Abstract
Molecular engineering of plant immunity to confer resistance against plant viruses holds great promise for mitigating crop losses and improving plant productivity and yields, thereby enhancing food security. Several approaches have been employed to boost immunity in plants by interfering with the transmission or lifecycles of viruses. In this review, we discuss the successful application of clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein (Cas) (CRISPR/Cas) systems to engineer plant immunity, increase plant resistance to viruses, and develop viral diagnostic tools. Furthermore, we examine the use of plant viruses as delivery systems to engineer virus resistance in plants and provide insight into the limitations of current CRISPR/Cas approaches and the potential of newly discovered CRISPR/Cas systems to engineer better immunity and develop better diagnostics tools for plant viruses. Finally, we outline potential solutions to key challenges in the field to enable the practical use of these systems for crop protection and viral diagnostics.
CRISPR-Cas systems unlock the potential of understanding the molecular basis of plant virus interactions, engineering immunity against plant viruses, and developing sensitive and specific diagnostics.
Introduction
The exceptional simplicity, efficacy, versatility, accuracy, and multifunctionality of the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated genes (Cas) (CRISPR/Cas) system has made it an unprecedented tool for genome editing, gene regulation, engineering virus resistance, nucleic acid-targeting, and diagnostics for eukaryotic systems (Jinek et al., 2012; Cong et al., 2013; Malina et al., 2013; Doudna and Charpentier, 2014; Ali et al., 2015a, 2020a; Piatek et al., 2015; Aman et al., 2020a). CRISPR/Cas has been successfully employed to boost immunity and to produce diagnostics for eukaryotic viruses, including plant viruses (Ali et al., 2015b; Ali et al., 2016; Pyott et al., 2016; Aman et al., 2018a; Ali et al., 2020a). Here, we discuss the unrivaled application of CRISPR/Cas systems (Box 1) against phytopathogenic viruses for viral interference and diagnostics. We also describe the development of plant viruses as tools to deliver the CRISPR/Cas machinery into plant cells (Box 2), for targeted genome editing, and thus enhance virus resistance.
Box 1.
CRISPR/Cas system and plant virology
The CRISPR/Cas system originated as a bacterial defense against viruses and therefore has the innate ability to facilitate virus resistance and interference, and it can be harnessed for diagnostics in plant virology. Moreover, the simplicity and diversity of the CRISPR/Cas systems make it an unrivaled toolbox to cope with all aspects of phytopathogenic viruses, such as the direct targeting of viral genomes. Even though they have not been studied thoroughly, both ZFNs and TALENs were shown to partially inhibit the replication of plant viruses, but in vivo viral genome cutting using these nucleases has never been reported (Sera, 2005; Cheng et al., 2015). Similarly, multiplexing for multisite targeting is almost impossible to perform with ZFNs and TALENs. Moreover, both ZFNs and TALENs can only be designed to target DNA sequences and are unable to target the genomes of plant RNA viruses. By comparison, multiple CRISPR/Cas modules can be used to target any genome (DNA or RNA), and it is very simple to perform multiplexing with these systems to target any plant virus or mixed virus infections (Ali et al., 2016; Aman et al., 2018a; Mahas et al., 2019).
Similarly, plant virus management requires an inexpensive, simple, specific, field-deployable virus diagnostic system. However, all currently used diagnostic tools are expensive, complicated, and cannot be deployed in the field by farmers. Even though CRISPR/Cas-based diagnostic tools are still in the development phase, they will soon fulfill all of the requirements of an agriculturally feasible diagnostic system (Aman et al., 2020a).
Virus-based delivery of genome editing machinery and the use of gene targeting reagents have severe consequences in animal systems (Chira et al., 2015). However, the short life spans of plants and the easy removal of viral vector remnants from plants in subsequent generations make plant viral vectors the best choice for delivering genome engineering reagents into plant cells.
Box 2.
Limitations of the current system and new perspectives
CRISPR/Cas9 has been successfully employed for genome editing and to enhance virus resistance. However, this system has some limitations, including its inherent inability to perform some of the required steps in virus resistance in plants and the additional steps or helper systems needed for particular tasks. For example, multisite targeting of the virus genome is required to avoid virus escape, but Cas9 cannot process its sgRNA. Therefore, several promoters or tRNA–sgRNA structures have been employed to target plant virus genomes at multiple sites. Similarly, the PAM requirement for Cas9 is very strict, and Cas9 cannot target ssDNA.
The newly discovered CRISPR/Cas systems offer precise and simple solutions to these problems (preventing viral escape, multiplexing DNA targeting, and even easy viral diagnostics). For example, CRISPR/Cas12 is widely used and much more versatile than CRISPR/Cas9. Specifically, the compact toolkit of Cas12 is well suited for plant virology. Cas12 requires only a short crRNA (making engineering easy), can process polycistronic crRNAs (making multiplexing possible, with no chance of virus escape, allowing multiple genomic loci to be edited at once), targets ssDNAs, and dsDNAs, and degrades ssDNAs via trans activity, Cas12 is comparatively small, and can easily be delivered via deconstructed viral vectors. Similarly, the Type I system can be used to cut the target DNA and to chop the entire viral genome into pieces or completely remove any susceptibility genes from the host genome with Cas3 enzyme.
Similar to viruses, viroids are causing severe diseases in plants. Based on the crRNA processing capability of the CRISPR/Cas13, a well-designed multiplex system to target ssRNA of viroid genome would greatly help to mitigate viroid-based diseases in plants.
Furthermore, the nonspecific degradation of ssDNAs or ssRNAs (reporters) upon recognition of a specific target by Cas12, Cas13, and Cas14 variants provides the opportunity to develop an efficient diagnostic system for deployment in the field. Coupling of the target specificity and nuclease activity of Cas variants with target enrichment (via isothermal amplification, LAMP, RPA) and signal amplification (CONAN or SENSR) has the potential to change the entire scope of plant virus detection and control measures.
Phytopathogenic viruses and viroids cause severe diseases in plants (Kovalskaya and Hammond, 2014; Sastry and Zitter, 2014). Plant viruses have small, highly compact genomes (2–20 kb genome) that encode only a few proteins (replication initiation factors, host immune system silencers, and coat proteins); these proteins allow the virus to use cellular resources to complete its lifecycle (Heinlein, 2015). Viroids, an even simpler type of small infectious phytopathogen, are composed of a 250–430 bases single-stranded circular RNA that encodes no protein but can cause plant diseases (Tsagris et al., 2008). Based on the genome organization, plant viruses are divided into six major types: double-stranded DNA (dsDNA) viruses, single-stranded DNA (ssDNA) viruses, double-stranded RNA (dsRNA) viruses, sense (positive) strand ssRNA (ssRNA+) viruses, and antisense (negative) strand ssRNA (ssRNA−) viruses (Roossinck, 2011). Similarly, plant viruses are named based on their visible disease symptoms and the host from which the virus was first isolated, e.g., Tobacco mosaic virus (TMV) and Tomato yellow leaf curl virus (TYLCV). The current list of disease-causing phytopathogenic viruses includes almost 1,000 viruses (Lefkowitz et al., 2018).
Despite tremendous efforts aimed at prevention, plant viruses cause 10–15% yield losses (60 billion USD) annually by infecting cucurbits like cucumbers (Cucumis sativus), melons (C. melo), cotton (Gossypium hirsutum), beet (Beta vulgaris), beans (Phaseolus vulgaris, P. coccineus, Vicia faba), tomato (Solanum lycopersicum), tobacco, cassava (Nicotiana tabacum), alfalfa (Medicago sativa), sugarcane (Saccharum officinarum), eggplants (S. melongena), flax (Linum usitatissimum) spinach (Spinacia oleracea), squash (Cucurbita maxima, C. pepo), barley (Hordeum vulgare), wheat (Triticum aestivum), maize (Zea mays), petunia (Petunia axillaris), pansy (Viola tricolor), geranium (Geranium aculeolatum), delphinium (Delphinium elatum), oranges (Citrus limon, C. reticulata, C. x paradise), peaches (Prunus persica), apple (Malus domestica), banana (Musa x paradisiaca), tulip (Tulipa x gesneriana), and lily (Lilium candidum; Loebenstein, 2008; Nicaise, 2014). Moreover, severe, uncontrolled viral attacks can cause 100% crop losses in tomato, cotton, beans, and cassava, with disastrous effects on agriculture and food security (Briddon and Markham, 2000; Dovas et al., 2002; Hanssen et al., 2010; Chikoti et al., 2019). Multiple factors including the movement of plants and plant products in the food trade, the use of unhealthy seeds, mixed crop culture practices, vector migration, unexpected vector outbreaks, and vector resistance to pesticides, viral diversity, host range break, the presence of a mixed population of different plant viruses in the field, and the rapid evolution of the viral genome increase the virus/viroid transmission and thus pose great risks to sustainable food production (Loebenstein and Katis, 2014; Moreno and López-Moya, 2020). Conventional methods, i.e., breeding resistant varieties and propagation of virus-free material in nurseries played a major role in food security, but these methods are time-consuming and require tremendous economic investment.
Antibiotics and other chemicals can limit the spread of some phytopathogens (such as bacteria and fungi), but no reagents are currently available to recover virus-infected plants or eliminate viral infection (Cao et al., 2020). Most of the conventional control strategies for plant viruses are based on controlling viral vectors (nematodes, mites, and insects) using pesticides, natural predators, physical barriers, replicative mulches, sticky pads, or UV-absorbing sheets (Bragard et al., 2013). Good agricultural practices, such as selecting virus-free stock/seeds, early sowing, and employing weed and crop management techniques are used to limit viral pathogenicity (Goldbach, 1998; Hilje et al., 2001; Singh and Srivastava, 2020). However, designing long-term strategies to manage plant viruses using conventional approaches remains challenging.
Fighting an invisible foe outside of the cell is challenging. Therefore, strategies to restrict pathogenic attack or target the pathogen inside the cell are good options to avoid diseases in plants. Different molecular approaches have been employed to boost plant immunity against viruses, including molecular breeding of resistance-related quantitative trait loci (QTLs), mutagenesis (Lellis et al., 2002), and the introduction of resistance genes (R genes; Seo et al., 2006) or RNA interference (RNAi) constructs via transgenics (Pooggin et al., 2003; Zhang et al., 2011). However, the lack of information about such QTLs in most crops, the rapid evolution, and diversity of viruses, and the labor-intensive, time-consuming methods needed to introduce plant resistance using these strategies limit their broad-scale application in modern agriculture (Dong and Ronald, 2019). Similarly, the introduction of dominant/recessive genes for viral resistance and RNAi is still in use in some crops (for detailed reviews, see Wang et al., 2012; Hashimoto et al., 2016). However, the inability to transform and regenerate many crop varieties, the emergence of new viral strains, resistance breaks, the lack of specificity, growth penalty issues for RNAi systems, and regulatory constraints limit the widespread use of these techniques in the field.
