Abstract
Objectives
We explore the importance of SARS-CoV-2 sentinel surveillance testing in primary care during a regional COVID-19 outbreak in Austria.
Design
Prospective cohort study.
Setting
A single sentinel practice serving 22 829 people in the ski-resort of Schladming-Dachstein.
Participants
All 73 patients presenting with mild-to-moderate flu-like symptoms between 24 February and 03 April, 2020.
Intervention
Nasopharyngeal sampling to detect SARS-CoV-2 using real-time reverse transcriptase-quantitative PCR (RT-qPCR).
Outcome measures
We compared RT-qPCR at presentation with confirmed antibody status. We split the outbreak in two parts, by halving the period from the first to the last case, to characterise three cohorts of patients with confirmed infection: early acute (RT-qPCR reactive) in the first half; and late acute (reactive) and late convalescent (non-reactive) in the second half. For each cohort, we report the number of cases detected, the accuracy of RT-qPCR, the duration and variety of symptoms, and the number of viral clades present.
Results
Twenty-two patients were diagnosed with COVID-19 (eight early acute, seven late acute and seven late convalescent), 44 patients tested SARS-CoV-2 negative and 7 were excluded. The sensitivity of RT-qPCR was 100% among all acute cases, dropping to 68.1% when including convalescent. Test specificity was 100%. Mean duration of symptoms for each group were 2 days (range 1–4) among early acute, 4.4 days (1–7) among late acute and 8 days (2–12) among late convalescent. Confirmed infection was associated with loss of taste. Acute infection was associated with loss of taste, nausea/vomiting, breathlessness, sore throat and myalgia; but not anosmia, fever or cough. Transmission clusters of three viral clades (G, GR and L) were identified.
Conclusions
RT-qPCR testing in primary care can rapidly and accurately detect SARS-CoV-2 among people with flu-like illness in a heterogeneous viral outbreak. Targeted testing in primary care can support national sentinel surveillance of COVID-19.
Keywords: primary care, COVID-19, public health, virology
Strengths and limitations of this study.
Our study was conducted in a state-of-the-art sentinel surveillance practice, participating in the Austrian National Influenza Screening Programme, covering the entire period of a regional COVID-19 outbreak.
Symptomatic patients received same-day appointments with a clinician for nasopharyngeal swabs, and people testing RT-qPCR reactive were notified within 24 hours.
Cases were confirmed using a combination of five different ELISA platforms and neutralising antibody assay.
The relatively small patient cohort from a single testing site limits conclusion on causality and generalisability.
Any difference in symptoms observed between study cohorts may be due to recall bias occurred, particularly among those people presenting late.
Introduction
The COVID-19 pandemic, caused by SARS-CoV-2, continues to spread globally with more than 192 million cases, and over four million deaths reported as of 26 July 2021. Undetected infection and delays in implementing an effective test-trace-isolate (TTI) strategy have contributed to the spread of the virus becoming a pandemic. SARS-CoV-2 has a wide spectrum of manifestations including no symptoms (asymptomatic infection), mild to moderate to severe flu-like illness, loss of taste or smell, pneumonia and acute respiratory distress syndrome, sepsis, multi-organ failure and death.1 In studies to date, the reported time for the infection to become symptomatic (incubation period) varies among different cohorts and settings, with a median incubation period around 5.1 days,2 infectivity starting 2.3 days before symptom onset, peaking 1–2 days before that and gradually declining over 7–10 days.3–6
SARS-CoV-2 has the potential for ‘superspreading’ events, resulting in clusters of disease outbreaks among a large number of people. Most infections remain isolated cases, but a small number of individuals (10%) may cause up to 80% of secondary transmissions.7 Although symptomatic infection is common (17%, range 4%–41%), the relative risk for symptomatic transmission may be up to six times higher than for asymptomatic infection.8–10 Undocumented infection may constitute the majority of cases (86%), causing more than half (55%) of all documented infections.11 Superspreading events have been reported from across the globe, and countries achieving early viral suppression took rapid and decisive action to implement comprehensive case identification and testing, combined with contact tracing and isolation.12 13 For epidemic control of COVID-19, the effective reproduction number, Re, needs to be less than 1; the presence of undetected and persistent infection within the population, even if very small, can increase Re and induce a secondary peak of infections. Therefore, rapid identification and containment of infection are key factors for the prevention of onward transmission and controlling the virus to protect the public.14
In Austria, the first two COVID-19 cases were reported among travellers from Italy in the city of Innsbruck on 25 February 2020.15 Multiple superspreading events then occurred among tourists visiting Austrian ski resorts, including the town of Ischgl, that are believed to have led to further outbreaks in the tourists’ home countries, including Germany, Denmark and Sweden.15 16 Austria was one of the first countries to adopt comprehensive lockdown measures on 16 March 2020, including protection of vulnerable groups, penalty fees for breaching self-isolation and a national health hotline to facilitate testing at acute care settings and via mobile units.17 The first death from COVID-19-associated complications occurred on 12 March 2020, and as of 26 July, 656 582 cases and 10 732 COVID-19 related deaths have been reported.
General practice (GP) is considered a key partner in case recording, managing high-risk groups and delivery of equitable care.18–20 The European Centre for Disease Prevention and Control (ECDC) recommended integration of ‘COVID-19 surveillance with sentinel surveillance of influenza-like illness or acute respiratory infection’.21 However, in some countries, like the UK and the USA, primary care has been largely excluded from the national TTI strategy.22 23 In contrast, Austria additionally offered SARS-CoV-2 real-time reverse transcriptase PCR (RT-qPCR) testing to people presenting with mild-to-moderate flu-like symptoms to any of the 92 sentinel surveillance sites (GPs and paediatric practices) beginning 24 February 2020.24 The new service supplemented the existing national health hotline for people at risk of COVID-19.25 RT-qPCR is an established technique to detect viral RNA from nasopharyngeal sampling used to diagnose COVID-19.26 Early detection of SARS-CoV-2 is essential for effective contact tracing,27 and whole genome sequencing may provide data on dynamics of transmission.28
The overall aim of this work is to test whether rapid early RT-qPCR testing in primary care can accurately and timely detect SARS-CoV-2 and inform outbreak surveillance. To attest this, we report the outcomes of SARS-CoV-2 RT-qPCR testing at a sentinel GP in the ski resort of Schladming-Dachstein, Austria. We report (a) the accuracy (via sensitivity and specificity) of rapidly deployed RT-qPCR testing in patients presenting with acute infection by comparing it to anti-SARS-CoV-2 antibody status during convalescence in the same geographically defined study cohort; (b) the earliness of viral RNA detection by comparing the duration, number and type of symptoms among patients presenting during the first half (early presenters) and the second half (late presenters) of the outbreak, measured by the number of days from the first to the last case detected and dividing that period by two; (c) the identification of key clinical symptoms of acute and convalescent disease and determine a correlation between these and (d) the number of SARS-CoV-2 clades implicated in the outbreak.
