Abstract
The trillions of microbes that make up the gut microbiome are an important contributor to health and disease. With respect to xenobiotics, particularly orally administered compounds, the gut microbiome interacts directly with drugs to break them down into metabolic products. In addition, microbial products such as bile acids interact with nuclear receptors on host drug-metabolizing enzyme machinery, thus indirectly influencing drug disposition and pharmacokinetics. Gut microbes also influence drugs that undergo enterohepatic recycling by reversing host enzyme metabolic processes and increasing exposure to toxic metabolites as exemplified by the chemotherapy agent irinotecan and non-steroidal anti-inflammatory drugs. Recent data with immune checkpoint inhibitors demonstrate the impact of the gut microbiome on drug pharmacodynamics. We summarize the clinical importance of gut microbe interaction with digoxin, irinotecan, immune checkpoint inhibitors, levodopa, and non-steroidal anti-inflammatory drugs. Understanding the complex interactions of the gut microbiome with xenobiotics is challenging; and highly sensitive methods such as untargeted metabolomics with molecular networking along with other in silico methods and animal and human in vivo studies will uncover mechanisms and pathways. Incorporating the contribution of the gut microbiome to drug disposition, pharmacokinetics, and pharmacodynamics is vital in this era of precision medicine.
Key Points
| The gut microbiome is emerging as an important contributor to drug pharmacokinetics and pharmacodynamics interacting with drugs by directly metabolizing them, indirectly affecting host drug-metabolizing enzymes, and modifying the response to drugs |
| We summarize clinically important interactions between the gut microbiota and several drugs such as digoxin, irinotecan, immune checkpoint inhibitors, levodopa, and non-steroidal anti-inflammatory drugs |
Introduction
The goal of precision medicine is to utilize an individual’s genetic, environmental, and lifestyle characteristics to ensure appropriate drug therapy and disease state management. Understanding the factors that contribute to the variability in pharmacokinetics and pharmacodynamics is paramount. For example, progress has been made in determining genetic variability in drug-metabolizing enzymes, drug transporters, and drug target genes, resulting in clinically actionable guidelines for select drugs [1]. The gut microbiome with its trillions of microbial cells including bacteria, viruses, fungi, and archaea has recently emerged as an important contributor to drug action and variability, particularly with orally administered compounds. Genes encoding organisms in the human gut microbiome in recent estimates number at 232 million [2], far outnumbering human germline genes of ~ 20,000 [3]. More than 90% of the gut microbiota are members of two bacterial phyla, Bacteriodetes and Firmicutes [4]. The enormous inter-patient diversity in human gut microbiomes and inter-related factors such as diet, circadian rhythms, and immune function are significant contributors to variability in drug disposition and response. Additionally, intra-individual variability across time and influences such as diet are also important considerations when determining relationships between the gut microbiome and drugs [5].
There is a bidirectional nature to the interaction between drugs and the microbiome. Antibiotics, particularly those that impact Gram-positive organisms and anaerobes, can profoundly alter the microbial composition. Some evidence suggests that antibiotic use in infants may change the microbiome ontogeny [6–8] and lead to long-term adverse immunological, neurological, and metabolic outcomes [6–8]. In addition, a significant number of non-antibiotic compounds can alter the gut microbiome with up to 240 drugs showing inhibition of at least one bacterial strain in vitro [9]. This may have implications on antibiotic resistance and dysbiosis-induced disease from traditionally categorized non-antibiotic drugs. Conversely, microbes in the gut can modify drugs prior to entering the systemic circulation, thus affecting oral relative bioavailability (F). Microbes and their products also play a role in the gut-liver crosstalk affecting enterohepatic recycling, intestinal and hepatic drug metabolism, and transporters affecting total body drug exposure.
While knowledge of direct microbial drug metabolism has been known, with an azo bond cleavage by colonic bacteria activating the antibiotic sulfasalazine [10], more recent investigations demonstrate that microbes indirectly influence drug action by altering host drug metabolism/transport [11–13]. Microbial products such as bile acids and indoles can alter drug-metabolizing enzyme and transporter activity [14]. Technological advances such as metagenomic sequencing of gut microbes have provided insight into the functions and variability of gut microbes; however, our knowledge is limited and data in humans are evolving. This review focuses on the impact of the gut microbiome on drug disposition and the pharmacokinetics and pharmacodynamics for select drugs. Digoxin, irinotecan, immune checkpoint inhibitors, levodopa, and nonsteroidal anti-inflammatory drugs affected by microbiome alterations with potential clinical impact will be highlighted.
Absorption
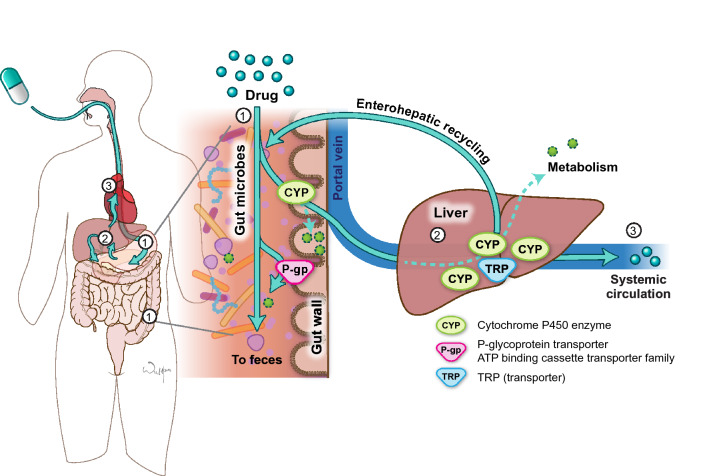
An orally administered compound must clear several hurdles on its way through the gastrointestinal tract into the portal vein and through the liver before ultimately reaching the systemic circulation. These barriers include physicochemical barriers, transporters, metabolizing enzymes, and bacteria (Fig. 1). Bioavailability variability as a result of these factors is a major contributor to therapeutic failure and/or toxicity. Drugs and other orally administered substances may be metabolized directly by bacteria and bacteria may interact with host-metabolizing enzymes and transporters to alter their activity and thus indirectly affect metabolism. Bacteria may metabolize drugs before absorption, after absorption through the intestinal epithelia, or after biliary excretion from the liver, which may then lead to reabsorption of the drug through enterohepatic recycling.
Fig. 1.
Pathway of an orally administered drug. When a drug is administered orally (1), it encounters gut microbes, cytochrome P450 (CYP) enzymes, and transporters (TRP) such as P-glycoprotein (P-gp) in the small and large intestine. Some drug will be lost in the feces in these processes. The drug that survives the small intestine will then travel through the portal vein to the liver where it encounters more CYPs and TRP and more drug may be lost to metabolism (2). Some drugs undergo enterohepatic recycling in which drug conjugates are transported from the liver back to the intestine where they encounter microbes, CYP, and TRP again. The amount of drug that enters the systemic circulation is a fraction of what was originally ingested (3). ATP adenosine triphosphate
Direct Metabolism of Drugs by Bacteria
Bacteria have distinct types of reactions compared to the human host. Bacteria metabolize drugs into more hydrophobic compounds potentially increasing toxicity, while the goal of host metabolism is to metabolize drugs into more hydrophilic compounds decreasing toxicity and facilitating excretion [15]. Modifications performed by bacteria include reduction, hydrolysis, hydroxylation, dihydroxylation, dealkylation, and rarely oxidation. They also can remove functional groups such as N-oxide cleavage, proteolysis, and deconjugation [16]. These bacterial modifications are distinct from those performed by the host cytochrome P450 (CYP) system, which traditionally include N-oxidation and S-oxidation, N-dealkylation and O-dealkylation, aromatic hydroxylation, deamination, and dehalogenation [17].
Gut bacteria directly metabolize a variety of drugs [18]. In many cases, the bacterial species responsible for the drug modification is unknown. In a few cases, extensive investigation has been conducted to determine the species and even strain level. Direct microbe modification of drugs that may be clinically relevant is displayed in Table 1. For example, the cardiac glycoside, digoxin, has been shown to be directly inactivated by the gut microbe Eggerthella lenta, leading to the potential for variability in drug concentrations and toxicity in this narrow therapeutic range drug [19]. Recently, another narrow therapeutic range drug, tacrolimus, an immunosuppressive agent used in transplantation, was shown to be linked to Faecalibacterium prausnitzii. Kidney transplant patients who required higher doses of tacrolimus had increased amounts of Faecalibacterium prausnitzii, a non-motile Gram-positive bacterium present in the gut microbiome [20]. Further investigation showed that incubation of tacrolimus with F. prausnitzii produces a keto-reduction product of tacrolimus that was not found when incubated in hepatic microsomes, suggesting a direct biotransformation by gut microbes [21].
Table 1.
