Abstract
Pattern recognition receptors (PRRs) are a class of receptors that can directly recognize the specific molecular structures on the surface of pathogens, apoptotic host cells, and damaged senescent cells. PRRs bridge nonspecific immunity and specific immunity. Through the recognition and binding of ligands, PRRs can produce nonspecific anti-infection, antitumor, and other immunoprotective effects. Most PRRs in the innate immune system of vertebrates can be classified into the following five types based on protein domain homology: Toll-like receptors (TLRs), nucleotide oligomerization domain (NOD)-like receptors (NLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), C-type lectin receptors (CLRs), and absent in melanoma-2 (AIM2)-like receptors (ALRs). PRRs are basically composed of ligand recognition domains, intermediate domains, and effector domains. PRRs recognize and bind their respective ligands and recruit adaptor molecules with the same structure through their effector domains, initiating downstream signaling pathways to exert effects. In recent years, the increased researches on the recognition and binding of PRRs and their ligands have greatly promoted the understanding of different PRRs signaling pathways and provided ideas for the treatment of immune-related diseases and even tumors. This review describes in detail the history, the structural characteristics, ligand recognition mechanism, the signaling pathway, the related disease, new drugs in clinical trials and clinical therapy of different types of PRRs, and discusses the significance of the research on pattern recognition mechanism for the treatment of PRR-related diseases.
Subject terms: Drug development, Innate immunity, Structural biology
Introduction
The first line of defense against pathogens that gradually evolved in organisms is innate immunity,1 which is divided into two levels: first, the skin, mucosal tissue, blood–brain barrier, and chemical barrier (e.g. fatty acid, pH, enzyme, and complement system) of the host can effectively resist the invasion of general pathogenic microorganisms;2–4 second, the innate immune system of vertebrates protects the organism through nonspecific immune defense and surveillance by innate immune cells. Innate immune cells mainly include monocytes, neutrophils, macrophages, dendritic cells, natural killer (NK) cells, mast cells, eosinophils, and basophils.5,6 Unlike T cells and B cells, which have high specificity, innate immune cells do not express specific antigen recognition receptors. Through the recognition and binding of some common molecules on the surface of pathogens, apoptotic host cells, and damaged senescent cells, pattern recognition receptors (PRRs) induce immunoprotective effects, such as anti-infection and antitumor effects, and participate in the initiation and effect process of specific immune response.7–9
In the 1990s, the hypothesis of pathogen-associated molecular patterns (PAMPs) and PRRs that recognize PAMPs was proposed by Janeway, which was of epoch-making significance and changed research on innate immunity.10 The main point of this hypothesis is the connection between the innate immune signal and the initiation of the adaptive immune response. Some unique and conserved components of pathogenic microorganisms can induce the second signal required to activate T cells, so as to control the adaptive immunity from being activated under normal conditions.11,12 In addition, there are a class of receptors in the host that can recognize pathogenic microorganisms and activate the second signal in time, which are independent of gene rearrangement. In vertebrates, innate immunity recognizes pathogenic microorganisms and assists in the activation and expression of second signals that activate the adaptive immunity.13
Toll-like receptors (TLRs) are one of the earliest PRRs discovered in the innate immune system, which plays an important role in inflammatory responses.14,15 Therefore, here is a brief description of the development history of PRRs with TLRs as a representative. TLRs were first found in Drosophila in the form of genes in 1994. Studies have shown that the function of this gene is related to the formation of the dorsal–ventral axis during the embryonic development of Drosophila.16 In 1988, Hashimoto et al. discovered that the Toll gene encodes a transmembrane protein and clarified the structure of the Toll protein.17 In 1991, Gay et al. found that Toll protein had structural homology with interleukin-1 (IL-1), a natural immune molecule in mammals, suggesting that the function of Toll may be related to immunity.18 In 1996, Hoffmann team found that Toll plays a role in the resistance of Drosophila to fungal infection. Toll-activated mutants persistently express antifungal peptides, while Toll-deletion mutants, on the contrary, lose their ability to arrest fungal infection. It has been found that Toll can recognize spatzle (an important protein in the development of the dorsal and abdomen of Drosophila) and initiate a series of signal transduction to activate the expression of antifungal peptide.19 In 1997, Janeway et al. cloned human TLR4. TLR4 can induce the activation of nuclear factor (NF)-κB and the expression of the co-stimulatory molecule CD80. This proves that innate immunity recognizes pathogenic microorganisms and activates the expression of the second signal, which is indispensable for the activation of adaptive immunity.20 Since the discovery of TLR4, many PRRs and their corresponding ligands have been discovered. PRRs can be divided into the following five types based on protein domain homology: TLRs, nucleotide oligomerization domain (NOD)-like receptors (NLRs), retinoic acid-inducible gene-I (RIG-I)-like receptors (RLRs), C-type lectin receptors (CLRs), and absent in melanoma-2 (AIM2)-like receptors (ALRs) (Table 1).21 PRRs are representative of immune receptors in innate immunity and exist in various forms. PRRs are not only expressed on the cell membrane but also widely distributed in intracellular compartment membranes and the cytoplasm.22 Membrane-bound PRRs and PRRs in the cytoplasm are basically composed of ligand recognition domains, intermediate domains, and effector domains.23,24 PRRs activate downstream signaling pathways through recognition of their ligands. The activation of downstream signaling pathways can produce many effects: recruiting and releasing cytokines, chemokines, hormones, and growth factors; inducing chronic inflammation; forming an inflammatory microenvironment; initiating innate immune killing and subsequent acquired immune response,9 maintaining the balance of host microecology; and eliminating dead or mutated cells.
Table 1.
