Transcriptome-wide association study offers accurate gene-level association mapping in several plant species and is reasonably robust to sources of expression data.
Abstract
A genome-wide association study (GWAS) is used to identify genetic markers associated with phenotypic variation. In contrast, a transcriptome-wide association study (TWAS) detects associations between gene expression levels and phenotypic variation. It has previously been shown that in the cross-pollinated species, maize (Zea mays), GWAS, and TWAS identify complementary sets of trait-associated genes, many of which exhibit characteristics of true positives. Here, we extend this conclusion to the self-pollinated species, Arabidopsis thaliana and soybean (Glycine max). Linkage disequilibrium (LD) can result in the identification, via GWAS, of false-positive associations. In all three analyzed plant species, most trait-associated genes identified via TWAS are well separated physically from other candidate genes. Hence, TWAS is less affected by LD than is GWAS, demonstrating that TWAS is particularly well suited for association studies in genomes with slow rates of LD decay, such as soybean. TWAS is reasonably robust to the plant organs/tissues used to determine expression levels. In summary, this study confirms that TWAS is a promising approach for accurate gene-level association mapping in plants that is complementary to GWAS, and established that TWAS can exhibit substantial advantages relative to GWAS in species with slow rates of LD decay.
Introduction
Identifying genes that contribute to phenotypic variation enhances our understanding of plant biology and can contribute crop improvement. Genome-wide association study (GWAS) detects associations between genetic variants and phenotypic variation in diversity panels by taking advantage of ancient recombination events to identify genetic markers that co-segregate with phenotypic variation (Hirschhorn and Daly, 2005). Benefiting from cost-efficient genotyping technologies and improved statistical methods, GWAS is widely used to dissect the genetic basis of complex traits in plants. Over the past decade, thousands of loci have been identified for hundreds of plant traits using variants of this method (Tian et al., 2020). However, due to linkage disequilibrium (LD), it is often not possible to unambiguously determine which of multiple genes linked to a genetic marker associated with a trait via GWAS is in fact the causal gene (Wallace et al., 2014).
Transcriptome-wide association study (TWAS) detects associations between variation in gene expression levels and phenotypic variation (GTEx Consortium et al., 2015; Gusev et al., 2016). In maize (Zea mays), a TWAS method termed expression read depth GWAS (eRD-GWAS; Lin et al., 2017) that employs Bayesian statistical methods was developed. Unlike some TWAS methods used in humans, where gene expression levels were predicted, eRD-GWAS used empirically measured gene expression levels as explanatory variables. It was performed on 13 agronomic traits of maize and many of the trait-associated genes have predicted functions consistent with the corresponding traits (Lin et al., 2017). One of the genes identified as being associated with flowering time, MADS TRANSCRIPTION FACTOR69 (ZmMADS69), has since been cloned and its role in flowering time is functionally characterized (Liang et al., 2019). More recently, eRD-GWAS has been used to identify genes associated with root traits and again, many of the trait-associated genes have been previously associated with root architecture (Zheng et al., 2020). A different approach to TWAS was applied to analyze multiple traits using expression data from seven diverse maize tissues (Kremling et al., 2019). These studies used different approaches to interpret their results. Because Lin et al. (2017) demonstrated that TWAS is complementary to GWAS, they recommended treating the union of the two sets of trait-associated genes as candidates. In contrast, Kremling et al. (2019) employed an ensemble approach that intersects results from GWAS and TWAS to prioritize causal genes. Lin et al. (2017) and Kremling et al. (2019) both obtained hundreds of associated genes but did not quantitatively evaluate the qualities of the resulting gene sets based on the published literature.
To assess the ability of TWAS to specifically identify causal genes in self-pollinated species, we analyzed several well-studied quantitative traits in Arabidopsis thaliana for which hundreds of genes have been identified, and in many cases characterized (Bouché et al., 2016). This body of prior research made it possible to evaluate the ability of TWAS to identify true positives. LD can complicate the interpretation of GWAS results (Atwell et al., 2010). Some plant species exhibit slow rates of LD decay, which reduces the utility of GWAS in these species. To test whether TWAS can overcome this challenge, we compared GWAS and TWAS results on the qualitative trait pubescence color in soybean (Glycine max), which has an average LD of ∼100 kb (Zhou et al., 2015). Similarly, we show that TWAS does not identify closely linked candidate genes.
Our results establish that in multiple plant species, TWAS provides results that are complementary to those from GWAS, and that TWAS can identify high-quality candidate genes even in species with low rates of LD decay.
Results
TWAS is complementary to GWAS
Lin et al. (2017) reported that in the cross-pollinated species maize, TWAS and GWAS identify complementary sets of trait-associated candidate genes. To extend these findings to a self-pollinated species, we conducted GWAS and TWAS independently for the same phenotype using the same Arabidopsis diversity panel. The data used for these comparisons are summarized in “Materials and methods”.
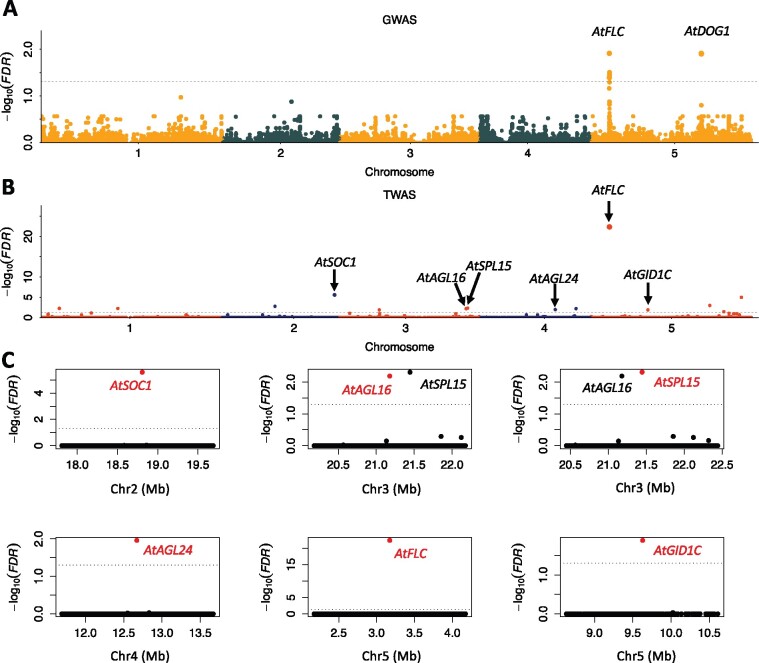
We repeated previously published single-nucleotide polymorphism (SNP)-based GWAS for flowering time at 16°C (FT16) in a panel of 970 Arabidopsis accessions, and detected the same two loci (Alonso-Blanco et al., 2016), FLOWERING LOCUS C (AtFLC) and DELAY OF GERMINATION1 (AtDOG1; Figure 1A). In parallel, TWAS was performed on the subset of accessions from this panel for which both leaf tissue RNA-seq data and FT16 phenotypes were available (N = 690 samples). This analysis identified 14 trait-associated genes (Figure 1B;Table 1), including only one of the two genes identified via GWAS, AtFLC. Although the other gene associated with FT16 via GWAS, AtDOG1, is expressed in leaf tissue, it was not associated with flowering time via TWAS. A total of 6 of the 14 trait-associated genes from TWAS are included in the FLOR-ID database (Bouché et al., 2016), which includes 306 hand-curated Arabidopsis flowering-related genes. This is substantially more overlap than expected by chance (one-sided Fisher’s exact test P-value 5E-8). The 2-Mb windows centered on four of these genes do not contain any additional trait-associated genes (Figure 1C). Two of the six genes (AGAMOUS-LIKE16 [AtAGL16] and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE15 [AtSPL15]) are separated by only 263 kb, but both are known to be related to flowering time (Figure 1C;Table 1). Five of these six genes encode transcription factors. Four of these exhibit regulatory interactions. SUPPRESSOR OF OVEREXPRESSION OF CO1 (AtSOC1; false-discovery rate [FDR] 2.6E-06) and AtSPL15 (FDR 4.9E-03) are regulated by AtFLC (FDR 4.5E-23), and AGAMOUS-LIKE24 (AtAGL24; FDR 1.1E-02) interacts with AtSOC1 (Supplemental Figure S1).