Direct metabolism of drugs by gut microbes associated with pharmacokinetic changes
| Drug | Animal/human | In vivo/in vitro | Organism | Effect on PK (intact gut microbiome) | Comments |
|---|---|---|---|---|---|
| Amlodipine [22] | Both | In vivo | N/A | ↓ F, ↓ Tmax, ↓ Cmax, ↓ AUC | Antibiotics may increase bioavailability by suppressing gut microbial metabolic activities |
| Aspirin [23, 24] | Animal |
Both In vivo |
Lysinibacillus sphaericus |
↓ Cmax and AUC ↓ F |
Reduced gut microbial ASA-metabolizing activity by 67% in rats ↑ F in microbiota-depleted mice |
| Diclofenac acyl glucuronide [25] | Animal | Both | Escherichia coli | ↓ Cmax | E. coli β-glucuronidase catalyzed the deconjugation of diclofenac acyl glucuronides in vitro |
| Digoxin [19] | Both | Both | Eggerthella lenta | ↑ F | E. lenta reduces Cgr 2 → less reduction of digoxin |
| Indomethacin [26] | Human | In vitro | Enterobacteriaceae | ↓ Concentrations | β-glucuronidase expression by human gut Enterobacteriaceae |
| Irinotecan [27] | Both | In vitro | Bacteroides uniformis and E. coli | ↑ Abundance (SN-38) | Gut microbial enzymes promote drug toxicity by hydrolyzing the inactive drug → active drug |
| Levodopa [28–30] | Both | In vivo | Enterococcus faecalis and E. lenta | ↓ Levodopa concentration | Microbes cause less levodopa to be available to cross the blood–brain barrier |
| Lovastatin [31] | Both | In vivo | N/A | May increase Cmax, Tmax, and AUC of M8 | The gut microbiota is involved in the metabolism of lovastatin to its bioactive metabolite (M8) |
| Nabumetone [32] | Animal | Both | E. coli | ↓ AUC, Cmax, and half-life | E. coli converts the NSAID to a reduced pharmacologically inactive metabolite |
| Tacrolimus [20] | Human | In vivo | Faecalibacterium prausnitzii | Positive correlation b/w F. prausnitzii abundance and tacrolimus dose | Study did not explore the potential mechanisms by which F. prausnitzii may have influenced tacrolimus metabolism |
| Tacrolimus [21] | Human | In vitro | F. prausnitzii and Clostridiales order |
Tacrolimus → M1 ↓ F |
F. prausnitzii may metabolize tacrolimus into M1 (five-fold less potent than tacrolimus as an immunosuppressant) |
ASA aspirin, AUC area under the concentration–time curve, b/w between, Cgr2 cardiac glycoside reductase operon, Cmax maximum concentration, F relative bioavailability, M1 tacrolimus metabolite, M8 lovastatin metabolite, N/A not available, NSAID non-steroidal anti-inflammatory drug, PK pharmacokinetics, SN-38 active metabolite of irinotecan, Tmax time to maximum concentration, ↓ decreased
Indirect Metabolism of Drugs by Gut Bacteria
Animal and human studies have provided preliminary evidence that the gut microbiome affects the regulation and activity of metabolizing enzymes. Animal studies have demonstrated that gene expression, protein levels, and activity of drug-metabolizing enzymes are altered in germ-free mice (i.e., mice without a microbiome). CYP3A gene expression was shown to be markedly downregulated in germ-free mice. This was correlated with decreased pregnane X receptor (PXR) binding, a known transcriptional upregulator of CYP3A in the liver [11]. Conventionalization of the germ-free mice restored CYP3A to near-normal levels [12]. Treatment of conventional mice with the quinolone antibiotic ciprofloxacin caused decreased hepatic CYP3A expression and decreased metabolic activity of the CYP3A substrate triazolam; however, no changes were seen when ciprofloxacin was given to germ-free mice [13]. This suggests that microbes or microbial products may bind to nuclear factors such as PXR to downregulate the expression of drug-metabolizing enzymes such as CYP3A.
There are sparse human data but the results are concordant with animal results (Table 2). A study in healthy volunteers showed decreased CYP1A2, CYP2C19, and CYP3A4 activity after a 7-day course of the second-generation cephalosporin, cefprozil [33]. Enzyme activities were measured using a modified Cooperstown cocktail [34] of caffeine for CYP1A2, omeprazole for CYP2C19, and midazolam for CYP3A4. Analysis of the microbial community showed decreased alpha diversity and a correlation between loss of alpha diversity and increased drug and metabolite formation for all three probe compounds [33]. Altering the microbiome with antibiotic therapy modestly decreased enzyme activity, which suggests that a healthy and diverse microbiome may be necessary for optimal functioning of drug-metabolizing enzymes. Future investigations into the mechanism of this effect as well as with other antibiotics will provide additional clinically actionable information.
Table 2.
Indirect metabolism of drugs by gut bacteria associated with pharmacokinetic changes
| Indirect metabolism | ||||||
|---|---|---|---|---|---|---|
| Drug | Animal/human | In vivo/in vitro | Effect on PK (intact gut microbiome) | Enzyme | Diversity | Comments |
| Acetaminophen [35] | Animal | In vivo | ↑ AUC and Cmax | SULT1A1 | N/A | P-cresol competes with acetaminophen binding to SULT1A1 → prevents host from detoxifying acetaminophen |
| Caffeine [33] | Human | In vivo | ↓ CL | CYP1A2 | ↓ α ↑ β | Decreased CYP activity when treated with cefprozil |
| Metformin [36] | Animal | In vivo | ↓ Cmax and ↑ half-life | Oct1 | N/A | Pharmacokinetic changes likely owing to ↓ Oct1 expression in the liver → altered hepatic uptake of metformin in vivo |
| Midazolam [33, 37] |
Animal Human |
In vitro In vivo |
↓ Cmax, AUC, and half-life four-fold ↓ CL |
CYP3A; UGT |
N/A ↓ α ↑ β |
Low levels of CYP3A activity in GF mice decrease drug metabolism in vivo Decreased CYP activity when treated with cefprozil |
| Omeprazole [33] | Human | In vivo | ↓ AUC metabolite ratio | CYP2C19 | ↓ α ↑ β | Decreased CYP activity when treated with cefprozil |
| Progestogens [38] | Human | In vivo | MPA had longest half-life | CYP450 | N/A | Hydroxylation of progestins are likely CYP450 mediated |
| Triazolam [13, 39] | Animal | In vivo | Increased metabolite-to-parent drug ratio in SPF vs GF mice |
CYP3A CYP3A11 CYP3A25 |
N/A |
CYP activity higher for the livers of SPF mice (Bacteroides and Escherichia coli) Ciprofloxacin administration to SPF mice → significant ↓ mRNA expression of CYP3A11 in the liver |
AUC area under the concentration–time curve, CL clearance, Cmax maximum concentration, CYP cytochrome P450, GF germ-free, mRNA messenger RNA, N/A not available, Oct1 organic cation transporter 1, PK pharmacokinetics, SPF specific pathogen-free, SULT1A1 sulfotransferase 1A1, UGT uridine diphosphate glucuronosyltransferase (animal), ↑ increased, ↓ decreased
Enterohepatic Recycling
Enterohepatic recycling occurs when xenobiotics or endogenous substances are absorbed through enterocytes, processed by hepatocytes, then secreted into the bile where they are then reabsorbed by intestinal cells. Enterohepatic recycling can often be accompanied by hepatic conjugation and intestinal deconjugation. This process can occur continuously and results in a longer mean residence time including multiple peaks during a single-dose concentration vs time profile. Many drugs and endogenous substances are modified by phase II enzymes such as UDP glucuronosyltransferases (UGT), which adds a glucuronic acid moiety to make a more water-soluble metabolite that is more easily excreted into urine or bile. These metabolites often undergo enterohepatic recycling, being secreted into the bile and transported back into the intestine. In the intestine, they can encounter bacterial enzymes such as β-glucuronidase, β-glucosidase, demethylase, desulfurase, and other enzymes with phase II reversing activity that cleave off the small molecules such as glucuronide and make them available again for reabsorption.
The clinical implications of gut bacterial involvement in enterohepatic recycling are discussed in Sects. 6.2 and 6.5 for irinotecan and nonsteroidal anti-inflammatory drugs (NSAIDs), respectively. In both cases, gut bacterial enzymes remove the glucuronide moiety from the drug, which causes the drug to become active again and available to exert toxicities such as diarrhea and enteropathy. Variability in gut microbial β-glucuronidase activity, in UGT activity, and antibiotics that reduce β-glucuronidase activity may be factors in whether an individual develops diarrhea or enteropathy with these agents. As more data are developed in humans, giving a β-glucuronidase inhibitor, or a pre-biotic or probiotic, may soon be a reality in order to manipulate the gut microbiota to mitigate undesirable effects resulting from β-glucuronidase activity.
Volume of Distribution
The distribution of a drug within the body is affected by drug properties (e.g., lipophilicity, molecular size) and its interactions with body constituents, including binding to plasma proteins and tissues. The relationship between the apparent volume of distribution, drug binding, and anatomical volumes is given by:
where VP is the plasma volume, VT is the tissue volume, and fu and fuT are the unbound fractions of drug in plasma and tissue, respectively. Additionally, although some drugs can passively distribute throughout body compartments, facilitated movement via transporters often governs distribution to and from various tissues. Transporters may also form physiological barriers such as the blood–brain barrier (BBB) and placental barrier and limit movement of drugs into tissues. Thus, microbiome effects on transporters, tissue binding, and plasma protein binding may alter the distribution of a drug within the body. These effects may have therapeutic consequences, for example, for a drug that must reach the brain to elicit a pharmacologic response.
