Abstract
Wastewater surveillance has shown to be a valuable and efficient tool to obtain information about the trends of COVID-19 in the community. Since the recent emergence of new variants, associated with increased transmissibility and/or antibody escape (variants of concern), there is an urgent need for methods that enable specific and timely detection and quantification of the occurrence of these variants in the community. In this study, we demonstrate the use of RT-ddPCR on wastewater samples for specific detection of mutation N501Y. This assay enabled simultaneous enumeration of lineage B.1.351 (containing the 501Y mutation) and Wild Type (WT, containing 501N) SARS-CoV-2 RNA. Detection of N501Y was possible in samples with mixtures of WT with low proportions of B.1.351 (0.5%) and could accurately determine the proportion of N501Y and WT in mixtures of SARS-CoV-2 RNA. The application to raw sewage samples from the cities of Amsterdam and Utrecht demonstrated that this method can be applied to wastewater samples. The emergence of N501Y in Amsterdam and Utrecht wastewater aligned with the emergence of B.1.1.7 as causative agent of COVID-19 in the Netherlands, indicating that RT-ddPCR of wastewater samples can be used to monitor the emergence of the N501Y mutation in the community. It also indicates that RT-ddPCR could be used for sensitive and accurate monitoring of current (like K417N, K417T, E484K, L452R) or future mutations present in SARS-CoV-2 variants of concern. Monitoring these mutations can be used to obtain insight in the introduction and spread of VOC and support public health decision-making regarding measures to limit viral spread or allocation of testing or vaccination.
Keywords: Sewage surveillance, COVID-19, RNA, N501Y, Public health, SARS-CoV-2, Coronavirus
Graphical abstract
1. Introduction
The first reports of gastrointestinal infection by SARS-CoV-2 and the detection of SARS-CoV-2 RNA in the feces of 30–60% of infected individuals (Jones et al., 2020; Wang et al., 2020; Wölfel et al., 2020; Xiao et al., 2020), sparked the idea that wastewater could be used to monitor the circulation of SARS-CoV-2 at community level. Methods for the detection of SARS-CoV-2 RNA in wastewater were developed and deployed already in the early stage of the pandemic (Lodder and de Roda Husman, 2020, Medema et al., 2020, Medema et al., 2020). The concentration of SARS-CoV-2 RNA in wastewater has shown to reflect and even precede the trends of the newly reported cases or COVID-19 hospitalizations (Medema et al., 2020a; Prado et al., 2021). The possibility to obtain objective information about virus circulation in a community via wastewater sampling, independent from diagnostic testing availability, willingness and awareness (i.e. asymptomatic virus carriers), is currently being used to support public health decisions. In situations with low SARS-CoV-2 circulation in the community, wastewater surveillance is being used as an early warning system (Betancourt et al., 2020; Medema et al., 2020, Medema et al., 2020; Ahmed et al., 2021) enabling rapid and targeted measures to limit viral spread. The added value of wastewater surveillance has led to worldwide implementation of methods for SARS-CoV-2 monitoring in sewage (Bivins et al., 2020).
The occurrence of new SARS-CoV-2 variants that have unusual numbers of mutations and are associated with increased transmissibility, antibody escape or both (“Variants of Concern”, VoC), has recently gained attention (Altmann et al., 2021; Challen et al., 2021; Eurosurveillance-editorial-team, 2021). Four VoC lineages are recognized since the end of 2020 and are currently emerging worldwide and additional variants are under consideration: Lineage B.1.1.7 (Alpha) was recognized first through the large scale genomics initiative in the UK, lineage B.1.351 (Beta) was first recognized in South Africa, lineage P.1 (Gamma) emerged in late 2020 in Manaus (Brazil) and lineage B.1.617.2 (Delta) was first documented in India. These VoC's appear to spread at higher efficiency (Davies et al., 2021; Faria et al., 2021; Volz et al., 2021). In addition, lineages Beta, Gamma and Delta appear to be partially resistant to neutralizing antibodies from infection, vaccination, or treatment (Bernal et al., 2021; Garcia-Beltran et al., 2021; Wang et al., 2021). Therefore, it is of great importance to monitor the spread of these VoC in the community to understand the dynamics of transmission of VoC and be able to take appropriate public health protection measures. Currently, VoC surveillance is mainly done through whole genome sequencing of patient samples. This approach is relatively expensive, labour intensive and time consuming and may not be achievable in many parts of the world. Variant surveillance via wastewater could be a more efficient and rapid method to monitor the emergence and spread of VoC in a community. However, there are currently no validated efficient methods available to quantify the presence of different VoC's in sewage. Sequence analysis of RNA isolated from sewage can specifically detect and identify SARS-CoV-2 RNA sequence variation (Izquierdo-Lara et al., 2020) and sequence analysis has the potential to identify VoC in sewage. However, it is expected that this needs deep sequencing efforts and intensive bioinformatics approaches to find rare mutations in sewage samples which likely contain mixtures of SARS-CoV-2 lineages. The limited sequence differences between VoC and the likely presence of mixtures of sequence variants in sewage makes it difficult to design VoC specific RT-qPCR assays for sewage. In droplet digital RT-PCR (RT-ddPCR) the sample and reagents are partitioned in a large number (~10.000 to 20.000) “water in oil” droplets and PCR-reactions are performed on single molecules in individual closed droplets. This makes it possible to detect rare mutations and to discriminate closely related sequences on the base of probe binding kinetics (Pekin et al., 2011). This makes RT-ddPCR potentially attractive to specifically detect the presence and proportion of mutations that are specific to VoC's (signature mutations) in wastewater. In this study we evaluated the use of RT-ddPCR to detect VoC signature mutations and to determine the proportion and concentration of signature mutations in wastewater. Mutation N501Y was selected to study the possibilities to apply RT-ddPCR to monitor VoC signature mutations. This N501Y mutation is present in lineages B.1.1.7, B.1.351 and P.1 and started to emerge rapidly at the end of 2020. The mutation leads to an amino acid change at position 501 in the receptor binding domain of the spike protein. The N501Y mutation appears to influence binding of the spike protein to the human ACE2 receptor (Liu, Zhang et al. 2021; Rynkiewicz et al., 2021) resulting in more efficient transmission (Liu, Liu et al. 2021). Validation was performed with RNA isolated from wild type virus and a B.1.351 stock and the applicability of the RT-ddPCR was studied on domestic wastewater samples from the Dutch cities of Amsterdam and Utrecht.
