Abstract
Parasitic plants reduce crop yield worldwide. Dodder (Cuscuta campestris) is a stem parasite that attaches to its host, using haustoria to extract nutrients and water. We analyzed the transcriptome of six C. campestris tissues and identified a key gene, LATERAL ORGAN BOUNDARIES DOMAIN 25 (CcLBD25), as highly expressed in prehaustoria and haustoria. Gene coexpression networks from different tissue types and laser-capture microdissection RNA-sequencing data indicated that CcLBD25 could be essential for regulating cell wall loosening and organogenesis. We employed host-induced gene silencing by generating transgenic tomato (Solanum lycopersicum) hosts that express hairpin RNAs to target and down-regulate CcLBD25 in the parasite. Our results showed that C. campestris growing on CcLBD25 RNAi transgenic tomatoes transited to the flowering stage earlier and had reduced biomass compared with C. campestris growing on wild-type (WT) hosts, suggesting that parasites growing on transgenic plants were stressed due to insufficient nutrient acquisition. We developed an in vitro haustorium system to assay the number of prehaustoria produced on strands from C. campestris. Cuscuta campestris grown on CcLBD25 RNAi tomatoes produced fewer prehaustoria than those grown on WT tomatoes, indicating that down-regulating CcLBD25 may affect haustorium initiation. Cuscuta campestris haustoria growing on CcLBD25 RNAi tomatoes exhibited reduced pectin digestion and lacked searching hyphae, which interfered with haustorium penetration and formation of vascular connections. The results of this study elucidate the role of CcLBD25 in haustorium development and might contribute to developing parasite-resistant crops.
Transcription factor CcLBD25 plays a pivotal role in haustorium initiation, regulating pectin digestion, and searching hyphae development during the haustorium penetration process.
Introduction
Parasitic plants are heterotrophic, reducing the yields of crops worldwide (Agrios, 2005; Yoder and Scholes, 2010). They parasitize host plants using specialized organs known as haustoria, which extract nutrients and water from the hosts. Cuscuta species (dodders) are stem holoparasites without functional roots and leaves. Stems of Cuscuta spp. coil counterclockwise around their host and then form a series of haustoria along stems to attach to the hosts (Furuhashi et al., 2011; Alakonya et al., 2012). Cuscuta campestris is one of the most widely distributed and destructive parasitic weeds (Parker, 2021). A better understanding of the underlying molecular mechanisms of C. campestris haustorium development will aid in applying parasitic weed control and producing parasitic plant-resistant crops.
Many previous studies have identified the key factors needed for seed germination, host recognition, and haustorium induction and growth in root parasites (Shen et al., 2006; López-Ráez et al., 2009; Yoder and Scholes, 2010). Focusing on haustorium development, a previous study indicated that root parasitic plants co-opted the mechanism of lateral root formation in haustorium organogenesis (Ichihashi et al., 2017). The LATERAL ORGAN BOUNDARIES DOMAIN (LBD) family of transcription factors (TFs) are reported to be crucial in both lateral root formation in nonparasitic plants and haustorium developmental programming in root parasites (Ichihashi et al., 2020). In nonparasitic model plants, like Arabidopsis, LBD genes are shown to be involved in auxin signaling, interact with AUXIN RESPONSE FACTORs, and promote lateral root formation (Mangeon et al., 2010; Porco et al., 2016). During the haustorium development stage, LBD orthologs are reported to be upregulated at attachment sites of root parasitic plants like Thesium chinense (Ichihashi et al., 2017) and Striga hermonthica (Yoshida et al., 2019). On the other hand, the molecular pathways regulating haustorium development in stem parasitic plants are still largely unexplored. Although a few gene orthologs that regulate auxin accumulation during lateral root development in nonparasitic plants are found to be expressed in Cuscuta seedlings and stems, whether these genes are also involved haustorium formation remains unknown (Ranjan et al., 2014). Our previous studies showed that SHOOT MERISTEMLESS-like plays a role in Cuscuta spp. haustorium development (Alakonya et al., 2012). These results suggest that Cuscuta spp. might have repurposed the shoot developmental programs into haustorium organogenesis, but a recent study indicates that some genes that are involved in root development were also expressed in Cuscuta australis prehaustoria and haustoria (Sun et al., 2018), suggesting that the lateral root programming system was co-opted into haustorium development.
In this study, we provide an insight into the gene regulatory mechanisms of haustorium organogenesis and identify one of the LBD TFs, CcLBD25, as a vital regulator of C. campestris haustorium development. This discovery supports the hypothesis that stem parasitic plants adapted both shoot and root molecular machinery into haustorium formation. Using detailed transcriptome analysis and gene coexpression networks (GCNs) coupled with cellular and developmental phenotype assays, we also show that CcLBD25 is not only involved in haustorium initiation through auxin signaling, but also participates in other aspects of haustorial developmental reprogramming, including cell wall loosening, searching hyphae development, and other phytohormone mediated signaling pathways. The results of this study will not only shed light on the field of haustorium development in stem parasitic plants but will also help develop a potentially universal parasitic weed-resistant system in crops to reduce economic losses caused by both root and stem parasites.
Results
Establishing genomic resources for C. campestris and constructing GCNs that regulate haustorium formation
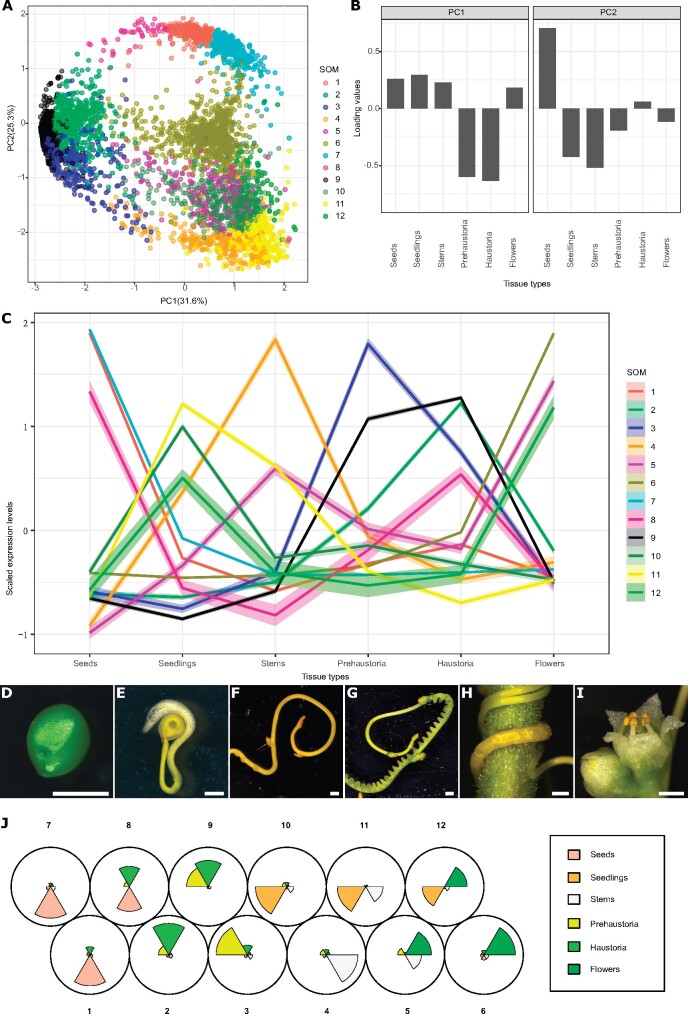
In this study, we analyzed the transcriptome of different C. campestris tissues, including seeds, seedlings, stems, prehaustoria, haustoria, and flowers, grown on the tomato (Solanum lycopersicum) Heinz 1706 (H1706) cultivar and Nicotiana benthamiana (Ranjan et al., 2014) by mapping reads to the recently available genome of C. campestris (Vogel et al., 2018). In general, seed tissues have distinctively different gene expression profiles compared to all other tissues (Supplemental Figure S1). In addition, the expression patterns in invasive tissues (prehaustoria and haustoria) and noninvasive tissues are also disparate (Supplemental Figure S1). We conducted principal component analysis (PCA) analysis and noticed that the genes that are highly expressed in invasive tissues can be separated from the genes that are highly expressed in noninvasive tissues on PC1 (Figure 1, A and B). To identify the genes that might be involved in haustorium development, we performed clustering analysis using self-organizing maps (SOMs) in R, and identified a cluster enriched with genes that are highly expressed in both prehaustoria and haustoria tissues (SOM9; Figure 1;Supplemental Figure S2; Supplemental Table S1).
Figure 1.
PCA and SOMs clustering of gene expression in C. campestris tissue type RNA-seq data mapped to C. campestris genome. A, PCA analysis based on gene expression across different C. campestris tissues. Each dot represents a gene and is in the color indicating its corresponding SOM group. The percentage next to each axis indicates the proportion of variance that can be explained by the PC. B, Loading values of PC1 and PC2. PC1 separates the genes that are specifically expressed in intrusive tissues (prehaustoria and haustoria) from those that are expressed in nonintrusive tissues. PC2 divides the seed-specific genes from other genes. C, Scaled expression levels of each SOM group across different C. campestris tissue types. Each line is colored based on the corresponding SOM groups. The highlighting around the lines indicates a 95% confidence interval. D–I, The six different tissue types that were used in this transcriptomic study. Scale bars = 1 mm. D, Seed. E, Seedling. F, Stem. G, Prehaustoria. H, Haustoria. White arrowheads indicate haustoria. I, Flowers. J, A code plot of SOM clustering based on gene expression in C. campestris tissue type RNA-seq data mapped to the C. campestris genome. Each sector represents a tissue type and is in the color indicating its corresponding tissue type. The size of each sector illustrates the amount of expression from each tissue type in SOM groups.