Microbial-induced changes in plasma proteins have been reported, which could theoretically affect drug distribution. For example, gut bacteria may be related to serum albumin levels. In one study, higher abundance of Sutterella was correlated with lower serum albumin levels in patients with colorectal cancer. However, the mechanistic basis underlying this finding is unclear [40]. Indirect relationships are also possible. To illustrate, phenolic fragments (e.g., hydroxybenzoic, hydroxyacetic and hydroxycinnamic acids, and hydroxybenzenes) are produced from flavonoids by bacterial microflora. Some of these metabolites form stable complexes with albumin in vitro [41] though appear to have limited potential to displace drugs from binding sites. Finally, several “uremic toxins” that are dependent on the presence of gut microflora, such as indoxyl sulfate, hippuric acid, and phenylacetic acid, are also highly bound in plasma to albumin [42, 43].
Relationships between the microbiome and the BBB have been identified. Germ-free mice show increased BBB permeability as compared with mice with a normal gut flora, with reduced expression of tight junction proteins persisting into adulthood [44]. Bacteria and bacterially released factors can reach the systemic circulation and affect immune cells to influence interactions with the BBB [45]. Other mechanisms that have been proposed include an indirect effect on cytokines, which then alters BBB transport sites and overall integrity [45].
Metabolism and Excretion
Determining the differential contributions of the intestine and liver to drug metabolism and excretion can be challenging and the contribution of the microbiome to these routes is emerging. It is increasingly evident that there is significant cross-talk between the intestine and liver and that bile acids, produced in the liver and modified by bacteria in the gut, are important signaling molecules that regulate host metabolism [46]. Bile acids achieve their signaling properties by binding to G-protein-coupled receptors such as the farnesoid X receptor and TGR5 [47]; and binding of bile acids to the farnesoid X receptor modulates CYP3A [48] and transporter activity [49, 50]. Other microbial products such as the secondary bile acid lithocholic acid (LCA), lipopolysaccharides produced from Gram-negative bacteria, and indole-3-propionic acid have also been shown to activate the nuclear receptor, PXR, another nuclear receptor involved in regulating drug metabolism and transport [51]. Animal data support the role of the gut microbiome in modifying host drug metabolism and transport. The protein expression of several CYPs and transporters such as Oatp and Bcrp1 were altered in germ-free and antibiotic-treated mice [52]; and ciprofloxacin-treated mice had significantly reduced LCA-producing bacteria in their feces. In germ-free mice given LCA, CYP3A expression was significantly elevated suggesting that LCA activated farnesoid X receptor and PXR [13]. Hepatic CYP3A and the activity of the CYP3A substrate midazolam were significantly lower in germ-free mice compared with conventional mice, suggesting that gut microbes may alter the metabolic activity of CYP3A [37].
One example of gut microbes altering host liver metabolism is with the analgesic acetaminophen (Table 2). Acetaminophen undergoes glucuronidation and bacterial glucuronidases can deconjugate the glucuronide metabolite allowing for reabsorption of the parent acetaminophen or further metabolism to sulfate and/or glucuronide conjugates. With antibiotic treatment, there is a decrease in the sulfate conjugate of acetaminophen [35]. In addition, gut bacteria produce a metabolite of aromatic amino acid metabolism, p-cresol, that competes with acetaminophen for binding to the enzyme sulfotransferase 1A1. Individuals who produce high levels of p-cresol were shown to have a low capacity for sulfonate acetaminophen [53]. Therefore, antibiotic therapy and/or high levels of the bacterially derived metabolite p-cresol could predispose individuals to the hepatotoxic effects of acetaminophen.
Drugs Affected by Microbiome Alterations with Clinical Significance
Digoxin
Digoxin is a cardiac glycoside for the treatment of atrial fibrillation and congestive heart failure [54]. Digoxin is a positive inotropic drug that inhibits the Na+/K+-ATPase pump, resulting in increased intracellular calcium in cardiac myocytes [55]. The narrow therapeutic window (target concentration range 0.5–2 mcg/L) of digoxin requires therapeutic drug monitoring [56]. Digoxin relative F is influenced by the formulation, with higher bioavailability in capsule formulations compared with tablets [57]. Digoxin F can also be influenced by malabsorption syndromes, gastrointestinal motility, and drug–drug or drug–food interactions [58]. Digoxin is a substrate for P-glycoprotein (P-gp) [59] and P-gp genetic polymorphisms impact digoxin pharmacokinetic (PK) variability as the partial area under the concentration–time curve (AUC) increased in subjects with the MDR1 3435TT genotype vs the MDR1 3435CC genotype (p ≤ 0.05) [60]. Conflicting evidence exists and suggests no difference in digoxin partial AUC across MDR1 3435TT, CC, and CT genotypes [61, 62]. Epigenetic effects, via methylation of the ABCB1 promoter region, also impact digoxin PK variability. Subjects with a high methylated epigenetic profile (n = 15) had higher digoxin partial AUC0–4 (5.12 ± 1.42 vs 4.31 ± 1.03 ng*h/mL, p ≤ 0.05) and Cmax (2.49 ± 0.18 vs 1.92 ± 0.26 ng/mL, p ≤ 0.05) compared with subjects with a low methylated epigenetic profile (n = 15) [63].
Studies using various digoxin formulations provide evidence of the effect of Eggerthella lenta (E. lenta, previously named Eubacterium lentum) on urinary digoxin and digoxin reduction products (DRP) excretion. Digoxin absorption is proportional not only to exposure but to urinary digoxin excretion [58]. Urinary DRP excretion varies inversely with oral digoxin F [64, 65]. In healthy adults (n = 4) administered an oral tablet digoxin 0.25 mg once daily, DRP urinary excretion (described as a percentage of total digoxin and DRP excretion) was 45–80% compared to elixir and intravenous formulations that had lower DRP of 20–40%. Upon co-administration of a 5-day course of erythromycin or tetracycline antibiotics, DRP urinary concentrations and DRP excretion percentages dramatically reduced while digoxin serum concentrations increased [64]. In another study, healthy subjects (n = 22) received 0.4 mg of oral digoxin formulated as an encapsulated liquid or a tablet [65]. Mean cumulative digoxin urinary excretion was higher with the encapsulated liquid compared with the tablet (195 ± 8.6 vs 137.5 ± 6.3 mcg). In contrast, DRP urinary excretion (60.8 ± 5.5 vs 102.7 ± 9.5 mcg) and percentage DRP (23.5 ± 1.8% vs 41.2 ± 2.7%) was also lower with the encapsulated liquid compared with the tablet [65]. These results have been confirmed elsewhere [66].
Eggerthella lenta was identified as the bacteria that metabolized digoxin to an inactive reduced dihydrodigoxin metabolite in human gut flora [67, 68]. However, the presence as well as concentrations of E. lenta in the gut flora did not always correlate with DRP production [68]. Haiser et al. [19] later determined that a two-gene cytochrome-encoding operon (now referred to as cardiac glycoside reductase) was upregulated > 100-fold in the presence of digoxin in certain E. lenta strains.
Conditions that decrease and/or eliminate E. lenta activity may have clinical implications given the narrow therapeutic window of digoxin and target concentration range. Exposure to antibiotics during co-administration of digoxin is one example of a microbiome–drug interaction. Studies provide evidence of such an effect [64, 69] whereby eliminating E. lenta results in little to no urinary DRP formation. One would then expect increased digoxin F and increased systemic concentrations, which may impact the target concentration range. Other groups have speculated that diet may be clinically impactful given in vitro and in vivo animal studies support that E. lenta exposure to arginine decreased cardiac glycoside reductase operon expression and prevented the conversion of digoxin to dihydrodigoxin [19]. Monitoring of an individual’s dietary protein intake may be needed during digoxin therapy as increased consumption of protein-rich foods, which contain arginine, would inhibit E. lenta-mediated digoxin reduction resulting in increased digoxin F.
Irinotecan
Irinotecan in combination with other agents is indicated for gastrointestinal carcinomas and small cell lung cancer. Irinotecan blocks DNA replication by inhibiting topoisomerase [70]. In hepatocytes, irinotecan is metabolized by carboxylesterase to SN-38, which is an active metabolite. SN-38 is then metabolized by UGT to form inactive SN-38G. Biliary excretion removes SN-38G into the intestinal lumen [70]. In Caucasian patients with cancer (n = 30), the UGT1A1*28 polymorphism and CYP3A4 phenotype (as measured by midazolam clearance) were statistically significant variables associated with irinotecan pharmacokinetics [71]. Midazolam clearance varied approximately four-fold during irinotecan therapy. Patients with the UGT1A1*28 polymorphism had higher SN-38 AUCs compared with patients without the UGT1A1*28 polymorphism [71].