2. Materials and methods
2.1. Wastewater samples
The inlet of the wastewater treatment plants of Amsterdam, serving 669,400 inhabitants, and of Utrecht, serving 267,900 inhabitants (data Netherlands Central Bureau of Statistics) were sampled. In both cases, this was approx. 75% of the population of the city. Composite, flow-dependent 24 h samples were collected by the operators of the wastewater treatment plants (WWTP) that serve the cities of Amsterdam and Utrecht in the Netherlands. Samples were stored at 4 °C during 24 h periods of sampling as previously described (Medema et al., 2020, Medema et al., 2020, Medema et al., 2020). Samples were taken weekly from March 2020 to March 1 2021. RT-qPCR was applied on all samples, RT-ddPCR analysis was applied on the samples from the period November 9 2020 to March 1 2021.
2.2. SARS-CoV-2 viral RNA
Reference genomic RNA from WT SARS-CoV-2 virus (WIV04/2019) and variants B.1.351 and B.1.1.7 containing the N501Y mutation was isolated from cell-cultured virus at passage 3 after inoculation of primary patient sample. RNA isolation from 0,2 TCID50 cell-cultured virus was performed by using the MagNA pure 96 (Roche diagnostics) and the total nucleic acid isolation kit (Roche Diagnostics). The sequence of the cell-cultured RNA was confirmed by whole-genome sequencing performed at Erasmus University Medical Center (Rotterdam, The Netherlands).
2.3. Virus concentration and nucleic acid extraction
Samples were transported to the laboratory and processed as previously described (Medema et al., 2020, Medema et al., 2020, Medema et al., 2020). In short, centrifugation was used to pellet larger particles, viral particles were concentrated from 50 ml supernatant by ultrafiltration through Centricon® Plus-70 centrifugal ultrafilters with a cut-off of 30 or 100 kDa, dependent on supplies (Millipore, Amsterdam, The Netherlands). RNA extraction was also performed on ultrafiltrated PCR grade and RNAse free distilled water (Applied Biosystems, Fisher Scientific, Landsmeer, The Netherlands) water to use as negative controls. Approximately 2 × 104 genomic RNA copies from the murine coronavirus Mouse Hepatitis Virus (MHV)-A59 (obtained from “Leids Universitair Medisch Centrum”) was spiked to each concentrate and co-isolated during the extraction procedure to monitor the possible presence of RT-PCR inhibitors and measure the recovery efficiency of the extraction procedure. Nucleic acid was extracted from the concentrate using the magnetic extraction reagents of the Biomerieux Nuclisens kit (Biomerieux, Amersfoort, the Netherlands) in combination with the semi-automated KingFisher mL (Thermo Scientific, Bleiswijk, The Netherlands) as previously described. Extracted nucleic acid was eluted in a volume of 100 μl.
2.4. RT-qPCR for coronavirus RNA-quantification in wastewater
The N2 assay targeting a fragment of the nucleocapsid gene, as published by US CDC (US-CDC, 2020), was used to quantify SARS-CoV-2 RNA in the sewage samples weekly in the period from. Reagents and reaction conditions were as previously described (Medema et al., 2020, Medema et al., 2020, Medema et al., 2020). All RT-PCR's were run as technical duplicates on 5 μl extracted nucleic acid. Duplicate RT-qPCR reactions on serial dilutions containing RT-ddPCR calibrated (five 10-fold concentration steps containing 2.101 to 2.105 RNA copies/reaction) EURM-019 single stranded RNA (provided by the Joint Research Centre) were used to construct calibration curves. The average PCR efficiency, based on all weekly performed RT-qPCR experiments (n = 26) presented in this paper, was 96.8% (Range: 93.2–103%; R2 0.999). These calibration curves were subsequently used to quantify N2 in RNA extracted from the sewage samples. Spiked MHV-A59 RNA was detected by performing a MHV-A59 specific RT-qPCR targeting the N-gene using the primers and reaction profile described by Raaben et al. (2007). Calibration curves for quantification were generated by performing RT-PCR assays on dilution series of a synthetic quantified gBlock gene fragment containing a partial sequence MHV-A59 N-gene (obtained from IDT, Leuven Belgium). The recovery efficiency was calculated by determining the ratio (%) between the MHV-A59 RNA concentration in the spiked sewage sample with and the concentration MHV-A59 suspension used to spike the samples. This recovery efficiency was used as quality control to monitor the performance of the RNA-isolation procedure and the possible presence of RT-PCR inhibitors. The recovery efficiencies (Tables S1 and S2) are used as process control (Kantor et al., 2021), only samples with a recovery efficiency of >5% are reported in this paper. Two types of negative controls were analyzed in every experiment: 1) RT-qPCR reactions performed on PCR grade and RNAse free distilled water and 2) and RT-qPCR reactions performed on RNA extracted from PCR grade and RNAse free distilled water.
2.5. qPCR for CrAssphage quantification in wastewater
A previously described, CrAssphage CPQ_064 specific PCR (Stachler et al., 2017) was used to quantify this DNA-virus that exclusively occurs in human intestinal tracts. Assays were performed in duplicate on 5 μl 1:10 diluted extracted nucleic acid. Quantification was performed using qPCR assays on dilution series of a synthetic quantified gBlock (obtained from IDT, Leuven Belgium) containing the CPQ_064 gene fragment. The CrAssphage concentrations were used to normalize measured viral loads to account for population dynamics and fluctuating non-human inputs (rain, industrial wastewater, groundwater) in the sewer network (Medema et al., 2020a).
