Abstract
Neoadjuvant Chemotherapy (NAC) is not frequently used in ER-positive/HER2-negative breast cancer (BC) because around 10% patients achieve pathological complete response (pCR). Since NAC can result in cancer downstaging both in the breast and axilla and prevent a morbid surgery, thus a score to predict pCR in this population will be crucial to identify patients who can benefit from this approach. A total of 4038 patients from cohorts; GSE25066, GSE20194, Hess, GSE20181, TCGA-BRCA and METBRIC were analyzed. The score was generated by the 5 most highly expressed genes in the Hallmark E2F targets gene set amongst patients in the GSE25066 cohort with ER-positive/HER2-negative BC who achieved pCR. The area under the curve was significantly higher in the score than that for the E2F targets score. High score ER-positive/HER2-negative BCs were significantly associated with higher Nottingham pathological grade, AJCC cancer stage, MKI67 expression levels, intratumor heterogeneity, homologous recombination defects, mutation burden, neoantigen load, and infiltration of anti-cancer immune cells (CD4+, T helper type1, plasmacytoid dendritic cells, M1 macrophages). They also expressed lower abundance of stromal cells including fibroblasts, lymphatic endothelial cells, pericytes and adipocytes consistently in GSE25066, TCGA and METABRIC cohorts. All cell proliferation-related gene sets, G2M checkpoint, E2F targets, MYC targets v1 and v2, Mitotic Spindle, were strongly enriched in high score BCs consistently in 3 cohorts. The gene score was significantly associated with high pCR rate consistently in the GSE25066 (38%, P < 0.001), GSE20194 (16%, P = 0.006), and Hess cohort (23%, P = 0.037). In conclusion, the 5-gene score reflects cancer cell proliferation and immune cell infiltration, and predicts pCR after NAC in ER-positive/HER2-negative breast cancer.
Keywords: 5-gene, ER-positive/HER2-negative breast cancer, predictive biomarker, neoadjuvant chemotherapy, tumor immune microenvironment
Introduction
Breast cancer is one of the most common types of cancer among women in the world [1]. The most abundant subtype is Estrogen receptor (ER)-positive/human epidermal growth factor receptor 2 (HER2)-negative accounting for approximately 70% of all breast cancers [2]. Compared to the other subtypes, this subtype is generally less aggressive. Given the mechanism that cytotoxic chemotherapy is effective against highly proliferative cells, around 10% of patients with this subtype are likely to achieve complete pathological response (pCR) after neoadjuvant chemotherapy (NAC) [3-5]. Attainment of pCR after NAC is important since it is currently considered to be a surrogate marker for survival [6]. Recently, tumor infiltrating lymphocytes (TIL) in the tumor microenvironment have been reported to correlate with NAC response [7]; however, currently no established universal predictive biomarker for NAC response exists that can be readily used in clinical practice. Certain tumor characteristics may decrease the likelihood of response to chemotherapy, in which case, attempting NAC even for bulky disease may not be the correct approach due to low chemosensitivity and would only result in unnecessary toxicities. Therefore, given poor response rate to chemotherapy, surgeons usually hesitate to consult medical oncology for NAC for this less aggressive ER-positive/HER2-negative subtype even when the tumor is relatively large. As expected, this results in these patients undergoing an extensive operation with significant morbidity. Therefore, identification of a reliable biomarker to predict the response of ER-positive/HER2-negative tumors to NAC will have a huge clinical benefit as it would emphasize the need for NAC to downstage patients with bulky disease prior to taking them for surgery, by converting lymph node-positive to lymph node-negative disease avoiding an axillary lymph node dissection, and resulting in higher rate for breast conserving surgery compared to mastectomy. This would be particularly true, especially when there is evidence that NAC would be clinically useful due to upfront knowledge that the tumor is chemo sensitive, an indication where currently other genomic tests like Mamma Print and Oncotype Dx that predict chemotherapy benefit are not approved.
Recent advances in comprehensive profiling of transcriptome by gene expression microarray or RNA-sequencing technologies have revolutionized how we understand the cancer biology. Many computational algorithms that analyze the entire gene expressions in the bulk tumor allow the researchers to understand not only the biology of the cancer cells but also every single cell that is transcribed in that tumor microenvironment. The researchers can investigate the clinical relevance of a gene expression when transcriptomic profile is associated with clinical parameters such as cancer staging, pathological results, and survival. Yet, it is a challenge to interpret the biological meaning of a single gene expression change, and it is known to often have limited reproducibility across independent cohorts. Cancer response to drugs are concerted maneuver of multiple gene expressions [8], and a score that captures multiple gene expressions can provide a more accurate value than that of a single gene [9]. Our group has utilized pathway based approaches that considered such coordination of genes, simplified the model, and improved the interpretation of the results [10,11]. We previously reported that E2F pathway score, calculated by gene set variation analysis (GSVA) algorithm, have a potential as not only a prognostic but also a predictive biomarker for treatment response after NAC in ER-positive/HER2-negative breast cancer [11]. The E2F pathway score consist of expression data of E2F target pathway-related 200 genes. Scoring by GSVA is a useful method for investigating the relationship between various pathways and clinical outcomes [12-16]; however, it requires comprehensive gene expression analysis and a score with single genes will be much practical from cost stand point.
In this study, we hypothesized that there are some genes among E2F gene sets that are strongly associated with pCR after NAC. Therefore, we aimed to develop a novel gene expression-based score utilizing those genes that strongly predict pCR after NAC in ER-positive/HER2-negative breast cancer patients, and examined its biological features.