Figure 1.
GWAS and TWAS of Arabidopsis FT16. Manhattan plots of GWAS (A) and TWAS (B). Horizontal dashed lines in each panel designate the 0.05 FDR significance cutoff. Each dot represents a single SNP in GWAS and a single gene in TWAS plots. The dots in (A) and (B) are in four different colors to alternate through the association studies and chromosomes. Six TWAS identified Genes included in the FLOR-ID database are labeled with black text with arrow (B). C, The 2-Mb windows centered on each of the six genes identified via TWAS that are included in the FLOR-ID database. Each dot represents a single gene. The centered genes are highlighted in red and labeled in red text, while other significant genes are labeled in black text.
Table 1.
Fourteen Arabidopsis genes associated with flowering time via TWAS
| Relationship to FT in Literature | ID | Gene Symbol | FT16a (690 Samples) |
FT16a (631 Samples) |
Gene Annotation | References |
|---|---|---|---|---|---|---|
| Known | AT2G45660 | AtSOC1 | 2.6E-06 | 5.3E-06 | MADS-box transcription factor | Bouché et al. (2016) |
| AT3G57230 | AtAGL16 | 6.5E-03 | NS | MADS-box transcription factor | Bouché et al. (2016) | |
| AT3G57920 | AtSPL15 | 4.9E-03 | NS | SBP-box transcription factor | Bouché et al. (2016) | |
| AT4G24540 | AtAGL24 | 1.1E-02 | 5.4E-03 | MADS-box transcription factor | Bouché et al. (2016) | |
| AT5G10140 | AtFLC | 4.5E-23 | 2.3E-20 | MADS-box transcription factor | Bouché et al. (2016) | |
| AT5G27320 | AtGID1C | 1.3E-02 | 2.8E-02 | GA INSENSITIVE DWARF1C. Encodes a soluble gibberellin receptor that targets DELLA proteins | Bouché et al. (2016) | |
| AT1G09530 | AtPIF3 | 5.5E-03 | 8.0E-03 | Transcription factor interacting with photoreceptors phyA and phyB | Oda et al. (2004) | |
| Weak Evidenceb | AT3G19150 | AtACK1 | 1.3E-02 | 4.8E-02 | Kip-related protein gene negatively affects plant development and fertility | Han et al. (2005); Mahrez et al. (2016) |
| AT5G63120 | AtRH30 | 1.0E-05 | 1.1E-05 | P-loop containing nucleoside triphosphate hydrolases superfamily protein | Szklarczyk et al. (2019) | |
| AT5G55120 | AtVTC5 | 3.7E-02 | NS | GDP-L-galactose phosphorylase | ||
| No Evidence | AT2G20440 | 1.7E-03 | 4.5E-03 | Ypt/Rab-GAP domain of gyp1p superfamily protein | ||
| AT1G35180 | 6.0E-03 | 7.8E-03 | TRAM, LAG1, and CLN8 (TLC) lipid-sensing domain-containing protein. | |||
| AT4G33625 | 6.5E-03 | NS | Vacuole protein | |||
| AT5G49360 | 1.0E-03 | 3.3E-03 | Putative beta-xylosidase gene involved in secondary cell wall metabolism and plant development |
FDR of TWAS; NS, not significant.
Genes are functionally related with flowering time but have no evidence that mutants alter flowering times.
Information about the other eight trait-associated genes identified via TWAS is summarized in Table 1. Antisense suppression of PHYTOCHROME INTERACTING FACTOR3 (AtPIF3) leads to early flowering (Oda et al., 2004). There is evidence that three of the other trait-associated genes (ARABIDOPSIS CDK INHIBITOR1 [AtACK1], RNA HELICASE30 [AtRH30], and VITAMIN C DEFECTIVE5 [AtVTC5]) are involved in flowering regulatory networks (Dowdle et al., 2007; Kotchoni et al., 2009; Duan et al., 2016; Mahrez et al., 2016; Szklarczyk et al., 2019). AtACK1 encodes a cyclin-dependent kinase inhibitor and is a negative regulator of cell division (Han et al., 2005). Its expression is altered in the early flowering mutant BRR2a-T895I (Mahrez et al., 2016). Based on the STRING database (Szklarczyk et al., 2019), AtRH30 has at least two predicted “functional partners” involved in flowering, SNW/SKI-INTERACTING PROTEIN and GLYCINE RICH PROTEIN2. AtVTC5 encodes a guanosine diphosphate (GDP)‐l‐galactose phosphorylase required for the biosynthesis of ascorbic acid (Dowdle et al., 2007). Ascorbic acid can affect flowering time in Arabidopsis (Kotchoni et al., 2009). Furthermore, AtVTC5 is a putative target of the flowering gene, AtFLC (Duan et al., 2016). We have not identified evidence linking the remaining four genes to flowering time.
To conduct a fair comparison between GWAS and TWAS, association studies were performed on the subset of the Arabidopsis samples for which genotypes, expression data, and phenotypic data were all available (N = 631). Using these data, GWAS and TWAS identified 1 locus and 10 genes associated with FT16, respectively (Supplemental Figure S2; Table 1). The finding that many (1/1 from GWAS and 7/10 from TWAS) of these trait-associated genes can be linked to flowering time by independent studies demonstrates that both methods identify true positives at high rates.
To extend these conclusions, additional TWASs were performed for five highly correlated developmental traits (three for flowering time and two for leaf number), using data from Grimm et al. (2017). The numbers of samples contributing phenotypic values, genotype, and expression levels range from 574 to 620 depending upon the trait (Supplemental Table S1). Across the five traits, TWASs identified 41 trait-associated genes, consisting of 16 unique genes, using an FDR cutoff of 0.05 (Supplemental Table S1). Ten of these 16 genes had also been identified by the FT16 TWAS. One of the remaining six genes is FLOWERING LOCUS T (AtFT; AT1G65480), which was associated with the trait rosette leaf (RL) number; AtFT mutants exhibit increased leaf number (Onouchi et al., 2000). The number of known flowering-related genes identified by TWAS ranged from 4 to 6/trait (Supplemental Table S1). It is not surprising that flowering-related genes were associated with leaf number because these two traits are highly and positively correlated with flowering time in Arabidopsis (Piñeiro and Coupland, 1998; Grimm et al., 2017). The corresponding GWAS (Supplemental Table S1) identified five unique loci, including AtDOG1. None of these candidate genes overlapped with those identified via TWAS (Supplemental Figure S3). Using a superset of samples (N = 860–936; those with genotype but not expression data), Grimm et al. (2017) identified via GWAS 30 candidate genes for these 5 traits. Only one of these, AtFLC, was also identified via TWAS. The very limited overlap between the trait-associated genes from GWAS and TWAS for these five developmental traits of Arabidopsis provides further support for the complementarity of these approaches in a self-pollinated species.