2.6. RT-ddPCR for the N501Y mutation
Digital droplet RT-PCR was used to quantify the N501Y and the wild-type (WIV04/2019, WT) sequence in one single tube multiplex mutation assay designed by BioRad (Assay ID: dMDS731762551). This assay uses primers that amplify an 80 bp fragment of the Spike gene including the area containing an A to T point mutation that leads to the N501Y amino acid change in the Spike protein (N501Y). Two probes are used to detect PCR-amplification in the droplets: one FAM-labeled probe which perfectly binds to the N501Y mutation and one HEX-labeled probe which perfectly binds to the wild-type SARS-CoV-2 sequence. The ability to perform the PCR-assay in discrete self-contained droplets makes it possible to discriminate between droplets containing SARS-CoV-2 mutant fragments at low frequencies in a background of wild-type fragments. Assays were performed in 20 μl reaction volumes containing the reagents from the One-Step Advance RT-ddPCR for probes: 5 μl RT-ddPCR One-Step Advanced Supermix, 2 μl Reverse Transcriptase, 1 μl DTT (300 mM) supplemented with 1 μl Single tube mutation assay, 6 μl PCR grade and RNAse free distilled and 5 μl sample-RNA. The BioRad QX200 droplet generator was used to partitionate sample-RNA and reagents in droplets. The temperature profile used for RT-ddPCR was as follows: 60 min. 50 °C, 10 min 50 °C, 40 cycles with 30 s. 95 °C and 1 min. 55 °C followed by 10 min. 98 °C, 30 min. 4 °C and hold at 12 °C. Samples were scanned using the QX200 system (BioRad) and analyzed using the QuantaSoft-Analysis software (BioRad). For each sample, the number of negative and WT or N501Y ddPCR positive droplets were recorded and used to determine the WT or N501Y concentrations. The proportion of Spike gene specific RNA fragments containing the N501Y mutation was calculated by the QuantaSoft-Analysis software as the concentration N501Y in the ddPCR reaction, divided by the sum of WT and N501Y concentrations in the ddPCR reaction. The 95% confidence intervals in the proportion of N501Y were calculated assuming a Poisson distribution of RNA molecules in the droplets. RT-ddPCR reactions performed on PCR grade and RNAse free distilled water and RNA extracted from PCR grade and RNAse free distilled water were used as negative controls. RT-ddPCR performed on 500–900 genome copies wild-type, B.1.351 and B.1.1.7 were used as positive controls.
2.7. Validation experiments
Two dilution series were analyzed to evaluate the ability of RT-ddPCR to detect WT and the N501Y variants simultaneously in mixtures of different concentration ratios of reference genomic RNA of SARS-CoV-2 lineage B.1.351 and WT. The approximate concentration of RNA from WT SARS-CoV-2 virus (Wuhan type) and variant B.1.351 was first quantified using the N2 specific RT-qPCR assay. The first dilution series consisted of a stable concentration of approximately 600 RNA copies of WT, mixed with 2-fold dilutions of lineage B.1.351. The second dilution series contained a stable concentration of approximately 700 RNA copies of variant B.1.351 mixed with 2-fold dilution series of RNA extracted from WT virus. The average concentrations measured with RT-ddPCR in the samples containing stable concentrations of WT or variant B.1.351 respectively were used as values, the dilution factors were subsequently used to calculate the expected proportions of WT and variant B.1.351 in the mixed samples.
3. Results
3.1. Method validation
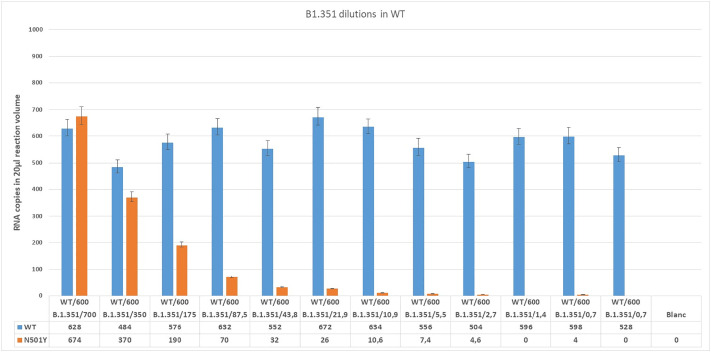
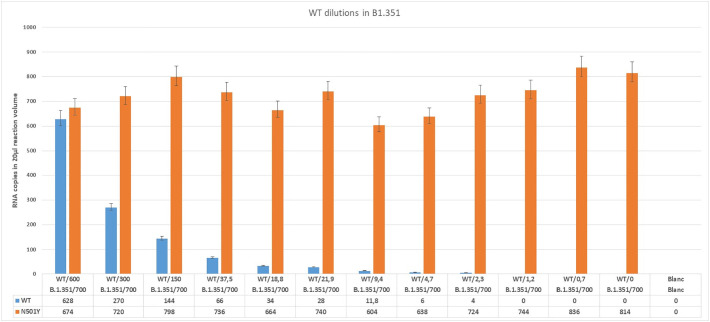
To study the ability of RT-ddPCR to differentiate between 501Y and 501N sequences and to detect low concentrations of SARS-CoV-2 N501Y mutant in the background of WT RNA two dilutions series were analyzed. For this, we isolated RNA from wild type virus and a B.1.351 strain containing the N501Y mutation. Next a two-fold dilution series of B.1.351 RNA was made in a stable background of WT RNA (Fig. 1 ) and vice versa (Fig. 2 ).
Fig. 1.
Detected copy numbers of WT and N501Y in mixtures containing ~600 copies WT and 2-fold dilution series of variant lineage B.1.351.
Fig. 2.
Detected copy numbers of WT and N501Y in mixtures containing ~700 copies variant lineage B.1.351WT and 2-fold dilution series of WT.
These results demonstrate the ability of the method to simultaneously detect and discriminate between the sequences of WT and the N501Y mutation in lineage B.1.351 from a mixed sample. It also shows that low concentrations of N501Y mutation can be detected in the presence of WT virus RNA and vice versa. Detection of N501Y was possible in a sample dilution containing a theoretical concentration of only 2.7 copies of B.1.351 or WT detection in a sample containing 2.3 copies of WT suggests the feasibility for specific detection of low concentrations of WT and N501Y.
