Abstract
The genomic shock of whole-genome duplication (WGD) and hybridization introduces great variation into transcriptomes, for both coding and noncoding genes. An altered transcriptome provides a molecular basis for improving adaptation during the evolution of new species. The allotetraploid cotton, together with the putative diploid ancestor species compose a fine model for study the rapid gene neofunctionalization over the genome shock. Here we report on Drought-Associated Non-coding gene 1 (DAN1), a long intergenic noncoding RNA (lincRNA) that arose from the cotton progenitor A-diploid genome after hybridization and WGD events during cotton evolution. DAN1 in allotetraploid upland cotton (Gossypium hirsutum) is a drought-responsive lincRNA predominantly expressed in the nucleoplasm. Chromatin isolation by RNA purification profiling and electrophoretic mobility shift assay analysis demonstrated that GhDAN1 RNA can bind with DNA fragments containing AAAG motifs, similar to DNA binding with one zinc finger transcription factor binding sequences. The suppression of GhDAN1 mainly regulates genes with AAAG motifs in auxin-response pathways, which are associated with drought stress regulation. As a result, GhDAN1-silenced plants exhibit improved tolerance to drought stress. This phenotype resembles the drought-tolerant phenotype of the A-diploid cotton ancestor species, which has an undetectable expression of DAN1. The role of DAN1 in cotton evolution and drought tolerance regulation suggests that the genomic shock of interspecific hybridization and WGD stimulated neofunctionalization of non-coding genes during the natural evolutionary process.
A hybridization- and polyploidization-activated lncRNA neofunctionalized into a regulator of drought responses by forming RNA–DNA triplexes targeting genes mainly in the auxin response pathway.
Introduction
Interspecific hybridization and whole-genome duplication (WGD), also known as polyploidization, can stimulate the formation of new species via dramatic alterations in expression of coding and noncoding genes, neofunctionalization of redundant genes, and other variations, which allow for fast adaptation to a new habitat (Adams and Wendel, 2005). Over a long span of evolution, redundant genes generated by WGD might remain functional due to their biological importance. Alternatively, some duplicated genes lose their function because of expressional suppression (defunctionalization), sequence mutation (pseudogene), or gene loss because of redundancy (Adams et al., 2004; Conrad and Antonarakis, 2007). Some genes, especially coding genes, might gain new functions (i.e. undergo neofunctionalization) by means of new expression patterns or gene structures (Conrad and Antonarakis, 2007; Semon and Wolfe, 2008). When comparing gene expression activity in the hybrid or allotetraploid genomes with that in the parental genomes, the differentially expressed genes (DEGs) can be classified as having additive expression or nonadditive expression (Chen, 2010). Genes with nonadditive expression are understood to be the major contributors to the heterosis of hybridization and polyploidization (Chen, 2010). Some epigenomic marks, such as small RNAs (Ha et al., 2009) and chromatin modifications (Ha et al., 2009; Zhang et al., 2016), are also believed to play critical roles as buffers in response to the “genomic shock” caused by hybridization and WGD (Mcclintock, 1984; Chen, 2007). Previously, we reported that lncRNA in the cotton lineage also proactively follows a nonadditive expression pattern in interspecies hybridization and polyploidization (Zhao et al., 2018a, 2018b). Unique expression patterns and functions of lncRNA are commonly reported in animal and plant genomes (Li et al., 2014; Necsulea et al., 2014; Deng et al., 2018; Zhao et al., 2018a, 2018b). Whether these nonadditive or newly stimulated and activated lncRNAs arising during genomic shock are biologically important for the hybrid and polyploid species remains largely unknown.
The putative diploid ancestors of the current allotetraploid Gossypium (cotton) lineage species are Gossypium herbaceum (A1) or Gossypium arboreum (A2) and Gossypium raimondii (D5) (Cronn et al., 1996; Zhang et al., 2015). The cotton diploid ancestors formed an allotetraploid by hybridization and polyploidization approximately one to two million years ago, and from these ancestors, seven current allotetraploid species were derived (Wendel and Grover, 2015; Wang et al., 2018). Two diploid species (G. herbaceum, A1 and G. arboreum, A2) and two allotetraploid species [Gossypium hirsutum, (Gh) AD1 and Gossypium barbadense, AD2] have been independently domesticated for farming. Diploid cultivars from G. arboreum (A2) are low in product yield and have a high tolerance to drought stress (Zhang et al., 2010; Chen et al., 2015; Jia et al., 2018). An allotetraploid species (upland cotton, G. hirsutum) is the dominant cultivated species of cotton worldwide; it has high productivity and accounts for 90% of the cotton grown. LncRNAs have been shown to play crucial regulatory roles in plant–environmental interactions, including seed dormancy (Fedak et al., 2016), drought and salt stress tolerance (Qin et al., 2017), cold sensing and flowering (Swiezewski et al., 2009), phosphate nutrition homeostasis (Franco-Zorrilla et al., 2007), pathogen interaction (Yang et al., 2019), pathogen resistance (Zhang et al., 2018), and so on. We employed cotton lineage as a model system, using functional tests of abiotic stress regulation to examine the evolutionary path from diploid to allotetraploid species in order to identify a lncRNA activated in hybridization and polyploidization. Here, we report on a long intergenic non-coding RNA (lincRNA) responsible for drought stress regulation, the Drought-Associated Non-coding gene 1 (DAN1).
Results
GhDAN1 is found to be associated with the drought tolerance trait
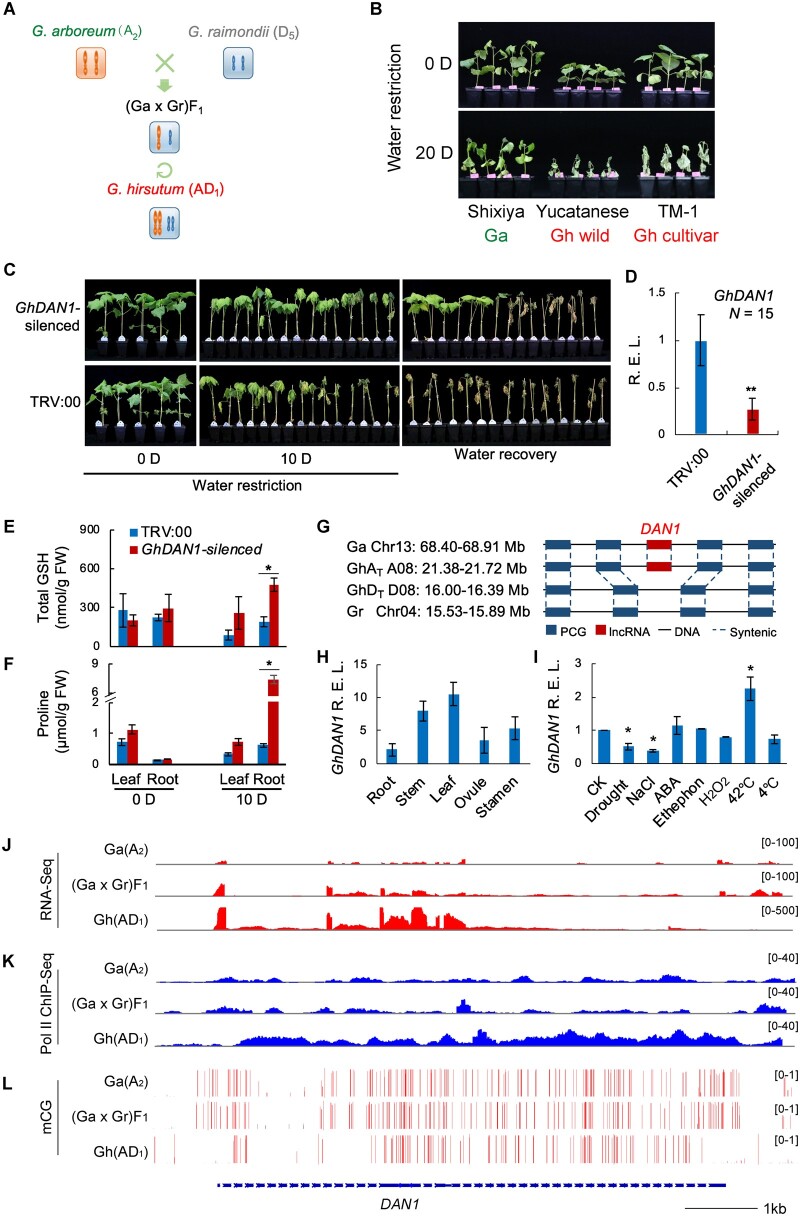
We employed a simplified model system with G. arboreum (Ga, A2), G. raimondii (Gr, D5), (Ga x Gr)F1, and G. hirsutum (Gh, AD1) to mimic cotton’s evolutionary lineage (Figure 1A) and demonstrate that lncRNA follows a nonadditive expression pattern from diploid to allotetraploid by interspecies hybridization (Zhao et al., 2018a, 2018b). Diploid cultivars from G. arboreum (A2) show high tolerance to drought stress in comparison to allotetraploid cotton (Chen et al., 2015; Figure 1B). To address the potential link between the lncRNA reprogramming and the alteration of the drought tolerance trait, we conducted a low-throughput functional screening of the lncRNAs in Gh with virus-induced gene silencing (VIGS) technology. A lincRNA responsive to drought stress, the DAN1 gene, was selected for further study because of its stable phenotyping result from at least five independent biological replicates (Supplemental Figure S1). The GhDAN1-silenced plants showed improved tolerance to drought stress (Figure 1, C and D), with increased glutathione (GSH; Figure 1E) and proline contents in drought-treated roots (Figure 1F), but without obvious changes in its photosynthesis traits (Supplemental Figure S2).
Figure 1.