Materials and methods
Data collection
The Gene Expression Omnibus (GEO) repository was used to access the tumor gene expression and clinical data from the Symmans et al. (GSE25066; n = 508) [17], Shi et al. (GSE20194; n = 248) [18], and Hess et al. (n = 133) [19] cohorts. GSE25066 cohort was chosen as the testing cohort because of large sample number. Other two cohorts were used as validation cohorts for the NAC response- predictive biomarker value of the 5-gene score. Furthermore, data of gene expression and clinical data of the TCGA-BRCA (n = 1069, female) [20] and METABRIC (n = 1904) [21] cohorts were also obtained as validation cohorts for the association of the 5-gene score with clinical and molecular features through cBio Cancer Genomics Portal [22] in late 2017. Interferon (IFN)-γ response, tumor infiltrating lymphocyte (TIL) regional fraction, T cell receptor (TCR) and B cell receptor (BCR) shannon, silent and non-silent mutation rate, fraction altered, single nucleotide variant (SNV) and indel neoantigens, intratumor heterogeneity, homologous recombination defects, and proliferation score were calculated as described by Thorsson et al. [23].
Gene expression analyses
For the analysis, we used limma package in R. False discovery rate (FDR) < 0.001 after adjustment for multi-testing was considered as differentially expressed. For multiple testing adjustments, the FDR < 0.001 was chosen as the cut-off to identify the candidate genes. For gene set enrichment analysis, GSEA software and MSigDB Hallmark gene set collections [24] were used. A nominal p value threshold of 0.05 and a FDR of 0.25, as recommended by the GSEA software, was used to deem significance.
Statistical analysis
Comparison between groups were performed using the Mann-Whitney U test or Kruskal test. Limma models were used to investigate the impact of various genes on the risk of residual disease (RD) after NAC. P values were considered significant when less than 0.05. All statistical analyses were performed by R software (version 4.0.1).
Others
The gene set variation analysis (GSVA) algorithm [25] was utilized with the GSVA Bioconductor package (version 3.10) to determine the E2F pathway score from the “HALLMARK_E2F targets” gene sets of the MSigDB Hallmark collection (Table S1) [24]. Gene set enrichment analysis were performed with Gene Set Enrichment Analysis (GSEA) software (Java version 4.0.1) with MSigDB Hallmark gene sets. Statistical significance was set with a false discovery rate (FDR) of 0.25, as recommended by the GSEA software. The fraction of infiltrating cells in bulk tumor was calculated by xCell algorithm [26], as we previously reported [27-30].
Results
Establishment of a novel 5-gene score to predict pathological complete response (pCR) after neoadjuvant chemotherapy (NAC) in ER-positive/HER2-negative breast cancer
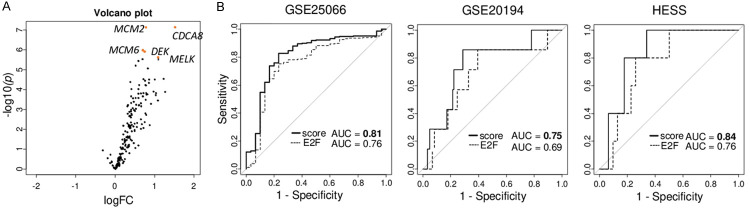
To identify the genes that associate the strongest with NAC response among genes in Hallmark E2F target gene set that we previously reported as a predictive biomarker in ER-positive/HER2-negative breast cancer [11], differential gene expression analysis (DGEA) that compared the gene expressions between groups that did and did not completely respond (pCR vs non-pCR) to NAC was conducted among ER-positive/HER2-negative breast cancer in GSE25066 cohort. Of the 200 genes in E2F target gene sets, expressions of 184 genes were identified in the GSE25066 cohort. We found that among those 184 genes, the expression of 20 genes was significantly higher with false discovery rate (FDR) < 0.001. No genes were highly expressed with FDR < 0.001 among the patients that did not respond to NAC. We identified five genes, CDCA8, MCM2, MCM6, MELK and DEK, that were highly associated with response to NAC (Figure 1A). We generated a multivariate 5-gene score using tumor gene expressions and log2 (fold change) was calculated as: 1.522 × (expressionCDCA8) + 0.780 × (expressionMCM2) + 0.708 × (expressionMCM6) + 1.089 × (expressionMELK) + 0.694 × (expressionDEK).
Figure 1.
Establishment and association of the 5-gene score with pathological complete response (pCR) after neoadjuvant chemotherapy (NAC) in ER-positive/HER2-negative breast cancer. A. Volcano plots illustrating the differentially expressed mRNAs between pCR (n = 30) and non-pCR groups (n = 248) of ER-positive/HER2-negative breast cancer in the GSE25066 cohort. X-axes; log2 (fold change), Y-axes; - log10 P-value from limma analysis. mRNA with adjusted p-value < 0.05 are marked in blue, and top five gene of p-vale are marked in red. B. Receiver operating characteristic (ROC) curve of 5-gene score and E2F targets score with area under the curve (AUC) in the GSE25066, GSE20194, and HESS cohorts. 5-gene score is in bold lines, and E2F targets score in dotted lines.
Next, we tested the predictive accuracy of the score by receiver operating characteristic-area under the curve (ROC-AUC) analysis. The AUC of 5-gene score was higher than any other 184 genes in the GSE25066 cohort (Table S2) as well as that of E2F pathway score itself consistently in three cohorts (Figure 1B; AUC = 0.813, and 0.759, respectively, P = 0.0002 in the GSE25066, AUC = 0.75 and 0.69, respectively, P = 0.0318 in the GSE20194, and AUC = 0.84 and 0.76, respectively, P = 0.2749 in the HESS cohort).
