Abstract
Tumor infiltrating immune cells plays a critical role in cancer progression. Apoptosis is an autonomous cell death that counteracts tumor growth. To this end, we hypothesized that increased apoptosis in breast cancer is associated with immune cell killing. Apoptosis score of MSigDB Hallmark collection was used to analyze METABRIC cohort (n=1904) and TCGA (n=1069) as validation cohort. High apoptosis tumors enriched cancer promoting signaling pathways; hypoxia, KRAS, TGF-β, PI3K signaling, and was associated with low MKI67 expression and less cell proliferation gene sets, less homologous recombination defects, and less altered fraction. High apoptosis tumors also enriched angiogenesis and high infiltration of vascular endothelial cells, pericytes and stromal cells and significantly enriched inflammation and immune response-related gene sets and high infiltration of CD8, CD4 memory, dendritic cells, M1 and M2 macrophages and significant elevation of cytolytic activity and immune checkpoint molecules, consistently in both cohorts. In conclusion, breast cancer patients with high apoptosis are associated with angiogenesis, immune response, high immune cell infiltration and cytolytic activity. To the best of our knowledge, this is the first study to utilize in silico translational approach to demonstrate the clinical relevance of apoptosis in breast cancer patients in large cohorts.
Keywords: Apoptosis, breast cancer, immune reaction, gene set, inflammatory response, cytolytic activity, tumor immune microenvironment
Introduction
Breast cancer (BC) is the most common cancer in women worldwide and the second leading cause of mortality due to cancer in women in the United States [1]. Despite the rapid advances in management of breast cancer, approximately 40,000 lives are lost every year to this disease in the United States. Thus, better understanding of the mechanisms how breast cancer worsens in patient population level, is essential to further improve clinical outcome. Apoptotic cell death is one of the most studied processes in biological research since its description in the 1970s [2].
Apoptosis is defined as programmed cell death which is finely regulated at the genetic level causing orderly and efficient removal of damaged cells such as that occur after DNA damage or excessive cell proliferation [3]. Since evasion of apoptosis is counted as one of the Hallmarks of cancer, apoptosis is considered one of the key mechanisms to control cancer [4]. The apoptosis mechanism is not only highly selective but also complex and involves many pathways such as inflammation, immune response, reactive oxygen species and p53 signaling pathway and is mediated in a cell through caspase mediated pathways [5].
Chronic inflammation plays a critical role in tumor development through persistent damage to cells and their components such as DNA [6]. Apoptosis is one of the main processes that respond to the consequences of damage caused by inflammation, through subsequent microenvironment stimulation [7]. Studies have shown that inflammation can evoke either favorable (such as high IFN-γ response and infiltration of CD8+ T cells as well as its cytolytic activity) or unfavorable immune reactions [8]. The inflammatory mediators directly affect both cancer and stromal cells and contribute to several hallmarks of cancer, such as promotion of the epithelial-to-mesenchymal transition (EMT) and metastasis of cancer cells. Its clinical impact depends on which immune reaction occurs in the tumor [8]. Localized inflammation and possibly hypoxia coincide to promote tumor angiogenesis. Angiogenesis (development of new blood vasculatures from preexisting ones) is considered as another hallmark of cancer, with blood vessels acting as a conduit not only for nutrient and oxygen supply to the cancer cells but also deliver cells to the tumor microenvironment (TME) [9]. Indeed, we have previously reported that intratumoral angiogenesis is associated with unfavorable inflammation but favorable immune response [10]. Studies have also demonstrated that high mutation tumors generate increased neoantigens and increased mutation burden evokes strong immunogenicity in TME [11] that recruits tumor infiltrating lymphocytes to attack the cancer cells. High mutation tumors also demonstrate high MKI67 gene expression and enriched cell cycle and cell proliferation pathways and hence more aggressive phenotypes [12]. Collectively all these studies have shown that altering components of the apoptosis machinery can affect the dynamics of not only tumor progression but also provide a rationale that inactivation of this machinery occurs during tumor development. Although theoretically, the above-mentioned studies do show an association between inflammation, immune response and apoptosis, whether it really is happening in patients remains unclear given that patient analyses with large cohorts are lacking.
Recently, our group has performed in silico translational research to identify biomarkers [13-15], clinically relevant immune cells [16-18], predictive genes [19], as well as microRNAs in breast cancers [20]. We employed computational algorithms, such as gene set enrichment analysis (GSEA), which enables us to simplify and understand the biological pathways between two distinct groups [21]. We also utilized xCell [22], which uses transcriptomic data to estimate the composition of immune cells within human tumors. Given the role of TME in apoptosis, in this study, we hypothesized that increased apoptosis in breast cancer patients is associated with increased angiogenesis, enhanced inflammatory and immune response, high infiltration of anti-cancer immune cells and high cytolytic activity.
Materials and methods
Genetic profiling and clinical information of breast cancer cohorts
The Cancer Genome Atlas Breast Cancer cohort (TCGA-BRCA) clinical information and RNA-sequencing data of 1069 female patients with confirmed pathological breast cancer, were obtained from Pan-Cancer Clinical Data Resource [23] and through cBio Cancer Genomic Portal [24]. The data of 1903 cases in the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) cohort [25] was also obtained from the cBioPortal. We used normalized tumoral genomic and clinical data provided by the Gene Expression Omnibus (GEO) repository of the US National Institutes of Health (http://www.ncbi.nlm.nih.gov/geo, accessed on 19 October 2020). For genes having multiple probes, average value was used. Gene expression data were transformed for log2 in all analyses. We used the study of Symmans et al. (GSE25066, n=467) [26].
Gene set expression analyses
Gene set variation analysis (GSVA) score of the “HALLMARK_APOPTOSIS” gene set of the MSigDB Hallmark [27] collection was used to measure apoptosis pathway score using the GSVA Bioconductor package (version 3.10) [28], similar to how we measured Angiogenesis signaling score [10], KRAS signaling score [29], G2M cell cycle pathway score [30] and E2F pathway score [31]. False discovery rate (FDR) of less than 0.25 was used for statistical significance in the GSEA analysis, as recommended by GSEA software (Lava version 4.0) as we previously reported [12,13,32-38].
Statistical analysis
The median value of the apoptosis pathway score was used to divide low and high apoptosis score groups in each cohort. Analysis of comparisons between groups used Mann-Whitney U test and Fischer’s exact tests. The xCell algorithm (version 16 for Windows; Microsoft, Redmond, WA, USA) was used to estimate cell fraction of a tumor from its mRNA gene expression data [22]. P value <0.05 was taken as statistically significant for all tests. Turkey type boxplots show median and interquartile level values. R software (version 4.0.1, R Project for Statistical Computing) and Excel (version 16 for Windows, Redmond, WA, USA) were used for mRNA data analysis and figures construction.