GhDAN1 is associated with the drought-tolerance trait in the cotton ancestor. A, a simplified model system that mimics interspecific hybridization and WGD in the cotton lineage. B, drought-tolerance phenotypes of Ga diploid and Gh allotetraploid cotton (wild and cultivar accessions). C and D, drought-tolerance phenotype of GhDAN1-silenced plants and GhDAN1 expression. Error bars represent the sd of 15 seedlings in each group of treatment. **P < 0.01, Student’s t test. Total GSH (E) and proline contents (F) before and after drought treatment. Error bars represent the sd of three biological replicates. *P < 0.05, Mann–Whitney U-test. G, syntenic analysis of DAN1 in diploid Ga and allotetraploid Gh genomes. H and I, GhDAN1 expression in different tissues and the profile in response to multiple abiotic treatments. Error bars represent the sd of three biological replicates. *P < 0.05, Mann–Whitney U-test. J–L, stack plots for transcript profile (RNA-seq) (J), Pol II ChIP-seq profile (K), and the DNA CG-methylation status (L) of DAN1 loci in Ga (A2), (Ga x Gr)F1, and Gh (AD1) genomes.
Allotetraploid DAN1 (GhDAN1) is a non-coding gene with a predicted length of 8,581 base pairs (bps). At least three different isoforms of transcripts were predicted from the GhDAN1 locus. The longest of these isoforms (6,779 nt) that had abundant transcripts was confirmed in the results of both the lncRNA-seq and reverse transcription quantitative PCR (RT-qPCR) assays, and was further analyzed in our study (Supplemental Figure S3). Like mRNA, GhDAN1 has a poly(A) tail (Supplemental Figure S4A). However, according to the coding potential calculation, GhDAN1 does not seem to contain an open reading frame encoding any known peptide or protein. This is also in agreement with the low frequency of fragments found in the ribosome profiling (Supplemental Figure S4B). Synteny analysis indicated that GhDAN1 is inherited from the A diploid genome progenitor (Figure 1G). GhDAN1 was predominantly expressed in leaf and stem tissues (Figure 1H) and was spontaneously suppressed by drought and salt treatments (Figure 1I). RNA-seq and RNA polymerase II (Pol II) chromatin immunoprecipitation (ChIP)-seq analysis showed that DAN1 was very probably transcribed by Pol II. Its transcription was only slightly activated in the F1 hybrid but was highly expressed in the allotetraploid Gh genome (Figure 1, J and K). Interestingly, the CG and CHG methylation levels on the promoter region and 5′-end region of the DAN1 gene were lower in Gh compared with those in Ga and (Ga x Gr)F1 (Figure 1L;Supplemental Figure S5), which was in agreement with our previous finding that the activation of lncRNA transcription is predominantly associated with DNA demethylation after hybridization and polyploidization (Zhao et al., 2018a, 2018b).
DAN1 is active in the allotetraploid cotton species
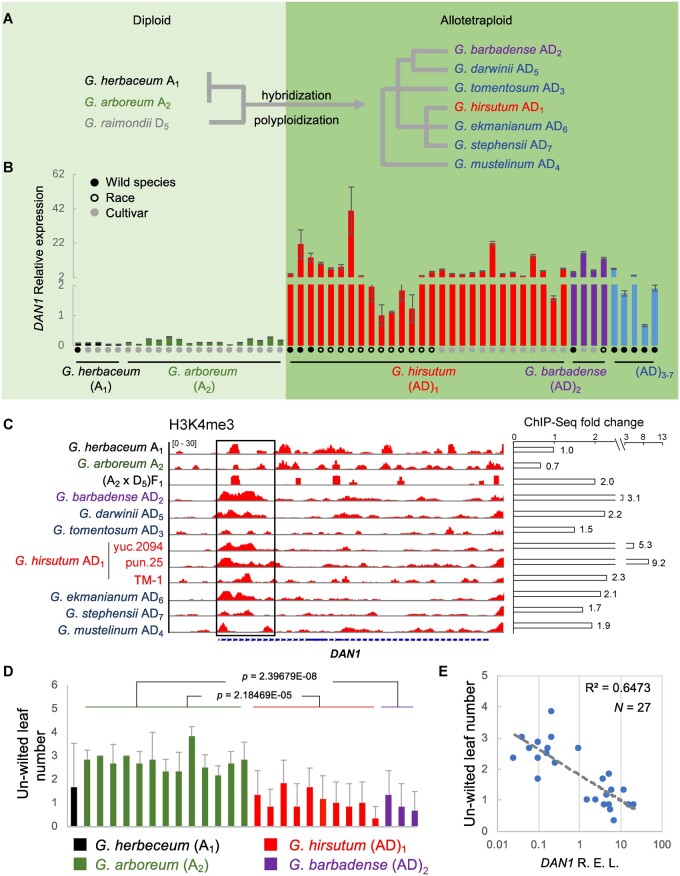
To further determine the behavior of DAN1 in the cotton lineage, we examined 5 accessions of G. herbaceum (A1), 17 accessions of G. arboreum (A2), 28 accessions of G. hirsutum (AD1), and 4 accessions of G. barbadense (AD2). We also examined one accession each of the other allotetraploid species, including Gossypium tomentosum (AD3), Gossypium mustelinum (AD4), Gossypium darwinii (AD5), Gossypium ekmanianum (AD6), and Gossypium stephensii (AD7) (Figure 2A;Supplemental Table S1). With this selection, we were able to examine a diverse cotton population that included wild species, primitive landraces, and cultivars of the A diploid species and allotetraploid species. The primary sequences of DAN1 were conserved in the A diploid species and allotetraploid species (Supplemental Figure S6). In all of the A diploid species examined, the transcription of DAN1 in leaf tissue was either below detection or had very low expression (Figure 2B). However, the expression of DAN1 in leaf tissue was commonly 10- to 80-fold higher in allotetraploid species than in the A diploid species (Figure 2B). To determine the variations in epigenetic modification underlying DAN1 activation, we further examined the histone modification profiles of H3K4me3 (active marker) and H3K27me3 (repressive marker) using ChIP-seq with the leaves harvested from the cotton lineage collection. Results indicated that the active expression of DAN1 in allotetraploid species was consistent with the presence of the active histone modification marker H3K4me3 at the 5′-end of the gene (Figure 2C); no obvious differences in the H3K27me3 signal were found (Supplemental Figure S7).
Figure 2.
DAN1 is active in the allotetraploid cotton species. A, schematic chart showing phylogenetic relationships of diploid ancestors and seven allotetraploid cotton species. B, DAN1 transcription activity in two diploid A species and seven allotetraploid species (accessions are listed in order in Supplemental Table S1). Error bars represent the se of three technical replicates. C, enrichment of H3K4me3 ChIP-seq signals (boxed region) near the DAN1 transcription starting site in the allotetraploid cotton lineage. Fold changes were calculated using normalized peak counts compared with their corresponding mock control. D, drought-tolerance phenotype of diploid and allotetraploid cotton cultivars. The number of unwilted leaves after water restriction was calculated (see the Supplemental Figure S8 for details); Error bars represent the sd of six seedlings from each group. P-value was indicated by comparing Ga (A2) diploid with Gh (AD1) and Gb (AD2) allotetraploid cultivars, respectively. Student’s t test. E, analysis of the correlation between DAN1 expression and the unwilted leaf count from (D).
We also compared the drought tolerance characteristics of diploid and allotetraploid cotton species with varying levels of DAN1 expression. Results showed a negative correlation (R2 = 0.6473) between the expression of DAN1 and the drought tolerance in the species we tested (Figure 2, D and E;Supplemental Figure S8). These data confirmed that DAN1 was activated after hybridization and polyploidization in the Gossypium lineage and is one of the factors that contributes to the valuable characteristic of drought tolerance in A diploid species, which most allotetraploid cultivated cotton species have lost.
GhDAN1 can form RNA–DNA triplexes to anchor the AAAG DNA target site and regulate gene expression in trans
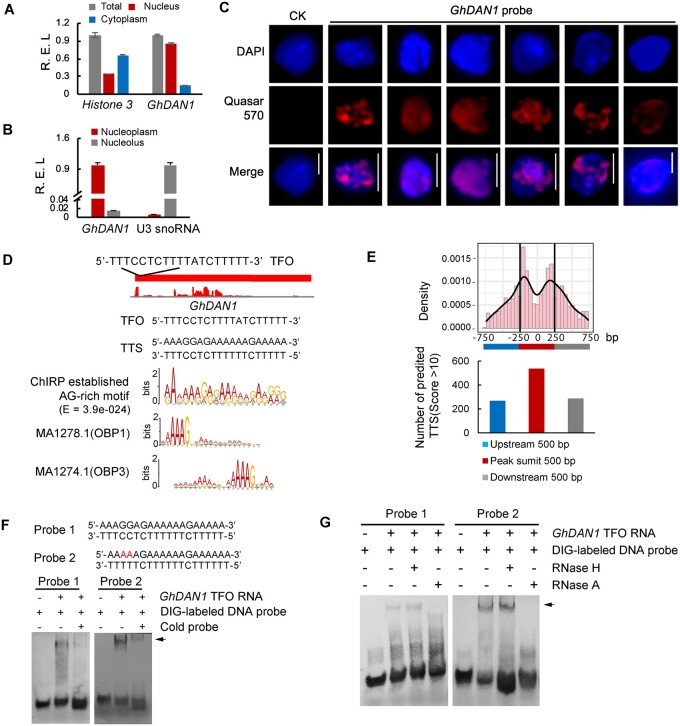
To determine the function of GhDAN1 in drought tolerance, the gene expression profiles of GhDAN1-silenced plants were examined using mRNA-seq and were compared with the VIGS control group. The silenced expression of GhDAN1 by VIGS did not significantly affect the activity of the 25 flanking genes of GhDAN1 (which covered ∼8 Mb of the flanking region) in the chromosome (Supplemental Figure S9), which suggests that GhDAN1 might not mediate the control of drought tolerance in cis. GhDAN1 transcripts were preferentially present in the nucleus (Figure 3A) and more specifically, in the nucleoplasm (Figure 3B). Fluorescence in situ hybridization assay with RNA probes (RNA-FISH) using isolated nuclei showed that the GhDAN1 signal was dispersed throughout the nucleus (Figure 3C). Our data are consistent with a model in which the GhDAN1 binding sites were concentrated in multiple specific spots on different chromosomes. The above analysis demonstrated that GhDAN1 is a nucleus-localized lincRNA, which might be required for the ability of a lncRNA to act in trans.
Figure 3.