Even though they have not been fully exploited to target plant viruses, the development of the sequence-specific nucleases zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs) have attracted the attention of plant virologists for their ability to confer direct immunity against phytopathogenic viruses (Sera, 2005; Mahfouz et al., 2011; Chen et al., 2014; Cheng et al., 2015). The use of ZFNs and TALENs provides only partial interference but opens the possibility for the use of any nucleic acid-targeting system against viruses (Sera, 2005; Cheng et al., 2015). The major problem with these systems is that both ZFNs and TALENs are protein-based, and must be assembled for each target independently. For an ever-changing foe such as viruses, designing ZFNs, and TALENs remains challenging.
CRISPR/Cas employs Cas9 as a guided endonuclease to cleave the target DNA in a site-specific manner. Cas9 is guided to the target DNA sequence by a short single guide RNA (sgRNA) containing a 20-nucleotide sequence complementary to the target site. A short NGG sequence (the adjacent protospacer motif, PAM) following the 20-nucleotide target DNA sequence is the only other requirement for the targeting of DNA by CRISPR/Cas9 (Jinek et al., 2012; Cong et al., 2013; Doudna and Charpentier, 2014). Plant scientists have taken advantage of the simplicity and robustness of the CRISPR/Cas9 system to study basic biological questions and have applied this revolutionary technology for crop improvement, including plant protection against pathogenic viruses (Ali et al., 2015a, 2015b, 2016, 2020b; Baltes et al., 2015; Beying et al., 2020).
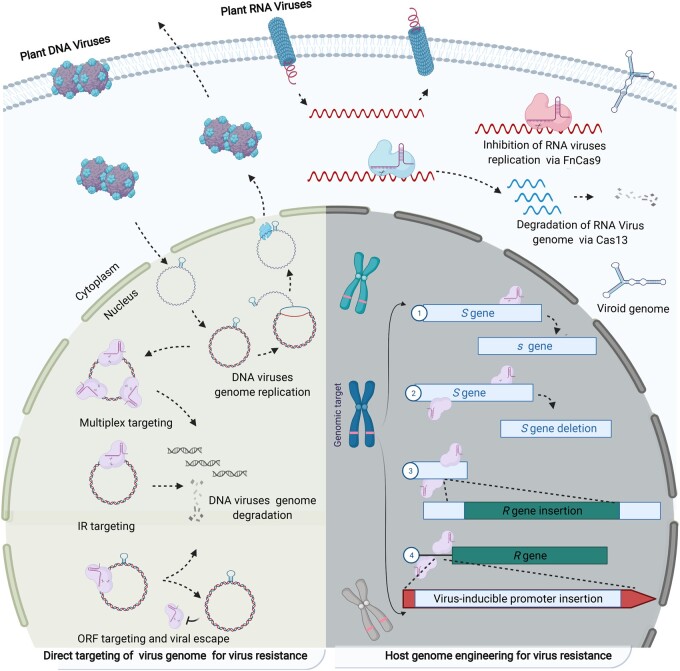
CRISPR/Cas reagents expressed inside the cell can target the viral genome and thus provide resistance against phytopathogenic viruses (Figure 1, Table 1). Even though viral infection is highly efficient and quick, the virus lifecycle provides several opportunities (Dangl et al., 2013) for the use of a CRISPR/Cas-based system to attack the viral genome or interrupt the virus’s lifecycle (Chaparro-Garcia et al., 2015; Chandrasekaran et al., 2016). For example, viruses replicate in the host cell, and their genomes are exposed to the cellular machinery. Moreover, the versatility of the CRISPR/Cas systems and recent discoveries have provided the CRISPR/Cas-based antivirus toolbox with all the gadgets needed to foster robust immunity against pathogenic viruses regardless of their genome composition.
Figure 1.
Schematic of CRISPR/Cas-mediated resistance to plant viruses. Left, CRISPR/Cas-based direct targeting and interference with the genome of phytopathogenic viruses. Multisite targeting or targeting the conserved inverted repeat (IR) region provides resistance by cutting the genome of DNA viruses into multiple pieces inside the plant nucleus. Targeting plant virus genomes at the open reading frame (ORF) sequence can lead to virus escape via evolution of new ORF sequence variants that can complete the virus life cycle but are not targeted by the existing CRISPR/Cas system. Similarly, inhibition of RNA virus genome replication or degradation of the RNA genome was achieved via FnCas9 and Cas13 variants in the cytoplasm. CRISPR/Cas systems can also be designed to target the RNA genomes of viroids. Right, genetic engineering of susceptibility (S) and resistance (R) genes in the host genome can provide durable resistance to multiple viruses. CRISPR/Cas editing of S genes mimics natural resistance (point repair). Removal of S genes and insertion of R genes into neutral, high-expression loci or under the control of strong promoters, including virus-inducible promoters, via homology-directed repair can result in over-expression of R genes for virus resistance.
Table 1.
Summary of CRISPR/Cas-mediated virus resistance
| CRISPR/Cas system used | Targeted plant virus | Plant used | Main observation of the study | References |
|---|---|---|---|---|
| Sp-CRISPR/Cas9 | TYLCV | Nicotiana benthamiana, Tomato (Lycopersicon esculentum) | CRISPR/Cas9 validation for plant virus resistance / Virus genome cutting | (Ali et al., 2015b; Tashkandi et al., 2018) |
| Sp-CRISPR/Cas9 | BSCTV | Nicotiana benthamiana | CRISPR/Cas9 validation for plant virus resistance / Virus genome cutting | (Ji et al., 2015) |
| Sp-CRISPR/Cas9 | BeYDV | Nicotiana benthamiana | CRISPR/Cas9 validation for plant virus resistance / Virus genome cutting | (Baltes et al., 2015) |
| Sp-CRISPR/Cas9 | MeMV | Nicotiana benthamiana | Multiple virus targeting with single sgRNA | (Ali et al., 2016) |
| Sp-CRISPR/Cas9 | BCTV. | Nicotiana benthamiana | Multiple virus targeting with single sgRNA | (Ali et al., 2016) |
| Sp-CRISPR/Cas9 | CLCuMV | Nicotiana benthamiana | Multiplex targeting and virus escape, Replication inhibition | (Ali et al., 2016; Khan et al., 2019; Yin et al., 2019) |
| Sp-CRISPR/dCas9 | ||||
| Sp-CRISPR/Cas9 | ChLCV | Nicotiana benthamiana | Multiplex targeting and virus escape | (Roy et al., 2019) |
| Sp-CRISPR/Cas9 | CaMV | Arabidopsis thaliana | Multiplex targeting and virus escape | (Liu et al., 2018) |
| Sp-CRISPR/Cas9 | endogenous eBSV | banana (Musa spp.) | Multiplex targeting and virus escape | (Tripathi et al., 2019) |
| Sp-CRISPR/Cas9 | WDV | barley (H. vulgare L. cv Golden promise) | Multiplex targeting of overlapping sequences of different genes in the virus genome | (Kis et al., 2019b) |
| Sp-CRISPR/Cas9 | ACMV | Nicotiana benthamiana | virus escape | (Mehta et al., 2019) |
| Fn-CRISPR/Cas9, Fn-CRISPR/dCas9 | TMV | Nicotiana benthamiana | Replication inhabitation of RNA virus | (Zhang et al., 2019) |
| Fn-CRISPR/Cas9, Fn-CRISPR/dCas9 | CMV | Nicotiana benthamiana | Replication inhabitation of RNA virus | (Zhang et al., 2019) |
| Lsh-CRISPR/Cas13a | TuMV | Nicotiana benthamiana, Arabidopsis thaliana | RNA virus genome targeting | (Aman et al., 2018a, 2018b, Zhan et al., 2019) |
| CRISPR/Cas13 | PVY | Potato (S. tuberosum) | RNA virus genome targeting, Multiple strains targeting, broad-spectrum resistance | (Zhan et al., 2019) |
| Lsh-CRISPR/Cas13a | Southern rice black-streaked draft virus (SRBsDV) | Rice (Oryza sativa) | RNA virus genome targeting | (Zhan et al., 2019) |
|
TuMV | Nicotiana benthamiana | Characterization for robust CRISPR/Cas system for RNA virus genome targeting | (Mahas et al., 2019) |
| Sp-CRISPR/Cas9 | Cucumber vein yellow virus | Cucumber (C. sativus L.) | Host genome (eIF4E) modification for broad-spectrum resistance to potyviruses | (Chandrasekaran et al., 2016) |
| Sp-CRISPR/Cas9 | CuVYV), Zucchini yellow mosaic virus (ZYMV) | Cucumber (C. sativus L.) | Host genome (eIF4E) modification for broad-spectrum resistance to potyviruses | (Chandrasekaran et al., 2016) |
| Sp-CRISPR/Cas9 | Papaya ring spot virus-W (PRSV-W) | Cucumber (C. sativus L.) | Host genome (eIF4E) modification for broad-spectrum resistance to potyviruses | (Chandrasekaran et al., 2016) |
| Sp-CRISPR/Cas9 | TuMV | Arabidopsis thaliana | Host genome (eIF4E) modification for resistance to RNA viruses | (Pyott et al., 2016) |
| Sp-CRISPR/Cas9 | Rice tungro spherical virus (RTSV) | Rice (O. sativa) | Host genome (eIFiso4E) modification for broad-spectrum resistance to RNA viruses | (Macovei et al., 2018) |
| Sp-CRISPR/nCas9-base editor | Clover yellow vein virus (ClYVV) | Arabidopsis thaliana | Mimicking natural resistance alleles elF4E (N176K; amino acid substitutions) | (Bastet et al., 2019) |
| Sp-CRISPR/Cas9 | Cassava brown streak virus (CBSV) | Cassava plant (Manihot esculenta Crantz) | Host genome (eIF4E isoforms nCBP‐1 and nCBP‐2) modification for broad-spectrum resistance to RNA viruses | (Gomez et al., 2019) |
| Sp-CRISPR/Cas9 | PVY | Potato (S. tuberosum) | Host genome (Coilin) modification for broad-spectrum resistance to RNA viruses | (Makhotenko et al., 2019) |
| Sp-CRISPR/Cas9 | Soya bean mosaic virus (SMV) | soya bean [Glycine max (L.) Merr.] | Host genome (GmF3H1, GmF3H2, and GmFNSII-1, members of the metabolites Isoflavonoids pathway) modification for broad-spectrum resistance to RNA viruses | (Zhang et al., 2020) |
Direct inhibition of plant DNA viruses
DNA viruses belonging to the Geminiviridae family infect many crops, including cotton, tomato, cassava, tobacco, potato, pepper, watermelon, melon, cowpea, soybean, common bean, mung bean, barley, and banana (Seal et al., 2006). The genomes of geminiviruses are very compact, contain genes encoding only six to seven proteins on both the sense and antisense strands, and are expressed from a bidirectional intergenic region (IR) promoter. Both the IR and open reading frames of protein-coding genes contain important conserved regions essential for the replication/lifecycle of the virus (Chatterji et al., 2000). Any alterations in these conserved regions can interfere with virus replication (Figure 1).
