Abstract
Bladder cancer is the ninth most diagnosed cancer in the world. This study aims to investigate the role and mechanisms of the taurine-upregulated gene 1 (TUG1)/miR-140-3p/annexin A8 (ANXA8) axis in bladder cancer. Western blotting and qRT-PCR determined the expression levels of ANXA8, miR-140-3p, TUG1, and epithelial-mesenchymal transition (EMT) markers. RNA immunoprecipitation (RIP), luciferase assay, and RNA pull-down assay validated the association among ANXA8, miR-140-3p, and TUG1. The biological functions were determined by colony formation, Annexin V-fluorescein isothiocyanate (FITC)/propidium (PI) staining, and transwell assays. Xenograft tumorigenesis detected tumor growth and metastasis in vivo. Pathological analysis was examined by hematoxylin and eosin (H&E) and immunohistochemistry (IHC) analyses. ANXA8 was elevated in bladder tumors and cells. Knockdown of ANXA8 suppressed cell growth, migration, invasion, and EMT in UMUC-3 and T24 cells. ANXA8 was determined as a miR-140-3p target gene. Overexpression of miR-140-3p suppressed cell proliferation, migration, invasion, and EMT via targeting ANXA8. TUG1 promoted ANXA8 expression via sponging miR-140-3p. Silencing of miR-140-3p or ANXA8 overexpression abrogated the tumor-suppressive effects of TUG1 silencing on bladder cancer cell growth and metastasis. The TUG1/miR-140-3p/ANXA8 axis was also implicated in tumor growth and lung metastasis in vivo. TUG1 promotes bladder cancer progression and metastasis through activating ANXA8 by sponging miR-140-3p, which sheds light on the mechanisms of bladder cancer pathogenesis.
Keywords: lncRNA-TUG1, miR-140-3p, ANXA8, tumor metastasis, bladder cancer
Graphical Abstract
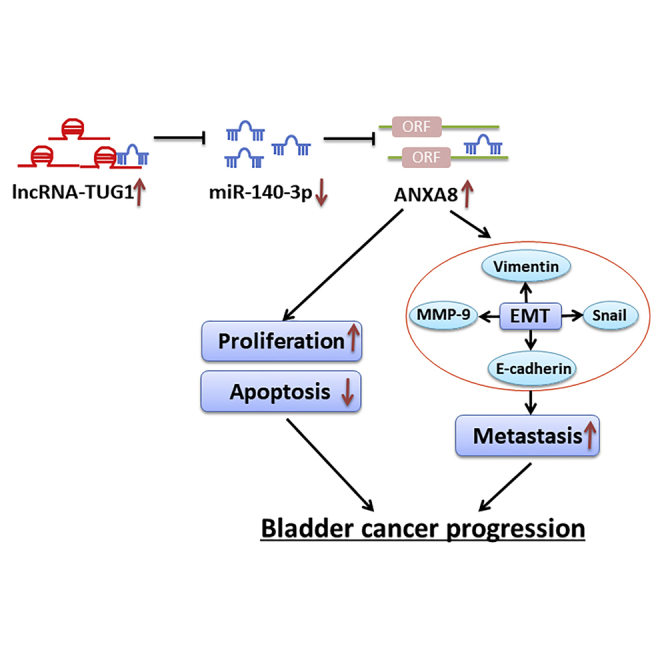
The current study first demonstrated the interactions among ANXA8, miR-140-3p, and long non-coding RNA (lncRNA)-TUG1 in bladder cancer. TUG1 promoted bladder cancer progression and metastasis through activating ANXA8 by sponging miR-140-3p. These findings illustrated the regulatory role of the TUG1/miR-140-3p/ANXA8 axis in bladder cancer.
Introduction
As the ninth most diagnosed cancer and the second most common of genitourinary malignancies worldwide, bladder cancer is associated with high recurrence or metastasis rates. In clinical practice, muscle-invasive bladder cancer (MIBC) is associated with increased incidence of distance metastasis.1 Even though neoadjuvant chemotherapy (NAC), antibodies, and immune checkpoint inhibitors have been introduced and shown to be beneficial for a subset of bladder cancer patients recently, the outcomes of advanced cases have not been significantly improved.2,3 Thus, it is necessary to explore the novel therapeutic targets and unravel the underlying mechanisms of bladder cancer development.
Annexins (ANXA1-ANXA11, ANXA13) belong to a superfamily of calcium-dependent, phospholipid-binding proteins. Annexins share a core domain containing four or more copies of annexin repeats responsible for Ca2+ and phospholipid-binding activity.4 Previous studies have illustrated the aberrant expression of annexins in cancer cells and their involvement in tumorigenesis and progression of different types of cancer, including bladder cancer.5,6 Annexin A8 (ANXA8) was first discovered as an anticoagulation factor,7 and growing evidence has illustrated that ANXA8 is elevated in various types of cancers, including pancreatic cancer, breast cancer, acute promyelocytic leukemia, and ovarian cancer.8, 9, 10, 11 As reported, upregulated ANXA8 in pancreatic cancer is positively correlated with tumor grade and decreased 5-year survival, and knockdown of ANXA8 impairs cell viability in pancreatic cancer cells,10 indicating its oncogenic role in pancreatic cancer. However, the biological roles of ANXA8 and its relevant mechanisms in bladder cancer remain elusive.
It is well established that only ∼2% of the transcripts are protein-coding transcripts, while the majority are transcribed non-coding RNAs (ncRNAs). ncRNAs can be divided into two major categories: long non-coding RNAs (lncRNAs; >200 nt in length) and small ncRNAs (microRNAs [miRNAs] or small interfering RNAs [siRNAs]; <50 nt).12 Compelling evidence has suggested that lncRNAs or miRNAs play crucial roles in a diversity of cellular and disease processes, including cell proliferation, metastasis, apoptosis, and epithelial-mesenchymal transition (EMT).13,14 The well-known competing endogenous RNA (ceRNA) hypothesis demonstrates that lncRNA serves as a miRNA sponge, thereby regulating miRNA target gene expression.15,16 miR-140-3p was found as either a poor prognostic marker or tumor suppressor in certain types of cancer.17, 18, 19 More importantly, a recent report has illustrated that miR-140-3p is markedly decreased in bladder cancer tissues, and it exerts tumor-suppressive functions in bladder cancer.20 Interestingly, our preliminary bioinformatics analysis indicated that lncRNA Taurine Upregulated Gene 1 (TUG1) might function as a miR-140-3p sponge. TUG1, which was originally identified as a taurine upregulated transcript, is aberrantly elevated and plays tumor-promoting effects in bladder cancer.21, 22, 23, 24 For instance, silencing of TUG1 improves responses for radiation therapy of bladder cancer by targeting HMGB1.23 However, knowledge is unavailable concerning the role of TUG1 and miR-140-3p in bladder cancer progression and metastasis.
In this study, we found that ANXA8 was elevated in bladder cancer, and silencing of ANXA8 inhibited cell growth, migration, invasion, and EMT in UMUC-3 and T24 cells. ANXA8 was determined as a miR-140-3p target gene. Overexpression of miR-140-3p suppressed cell growth, migration, invasion, and EMT, but promoted apoptosis via targeting ANXA8. Moreover, silencing of TUG1 suppressed bladder cancer cell growth and metastasis by targeting miR-140-3p to inhibit ANXA8 signaling. The TUG1/miR-140-3p/ANXA8 axis also regulated tumor growth and lung metastasis in vivo. These findings indicated that TUG1, miR-140-3p, and ANXA8 could be potential targets for bladder cancer treatment.
