Abstract
Evaluation of the functional aspects if the tumor immune microenvironment (TIME), such as the recently introduced cytolytic activity score (CYT) index have been under the spotlight in cancer research; however, clinical relevance of immune cell killing activity in breast cancer has never been analyzed in large patient cohorts. We hypothesized that CYT reflects the immune activity of TIME and can predict patient survival. A total of 7533 breast cancer patients were analyzed as both discovery and validation cohorts. We found that high CYT was associated with advanced histological grade and triple-negative breast cancer (TNBC). High CYT in tumors was significantly associated with better survival in TNBC, but unexpectedly, not in other breast cancer subtypes. High CYT TNBC included both favorable immune-related, as well as unfavorable (suppressive) inflammation-related gene sets, and characterized by high infiltration with T cells and B cells. High CYT TNBC was associated with high homologous recombination deficiency and low somatic copy number alteration score and less mutant allele tumor heterogeneity, but not with tumor mutation burden (TMB). Although CYT was not associated with pathological complete response after neoadjuvant chemotherapy, it was significantly associated with high expression of multiple immune checkpoint molecules. In conclusion, CYT of TNBC is associated with enhanced anti-cancer immunity, less intra-tumoral heterogeneity, and with better survival.
Keywords: Breast cancer, CYT, cytolytic activity, heterogeneity, immune cells, immune checkpoint inhibitor, mutation, TNBC, transcriptome
Introduction
Tumor immune microenvironment (TIME) plays a critical role in tumor progression, response to therapeutics, and prognosis in breast cancer [1,2]. Tumor-infiltrating lymphocytes (TILs) are one of the major components of TIME, and the density and types of lymphocytes in the TIL fraction of a tumor have marked prognostic associations in breast cancer [3,4].
Breast cancer has long been considered to be a non-immunogenic malignancy, also known as “cold” or “immune desert” tumors [5,6]. However, recently TILs were found to have a clinical impact on treatment response and patient outcomes in some types of breast cancer including biologically aggressive triple negative breast cancer (TNBC) [7,8]. Immunogenic breast cancer with high mutation burden and neoepitope load attracts effector CD8+ T cells that induce cancer immunoediting [9-11]. Cancer immunoediting is the process whereby the immune system can both constrain and promote tumor development, which proceeds through three phases termed elimination, equilibrium, and escape [12]. Rare tumor subclones capable of surviving elimination can progress into the equilibrium phase, in which net tumor mass is sustained over time. However, the constant pressure from the adaptive immune system coupled with the genetic instability of tumor cells can select for tumor subclones with reduced immunogenicity that can evade immune recognition and destruction [13]. In particular, the importance of CD8+ T cells in cancer immunoediting has been shown, and more broadly in those tumors with an adaptive immune resistance phenotype [14]. TNBC patients with high infiltration of TILs are reported to correlate with a higher response to neoadjuvant therapy [7,8,15,16]. Given the mechanism that not only the existence of CD8+ T cells, but its immune cell killing activity is linked to treatment response, we speculated that cytolytic activity may correlate with clinical outcomes stronger than the number of TILs, which was the measure used in previous studies.
A recent study identified that immune cytolytic activity (CYT), which reflects the cell killing function by a geometric mean of gene expressions of GZMA and PRF1, can be used to assess immune-mediated attack against cancer cells [17], providing an attractive, easy to widely implement index for prognosis of cancer and guide therapeutic decisions. CYT and its related genes have been demonstrated to represent effector function of CD8+ T-cells with the resistance or responsiveness to immunotherapy in melanoma [18]. CYT was also shown to be inversely linked to genomic alterations compared to benign tissue suggesting that intrinsic oncogenic processes drive immune inactivity in pancreatic cancer [19]. In colorectal cancer, our group and others determined that CYT is associated with the mutational burden and immune microenvironment [20,21]. We also reported that CD8+ T cells estimated by xCell algorithm from bulk tumor transcriptome was highly correlated with CYT in breast cancer patients [4]. However, less is known on the clinical impact of CYT and its interaction with cancer cells in TIME of breast cancer patients. In the present study, we hypothesized that CYT is associated with immune-mediated antitumor elimination and with alteration of intra-tumor genetic heterogeneity, which impacts clinical outcomes including survival and treatment response in breast cancer. We test this hypothesis using CYT score in multiple large breast cancer cohorts; The Cancer Genome Atlas (TCGA), Molecular Taxonomy of Breast Cancer International Consortium (METABRIC), and multiple Gene Expression Omnibus (GEO) cohorts.
Materials and methods
Patient cohort and genomic data processing
Transcriptome data was acquired from The Cancer Genome Atlas (TCGA) Breast cancer cohort through cBioportal [22]. Survival endpoints for TCGA cases were used the Pan-Cancer Clinical Data Resource [23]. Female samples with gene expression data were used in this analysis (n = 1069), as we previously reported [24,25]. Regarding sample processing in TCGA project, frozen samples were collected from patients with untreated breast cancers and processed by the Biospecimen Core Resource [26,27]. The RNA was processed and sent to Genome Characterization Centers (GSCs) and Genome Sequencing Centers (GSCs) where they were sequenced. These results were then sent to the TCGA Research network and further analyzed, interpreted, and made publicly available [26,27]. The data set derived from Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) (n = 1904) was utilized to validate survival analysis [28]. Gene Expression Omnibus (GEO) data sets from GSE96058 (n = 3723) studied by Brueffer et al. [29] was also utilized as validation cohort. GEO data sets from studies by Shi et al. (GSE20194 (n = 248)) [30], Hatzis et al. (GSE25066 (n = 508)) [31], and Massarweh et al. (GSE33658 (n = 81)) [32] were utilized to correlate pre-treatment CYT with therapeutic response to chemotherapy in breast cancer patients. This study was deemed exempt from Roswell Park Cancer Institution Institutional Review Board evaluation because all genomic and clinical information within TCGA is publicly accessible and de-identified [33,34].
Gene expression data and analysis based on RNA-Seq
Gene expression data were obtained in RSEM format and converted to Transcripts Per Million (TPM) by a given gene’s estimated fraction of transcripts and multiplying with 10^6. CYT was defined as the geometric mean of GZMA and PRF1 expression values in TPM [17,19]. The threshold of dichotomization of CYT high and low groups was determined by comparing differences in the overall survival between the two groups and the cutoff point that gave the least p-value was chosen.
Determination of tumor infiltrating immune cell composition utilizing xCell
xCell algorithm [35] was used to estimate the fraction of sixty-four infiltrating immune cell types as well as stromal cells in each tumor tissue to evaluate intra-tumor cell composition. The sixty-four cell fractions were calculated via their online calculator, as we previously reported [36,37].
Determination of mutant-allele tumor heterogeneity (MATH), tumor clonal analysis, T-cell receptor (TCR) diversity and inhibitory checkpoint molecule (ICM) index
Mutant-allele tumor heterogeneity (MATH) score, a measure of intra-tumor heterogeneity, was calculated through R/Bioconductor package “maftools”; efficient analysis, visualization and summarization of (MAF) files from large-scale cohort-based cancer studies (https://www.biorxiv.org/content/early/2016/05/11/052662) [38-40]. PyClone was utilized to infer tumor clonality by clustering variants of similar MAF from tumor samples in TCGA [41,42]. ICM index was created by Balli et al., using six gene expression [19].