Common PRRs in innate immunity
| Items | PRR | Domains | Cellular distribution | PAMP | Sources | Signaling pathways |
|---|---|---|---|---|---|---|
| Toll-like receptors (TLRs) | TLR1 (TLR1–TLR2) | LRR domain–transmembrane domain–TIR domain (extracellular to intracellular) | Mo, DC, Ma, Eo, Ba | Triacyl lipopeptide | Bacteria | Most TLRs: MyD88-dependent pathways; TLR3: TRIF-dependent pathways; TLR4: MyD88-dependent pathways and TRIF-dependent pathways |
| TLR2 (TLR1–TLR2, TLR2–TLR6) | Mo, DC, Ma, Eo, Ba | Lipoteichoic acid | Bacteria | |||
| Arabinomannan | Mycobacterium | |||||
| Peptidoglycan | Bacteria | |||||
| Zymosan | Fungi | |||||
| Lipoprotein | Mycoplasma | |||||
| Pore protein | Neisseria | |||||
| TLR3 | Mφ, DC, IEC | dsRNA | Virus | |||
| TLR4 (MD-2/CD14) | Mφ, DC, Ma, Eo | Lipopolysaccharides | Bacteria | |||
| Heat-shock proteins | Host | |||||
| TLR5 | IEC | Flagellin | Bacteria | |||
| TLR6 (TLR2–TLR6) | Mo, DC, Ma, Eo, Ba | Lipoteichoic acid | Bacteria | |||
| Peptidoglycan | Bacteria | |||||
| TLR7 | pDC, Mφ, Eo | ssRNA | Virus | |||
| Imidazoquinoline | Artificially synthesized | |||||
| TLR8 | Mφ, N | ssRNA | Virus | |||
| TLR9 | pDC, Eo, Ba | Non-methylated CpG DNA | Bacteria, Virus | |||
| TLR10 (human) | pDC, Eo, Ba | dsRNA | Virus | |||
| TLR11 (mouse) | Mφ, DC | Profilin and related proteins | Toxoplasma gondii | |||
| TLR12 (mouse) | DC | Profilin and related proteins | Toxoplasma gondii | |||
| TLR13 (mouse) | Unknown | 23s ribosomal RNA | Bacteria | |||
| Nucleotide-binding oligomerization domain-like receptors (NLRs) | NOD1 | LRR domain–NBD–effector domains | IEC, cytosol of Mφ | iE-DAP | Gram negative bacteria | RIP2-TAK1-NF-κB pathways |
| NOD2 | MDP | Gram-negative bacteria, Gram-positive bacteria | ||||
| RIG-I-like receptors (RLRs) | RIG-I | (RD)-CTD-DexD/H helicase domain–CARD | Cytosol | 5’-triphosphorylated RNA, short-chain dsRNA | Virus | MAVS-TRAF6-NF-κB/TBK1 pathways |
| MDA5 | poly IC, long-chain dsRNA | Virus | ||||
| LGP2 | dsRNA | Virus | ||||
| C-type lectin receptors (CLRs) | Dectin-1 | CTLD–ITAM | DC, Mφ | β-Glucan | Fungus | Tyrosine kinase-dependent and non-tyrosine kinase-dependent pathways |
| Dectin-2 | α-Mannan | Fungus | ||||
| Absent in melanoma-2-like receptors (ALRs) | ALRs | HIN-200-PYD | Cytosol | dsDNA | Bacteria | Inflammasome–pyroptosis |
LRR leucine-rich repeat, TIR Toll/IL-1R domain, NBD nucleotide-binding domain, RD repressor domain, CTD C-terminal domain, CARD caspase activation and recruitment domain, CTLD C-type lectin-like domains, ITAM immunoreceptor tyrosine-based activation motif, PYD pyrin domain, Mo monocyte, DC dendritic cell, Ma mastocyte, Eo eosinophils, Ba basophils, pDC plasmacytoid dendritic cell, IEC intestinal epithelial cell, N neutrophil, dsRNA double-stranded RNA, ssRNA single-stranded RNA, iE-DAP γ-D-glu-meso-diaminopimelic acid, MDP muramyl dipeptide, MyD88 myeloid differentiation factor 88, TRIF TIR domain-containing adaptor protein-inducing interferon β, RIP2 receptor-interacting serine–threonine protein 2, TAK1 transforming growth factor-β-activated kinase 1, NF-κB nuclear factor κB, MAVS mitochondrial antiviral signaling protein, TRAF6 tumor necrosis factor receptor-associated factor, TBK1 TANK-binding kinase 1
PAMPs are the specific and highly conserved molecular structures shared by the same kind of pathogenic microorganisms,25,26 including lipids, proteins, and nucleic acids, such as lipopolysaccharides (LPS), lipoteichoic acid (LTA), and bacterial DNA.27,28 PAMPs are essential for pathogen survival and usually have unique molecular or subcellular characteristics that are not found in host cells. Therefore, innate immune cells can recognize PAMPs via PRRs, distinguish “self” and “non-self,”29 and respond to pathogens and their products. However, the host will produce some proteins and metabolites after being stimulated by its own tissue damage, cell necrosis, and other factors.30 These molecules are called damage-associated molecular pattern (DAMP).31 PRRs can also recognize such molecules, activate natural immunity, and cause inflammation.32
With advances in research on PRRs structure and distribution at different levels, the role of PRRs in the innate immune regulatory network has become clearer. The recognition and binding of PRRs to their ligands is critical in initiating the innate immune response. Therefore, the study of PRR-mediated pattern recognition mechanisms will help to elucidate the signaling pathways and mechanisms of disease and provide new targets and methods for the treatment of diseases. In this review, we describe the structural characteristics, ligand recognition mechanism, the signaling pathway, the related disease, new drugs in clinical trials, and clinical therapy of different types of PRRs in detail. We focus on the different domains and ligand recognition mechanisms between PRRs, which can not only provide new ideas for the definition, role, and clinical application of PRRs but also promote the study of the role of the innate immune system in related diseases and even tumors.