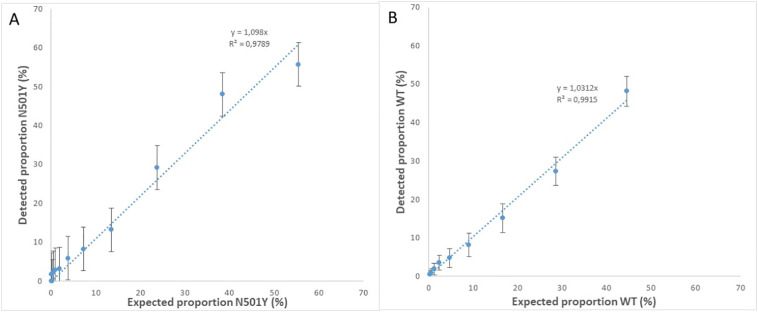
Comparing the measured versus expected proportion of the 2-fold dilutions of B.1.351 (Fig. 3 A) or the diluted WT (Fig. 3B) shows the high linear correlation between the expected proportion and detected proportion of N501Y and WT respectively.
Fig. 3.
The expected and detected proportion of N501Y (A) and WT (B) in artificial mixtures of WT and lineage B.1.351 as detected by ddPCR.
These results demonstrate that the assay can be used to discriminate between WT SARS-CoV-2 RNA and variants containing the N501Y mutation and simultaneous detection and quantification of these sequences. The specificity of the assay to detect the N501Y mutation was confirmed on RNA from SARS-CoV-2 lineage B.1.1.7, the specificity of the N501Y assay for RNA from B.1.1.7 and B.1.351 VOC's is demonstrated in Supplemental Fig. S1. It is expected that the assay can be used to detect low concentrations of WT and N501Y and to obtain reliable insight in the proportion of N501Y in mixtures of viruses, and therefore has the potency to collect information about the spread of variants.
3.2. SARS-CoV-2 VoC surveillance in wastewater of Amsterdam and Utrecht
Wastewater samples from the cities of Amsterdam and Utrecht were analyzed in the period between September 2020 and March 2021 to evaluate the applicability of sewage surveillance to monitor the emergence of VoC's. The concentration SARS-CoV-2 in these samples was first determined using RT-qPCR and compared to COVID-19 incidence and the emergence of N501Y was subsequently studied on the same set of samples.
3.2.1. SARS-CoV-2 surveillance
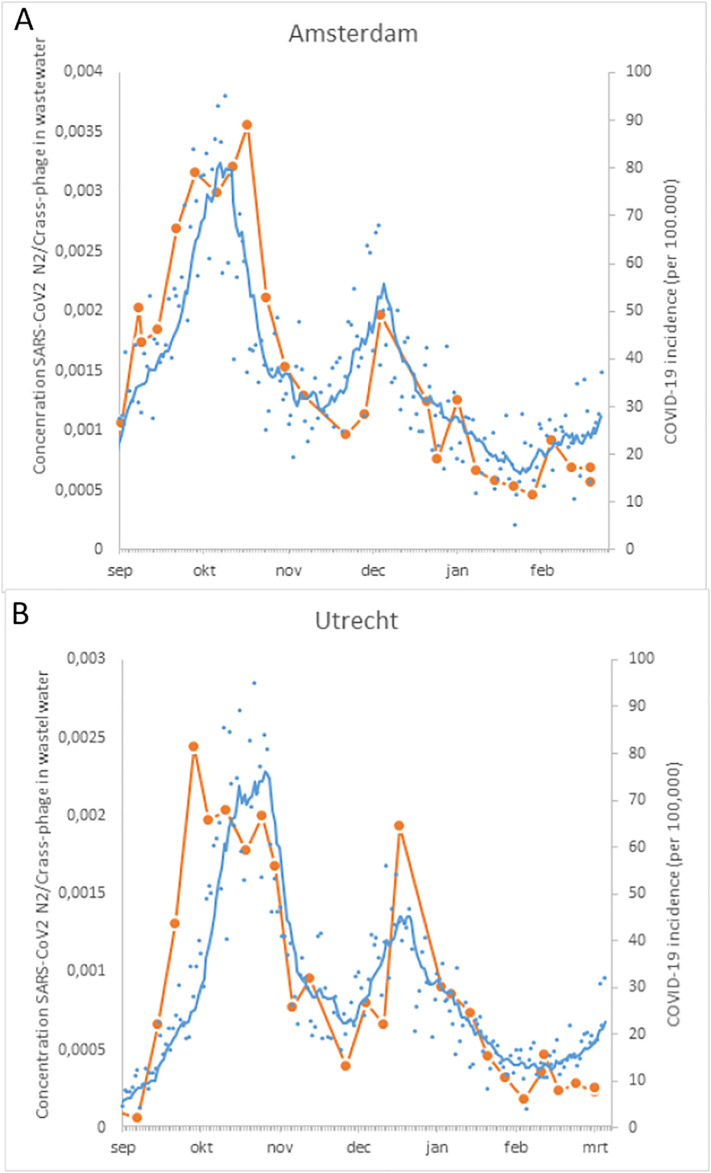
The concentrations of SARS-CoV-2 N2 (determined with RT-qPCR and normalized for the concentration of CrAssphage in the same sample) are compared with the number of newly reported cases (Fig. 4 ). The results demonstrate that the concentrations of SARS-CoV-2 RNA in wastewater reflected the trends in the newly reported positive COVID-19 diagnostic tests in the population of these cities. Both cities experienced a peak in COVID-19 cases in October and December 2020.
Fig. 4.
The concentration of SARS-CoV-2 N2 gene determined with RT-qPCR (normalized for the concentration of Crass-phage) in wastewater of Amsterdam (A) and Utrecht (B) from Aug 26 2020 to March 8 2021 (orange circles) and the newly reported COVID-19 cases in the city population (blue dots) and the 7-day moving average (blue line). COVID-19 data: National Institute of Public Health and the Environment, data source ESRI NL COVID-19 Hub (Esri-Nederland, 2021). The wastewater data covered 75% of the population in both cities.