TWAS is less affected by LD than GWAS
GWAS exploits LD between markers and functional variation. In species with high LD, markers are often tightly linked to multiple genes. In such instances, it is often difficult to uniquely associate a single causal gene to a trait (Atwell et al., 2010). To test whether TWAS can overcome this challenge in a species with high levels of LD, we conducted GWAS and TWAS for pubescence color in soybean, which has an average LD of ∼100 kb (Zhou et al., 2015).
Pubescence color trait data were available for 75 of 102 soybean lines for which both expression data and SNP genotypes were available (Supplemental Table S2). These lines had either tawny (N = 34) or gray (N = 41) pubescence color. The T locus (Glyma.06g202300), which controls pubescence color (Toda et al., 2002), encodes a flavonoid 3′- hydroxylase (F3′H). The dominant (T) and recessive (t) alleles confer tawny and gray color, respectively. GWAS (Figure 2A) and TWAS (Figure 2B) were conducted separately for pubescence color using comparable statistical methods.
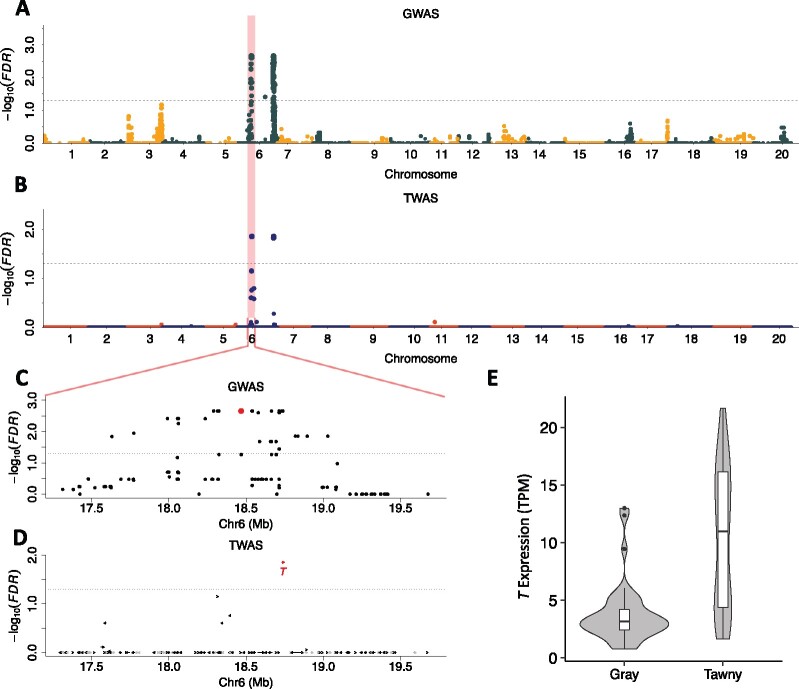
Figure 2.
Analysis of soybean pubescence color via GWAS and TWAS. Manhattan plots of GWAS (A) and TWAS (B). Horizontal dashed lines designate the 0.05 FDR significance cutoff. Each dot represents a single SNP in GWAS and a single gene in TWAS plots. The known causal locus (T) is highlighted. The regions surrounding the T gene from the GWAS and TWAS analyses are magnified in parts (C) and (D), respectively. The red dots in these panels designate the most significant trait-associated SNP (C) and gene (D) in each analysis. In (D), the causal gene, T, is labeled and arrows indicate the directions in which genes are transcribed. Gray arrows represent genes that are not expressed in the RNA samples used in the TWAS. E, violin plots and boxplots of expression of the T gene in soybean lines with gray and tawny pubescence. The unit of expression is TPM. Violin plots show the probability density curves of T expression in two groups of soybean lines, the width of the curve corresponds with the approximate frequency of expression values in each region. The box in the boxplot shows 25–75th percentiles; black line within the box shows the median; whiskers represent the 1.5 times interquartile range; points represent outliers.
The GWAS detected 80 trait-associated SNPs spanning a ∼1.4 Mb interval (Chr06: 17,632,002–19,029,221) that includes 68 annotated genes, one of which is the T locus. Within this interval, the SNP with the smallest FDR (Chr06-18468010) is located 263-kb upstream of the T locus (Glyma.06g202300; Figure 2C). Similarly, three previous GWASs reported the most significant SNP as having distances of ∼100–600 kb away from the T locus, and associated SNPs spanned intervals of 2–4 Mb using diversity panels from 139 to 12,360 lines (Sonah et al., 2015; Wen et al., 2015; Bandillo et al., 2017). Another trait-associated signal detected in our GWAS (Chr06:165–167 Mb) has not previously been associated with this well-studied trait.
In contrast to the results obtained via GWAS, the T gene was the only trait-associated gene identified by TWAS within the 1.4-Mb interval defined by GWAS hits (Figure 2D). T exhibits significant differential gene expression (Welch two Sample t test, P-value 8E-7) between gray versus tawny lines (Figure 2E). Hence, despite the existence of extensive local LD (the average pairwise LD among these 80 trait-associated SNPs is 0.8), TWAS correctly identified the causal gene (T) associated with pubescence color.
As discussed above, the pattern of LD decay affects the resolving power of SNP-based GWAS. Similarly, the resolving power of TWAS would be limited if the expression patterns of neighboring genes were highly correlated, which could result in false-positive signals, as has been reported in human TWAS (Wainberg et al., 2019; Mancuso et al. 2019). We (Lin et al., 2017; Zheng et al., 2020) and others (Kremling et al., 2019) have not observed this problem in maize. If expression patterns of neighboring Arabidopsis genes were highly correlated, we would have expected to identify many closely linked genes associated with the newly analyzed developmental traits of this species (i.e. flowering time and leaf number). But this was not the case. These data, therefore, indicate that TWAS is less affected by LD than is GWAS, even in high LD species.
Effect of tissue selection on TWAS
Gene expression patterns differ across organs, tissues, environments, and developmental stages, which leads to the question of the importance of identifying the appropriate RNA-seq source to conduct TWAS for a given trait. To address this question, we used RNA-seq data from seven maize tissues derived from a diversity panel of maize inbreds (Kremling et al., 2018) to conduct TWASs for qualitative endosperm color. Endosperm color phenotypes are available for 229 of the 300 inbreds in this panel (Supplemental Tables S3 and S4). The yellow endosperm1 (y1) and white cap1 (wc1) genes, which are known to regulate endosperm color (Buckner et al., 1996; Tan et al., 2017), have diverse expression patterns across the genotypes and tissues analyzed in this study (Figure 3).