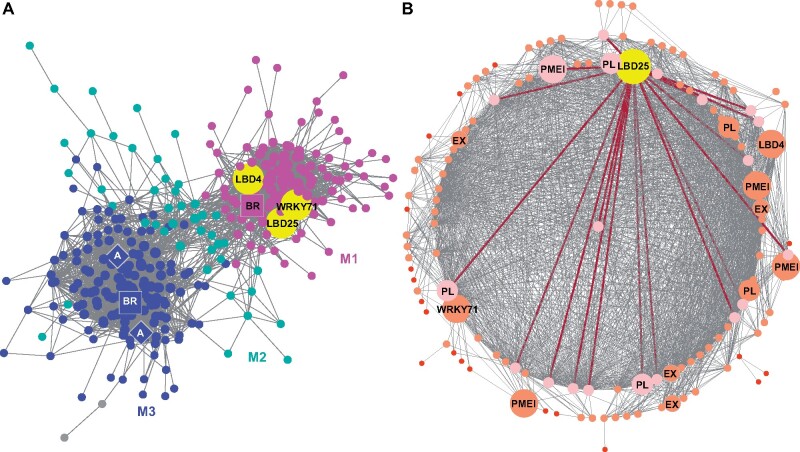
We focused on the genes contained in this SOM9 cluster and constructed a GCN. Using the fast greedy modularity optimization algorithm to analyze the GCN community structure (Clauset et al., 2004) and visualizing the network using Cytoscape (Cline et al., 2007), we noticed this SOM9 GCN is composed of three major modules (Figure 2A;Supplemental Table S2). Since the current gene annotation of C. campestris genome is not as complete as that of most model organisms, we used BLAST to combine our previously annotated transcriptome with current C. campestris genome gene IDs (Supplemental Table S3). With this more comprehensive annotation profile, we conducted Gene Ontology (GO) enrichment analysis using the TAIR ID for each C. campestris gene in the network to identify the major GO term for each module (Supplemental Table S4). Based on our GO enrichment results, the major biological process of Module 1 can be classified as “plant-type cell wall loosening,” and the cellular component of Module 1 is “extracellular region and intracellular membrane-bounded organelle” (Figure 2; Supplemental Table S4). This result indicates the genes contained in Module 1 are mostly involved in cell wall loosening, which is needed for the haustorium to penetrate through the host tissue. On the other hand, the major biological processes of Module 3 include “transport, response to hormones, secondary metabolite biosynthetic process, and regulation of lignin biosynthetic process.” The molecular function of Module 3 is “transmembrane transporter activity,” and the cellular component of Module 3 is “plasma membrane” (Figure 2A;Supplemental Table S4). This analysis suggests that these genes might be involved in later stages of development and nutrient transport from the host to the parasite once a connection is established between the host and the parasite.
Figure 2.
SOM9 GCNs from C. campestris tissue type RNA-seq data. A, GCN of genes that are classified in SOM9, which includes genes that are highly expressed in both prehaustoria and haustoria. This SOM9 GCN is composed of three major modules. Magenta indicates genes in Module 1, which has enriched biological process GO term “plant-type cell wall loosening.” Cyan indicates genes in Module 2. Blue indicates genes in Module 3, which has enriched biological process GO terms “transport, response to hormones, secondary metabolite biosynthetic process, and regulation of lignin biosynthetic process.” The only three TFs in Module 1 are enlarged and labeled in yellow. The genes involved in auxin transport are labeled in diamonds. The genes involved in brassinosteroid signaling are labeled in squares. B, GCN of genes that are classified in SOM9 Module 1. Dark red lines indicate the connection between CcLBD25 (yellow) and its first layer neighbors. The genes that are first layer neighbors of CcLBD25 are labeled in pink. The genes that are second layer neighbors of CcLBD25 are labeled in orange with medium size dots. The genes that are outside the second layer neighbors of CcLBD25 are labeled in red with small size dots. The only three TFs as well as cell wall loosening related genes are enlarged, highlighted and labeled in the network. A and B, PL. A, auxin efflux carrier-like protein. BR, brassinosteroid insensitive 1-associated receptor kinase 1-like.
To identify the key regulators in the haustorium penetration process, we focused on genes in Module 1 and calculated the degree centrality and betweenness centrality scores of each gene within this group. Many central hub genes in Module 1 are proteins or enzymes involved in cell wall modifications, like pectin lyases (PLs), pectinesterase inhibitors, and expansins (EXs; Figure 2B;Supplemental Table S2). To find the upstream regulators of these pathways, we focused on TFs that are classified in Module 1. Intriguingly, only three TFs are included in Module 1: LBD gene 25 (CcLBD25; Cc019141), CcLBD4 (Cc017015), and CcWRKY71 (Cc004070; Figure 2B;Supplemental Table S2). According to the GCN of SOM9, we noticed that CcLBD25, CcLBD4, and CcWRKY71 share several common first-layer neighbors (Figure 2). Based on previous reports, AtLBD25 regulates lateral root development in Arabidopsis by promoting auxin signaling (Dean et al., 2004; Mangeon et al., 2010). Furthermore, an LBD25 ortholog (TcLBD25) in T. chinense, a root parasitic plant in the Santalaceae family, was also detected to be upregulated during the haustorium development process (Ichihashi et al., 2017). Using CcLBD25 nucleotide sequence alignments and a LBD25 phylogenetic tree constructed using MEGA X (Kumar et al., 2018; Supplemental Figure S3; Supplemental Dataset S1), we confirmed orthology between the TcLBD25, AtLBD25, and CcLBD25 genes. Based on these serendipitous pieces of evidence, we suspected that CcLBD25 may also regulate haustorium formation and the parasitism process in C. campestris.
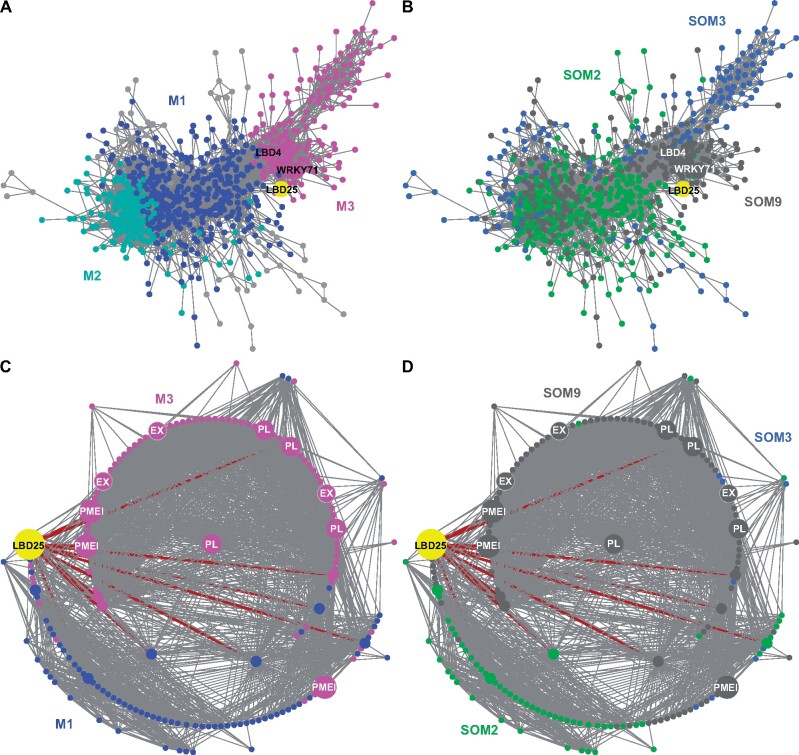
To understand the role of CcLBD25 and the potential connection with other genes, we included genes in SOM2 (genes that are only highly expressed in haustoria) and SOM3 (genes that are only highly expressed in prehaustoria) to build a more comprehensive GCN (Figure 1;Supplemental Figure S2). Based on the community structure analysis, this comprehensive network is composed of three major modules (Figure 3A;Supplemental Table S5). Based on our GO enrichment results, the major biological process of Module 3 is plant-type cell wall loosening and the major biological processes of Module 1 include morphogenesis of a branching structure, plant organ formation, and several hormone responses and biosynthetic processes (Supplemental Table S6). CcLBD25 itself is placed in Module 1 (Supplemental Table S5), but CcLBD25 has many first layer connections with genes that are classified in Modules 1 or 3 (Figure 3, A and C). This result indicates that CcLBD25 might play a role in connecting genes involved in different pathways or aspects of haustorium development. Furthermore, by coloring the network with their corresponding SOM groups, we noticed that even though CcLBD25 itself is in SOM9, many of the CcLBD25 first and second layer neighbors are in SOM2 and SOM3 (Figure 3, B and D). Thus, CcLBD25 might be a key regulator of the haustorium development process in both early and late stages of haustorium development and may also play a critical role in coordinating the function of genes that are expressed only during discrete developmental stages.
Figure 3.
GCNs of SOM2, 3, 9 genes based on C. campestris tissue type RNA-seq data. A, GCN of genes that are in SOM2, SOM3, and SOM9 with colors based on network modules. This SOM2+SOM3+SOM9 GCN is composed of three major modules. Blue indicates genes in Module 1, which has the biological process GO enrichment “morphogenesis of a branching structure, plant organ formation, strigolactone responses, and biosynthetic processes.” Cyan indicates genes in Module 2, which has the biological process GO enrichment “response to karrikin, hormone-mediated signaling pathway and defense response.” Magenta indicates genes in Module 3, which has the biological process GO enrichment “plant-type cell wall loosening.” Light grey indicates genes that are not included in Modules 1, 2, or 3. B, GCN of genes that are in SOM2, SOM3, and SOM9, with colors based on SOM clustering groups. Green indicates genes in SOM2. Blue indicates genes in SOM3. Grey indicates genes in SOM9, which includes the genes that are highly expressed in both prehaustoria and haustoria. SOM2 includes genes that are only highly expressed in haustoria and SOM3 includes genes that are only highly expressed in prehaustoria. C, GCN of CcLBD25 and its first and second layer neighbors with colors based on network modules as in (A). D, GCN of CcLBD25 and its first and second layer neighbors with colors based on SOM clustering groups as in (B). C and D, Dark red lines indicate the connection between CcLBD25 and its first layer neighbors. The genes that are second-layer neighbors of CcLBD25 are labeled with medium size dots. The genes that are outside the second layer neighbors of CcLBD25 are labeled with small size dots. CcLBD25 and cell wall loosening related genes are enlarged, highlighted, and labeled in the network.