Irinotecan-related toxicities such as neutropenia and diarrhea are dose limiting, potentially life threatening, and can be partially attributed to SN-38. Escherichia coli is a pathogen that produces β-glucuronidase, which converts SN-38G back to SN-38 in the intestinal lumen. Consequently, higher SN-38 intestinal lumen concentrations may increase the risk of diarrhea and localized enteric injury [72, 73] (Table 3). One animal study provided an initial glimpse of potential clinical implications. In this study, oral administration of a bacterial β-glucuronidase inhibitor protected mice from irinotecan-related toxicity [73], thus suggesting specificity of the β-glucuronidase inhibitor against bacterial, but not against mammalian-specific cells.
Table 3.
Gut bacteria associated with pharmacodynamic effects of drugs
| Drug | Animal/human | In vivo/in vitro | Organism or phyla | Effect on PD | Comments |
|---|---|---|---|---|---|
| Anti-PD-1 therapy [74] | Both | In vivo | Faecalibacterium and Ruminococcaceae | ↑ Systemic and anti-tumor immunity | Enriched gut microbiome increased antigen presentation and improved effector T-cell function in the periphery and the tumor environment |
| Ipilimumab [75] | Both | In vivo | B. fragilis and/or B. thetaiotaomicron and Burkholderiales | ↓ Tumor size | Antitumor effects of the CTLA-4 monoclonal antibody, ipilimumab depend on Bacteroides spp. |
| Irinotecan [27, 76] | Both | In vitro | B. uniformis and Escherichia coli | ↑ GI toxicity | Variability of GI toxicity by anti-cancer drug, irinotecan, is a result of differences in types and levels of gut bacterial β-glucuronidases |
| NSAIDs [77, 78] | Both | In vivo | β-Glucoronidases | Enteropathy | Various gut microbes have β-glucuronidases that de-glucuronidate NSAIDs in the intestine causing increased susceptibility for enteropathy |
| Metformin [79, 80] | Both | In vivo |
E. coli Intestinibacter |
Regulation of glucose homeostasis | Metformin-treated microbiota transferred to germ-free mice improved glucose metabolism |
| Methotrexate [81] | Human | In vivo | Higher Firmicutes/Bacteroidetes ratio in patients with RA who were non-responsive to methotrexate | Response to methotrexate in patients with RA | Methotrexate-responsive patients had lower microbial diversity; methotrexate responders vs non-responders differed significantly in microbial gene abundance reflecting different microbial pathways |
CTLA-4 cytotoxic lymphocyte antigen 4, GI gastrointestinal, NSAID non-steroidal anti-inflammatory drug, PD pharmacodynamics, PD-1 programmed cell death receptor, RA rheumatoid arthritis, ↑ increased, ↓ decreased
ICIs
Immune checkpoint inhibitors (ICIs) are indicated for a variety of solid tumor and hematological malignancies and induce an immune response by suppressing pathways involved in the negative regulation of the immune system. On the surface of T lymphocytes, cemiplimab, nivolumab, and pembrolizumab bind to programmed death receptor 1 (PD-1), while ipilimumab binds to the cytotoxic lymphocyte antigen 4 (CTLA-4) receptor. Atezolizumab, avelumab, and durvalumab target PD-1 ligands (PD-L1). Population PK analyses have identified intrinsic and extrinsic covariates having a modest effect on ICI PK variability. For most ICIs, statistically significant covariates on clearance include sex, body weight, estimated glomerular filtration rate, and immunogenicity [82–84]. Some have suggested that the modest influence of sex, renal function, and hepatic impairment on ICI clearance is due to various physiological mechanisms involved in clearance for monoclonal antibodies, specifically proteolytic catabolism in plasma and peripheral tissues and receptor-mediated endocytosis via target-mediated drug disposition [85, 86].
Gut/intestinal microbiome composition affects ICI activity, efficacy, and toxicity. Intestinal recolonization of antibiotic-treated or germ-free mice with a combination of Bacteriodes fragilis, B. thetaiotaomicron, and Burkholderia cepacia restores the cytotoxic lymphocyte antigen 4-mediated anticancer response via induction of the interleukin-12-dependent, T-helper-1 immune response [87] (Table 3). In another mouse study, oral administration of Bifidobacterium species restored anti-PD-L1 antitumor activity resulting in dendritic cell maturation and increasing tumor-specific T-cell activity [88]. In humans, Faecalibacterium genus [89] and Akkermansia muciniphila [90] impact ICI activity and/or efficacy. In one study, patients with melanoma receiving anti-PD-1 immunotherapy (n = 112) were separated into responders (e.g., complete or partial response or stable disease for at least 6 months, n = 30) and non-responders (e.g., disease progression or stable disease less than 6 months, n = 13) [74]. An abundance of Faecalibacterium was observed in responders and was a strong predictor to anti-PD-L1 immunotherapy (hazard ratio = 2.92, 95% confidence interval 1.08–7.89). In another study, patients with metastatic melanoma treated with ipilimumab and whom had Faecalibacterium and other Firmicutes phylum composition (n = 12) had increased progression-free survival (PFS) and increased overall survival [89].
Immune checkpoint inhibitor activity and/or efficacy is also affected by microbiome diversity. Higher microbial fecal diversity was observed in patients with melanoma who responded to anti-PD-1 immunotherapy [74]. In other studies, antibiotic exposure resulting in a loss of microbial diversity (dysbiosis) decreased PFS and OS in patients with cancer receiving ICI immunotherapy [90, 91]. Derosa et al. [91] retrospectively analyzed patients with advanced renal cell carcinoma (RCC, n = 121) and patients with non-small-cell lung cancer (n = 239) receiving antibiotic therapy within 30 days (for oral) or 60 days (for intravenous administration) prior to starting PD-1/PD-L1 alone or in combination. An increased rate of primary progressive disease (75% vs 33%, p < 0.01) was observed in patients with RCC receiving antibiotic therapy (n = 16). Median PFS (1.9 vs 7.4 months, hazard ratio = 3.1, 95% confidence interval 1.4–6.9, p < 0.05) and median OS (17.3 vs 30.6 months, hazard ratio = 3.5, 95% confidence interval 1.1–10.8, p < 0.05) were also shorter compared with patients with RCC who were not receiving antibiotic therapy (n = 105). Progression-free survival and OS were also significantly shorter in patients with non-small-cell lung cancer receiving antibiotic therapy. The results of a shorter PFS and OS are consistent with another study in patients with non-small-cell lung cancer, RCC, and urothelial carcinoma receiving antibiotic therapy before or during PD-1/PD-L1 immunotherapy [90].
L-dopa
Levodopa (L-dopa) plus carbidopa is indicated to treat motor symptoms of Parkinson’s disease such as tremors, stiffness, and gait. Levodopa is a prodrug requiring passage through the BBB and metabolism to dopamine by the human enzyme aromatic amino acid. Carbidopa is an aromatic amino acid inhibitor and is given in combination to reduce peripheral (non-central nervous system) metabolism of L-dopa to dopamine. Peripheral dopamine is unable to cross the BBB and is believed to mediate side effects [92]. Upon oral administration, there is known L-dopa serum concentration variability potentially impacting the pharmacodynamic and/or therapeutic response in patients with Parkinson’s disease [93]. There is suggestion that Helicobacter pylori infection affects L-dopa F via duodenal mucosa disruption [94]. There is limited supportive evidence from studies that upon H. pylori eradication in patients with Parkinson’s disease, L-dopa absorption increased and was associated with an improvement in motor symptoms [95, 96].
In 2019, a gut bacterial pathway involved in L-dopa metabolism was discovered [28, 29]. This pathway is distinct from the aforementioned host aromatic amino acid-mediated pathway of L-dopa metabolism. Enterococcus faecalis was identified as the strain that possessed a conserved tyrosine decarboxylase with the ability to metabolize L-dopa to dopamine. Eggerthella lenta was also identified as the strain mediating metabolism of dopamine to m-tyramine. In addition, carbidopa had little impact on the tyrosine decarboxylase-mediated E. faecalis pathway [28]. In October 2020, Clostridium sporogenes was identified in deaminating L-dopa in the gut whereby the 3-(3,4-dihydroxyphenol)propionic acid metabolite formed inhibits ileal motility in an ex vivo model. In addition, stool samples of patients with Parkinson’s disease receiving L-dopa therapy contain this metabolite, thus suggesting active production by the gut microbiota [97]. The potential clinical impact is increased L-dopa dosage requirements in patients with Parkinson’s disease given that several strains metabolize L-dopa prior to reaching the BBB for penetration into the brain [26].
NSAIDs
Nonsteroidal anti-inflammatory drugs are used to treat pain, inflammation, and fever. Non-steroidal anti-inflammatory drugs inhibit the enzyme cyclooxygenase, which is involved in the breakdown of arachidonic acid into prostaglandin, prostacyclin, and thromboxane. The majority of NSAIDs (e.g., aspirin, ibuprofen, indomethacin, and naproxen) are non-selective cyclooxygenase inhibitors, while celecoxib is a selective cyclooxygenase-2 inhibitor. Age, sex, and disease state contribute to NSAID PK variability [98–100]. In addition, CYP2C9 genetic polymorphisms are also another contributory factor whereby meloxicam AUC was 2.4-fold higher in CYP2C9*1/*13 vs CYP2C9*1/*1 genotyped adults [100].