Results
The oncogenic role of ANXA8 in the progression and metastasis of bladder cancer
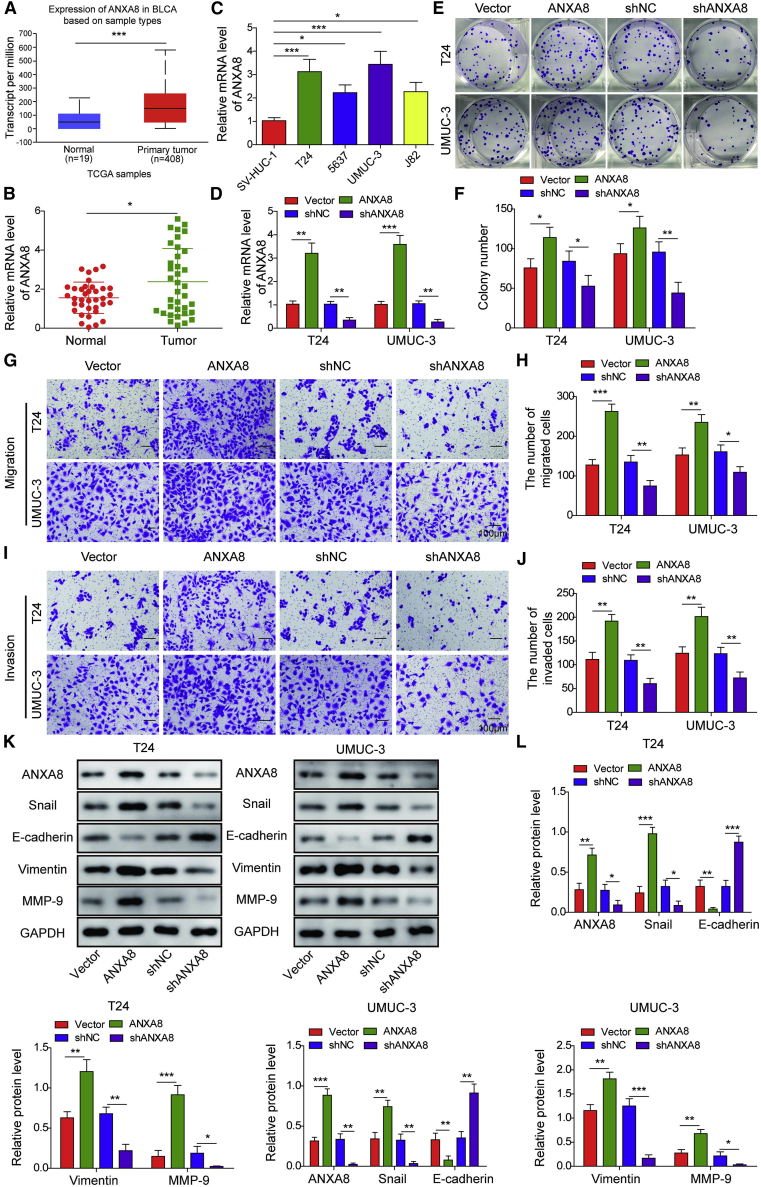
To study the expression profile of ANXA8 in bladder cancer, we evaluated the transcriptional level of ANXA8 between the bladder tumor and paired adjacent normal urinary bladder tissue by the UALCAN database (http://ualcan.path.uab.edu/). As presented in Figure 1A, the transcriptional level of ANXA8 was remarkably elevated in bladder cancer tissues (n = 408) in comparison with normal tissues (n = 19). We further examined the mRNA level of ANXA8 in clinical specimen and bladder carcinoma cell lines. Consistently, ANXA8 was remarkably elevated in bladder cancer tissues and cell lines 5637, T24, UMUC-3, and J82 cells compared with paired normal tissues and normal uroepithelial SV-HUC-1 cells, respectively (Figures 1B and 1C). UMUC-3 and T24 cells were selected for the subsequent experiments because of the high ANXA8 expression in these cells. To study the functional role of ANXA8, we carried out gain- and loss-of-function experiments. Transfection of pcDNA3.1-ANXA8 or shANXA8 successfully upregulated or downregulated the ANXA8 mRNA level in both UMUC-3 and T24 cells, respectively (Figure 1D). Colony formation assays revealed that ANXA8 overexpression markedly enhanced the clonogenic ability, whereas knockdown of ANXA8 exerted the opposite effect on colony formation in both UMUC-3 and T24 cells (Figures 1E and 1F). To test the effect of ANXA8 on cell migration and invasion, we performed transwell assays. As presented in Figures 1G–1J, the migrated and invaded cell numbers were remarkably higher in UMUC-3 or T24 cells overexpressing ANXA8, whereas knockdown of ANXA8 notably impaired the migratory and invasive capacities of UMUC-3 and T24 cells. These findings indicate that ANXA8 may be correlated with bladder cancer metastasis. It is well accepted that EMT is often involved in cell migration and invasion. As expected, overexpression of ANXA9 dramatically decreased epithelial marker E-cadherin expression but resulted in a significant induction of mesenchymal markers Snail, Vimentin, and MMP-9 (Figures 1K and 1L). Knockdown of ANXA8 exerted the opposite effects on EMT marker expression, suggesting that ANXA8 promotes EMT in bladder cancer cells (Figures 1K and 1L). Taken together, upregulated ANXA8 promotes cell proliferation and metastasis in UMUC-3 and T24 cells.
Figure 1.
The oncogenic role of ANXA8 in bladder cancer
(A) Comparison of ANXA8 mRNA expression based on data mining via the UALCAN database. (B) The mRNA levels of ANXA8 in different tissues were assessed by qRT-PCR. (C) The ANXA8 mRNA levels in different cells were detected by qRT-PCR. pcDNA3.1 alone, pcDNA3.1-ANXA8, shNC, or shANXA8 was transfected into T24 and UMUC-3 cells. (D) The ANXA8 mRNA levels in ANXA8-overexpressing and knockdown cells were determined by qRT-PCR. (E and F) Colony-forming ability was examined by colony formation assay with quantitative analysis. (G–J) The migratory (G and H) and invasive (I and J) capacities were examined by transwell migration and invasion assays with quantitative analysis. Scale bars, 100 μm. (K and L) The ANXA8 and EMT markers expressions were assessed by western blotting. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
ANXA8 is a direct target gene of miR-140-3p in bladder cancer cells
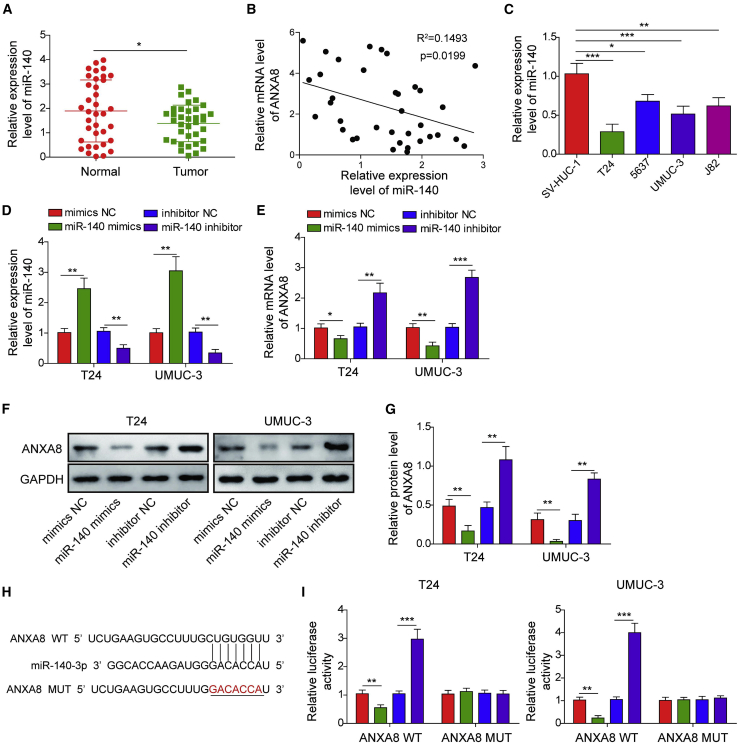
Bioinformatics analysis predicted that miR-140-3p might serve as an upstream molecule of ANXA8. To validate bioinformatics analyses, we next checked the miR-140-3p level in bladder cancer tissues. Interestingly, miR-140-3p was markedly decreased in bladder tumors compared with their normal tissue counterparts (Figure 2A). Additionally, there was a negative correlation between ANXA8 and miR-140-3p in bladder tumors (Figure 2B). In line with these findings, miR-140-3p was remarkably decreased in 5637, T24, UMUC-3, and J82 cells compared with normal uroepithelial SV-HUC-1 cells (Figure 2C). Overexpression and knockdown experiments were further conducted to study the effect of miR-140-3p on ANXA8 expression. As presented in Figure 2D, miR-140-3p level was induced or reduced by miR-140-3p mimics or inhibitor in both T24 and UMUC-3 cells, respectively. Transfection of miR-140-3p mimics resulted in a dramatic reduction of ANXA8, whereas miR-140-3p inhibitor led to an induction of ANXA8 in both bladder cancer cell lines (Figure 2E). A similar observation was found at ANXA8 protein levels by western blotting (Figures 2F and 2G). The putative binding sites between ANXA8 3′ UTR and miR-140-3p were predicted using TargetScan (Figure 2H). In UMUC-3 and T24 cells, co-transfection of miR-140-3p mimics and wild-type (WT)-ANXA8 caused a marked reduction of luciferase activity, while co-transfection of miR-140-3p inhibitor and WT-ANXA8 led to a dramatic increase of luciferase activity in comparison with control. By contrast, the mutant of ANXA8 (MUT-ANXA8) abrogated the effects of miR-140-3p mimics or inhibitor on luciferase activity in both UMUC-3 and T24 cells (Figure 2I). Collectively, these data indicate that ANXA8 is a target gene of miR-140-3p in bladder cancer cells.
Figure 2.