Methods
Setting
This study was set in a sentinel GP participating in the National Influenza Surveillance Network in the ski resort of Schladming-Dachstein, political subdistrict of Groebming (population 22 829), Austria. The study was conducted during a local COVID-19 outbreak in March and April 2020, during which 29 cases were detected by RT-qPCR locally. The bulk of the outbreak occurred after a 3-day party (March 13–15) prior to implementation of the national lockdown policy on 16 March, which led to premature termination of the skiing season. All patients presenting with mild-to-moderate flu-like illness were included. Following the report of the first cases in Austria, people with flu-like symptoms were advised to call the national health hotline instead of directly presenting to the hospital or GP. Patients were advised to phone the GP or receive in-home testing by mobile testing units, and home self-isolate and self-care. Asymptomatic people were excluded from this study.
Design
We conducted a longitudinal evaluation comprising a prospective cohort to examine the impact of SARS-CoV-2 RT-qPCR testing on COVID-19 case detection. Between 24 February and 03 April 2020, RT-qPCR testing and seropositivity data were collected to compare two groups within this cohort of patients:
Patients testing RT-qPCR reactive at presentation with acute disease.
Patients confirmed anti-SARS-CoV-2 antibody positive during the convalescence phase (confirmed infection).
We define acute disease as the presence of flu-like symptoms combined with reactive SARS-CoV-2 RT-qPCR and positive serostatus; and confirmed infection as the presence of convalescent anti-SARS-CoV-2 antibody 3–6 weeks after the acute illness, irrespective of the RT-qPCR result.
Intervention
On 24 February 2020, 1 day before the first two cases were reported in Austria, the National Influenza Screening Network was enhanced to include SARS-CoV-2 RT-qPCR testing.
Patients with mild-to-moderate flu-like symptoms calling the study sentinel GP were offered same-day appointments for SARS-CoV-2 RT-qPCR testing. RT-qPCR results were available within 24 hours, and those patients with a reactive outcome were immediately notified by a clinician and advised to self-isolate for a minimum of 2 weeks following national policy at that time. Repeat follow-up RT-qPCR was arranged by the local public health authority (District Commissioner of Liezen, Austria), and people testing non-reactive on repeat RT-qPCR were released from self-isolation. After 3–6 weeks, venous blood was obtained to confirm SARS-CoV-2 infection using ELISA IgG and neutralising antibody assay. We defined the period of the outbreak as the number of days from the first patient to the last patient testing RT-qPCR reactive at the GP.
Since the winter season 2000/2001, the National Influenza Screening Network has conducted influenza screening for patients attending sentinel GPs and paediatric practices. Between November and March of each year, participating practices routinely collect nasopharyngeal swabs from patients presenting with flu-like symptoms. Specimens are sent to the Center for Virology, Medical University of Vienna, Austria, for virus isolation on tissue cultures and PCR detection. This surveillance programme allows for near real-time recording of seasonal influenza virus activity in the country.
Clinical data
We obtained anonymous patient data held within the GP computer system. The practice lead clinician (OL) generated a clinical master case report form before extracting pseudonymised patient records into an Excel spreadsheet. E-MH and CH verified the accuracy of the data extraction for all patients. Data were stored on a secure computer at the Institute of General Practice and Evidence-based Health Services Research, University of Graz, Austria, before sharing it with the study statistician (JP-G) using encrypted email and secure storage at the University of Oxford, UK.
Testing
Reverse transcriptase-quantitative PCR
SARS-CoV-2 RT-qPCR was performed in scope of the routine surveillance at the Center for Virology, Medical University of Vienna on a Roche LightCycler (http://www.roche.com; Switzerland) using a primer set provided by TIB MOLBIOL (https://www.tib-molbiol.com/; Germany).26 RT-qPCR targeting the E-gene was considered reactive at a cycle threshold (Ct) value of less than 40, and Ct values above 32 were confirmed by RNA-dependent RNA polymerase (RdRp) gene detection.
Enzyme-linked immune assays
IgG serostatus assays were performed according to the manufacturers’ protocol using five different commercial test kits of anti-SARS-CoV-2 IgG ELISA provided by the following companies: EUROIMMUN (EUROIMMUN Medizinische Labordiagnostika AG, www.euroimmun.com)29 and EPITOPE DIAGNOSTICS (Immunodiagnostik AG, www.euroimmun.com), respectively.30 Reagent wells of the anti-SARS-CoV-2 IgG ELISA are coated with recombinant antigen derived from the spike protein (S1 domain) of SARS-CoV-2. Reagent wells of the EDI Novel Coronavirus COVID-19 IgG ELISA are coated with COVID-19 recombinant full-length nucleocapsid protein. ABBOTT performed on the Architect platform (ABBOTT LABORATORIES, www.abbott.com), DIASORIN (DIASORIN S.p.A., https://www.diasorin.com/home) performed on the LIAISON platform and ROCHE performed on the cobas e 801 analyzer. The Abbott SARS-CoV-2 IgG assay is a chemiluminescent microparticle immunoassay for the qualitative detection of IgG against a recombinant SARS-CoV-2 nucleoprotein. Results are reported in the form of an index value (S/C). LIAISON SARS-CoV-2 S1/S2 IgG assay is a chemiluminescence immunoassay for the quantitative detection of IgG against the recombinant S1 and S2 domain of the spike protein. Results are reported in arbitrary units (AU/mL). Elecsys anti-SARS-CoV-2 assay (Roche Diagnostics) is a electrochemiluminescence immunoassay for qualitative detection of SARS-CoV-2 antibodies in human serum against a recombinant nucleocapsid protein of SARS-CoV-2. It is a total antibody assay not differentiating between IgA, IgM or IgG but detecting IgG predominantly. Results are reported as numeric values in the form of signal sample/cut-off (COI).