GhDAN1 forms RNA–DNA triplexes to anchor the AAAG DNA target site and regulate gene expression in trans. A and B, RT-qPCR analysis of GhDAN1 expression in the nucleus versus cytoplasm fractions, and in the nucleolus versus nucleoplasm. Histone3 and U3 snoRNA were used as controls. Error bars represent the se of three technical replicates. C, RNA-FISH assay showing the GhDAN1 distribution in the nucleus. Bars = 5 µm. D, alignment of the Triplexator software-predicted GhDAN1 TTSs with ChIRP-established AG-rich motif and Dot TFs binding motifs. E, number and density of TTSs over the gene-associated GhDAN1 peak summits and neighboring regions. The TTS motif was predicted by FIMO in the peaks (±250 bp from the peak summits) and the flanking control regions (500-bp upstream and downstream from the peaks). The density of the TTS motif was generated by ggplot2 in R using the distances between the predicted TTS motif and the peak summit. F and G, EMSA assay showed triplex formation of GhDAN1 RNA with DIG-labeled TTS DNA probes in vitro. The nucleotide in red color in Probe 2 indicated two mismatches compared with predicted TTS motif. The arrows indicated the RNA–DNA triplexes.
Generally, the lncRNA affects the transcription in trans by recruiting a regulatory ribonucleoprotein (RNP) complex to the target gene. In such cases, lncRNA acts as a partner of the RNPs (Rinn and Chang, 2012). LncRNA can also act in trans by inhibiting the binding of a transcriptional regulatory factor by acting as a decoy or can inhibit its activity by direct active-site occlusion to prevent its activity (Rinn and Chang, 2012; Long et al., 2017). To investigate the possible molecular mechanism of GhDAN1 in trans, chromatin isolation by RNA purification (ChIRP) assays were performed to map the regions of chromatin interacting with the GhDAN1 transcripts. We set up the ChIRP reactions using anti-sense oligonucleotide probes that separated into two groups (even and odd), which was followed by DNA-seq analysis. Only those peaks shared by both the odd and even probe sets were considered to be valid peaks (see the section “Materials and methods” for a detailed ChIRP-seq workflow). Based on the filter parameters, 12,142 valid peaks were identified, with 10,941 (90.1%) peaks located intergenically, and 1,201 (9.9%) peaks located near 945 coding genes (including the 3-kb 5′-flanking region, gene body, and 3-kb 3′-flanking region, designated as the gene-associated DAN1 ChIRP peaks; Supplemental Figure S10A). An AG-rich motif similar to the binding motif of DNA binding with one finger (Dof)-type plant-specific transcription factors (TFs) was established by MEME (https://meme-suite.org/meme/; Bailey et al., 2009) using the sequences of the top 1,214 valid peaks (i.e. the top 10% of the total valid peaks as ranked by fold enrichment; Figure 3D). To determine that this established motif resulted from the ChIRP binding but not from random sequences, the occurrence of the motif in the 1,201 gene-associated DAN1 ChIRP peaks was analyzed by FIMO scanning. Results indicated that this AG-rich motif was enriched in the peaks (±250 bp from the peak summits) compared with the control flanking regions (500-bp upstream and downstream from the peaks; Figure 3E). The latest studies have revealed that multiple mammalian lncRNAs (e.g. MEG3, HOTAIR, and PARTICLE; Mondal et al., 2015; Oleary et al., 2015; Kalwa et al., 2016) are capable of regulating gene expression by forming RNA–DNA triplexes. We, therefore, scanned the GhDAN1 RNA sequence for possible triplex-forming oligonucleotides (TFOs) using Triplexator software (Buske et al., 2012). High-score TFOs were obtained (Supplemental Table S2) with TC-rich sequences. Among these TFOs, GhDAN1 TFO motif 1, which was a 19-bp TC-rich motif at the 5′-end of GhDAN1 (from positions +118 to +137), exhibited the triplex target site (TTS) sequence that was most similar to the ChIRP-established AG-rich motif (Figure 3D). Accordingly, we speculated that the TC-rich GhDAN1 RNA might be recruited to the AG-rich genomic DNA targeting sites through RNA–DNA triplex formation following the Hoogsteen configuration (Li et al., 2016).
To validate the ability of DAN1 lincRNA to form triplex structures, an electrophoretic mobility shift assay (EMSA) was performed. It examined the triplex formation of a 178-nt GhDAN1 single-stranded RNA fragment (+9 to +186) containing GhDAN1 TFO (+118 to +137) with the digoxigenin (DIG)-labeled AG-rich double-stranded DNA TTSs. The results indicated that GhDAN1 RNA can bind to the double-stranded DNA probe and has a sequence identical to that of the predicted DAN1 TTS motif (probe 1). To evaluate the sequence discrepancy of the RNA–DNA triplex formation, the probe with mismatches of the predicted TTS motif was also examined with EMSA. We observed that an alternative probe with two mismatches (probe 2) preserved the triplex formation (Figure 3F). Therefore, the formation of the RNA–DNA triplex exhibited a mild tolerance to sequence variation. In the control, RNA–DNA triplex binding was retarded upon the addition of a corresponding unlabeled cold probe serving as a competitor (Figure 3F). This RNA–DNA triplex structure was sensitive to RNase A but resistant to RNase H (Figure 3G). Because the RNA strand of an RNA–DNA hybrid can be specifically degraded by RNase H, these results suggested that the EMSA shift band detected in vitro was not due to the RNA–DNA hybrid formed by Watson–Crick pairing. Because GhDAN1 recognizes DNA sites similar to the Dof TFs binding motif, we speculated that the pathway corresponding to Dof TFs in plants may be coordinated by GhDAN1 regulation.
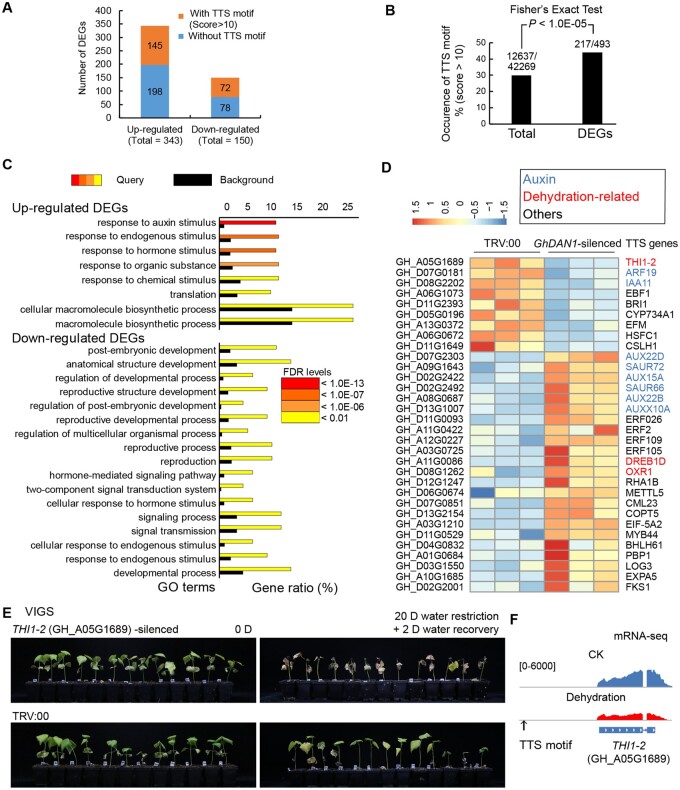
Genes with the GhDAN1 TTS motif were sensitive to the silencing of GhDAN1 and significantly enriched in the auxin response pathway
The mRNA-seq analysis with leaves of VIGS-silenced plants and tobacco rattle virus (TRV) control plants before drought treatment was performed in order to determine the early response genes under GhDAN1 regulation. As a result, 493 DEGs [P < 0.001, false discovery rate (FDR) < 0.05] were identified in GhDAN1-silenced plants in comparison to the control group, including 343 upregulated and 150 downregulated genes (Figure 4A;Supplemental Figure S11; Supplemental Table S3). Gene ontology (GO) enrichment analysis showed that the DEGs were significantly enriched in the category of “response to hormone stimulus” (FDR < 1.0E-07), especially in the “response to auxin stimulus” (FDR < 1.0E-13) for the upregulated DEGs (Figure 4C). The downregulated DEGs were enriched in the developmental and reproductive processes (Figure 4C). We further validated the main auxin biosynthesis pathway in plants, in which tryptophan is first converted to indole-3-pyruvate (IPA) by the TAA/TAR family of amino transferases, and IAA is subsequently produced from IPA by the YUC family of flavin monooxygenases (Mano and Nemoto, 2012), and the results indicated that none of the TAA/TAR or YUC genes were differentially expressed in GhDAN1-silenced plants in comparison to the control group (Supplemental Figure S12A). Considering the importance of abscisic acid (ABA) in drought stress responses, the expression profile of genes in ABA biosynthesis and catabolism pathways were summarized, and the results showed that none of these genes were differentially expressed (Supplemental Figure S12B). Thus, GhDAN1 was unlikely to affect phytohormone biosynthesis, including auxin and ABA.
Figure 4.
Genes with GhDAN1 the TTSs motif were sensitive to silencing of GhDAN1 and significantly enriched in the auxin response pathway. A, the column chart shows the number of DEGs in GhDAN1-silenced plants and the distribution of genes in each up and down-regulated groups that were predicted with or without the GhDAN1 TTS motif in the 2-kb promoters using FIMO scanning (score >10). B, occurrence of GhDAN1 TTS motif in DEGs and the total coding genes. C, GO analysis of upregulated and downregulated genes in GhDAN1-silenced plants showing significant enrichment in the auxin response pathway. D, Heatmap showing the expression profiles of the representative DEGs with the GhDAN1 TTS motif. E, cotton seedlings were sensitive to drought stress after silencing of the GhDAN1 downstream gene GhTHI1-2. F, stack plots of mRNA-seq on GhTHI1-2 transcription activity during dehydration treatment. The predicted GhDAN1 TTS motif in the promoter was indicated by the black arrow.