Non-steroidal anti-inflammatory drugs are glucuronidated in the liver and undergo enterohepatic circulation. In the intestine, NSAID-glucuronide conjugates are cleaved by microbiome-encoded β-glucuronidases resulting in reformation of NSAIDs in the parent form and increased aglycones. In theory, reformed NSAIDs and aglycones in the intestine contribute to mucosa damage and enteropathy. Evidence to support this mechanism is from a mice study, whereby β-glucuronidase inhibition protected against NSAID-induced enteropathy by indomethacin, ketoprofen, and/or diclofenac [77]. As discussed earlier with irinotecan, an area of potential clinical implication and/or development is to target bacterial β-glucuronidase inhibition to minimize NSAID-induced enteropathy. In addition, restoration of an altered intestinal mucosa caused by NSAIDs is another area of clinical intervention. Supplementation with probiotics such as several Lactobacillus strains, as well as use of a mucosal protective agent have shown decreased NSAID-associated small intestinal injury and inflammation in humans [78, 101–103].
What is the Future for Understanding Complex Drug–Microbe Interactions?
Modern and emerging chemical analysis methods, such as untargeted mass spectrometry-based metabolomics, are crucial tools necessary to untangle the complex influence of the gut microbiome on drug metabolism. Mass spectrometry is an ideal complement, revealing the chemical transformations and chemical changes associated with the gut microbiota, to genetic sequencing (e.g., next-generation sequencing such as 16S rRNA sequencing, shotgun metagenomic sequencing, microbial metatranscriptomics), which measures what microbes are present and measures alterations at the genetic level. Integration of these techniques will provide immensely different insights than currently possible.
One challenge, specific to mass spectrometry, is the amount of data which cannot be assigned a chemical name. While the annotation rate varies by experiment and sample type, it is not uncommon for the vast majority (> 90%) of all possible chemicals detected to go unannotated. The ability to detect and annotate parent drugs is strikingly good compared to other classes of chemicals as it is often relatively easy to purchase or acquire a genuine chemical standard. Conversely, drug metabolites are often poorly annotated, while generally amenable to detection, owing to the difficulty in acquiring genuine chemical standards. In silico methods to predict and identify metabolites (e.g., BioTransformer) [104] are likely to help overcome some of these aspects. Another data analysis solution is to utilize molecular networking [105], which aims to connect structurally related chemicals via similarity in mass spectrometry/mass spectrometry fragmentation. It is not uncommon for a parent drug to be connected to a desmethyl metabolite as well as other related unannotated compounds. A recently reported approach is to use a repository-scale data analysis as illustrated in Jarmusch et al. [106] in which clindamycin metabolites were connected via molecular networking of mass spectrometry/mass spectrometry data across multiple disparate metabolomics datasets. Regardless of the approaches, substantial progress in the ability to annotate chemicals in mass spectrometry data will enhance our understanding of microbial influence on drug metabolism. When elucidating interactions, it helps to have a chemical identification to connect with genetic sequencing information.
The most pronounced challenge is the difficulty in integrating microbiome and mass spectrometry (e.g., metabolomics) data in order to derive meaningful insights. Experimentally, one can explore the interactions by the quantitation of parent and known metabolites in multiple matrices, such as blood and feces, and model the contribution as reported by Zimmermann et al. [107] Computationally, there have been a few reported methods specifically intended to address this challenge that go beyond correlation measures such as Pearson, Spearman, and Kendall. Note, that correlation methods can be used to color molecular networks to enhance interpretation [108]. Microbe-metabolite vectors (mmvec), reported in Morton et al. [109] utilize the probability of metabolites and microbes co-occurring rather than a measure of correlation. The illustrative use reported in that paper connected Pseudomonas aeruginosa-associated molecules detected by mass spectrometry with taxa assignments from sequencing including P. aeruginosa. Certainly, one could imagine an analogous use in the study of the co-occurrence between drug metabolites and microbes. Last, visualization approaches, procrustes analysis [110], and molecular cartography [111, 112], will help link microbes to chemistry and inform their influences. In summary, one must carefully review the results and subsequent validation experiments should be the norm as comparisons between microbiome and metabolomics are bound to contain false discoveries.
Conclusions
The role of the gut microbiome and its clinical impact on pharmacokinetics and pharmacodynamics is still evolving. While there are examples of direct metabolism by gut microbes affecting drug pharmacokinetics, questions remain such as the specific species and strains involved, the redundancy and variability of the microbial community to metabolize these drugs, as well as other influences such as diet, other drugs, immunity, and circadian rhythms that may affect these activities. There is also evidence of indirect effects of the gut microbiome on drug metabolism involving gut microbial products such as bile acids interacting with host drug-metabolizing machinery. Further studies in humans to determine the clinical significance of these interactions as well as animal and/or in silico studies to investigate the mechanisms behind these effects are paramount. We have reviewed several examples of compounds in which alterations in gut microbial taxa can cause significant perturbations in pharmacokinetics and/or pharmacodynamics. The importance of assessing the role that the gut microbiome plays in the variability of xenobiotic metabolism and the resulting clinical effect in humans cannot be underestimated. As with other factors influencing individual variability, accounting for the influence of the microbiome is even more critical with narrow therapeutic range drugs. Currently, the available data on the gut microbiome’s influence on drug pharmacokinetics and pharmacodynamics are not robust enough to translate to clinically actionable guidance. As the field advances, we anticipate that the gut microbiome’s impact and the variability within and between individuals will be an important component in addition to genetics, diet, and other drugs in determining dose and response to drugs. Studying this highly variable and complex system will require a multi-pronged approach with animal, human, and systems biology models. There are immense challenges that remain in understanding the impact of the microbiome on drug metabolism; however, highly sensitive techniques such as mass spectrometry coupled with advanced in silico methods will certainly play a future role in revealing direct chemical transformation performed by microbes as well as the microbiota’s indirect influences.
Declarations
Funding
No funding was received for the preparation of this article.
Conflicts of interest
Shirley M. Tsunoda, Christopher Gonzales, Alan K. Jarmusch, Jeremiah D. Momper, and Joseph D. Ma have no conflicts of interest that are directly relevant to the content of this article.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Availability of data and material
Not applicable.
Code availability
Code available on reasonable request.
Authors' contributions
SMT, AKJ, JDM, and JDM wrote and edited the manuscript. CG created the tables in the manuscript.