Based on their economic importance, the pioneering work of engineering CRISPR/Cas-mediated virus interference in plants began with virus inhibition strategies against geminiviruses (genus Begomoviruses; Ali et al., 2015b, 2016; Baltes et al., 2015; Ji et al., 2015). The CRISPR/Cas9 system was programmed to boost the molecular immunity of Arabidopsis thaliana and N. benthamiana against TYLCV, Beet severe top curly virus (BSCTV), and Bean yellow dwarf virus (BeYDV), belonging to three different genera: Begomovirus, Curtovirus, and Mastervirus. All three studies focused on the same important regions of the virus: Rep, coat proteins, and the IR harboring the conserved nine-nucleotide (TAATATTAC) replication initiation sequence. Baltes et al. (2015) and Ji et al. (2015) both used T-DNA plasmids to express Cas9 and sgRNA to target BSCTV and BeYDV. Virus targeting was first confirmed transiently by infiltrating the plants with Agrobacterium containing the CRISPR/Cas9 expression cassette. Next, CRISPR/Cas9-expressing transgenic plants were regenerated and progeny plants were challenged with BSCTV and BeYDV (Baltes et al., 2015; Ji et al., 2015, 2018). Plants expressing CRISPR/Cas9 against BSCTV and BeYDV developed no viral symptoms, and the accumulation of the viral genome decreased by 80%. A modified version of a virus-inducible virus targeting system was subsequently developed using a viral promoter to express the CRISPR/Cas9 machinery against BSCTV. This system provides highly efficient virus resistance, as any replication of BSCTV inside the cell simultaneously induces the CRISPR/Cas9 machinery against BSCTV (Ji et al., 2018).
Ali et al. (2015b) employed another interesting combinatorial approach to boost plant immunity against plant viruses via CRISPR/Cas9. In this approach, the sgRNA targeting TYLCV was systemically delivered via the well-established Tobacco rattle virus (TRV) into Cas9-expressing plants. After establishing CRISPR/Cas9-mediated immunity (7 d), plants were challenged with TYLCV. Sequencing of the targeted TYLCV genomic fragments (IR, RCRII, and CP [encoding coat protein]) revealed typical Cas9 cutting signatures. However, the robust inhibition of TYLCV genome accumulation and reduced TYLCV symptoms were observed only by targeting the conserved regions of the IR and Rep (RCRII, metal ion binding motif) compared to CP-targeted plants. In plants, the cutting of target DNA by CRISPR/Cas9 is generally followed by nonhomologous end joining-based DNA repair, leading to random insertions and deletions (indels) at the DNA target site (Ali et al., 2015b). Ali et al. (2015b) reasoned that indel formation in the essential conserved regions, IR, RCRII, and RCRIII (Rep), renders the TYLCV genome unable to replicate. However, indels in the CP sequence lead to variants that replicate properly.
Indels in a nonconserved sequence generated via CRISPR/Cas9 may lead to the escape of mutated virus from the existing CRISPR/Cas9, as the newly formed sequence (with indels) is no longer complementary to the sgRNA and cannot be digested by Cas9. In a follow-up study, the viral escape mechanism was evaluated in detail and demonstrated that CRISPR/Cas9-mediated targeting of the CP induces indels in the CP sequence that can create variants of intact CP still capable of packing viral DNA (Ali et al., 2016). Also, it was observed that an sgRNA targeting the conserved stem-loop nine-nucleotide (TAATATTAC) sequence of TYLCV was capable of targeting multiple geminiviruses, including Merremia mosaic virus (MeMV) and Beet curly top virus (BCTV; Ali et al., 2016).
Targeting the viral genome at multiple loci by CRISPR/Cas9 can eliminate the possibility of virus replication (Figure 1). Ali et al. (2016) coupled tRNA-sgRNA (Xie et al., 2015) with the TRV system to express three independent sgRNAs targeting the virus genome at three different loci. This multiplex approach led to enhanced resistance in the targeted plants, which were free of any TYLCV symptoms (Ali et al., 2015b, 2016). Later effectiveness of the multiplex targeting (IR, Rep, and CP) approach was confirmed by increase resistance against Cotton leaf curl Multan virus (CLCuMV; Yin et al., 2019), Chilli leaf curl virus (ChLCV; Roy et al., 2019), and against dsDNA of the pararetrovirus Cauliflower mosaic virus (CaMV; Liu et al., 2018).
ZFNs and TALENs inhibit viral replication without cutting the viral genome (Sera, 2005; Cheng et al., 2015). Inhibition of plant viruses without cutting the genome is also quite feasible using the CRISPR/Cas system. A mutation in the nuclease domain renders Cas9 unable to cut DNA, but dCas9-sgRNA still possesses a high binding affinity to the target site. Khan et al. (2019) produced transgenic plants expressing dCas9-sgRNA to demonstrate the inhibition of CLCuMV replication without genome cutting. However, compared to the efficient inhibition by wild-type Cas9, targeting of the dCas9-sgRNA to the IR provides only partial protection against CLCuMV (Khan et al., 2019).
After successfully producing CRISPR/Cas9-based immunity against phytopathogenic viruses in model plants, various groups used CRISPR/Cas9 to generate viral immunity in crop plants, including tomato, cassava, barley, and banana. Tashkandi et al. (2018) produced transgenic tomato plants expressing CRISPR/Cas9 machinery against TYLCV (Tashkandi et al., 2018). Similarly, multiplex CRISPR/Cas9-based immunity was engineered in barley against Wheat dwarf virus (WDV; Kis et al., 2019a). CRISPR/Cas9 efficiently targeted the IR sequences of TYLCV and WDV in tomato and barley, and the transgenic plants were free of viral symptoms. Mehta et al. (2019) generated transgenic cassava plants expressing the CRISPR/Cas9 machinery to target the coding regions of transcription activator protein (AC2) and replication enhancer protein (AC3) in African cassava mosaic virus (ACMV). Similar to observations by Ali et al. (2016) for CLCuMV and Liu et al. (2018) for CaMV, targeting of the coding sequences of ACMV by CRISPR/Cas9 resulted in the generation of ACMV variants that were capable of replication and infection (Mehta et al., 2019). These findings (Table 1) are critical for designing virus resistance strategies and applying the CRISPR/Cas system to plant virology.
Plant viruses genome rarely integrate into the host plant genome. However, the genomes of pararetroviruses such as Banana streak virus (BSV) integrate into the genome of banana, generating an endogenous BSV called eBSV. These integrated viral genomes remain a continuous threat, as eBSV can be activated at any stage in the plant lifecycle. Tripathi et al. (2019) designed a CRISPR/Cas9 system to inactivate the eBSV genome present in the banana genome. Regenerated banana plants expressing CRISPR/Cas9 against the eBSV genome were produced, demonstrating the efficient targeting of the eBSV genome and mitigating the potential risks of eBSV activation (Tripathi et al., 2019).
Direct inhibition of plant RNA viruses
RNA viruses are the most abundant phytopathogens worldwide, causing devastating diseases in cultivated plants. The genomes of plant RNA viruses are composed of positive-strand ssRNA, negative-strand ssRNA, or dsRNA. Multiple CRISPR/Cas system variants (Fn-CRISPR/Cas9 and CRISPR/Cas13) have the potential to increase plant immunity against RNA viruses (Figure 1, Table 1). Zhang et al. (2018) produced N. benthamiana and Arabidopsis plants expressing FnCas9, FndCas9 (nuclease inactive), and multiple sgRNAs to target the RNA genomes of TMV and Cucumber mosaic virus (CMV). Interestingly, both FnCas9 and its modified dead version FndCas9 provided efficient resistance against both TMV and CuMV, suggesting that the binding of FnCas9 or FndCas9 to the RNA genomes of TMV and CuMV is sufficient to block viral replication in plant cells (Zhang et al., 2018). Aman et al. (2018a, 2018b) engineered a multiplexed CRISPR/Cas13 system to cut viral genome and provide resistance against Turnip mosaic virus (TuMV) and TMV in N. benthamiana and Arabidopsis plants. The successful targeting of the RNA genomes of viruses in model plants (Aman et al., 2018a, 2018b) paved the way for applying this powerful technology to crop plants to increase resistance to viruses. Zhan et al. (2019) successfully used the CRISPR/Cas13 system to efficiently inhibit the replication of Potato virus Y (PVY) in potato (Zhan et al., 2019). Similarly, CRISPR/Cas13 was used to generate transgenic rice resistance to Southern rice black-streaked draft virus and Rice stripe mosaic virus (Zhang et al., 2019).
Plant RNA viruses are highly diverse, and their genomes evolve rapidly. Thus, it is crucial to develop a CRISPR/Cas system capable of providing efficient, broad-spectrum resistance to RNA viruses. The Type VI CRISPR/Cas13 system is divided into four subtypes (A–D) based on the phylogeny of Cas13 variants (O'connell, 2019). Some Cas13 variants have more robust catalytic activity and specificity than the well-known LshCas13a (Abudayyeh et al., 2017; Cox et al., 2017; Gootenberg et al., 2017; Konermann et al., 2018; Yan et al., 2018). Mahas et al. (2019) characterized a set of Cas13 family members (LwaCas13, LwaCas13a, PspCas13b, and Cas13d) in planta to identify a robust system for RNA virus interference in plants. Most of the Cas13 variants provided resistance against plant RNA viruses, but CasRx (CRISPR/Cas13d) conferred robust resistance against the RNA virus TuMV (Mahas et al., 2019). Interestingly, nuclease-inactive (dCasRx)-based targeting did not mediate virus interference, indicating that in contrast to dCas9-sgRNA and FnCas9-sgRNA (Sera, 2005; Zhang et al., 2018), binding of only dCasRx-crRNA complex to viral genomic RNA is not sufficient to interfere with virus replication. In the same study, multiplexing of CasRx-crRNAs to target two different RNA viruses was successfully used to restrict the accumulation of the genomes of both viruses in plant cells (Mahas et al., 2019).