To establish the link between the ChIRP peak-associated genes and the DEGs in GhDAN1-silenced plants, we compared the ChIRP peaks and mRNA-seq data. Of the 945 genes with DAN1 ChIRP peaks, 3 overlapped with the DEGs. They were (1) a 50S ribosomal protein L9 gene encoding one of the primary rRNA-binding proteins, (2) a copper transporter five gene that responded to ABA and was involved in the transport of copper for protein maturation and copper homeostasis in Arabidopsis thaliana (Sancenon et al., 2003), and (3) a germin-like protein subfamily 1 member 13, the GLP6 gene that is involved in manganese and iron-binding and nutrient reservoir activity, and which may play a role in plant defense (Davidson et al., 2010; Supplemental Figure S10B). These three genes were all significantly upregulated in GhDAN1-silenced plants (Supplemental Table S3).
Because of the low resolution of the genes that were associated with ChIRP peaks and were also differentially expressed in GhDAN1-silenced plants, we evaluated, as an alternative, the effects of the GhDAN1 RNA–DNA triplex on the transcriptional activity of the potential target-coding genes. This was done by examining the presence of the GhDAN1 TTS motif in the promoters of the DEGs. The promoters of the total 42,269 expressed coding genes (FPKM > 1) in the Gh genome were used as background controls. Results indicated that the TTS motif was significantly enriched in the promoter regions of the DEGs (Figure 4B;P < 1.0E-05, Fisher’s exact test), indicating that the genes containing the GhDAN1 TTS motif were dominant among the DEGs that were responsive in the early stages following GhDAN1 regulation. Multiple DEGs related to the auxin response pathway, ethylene response pathway, brassinosteroid response and biosynthesis pathway, dehydration response/oxidation resistance, and cell wall biosynthesis contained the GhDAN1 TTS motif in their promoters, suggesting that they are the primary targets of GhDAN1 (Figure 4D). Orthologs of 13 TTS motif-containing DEGs in other plants were reported to be involved in drought stress tolerance (Supplemental Table S4). Based on all of this evidence, it can be assumed that GhDAN1 preferentially regulates genes containing the GhDAN1 TTS motif, and that these genes are significantly enriched in the auxin response pathway.
To determine whether the DEGs with the GhDAN1 TTS motif in GhDAN1-silenced plants are subject to drought stress regulation, we examined the transcriptome analysis of drought treatment in upland cotton TM-1. Among the 145 TTS-containing genes up-regulated in GhDAN1-silenced plants, 41 (28.3%) and 25 (17.2%) showed significantly increased and suppressed expression in response to dehydration treatment, respectively (Supplemental Figure S13A; Supplemental Table S5). Among the 72 TTS-containing genes downregulated in GhDAN1-silenced plants, 15 (20.8%) and 8 (11.1%) showed significantly increased and suppressed expression in response to dehydration treatment, respectively (Supplemental Figure S13A; Supplemental Table S5). For comparison, in the total 42,269 expressed coding genes (FPKM > 1) in the Gh genome, 4,441(10.5%), and 4,443(10.5%) were significantly up- and downregulated, respectively, in response to dehydration treatment. Statistical analysis indicated that the TTS motif-containing DEGs that were upregulated in both GhDAN1-silenced plants and dehydration-treated Gh plants were significantly enriched compared with dehydration upregulated genes at the whole genome background (Supplemental Figure S13A; P < 1.0E-05, Fisher’s exact test), indicating that the major effect of GhDAN1 is targeting these up-regulated genes. Totally 89 of the TTS motif-containing DEGs in GhDAN1-silenced plants were dehydration-responsive, which are enriched in the GO term “response to auxin stimulus” (FDR < 0.05; Supplemental Figure S13B). We found that 6 out of 13 documented drought tolerance-related genes are both up- or downregulated in GhDAN1-silenced plants and drought-treated, Gh wild-type plants (Supplemental Table S4). We further selected 11 candidate genes from the DEGs with the GhDAN1 TTS motif to examine their functions under drought stress using the VIGS assay (Supplemental Figure S14). A representative gene encoding thiamine thiazole synthase 2 (THI1-2, GH_A05G1689), which was responsive to both GhDAN1 silencing and drought treatment, was demonstrated to be involved in drought tolerance regulation in the VIGS and dehydration assays (Figure 4, E and F). These findings confirmed that GhDAN1 can regulate drought stress regulation by targeting genes with the TTS motif.
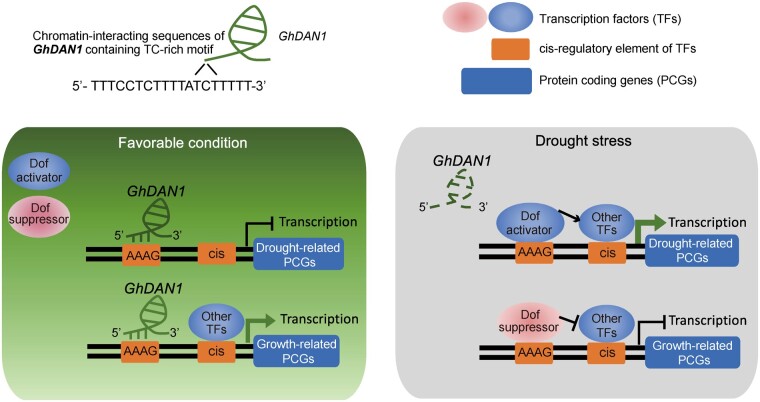
Working model of DAN1-mediated drought-response dynamic
Given all the data we obtained for DAN1, a most likely working model for DAN1 evolution and its role in drought tolerance regulation is proposed. DAN1 originated from an A diploid ancestor genome after polyploidization due to DNA CG demethylation and H3k4me3 modification at the 5′-end of the gene (Figures 1L and 2C). It obtained new functions associated with stress adaptation over the evolution of the cotton allotetraploid. In favorable conditions, GhDAN1 is transcribed at a sufficient level and engages with genomic regions with AAAG motifs by forming RNA–DNA triplexes. When plants encounter conditions of drought stress, the GhDAN1 transcript levels are reduced (Figure 1I), thus releasing the AAAG motifs, possibly for the Dof TFs. Because members of the Dof TFs are characterized as being activators and suppressors in plants (Noguero et al., 2013), the expression of growth/development-related genes (downregulated DEGs in Figure 4, C and D) could be decreased by possible Dof suppressors, and the expression of drought defense-related genes (upregulated DEGs in Figure 4, C and D) could be promoted by possible Dof activators. In an orchestrated result, plants could successfully adapt to environmental dynamics (Figure 5). This allows plants to achieve a balance between the energy they expend in growth and the stress tolerance they gain. GhDAN1 activation is a remarkable case of a gene that, during hybridization and WGD, was selected for and fixed in such a way that it could maintain its growth and fitness when encountering environmental stress.
Figure 5.
Working model of DAN1-mediated drought-response dynamics.
Discussion
WGD-induced GhDAN1 plays a regulatory role in drought stress tolerance
WGD in plants that is led by polyploidization and interspecies hybridization usually leads to dramatic changes, both genetically and physically (Chen, 2013). The encounter of two genomes in one nucleus proactively stimulates the noncoding region of the genome to transcribe RNAs (Zhao et al., 2018a, 2018b). As has been previously reported, these activated RNAs are mainly transcribed by Pol II (Zhao et al., 2018a, 2018b). These newly transcribed regions of the genome are also highly associated with markers of epimodification, such as DNA methylation (Zhao et al., 2018a, 2018b). We proposed that the total effects of the epimodifications to chromatin and the transcriptome provided a reservoir for variations in heredity. The transcriptional activity and function of DAN1 in the cotton lineage support this hypothesis. The expression of DAN1 in diploid species was almost undetectable. In all allotetraploid species, however, DAN1 expression is active. DAN1 activation in the synthetic hybrid of the A2 and D5 diploids also provided strong evidence to suggest that the DAN1 locus is sensitive to genomic shock. Meanwhile, the primary sequence of DAN1 was conserved in the cotton lineage from the A diploid to the A subgenome in the allotetraploid over one to two million years. Therefore, DAN1 displays a unique evolutionary pattern wherein RNA expression is activated with no fundamental changes to the sequence. The activation and function of GhDAN1 in allotetraploid cotton species demonstrate that the noncoding transcripts activated by genomic shock can cause the emergence of new functional and beneficial genes, which can be fixed in the natural population during evolution.
In recent reports, there is controversy about the sequence and functional conservation of lncRNAs. The primary sequences of lncRNAs evolved faster relative to mRNAs, both in tetrapod animals (Necsulea et al., 2014), and the relatively closely related plant species (Zhao et al., 2018a, 2018b; Barrera-Redondo et al., 2019). The functional characterization of the known lncRNAs in one species is rarely identified in other relatively close species, suggesting that lncRNA has very strong species specificity. A recent report in Drosophila found that functional tests of 30 relatively conserved lncRNAs using CRISPR-CAS9 technology resulted in no observed developmental functions for any of them (Goudarzi et al., 2019). Here we argue that according to its primary sequence, DAN1 is a conserved gene in the cotton lineage. In terms of transcriptional activity, however, DAN1 is a novel gene in the allotetraploid, harbored in a noncoding region. The activity of DAN1 we are reporting here demonstrates the flexibility of the genome in employing the noncoding region in response to environmental stimuli.
The molecular basis of DAN1-mediated gene expression regulation in trans
Previous studies have shown that multiple mammalian lncRNAs (e.g. MEG3, HOTAIR, and PARTICLE; Mondal et al., 2015; Oleary et al., 2015; Kalwa et al., 2016) can regulate gene expression by RNA–DNA triplex formation, indicating that this type of formation may be a general mechanism of target site recognition by lncRNAs. GhDAN1 could bind to an AAAG-rich DNA motif corresponding to the Dof TFs’ core binding sites in chromatin by forming an RNA–DNA triplex. The triplex can engage cis-elements on the promoters of stress-responsive and developmentally related genes in order to prevent gene transcription under certain conditions. Dof TFs are plant-specific, and their involvement in a wide range of processes is suggested by their binding to the AAAG consensus sequence in target promoters; this implies that they have important roles, such as regulating stress responses, photosynthesis, or flower induction. Dof TFs can act as either positive or negative regulators of gene expression (Noguero et al., 2013). It was documented that a Dof protein DNA binding sequence (ACTTTA), which is targeted by a tobacco Dof protein NtBBF1, acts as the cis-regulatory element required in auxin induction of rolB (Baumann et al., 1999), which linked the relationship between Dof TFs and the auxin response pathway. More transcriptional association between Dof TFs and their potential target genes involved in auxin signaling has been established (Pattison et al., 2015). As the first plant hormone to be identified, auxin and its response pathway not only play important roles in plant growth regulation, but also interconnect and stimulate different metabolic pathways under abiotic stresses (Tognetti et al., 2012; Du et al., 2013; Tiwari et al., 2017; Wang et al., 2019). Recently, auxin-sensitive Aux/IAA proteins were found to mediate drought tolerance in Arabidopsis by regulating glucosinolate levels, which provide a connection between auxin signaling and drought response (Salehin et al., 2019). GhDAN1 has the ability to “sequester” the core binding sites of Dof TFs, thus retarding positive or negative regulation by TFs and coordinating proper gene expression during drought stress. Accordingly, the genes in auxin response pathway corresponding to Dof TFs in plants were coordinated by GhDAN1 regulation. We infer that by coordinating TFs such as Dof for the binding of a specific DNA motif, GhDAN1 forms a negative feedback loop in response to abiotic stress.