Zooming into tissue-specific expression using laser-capture microdissection coupled with RNA-seq
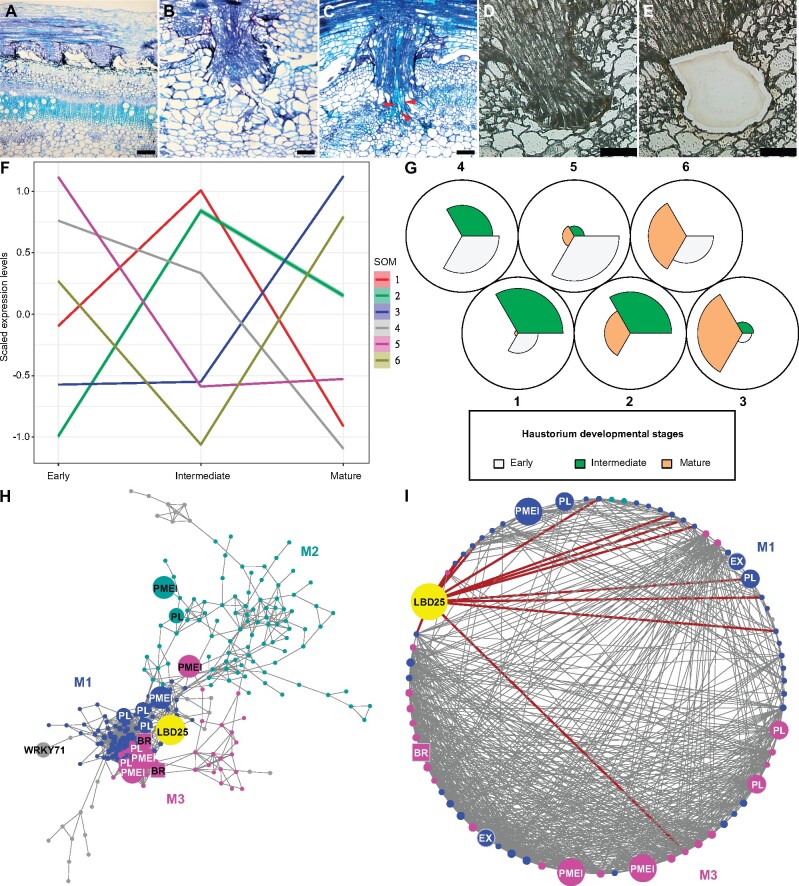
Our first transcriptome data came from hand-collected tissue samples. To further dissect Cuscuta haustorium developmental stages, we used laser-capture microdissection (LCM) with RNA-sequencing (RNA-seq) to analyze only pure haustorial tissues from three different haustorium developmental stages (Figure 4, A–E). Based on a previous study, changes in the levels of jasmonic acid and salicylic acid are observed ∼36–48 h after first haustorial swelling, which is ∼4 d postattachment (DPA; Runyon et al., 2010). We also noticed that haustorium growth is a continuous process for C. campestris, so all developmental stages of haustoria can be found on the same strand at the 4 DPA time point. Therefore, we focused on 4 DPA and defined three developmental stages based on their haustorium structure: early (the haustorium has just contacted the host), intermediate (the haustorium has developed searching hyphae but has not formed vascular connections), and mature (a mature haustorium with continuous vasculature between host and parasite; Figure 4, A–C). Cuscuta campestris haustorium tissues, especially the protruding region of haustoria, were collected from C. campestris using LCM at these three developmental stages attached to H1706 and subjected to RNA-seq (Figure 4, D–E).
Figure 4.
SOM clustering and GCNs of gene expression in C. campestris haustoria across three developmental stages (from the LCM RNA-seq data). A–E, Sections of developmental stages for LCM RNA-seq. A–C, Three developmental stages for LCM RNA-seq. Paraffin sections stained with Toluidine Blue. A, Early stage. B, Intermediate stage. C, Mature stage. The red arrowheads indicate vascular connections between host and C. campestris. D–E, The C. campestris haustorium tissues were collected using LCM. A and C, Scale bars = 250 µm. B, D, and E, Scale bars = 100 µm. F, Scaled expression levels of each SOM group across three haustorium developmental stages. Each line is colored based on the corresponding SOM group. The highlighting around the lines indicates a 95% confidence interval. G, A code plot of SOM clustering illustrating which developmental stages are highly represented in each SOM group by the size of sectors. Each sector represents a developmental stage and is in the color indicating its corresponding developmental stage. H, GCN based on LCM RNA-seq expression profiles with genes in tissue type RNA-seq SOM9. Blue indicates genes in Module 1, which has enriched biological process GO term “plant-type cell wall loosening.” Cyan indicates genes in Module 2, which has enriched biological process GO term “respiratory burst.” Magenta indicates genes in Module 3, which has enriched biological process GO term “brassinosteroid mediated signaling pathway.” Light grey indicates genes that are not included in Modules 1, 2, or 3. CcLBD25, CcWRKY71, and pectin degradation-related genes are enlarged, highlighted, and labeled in the network. The genes involved in brassinosteroid signaling are labeled in squares. I, GCN of CcLBD25 and its first and second layer neighbors with colors based on network modules as in (H). Dark red lines indicate the connection between CcLBD25 and its first layer neighbors. CcLBD25 and cell wall loosening related genes are enlarged, highlighted, and labeled in the network. H–I, PL.
Next, we mapped our LCM RNA-seq data to the C. campestris genome. Visualizing the gene expression changes using multidimensional scaling (MDS) showed that the expression profile of the mature stage is distinct from the early and intermediate stages (Supplemental Figure S4). We then conducted clustering analyses using SOM to group genes based on their expression patterns at these three different developmental stages (Figure 4, F–G). According to our PCA analysis, PC1 obviously separated genes that are specifically expressed in the mature stage from those expressed in the other two stages, and PC2 distinguished the genes expressed in the early stage from those expressed in the intermediate stage (Supplemental Figure S5). Interestingly, and similar to what was seen in our tissue type transcriptome data, CcLBD25 is grouped in SOM6, which is the cluster of genes that are relatively highly expressed in both early and mature stages (Figure 4, F–G;Supplemental Figure S6; Supplemental Table S7). Notably, CcLBD25 expression in intermediate-stage is relatively reduced compared with early-stage or mature-stage but is still much higher compared with other noninvasive tissues like seed, seedlings, stems, and flowers. Genes with low expression levels were often not detectable in LCM RNA-seq data, which might be caused by the preparation process of LCM tissues, including fixation, sectioning, and dissection processes, which are likely to lead to loss of some RNAs due to unpreventable degradation. To investigate gene regulatory dynamics within the haustorium developmental process, we used the same gene list from tissue type RNA-seq SOM9 and constructed another GCN of these genes that was based on the LCM RNA-seq expression profiles (Figure 4H). By using the same gene list, but the expression dataset from samples of precisely collected haustorial cells, we obtained detailed regulatory connections between genes by comparing the tissue type GCN and LCM GCN (Figures 2A, 4H). Based on the fast greedy community structure analysis, this LCM GCN is composed of three major modules with CcLBD25 in Module 1 (Figure 4H;Supplemental Table S8). According to our GO enrichment results, the major biological process for Module 1 is plant-type cell wall loosening, and for Module 3, is brassinosteroid mediated signaling pathway (Figure 4H;Supplemental Table S9). In addition to cell wall loosening related enzyme encoding genes forming central hubs, we noticed CcLBD25 is the TF with the highest number of connections in Module 1. CcLBD25 has 13 first layer neighbors and 70 second-layer neighbors, including many cell wall loosening-related genes (Figure 4, H–I;Supplemental Table S8). Zooming in to focus on CcLBD25, we noticed that the CcLBD25 first and second layer neighbors are genes classified in Modules 1 or 3, indicating that CcLBD25 might play a role in connecting these two pathways. Many of the CcLBD25 first and second layer neighbors are pectin degradation-related genes like PL and PMEI. On the other hand, CcLBD4 is not in the LCM GCN, and CcWRKY71 is at a marginal location with only one connection. This result provided further support for our hypothesis that CcLBD25 is the major TF regulating cell wall modification in the haustorium penetration process. CcLBD4 and CcWRKY71 might also be key regulators but are likely involved in a different aspect of haustorium development. Thus, we focused our attention on understanding the function of CcLBD25 in haustorium development.
