Abstract
Microfluidic devices can provide a versatile, cost-effective platform for disease diagnostics and risk assessment by quantifying biomarkers. In particular, simultaneous testing of several biomarkers can be powerful. Here, we critically review work from the previous 4 years up to February 2021 on developing microfluidic devices for multiplexed detection of biomarkers from samples. We focus on two principal approaches: electrical and optical detection methods that can distinguish and quantify biomarkers. Both electrical and spectroscopic multiplexed detection strategies are being employed to reach limits of detection below clinical sample levels. Some of the most promising strategies for point-of-care assays involve inexpensive materials such as paper-based microfluidic devices, or portable and accessible detectors such as smartphones. This review does not comprehensively cover all multiplexed microfluidic biomarker studies, but rather provides a critical evaluation of key work and suggests promising prospects for future advancement in this field.
Graphical abstract
Electrical and optical multiplexing are powerful approaches for microfluidic biomarker analysis.
Keywords: Multiplexing, Paper microfluidics, Immunoassays, Electrical detection, Spectroscopy
Introduction
Biomarkers are analytes such as proteins or other molecules in the body that can be measured to give insight into physiological conditions or diseases. Detecting and quantifying biomarkers enables disease monitoring, diagnostics, and risk assessment. Recent studies have involved specific biomarkers for assessing preterm birth risk [1], diagnosing cardiovascular disease [2], monitoring drug side effects [3], and so on. Many diseases and conditions require rapid treatment, so researchers are developing tests for biomarkers that are usable at the point-of-care (POC) [4], where the test can be performed on-site with sometimes limited resources. The World Health Organization recommends POC tests that are Affordable, Sensitive, Specific, User-friendly, Rapid and robust, Equipment-free and Deliverable to end-users (ASSURED) [5]. Although traditional methods for detecting biomarkers, such as immunoassays, mass spectrometry, and HPLC, are highly sensitive and specific, they are not optimal for POC testing because they often require costly equipment or extensive training [3]. Development of rapid, inexpensive methods can make biomarker tests more accessible for POC and limited-resource locations.
Microfluidics provide a versatile platform for POC detection of biomarkers [6]. Microfluidic devices have small channels that carry micro- to nanoliter volumes of fluids. They are portable and relatively cheap, and use small sample and reagent volumes, making them advantageous for miniaturized sample preparation or integration into a “lab-on-a-chip” system. On-chip sample processing also potentially eliminates the need for bulky or expensive robotic or other equipment that can require considerable space as well as operator technical training [7]. These benefits open possibilities for remote or resource-limited locations to use the devices for detecting biomarkers, while maintaining the necessary selectivity and specificity obtained with traditional methods [8].
Sonker et al. [6] previously reviewed advances in on-chip sample preparation and separation. A key concern they raised was that more work was needed in multiplexing, or the simultaneous detection of multiple biomarkers. Analyzing multiple biomarkers at the same time can give a more complete understanding of conditions, enabling more accurate medical diagnostics. In the years since Sonker’s publication, important progress has been made in multiplex detection of biomarkers [9], improving microfluidic device usability [4, 10] and developing microfluidic devices for other medical uses [11–14]. This review discusses advances in detection in multiplexed microfluidic devices for the simultaneous detection of biomarkers. We review papers demonstrating microfluidic electrical or spectroscopic detection of multiple biomarkers published between March 1, 2017, and February 1, 2021.
Electrical detection
Electrical-based detection techniques find use in bioanalysis because they can provide information on a broad range of species with or without labeling prior to analysis. In this section, we review common electrical-based detection techniques, which include electrochemistry, giant magnetoresistance, field effect transistors, electrophoresis, and Coulter counting. We discuss recent multiplexing work with these techniques and cover their pros and cons.
Electrochemical detection
Electrochemical detection is based on measuring electrical currents produced by redox reactions for the compounds under analysis. It utilizes electrodes (which are sometimes referred to as sensors) in contact with an electrolyte for analysis. Due to its high selectivity, high sensitivity, low limit of detection, and low power requirement, electrochemical detection is useful in biochemical analysis. Also, because electrochemical sensors can be easily miniaturized, they can be readily integrated into microfluidic devices, making them potentially economical and suitable for POC analysis. Several electrochemical detection methods are available for multiplexing; we first discuss two commonly used approaches—voltammetry and amperometry.
Voltammetry
In voltammetry, a three-electrode system is typically used with reference, working, and counter electrodes. Fundamentally, voltammetry involves applying a voltage as a function of time, and then measuring the resulting current through the circuit. This measured current is then plotted against the applied voltage, yielding insights into the properties of a target analyte.
Pursey et al. [15] developed a microfluidic device integrated with porphyrin-based electrochemical sensors for the multiplex detection in 20 min of three bladder cancer biomarkers, a tumor suppressor (E. Cad), a mediator of cell death (DAPK), and a mediator of cell growth (RARβ). A molecular beacon modified with a free-base porphyrin tag was immobilized on 20 gold sensors. The molecular beacon facilitated a large change in current when the DNA sequence on the end of its probe hybridized with the matched targets, compared to exposure to mismatched targets. With 18 sensor pads, multiple parallel runs were done for each of the matched and mismatched targets to ensure reproducibility and accuracy, while two other sensor pads were used for background measurement. This electrochemical integrated microfluidic system had a dynamic range of about six orders of magnitude for detecting these bladder cancer biomarkers. Also, a limit of detection (LOD) of 250 fM was obtained for this measurement, which is below the typical concentrations found in a patient’s urine sample. Although the dynamic range and LOD are favorable, this is not a label-free electrochemical detection method, which makes operational procedures and biosensor development less simple. Also, off-chip methylation-specific polymerase chain reaction is crucial for this method to distinguish the matched target from the mismatched target, which results in an increase in analysis time. Integrating the methylation-specific PCR on-chip could help in reducing the total analysis time.
Using label-free detection simplifies operational procedures and reduces analysis time, for example, as described by Wang et al. [16]. Label-free aptasensors were integrated into a paper-based microfluidic device for the simultaneous detection of two lung cancer protein biomarkers, carcinoembryonic antigen (CEA) and neuron-specific enolase (NSE). In this approach, the sample flowed by capillary action through the microchannel onto the screen-printed electrodes that had been modified with their corresponding DNA aptamers. Once the target analyte was recognized, there was a drop in current due to the aptamer-biomarker complex formed on the electrode surface, which hindered electron transfer, as probed by cyclic voltammetry and differential pulse voltammetry. LODs obtained were 2 pg/mL and 10 pg/mL for CEA and NSE, respectively, which are well below the minimum level to diagnose risk for lung cancer in humans. Also, data showed that the biosensors had good specificity and a 5 order of magnitude dynamic range for both CEA and NSE.
In another work, Zhu et al. [3] detected four leukemia biomarkers (methotrexate, lactate dehydrogenase, uric acid, and urea) with screen-printed electrodes in four different sections of a device. They flowed a blood sample through a filtering film to remove components >1000 Da, then divided the sample into four detection cells. Each cell contained an electrode with different modifying groups specific to one of the biomarkers, with detection by cyclic voltammetry, electrochemical impedance spectroscopy, and differential pulse voltammetry. Biomarkers were detected from a spiked rabbit blood sample using the on-chip electrodes; similar recovery efficiencies were found for all four targets compared to the standard HPLC method, and the LODs were below levels found in human blood serum.
One difference between this method and that of Wang et al. [16] is that this microfluidic chip used a photosensitive resin that required a film for sample filtration, whereas Wang et al.’s used cellulose filter paper which allowed for functions such as sample filtration and auto-injection without the need for external systems. The work done by Wang et al. uses a label-free method, but unlike in the work done by Pursey et al. [15], the devices are not readily reusable.
Amperometry
In amperometry, the redox potential applied to the working electrode is kept constant, while the current in the circuit is measured to detect the concentration of the oxidized or reduced analyte of interest [17]. Amperometry finds numerous applications in miniaturized biochemical analysis because it is sensitive and selective and can be easily integrated into microfluidic devices.