ANXA8 was a direct target gene of miR-140-3p in T24 and UMUC-3 cells
(A) The miR-140-3p level in bladder tumors and control was determined by qRT-PCR. (B) Correlation analysis between ANXA8 and miR-140-3p. (C) The miR-140-3p levels in SV-HUC-1 cells and bladder carcinoma cell lines were assessed by qRT-PCR. T24 and UMUC-3 cells were transfected with miR-140-3p mimics/inhibitor or corresponding controls. (D and E) The levels of miR-140-3p (D) and ANXA8 (E) in miR-140-3p-overexpressing and knockdown cells were detected by qRT-PCR. (F and G) The ANXA8 expression was assessed by western blotting with quantitative analysis. (H) Putative binding sites between ANXA8 and miR-140-3p. (I) Relative luciferase activity was measured by luciferase assay. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
miR-140-3p inhibits cell growth, migration, invasion, and EMT but enhances apoptosis via targeting ANXA8
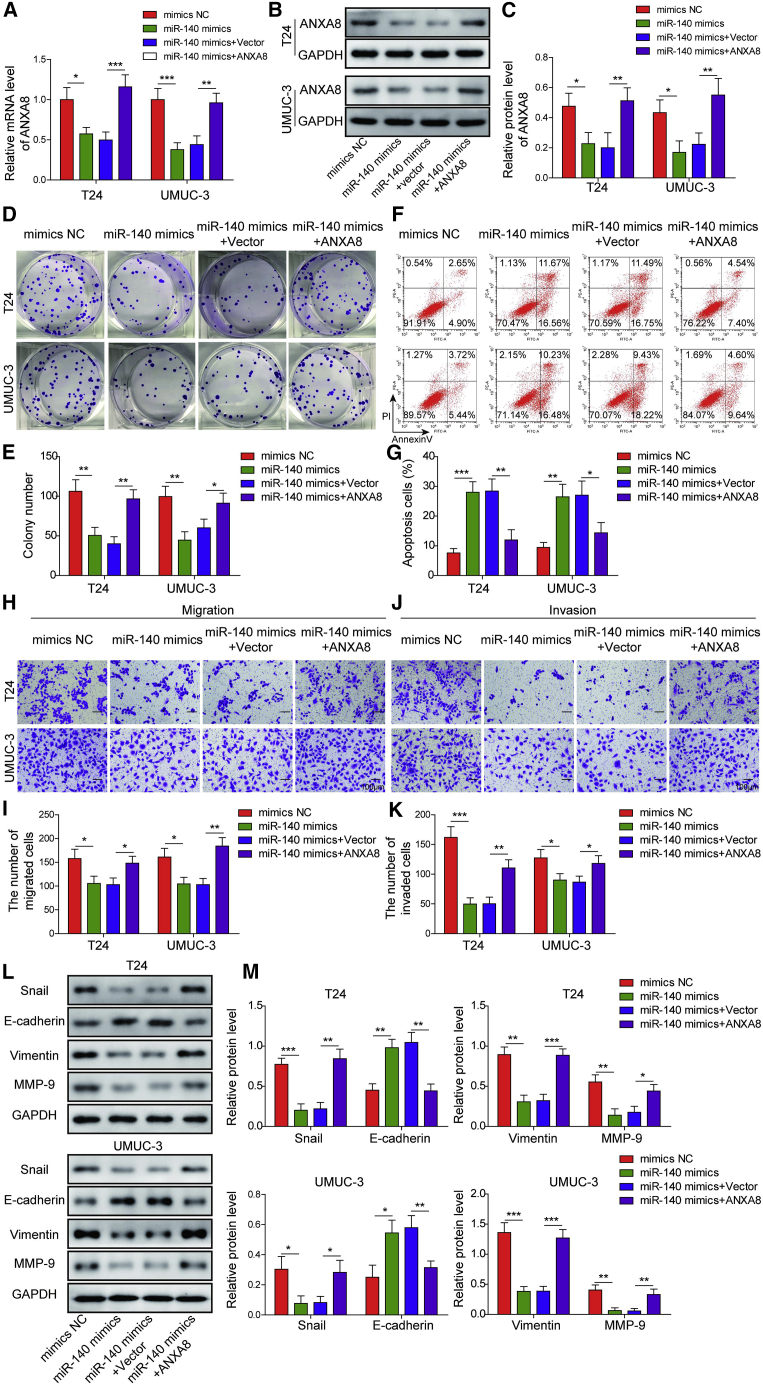
To validate the role of the miR-140-3p/ANXA8 axis, we transfected UMUC-3 and T24 cells with mimics NC, miR-140-3p mimics, miR-140-3p mimics+pcDNA3.1, or miR-140-3p+pcDNA3.1-ANXA8. As presented in Figures 3A–3C, transfection of miR-140-3p mimics significantly decreased ANXA8 expression at both mRNA and protein levels, whereas co-transfection of miR-140-3p mimics and pcDNA3.1-ANXA8 reversed this effect on ANXA8 expression. Overexpression of miR-140-3p markedly impaired clonogenic abilities of bladder cells, whereas exogenous expression of ANXA8 caused a rebound in colony number in comparison with the miR-140-3p mimics group (Figures 3D and 3E). In contrast, miR-140-3p mimics promoted apoptosis, while co-transfection of miR-140-3p and pcDNA3.1-ANXA8 markedly attenuated miR-140-3p-induced apoptosis in bladder cancer cells (Figures 3F and 3G). Moreover, impaired migration and invasion were observed in miR-140-3p mimics-transfected UMUC-3 and T24 cells, whereas overexpression of ANXA8 attenuated the miR-140-3p-inhibited migration and invasion (Figures 3H–3K). In accordance with the results of transwell assays, overexpression of miR-140-3p remarkably induced E-cadherin expression, accompanied by decreased expression of Snail, Vimentin, and MMP-9, whereas ANXA8 overexpression reversed the miR-140-3p-mediated changes on the protein levels of E-cadherin, Snail, Vimentin, and MMP-9 (Figures 3L and 3M). Together, these results suggest that miR-140-3p plays as a tumor suppressor via targeting ANXA8.
Figure 3.
miR-140-3p inhibited cell proliferation, migration, invasion, and EMT via targeting ANXA8
Mimics NC, miR-140-3p mimics, miR-140-3p mimics+pcDNA3.1, or miR-140-3p mimics+pcDNA3.1-ANXA8 were transfected into T24 and UMUC-3 cells. (A) The mRNA level of ANXA8 was determined by qRT-PCR. (B and C) The ANXA8 expression was assessed by western blotting with quantitative analysis. (D and E) Clonogenic ability was assessed by colony formation assay with quantitative analysis. (F and G) Cell apoptotic rate was assessed by flow cytometry with quantitative analysis. (H–K) The migratory (H and I) and invasive (J and K) capacities were examined by transwell migration and invasion assays with quantitative analysis. Scale bar, 100 μm. (L and M) The expressions of EMT markers were assessed by western blotting with quantitative analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
TUG1 promotes ANXA8 expression via sponging miR-140-3p in bladder cancer cells
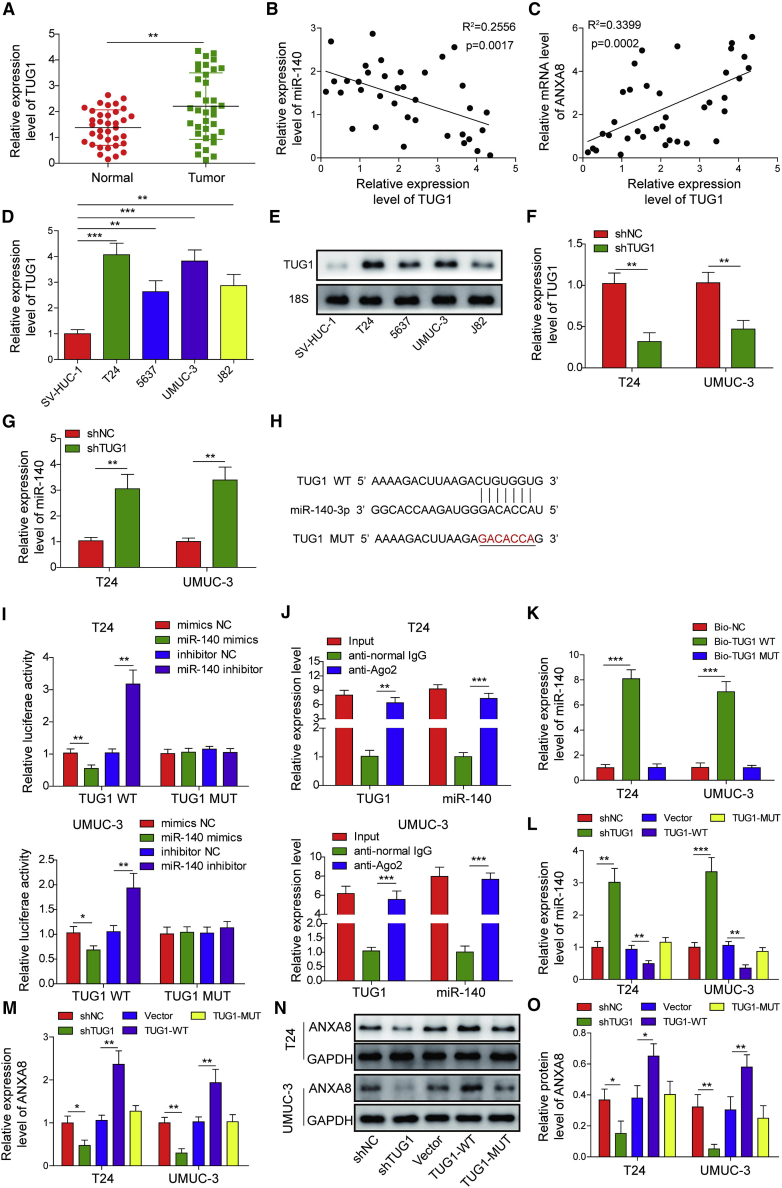
To further validate the TUG1/miR-140-3p/ANXA8 ceRNA network in bladder cancer, we next examined TUG1 level in bladder tumors. Consistent with previous study, TUG1 was markedly elevated in bladder tumors in comparison with their normal counterparts (Figure 4A). As expected, TUG1 was negatively correlated with miR-140-3p and positively correlated with ANXA8 in bladder tumors (Figures 4B and 4C). Compared with normal uroepithelial SV-HUC-1 cells, TUG1 was remarkably elevated in bladder cancer cell lines T24, 5637, UMUC-3, and J82 cells as detected by qRT-PCR and northern blot (Figures 4D and 4E). Gain- and loss-of-function experiments were then carried out to reveal the interaction between TUG1 and miR-140-3p. As shown in Figures 4F and 4G, shTUG1 successfully decreased TUG1 expression level and increased miR-140-3p level in both T24 and UMUC-3 cells. The putative binding sites between TUG1 and miR-140-3p were predicted by starBase (http://starbase.sysu.edu.cn/index.php), and the mutant of TUG1 (TUG1-MUT) was generated by inserting the mismatched sequences in the potential binding site for miR-140-3p (Figure 4H). Luciferase assay revealed that co-transfection of miR-140-3p mimics and wild-type TUG1 (TUG1-WT) dramatically decreased luciferase activity, whereas miR-140-3p inhibitor exerted an opposite effect. However, TUG1-MUT abrogated the effects of miR-140-3p mimics/inhibitor in both UMUC-3 and T24 cells (Figure 4I). Additionally, the results of RNA immunoprecipitation (RIP) showed that both TUG1 and miR-140-3p were remarkably enriched in the Ago2-immunoprecipitation fractions of T24 and UMUC-3 cells, as compared with normal immunoglobulin G (IgG) (Figure 4J). RNA pull-down assay further confirmed that miR-140-3p was successfully pulled down by TUG1-WT rather than TUG1-MUT (Figure 4K). Silencing of TUG1 caused a significant increase of miR-140-3p (Figure 4L). By contrast, overexpression of TUG1-WT led to a remarkable reduction of miR-140-3p, whereas TUG1-MUT abolished this effect (Figure 4L). Furthermore, ANXA8 mRNA and protein levels were dramatically decreased, which were correlated with TUG1 downregulation (Figures 4M–4O). ANXA8 was upregulated in TUG1-WT-transfected T24 and UMUC-3 cells, whereas no significant change of ANXA8 was observed in TUG1-MUT-overexpressing cells (Figures 4M–4O). These findings indicate that TUG1 promotes ANXA8 expression via sponging miR-140-3p in bladder cancer cells.