Results
High apoptosis breast cancer significantly enriched cancer promoting signaling pathways whereas low tumors enriched cell proliferation and associated with increased MKI67 expression
In order to understand the role of apoptosis in the TME we utilized the apoptosis score defined by the Hallmark collection of Gene set variation analysis (GSVA) algorithm in molecular signatures database (MSigDB) [27] (included genes are presented in Table S1) as we have done with the other scores previously [8,29,31,39-42]. We performed GSEA of Hallmark gene sets using apoptosis score in the TCGA and METABRIC cohorts to investigate what cancer biology is associated with apoptosis in TME. The median was used as cut-off to divide into high and low score groups within each cohort. We found that high apoptosis tumors significantly enriched cancer promoting signaling pathways-related Hallmark gene sets such as-Hypoxia, KRAS signaling up, TGFβ signaling and P13K-AKT-mTOR signaling, consistently in both cohorts [Figure 1A, all normalized enrichment score (NES) >1.44 with a false discovery rate (FDR) <0.25]. Further, we found that low apoptosis tumors enriched multiple cell proliferation pathways such as E2F targets, MYC Targets V1 and MYC Targets V2, but significantly only in TCGA and not in METABRIC cohort (Figure 1B, NES >-1.4, FDR <0.25). In agreement, MKI67 expression, being a cellular marker for proliferation was also high in low apoptosis tumors consistently in both TCGA and METABRIC cohorts (Figure 1C, P=0.034 and P<0.001 respectively). Proliferation score determined by Thorsson et al. [43] was also high in low apoptosis tumors in the TCGA cohort (Figure 1D, P<0.001). These results suggest that high apoptosis tumors are associated with cancer promoting signaling pathways, but less cell proliferation.
Figure 1.
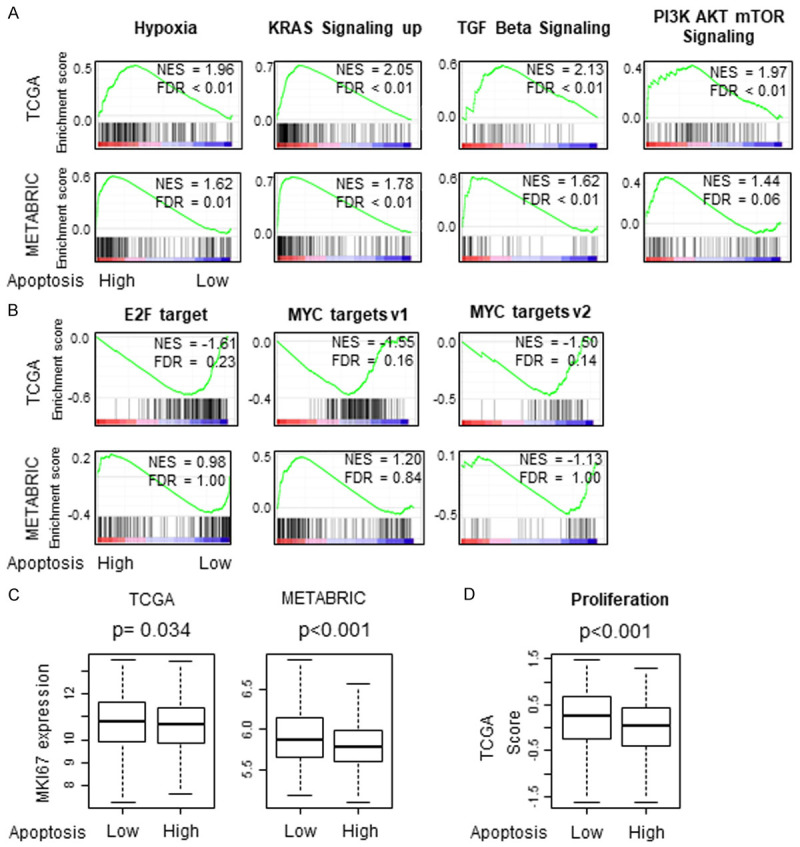
Gene Set Enrichment Assay (GSEA) of apoptosis score in TCGA and METABRIC cohorts, (A) High apoptosis tumors enrich Signaling pathway gene sets; Hypoxia, KRAS signaling up, TGFβ signaling and P13K-AKT-mTOR signaling (B) low apoptosis tumors enrich E2F Targets, MYC Targets V1 and MYC Targets V2. (C) Boxplots of MKI67 expression of low and high apoptosis score groups in TCGA and METABRIC cohorts (D) Boxplot of Proliferation score of low and high apoptosis tumors in TCGA cohort. The median was used as cut-off to divide into high and low score groups within each cohort. Normalized enrichment score (NES) and false discovery rate (FDR) <0.25 was considered significant following the definition by the developer. Mann-Whitney U test was used to calculate P values. Turkey type box plots show median and inter-quartile level values.
Low apoptosis tumor is associated with high homologous recombination deficiency (HRD) and altered fractions
Carcinogenesis is known to be driven by an accumulation of somatic mutations in cancer cells [44] and different cancers have varying degrees of mutations and this variability in mutation rate is likely due to differences in the etiology of mutagenesis [45]. Based upon previous reports that highly proliferative cancers are aggressive and highly mutative [12], we expected that low apoptosis tumors would have more mutations. We found that homologous recombinant deficiency (HRD) and altered fractions were significantly elevated in low apoptosis tumors (both P<0.001) but there was no increase in silent and non-silent mutations, single nucleotide variant (SNV) and Insertion and deletion (Indel) neoantigens (Figure 2).
Figure 2.
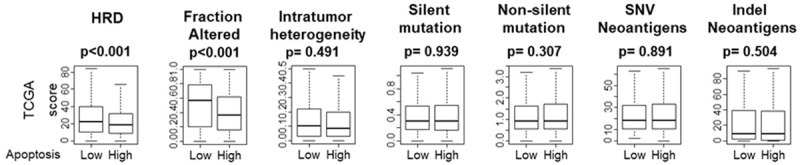
Association between mutation and tumor apoptosis score in TCGA breast cancer cohort. Boxplots of the apoptosis score by Homologous recombination defects (HRD), Fraction altered, Intratumor heterogeneity, silent mutation rate, Non-silent mutation rate, Single nucleotide variant (SNV) neoantigens and Indel neoantigens in the TCGA cohort. The median was used as a cut off to divide patients into high and low score groups within the cohort. Mann-Whitney U test was used to calculate P values. Turkey box plots show median and inter-quartile level values.