Homologous Recombination Deficiency (HRD) determination
Homologous recombination deficiency (HRD) has emerged as a biomarker representing genomic scar or genomic instability in cancers [43,44]. In the present study, HRD score is defined using large-scale state transitions (LST) score based on the previous reports [45].
Gene Set Enrichment Analysis (GSEA) of hallmark gene sets
Gene Set Enrichment Analysis (GSEA) was performed using software provided by the Broad Institute (http://software.broadinstitute.org/gsea/index.jsp) [46], as we previously reported [47-51]. A collection of annotated gene sets for use with GSEA software can be found in the Molecular Signatures Database (MSigDB) (http://software.broadinstitute.org/gsea/msigdb). The result of the GSEA is the enrichment score (ES), which reflects the degree to which a gene set is overrepresented at the top or bottom of a ranked list of genes. Statistical analyses were performed using R software (http://www.r-project.org/). Statistical significance was defined by false discovery rate (FDR) less than 25%, as GSEA software recommended.
Statistical analysis
All statistical analyses were performed using R software (https://www.r-project.org/) and Bioconductor (https://www.bioconductor.org/). Kaplan-Meier method with log-rank test was used to compare survival curves between groups. For continuous variables, the differences between two groups were assessed by Mann-Whitney test; for discrete variables, Fisher’s exact test was used to evaluate the association between factors. Spearman correlation was used to describe the relationship between gene expressions and CYT. Association between variables (e.g. gene expression, mutation load and immune checkpoint molecule index) was determined using the Mann-Whitney U test. In all analyses, a two-sided P < 0.05 was considered statistically significant and this “prognostic marker” study is conducted according to the REMARK guidelines [52].
Results
High cytolytic activity score (CYT) is signficantly associated with Nottingham histologic grade in breast cancer
We reported that breast cancer with high mutation load was associated with both aggressive phenotype and infiltration of anti-cancer immune cells [53]. To this end, we expected breast cancer with high CYT to associate with a more aggressive phenotype. We used median value to divide into low and high CYT groups within each cohort. Disease specific survival (DSS) in The Cancer Genome Atlas (TCGA; n = 1069) and the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC; n = 1904) cohort, as well as overall survival (OS) in the GSE96058 (n = 3273) cohort was analyzed, and high CYT was not associated with worse survival in any cohorts (Figure 1A). We next examined whether there was an association between the CYT and clinical aggressiveness in breast cancer. The CYT was significantly higher in advanced American Joint Committee on Cancer (AJCC) staging in METABRIC, but not in the TCGA cohort (Figure 1B). On the other hand, CYT was high in advanced Nottingham histological grade consistently in all three cohorts, TCGA, METABRIC and GSE96058 (Figure 1B; all P < 0.001). These results suggest that CYT is associated with cancer cell proliferation, but does not translate to cancer stage or survival in the whole cohorts.
Figure 1.
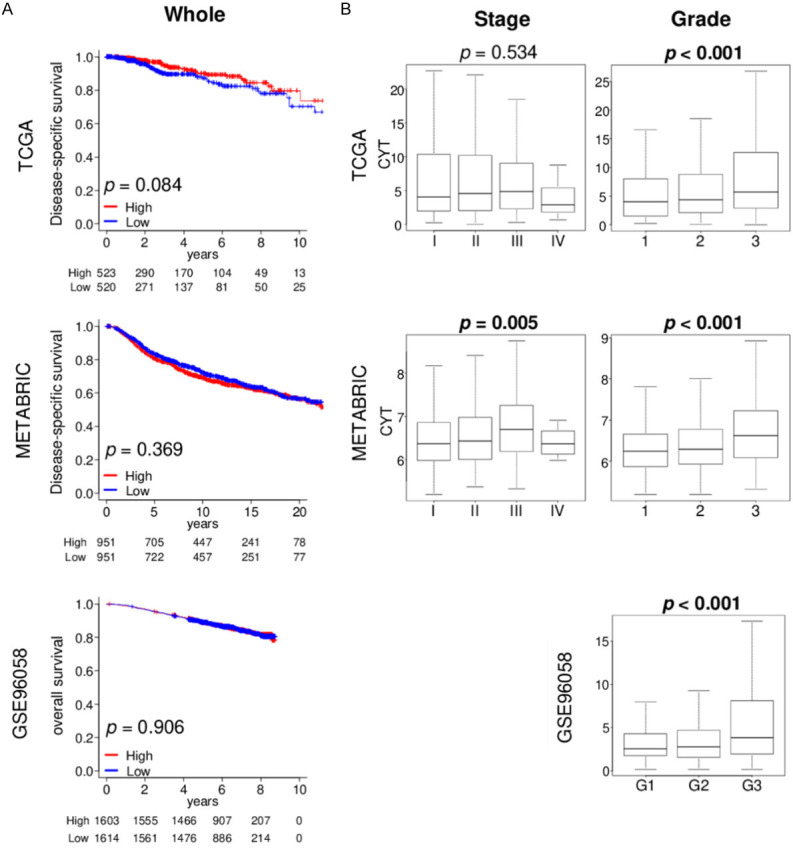
Association of the cytolytic activity score (CYT) with clinical cancer aggressiveness in the TCGA, METABRIC, and GSE96058 cohorts. A. Kaplan-Meier plots of disease-specific survival in the TCGA and METABRIC cohorts, and overall survival in the GSE96058 cohort by CYT low (blue line) and high (red line) within whole breast cancer samples. Median cut-off was used to divide two groups. Log-rank test was used to calculate p value. B. Boxplots of the CYT by AJCC stage in the TCGA and METABRIC cohort, and Nottingham histological grade in the TCGA, METABRIC, and GSE96058 cohorts. Kruskal-Wallis test was used to calculate p value. AJCC, American Joint Committee on Cancer.