Neutralising antibody assay
Samples with discordant antibody results (see below) were further evaluated using an in-house neutralising antibody assay as follows: serial dilutions of heat-inactivated serum samples were incubated with 50–100 TCID50 SARS-CoV-2 (hCoV-19/Austria/CeMM0360/2020; GISAID EPI_ISL: 438123) for 1 hour at 37°C. The mixture was added to Vero E6 (ATCC CRL-1586) cell monolayers and incubation was continued for 2–3 days. Neutralisation titres (NT) were expressed as the reciprocal of the serum dilution that protected against virus-induced cytopathic effects. NT ≥10 were considered positive. The study has been reported in accordance with STARI reporting guidelines for implementation studies.31
Outcome measures and statistical analysis
We present a descriptive statistics of patient demographics including age, gender and ethnicity; and the following four testing, viral and genomic outcomes:
Outcome A
The diagnostic accuracy (using sensitivity and specificity) of SARS-CoV-2 RT-qPCR among patients with mild-to-moderate flu-like symptoms at presentation by comparing molecular diagnosis with anti-SARS-CoV-2 antibody testing during convalescence, and hospital admission and death, including any alternative diagnoses for patients testing SARS-CoV-2 negative. To determine the accuracy of RT-qPCR, we stratified RT-qPCR results in four groups: true reactive (RT-qPCR reactive and confirmed antibody positive); false reactive (RT-qPCR reactive, antibody negative); true non-reactive (RT-qPCR non-reactive, antibody negative) and false non-reactive (RT-qPCR non-reactive, antibody positive).
Outcome B
The earliness of RT-qPCR testing by comparing the duration and number of symptoms during the first half of the outbreak (early presenters) and during the second half of the outbreak (late presenters). We calculated the earliness of RT-qPCR testing by determining the mean duration of symptoms, in days (range), and mean number of symptoms (range), across the three cohorts of patients with confirmed infection: early acute, late acute and late convalescent. The three cohorts were obtained by stratifying people with confirmed infection according to the date of presentation to the GP during the outbreak as follows: people presenting with acute infection (RT-qPCR reactive, confirmed antibody positive) during the first half of the outbreak (early acute disease) versus those presenting during the second half of the outbreak (late acute) and those presenting with previous disease (RT-qPCR non-reactive but confirmed antibody positive) in the second half of the outbreak (late convalescent).
Outcome C
The key clinical symptoms associated with RT-qPCR reactivity (acute infection) and convalescent seropositivity (confirmed infection) to determine any potential correlation between these stages of disease. We used multivariate logistic regression to test the association of 15 clinical symptoms with RT-qPCR reactivity at presentation and among all patients with confirmed infection. We reported the ORs and the significance value (p) of each covariate on testing RT-qPCR reactive and confirmed positive antibody status, respectively. We quantified the association between patients with reactive RT-qPCR (and confirmed antibody positive) and all patients with confirmed infection by calculating the correlation coefficient r and estimating the 95% CI.
Outcome D
The number of viral clades implicated in the outbreak. To do this, SARS-CoV-2 full genome sequencing was undertaken as part of a wider study covering the whole of Austria.28 The full-length sequences were matched to patient records by an anonymised unique identifier and uploaded to the Global Initiative on Sharing All Influenza Data (GISAID) database (http://gisaid.org).32 Sequences were aligned in MEGA7 and non-synonymous nucleotide variants were identified to determine the respective clades, following the GISAID classification scheme for lineages.33
Patient and public involvement
Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Results
Overall testing results
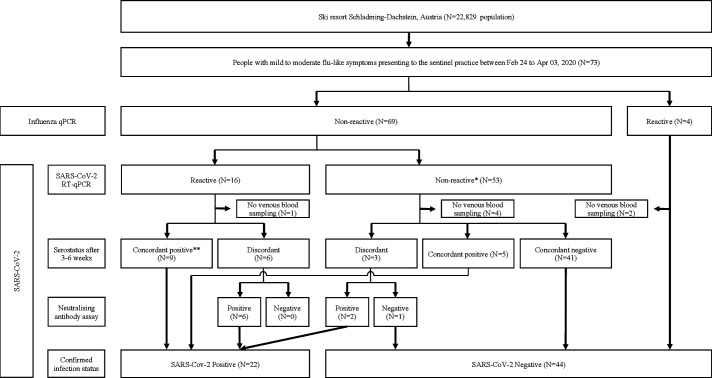
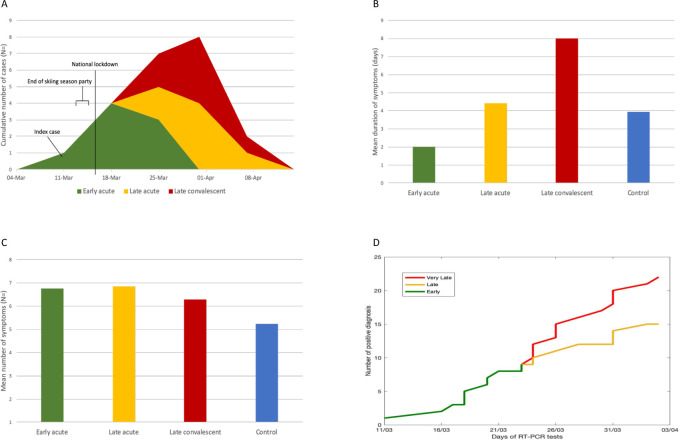
Baseline characteristics for confirmed cases were similar for sex, age and ethnic origin (table 1). All patients were local residents and no endemic cases were documented among tourists. Figure 1 shows the flowchart for the patient cohorts of this study. Seventy-three patients presented with mild-to-moderate flu-like illness, all of whom received SARS-CoV-2 RT-qPCR (and influenza qPCR). Of those, 16 (21.9%) tested RT-qPCR reactive and 57 (78.1%) tested non-reactive, including 4 that tested influenza PCR reactive. Due to lack of venous blood sampling (obtained 3–6 weeks after initial presentation), antibody data was not available for 7 patients (1 RT-qPCR reactive vs 6 non-reactive) that were excluded from this analysis. Therefore, of the 66 patients included in this analysis, 22 patients (33.3%) had SARS-CoV-2 infection confirmed by antibody testing and 44 (66.7%) patients were confirmed seronegative. Of the former, 8 patients (early acute presenters) presented in the first half of the outbreak (12 days from 11 to 22 March 2020) and 14 patients presented in the second half (23 March to 03 April 2020); of the latter, 7 patients were late acute and 7 late convalescent (figure 2A). Alternative diagnoses of the 44 patients who tested SARS-CoV-2 negative included: influenza and infectious mononucleosis (N=2, each); bacterial tonsillitis, bacterial pneumonia, bronchitis and exacerbation of chronic obstructive pulmonary disease (N=1, each) (see flowchart, figure 1). No hospital admissions or deaths were reported.