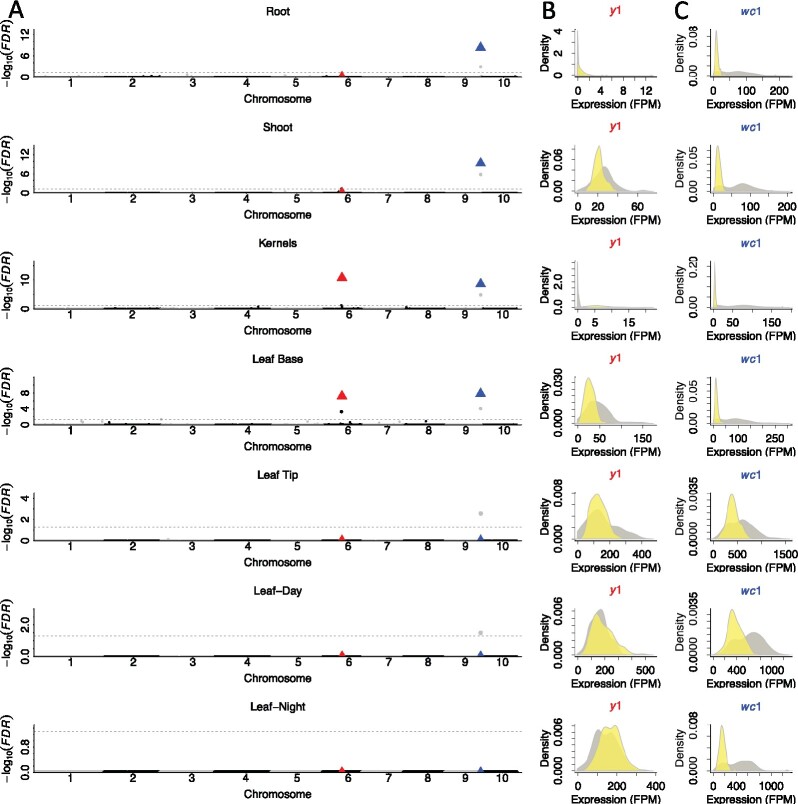
Figure 3.
Effects of tissue source on TWAS results for maize endosperm color. A, Manhattan plots of TWAS conducted for maize endosperm color on each of seven tissues; each dot represents a single gene. The gray points adjacent to wc1 designate GRMZM2G089421. Horizontal dashed lines designate the 0.05 FDR significance cutoff. The red and blue triangles represent the known causal loci, y1 and wc1, respectively. The distributions of expression levels of the y1 (B) and wc1 (C) genes in inbred lines with yellow (yellow) and white (gray) endosperms. The density plots show the probability density across each bandwidth. Sum of densities × bandwidth equals to 1.
Both the y1 and wc1 genes were associated with endosperm color via TWAS using expression data from kernels (Figure 3A). Similar results were obtained using expression data from the leaf base. Additionally, the wc1 gene, but not the y1 gene, was associated with endosperm color when using expression data from two additional tissues, shoot, and root. Interestingly, our ability to detect an association of the y1 gene with endosperm color using expression data from leaf base was due to the fact that, in contrast to the situation in kernels, inbred lines having white endosperms accumulated higher levels of y1 transcript in leaf base tissue than did those with yellow endosperms (Figure 3B).
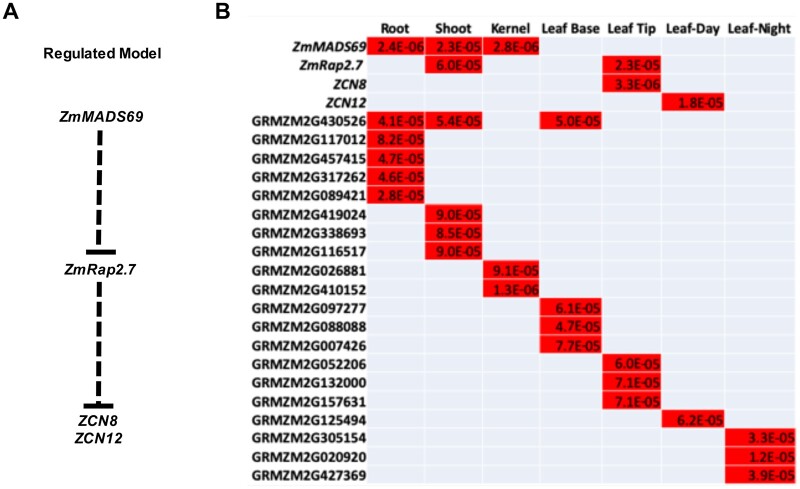
To extend this exploration, we used the same seven sets of expression data to conduct TWASs on the flowering time trait, days to anthesis (DTA; Peiffer et al., 2014). The number of inbreds for which both expression data and phenotypes varied from 191 to 258, with an average of 238 (Supplemental Table S5). A total of 24 unique genes were associated with flowering time across the 7 TWASs with the more relaxed P-value cutoff of 1E-04 (Figure 4; Supplemental Table S5). Four genes known to function in flowering (Liang et al., 2019; Castelletti et al., 2020) were identified in one or more of the seven tissues. MADS TRANSCRIPTION FACTOR69 (ZmMADS69) was identified in three of seven tissues. Interestingly, although the ZmMADS69-regulated genes, RELATED TO APETALA2.7 and ZEA CENTRORADIALIS8, were both identified using expression data from leaf tip, ZmMADS69 per se was not identified using this dataset. At least one of the four known flowering-related genes was detected in five of the seven tissues, including root and kernels, which are not obviously associated with the DTA trait. Two of the 24 genes (ZmMADS69 and GRMZM2G430526) were identified in a set of five DTA-associated genes from another TWAS that relied on the same phenotypic data, but independent expression data (Lin et al., 2017). This is more overlap than expected by chance (one-sided Fisher’s exact test, P-value 2E-5). The analysis of expression data from multiple tissues, even some of which would appear unrelated to flowering, detected additional loci associated with the DTA trait.
Figure 4.
Maize genes associated with DTA via TWAS. A, Regulatory relationships among four known flowering genes (Liang et al., 2019; Castelletti et al., 2020); (B) the 24 genes identified via TWAS identified using expression data from seven tissues. Red cells represent genes that were significantly associated with the DTA trait in the indicated source of expression data. Significant P-values are indicated. Gray cells represent tests that were not significant.
To further explore the effects of using expression data from unrelated tissues, TWASs were performed for 24 carotenoid-related traits in kernels (Owens et al., 2014) using expression data from maize seedlings (Hirsch et al., 2014). Although Owens et al. data set contains carotenoid concentrations for over 200 inbreds, expression data from Hirsch et al. were available for only about half of these samples (Supplemental Table S6). Using a somewhat relaxed P-value cutoff of 1E-04 data (refer to “Materials and methods”) to control for the limited number of samples (∼100), 16 unique trait-associated genes were identified for the 24 carotenoid-related traits (Supplemental Table S6). GRMZM2G143202 (LUTEIN DEFICIENT1) had the most significant P-value (i.e. 5E-6) and encodes a cytochrome P450 protein required for the biosynthesis of lutein (Tian et al., 2004). Another cytochrome P450 family gene, GRMZM2G013357 was associated with two carotenoid traits, “β-Cryptoxanthin/Zeaxanthin” and “Provitamin A.” GRMZM2G087207 is involved in hydroxymethylglutaryl-CoA synthase activity, which takes part in supplying a precursor of β-carotene biosynthesis (Qiang et al., 2020). Only 1 of the 16 candidate genes, that is, LUTEIN DEFICIENT1, identified via TWAS was also detected via a GWAS based on the same phenotypic dataset but using data from nearly twice as many inbreds (N = 210), despite the fact that this GWAS identified 58 candidate genes (Owens et al., 2014). The success of using seedling expression data to identify presumptively true positives for carotenoid traits in kernels lends further support to our conclusion that it is possible to use expression data from unrelated tissues to identify causal genes via TWAS.