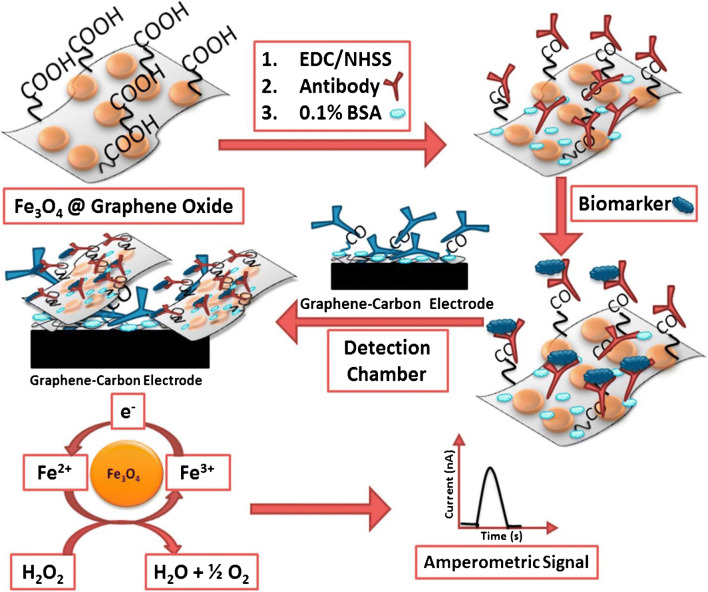
Sharafeldin et al. [18] used a combination of amperometry and a microfluidic immunoarray with 8 sensors for the multiplex detection of prostate cancer biomarkers. In this approach, off-chip capture of prostate-specific antigen (PSA) and prostate-specific membrane antigen (PSMA) on iron oxide nanoparticles assembled onto graphene oxide nanosheets and decorated with detection antibodies was first done. Next, the analytes were delivered into the microfluidic device containing the amperometric detection chamber, having a constant applied potential and 8 sensors modified with capture antibodies for analysis. The amperometric peak current obtained for the measurement was dependent on the amounts of antibody-linked nanoparticles bound to the sensor, which in turn was proportional to the concentration of the captured antigen of interest. Figure 1 shows a schematic of the protein capture and detection process. LODs of 15 and 4.8 fg/mL were obtained for PSA and PMSA, respectively, which are lower than the levels found in human blood serum. Furthermore, the accuracy of this method was validated by comparing the data obtained with ELISA results. One limitation of this work was that analyte capture was done off-chip. With improvements to the design of the microfluidic device, this method could allow on-chip capture of the analytes of interest, which would in turn improve portability of the device and make it suitable for POC application.
Fig. 1.
Process flow of biomarker immobilization and amperometric detection mediated by iron oxide nanoparticles assembled onto graphene oxide nanosheets. Adapted with permission from ref. [18]. Copyright 2017 Elsevier
Giant magnetoresistance
Giant magnetoresistance (GMR) is a surface-based biosensing method that leverages the large change in electrical resistance when the surface of a GMR material experiences a magnetic field. Due to the sensitive nature of this method, it can find use in the detection of biological molecules, such as DNA and proteins.
Gao et al. [19] demonstrated the use of GMR for the detection of 12 tumor biomarkers. In this method, a GMR chip comprising multilayer structures of different magnetic properties was utilized, and the surface was modified with polyvinyl chloride to improve its adsorption characteristics. A change in analyte concentration influenced the number of magnetic nano-beads (captured by biotinylated antibodies) attached to the GMR surface, which resulted in a decrease in the applied alternating current magnetic field. This led to a change in resistance of the GMR sensor surface, which was used to determine the analyte concentration. The LODs (<1 ng/mL) obtained for this experiment may not give a true reflection of the capabilities of this technology because nonspecific adsorption limited the precision at low biomarker concentrations. GMR technology for bioanalysis is expensive because of the sensor cost and the effort needed to functionalize the GMR biosensor surface and immobilize antibodies. Additionally, the magnetic nature of GMR-based sensors makes hysteresis a common concern that can reduce performance.
Field-effect transistor chemiresistor
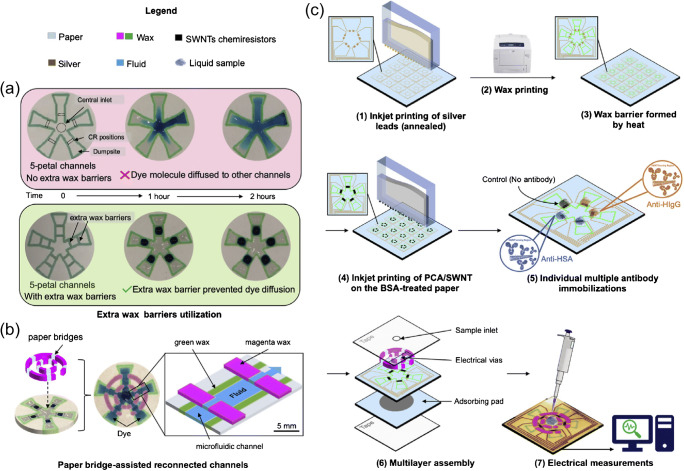
Another label-free technology is a field-effect transistor chemiresistor-based biosensor, whose electrical resistance varies with changes in the chemical environment. An origami-paper-based microfluidic device integrated with chemiresistor biosensors was developed by Shen et al. [20] for the multiplex detection of human serum albumin (HSA) and human immunoglobulin G (IgG) in fluids. The fabrication method for the multiplexed paper microfluidic device used wax printing, and the biosensors were made from single-walled carbon nanotubes (SWNTs). Figure 2 shows a schematic of the microfluidic channel design and an overview of sensor fabrication and testing. Detection occurred upon sample transport by the microfluidic channels to biosensors functionalized with monoclonal anti-HSA and anti-IgG, with an applied source-drain voltage of 0.1 V. The change in resistance of the chemiresistor was monitored and used to compute the analyte concentration. Concentrations as low as 1.5 pM were detected for the target analytes, demonstrating good sensitivity for a paper-based microfluidic device.
Fig. 2.
Five-petal channel design using extra wax barriers and paper bridges for channel reconnections. a Top: dye molecules spreading to other channels without the extra wax barriers. Bottom: spread of dye molecules prevented by the extra wax barriers. b Paper bridges assembly. c An overview of the fabrication process. Adapted with permission from ref. [20]. Copyright 2021 Elsevier
Electrophoresis
Microchip electrophoresis (μCE) is a technique that separates ions based on their electrophoretic mobility using an applied voltage. It provides high resolution and requires a small amount of sample, which, in combination with quantitation capabilities, makes μCE effective for multiplexing.
Sahore et al. [21] described a device with microvalves and porous polymer monoliths that facilitated the analysis of the preterm birth (PTB) risk biomarkers, ferritin and corticotropin-releasing factor. Their device first preconcentrated the biomarkers on a reverse-phase monolith in the channel; they then attached fluorescent labels and eluted the labeled biomarkers from the monolith. The biomarkers were separated using μCE, and peaks were detected using laser-induced fluorescence. This integrated system provided automated labeling and separation of PTB biomarkers on-chip. However, the multilayer fabrication process used is slow and scales poorly for making many devices.
Nielsen et al. [1] carried out μCE separation of six of the nine PTB biomarkers in the panel with sizes that ranged from 2 to 470 kDa. Their LODs were still above those needed for PTB risk assessment; further work with upstream sample preparation could improve the LODs.
Xie et al. [22] reported the separation and detection of ɑ-fetoprotein (AFP), carbohydrate antigen 125 (CA125), and CEA using μCE and fluorometry. They used aptamers, attached to gold nanoparticles on iron oxide magnetic particles, as capture probes. Different biomarkers captured on these particles allowed generation of different sizes of DNA that were then separated by μCE with fluorescence detection. LODs obtained for these three biomarkers were 0.1–0.2 pg/mL. A notable feature of this method is that the aptamer-functionalized magnetic beads are reusable.