Table 1.
Summary of the demographic characteristics of COVID-19 cases
| People with confirmed infection (seropositive, any RT-qPCR result) (N=22) | People with acute infection (RT-qPCR reactive and seropositive) (N=15) | |
| Sex | ||
| Female | 14 (63.6%) | 9 (60%) |
| Male | 8 (36.4%) | 6 (40%) |
| Age (years) | ||
| 16–24 | 4 (26.7%) | 3 (20%) |
| 25–34 | 4 (26.7%) | 2 (13.3%) |
| 35–49 | 6 (40%) | 4 (26.7%) |
| >50 | 8 (36.4%) | 6 (40%) |
| Ethnic origin | ||
| White | 22 (100%) | 15 (100%) |
RT-qPCR, real-time reverse transcriptase-quantitative PCR.
Figure 1.
Flowchart. Twenty-two patients had COVID-19 infection confirmed by antibody testing, including 15 patients diagnosed with acute disease (reactive RT-qPCR) and 7 with convalescent disease (non-reactive RT-qPCR); among the former, 9 patients tested concordant antibody positive and 6 patients tested neutralising antibody positive following discordant ELISA result; and among the latter, 5 patients tested concordant antibody positive and 2 patients tested neutralising antibody positive following discordant ELISA result. Forty-four patients with non-reactive RT-qPCR tested antibody negative, including 41 with concordant negative ELISA, 1 patient with negative neutralising antibody after discordant ELISA result and 2 patients diagnosed with influenza. Antibody status was not available for 7 patients. *Final clinical diagnoses included infectious mononucleosis (N=2); bacterial tonsillitis, bacterial pneumonia and bronchitis and exacerbation of chronic obstructive pulmonary disease (N=1, each). **No concordant negatives. RT-qPCR, real-time reverse transcriptase-quantitative PCR.
Figure 2.
(A) Cumulative COVID-19 diagnosis in the ski resort Schladming-Dachstein over time. The main outbreak occurred after a 3-day party event (13 to 15 March) celebrating the early termination of the skiing season due to national lockdown commencing on 16 March. Between 11 March (index case) and 03 April (last endemic case), eight people were diagnosed with acute infection (RT-qPCR-reactive, confirmed antibody positive) in the first half (12 days from 11 to 22 March 2020) of the outbreak (green colour), and seven people with late acute infection (amber) and seven people with convalescent infection (red) were detected during the second half; (B) mean duration of symptoms; (C) mean number of symptoms and (D) cumulative weekly numbers of confirmed COVID-19 cases during the outbreak. RT-qPCR was 100% sensitive among all early acute and late acute presenters. RT-qPCR did not detect any of the late convalescent presenters. RT-qPCR, real-time reverse transcriptase PCR.
Specificity and sensitivity of RT-qPCR
In the absence of a gold standard, we used a consensus statement on serostatus, irrespective of RT-qPCR outcomes, to establish whether an infection had occurred. We considered an infection as confirmed in any patient who tested IgG ELISA positive on all five screening platforms (concordant results) or in any patient with mismatch between ELISA test results (discordant results) but positive neutralising antibody assay (see flowchart, figure 1). Of the 15 patients with reactive RT-qPCR, sera from 9 patients were concordant positive and 6 were discordant; and of the 53 patients with non-reactive RT-qPCR, sera from 41 patients were concordant negative, 5 were concordant positive and 3 were discordant. Sera from 2 patients diagnosed with influenza who tested RT-qPCR non-reactive were concordant negative and included in this analysis. For the 9 patients with discordant results, we used neutralising antibody assay to confirm infection status. All patients (N=6) with reactive RT-qPCR were neutralising antibody positive; and of the 3 patients with non-reactive RT-qPCR, 2 were neutralising antibody positive and 1 was negative. Therefore, overall, when combining ELISA and neutralising antibody assay, 22 patients had confirmed infection, of whom 15 patients were RT-qPCR reactive (true reactive) and 7 were non-reactive (false non-reactive). There were no false reactive RT-qPCR results. Therefore, RT-qPCR correctly identified infection in 15/22 patients (overall sensitivity of 68.1%). Sensitivity of RT-qPCR among all acute (early and late) presenters and during the first half of the outbreak was high (100%), but dropped to 50% in the second half of the outbreak. RT-qPCR correctly identified absence of infection for all 44 patients testing antibody negative (true non-reactive) indicating specificity of 100%.
Earliness of RT-qPCR testing
The mean duration of symptoms was 2 days (range 1–4) among early acute presenters, 4.4 days (range 1–7) among late acute presenters, 8 days (range 2–12) among people with late convalescent infection and 3.9 days (range 1–14) among non-COVID-19 controls (figure 2B). The mean number of symptoms was 6.75 (range 4–9) among early acute presenters, 6.86 (3–12) among late acute presenters, 6.3 (1–11) among people with convalescent infection and 5.23 (range 2–11) among non-COVID-19 controls (figure 2C).
Regression analysis on confirmed infection
Multivariate regression on all 66 patients, including 22 (31.9%) with confirmed infection, suggested that loss of taste, but not loss of smell, was the key covariate significantly associated with positive serostatus (ORs=6.03; p=0.047) (table 2). Breathlessness (OR=6.9, p=0.054) and cough (OR=0.12, p=0.053) were also possible covariates of confirmed infection.
Table 2.
Regression analysis on symptoms reported by patients diagnosed with COVID-19
| Clinical symptom | People with confirmed infection (seropositive, any RT-qPCR result) (N=22) | People with acute disease (RT-qPCR reactive and seropositive) (N=15) | ||||
| OR | 95% CI | P value | OR | 95% CI | P value | |
| Change in taste | 6.02 | (1.02 to 35.51) | 0.047 | 571.72 | (1.92 to 170 629.2) | 0.029 |
| Nausea/vomiting | 4.42 | (0.748 to 26.09) | 0.101 | 370.11 | (2.71 to 50 429.42) | 0.018 |
| Sore throat | 0.36 | (0.067 to 1.93) | 0.233 | 0.002 | (0.000006 to 0.74) | 0.039 |
| Myalgia | 1.15 | (0.24 to 5.51) | 0.865 | 121.82 | (1.52 to 9749.08) | 0.032 |
| Breathlessness | 6.90 | (0.96 to 49.40) | 0.054 | 134.46 | (1.02 to 17 796.87) | 0.049 |
| Change in smell | 0.77 | (0.098 to 6.15) | 0.811 | 0.37 | (0.008 to 15.87) | 0.607 |
| Fever | 2.97 | (0.44 to 20.35) | 0.266 | 1.44 | (0.057 to 36.66) | 0.825 |
| Cough | 0.12 | (0.014 to 1.03) | 0.053 | 0.011 | (0.00008 to 1.42) | 0.069 |
Symptoms associated with confirmed SARS-CoV-2 infection (antibody confirmed positive, irrespective of RT-qPCR result) among 22 patients and with acute infection (RT-qPCR reactive, antibody confirmed positive) among 15 patients, respectively.