3.2.2. Emergence of the SARS-CoV-2 N501Y mutation in sewage
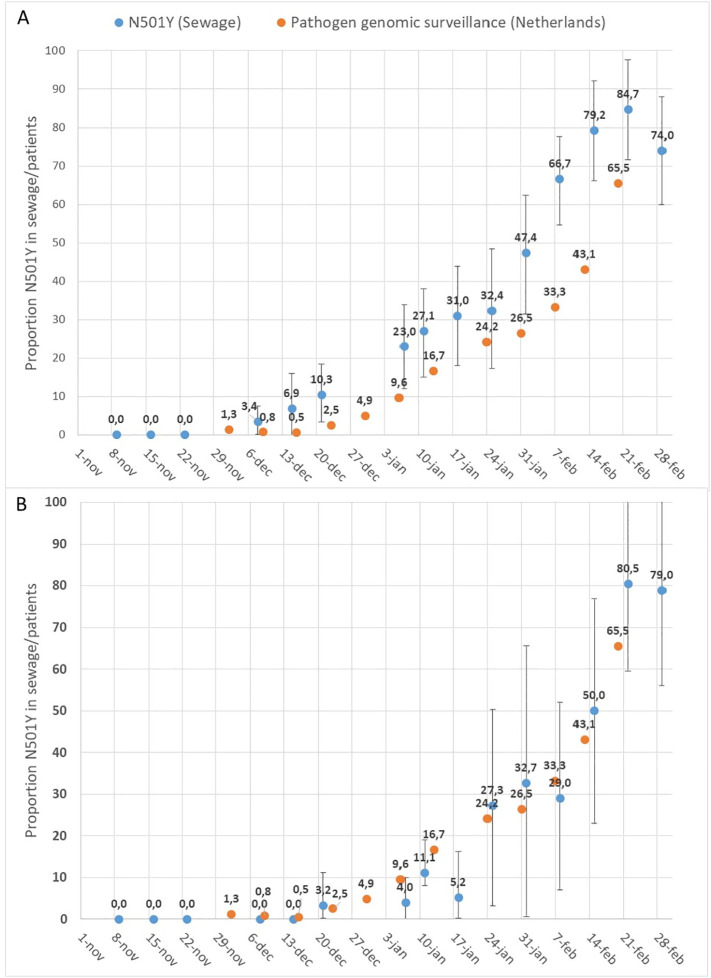
Analysis of the sewage samples was performed and the results are summarized in Table S1 (Amsterdam) and Table S2 (Utrecht). The proportion of N501Y to WT sequences in wastewater is determined and compared with publicly available data from the Dutch National Institute for Public Health and the Environment (RIVM). The available data is based on surveillance of variants by sequencing a random selection of SARS-COV-2 isolates from several hundreds of new cases in the Netherlands every week (Table S3, “Pathogen Genomic surveillance”: https://www.rivm.nl/coronavirus-covid-19/virus/varianten, accessed March 09 2021). The results of these comparisons are shown in Fig. 5A (Amsterdam) and B (Utrecht).
Fig. 5.
Proportion of Spike gene RNA fragments containing the N501Y mutation in wastewater from Amsterdam (A) and Utrecht (B) calculated as relative concentration of N501Y containing Spike gene fragments divided by the total concentration of S-gene fragments (WT + N501Y). Error bars are calculated assuming Poisson distribution of RNA molecules in droplets. The proportion of newly COVID-19 patients infected with N501Y containing variants (Pathogen genomic surveillance) are shown as (the proportional sum of lineages B.1.1.7, B.1.351 and P.1).
The earliest detection of N501Y was in the wastewater sample of December 8 2020 in Amsterdam and in the wastewater sample of December 21 2020 in Utrecht. The proportion of N501Y gradually increased to 85% on February 22 2021 in Amsterdam and 81% on February 22 2021 in Utrecht. The total SARS-CoV-2 RNA concentrations were higher in wastewater from Amsterdam compared to Utrecht (Tables S1 and S2), yielding a more accurate assessment of the N501Y proportion, as indicated by the error-bars.
The first detection of the British variant (lineage B.1.1.7) in patients in the Netherlands was in the first week of December 2020. This corresponds to the first detection of N501Y in wastewater from Amsterdam. The first detection of N501Y in wastewater from Utrecht was two weeks later. After the emergence of variants containing the N501Y mutation, the increase in the proportion of this marker in wastewater follows a trend that is comparable with the increase of N501Y containing variants in patients. The percentage of N501Y containing variants appears to be higher in Amsterdam wastewater in the complete period suggesting earlier introduction in Amsterdam than in Utrecht. The introduction and emergence of SARS-CoV-2 variants containing the N501Y mutation in Utrecht wastewater aligns more in time with the national ‘average’ emergence as obtained through the genomic pathogen surveillance data.
3.2.3. Comparison N2 gene RT-qPCR with S gene RT-ddPCR
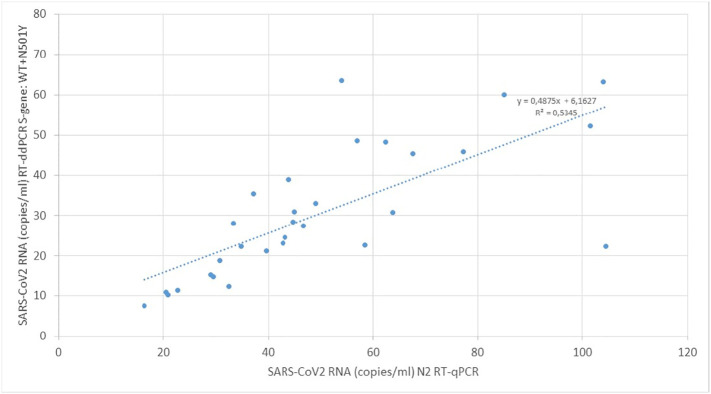
For the period of Nov 9, 2020 to Mar 1, 2021 concentrations of SARS-CoV-2 RNA was measured in sewage from WWTP Amsterdam and Utrecht with RT-qPCR directed to the N2 amplicon of the N-gene and with RT-ddPCR as the sum of WT and N501Y sequences of the Spike gene. Although these are two different targets and assays, we compared the concentrations obtained in the same samples with both assays in Fig. 6 . This comparison shows that there is a linear relation between the values measured with both methods. However, concentrations measured with RT-qPCR are consistently higher (61% on average) than concentrations measured with RT-ddPCR. On basis of current knowledge of SARS-CoV-2 sequences in the Dutch population, it is unlikely that there are other sequence variants in the populations that were detected by RT-qPCR but missed with the RT-ddPCR assay. The higher RT-qPCR results could be the result of targeting different gene fragments with different assays. The different RNA-targets can have different degradation rates in sewage but it can also be the result of higher expression levels of the N-gene compared to the S-gene in coronavirus infected cells and the presence of the N-gene in every subgenomic mRNA (Kim et al., 2020) making it likely that higher levels of N-sequences will be present in infected intestinal cells and eventually may lead to higher N-sequence concentrations in sewage.