Triplex-forming motifs identified in the mammalian lncRNAs MEG3, HOTAIR, and PARTICLE transcripts were seen to be mostly AG-rich sequences, which formed antiparallel triplexes with their AG-rich DNA target sequences by forming antiparallel reverse Hoogsteen hydrogen bonds with the purines of the DNA duplex (Mondal et al., 2015; Oleary et al., 2015; Kalwa et al., 2016). The lncRNAs Fendrr and Khps1, with TC-rich TFOs in their transcripts, bind to AG-rich DNA target sequences by means of parallel Hoogsteen hydrogen bonds with the purines of the DNA duplex (Grote et al., 2013; Postepska-Igielska et al., 2015). Our example of GhDAN1 forms an RNA–DNA triplex according to the rules of parallel Hoogsteen configuration, and this indicates a common functional strategy of lncRNA in both plant and animal genomes. Interestingly, a new AG-rich motif in a plant with a sequence of AAAG, rather than simple AG repeats, might represent a divergence in patterns of RNA–DNA triplex formation between plant and animal genomes.
Triplex-forming lnRNAs are also reported to recruit specific proteins (e.g. PRC2) to the correct locus on chromatin in response to stimuli or oncogene expression (Mondal et al., 2015; Oleary et al., 2015; Kalwa et al., 2016). The RNA–DNA triplex formed by DAN1 could also be involved in site-specific protein binding in response to environmental signals or abiotic stress. Further examination of DAN1-binding proteins is the next critical question to address.
The potential of noncoding genes in abiotic stress
Drought causes severe losses in global crop production (Comas et al., 2013). It is a major unsolved issue in modern agriculture. The study of mechanisms of drought tolerance is needed in order to improve crop adaptability under drought stress. Knowledge of drought tolerance variations among Gossypium species may help us to understand how genomic shock leads to the activation and fixation of specific functional genes, and it may also shed light on our progress in the genetic breeding of cultivated cotton species with altered characteristics that would improve stress tolerance. According to the phenotypic analysis, the A diploid cotton species we tested showed high drought tolerance, whereas most cultivated allotetraploids seemed to have lost this valuable characteristic (this change has often been recognized in previous reports; Chen et al., 2015; Jia et al., 2018). Plants might take on defensive strategies that sacrifice their biomass accumulation and developmental processes in stress conditions, a tradeoff we have called fitness cost (Bostock et al., 2014; Cipollini et al., 2017). According to our data, GhDAN1 may act as a modulator of the auxin pathways, thereby adjusting the balance between biomass accumulation and stress tolerance processes. To optimize the balance between stress tolerance and biomass accumulation after hybridization and polyploidization, the allotetraploid genome could activate noncoding genes such as DAN1 to create a novel regulatory feedback loop in response to drought. Considering the evolutionary profile of DAN1 and its mode of action, we have inferred that GhDAN1 activation is a remarkable case of a gene that, during hybridization and WGD, was selected for and fixed to maintain the balance between its growth and fitness costs when encountering environmental stress.
Materials and methods
Cotton species and plant culture
Seed and leaf sources of the varieties of diploid and allotetraploid cotton used in this study, including wild species, landraces, and cultivars, are listed in Supplemental Table S1. Plant material of the interspecies hybrid F1 of G. arboreum (A2; 2n = 2x = 26) and G. raimondii (D5; 2n = 2x = 26) was used as previously described (Zhao et al., 2018a, 2018b). All leaf samples were collected in identical field growth conditions, frozen in liquid nitrogen immediately, and stored at −70°C until they were prepared for RNA isolation and DAN1 gene expression analysis. For the VIGS assay, cotton seedlings were grown in pots at 22 °C in a greenhouse in a 16-/8-h light/dark cycle with 60% humidity. Seedlings with mature cotyledons but without visible true leaves (7 d after germination) were infiltrated in the VIGS assay. For drought treatment, abiotic treatment, and leaf collection for the RNA-FISH, ChIP, and ChIRP assays, seedlings were grown in the same greenhouse conditions as for the VIGS, and the treatment or sample collection was performed when the seedlings had two or three true leaves (i.e. they were 4-week-old seedlings).
Virus-induced gene silencing
The preparation of TRV vectors and Agrobacterium tumefaciens bacteria for our experiments in cotton plants was conducted as previously reported (Zhao et al., 2018a, 2018b). Fragments of candidate genes were amplified and separately cloned into a pTRV2 vector by the infusion method to construct recombinant pTRV2 vectors. Of these, the recombinant pTRV2–GhCLA vector was used as a positive control in order to monitor the effect of the VIGS. The primer pairs used to construct recombinant pTRV2 vectors are listed in Supplemental Table S6. The vectors, including pTRV1, recombinant pTRV2, and pTRV2 empty vectors, were then separately introduced into specimens of A. tumefaciens strain GV3101 by electroporation. The specimens of A. tumefaciens GV3101 containing pTRV1 and recombinant pTRV2 were cultured overnight at 28 °C in an antibiotic selection medium with rifampicin (25 mg/L) and kanamycin (50 mg/L). Agrobacterium cells were then centrifuged and resuspended in an infiltration buffer (10 mM MgCl2, 10 mM MES, and 200 mM acetosyringone) adjusted to an OD600 = 0.4-0.8. Specimens of A. tumefaciens GV3101 with pTRV1 vectors were mixed individually with either recombinant pTRV2 vectors containing candidate genes or a positive marker gene or an empty vector pTRV2 at a ratio of 1:1. The mixture was left in darkness at 28 °C for 3 h. Finally, 7-d-old seedlings were infiltrated by inserting the Agrobacterium suspension into the cotyledons with a syringe. Four weeks after infiltration, the second true leaves from each seedling were collected for gene expression analysis, or the intact seedlings were subsequently subjected to drought treatment. For each vector or gene, 15–30 Gh TM-1 seedlings were infiltrated for subsequent phenotypical analysis. The VIGS assay for GhDAN1 was performed with at least five biological replicates.
Drought treatment
For each independent biological experiment, 12–15 4-week-old cotton seedlings (having at least two fully expanded true leaves) were subjected to a cessation in watering. After 10–20 d of water deprivation, the phenotype was observed, and a sample collection or re-watering treatment for 2–3 d was performed when needed.
Abiotic treatment
Twelve to 15 4-week-old cotton seedlings (having at least two fully expanded true leaves) were used in each abiotic treatment group. Different foliar sprays composed of 150 mM NaCl, 1 μM ABA, 1 mM ethephon, or 2 mM H2O2 were applied, and the leaves were collected 6 h later for gene expression analysis. Plants sprayed with distilled water were used as controls. Seedlings were treated in a growth chamber at 42 °C or 4 °C for 6 h before analysis.
LncRNA-seq and pipeline for lncRNA identification
LncRNA sequencing and the pipeline for lncRNA identification in the cotton lineage were described previously (Zhao et al., 2018a, 2018b). The total RNA was isolated from the plant tissues using the Spectrum Plant Total RNA Kit (Sigma-Aldrich, St Louis, MO, USA). Ribosomal RNA was removed using the Epicentre Ribo-Zero rRNA Removal Kit (Epicentre, Madison, WI, USA). LncRNA libraries were generated from rRNA-depleted RNA using the NEBNext® Ultra Directional RNA Library Prep Kit for Illumina® (New England Biolabs, Inc., Beverly, MA, USA). Strand-specific sequencing was performed with the Illumina HiSeq 2000 system (paired-end 125-bp reads; Illumina, Inc., San Diego, CA, USA). The quality control of strand-specific RNA-seq reads, the mapping of reads to the reference genomes, and the assembly of all of the transcripts was as described (Zhao et al., 2018a, 2018b). The coding potential of the transcripts was calculated using the Coding Potential Calculator (CPC) software (Kong et al., 2007). All transcripts with CPC scores above zero were discarded. The remaining transcripts were subjected to HMMER version 3.1b2 to exclude the transcripts containing known protein domains (cutoff <0.001; Finn et al., 2011). The remaining transcripts were candidate lncRNAs.
Analysis of whole-genome MethylC-seq reads
MethylC-seq data for diploid G. arboreum, (G. arboreum × G. raimondii)F1, and G. hirsutum were retrieved from a previous study (Song et al., 2017). Methylation levels in the DAN1 promoter and gene body regions were calculated using in-house Perl and R scripts.
Physiological trait measurements
The total GSH content was measured using a GSH assay kit (Sigma-Aldrich; cat. no. CS0260). For the sample preparation, 50 mg of ground tissue was homogenized with 0.5 mL 5% (w/v) 5-sulfosalicylic acid (SSA) solution to obtain the supernatant. The reaction procedure was performed according to the user manual. After the reaction, a plate reader was set to 412 nm with a kinetic read at 1-min intervals for 10 min. The standard curve was established by using a series of concentrations of GSH (3.125, 6.25, 12.5, 15, and 50 μM). Based on the standard, the total GSH content of the samples was calculated. GSH content values from three independent biological replicates were collected for each treatment.