A 5-gene score was significantly associated with clinical aggressiveness in ER+/HER2- breast cancer
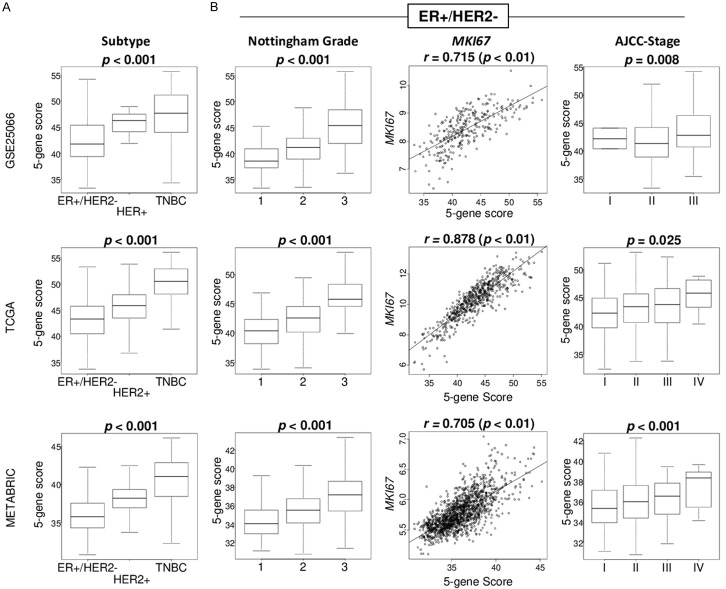
We expected that the 5-gene score is associated with aggressive cancer biology because the score is generated by the genes included in cell proliferation-related E2F targets gene set. We found that the 5-gene score was highest in triple-negative breast cancer (TNBC), which is the most aggressive subtype, consistently in all GSE25066, TCGA, and METABRIC cohorts (Figure 2A; all P < 0.001). Within ER-positive/HER2-negative breast cancer, higher cancer stage by American Joint Committee on Cancer (AJCC) cancer staging and Nottingham histological grade were both significantly associated with elevated 5-gene score consistently in all the three cohorts (Figure 2B; all P < 0.03). Further, the 5-gene score strongly correlated with MKI67 gene expression, which is the most commonly used marker for cell proliferation in clinical practice, consistently in three cohorts (Spearman rank correlation (r) = 0.715, 0.878, and 0.705, respectively, all P < 0.01). These results suggest that the 5-gene score was significantly associated with clinical parameters of cancer aggressiveness in ER-positive/HER2-negative breast cancer.
Figure 2.
Association of the 5-gene score level with clinical characteristics in the GSE25066, TCGA, and METABRIC cohorts. A. Boxplots of the 5-gene score level by subtype in whole cohort. B. Boxplots of the 5-gene score level by AJCC cancer staging and Nottingham histological grade, and correlation plots between 5-gene score and MKI67 expression in ER-positive/HER2-negative breast cancer. Kruskal-Wallis test and spearman correlation test were used accordingly.
High 5-gene score ER-positive/HER2-negative breast cancer enriched cell proliferation-related gene sets and other pro-cancer-related gene sets
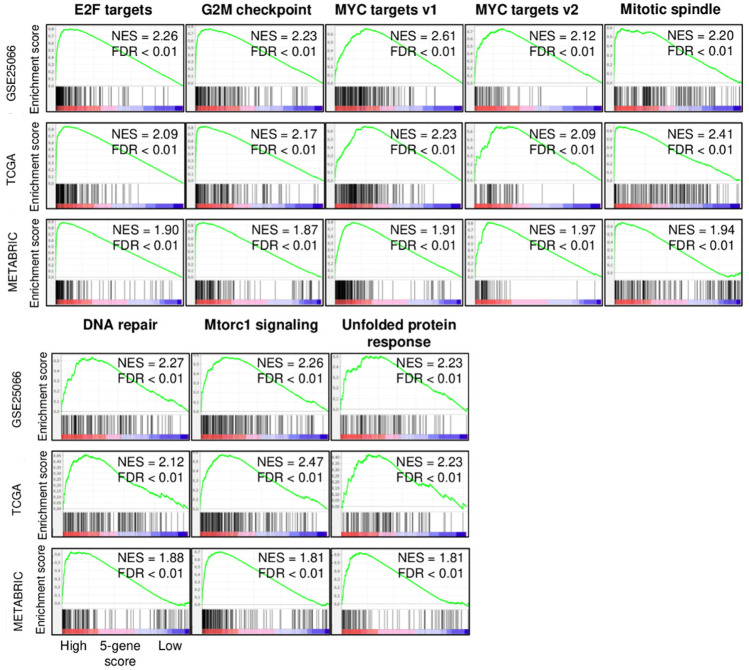
Using Gene Set Enrichment Analysis (GSEA) with MSigDB hallmark gene, we investigated the association of the 5-gene score with cancer biology of ER-positive/HER2-negative breast cancer in three independent cohorts (GSE25066, TCGA, and METABRIC). The top one-third was defined as high and the rest to be low score. High 5-gene score ER-positive/HER2-negative breast cancer significantly enriched all of the cell proliferation-related gene sets in Hallmark collection (E2F targets, G2M checkpoint, MYC targets v1, MYC targets v2, and Mitotic spindle), and other pro-cancer-related gene sets (DNA repair, mtorc1 signaling, and unfolded protein response) consistently in all three cohorts (Figure 3). These results suggest that the 5-gene score strongly reflects cell proliferation, which agrees with the notion that highly proliferative cancer responds to neoadjuvant cytotoxic chemotherapy better than the ones that do not.