Fig. 6.
Comparison of the wastewater concentrations of SARS-CoV-2 S-gene fragment, measured with RT-ddPCR, and the N2 gene fragment, measured with RT-qPCR. Wastewater samples from Amsterdam and Utrecht Nov 9 2020 – Mar 1 2021.
4. Discussion and conclusions
This research describes the first use of RT-ddPCR to specifically detect and quantify SARS-CoV-2 mutations associated with VoC's in wastewater. This assay can be used for sensitive and simultaneous detection of WT and the N501Y mutation in sewage. Although more extensive research is needed to determine the limit of detection accurately, experiments on SARS-CoV-2 RNA from lineage B.1.351 (with mutation N501Y) and WT in different proportional mixtures demonstrated that proportions as low as 0,5% of lineage B.1.351 can be detected in a background of 700 RNA-copies of WT SARS-CoV-2. These experiments also demonstrated the linearity of the expected proportion and the measured proportion of B.1.351 in the mixtures and vice versa, indicating that this N501Y assay can be used to accurately determine the ratio between SARS-CoV-2 genomes containing N501Y and those containing the WT sequence. The weekly analysis of wastewater from the cities of Amsterdam and Utrecht using RT-qPCR targeting the N-gene (CDC N2 target) clearly demonstrated the relationship between SARS-CoV-2 RNA levels and the Covid-19 incidence in the communities connected to the respective WWTP's. N501Y containing B 1.1.7 variants were first detected on December 82,020 in Amsterdam and two weeks later in Utrecht. The proportion of N501Y variants in the weekly samples increased to approximately 80% on March 1, 2021.
Concentrations measured in sewage with RT-qPCR (N2 amplicon) and RT-ddPCR (S-gene specific amplicon) showed consistently higher concentrations measured with RT-ddPCR then RT-ddPCR. On basis of current knowledge of SARS-CoV-2 sequences in the Dutch population, it is unlikely that there are other sequence variants in the populations that were detected by RT-qPCR but missed with the RT-ddPCR assay. The higher RT-qPCR values could be the result of targeting different gene fragments with potential differences in detected concentrations in wastewater or the use of different detection techniques. The different RNA-targets can have different degradation rates in sewage but it can also be the result of higher expression levels of the N-gene compared to the S-gene in coronavirus infected cells and the presence of the N-gene in every subgenomic mRNA (Kim et al., 2020) making it likely that higher levels of N-sequences will be present in infected intestinal cells and eventually may lead to higher N-sequence concentrations in sewage. Comparing concentrations using RT-qPCR targeting the (S-gene specific) RT-ddPCR fragment will give insight in the influence of the detection technique used on the measured concentrations.
These results demonstrate the applicability of this RT-ddPCR assay to monitor the absolute quantities and ratios of WT viral sequences and sequences containing the N501Y mutation. This makes the use of RT-ddPCR on wastewater samples an attractive, rapid and efficient method to follow the emergence of (mutations associated with) VoC's in the community, that can generate an early warning of the emergence of VoC's on the basis of one “population-sample” via domestic wastewater. This proof-of-principle demonstrates the value of RT-ddPCR to detect N501Y, and suggests RT-ddPCR can also be used for the detection of multiple signature mutations (like f.e. E484K, K417T, K417N or L452R) characteristic for various epidemiologically or immunologically relevant VoC's. At the onset of this study in January 2021, the threat of emergence of B 1.1.7 (the Alpha VOC) in the Netherlands was most imminent and our data show that the emergence of this VOC was detectable via N501Y monitoring of wastewater. In the situation of June 2021, emergence of the Beta, Gamma or Delta variant is imminent. To use wastewater surveillance to monitor for these VOC, specific primer/probe sets for other signature mutations are needed and under development. Whole genome sequencing of SARS-CoV-2 from clinical samples from infected individuals is commonly used to identify VoC's (Tegally et al., 2021). Although whole genome sequencing has shown to be feasible to give insight in the sequence variations of SARS-CoV-2 genomes in sewage (Izquierdo-Lara et al., 2020) and has been used to detect mutations associated with variants of concern in wastewater (Jahn et al., 2021). This approach requires deep sequencing and intensive bioinformatics analyses to determine the presence of mutations associated with VoC's in wastewater. An advantage of whole genome sequencing of wastewater samples is that it can provide more extensive information about the co-occurrence of the range of mutations and deletions associated with VoC. It would be valuable to compare the results of RT-ddPCR with whole genome sequencing on the same wastewater samples and on samples of the COVID-19 cases in the population that contributes to this wastewater.
CRediT authorship contribution statement
Leo Heijnen: Conceptualization, Methodology, Writing – original draft. Goffe Elsinga: Writing – original draft, Methodology. Miranda de Graaf: Writing – review & editing, Resources. Richard Molenkamp: Writing – review & editing, Resources. Marion P.G. Koopmans: Writing – review & editing, Supervision. Gertjan Medema: Writing – review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are very grateful for the assistance of the Water Authorities and WWTP operators of Waternet and Hoogheemraadschap de Stichtse Rijnlanden in the Netherlands who organized the sampling and provided the sewage samples. We are also grateful to Meindert de Graaf for sample transport. Eddy van Collenburg (BioRAD) is acknowledged for technical advice and providing the test kits. Coronavirus MHV-A59 was a kind gift from Prof. Eric Snijder (Leids Universitair Medisch Centrum) and provided by Linda Boomaars. This study was financed by “TKI Health Holland” in EUREKA project WASTEWATER4COVID.
Editor: Adrian Covaci
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.149456.