All Cas13 variants upon digestion of the specific viral RNA target, the activated Cas13 nonspecifically degrades cellular RNA transcripts, representing an essential component of bacterial immunity to restrict further viral spread via programmed cell death (Meeske et al., 2019; O'connell, 2019). Interestingly, such nonspecific promiscuous degradation of cellular RNA transcripts in trans has not been observed in planta. Still, the underlying molecular mechanism must be experimentally confirmed. Perhaps accessory/helper factors or structures with a specific confirmation enable Cas13 variants to promiscuously digest their nonspecific targets in bacterial cells in trans, but such factors are absent in plants, and Cas13 variants only target viral genomes for degradation (Mahas et al., 2019). The ability of CRISPR/Cas13 to precisely cut the target viral RNA while not disturbing cellular transcripts are of great importance for providing crops with an efficient immune system against RNA viruses.
CRISPR/Cas-mediated host genome editing for virus resistance
The genomes of plant viruses encode only a few proteins, including proteins important for suppressing the host immune system and initiating viral replication, coat proteins for genome encapsidation, and movement protein for systemic infection of the plant body. To replicate and complete their lifecycles, viruses hijack the host’s cellular machinery to facilitate the infection process (Dong and Ronald, 2019). Most of these host factors function as susceptibility factors (S factors) for plant viruses (Van Schie and Takken, 2014). Modifying these S factors (Figure 1) to limit their availability or to prevent them from interacting with the viral lifecycle can mitigate the pathogenicity of viruses in plants (Dong and Ronald, 2019). The first recessive virus-resistant mutants were discovered in Arabidopsis and demonstrated that the potyvirus Tobacco etch virus could not infect plants containing the eIFiso4E (Eukaryotic initiation factor) variant (Lellis et al., 2002). Such recessive resistance to different plant RNA viruses was subsequently observed in many resistant cultivars of pepper, melon, lettuce, the wild tomato S. habrochaites, rice, and barley. For a detailed review, see Hashimoto et al. (2016). These observations prompted all of the initial efforts to modify the host genome for virus resistance to focus on eIF4E, eIFiso4E, and eIF4G. Chandrasekaran et al. (2016) used a transgene-free approach to deliver CRISPR/Cas9 to cucumber targeting eIF4E. The eIF4E-mutated (elf4e) cucumber plants showed efficient resistance to Cucumber vein yellow virus, Zucchini yellow mosaic virus, and Papaya ring spot virus-W (Chandrasekaran et al., 2016). Using the same approach, elF4E in Arabidopsis and eIFiso4E in rice were mutated for resistance against TuMV and Rice tungro spherical virus respectively (Pyott et al., 2016; Macovei et al., 2018). Similarly, the inactivation of the eIF4E isoforms nCBP‐1 and nCBP‐2 by CRISPR/Cas9 reduced susceptibility to Cassava brown streak virus in cassava plants (Gomez et al., 2019).
Mimicking natural resistance alleles (amino acid substitutions) where the original physiological functionality of elF4E is not affected but the engineered elF4E (N176K) allele (Gao et al., 2004) can provide broad-spectrum virus resistance, are preferred in plant biotechnology (Zaidi et al., 2020). Bastet et al. (2019) applied the CRISPR/Cas9-coupled base editing approach to convert the wild-type susceptible elF4E allele to the virus-resistant elF4E (N176K) allele. The specific conversion of N176K conferred resistance against Clover yellow vein virus in Arabidopsis (Bastet et al., 2019).
In addition to eIF4E, several other host factors were identified as recessive susceptibility factors for plant viruses. Coilin, encoding a signature protein of subnuclear structures Cajal bodies mediate susceptibility to PVY (Shaw et al., 2014; Love et al., 2017). Makhotenko et al. (2019) delivered ribonucleoprotein complex (Cas9-sgRNA) to the apical meristem of potato via biolistics, to mutate the Coilin locus in the genome. Interestingly, editing of at least one allele of Coilin in the tetraploid genome of potato was sufficient to provide resistance against PVY (Makhotenko et al., 2019).
Secondary metabolites play a critical role in plant-microbe interactions, including virus resistance (Mishra et al., 2020). Particularly, accumulation of isoflavones contents resulting from the Glycine max isoflavone synthase (GmIFS’s) polymorphisms is considered to be associated with Soybean mosaic virus (SMV) resistance in soya bean (Cheng et al., 2010). To demonstrate a direct connection of isoflavones content and SMV resistance in soybean, Zhang et al. (2020) employed CRISPR/Cas9‐mediated multiplex gene‐editing. Simultaneous targeting of GmF3H1, GmF3H2, and GmFNSII‐1 doubled the leaf isoflavone content, and reduced the SMV by one‐third, confirming the direct relationship of the increased isoflavone content to SMV resistance in soybean (Zhang et al., 2020).
CRISPR/Cas and viral diagnostics in plants
Early detection of a pathogen can give growers enough time to devise a disease prevention strategy. Molecular techniques such as PCR and RT-PCR and field-based isothermal techniques such as LAMP, RT-LAMP, RPA, and NASBA are commonly used to detect phytopathogens (Szemes et al., 2002; Lu et al., 2018; Wilisiani et al., 2019; Edgü et al., 2020; Wang et al., 2020b). However, these techniques are time-consuming, expensive, have a high ratio of nonspecificity or low sensitivity, and cannot be applied to bulk plant samples in the field. Simple, precise, inexpensive, rapid, large-scale, field-deployable diagnostic tools are still needed to detect phytopathogens in crops in the field.
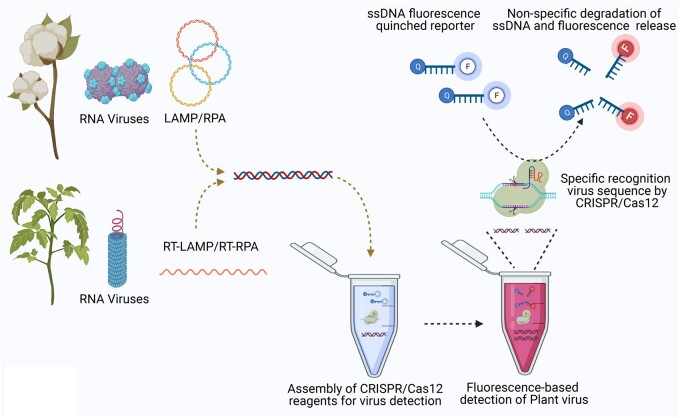
The use of CRISPR/Cas systems as sequence-specific nucleases revolutionized the field of genome editing (Doudna and Charpentier, 2014), but also, techniques based on the ability of CRISPR/Cas variants to promiscuously degrade any ssDNA or ssRNA in the vicinity following its specific target recognition and digestion (Chertow, 2018) brought the field of molecular diagnostics to the next level (Wang et al., 2020a). In the past 5 years, more than 100 different CRISPR/Cas-based molecular diagnostic strategies were reported to detect nucleic acids, including plant viruses. After cutting the target DNA, Cas12a nonspecifically degrades the reporter ssDNA (Chen et al., 2018). In most cases, ssDNA (fluorescence quenched) reporter upon nonspecific degradation by Cas12, releases the fluorescent signal for visual detection (Kellner et al., 2019; Ali et al., 2020a). Like Cas12, most Cas13, and Cas14 variants possess collateral activity against ssDNA or ssRNA in trans. Variants of Cas12, Cas13, and Cas14 have been harnessed to develop efficient nucleic acid diagnostic platforms such as DETECTR, SHERLOCK, HOLMES, HOLMESv2, CaT-SMelor, iSCAN, and SHINE for specific and highly sensitive (up to attomole) detection of nucleic acids and small molecules (Gootenberg et al., 2017; Li et al., 2018; Kellner et al., 2019; Li et al., 2019; Liang et al., 2019; Ali et al., 2020a; Arizti-Sanz et al., 2020; Broughton et al., 2020). The impressive advancements in molecular diagnostics via CRISPR/Cas prompted plant scientists to apply these techniques as specific and sensitive diagnostic platforms for plant virus detection. Specifically, the coupling of the isothermal amplification assays LAMP and RPA and reverse transcription techniques to enrich target and visual detection systems (lateral flow strips, florescence detection systems) with CRISPR/Cas can provide an inexpensive, field-deployable diagnostic system (Figures 2 and 3) for plant viruses (Mahas et al., 2021).
Figure 2.
Field-deployable CRISPR/Cas-based detection of plant viruses. Nucleic acid (DNA, or RNA) isolated from plants is subjected to isothermal amplification (loop-mediated isothermal amplification, LAMP or recombinase polymerase amplification, RPA for DNA viruses or reverse transcriptase coupled loop-mediated isothermal amplification, RT-LAMP, reverse transcriptase coupled recombinase polymerase amplification, RT-RPA for RNA viruses). Upon specific recognition and digestion of virus nucleic acid, Cas12a nonspecifically degrades an ssDNA reporter and releases the quenched fluorescence. End-point fluorescence (color change) is visualized with the naked eye using a P51 fluorescence lightbox.
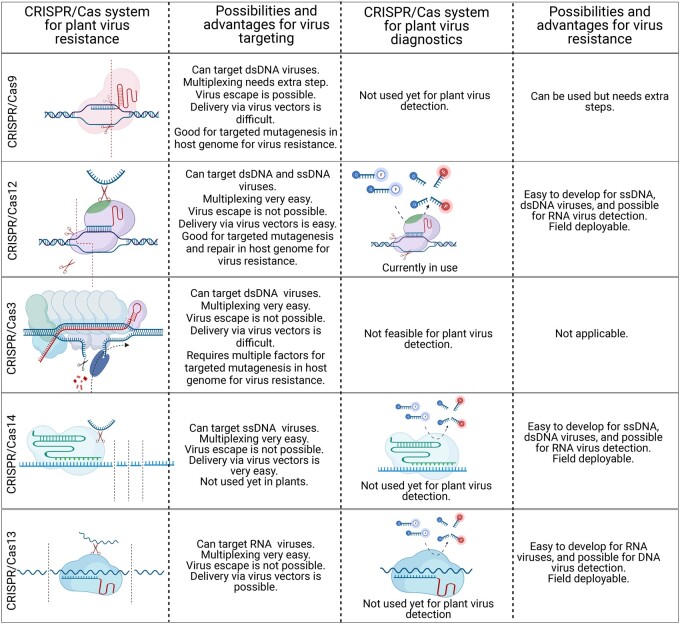
Figure 3.