The proline concentration was measured based on its reaction with ninhydrin according to the following method (Lee et al., 2018). Fresh tissue samples (∼0.1 g) were homogenized in 1 mL of extracting solution (3% (w/v) SSA) on ice, followed by incubation at 100°C for 10 min. Samples were then centrifuged at 25 °C at 1500 g for 10 min. An aliquot of 0.25 mL of the supernatant was transferred to a new tube and combined with 0.25 mL glacial acetic acid and 0.25 mL 2.5% (w/v) acid ninhydrin. The reaction was inoculated at 100 °C for 0.5 h and terminated in an ice bath. An amount of 0.5 mL of toluene was added to the reaction, and the solution was vortexed for 30 s. The absorbance at 520 nm was recorded. The standard curve was established by using a series of concentrations of proline (0, 1, 2, 4, 6, 8, 10, and 15 μg/mL). Based on the standard, the proline content of the samples was calculated. The proline content values were collected from three independent biological replicates for each treatment.
Photosynthetic parameters were determined using intact, fully expanded, functional leaves (the third or fourth topmost leaves). Measurements of the net photosynthetic rate (Pn), stomatal conductance (Gs), transpiration rate (Tr), and intercellular CO2 concentration (Ci) were gathered with an LI-6400 portable photosynthesis system (LI-COR Biosciences, Lincoln, NE, USA) according to instructions in the user manual. Photosynthetic parameters of 15–30 seedlings were collected for each treatment.
Fractionation of RNA
Isolation of nuclei, cytoplasm, nucleoplasm, and nucleoli from the cotton leaves was performed as previously described (Xing et al., 2017) with modifications. One gram of cotton leaves was ground into a fine powder using liquid nitrogen. The ground powder was transferred into a 50-mL ice-cold centrifuge tube containing 30 mL ice-cold Tris buffer (10 mM Tris pH 8.0, 10 mM MgCl2, 0.1 mM PMSF, 5 mM β-mercaptoethanol). After being agitated gently, the mixture was immediately filtered through four layers of Miracloth into a new 50-mL tube and centrifuged at 100 g for 5 min at 4 °C to derive a pellet. Two milliliters of a nuclei isolation buffer (10 mM Tris pH 8.0, 10 mM MgCl2, 0.1 mM PMSF, 5 mM β-mercaptoethanol, 1% (v/v) Triton X-100, 1× proteinase inhibitors, 5 U/μL RNAse inhibitors) was added to resuspend the pellet. The suspension was incubated on ice for 10 min and then centrifuged at 100 g for 3 min at 4 °C to form a pellet of the nuclei. An aliquot of 0.5 mL supernatant (the cytoplasmic fraction) was transferred to a new 2-mL tube for cytoplasmic RNA extraction. The nuclei pellet was resuspended with 1 mL 340 mM sucrose solution containing 5 mM MgCl2, and 250 μL of the lysate was transferred to a new 2-mL tube for RNA extraction of the nuclei. The rest of the lysate of the nuclei was broken by sonication until intact nuclei could not be detected in the suspension by a microscope. A solution of 750 μL 880 mM sucrose containing 5 mM MgCl2 was gently added to the sonicated nuclei and then centrifuged for 20 min at 200 g at 4°C to form a pellet of the nucleoli. The supernatant was the nucleoplasmic fraction. Fractionated RNAs of the nuclei, cytoplasm, nucleoplasm, and nucleoli were extracted using the PureLink Plant RNA Reagent (Invitrogen Carlsbad, CA, USA; cat. no. 12322012; Thermo Fisher Scientific, Waltham, MA, USA). Fractionated RNAs from the same amount of cells were used for cDNA synthesis and RT-qPCR. Each fractionated RNA had three biological replicates for analysis.
RNA-FISH assay
The RNA-FISH assay was performed as previously described (Zhu et al., 2016) using isolated nuclei. For plant material cross-linking, 1 g of fresh cotton leaves was cut into pieces and transferred to a 50-mL RNase/DNase-free tube and cross-linked with 30 mL of fixation buffer (10 mM Tris pH 8.0, 1% w/v formaldehyde, 0.1 mM PMSF, 5 mM β-mercaptoethanol) for 15 min by vacuum infiltration in an exicator at room temperature. After cross-linking, 2.5 mL 2 M glycine was added, followed by vacuum infiltration for 5 min to quench the reaction. The tissue was then washed 3 times with plenty of ddH2O and briefly dried between sheets of clean absorbent paper for 30 s to remove most of the water. For nuclei isolation, tissue reserved from the cross-linking step was ground using liquid nitrogen to form a fine-grained dry powder. Nuclei were isolated as described above in the section entitled Fractionation of RNA. A Quasar 570-labeled GhDAN1-specific probe set was ordered. A hybridization assay was performed with 2 mL of the nuclei suspension using Stellaris RNA-FISH Hybridization Buffer, Wash Buffer A, and Wash Buffer B (Biosearch Technologies, Beverly, MA, USA; cat. nos. SMF-HB1-10, SMF-WA1-60, and SMF-WB1-20) strictly following the protocol for hybridization, washing, and DAPI staining according to the Stellaris RNA-FISH kit before proceeding to image. For probes labeled with Quasar 570, an excitation line of 561 nm was applied and signals were detected at 570–640 nm; for DAPI (4′,6-diamidino-2-phenylindole), an excitation line of 405 nm was applied and signals were detected at 420–480 nm.
ChIRP The ChIRP assay was performed as described (Chu et al., 2012; Ariel et al., 2020) using the Magna ChIRP RNA Interactome Kit (Millipore Sigma, Burlington, MA, USA; cat. no. 17-10495), with minor modifications. Antisense oligo probes for GhDAN1 were designed using the online probe designer (https://www.biosearchtech.com/support/tools/design-software/chirp-probe-designer). The probes were designed against every 100 bp over the 2 kb region on GhDAN1 (as indicated in Supplemental Figure S3) and numbered based on their position along the RNA from 5′ to 3′. GhDAN1 ChIRP reactions were performed using “even” numbered probe pool and “odd” numbered probe pool. Real RNA-dependent signals will be present in both “even” and “odd” pools as shared DNA peaks, while the random noises will be unique to each pool . Two grams of fresh cotton leaves was sufficient for the ChIRP reaction with the gene-specific probes (odd and even probe sets) and input control. The plant material cross-linking and nuclei isolation was performed as described above in the section entitled RNA-FISH Assay. Next, the pellet of nuclei from each 1 g of tissue was resuspended in 2 mL of complete lysis buffer (from the Millipore ChIRP Kit), followed by immediate sonicating in a Bioruptor (Diagenode, Denville, NJ, USA) to shear the DNA (aliquot 350 μL in each tube for sonication) to 100–500 bp in length. The sonicated chromatin was centrifuged for 10 min at 12,000 g at 4°C to collect the supernatant. Ultimately, ∼2 mL of the sonicated chromatin was obtained from each 1 g of fresh leaves. The tubes were snap frozen in liquid nitrogen and stored at −80°C. For ChIRP, each 1-mL sample of sonicated chromatin was transferred to a 15-mL RNase/DNase-free tube to be used in one ChIRP reaction. The Millipore ChIRP Kit user manual guidelines were followed carefully for completion of the ChIRP reaction assay and DNA isolation, followed by next-generation sequencing (NGS). Magna ChIRP Negative Control Probe Set (LacZ) in the Magna ChIRP RNA Interactome Kit was used as negative control. For the ChIRP-seq bioinformatic analysis, the first step after quality control and trimming of the adaptor was to map the clean data of the NGS to the reference cotton genome (TM-1; Hu et al., 2019) using hierarchical indexing for spliced alignment of transcripts 2 (HISAT2; the no-spliced-alignment parameter; Pertea et al., 2016). Second, peaks were called separately using data from the odd and even probe sets by MACS2 (using the P-value 1 × 10−5 parameter; Feng et al., 2011). Third, the peaks shared by the odd and even pools (designated as ChIRP valid peaks) were generated using intersectBed in the BEDTools suite (Quinlan and Hall, 2010). The peaks with a fold enrichment ≥two were selected as the final valid ChIRP peaks. The motif discovery was performed using MEME Suite version 5.0.5 (Bailey et al., 2009) with the top 10% most-enriched valid peaks. (MEME Top1000validpeaks.fasta -dna -nmotifs 20 -minsites 50 -minw 6 -maxw 20 -maxsize 6000000); motif scanning was performed using FIMO in MEME Suite version 5.0.5 (using default settings); and motif comparison was performed using Tomtom in MEME Suite version 5.0.5 (using default settings). Two biological replicates for ChIRP reaction were performed, and similar AAAG rich motif was repetitively established by MEME software.
Triplex prediction
Prediction of the TFOs in GhDAN1 RNA was performed using Triplexator software (Buske et al., 2012).
EMSA
EMSA for lncRNA and DNA interaction was performed as previously described, with modifications (Postepska-Igielska et al., 2015). A GhDAN1 fragment from +9 to +186 bp was first cloned into the pMD19-T vector to construct pMD19-T-GhDAN1 for sequence confirmation. PCR products containing a T7 promoter in the correct orientation were then prepared and purified by phenol–chloroform extraction and ethanol precipitation. RNA synthesis was performed using the HiScribe T7 High Yield RNA Synthesis Kit (NEB; cat. no. E040S), with the above PCR products used as templates, according to the user manual. As a result, ∼8 μg/μL (total 30 μL) GhDAN1 RNA was generated from each reaction. The 3′-DIG-labeled single-stranded oligonucleotide was synthesized and stored in ddH2O at a concentration of 100 pmol/μL. DIG-labeled DNA double-stranded probes (4 pmol) and cold probes (40 pmol) were annealed in a 50 μL reaction containing 50 mM NaCl following the prescribed program (95 °C for 10 min, 75 °C for 15 min, 55 °C for 15 min, 45 °C for 15 min, and 35°C for 15 min). In the EMSA reaction, 4 pmol (1 μL) of DIG-labeled double-stranded oligonucleotide, comprised nucleotides containing the GhDAN1 TTS motif that were predicted within the gene promoter (see Supplemental Table S6 for detailed sequences) was incubated for 2 h at 37 °C with 16 μg (∼320 pmol) synthetic GhDAN1 fragment in 40 mM Tris–acetate (pH 7.5), 20 mM KCl, 10 mM Mg (CH3COO)2, and 10% (v/v) glycerol. Cold probes (40 pmol) were added for a competitive reaction. RNaseA or RNaseH was added after the reaction when needed to confirm the triplex formation. Triplex formation was confirmed by EMSA using 12% (w/v) polyacrylamide gels containing 40 mM Tris–acetate (pH 7.5) and 10 mM MgCl2. After electrophoresis in 1× TBE, the DNA/RNA was transferred from the gel to an Amersham Hybond −N+ membrane in 1× TBE at 400 mA for 1 h. The probe signal was detected using the DIG Nucleic Acid Detection Kit (Roche, Basel, Switzerland; cat. no. 11175041910).
mRNA-seq
The total RNA was extracted using the PureLink Plant RNA Reagent (Invitrogen, Carlsbad, CA, USA; cat. no. 12322012). Libraries of mRNA were prepared and sequenced using the HiSeq 150-bp paired-end Illumina RNA-seq protocol, with three biological replicates for each treatment. Reads were quality controlled and trimmed using fastp (Chen et al., 2018). The clean reads were mapped onto the cotton genome using HISAT2 (Pertea et al., 2016). Gene expression was calculated using StringTie with parameter (-A) (Pertea et al., 2016). For differential expression analysis, each gene count was calculated using HTseq-count (Anders et al., 2015). DEGs were tested using the edgeR package (Robinson et al., 2010).