PRRs and ligand-recognition mechanisms
Toll-like receptors
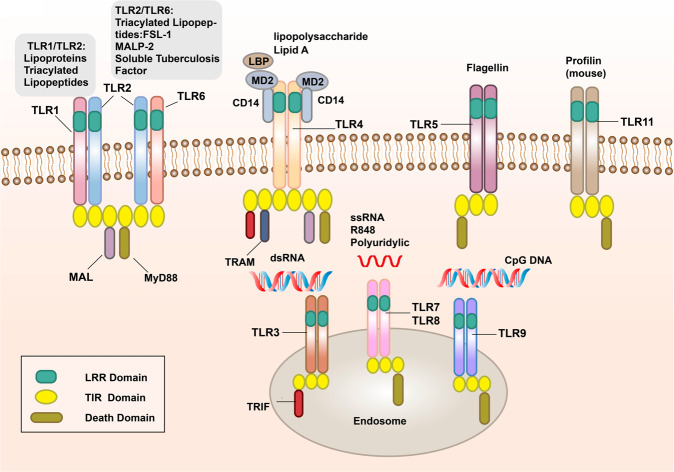
TLRs are membrane-bound signal receptors and are important PRRs in the innate immune system of vertebrates.15,33 Such receptor molecules usually have two functions, one is to bind specifically to the ligand, and the other is to transmit signals. The corresponding signal transduction will amplify the effect of anti-pathogen infection, so that the immune cells active in the inflammatory response can be activated through the transcription of genes, and produce and secrete a variety of pro-inflammatory and antiviral factors.34–36 Up to now, 10 functional TLRs (TLR1–10) have been found in humans and 12 (TLR1–9 and TLR11–13) in mice.37–41 TLR10 in mice is not functional due to the insertion of reverse transcriptase.42 TLRs recognize PAMPs in different subcellular structures. The cellular localization of TLRs determines the types of ligands and the recognition mechanism. Some TLRs (TLR1, 2, 4, 5, 6, 10) are expressed on the surface of immune cells in the form of heterodimers or homodimers, mainly recognizing the membrane components of pathogenic microorganisms, such as lipids, lipoproteins, and proteins; others (TLR3, 7, 8, 9) are expressed in the form of homodimers, which mainly recognize the nucleic acids of microorganisms (Fig. 1).43
Fig. 1.
The signal transduction pathways and structure of TLR-binding ligand complex. TLRs can recognize one or more PAMPs through LRR domain. They usually dimerize themselves and recruit adaptor molecules with the same TIR domain to transmit signals
TLRs are type I transmembrane glycoproteins and are composed of an extracellular region, a transmembrane region, and an intracellular region.44 The extracellular region contains leucine-rich repeats (LRRs), which are responsible for the recognition of specific ligands and perform extracellular pattern recognition. The intracellular domain contains the same Toll/IL-1R (TIR) domain as IL-1R, which plays a role in signal transduction. The extracellular region of TLRs contains LRRs, which mediate the pattern recognition of TLRs (Fig. 1).45 In 2007, researchers used X-ray crystal diffraction to analyze and determine the structure of the TLR–ligand complex,46 which provided a deeper understanding of the LRR domain. The LRR domain is shaped like a horseshoe, and each module consists of a conserved leucine motif and a variable region. The “LxxLxLxxN (L leucine, x any amino acid, N asparagine)” motif is composed of 20–30 amino acids and is on the concave surface of the horseshoe-like structure.47–50 The horseshoe-shaped N-terminus and C-terminus contain disulfide bridges formed by cysteine clusters51,52 to protect the hydrophobic core. After TLRs recognize and bind the corresponding PAMPs and endogenous ligands, the TIR domains conduct signals by binding to different receptor adaptor proteins in the cytoplasmic region.53,54 The TIR domain has three conserved amino acid sequences, which are called 1,2,3 cassettes. Depending on the different adaptor proteins, TLRs signaling can be divided into myeloid differentiation factor 88 (MyD88)-dependent and MyD88-independent pathways (Fig. 1).55,56
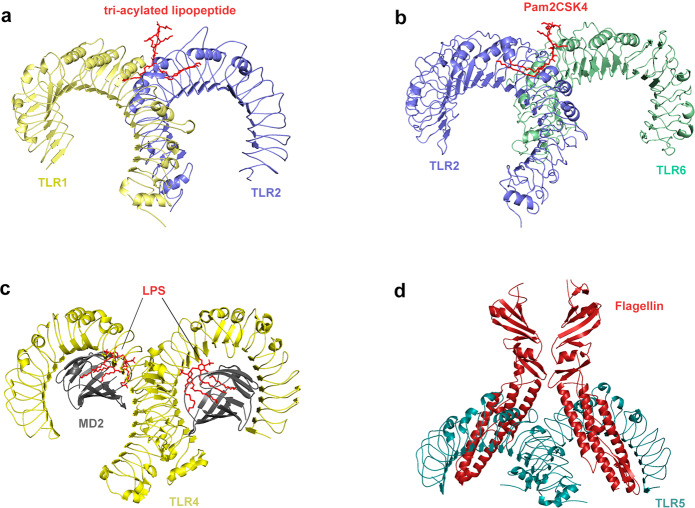
Exploring the pattern recognition mechanisms of TLRs is very valuable for understanding innate immunity and some tumorigenesis mechanisms. Therefore, researchers used X-ray crystal diffraction to determine the crystallographic structure of the extracellular domain of TLRs and the ligand complex. Although the ligand complexes have different structures, all these complexes have similar M-type crystal structures (Fig. 2).50,51 TLR1 or TLR6 can form TLR1/TLR2 and TLR6/TLR2 heterodimers with TLR2 to recognize tri-acylated lipopeptide and di-acylated lipopeptide,57 respectively. After recognizing the appropriate ligands, TLR2 can form an M-type