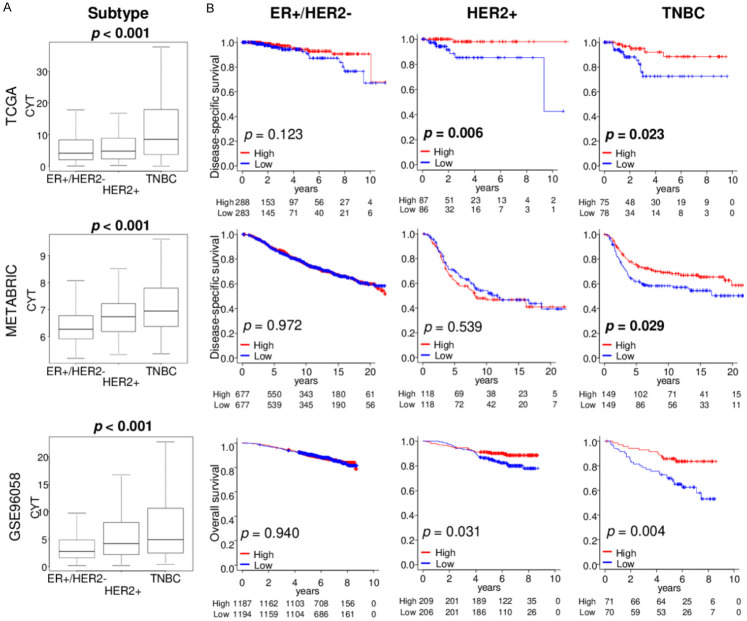
High CYT in triple-negative breast cancer (TNBC) is signficantly associated with better survival
Since it is well known that the amount of immune cell infiltration differs by each subtype, where TNBC is the most abundant, we studied the relationship of CYT with patient survival in each subtype. We found that CYT was significantly higher in TNBC compared to the other subtypes consistently in all three cohorts, TCGA, METABRIC and GSE96058 (Figure 2A; all P < 0.001). High CYT was significantly associated with better patient survival in TNBC consistently in all three cohorts (Figure 2B; DSS: P = 0.023 and 0.029 in TCGA and METABRIC, and OS: P = 0.004 in GSE96058, respectively), and in HER2 in TCGA and GSE96058 cohorts (Figure 2B; DSS: P = 0.006 in TCGA and OS: P = 0.031 in GSE96058, respectively). In HER2-positive breast cancer group, high CYT was significantly associated with better survival in two cohort (DSS: P = 0.006 in TCGA and p = 0.539 in METABRIC, and P = 0.031 in GSE96058), but there were no significance in ER-positive/HER2-negative breast cancer cohorts. These findings suggest that high CYT was associated with favorable patient survival in aggressive immunogenic TNBC.
Figure 2.
CYT levels by subtype and its association with patient survival by subtype in TCGA, METABRIC, and GSE96058 cohorts. A. Boxplots of the CYT by breast cancer subtypes; ER+/HER2-, HER2+, and TNBC. Kruskal-Wallis test was used to calculate p value. B. Kaplan-Meier plots of disease-specific survival in the TCGA and METABRIC cohorts, and overall survival in the GSE96058 cohort by CYT low (blue line) and high (red line) within each breast cancer subtype groups. Median cut-off was used to divide two groups. Log-rank test was used to calculate p value.
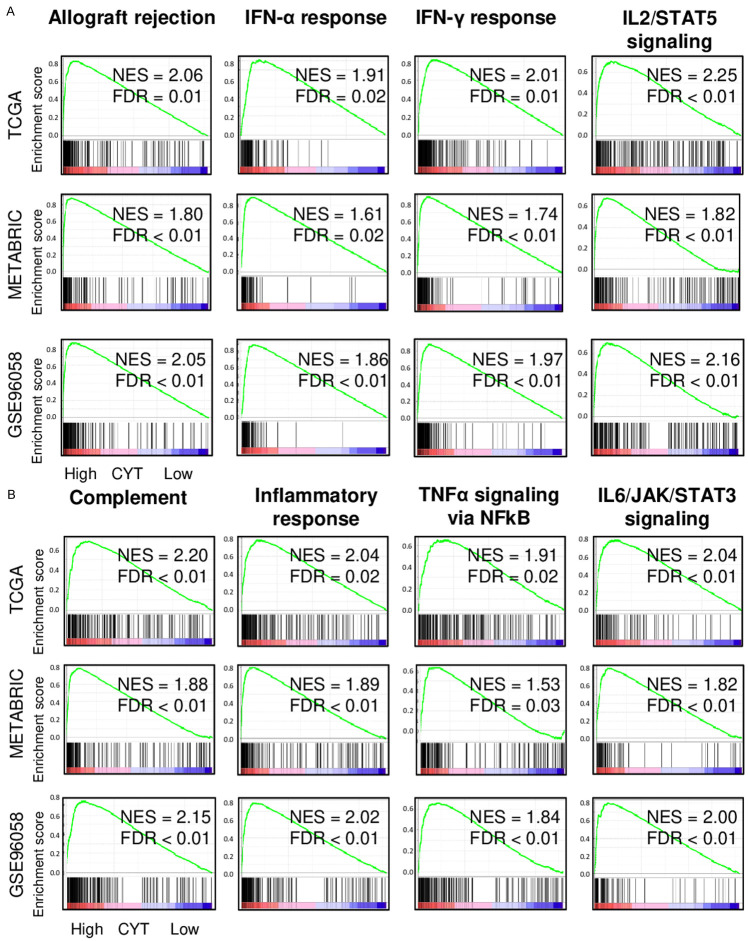
High CYT TNBC significantly enriched both favorable immune-relaated and unfavorable inflammation-related gene sets
TNBC with a high CYT was expected to have favorable tumor immune microenvironment since it was associated with better survival. To test whether this is the case, gene set enrichment analyses (GSEA) of the Hallmark gene sets was performed in TNBC of TCGA, METABRIC, and GSE96058 cohorts. High CYT TNBC significantly enriched many favorable immune-related Hallmark gene sets, such as allograft rejection, interferon (IFN)-α response and IFN-γ response, IL2/STAT5 signaling, and complement consistently in all three cohorts (Figure 3A). Interestingly, high CYT TNBC enriched unfavorable inflammation-related gene sets as well, such as inflammatory response, TNF-α signaling, and IL6/JAK/STAT3 signaling consistently in all three cohort (Figure 3B). These results suggest that CYT is associated with both favorable immune reaction and unfavorable inflammation which are most likely intricately intertwined and led to context-dependent clinical outcomes in TNBC.
Figure 3.
Gene Set Enrichment Assay (GSEA) of high CYT TNBC in the TCGA, METABRIC and GSE96058 cohort. Enrichment plots are shown for (A) favorable immune-related and (B) unfavorable inflammation-related Hallmark gene sets. CYT was compared from high (left) to low (right), along with normalized enrichment score (NES) and false discovery rate (FDR). FDR of 0.25 was used as statistical significance of GSEA. Median cut-off was used to divide two groups.
High CYT TNBC is significantly associated with high fraction of favorable anti-cancer immune cells
Since high CYT TNBC enriched immune-related gene sets, it was of interest to determine which types of immune cells are infiltrated in these tumors. xCell algorithm, which estimates immune cell composition by gene expression data of a bulk tumor, was utilized. We found that a significantly higher fraction of anti-cancer immune cells, CD8+ T cells, CD4+ memory T cells, M1 macrophages, and dendritic cells, as well as B cells, were infiltrated in the high CYT TNBC consistently in TCGA, METABRIC, and GSE96058 cohorts (Figure 4A; all P < 0.001). High CYT significantly correlated with higher T cell receptor (TCR) diversity, which is calculated by Thorsson et al. in the TCGA cohort (Figure 4B; P < 0.001). Furthermore, high CYT score was significantly associated with high TCR Shannon score as well as B cell receptor (BCR) Shannon score, which represent the TCR and BCR diversity that is thought to enhance their anti-cancer potency (Figure 4C; all P < 0.001). On the other hand, high CYT TNBC was not associated with fraction of pro-cancer immune cells; regulatory T cells, T helper type 2 cells (Th2), and M2 macrophages in the TCGA, whereas they did in the METABRIC and GSE96058 cohorts (Figure 4D). These results suggest that high CYT TNBC was associated with enhanced immune response and favorable anti-cancer immune cells compared to low CYT.
Figure 4.