High apoptosis tumor significantly enriched angiogenesis and is associated with high infiltration of vascular cells (microvascular endothelial cells (mvE), lymphatic endothelial cells (lyE) and pericytes) and stromal cells (fibroblasts, endothelial cells, adipocytes)
Based upon our finding that high apoptosis tumors are associated with hypoxia, we expected high apoptosis tumors to be associated with enhanced angiogenesis. Indeed, high apoptosis tumors significantly enriched Angiogenesis gene set in GSEA consistently in both TCGA and METABRIC cohorts (Figure 3A, both NES >1.5 and FDR <0.25). Given that angiogenesis often results in generation of immature pathological vessels, it was somewhat surprising to find high apoptosis tumors were associated with high infiltration of microvascular endothelial cells (mvE), lymphatic endothelial cells (lyE) and pericytes, which are components of mature vessels, consistently in both cohorts (Figure 3B, all P<0.001 except mvE in METABRIC).
Figure 3.
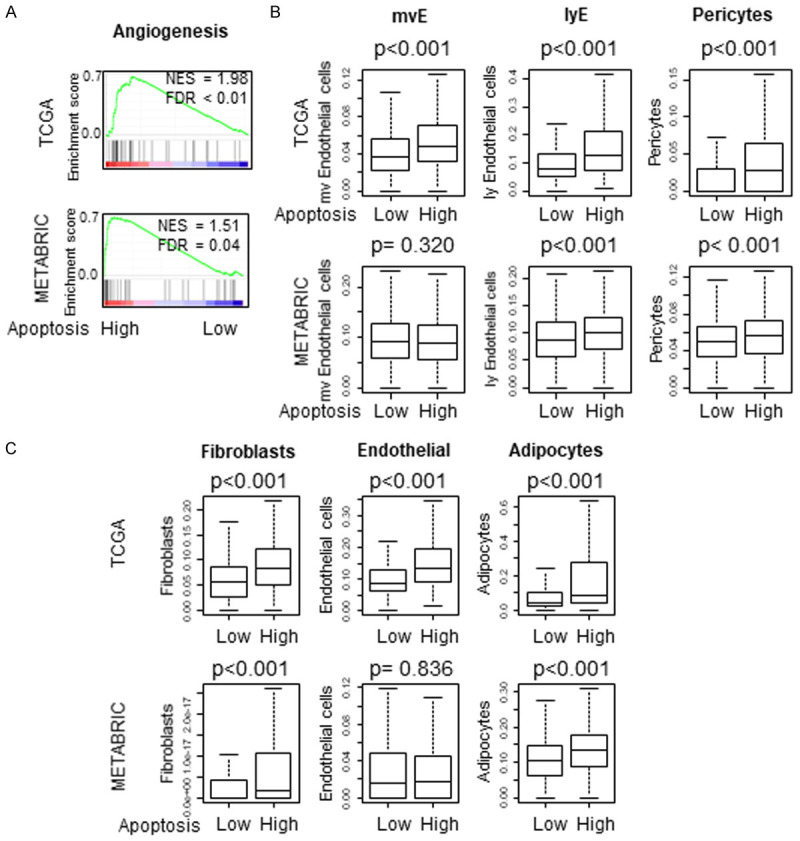
Association between tumor apoptosis and angiogenesis, cell infiltration of vascular and stromal cells (A) Gene Set Enrichment Assay (GSEA) of apoptosis score and angiogenesis pathway in TCGA and METABRIC cohorts (B) Vascular cells like microvascular endothelial cells (mvE), Lymphatic endothelial cells (lyE) and pericytes in TCGA and METABRIC cohorts. (C) Stromal infiltration of fibroblasts, endothelial cells and adipocytes. Normalized enrichment score (NES) and false discovery rate (FDR) are demonstrated. The median was used as cut-off to divide into high and low score groups within each cohort. Mann-Whitney U test was used to calculate P values. Turkey type box plots show median and inter-quartile level values.
We also reported that highly proliferative tumors are associated with less adipocytes in TME using xCell computational algorithm [16]. In agreement, we found that high apoptosis tumors which are less proliferative, are associated with high infiltration of stromal cells such as fibroblasts, endothelial cells and adipocytes in the TCGA cohort (all P<0.001) and fibroblasts and adipocytes in the METABRIC cohort (Figure 3C).
High apoptosis tumor significantly enriched inflammation and immune response gene sets and increased immune response score
Inflammation and immune cell infiltration to the TME are known to play a critical role in cancer biology. It has been shown that local injury and cell death induce inflammation and cytokine production and impact tumor promotion [46,47]. To this end, it was of interest to see the association of apoptosis and immune response. As expected, high apoptosis tumors significantly enriched inflammatory response and IL6-JAK-STAT3 signaling gene sets consistently in both TCGA and METABRIC cohorts (Figure 4A). High apoptosis tumors also significantly enriched immune response related Hallmark gene sets, Allograft rejection, Complement, Interferon (IFN)-α response and Interferon (IFN)-γ response (Figure 4B). Furthermore among the scores reported in Thorsson’s publication [43], we found that high apoptosis tumors are associated with high (IFN)-γ response, Tumor infiltrating lymphocyte (TIL) regional fraction, Lymphocyte infiltration signature, Leukocyte fraction, T cell receptor (TCR) richness and B cell receptor (BCR) richness in the TCGA cohort (Figure 4C).
Figure 4.