structure with the extracellular region of TLR1 and TLR6, and the pocket structure formed binds to the ligand.58–61 The crystal structure of TLR1–TLR2–tri-acylated lipopeptide complex is similar to that of TLR2–TLR6–di-acylated lipopeptide complex, but there are important structural differences between TLR1 and TLR6 in the ligand-binding site and dimerization surface. The ligand-binding pocket of TLR1–TLR2 is located in the interface between the central and C-terminal domain, and TLR1–TLR2–tri-acylated lipopeptide is stabilized by non-covalent bonds such as hydrogen bonds, hydrophobic interactions, and ionic interactions near the ligand-binding pocket.62 In TLR6, the side chains of amino acid residues block the ligand-binding pocket, resulting in a pocket less than half the length of TLR1. In addition, the TLR2–TLR6 heterodimer is mainly regulated by the surface exposed residues of the LRR11–14 module (Fig. 2a, b).63 Researches on the structure of ligand complexes can significantly promote the discovery of small molecule agonists/antagonists targeting PRRs. A recent study revealed the activation mechanism of atypical agonists for TLR1–TLR2. Diprovocim is a recently found small molecule activator for TLR1–TLR2, but it has no structural similarity with the tri-acylated lipopeptide complex. It also interacts with TLR1–TLR2 in the same binding pocket as typical lipopeptide ligand.64 Crystal structure analysis revealed that double-stranded RNA (dsRNA) binds to the LRR domains of the N-terminus and C-terminus of TLR3.65,66 Different from the way that other TLRs directly recognize ligands,67–69 TLR4 specifically recognizes LPS in combination with two auxiliary molecules, myeloid differentiation factor 2 (MD2) and the LRR structural protein CD14. LPS is transported by LPS-binding protein to CD14 on the cell membrane of monocytes and macrophages to form a complex and then interacts with TLR4/MD2.70 After LPS binds with the TLR4/MD2 complex, the hydrophobic pocket of MD2 is used to bridge the two TLR4–MD2–LPS complexes to form a spatially symmetrical M-type TLR4–MD2–LPS dimer,71 and then conformational changes affect their respective functional domains and transmit signals (Fig. 2c). In addition to binding to LPS, TLR4 is also involved in the recognition of natural products (carnosic acid, paclitaxel) and pneumolysin.72–74 TLR5 is the most conserved and important PRR, which is usually stimulated by bacterial flagellin. In the form of homodimer, TLR5 plays a major role in the primary defense of invasive pathogens and immune homeostasis regulation.75 Although the heterodimeric structure of TLR5a–TLR5b in zebrafish and the crystallographic structure of TLR5–flagellin complex has been clearly reported, the lack of biochemical and structural information of fish TLR hinders the understanding of flagellin-based therapies. In the future, experimental TLR5–flagellin complex structure modeling and computational simulation should be used to study flagellin-mediated interactions between various pathogens and host immune receptors (Fig. 2d).76 It has been reported that TLR1–6 each exist as monomers in solution, and dimerization occurs only when the ligand is bound; in contrast, TLR8 and TLR9 exist as preformed dimers, and the binding of ligands induces conformational changes in preformed dimers (Fig. 1).49,77,78 Lee revealed that TLR10 binds dsRNA in vitro at endosomal pH, indicating that dsRNA is a ligand of TLR10. The recognition of dsRNA by TLR10 recruits MyD88, thereby transducing signals and inhibiting interferon regulatory factor 7 (IRF7)-dependent type I interferon (IFN) production.79 In mice, TLR11 and TLR12 are the main effector molecules to recognize Toxoplasma gondii. The recognition of T. gondii profilin by TLR11 depends on the parasite-specific, surface-exposed motif in TgPRF consisting of an acidic loop and a β-hairpin.80–82
Fig. 2.