Coulter counter
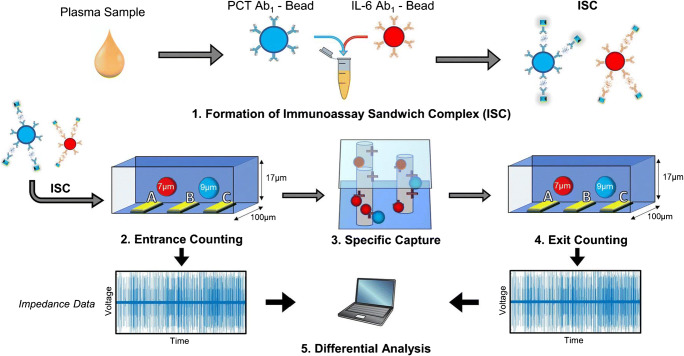
A typical Coulter counter has entrance and exit openings connected by one or more microchannels; it works by recording the change in impedance that is proportional to the volume of the particle passing through the entrance/exit. Coulter counting finds use in numerous applications, like determining cell count and size, particle characterization, and hematology. Recent research has utilized Coulter counters to successfully analyze biomarkers. For example, Berger et al. [23] demonstrated the detection of two sepsis biomarkers, interleukin-6 (IL-6) and procalcitonin (PCT). The microfluidic platform utilized entrance and exit electrodes, and a streptavidin-functionalized chamber captured protein-specific microbeads having a biotinylated secondary antibody. Figure 3 shows an overview of the detection of the biomarkers using this method. After loading, the beads were counted, with ones of interest captured in the chamber. Following that, the exit counter identified the non-captured population, with the difference between the entrance and exit count used to calculate the target analyte concentration. The LODs were 130–150 pg/mL; the use of different microbead sizes provided distinguishable signals for identifying the different biomarker complexes with a single set of Coulter electrodes. The method has limitations in capture efficiency and nonspecific capture; improvements by developing new types of immunocapture microbeads that are of similar diameter could lower the LOD.
Fig. 3.
Process flow for the multiplexed detection of PCT and IL-6 using electrical differential counting: (1) incubation of plasma with microbeads. (2) Enumeration of the sandwich complex for PCT and IL-6 by the entrance counter. (3) A population of the beads is captured based on the level of the biomarker in the plasma sample. (4) An exit counter enumerates the exit population. (5) Differential analysis of the entrance and exit populations. Adapted from ref. [23]. Copyright 2020 Springer
Cowell et al. [24] developed a microfluidic platform similar to that of Berger et al. [23] for the detection of PCT and IL-6. Their method utilized polyacrylamide-based hydrogel beads having the same size. Using droplet microfluidics, the hydrogel beads were synthesized by varying the size and density of hydrogel microparticles, resulting in beads with distinct solid volumes. Thus, bead populations had distinct impedance signatures during Coulter counting for easy multiplex analysis, with comparable LODs to those of Berger et al. [23]. One limitation of this approach is the device fabrication time.
Summary of electrical methods
Electrical detection shows promise in multiplexing because it is scalable, easy to miniaturize, and often straightforward. Table 1 shows a summary of different multiplexed electrical analysis methods. Electrical methods for multiplexed analysis work for a range of biomarkers indicative of different disease states, and can have excellent LODs. While electrical detection has numerous advantages, issues remain, such as nonspecific adsorption, electromagnetic noise, and chemical interferences. Emerging developments involving micro- and nano-fabrication can miniaturize electro-biosensor elements to improve results.
Table. 1.
Biomarker types, their relevance, and the immobilization and detection methods used for multiplexed electrical analysis
| Biomarker type | Size range (kDa) | Disease or condition | Immobilization | Detection method | LOD | Ref. |
|---|---|---|---|---|---|---|
| DNA | 0.65–0.66 | Bladder cancer | Electrode surface | Cyclic voltammetry | 200–250 fM* | [15] |
| Proteins | 45–200 | Lung cancer | Electrode surface | Differential pulse voltammetry | 2–10 pg/mL* | [16] |
| Proteins | 30–84 | Prostate cancer | Electrode surface | Amperometry | 5–15 fg/mL* | [18] |
| Proteins | 23–660 | Cancer | Device surface | Giant magnetoresistance | 0.02–1 ng/mL* | [19] |
| Proteins | 67–150 | Blood biomarkers | Device surface | Chemiresistor | 1.5–2 pM* | [20] |
| Peptides and proteins | 2–470 | Preterm birth | N/A | Electrophoresis with fluorescence | 1–200 nM** | [1] |
| Glycoproteins | 67–1500 | Ovarian cancer | Magnetic particles | Electrophoresis with fluorescence | 0.1–0.2 pg/mL* | [22] |
| Proteins | 14–21 | Sepsis | Microbead surface | Coulter counter | 130–150 pg/mL** | [23, 24] |
*Meets clinical threshold
**Does not meet clinical threshold
Spectroscopic detection
Spectroscopic detection in microfluidic devices utilizes wavelength-specific emission or absorption of light for biomarker detection. Fluorescence, chemiluminescence (CL), and colorimetric immunoassays are some of the more common multiplexing strategies. Geometric multiplexing, a common approach for spectroscopic multiplexing, entails dividing a sample across multiple sections of a device and then detecting individual biomarkers in each region. Partitioning samples in this way simplifies detection because researchers apply similar methods for detecting each biomarker in different sections of the device.
Microfluidic paper analytical devices
Researchers are developing microfluidic assays to match the detection limits and accuracy of ELISA and other methods while improving portability, cost, automation, etc. Microfluidic paper analytical devices (μPADs) provide an inexpensive platform with minimal assembly for these assays, making them a popular spectroscopy-based multiplexing strategy [25]. Figure 4 shows a typical μPAD layout.
Fig. 4.
An example μPAD design. The sample and additional reagents are added to the top of the device and capillary action causes them to flow through channels to detection regions for each biomarker. A sandwich immunoassay is created with immobilized antibodies, target antigen, and optically detectable secondary antibodies. The detection region is then accessed, and biomarkers are spectroscopically measured using a smartphone, scanner, or spectrometer
A simple μPAD design was created by Guo et al. [26]. They used a single-layer device and added horseradish peroxidase (HRP)-bound antibodies followed by luminol-H2O2, similar to ELISA. For three cancer biomarkers, they had LODs ranging from 0.2 to 2 ng/mL with a CL detector. Reaching lower LODs with microfluidic immunoassays requires improvements in amplification, for example, by adding CL-amplifying nanoparticles or utilizing biotin-streptavidin to increase HRP to biomarker ratio. Jiao et al. [27] used a 3D μPAD that included a negative control and pre-coated each of the sample-dividing channels with fluorescently labeled antibodies specific to that detection region, reducing antibody crossover. Pre-separating the mobile antibodies and including a negative control helped them achieve LODs of 30–50 pg/mL for cancer biomarkers using a smartphone detector and UV excitation. This design is promising for POC work; the assays take ~5 min, the materials are inexpensive, the detectors are portable, and the detection ranges are clinically relevant.
Nanowires and nanoparticles have a variety of uses in spectroscopic multiplexing, including surface-enhanced Raman spectroscopy (SERS), and CL or fluorescence amplification. Pinheiro et al. [28] created a μPAD that used plasmonic properties of gold nanoparticles (AuNPs) for colorimetric detection. They used in situ AuNP synthesis for glucose measurement, relying on the ability of glucose to reduce Au3+ to Au0, creating nucleation sites proportional to the glucose concentration. For uric acid and cholesterol, they relied on aggregative properties of functionalized nanoparticles; both nucleation and aggregation created shifts in solution color. They obtained LODs of 1 mM, 70 μM, and 80 μM for glucose, uric acid, and cholesterol, respectively. Although these devices have promising capabilities, the methods may not be generalizable to broad classes of biomarkers.
Lim et al. [29] used antibody-tagged nanoparticles having different shapes and composed of different metals as labels for a colorimetric μPAD. Although they achieved LODs of 500 pg/mL for three different cardiac biomarkers, they found that the calibration curve was nonlinear at high concentrations and the LOD varied depending on the detector. In subsequent work [30], they added Raman reporters to the nanoparticles and used SERS to create a calibration-free device with lower LODs. The improved LODs for the same cardiac biomarkers were 1–10 pg/mL, similar to their clinical cutoff values. The tradeoff for the lower LOD is that the SERS approach required a Raman microscope, whereas the initial method only required a smartphone for detection.