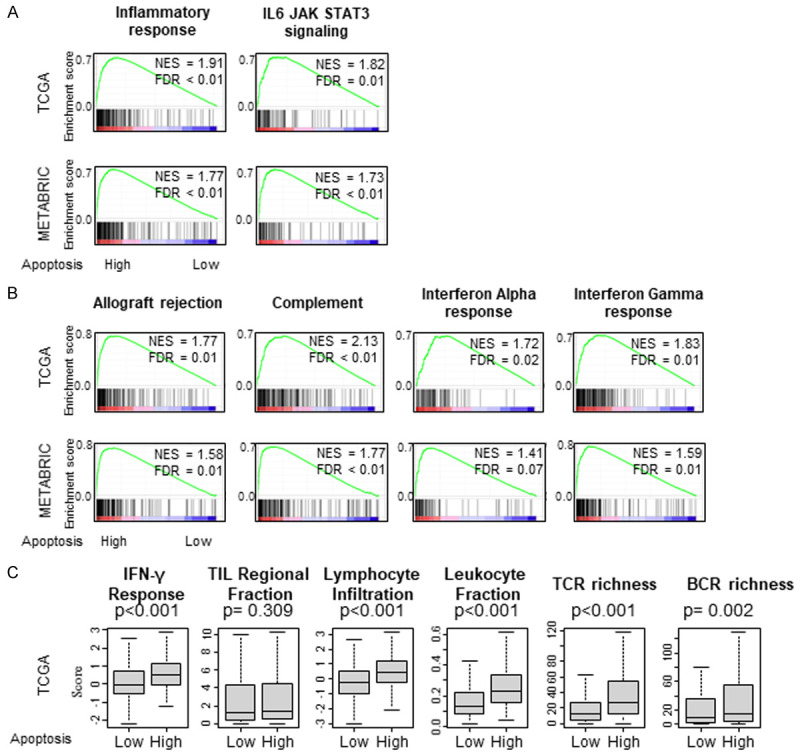
Gene Set Enrichment Assay (GSEA) apoptosis score in TCGA and METABRIC cohorts. Correlation plot of (A) Inflammation-related gene sets; Inflammatory response, IL6-JAK-STAT3 signaling, (B) Immune response gene sets; Allograft rejection, Complement, Interferon (IFN)-α response and Interferon (IFN)-γ response, with normalized enrichment score (NES) and false discovery rate (FDR). (C) Comparison of Thorsson’s score between low and high apoptosis tumors in the TCGA cohort. Boxplots of the comparison with Interferon (IFN)-γ response, Tumor infiltrating lymphocyte (TIL) regional fraction, Lymphocyte infiltration signature, Leukocyte fraction, T cell receptor (TCR) richness and B cell receptor (BCR) richness. The median was used as cut-off to divide into high and low score groups within the TCGA cohort. Mann-Whitney U test was used to calculate P values. Turkey type box plots show median and inter-quartile level values.
High apoptosis tumor is associated with high infiltration of anti-cancer dominant immune cells and elevated cytolytic activity
Given that immune response was associated with high apoptosis in TME, we expected anti-cancer (favorable) immune cells to be infiltrated in high apoptosis tumors. To examine this, we estimated the abundance of different fractions of infiltrating immune cells using the xCell algorithm [22]. We found that high apoptosis tumors had significantly high infiltration of anti-cancer immune cells CD8, CD4 memory, T helper type 1 (Th1) cells, M1 macrophages and Dendritic cell (DC) uniformly in both TCGA and METABRIC cohorts (Figure 5A, all P<0.001). High infiltration of pro-cancer immune cells, such as regulatory T cell (Treg), T helper type 2 (Th2) cells, M2 macrophage, B cell and Plasma cells were observed, however not consistently in the two cohorts (Figure 5B). Cytolytic activity that reflects the overall immune cell killing in the TME was significantly higher in high apoptosis tumors consistently in both cohorts (Figure 5C, all P<0.001). These results demonstrate that high apoptosis in the TME is strongly associated with infiltration of immune cells and immune cell killing.
Figure 5.
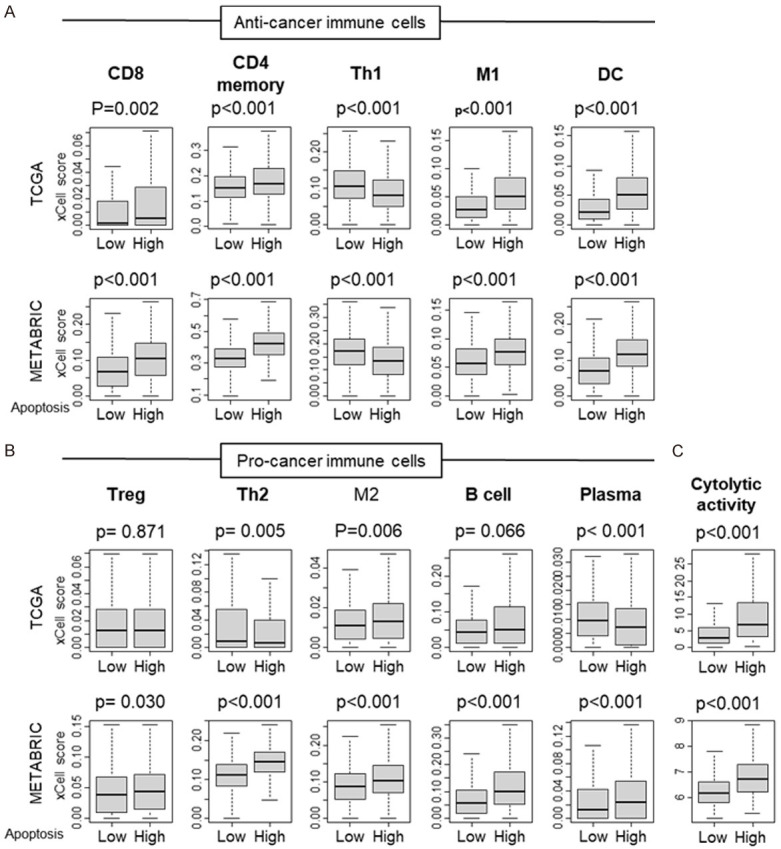
Comparison of tumor infiltrating immune cells between low and high apoptosis score tumors. Boxplots of comparison of Apoptosis score with (A) Anti-cancer immune cells: CD8, CD4 memory, T helper type 1 (Th1) cells, M1 macrophages and Dendritic cell (DC); and (B) Pro-cancer immune cells: regulatory T cell (Treg), T helper type 2 (Th2) cells, M2 macrophage, B cell and Plasma cells by low and high apoptosis score in the TCGA and METABRIC cohorts. (C) Comparison of low and high apoptosis scores with cytolytic activity in the TCGA and METABRIC cohorts. The median was used as cut-off to divide into high and low score groups within each cohort. Mann-Whitney U test was used to calculate P values. Turkey type box plots show median and inter-quartile level values.