References
- 1.Relling MV, Klein TE. CPIC: clinical pharmacogenetics implementation consortium of the pharmacogenomics research network. Clin Pharmacol Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tierney BT, Yang Z, Luber JM, Beaudin M, Wibowo MC, Baek C, et al. The landscape of genetic content in the gut and oral human microbiome. Cell Host Microbe. 2019;26(283–95):e8. doi: 10.1016/j.chom.2019.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449:804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flores GE, Caporaso JG, Henley JB, Rideout JR, Domogala D, Chase J, et al. Temporal variability is a personalized feature of the human microbiome. Genome Biol. 2014;15:531. doi: 10.1186/s13059-014-0531-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koleva PT, Bridgman SL, Kozyrskyj AL. The infant gut microbiome: evidence for obesity risk and dietary intervention. Nutrients. 2015;7:2237–2260. doi: 10.3390/nu7042237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cho I, Yamanishi S, Cox L, Methé BA, Zavadil J, Li K, et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature. 2012;488:621–626. doi: 10.1038/nature11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nobel YR, Cox LM, Kirigin FF, Bokulich NA, Yamanishi S, Teitler I, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun. 2015;6:7486. doi: 10.1038/ncomms8486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sousa T, Yadav V, Zann V, Borde A, Abrahamsson B, Basit AW. On the colonic bacterial metabolism of azo-bonded prodrugs of 5-aminosalicylic acid. J Pharm Sci. 2014;103:3171–3175. doi: 10.1002/jps.24103. [DOI] [PubMed] [Google Scholar]
- 11.Björkholm B, Bok CM, Lundin A, Rafter J, Hibberd ML, Pettersson S. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE. 2009;4:e6958. doi: 10.1371/journal.pone.0006958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Selwyn FP, Cheng SL, Klaassen CD, Cui JY. Regulation of hepatic drug-metabolizing enzymes in germ-free mice by conventionalization and probiotics. Drug Metab Dispos. 2016;44:262–274. doi: 10.1124/dmd.115.067504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toda T, Ohi K, Kudo T, Yoshida T, Ikarashi N, Ito K, et al. Ciprofloxacin suppresses Cyp3a in mouse liver by reducing lithocholic acid-producing intestinal flora. Drug Metab Pharmacokinet. 2009;24:201–208. doi: 10.2133/dmpk.24.201. [DOI] [PubMed] [Google Scholar]
- 14.Staudinger JL, Woody S, Sun M, Cui W. Nuclear-receptor-mediated regulation of drug- and bile-acid-transporter proteins in gut and liver. Drug Metab Rev. 2013;45:48–59. doi: 10.3109/03602532.2012.748793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheweita SA. Drug-metabolizing enzymes: mechanisms and functions. Curr Drug Metab. 2000;1:107–132. doi: 10.2174/1389200003339117. [DOI] [PubMed] [Google Scholar]
- 16.Sousa T, Paterson R, Moore V, Carlsson A, Abrahamsson B, Basit AW. The gastrointestinal microbiota as a site for the biotransformation of drugs. Int J Pharm. 2008;363:1–25. doi: 10.1016/j.ijpharm.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 17.Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138:103–141. doi: 10.1016/j.pharmthera.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 18.Haiser HJ, Turnbaugh PJ. Developing a metagenomic view of xenobiotic metabolism. Pharmacol Res. 2013;69:21–31. doi: 10.1016/j.phrs.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP, Turnbaugh PJ. Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science. 2013;341(6143):295–298. doi: 10.1126/science.1235872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JR, Muthukumar T, Dadhania D, Taur Y, Jenq RR, Toussaint NC, et al. Gut microbiota and tacrolimus dosing in kidney transplantation. PLoS ONE. 2015;10:e0122399. doi: 10.1371/journal.pone.0122399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Y, Crnkovic CM, Won K-J, Yang X, Lee JR, Orjala J, et al. Commensal gut bacteria convert the tmmunosuppressant tacrolimus to less potent metabolites. Drug Metab Dispos. 2019;47:194–202. doi: 10.1124/dmd.118.084772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo HH, Kim IS, Yoo D-H, Kim D-H. Effects of orally administered antibiotics on the bioavailability of amlodipine: gut microbiota-mediated drug interaction. J Hypertens. 2016;34:156–162. doi: 10.1097/HJH.0000000000000773. [DOI] [PubMed] [Google Scholar]
- 23.Kim IS, Yoo D-H, Jung I-H, Lim S, Jeong J-J, Kim K-A, et al. Reduced metabolic activity of gut microbiota by antibiotics can potentiate the antithrombotic effect of aspirin. Biochem Pharmacol. 2016;122:72–79. doi: 10.1016/j.bcp.2016.09.023. [DOI] [PubMed] [Google Scholar]
- 24.Zhao R, Coker OO, Wu J, Zhou Y, Zhao L, Nakatsu G, et al. Aspirin reduces colorectal tumor development in mice and gut microbes reduce its bioavailability and chemopreventive effects. Gastroenterology. 2020;159(969–83):e4. doi: 10.1053/j.gastro.2020.05.004. [DOI] [PubMed] [Google Scholar]
- 25.LoGuidice A, Wallace BD, Bendel L, Redinbo MR, Boelsterli UA. Pharmacologic targeting of bacterial β-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J Pharmacol Exp Ther. 2012;341:447–454. doi: 10.1124/jpet.111.191122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little MS, Pellock SJ, Walton WG, Tripathy A, Redinbo MR. Structural basis for the regulation of β-glucuronidase expression by human gut Enterobacteriaceae. Proc Natl Acad Sci USA. 2018;115:E152–E161. doi: 10.1073/pnas.1716241115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jariwala PB, Pellock SJ, Goldfarb D, Cloer EW, Artola M, Simpson JB, et al. Discovering the microbial enzymes driving drug toxicity with activity-based protein profiling. ACS Chem Biol. 2020;15:217–225. doi: 10.1021/acschembio.9b00788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maini Rekdal V, Bess EN, Bisanz JE, Turnbaugh PJ, Balskus EP. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364(6445):eaau6323. doi: 10.1126/science.aau6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Kessel SP, Frye AK, El-Gendy AO, Castejon M, Keshavarzian A, van Dijk G, et al. Gut bacterial tyrosine decarboxylases restrict the bioavailability of levodopa, the primary treatment in Parkinson’s disease. Nat Commun. 2019;10:310. doi: 10.1038/s41467-019-08294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulroy E, Bhatia KP. The gut microbiome: a therapeutically targetable site of peripheral levodopa metabolism. Mov Disord Clin Pract. 2019;6:547–548. doi: 10.1002/mdc3.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoo D-H, Kim IS, Van Le TK, Jung I-H, Yoo HH, Kim D-H. Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug Metab Dispos. 2014;42:1508–1513. doi: 10.1124/dmd.114.058354. [DOI] [PubMed] [Google Scholar]
- 32.Jourova L, Anzenbacher P, Matuskova Z, Vecera R, Strojil J, Kolar M, et al. Gut microbiota metabolizes nabumetone in vitro: consequences for its bioavailability in vivo in the rodents with altered gut microbiome. Xenobiotica. 2019;49:1296–1302. doi: 10.1080/00498254.2018.1558310. [DOI] [PubMed] [Google Scholar]
- 33.Jarmusch AK, Vrbanac A, Momper JD, Ma JD, Alhaja M, Liyanage M, et al. Enhanced characterization of drug metabolism and the influence of the intestinal microbiome: a pharmacokinetic, microbiome, and untargeted metabolomics study. Clin Transl Sci. 2020;13:972–984. doi: 10.1111/cts.12785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streetman DS, Bleakley JF, Kim JS, Nafziger AN, Leeder JS, Gaedigk A, et al. Combined phenotypic assessment of CYP1A2, CYP2C19, CYP2D6, CYP3A, N-acetyltransferase-2, and xanthine oxidase with the “Cooperstown cocktail”. Clin Pharmacol Ther. 2000;68:375–383. doi: 10.1067/mcp.2000.109519. [DOI] [PubMed] [Google Scholar]
- 35.Malfatti MA, Kuhn EA, Murugesh DK, Mendez ME, Hum N, Thissen JB, et al. Manipulation of the gut microbiome alters acetaminophen biodisposition in mice. Sci Rep. 2020;10:4571. doi: 10.1038/s41598-020-60982-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu B, Chen M, Gao Y, Hu J, Liu M, Zhang W, et al. In vivo pharmacodynamic and pharmacokinetic effects of metformin mediated by the gut microbiota in rats. Life Sci. 2019;226:185–192. doi: 10.1016/j.lfs.2019.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Togao M, Kawakami K, Otsuka J, Wagai G, Ohta-Takada Y, Kado S. Effects of gut microbiota on in vivo metabolism and tissue accumulation of cytochrome P450 3A metabolized drug: midazolam. Biopharm Drug Dispos. 2020;41:275–282. doi: 10.1002/bdd.2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coombes Z, Yadav V, McCoubrey LE, Freire C, Basit AW, Conlan RS, et al. Progestogens are metabolized by the gut microbiota: implications for colonic drug delivery. Pharmaceutics. 2020;12(8):760. doi: 10.3390/pharmaceutics12080760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Toda T, Saito N, Ikarashi N, Ito K, Yamamoto M, Ishige A, et al. Intestinal flora induces the expression of Cyp3a in the mouse liver. Xenobiotica. 2009;39:323–334. doi: 10.1080/00498250802651984. [DOI] [PubMed] [Google Scholar]
- 40.Shuwen H, Wei W, Miao D, Qing Z, Jiamin X, Jing Z, et al. Gut microbes and microbial metabolites in colorectal cancer complicated with different serum albumin levels. Research Square. 2020. https://www.researchsquare.com/article/rs-33532/v1. Accessed 28 Apr 2021.