Figure 3.
Gene set enrichment analysis (GSEA) of high 5-gene score ER-positive/HER2-negative breast cancer in the GSE25066, TCGA, and METABRIC cohorts. Enrichment plots with normalized enrichment score (NES) and false discovery rate (FDR) of Hallmark gene sets, which were significantly enriched in high 5-gene score ER-positive/HER2-negative breast cancer consistently in three cohorts; E2F targets, G2M checkpoint, MYC targets v1 and v2, mitotic spindle, DNA repair, Mtorc1 signaling, and unfolded protein response gene sets. NES and FDR were determined with the classical GSEA method, where FDR < 0.25 is considered significant.
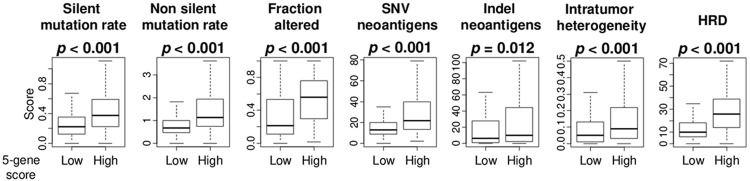
High 5-gene score was significantly associated with high mutation load, intratumor heterogeneity, homologous recombination deficiency (HRD) as well as proliferation
Previously we have shown that breast cancer with high mutation is highly proliferative [31], and vice versa [11,32]. Taken together with the above result that high 5-gene score cancer enriched DNA repair gene set, we expected the 5-gene score to associate with mutation rate, intratumor heterogeneity, and HRD. We utilized several scores previously reported on TCGA cohort by Thorsson et al. High score was significantly associated with high level of mutation-related scores (silent and non-silent mutation rate, fraction altered, SNV and indel neoantigens), intratumor heterogeneity, and HRD score (Figure 4, all P < 0.001).
Figure 4.
Association of the 5-gene score with mutation rates, intratumor heterogeneity, and homologous recombination defects (HRD) in ER-positive/HER2-negative breast cancer. Boxplots of level of mutation-related score; silent and non-silent mutation load, fraction altered, single-nucleotide variant (SNV) and indel neoantigens, intratumor heterogeneity, and HRD, by high and low 5-gene score in the TCGA cohorts. Mann-Whitney U test was used to calculate p values.
High 5-gene score tumors were infiltrated with high fractions of anti-cancer immune cells
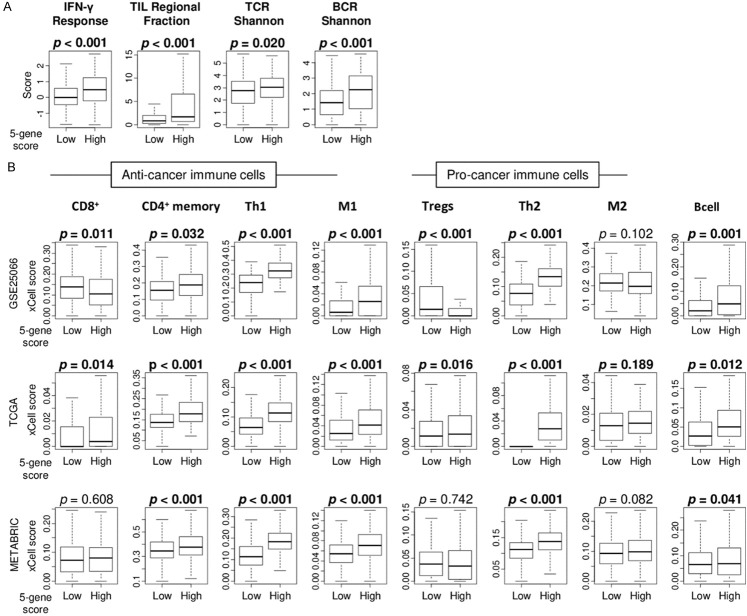
Since it has been reported that several type of cells in the tumor microenvironment (TME) are strongly associated with drug response in breast cancer [33,34], we examined the association of the 5-gene score with immune response in ER-positive/HER2-negative breast cancer in the GSE25066, TCGA, and METABRIC cohorts. High 5-gene score was significantly associated with high level of immune-related scores; interferon (IFN)-γ response, tumor infiltrating lymphocyte (TIL) regional fraction, and T cell receptor (TCR) as well as B cell receptor (BCR) Shannon, calculated by Thorsson et al. in the TCGA cohort (Figure 5A; P < 0.001, < 0.001, = 0.020, and < 0.001, respectively). Next, we examined the association of the 5-gene score with fraction of each immune cell in TME using xCell algorithm. High score tumors were significantly associated with high infiltration of anti-cancer immune cells, including CD4+ memory T cells, T helper type 1 (Th1) cells, and M1 macrophages consistently in three cohorts (Figure 5B). Th2 cells and B cells were also highly infiltrated in high score tumors consistently in three cohorts (Figure 5B).
Figure 5.