Figure 4.
TUG1 promoted ANXA8 expression via sponging miR-140-3p in bladder cancer cells
(A) The TUG1 levels in bladder tumors and normal counterparts were examined by qRT-PCR. (B) Correlation analysis between miR-140-3p and TUG1. (C) Correlation analysis between ANXA8 and TUG1. (D) The TUG1 levels in SV-HUC-1 cells and bladder carcinoma cell lines were assessed by qRT-PCR. (E) The TUG1 levels in SV-HUC-1 cells and bladder carcinoma cell lines were assessed by northern blot. 18S acted as a loading control. (F and G) The TUG1 (F) and miR-140-3p (G) levels in shNC or shTUG1-transfected cells were detected by qRT-PCR. (H) Putative binding sites between TUG1 and miR-140-3p. (I) Relative luciferase activity was measured by dual-luciferase reporter assay. (J) Relative enrichment of TUG1 and miR-140-3p after RIP was determined by qRT-PCR. (K) The directed interaction between TUG1 and miR-140-3p was validated by RNA pull-down assay. (L) The miR-140-3p level in different cells was detected by qRT-PCR. (M) The ANXA8 mRNA level in different cells was detected by qRT-PCR. (N and O) The ANXA8 expression in different cells was assessed by western blotting with quantitative analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
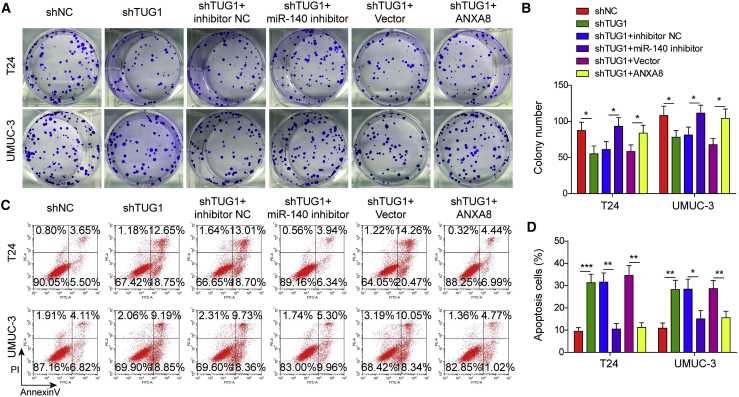
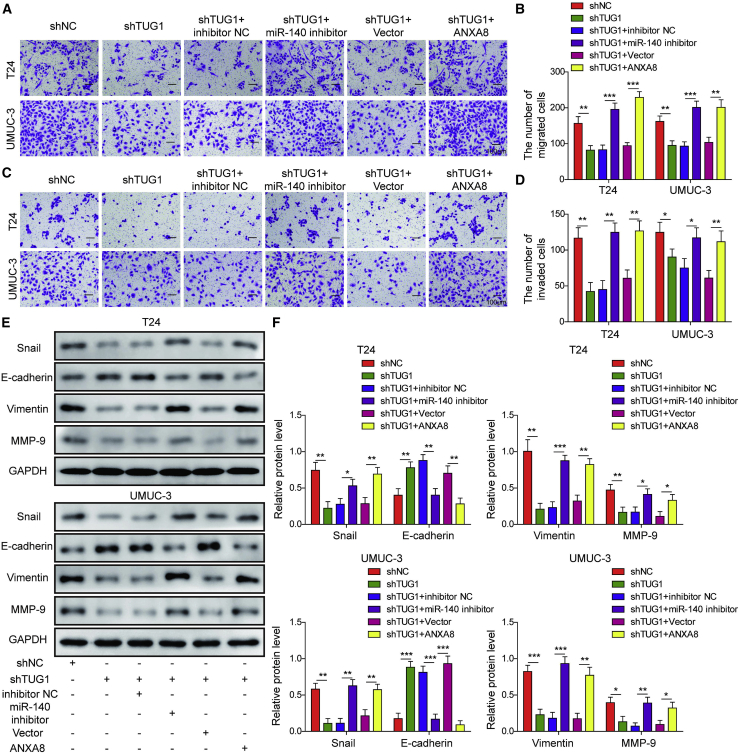
Silencing of miR-140-3p or ANXA8 overexpression reverses the effects of TUG1 knockdown on bladder cell proliferation and metastasis
We next examined the function of the TUG1/miR-140-3p/ANXA8 axis in T24 and UMUC-3 cells. As presented in Figures 5A and 5B, knockdown of TUG1 significantly impaired the colony-forming abilities of T24 and UMUC-3 cells, whereas silencing of miR-140-3p or ANXA8 overexpression led to a marked rebound of the clonogenic abilities compared with the corresponding control. Cell apoptosis assay showed that silencing of TUG1 induced apoptosis in bladder cancer cells, whereas miR-140-3p silencing or ANXA8 overexpression extenuated shTUG1-induced apoptosis (Figures 5C and 5D). Moreover, miR-140-3p silencing or ANXA8 overexpression abolished the negative effects of shTUG1 on migratory and invasive capacities of bladder cancer cells (Figures 6A–6D). Consistent with the results of transwell assays, transfection of miR-140-3p inhibitor or ANXA8 overexpression construct caused a rebound of shTUG1-mediated repression of Snail, Vimentin, and MMP-9 and also attenuated shTUG1-mediated induction of E-cadherin in both UMUC-3 and T24 cells (Figures 6E and 6F). Together, the TUG1/miR-140-3p/ANXA8 axis plays crucial roles in cell growth, apoptosis, migration, invasion, and EMT in bladder cancer cells.
Figure 5.
Silencing of miR-140-3p or ANXA8 overexpression reversed the effects of TUG1 knockdown on bladder cell proliferation and apoptosis
T24 and UMUC-3 cells were transfected with shNC, shTUG1, shTUG1+inhibitor NC, shTUG1+miR-140-3p inhibitor, shTUG1+pcDNA3.1, or shTUG1+pcDNA3.1-ANXA8. (A and B) Colony-forming ability was determined by colony formation assay with quantitative analysis. (C and D) Cell apoptotic rate was assessed by flow cytometry with quantitative analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Figure 6.
Silencing of miR-140-3p or ANXA8 overexpression reversed the effects of TUG1 knockdown on bladder cell migration, invasion, and the EMT process
T24 and UMUC-3 cells were transfected with shNC, shTUG1, shTUG1+inhibitor NC, shTUG1+miR-140-3p inhibitor, shTUG1+pcDNA3.1, or shTUG1+pcDNA3.1-ANXA8. (A–D) The migratory (A and B) and invasive (C and D) capacities were examined by transwell migration and invasion assays with quantitative analysis. Scale bar, 100 μm. (E and F) The EMT marker level was assessed by western blotting with quantitative analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
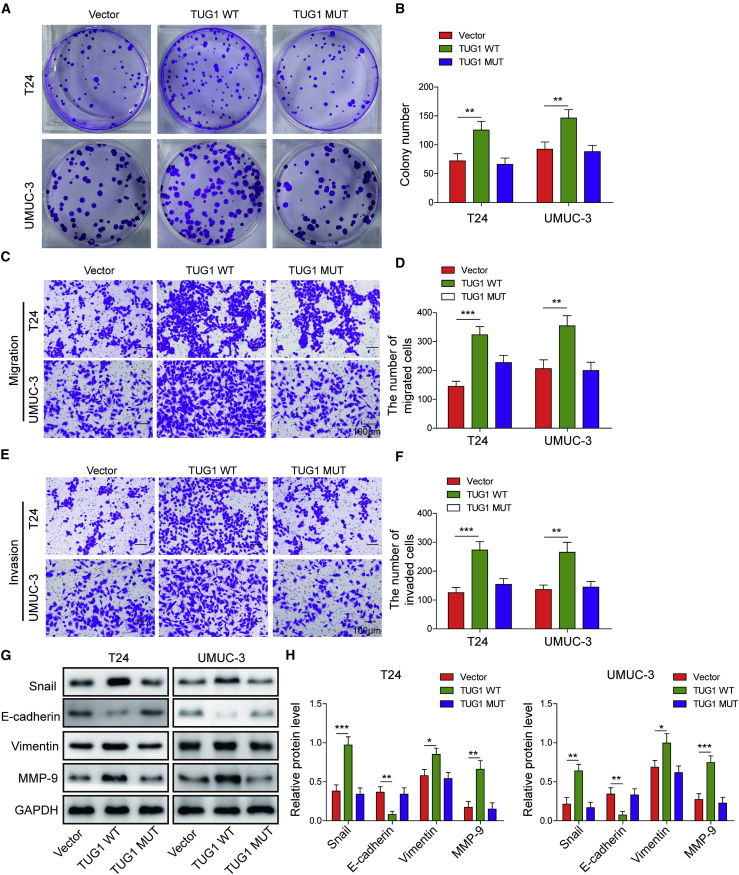
Overexpression of TUG1 promotes colony formation, cell migration, invasion, and EMT in bladder cancer cells by sponging miR-140-3p
To further delineate the biological function of TUG1 in bladder cancer cells, we conducted gain-of function experiments in T24 and UMUC-3 cells. The colony numbers significantly increased by TUG1-WT in bladder cancer cells, whereas TUG1-MUT containing the mutated miR-140-3p binding sites exhibited no effect on colony-forming abilities of T24 and UMUC-3 cells (Figures 7A and 7B). Moreover, overexpression of TUG1-WT promoted cell migration and invasion as detected by transwell assays, while TUG1-MUT abrogated these effects (Figures 7C–7F). In line with these findings, western blot revealed that TUG1-WT overexpression resulted in induction of Snail, Vimentin, and MMP-9, accompanied by the reduction of E-cadherin in T24 and UMUC-3 cells. As expected, TUG1-MUT caused no significant changes on EMT marker expression compared with the control group (Figures 7G and 7H). These data indicate that overexpression of TUG1 promotes colony formation, cell migration, invasion, and EMT in bladder cancer cells, and lack of miR-140-3p binding site attenuated the oncogenic function of TUG1 in T24 and UMUC-3 cells.