- 41.Mohos V, Fliszár-Nyúl E, Lemli B, Zsidó BZ, Hetényi C, Mladěnka P, et al. Testing the pharmacokinetic interactions of 24 colonic flavonoid metabolites with human serum albumin and cytochrome P450 enzymes. Biomolecules. 2020;10:409. 10.3390/biom10030409. [DOI] [PMC free article] [PubMed]
- 42.Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, et al. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc Natl Acad Sci USA. 2009;106:3698–3703. doi: 10.1073/pnas.0812874106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramezani A, Massy ZA, Meijers B, Evenepoel P, Vanholder R, Raj DS. Role of the gut microbiome in uremia: a potential therapeutic target. Am J Kidney Dis. 2016;67:483–498. doi: 10.1053/j.ajkd.2015.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braniste V, Al-Asmakh M, Kowal C, Anuar F, Abbaspour A, Tóth M, et al. The gut microbiota influences blood-brain barrier permeability in mice. Sci Transl Med. 2014;6:263ra158. doi: 10.1126/scitranslmed.3009759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Logsdon AF, Erickson MA, Rhea EM, Salameh TS, Banks WA. Gut reactions: how the blood-brain barrier connects the microbiome and the brain. Exp Biol Med. 2018;243:159–165. doi: 10.1177/1535370217743766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wahlström A, Kovatcheva-Datchary P, Ståhlman M, Bäckhed F, Marschall H-U. Crosstalk between bile acids and gut microbiota and its impact on farnesoid X receptor signalling. Digest Dis. 2017;35(3):246–250. doi: 10.1159/000450982. [DOI] [PubMed] [Google Scholar]
- 47.Pols TWH, Noriega LG, Nomura M, Auwerx J, Schoonjans K. The bile acid membrane receptor TGR5: a valuable metabolic target. Dig Dis. 2011;29:37–44. doi: 10.1159/000324126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gnerre C, Blättler S, Kaufmann MR, Looser R, Meyer UA. Regulation of CYP3A4 by the bile acid receptor FXR: evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics. 2004;14:635–645. doi: 10.1097/00008571-200410000-00001. [DOI] [PubMed] [Google Scholar]
- 49.Ananthanarayanan M, Balasubramanian N, Makishima M, Mangelsdorf DJ, Suchy FJ. Human bile salt export pump promoter is transactivated by the farnesoid X receptor/bile acid receptor. J Biol Chem. 2001;276:28857–28865. doi: 10.1074/jbc.M011610200. [DOI] [PubMed] [Google Scholar]
- 50.Plass JRM, Mol O, Heegsma J, Geuken M, Faber KN, Jansen PLM, et al. Farnesoid X receptor and bile salts are involved in transcriptional regulation of the gene encoding the human bile salt export pump. Hepatology. 2002;35:589–596. doi: 10.1053/jhep.2002.31724. [DOI] [PubMed] [Google Scholar]
- 51.Staudinger JL, Goodwin B, Jones SA, Hawkins-Brown D, MacKenzie KI, LaTour A, et al. The nuclear receptor PXR is a lithocholic acid sensor that protects against liver toxicity. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuno T, Hirayama-Kurogi M, Ito S, Ohtsuki S. Effect of intestinal flora on protein expression of drug-metabolizing enzymes and transporters in the liver and kidney of germ-free and antibiotics-treated mice. Mol Pharm. 2016;13:2691–2701. doi: 10.1021/acs.molpharmaceut.6b00259. [DOI] [PubMed] [Google Scholar]
- 53.Clayton TA, Baker D, Lindon JC, Everett JR, Nicholson JK. Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc Nat Acad Sci USA. 2009;2019:14728–14733. doi: 10.1073/pnas.0904489106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Digitalis Investigation Group The effect of digoxin on mortality and morbidity in patients with heart failure. N Engl J Med. 1997;336:525–533. doi: 10.1056/NEJM199702203360801. [DOI] [PubMed] [Google Scholar]
- 55.Orrego F. Calcium and the mechanism of action of digitalis. Gen Pharmacol. 1984;15(4):1273–1280. doi: 10.1016/0306-3623(84)90001-6. [DOI] [PubMed] [Google Scholar]
- 56.Cañas F, Tanasijevic MJ, Maluf N, Bates DW. Evaluating the appropriateness of digoxin level monitoring. Arch Intern Med. 1999;159:363–368. doi: 10.1001/archinte.159.4.363. [DOI] [PubMed] [Google Scholar]
- 57.Binnion PF. A comparison of the bioavailability of digoxin in capsule, tablet, and solution taken orally with intravenous digoxin. J Clin Pharmacol. 1976;16:461–467. [PubMed] [Google Scholar]
- 58.Greenblatt DJ, Smith TW, Koch-Weser J. Bioavailability of drugs: the digoxin dilemma. Clin Pharmacokinet. 1976;1:36–51. doi: 10.2165/00003088-197601010-00004. [DOI] [PubMed] [Google Scholar]
- 59.Bodor M, Kelly EJ, Ho RJ. Characterization of the human MDR1 gene. AAPS J. 2005;7:E1–5. doi: 10.1208/aapsj070101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verstuyft C, Strabach S, El Morabet H, Kerb R, Brinkmann U, Dubert L, et al. Dipyridamole enhances digoxin bioavailability via P-glycoprotein inhibition. Clin Pharmacol Ther. 2003;73:51–60. doi: 10.1067/mcp.2003.8. [DOI] [PubMed] [Google Scholar]
- 61.Sakaeda T, Nakamura T, Horinouchi M, Kakumoto M, Ohmoto N, Sakai T, et al. MDR1 genotype-related pharmacokinetics of digoxin after single oral administration in healthy Japanese subjects. Pharm Res. 2001;18:1400–1404. doi: 10.1023/A:1012244520615. [DOI] [PubMed] [Google Scholar]
- 62.Gerloff T, Schaefer M, Johne A, Oselin K, Meisel C, Cascorbi I, et al. MDR1 genotypes do not influence the absorption of a single oral dose of 1 mg digoxin in healthy white males. Br J Clin Pharmacol. 2002;54:610–616. doi: 10.1046/j.1365-2125.2002.01691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu L-X, Wen C-J, Li Y, Zhang X, Shao Y-Y, Yang Z, et al. Interindividual epigenetic variation in ABCB1 promoter and its relationship with ABCB1 expression and function in healthy Chinese subjects. Br J Clin Pharmacol. 2015;80:1109–1121. doi: 10.1111/bcp.12675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lindenbaum J, Rund DG, Butler VP, Jr, Tse-Eng D, Saha JR. Inactivation of digoxin by the gut flora: reversal by antibiotic therapy. N Engl J Med. 1981;305:789–794. doi: 10.1056/NEJM198110013051403. [DOI] [PubMed] [Google Scholar]
- 65.Rund DG, Lindenbaum J, Dobkin JF, Butler VP, Jr, Saha JR. Decreased digoxin cardioinactive-reduced metabolites after administration as an encapsulated liquid concentrate. Clin Pharmacol Ther. 1983;34:738–743. doi: 10.1038/clpt.1983.243. [DOI] [PubMed] [Google Scholar]
- 66.Lindenbaum J, Tse-Eng D, Butler VP, Rund DG. Urinary excretion of reduced metabolites of digoxin. Am J Med. 1981;1981:67–74. doi: 10.1016/0002-9343(81)90260-6. [DOI] [PubMed] [Google Scholar]
- 67.Dobkin JF, Saha JR, Butler VP, Jr, Neu HC, Lindenbaum J. Inactivation of digoxin by Eubacterium lentum, an anaerobe of the human gut flora. Trans Assoc Am Physicians. 1982;95:22–29. [PubMed] [Google Scholar]
- 68.Saha JR, Butler V, Jr, Neu H, Lindenbaum J. Digoxin-inactivating bacteria: identification in human gut flora. Science. 1983;220:325–327. doi: 10.1126/science.6836275. [DOI] [PubMed] [Google Scholar]
- 69.Bennett RG, Beamer BA, Greenough WB, 3rd, Lindenbaum J, Bartlett JG. Colonisation with digoxin-reducing strains of Eubacterium lentum and Clostridium difficile infection in nursing home patients. J Diarrhoeal Dis Res. 1992;10:87–92. [PubMed] [Google Scholar]
- 70.Robert J, Rivory L. Pharmacology of irinotecan. Drugs Today (Barc). 1998;34:777–803. doi: 10.1358/dot.1998.34.9.485276. [DOI] [PubMed] [Google Scholar]
- 71.Mathijssen RHJ, de Jong FA, van Schaik RHN, Lepper ER, Friberg LE, Rietveld T, et al. Prediction of irinotecan pharmacokinetics by use of cytochrome P450 3A4 phenotyping probes. J Natl Cancer Inst. 2004;96:1585–1592. doi: 10.1093/jnci/djh298. [DOI] [PubMed] [Google Scholar]
- 72.Gupta E, Lestingi TM, Mick R, Ramirez J, Vokes EE, Ratain MJ. Metabolic fate of irinotecan in humans: correlation of glucuronidation with diarrhea. Cancer Res. 1994;54:3723–3725. [PubMed] [Google Scholar]
- 73.Wallace BD, Wang H, Lane KT, Scott JE, Orans J, Koo JS, et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science. 2010;330:831–835. doi: 10.1126/science.1191175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science. 2018;359:97–103. doi: 10.1126/science.aan4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vétizou M, Pitt JM, Daillère R, Lepage P, Waldschmitt N, Flament C, et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science. 2015;350:1079–1084. doi: 10.1126/science.aad1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vanlancker E, Vanhoecke B, Stringer A, Van de Wiele T. 5-Fluorouracil and irinotecan (SN-38) have limited impact on colon microbial functionality and composition in vitro. PeerJ. 2017;5:e4017. doi: 10.7717/peerj.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saitta KS, Zhang C, Lee KK, Fujimoto K, Redinbo MR, Boelsterli UA. Bacterial β-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica. 2014;44:28–35. doi: 10.3109/00498254.2013.811314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suzuki T, Masui A, Nakamura J, Shiozawa H, Aoki J, Nakae H, et al. Yogurt containing Lactobacillus gasseri mitigates aspirin-induced small bowel injuries: a prospective, randomized, double-blind, placebo-controlled trial. Digestion. 2017;95:49–54. doi: 10.1159/000452361. [DOI] [PubMed] [Google Scholar]