RT-qPCR
The total RNA was extracted using the PureLink Plant RNA Reagent (Invitrogen; cat. no. 12322012). cDNAs were synthesized from the total RNA using HiScript II Q RT SuperMix for qPCR (+gDNA wiper; Vazyme, Nanjing, China; cat. no. R233). RT-qPCR was performed on the ABI 7900HT Fast Real-Time PCR System using histone h3 (AF024716) as a reference gene. Primers used in this study are presented in Supplemental Table S6.
ChIP
For the ChIP assay, a procedure similar to that of the ChIRP assay was followed to complete the cross-linking of the plant material, nuclei isolation, cell lysis, and chromatin sonication (Haring et al., 2007). The immunoprecipitation of the cross-linked protein/DNA was performed using the Magna ChIP A/G ChIP Kit (Millipore; cat. no. 17-10085) according to the user manual instructions. Anti-histone H3 (trimethyl K4) antibodies (ab8580) and anti-histone H3 (trimethyl K27) antibodies (ab6002) were used in the assay. NGS reads were mapped to the cotton genome using Bowtie, allowing up to two mismatches (Langmead et al., 2009). The reference genome for G. herbaceum subs. Africanum (A1-a), G. arboreum (A2, accession Shixiya 1), and (G. arboreum x G. raimondii)F1 was G. arboreum (A2, accession Shixiya 1; Du et al., 2018); the reference genome for G. barbadense (AD2, accession H7124) and G. darwinii (AD5) was G. barbadense (AD2, accession H7124; Hu et al., 2019); and the reference genome for G. hirsutum (AD1, accessions yucatanense2094, punctatum25 and TM-1), G. tomentosum (AD3), G. mostelinum (AD4), G. ekmanianum (AD6), and G. stephensii (AD7) was G. hirsutum (AD1, accession TM-1; Hu et al., 2019). For the uniquely mapped reads, peak calling of the ChIP-seq data was performed using MACS2 (Zhang et al., 2008).
Phylogenetic analysis
Phylogenetic tree depicting the evolutionary relationship of DAN1 in cotton lineage was performed using MEGA software (Tamura et al., 2007).
Statistical analyses
All statistical analysis was done with the software Graphpad Prism (GraphPad Software, La Jolla, CA, USA). The Student’s t test, Mann–Whitney U-test, or Fisher’s exact test was used whenever appropriate. The P ˂0.05 were considered statistically significant.
Accession numbers
Novel data generated in this study, including raw data of mRNA-seq and ChIRP-seq, have been deposited in the National Center for Biotechnology Information under the accession number PRJNA637977. Pol II ChIP-seq data are under the accession number PRJNA373801, as previously reported (Zhao et al., 2018a, 2018b).
Supplemental data
The following supplemental materials are available in the online version of this article.
Supplemental Figure S1. Phenotypes of GhDAN1-, CLA1-, and GoPGF-silenced plants.
Supplemental Figure S2. Analysis of physiological traits associated with photosynthesis.
Supplemental Figure S3. Alternative splicing of GhDAN1.
Supplemental Figure S4. The lincRNA GhDAN1 has a poly(A) tail and does not encode any peptide/protein.
Supplemental Figure S5. DNA CHG and CHH methylation status of the DAN1 gene body and promoter region.
Supplemental Figure S6. Sequence alignment of DAN1 genomic DNA in 13 cotton lineages.
Supplemental Figure S7. Stack plots of ChIP-seq showing the H3K27me3 profile in DAN1 loci in 12 cotton lineages.
Supplemental Figure S8. Phenotype of A diploid and AD allotetraploid plants before and after drought treatment lasting 10 d.
Supplemental Figure S9. GhDAN1 is not a regulator in cis.
Supplemental Figure S10. Analysis of ChIRP-seq peaks and peak-associated genes.
Supplemental Figure S11. mRNA-seq results of selected DEGs and RT-qPCR confirmation.
Supplemental Figure S12. GhDAN1 does not affect genes in the auxin biosynthesis or the ABA biosynthesis/catabolism pathways.
Supplemental Figure S13. Profile of the TTS motif-containing DEGs from GhDAN1-silenced plants under dehydration treatment.
Supplemental Figure S14. Phenotypes of plants that have undergone VIGS before and after drought treatment.
Supplemental Table S1. Information of cotton accessions used in this study.
Supplemental Table S2. TFOs in GhDAN1 predicted by Triplexator software.
Supplemental Table S3. List of DEGs in GhDAN1-silenced plants.
Supplemental Table S4. List of orthologous genes of TTS motif-containing DEGs in other plants that were involved in drought stress tolerance.
Supplemental Table S5. Shared DEGs with the TTS motif between GhDAN1-silenced plants and dehydration-treated Gh plants.
Supplemental Table S6. Primers used in this study.
Supplementary Material
Acknowledgment
We would like to thank Professor Kunbo Wang from the National Wild Cotton Nursery, Sanya, and Professor Xiongming Du from the National Medium Term Gene Bank of Cotton in China, the Institute of Cotton Research of the Chinese Academy of Agricultural Sciences, for providing the seed and leaf tissues of diploid cotton and wild allotetraploid cotton germplasm. We would also like to thank Professor Liang Wu for providing insightful comments on the manuscript.
Funding
This work was financially supported in part by grants from the National Key Research and Development Program (2016YFD0100605), National Natural Science Foundation of China (NSFC, 31971985, 31900395, and 31901430), Fundamental Research Funds for the Central Universities, and the funding of “Leading Innovative and Entrepreneur Team Introduction Program of Zhejiang” (2019R01002).
Conflict of interest statement. The authors declare no conflicts of interest.
X.G. and X.T. conceptualized this project and designed the experiments. X.T., M.L., and H.Z. performed the experiments and generated the data. T.Z., X.T., and S.F. carried out the bioinformatic analysis. X.G., X.T., M.L., H.Z., Q.C., B.Z., and L.W. analyzed the data. M.L., L.W., J.H., M.G., and K.L. conducted the functional lncRNA screening. B.Z. provided the (G. arboreum × G. raimondii) F1 leaves for the study. X.G., X.T., and M.L. wrote the manuscript. All authors discussed results and commented on the manuscript. All authors read and approved the final manuscript.
The author responsible for the distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (https://academic.oup.com/plphys/pages/general-instructions) is Xueying Guan (xueyingguan@zju.edu.cn).