Appendix A. Supplementary data
Supplementary material
References
- Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., O'Brien J.W., Simpson S.L., Sherman P., Thomas K.V., Verhagen R., Zaugg J., Mueller J.F. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761 doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann D.M., Boyton R.J., Beale R. Immunity to SARS-CoV-2 variants of concern. Science. 2021;371(6534):1103–1104. doi: 10.1126/science.abg7404. [DOI] [PubMed] [Google Scholar]
- Bernal J.L., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., Stowe J., Tessier E., Groves N., Dabrera G., Myers R., Campbell C., Amirthalingam G., Edmunds M., Zambon M., Brown K., Hopkins S., Chand M., Ramsay M. Effectiveness of COVID-19 vaccines against the B.1.617.2 variant. medRxiv 2021.05.22.21257658. 2021 doi: 10.1101/2021.05.22.21257658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betancourt W.W., Schmitz B.W., Innes G.K., Brown K.M.Pogreba, Prasek S.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. Wastewater-based epidemiology for averting COVID-19 outbreaks on the University of Arizona Campus. medRxiv 2020.11.13.20231340. 2020 doi: 10.1101/2020.11.13.20231340. [DOI] [Google Scholar]
- Bivins A., North D., Ahmad A., Ahmed W., Alm E., Been F., Bhattacharya P., Bijlsma L., Boehm A.B., Brown J., Buttiglieri G., Calabro V., Carducci A., Castiglioni S., Gurol Z.Cetecioglu, Chakraborty S., Costa F., Curcio S., de Los Reyes F.L., 3rd, Vela J.Delgado, 3rd, Farkas K., 3rd, Fernandez-Casi X., 3rd, Gerba C., 3rd, Gerrity D., 3rd, Girones R., 3rd, Gonzalez R., 3rd, Haramoto E., 3rd, Harris A., 3rd, Holden P.A., 3rd, Islam M.T., 3rd, Jones D.L., 3rd, Kasprzyk-Hordern B., 3rd, Kitajima M., 3rd, Kotlarz N., 3rd, Kumar M., 3rd, Kuroda K., 3rd, Rosa G.La, 3rd, Malpei F., 3rd, Mautus M., 3rd, McLellan S.L., 3rd, Medema G., 3rd, Meschke J.S., 3rd, Mueller J., 3rd, Newton R.J., 3rd, Nilsson D., 3rd, Noble R.T., 3rd, van Nuijs A., 3rd, Peccia J., 3rd, Perkins T.A., 3rd, Pickering A.J., 3rd, Rose J., 3rd, Sanchez G., 3rd, Smith A., 3rd, Stadler L., 3rd, Stauber C., 3rd, Thomas K., 3rd, van der Voorn T., 3rd, Wigginton K., 3rd, Zhu K., 3rd, Bibby K., 3rd Wastewater-based epidemiology: global collaborative to maximize contributions in the fight against COVID-19. Environ. Sci. Technol. 2020;54(13):7754–7757. doi: 10.1021/acs.est.0c02388. [DOI] [PubMed] [Google Scholar]
- Challen R., Brooks-Pollock E., Read J.M., Dyson L., Tsaneva-Atanasova K., Danon L. Risk of mortality in patients infected with SARS-CoV-2 variant of concern 202012/1: matched cohort study. BMJ. 2021;372 doi: 10.1136/bmj.n579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies N.G., Abbott S., Barnard R.C., Jarvis C.I., Kucharski A.J., Munday J.D., Pearson C.A.B., Russell T.W., Tully D.C., Washburne A.D., Wenseleers T., Gimma A., Waites W., Wong K.L.M., van Zandvoort K., Silverman J.D., Diaz-Ordaz K., Keogh R., Eggo R.M., Funk S., Jit M., Atkins K.E., Edmunds W.J. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science. 2021;372(6538):eabg3055. doi: 10.1126/science.abg3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esri-Nederland Website: COVID-19 - Historische gegevens RIVM. 2021. https://nlcovid-19-esrinl-content.hub.arcgis.com/
- Eurosurveillance-editorial-team Updated rapid risk assessment from ECDC on the risk related to the spread of new SARS-CoV-2 variants of concern in the EU/EEA – first update. Eurosurveillance. 2021;26(3):2101211. doi: 10.2807/1560-7917.ES.2021.26.3.2101211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C., Hawryluk I., McCrone J.T., Hulswit R.J.G., Franco L.A.M., Ramundo M.S., de Jesus J.G., Andrade P.S., Coletti T.M., Ferreira G.M., Silva C.A.M., Manuli E.R., Pereira R.H.M., Peixoto P.S., Kraemer M.U., Gaburo N., Camilo C.D.C., Hoeltgebaum H., Souza W.M., Rocha E.C., de Souza L.M., de Pinho M.C., Araujo L.J.T., Malta F.S.V., de Lima A.B., Silva J.D.P., Zauli D.A.G., Ferreira A.C.S., Schnekenberg R.P., Laydon D.J., Walker P.G.T., Schlüter H.M., Santos A.L.P.Dos, Vidal M.S., Caro V.S.Del, Filho R.M.F., Santos H.M.Dos, Aguiar R.S., Modena J.L.P., Nelson B., Hay J.A., Monod M., Miscouridou X., Coupland H., Sonabend R., Vollmer M., Gandy A., Suchard M.A., Bowden T.A., Pond S.L.K., Wu C.H., Ratmann O., Ferguson N.M., Dye C., Loman N.J., Lemey P., Rambaut A., Fraiji N.A., Carvalho M., Pybus O.G., Flaxman S., Bhatt S., Sabino E.C. Genomics and epidemiology of a novel SARS-CoV-2 lineage in Manaus, Brazil. medRxiv. 2021 doi: 10.1101/2021.02.26.21252554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Beltran W.F., Lam E.C., Denis K.S., Nitido A.D., Garcia Z.H., Hauser B.M., Feldman J., Pavlovic M.N., Gregory D.J., Poznansky M.C., Sigal A., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. Circulating SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. medRxiv. 2021 doi: 10.1101/2021.02.14.21251704. [DOI] [Google Scholar]
- Izquierdo-Lara R., Elsinga G., Heijnen L., Munnink B.B.O., Schapendonk C.M.E., Nieuwenhuijse D., Kon M., Lu L., Aarestrup F.M., Lycett S., Medema G., Koopmans M.P.G., de Graaf M. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg. Infect. Dis. 2021;27(5):1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., Bänziger C., Stachler E., Fuhrmann L., Jablonski K.P., Chen C., Aquino C., Stadler T., Ort C., Kohn T., Julian T.R., Beerenwinkel N. Detection of SARS-CoV-2 variants in Switzerland by genomic analysis of wastewater samples. medRxiv 2021.2001.2008.21249379. 2021 doi: 10.1101/2021.01.08.21249379. [DOI] [Google Scholar]