RT-qPCR, real-time reverse transcriptase-quantitative PCR.
Regression analysis on acute disease
All 15 patients with acute disease reported fatigue and therefore this covariate was removed from the analysis; and observations from two patients with non-reactive RT-qPCR, who did not report fatigue, were also removed (table 2). The multivariate logistic regression on the remaining 66 patients showed that the following covariates were associated with acute disease: loss of taste (OR=571.72; p=0.029), nausea and vomiting (OR=370.11; p=0.018), breathlessness (OR=134.46; p=0.049), myalgia (OR=121.82; p=0.032) and sore throat (OR=0.002, p=0.039), but not loss of smell (OR=0.37, p=0.607), fever (OR=1.44, p=0.825) or cough (OR=0.01, p=0.069).
Correlation between acute and confirmed infection
Testing RT-qPCR reactive was correlated with testing seropositive for COVID-19 infection (r=0.77, 95% CI 0.65 to 0.89). Among early and acute presenters, the correlation between the two tests was perfect (green and amber in figure 2D), irrespective of the stage of the outbreak; whereas in the second half of the outbreak, RT-qPCR did not detect any case with convalescent infection (red curve on figure 2D).
Viral clade analysis
Thirteen of 15 full-length genome sequences were available for clade analysis via GISAID (table 3); and two sequences were not available at the time of analysis. Lineages of SARS-CoV-2 have been identified based on mutations in key amino acid positions.33 Clade G is defined by the mutations D614G, C241T, C3037T and A23403G in the Spike protein; and clade GR by additional RG203KR mutations in the Nucleocapsid protein N; clade L is most closely related to the Wuhan reference strain (NC_045512.2).34 Accordingly, among the 13 viral isolates, three different clades were identified, including clade L (N=2), GR (N=4) and L (N=7).
Table 3.
Genomic sequences accessed via GISAID listing key amino acid locations used for SARS-CoV-2 classification
| Disease classification | Virus name (GISAID) | EPI_ISL_# | Date of RT-qPCR | Lineage | ORF8: 84 | ORF3a: 57 | S:614* | N:203** | N:204** |
| Early acute | hCoV-19/Austria/CeMM0191/2020 | 438 032 | 13/03/2020 | B(L) | L | Q | D | R | G |
| Early acute | hCoV-19/Austria/CeMM0248/2020 | 438 078 | 21/03/2020 | B (L) | L | Q | D | R | G |
| Early acute | hCoV-19/Austria/CeMM0018/2020 | 419 671 | 19/03/2020 | B.1.1 (GR) | L | Q | G | K | R |
| Early acute | hCoV-19/Austria/CeMM0228/2020 | 438 061 | 18/03/2020 | B.1.1 (GR) | L | Q | G | K | R |
| Early acute | hCoV-19/Austria/CeMM0235/2020 | 438 066 | 19/03/2020 | B.1.1 (GR) | L | Q | G | K | R |
| Early acute | hCoV-19/Austria/CeMM0250/2020 | 438 080 | 21/03/2020 | B.1.1 (GR) | L | Q | G | K | R |
| Early acute | hCoV-19/Austria/CeMM0222/2020 | 438 056 | 17/03/2020 | B.1.8 (G) | L | Q | G | R | G |
| Early acute | hCoV-19/Austria/CeMM0249/2020 | 438 079 | 21/03/2020 | B.1.8 (G) | L | Q | G | R | G |
| Late acute | hCoV-19/Austria/CeMM0267/2020 | 438 096 | 24/03/2020 | B.1.8 (G) | L | Q | G | R | G |
| Late acute | hCoV-19/Austria/CeMM0276/2020 | 438 103 | 25/03/2020 | B.1.8 (G) | L | Q | G | R | G |
| Late acute | hCoV-19/Austria/CeMM0303/2020 | 475 778 | 29/03/2020 | B.1.8 (G) | L | Q | G | R | G |
| Late acute | hCoV-19/Austria/CeMM0324/2020 | 475 794 | 01/04/2020 | B.1.8 (G) | L | Q | G | R | G |
| Late acute | hCoV-19/Austria/CeMM0337/2020 | 475 800 | 03/04/2020 | B.1.8 (G) | L | Q | G | R | G |
SARS-CoV-2 clades are classified by The Global Initiative on Sharing All Influenza Data (GISAID) using specific non-synonymous mutations in the viral genome. Clade G is defined by the mutations D614G, C241T, C3037T and A23403G in the Spike protein; and clade GR by additional RG203KR mutations in the Nucleocapsid protein N; clade L is most closely related to the Wuhan reference strain (NC_045512.2).34 Whole genome data were available for 13/15 sequences; data for two sequences were not available at the time of analysis. Accordingly, among the 13 sequences analysed, three different clades were identified, including clades L (N=2), GR (N=4) and G (N=7). All three clades were detected in early acute infection, and clade G was additionally detected in late acute infection. *For simplicity reasons, only mutation D614G (grey background) in the Spike protein defining clade G is shown. **Additional mutations R203K and G204R in the Nucleocapsid protein N defining clade GR are also shown in grey.
ORF, open reading frame.
Discussion
Our results demonstrate that SARS-CoV-2 RT-qPCR testing, when added to a national influenza surveillance programme in primary care, can rapidly, early and accurately diagnose COVID-19 during an outbreak. Of the 73 patients presenting to the sentinel GP, 22 were diagnosed with COVID-19, including 15 patients with acute disease and 7 with late convalescent infection, respectively. The sensitivity and specificity of RT-qPCR were 68.1% and 100%. Testing RT-qPCR reactive showed perfect correlation with seropositivity during the first half of the outbreak and among early acute (N=8 patients) and late acute presenters (N=7). Strikingly, the mean duration of symptoms of early presenters (2 days) was less than half of late acute presenters (4.4 days) and a quarter of late convalescent presenters (8 days). These findings highlight the need to undertake RT-qPCR testing rapidly and early as soon as symptoms occur. Acute infection was strongly associated with multiple symptoms, including loss of taste, nausea and vomiting, breathlessness, myalgia and sore throat; but loss of smell, fever and cough were not. Surprisingly, loss of taste, but not any other clinical symptom, was significantly associated with convalescent infection. Finally, viral genome analysis demonstrated the presence of three major SARS-CoV-2 clades during the outbreak, suggesting that the outbreak was the result of independent transmission chains.