Crystal structure of TLRs with ligands. a Crystal structure of the TLR1–TLR2 heterodimer induced by binding of a tri-acylated lipopeptide (PDB 2Z7X). TLR2 initiates immune responses by recognizing di-acylated and tri-acylated lipopeptides. The ligand specificity of TLR2 is controlled by whether it heterodimerizes with TLR1 or TLR6. Binding of the tri-acylated lipopeptide (red) induced the formation of M-type crystal structures of the TLR1 (pale yellow) and TLR2 (slate) ectodomains. b Crystal structure of TLR2–TLR6–Pam2CSK4 complex (PDB 3A79). Binding of the di-acylated lipopeptide, Pam2CSK4 (red), induced the formation of M-type crystal structures of the TLR2 (slate) and TLR6 (pale green) ectodomains. c Crystal structure of mouse TLR4/MD2/LPS complex (PDB 3VQ2). After LPS (red) binds with the TLR4 (yellow)/MD2 (gray) complex, the hydrophobic pocket of MD2 is used to bridge the two TLR4–MD2–LPS complexes to form a spatially symmetrical M-type structure. Mouse TLR4/MD2/LPS exhibited an complex similar to the human TLR4/MD2/LPS complex. d Crystal structure of the N-terminal fragment of zebrafish TLR5 in complex with Salmonella flagellin (PDB 3V47). Two TLR5 (cyan)–flagellin (firebrick) 1:1 heterodimers assemble into a 2:2 tail-to-tail signaling complex to function
NOD-like receptors
The growth cycle of some pathogenic microorganisms involves infection of the cytoplasm. For example, viral genes are often transcribed and translated in the cytoplasm, and virus particles are assembled. In addition, some bacteria and parasites have a series of escape mechanisms, such as making holes in the phagosome membrane and entering the cytoplasm. Therefore, pathogens and their components, as well as other components produced by infection and injury, will appear in the cytoplasm,83 which requires the recognition of PRRs in the body. NLRs are intracellular PRRs, composed of three domains:84,85 one is the central nucleotide-binding domain (NBD), also known as the NACHT domain (synthesized by the abbreviations of the following four kinds of NLR members: NAIP, CIITA, HETE, TP1), which is shared by the NLR family and is very important for nucleic acid binding and oligomerization of NLRs (Fig. 3); LRRs at the C-terminus, which are used to identify ligands; and the N-terminal effector domain, which is the protein interaction domain, such as the caspase activation and recruitment domain (CARD) or the pyrin domain (PYD).86–89 According to the different N-terminal effector domains, the NLRs family can be divided into five subfamilies: the NLRC subfamily, which contain CARDs; the NLRP subfamily, which contain PYDs; the NLRB subfamily, which contain baculovirus inhibitor of apoptosis protein repeats; the NLRA subfamily, which contain acidic activation domains; and the NLRX subfamily containing other NLR effector domains.85
Fig. 3.
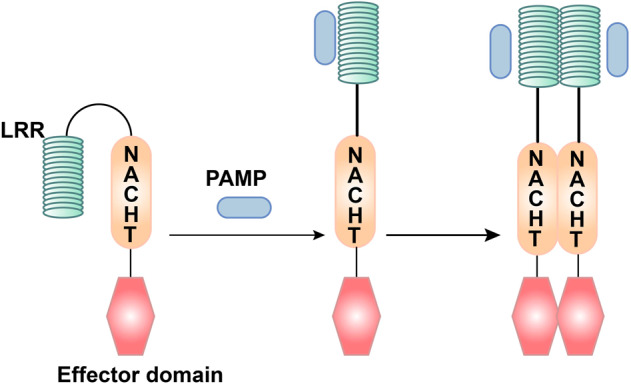
The ligand recognition mechanism of NLRs. The combination of PAMP and LRR changes the conformation of NLRs from self-inhibition to activation
Among the NLRs family, the most in-depth study has focused on NOD1 and NOD2 proteins. NOD1 mainly recognizes the diaminopimelic acid (γ-D-glu-meso-diaminopimelic acid (iE-DAP)) of the cell wall of Gram-negative bacteria.90,91 In addition to recognizing muramyl dipeptide (MDP) in all bacterial cell walls, NOD2 can also recognize single-stranded RNA (ssRNA) of the virus, but it must be a complete viral ssRNA.92 The basic process of NOD2 activation and signal transduction is as follows: after pathogenic bacteria are phagocytosed by macrophages, they first form phagosomes, and then fuse with lysosomes to become phagolysosomes. Under the action of lysosomal enzymes, bacterial cell wall components are decomposed into peptidoglycan, which can be degraded into a cell wall peptide with immunomodulatory activity and enter the cytosol, thereby activating NOD2.93 In general, the LRR domain of the NLR molecule folds to form a U-shaped configuration with the central NACHT domain, which inhibits its multimerization and makes the NLRs inactive.94 Once PAMPs directly or indirectly bind to the LRRs, the NLR molecule change their conformation, exposing the NACHT oligomerization domain, which triggers oligomerization, and the NLR molecule is activated.95 At the same time, the N-terminal effector domain is exposed, and through homotypic interactions, downstream adaptor molecules and signaling proteins with the same structure are recruited to initiate the corresponding signal transduction (Fig. 3).96 Although NOD1 and NOD2 do not have transmembrane domains, studies have shown that they are recruited into the plasma membrane and endosomal membrane, which is necessary for signal transduction.97 In this process, palmitoylation plays a vital role. The modification of NOD1/2 protein under the action of palmitoyltransferase ZDHHC5, which makes NOD1/2, possess the characteristics of rapid and reversible localization changes, which is necessary for membrane recruitment and inflammatory signal transduction.98 This study gives us a good enlightenment that the modification of PRRs may play a key role in the regulation of host innate immune signal.
RIG-I-like receptors
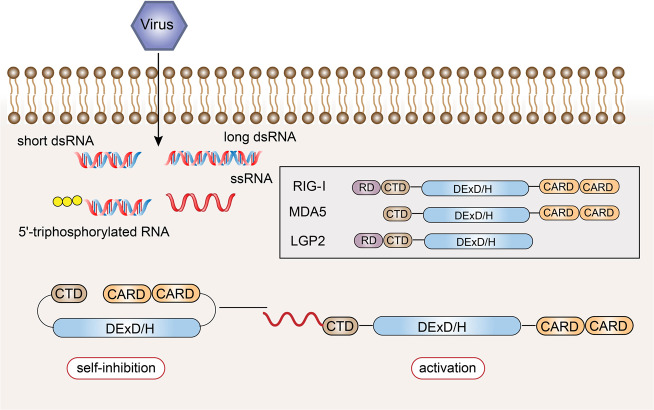
RLRs are also intracellular PRRs. In innate antiviral immunity, in addition to the recognition of viral nucleic acids by TLR7 and TLR9, most other types of cells recognize viral nucleic acids through RLRs to induce antiviral immune responses.99,100 The currently discovered RLR family members mainly include three: RIG-I, melanoma differentiation-associated gene 5 (MDA5), and laboratory of genetics and physiology 2 (LGP2) (Fig. 4).101
Fig. 4.