Association of the 5-gene score with tumor infiltrating immune cells. A. Boxplots of level of immune-related scores; interferon (IFN)-γ response, tumor infiltrating lymphocyte (TIL) regional fraction, T cell receptor (TCR) and B cell receptor (BCR) shannon, by high and low 5-gene score in the TCGA cohorts. B. Boxplots of the fraction of anti-cancer immune cells; CD8+ T cells, CD4+ T cells, type 1 T helper (Th1) cells, M1 macrophages, and pro-cancer immune cells; Regulatory T cells (Tregs), type 2 T helper (Th2) cells, M2 macrophages, and B cells by high and low and 5-gene scores ER-positive/HER2-negative breast cancer in the GSE25066, TCGA, and METABRIC cohorts. Mann-Whitney U test was used to calculate p values.
High 5-gene score tumors were infiltrated with low fraction of several stromal cells
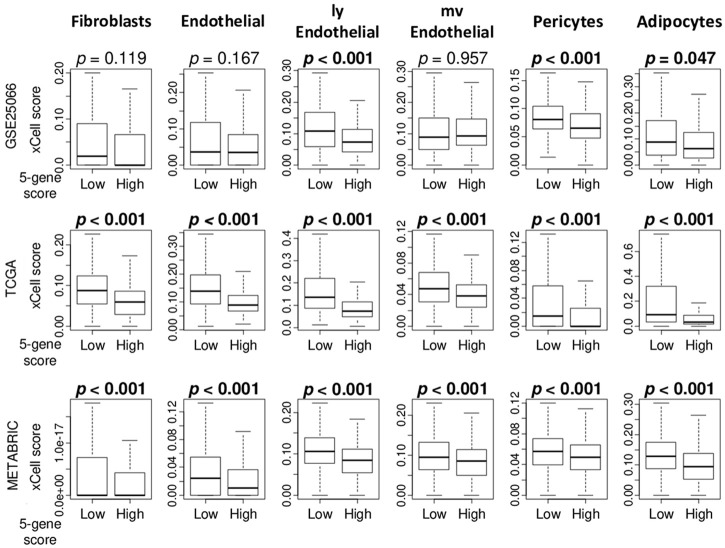
Given our previous studies that demonstrated that infiltrations of stromal cells reflect less aggressive cancer [35-37], we investigated the association of the score with stromal cells in TME in the GSE25066, TCGA and METABRIC cohorts. We found that high 5-gene score tumors were significantly associated with low fraction of several stromal cells; lymph endothelial (lyE) cells, pericytes, and adipocytes consistently in three cohorts (Figure 6; P < 0.05). High 5-gene score tumors were also significantly associated with low fraction of fibroblasts, endothelial cells, and micro vessel endothelial (mvE) cells consistently in the two cohorts.
Figure 6.
Association of the MELK expression with the fraction of stromal cells in the tumor microenvironment in ER-positive/HER2-negative breast cancer. Boxplots of the fraction of several stromal cells, including fibroblasts, endothelial cells, lymphatic endothelial (lyE) cells, micro vessel endothelial (mvE) cells, pericytes cells, and adipocytes cells by high and low 5-gene ER-positive/HER2-negative breast cancer groups in the GSE25066, TCGA, and METABRIC cohorts. P values were calculated by Mann-Whitney U test.
A high score was significantly associated with high rate of pCR for neoadjuvant chemotherapy in ER+/HER2- breast cancer
Finally, we tested the utility of the score as predictive biomarker for drug treatment therapy. High score was significantly associated with high rate of pCR after NAC compared to low score group (Figure 7; n = 289, pCR rate = 37.3% and 2.8%, respectively, P < 0.001) in the GSE25066 cohort. The result was validated by two other cohorts, GSE20194 (Figure 7; n = 129, pCR rate = 16.2% and 1.2% respectively, P = 0.006), and HESS (Figure 7; n = 67, pCR rate = 22.2% and 2.3%, respectively, P = 0.037).
Figure 7.
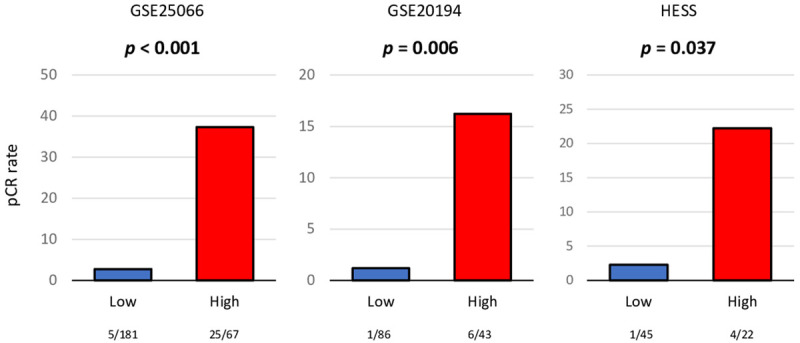
Association of the 5-gene score with pathological complete response (pCR) after neoadjuvant chemotherapy (NAC) for ER-positive/HER2-negative breast cancer patients. Bar plots of the comparison of pCR rate after NAC between the 5-gene score low (blue) and high (red) ER-positive/HER2-negative breast cancer groups in the GSE25066 (n = 278), GSE20194 (n = 129), and HESS (n = 67) cohorts. Fisher’s exact test was used for the analysis. Group sizes are shown underneath the bar.