Overall, our findings help untangle COVID-19 infection during an outbreak in a ski resort in Austria. Our results suggest that acute COVID-19 may be associated with a spectrum of symptoms and presence of multiple strains within one setting. This highlights the heterogeneity of COVID-19 and the importance in containing outbreaks early before spread. While effective TTI strategies have been suggested as the key to containing the outbreak without intermittent lockdowns,35 we suggest that systemic changes may also be needed. For example, behavioural changes such as large-scale gathering of people in closed spaces have to be avoided as they may trigger emergence of individual clusters to form a superspreading event. Keeping a level of compliance to social distancing and reduced physical contacts are necessary to prevent any future wave. Enhanced testing is an important factor, and our study suggests that testing in primary care at symptom onset is highly accurate and should be something that governments should consider as an additional strategy.
Loss of taste of smell has been recognised as an important marker of COVID-19;36 37 however, more than half of patients reported olfactory dysfunction after the onset of other symptoms when sensitivity of RT-qPCR may be reduced.38 Furthermore, loss of taste could not be objectively confirmed in one-third of people,38 suggesting that self-assessment using a mobile phone application may not be as accurate as clinician-initiated RT-qPCR testing of people presenting with acute disease.39 Timely and accurate testing is also a prerequisite for effective contact tracing.27
The outbreak we explored occurred after a 3-day party (13–15 March) just before the skiing season was brought to a premature end due to the Austrian national lockdown measures on 16 March. The index case was diagnosed on 11 March and the first secondary cases were reported 2 days after the celebrations. Therefore, it is possible that the outbreak we are describing here could be a possible superspreading event. Superspreading events have been associated with high-intensity aerosol producing activities (shouting, singing) in confined spaces and potentially, the lockdown party might have triggered the local outbreak. The two acute disease clusters observed in this study may represent different types of viral exposure. First, inhalation of high-density aerosols at the party causing acute illness among early presenters and second, low-level home transmission of party goers to (late presenting) friends and family during the lockdown. In our study, no COVID-19 cases were observed among children (persons <18 years of age), suggesting that any infected children may have remained asymptomatic or did not attend the practice because of mild disease.40 No further endemic cases were detected after the outbreak. This suggests that combination prevention including rapid testing and case notification in primary care, contact tracing and isolation, and lockdown measures can effectively terminate an outbreak. To our knowledge, our study is the first to demonstrate that the ECDC policy of additional COVID-19 screening at national influenza screening sites can effectively detect and control a regional outbreak.21
Our study has many strengths. Our study was enabled by data from a well-established sentinel GP, participating in the National Influenza Screening Programme, covering the entire area of the outbreak. Importantly, national SARS-CoV-2 screening was adopted early, starting the day before the first two cases were reported in Austria; and 16 of 29 (55.1%) cases documented in the Schladming-Dachstein region, including the first and the last case, were detected at the sentinel GP. RT-qPCR testing was rapidly deployed by offering same-day GP appointments, and result reporting and case notification within 24 hours. Rapid adoption of new commercial antibody platforms (Lab Mustafa, Salzburg) and in-house neutralising antibody testing assay (Medical University of Vienna) enabled accurate interpretation of RT-qPCR results.
There are some limitations of our study. We used a relatively small patient cohort from a single sentinel GP, potentially limiting conclusions on causality and generalisability of our finding to other areas excluding seven patients for whom COVID-19 serostatus were not available. Lack of association with high fever and cough in our COVID-19 cohort may be due to the national health hotline directing patients with more severe disease to attend emergency service. Therefore, people with these symptoms might have preferred to attend acute services rather than the GP. Although we collected data prospectively, recall bias cannot be excluded. This could be suggested by the lack of association of symptoms of acute infection (nausea and vomiting, breathless and myalgia) among all people confirmed with infection (when including those with negative RT-qPCR), compared with those people presenting early (reactive RT-qPCR). Specific recall bias of taste is less likely, as it featured in both groups and data collection was completed prior to publication of the first systematic review of altered taste and smell in the media.41 However, change or loss in smell/taste was not quantified using an established tool such as the Visual Analogue Scale,42 43 but rather assessed by simple ‘yes’ and ‘no’ answers using a standard clinical questionnaire, potentially leading to response style bias. Although asymptomatic infection is common,9 asymptomatic people were excluded from this study as we were focusing on symptom-driven presentation. This potentially excludes an important segment of the infected population and future studies will focus on exploring this further. The presence of three viral clades within the outbreak suggests heterogeneity of the virus, but we have not explored this aspect in great details in this study, as this was beyond the scope of this work. In fact, the data presented here is part of the ongoing work untangling the phylogeny of SARS-CoV-2 clades in Austria and their worldwide spread.28
To our knowledge, this is the first study to show that primary care can contribute to early case detection and termination of a SARS-CoV-2 outbreak in the community. Our study has important implications for patients, public health and health systems; nationally and internationally for outbreak epidemiology and control. As countries enter the viral suppression phase, early detection will be crucial in the prevention and control of the disease. Early testing at onset of disease, followed by timely contact tracing and case isolation of secondary cases should prevent onward transmission and reduce the reproduction number Re below 1. Austria has increased the number of its sentinel sites from 91 to 231 due to COVID-19, indicating that primary care has become an essential partner in a comprehensive surveillance strategy for disease prevention and control. Clade analysis could greatly enhance public health surveillance in the UK where only three quarters of contact tracing is being completed.44 Key priorities for future research include systematic prospective quantitative and qualitative evaluation of the Austrian National SARS-CoV-2 screening programme during the seasonal influenza season and generalisability of the intervention in multi-ethnic inner-city settings including genomic analysis using deep viral genome sequencing to support complex contact tracing and adaption of the REAP-1 protocol to include SARS-CoV-2 lateral flow antigen testing.
Conclusions
RT-qPCR testing in primary care can rapidly and accurately detect SARS-CoV-2 among people presenting with mild-to-moderate illness in a heterogenous viral community outbreak. This study demonstrates high rates of accurate and early viral detection associated with symptomatic testing in primary care during a COVID-19 outbreak, which is required for an effective TTI strategy. Targeted testing in primary care can support national sentinel surveillance of COVID-19.