Structural features and ligand recognition mechanism of RLRs. The structure and functions of MDA5 are similar to those of RIG-I. However, MDA5 lacks the repressor domain, so it does not have self-inhibitory functions. LGP2 does not have CARD, and so it cannot transmit signals. The combination of viral RNA and CTD changes the conformation of RLRs
RIG-I was first discovered in acute promyelocytic leukemia cells induced by retinoic acid. In 2004, it was found that RIG-I could induce the expression of a reporter gene in the IFN-β promoter region, which confirmed its antiviral activity.102 The structure of the RIG-I protein consists of three parts.103–105 The middle part is the DexD/H helicase domain, which is the common domain of the RLR family, and has ATPase and helicase activities.106–108 The N-terminus of the RIG-I protein is composed of two caspase activation and recruitment domains in series,109 which are responsible for transmitting signals downstream.110 The C-terminus is composed of the repressor domain (RD) and the C-terminal domain (CTD), which can regulate its own state.106,111 The former can inhibit the activation of the receptor, and the latter is responsible for the recognition of viral RNA.112,113 In the resting state, CARD, CTD, and the helicase domain are folded, and RIG-I is in a self-inhibited state. During viral infection, the CTD of RIG-I recognizes viral RNA and undergoes a conformational change.114 RIG-I uses ATP hydrolase activity to expose and activate the CARD and multimerize, thereby recruiting downstream signaling linker molecules (Fig. 4).115–117
The structure and functions of MDA5 are similar to those of RIG-I, with the DexD/H helicase domain in the middle, two CARD at the N-terminus, and a CTD at the C-terminus; however, MDA5 lacks the RD, and so it does not have self-inhibitory functions. In contrast to other RLRs, LGP2 does not have CARD,118,119 and so it cannot recruit molecules of the same structure to transmit signals, but it can regulate the recognition of viral nucleic acids by RIG-I and MDA5, thereby preventing RLR-mediated resistance.120–123 LGP2 can negatively regulate RIG-I-mediated recognition of viral dsRNA, reduce the production of IFNs and inflammatory factors, and ultimately inhibit the antiviral innate immune response.124 LGP2 is also critical in the antiviral response mediated by MDA5.125 LGP2 exhibits a concentration-dependent conversion between MDA5-specific enhancement and interference.126 The latest research revealed a mechanistic basis for LGP2-mediated regulation of MDA5 antiviral innate immune responses. LGP2 facilitates MDA5 fiber assembly and is incorporated into the fibers, forming hetero-oligomers with MDA5.127 In addition, LGP2 can significantly induce the exposure of the CARD domain of MDA5.128 Under bacterial infection of the Indian major carp Labeo rohita, LGP2 gene expression was significantly increased after dsRNA and various PAMPs were stimulated, indicating that LGP2 can act as an antiviral and antibacterial cytoplasmic receptor.129
Although the RLR family members have similar structures, they recognize the RNA of different viruses through ligand-recognition domains.130 Both RIG-I and MDA5 can recognize viral dsRNA, but their recognition depends on the length of the dsRNA.131 RIG-I mainly recognizes viruses with relatively short dsRNA (<1000 bp), while MDA5 tends to recognize long-chain dsRNA (>1000 bp).132 Additionally, RIG-I mediates the antiviral response by recognizing the 5’-triphosphate RNA of viruses.133 The 5’-terminal triphosphate group can be recognized by RIG-I as a non-self component, but after posttranslational modification, this molecule cannot be recognized by RIG-I.134 Because host cell RNA needs to undergo different degrees of processing and modification after synthesis in the nucleus, these results indicate that RIG-I can distinguish viral dsRNA from endogenous RNA. In the cell, RIG-I mainly recognizes influenza virus,135 vesicular stomatitis virus,136 Sendai virus, and Japanese encephalitis virus,137,138 while MDA5 mainly recognizes small RNA viruses, such as poliovirus.139,140 MDA5 also participates in the synthesis of the dsRNA analog polycytidylic acid (poly I:C). Previous studies have shown that filamentous fibers are formed during the recognition of ligands by RIG-I and MDA5, and signaling pathways are initiated from the tail and inside of the viral dsRNA, respectively.141
Although it is mentioned in the “Toll-like receptors” section that TLR3, TLR7, TLR8, and TLR9 specifically recognize virus-derived nucleic acid molecules and bacterial nuclear components, they mainly appear in the endosomal membrane. RLRs can not only be expressed in cells infected by various viruses but also can directly recognize and perceive the virus products and virus particles that exist in the cytosol. Its antiviral significance cannot be ignored.142
C-type lectin receptors
CLRs, which belong to phagocytic PRRs, are also a popular type of receptor under study.143 The function of phagocytic receptor is different from the receptor that activates cells by signal transduction. It recognizes and binds to PAMPs through PRRs and places pathogens in cytoplasmic vesicles for direct digestion and elimination to control infection.144 CLRs are a class of receptors that recognize carbohydrates on the surface of pathogenic microorganisms with the participation of Ca+.145 It is expressed on macrophages, dendritic cells (DCs), and certain tissue cells. The ability of CLRs to recognize carbohydrates existing on self and non-self structures is mediated by carbohydrate recognition domain (CRD).146 The CRD of CLRs is a compact spherical structure, and this region is called C-type lectin-like domain (CTLD).147,148 Depending on the location of the protein on the cell membrane, CLRs are divided into transmembrane receptors and secretory receptors.146,149,150 The main representative of secretory receptors is collagen lectin family (under the “Extracellular pattern recognition molecules”).151 Transmembrane receptors can be divided into type I and type II according to their topological structure.152,153 The N-terminal of type I receptors points to extracellular and contains multiple CRDs, while the N-terminal of type II receptors points to intracellular and contains only one CRD.154,155 It has been shown that the vast majority of CLRs are involved in the presentation of antigens as active membrane-associated receptors, and CLRs are mainly expressed on antigen-presenting cells such as DCs and macrophages.145 CLRs are circular structures connected by two disulfide bonds.156 CLRs contain at least one CTLD outside the cell, while the intracellular domain is different.