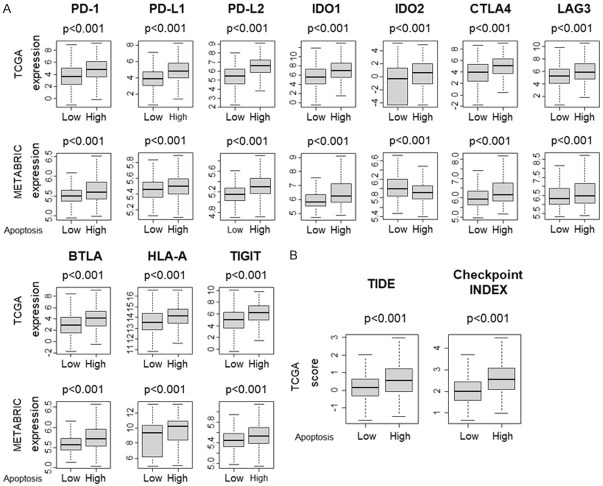
High apoptosis tumor is associated with uniformly elevated immune checkpoint molecules
We hypothesized that high apoptosis tumors would express major T cell exhaustion markers to counterbalance the enhanced immune reaction. As expected, gene expression of major T cell markers such as programmed cell death 1 (PD-1), programmed death ligand 1 (PD-L1), programmed death ligand 2 (PD-L2), indoleamine 2,3-dioxygenase 1 (IDO1), indoleamine 2,3-dioxygenase 2 (IDO2), cytotoxic T-lymphocyte associated antigen 4 (CTLA4), lymphocyte activating 3 (LAG3), B and T lymphocyte attenuator (BTLA), Human Leukocyte Antigen-A (HLA-A) and T-cell immunoglobulin and ITIM domain (TIGIT), was significantly elevated in high apoptosis tumors (Figure 6A). Further, high apoptosis tumors were associated with significantly high Tumor Immune Dysfunction and Exclusion (TIDE) scores and Immune checkpoint index (Figure 6B, both P<0.001). With regards to checkpoint molecules, on further analysis we found that high Immune check point molecule scores was associated with increased apoptosis except IDO2 in the METABRIC cohort where lower IDO2 score was associated with increased apoptosis (Figure S2).
Figure 6.
Comparison of immune check point molecules between low and high apoptosis score tumors. Boxplots of the comparison with (A) T cell exhaustion markers; programmed cell death 1 (PD-1), programmed death ligand 1 (PD-L1), programmed death ligand 2 (PD-L2), indoleamine 2,3-dioxygenase 1 (IDO1), indoleamine 2,3-dioxygenase 2 (IDO2), cytotoxic T-lymphocyte associated antigen 4 (CTLA4), lymphocyte activating 3 (LAG3), B and T lymphocyte attenuator (BTLA), Human Leukocyte Antigen-A (HLA-A) and T-cell immunoglobulin and ITIM domain (TIGIT) in the TCGA and METABRIC cohorts. (B) Comparison of low and high apoptosis scores with Tumor Immune Dysfunction and Exclusion (TIDE) score and immune Checkpoint Index in the TCGA cohort. The median was used as a cut-off to divide into high and low groups within each cohort. Mann-Whitney U test was used to calculate P values. Turkey type box plots show median and inter-quartile level values.
Discussion
Apoptosis is a fundamental phenomenon that occurs in cancer. Cancer cells exhibit more apoptosis than normal cells because cancer cells being highly proliferative, that require more DNA repair, are more amenable to cell death. In the current study, we investigated the clinical relevance of apoptosis in breast cancer patients. We found that high apoptosis tumors significantly enriched Hypoxia, KRAS signaling up, TGFβ signaling and P13K-AKT-mTOR signaling pathways. Low apoptosis tumors enriched cell proliferation pathways such as E2F targets, MYC Targets V1 and MYC Targets V2, as well as increased MKI67 expression and high proliferation score. Low apoptosis tumors were associated with a higher homologous recombination deficiency and Fraction altered but not with mutation rates. High apoptosis tumors enriched angiogenesis pathway and were associated with high infiltration of vascular cells (mvE, lyE and pericytes) and stromal cells (fibroblasts, endothelial cells, adipocytes). High apoptosis tumors significantly enriched Inflammation-related Hallmark gene sets, inflammatory response, IL6-JAK-STAT3 signaling, as well as immune response-related Hallmark gene sets. High apoptosis tumors had significantly high infiltration of anti-cancer immune cells CD8, CD4 memory, Th1, M1 macrophages and DC uniformly, as well as high cytolytic activity that reflects immune cell killing. Lastly, gene expression of major T cell markers was significantly elevated in high apoptosis tumors; and expression of high TIDE scores and Checkpoint index. We found that although high apoptosis score was seen in HER 2-overexpressing subtype, it was not statistically significant (Figure S1).
Breast cancer is a complex multistage disease involving the deregulation of a number of different signaling cascades [48]. Despite improvements in breast cancer treatment, mortality is principally a result of distant metastases that has become resistant to treatment. Many studies have now implicated that apoptosis is a common mechanism through which chemotherapy agents exert their cytotoxicity [49]. Deregulation of the apoptosis process is considered one of the hallmarks of cancer and therapeutic strategies targeting molecules involved in apoptosis resistance represent a valid reason to investigate, in order to restore sensitivity of these tumor cells to apoptosis and overcome their resistance.
Hypoxia is a result of worse circulation. Angiogenesis is neovascularization often with immature pathologic vessels. What we discovered was that high apoptosis was not only associated with enrichment of the angiogenesis pathway with increased immature cells (mvE, lyE) of neovascularization, but also exhibited elevation of mature cells (pericytes). These findings are important as pathologically determined higher angiogenesis state is known to associate with occurrence of metastasis and poorer breast cancer outcome [50,51].
We utilized the apoptosis pathway score that analyzed 161 gene expressions to capture the complex mechanism of apoptosis (Table S1) and using a similar approach, our lab has previously investigated an angiogenesis score, and our present findings are in agreement with our angiogenesis study which demonstrated that intra tumoral angiogenesis is associated with immune reaction, inflammation and metastasis related pathway in breast cancer [10]. Thus, there appears to be two different mechanisms here - first is that high apoptosis is associated with mature vessels that deliver immune cells and the second is - yet high apoptosis tumors are associated with more immature cells that cause hypoxia and thus both occurrences are true.
It has been proven that increased tumor burden evokes strong immune reaction in TME [11]. We have previously analyzed tumors with high mutation rates and neoantigens and demonstrated aggressive clinical features, such as elevated MKI-67 expression and enriched cell cycle and cell proliferation related gene sets on GSEA [12]. In agreement, in the present study we found that low apoptosis tumors were associated with increased mutations in HRD and Fraction altered. These findings are important as HRD status provides significant improvement over clinical parameters or BRCA mutations status in identifying tumors with TNBC who could potentially respond to platinum-based chemotherapy [52].