Supplementary Material
Acknowledgments
We thank Evelyn Marktl for daily updates on the Christian Drosten’s COVID-19 podcast (https://www.ndr.de/nachrichten/info/podcast4684.html). We are grateful to the team of Praxis Dr Lammel for their contributions, and in particular to the nurse Sabine Roiderer for providing direct patient care and help with administration. We thank the patients of Schladming-Dachstein for participating in the study. We thank the Styrian Academy for General Practice (STAFAM) for carrying the publication costs.
Footnotes
Twitter: @WernerLeber
WL and JP-G contributed equally.
Contributors: WL, OL, MR-F, MEM-K, E-MH, CH and JP-G contributed to the design of the study; OL and E-MH took nasopharyngeal swabs; OL, E-MH and CH maintained the clinical data base; AS and RCG submitted the ethics application; MR-F provided RT-qPCR data; BA, AL, AP, J-WG, TP, SA, CB, AB and JC conducted clade analysis; MEM-K produced ELISA data; KS performed the neutralising antibody assay; JP-G and WL conducted the statistical analysis; WL and JP-G wrote the manuscript with contributions from OL, MR-F, MEM-K, RCG, JC, CB, AB, KS, E-MH, CH, AS and CG. All authors read and approved the final version.
Funding: This research was self-funded by each individual co-author. This project received funding from the Vienna Science and Technology Fund (WWTF) as part of the WWTF COVID-19 Rapid Response Funding 2020. Award/Grant number is not applicable.
Competing interests: None declared.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement
Data are available upon reasonable request. All data relevant to the study are included in the article. Raw data are stored on a secure computer at the Institute of General Practice and Evidence-based Health Services Research, University of Graz, Austria.
Ethics statements
Patient consent for publication
Not required.
Ethics approval
The Medical University of Graz Research Ethics Committee (reference number: 32–429 ex 19/20) approved collection of anonymised RT-qPCR and antibody status data, and the Medical University of Vienna Research Ethics Committee (reference number: EK1339/2017) additionally approved usage of anonymised RT-qPCR data collected as part of the National Influenza Surveillance Network including generation of secondary genomic data. Written consent was obtained from all participating patients agreeing on anonymised data collection for data validation, quality control and research purposes.
References
- 1.World Health Organization (WHO) . Clinical management of severe acute respiratory infection when COVID-19 is suspected, 2020. Available: https://www.who.int/publications-detail/clinical-management-of-severe-acute-respiratory-infection-when-novel-coronavirus-(ncov)-infection-is-suspected [Accessed 05 May 2021].
- 2.Lauer SA, Grantz KH, Bi Q, et al. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann Intern Med 2020;172:577–82. 10.7326/M20-0504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature 2020;581:465–9. 10.1038/s41586-020-2196-x [DOI] [PubMed] [Google Scholar]
- 4.Cevik M, Tate M, Lloyd O, et al. SARS-CoV-2, SARS-CoV-1 and MERS-CoV viral load dynamics, duration of viral shedding and infectiousness: a living systematic review and meta-analysis. SSRN Journal 2020:2020.07.25.20162107. 10.2139/ssrn.3677918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byrne AW, McEvoy D, Collins AB, et al. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open 2020;10:e039856. 10.1136/bmjopen-2020-039856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis 2020;71:2663–6. 10.1093/cid/ciaa638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Endo A, Abbott S, Kucharski AJ, et al. Estimating the overdispersion in COVID-19 transmission using outbreak sizes outside China. Wellcome Open Res 2020;5:67. 10.12688/wellcomeopenres.15842.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sayampanathan AA, Heng CS, Pin PH, et al. Infectivity of asymptomatic versus symptomatic COVID-19. Lancet 2021;397:93–4. 10.1016/S0140-6736(20)32651-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Byambasuren O, Cardona M, Bell K, et al. Estimating the extent of asymptomatic COVID-19 and its potential for community transmission: systematic review and meta-analysis. Official Journal of the Association of Medical Microbiology and Infectious Disease Canada 2020;5:223–34. 10.3138/jammi-2020-0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bi Q, Wu Y, Mei S, et al. Epidemiology and transmission of COVID-19 in 391 cases and 1286 of their close contacts in Shenzhen, China: a retrospective cohort study. Lancet Infect Dis 2020;20:911–9. 10.1016/S1473-3099(20)30287-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li R, Pei S, Chen B, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 2020;368:489–93. 10.1126/science.abb3221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.European Centre for Disease Prevention and Control (ECDC) . Rapid risk assessment: coronavirus disease 2019 (COVID-19) in the EU/EEA and the UK– ninth update, 2020. Available: https://www.ecdc.europa.eu/en/publications-data/rapid-risk-assessment-coronavirus-disease-2019-covid-19-pandemic-ninth-update [Accessed 05 May 2020].
- 13.Koo JR, Cook AR, Park M, et al. Interventions to mitigate early spread of SARS-CoV-2 in Singapore: a modelling study. Lancet Infect Dis 2020;20:678–88. 10.1016/S1473-3099(20)30162-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frieden TR, Lee CT. Identifying and interrupting Superspreading Events—Implications for control of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 2020;26:1059–66. 10.3201/eid2606.200495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kreidl P, Schmid D, Maritschnik S, et al. Emergence of coronavirus disease 2019 (COVID-19) in Austria. Wien Klin Wochenschr 2020;132:645–52. 10.1007/s00508-020-01723-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Popa A, Genger J-W, Nicholson M. Mutational dynamics and transmission properties of SARS-CoV-2 superspreading events in Austria. bioRxiv 2020:2020.07.15.204339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The independent, 2020. Available: https://www.independent.co.uk/news/world/europe/coronavirus-austria-cases-covid-19-hospital-lockdown-latest-a9466281.html [Accessed 05 May 2020].
- 18.European Centre for Disease Prevention and Control (ECDC) . Coronavirus disease 2019 (COVID-19) in the EU/EEA and the UK –ninth update, 2020. Available: https://www.ecdc.europa.eu/sites/default/files/documents/covid-19-rapid-risk-assessment-coronavirus-disease-2019-ninth-update-23-april-2020.pdf [Accessed 02 Jun 2020].