- Jones D.L., Baluja M.Q., Graham D.W., Corbishley A., McDonald J.E., Malham S.K., Hillary L.S., Connor T.R., Gaze W.H., Moura I.B., Wilcox M.H., Farkas K. Shedding of SARS-CoV-2 in feces and urine and its potential role in person-to-person transmission and the environment-based spread of COVID-19. Sci. Total Environ. 2020;749 doi: 10.1016/j.scitotenv.2020.141364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantor R.S., Nelson K.L., Greenwald H.D., Kennedy L.C. Challenges in measuring the recovery of SARS-CoV-2 from wastewater. Environ. Sci. Technol. 2021;51(16):9146–9154. doi: 10.1021/acs.est.0c08210. [DOI] [PubMed] [Google Scholar]
- Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181(4):914–921. doi: 10.1016/j.cell.2020.04.011. e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Zhang Q., Wei P., Chen Z., Aviszus K., Yang J., Downing W., Peterson S., Jiang C., Liang B., Reynoso L., Downey G.P., Frankel S.K., Kappler J., Marrack P., Zhang G. The basis of a more contagious 501Y.V1 variant of SARS-COV-2. bioRxiv. 2021 doi: 10.1101/2021.02.02.428884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Liu J., Plante K.S., Plante J.A., Xie X., Zhang X., Ku Z., An Z., Scharton D., Schindewolf C., Menachery V.D., Shi P.-Y., Weaver S.C. The N501Y spike substitution enhances SARS-CoV-2 transmission. bioRxiv. 2021 doi: 10.1101/2021.03.08.434499. The preprint server for biology: 2021.03.08.434499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet Gastroenterol. Hepatol. 2020;5(6):533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 in sewage. medRxiv 2020.03.29.20045880. 2020 doi: 10.1101/2020.03.29.20045880. [DOI] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020;7(7):511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Pekin D., Skhiri Y., Baret J.C., Le Corre D., Mazutis L., Salem C.B., Millot F., El Harrak A., Hutchison J.B., Larson J.W., Link D.R., Laurent-Puig P., Griffiths A.D., Taly V. Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip. 2011;11(13):2156–2166. doi: 10.1039/c1lc20128j. [DOI] [PubMed] [Google Scholar]
- Prado T., Fumian T.M., Mannarino C.F., Resende P.C., Motta F.C., Eppinghaus A.L.F., do Vale V.H.Chagas, Braz R.M.S., Maranhão A.G., Miagostovich M.P., de Andrade J.D.S.R. Wastewater-based epidemiology as a useful tool to track SARS-CoV-2 and support public health policies at municipal level in Brazil. Water Res. 2021;191:116810. doi: 10.1016/j.watres.2021.116810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaben M., Einerhand A.W.C., Taminiau L.J.A., van Houdt M., Bouma J., Raatgeep R.H., Büller H.A., de Haan C.A.M., Rossen J.W.A. Cyclooxygenase activity is important for efficient replication of mouse hepatitis virus at an early stage of infection. Virol. J. 2007;4:55. doi: 10.1186/1743-422X-4-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rynkiewicz P., Babbitt G.A., Cui F., Hudson A.O., Lynch M.L. A comparative survey of betacoronavirus binding dynamics relevant to the functional evolution of the highly transmissible SARS-CoV-2 variant N501Y. bioRxiv. 2021 doi: 10.1101/2020.09.11.293258. [DOI] [Google Scholar]
- Stachler E., Kelty C., Sivaganesan M., Li X., Bibby K., Shanks O.C. Quantitative CrAssphage PCR assays for human fecal pollution measurement. Environ. Sci. Technol. 2017;51(16):9146–9154. doi: 10.1021/acs.est.7b02703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., Mlisana K., von Gottberg A., Walaza S., Allam M., Ismail A., Mohale T., Glass A.J., Engelbrecht S., Van Zyl G., Preiser W., Petruccione F., Sigal A., Hardie D., Marais G., Hsiao M., Korsman S., Davies M.A., Tyers L., Mudau I., York D., Maslo C., Goedhals D., Abrahams S., Laguda-Akingba O., Alisoltani-Dehkordi A., Godzik A., Wibmer C.K., Sewell B.T., Lourenço J., Alcantara L.C.J., Kosakovsky Pond S.L., Weaver S., Martin D., Lessells R.J., Bhiman J.N., Williamson C., de Oliveira T. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021 doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- US-CDC 2019-Novel Coronavirus (2019-nCoV) Real-Time rRT-PCR Panel Primers and Probes. 2020. https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html
- Volz E., Mishra S., Chand M., Barrett J.C., Johnson R., Geidelberg L., Hinsley W.R., Laydon D.J., Dabrera G., O’Toole Á., Amato R., Ragonnet-Cronin M., Harrison I., Jackson B., Ariani C.V., Boyd O., Loman N.J., McCrone J.T., Gonçalves S., Jorgensen D., Myers R., Hill V., Jackson D.K., Gaythorpe K., Groves N., Sillitoe J., Kwiatkowski D.P., Flaxman S., Ratmann O., Bhatt S., Hopkins S., Gandy A., Rambaut A., Ferguson N.M. Transmission of SARS-CoV-2 Lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv 2020.12.30.20249034. 2021 doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S., Mascola J.R., Chang J.Y., Yin M.T., Sobieszczyk M., Kyratsous C.A., Shapiro L., Sheng Z., Nair M.S., Huang Y., Ho D.D. Increased resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7 to antibody neutralization. bioRxiv. 2021 doi: 10.1101/2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809):465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158(6):1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material