Cross-species RNAi (host-induced gene silencing) CcLBD25 affects whole-plant phenotypes and reduces parasite fitness
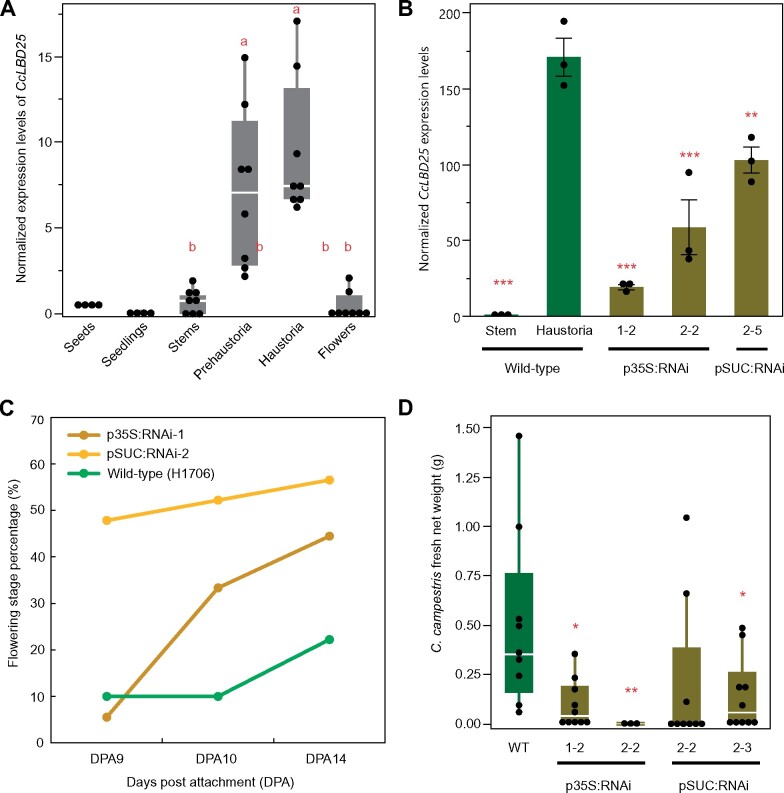
In our previous studies, we found cross-species transport of mRNAs and siRNAs between C. campestris and their hosts (when the initial haustoria are successfully connected with host vascular tissue), and demonstrated host-induced gene silencing (HIGS; Runo et al., 2011; Alakonya et al., 2012). Many previous studies have also shown that large-scale mRNA and small RNAs are transported through the haustorium connections in Cuscuta species (Kim et al., 2014; Johnson et al., 2019). Therefore, we generated transgenic host tomatoes with hairpin RNAs that target and down-regulate CcLBD25 after the parasite forms the first attachment and takes up RNAs from the host (Supplemental Figure S7). When C. campestris grows on wild-type (WT) tomato hosts, CcLBD25 is highly expressed in invasive tissues (Figure 5, A and B). However, CcLBD25 expression levels are significantly knocked down in the tissues on and near the initial functional attachment sites of C. campestris plants that are growing on CcLBD25 RNAi transgenic plants (Figure 5B). Based on our results, expressing CcLBD25 RNAi constructs has no effect on tomato plant growth and, with one exception, transgenic CcLBD25 RNAi tomato plants have the same phenotype as WT plants (Supplemental Figure S8). Notably, CcLBD25 and SlLBD25 nucleotide sequences shared low similarity based on results of Blastn with the most recently published tomato genome (ITAG 4.0; Supplemental Data S1). This also explains why using CcLBD25 RNAi constructs do not influence tomato growth (Supplemental Figure S8). We suspect that the p35S:RNAi line 2-2 might be different from WT due to tissue culture or insertion effects. Nevertheless, pSUC:RNAi would be a better choice for agricultural application due to the specific expression of the RNAi construct in phloem tissues, which would allow efficient transfer of siRNAs to C. campestris after establishment of vascular connection. However, for the purpose of verifying the function of CcLBD25 in C. campestris, both p35S:RNAi lines and pSUC:RNAi lines worked well and successfully downregulated the expression of CcLBD25 in C. campestris (Figure 5B). If CcLBD25 is important in haustorium development and parasitism, then downregulating CcLBD25 should influence haustorium structure or formation, and might also affect nutrient transport. To verify our hypothesis, we measured flowering time in C. campestris growing on various tomato hosts. The result showed that parasites growing on CcLBD25 RNAi transgenic tomatoes transitioned to the flowering stage and subsequently senesced earlier than those growing on WTs (Figure 5C). Based on previous studies, many plant species respond to environmental stress factors by inducing flowering (Wada and Takeno, 2010; Riboni et al., 2014). This early transition to the reproductive stage and senescence in C. campestris grown on CcLBD25 RNAi plants suggests that C. campestris was growing under stress, likely because of nutrient deficiency.
Figure 5.
Gene expression levels and whole-plant phenotypes of C. campestris growing on HIGS CcLBD25 RNAi transgenic plants. A, The normalized expression level of CcLBD25 in six different tissue types of C. campestris (from RNA-seq data). Data presented are assessed using pair-wise comparisons with the Tukey’s test. P-values of the contrasts between “a” and “b” are ˂0.001. B, Expression levels of CcLBD25 in C. campestris haustoria grown on WT tomatoes and T2 CcLBD25 RNAi transgenic plants (by RT-qPCR). Data presented are assessed using one-tailed Welch’s t test with WT haustoria as control. *P-value < 0.05; **P-value < 0.01; ***P-value < 0.005. The error bars indicate standard errors of the data. All data points are plotted as black dots. C, The flowering time of C. campestris growing on WT tomatoes and T1 CcLBD25 RNAi transgenic plants. The early transition to the flowering stage indicates that C. campestris may be growing under stress conditions because they might not obtain sufficient nutrients from their host. Quantification was assessed with the whole plant as a unit. Sample size: WT, 9 biological replicates; p35S:RNAi-1, 18 biological replicates; pSUC:RNAi-2, 23 biological replicates. D, Biomass of C. campestris growing on WT tomatoes and T2 CcLBD25 RNAi transgenic plants. Fresh net weights of C. campestris were measured in grams. Data presented are assessed using one-tailed Welch’s t test with WT as control. *P-value < 0.05; **P-value < 0.01. B–D, p35S:RNAi indicates the transgenic plants with the 35S promoter driving the CcLBD25 RNAi construct. pSUC:RNAi indicates the transgenic plants with the SUC2 promoter driving the CcLBD25 RNAi construct. A and D, The centerline in the box indicates the median. The bottom and top of the box indicate the 25th and 75th quantiles. The whiskers represent the expected variation of the data. The whiskers extend 1.5 times the interquartile range from the top and bottom of the box. All measured data points are plotted as black dots.
To verify if downregulating CcLBD25 affects the ability of the parasite to acquire resources from the host, we also measured the biomass of C. campestris grown on WT H1706 and CcLBD25 RNAi transgenic plants. At 14 DPA, we noticed that C. campestris plants grown on CcLBD25 RNAi transgenic tomatoes had less biomass compared with the C. campestris plants grown on WT H1706 (Figure 5D). Both whole-plant level phenotypes suggest that CcLBD25 might be involved in haustorium development and knocking down the expression level of CcLBD25 influences the ability of C. campestris to establish connections with hosts and interferes with parasite nutrient acquisition.
Using an in vitro haustoria system to investigate the impact of CcLBD25 on early-stage haustorium development
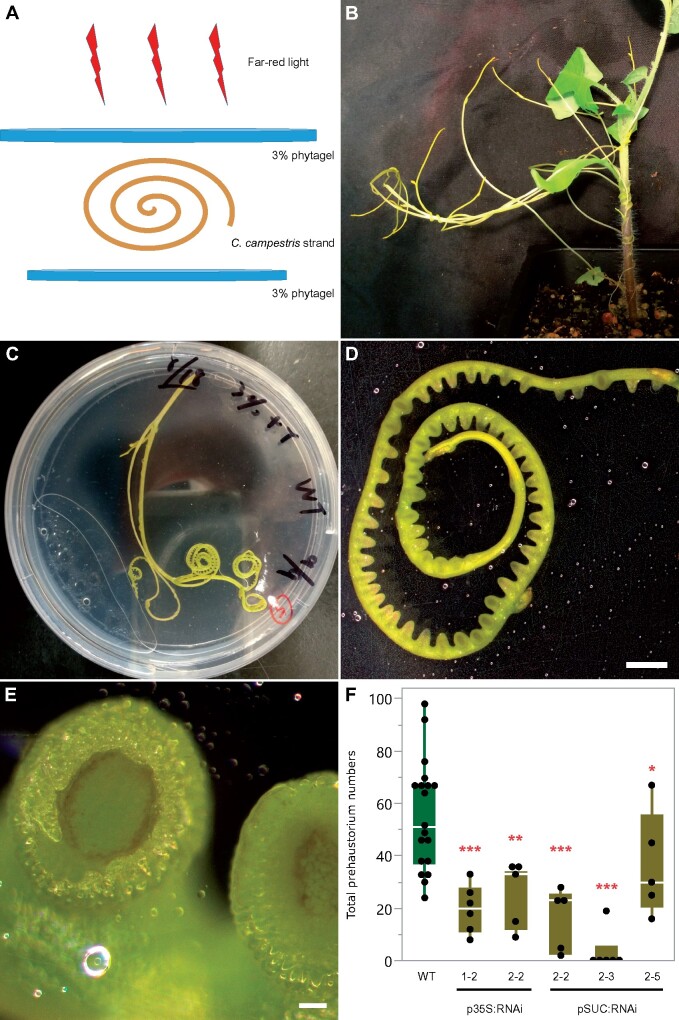
Previous studies indicate that several auxin-inducible LBD genes function in lateral root initiation (Goh et al., 2012). We noticed that auxin efflux carriers and auxin-responsive genes are also in the SOM9 GCN (Figure 2). Therefore, we proposed that CcLBD25 might regulate early-stage haustorium development in C. campestris. In order to assay the role of CcLBD25 in C. campestris haustorium initiation, we developed an in vitro haustorium (IVH) system coupled with HIGS (Figure 6A). This method is inspired by the previous discovery that Cuscuta haustoria can be induced by physical contact and far-red light signals (Tada et al., 1996) and many studies confirmed that small RNAs and mRNAs can move cross-species through the haustorial phloem connection (David-Schwartz et al., 2008; Alakonya et al., 2012; Kim et al., 2014; Johnson et al., 2019). Therefore, we took the C. campestris strands growing near the haustorium attachment sites on WT and CcLBD25 RNAi transgenic tomato (Figure 6B) and sandwiched these strands between two layers of agar to provide sufficient physical contact (Figure 6, A and C). We then illuminated these plates under far-red light for 5 d, at which point prehaustoria are readily visible (Figure 6, D–E). Since the IVH induction is rapid and these prehaustoria can easily be separated from the agar, this method allowed us to count prehaustoria numbers under the microscope and validate the effect of CcLBD25 RNAi on haustorium initiation. The strands from the C. campestris grown on CcLBD25 RNAi transgenic tomatoes produced much fewer prehaustoria than the strands from those grown on WT (Figure 6F). This result indicates that reduced CcLBD25 expression impeded haustorium initiation and confirms that CcLBD25 is a key regulator of early-stage haustorium development, as suggested by our LCM RNA-seq analysis results.
Figure 6.
Far-red light-induced IVH phenotypes of C. campestris growing on T2 HIGS CcLBD25 RNAi transgenic plants. A, An illustration of the setup for the IVH system. Each C. campestris strand that was used in the IVH system is ∼10 cm long. B, Cuscuta campestris strands near the haustorium attachment sites. C, An IVH plate with a C. campestris strand sandwiched in between two layers of agar to provide sufficient physical contact signals. D and E, After illuminating these plates under far-light for 5 d, prehaustoria is readily visible. D, Scale bar = 2 mm. E, Scale bar = 100 µm. F, C. campestris strands were detached and subjected to IVH, and the numbers of prehaustoria were counted. Data presented are assessed using one-tailed Welch’s t test with WT as control. *P-value < 0.06; **P-value < 0.001; ***P-value < 0.0005. p35S:RNAi indicates the transgenic plants with the 35S promoter driving the CcLBD25 RNAi construct. pSUC:RNAi indicates the transgenic plants with the SUC2 promoter driving the CcLBD25 RNAi construct. The centerline in the box indicates the median. The bottom and top of the box indicate the 25th and 75th quantiles. The whiskers represent the expected variation of the data. The whiskers extend 1.5 times the interquartile range from the top and bottom of the box. All measured data points are plotted as black dots.