Discussion
It has previously (Lin et al., 2017; Zheng et al., 2020) been shown that at least in the cross-pollinated species, maize, TWAS is complementary to GWAS, in that these two types of analyses identify only partially overlapping sets of trait-associated genes that exhibit characteristics of true positives. This study extends this conclusion to the self-pollinated species, Arabidopsis. Our TWAS of development-related traits in Arabidopsis identified distinct trait-associated genes as compared to those identified via GWAS using the same genotypes and the same phenotypic data. Importantly, half of the identified TWAS-specific genes associated with Arabidopsis traits have been functionally associated with flowering time by independent studies.
Lin et al. (2017) recommended treating all genes in the union of results from GWAS and TWAS as candidates to better explain the genetic basis of the traits. In contrast, Kremling et al. (2019) intersected the results from GWAS and TWAS to achieve higher predictive ability than genes identified by either method alone. This is presumably because the frequency of true positives in the intersection of the gene sets identified via GWAS and TWAS is higher than among the method-specific trait-associated genes. We note, however, that many of the true positives detected in our Arabidopsis TWASs were not identified by the comparable GWAS and that the estimate of true positives, that is, the percentage of known genes and evidence supported genes detected, identified via TWAS ranged from 50% to 71% for each of the Arabidopsis traits (Table 1; Supplemental Table S1). Hence, there may be substantial lost opportunities if candidate genes detected only by TWAS are ignored.
How might TWAS identify trait-associated genes missed by GWAS? If a mapping population does not contain a polymorphic genetic marker near a causal gene, GWAS cannot associate that gene with phenotypic variation. But this gene–trait association could potentially be detected via TWAS if variation in the expression of this gene contributes to phenotypic variation. This situation could arise in several ways. First, absent full genome sequences, the failure to detect a polymorphic marker near the trait-associated gene does not indicate the absence of cis-expression quantitative loci (eQTL). Second, differences in expression could be the result of heritable epigenetic variation in or near the trait-associated gene that would not be detected via sequence analyses. Third, the differences in the expression of the trait-associated gene could be due to the segregation of trans-eQTL. In this case, the detected trait-associated gene is serving as a read-out for variation in the trans-eQTL. Finally, because TWAS involves fewer statistical tests than a typical SNP-based GWAS and the need to control for multiple testing, it is expected that all other things being equal, TWAS will be capable of detecting gene–trait associations with smaller effect sizes than GWAS.
Just as GWAS can fail to identify candidate genes identified via TWAS, TWAS can fail to identify candidate genes identified via GWAS. At least some of these could be false-negative results (others may have been false-positive results from the corresponding GWAS). False-negative results could occur because a gene is not expressed (or not expressed at levels sufficient to provide statistical significance) and/or because the functional variation segregating in the association panel is not associated with variation in expression levels. For example, an allele that encodes an altered protein might be detected via GWAS but not by TWAS.
LD can result in false-positive gene–trait associations during GWAS. This is particularly challenging in species that exhibit low rates of LD decay. In such species, a single trait-associated marker may be in LD with dozens of candidate genes. It is often difficult to prioritize among these candidate genes to select those for functional analyses. Fortunately, we have demonstrated that TWAS can accurately identify trait-associated genes even in species with high levels of local LD. For example, in soybean, the causal gene for pubescence color, T, was uniquely identified via TWAS, whereas GWAS-associated markers across a 1.4-Mb interval that includes 68 genes with the T.
These results with soybean represent a single case in which TWAS correctly identified the causal gene despite high levels of local LD. If LDs were resulting in many false-positive trait associations in TWAS, we would expect to observe sets of closely linked candidate genes, as occurs in GWAS and in human TWAS, which rely upon imputed expression data (Wainberg et al., 2019; Mancuso et al. 2019). In contrast, the minimum physical distance between the trait-associated genes for the five Arabidopsis developmental traits (flowering time and leaf number) is 813 kb (Supplemental Table S1). Because Arabidopsis exhibits a global LD of <10 kb (Kim et al., 2007), our results are inconsistent with TWAS generating high levels of LD-induced false-positive associations.
Our TWAS for endosperm color identified the true positive, wc1 (Tan et al., 2017). The same TWAS also associated GRMZM2G089421 with endosperm color (Figure 3A) despite the fact that no prior literature links GRMZM2G089421 with this trait. Interestingly this gene is only 39 kb from wc1 and the two genes share a common cis-eQTL (Wang et al., 2018). Consequently, GRMZM2G089421 may represent a false-positive signal that arose as a consequence of co-regulation. Although we cannot rule out the possibility that GRMZM2G089421 has an as yet undescribed role in regulating endosperm color, it is certainly possible that a transcription factor that co-regulates two closely linked loci could result in the two genes exhibiting correlated expression patterns, and thereby being co-discovered via TWAS. If only one of these genes actually contributes to phenotypic variation in the trait of interest, the second gene would represent a false positive. But our empirical data suggest that this situation with linked co-regulated loci does not often occur; otherwise, we would have observed linked candidate genes in the Arabidopsis TWAS. More generally, however, false positives will be detected via TWAS if a gene’s expression patterns are correlated with phenotypic variation, but that gene is not causative (Wainberg et al., 2019). This situation might be more common when using expression data from tissues unrelated to the trait of interest.
Because it is possible to identify SNPs from RNA-seq data, it is possible to conduct both GWAS and TWAS using RNA-seq data. This combined approach is particularly attractive for species with large genomes, in which genome re-sequencing of large diversity panels remains expensive. Practitioners of TWAS are faced with the selecting tissue samples from which to analyze gene expression. Our results demonstrate that although expression data from tissues associated with the trait of interest may be optimal, even tissues that would not be expected to be associated with the trait can be used to identify gene–trait associations. For example, although two maize genes known to affect endosperm color (i.e. y1 and wc1) were successfully identified via TWAS using expression data from kernel, these genes were also identified using expression data from some (but not all) other tissues. Similarly, we used previously published sets of expression data from diverse tissues to conduct TWASs on a maize flowering time trait (DTA). At least one of four known flowering-related genes was detected using expression data from five of seven tissues. Finally, genes known to be associated with development-related traits in Arabidopsis at 16°C were identified via TWAS using expression data from plants grown at 20°C. Hence, our results indicate that although expression data from a trait-related tissue or environment is ideal for TWAS, even expression data from less obviously trait-related tissue samples or from plants grown in different environments can enable the discovery of trait-associated genes. This demonstrates the feasibility of conducting TWAS using existing gene expression data sets. Finally, our results also suggest that trait-associated genes identified via TWAS using different sources of expression data can be complementary.