Our previous studies have reported that high inflammatory pathway tumors are associated with not only apoptosis enriched pathways but also favorable immune response with mainly favorable anti-cancer cell infiltration [8]. In agreement our present study found the high apoptosis tumors not only enriched inflammatory response but also other immune related scores such as TIL regional fraction, IFN-γ response, Lymphocyte infiltration and Leukocyte fraction as well as cytolytic scores and thus shows that inflammatory pathway is critical in breast cancer.
Immune check point inhibition constitutes a promising modality in the anticancer therapeutic approach. Immune checkpoint inhibitors such as anti-PD-1/PD-L1 and CTLA-4 antibodies have added significant elevation to the immunotherapy modality of breast cancer treatment armamentarium [53]. Patient selection is a big challenge in immunotherapy. Although we do not have data, given the high gene expression of all major T cell markers, we cannot help but speculate that high apoptosis breast cancer patients may be a candidate for immune check point inhibitors.
Several basic science papers have studied the mechanism of how apoptosis can drive immune cell infiltration and anti-cancer immunity. Wu et al. demonstrated that immunogenic tumor apoptosis enhanced antitumor immunity and increased IFN-γ and IL-12 levels in triple negative breast cancer [54]. Role of Sphingosine-1-phosphate (S1P) released from apoptotic cells cause migration and chemo-attraction of phagocytes as well as mitogens leading to apoptosis-induced compensatory proliferation and alteration of extracellular matrix and vascular network, was studied by Gadiyar V et al. [55]; while Horton BL et al. demonstrated that prevention of CD8+ TIL apoptosis by Bcl-xl overexpression resulted in improved tumor control [56] and Frank AC et al. demonstrated miR-375 transfer mechanism from apoptotic breast cancer cells to tumor associated macrophages involving CD36, which may pave the way for identifying new drug targets in breast cancer in the future [57]. However, these results are mainly highlighted in mouse tumor models and none of these papers have looked at patient samples. Using a similar gene expression approach, our lab studies have also demonstrated that a KRAS score high in TNBC is associated with favorable TME and better survival [29], G2M cell cycle pathway score is a prognostic biomarker for metastasis in ER positive tumors [30], and E2F pathway score is a predictive marker of neoadjuvant treatment response in ER positive/HER2 negative breast cancer [31]. Despite these strengths, our study is not free from limitations. First these cohorts are retrospective, and are limited by clinical parameters; however, our assessment is objective, and we understand our findings and conclusion would have been stronger with larger cohorts. Our study does not include in vitro or in vivo experiments and hence all our findings were observational associations and do not prove causal mechanisms. Our analyses are limited to measurement of gene expression whereas apoptosis is a process that is ongoing throughout the life of the tumor just like inflammation. Lastly, although we have validated the apoptosis score using the largest publicly available breast cancer cohorts, the apoptosis score should be used in a prospective cohort to be meaningfully utilized for clinical application.
Conclusions
High apoptosis in breast cancer patient is associated with decreased cell proliferation and increased angiogenesis, immune response, immune cell infiltrations, global cytolytic activity that reflect immune cell killing and elevated immune check point molecules.
Acknowledgements
This work was supported by US National Institutes of Health/National Cancer Institute grant 5T32CA108456 to V.M., R01CA160688, R01CA250412, R37CA248018, US Department of Defense BCRP grant W81XWH-19-1-0674, as well as the Edward K. Duch Foundation and Paul & Helen Ellis Charitable Trust to K.T., and US National Cancer Institute cancer center support grant P30-CA016056 to Roswell Park Comprehensive Cancer Center.
Disclosure of conflict of interest
None.
Abbreviations
- AJCC
American Joint Committee on Cancer
- ER
Estrogen receptor
- FDR
False discovery rate
- GSVA
Gene set variation analysis
- HER2
Human Epidermal growth factor receptor 2
- METABRIC
Molecular Taxonomy of Breast Cancer International Consortium
- NES
Normalized enrichment score
- TCGA
The Cancer Genome Atlas
Supporting Information
References
- 1.American cancer society facts and figures 2020. 2020. https://www.cancer.org/content/dam/cancer-org/research/cancer-facts-and-statistics/annual-cancer-facts-and-figures/2020/cancer-facts-and-figures-2020.pdf. [DOI] [PMC free article] [PubMed]
- 2.Kerr JF, Wyllie AH, Currie AR. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972;26:239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pistritto G, Trisciuoglio D, Ceci C, Garufi A, D’Orazi G. Apoptosis as anticancer mechanism: function and dysfunction of its modulators and targeted therapeutic strategies. Aging (Albany NY) 2016;8:603–619. doi: 10.18632/aging.100934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 5.Wong RS. Apoptosis in cancer: from pathogenesis to treatment. J Exp Clin Cancer Res. 2011;30:87. doi: 10.1186/1756-9966-30-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shacter E, Weitzman SA. Chronic inflammation and cancer. Oncology (Williston Park) 2002;16:217–226. 229. discussion 230-232. [PubMed] [Google Scholar]
- 7.Schuetz JM, Grundy A, Lee DG, Lai AS, Kobayashi LC, Richardson H, Long J, Zheng W, Aronson KJ, Spinelli JJ, Brooks-Wilson AR. Genetic variants in genes related to inflammation, apoptosis and autophagy in breast cancer risk. PLoS One. 2019;14:e0209010. doi: 10.1371/journal.pone.0209010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. Inflammation is associated with worse outcome in the whole cohort but with better outcome in triple-negative subtype of breast cancer patients. J Immunol Res. 2020;2020:5618786. doi: 10.1155/2020/5618786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Katsuta E, Rashid OM, Takabe K. Clinical relevance of tumor microenvironment: immune cells, vessels, and mouse models. Hum Cell. 2020;33:930–937. doi: 10.1007/s13577-020-00380-4. [DOI] [PubMed] [Google Scholar]