Down-regulation of CcLBD25 leads to structural changes in haustoria
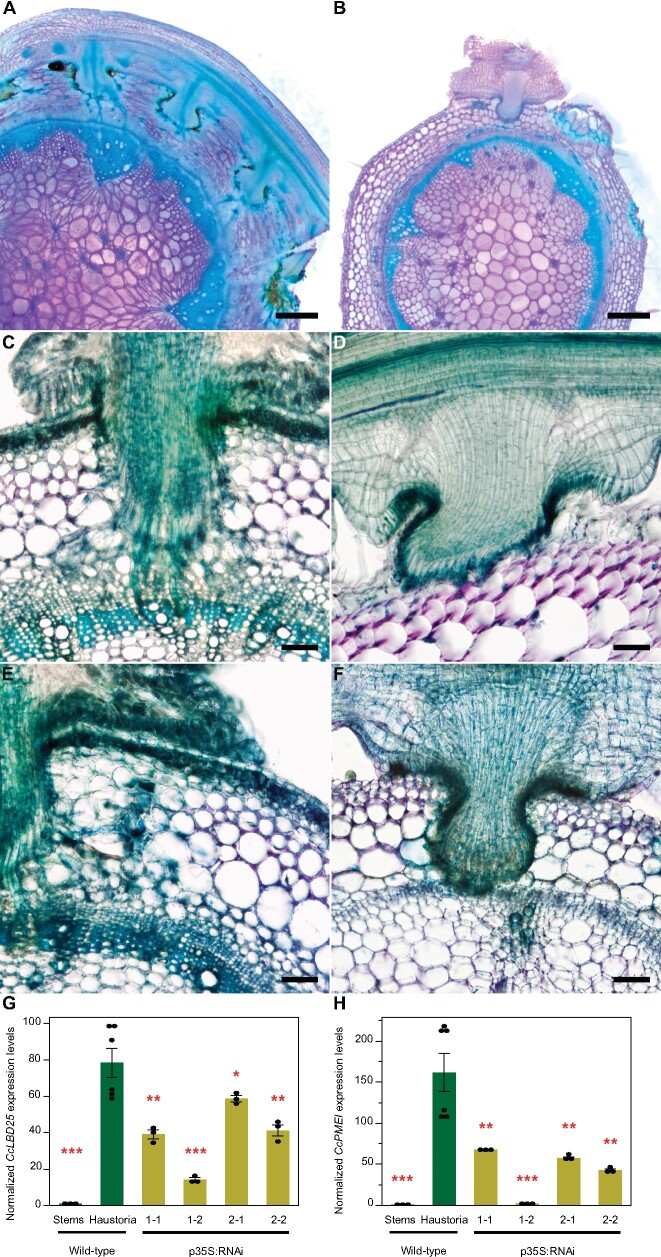
To verify the crucial role CcLBD25 plays in haustorium development and to investigate how down-regulating CcLBD25 affects haustorium structure and the parasitism process, we prepared 100 µm-thick fresh haustorium sections using a vibratome, and stained them with Toluidine Blue O (O’Brien et al., 1964). In sections of haustoria growing on WT plants, we could observe searching hyphae that had penetrated the host cortex region and transformed into xylic or phloic hyphae as they had connected to host xylem and phloem (Figure 7, A, C, and E). However, we observed that many haustoria growing on CcLBD25 RNAi transgenic tomatoes had formed a dome shape structure and lacked searching hyphae (Figure 7, B, D, and F; Supplemental Figure S9; Supplemental Tables S10 and S11). This result indicates that CcLBD25 might be involved in development of searching hyphae. Therefore, knocking down of CcLBD25 affects the ability of C. campestris to establish connections with the host vascular system and leads to nutrient deficiency, as observed in the whole-plant level phenotypes.
Figure 7.
Haustorium phenotypes and gene expression levels of C. campestris growing on HIGS CcLBD25 RNAi transgenic plants. A, C, and E, Cuscuta campestris haustoria growing on a WT H1706 host. B, D, and F, Cuscuta campestris haustoria growing on T2 CcLBD25 RNAi transgenic tomato plants. B, p35S:RNAi line 1-1. D, pSUC:RNAi line 1-2. F, p35S:RNAi line 1-2. A, B, Scale bars = 500 µm. C–F, Scale bars = 100 µm. A–F, 100 µm thick vibratome sections of fresh haustorium stained with Toluidine Blue O. G, H, Expression levels of CcLBD25 and CcPMEI in C. campestris haustoria grown on WT tomatoes and CcLBD25 RNAi transgenic plants. Data presented are assessed using one-tailed Welch’s t test with WT haustoria as control. *P-value < 0.05; **P-value < 0.005; ***P-value < 0.001. p35S:RNAi indicates the transgenic plants with the 35S promoter driving CcLBD25 RNAi construct. The error bars indicate standard errors of the data.
We also noticed the down-regulation of CcLBD25 influenced the parasite penetration process. Fresh tissue sections of the haustoria growing on WT showed a clear zone in tomato cortex tissues near haustorium tissues (Figure 7, A, C, and E). Since the metachromatic staining of Toluidine Blue O is based on cell wall composition and pH values and a pink to purple color indicates pectin presence, this result indicates that the pectins in tomato cortex tissues may have been digested or the pH condition in the cell wall had been changed in the haustorium penetration process (Figure 7, A, C, and E). On the other hand, the C. campestris growing on CcLBD25 RNAi transgenic tomatoes still showed pink to purple color in the cortex near the haustorium attachment sites (Figure 7, B and D). Hence, less pectin digestion or cell wall modification happened in tomato cortex tissues near these CcLBD25 downregulated haustorium tissues compared to the haustoria growing on WT. These haustorium structural phenotypes correspond well with our SOM9 GCN from the C. campestris tissue transcriptome. CcLBD25 is one of the TF central hub genes in Module 1, with many first and second layers of connection to genes involved in cell wall modification, including PLs and pectin methyl-esterase (PME) inhibitors (PMEIs). Based on many previous studies, the interplay between PME and PMEI is an important determinant of cell wall loosening, strengthening, and organ formation (Wormit and Usadel, 2018). Therefore, we hypothesized that PMEIs might be one of the key regulators that cause the haustorium phenotype in CcLBD25-downregulated haustorium tissues. To test if the downregulation of CcLBD25 would affect PMEI expression levels, we conducted qPCR to detect CcPMEI expression levels in the tissues of C. campestris plants that are growing on CcLBD25 RNAi transgenic plants. Our results show that CcPMEI expression levels are also significantly reduced when CcLBD25 is knocked down (Figure 7, G and H). Thus, CcLBD25 might directly or indirectly regulate CcPMEI at the transcriptional level. These results verify the hypothesis that CcLBD25 plays an important role in haustorium development and might regulate cell wall modification.
Discussion
In this study, we demonstrate that CcLBD25 is a crucial regulator of several aspects of C. campestris haustorium development, including haustorium initiation, cell-wall loosening, and searching hyphae growth. We use transcriptome of six C. campestris tissue types and RNA-seq data of LCM captured haustoria at three developmental stages to reveal the potential molecular mechanisms and the complexity of gene networks that are regulated during the haustorium formation process. Our results provide a comprehensive analysis of the CcLBD25 centered regulatory system and illustrate that CcLBD25 might directly or indirectly coordinate different groups of genes that are expressed only at the early or mature stage during haustorium development.
Lateral root development and haustorium development
In nonparasitic plants, like Arabidopsis, AtLBD25 was also named DOWN IN DARK AND AUXIN1 because lbd25 mutant plants exhibited reduced sensitivity to auxin and reduced number of lateral roots (Mangeon et al., 2010). These phenotypes indicate that AtLBD25 functions in lateral root formation by promoting auxin signaling (Mangeon et al., 2010). In the root parasitic plant, T. chinense, TcLBD25 was highly expressed during haustorium formation (Ichihashi et al., 2017). This supports the hypothesis that root parasitic plants co-opted the lateral root formation machinery into haustorium organogenesis. However, whether rootless stem parasitic plants Cuscuta spp. also followed the same path to generating haustoria was unknown. In this study, we identified CcLBD25 as playing a key role in Cuscuta haustorium development. Our SOM9 GCN shows that auxin efflux carriers and auxin-responsive genes are also remotely connected with CcLBD25, but not in the first or second layers of neighbors (Figure 2). Our hypothesis is that the increased expression of CcLBD25 might induce the genes that are involved in auxin signaling, which was observed in the Arabidopsis lateral root development system (Dean et al., 2004; Mangeon et al., 2010). These pieces of evidence suggest that Cuscuta spp. adapted not only the shoot developmental programs (Alakonya et al., 2012), but also the lateral root programming system, into haustorium organogenesis. According to our LBD25 gene phylogenetic tree, CcLBD25 and TcLBD25 likely evolved convergently to function in haustorium development (Supplemental Figure S3; Supplemental Data S1).
Development of searching hyphae
Down-regulating CcLBD25 reduced searching hyphae formation (Figure 7, B, D, and F; Supplemental Figure S9), indicating that CcLBD25 is involved in searching hyphae development. Surprisingly, AtLBD25 is not only expressed in roots but is also expressed in pollen (Mangeon et al., 2010). Previous reports indicate that AtLBD25 is especially highly expressed during the pollen late developmental stage (Kim et al., 2016). Intriguingly, many genes that are involved in haustoria development also play important roles in flower and pollen development (Yang et al., 2015; Yoshida et al., 2019). Recent research on haustoria 3D structures also indicates that the growth pattern of intrusive cells is similar to the rapid polar growth of pollen tubes (Masumoto et al., 2021). Taken together with our results in this study and previous findings in other organisms, we suggest that the genes that regulate pollen development or pollen tube growth, like LBD25, might be adapted by parasitic plants for development of haustorium intrusive cells and searching hyphae. This discovery also confirmed the hypothesis that parasitic plants co-opted the developmental reprogramming process from multiple sources instead of just a single organ.