Discussion
In the current study, we established a novel 5-gene score as a strong predictive biomarker for pCR after NAC in ER-positive/HER2-negative breast cancer. The score is calculated by the expressions of CDCA8, MCM2, MCM6, MELK, and DEK genes that were most elevated among the 200 genes in E2F target gene sets in ER-positive/HER2-negative breast cancer patients that achieved pCR compared to those who did not in the GSE25066 cohort. AUC of the 5-gene score was significantly higher than that for the E2F pathway score as well as any genes in the GSE25066 cohort, and was also higher than the E2F pathway score in other two cohorts (GSE20194 and HESS). Among breast cancer subtypes, the score of ER-positive/HER2-negative breast cancer was lowest compared to other subtypes in the GSE25066, TCGA, and METABRIC cohorts. In ER-positive/HER2-negative tumors, the 5-gene score was significantly associated with higher Nottingham pathological grade, AJCC cancer stage, and highly correlated with MKI67 expression in three cohorts. Biologically, high-score ER-positive/HER2-negative cancer enriched all 5 Hallmark cell proliferation-related gene sets (E2F targets, G2M checkpoint, MYC targets v1 and v2, and Mitotic spindle) as well as DNA repair, Mtorc1 signaling, and unfolded protein response gene sets, consistently in three cohorts. The high-score tumors also had greater mutation rates, neoantigen load, intratumor heterogeneity, and HRD, compared to low-score tumors in the TCGA cohort. They also had higher immune response, and had more abundance of anti-cancer immune cells, including CD4+ memory T cells, Th1 cells, and M1 macrophages, as well as Th2 cells and B cells, and less abundance of stromal cells (fibroblasts, lymphatic endothelial cells, pericytes, and adipocytes) consistently in three cohorts. Finally, the 5-gene score was significantly associated with high pCR rate after NAC consistently in three cohorts.
Among the 5 genes that consist the score, CDCA8 was reported to be a key mediator of estrogen-stimulated breast cancer cell growth. And some suggested it to be a therapeutic target in breast cancer [38]. Gene silencing of CDCA8 suppressed cancer cell growth by promoting G1 arrest of the cell cycle that coordinated with a decrease in E2-induced molecules, Cyclin D1 (CCND1) and B-Cell CLL/Lymphoma 2 (BCL2) [38]. Minichromosome maintenance protein 2 (MCM2) and MCM6 are members of the MM protein family that plays an important role in DNA replication and in cell cycle progression. MCM2 expression was reported to have a significant correlation with worse patient survival, and MCM6 was also reported to associate with MKI67 expression and prognosis in breast cancer. Maternal and embryonic leucine zipper kinase (MELK) is overexpressed in breast cancer [39] and was reported to suppress breast cancer cell proliferation by arresting different cell cycle phases that is mediated by different mediators, which may be involved in the crosstalk between MELK signaling and the estrogen receptor signaling pathway [40]. DEK is an oncogene and its expression in breast cancer creates an immune suppressed tumor microenvironment by inducing M2 tumor associated macrophage polarization [41]. DEK expression was reported to be associated with pCR [42].
Although initial intention of NAC was to decrease the size and extent of locally advanced cancer to become operable, currently it is used to predict the ultimate course of cancer progression. The extent of response to NAC not only reveals tumor response to a given therapy independent of other prognostic features of cancer, but it also is a surrogate marker for survival [17,43]. It is important to remember that NAC is not always helpful for patients with ER positive breast cancer where tumors have low chemosensitivity, on the contrary, use of anthracyclines and taxanes, the standard chemotherapy used for ER positive breast cancer, may result in unnecessary immediate and long-term toxicities like, increased risk of infections, cardiac morbidity, debilitating neuropathy, and in rare cases leukemia several years later without any clinical benefit. Thus, a predictive biomarker is expected to maximize the treatment benefit, minimize the physical and financial toxicities, and improve quality of life by precise patient selection for NAC. At the same time, bulky disease where the biomarker suggests poor response to chemotherapy may help us prioritize clinical trials for this patient population with novel agents with the ultimate aim to improve responses. Given that NAC is most effective against highly proliferative cells [42], we have previously generated scores that reflect cell proliferation and predict NAC response to breast cancer as biomarkers. We found that the E2F target genes play a critical role in the cell cycle and the score predicted NAC response in ER-positive/HER2-negative breast cancer patients [11]. In the current study, we investigated the key genes in the E2F targets gene set that is most relevant to NAC response. We found that 5-gene score reflected the cell proliferation activity, which also showed correlation with clinical aggressiveness and MKI67, as well as gene set enrichment analysis in ER-positive/HER2-negative breast cancer. Furthermore, high score was associated with presence of more anti-cancer immune cells as well as low fraction of stromal cells in TME. Many studies have been reported on the association between tumor immunity and NAC response [33,34]. We have previously reported that tumor immune cells, especially Tregs, are involved in NAC response [44], but the 5-gene score had even higher predictive value. In fact, the 5-gene score has stronger predictive value than the E2F pathway score (using 200 genes) as well as other several single gene expressions. Genomic signature profiling, such as Oncotype Dx and MammaPrint, has been used in the clinical practice to predict the benefit of adjuvant chemotherapy in hormone-positive breast cancers, but are not yet approved to predict NAC response prior to definitive surgery. Since the 5-gene score predicts the NAC response, it does not overlap with the current setting in which the existing genomic signature profiling is utilized. Further, it only uses 5 genes, which is far more clinically appreciable in terms of cost and simplicity. We cannot help but speculate that the 5-gene score may have a clinical utility to be used for patient selection and as a predictive biomarker for NAC in ER-positive/HER2-negative breast cancer patients Knowledge of this predictive biomarker in the upfront setting would let clinicians confidently treat high risk ER positive breast cancer patients in the neoadjuvant setting with chemotherapy with the expectation of a good treatment response, downstaging of tumor which would eventually lead to a less morbid surgery.