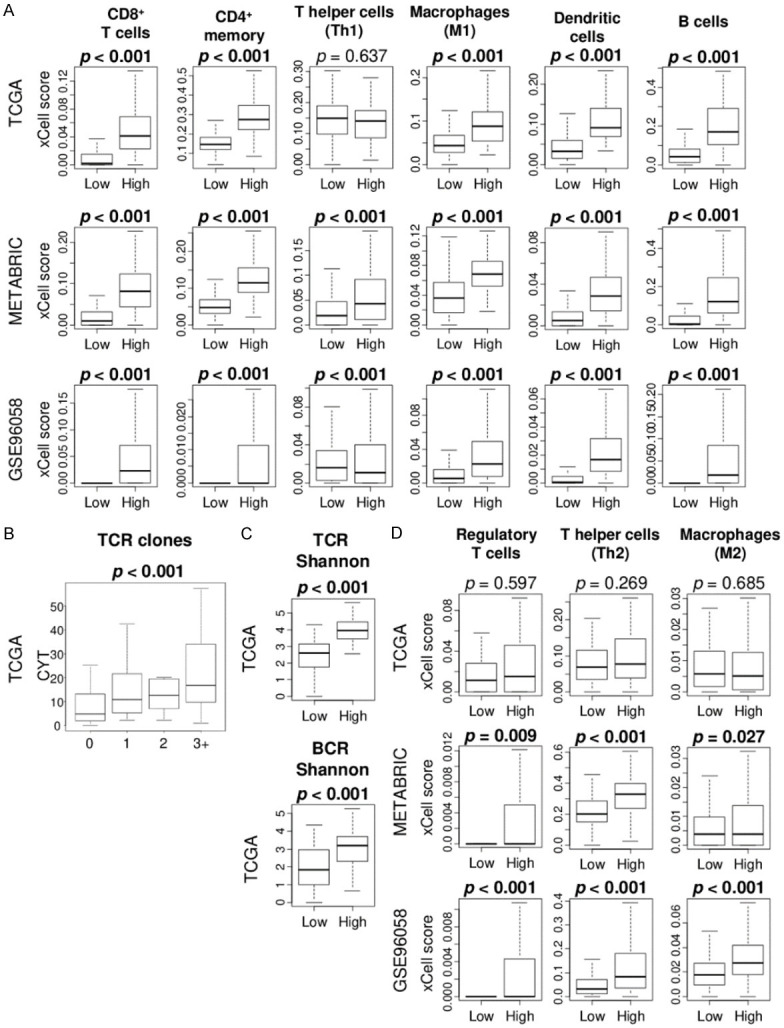
Association of CYT with fraction of infiltrating immune cells in the TCGA, METABRIC, and GSE96058 cohorts. A. Boxplots of the fraction of anti-cancer immune cells; CD8+ T cells, CD4+ memory T cells, T helper type1 cells (Th1), M1 macrophages, and dendritic cells, and B cells were compared by low and high CYT group. Mann Whitney U test was used to calculate p value. B. Boxplots of the CYT by T cell receptor (TCR) clones score in the TCGA cohort. Kruskal-Wallis test was used to calculate p value. C. Boxplots of the comparison of TCR richness and BCR richness score between low and high CYT groups in the TCGA cohort. D. Boxplots of the fraction of pro-cancer immune cells; regulatory T cells, T helper type 2 (Th2) cells, and M2 macrophages were compared by low and high CYT. Mann Whitney U test was used to calculate p value. Median cut-off was used to divide two groups.
High CYT TNBC are significantly associated with high homologous recombination deficiency (HRD) but with less intra-tumoral heterogeneity
The concept of immunoediting is that the immune system can both constrain and promote tumor development, where the constant pressure from the adaptive immune system coupled with the genetic instability of tumor cells can select for aggressive tumor subclones that can evade immune recognition and destruction [54]. Together with the notion that high mutation burden and neoepitope load attracts and is attacked by effector CD8+ T cells, we expected that high CYT TNBC is associated with mechanisms to generate mutations, but highly mutated cells may be eliminated which results in less intra-tumoral heterogeneity. Indeed, high CYT TNBC was associated with high homologous recombination deficiency (HRD), but with low somatic copy number alteration (SCNA) score and mutant allele tumor heterogeneity (MATH) index (Figure 5A and 5B). CYT did show the trend to be lower in high number of clones by Pyclone although there was no statistical difference, and there was no trend in intratumor heterogeneity score in the TCGA cohort (Figure 5C and 5D). CYT was not associated with mutation-related score, including single-nucleotide variants (SNV) and indel neoantigens, silent and non-silent mutation rate in the TCGA cohort (Figure 5D). These results are in alignment with the notion that anti-cancer immunity represented by CYT enhances selection pressure to clonability and less heterogeneity which is associated with less aggressive biological phenotype of TNBC.
Figure 5.
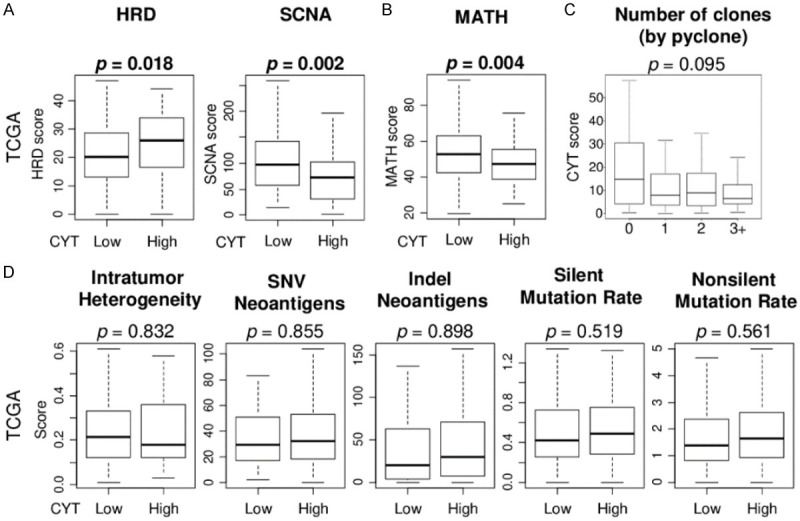
Association of the CYT score with intra-tumor genetic heterogeneity and mutations in the TCGA TNBC cohort. (A) Box plots of the homologous recombination deficiency (HRD) and somatic copy number alteration (SCNA), and (B) mutant allele tumor heterogeneity (MATH) index by low and high CYT score groups. (C) Box plots of the CYT score by number of clones by Pyclone. (D) Box plots of the intratumor heterogeneity and mutation-related score; single-nucleotide variants (SNV) and indel neoantigens, silent and non-silent mutation rate, by low and high CYT score groups. Mann Whitney U test and Kruskal-Wallis test were used to calculate p value. Median cut-off was used to divide two groups.