Rather than use nanoparticles as labels, Guo et al. [31] enhanced fluorescence signal using ZnO nanowires in a sandwich immunoassay. They formed the nanowires directly on μPADs and attached capturing antibodies to the nanowires. The target cardiac biomarker antigens and fluorophore-labeled secondary antibodies bound to the immobilized antibodies, bringing the labels close to ZnO and amplifying their signal. Compared to a control device, the ZnO nanowire device modestly lowered LODs for three cardiac biomarkers from 5–8 to 1–2 ng/mL with UV light excitation and smartphone detection.
An advantage of spectroscopic designs is their compatibility with smartphone detection. Eliminating the use of microscopes or bulky optical setups enables these devices to be implemented in more remote settings. Smartphone detection is aided by devices that include built-in negative controls or calibration curves. Colorimetry is especially sensitive to changes in temperature, humidity, lighting, etc., so integrating a calibration curve on-chip improves quantitative analysis by accounting for these environmental factors. Kim et al. [32] prepared four additional chambers per biomarker in a paper-based ELISA device to create a calibration curve from pre-loaded standards. They used colorimetric detection with a flatbed scanner or smartphone for readout. The LODs were 0.30 mM for glucose and lactate, which is below the levels in blood in healthy individuals, and only 0.4 μL of sample was needed. A disadvantage to this approach was that it required five chambers per biomarker, making it more challenging to multiplex beyond two biomarkers.
One strategy for CL multiplexing is to add reagents to each detection region sequentially, giving the signal temporal resolution. A challenge with temporally resolved CL is that it requires a slower flow rate, which lengthens the assay time. Li et al. [33] addressed this issue by creating a μPAD with larger detection regions connected in series by smaller channels, which allowed them to detect CL peaks with a single sensor and maintain a continuous flow rate. They achieved LODs of 0.7–15 μM for small biomolecules such as glucose, but generating H2O2 with biomarker-specific oxidases limits the method to biomolecules with available oxidases. Li et al. [34] later showed that they could amplify signal by switching from target-specific oxidases to AuNPs with associated Co(II), luminol, and antibodies. This process was easily adaptable to different proteins by changing the antibody conjugated to the AuNPs, and they lowered the LODs to 0.06–0.4 pg/mL with a CL detector. Li et al. [35] also demonstrated color-resolved CL using resonance energy transfer. They paired the CL reaction with fluorophores having different spectral properties, giving each biomarker a different location, time, and color. This combined strategy facilitated the use of smartphone detection and gave detection limits of 0.2 pg/mL, 3.2 pg/mL, and 4.2 fg/mL for CEA, AFP, and PSA, respectively. Of these three designs by Li et al., the color-resolved one is the best suited for POC use with smartphone detection.
μPADs use inexpensive materials and rely on capillary action instead of complicated pump systems, which is desirable for POC applications. With signal amplification strategies, many μPADs can even use smartphone detection. An issue with μPADs is that they are often limited to multiplexes of three to four protein or organic molecule biomarkers.
Microfluidic immunoassays
Immunoassays rely on antibody-antigen interactions to selectively capture and/or label biomarkers. Although traditional ELISA is considered the gold standard because of its commercial availability and low LOD, it requires several hours for analysis [9] and is most effective in clinical laboratories. Analysis time can potentially be reduced by miniaturizing immunoassay methods and automating processes.
Increased concentration of peroxidase per antibody can improve CL signal. Wu et al. [36] amplified CL by attaching biotin to the secondary antibody and then flowing streptavidin with biotin-bound HRP, allowing several HRP molecules to bind to each antibody. They performed a microfluidic immunoassay in a PDMS device by adding reagents through multiple perpendicular channels, creating a grid of detection regions. They obtained LODs of 1.0 and 44 pg/mL for IL-6 and PCT, a 2–4-fold improvement from traditional ELISA. This approach also reduced the analysis time > 2-fold relative to ELISA; this is still slower than other microfluidic assays, but some of the operation time could be eliminated by automating on-chip processes.
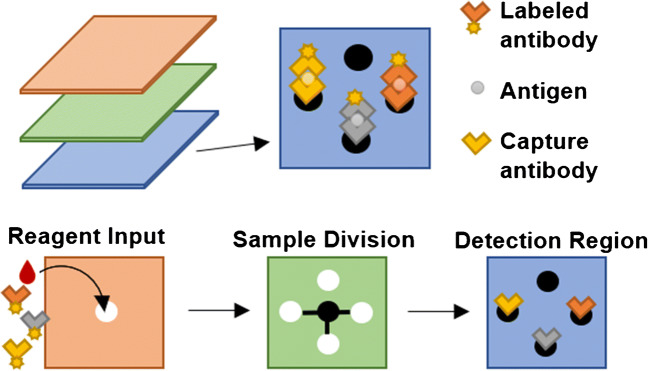
Many miniaturized methods use a different detection region for each biomarker. Alternatively, all biomarkers can be detected in the same location if there is a unique label or signal for each. In this format, the number of distinct signals that can be created/detected limits the level of multiplexing. Dinter et al. [37] distinguished fluorescently labeled biomarkers in an immunoassay using physically and spectroscopically different functionalized microbeads. Each bead type had a distinct diameter between 11.0 and 15.4 μm and a different ratio of rhodamine 6G to coumarin 334 to create a unique code, as represented in Fig. 5. They pooled all the beads to create a sandwich immunoassay using Cy5-conjugated antibodies and a hybridization assay with Cy5-conjugated DNA probes. With commercial video imaging, the size, fluorescent coding, and Cy5 signal were used to quantify cardiovascular disease biomarkers with LODs of 0.2–3 μg/mL, but major LOD improvements will be needed to access clinically relevant levels. Although they detected four biomarkers in this study, this design can allow multiplexing of up to eighteen biomarkers and can simultaneously detect both protein and DNA analytes.
Fig. 5.
Schematic of bead coding. a Each coded bead population is functionalized with antigens or probes for one biomarker. Target antibodies and secondary probes are prelabeled with Cy5. b As illustrated with false colors, microbead populations are distinguished by fluorescence and size. c Fluorescence coding is visible through a red corona created from Cy5. Reprinted from ref. [37]. Copyright 2019 Springer
Chang et al. [38] demonstrated a multilayer microfluidic chip fabricated with polymer sheets, and containing silica-based photonic crystal beads (PCBs) for the simultaneous detection of human IgG, CEA, and AFP. Antibodies to the biomarkers were attached to the PCBs to achieve selectivity. The microfluidic device leveraged the reflection spectra coding of the PCBs for the detection of the biomarkers, such that the analytes of interest were uniquely identified by the colors of their immobilized PCBs while their concentrations were determined from the PCB fluorescent intensity. The LOD for AFP was 20 ng/mL, which is about at clinical levels, such that further improvements would be needed for medical applications.
Yuan et al. [39] developed a portable imaging setup comprising microfluidic chips with an objective lens, prism, filters, and camera for the multiplex detection and quantification of a protein and human IgG related to dengue. Carboxyl-modified magnetic beads encoded with different concentrations of allophycocyanin dye provided unique spectral signatures. To capture dengue virus non-structural protein (DENV-NS1), the magnetic beads with higher dye concentration were coupled with the anti-DENV-NS1 specific antibody, while the beads with lower dye concentration were loaded with DENV-NS1 to capture anti-DENV-NS1 antibodies. The coupled beads were then introduced into microfluidic channels for analysis using a laser and portable imaging setup. The biomarker concentration was calculated from the fluorescence intensity, and LODs for DENV-NS1 and anti-DENV-NS1 were 8 ng/mL and 16 ng/mL, respectively. This method showed suboptimal repeatability in measuring DENV-NS1, so optimizing the bead coupling procedure and laser illumination source could improve the repeatability of this biosensing method. Modifying the imaging setup or using alternative dyes less susceptible to photobleaching could further improve the sensitivity of this method.
Dai et al. [40] developed a portable imaging device for PDMS microfluidic chips having channels connecting successive teardrop-shaped detection regions. They attached antibodies in the detection regions and quantified four colorectal cancer biomarkers using an ELISA-based reaction. The teardrop shape reduced the flux through the detection region while maintaining a continuous flow rate throughout the device. The pump-free portable analysis platform had filter paper that drove the fluid flow through capillary action and used a CCD camera for CL imaging, resulting in LODs of 0.9–1.8 ng/mL. Although their detector was more portable, this method had higher LODs than others that used temporally resolved CL [34, 35].