Applications of CRISRP/Cas systems comparison of CRISPR/Cas systems for virus resistance and diagnostics. Targeting and digestion site pattern in the dsDNA or ssDNA and RNA are shown by small dashed lines. For each system, the possibilities and advantages are described. For plant virus diagnostics, the fluorescence detection method is presented.
Early detection of RNA viruses and viroids in apple can reduce financial losses when combined with viral spread prevention strategies. Jiao et al. (2021) developed the first inexpensive, field-deployable CRISPR/Cas12a based diagnostic system to detect the prevalent RNA viruses/viroids of apple. A CRISPR/Cas12a‐based platform was designed to detect multiple apple viruses, including Apple necrotic mosaic virus, Apple stem pitting virus, Apple stem grooving virus, Apple chlorotic leaf spot virus, and Apple scar skin viroid (Cho et al., 2016). Using reverse transcription‐recombinase polymerase amplification (RT‐RPA), the target viral RNA is first converted to DNA, and the amplified DNA is then detected using the CRISPR/Cas12a detection system. This visual detection system is based on a linear linker ssDNA and oligonucleotide‐conjugated gold nanoparticles. In a positive sample, upon target recognition, Cas12a nonspecifically degrades the linker‐ssDNA to prevent aggregation of DNA1/2‐AuNPs (there is no possibility of cross-linked hybridization), and the solution remains red (positive result). However, in the absence of the viral target (no activity of Cas12a and no degradation of linker‐ssDNA), the intact linker-ssDNA crosslinks with the complementary ssDNA conjugated to gold nanoparticles. This results in the aggregation of DNA and gold nanoparticles, and solution becomes colorless (negative result; Jiao et al., 2021). The only drawback of this system is its relatively low sensitivity (250 viral copies per reaction), as it would not be possible to detect viral attack before a certain incubation period in the field.
To make the plant virus detection easier, recently Aman et al. (2020b) developed a much simpler but sensitive CRISPR/Cas12a-based assay to detect plant RNA viruses. The original in vitro Specific CRISPR-based Assay for Nucleic acids detection (iSCAN; Ali et al., 2020a) protocol was modified by replacing RT-LAMP with a sensitive RT-RPA technique to develop a one-pot assay named iSCAN one-pot (iSCAN-OP) for the detection of Potato virus X (PVX), PVY, and TMV. With iSCAN-OP a very low titer of the virus (up to 10 pM) can be easily detected within 15–20 min in a single tube. Moreover, the presence of the virus (positive read-out) is visible to the naked eye (Aman et al., 2020b). The iSCAN-OP detection system can be used for large-scale, reliable, affordable, and rapid screening of plant viruses in the field.
Selection of disease-free seeds and early detection of virus at nursery level is important for sustainable agriculture. Very recently, Mahas et al. (2021) developed an inexpensive LAMP-coupled CRISPR–Cas12a module for rapid and sensitive detection of plant DNA viruses. This in field, easy-to-interpret visual readout CRISPR–Cas12a based detection module was shown to detect TYLCV and Tomato leaf curl New Delhi virus (ToLCNDV) in less than 1 h (Mahas et al., 2021). These molecular diagnostics systems are low-cost, field suitable, and can be used in any point-of-use applications.
The development of such systems is just the beginning (Figure 3). Fozouni et al. (2021) recently developed a new CRISPR/Cas13a diagnostic method that does not require pre-amplification of the target sequence, and the results can be evaluated using a smartphone (Fozouni et al., 2021). Adopting, the amplification-free method could make a truly field-deployable plant virus detection system.
Plant viruses as CRISPR/Cas delivery tools for virus targeting and resistance
Apart from their devastating effects on plant health, the programmability of plant virus nucleoprotein nanostructures allows these natural foes to be converted into efficient tools to deliver DNA, RNA, proteins, medicines, and reagents of agricultural importance into living cells (Chira et al., 2015; Abrahamian et al., 2020; Kalinina et al., 2020; Varanda et al., 2021; DeHart and Kamrud, 2018). In a scenario where antibiotics and chemicals cannot be used against plant viruses, but CRISPR/Cas machinery can confer virus resistance in plants, the delivery of the CRISPR/Cas system via viruses has several advantages. Beginning with the work of Baltes et al. (2014; DNA replicons) and Ali et al. (2015a), (TRV, RNA virus) now more than 30 different viral vectors (Table 2 and Box 3) have been designed for the delivery of CRISPR/Cas machinery into plant cell for multiple purposes (for detailed reviews, see Puchta, 2002; Wigge, 2011; Fauser et al., 2012; Puchta and Fauser, 2013; Gover et al., 2014; Lozano-Durán, 2016; Zhang et al., 2016; Schmidt et al., 2019; Van Vu et al., 2019; Liu and Zhang, 2020; Ma et al., 2020; Varanda et al., 2021). Here, we discuss only the pioneering studies, major milestones, and potential applications of some of these vector systems (for viral vectors expressing the whole CRISPR/Cas machinery see Box 3 and Table 2) to deliver the CRISPR/Cas machinery into plant cells for virus resistance in plants.
Table 2.
Summary of virus vectors for CRISPR/Cas machinery delivery into plant
| Plant virus vector | Genome | Target plant | Main goal of virus-based delivery | Reference/s |
|---|---|---|---|---|
| BeYDV replicons | DNA |
|
Virus replicons development for CRISRP/Cas system delivery, for host genome editing and HDR-based repair, gene targeting | (Baltes et al., 2014; Butler et al., 2016; Čermák et al., 2017; Dahan-Meir et al., 2018) |
| Cabbage leaf curl virus (CaLCuv) | DNA | Nicotiana benthamiana | VIGE (Virus induce genome editing), systemic delivery of sgRNA to increase the recovery of edited regenerated plants | (Yin et al., 2015) |
| WDV | DNA |
|
Virus replicons development for CRISRP/Cas system delivery and HDR based repair | (Čermák et al., 2017; Wang et al., 2017) |
| Cotton leaf crumple virus (CLCrV) | DNA | Arabidopsis thaliana | sgRNAs delivery stem apical meristem and germline cells | (Lei et al., 2021) |
| TRV | RNA | Nicotiana benthamiana | Systemic delivery of sgRNA, sgRNA-FT, sgRNA-tRNA to invade germline cells to increase the recovery of edited plants, to obviate tissue culture | (Ali et al., 2015a, 2015b, 2016; Aman et al., 2018a, 2018b; Mahas et al., 2019; Ellison et al., 2020) |
| PEBV | RNA | Nicotiana benthamiana, Arabidopsis thaliana | Systemic delivery of sgRNA to increase the recovery of edited plants | (ALi et al., 2018) |
| TMV | RNA | Nicotiana benthamiana | To enhance systemic delivery of sgRNA to increase the recovery of edited plants, sgRNAs processing | (Cody and Scholthof, 2020) |
| Beet necrotic yellow vein virus (BNYVV) | RNA | Nicotiana benthamiana | Multiple sgRNAs delivery | (Jiang et al., 2019) |
| Foxtail mosaic virus | RNA | Foxtail (Setaria viridis), Maize (Z. mays) | sgRNAs delivery to monocots | (Mei et al., 2019) |
| Barley yellow striate mosaic virus (BYSMV) | RNA | Nicotiana benthamiana | Delivery of the whole CRISPR/Cas system to plant tissue to increase the recovery of foreign DNA free genome-edited plants | (Ma et al., 2020) |
| Sonchus yellow net rhabdovirus (SYNV) | RNA | Nicotiana benthamiana | Delivery of the whole CRISPR/Cas system to plant tissue to increase the recovery of foreign DNA free genome-edited plants | (Ma et al., 2020) |
| PVX | RNA | Nicotiana benthamiana | Delivery of tandemly arranged sgRNAs for simultaneous multisite genome editing and the whole CRISPR/Cas system to plant tissue to increase the recovery of foreign DNA free genome-edited plants | (Ariga et al., 2020; Uranga et al., 2021) |
Box 3.
Virus vectors for CRISPR/Cas system delivery into plants
Plant virus-based vectors known as replicons (Gover et al., 2014; Lozano-Durán, 2016; Liu and Zhang, 2020; Ma et al., 2020) were developed to deliver whole CRISPR/Cas9 machinery or only sgRNA (Cabbage leaf curl virus) machinery into tobacco and potato, leading to high rates (75%–85%) of targeted mutagenesis and herbicide resistance (Baltes et al., 2014; Yin et al., 2015; Butler et al., 2016).
Mimicing natural resistance (s alleles, loss-of-function mutations in susceptibility factors) and to avoid silencing, insertion of R factors or virus resistance QTL into safe harbors via gene targeting (homology-directed repair) is important to create elite crop varieties (Puchta, 2002; Puchta and Fauser, 2013; Van Vu et al., 2019). Geminivirus replicons were developed to enhance the frequency of repair 1.5- to 9-fold in tomato, tobacco, M. truncatula, wheat, and barley (Fauser et al., 2012; Čermák et al., 2017; Schmidt et al., 2019). Similarly, the BeYDV replicon system (Baltes et al., 2014) was modified to make freely available repair template (Fauser et al., 2012), this CRISPR/Cas9-based recovered gene targeting events at the CRTISO and PSY1 loci in 25% of T0 tomato plants (Dahan-Meir et al., 2018).
Germline cells genome engineering can increase the recovery of modified plants in the next generation. Ellison et al. (2020) modified the TRV-based sgRNA expression system (Ali et al., 2015a; Ellison et al., 2020. Lei et al. (2021) modified Cotton leaf crumple virus replicons by fusing an mRNA FT-translocation signaling motif (Wigge, 2011) or tRNA (Zhang et al., 2016) to the 3' end of sgRNA to enhance its translocation into germline cells (Lei et al., 2021).