Figure 7.
Overexpression of TUG1 promotes colony formation, cell migration, invasion, and EMT in bladder cancer cells by sponging miR-140-3p
T24 and UMUC-3 cells were transfected with vector, TUG1-WT, or TUG1-MUT. (A and B) Colony-forming ability was determined by colony formation assay with quantitative analysis. (C–F) The migratory (C and D) and invasive (E and F) capacities were examined by transwell migration and invasion assays with quantitative analysis. Scale bar, 100 μm. (G and H) The EMT marker level was assessed by western blotting with quantitative analysis. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
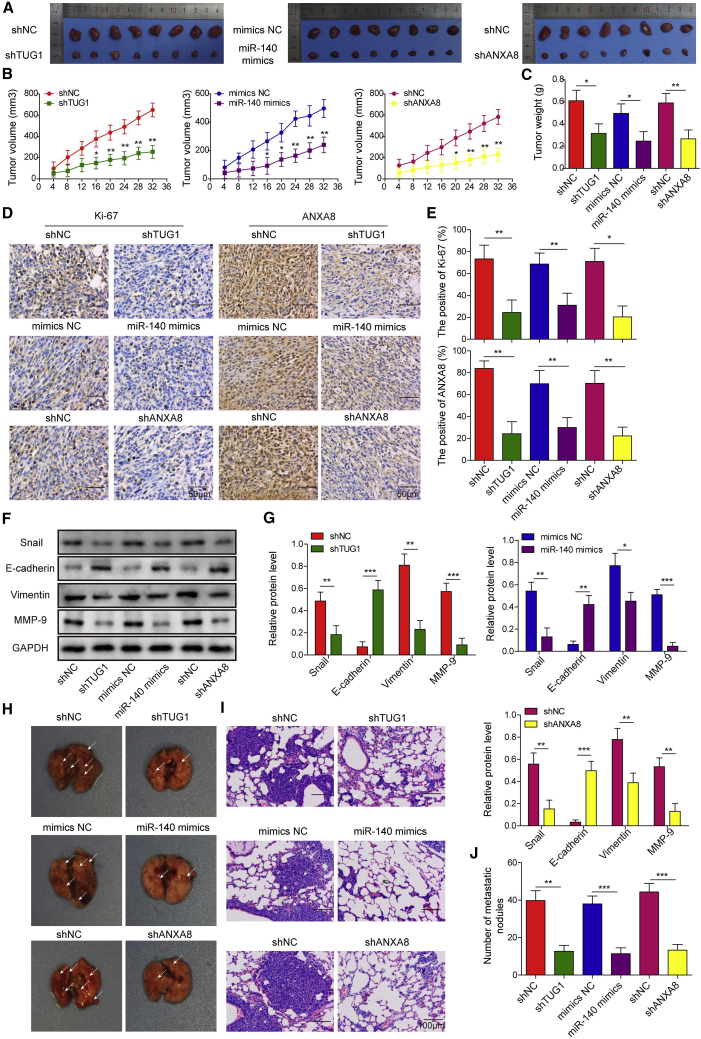
The TUG1/miR-140-3p/ANXA8 axis regulates tumor growth and lung metastasis in vivo
In order to further validate the function of the TUG1/miR-140-3p/ANXA8 axis, we performed in vivo xenograft tumorigenesis and metastasis studies. Interestingly, tumors derived from TUG1 knockdown, miR-140-3p overexpression, or ANXA8 knockdown cells grew significantly slower than that of the corresponding control group (Figures 8A and 8B). Tumors were dissected and weighed on day 30. As presented in Figure 8C, silencing of TUG1 or ANXA8 and miR-140-3p overexpression resulted in a remarkable decrease of tumor weight. In addition, tumors were subjected to immunohistochemistry (IHC) analysis. The cell proliferation markers Ki-67 and ANXA8 were significantly decreased by TUG1 knockdown, miR-140-3p overexpression, or ANXA8 knockdown (Figures 8D and 8E), suggesting that the TUG1/miR-140-3p/ANXA8 axis might be involved in cell proliferation in bladder cancer. Western blotting confirmed that silencing of TUG1 or ANXA8 and overexpression of miR-140-3p reduced Snail, Vimentin, and MMP-9 protein levels but induced E-cadherin expression (Figures 8F and 8G). Furthermore, a lung metastasis in vivo model was generated using lateral tail vein injection of transfected cells. After 6 weeks, the lung tissues were harvested and subjected to histological analysis. Knockdown of TUG1 or ANXA8 and overexpression of miR-140-3p suppressed lung metastasis of bladder cancer (Figures 8H and 8I) and also decreased the number of pulmonary nodules (Figure 8J). Taken together, the in vivo studies suggest that the TUG1/miR-140-3p/ANXA8 axis regulates tumor growth and lung metastasis in bladder cancer.
Figure 8.
The TUG1/miR-140-3p/ANXA8 axis regulated tumor growth and lung metastasis in vivo
(A) Images of tumor size on day 30. (B and C) Tumor volumes and weights were examined every 5 days or after 30 days, respectively. (D and E) The Ki-67 and ANXA8 expressions in xenograft tumor tissues were determined by IHC with quantitative analysis. Ki-67 and ANXA8-positive cells were counted in the five random fields. Scale bars, 50 μm. (F) The EMT marker protein levels in tumors were assessed by western blotting. (G) Quantitative analysis of western blotting. (H) Images of lungs in different groups. (I) H&E analysis was used to visualize metastasis colonization. Scale bar, 100 μm. (J) The number of pulmonary nodules was counted. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Currently, transurethral resection is the gold standard for the initial treatment of non-muscle-invasive bladder cancer (NMIBC), followed by intravesical chemotherapy and immunotherapy. For the patients with MIBC, radical cystectomy and chemotherapy are standard treatments.3 Despite the advances in clinical treatments, ∼50%–70% of patients with bladder cancer experienced a relapse within 5 years.25 Distant metastases and lymph node involvement are correlated with poor prognosis.26,27 Thus, it is critical to identify the novel therapeutic target to improve the clinical outcomes. In the current study, our data showed that ANXA8 and TUG1 were elevated, whereas miR-140-3p was downregulated in bladder cancer tissues and cells. ANXA8 was a miR-140-3p target gene, and TUG1 acted as ceRNA of miR-140-3p in bladder cancer cells. TUG1 promoted cell proliferation, migration, invasion, and EMT but inhibited apoptosis through regulating ANXA8 by sequestering miR-140-3p in bladder cancer cells, and this ceRNA network was also involved in tumor growth and lung metastasis in vivo models, indicating that the TUG1/miR-140-3p/ANXA8 axis may provide new insight into the prognosis of bladder cancer.
Compelling evidence has demonstrated that dysregulated lncRNAs are implicated in a diversity of cellular processes in bladder cancer, such as cell apoptosis, proliferation, migration, and invasion.25 Meta-analyses have revealed that lncRNAs function as diagnostic or prognostic markers for bladder cancer.28,29 For instance, elevated lncRNA-UCA1 was positively correlated with tumor grade and is expected to be a diagnostic marker for bladder cancer.30 In the current study, we reported that TUG1 was significantly upregulated in bladder cancer tissues and cells. This observation was in agreement with previous studies that reported the aberrant upregulation of TUG1 in bladder cancer.22, 23, 24 TUG1 promotes radioresistance by targeting ZEB2 in bladder cancer cells.22 A more recent report has illustrated that silencing of TUG1 improves response for radiation treatment for bladder cancer by repressing HMGB1.23 Both of these studies mainly focused on the effect of TUG1 in radiosensitivity in bladder cancer; however, the biological role and mechanism of TUG1 during bladder cancer progression have not been fully disclosed. It is worthy to note that knockdown of TUG1 inhibits EMT and cell invasion by targeting the miR-145/ZEB2 axis in T24 cells and suppresses tumor growth in vivo.22 The present data support this by demonstrating that silencing of TUG1 suppressed cell growth, migration, invasion, and EMT but enhanced apoptosis by regulating the miR-140-3p/ANXA8 axis in bladder cancer cells. In vivo study also illustrated that knockdown of TUG1 significantly inhibited tumor growth, EMT, and lung metastasis. A novel ceRNA network of TUG1 has been identified in bladder cancer cells.