One of the downsides of parallel detection is that the flow systems can become complicated; automating devices makes it possible to use intricate flow systems to process and preconcentrate samples on-chip while maintaining usability. Sharafeldin et al. [41] used a system of micropumps and an Arduino microcontroller to create an automated 3D-printed device that lysed cells and performed a sandwich immunoassay for four metastasis biomarkers. They detected proteins at the single-cell level with LODs ranging from 0.1 to 0.2 fg/mL. Their low LOD was attributed to using streptavidin-biotin to increase the amount of HRP and using a chitosan hydrogel to maximize interactions in a small volume in their detection region.
Some multiplexed microfluidic immunoassays demonstrated LODs comparable to ELISA, but many need improved LODs before potential application for POC detection. Additionally, these methods should be more automated for convenient POC use and need to have competitive selectivity with ELISA.
Microfluidic nucleic acid analysis
For nucleic acid analysis, selectivity is typically achieved through hybridization conditions. Because of the availability of nucleic acid synthesis, these methods are readily modified for different targets by changing the probe sequence.
Surface plasmon resonance (SPR) is an optical technique that measures the change in refractive index at the surface of a material due to molecular interactions between a mobile analyte and a receptor molecule immobilized to the surface. Yeung et al. [42] demonstrated an automated microfluidic device for the multiplex detection of urinary miRNA biomarkers by using label-free transmission SPR. They used AuNPs and capped gold nanoslits for signal amplification. The AuNPs and nanoslits were derivatized with oligonucleotides, each of which was complementary to half of the target sequence. The AuNPs conveyed the partially hybridized target molecules to the nanoslit surface, where the target AuNP complex further hybridized to the nanoslit probes, changing the refractive index in a target concentration-dependent fashion. For an analysis time of 20 min, a LOD of 30 fM was obtained, with limited nonspecific binding in a urine sample.
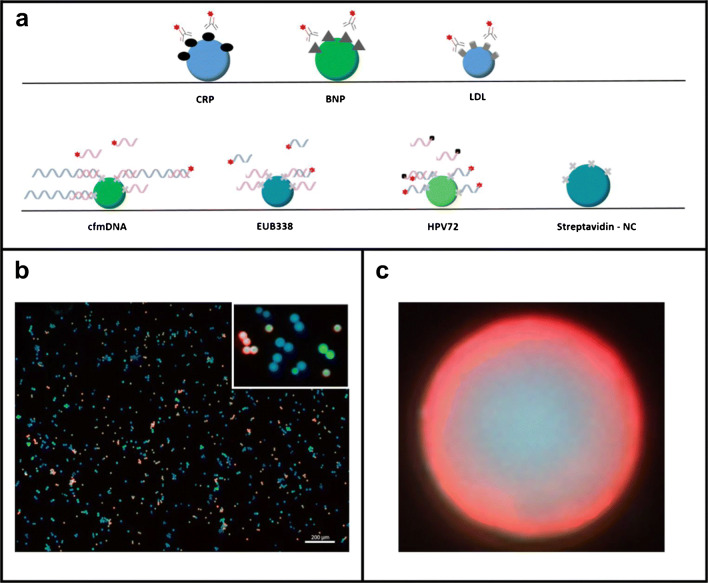
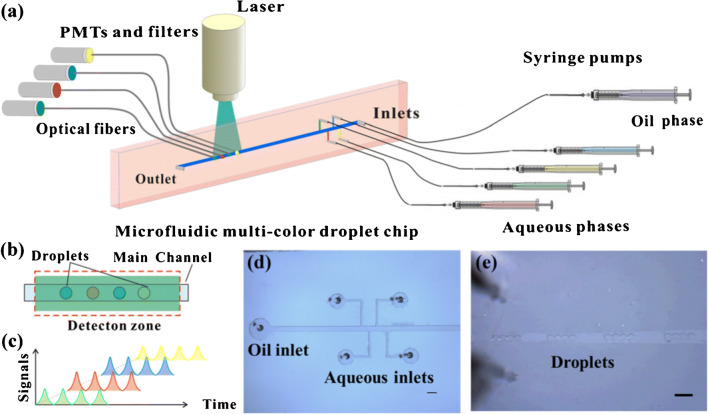
Ye et al. [43] used droplet microfluidics and quantum dots with different spectroscopic properties to distinguish four miRNAs. They attached DNA probes for each miRNA target to quantum dots and used a second DNA probe complementary to the miRNA target with a quencher for quantification. The quantum dot populations were separately mixed with the sample and quencher, loaded into the channel via 10-nL droplet generators, and optically detected, as shown in Fig. 6. They used the average fluorescence for 65 sets of droplets in 50 s to determine the concentration for each miRNA, with a LOD of 40 pM. However, this approach is currently limited to four biomarkers because of the quantum dot emission spectra breadth, and the method involves off-chip sample preparation steps. Integrating these sample labeling processes on-chip could simplify work for POC usage.
Fig. 6.
Schematic of multiplexed droplet microfluidic miRNA analysis. a Syringe pumps create 10-nL droplets of labeled sample that flow to a laser-induced fluorescence setup with four sets of optical fibers, filters, and photomultiplier tubes (PMTs). b One droplet from each sample is formed in sequence. c Individual signals are measured for each sample. d Droplet creation zone. e Droplets generated in a channel; scale bar is 500 μm. Adapted from ref. [43]. Copyright 2020 Springer
Devices requiring minimal training and time from personnel, small reagent volumes, and cheap, portable equipment are desirable for POC usage. Integrating sample processing and preconcentration on-chip can address these issues. Meena et al. [44] used on-chip preconcentration followed by labeling and single-particle counting to quantify specific DNA sequences that encode antibiotic resistance in bacterial infections. They used one device to preconcentrate, isolate, and fluorescently label a DNA target from mixed samples using a monolith column with affixed capture probes. Three target DNAs isolated using different versions of the first device were then loaded into one of three parallel channels in a second device. This device used anti-resonant reflecting optical waveguides with different, spatially dependent fluorescence patterns in each channel to simultaneously detect all three DNA targets. Single-molecule sensitivity was attributed to the combination of preconcentrating the targets and the ultra-small detection volume. Future work toward processing all targets on a single monolith and simultaneous detection within a single channel should further simplify the process.
Multiplexed nucleic acid detection in microfluidics has excellent promise for low LODs, even with complex samples. High levels of automation and integration are still needed for these devices to become prevalent in POC work.
Microfluidic cell and subcellular component analysis
Chemical information present in cells and subcellular structures can serve as a biomarker for medical conditions. For example, cells can be distinguished by identifying membrane proteins; such information gathered from cells or subcellular components can then be used to diagnose or monitor progression of different cancers.
Zhang et al. [45] isolated circulating tumor cells (CTCs) from blood samples on-chip before profiling the cells using multiplex SERS detection of membrane proteins. Because of the large size of CTCs relative to leukocytes and red blood cells, a filter between two channels on the device trapped CTCs in the first channel while allowing other cells to pass through. With aptamers linked to nanoparticles, three proteins could be distinguished using SERS. This approach allowed them to determine four different types of CTCs with 88–97% sensitivity and 96–98% specificity. White blood cells were filtered out before adding sample to the device, but this step could potentially be carried out on-chip using similar filter technology to what was used for CTCs.
A microfluidic technology that involves templated gold plating on exosomes for plasmonic detection was developed by Wu et al. [46]. In their approach, exosomes with associated proteins were fluorescently labeled with DNA aptamers and incubated with AuNPs. The role of the AuNPs was to facilitate in situ nanomaterial growth, which occurred on the exosome surface and induced a strong localized plasmonic resonance shift toward the red. This disparity in the plasmonic properties was used as the basis for the analysis of the cancer markers in exosomes. This platform showed promise in clinical validation, and could be expanded by incorporating other fluorescent probes or different molecular recognition mechanisms.
Present methods for multiplexing cell and subcellular component analysis require some advance, off-chip sample preparation (e.g., culture, centrifugation to isolate exosomes), offering opportunities for automation to provide improvements. Further work to expand these approaches to additional cell types and subcellular components is also needed.