- 19.de Sutter A, Llor C, Maier M, et al. Family medicine in times of 'COVID-19': A generalists' voice. Eur J Gen Pract 2020;26:58–60. 10.1080/13814788.2020.1757312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hull SA, Williams C, Ashworth M. Suspected COVID-19 in primary care: how GP records contribute to understanding differences in prevalence by ethnicity. medRxiv 2020:2020.05.23.20101741. [Google Scholar]
- 21.European Centre for Disease Prevention and Control (ECDC) . Strategies for the surveillance of COVID-19, 2020. Available: https://www.ecdc.europa.eu/sites/default/files/documents/COVID-19-surveillance-strategy-9-Apr-2020.pdf [Accessed 11 Jul 2020].
- 22.Roehr B. Covid-19 is threatening the survival of US primary care. BMJ 2020;369:m2333. 10.1136/bmj.m2333 [DOI] [PubMed] [Google Scholar]
- 23.Harding-Edgar L, McCartney M, Pollock AM. Test and trace strategy has overlooked importance of clinical input, clinical oversight and integration. J R Soc Med 2020;113:428–32. 10.1177/0141076820967906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zentrum für Virologie Medizinische Universität Wien. Projekt Diagnostisches Influenzanetzwerk Österreich (DINÖ). Available: https://www.virologie.meduniwien.ac.at/wissenschaft-forschung/virus-epidemiologie/influenza-projekt-diagnostisches-influenzanetzwerk-oesterreich-dinoe/ [Accessed 28 May 2020].
- 25.Federal Ministry of Social Affairs H, Care and Consumer Protection, Republic of Austria . National health Hotline 1450, 2019. Available: https://www.1450.at/1450-die-gesundheitsnummer/ [Accessed 28 May 2020].
- 26.Corman V, Bleicker T, Brünink S. Diagnostic detection of 2019-nCoV by real-time RT-PCR, 2020. Available: https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf?sfvrsn=a9ef618c_2 [Accessed 02 Jul 2020]. [DOI] [PMC free article] [PubMed]
- 27.Kretzschmar ME, Rozhnova G, Bootsma MCJ, et al. Impact of delays on effectiveness of contact tracing strategies for COVID-19: a modelling study. Lancet Public Health 2020;5:e452–9. 10.1016/S2468-2667(20)30157-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Popa A, Genger J-W, Nicholson MD, et al. Genomic epidemiology of superspreading events in Austria reveals mutational dynamics and transmission properties of SARS-CoV-2. Sci Transl Med 2020;12:eabe2555. 10.1126/scitranslmed.abe2555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stadlbauer D, Amanat F, Chromikova V, et al. SARS‐CoV‐2 seroconversion in humans: a detailed protocol for a serological assay, antigen production, and test setup. Curr Protoc Microbiol 2020;57:e100. 10.1002/cpmc.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn JY, Sohn Y, Lee SH, et al. Use of convalescent plasma therapy in two COVID-19 patients with acute respiratory distress syndrome in Korea. J Korean Med Sci 2020;35:e149. 10.3346/jkms.2020.35.e149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinnock H, Epiphaniou E, Sheikh A, et al. Developing standards for reporting implementation studies of complex interventions (STARI): a systematic review and e-Delphi. Implement Sci 2015;10:42. 10.1186/s13012-015-0235-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu Y, McCauley J. GISAID: global initiative on sharing all influenza data – from vision to reality. Eurosurveillance 2017;22. 10.2807/1560-7917.ES.2017.22.13.30494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mercatelli D, Giorgi FM. Geographic and genomic distribution of SARS-CoV-2 mutations. Front Microbiol 2020;11:1800. 10.3389/fmicb.2020.01800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Global Initiative on Sharing All Influenza Data (GISAID) . Clade and lineage nomenclature AIDS in genomic epidemiology studies of active hCoV-19 viruses, 2020. Available: https://www.gisaid.org/references/statements-clarifications/clade-and-lineage-nomenclature-aids-in-genomic-epidemiology-of-active-hcov-19-viruses/ [Accessed September 05, 2020].
- 35.Panovska-Griffiths J, Kerr CC, Stuart RM, et al. Determining the optimal strategy for reopening schools, the impact of test and trace interventions, and the risk of occurrence of a second COVID-19 epidemic wave in the UK: a modelling study. Lancet Child Adolesc Health 2020;4:817–27. 10.1016/S2352-4642(20)30250-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Aziz M, Goyal H, Haghbin H, et al. The Association of "Loss of Smell" to COVID-19: A Systematic Review and Meta-Analysis. Am J Med Sci 2021;361:216–25. 10.1016/j.amjms.2020.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Bartheld CS, Hagen MM, Butowt R. Prevalence of chemosensory dysfunction in COVID-19 patients: a systematic review and meta-analysis reveals significant ethnic differences. ACS Chem Neurosci 2020;11:2944–61. 10.1021/acschemneuro.0c00460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lechien JR, Chiesa-Estomba CM, Hans S, et al. Loss of smell and taste in 2013 European patients with mild to moderate COVID-19. Ann Intern Med 2020;173:672–5. 10.7326/M20-2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menni C, Valdes AM, Freidin MB, et al. Real-Time tracking of self-reported symptoms to predict potential COVID-19. Nat Med 2020;26:1037–40. 10.1038/s41591-020-0916-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maltezou HC, Vorou R, Papadima K, et al. Transmission dynamics of SARS-CoV-2 within families with children in Greece: a study of 23 clusters. J Med Virol 2021;93:1414–20. 10.1002/jmv.26394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lovato A, de Filippis C. Clinical presentation of COVID-19: a systematic review focusing on upper airway symptoms. Ear Nose Throat J 2020;99:569–76. 10.1177/0145561320920762 [DOI] [PubMed] [Google Scholar]
- 42.Sung Y-T, Wu J-S. The visual analogue scale for rating, ranking and Paired-Comparison (VAS-RRP): a new technique for psychological measurement. Behav Res Methods 2018;50:1694–715. 10.3758/s13428-018-1041-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rojas-Lechuga MJ, Izquierdo-Domínguez A, Chiesa-Estomba C, et al. Chemosensory dysfunction in COVID-19 out-patients. Eur Arch Otorhinolaryngol 2021;278:695–702. 10.1007/s00405-020-06266-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.NHS England . Nhs test and trace – week 4 of contact tracing, England: 18 to 24 June 2020, 2020. Available: https://www.gov.uk/government/publications/nhs-test-and-trace-statistics-england-18-june-to-24-june-2020/weekly-nhs-test-and-trace-bulletin-england-18-24-june-2020 [Accessed 05 May 2021].
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available upon reasonable request. All data relevant to the study are included in the article. Raw data are stored on a secure computer at the Institute of General Practice and Evidence-based Health Services Research, University of Graz, Austria.