Conclusions
We extended TWAS from cross-pollinated maize to self-pollinated Arabidopsis and soybean and confirmed that TWAS is also complementary with GWAS (Lin et al., 2017) in self-pollinating species. By studying well-characterized traits, we demonstrated that TWAS offers high rates of true positive results and is less affected by LD than GWAS, which makes it a promising tool in species with high LD. Although trait-related tissue is preferred as the source of expression data, our results show that expression data from other tissues can also be used to identify trait-associated genes. In summary, by addressing these important open questions about TWAS and making its implementation more accessible, this study promises to encourage more plant scientists to exploit this complementary approach to identifying gene–trait associations.
Materials and methods
Data sources
Arabidopsis thaliana
SNPs from The 1001 Genome Project (Alonso-Blanco et al., 2016) were downloaded and filtered for those with a minor allele frequency (MAF) cutoff of ≥5% and site-level missingness of <20%. The 1,064,218 remaining SNPs were further imputed using Beagle version 4.1 (Browning and Browning, 2007, 2016), with default parameters. Read counts of RNA-seq data of 728 accessions from RLs grown at 20°C (Kawakatsu et al., 2016) were downloaded and normalized with transcripts per kilobase million (TPM) method. Then 22,708 annotated protein-coding genes with an average TPM > 0.1 were used for subsequent TWAS. Flowering time data of plants grown at 16°C with 4 replicates (blocks) in a random block design were from Alonso-Blanco et al. (2016). Data for three additional flowering time traits and two leaf number traits scored at 16°C with four replicates (blocks) in a random block design were from Grimm et al. (2017).
Expression data from seven maize (Z. mays) tissues
Fragments per million (FPM) normalized expression data from seven tissues, viz., germinating shoot (Shoot), germinating root (Root), third leaf base (Leaf Base), third leaf tip (Leaf Tip), adult leaf collected during the day (Leaf-Day), kernels 350 growing degree days after pollination (Kernels), and adult leaf collected at night (Leaf-Night) were collected from a maize diversity panel with an average of 255 inbreds (Supplemental Table S4; Kremling et al., 2018) and were used directly for TWAS because the 3′-RNA-seq method used to collect these data negates the impact of gene length (Ma et al., 2019). And annotated protein-coding genes with average FPM values of >0.1 were used for TWAS for each tissue (Supplemental Table S4).
Soybean (G. max) and maize phenotypic data
Pubescence color phenotypes of soybean lines and endosperm color phenotypes of maize lines were obtained from the US National Plant Germplasm System Website (https://npgsweb.ars-grin.gov/gringlobal/search.aspx; Supplemental Tables S2 and S3). Phenotypic data associated with flowering time (DTA) and kernel carotenoid composition (24 traits) in maize were obtained from Peiffer et al. (2014) and Owens et al. (2014), respectively. Peiffer et al. (2014) evaluated the DTA for each inbred with 12–15 plants in each of the three different environments in 2010. Owens et al. (2014) measured carotenoid levels of ≥4 ears/year grown in two different years (2009 and 2010) but at the same location.
Trimming and alignment of soybean and maize RNA-seq reads
Soybean RNA-seq data generated from V2 (18 d old) leaves of the 41 soyNAM parental lines and 61 milestone cultivars (El Baidouri et al., 2018) were aligned to the reference genome of G. max Williams 82 V2 (Schmutz et al., 2010). Maize RNA-seq data generated from whole seedlings of 503 inbreds (Hirsch et al., 2014) were aligned to the B73 reference genome Z. mays B73 AGPv3 (Schnable et al., 2009).
Prior to alignment, raw sequence reads were trimmed using Trimmomatic version 0.36 (Bolger et al., 2014) using following parameters: LEADING:3, TRAILING:3, SLIDINGWINDOW:4:15, and MINLEN:40. Trimmed reads were aligned to the corresponding reference genome with GSNAP (Wu and Watanabe, 2005) according to the method of Liu et al. (2012). Only uniquely qualified aligned reads were retained for subsequent SNP calling and gene expression analyses. Trimming and alignment results for each line are summarized in Supplemental Tables S2 and S3.
SNP calling from aligned soybean RNA-seq reads
Bi-allelic SNPs were identified within each sample using custom scripts (Liu et al., 2012; Li et al., 2019). For each sample, uniquely aligned reads were used while ignoring the first and last 3 bp of each read and only considering sites with PHRED scores ≥20. SNPs sites were required to have at least five covered reads and a combined overall allele frequency of ≥80%. Subsequently, the genotypes of those identified SNPs sites were determined for each sample using the following parameters. Homozygous SNP sites were defined as having ≥5 reads of the major allele, and overall major allele reads account ≥90%. Heterozygous SNP sites were defined as having ≥2 reads for each of the two alleles and the sum of the reads from both alleles being ≥5, each allele accounting for 20% of all reads and together accounting for ≥90% of all reads. For all the other situations, a missing genotype was assigned to the site in a given sample.
The SNPs were further filtered. First, SNPs were required to have a MAF of ≥5%, the number of samples homozygous for the minor allele was required to be ≥5 and the missing data rate was required to be ≤50%. Finally, 75,289 SNPs were retained for the 102 soybean lines. Those SNPs have a median coverage of 32 reads for genotyped samples, and the median missing rate of SNPs per sample is only 7.8% (Supplemental Figure S4).
Next, imputation was performed among those remaining SNPs using Beagle version 4.1 with default parameters. The imputed genotypes were used for GWAS.
Read counting from aligned RNA-seq reads and conversion for TWAS
The number of reads uniquely aligned to each gene in each individual was determined. TPM was used to normalize read counts, and only annotated protein-coding genes with an overall average TPM of >0.1 were defined as expressed and used for subsequent TWAS.
Normalized expression values were transformed for use in GAPIT (Lipka et al., 2012), which requires a numeric range from 0 to 2 for each gene. To handle extreme expression values (outliers), first, expression values smaller than quantile 5 were converted to 0 and values larger than quantile 95 were converted to 2. The remaining expression values were linearly transformed into values between 0 and 2.
GWAS and TWAS
The GWAS and TWAS were conducted using the Compressed Mixed Linear Model implemented in the R package GAPIT (Zhang et al., 2010; Lipka et al., 2012). A further MAF cutoff of ≥5% on samples with a phenotype was applied for GWAS. The first three components of principal component analysis derived from input SNPs (GWAS) and gene expression (TWAS) were used to separately control for population structure. The resulting P-values were adjusted to control for multiple testing (Benjamini and Hochberg, 1995). SNPs or genes that exhibited an FDR of ˂0.05 were defined as trait associated if not otherwise specified. To enhance the probability of identifying true positives given the reduced sample numbers in the maize DTA and maize carotenoid studies, for only these analyses we used a relaxed significance cut-off of 1E-04 (which is close to 1/genes number [i.e. 4E-05]) to define trait-associated genes. The LD R2 between SNPs in the soybean GWAS was estimated using Plink version 1.9 (Gaunt et al., 2007). The LD heatmap analysis was conducted with LDheatmap version 0.99-8 (Shin et al., 2006).
Statistical analyses
The difference between the average expression levels of the T gene in the two soybean pubescence color groups (tawny and gray) was analyzed using the Welch two-sample t test (Welch, 1947). A one-sided Fisher’s exact test (Fisher, 1922) was used for whether there is more overlap than expected by chance between two groups of genes. P <0.01 from those tests was considered significant.