Summary of spectroscopic methods
Table 2 summarizes the results of the various microfluidic spectroscopic multiplexing methods. For protein analysis, most methods have LODs below the low nanogram per milliliter range, and the best LODs reach even to femtogram per milliliter concentrations. Most devices with LODs <1 ng/mL rely on CL signal amplified by increasing the enzyme-to-analyte ratio or adding nanoparticles. Many of the optical multiplexing approaches discussed have LODs below clinical levels and use cheap, portable devices. Improvements to automation to eliminate off-chip sample preparation and maintain excellent LODs will help to make spectroscopically multiplexed systems worthwhile options for POC analyses.
Table. 2.
Biomarker types, their relevance, and the immobilization and detection methods used for multiplexed spectroscopic analysis
| Biomarker type | Size range (kDa) | Disease or condition | Immobilization | Detection method | LOD | Ref. |
|---|---|---|---|---|---|---|
| Proteins | 34–180 | Cancer | Channel surface | CL | 0.02–2.0 ng/mL* | [26] |
| Proteins | 70–180 | Cancer | Channel surface | Fluorescence | 0.03–0.05 ng/mL* | [27] |
| Small molecules | 0.18–0.39 | Diabetes | N/A | SERS | 70 μM–1 mM* | [28] |
| Proteins | 35–97 | AMI | Channel surface | SERS | 1–500 pg/mL* | [29, 30] |
| Proteins | 14–24 | AMI | Channel surface | Fluorescence | 1.00–2 ng/mL† | [31] |
| Organic molecules | 0.09–0.18 | None Tested | Channel surface | Colorimetry | 0.3 mM* | [32] |
| Organic molecules | 0.09–0.39 | None Tested | Channel surface | CL | 0.07–15 μM* | [33] |
| Proteins | 5–24 | AMI | Channel surface | CL | 0.06–0.4 pg/mL* | [34] |
| Proteins | 33–180 | Cancer | Channel surface | CL | 4 fg/mL–3 ng/mL* | [35] |
| Proteins | 13–80 | Inflammation | Channel surface | CL | 1.0–44 pg/mL* | [36] |
| Proteins and DNA | 110–150 | CVD | Microbeads | Fluorescence | 0.2–3 ng/μL* | [37] |
| Proteins | 48–150 | Dengue | Microbeads | Fluorescence | 8–16 ng/mL* | [39] |
| Proteins | 10–400 | Cancer | Channel surface | CL | 0.9–1.8 ng/mL* | [40] |
| Proteins | 46–107 | Cancer | Chitosan hydrogel | CL | 0.10–0.20 fg/mL* | [41] |
| miRNAs | 7 | None Tested | Capped gold nanoslits | SPR | 30 fM‡ | [42] |
| miRNAs | 7 | Cancer | In-solution | Fluorescence | 40 pM‡ | [43] |
Abbreviations: CVD cardiovascular disease, AMI acute myocardial infarction
*Meets clinical threshold
†Partially meets clinical threshold
‡Unknown clinical threshold
Trends in biomarker detection
As public health concerns shift over time, the applications for biomarker detection follow suit. Beginning in early 2020, the SARS-CoV-2 pandemic led to the rapid development of diagnostic tests [47], resulting in new technologies that can now be adapted for other diseases. However, none of the works discussed in this review addresses SARS-CoV-2. As shown in Fig. 7a, multiplexed biomarker detection has largely focused on cancer and heart disease in recent studies. These diseases have multiple indicators, such that multiplexed detection improves diagnostic accuracy, whereas virus detection often only involves one biomarker.
Fig. 7.
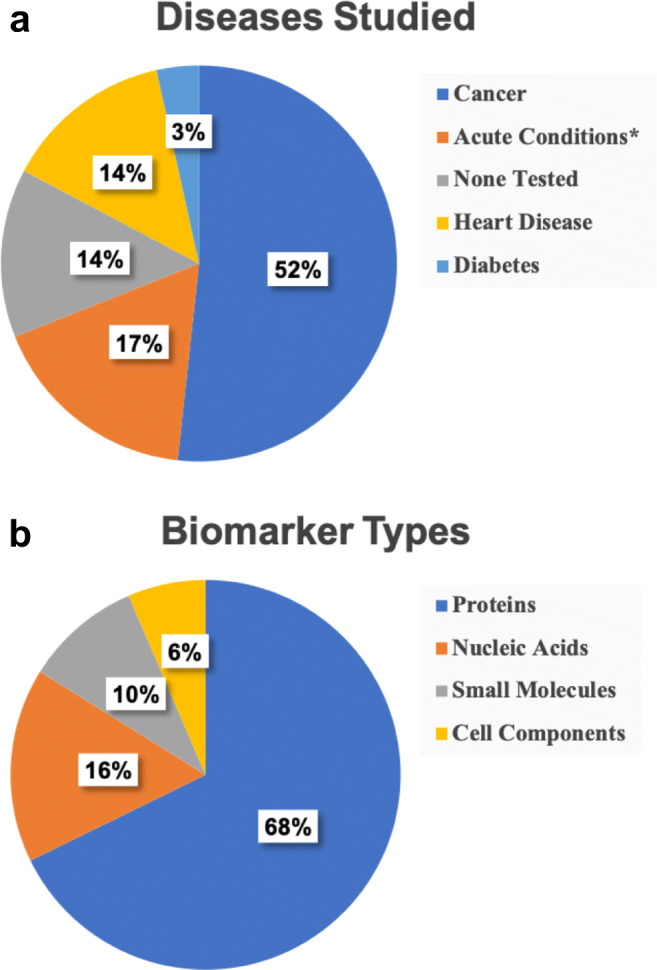
Pie charts showing the distributions of diseases and biomarker types studied in multiplexed microfluidic analysis. a Percentages of papers on diagnosis or monitoring of indicated diseases. b Percentages of papers on detection of various biomarker types. *Includes preterm birth, sepsis, inflammation, and dengue virus
Figure 7b shows that nearly 70% of the papers discussed in this review involved multiplexed detection of proteins. This can be attributed to two factors: (1) cancer and heart disease are often diagnosed by measuring protein levels, and (2) protein detection is more advanced than that for some biomarker types. Antibodies can be immobilized in microfluidic devices or modified with fluorescent or electrochemical indicators, providing highly specific interaction with proteins, as well as facile adaptation to different protein targets. Nucleic acid analysis can be advantageous for detecting other conditions, including viral infection and sepsis. As public health concerns change, emphasis on nucleic acid or other types of biomarker analysis may increase.
Conclusions
In this review are described advances in methodologies for multiplexed microfluidic biomarker analysis. Important areas of improvement have been in sample labeling and preconcentration, analyte capture, device portability, and device automation. In many instances, data obtained using these novel methods were comparable to those acquired with conventional approaches like ELISA, supporting suitability for use in POC assays.
Important innovations have also emerged for multiplex detection of biomarkers. For example, novel modifications to microbeads produced unique signals without requiring physical separation of biomarkers, enabling multiplexing. Cheaper, more portable detectors, such as screen-printed electrodes and smartphones, are also being incorporated into assays, further facilitating the development of POC applications. New designs and devices can detect analytes in complex mixtures, with fewer sample preparation steps, improving adaptability to specific diagnostic tests. Detecting increased combinations of biomarkers makes these devices even better candidates for future POC or clinical use.
Most of the studies described herein report detection of two to four biomarkers, but many tests require higher-level multiplexing. Challenges with detecting more biomarkers include the push toward larger devices, more reagents, or more complicated detection methods, while still maintaining acceptable LODs. The multichannel devices discussed generally have lower LODs, but are also physically limited by the number of channels that can fit on a device. Single-channel layouts avoid this issue, but often require more sample processing steps or more complex assays to decrease the LOD. Overcoming these barriers of lowering the LOD while increasing the number of biomarkers detected will be critical in transitioning these devices to POC use.
Multiplex detection technologies can be advanced further with improvements in microfluidics; for example, analysis times could be reduced through sample preparation on-chip. Also, device fabrication times and costs could be reduced if enabling technologies are developed and implemented. One promising future direction to make this a reality is 3D printing, which can provide direct integration of many components in a single, rapidly constructed microdevice [48, 49]. Finally, to the transition to future POC and clinical analysis, electrical and spectroscopic technologies should be further miniaturized and integrated into microfluidic devices for compatibility and ease of use.