High CYT was not associated with pathological complete response (pCR) after neoadjuvant chemotherapy (NAC), but was significanly associated with high expression of immune checkpoint genes in both ER-positive/HER2-negative and TNBC
Since TIL has been reported as a predictive biomarker for pCR after NAC [55], we expected that high cytolytic activity in breast cancer would associate with response to NAC particularly in TNBC. However, CYT was not associated with pCR after NAC in ER-positive/HER2-negativie breast cancer nor TNBC in any of the cohort examined (GSE20194, GSE25066, and GSE32646) (Figure 6A). Recently there have been number of reports on the use of immune checkpoint inhibitors for breast cancer [56]. Since high CYT tumors attract effector T cells, we expected the immune checkpoint molecule expressions to be elevated. We found that high CYT was significantly associated with high expression of numerous immune checkpoint molecules in the TCGA, METABRIC and GSE96058 cohorts. Specifically, programmed cell death 1 (PDCD1 (PD-1)), programmed cell death 1 ligand 1 (CD274 (PD-L1)), programmed cell death 1 ligand 2 (PDCD1LG2 (PD-L2)), cytotoxic T-lymphocyte-associated protein 4 (CTLA4), indoleamine 1 (IDO1), IDO2, lymphocyte activation gene 3 (LAG3), and T cell immunoreceptor with Ig and ITIM domains (TIGIT) were significantly highly expressed in high CYT TNBC, as well as ER-positive/HER2-negative, and HER2-positive breast cancer (Figure S1). Furthermore, we found a strong correlation between CYT and the inhibitory checkpoint molecule (ICM) index using six gene expressions, in the TCGA and GSE96058 cohorts (Figure 6B; Spearman’s rank correlation (r) = 0.842 and 0.755 in ER-positive/HER2-negative, r = 0.848 and 0.784 in TNBC, respectively, all P < 0.01). These results suggest that CYT reflects global expression of immune checkpoint molecules.
Figure 6.
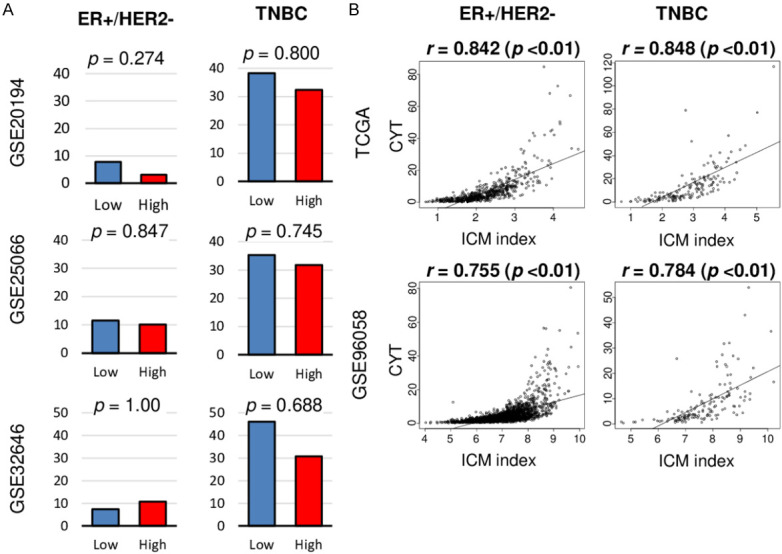
Association of the CYT score with drug treatment response for breast cancer. A. Bar plots of comparison of the pathological complete response (pCR) rate by low and high CYT group in ER+/HER2- and TNBC in the GSE20194, GSE25066, and GSE33658 cohorts. Fisher’s exact test was used to calculate p value. Median cut-off was used to divide two groups. Fisher’s exact test was used to calculate p value. B. Correlation plots between CYT and inhibitory checkpoint molecule (ICM) index in ER+/HER2- and TNBC in the TCGA and GSE96058 cohorts. Spearman’s rank correlation was used for the analysis.
Discussion
In this study, we found that high CYT was significantly associated with advanced histological grade and TNBC subtype. High CYT TNBC was significantly associated with better survival, but the other subtypes were not. High CYT TNBC enriched both favorable immune-related and unfavorable inflammation-related gene sets. High CYT TNBC was associated with high infiltration of anti-cancer immune cells (CD8+ T cells, CD4+ memory T cells, M1 macrophages, and dendritic cells, and B cells). High CYT TNBC was also associated with high level of T cell receptor (TCR) clones and TCR and B cell receptor (BCR) richness. High CYT TNBC was associated with high HRD and less intra-tumor heterogeneity (SCNA and MATH) but not with mutation load. Although CYT was not associated with NAC response in both ER-positive/HER2-negative nor TNBC, high CYT strongly correlated with immune checkpoint molecule index and was associated with high expression of multiple immune checkpoint molecule genes in both ER-positive/HER2-negative and TNBC.
Higher number of infiltrating lymphocytes into the tumor microenvironment is known to associate with favorable outcomes in several cancers [57]. We speculated that improvement in patient outcomes with TILs are related to the increased anti-tumor CYT rather than the number of cells. In the present study, using publicly available cohorts with total of 7533 primary breast cancer patients, we profiled the genomic and transcriptional data in the context of the tumor immune microenvironment. We utilized CYT that is calculated by the gene expression signature of granzyme A (GZMA) and perforin-1 (PRF1) to assess immune cytolytic activity in a bulk tumor [17]. GZMA is a tryptase that induces caspase-independent programmed cell death and PRF1 is a pore-forming enzyme that mediates entry of granzymes into target cells through perforin-polymer, both produced by activated cytolytic CD8+ T-cells or macrophages and upregulated following immune reaction or response to immunotherapy [58-60]. Granzyme B is also secreted by cytotoxic CD8+ T-cells same as granzyme A [58], however it was not included in the original paper that established CYT by Rooney et al. [17]. Additionally, recent data suggest that the relative contributions of granzymes A and B depend on properties of the target cell [61]. We previously reported that CYT was strongly correlated with fraction of CD8+ T cell in breast cancer using xCell algorithm [4], and is associated with tumor immune microenvironment of hepatocellular carcinoma [62] and colorectal cancer [21]. In the current study GSEA showed that both immune response-related and inflammation-related gene sets were enriched in the high CYT tumors. Of note, several immune response-related gene sets had higher NES compared to inflammation-related gene sets consistently in all three cohorts. In addition, cytolytic activity has been shown to be associated with high infiltration of many anti-cancer immune cells, which has been linked to its association with better prognosis in TNBC patients.
Recently, multiregional genome sequencing has allowed us to elucidate intra-tumor cancer evolution [63]. Some claim that tumors commonly show two types of the cancer evolution; branched evolution [64-66] and neutral evolution [67-69], which depend on various selective pressure during its evolution, including host microenvironment, intra-tumor inter-cellular cooperation, therapeutics, and immunoediting [63]. Namely, lower selective pressure allows tumors to grow heterogeneously (neutral evolution) and higher selective pressure leads to less tumor heterogeneity (branched evolution). In the present study, our results, which indicated that lower CYT is associated with higher intra-tumor heterogeneity, is in agreement with this concept.
KEYNOTE-522 is a phase III study which shows that achievement of pCR was significantly higher among those who received anti-PD-1 antibody (pembrolizumab) plus NAC than among those who received placebo plus NAC in TNBC [70]. Immune checkpoint inhibitors have been shown to be effective in metastatic breast cancer as well, but the overall response rate remains low [71]. Approximately 10% of patients receiving anti-PD-1 antibodies report more than 3 adverse effects [72]. Although CYT did not predict pCR after NAC, we found that tumors with high CYT were associated with significant increase in expression of multiple immune checkpoint genes. We cannot help but speculate that CYT may be a useful tool as a biomarker of immune checkpoint inhibitors and this warrants further prospective studies to prove its utility in the future.