Cell adhesion and cell wall loosening in parasitism
The mechanical properties and chemical conditions of cell walls have been reported to be critical for regulating plant organ morphogenesis (Chebli and Geitmann, 2017; Zhao et al., 2018). By remodeling cell wall composition or extracellular environments, plants generate local cell wall loosening and strengthening, which allows anisotropic growth processes to occur (Chebli and Geitmann, 2017). Recent studies also indicate that the interaction between pectin and other cell wall components is an important determinant for plant organogenesis (Chebli and Geitmann, 2017; Saffer, 2018), and the interplay between PME and PMEI plays a vital role in regulating physical properties of the cell wall (Wormit and Usadel, 2018). In the root parasitic plant, Orobanche cumana, a PME is shown to be present at the host and parasite interface and to have pectolytic activity (Losner-Goshen et al., 1998). These results suggest that parasitic plants produce PME to degrade pectin in the host cell wall and help with haustorium penetration. Our SOM9 GCNs shows that CcLBD25 is coexpressed with many PLs and PMEIs (Figures 2B; 3, C–D; 4, H–I), implying that CcLBD25 might be the key TF regulating expression of the enzymes involved in pectin remodeling. The haustoria grown on CcLBD25 RNAi transgenic plants failed to penetrate host tissues and were unable to create a clear zone at the host and parasite interface (Figure 7, A–F), supporting the existence of a tight connection between CcLBD25 and pectin-modifying enzymes. CcLBD25 and PMEIs were coexpressed in the mature stage of haustorium (Figure 4, H–I), when cell wall loosening occurs for haustorium penetration.
On the other hand, since the patterns of demethylesterification on homogalacturonans determine cell wall loosening or strengthening, pectin properties also play a role in cell adhesion, which is regulated by PME and PMEI (Wormit and Usadel, 2018). Previous studies also indicate that Cuscuta spp. secrete pectin-rich adhesive materials to help with adhesion and allow attachment to their hosts (Vaughn, 2002; Shimizu and Aoki, 2019). This is consistent with our discovery that both CcLBD25 and PMEIs are highly expressed in the early stage of haustorium development, which would be responsible for the adhesion process in C. campestris (Figure 4, H–I).
Conclusions
Our detailed bioinformatic analysis on previously published C. campestris tissue type transcriptome coupled with LCM of RNA-seq data from three haustorium developmental stages helped us discern the molecular mechanism of parasitic plant haustorium development. The discovery that CcLBD25 plays a pivotal role in many aspects of haustorium formation shows that the regulatory machinery of haustorium development is potentially shared by both root and stem parasites. Although previous studies have indicated that parasitic plants evolved independently in about 13 different families, this conserved molecular mechanism supports the hypothesis that stem parasitic plants also adapted the lateral root formation programming of nonparasitic plants into haustorium development. The results of this study not only provide an insight into molecular mechanisms by which LBD25 may regulate parasitic plant haustorium development, but also raise potential for developing a universal parasitic weed-resistant crop that can defend against both stem and root parasitic plants at the same time.
Materials and methods
Cuscuta campestris materials
We thank W. Thomas Lanini for providing dodder seeds collected from tomato field in California. These dodder materials were previously identified as Cuscuta pentagona (Yaakov et al., 2001), a species closely related to C. campestris (Costea et al., 2015). We used molecular phylogenetics of plastid trnL-F intron / spacer region, plastid ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit, nuclear internal transcribed spacer, and nuclear large-subunit ribosomal DNA sequences (Stefanović et al., 2007; García et al., 2014; Costea et al., 2015) to verify our dodder isolate is the same as C. campestris 201, Rose 46281 (WTU ) from California, USA (Jhu et al., 2020) by comparing with published sequences (Costea et al., 2015).
RNA-seq data mapping and processing
For C. campestris tissue type RNA-seq analysis, we used the raw data previously published (Ranjan et al., 2014). This RNA-seq data contain six different C. campestris tissues, including seeds, seedlings, stems, prehaustoria, haustoria, and flowers, grown on the tomato (Solanum lycopersicum) H1706 cultivar and N. benthamiana. We mapped both C. campestris tissue type and LCM RNA-seq data to the genome of C. campestris (Vogel et al., 2018) with Bowtie 2 (Langmead and Salzberg, 2012) and used EdgeR (Robinson et al., 2009) to get normalized trimmed mean of M values for further analysis.
MDS and PCA with SOM clustering
After normalization steps, we used cmdscale in R stats package to create MDS data matrix and then generate MDS plots. For C. campestris tissue types RNA-seq data, we selected genes with coefficient of variation >0.85 for PCA analysis. We calculated PC values using prcomp function in R stats package. Selected genes are clustered for multilevel six-by-two hexagonal SOM using som function in the Kohonen package (Wehrens and Buydens, 2007). We visualized the SOM clustering results in PCA plots. The complete gene lists for all SOM units in C. campestris tissue type RNA-seq data with SOM distances and PCA PC values are included in Supplemental Table S1. For C. campestris LCM RNA-seq data, genes in the upper 50% quartile of coefficient of variation were selected for further analysis. Selected genes were then clustered for multilevel three-by-two hexagonal SOM. The complete gene lists for all SOM units in LCM RNA-seq data with SOM distances and PCA PC values are included in Supplemental Table S7.
Construct GCNs
We used the genes that are classified in selected SOM groups to build GCNs. The R script is modified from our previously published method (Ichihashi et al., 2014) and the updated script is uploaded to GitHub and included in code availability. The SOM9 GCNs for C. campestris tissue type data was constructed with normal quantile cutoff = 0.93. The SOM2 + 3 + 9 GCNs for C. campestris tissue type data was constructed with normal quantile cutoff = 0.94. For the GCN of C. campestris LCM data, we used the SOM9 gene list from tissue type RNA-seq and constructed the GCN of these genes based on the expression profiles in LCM RNA-seq data with normal quantile cutoff = 0.94. These networks were then visualized using Cytoscape version 3.8.0.
Functional annotation and GO enrichment analysis of RNA-seq data
Since many genes are not functionally annotated in the recently published C. campestris genome (Vogel et al., 2018), we used BLASTN with 1e-5 as an e-value threshold to compare our previously annotated transcriptome final contigs with current C. campestris genome genes and only kept the highest scored hit for each gene (Supplemental Table S3). After we obtained this master list, we combined the functional annotation of our published transcriptome based on the NCBI nonredundant database and TAIR10 (Ranjan et al., 2014) with the C. campestris genome gene IDs to create a more complete functional annotation (Supplemental Table S9). TAIR ID hits were used for GO Enrichment Analysis on http://geneontology.org/ for gene clusters and modules.
LCM RNA-seq library preparation and sequencing
We infested approximately four-leaves-stage H1706 tomato plants with C. campestris strands. Tomato stems with haustoria were collected at 4 DPA and fixed in formaldehyde–acetic acid–alcohol (FAA). These samples were dehydrated by the ethanol series and embedded in paraffin (Paraplast X-Tra; Thermo Fisher Scientific, Waltham, MA, USA). We prepared 10 μm thick sections on a Leica RM2125RT rotary microtome. Approximately 30 regions of 10 um thickness each were cut from each slide, and three to four slides used per library preparation. Tissue was processed within one month of fixation to ensure RNA quality. Haustorial tissues of the three defined developmental stages were dissected on a Leica LMD6000 Laser Microdissection System. Tissue was collected in lysis buffer from RNAqueous-Micro Total RNA Isolation Kit (Ambion, Austin, TX, USA) and stored at −80°C. RNA was extracted using RNAqueous-Micro Total RNA Isolation Kit (Ambion) and amplified using WT-Ovation Pico RNA Amplification System version 1.0 (NuGEN Technologies Inc., San Carlos, CA, USA) following manufacturer instructions. RNA-seq libraries for Illumina sequencing were constructed following a previously published method (Kumar et al., 2012) with slight modifications. Libraries were quantified, pooled to equal amounts, and their quality was checked on a Bioanalyzer 2100 (Agilent, Santa Clara, CA, USA). Libraries were sequenced on a HiSeq2000 Illumina Sequencer at the Vincent J Coates Genomics Sequencing Laboratory at UC Berkeley.
CcLBD25 RNAi transgenic plants and HIGS efficiency verification
We used the pTKO2 vector (Snowden et al., 2005; Brendolise et al., 2017), which enables streamlined cloning by using two GATEWAY cassettes positioned at opposite directions, separated by an Arabidopsis ACT2 intron, and under the control of the 35S constitutive promoter. We have previously shown that producing the RNAi construct in phloem cells specifically using the SUC2 promoter was effective for dodder HIGS (Alakonya et al., 2012). Therefore, we replaced the 35S promoter with the SUC2 promoter and generated pTKOS (Supplemental Figure S7). We used BLAST to identify a 292 bp fragment that was specific to CcLBD25 and different from tomato genes (Supplemental Dataset S2). This RNAi fragment was amplified from C. campestris gDNA, TOPO cloned into pCR8/GW-TOPO (Life Technologies, Carlsbad, CA, USA) and LR recombined into pTKO2 and pTKOS (Supplemental Dataset S3). These constructs were then sent to the UC Davis Plant Transformation Facility to generate CcLBD25 RNAi transgenic tomato plants.
All T0 transgenic plants were selected by kanamycin resistance and their gDNAs were extracted and PCR performed to verify they contained CcLBD25 RNAi constructs. To validate HIGS efficiency and quantify the expression level of CcLBD25 and CcPMEI in C. campestris, dodder tissues were harvested from both C. campestris grown on WT plants and T2 CcLBD25 RNAi transgenic plants. For validating the downregulation of CcLBD25, we collected the stem segment with haustoria and prehaustoria at the initial attachment site from C. campestris grown on WT and RNAi transgenic plants. About 100 mg tissues were used for each RNA extraction for each genotype sample. We froze tissues in liquid nitrogen and ground them in extraction buffer using a bead beater (Mini Beadbeater 96; BioSpec Products). Following our previously published poly-A-based RNA extraction method (Townsley et al., 2015), we obtained total mRNA from C. campestris and then used Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA) for reverse transcription to synthesize cDNA as described by the manufacturer instructions. Real-time qPCR was performed using a Bio-Rad iCycler iQ real-time thermal cycler with Bio-Rad IQ SYBR Green super mix. The sequences of qPCR primer pairs are included in Supplemental Table S12.
Whole-plant phenotype assays
Based on previous studies, many plant species are reported to have early flowering phenotypes in response to environmental stresses (Wada and Takeno, 2010; Riboni et al., 2014). Therefore, we grew C. campestris on WT H1706 tomatoes and CcLBD25 RNAi T1 transgenic tomato plants and then quantified how fast these C. campestris plants transitioned to their reproductive stage. The number of C. campestris plants that transitioned to the flowering stage was counted at 9, 10, and 14 DPA to test whether a stress-induced flowering phenotype could be observed.