- 79.Wu H, Esteve E, Tremaroli V, Khan MT, Caesar R, Mannerås-Holm L, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–858. doi: 10.1038/nm.4345. [DOI] [PubMed] [Google Scholar]
- 80.Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528:262–266. doi: 10.1038/nature15766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Artacho A, Isaac S, Nayak R, Flor-Duro A, Alexander M, Koo I, et al. The pre-treatment gut microbiome is associated with lack of response to methotrexate in new onset rheumatoid arthritis. Arthritis Rheumatol. 2020 doi: 10.1002/art.41622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Feng Y, Masson E, Dai D, Parker SM, Berman D, Roy A. Model-based clinical pharmacology profiling of ipilimumab in patients with advanced melanoma: clinical pharmacology profiling of ipilimumab in advanced melanoma. Br J Clin Pharmacol. 2014;78:106–117. doi: 10.1111/bcp.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahamadi M, Freshwater T, Prohn M, Li CH, De Alwis DP, De Greef R, et al. Model-based characterization of the pharmacokinetics of pembrolizumab: a humanized anti-PD-1 monoclonal antibody in advanced solid tumors. CPT Pharmacometrics Syst Pharmacol. 2017;6:49–57. doi: 10.1002/psp4.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bajaj G, Wang X, Agrawal S, Gupta M, Roy A, Feng Y. Model-based population pharmacokinetic analysis of nivolumab in patients with solid tumors. CPT Pharmacometrics Syst Pharmacol. 2017;6(1):58–66. doi: 10.1002/psp4.12143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Keizer RJ, Huitema ADR, Schellens JHM, Beijnen JH. Clinical pharmacokinetics of therapeutic monoclonal antibodies. Clin Pharmacokinet. 2010;49:493–507. doi: 10.2165/11531280-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 86.Centanni M, Moes DJAR, Trocóniz IF, Ciccolini J, van Hasselt JGC. Clinical pharmacokinetics and pharmacodynamics of immune checkpoint inhibitors. Clin Pharmacokinet. 2019;58:835–857. doi: 10.1007/s40262-019-00748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hannani D, Vétizou M, Enot D, Rusakiewicz S, Chaput N, Klatzmann D, et al. Erratum: anticancer immunotherapy by CTLA-4 blockade: obligatory contribution of IL-2 receptors and negative prognostic impact of soluble CD25. Cell Res. 2015;25:399–400. doi: 10.1038/cr.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dubin K, Callahan MK, Ren B, Khanin R, Viale A, Ling L, et al. Intestinal microbiome analyses identify melanoma patients at risk for checkpoint-blockade-induced colitis. Nat Commun. 2016;7:10391. doi: 10.1038/ncomms10391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chaput N, Lepage P, Coutzac C, Soularue E, Le Roux K, Monot C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol. 2017;28:1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 90.Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillère R, et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science. 2018;359:91–97. doi: 10.1126/science.aan3706. [DOI] [PubMed] [Google Scholar]
- 91.Derosa L, Hellmann MD, Spaziano M, Halpenny D, Fidelle M, Rizvi H, et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann Oncol. 2018;29:1437–1444. doi: 10.1093/annonc/mdy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Koller WC, Rueda MG. Mechanism of action of dopaminergic agents in Parkinson’s disease. Neurology. 1998;50(6 Suppl. 6):S11–S14. doi: 10.1212/wnl.50.6_suppl_6.s11. [DOI] [PubMed] [Google Scholar]
- 93.Bergmann S, Curzon G, Friedel J, Godwin-Austen RB, Marsden CD, Parkes JD. The absorption and metabolism of a standard oral dose of levodopa in patients with parkinsonism. Br J Clin Pharmacol. 1974;1:417–424. doi: 10.1111/j.1365-2125.1974.tb00280.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hamlet A, Thoreson AC, Nilsson O, Svennerholm AM, Olbe L. Duodenal Helicobacter pylori infection differs in cagA genotype between asymptomatic subjects and patients with duodenal ulcers. Gastroenterology. 1999;116:259–268. doi: 10.1016/S0016-5085(99)70121-6. [DOI] [PubMed] [Google Scholar]
- 95.Pierantozzi M, Pietroiusti A, Brusa L, Galati S, Stefani A, Lunardi G, et al. Helicobacter pylori eradication and l-dopa absorption in patients with PD and motor fluctuations. Neurology. 2006;66:1824–1829. doi: 10.1212/01.wnl.0000221672.01272.ba. [DOI] [PubMed] [Google Scholar]
- 96.Hashim H, Azmin S, Razlan H, Yahya NW, Tan HJ, Manaf MRA, et al. Eradication of Helicobacter pylori infection improves levodopa action, clinical symptoms and quality of life in patients with Parkinson’s disease. PLoS ONE. 2014;9:e112330. doi: 10.1371/journal.pone.0112330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Kessel SP, de Jong HR, Winkel SL, van Leeuwen SS, Nelemans SA, Permentier H, et al. Gut bacterial deamination of residual levodopa medication for Parkinson’s disease. BMC Biol. 2020;18:137. doi: 10.1186/s12915-020-00876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rugstad HE, Hundal Ø, Holme I, Herland OB, Husby G, Giercksky K-E. Piroxicam and naproxen plasma concentrations in patients with osteoarthritis: relation to age, sex, efficacy and adverse events. Clin Rheumat. 1986;5(3):389–398. doi: 10.1007/bf02054259. [DOI] [PubMed] [Google Scholar]
- 99.Tan SC, Patel BK, Jackson SHD, Swift CG, Hutt AJ. Influence of age on the enantiomeric disposition of ibuprofen in healthy volunteers. Br J Clin Pharmacol. 2003;55:579–587. doi: 10.1046/j.1365-2125.2003.01806.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bae J-W, Choi C-I, Jang C-G, Lee S-Y. Effects of CYP2C9*1/*13 on the pharmacokinetics and pharmacodynamics of meloxicam. Br J Clin Pharmacol. 2011;71(4):550–555. doi: 10.1111/j.1365-2125.2010.03853.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kurokawa S, Katsuki S, Fujita T, Saitoh Y, Ohta H, Nishikawa K, et al. A randomized, double-blinded, placebo-controlled, multicenter trial, healing effect of rebamipide in patients with low-dose aspirin and/or non-steroidal anti-inflammatory drug induced small bowel injury. J Gastroenterol. 2014;49(2):239–244. doi: 10.1007/s00535-013-0805-2. [DOI] [PubMed] [Google Scholar]
- 102.Watanabe T, Takeuchi T, Handa O, Sakata Y, Tanigawa T, Shiba M, et al. A multicenter, randomized, double-blind, placebo-controlled trial of high-dose rebamipide treatment for low-dose aspirin-induced moderate-to-severe small intestinal damage. PLoS ONE. 2015;10:e0122330. doi: 10.1371/journal.pone.0122330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Mortensen B, Murphy C, O’Grady J, Lucey M, Elsafi G, Barry L, et al. Bifidobacteriumbreve Bif195 protects against small-intestinal damage caused by acetylsalicylic acid in healthy volunteers. Gastroenterology. 2019;157(637–46):e4. doi: 10.1053/j.gastro.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 104.Djoumbou-Feunang Y, Fiamoncini J, Gil-de-la-Fuente A, Greiner R, Manach C, Wishart DS. BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. J Cheminform. 2019;11:2. doi: 10.1186/s13321-018-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Aron AT, Gentry EC, McPhail KL, Nothias L-F, Nothias-Esposito M, Bouslimani A, et al. Reproducible molecular networking of untargeted mass spectrometry data using GNPS. Nat Protoc. 2020;15:1954–1991. doi: 10.1038/s41596-020-0317-5. [DOI] [PubMed] [Google Scholar]
- 106.Jarmusch AK, Wang M, Aceves CM, Advani RS, Aguirre S, Aksenov AA, et al. ReDU: a framework to find and reanalyze public mass spectrometry data. Nat Methods. 2020;17:901–904. doi: 10.1038/s41592-020-0916-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, Goodman AL. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363(6427):eaat9931. doi: 10.1126/science.aat9931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Basu S, Duren W, Evans CR, Burant CF, Michailidis G, Karnovsky A. Sparse network modeling and metscape-based visualization methods for the analysis of large-scale metabolomics data. Bioinformatics. 2017;33:1545–1553. doi: 10.1093/bioinformatics/btx012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Morton JT, Aksenov AA, Nothias LF, Foulds JR, Quinn RA, Badri MH, et al. Learning representations of microbe–metabolite interactions. Nat Methods. 2019;16(12):1306–1314. doi: 10.1038/s41592-019-0616-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Gower JC. Generalized procrustes analysis. Psychometrika. 1975;40:33–51. doi: 10.1007/BF02291478. [DOI] [Google Scholar]
- 111.Protsyuk I, Melnik AV, Nothias L-F, Rappez L, Phapale P, Aksenov AA, et al. 3D molecular cartography using LC-MS facilitated by Optimus and ’ili software. Nat Protoc. 2018;13:134–154. doi: 10.1038/nprot.2017.122. [DOI] [PubMed] [Google Scholar]
- 112.Jarmusch AK, Elijah EO, Vargas F, Bouslimani A, da Silva RR, Ernst M, et al. Initial development toward non-invasive drug monitoring via untargeted mass spectrometric analysis of human skin. Anal Chem. 2019;91:8062–8069. doi: 10.1021/acs.analchem.8b05854. [DOI] [PubMed] [Google Scholar]