Our study is not without limitations. This is a retrospective study that utilized multiple cohorts with clinical and genetic data, however, data on co-morbidity and therapeutic intervention are missing. The largest limitation of this study from its clinical application standpoint is that we utilized curated gene expression data without any direct quantification of tumoral CYT with more affordable measures such as quantitative RT-PCR. We defined immune cells by the transcriptomic profile determined by the xCell algorithm, which may or may not capture all the cells defined by the gold standard, since not all gene expressions are translated to proteins. We should also recognize the limitation that the present study demonstrated immunogenomic landscape utilizing TCGA cohort, thus we demonstrate only association and not causality with CYT and the other factors we analyzed. To this end, the results of this study should be validated with a prospective cohort with generalizable methods to translate our result in order to have a higher clinical impact.
To our knowledge, this is the first report to elucidate that cytolytic activity in the bulk tumor is associated with enhanced anti-cancer immunity, less intra-tumoral heterogeneity, and with better survival in TNBC patients, suggesting that intrinsic oncogenic processes strongly correlate with intra-tumoral immune landscape.
Acknowledgements
This work was supported by US National Institutes of Health/National Cancer Institute grant R01CA160688, R01CA250412, R37CA248018, US Department of Defense BCRP grant W81XWH-19-1-0674, as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to K.T., and US National Cancer Institute cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ogino S, Galon J, Fuchs CS, Dranoff G. Cancer immunology--analysis of host and tumor factors for personalized medicine. Nat Rev Clin Oncol. 2011;8:711–719. doi: 10.1038/nrclinonc.2011.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid Dendritic Cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in Triple Negative Breast Cancer (TNBC) more strongly than Conventional Dendritic Cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speiser DE, Ho PC, Verdeil G. Regulatory circuits of T cell function in cancer. Nat Rev Immunol. 2016;16:599–611. doi: 10.1038/nri.2016.80. [DOI] [PubMed] [Google Scholar]
- 4.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 6.Cesano A, Warren S. Bringing the next generation of immuno-oncology biomarkers to the clinic. Biomedicines. 2018;6:14. doi: 10.3390/biomedicines6010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denkert C, von Minckwitz G, Brase JC, Sinn BV, Gade S, Kronenwett R, Pfitzner BM, Salat C, Loi S, Schmitt WD, Schem C, Fisch K, Darb-Esfahani S, Mehta K, Sotiriou C, Wienert S, Klare P, André F, Klauschen F, Blohmer JU, Krappmann K, Schmidt M, Tesch H, Kümmel S, Sinn P, Jackisch C, Dietel M, Reimer T, Untch M, Loibl S. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 2015;33:983–991. doi: 10.1200/JCO.2014.58.1967. [DOI] [PubMed] [Google Scholar]
- 8.Denkert C, Loibl S, Noske A, Roller M, Müller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, von Törne C, Weichert W, Engels K, Solbach C, Schrader I, Dietel M, von Minckwitz G. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 9.Green AR, Aleskandarany MA, Ali R, Hodgson EG, Atabani S, De Souza K, Rakha EA, Ellis IO, Madhusudan S. Clinical impact of tumor DNA repair expression and T-cell infiltration in breast cancers. Cancer Immunol Res. 2017;5:292–299. doi: 10.1158/2326-6066.CIR-16-0195. [DOI] [PubMed] [Google Scholar]
- 10.Mahmoud SM, Paish EC, Powe DG, Macmillan RD, Grainge MJ, Lee AH, Ellis IO, Green AR. Tumor-infiltrating CD8+ lymphocytes predict clinical outcome in breast cancer. J. Clin. Oncol. 2011;29:1949–1955. doi: 10.1200/JCO.2010.30.5037. [DOI] [PubMed] [Google Scholar]
- 11.Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, Earl HM, Poole CJ, Hiller L, Dunn JA, Bowden SJ, Twelves C, Bartlett JM, Mahmoud SM, Rakha E, Ellis IO, Liu S, Gao D, Nielsen TO, Pharoah PD, Caldas C. Association between CD8+ T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014;25:1536–1543. doi: 10.1093/annonc/mdu191. [DOI] [PubMed] [Google Scholar]
- 12.O’Donnell JS, Teng MWL, Smyth MJ. Cancer immunoediting and resistance to T cell-based immunotherapy. Nat Rev Clin Oncol. 2019;16:151–167. doi: 10.1038/s41571-018-0142-8. [DOI] [PubMed] [Google Scholar]
- 13.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, Bhatia S, Hwu WJ, Gajewski TF, Slingluff CL Jr, Chowell D, Kendall SM, Chang H, Shah R, Kuo F, Morris LGT, Sidhom JW, Schneck JP, Horak CE, Weinhold N, Chan TA. Tumor and Microenvironment evolution during immunotherapy with Nivolumab. Cell. 2017;171:934–949. e916. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teng MW, Galon J, Fridman WH, Smyth MJ. From mice to humans: developments in cancer immunoediting. J Clin Invest. 2015;125:3338–3346. doi: 10.1172/JCI80004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Melichar B, Študentova H, Kalábová H, Vitásková D, Čermáková P, Hornychová H, Ryška A. Predictive and prognostic significance of tumor-infiltrating lymphocytes in patients with breast cancer treated with neoadjuvant systemic therapy. Anticancer Res. 2014;34:1115–1125. [PubMed] [Google Scholar]
- 16.Ono M, Tsuda H, Shimizu C, Yamamoto S, Shibata T, Yamamoto H, Hirata T, Yonemori K, Ando M, Tamura K, Katsumata N, Kinoshita T, Takiguchi Y, Tanzawa H, Fujiwara Y. Tumor-infiltrating lymphocytes are correlated with response to neoadjuvant chemotherapy in triple-negative breast cancer. Breast Cancer Res Treat. 2012;132:793–805. doi: 10.1007/s10549-011-1554-7. [DOI] [PubMed] [Google Scholar]