Mannose receptors (MRs) belong to membrane CLRs, which are single-chain transmembrane molecules.157–159 The extracellular segment of MR consists of two parts: one is the proximal membrane end with eight consecutive CTLDs, which is responsible for the endocytosis and transport of the ligand; the other is the distal membrane end of the cysteine-rich lectin domain, which recognizes sulfation of carbohydrate conjugates.160 The endogenous ligands of MR are lysosomal hydrolase and myeloperoxidase, as well as the mannan-rich structure expressed by pathogens.161,162
Dendritic cell-associated C-type lectin (Dectin)-1 and Dectin-2 are typical representatives of the CLR family.163,164 Dectin-1 is a type II transmembrane protein expressed in DCs, macrophages, neutrophils, and monocytes.165 The extracellular region is a CTLD. The intracellular tail is connected to an immunoreceptor tyrosine-based activation motif (ITAM),166 indicating that the receptor also has a signal transduction function. Dectin-1 can identify a variety of fungi,167 including yeast,168 Candida albicans,169,170 Pneumocystis carinii,171,172 Cryptococcus,173,174 and Aspergillus.175,176 The ligand of Dectin-1 is β-1,3-glucan, which can activate downstream signals through tyrosine kinase-dependent and tyrosine kinase-independent pathways after recognition and binding of the ligand.177,178 Glycosylation is an important modification of the posttranslational modification of proteins (including antibodies),179 which can significantly change the structure and function of proteins or antibodies, so it is also a key mechanism for the immune system to regulate biological activity.180 Abnormal glycosylation is usually associated with malignant tumors.179 Therefore, the identification of molecules that bind glycosylated glycans can provide a new way for the treatment of human infectious and malignant diseases. Studies have found that Dectin-1 can recognize aromatic amino acids adjacent to the N-terminal asparagine at the glycosylation site as well as the core fucose on IgG antibodies, which do not compete for the same protein binding site for β-glucan, so Dectin-1 can regulate the immune response induced by IgG by combining with core fucose.181
Dectin-2, which is different from Dectin-1, does not contain the ITAM sequence and has no signal transduction function.182 Dectin-2 mainly recognizes α-mannan in the fungal cell wall and recognizes the Schistosoma mansoni egg antigen.183,184 The molecular mechanism by which Dectin-2 recognizes the binding ligand has always been the focus of research. Decout et al.185 found that the stimulation of Dectin-2 by purified Mycobacterium tuberculosis mannose-capped lipoarabinomannan requires the (α1 → 2)-linked mannosides forming the cap. Besides, Dectin-2 can also recognize lipoglycans from other bacterial species.185,186 From the perspective of the relationship between the structure and function of the above two ligands, dimannoside caps and multivalent interaction are necessary for Dectin-2 to recognize binding ligands and conduct signals.187
AIM2-like receptors
ALRs are a new type of PRRs that can recognize intracellular DNA.188,189 The C-terminus is the DNA-binding domain HIN-200, and the N-terminus is the PYD.189–192 The HIN-200 domain recognizes double-stranded DNA and binds to it. The N-terminal PYD binds to the PYD of apoptosis-associated speck-like protein containing CARD (ASC),193,194 thereby promoting the formation of inflammasomes and the maturation and release of IL-1β and IL-18.195 Both the DNA-binding affinity of AIM2 and the activity of its inflammasome depend on dsDNA, and it can assemble into filamentous structures along dsDNA. However, without dsDNA, it can also form filaments at high protein concentrations.196–198 ALRs can not only participate in the innate immune response but also regulate apoptosis, which is related to the occurrence and development of tumors.199
Extracellular soluble pattern recognition molecules
The initiation of the innate immune response depends on the recognition of PAMPs by pattern recognition molecules (PRMs), including cell PRRs and extracellular soluble PRMs. They are a class of free receptors that can play an antibacterial effect in serum.200 Although the pattern recognition of innate immunity does not have the antigen specificity of the adaptive immune response, some PRMs produced by the body after infection by pathogenic microorganisms will exist in the serum. Once the new pathogens invade, they can also bind to the pathogen like an antibacterial molecule and play an effective function. Unlike cell-related PRRs, extracellular soluble PRMs are an important part of non-specific humoral immunity.201 Extracellular soluble PRMs are composed of different molecular families, mainly including pentraxin,202 collectin, and ficolin.203 They generally function in two ways: one is that they recognize various pathogenic factors and eliminate them through complement activation,204,205 opsonization,206 aggregation, and neutralization of inflammatory regulation; the other is that they interact with cell-related PRRs and regulate their functions to jointly regulate innate immune response.207