References
- Adams KL, Percifield R, Wendel JF (2004) Organ-specific silencing of duplicated genes in a newly synthesized cotton allotetraploid. Genetics 168: 2217–2226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8: 135–141 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F, Lucero L, Christ A, Mammarella MF, Jegu T, Veluchamy A, Mariappan K, Latrasse D, Blein T, Liu C, et al. (2020) R-loop mediated trans action of the APOLO long noncoding RNA. Mol Cell 77: 1055–1065 e1054 [DOI] [PubMed] [Google Scholar]
- Chu C, Quinn J, Chang HY (2012) Chromatin isolation by RNA purification (ChIRP). J Vis Exp 61: 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acid Res 37: W202–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrera-Redondo J, Ibarra-Laclette E, Vázquez-Lobo A, Gutiérrez-Guerrero YT, Sánchez de la Vega G, Piñero D, Montes-Hernández S, Lira-Saade R, Eguiarte LE (2019) The genome of Cucurbita argyrosperma (silver-seed gourd) reveals faster rates of protein-coding gene and long noncoding RNA turnover and neofunctionalization within cucurbita. Mol Plant 12: 506–520 [DOI] [PubMed] [Google Scholar]
- Baumann K, De Paolis A, Costantino P, Gualberti G (1999) The DNA binding site of the D of protein NtBBF1 is essential for tissue-specific and auxin-regulated expression of the rolB oncogene in plants. Plant Cell 11: 323–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostock RM, Pye MF, Roubtsova TV (2014) Predisposition in plant disease: exploiting the nexus in abiotic and biotic stress perception and response. Ann Rev Phytopathol 52: 517–549 [DOI] [PubMed] [Google Scholar]
- Buske FA, Bauer DC, Mattick JS, Bailey TL (2012) Triplexator: detecting nucleic acid triple helices in genomic and transcriptomic data. Genome Res 22: 1372–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J (2018) fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34: i884–i890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Y, Zhao T, Yang J, Feng S, Nazeer W, Zhang T, Zhou B (2015) A new synthetic amphiploid (AADDAA) between Gossypium hirsutum and G. arboreum lays the foundation for transferring resistances to verticillium and drought. PLoS One 10(6): e0128981 doi:10.1371/journal.pone.0128981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2007) Genetic and epigenetic mechanisms for gene expression and phenotypic variation in plant polyploids. Ann Rev Plant Biol 58: 377–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2010) Molecular mechanisms of polyploidy and hybrid vigor. Trends Plant Sci 15: 57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ (2013) Genomic and epigenetic insights into the molecular bases of heterosis. Nat Rev Genet 14: 471–482 [DOI] [PubMed] [Google Scholar]
- Cipollini D, Walters D, Voelckel C, editors (2017) Costs of resistance in plants: from theory to evidence. InAnnual Plant Reviews Online: Insect‐Plant Interactions, Vol. 47, John Wiley & Sons, Ltd., pp 263–307 [Google Scholar]
- Comas LH, Becker SR, Cruz VM, Byrne PF, Dierig DA (2013) Root traits contributing to plant productivity under drought. Front Plant Sci 4: 442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad B, Antonarakis SE (2007) Gene duplication: a drive for phenotypic diversity and cause of human disease. Ann Rev Genom Hum Genet 8: 17–35 [DOI] [PubMed] [Google Scholar]
- Cronn RC, Zhao X, Paterson AH, Wendel JF (1996) Polymorphism and concerted evolution in a tandemly repeated gene family: 5S ribosomal DNA in diploid and allopolyploid cottons. J Mol Evol 42: 685–705 [DOI] [PubMed] [Google Scholar]
- Davidson RM, Manosalva P, Snelling J, Bruce M, Leung H, Leach JE (2010) Rice germin-like proteins: allelic diversity and relationships to early stress responses. Rice 3: 43–55 [Google Scholar]
- Deng P, Liu S, Nie X, Weining S, Wu L (2018) Conservation analysis of long non-coding RNAs in plants. Sci China-life Sci 61: 190–198 [DOI] [PubMed] [Google Scholar]
- Du H, Liu H, Xiong L (2013) Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci 4: 397–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Huang G, He S, Yang Z, Sun G, Ma X, Li N, Zhang X, Sun J, Liu M (2018) Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat Genet 50: 796–802 [DOI] [PubMed] [Google Scholar]
- Fedak H, Palusinska M, Krzyczmonik KLien Brzezniak L,Yatusevich R, Pietras Z, Kaczanowski S,Swiezewski S (2016) Control of seed dormancy in Arabidopsis by a cis-acting noncoding antisense transcript. Proc Natl Acad Sci U S A 113: E7846–E7855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Zhang Y (2011) Using MACS to identify peaks from ChIP-Seq data. Curr Protocol Hum Genet 34: 2.14.1–2.14.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR (2011) HMMER web server: interactive sequence similarity searching. Nucleic Acid Res 39: W29–W37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Valli A, Todesco M, Mateos I, Puga MI, Rubio-Somoza I, Leyva A, Weigel D, Garcia JA, Paz-Ares J (2007) Target mimicry provides a new mechanism for regulation of microRNA activity. Nat Genet 39: 1033–1037 [DOI] [PubMed] [Google Scholar]
- Goudarzi M, Berg K, Pieper LM, Schier AF (2019) Individual long non-coding RNAs have no overt functions in zebrafish embryogenesis, viability and fertility. Elife 8: e40815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix DA, Koch F, Wahrisch S, Beisaw A, Macura K, Blass G, Kellis M, Werber M (2013) The tissue-specific lncRNA fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha M, Lu J, Tian L, Ramachandran V, Kasschau KD, Chapman EJ, Carrington JC, Chen X, Wang XJ, Chen ZJ (2009) Small RNAs serve as a genetic buffer against genomic shock in Arabidopsis interspecific hybrids and allopolyploids. Proc Natl Acad Sci USA 106: 17835–17840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haring M, Offermann S, Danker T, Horst I, Peterhansel C, Stam M (2007) Chromatin immunoprecipitation: optimization, quantitative analysis and data normalization. Plant Methods 3: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, et al. (2019) Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet 51: 739–748 [DOI] [PubMed] [Google Scholar]
- Jia Y, Pan Z, He S, Gong W, Geng X, Pang B, Wang L, Du X (2018) Genetic diversity and population structure of Gossypium arboreum L. collected in China. J Cotton Res 1: 11 [Google Scholar]
- Kalwa M, Hanzelmann S, Otto S, Kuo C, Franzen J, Joussen S, Fernandezrebollo E, Rath B, Koch CM, Hofmann A (2016) The lncRNA HOTAIR impacts on mesenchymal stem cells via triple helix formation. Nucleic Acids Res 44: 10631–10643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G (2007) CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res 35: W345–W349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MR, Kim CS, Park T, Choi Y, Lee K (2018) Optimization of the ninhydrin reaction and development of a multiwell plate-based high-throughput proline detection assay. Anal Biochem 556: 57–62 [DOI] [PubMed] [Google Scholar]
- Li L, Eichten SR, Shimizu R, Petsch K, Yeh CT, Wu W, Chettoor AM, Givan SA, Cole RA, Fowler JE, et al. (2014) Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol 15: R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Syed J, Sugiyama H (2016) RNA-DNA triplex formation by long noncoding RNAs. Cell Chem Biol 23: 1325–1333 [DOI] [PubMed] [Google Scholar]
- Long Y, Wang X, Youmans DT, Cech TR (2017) How do lncRNAs regulate transcription? Sci Adv 3: eaao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano Y, Nemoto K (2012) The pathway of auxin biosynthesis in plants. J Exp Bot 63: 2853–2872 [DOI] [PubMed] [Google Scholar]
- Mcclintock B (1984) The significance of responses of the genome to challenge. Science 226: 792–801 [DOI] [PubMed] [Google Scholar]
- Mondal T, Subhash S, Vaid R, Enroth S, Uday S, Reinius B, Mitra S, Mohammed A, James AR, Hoberg E, et al. (2015) MEG3 long noncoding RNA regulates the TGF-beta pathway genes through formation of RNA-DNA triplex structures. Nat Commun 6: 7743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, Baker JC, Grutzner F, Kaessmann H (2014) The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature 505: 635–640 [DOI] [PubMed] [Google Scholar]
- Noguero M, Atif RM, Ochatt S, Thompson RD (2013) The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci 209: 32–45 [DOI] [PubMed] [Google Scholar]
- Oleary VB, Ovsepian SV, Carrascosa LG, Buske FA, Radulovic V, Niyazi M, Moertl S, Trau M, Atkinson MJ, Anastasov N (2015) PARTICLE, a triplex-forming long ncRNA, regulates locus-specific methylation in response to low-dose irradiation. Cell Rep 11: 474–485 [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Csukasi F, Zheng Y, Fei Z, van der Knaap E, Catalá C (2015) Comprehensive tissue-specific transcriptome analysis reveals distinct regulatory programs during early tomato fruit development. Plant Physiol 168: 1684–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protocol 11: 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postepska-Igielska A, Giwojna A, Gasri-Plotnitsky L, Schmitt N, Dold A, Ginsberg D, Grummt I (2015) LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol Cell 60: 626–636 [DOI] [PubMed] [Google Scholar]
- Qin T, Zhao H, Cui P, Albesher N, Xiong L (2017) A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol 175: 1321–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Ann Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehin M, Li B, Tang M, Katz E, Song L, Ecker JR, Kliebenstein DJ, Estelle M (2019) Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat Commun 10: 4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancenon V, Puig S, Mira H, Thiele DJ, Penarrubia L (2003) Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol Biol 51: 577–587 [DOI] [PubMed] [Google Scholar]
- Semon M, Wolfe KH (2008) Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis. Proc Natl Acad Sci USA 105: 8333–8338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Zhang T, Stelly DM, Chen ZJ (2017) Epigenomic and functional analyses reveal roles of epialleles in the loss of photoperiod sensitivity during domestication of allotetraploid cottons. Genome Biol 18: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley JT, Nei M, Kumar S (2007) MEGA4: molecular evolu-tionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tiwari S, Lata C, Chauhan PS, Prasad V, Prasad M (2017) A functional genomic perspective on drought signalling and its crosstalk with phytohormone-mediated signalling pathways in plants. Curr Genom 18: 469–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tognetti VB, Muhlenbock P, Van Breusegem F (2012) Stress homeostasis - the redox and auxin perspective. Plant Cell Environ 35: 321–333 [DOI] [PubMed] [Google Scholar]
- Wang K, Wendel JF, Hua J (2018) Designations for individual genomes and chromosomes in Gossypium. J Cotton Res 1: 3 [Google Scholar]
- Wang P, Shen L, Guo J, Jing W, Qu Y, Li W, Bi R, Xuan W, Zhang Q, Zhang W (2019) Phosphatidic acid directly regulates PINOID-dependent phosphorylation and activation of the PIN-FORMED2 auxin efflux transporter in response to salt stress. Plant Cell 31: 250–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendel JF, Grover CE (2015) Taxonomy and evolution of the cotton genus, Gossypium. Cotton 2: 1–20. [Google Scholar]
- Xing Y, Yao R, Zhang Y, Guo C, Jiang S, Xu G, Dong R, Yang L, Chen L (2017) SLERT regulates DDX21 rings associated with pol I transcription. Cell 169: 664–678 [DOI] [PubMed] [Google Scholar]
- Yang Y, Liu T, Shen D, Wang J, Ling X, Hu Z, Chen T, Hu J, Huang J, Yu W, et al. (2019) Tomato yellow leaf curl virus intergenic siRNAs target a host long noncoding RNA to modulate disease symptoms. PLoS Pathogens 15: e1007534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wang M, Li N, Wang H, Qiu P, Pei L, Xu Z, Wang T, Gao E, Liu J (2018) Long noncoding RNAs involve in resistance to Verticillium dahliae, a fungal disease in cotton. Plant Biotechnol J 16: 1172–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang D, Lang Z, He L, Yang L, Zeng L, Li Y, Zhao C, Huang H, Zhang H, Zhang H, Zhu JK (2016) Methylation interactions in Arabidopsis hybrids require RNA-directed DNA methylation and are influenced by genetic variation. Proc Natl Acad Sci USA 113: E4248– E–4256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Hu Y, Jiang W, Fang L, Guan X, Chen J, Zhang J, Saski CA, Scheffler BE, Stelly DM, et al. (2015) Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol 33: 531–537 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Li Fu, Liu C, Zhang C, Wu Z (2010) Isolation and analysis of drought-related gene from cotton (Gossypium arboreum L.) SSH library. Cotton Sci 22: 110–114 [Google Scholar]
- Zhao T, Tao X, Feng S, Wang L, Hong H, Ma W, Shang G, Guo S, He Y, Zhou B, et al. (2018a) LncRNAs in polyploid cotton interspecific hybrids are derived from transposon neofunctionalization. Genome Biol 19: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y (2018b) Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun 9: 5056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.