Acknowledgements
We thank the National Institutes of Health for funding this work (R01 EB027096). We also thank Brigham Young University for an Undergraduate Research Award to Kaitlynn Mitchell.
Code availability
Not applicable.
Author contribution
Conceptualization: Kaitlynn R. Mitchell. Literature search and data analysis: Joule E. Esene, Kaitlynn R. Mitchell. Drafting and critical revision: Joule E. Esene, Kaitlynn R. Mitchell, Adam T. Woolley.
Funding
This research was funded by the National Institutes of Health (R01 EB027096) and Brigham Young University.
Data availability
Not applicable.
Declarations
Conflict of interest
The authors declare that they have no conflicts of interest. ATW is Chair Editor for ABC, but was not involved in the peer review of this work.
Footnotes
Published in the topical collection celebrating ABCs 20th Anniversary.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Nielsen AV, Nielsen JB, Sonker M, Knob R, Sahore V, Woolley AT. Microchip electrophoresis separation of a panel of preterm birth biomarkers. Electrophoresis. 2018;39(18):2300–2307. doi: 10.1002/elps.201800078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ouyang M, Tu D, Tong L, Sarwar M, Bhimaraj A, Li C, et al. A review of biosensor technologies for blood biomarkers toward monitoring cardiovascular diseases at the point-of-care. Biosens Bioelectron. 2021;171:112621. doi: 10.1016/j.bios.2020.112621. [DOI] [PubMed] [Google Scholar]
- 3.Zhu L, Liu X, Yang J, He Y, Li Y. Application of multiplex microfluidic electrochemical sensors in monitoring hematological tumor biomarkers. Anal Chem. 2020;92(17):11981–11986. doi: 10.1021/acs.analchem.0c02430. [DOI] [PubMed] [Google Scholar]
- 4.Park J, Han DH, Park J-K. Towards practical sample preparation in point-of-care testing: user-friendly microfluidic devices. Lab Chip. 2020;20(7):1191–1203. doi: 10.1039/D0LC00047G. [DOI] [PubMed] [Google Scholar]
- 5.Kosack CS, Page A-L, Klatser PR. A guide to aid the selection of diagnostic tests. Bull World Health Organ. 2017;95(9):639. doi: 10.2471/BLT.16.187468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sonker M, Sahore V, Woolley AT. Recent advances in microfluidic sample preparation and separation techniques for molecular biomarker analysis: a critical review. Anal Chim Acta. 2017;986:1–11. doi: 10.1016/j.aca.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Negou JT, Hu J, Li X, Easley CJ. Advancement of analytical modes in a multichannel, microfluidic droplet-based sample chopper employing phase-locked detection. Anal Methods. 2018;10(28):3436–3443. doi: 10.1039/C8AY00947C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Du K, Cai H, Park M, Wall TA, Stott MA, Alfson KJ, et al. Multiplexed efficient on-chip sample preparation and sensitive amplification-free detection of Ebola virus. Biosens Bioelectron. 2017;91:489–496. doi: 10.1016/j.bios.2016.12.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones A, Dhanapala L, Kankanamage RNT, Kumar CV, Rusling JF. Multiplexed immunosensors and immunoarrays. Anal Chem. 2020;92(1):345–362. doi: 10.1021/acs.analchem.9b05080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murray LP, Mace CR. Usability as a guiding principle for the design of paper-based, point-of-care devices - a review. Anal Chim Acta. 2020;1140:236–249. doi: 10.1016/j.aca.2020.09.063. [DOI] [PubMed] [Google Scholar]
- 11.Zhang K, Li H, Wang W, Cao J, Gan N, Han H. Application of multiplexed aptasensors in food contaminants detection. ACS Sens. 2020;5(12):3721–3738. doi: 10.1021/acssensors.0c01740. [DOI] [PubMed] [Google Scholar]
- 12.Pei H, Li L, Han Z, Wang Y, Tang B. Recent advances in microfluidic technologies for circulating tumor cells: enrichment, single-cell analysis, and liquid biopsy for clinical applications. Lab Chip. 2020;20(21):3854–3875. doi: 10.1039/D0LC00577K. [DOI] [PubMed] [Google Scholar]
- 13.Davaran S, Sadeghinia M, Jamalpoor Z, Raeisdasteh Hokmabad V, Doosti-Telgerd M, Karimian A, et al. Multiple functions of microfluidic platforms: characterization and applications in tissue engineering and diagnosis of cancer. Electrophoresis. 2020;41(12):1081–1094. doi: 10.1002/elps.201900341. [DOI] [PubMed] [Google Scholar]
- 14.Zhu H, Fohlerova Z, Pekarek J, Basova E, Neuzil P. Recent advances in lab-on-a-chip technologies for viral diagnosis. Biosens Bioelectron. 2020;153:112041. doi: 10.1016/j.bios.2020.112041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pursey JP, Chen Y, Stulz E, Park MK, Kongsuphol P. Microfluidic electrochemical multiplex detection of bladder cancer DNA markers. Sensors Actuators B Chem. 2017;251:34–39. doi: 10.1016/j.snb.2017.05.006. [DOI] [Google Scholar]
- 16.Wang Y, Luo JP, Liu JT, Sun S, Xiong Y, Ma YY, et al. Label-free microfluidic paper-based electrochemical aptasensor for ultrasensitive and simultaneous multiplexed detection of cancer biomarkers. Biosens Bioelectron. 2019;136:84–90. doi: 10.1016/j.bios.2019.04.032. [DOI] [PubMed] [Google Scholar]
- 17.Dhanapala L, Jones AL, Czarnecki P, Rusling JF. Sub-zeptomole detection of biomarker proteins using a microfluidic immunoarray with nanostructured sensors. Anal Chem. 2020;92(12):8021–8025. doi: 10.1021/acs.analchem.0c01507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharafeldin M, Bishop GW, Bhakta S, El-Sawy A, Suib SL, Rusling JF. Fe3O4 nanoparticles on graphene oxide sheets for isolation and ultrasensitive amperometric detection of cancer biomarker proteins. Biosens Bioelectron. 2017;91:359–66. [DOI] [PMC free article] [PubMed]
- 19.Gao Y, Huo W, Zhang L, Lian J, Tao W, Song C, et al. Multiplex measurement of twelve tumor markers using a GMR multi-biomarker immunoassay biosensor. Biosens Bioelectron. 2019;123:204–210. doi: 10.1016/j.bios.2018.08.060. [DOI] [PubMed] [Google Scholar]
- 20.Shen Y, Modha S, Tsutsui H, Mulchandani A. An origami electrical biosensor for multiplexed analyte detection in body fluids. Biosens Bioelectron. 2021;171:112721. doi: 10.1016/j.bios.2020.112721. [DOI] [PubMed] [Google Scholar]
- 21.Sahore V, Sonker M, Nielsen AV, Knob R, Kumar S, Woolley AT. Automated microfluidic devices integrating solid-phase extraction, fluorescent labeling, and microchip electrophoresis for preterm birth biomarker analysis. Anal Bioanal Chem. 2018;410(3):933–941. doi: 10.1007/s00216-017-0548-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xie LS, Cao YT, Hu FT, Li TH, Wang QQ, Gan N. Microfluidic chip electrophoresis for simultaneous fluorometric aptasensing of alpha-fetoprotein, carbohydrate antigen 125 and carcinoembryonic antigen by applying a catalytic hairpin assembly. Microchim Acta. 2019;186(8):547. doi: 10.1007/s00604-019-3594-3. [DOI] [PubMed] [Google Scholar]
- 23.Berger J, Valera E, Jankelow A, Garcia C, Akhand M, Heredia J, et al. Simultaneous electrical detection of IL-6 and PCT using a microfluidic biochip platform. Biomed Microdevices. 2020;22(2):36. doi: 10.1007/s10544-020-00492-6. [DOI] [PubMed] [Google Scholar]