- 10.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Nagahashi M, Takabe K. Intra-tumoral angiogenesis is associated with inflammation, immune reaction and metastatic recurrence in breast cancer. Int J Mol Sci. 2020;21:6708. doi: 10.3390/ijms21186708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science. 2016;351:1463–1469. doi: 10.1126/science.aaf1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi H, Asaoka M, Yan L, Rashid OM, Oshi M, Ishikawa T, Nagahashi M, Takabe K. Biologically aggressive phenotype and anti-cancer immunity counterbalance in breast cancer with high mutation rate. Sci Rep. 2020;10:1852. doi: 10.1038/s41598-020-58995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oshi M, Katsuta E, Yan L, Ebos JML, Rashid OM, Matsuyama R, Endo I, Takabe K. A novel 4-gene score to predict survival, distant metastasis and response to neoadjuvant therapy in breast cancer. Cancers (Basel) 2020;12:1148. doi: 10.3390/cancers12051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oshi M, Tokumaru Y, Asaoka M, Yan L, Satyananda V, Matsuyama R, Matsuhashi N, Futamura M, Ishikawa T, Yoshida K, Endo I, Takabe K. M1 Macrophage and M1/M2 ratio defined by transcriptomic signatures resemble only part of their conventional clinical characteristics in breast cancer. Sci Rep. 2020;10:16554. doi: 10.1038/s41598-020-73624-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oshi M, Asaoka M, Tokumaru Y, Yan L, Matsuyama R, Ishikawa T, Endo I, Takabe K. CD8 T cell score as a prognostic biomarker for triple negative breast cancer. Int J Mol Sci. 2020;21:6968. doi: 10.3390/ijms21186968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tokumaru Y, Oshi M, Katsuta E, Yan L, Huang JL, Nagahashi M, Matsuhashi N, Futamura M, Yoshida K, Takabe K. Intratumoral adipocyte-high breast cancer enrich for metastatic and inflammation-related pathways but associated with less cancer cell proliferation. Int J Mol Sci. 2020;21:5744. doi: 10.3390/ijms21165744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Kalinski P, Endo I, Takabe K. Plasmacytoid dendritic cell (pDC) infiltration correlate with tumor infiltrating lymphocytes, cancer immunity, and better survival in triple negative breast cancer (TNBC) more strongly than conventional dendritic cell (cDC) Cancers (Basel) 2020;12:3342. doi: 10.3390/cancers12113342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oshi M, Asaoka M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Zsiros E, Ishikawa T, Endo I, Takabe K. Abundance of regulatory T cell (Treg) as a predictive biomarker for neoadjuvant chemotherapy in triple-negative breast cancer. Cancers (Basel) 2020;12:3038. doi: 10.3390/cancers12103038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oshi M, Newman S, Murthy V, Tokumaru Y, Yan L, Matsuyama R, Endo I, Takabe K. ITPKC as a prognostic and predictive biomarker of neoadjuvant chemotherapy for triple negative breast cancer. Cancers (Basel) 2020;12:2758. doi: 10.3390/cancers12102758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokumaru Y, Katsuta E, Oshi M, Sporn JC, Yan L, Le L, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of miR-34a associated with less aggressive cancer biology but not with survival in breast cancer. Int J Mol Sci. 2020;21:3045. doi: 10.3390/ijms21093045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, Omberg L, Wolf DM, Shriver CD, Thorsson V, Hu H. An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell. 2018;173:400–416. e411. doi: 10.1016/j.cell.2018.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Gräf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S METABRIC Group. Langerød A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Børresen-Dale AL, Brenton JD, Tavaré S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hatzis C, Pusztai L, Valero V, Booser DJ, Esserman L, Lluch A, Vidaurre T, Holmes F, Souchon E, Wang H, Martin M, Cotrina J, Gomez H, Hubbard R, Chacón JI, Ferrer-Lozano J, Dyer R, Buxton M, Gong Y, Wu Y, Ibrahim N, Andreopoulou E, Ueno NT, Hunt K, Yang W, Nazario A, DeMichele A, O’Shaughnessy J, Hortobagyi GN, Symmans WF. A genomic predictor of response and survival following taxane-anthracycline chemotherapy for invasive breast cancer. JAMA. 2011;305:1873–1881. doi: 10.1001/jama.2011.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tokumaru Y, Oshi M, Katsuta E, Yan L, Satyananda V, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. KRAS signaling enriched triple negative breast cancer is associated with favorable tumor immune microenvironment and better survival. Am J Cancer Res. 2020;10:897–907. [PMC free article] [PubMed] [Google Scholar]
- 30.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Matsuyama R, Endo I, Takabe K. G2M cell cycle pathway score as a prognostic biomarker of metastasis in estrogen receptor (ER)-positive breast cancer. Int J Mol Sci. 2020;21:2921. [Google Scholar]
- 31.Oshi M, Takahashi H, Tokumaru Y, Yan L, Rashid OM, Nagahashi M, Matsuyama R, Endo I, Takabe K. The E2F pathway score as a predictive biomarker of response to neoadjuvant therapy in ER+/HER2- breast cancer. Cells. 2020;9:1643. doi: 10.3390/cells9071643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okano M, Oshi M, Butash AL, Katsuta E, Tachibana K, Saito K, Okayama H, Peng X, Yan L, Kono K, Ohtake T, Takabe K. Triple-negative breast cancer with high levels of annexin A1 expression Is associated with mast cell infiltration, inflammation, and angiogenesis. Int J Mol Sci. 2019;20:4197. doi: 10.3390/ijms20174197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katsuta E, Rashid OM, Takabe K. Fibroblasts as a biological marker for curative resection in pancreatic ductal adenocarcinoma. Int J Mol Sci. 2020;21:3890. doi: 10.3390/ijms21113890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katsuta E, Yan L, Takeshita T, McDonald KA, Dasgupta S, Opyrchal M, Takabe K. High MYC mRNA expression is more clinically relevant than MYC DNA amplification in triple-negative breast cancer. Int J Mol Sci. 2019;21:217. doi: 10.3390/ijms21010217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gandhi S, Elkhanany A, Oshi M, Dai T, Opyrchal M, Mohammadpour H, Repasky EA, Takabe K. Contribution of immune cells to glucocorticoid receptor expression in breast cancer. Int J Mol Sci. 2020;21:4635. doi: 10.3390/ijms21134635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tokumaru Y, Asaoka M, Oshi M, Katsuta E, Yan L, Narayanan S, Sugito N, Matsuhashi N, Futamura M, Akao Y, Yoshida K, Takabe K. High expression of microRNA-143 is associated with favorable