To quantify the effect of CcLBD25 downregulation on C. campestris growth, we infested 3-week-old tomato plants with ∼10 cm stem segments C. campestris, which were originally grown on WT H1706. We harvested all C. campestris tissues grown on WT H1706 and CcLBD25 RNAi T2 transgenic plants at 14 DPA. These C. campestris tissues were then carefully separated from their host plant stems by hand, and their fresh weights were measured using chemical weighing scales.
IVH system
Inspired by the previous discovery that Cuscuta haustoria can be induced by physical contact and far-red light signals (Tada et al., 1996), we developed an (IVH system for haustorium induction without hosts. In this method, we detached Cuscuta stem segments, which were right next to a stable haustorium attachment, from the C. campestris grown on WT plants and T2 CcLBD25 RNAi transgenic plants. Cuscuta strands with shoot apices detached from a host plant were sandwiched between 3% Phytagel agar containing 0.5× Murashige and Skoog medium to provide tactile stimuli (Figure 6, A–C). These combined plates were then irradiated with far-red light for 2 h. After 5 d of growth in darkness in a 22°C growth chamber, prehaustoria were readily visible (Figure 6, D–E). We then counted the number of prehaustoria under a Zeiss SteREO Discovery, V12 microscope for quantification. Since the RNAi silencing signal is systemic (David-Schwartz et al., 2008; Alakonya et al., 2012)and IVH induction is rapid, we could validate the effect of CcLBD25 RNAi on haustoria development. We are aware of a similar system that was reported recently (Kaga et al., 2020). However, we used two layers of agar gel instead of one layer of gel with one glass slide. This prevented prehaustoria from attaching to the glass slide, making it easier to detach prehaustoria for further analysis without damaging their structure. Second, we used far-red light instead of blue light irradiation. Both methods seem to be effective.
Fresh tissue sectioning and histology
For fresh vibratome sections of haustoria attached to WT and CcLBD25 RNAi host stems, we collected samples and embedded them in 7% Plant Tissue Culture Agar. We then fixed these agar blocks in FAA (final concentration: 4% formaldehyde, 5% glacial acetic acid, and 50% ethanol) overnight, 50% ethanol for 1 h, and then transferred the samples to 70% ethanol for storage. These agar blocks were then sectioned using Lancer Vibratome Series 1000 to prepare 100 μm sections. We kept these sections in 4°C water and then conducted Toluidine Blue O Staining. We followed the published protocol (O'Brien et al., 1964) with some modifications. The sections were immersed in the stain for 30 s, and then washed with water three times for 30 s each wash. After removing the agar from around the sections using forceps, we mounted the sections with water on a slide and imaged using a Zeiss SteREO Discovery, V12 microscope, and a Nikon Eclipse E600 microscope.
Code availability
Updated R scripts for MDS, PCA, and SOM analysis and GCN analysis are all deposited on GitHub (Link: https://github.com/MinYaoJhu/CcLBD25_project.git).
Data availability
All data are available in the main text or the supplemental materials. LCM RNA-seq raw data are deposited on NCBI Sequence Read Archive (SRA) PRJNA687611.
Accession numbers
LCM RNA-seq raw reads from this article can be found in the NCBI SRA data under accession number PRJNA687611.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . MDS plot of expression profiles of all libraries from six different C. campestris tissue types mapped to the C. campestris genome.
Supplemental Figure S2 . Heatmaps of gene expression profiles in z-scores for SOM2, SOM3, and SOM9 from C. campestris tissue type RNA-seq data mapped to C. campestris genome.
Supplemental Figure S3 . LBD25 phylogenetic tree with top significant sequences aligned with CcLBD25, TcLBD25, AtLBD25, and SlLBD25.
Supplemental Figure S4 . MDS plot of RNA expression profile in all libraries from LCM of three different C. campestris developmental stages mapped to C. campestris genome.
Supplemental Figure S5 . PCA analysis with SOM clustering and GCNs of gene expression in C. campestris haustoria across three developmental stages in LCM RNA-seq data.
Supplemental Figure S6 . Heatmap of gene expression profiles in z-scores for SOM6 from C. campestris LCM RNA-seq data mapped to C. campestris genome.
Supplemental Figure S7 . CcLBD25 RNAi constructs for HIGS.
Supplemental Figure S8 . Whole-plant phenotypes of CcLBD25 RNAi transgenic tomato plants without C. campestris infestation treatment.
Supplemental Figure S9 . Quantification of haustorium status on CcLBD25 RNAi HIGS and WT plants.
Supplemental Table S1 . The SOM clustering gene list in C. campestris tissue type RNA-seq data and results of PCA analysis and multilevel SOM clustering using selected genes with coefficient of variation >0.85.
Supplemental Table S2 . The gene list of SOM9 GCN modules from C. campestris tissue type RNA-seq data.
Supplemental Table S3 . Combined annotation of C. campestris genes and transcriptome.
Supplemental Table S4 . The GO enrichment results and statistics of SOM9 GCN modules from C. campestris tissue type RNA-seq data.
Supplemental Table S5 . The gene list of SOM2, 3, 9 combined GCN modules from C. campestris tissue type RNA-seq data.
Supplemental Table S6 . The GO enrichment results and statistics of SOM2, 3, 9 combined GCN modules from C. campestris tissue type RNA-seq data.
Supplemental Table S7 . The SOM clustering gene list in LCM RNA-seq data and results of PCA analysis and multilevel SOM clustering using selected genes in the upper 50% quartile of coefficient of variation.
Supplemental Table S8 . The gene list in the modules of the GCN based on LCM RNA-seq expression with genes in tissue type RNA-seq SOM9.
Supplemental Table S9 . The GO enrichment results and statistics of the modules of the GCN based on LCM RNA-seq expression with genes in tissue type RNA-seq SOM9.
Supplemental Table S10 . Quantification and statistics of haustorium status on CcLBD25 RNAi HIGS and WT plants by section ID.
Supplemental Table S11 . Quantification and statistics of haustorium status on CcLBD25 RNAi HIGS and WT plants by sample ID.
Supplemental Table S12 . The primer pairs that are used for making the construct and quantifying expression level of CcLBD25 by quantitative reverse transcriptase PCR (RT-qPCR).
Supplemental Dataset S1 . Nucleotide sequence alignments with CcLBD25, TcLBD25, AtLBD25, SlLBD25, and their top 10 significant aligned sequences.
Supplemental Dataset S2 . Sequence of the CcLBD25 fragment that is used for making CcLBD25 RNAi construct.
Supplemental Dataset S3 . Sequence of the SUC2 promoter that is used for driving CcLBD25 RNAi construct in pTKOS.
Supplementary Material
Acknowledgments
We are grateful to the UC Davis Plant Transformation Facility for generating CcLBD25 RNAi transgenic tomato plants. We thank Kristina Zumstein for help in maintaining transgenic tomato seed stocks, and Richard Philbrook, Kaiwen Zhang, and Junqi Lu for helping with some parts of experiments and vibratome sectioning. We also thank Aaron Leichty, Steven Rowland, and Karo Czarnecki for their input on bioinformatics analyses.
Funding
This work was funded by United States Department of Agriculture-National Institute of Food and Agriculture (USDA-NIFA) (grant no. 2013-02345). M.-Y.J. was supported by Yen Chuang Taiwan Fellowship, Taiwan Government Scholarship to Study Abroad (GSSA), Elsie Taylor Stocking Memorial Fellowship, Katherine Esau Summer Graduate Fellowship, Loomis Robert S. and Lois Ann Graduate Fellowship in Agronomy, and the University of California Davis Graduate Research Award. Y.I. was supported by Grant-in-Aid for Young Scientists from the Ministry of Education, Culture, Sports, Science, and Technology, Japan (B; grant no. 15K18589 to Y.I.).
Conflict of interest statement: The authors declare that they have no conflicts of interest.
Y.I. initiated the project and all work was done under consultation and supervision from N.R.S.; Y.I. wrote the original R scripts for MDS, PCA, and SOM analysis, and M.-Y.J. edited the scripts and applied them on further analyses; M.-Y.J. mapped the transcriptome data to C. campestris genome and performed SOM clustering and coexpression network analysis; M.F. made CcLBD25 RNAi constructs and used the UC Davis transformation facility to generate transgenic plants; M.-Y.J. performed qPCR, functional characterization, and phenotyping of CcLBD25 transgenic plants; M.-Y.J. designed the IVH system and conducted HIGS experiments and analyzed the data; M.F. conducted LCM to capture tissues and prepared libraries for RNA-seq; M.-Y.J. mapped the LCM RNA-seq data to C. campestris genome and performed SOM clustering and GCN analysis; M.Y.J. conducted phylogenetic analysis and verified Cuscuta species; C.W. analyzed haustorium connection status; M.-Y.J. wrote the initial manuscript and made figures and tables with primary editing from N.R.S.; N.R.S. supervised this project and serves as the author responsible for contact and communication.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is: Neelima R. Sinha (nrsinha@ucdavis.edu).