It is well established that lncRNAs serve as molecular sponges of miRNA to modulate their functions.31,32 For instance, lncRNA-BCRT1 acts as a miR-1303 molecular sponge to regulate PTBP3 expression, thereby promoting breast cancer progression.33 Bioinformatics analyses, luciferase assay, RIP, and RNA pull-down assay unequivocally illustrated that TUG1 acted as an endogenous sponge of miR-140-3p in bladder cancer cells. The function of miR-140-3p is complicated, and it has been shown to be either a poor prognostic marker or tumor suppressor depending on tumor types.17, 18, 19 For example, miR-140-3p is involved in recurrence and tumor invasion in spinal chordoma.17 By contrast, miR-140-3p inhibits cell growth and invasion by directly targeting ATP6AP2 or ATP8A1 in non-small cell lung cancer (NSCLC).18,19 A recent study has illustrated that miR-140-3p overexpression remarkably suppressed cell growth, migration, and invasion in bladder cancer cells.20 In the present study, we found that T24 and UMUC-3 cells showed higher levels of ANXA8 compared with that in 5637 and J82 cells, while lower levels of miR-140-3p were observed in T24 and UMUC-3 cells. These data suggest that decreased expression of miR-140-3p led to high expression of its target gene ANXA8, thereby promoting cell proliferation, migration, and invasion. Even though the miR-140-3p level in UMUC-3 was higher than that in T24 cells, miR-140-3p still contributed to tumor-suppressive effects because of the reduction by >50% in both cells, in comparison with that in SV-HUC-1 cells. However, these findings have not been validated in vivo, and the regulatory mechanism by which miR-140-3p exerted its tumor-suppressive role remains unclear. In agreement with the previous study, in vitro and in vivo studies suggested that miR-140-3p functioned as a tumor suppressor in bladder cancer. Our findings provided mechanistic insight into the understanding of the tumor-suppressive role of miR-140-3p in bladder cancer.
Previous study has illustrated that lncRNA that contains multiple seed target sites of a specific miRNA is more potent to work as a competitive inhibitor.34 This study indicated that TUG1 contains only one target site of miR-140-3p, which has partial sequence complementarity to 7-mer seed region (position 2–8). Furthermore, TUG1 could inhibit miR-140-3p expression by directly binding to miR-140-3p, suggesting that TUG1 functioned as a ceRNA to negatively regulate miR-140-3p. Therefore, we searched and read a large number of reports about the mechanism of ceRNA, which showed that miRNA was negatively regulated by lncRNA overexpression or knockdown.35, 36, 37, 38 However, the specific mechanisms by which lncRNA negatively regulated miRNA have not been described in detail, which may be a meaningful research direction in the future. In contrast, although the seed target site “ACCACAG” on miR-140-3p could serve as a potential binding site for a number of lncRNAs or circular RNAs (circRNAs) based on bioinformatics analyses, gain- and loss-of function experiments supported that TUG1 acted as a miR-140-3p sponge to regulate bladder cancer progression. Moreover, TUG1-MUT containing the mismatched sequences in the putative miR-140-3p binding site had no effect on miR-140-3p level and the proliferation and metastasis of bladder cancer cells. Function experiments revealed that the miR-140-3p inhibitor abrogated the tumor-suppressive effects of TUG1 silencing on bladder cancer cell growth and metastasis, indicating that TUG1 acted as an oncogene by sponging miR-140-3p. Although several lncRNAs can interact with the same miRNA, whether other lncRNAs function as miR-140-3p sponges in bladder cancer needs further investigation.
Annexins are involved in various cellular processes associated with membrane transport and interaction, including cell proliferation, differentiation, and apoptosis.39 In recent years, the roles of Annexins in tumorigenesis and progression have gained more attention. In urinary bladder cancer, overexpression of ANXA1 is associated with tumorigenesis and differentiation.5 A recent study also demonstrated that ANXA2 is correlated to drug resistance and recurrence of bladder cancer.40 Previous studies have confirmed high expression of ANXA8 and its prognostic role in acute promyelocytic leukemia, high-grade breast cancer, pancreatic cancer, and ovarian cancer.8, 9, 10, 11 For instance, prognostic analysis has revealed that high expression of ANXA8 is closely associated with poor prognosis in ovarian cancer.11 However, the function and miRNA-mediated mechanisms of ANXA8 in bladder cancer remain poorly understood. The current study has illustrated that ANXA8 was remarkably elevated in bladder cancer tissues and cells, and ANXA8 exerted an oncogenic role on cell growth and metastasis in vivo and in vitro. Interestingly, we showed that knockdown of ANXA8 inhibited EMT in bladder cancer cells. A study in cholangiocarcinoma has reported that ANXA8 is downregulated by epidermal growth factor (EGF) transcriptionally, thus leading to the morphologic changes of EMT.41 Future study is needed to investigate whether EGF also acts as an upstream molecule of ANXA8 in bladder cancer cells. Moreover, we also identified ANXA8 as a direct target of miR-140-3p in bladder cancer cells, and TUG1 regulated ANXA8 expression via sponging miR-140-3p. Functional experiments further showed that ANXA8 overexpression attenuated the tumor-suppressive effects of TUG1 knockdown in bladder cancer cells, indicating that the TUG1/miR-140-3p/ANXA8 axis plays critical roles in bladder cancer progression and metastasis.
In conclusion, we first demonstrated that ANXA8 was markedly elevated in bladder cancer tissues and cells and served as an oncogene in bladder cancer. TUG1 promoted bladder cancer progression and metastasis through regulating ANXA8 expression by sponging miR-140-3p. Unraveling the novel ceRNA network provided in-depth understanding of the mechanism in the pathogenesis of bladder cancer, as well as candidates for targeted therapy of bladder cancer.
Materials and methods
Clinical specimen collection
A cohort of 36 bladder tumors and their normal counterparts was collected from bladder cancer patients post-operatively in the Xiangya Hospital of Central South University. The study was approved by the Xiangya Hospital of Central South University. Informed consents were obtained from patients recruited to this study.
Cell culture
Human bladder carcinoma cell lines and normal uroepithelial SV-HUC-1 cells were purchased from the Chinese Academy of Science (Shanghai, China). T24 and J82 cells were grown in DMEM-F12 supplemented with 10% fetal bovine serum (FBS; GIBCO, Thermo Fisher Scientific, Waltham, MA, USA). 5637, UMUC-3, and SV-HUC-1 cells were grown in RPMI 1640, DMEM, or F-12K containing 10% FBS (GIBCO), respectively. All cells were grown at 37°C/5% CO2.
Cell transfection
The full length of ANXA8 or TUG1 was constructed into pcDNA3.1 (Invitrogen, Thermo Fisher Scientific). The binding site sequence of miR-140-3p on TUG1 was mutated by QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent, Palo Alto, CA, USA). miR-140-3p mimics/inhibitor, mimics negative control (mimics NC), inhibitor negative control (inhibitor NC), short hairpin RNA (shRNA) against ANXA8 and TUG1, as well as scramble shRNA (shNC) were obtained from Genechem (Shanghai, China). Cells were transfected with shRNA, miRNA, or overexpression constructs using Lipofectamine 2000 transfection reagent (Invitrogen). The sequences of miRNAs used in this study are as follows: hsa-miR-140-3p mimics forward (F), 5′-UACCACAGGGUAGAACCACGG-3′ and reverse (R), 5′-GUGGUUCUACCCUGUGGUAUU-3′; hsa-miR-140-3p inhibitor, 5′-CCGUGGUUCUACCCUGUGGUA-3′; mimic NC F, 5′-UUCUCCGAACGUGUCACGUTT-3′ and R 5′-ACGUGACACGUUCGGAGAATT-3′; inhibitor NC, 5′-CAGUACUUUUGUGUAGUACAA-3′.
Total RNA isolation and real-time PCR
Total RNA was extracted from bladder tumors or cells using TRIzol (Sigma-Aldrich). cDNA was reverse transcribed using SuperScript III (Invitrogen). qRT-PCR was conducted using FastStart SYBR Green Mastermix (Roche, Germany). The miR-140-3p primers contain a stable stem-loop structure. The F primer adds additional nucleotides to optimize GC contents, melting temperature (Tm), and appropriate length, as well as enhance specificity. The specificity of miR-140-3p was confirmed by gel electrophoresis and sequencing of the single band. miRNA expression was assessed using TaqMan MicroRNA Assay (Applied Biosystems). cDNA was synthesized using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems), and qRT-PCR was performed using TaqMan miRNA Assays Human Panel sequence-specific primers in an ABI7900 Fast Real-Time PCR systems (Applied Biosystems). The following primers were used in this study: TUG1: 5′-ACGACTGAGCAAGCACTACC-3′ (F) and 5′-CTCAGCAATCAGGAGGCACA-3′ (R); miR-140-3p: 5′-CGGCTACCACAGGGTAGAA-3′ (F) and 5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCGTGG-3′ (R); ANXA8: 5′-TGACCCAAGGACACTGTGTT-3′ (F) and 5′-AGGTACCCAGTCTCAGTTGC-3′ (R); U6: 5′-CTCGCTTCGGCAGCACA-3′ (F) and 5′-AACGCTTCACGAATTTGCGT-3′ (R); GAPDH: 5′-CCAGGTGGTCTCCTCTGA-3′ (F) and 5′-GCTGTAGCCAAATCGTTGT-3′ (R). The relative quantification of target genes was carried out using the 2−ΔΔCt method. The specificity was confirmed by agarose gel electrophoresis and sequencing.
Colony formation assay
Cells were trypsinized and reseeded on six-well plates (5,000 cells/well). On day 14, the formaldehyde-fixed colonies were stained with crystal violet solution. The visible colonies were counted and used to calculate the colony formation rate.