tumor immune microenvironment and better survival in estrogen receptor positive breast cancer. Int J Mol Sci. 2020;21:3213. doi: 10.3390/ijms21093213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi H, Kawaguchi T, Yan L, Peng X, Qi Q, Morris LGT, Chan TA, Tsung A, Otsuji E, Takabe K. Immune cytolytic activity for comprehensive understanding of immune landscape in hepatocellular carcinoma. Cancers (Basel) 2020;12:1221. doi: 10.3390/cancers12051221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okano M, Oshi M, Butash AL, Asaoka M, Katsuta E, Peng X, Qi Q, Yan L, Takabe K. Estrogen receptor positive breast cancer with high expression of androgen receptor has less cytolytic activity and worse response to neoadjuvant chemotherapy but better survival. Int J Mol Sci. 2019;20:2655. doi: 10.3390/ijms20112655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oshi M, Kim TH, Tokumaru Y, Yan L, Matsuyama R, Endo I, Cherkassky L, Takabe K. Enhanced DNA repair pathway is associated with cell proliferation and worse survival in hepatocellular carcinoma (HCC) Cancers (Basel) 2021;13:323. doi: 10.3390/cancers13020323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oshi M, Tokumaru Y, Angarita FA, Yan L, Matsuyama R, Endo I, Takabe K. Degree of early estrogen response predict survival after endocrine therapy in primary and metastatic ER-positive breast cancer. Cancers (Basel) 2020;12:3557. doi: 10.3390/cancers12123557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze A, Oshi M, Endo I, Takabe K. MYC targets scores are associated with cancer aggressiveness and poor survival in ER-positive primary and metastatic breast cancer. Int J Mol Sci. 2020;21:8127. doi: 10.3390/ijms21218127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oshi M, Newman S, Tokumaru Y, Yan L, Matsuyama R, Endo I, Katz MHG, Takabe K. High G2M pathway score pancreatic cancer is associated with worse survival, particularly after margin-positive (R1 or R2) resection. Cancers (Basel) 2020;12:2871. doi: 10.3390/cancers12102871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH, Porta-Pardo E, Gao GF, Plaisier CL, Eddy JA, Ziv E, Culhane AC, Paull EO, Sivakumar IKA, Gentles AJ, Malhotra R, Farshidfar F, Colaprico A, Parker JS, Mose LE, Vo NS, Liu J, Liu Y, Rader J, Dhankani V, Reynolds SM, Bowlby R, Califano A, Cherniack AD, Anastassiou D, Bedognetti D, Mokrab Y, Newman AM, Rao A, Chen K, Krasnitz A, Hu H, Malta TM, Noushmehr H, Pedamallu CS, Bullman S, Ojesina AI, Lamb A, Zhou W, Shen H, Choueiri TK, Weinstein JN, Guinney J, Saltz J, Holt RA, Rabkin CS Cancer Genome Atlas Research Network. Lazar AJ, Serody JS, Demicco EG, Disis ML, Vincent BG, Shmulevich I. The immune landscape of cancer. Immunity. 2019;51:411–412. doi: 10.1016/j.immuni.2019.08.004. [DOI] [PubMed] [Google Scholar]
- 44.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson IR, Takahashi K, Futreal PA, Chin L. Emerging patterns of somatic mutations in cancer. Nat Rev Genet. 2013;14:703–718. doi: 10.1038/nrg3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuraishy A, Karin M, Grivennikov SI. Tumor promotion via injury- and death-induced inflammation. Immunity. 2011;35:467–477. doi: 10.1016/j.immuni.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grivennikov SI, Karin M. Inflammatory cytokines in cancer: tumour necrosis factor and interleukin 6 take the stage. Ann Rheum Dis. 2011;70(Suppl 1):i104–108. doi: 10.1136/ard.2010.140145. [DOI] [PubMed] [Google Scholar]
- 48.Hamed EA, Zakhary MM, Maximous DW. Apoptosis, angiogenesis, inflammation, and oxidative stress: basic interactions in patients with early and metastatic breast cancer. J Cancer Res Clin Oncol. 2012;138:999–1009. doi: 10.1007/s00432-012-1176-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomadaki H, Scorilas A. Molecular profile of the BCL2 family of the apoptosis related genes in breast cancer cells after treatment with cytotoxic/cytostatic drugs. Connect Tissue Res. 2008;49:261–264. doi: 10.1080/03008200802147829. [DOI] [PubMed] [Google Scholar]
- 50.Vermeulen PB, Gasparini G, Fox SB, Colpaert C, Marson LP, Gion M, Beliën JA, de Waal RM, Van Marck E, Magnani E, Weidner N, Harris AL, Dirix LY. Second international consensus on the methodology and criteria of evaluation of angiogenesis quantification in solid human tumours. Eur J Cancer. 2002;38:1564–1579. doi: 10.1016/s0959-8049(02)00094-1. [DOI] [PubMed] [Google Scholar]
- 51.Weidner N, Semple JP, Welch WR, Folkman J. Tumor angiogenesis and metastasis--correlation in invasive breast carcinoma. N Engl J Med. 1991;324:1–8. doi: 10.1056/NEJM199101033240101. [DOI] [PubMed] [Google Scholar]
- 52.Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC, Szallasi Z, Barry WT, Winer EP, Tung NM, Isakoff SJ, Ryan PD, Greene-Colozzi A, Gutin A, Sangale Z, Iliev D, Neff C, Abkevich V, Jones JT, Lanchbury JS, Hartman AR, Garber JE, Ford JM, Silver DP, Richardson AL. Homologous recombination deficiency (HRD) score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res. 2016;22:3764–3773. doi: 10.1158/1078-0432.CCR-15-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokumaru Y, Joyce D, Takabe K. Current status and limitations of immunotherapy for breast cancer. Surgery. 2020;167:628–630. doi: 10.1016/j.surg.2019.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu S, Liu D, Li W, Song B, Chen C, Chen D, Hu H. Enhancing TNBC Chemo-immunotherapy via combination reprogramming tumor immune microenvironment with immunogenic cell death. Int J Pharm. 2021;598:120333. doi: 10.1016/j.ijpharm.2021.120333. [DOI] [PubMed] [Google Scholar]
- 55.Gadiyar V, Lahey KC, Calianese D, Devoe C, Mehta D, Bono K, Desind S, Davra V, Birge RB. Cell death in the tumor microenvironment: implications for cancer immunotherapy. Cells. 2020;9:2207. doi: 10.3390/cells9102207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Horton BL, Williams JB, Cabanov A, Spranger S, Gajewski TF. Intratumoral CD8(+) T-cell apoptosis is a major component of T-cell dysfunction and impedes antitumor immunity. Cancer Immunol Res. 2018;6:14–24. doi: 10.1158/2326-6066.CIR-17-0249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frank AC, Ebersberger S, Fink AF, Lampe S, Weigert A, Schmid T, Ebersberger I, Syed SN, Brüne B. Apoptotic tumor cell-derived microRNA-375 uses CD36 to alter the tumor-associated macrophage phenotype. Nat Commun. 2019;10:1135. doi: 10.1038/s41467-019-08989-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.