Although we found a novel 5-gene score in ER-positive/HER2-negative breast cancer using multiple large human sample data, our study has limitations. This is a retrospective study, and due to the lack of a per-regiment cohort with enough sample numbers, the association between each regimen-specific response and the 5-gene score has not been investigated. For clinical application, appropriate cut-off values need to be evaluated under a prospective study. Furthermore, the impact of the 5-gene score on neoadjuvant treatment deserves further studies, with a specific evaluation in a prospective setting.
In conclusion, the 5-gene score reflects cell proliferation and has the potential to predict pCR after NAC in ER-positive/HER2-negative breast cancer.
Acknowledgements
This work was supported by US National Institutes of Health/National Cancer Institute grant R01CA160688, R01CA250412, R37CA248018, US Department of Defense BCRP grant W81XWH-19-1-0674, as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to K.T., and US National Cancer Institute cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center. Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award Number KL2TR001413 and UL1TR001412. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69:7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 2.Spring LM, Gupta A, Reynolds KL, Gadd MA, Ellisen LW, Isakoff SJ, Moy B, Bardia A. Neoadjuvant endocrine therapy for estrogen receptor-positive breast cancer: a systematic review and meta-analysis. JAMA Oncol. 2016;2:1477–1486. doi: 10.1001/jamaoncol.2016.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Esserman LJ, Berry DA, Cheang MC, Yau C, Perou CM, Carey L, DeMichele A, Gray JW, Conway-Dorsey K, Lenburg ME, Buxton MB, Davis SE, van’t Veer LJ, Hudis C, Chin K, Wolf D, Krontiras H, Montgomery L, Tripathy D, Lehman C, Liu MC, Olopade OI, Rugo HS, Carpenter JT, Livasy C, Dressler L, Chhieng D, Singh B, Mies C, Rabban J, Chen YY, Giri D, Au A, Hylton N. Chemotherapy response and recurrence-free survival in neoadjuvant breast cancer depends on biomarker profiles: results from the I-SPY 1 TRIAL (CALGB 150007/150012; ACRIN 6657) Breast Cancer Res Treat. 2012;132:1049–1062. doi: 10.1007/s10549-011-1895-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boughey JC, McCall LM, Ballman KV, Mittendorf EA, Ahrendt GM, Wilke LG, Taback B, Leitch AM, Flippo-Morton T, Hunt KK. Tumor biology correlates with rates of breast-conserving surgery and pathologic complete response after neoadjuvant chemotherapy for breast cancer: findings from the ACOSOG Z1071 (Alliance) Prospective Multicenter Clinical Trial. Ann Surg. 2014;260:608–614. doi: 10.1097/SLA.0000000000000924. discussion 614-606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Minckwitz G, Untch M, Blohmer JU, Costa SD, Eidtmann H, Fasching PA, Gerber B, Eiermann W, Hilfrich J, Huober J, Jackisch C, Kaufmann M, Konecny GE, Denkert C, Nekljudova V, Mehta K, Loibl S. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012;30:1796–1804. doi: 10.1200/JCO.2011.38.8595. [DOI] [PubMed] [Google Scholar]
- 6.Asaoka M, Gandhi S, Ishikawa T, Takabe K. Neoadjuvant chemotherapy for breast cancer: past, present, and future. Breast Cancer (Auckl) 2020;14:1178223420980377. doi: 10.1177/1178223420980377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loi S, Sirtaine N, Piette F, Salgado R, Viale G, Van Eenoo F, Rouas G, Francis P, Crown JP, Hitre E, de Azambuja E, Quinaux E, Di Leo A, Michiels S, Piccart MJ, Sotiriou C. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. 2013;31:860–867. doi: 10.1200/JCO.2011.41.0902. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Sun Z, Zimmermann MT, Bugrim A, Kocher JP. Predict drug sensitivity of cancer cells with pathway activity inference. BMC Med Genomics. 2019;12:15. doi: 10.1186/s12920-018-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi W, Jiang T, Nuciforo P, Hatzis C, Holmes E, Harbeck N, Sotiriou C, Peña L, Loi S, Rosa DD, Chia S, Wardley A, Ueno T, Rossari J, Eidtmann H, Armour A, Piccart-Gebhart M, Rimm DL, Baselga J, Pusztai L. Pathway level alterations rather than mutations in single genes predict response to HER2-targeted therapies in the neo-ALTTO trial. Ann Oncol. 2017;28:128–135. doi: 10.1093/annonc/mdw434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. [Google Scholar]