Transwell assays
Transwell assays were conducted as previously described.42 For invasion assay, cells were grown in the upper chambers (Corning, Corning, NY, USA) with Matrigel (Corning) in serum-free medium. The lower reservoirs were filled with complete medium. Forty-eight hours later, 4% paraformaldehyde (PFA)-fixed invading cells in the bottom chambers were stained with crystal violet and calculated. Transwell migration assay was conducted without Matrigel coating.
Western blotting
Protein extracts were lysed and estimated using a BCA protein assay kit (Beyotime, China). Protein lysates were separated and transferred onto the polyvinylidene difluoride (PVDF) membrane. After blocking, the blots were incubated with specific antibody at 4°C overnight, followed by the incubation of secondary antibody (Abcam). The results were visualized using ECL substrate (Applygen, Beijing, China).
Northern blot
Northern blot probes were synthesized by GenScript (Nanjing, China). The sequences of probes were as follows: TUG1, 5′-GAGGCACCAGCTTCAAAACCCGCTTGCTGA-3′; and 18S, 5′-CGGAACTACGACGGTATCTG-3′. Total RNA was prepared using TRIzol (Sigma-Aldrich). Northern blot was conducted using NorthernMax Kit (Invitrogen) following the manufacturer’s instructions. In brief, total RNAs were subjected to gel electrophoresis and transferred onto nylon membranes, followed by hybridization with biotin-labeled probes. The signals were detected using Biotin Chromogenic Detection Kit (Thermo Fisher Scientific).
Dual-luciferase reporter assay
Luciferase assay was performed using Dual-Luciferase Assay System (Promega, Madison, WI, USA).43 The wild-type TUG1 or ANXA8 3′ UTR containing the putative miR-140-3p binding site “CUGUGGU” was amplified by PCR, and mutants were synthesized using QuikChange Lightning Multi Site-Directed Mutagenesis Kit (Agilent, Palo Alto, CA, USA). The TUG1-WT/TUG1-MUT, as well as the WT-ANXA8/MUT-ANXA8 3′ UTRs were cloned into psiCHECK-2 vector (Promega). The construct was co-transfected with miR-140-3p mimics/inhibitor or corresponding control into T24 and UMUC-3 cells using Lipofectamine 2000 transfection reagent (Invitrogen).
Annexin V-fluorescein isothiocyanate (FITC)/PI staining
Cell apoptosis was detected using Annexin V-FITC Kit (Beyotime). In brief, resuspended cells were stained with Annexin V-FITC and PI at room temperature. The samples were then subjected to flow cytometry analysis.
RIP assay
RIP was carried out using Magna RIP Kit (Millipore). In brief, anti-Ago2 antibody (cat. no. 03-110; Millipore) or isotype control conjugated magnetic beads were incubated with cell lysates at 4°C overnight. RNA was purified, and TUG1 or miR-140-3p levels were examined by qRT-PCR.
RNA pull-down assay
RNA pull-down assay was performed using Pierce RNA pull-down kit (Pierce, Thermo Fisher Scientific). In brief, WT-TUG1 or MUT-TUG1 was labeled and purified using Pierce RNA 3′ End Desthiobiotinylation Kit. The biotinylated wild-type TUG1 (Bio-TUG1-WT), mutated TUG1 (Bio-TUG1-MUT) was then conjugated to streptavidin beads. After incubation with protein lysates, the RNA complex was extracted and purified using TRIzol reagent (Sigma-Aldrich). miR-140-3p level was examined by qRT-PCR.
In vivo study
For xenograft tumorigenesis study, BALB/c nude mice (6 weeks old, n = 48) were from SJA Laboratory Animal (Hunan, China). All animal studies were approved by the Xiangya Hospital of Central South University. Stable transfected UMUC-3 cells (5 × 106 cells) were subcutaneously implanted under the dorsa side of each mouse. The tumors were dissected and weighted on day 30. The lung metastasis study was conducted as previously described.22 In six groups, stable transfected cells were implanted by tail vein injection. After 6 weeks, the lungs were harvested and fixed with picric acid and subjected to pathological analyses.
Hematoxylin and eosin (H&E) staining
Formalin-fixed xenograft tumors were subjected to paraffin embedding and sectioning (5 μm thickness). The sections were deparaffinized and rehydrated as previously described44 and subsequently stained with hematoxylin solution, followed by counterstaining in eosin-phloxine solution. The images were acquired using a Nikon microscope (Nikon. Japan).
IHC
IHC was conducted as described previously.45 In brief, xenograft tumors were fixed in formalin, followed by paraffin embedding and serial sectioning. A paraffin-embedded section (5 μm in thickness) was then dewaxed, rehydrated, and subjected to epitope retrieval. After blocking, slides were incubated with specific antibody at 4°C overnight, followed by the incubation with secondary antibody. The results were visualized by AEC solution (Invitrogen) and acquired using a Nikon microscope (Nikon instruments).
Statistical analysis
Data are shown as the means ± SD. For comparison between two groups, unpaired two-tailed Student’s t test was used. One-way ANOVA was used for comparison among multiple groups using PRISM (GraphPad, San Diego, CA, USA). p <0.05 was statistically significant.
Availability of data and materials
All data generated or analyzed during this study are included in this article.
Acknowledgments
We would like to give our sincere gratitude to the reviewers for their constructive comments. This study was supported partially by Natural Science Foundation of Hunan Province (2018JJ3808 and 2019JJ40488), National Natural Science Foundation of China 82002098.
Author contributions
J.-B.Y. and B.-Y.F. contributed to the conception of the study and wrote the paper. J.-B.Y., L.G., and L.C. conducted the experiment and collected data. Y.Y. analyzed the data. All authors reviewed and approved the manuscript.
Declaration of interests
The authors declare no competing interests.
References
- 1.Prout G.R., Jr., Wesley M.N., Greenberg R.S., Chen V.W., Brown C.C., Miller A.W., Weinstein R.S., Robboy S.J., Haynes M.A., Blacklow R.S., Edwards B.K. Bladder cancer: race differences in extent of disease at diagnosis. Cancer. 2000;89:1349–1358. doi: 10.1002/1097-0142(20000915)89:6<1349::aid-cncr20>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 2.Sanli O., Dobruch J., Knowles M.A., Burger M., Alemozaffar M., Nielsen M.E., Lotan Y. Bladder cancer. Nat. Rev. Dis. Primers. 2017;3:17022. doi: 10.1038/nrdp.2017.22. [DOI] [PubMed] [Google Scholar]
- 3.Lemke E.A., Shah A.Y. Management of Advanced Bladder Cancer: An Update. J. Adv. Pract. Oncol. 2018;9:410–416. [PMC free article] [PubMed] [Google Scholar]
- 4.Gerke V., Creutz C.E., Moss S.E. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- 5.Kang W.Y., Chen W.T., Huang Y.C., Su Y.C., Chai C.Y. Overexpression of annexin 1 in the development and differentiation of urothelial carcinoma. Kaohsiung J. Med. Sci. 2012;28:145–150. doi: 10.1016/j.kjms.2011.10.004. [DOI] [PubMed] [Google Scholar]
- 6.Christensen M.V., Høgdall C.K., Jochumsen K.M., Høgdall E.V.S. Annexin A2 and cancer: A systematic review. Int. J. Oncol. 2018;52:5–18. doi: 10.3892/ijo.2017.4197. [DOI] [PubMed] [Google Scholar]
- 7.Hauptmann R., Maurer-Fogy I., Krystek E., Bodo G., Andree H., Reutelingsperger C.P. Vascular anticoagulant beta: a novel human Ca2+/phospholipid binding protein that inhibits coagulation and phospholipase A2 activity. Its molecular cloning, expression and comparison with VAC-alpha. Eur. J. Biochem. 1989;185:63–71. doi: 10.1111/j.1432-1033.1989.tb15082.x. [DOI] [PubMed] [Google Scholar]
- 8.Liu J.H., Stass S.A., Chang K.S. Expression of the annexin VIII gene in acute promyelocytic leukemia. Leuk. Lymphoma. 1994;13:381–386. doi: 10.3109/10428199409049626. [DOI] [PubMed] [Google Scholar]
- 9.Stein T., Price K.N., Morris J.S., Heath V.J., Ferrier R.K., Bell A.K., Pringle M.A., Villadsen R., Petersen O.W., Sauter G. Annexin A8 is up-regulated during mouse mammary gland involution and predicts poor survival in breast cancer. Clin. Cancer Res. 2005;11:6872–6879. doi: 10.1158/1078-0432.CCR-05-0547. [DOI] [PubMed] [Google Scholar]