Data accession numbers
All of the data used in this study were previously published or obtained from public databases (Supplemental Table S7). Maize seedling RNA-seq raw sequenced data (SRP018753) and soybean leaf RNA-seq raw sequenced data (SRP108748) were obtained from Sequence Read Archive, NCBI. The expression data from seven maize tissues were downloaded from Data Commons; Arabidopsis genotype data came from 1,001 Genome and expression data (GSE80744) came from Gene Expression Omnibus.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Interaction network of four Arabidopsis flowering time genes.
Supplemental Figure S2. GWAS and TWAS for Arabidopsis flowering time (FT16) on the subset of samples.
Supplemental Figure S3. GWAS and TWAS for five Arabidopsis developmental traits (three for flowering time and two for leaf number).
Supplemental Figure S4. Summary of soybean SNPs identified from leaf RNA-seq data.
Supplemental Table S1. Genes or loci identified via TWAS and GWAS for Arabidopsis flowering time and leaf number traits using data from Grimm et al. (2017).
Supplemental Table S2. Summary of leaf RNA-seq data and processing and pubescence colors of component genotypes from a soybean diversity panel.
Supplemental Table S3. Summary of seedling RNA-seq data and processing and endosperm colors of component genotypes from a maize diversity panel.
Supplemental Table S4. Numbers of genes expressed in seven tissues of a maize diversity panel and the frequencies of yellow and white endosperm phenotypes in subsets of this panel.
Supplemental Table S5. DTA-associated maize genes identified via TWAS using expression data from seven tissues.
Supplemental Table S6. Carotenoid-associated maize genes identified via TWAS using expression data from maize seedling.
Supplemental Table S7. Data used.
Supplementary Material
Acknowledgments
We thank Data2Bio LLC for access to computational resources, Cheng‐Ting Yeh for technical assistance, Aaron Kusmec and Hung-Ying Lin (all of, or formerly of the Schnable Lab at ISU) and James Schnable (University of Nebraska-Lincoln), and three anonymous reviewers for fruitful discussions and/or suggestions, and An-Ping Hsia and Lakshmi Attigala (Schnable Lab at ISU) for assistance editing the manuscript.
Funding
This work is partially supported by Hatch Project No. IOW04714 from the National Institute of Food and Agriculture of United States Department of Agriculture (to P.S.S.).
Conflict of interest statement. During much of this study DL was an employee of Data Biotech (Beijing) Co., Ltd., Beijing, China, which was a wholly owned subsidiary of Data2Bio LLC. PSS is a co-lead of the Genomes to Fields Initiative and a Changjiang Scholar at China Agriculture University. He is co-founder and managing partner of Data2Bio LLC; Dryland Genetics, LLC; EnGeniousAg, LLC; and LookAhead Breeding LLC. He is a member of the scientific advisory board and a shareholder of Hi-Fidelity Genetics, Inc. and a member of the scientific advisory boards of Kemin Industries and Centro de Tecnologia Canavieira.
Conceived and designed the experiments: D.L. and P.S.S. Analyzed the data: D.L., Q.L., and P.S.S. Wrote the manuscript: D.L. and P.S.S. All authors read and approved the final manuscript.
The author responsible for distribution of materials and data related to the findings presented in this study in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Patrick S. Schnable (schnable@iastate.edu).
References
- Alonso-Blanco C, Andrade J, Becker C, Bemm F, Bergelson J, Borgwardt KM, Cao J, Chae E, Dezwaan TM, Ding W, et al. (2016) 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandillo NB, Lorenz AJ, Graef GL, Jarquin D, Hyten DL, Nelson RL, Specht JE (2017) Genome-wide association mapping of qualitatively inherited traits in a germplasm collection. Plant Genome 10: 1–18 [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57: 289–300 [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouché F, Lobet G, Tocquin P, Périlleux C (2016) FLOR-ID: an interactive database of flowering-time gene networks in Arabidopsis thaliana. Nucleic Acids Res 44: D1167–D1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR (2016) Genotype imputation with millions of reference samples. Am J Hum Genet 98: 116–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL (2007) Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81: 1084–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner B, Miguel P, Janick-Buckner D, Bennetzen J (1996) The y1 gene of maize codes for phytoene synthase. Genetics 143: 479–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelletti S, Coupel-Ledru A, Granato I, Palaffre C, Cabrera-Bosquet L, Tonelli C, Nicolas SD, Tardieu F, Welcker C, Conti L (2020) Maize adaptation across temperate climates was obtained via expression of two florigen genes. PLoS Genet 16: e1008882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowdle J, Ishikawa T, Gatzek S, Rolinski S, Smirnoff N (2007) Two genes in Arabidopsis thaliana encoding GDP-l-galactose phosphorylase are required for ascorbate biosynthesis and seedling viability. Plant J 52: 673–689 [DOI] [PubMed] [Google Scholar]
- Duan W, Ren J, Li Y, Liu T, Song X, Chen Z, Huang Z, Hou X, Li Y (2016) Conservation and expression patterns divergence of ascorbic acid d-mannose/l-galactose pathway genes in Brassica rapa. Front Plant Sci. 7: 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M, Kim KD, Abernathy B, Li YH, Qiu LJ, Jackson SA (2018) Genic C-methylation in soybean is associated with gene paralogs relocated to transposable element-rich pericentromeres. Mol Plant 11: 485–495 [DOI] [PubMed] [Google Scholar]
- Fisher RA (1922) On the interpretation of χ2 from contingency tables, and the calculation of P. J Royal Stat Soc 85: 87–94 [Google Scholar]
- Gaunt TR, Rodríguez S, Day IN (2007) Cubic exact solutions for the estimation of pairwise haplotype frequencies: implications for linkage disequilibrium analyses and a web tool “CubeX”. BMC Bioinformatics 8: 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm DG, Roqueiro D, Salomé PA, Kleeberger S, Greshake B, Zhu W, Liu C, Lippert C, Stegle O, Schölkopf B, et al. (2017) easyGWAS: a cloud-based platform for comparing the results of genome-wide association studies. Plant Cell 29: 5–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- GTEx Consortium, Gamazon ER, Wheeler HE, Shah KP, Mozaffari SV, Aquino-Michaels K, Carroll RJ, Eyler AE, Denny JC, Nicolae DL, et al. (2015) A gene-based association method for mapping traits using reference transcriptome data. Nat Genet 47: 1091–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, Bhatia G, Chung W, Penninx BWJH, Jansen R, de Geus EJC, Boomsma DI, Wright FA, et al. (2016) Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet 48: 245–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W, Rhee HI, Cho JW, Ku MSB, Song PS, Wang MH (2005) Overexpression of Arabidopsis ACK1 alters leaf morphology and retards growth and development. Biochem Biophys Res Commun 330: 887–890 [DOI] [PubMed] [Google Scholar]
- Hirsch CN, Foerster JM, Johnson JM, Sekhon RS, Muttoni G, Vaillancourt B, Penagaricano F, Lindquist E, Pedraza MA, Barry K, et al. (2014) Insights into the maize pan-genome and pan-transcriptome. Plant Cell 26: 121–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschhorn JN, Daly MJ (2005) Genome-wide association studies for common diseases and complex traits. Nat Rev Genet 6: 95–108 [DOI] [PubMed] [Google Scholar]