- 17.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160:48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, Merchant AS, Mehta GU, Chichura A, Shalem O, Tran E, Eil R, Sukumar M, Guijarro EP, Day CP, Robbins P, Feldman S, Merlino G, Zhang F, Restifo NP. Identification of essential genes for cancer immunotherapy. Nature. 2017;548:537–542. doi: 10.1038/nature23477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Balli D, Rech AJ, Stanger BZ, Vonderheide RH. Immune cytolytic activity stratifies molecular subsets of human pancreatic cancer. Clin Cancer Res. 2017;23:3129–3138. doi: 10.1158/1078-0432.CCR-16-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zaravinos A, Roufas C, Nagara M, de Lucas Moreno B, Oblovatskaya M, Efstathiades C, Dimopoulos C, Ayiomamitis GD. Cytolytic activity correlates with the mutational burden and deregulated expression of immune checkpoints in colorectal cancer. J Exp Clin Cancer Res. 2019;38:364. doi: 10.1186/s13046-019-1372-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Narayanan S, Kawaguchi T, Yan L, Peng X, Qi Q, Takabe K. Cytolytic activity score to assess anticancer immunity in colorectal cancer. Ann Surg Oncol. 2018;25:2323–2331. doi: 10.1245/s10434-018-6506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oshi M, Newman S, Murthy V, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. ITPKC as a prognostic and predictive biomarker of neoadjuvant chemotherapy for triple negative breast cancer. Cancers (Basel) 2020;12:2758. doi: 10.3390/cancers12102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brueffer C, Vallon-Christersson J, Grabau D, Ehinger A, Häkkinen J, Hegardt C, Malina J, Chen Y, Bendahl PO, Manjer J, Malmberg M, Larsson C, Loman N, Rydén L, Borg Å, Saal LH. Clinical value of RNA sequencing-based classifiers for prediction of the five conventional breast cancer biomarkers: a report from the population-based multicenter Sweden cancerome analysis network-breast initiative. JCO Precis Oncol. 2018;2 doi: 10.1200/PO.17.00135. PO.17.00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shi L, Campbell G, Jones WD, Campagne F, Wen Z, Walker SJ, Su Z, Chu TM, Goodsaid FM, Pusztai L, Shaughnessy JD Jr, Oberthuer A, Thomas RS, Paules RS, Fielden M, Barlogie B, Chen W, Du P, Fischer M, Furlanello C, Gallas BD, Ge X, Megherbi DB, Symmans WF, Wang MD, Zhang J, Bitter H, Brors B, Bushel PR, Bylesjo M, Chen M, Cheng J, Cheng J, Chou J, Davison TS, Delorenzi M, Deng Y, Devanarayan V, Dix DJ, Dopazo J, Dorff KC, Elloumi F, Fan J, Fan S, Fan X, Fang H, Gonzaludo N, Hess KR, Hong H, Huan J, Irizarry RA, Judson R, Juraeva D, Lababidi S, Lambert CG, Li L, Li Y, Li Z, Lin SM, Liu G, Lobenhofer EK, Luo J, Luo W, McCall MN, Nikolsky Y, Pennello GA, Perkins RG, Philip R, Popovici V, Price ND, Qian F, Scherer A, Shi T, Shi W, Sung J, Thierry-Mieg D, Thierry-Mieg J, Thodima V, Trygg J, Vishnuvajjala L, Wang SJ, Wu J, Wu Y, Xie Q, Yousef WA, Zhang L, Zhang X, Zhong S, Zhou Y, Zhu S, Arasappan D, Bao W, Lucas AB, Berthold F, Brennan RJ, Buness A, Catalano JG, Chang C, Chen R, Cheng Y, Cui J, Czika W, Demichelis F, Deng X, Dosymbekov D, Eils R, Feng Y, Fostel J, Fulmer-Smentek S, Fuscoe JC, Gatto L, Ge W, Goldstein DR, Guo L, Halbert DN, Han J, Harris SC, Hatzis C, Herman D, Huang J, Jensen RV, Jiang R, Johnson CD, Jurman G, Kahlert Y, Khuder SA, Kohl M, Li J, Li L, Li M, Li QZ, Li S, Li Z, Liu J, Liu Y, Liu Z, Meng L, Madera M, Martinez-Murillo F, Medina I, Meehan J, Miclaus K, Moffitt RA, Montaner D, Mukherjee P, Mulligan GJ, Neville P, Nikolskaya T, Ning B, Page GP, Parker J, Parry RM, Peng X, Peterson RL, Phan JH, Quanz B, Ren Y, Riccadonna S, Roter AH, Samuelson FW, Schumacher MM, Shambaugh JD, Shi Q, Shippy R, Si S, Smalter A, Sotiriou C, Soukup M, Staedtler F, Steiner G, Stokes TH, Sun Q, Tan PY, Tang R, Tezak Z, Thorn B, Tsyganova M, Turpaz Y, Vega SC, Visintainer R, von Frese J, Wang C, Wang E, Wang J, Wang W, Westermann F, Willey JC, Woods M, Wu S, Xiao N, Xu J, Xu L, Yang L, Zeng X, Zhang J, Zhang L, Zhang M, Zhao C, Puri RK, Scherf U, Tong W, Wolfinger RD. The MicroArray Quality Control (MAQC)-II study of common practices for the development and validation of microarray-based predictive models. Nat Biotechnol. 2010;28:827–838. doi: 10.1038/nbt.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, Martin M, Cotrina J, Gomez H, Hubbard R, Chacón JI, Ferrer-Lozano J, Dyer R, Buxton M, Gong Y, Wu Y, Ibrahim N, Andreopoulou E, Ueno NT, Hunt K, Yang W, Nazario A, DeMichele A, O’Shaughnessy J, Hortobagyi GN, Symmans WF. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massarweh S, Tham YL, Huang J, Sexton K, Weiss H, Tsimelzon A, Beyer A, Rimawi M, Cai WY, Hilsenbeck S, Fuqua S, Elledge R. A phase II neoadjuvant trial of anastrozole, fulvestrant, and gefitinib in patients with newly diagnosed estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2011;129:819–827. doi: 10.1007/s10549-011-1679-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ramanathan R, Olex AL, Dozmorov M, Bear HD, Fernandez LJ, Takabe K. Angiopoietin pathway gene expression associated with poor breast cancer survival. Breast Cancer Res Treat. 2017;162:191–198. doi: 10.1007/s10549-017-4102-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stickeler E, Pils D, Klar M, Orlowsk-Volk M, Zur Hausen A, Jäger M, Watermann D, Gitsch G, Zeillinger R, Tempfer CB. Basal-like molecular subtype and HER4 up-regulation and response to neoadjuvant chemotherapy in breast cancer. Oncol Rep. 2011;26:1037–1045. doi: 10.3892/or.2011.1392. [DOI] [PubMed] [Google Scholar]
- 35.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.PLOS Medicine Staff. Correction: intra-tumor genetic heterogeneity and mortality in head and neck cancer: analysis of data from the cancer genome atlas. PLoS Med. 2015;12:e1001844. doi: 10.1371/journal.pmed.1001844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mroz EA, Rocco JW. MATH, a novel measure of intratumor genetic heterogeneity, is high in poor-outcome classes of head and neck squamous cell carcinoma. Oral Oncol. 2013;49:211–215. doi: 10.1016/j.oraloncology.2012.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rocco JW. Mutant allele tumor heterogeneity (MATH) and head and neck squamous cell carcinoma. Head Neck Pathol. 2015;9:1–5. doi: 10.1007/s12105-015-0617-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roth A, Khattra J, Yap D, Wan A, Laks E, Biele J, Ha G, Aparicio S, Bouchard-Côté A, Shah SP. PyClone: statistical inference of clonal population structure in cancer. Nat Methods. 2014;11:396–398. doi: 10.1038/nmeth.2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris LG, Riaz N, Desrichard A, Şenbabaoğlu Y, Hakimi AA, Makarov V, Reis-Filho JS, Chan TA. Pan-cancer analysis of intratumor heterogeneity as a prognostic determinant of survival. Oncotarget. 2016;7:10051–10063. doi: 10.18632/oncotarget.