- 24.Cowell TW, Valera E, Jankelow A, Park J, Schrader AW, Ding R, et al. Rapid, multiplexed detection of biomolecules using electrically distinct hydrogel beads. Lab Chip. 2020;20(13):2274–2283. doi: 10.1039/D0LC00243G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noviana E, Carrao DB, Pratiwi R, Henry CS. Emerging applications of paper-based analytical devices for drug analysis: a review. Anal Chim Acta. 2020;1116:70–90. doi: 10.1016/j.aca.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 26.Guo X, Guo Y, Liu W, Chen Y, Chu W. Fabrication of paper-based microfluidic device by recycling foamed plastic and the application for multiplexed measurement of biomarkers. Spectrochim Acta A Mol Biomol Spectrosc. 2019;223:117341. doi: 10.1016/j.saa.2019.117341. [DOI] [PubMed] [Google Scholar]
- 27.Jiao Y, Du C, Zong L, Guo X, Han Y, Zhang X, et al. 3D vertical-flow paper-based device for simultaneous detection of multiple cancer biomarkers by fluorescent immunoassay. Sensors Actuators B Chem. 2020;306:127239. doi: 10.1016/j.snb.2019.127239. [DOI] [Google Scholar]
- 28.Pinheiro T, Marques AC, Carvalho P, Martins R, Fortunato E. Paper microfluidics and tailored gold nanoparticles for nonenzymatic, colorimetric multiplex biomarker detection. ACS Appl Mater Interfaces. 2021;13(3):3576–3590. doi: 10.1021/acsami.0c19089. [DOI] [PubMed] [Google Scholar]
- 29.Lim WY, Thevarajah TM, Goh BT, Khor SM. Paper microfluidic device for early diagnosis and prognosis of acute myocardial infarction via quantitative multiplex cardiac biomarker detection. Biosens Bioelectron. 2019;128:176–185. doi: 10.1016/j.bios.2018.12.049. [DOI] [PubMed] [Google Scholar]
- 30.Lim WY, Goh C-H, Thevarajah TM, Goh BT, Khor SM. Using SERS-based microfluidic paper-based device (mu PAD) for calibration-free quantitative measurement of AMI cardiac biomarkers. Biosens Bioelectron. 2020;147:111792. doi: 10.1016/j.bios.2019.111792. [DOI] [PubMed] [Google Scholar]
- 31.Guo XY, Zong LJ, Jiao YC, Han YF, Zhang XP, Xu J, et al. Signal-enhanced detection of multiplexed cardiac biomarkers by a paper-based fluorogenic immunodevice integrated with zinc oxide nanowires. Anal Chem. 2019;91(14):9300–9307. doi: 10.1021/acs.analchem.9b02557. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Kim D, Kim S. Simultaneous quantification of multiple biomarkers on a self-calibrating microfluidic paper-based analytic device. Anal Chim Acta. 2020;1097:120–126. doi: 10.1016/j.aca.2019.10.068. [DOI] [PubMed] [Google Scholar]
- 33.Li F, Liu J, Guo L, Wang J, Zhang K, He J, et al. High-resolution temporally resolved chemiluminescence based on double-layered 3D microfluidic paper-based device for multiplexed analysis. Biosens Bioelectron. 2019;141:111472. doi: 10.1016/j.bios.2019.111472. [DOI] [PubMed] [Google Scholar]
- 34.Li F, Guo L, Hu Y, Li Z, Liu J, He J, et al. Multiplexed chemiluminescence determination of three acute myocardial infarction biomarkers based on microfluidic paper-based immunodevice dual amplified by multifunctionalized gold nanoparticles. Talanta. 2020;207:120346. doi: 10.1016/j.talanta.2019.120346. [DOI] [PubMed] [Google Scholar]
- 35.Li F, Guo L, Li Z, He J, Cui H. Temporal-spatial-color multiresolved chemiluminescence imaging for multiplex immunoassays using a smartphone coupled with microfluidic chip. Anal Chem. 2020;92(10):6827–6831. doi: 10.1021/acs.analchem.0c01405. [DOI] [PubMed] [Google Scholar]
- 36.Wu J, Chen Y, Yang M, Wang Y, Zhang C, Yang M, et al. Streptavidin-biotin-peroxidase nanocomplex-amplified microfluidics immunoassays for simultaneous detection of inflammatory biomarkers. Anal Chim Acta. 2017;982:138–147. doi: 10.1016/j.aca.2017.05.031. [DOI] [PubMed] [Google Scholar]
- 37.Dinter F, Burdukiewicz M, Schierack P, Lehmann W, Nestler J, Dame G, et al. Simultaneous detection and quantification of DNA and protein biomarkers in spectrum of cardiovascular diseases in a microfluidic microbead chip. Anal Bioanal Chem. 2019;411(29):7725–7735. doi: 10.1007/s00216-019-02199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chang N, Zhai J, Liu B, Zhou J, Zeng Z, Zhao X. Low cost 3D microfluidic chips for multiplex protein detection based on photonic crystal beads. Lab Chip. 2018;18(23):3638–3644. doi: 10.1039/C8LC00784E. [DOI] [PubMed] [Google Scholar]
- 39.Yuan X, Garg S, De Haan K, Fellouse FA, Gopalsamy A, Tykvart J, et al. Bead-based multiplex detection of dengue biomarkers in a portable imaging device. Biomed Opt Express. 2020;11(11):6154–6167. doi: 10.1364/BOE.403803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dai B, Yin C, Wu J, Li W, Zheng L, Lin F, et al. A flux-adaptable pump-free microfluidics-based self-contained platform for multiplex cancer biomarker detection. Lab Chip. 2021;21(1):143–153. doi: 10.1039/D0LC00944J. [DOI] [PubMed] [Google Scholar]
- 41.Sharafeldin M, Chen T, Ozkaya GU, Choudhary D, Molinolo AA, Gutkind JS, et al. Y detecting cancer metastasis and accompanying protein biomarkers at single cell levels using a 3D-printed microfluidic immunoarray. Biosens Bioelectron. 2021;171:112681. doi: 10.1016/j.bios.2020.112681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yeung WK, Chen H-Y, Sun J-J, Hsieh T-H, Mousavi MZ, Chen H-H, et al. Multiplex detection of urinary miRNA biomarkers by transmission surface plasmon resonance. Analyst. 2018;143(19):4715–4722. doi: 10.1039/C8AN01127C. [DOI] [PubMed] [Google Scholar]
- 43.Ye W-Q, Wei Y-X, Zhang Y-Z, Yang C-G, Xu Z-R. Multiplexed detection of micro-RNAs based on microfluidic multi-color fluorescence droplets. Anal Bioanal Chem. 2020;412(3):647–655. doi: 10.1007/s00216-019-02266-3. [DOI] [PubMed] [Google Scholar]
- 44.Meena GG, Hanson RL, Wood RL, Brown OT, Stott MA, Robison RA, et al. 3x multiplexed detection of antibiotic resistant plasmids with single molecule sensitivity. Lab Chip. 2020;20(20):3763–3771. doi: 10.1039/D0LC00640H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang YZ, Wang ZY, Wu L, Zong SF, Yun BF, Cui YP. Combining multiplex SERS nanovectors and multivariate analysis for in situ profiling of circulating tumor cell phenotype using a microfluidic chip. Small. 2018;14(20):1704433. doi: 10.1002/smll.201704433. [DOI] [PubMed] [Google Scholar]
- 46.Wu XJ, Zhao HT, Natalia A, Lim CZJ, Ho NRY, Ong CAJ, et al. Exosome-templated nanoplasmonics for multiparametric molecular profiling. Sci Adv. 2020;6(19):eaba2556. doi: 10.1126/sciadv.aba2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen LZ, Zhang GL, Liu LQ, Li ZD. Emerging biosensing technologies for improved diagnostics of COVID-19 and future pandemics. Talanta. 2021;225:121986. doi: 10.1016/j.talanta.2020.121986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beauchamp MJ, Nordin GP, Woolley AT. Moving from millifluidic to truly microfluidic sub-100-µm cross-section 3D printed devices. Anal Bioanal Chem. 2017;409(18):4311–9. [DOI] [PMC free article] [PubMed]
- 49.Nielsen AV, Beauchamp MJ, Nordin GP, Woolley AT. 3D printed microfluidics. Annu Rev Anal Chem. 2020;13:45–65. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.