- 10.Pimiento J.M., Chen D.T., Centeno B.A., Davis-Yadley A.H., Husain K., Fulp W.J., Wang C., Zhang A., Malafa M.P. Annexin A8 Is a Prognostic Marker and Potential Therapeutic Target for Pancreatic Cancer. Pancreas. 2015;44:122–127. doi: 10.1097/MPA.0000000000000218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gou R., Zhu L., Zheng M., Guo Q., Hu Y., Li X., Liu J., Lin B. Annexin A8 can serve as potential prognostic biomarker and therapeutic target for ovarian cancer: based on the comprehensive analysis of Annexins. J. Transl. Med. 2019;17:275. doi: 10.1186/s12967-019-2023-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brosnan C.A., Voinnet O. The long and the short of noncoding RNAs. Curr. Opin. Cell Biol. 2009;21:416–425. doi: 10.1016/j.ceb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 13.Schmitt A.M., Chang H.Y. Long Noncoding RNAs in Cancer Pathways. Cancer Cell. 2016;29:452–463. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gugnoni M., Ciarrocchi A. Long Noncoding RNA and Epithelial Mesenchymal Transition in Cancer. Int. J. Mol. Sci. 2019;20:1924. doi: 10.3390/ijms20081924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tay Y., Rinn J., Pandolfi P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmena L., Poliseno L., Tay Y., Kats L., Pandolfi P.P. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146:353–358. doi: 10.1016/j.cell.2011.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou M.X., Huang W., Wang X.B., Lv G.H., Li J., Deng Y.W. Identification of miR-140-3p as a marker associated with poor prognosis in spinal chordoma. Int. J. Clin. Exp. Pathol. 2014;7:4877–4885. [PMC free article] [PubMed] [Google Scholar]
- 18.Kong X.M., Zhang G.H., Huo Y.K., Zhao X.H., Cao D.W., Guo S.F., Li A.M., Zhang X.R. MicroRNA-140-3p inhibits proliferation, migration and invasion of lung cancer cells by targeting ATP6AP2. Int. J. Clin. Exp. Pathol. 2015;8:12845–12852. [PMC free article] [PubMed] [Google Scholar]
- 19.Dong W., Yao C., Teng X., Chai J., Yang X., Li B. MiR-140-3p suppressed cell growth and invasion by downregulating the expression of ATP8A1 in non-small cell lung cancer. Tumour Biol. 2016;37:2973–2985. doi: 10.1007/s13277-015-3452-9. [DOI] [PubMed] [Google Scholar]
- 20.Yamada Y., Kato M., Arai T., Sanada H., Uchida A., Misono S., Sakamoto S., Komiya A., Ichikawa T., Seki N. Aberrantly expressed PLOD1 promotes cancer aggressiveness in bladder cancer: a potential prognostic marker and therapeutic target. Mol. Oncol. 2019;13:1898–1912. doi: 10.1002/1878-0261.12532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han Y., Liu Y., Gui Y., Cai Z. Long intergenic non-coding RNA TUG1 is overexpressed in urothelial carcinoma of the bladder. J. Surg. Oncol. 2013;107:555–559. doi: 10.1002/jso.23264. [DOI] [PubMed] [Google Scholar]
- 22.Tan J., Qiu K., Li M., Liang Y. Double-negative feedback loop between long non-coding RNA TUG1 and miR-145 promotes epithelial to mesenchymal transition and radioresistance in human bladder cancer cells. FEBS Lett. 2015;589(20 Pt B):3175–3181. doi: 10.1016/j.febslet.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 23.Jiang H., Hu X., Zhang H., Li W. Down-regulation of LncRNA TUG1 enhances radiosensitivity in bladder cancer via suppressing HMGB1 expression. Radiat. Oncol. 2017;12:65. doi: 10.1186/s13014-017-0802-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Q., Liu H., Cheng H., Li Y., Li X., Zhu C. Downregulation of long noncoding RNA TUG1 inhibits proliferation and induces apoptosis through the TUG1/miR-142/ZEB2 axis in bladder cancer cells. OncoTargets Ther. 2017;10:2461–2471. doi: 10.2147/OTT.S124595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Terracciano D., Ferro M., Terreri S., Lucarelli G., D’Elia C., Musi G., de Cobelli O., Mirone V., Cimmino A. Urinary long noncoding RNAs in nonmuscle-invasive bladder cancer: new architects in cancer prognostic biomarkers. Transl. Res. 2017;184:108–117. doi: 10.1016/j.trsl.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Mar N., Dayyani F. Management of Urothelial Bladder Cancer in Clinical Practice: Real-World Answers to Difficult Questions. J. Oncol. Pract. 2019;15:421–428. doi: 10.1200/JOP.19.00215. [DOI] [PubMed] [Google Scholar]
- 27.Li J., Yang Z., Zou Q., Yuan Y., Li J., Liang L., Zeng G., Chen S. PKM2 and ACVR 1C are prognostic markers for poor prognosis of gallbladder cancer. Clin. Transl. Oncol. 2014;16:200–207. doi: 10.1007/s12094-013-1063-8. [DOI] [PubMed] [Google Scholar]
- 28.Quan J., Pan X., Zhao L., Li Z., Dai K., Yan F., Liu S., Ma H., Lai Y. LncRNA as a diagnostic and prognostic biomarker in bladder cancer: a systematic review and meta-analysis. OncoTargets Ther. 2018;11:6415–6424. doi: 10.2147/OTT.S167853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su G., He Q., Wang J. Clinical Values of Long Non-coding RNAs in Bladder Cancer: A Systematic Review. Front. Physiol. 2018;9:652. doi: 10.3389/fphys.2018.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Srivastava A.K., Singh P.K., Rath S.K., Dalela D., Goel M.M., Bhatt M.L. Appraisal of diagnostic ability of UCA1 as a biomarker of carcinoma of the urinary bladder. Tumour Biol. 2014;35:11435–11442. doi: 10.1007/s13277-014-2474-z. [DOI] [PubMed] [Google Scholar]
- 31.Wang P., Ning S., Zhang Y., Li R., Ye J., Zhao Z., Zhi H., Wang T., Guo Z., Li X. Identification of lncRNA-associated competing triplets reveals global patterns and prognostic markers for cancer. Nucleic Acids Res. 2015;43:3478–3489. doi: 10.1093/nar/gkv233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang M., Mao C., Ouyang L., Liu Y., Lai W., Liu N., Shi Y., Chen L., Xiao D., Yu F. Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2019;26:2329–2343. doi: 10.1038/s41418-019-0304-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang Y., Song X., Li Y., Chen B., Zhao W., Wang L. LncRNA BCRT1 promotes breast cancer progression by targeting miR-1303/PTBP3 axis. Mol. Cancer. 2020;19:85. doi: 10.1186/s12943-020-01206-5. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Denzler R., McGeary S.E., Title A.C., Agarwal V., Bartel D.P., Stoffel M. Impact of MicroRNA Levels, Target-Site Complementarity, and Cooperativity on Competing Endogenous RNA-Regulated Gene Expression. Mol. Cell. 2016;64:565–579. doi: 10.1016/j.molcel.2016.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D.L., Lu Y.X., Zhang J.X., Wei X.L., Wang F., Zeng Z.L., Pan Z.Z., Yuan Y.F., Wang F.H., Pelicano H. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding J., Yeh C.R., Sun Y., Lin C., Chou J., Ou Z., Chang C., Qi J., Yeh S. Estrogen receptor β promotes renal cell carcinoma progression via regulating LncRNA HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene. 2018;37:5037–5053. doi: 10.1038/s41388-018-0175-6. [DOI] [PubMed] [Google Scholar]
- 37.He Z.Y., Wei T.H., Zhang P.H., Zhou J., Huang X.Y. Long noncoding RNA-antisense noncoding RNA in the INK4 locus accelerates wound healing in diabetes by promoting lymphangiogenesis via regulating miR-181a/Prox1 axis. J. Cell. Physiol. 2019;234:4627–4640. doi: 10.1002/jcp.27260. [DOI] [PubMed] [Google Scholar]
- 38.Liu P., Jia S.B., Shi J.M., Li W.J., Tang L.S., Zhu X.H., Tong P. LncRNA-MALAT1 promotes neovascularization in diabetic retinopathy through regulating miR-125b/VE-cadherin axis. Biosci. Rep. 2019;39 doi: 10.1042/BSR20181469. BSR20181469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fatimathas L., Moss S.E. Annexins as disease modifiers. Histol. Histopathol. 2010;25:527–532. doi: 10.14670/HH-25.527. [DOI] [PubMed] [Google Scholar]
- 40.Hu H., Zhao J., Zhang M. Expression of Annexin A2 and Its Correlation With Drug Resistance and Recurrence of Bladder Cancer. Technol. Cancer Res. Treat. 2016;15:NP61–NP68. doi: 10.1177/1533034615617078. [DOI] [PubMed] [Google Scholar]
- 41.Lee M.J., Yu G.R., Yoo H.J., Kim J.H., Yoon B.I., Choi Y.K., Kim D.G. ANXA8 down-regulation by EGF-FOXO4 signaling is involved in cell scattering and tumor metastasis of cholangiocarcinoma. Gastroenterology. 2009;137:1138–1150, 1150.e1–9. doi: 10.1053/j.gastro.2009.04.015. [DOI] [PubMed] [Google Scholar]
- 42.Xiao P., Liu W., Zhou H. miR-200b inhibits migration and invasion in non-small cell lung cancer cells via targeting FSCN1. Mol. Med. Rep. 2016;14:1835–1840. doi: 10.3892/mmr.2016.5421. [DOI] [PubMed] [Google Scholar]
- 43.Liu L., Zhao X., Zhu X., Zhong Z., Xu R., Wang Z., Cao J., Hou Y. Decreased expression of miR-430 promotes the development of bladder cancer via the upregulation of CXCR7. Mol. Med. Rep. 2013;8:140–146. doi: 10.3892/mmr.2013.1477. [DOI] [PubMed] [Google Scholar]
- 44.Sun W., Li S., Yu Y., Jin H., Xie Q., Hua X., Wang S., Tian Z., Zhang H., Jiang G. MicroRNA-3648 Is Upregulated to Suppress TCF21, Resulting in Promotion of Invasion and Metastasis of Human Bladder Cancer. Mol. Ther. Nucleic Acids. 2019;16:519–530. doi: 10.1016/j.omtn.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B., Shen E.D., Liao M.M., Hu Y.B., Wu K., Yang P., Zhou L., Chen W.D. Expression and mechanisms of long non-coding RNA genes MEG3 and ANRIL in gallbladder cancer. Tumour Biol. 2016;37:9875–9886. doi: 10.1007/s13277-016-4863-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this article.