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies H, Glodzik D, Morganella S, Yates LR, Staaf J, Zou X, Ramakrishna M, Martin S, Boyault S, Sieuwerts AM, Simpson PT, King TA, Raine K, Eyfjord JE, Kong G, Borg Å, Birney E, Stunnenberg HG, van de Vijver MJ, Børresen-Dale AL, Martens JW, Span PN, Lakhani SR, Vincent-Salomon A, Sotiriou C, Tutt A, Thompson AM, Van Laere S, Richardson AL, Viari A, Campbell PJ, Stratton MR, Nik-Zainal S. HRDetect is a predictor of BRCA1 and BRCA2 deficiency based on mutational signatures. Nat Med. 2017;23:517–525. doi: 10.1038/nm.4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Polak P, Kim J, Braunstein LZ, Karlic R, Haradhavala NJ, Tiao G, Rosebrock D, Livitz D, Kübler K, Mouw KW, Kamburov A, Maruvka YE, Leshchiner I, Lander ES, Golub TR, Zick A, Orthwein A, Lawrence MS, Batra RN, Caldas C, Haber DA, Laird PW, Shen H, Ellisen LW, D’Andrea AD, Chanock SJ, Foulkes WD, Getz G. A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer. Nat Genet. 2017;49:1476–1486. doi: 10.1038/ng.3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Riaz N, Blecua P, Lim RS, Shen R, Higginson DS, Weinhold N, Norton L, Weigelt B, Powell SN, Reis-Filho JS. Pan-cancer analysis of bi-allelic alterations in homologous recombination DNA repair genes. Nat Commun. 2017;8:857. doi: 10.1038/s41467-017-00921-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21:6708. doi: 10.3390/ijms21186708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. [Google Scholar]
- 50.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McShane LM, Altman DG, Sauerbrei W, Taube SE, Gion M, Clark GM. REporting recommendations for tumor MARKer prognostic studies (REMARK) Nat Clin Pract Urol. 2005;2:416–422. [PubMed] [Google Scholar]
- 53.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 55.Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, Kellokumpu-Lehtinen PL, Bono P, Kataja V, Desmedt C, Piccart MJ, Loibl S, Denkert C, Smyth MJ, Joensuu H, Sotiriou C. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014;25:1544–1550. doi: 10.1093/annonc/mdu112. [DOI] [PubMed] [Google Scholar]
- 56.Tokumaru Y, Joyce D, Takabe K. Current status and limitations of immunotherapy for breast cancer. Surgery. 2020;167:628–630. doi: 10.1016/j.surg.2019.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: impact on clinical outcome. Nat Rev Cancer. 2012;12:298–306. doi: 10.1038/nrc3245. [DOI] [PubMed] [Google Scholar]
- 58.Johnson BJ, Costelloe EO, Fitzpatrick DR, Haanen JB, Schumacher TN, Brown LE, Kelso A. Single-cell perforin and granzyme expression reveals the anatomical localization of effector CD8+ T cells in influenza virus-infected mice. Proc Natl Acad Sci U S A. 2003;100:2657–2662. doi: 10.1073/pnas.0538056100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, Kohrt HE, Horn L, Lawrence DP, Rost S, Leabman M, Xiao Y, Mokatrin A, Koeppen H, Hegde PS, Mellman I, Chen DS, Hodi FS. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515:563–567. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chowdhury D, Lieberman J. Death by a thousand cuts: granzyme pathways of programmed cell death. Annu Rev Immunol. 2008;26:389–420. doi: 10.1146/annurev.immunol.26.021607.090404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pardo J, Balkow S, Anel A, Simon MM. The differential contribution of granzyme A and granzyme B in cytotoxic T lymphocyte-mediated apoptosis is determined by the quality of target cells. Eur J Immunol. 2002;32:1980–1985. doi: 10.1002/1521-4141(200207)32:7<1980::AID-IMMU1980>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 62.Takahashi H, Kawaguchi T, Yan L, Peng X, Qi Q, Morris LGT, Chan TA, Tsung A, Otsuji E, Takabe K. Immune cytolytic activity for comprehensive understanding of immune landscape in hepatocellular carcinoma. Cancers (Basel) 2020;12:1221. doi: 10.3390/cancers12051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McGranahan N, Swanton C. Clonal heterogeneity and tumor evolution: past, present, and the future. Cell. 2017;168:613–628. doi: 10.1016/j.cell.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 64.Murugaesu N, Wilson GA, Birkbak NJ, Watkins T, McGranahan N, Kumar S, Abbassi-Ghadi N, Salm M, Mitter R, Horswell S, Rowan A, Phillimore B, Biggs J, Begum S, Matthews N, Hochhauser D, Hanna GB, Swanton C. Tracking the genomic evolution of esophageal adenocarcinoma through neoadjuvant chemotherapy. Cancer Discov. 2015;5:821–831. doi: 10.1158/2159-8290.CD-15-0412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gerlinger M, Rowan AJ, Horswell S, Math M, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Francis JM, Zhang CZ, Maire CL, Jung J, Manzo VE, Adalsteinsson VA, Homer H, Haidar S, Blumenstiel B, Pedamallu CS, Ligon AH, Love JC, Meyerson M, Ligon KL. EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov. 2014;4:956–971. doi: 10.1158/2159-8290.CD-13-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Identification of neutral tumor evolution across cancer types. Nat Genet. 2016;48:238–244. doi: 10.1038/ng.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sottoriva A, Kang H, Ma Z, Graham TA, Salomon MP, Zhao J, Marjoram P, Siegmund K, Press MF, Shibata D, Curtis C. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shi H, Hugo W, Kong X, Hong A, Koya RC, Moriceau G, Chodon T, Guo R, Johnson DB, Dahlman KB, Kelley MC, Kefford RF, Chmielowski B, Glaspy JA, Sosman JA, van Baren N, Long GV, Ribas A, Lo RS. Acquired resistance and clonal evolution in melanoma during BRAF inhibitor therapy. Cancer Discov. 2014;4:80–93. doi: 10.1158/2159-8290.CD-13-0642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schmid P, Cortes J, Pusztai L, McArthur H, Kümmel S, Bergh J, Denkert C, Park YH, Hui R, Harbeck N, Takahashi M, Foukakis T, Fasching PA, Cardoso F, Untch M, Jia L, Karantza V, Zhao J, Aktan G, Dent R, O’Shaughnessy J. Pembrolizumab for early triple-negative breast cancer. N Engl J Med. 2020;382:810–821. doi: 10.1056/NEJMoa1910549. [DOI] [PubMed] [Google Scholar]
- 71.Zou Y, Zou X, Zheng S, Tang H, Zhang L, Liu P, Xie X. Efficacy and predictive factors of immune checkpoint inhibitors in metastatic breast cancer: a systematic review and meta-analysis. Ther Adv Med Oncol. 2020;12:1758835920940928. doi: 10.1177/1758835920940928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martins F, Sofiya L, Sykiotis GP, Lamine F, Maillard M, Fraga M, Shabafrouz K, Ribi C, Cairoli A, Guex-Crosier Y, Kuntzer T, Michielin O, Peters S, Coukos G, Spertini F, Thompson JA, Obeid M. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat Rev Clin Oncol. 2019;16:563–580. doi: 10.1038/s41571-019-0218-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.