Very recently, simple mechanical inoculation of PYX and negative-strand RNA viruses vectors were used to deliver sgRNAs for simultaneous multisite genome editing or the whole CRISPR/Cas machinery into plant tissue to increase (up to 90%) the recovery of foreign DNA free genome-edited plants (Ariga et al., 2020; Ma et al., 2020; Uranga et al., 2021). Mechanical inoculation could be used to directly introduce constructs harboring new S and R genes for virus resistance into mature plants such as fruit trees to regenerate foreign DNA-free plants. The direct recovery of genome-edited seeds will eliminate the need for time-consuming tissue culture and can enhance the speed of crop breeding for virus resistance and could potentially reshape agriculture.
The RNA virus, TRV is widely used to deliver (200–500) stretches nucleic acid into plant cells (for detailed reviews, see Creager et al., 1999; Ratcliff et al., 2001; Senthil-Kumar and Mysore, 2011; Ali et al., 2015a). The delivery of short RNA, such as sgRNAs systemically to the entire plant provides multiple opportunities. In their pioneering study, Ali et al. (2015a) successfully engineered TRV2 using a short RNA-dependent RNA polymerase promoter from the related RNA virus Pea early browning virus (PEBV) to express sgRNA with a proper 5' end to target the TYLCV genome. The systemic targeting of TYLCV by Cas9-sgRNA resulted in a high rate of TYLCV genome editing, blocked the spread and accumulation of virus genome, and led to the recovery of N. benthamiana plants devoid of any TYLCV, CLCuMV, MeMV, and BCTV symptoms (Ali et al., 2015b, 2016).
Next, TRV-based sgRNA expression for CRISPR/Cas13-based was engineered to restrict the replication of RNA viruses like TuMV in N. benthamiana (Aman et al., 2018a). Similarly, TRV-based sgRNA system was adopted to identify a robust Cas13 ortholog for RNA virus targeting. The use of TRV vectors readily identified Cas13d from Ruminococcus flavefaciens XPD3002 (CasRx) as an efficient RNA virus targeting protein (Mahas et al., 2019) compared to Cas13a from Leptotrichia wadei (LwaCas13a), Cas13b from Bergeyella zoohelcum (BzCas13b), and Cas13b from Prevotella sp. P5-125 (PspCas13b).
Conclusion
The CRISPR/Cas system is currently the only molecular tool used to cut the viral genome inside plant cells without perturbing the biochemical functionality of the cell (Ali et al., 2015a; Baltes et al., 2015; Ji et al., 2015). Plant replicons have the potential to deliver CRISPR/Cas-based machinery into mature plants to fight against pathogenic viruses. As an alternative to the direct targeting of viral genomes, the CRISPR/Cas system can be used to modify host factors to provide cellular immunity against plant viruses. The recently discovered CRISPR/Cas variants, i.e., CRISPR (Cas3, 12, 13, 14) are offering the accurate tools that can cover all aspects including, virus resistance and field-deployable diagnostics. Similarly, modifications (base editing, EvolvR, prime editing) to the existing CRISPR/Cas9 system, and Replicon-based delivery of CRISPR/Cas to cut and repair a host factor to mimic the natural resistance allele or to insert an R factor into a highly expressing locus in the genome, can provide efficient resistance against plant viruses.
As a whole, CRISPR/Cas system-based nucleic acid-targeting and detection represents the best toolbox in the field of plant virology. In the Outstanding Questions Box, we describe some challenges faced when using the CRISPR/Cas system in the field. Successfully overcoming these challenges could lead to the development of efficient resistance against plant viruses in the field.
Advances
CRISPR/Cas systems can boost plant immunity by directly targeting the viral genome (DNA or RNA) and have the potential to provide resistance against multiple pathogenetic viruses (mixed population in the field).
CRISPR/Cas systems can be used to help fully grown plants recover from virus symptoms and pathogenicity.
CRISPR/Cas systems can modify host genes to increase recessive resistance against pathogenic viruses.
A CRISPR/Cas-based field-deployable plant virus diagnostic system is now feasible.
Plant virus-based vectors can deliver genome modification reagents for efficient mutagenesis and gene targeting and delivery through RNA virus vectors can generate nontransgenic genetically modified elite crops.
Outstanding questions
How can CRISPR/Cas systems be delivered into mature plants (vaccination) such as field crops or fruit trees to mitigate virus attack?
What is the best CRISPR/Cas system for providing efficient, broad-spectrum resistance to viruses in the field?
How can viral escape be controlled?
How can CRISPR/Cas systems be used to identify additional susceptibility (S) factors to be modified for broad-spectrum viral resistance?
How can resistance genes (R) be created or mimicked to produce virus-resistant elite crop varieties?
How can genome-modified, virus-resistant elite crop varieties be created without the need for tissue culture?
How should genome editing, field trails, and innovations be regulated?
Acknowledgments
We would like to thank members of the genome engineering and synthetic biology laboratory for insightful discussions and technical support. This work was supported by baseline funding from the KAUST to M.M.M.
Funding
This work was supported by CRG # URF/1/4029-01-01 from KAUST.
Conflict of interest statement. The authors declare no conflict of interest.
Senior author. Z.A. and M.M.M. wrote the article.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Magdy M. Mahfouz (magdy.mahfouz@kaust.edu.sa).
References
- Abrahamian P, Hammond RW, Hammond J (2020) Plant virus-derived vectors: applications in agricultural and medical biotechnology. Annu Rev Virol 7: 513–535 [DOI] [PubMed] [Google Scholar]
- Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. (2017) RNA targeting with CRISPR-Cas13. Nature 550: 280–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Abul-Faraj A, Li L, Ghosh N, Piatek M, Mahjoub A, Aouida M, Piatek A, Baltes NJ, Voytas DF, et al. (2015a) Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol Plant 8: 1288–1291 [DOI] [PubMed] [Google Scholar]
- Ali Z, Abulfaraj A, Idris A, Ali S, Tashkandi M, Mahfouz MM (2015b) CRISPR/Cas9-mediated viral interference in plants. Genome Biol 16: 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Ali S, Tashkandi M, Zaidi SS-E-A, Mahfouz MM (2016) CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Sci Rep 6: 26912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Aman R, Mahas A, Rao GS, Tehseen M, Marsic T, Salunke R, Subudhi AK, Hala SM, Hamdan SM, et al. (2020a) iSCAN: An RT-LAMP-coupled CRISPR-Cas12 module for rapid, sensitive detection of SARS-CoV-2. Virus Res 288: 198129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Z, Eid A, Ali S, Mahfouz M (2018) Pea early-browning virus-mediated genome editing via the CRISPR/Cas9 system in Nicotiana benthamiana and Arabidopsis. Virus Res 244: 333–337 [DOI] [PubMed] [Google Scholar]
- Ali Z, Shami A, Sedeek K, Kamel R, Alhabsi A, Tehseen M, Hassan N, Butt H, Kababji A, Hamdan SM, et al. (2020b) Fusion of the Cas9 endonuclease and the VirD2 relaxase facilitates homology-directed repair for precise genome engineering in rice. Commun Biol 3: 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R, Ali Z, Butt H, Mahas A, Aljedaani F, Khan MZ, Ding S, Mahfouz M (2018a) RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol 19: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R, Mahas A, Butt H, Aljedaani F, Mahfouz M (2018b) Engineering RNA Virus Interference via the CRISPR/Cas13 Machinery in Arabidopsis. Viruses 10: 732; doi: 10.3390/v10120732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aman R, Mahas A, Mahfouz M (2020a) Nucleic Acid Detection Using CRISPR/Cas Biosensing Technologies. ACS Synth Biol 9: 1226–1233 [DOI] [PubMed] [Google Scholar]
- Aman R, Mahas A, Marsic T, Hassan N, Mahfouz MM (2020b) Efficient, Rapid, and Sensitive Detection of Plant RNA Viruses With One-Pot RT-RPA-CRISPR/Cas12a Assay. Front Microbiol 11: 610872–610872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariga H, Toki S, Ishibashi K (2020) Potato Virus X Vector-Mediated DNA-Free Genome Editing in Plants. Plant Cell Physiol 61: 1946–1953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arizti-Sanz J, Freije CA, Stanton AC, Petros BA, Boehm CK, Siddiqui S, Shaw BM, Adams G, Kosoko-Thoroddsen T-SF, Kemball ME, et al. (2020) Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2. Nat Commun 11: 5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26: 151–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltes NJ, Hummel AW, Konecna E, Cegan R, Bruns AN, Bisaro DM, Voytas DF (2015) Conferring resistance to geminiviruses with the CRISPR–Cas prokaryotic immune system. Nat Plants 1: 15145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastet A, Zafirov D, Giovinazzo N, Guyon-Debast A, Nogué F, Robaglia C, Gallois J-L (2019) Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol J 17: 1736–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beying N, Schmidt C, Pacher M, Houben A, Puchta H (2020) CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat Plants 6: 638–645 [DOI] [PubMed] [Google Scholar]
- Bragard C, Caciagli P, Lemaire O, Lopez-Moya JJ, Macfarlane S, Peters D, Susi P, Torrance L (2013) Status and prospects of plant virus control through interference with vector transmission. Annu Rev Phytopathol 51: 177–201 [DOI] [PubMed] [Google Scholar]
- Briddon RW, Markham PG (2000) Cotton leaf curl virus disease. Virus Res 71: 151–159 [DOI] [PubMed] [Google Scholar]
- Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, Miao X, Streithorst JA, Granados A, Sotomayor-Gonzalez A, et al. (2020) CRISPR–Cas12-based detection of SARS-CoV-2. Nat Biotechnol 38: 870–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler NM, Baltes NJ, Voytas DF, Douches DS (2016) Geminivirus-Mediated Genome Editing in Potato (Solanum tuberosum L.) Using Sequence-Specific Nucleases. Front Plant Sci 7: 1045–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Zhou H, Zhou X, Li F (2020) Control of Plant Viruses by CRISPR/Cas System-Mediated Adaptive Immunity. Front Microbiol 11: 593700–593700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, et al. (2017) A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 29: 1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekaran J, Brumin M, Wolf D, Leibman D, Klap C, Pearlsman M, Sherman A, Arazi T, Gal-On A (2016) Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol :17: 1140–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro-Garcia A, Kamoun S, Nekrasov V (2015) Boosting plant immunity with CRISPR/Cas. Genome Biol 16: 254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterji A, Chatterji U, Beachy RN, Fauquet CM (2000) Sequence parameters that determine specificity of binding of the replication-associated protein to its cognate site in two strains of tomato leaf curl virus-New Delhi. Virology 273: 341–350 [DOI] [PubMed] [Google Scholar]
- Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, Doudna JA (2018) CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science 360: 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Qian Y, Wu X, Sun Y, Wu X, Cheng X (2014) Inhibiting replication of begomoviruses using artificial zinc finger nucleases that target viral-conserved nucleotide motif. Virus Genes 48: 494–501 [DOI] [PubMed] [Google Scholar]
- Cheng H, Yang H, Zhang D, Gai J, Yu D (2010) Polymorphisms of soybean isoflavone synthase and flavanone 3-hydroxylase genes are associated with soybean mosaic virus resistance. Mol Breeding 25: 13–24 [Google Scholar]
- Cheng X, Li F, Cai J, Chen W, Zhao N, Sun Y, Guo Y, Yang X, Wu X (2015) Artificial TALE as a Convenient Protein Platform for Engineering Broad-Spectrum Resistance to Begomoviruses. Viruses 7: 4772–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chertow DS (2018) Next-generation diagnostics with CRISPR. Science 360: 381. [DOI] [PubMed] [Google Scholar]
- Chikoti PC, Mulenga RM, Tembo M, Sseruwagi P (2019) Cassava mosaic disease: a review of a threat to cassava production in Zambia. J Plant Pathol 101: 467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chira S, Jackson CS, Oprea I, Ozturk F, Pepper MS, Diaconu I, Braicu C, Raduly LZ, Calin GA, Berindan-Neagoe I (2015) Progresses towards safe and efficient gene therapy vectors. Oncotarget 6: 30675–30703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho I-S, Igori D, Lim S, Choi G-S, Hammond J, Lim H-S, Moon JS (2016) Deep sequencing analysis of apple infecting viruses in Korea. Plant Pathol J 32: 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cody WB, Scholthof HB (2020) Native processing of single guide RNA transcripts to create catalytic Cas9/single guide RNA complexes in planta. Plant Physiol 184: 1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F (2017) RNA editing with CRISPR-Cas13. Science 358: 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creager AN, Scholthof KB, Citovsky V, Scholthof HB (1999) Tobacco mosaic virus. Pioneering research for a century. Plant Cell 11: 301–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J 95: 5–16 [DOI] [PubMed] [Google Scholar]
- Dangl JL, Horvath DM, Staskawicz BJ (2013) Pivoting the plant immune system from dissection to deployment. Science 341: 746–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehart JL, Kamrud KI (2018) From Foe to Friend: Therapeutic and Commercial Applications of Engineered Viruses. eLS, John Wiley & Sons, Ltd (Ed.), 1–10 [Google Scholar]
- Dong OX, Ronald PC (2019) Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol 180: 26–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna JA, Charpentier E (2014) Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 346: 1258096. [DOI] [PubMed] [Google Scholar]
- Dovas CI, Katis NI, Avgelis AD (2002) Multiplex detection of criniviruses associated with epidemics of a yellowing disease of tomato in Greece. Plant Dis 86: 1345–1349 [DOI] [PubMed] [Google Scholar]
- Edgü G, Freund LJ, Hartje S, Tacke E, Hofferbert HR, Twyman RM, Noll GA, Muth J, Prüfer D (2020) Fast, precise, and reliable multiplex detection of potato viruses by loop-mediated isothermal amplification. Int J Mol Sci 21: 8741; doi: 10.3390/ijms21228741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison EE, Nagalakshmi U, Gamo ME, Huang P-J, Dinesh-Kumar S, Voytas DF (2020) Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants 6: 620–624 [DOI] [PubMed] [Google Scholar]
- Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, Biesgen C, Puchta H (2012) In planta gene targeting. Proc Natl Acad Sci USA 109: 7535–7540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fozouni P, Son S, Díaz De León Derby M, Knott GJ, Gray CN, D'ambrosio MV, Zhao C, Switz NA, Kumar GR, Stephens SI, et al. (2021) Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell 184: 323–333.e9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Eyers S, Thomas C, Ellis N, Maule A (2004) Identification of markers tightly linked to sbm recessive genes for resistance to Pea seed-borne mosaic virus. Theor Appl Genet 109: 488–494 [DOI] [PubMed] [Google Scholar]
- Goldbach R (1998) All you wanted to know about plant virus control. Trends Plant Sci 3: 490 [Google Scholar]
- Gomez MA, Lin ZD, Moll T, Chauhan RD, Hayden L, Renninger K, Beyene G, Taylor NJ, Carrington JC, Staskawicz BJ, et al. (2019) Simultaneous CRISPR/Cas9-mediated editing of cassava eIF4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol J 17: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, Verdine V, Donghia N, Daringer NM, Freije CA, et al. (2017) Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 356: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gover O, Peretz Y, Mozes-Koch R, Maori E, Rabinowitch HD, Sela I (2014) Only minimal regions of tomato yellow leaf curl virus (TYLCV) are required for replication, expression and movement. Arch Virol 159: 2263–2274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanssen IM, Lapidot M, Thomma BP (2010) Emerging viral diseases of tomato crops. Mol Plant Microbe Interact 23: 539–548 [DOI] [PubMed] [Google Scholar]
- Hashimoto M, Neriya Y, Yamaji Y, Namba S (2016a) Recessive resistance to plant viruses: potential resistance genes beyond translation initiation factors. Front Microbiol 7: 1695–1695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinlein M (2015) Plant virus replication and movement. Virology 479–480: 657–671 [DOI] [PubMed] [Google Scholar]
- Hilje L, Costa HS, Stansly PA (2001) Cultural practices for managing Bemisia tabaci and associated viral diseases. Crop Prot 20: 801–812 [Google Scholar]
- Ji X, Si X, Zhang Y, Zhang H, Zhang F, Gao C (2018) Conferring DNA virus resistance with high specificity in plants using virus-inducible genome-editing system. Genome Biol 19: 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Zhang H, Zhang Y, Wang Y, Gao C (2015) Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat Plants 1: 15144. [DOI] [PubMed] [Google Scholar]
- Jiang N, Zhang C, Liu JY, Guo ZH, Zhang ZY, Han CG, Wang Y (2019) Development of Beet necrotic yellow vein virus-based vectors for multiple-gene expression and guide RNA delivery in plant genome editing. Plant Biotechnol J 17: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao J, Kong K, Han J, Song S, Bai T, Song C, Wang M, Yan Z, Zhang H, Zhang R, et al. (2021) Field detection of multiple RNA viruses/viroids in apple using a CRISPR/Cas12a-based visual assay. Plant Biotechnol J 19: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinina NO, Khromov A, Love AJ, Taliansky ME (2020) CRISPR applications in plant virology: virus resistance and beyond. Phytopathology 110: 18–28 [DOI] [PubMed] [Google Scholar]
- Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F (2019) SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Prot 14: 2986–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan Z, Khan SH, Ahmad A, Aslam S, Mubarik MS, Khan S (2019) CRISPR/dCas9-mediated inhibition of replication of Begomoviruses. Int J Agri Biol 21: 711–718 [Google Scholar]
- Kis A, Hamar É, Tholt G, Bán R, Havelda Z (2019a) Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol J 17: 1004–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kis A, Hamar É, Tholt G, Bán R, Havelda Z (2019b) Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system. Plant Biotechnol J 171004–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD (2018) Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173: 665–676.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovalskaya N, Hammond RW. (2014) Molecular biology of viroid–host interactions and disease control strategies. Plant Sci 228: 48–60 [DOI] [PubMed] [Google Scholar]
- Lefkowitz EJ, Dempsey DM, Hendrickson RC, Orton RJ, Siddell SG, Smith DB (2018) Virus taxonomy: the database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res 46: D708–d717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei J, Dai P, Li Y, Zhang W, Zhou G, Liu C, Liu X (2021) Heritable gene editing using FT mobile guide RNAs and DNA viruses. Plant Methods 17: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lellis AD, Kasschau KD, Whitham SA, Carrington JC (2002) Loss-of-susceptibility mutants of Arabidopsis thaliana reveal an essential role for eIF(iso)4E during potyvirus infection. Curr Biol 121046–1051 [DOI] [PubMed] [Google Scholar]
- Li L, Li S, Wu N, Wu J, Wang G, Zhao G, Wang J (2019) HOLMESv2: a CRISPR-Cas12b-assisted platform for nucleic acid detection and DNA methylation quantitation. ACS Synth Biol 8: 2228–2237 [DOI] [PubMed] [Google Scholar]
- Li S-Y, Cheng Q-X, Wang J-M, Li X-Y, Zhang Z-L, Gao S, Cao R-B, Zhao G-P, Wang J (2018) CRISPR-Cas12a-assisted nucleic acid detection. Cell Discovery 4: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang M, Li Z, Wang W, Liu J, Liu L, Zhu G, Karthik L, Wang M, Wang K-F, Wang Z, et al. (2019) A CRISPR-Cas12a-derived biosensing platform for the highly sensitive detection of diverse small molecules. Nat Commun 10: 3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Soyars CL, Li J, Fei Q, He G, Peterson BA, Meyers BC, Nimchuk ZL, Wang X (2018) CRISPR/Cas9-mediated resistance to cauliflower mosaic virus. Plant Direct 2: e00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Zhang B (2020) Virus-based CRISPR/Cas9 genome editing in plants. Trends Genet 36: 810–813 [DOI] [PubMed] [Google Scholar]
- Loebenstein G (2008) Plant virus diseases: economic aspects. InMahy BWJ, Van Regenmortel MHV, eds. Encyclopedia of Virology, Ed 3. Academic Press, Oxford, pp 197–201 [Google Scholar]
- Loebenstein G, Katis N (2014) Control of plant virus diseases seed-propagated crops. Preface. Adv Virus Res 90: xi. [DOI] [PubMed] [Google Scholar]
- Love AJ, Yu C, Petukhova NV, Kalinina NO, Chen J, Taliansky ME (2017) Cajal bodies and their role in plant stress and disease responses. RNA Biol 14: 779–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R (2016) Geminiviruses for biotechnology: the art of parasite taming. New Phytol 210: 58–64 [DOI] [PubMed] [Google Scholar]
- Lu Y, Yao B, Wang G, Hong N (2018) The detection of ACLSV and ASPV in pear plants by RT-LAMP assays. J Virol Methods 252: 80–85 [DOI] [PubMed] [Google Scholar]
- Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat Plants 6: 773–779 [DOI] [PubMed] [Google Scholar]