References
- Agrios GN (2005) Chapter 13—Plant diseases caused by parasitic higher plants, invasive climbing plants, and parasitic green algae. Plant Pathology, Ed 5.Academic Press, San Diego, CA, pp 705–722 [Google Scholar]
- Alakonya A, Kumar R, Koenig D, Kimura S, Townsley B, Runo S, Garces HM, Kang J, Yanez A, David-Schwartz R, et al. (2012) Interspecific RNA interference of SHOOT MERISTEMLESS-like disrupts Cuscuta pentagona plant parasitism. Plant Cell 24:3153–3166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendolise C, Montefiori M, Dinis R, Peeters N, Storey RD, Rikkerink EH (2017) A novel hairpin library-based approach to identify NBS–LRR genes required for effector-triggered hypersensitive response in Nicotiana benthamiana. Plant Methods 13:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chebli Y, Geitmann A (2017) Cellular growth in plants requires regulation of cell wall biochemistry. Curr Opin Cell Biol 44:28–35 [DOI] [PubMed] [Google Scholar]
- Clauset A, Newman MEJ, Moore C (2004) Finding community structure in very large networks. Phys Rev E 70:066111. [DOI] [PubMed] [Google Scholar]
- Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. (2007) Integration of biological networks and gene expression data using Cytoscape. Nat Protocol 2:2366–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costea M, García MA, Baute K, Stefanović S (2015) Entangled evolutionary history of Cuscuta pentagona clade: a story involving hybridization and Darwin in the Galapagos. Taxon 64:1225–1242 [Google Scholar]
- David-Schwartz R, Runo S, Townsley B, Machuka J, Sinha N (2008) Long-distance transport of mRNA via parenchyma cells and phloem across the host–parasite junction in Cuscuta. New Phytol 179:1133–1141 [DOI] [PubMed] [Google Scholar]
- Dean G, Casson S, Lindsey K (2004) KNAT6 gene of Arabidopsis is expressed in roots and is required for correct lateral root formation. Plant Mol Biol 54:71–84 [DOI] [PubMed] [Google Scholar]
- Furuhashi T, Furuhashi K, Weckwerth W (2011) The parasitic mechanism of the holostemparasitic plant Cuscuta. J Plant Interact 6:207–219 [Google Scholar]
- García MA, Costea M, Kuzmina M, Stefanović S (2014) Phylogeny, character evolution, and biogeography of Cuscuta (dodders; Convolvulaceae) inferred from coding plastid and nuclear sequences. Am J Bot 101:670–690 [DOI] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, Fukaki H (2012) The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139:883–893 [DOI] [PubMed] [Google Scholar]
- Ichihashi Y, Aguilar-Martínez JA, Farhi M, Chitwood DH, Kumar R, Millon LV, Peng J, Maloof JN, Sinha NR (2014) Evolutionary developmental transcriptomics reveals a gene network module regulating interspecific diversity in plant leaf shape. Proc Natl Acad Sci 111:E2616–E2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Hakoyama T, Iwase A, Shirasu K, Sugimoto K, Hayashi M (2020) Common mechanisms of developmental reprogramming in plants—lessons from regeneration, symbiosis, and parasitism. Front Plant Sci 11: 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi Y, Kusano M, Kobayashi M, Suetsugu K, Yoshida S, Wakatake T, Kumaishi K, Shibata A, Saito K, Shirasu K (2017) Transcriptomic and metabolomic reprogramming from roots to haustoria in the parasitic plant, Thesium Chinense. Plant Cell Physiol 59:729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhu MY, Farhi M, Wang L, Philbrook RN, Belcher MS, Nakayama H, Zumstein KS, Rowland SD, Ron M, Shih PM, et al. (2020) Lignin-based resistance to Cuscuta campestris parasitism in Heinz resistant tomato cultivars. bioRxiv:706861. [DOI] [PMC free article] [PubMed]
- Johnson NR, dePamphilis CW, Axtell MJ (2019) Compensatory sequence variation between trans-species small RNAs and their target sites. eLife 8:e49750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaga Y, Yokoyama R, Sano R, Ohtani M, Demura T, Kuroha T, Shinohara N, Nishitani K (2020) Interspecific signaling between the parasitic plant and the host plants regulate xylem vessel cell differentiation in haustoria of Cuscuta campestris. Front Plant Sci 11: 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim G, LeBlanc ML, Wafula EK, dePamphilis CW, Westwood JH (2014) Genomic-scale exchange of mRNA between a parasitic plant and its hosts. Science 345:808–811 [DOI] [PubMed] [Google Scholar]
- Kim M, Kim MJ, Pandey S, Kim J (2016) Expression and protein interaction analyses reveal combinatorial interactions of LBD transcription factors during Arabidopsis pollen development. Plant Cell Physiol 57:2291–2299 [DOI] [PubMed] [Google Scholar]
- Kumar R, Ichihashi Y, Kimura S, Chitwood D, Headland L, Peng J, Maloof J, Sinha N (2012) A high-throughput method for illumina RNA-Seq library preparation. Front Plant Sci 3: 202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ráez JA, Matusova R, Cardoso C, Jamil M, Charnikhova T, Kohlen W, Ruyter-Spira C, Verstappen F, Bouwmeester H (2009) Strigolactones: ecological significance and use as a target for parasitic plant control. Pest Manag Sci 65:471–477 [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losner-Goshen D, Portnoy VH, Mayer AM, Joel DM (1998) Pectolytic activity by the haustorium of the parasitic plant Orobanche L. (Orobanchaceae) in host roots. Ann Bot 81:319–326 [Google Scholar]
- Mangeon A, Bell EM, Lin W-c, Jablonska B, Springer PS (2010) Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J Exp Bot 62:221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masumoto N, Suzuki Y, Cui S, Wakazaki M, Sato M, Kumaishi K, Shibata A, Furuta KM, Ichihashi Y, Shirasu K, et al. (2021) Three-dimensional reconstructions of haustoria in two parasitic plant species in the Orobanchaceae. Plant Physiol 185: 1429–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien TP, Feder N, McCully ME (1964) Polychromatic staining of plant cell walls by toluidine blue O. Protoplasma 59:368–373 [Google Scholar]
- Parker C (2021) Cuscuta Campestris (Field Dodder) in Invasive Species Compendium. CAB International, Wallingford, UK [Google Scholar]
- Porco S, Larrieu A, Du Y, Gaudinier A, Goh T, Swarup K, Swarup R, Kuempers B, Bishopp A, Lavenus J, et al. (2016) Lateral root emergence in Arabidopsis is dependent on transcription factor LBD29 regulation of auxin influx carrier LAX3. Development 143:3340–3349 [DOI] [PubMed] [Google Scholar]
- Ranjan A, Ichihashi Y, Farhi M, Zumstein K, Townsley B, David-Schwartz R, Sinha NR (2014) De novo assembly and characterization of the transcriptome of the parasitic weed dodder identifies genes associated with plant parasitism. Plant Physiol 166:1186–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riboni M, Robustelli Test A, Galbiati M, Tonelli C, Conti L (2014) Environmental stress and flowering time: the photoperiodic connection. Plant Signal Behav 9:e29036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runo S, Alakonya A, Machuka J, Sinha N (2011) RNA interference as a resistance mechanism against crop parasites in Africa: a ‘Trojan horse’ approach. Pest Manag Sci 67:129–136 [DOI] [PubMed] [Google Scholar]
- Runyon JB, Mescher MC, Felton GW, De Moraes CM (2010) Parasitism by Cuscuta pentagona sequentially induces JA and SA defence pathways in tomato. Plant Cell Environ 33:290–303 [DOI] [PubMed] [Google Scholar]
- Saffer AM (2018) Expanding roles for pectins in plant development. J Integr Plant Biol 60:910–923 [DOI] [PubMed] [Google Scholar]
- Shen H, Ye W, Hong L, Huang H, Wang Z, Deng X, Yang Q, Xu Z (2006) Progress in parasitic plant biology: host selection and nutrient transfer. Plant Biol 8:175–185 [DOI] [PubMed] [Google Scholar]
- Shimizu K, Aoki K (2019) Development of parasitic organs of a stem holoparasitic plant in genus cuscuta. Front Plant Sci 10:1435–1435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ (2005) The decreased apical dominance1/Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. Plant Cell 17:746–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanović S, Kuzmina M, Costea M (2007) Delimitation of major lineages within Cuscuta subgenus Grammica (Convolvulaceae) using plastid and nuclear DNA sequences. Am J Bot 94:568–589 [DOI] [PubMed] [Google Scholar]
- Sun G, Xu Y, Liu H, Sun T, Zhang J, Hettenhausen C, Shen G, Qi J, Qin Y, Li J, et al. (2018) Large-scale gene losses underlie the genome evolution of parasitic plant Cuscuta australis. Nat Commun 9: 2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada Y, Sugai M, Furuhashi K (1996) Haustoria of Cuscuta japonica, a holoparasitic flowering plant, are induced by the cooperative effects of far-red light and tactile stimuli. Plant Cell Physiol 37:1049–1053 [Google Scholar]
- Townsley BT, Covington MF, Ichihashi Y, Zumstein K, Sinha NR (2015) BrAD-seq: breath adapter directional sequencing: a streamlined, ultra-simple and fast library preparation protocol for strand specific mRNA library construction. Front Plant Sci 6: 366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughn KC (2002) Attachment of the parasitic weed dodder to the host. Protoplasma 219:227–237 [DOI] [PubMed] [Google Scholar]
- Vogel A, Schwacke R, Denton AK, Usadel B, Hollmann J, Fischer K, Bolger A, Schmidt MHW, Bolger ME, Gundlach H, et al. (2018) Footprints of parasitism in the genome of the parasitic flowering plant Cuscuta campestris. Nat Commun 9:2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada KC, Takeno K (2010) Stress-induced flowering. Plant Signal Behav 5:944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrens R, Buydens L (2007) Self- and super-organizing maps in R: the kohonen package. J Stat Softw 21:1–19 [Google Scholar]
- Wormit A, Usadel B (2018) The multifaceted role of pectin methylesterase inhibitors (PMEIs). Int J Mol Sci 19:2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaakov G, Lanini WT, Wrobel RL (2001) Tolerance of tomato varieties to lespedeza dodder. Weed Sci 49:520–523 [Google Scholar]
- Yang Z, Wafula EK, Honaas LA, Zhang H, Das M, Fernandez-Aparicio M, Huang K, Bandaranayake PCG, Wu B, Der JP, et al. (2015) Comparative transcriptome analyses reveal core parasitism genes and suggest gene duplication and repurposing as sources of structural novelty. Mol Biol Evol 32:767–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JI, Scholes JD (2010) Host plant resistance to parasitic weeds; recent progress and bottlenecks. Curr Opinion Plant Biol 13:478–484 [DOI] [PubMed] [Google Scholar]
- Yoshida S, Kim S, Wafula EK, Tanskanen J, Kim YM, Honaas L, Yang Z, Spallek T, Conn CE, Ichihashi Y, et al. (2019) Genome sequence of Striga asiatica provides insight into the evolution of plant parasitism. Curr Biol 29:3041–3052.e3044 [DOI] [PubMed] [Google Scholar]
- Zhao F, Chen W, Traas J (2018) Mechanical signaling in plant morphogenesis. Curr Opin Genet Dev 51:26–30 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplemental materials. LCM RNA-seq raw data are deposited on NCBI Sequence Read Archive (SRA) PRJNA687611.