- 11.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res. 2020;2020:5618786. doi: 10.1155/2020/5618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21:6708. doi: 10.3390/ijms21186708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC) Cancers (Basel) 2021;13:323. doi: 10.3390/cancers13020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, Martin M, Cotrina J, Gomez H, Hubbard R, Chacón JI, Ferrer-Lozano J, Dyer R, Buxton M, Gong Y, Wu Y, Ibrahim N, Andreopoulou E, Ueno NT, Hunt K, Yang W, Nazario A, DeMichele A, O’Shaughnessy J, Hortobagyi GN, Symmans WF. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, Shaughnessy JD Jr, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M, Chen M, Cheng J, Cheng J, Chou J, Davison TS, Delorenzi M, Deng Y, Devanarayan V, Dix DJ, Dopazo J, Dorff KC, Elloumi F, Fan J, Fan S, Fan X, Fang H, Gonzaludo N, Hess KR, Hong H, Huan J, Irizarry RA, Judson R, Juraeva D, Lababidi S, Lambert CG, Li L, Li Y, Li Z, Lin SM, Liu G, Lobenhofer EK, Luo J, Luo W, McCall MN, Nikolsky Y, Pennello GA, Perkins RG, Philip R, Popovici V, Price ND, Qian F, Scherer A, Shi T, Shi W, Sung J, Thierry-Mieg D, Thierry-Mieg J, Thodima V, Trygg J, Vishnuvajjala L, Wang SJ, Wu J, Wu Y, Xie Q, Yousef WA, Zhang L, Zhang X, Zhong S, Zhou Y, Zhu S, Arasappan D, Bao W, Lucas AB, Berthold F, Brennan RJ, Buness A, Catalano JG, Chang C, Chen R, Cheng Y, Cui J, Czika W, Demichelis F, Deng X, Dosymbekov D, Eils R, Feng Y, Fostel J, Fulmer-Smentek S, Fuscoe JC, Gatto L, Ge W, Goldstein DR, Guo L, Halbert DN, Han J, Harris SC, Hatzis C, Herman D, Huang J, Jensen RV, Jiang R, Johnson CD, Jurman G, Kahlert Y, Khuder SA, Kohl M, Li J, Li L, Li M, Li QZ, Li S, Li Z, Liu J, Liu Y, Liu Z, Meng L, Madera M, Martinez-Murillo F, Medina I, Meehan J, Miclaus K, Moffitt RA, Montaner D, Mukherjee P, Mulligan GJ, Neville P, Nikolskaya T, Ning B, Page GP, Parker J, Parry RM, Peng X, Peterson RL, Phan JH, Quanz B, Ren Y, Riccadonna S, Roter AH, Samuelson FW, Schumacher MM, Shambaugh JD, Shi Q, Shippy R, Si S, Smalter A, Sotiriou C, Soukup M, Staedtler F, Steiner G, Stokes TH, Sun Q, Tan PY, Tang R, Tezak Z, Thorn B, Tsyganova M, Turpaz Y, Vega SC, Visintainer R, von Frese J, Wang C, Wang E, Wang J, Wang W, Westermann F, Willey JC, Woods M, Wu S, Xiao N, Xu J, Xu L, Yang L, Zeng X, Zhang J, Zhang L, Zhang M, Zhao C, Puri RK, Scherf U, Tong W, Wolfinger RD. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, Rouzier R, Sneige N, Ross JS, Vidaurre T, Gómez HL, Hortobagyi GN, Pusztai L. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J. Clin. Oncol. 2006;24:4236–4244. doi: 10.1200/JCO.2006.05.6861. [DOI] [PubMed] [Google Scholar]
- 20.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS, Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2018;48:812–830. e814. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oshi M, Newman S, Murthy V, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. ITPKC as a prognostic and predictive biomarker of neoadjuvant chemotherapy for triple negative breast cancer. Cancers (Basel) 2020;12:2758. doi: 10.3390/cancers12102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Loi S, Drubay D, Adams S, Pruneri G, Francis PA, Lacroix-Triki M, Joensuu H, Dieci MV, Badve S, Demaria S, Gray R, Munzone E, Lemonnier J, Sotiriou C, Piccart MJ, Kellokumpu-Lehtinen PL, Vingiani A, Gray K, Andre F, Denkert C, Salgado R, Michiels S. Tumor-infiltrating lymphocytes and prognosis: a pooled individual patient analysis of early-stage triple-negative breast cancers. J. Clin. Oncol. 2019;37:559–569. doi: 10.1200/JCO.18.01010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, Piccart MJ, Loibl S, Denkert C, Smyth MJ, Joensuu H, Sotiriou C. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 35.Katsuta E, Qi Q, Peng X, Hochwald SN, Yan L, Takabe K. Pancreatic adenocarcinomas with mature blood vessels have better overall survival. Sci Rep. 2019;9:1310. doi: 10.1038/s41598-018-37909-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katsuta E, Rashid OM, Takabe K. Fibroblasts as a biological marker for curative resection in pancreatic ductal adenocarcinoma. Int J Mol Sci. 2020;21:3890. doi: 10.3390/ijms21113890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21:5744. doi: 10.3390/ijms21165744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bu Y, Shi L, Yu D, Liang Z, Li W. CDCA8 is a key mediator of estrogen-stimulated cell proliferation in breast cancer cells. Gene. 2019;703:1–6. doi: 10.1016/j.gene.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Li BB, Li J, Roberts TM, Zhao JJ. A conditional dependency on MELK for the proliferation of triple-negative breast cancer cells. iScience. 2018;9:149–160. doi: 10.1016/j.isci.2018.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li G, Yang M, Zuo L, Wang MX. MELK as a potential target to control cell proliferation in triple-negative breast cancer MDA-MB-231 cells. Oncol Lett. 2018;15:9934–9940. doi: 10.3892/ol.2018.8543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pease NA, Shephard MS, Sertorio M, Waltz SE, Vinnedge LMP. DEK expression in breast cancer cells leads to the alternative activation of tumor associated macrophages. Cancers (Basel) 2020;12:1936. doi: 10.3390/cancers12071936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Witkiewicz AK, Balaji U, Knudsen ES. Systematically defining single-gene determinants of response to neoadjuvant chemotherapy reveals specific biomarkers. Clin Cancer Res. 2014;20:4837–4848. doi: 10.1158/1078-0432.CCR-14-0885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.von Minckwitz G, Martin M. Neoadjuvant treatments for triple-negative breast cancer (TNBC) Ann Oncol. 2012;23(Suppl 6):vi35–39. doi: 10.1093/annonc/mds193. [DOI] [PubMed] [Google Scholar]
- 44.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.