- Macovei A, Sevilla NR, Cantos C, Jonson GB, Slamet-Loedin I, Čermák T, Voytas DF, Choi IR, Chadha-Mohanty P (2018) Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol J 16: 1918–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahas A, Aman R, Mahfouz M (2019) CRISPR-Cas13d mediates robust RNA virus interference in plants. Genome Biol 20: 263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahas A, Hassan N, Aman R, Marsic T, Wang Q, Ali Z, Mahfouz MM (2021) LAMP-coupled CRISPR–Cas12a module for rapid and sensitive detection of plant DNA viruses. Viruses 13: 466; doi: 10.3390/v13030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu J-K (2011) De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci 108: 2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhotenko AV, Khromov AV, Snigir EA, Makarova SS, Makarov VV, Suprunova TP, Kalinina NO, Taliansky ME (2019) Functional analysis of coilin in virus resistance and stress tolerance of potato Solanum tuberosum using CRISPR-Cas9 editing. Dokl Biochem Biophys 484: 88–91 [DOI] [PubMed] [Google Scholar]
- Malina A, Mills JR, Cencic R, Yan Y, Fraser J, Schippers LM, Paquet M, Dostie J, Pelletier J (2013) Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev 27: 2602–2614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeske AJ, Nakandakari-Higa S, Marraffini LA (2019) Cas13-induced cellular dormancy prevents the rise of CRISPR-resistant bacteriophage. Nature 570: 241–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta D, Stürchler A, Anjanappa RB, Zaidi SS-E-A, Hirsch-Hoffmann M, Gruissem W, Vanderschuren H (2019) Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol 20: 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Y, Beernink BM, Ellison EE, Konečná E, Neelakandan AK, Voytas DF, Whitham SA (2019) Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct 3: e00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra J, Srivastava R, Trivedi PK, Verma PC (2020) Effect of virus infection on the secondary metabolite production and phytohormone biosynthesis in plants. Biotech 10: 547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno AB, López-Moya JJ (2020) When viruses play team sports: mixed infections in plants. Phytopathology 110: 29–48 [DOI] [PubMed] [Google Scholar]
- Nicaise V (2014) Crop immunity against viruses: outcomes and future challenges. Front Plant Sci 5: 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'connell MR (2019) Molecular mechanisms of RNA targeting by Cas13-containing type VI CRISPR–Cas systems. J Mol Biol 431: 66–87 [DOI] [PubMed] [Google Scholar]
- Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM (2015) RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J 13: 578–589 [DOI] [PubMed] [Google Scholar]
- Pooggin M, Shivaprasad PV, Veluthambi K, Hohn T (2003) RNAi targeting of DNA virus in plants. Nat Biotechnol 21: 131–132 [DOI] [PubMed] [Google Scholar]
- Puchta H (2002) Gene replacement by homologous recombination in plants. Plant Mol Biol 48: 173–182 [PubMed] [Google Scholar]
- Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57: 629–637 [DOI] [PubMed] [Google Scholar]
- Pyott DE, Sheehan E, Molnar A (2016) Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol 17: 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F, Martin-Hernandez AM, Baulcombe DC (2001) Technical Advance. Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J 25: 237–245 [DOI] [PubMed] [Google Scholar]
- Roossinck MJ (2011) The big unknown: plant virus biodiversity. Curr Opin Virol 1: 63–67 [DOI] [PubMed] [Google Scholar]
- Roy A, Zhai Y, Ortiz J, Neff M, Mandal B, Mukherjee SK, Pappu HR (2019) Multiplexed editing of a begomovirus genome restricts escape mutant formation and disease development. PLoS One 14: e0223765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastry KS, Zitter TA (2014) Management of virus and viroid diseases of crops in the tropics. InSastry KS, Zitter TA, eds. Plant Virus and Viroid Diseases in the Tropics: Volume 2: Epidemiology and Management. Springer Netherlands, Dordrecht, pp 149–480 [Google Scholar]
- Schmidt C, Pacher M, Puchta H (2019) DNA break repair in plants and its application for genome engineering. Methods Mol Biol 1864: 237–266 [DOI] [PubMed] [Google Scholar]
- Seal SE, Jeger MJ, Van Den Bosch F (2006) Begomovirus evolution and disease management. Adv Virus Res 67: 297–316 [DOI] [PubMed] [Google Scholar]
- Senthil-Kumar M, Mysore KS (2011) New dimensions for VIGS in plant functional genomics. Trends Plant Sci 16: 656–665 [DOI] [PubMed] [Google Scholar]
- Seo YS, Rojas MR, Lee JY, Lee SW, Jeon JS, Ronald P, Lucas WJ, Gilbertson RL (2006) A viral resistance gene from common bean functions across plant families and is up-regulated in a non-virus-specific manner. Proc Natl Acad Sci USA 103: 11856–11861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera T (2005) Inhibition of virus DNA replication by artificial zinc finger proteins. J Virol 79: 2614–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw J, Love AJ, Makarova SS, Kalinina NO, Harrison BD, Taliansky ME (2014) Coilin, the signature protein of Cajal bodies, differentially modulates the interactions of plants with viruses in widely different taxa. Nucleus 5: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Srivastava A (2020) Chapter 41 - prevention and control of viral diseases of crops. InAwasthi LP, ed. Applied Plant Virology. Academic Press, Lucknow, India, pp 593–599 [Google Scholar]
- Szemes M, Klerks MM, Van Den Heuvel JF, Schoen CD (2002) Development of a multiplex AmpliDet RNA assay for simultaneous detection and typing of potato virus Y isolates. J Virol Methods 100: 83–96 [DOI] [PubMed] [Google Scholar]
- Tashkandi M, Ali Z, Aljedaani F, Shami A, Mahfouz MM (2018) Engineering resistance against Tomato yellow leaf curl virus via the CRISPR/Cas9 system in tomato. Plant Signal Behav 13: e1525996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi JN, Ntui V, Ron M, Muiruri SK, Britt A, Tripathi L (2019) CRISPR/Cas9 editing of endogenous banana streak virus in the B genome of Musa spp. overcomes a major challenge in banana breeding. Commun Biol 2: 46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagris EM, Martínez De Alba AE, Gozmanova M, Kalantidis K (2008) Viroids. Cell Microbiol 10: 2168–2179 [DOI] [PubMed] [Google Scholar]
- Uranga M, Aragonés V, Selma S, Vázquez-Vilar M, Orzáez D, Daròs JA (2021) Efficient Cas9 multiplex editing using unspaced sgRNA arrays engineering in a Potato virus X vector. The Plant Journal, 10.1111/tpj.15164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Schie CC, Takken FL (2014) Susceptibility genes 101: how to be a good host. Annu Rev Phytopathol 52: 551–581 [DOI] [PubMed] [Google Scholar]
- Van Vu T, Sung YW, Kim J, Doan DTH, Tran MT, Kim J-Y (2019) Challenges and perspectives in homology-directed gene targeting in monocot plants. Rice 12: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varanda CM, Félix MDR, Campos MD, Patanita M, Materatski P (2021) Plant viruses: from targets to tools for CRISPR. Viruses 13: 141; doi: 10.3390/v13010141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu JK (2017) Gene targeting by homology-directed repair in rice using a geminivirus-based CRISPR/Cas9 system. Mol Plant 10: 1007–1010 [DOI] [PubMed] [Google Scholar]
- Wang M, Zhang R, Li J (2020a) CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens Bioelect 165: 112430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MB, Masuta C, Smith NA, Shimura H (2012) RNA silencing and plant viral diseases. Mol Plant Microbe Interact 25: 1275–1285 [DOI] [PubMed] [Google Scholar]
- Wang Y, Chen R, Nie X, Zhong Z, Li C, Li K, Huang W, Fu X, Liu J, Nie B (2020b) Rapid and sensitive detection of potato virus Y by isothermal reverse transcription-recombinase polymerase amplification assay in potato. Mol Cell Probes 50: 101505. [DOI] [PubMed] [Google Scholar]
- Wigge PA (2011) FT, a mobile developmental signal in plants. Curr Biol 21: R374–R378 [DOI] [PubMed] [Google Scholar]
- Wilisiani F, Tomiyama A, Katoh H, Hartono S, Neriya Y, Nishigawa H, Natsuaki T (2019) Development of a LAMP assay with a portable device for real-time detection of begomoviruses under field conditions. J Virol Methods 265: 71–76 [DOI] [PubMed] [Google Scholar]
- Xie K, Minkenberg B, Yang Y (2015) Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc Natl Acad Sci USA 112: 3570–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, Scott DA (2018) Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell 70: 327–339.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Han T, Liu G, Chen T, Wang Y, Yu AY, Liu Y (2015) A geminivirus-based guide RNA delivery system for CRISPR/Cas9 mediated plant genome editing. Sci Rep 5: 14926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Han T, Xie K, Zhao J, Song J, Liu Y (2019) Engineer complete resistance to cotton leaf curl multan virus by the CRISPR/Cas9 system in Nicotiana benthamiana. Phytopathol Res 1: 9 [Google Scholar]
- Zaidi SS, Mahas A, Vanderschuren H, Mahfouz MM (2020) Engineering crops of the future: CRISPR approaches to develop climate-resilient and disease-resistant plants. Genome Biol 21: 289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan X, Zhang F, Zhong Z, Chen R, Wang Y, Chang L, Bock R, Nie B, Zhang J (2019) Generation of virus-resistant potato plants by RNA genome targeting. Plant Biotechnol J 17: 1814–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Du H, Wang J, Pu Y, Yang C, Yan R, Yang H, Cheng H, Yu D (2020) Multiplex CRISPR/Cas9-mediated metabolic engineering increases soya bean isoflavone content and resistance to soya bean mosaic virus. Plant Biotechnol J 18: 1384–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zhao Y, Ye J, Cao X, Xu C, Chen B, An H, Jiao Y, Zhang F, Yang X, et al. (2019) Establishing CRISPR/Cas13a immune system conferring RNA virus resistance in both dicot and monocot plants. Plant Biotechnol J 17: 1185–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Zheng Q, Yi X, An H, Zhao Y, Ma S, Zhou G (2018) Establishing RNA virus resistance in plants by harnessing CRISPR immune system. Plant Biotechnol J 16: 1415–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Thieme CJ, Kollwig G, Apelt F, Yang L, Winter N, Andresen N, Walther D, Kragler F (2016) tRNA-related sequences trigger systemic mRNA transport in plants. Plant Cell 28: 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Sato S, Ye X, Dorrance A, Morris TJ, Clemente TE, Qu F (2011) Robust RNAi-based resistance to mixed infection of three viruses in soybean plants expressing separate short hairpins from a single transgene. Phytopathology 101: 1264–1269 [DOI] [PubMed] [Google Scholar]