- Kawakatsu T, Huang SC, Jupe F, Sasaki E, Schmitz RJ, Urich MA, Castanon R, Nery JR, Barragan C, He Y, et al. (2016) Epigenomic diversity in a global collection of Arabidopsis thaliana accessions. Cell 166: 492–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M (2007) Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat Genet 39: 1151–1155 [DOI] [PubMed] [Google Scholar]
- Kotchoni SO, Larrimore KE, Mukherjee M, Kempinski CF, Barth C (2009) Alterations in the endogenous ascorbic acid content affect flowering time in Arabidopsis. Plant Physiol 149: 803–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremling KAG, Chen SY, Su MH, Lepak NK, Romay MC, Swarts KL, Lu F, Lorant A, Bradbury PJ, Buckler ES (2018) Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555: 520–523 [DOI] [PubMed] [Google Scholar]
- Kremling KAG, Diepenbrock CH, Gore MA, Buckler ES, Bandillo NB (2019) Transcriptome-wide association supplements genome-wide association in Zea mays. G3 9: 3023–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li D, Jiao Y, Schnable JC, Li Y, Li H, Chen H, Hong H, Zhang T, Liu B, et al. (2019) Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnol J 18: 389–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Liu Q, Wang X, Huang C, Xu G, Hey S, Lin H, Li C, Xu D, Wu L, et al. (2019) ZmMADS69 functions as a flowering activator through the ZmRap2.7‐ZCN8 regulatory module and contributes to maize flowering time adaptation. New Phytol 221: 2335. [DOI] [PubMed] [Google Scholar]
- Lin H, Liu Q, Li X, Yang J, Liu S, Huang Y, Scanlon MJ, Nettleton D, Schnable PS (2017) Substantial contribution of genetic variation in the expression of transcription factors to phenotypic variation revealed by eRD-GWAS. Genome Biol 18: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Tian F, Wang Q, Peiffer J, Li M, Bradbury PJ, Gore MA, Buckler ES, Zhang Z (2012) GAPIT: genome association and prediction integrated tool. Bioinformatics 28: 2397–2399 [DOI] [PubMed] [Google Scholar]
- Liu S, Yeh CT, Tang HM, Nettleton D, Schnable PS (2012) Gene mapping via bulked segregant RNA-Seq (BSR-Seq). PLoS One 7: e36406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F, Fuqua BK, Hasin Y, Yukhtman C, Vulpe CD, Lusis AJ, Pellegrini M (2019) A comparison between whole transcript and 3’ RNA sequencing methods using Kapa and Lexogen library preparation methods. BMC Genomics 20: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrez W, Shin J, Muñoz-Viana R, Figueiredo DD, Trejo-Arellano MS, Exner V, Siretskiy A, Gruissem W, Köhler C, Hennig L (2016) BRR2a affects flowering time via FLC splicing. PLoS Genet 12: e1005924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso N, Freund MK, Johnson R, Shi H, Kichaev G, Gusev A, Pasaniuc B (2019) Probabilistic fine-mapping of transcriptome-wide association studies. Nat Genet 51: 675–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda A, Fujiwara S, Kamada H, Coupland G, Mizoguchi T (2004) Antisense suppression of the Arabidopsis PIF3 gene does not affect circadian rhythms but causes early flowering and increases FT expression. FEBS Lett 557: 259–264 [DOI] [PubMed] [Google Scholar]
- Onouchi H, Igeño MI, Périlleux C, Graves K, Coupland G (2000) Mutagenesis of plants overexpressing CONSTANS demonstrates novel interactions among Arabidopsis flowering-time genes. Plant Cell 12: 885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owens BF, Lipka AE, Magallanes-Lundback M, Tiede T, Diepenbrock CH, Kandianis CB, Kim E, Cepela J, Mateos-Hernandez M, Buell CR, et al. (2014) A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CAC, McMullen MD, Holland JB, Bradbury PJ, et al. (2014) The genetic architecture of maize height. Genetics 196: 1337–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piñeiro M, Coupland G (1998) The control of flowering time and floral identity in Arabidopsis. Plant Physiol 117: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang S, Wang J, Xiong XC, Qu YL, Liu L, Hu CY, Meng YH (2020) Promoting the synthesis of precursor substances by overexpressing hexokinase (Hxk) and hydroxymethylglutaryl-CoA synthase (Erg13) to elevate β-carotene production in engineered Yarrowia lipolytica. Front Microbiol 11: 1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. (2010) Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183 [DOI] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Shin JH, Blay S, McNeney B, Graham J (2006) LDheatmap: an R function for graphical display of pairwise linkage disequilibria between single nucleotide polymorphisms. J Stat Softw Code Snippets 16: 1–9 [Google Scholar]
- Sonah H, O’Donoughue L, Cober E, Rajcan I, Belzile F (2015) Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol J 13: 211–221 [DOI] [PubMed] [Google Scholar]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, et al. (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47: D607–D613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BC, Guan JC, Ding S, Wu S, Saunders JW, Koch KE, McCarty DR (2017) Structure and origin of the white cap locus and its role in evolution of grain color in maize. Genetics 206: 135–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian D, Wang P, Tang B, Teng X, Li C, Liu X, Zou D, Song S, Zhang Z (2020) GWAS Atlas: a curated resource of genome-wide variant-trait associations in plants and animals. Nucleic Acids Res 48: D927–D932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Musetti V, Kim J, Magallanes-Lundback M, DellaPenna D (2004) The Arabidopsis LUT1 locus encodes a member of the cytochrome P450 family that is required for carotenoid ε-ring hydroxylation activity. Proc Natl Acad Sci 101: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toda K, Yang D, Yamanaka N, Watanabe S, Harada K (2002) A single-base deletion in soybean flavonoid 3 -hydroxylase gene is associated with gray pubescence color. Plant Mol Biol 50: 187–196 [DOI] [PubMed] [Google Scholar]
- Wainberg M, Sinnott-Armstrong N, Mancuso N, Barbeira AN, Knowles DA, Golan D, Ermel R, Ruusalepp A, Quertermous T, Hao K, et al. (2019) Opportunities and challenges for transcriptome-wide association studies. Nat Genet 51: 592–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JG, Bradbury PJ, Zhang N, Gibon Y, Stitt M, Buckler ES (2014) Association mapping across numerous traits reveals patterns of functional variation in maize. PLoS Genet 10: e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Chen Q, Wu Y, Lemmon ZH, Xu G, Huang C, Liang Y, Xu D, Li D, Doebley JF, et al. (2018) Genome-wide analysis of transcriptional variability in a large maize-teosinte population. Mol Plant 11: 443–459 [DOI] [PubMed] [Google Scholar]
- Welch BL (1947) The generalization of ‘Student’s’ problem when several different population variances are involved. Biometrika 34: 28–35 [DOI] [PubMed] [Google Scholar]
- Wen Z, Boyse JF, Song Q, Cregan PB, Wang D (2015) Genomic consequences of selection and genome-wide association mapping in soybean. BMC Genomics 16: 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK (2005) GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Z, Hey S, Jubery T, Liu H, Yang Y, Coffey L, Miao C, Sigmon B, Schnable JC, Hochholdinger F, et al. (2020) Shared genetic control of root system architecture between Zea mays and Sorghum bicolor. Plant Physiol 182: 977–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Jiang Y, Wang Z, Gou Z, Lyu J, Li W, Yu Y, Shu L, Zhao Y, Ma Y, et al. (2015) Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol 33: 408–414 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.