Pentraxin is characterized by the aggregation of five molecules and is highly conserved in evolution, including two families of short molecules and long molecules.208–211 The family of short molecules is called acute phase proteins, which is represented by C-reactive protein (CRP)212–214 and serum amyloid P component215,216 in humans and mice, respectively. These molecules are mainly produced by the liver under the stimulation of inflammatory signals and interleukins. They are non-specific proteins that reflect the systemic inflammatory response. Serum levels increase rapidly after the body is infected or injured. CRP generally binds to phosphocholine expressed on the surface of pathogenic microorganisms in a Ca+-dependent manner.217 SAA can bind to the outer membrane protein A of bacteria and interact with TLRs.218,219 In clinic, SAA and CRP are usually used as auxiliary diagnostic indicators for infectious diseases, but studies have shown that they also have diagnostic value in non-infectious diseases and can be used as disease classification markers.220,221 The representative of the pentraxin long molecule family is PTX3,222 which is unique in that it has a long N-terminal domain. PTX3 is produced by dendritic cells, monocyte macrophages, epithelial cells, smooth muscle cells (SMCs), and endothelial cells under the regulation of a variety of inflammatory factors.223 PTX3 is involved in the defense of selected pathogens and the regulation of inflammation.224–226 Due to its expression increases sharply under the conditions of inflammatory stimulation, PTX3 can become a biomarker of general acute inflammation and a variety of tumors.227 In coronavirus disease 2019 (COVID-19) patients, circulating and lung bone marrow monocytes and endothelial cells express high levels of PTX3, and PTX3 plasma concentration can serve as an independent strong prognostic indicator of short-term mortality in COVID-19.228,229
Collectin mainly includes mannose-binding lectin (MBL) and surfactant protein (SP).151,230 MBL is formed by connecting multiple homotrimers. Each component of the trimer includes a CRD, an alpha helix, and a main stem formed by spirals of collagen.231,232 The main stem of collagen gathers each trimer into bundles. MBL is composed of six CRDs.151 The end of CRD can identify the sugar structure on the surface of various pathogens, such as mannose, fucose, glucose, etc.233–235 The pathogens involved include yeast, parasites, Gram bacteria, and so on.236–240 When the distance of each CRD between the same trimer or adjacent trimers is 45 Å, it is most conducive to ligand binding.241 The other family members include A and D,242 which exist on the surface of the alveoli and are important innate immune defense molecules in the lungs. Both of them are composed of N-terminal region, CRD, neck region, collagen-like region, and other parts.243 CRD recognizes and binds glycosyl groups. The biological significance is that they can selectively identify microbial carbohydrate structures that are harmful to themselves.244,245
The domain of ficolin is similar to collectin, but it recognizes a variety of bacteria with a fibrinogen-type carbohydrate recognition structure.246,247 Its ligands are N-acetylglucosamine and LTA, a cell wall component of Gram-positive bacteria.248,249
Signaling pathways of PRRs
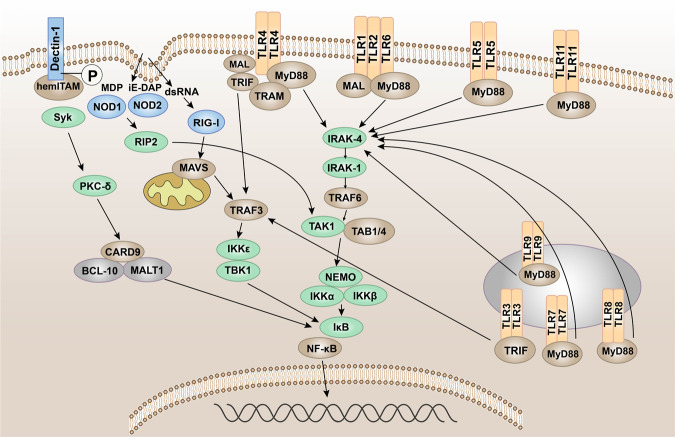
There are three main types of molecules involved in signal transduction: protein kinases, adaptor proteins, and transcription factors. Although PRRs are activated by their respective ligands in different subcellular structures with different mechanisms, the three main types of molecules involved in signal transduction have similar structures and functions, and the signals they transmit are cross-talking, which can converge into several common signaling pathways.
The NF-κB signaling
The transcription factor NF-κB is named after it was first discovered to be involved in the transcription of B cell κ chain genes.250 NF-κB is a heterodimer composed of two molecules, p50 and p65, and is inactive due to binding to the inhibitory protein IκB under normal conditions. NF-κB plays a key role in the process of cellular inflammation and immune response,251,252 and its mediated signal pathways are commonly seen in the activation of various immune cells, including signal transduction initiated by PRRs in innate immunity (Fig. 5).253
Fig. 5.
Pattern recognition receptor-mediated NF-κB signaling. The NF-κB protein can regulate gene expression and affect various biological processes, including innate and adaptive immunity, inflammation, stress response, B cell development, and lymphoid organ formation. TLRs, NLRs, RLRs, and CLRs can generally phosphorylate IκB protein, which inhibits the activation of NF-κB protein, thereby promoting the transcription and activation of inflammatory genes
In the signal transduction initiated by TLRs,45 after TLRs recognize and bind the corresponding PAMPs and DAMPs, the TIR domains conduct signals by binding to different receptor adaptor proteins in the cytoplasmic region.254,255 Depending on the different adaptor proteins, TLR signaling can be divided into MyD88-dependent and MyD88-independent pathways.256 MyD88 has a TIR domain at the C-terminus and a death domain at the N-terminus and is the linker molecule in most TLR signal transduction pathways.257 The current research indicated that, in the MyD88-dependent pathway, MyD88 signaling mainly leads to the production of pro-inflammatory cytokines, such as tumor necrosis factor (TNF), IL-6, IL-1, and chemokines.258–260 The C-terminus of MyD88 binds to the intracellular TIR domain of TLRs, and the N-terminus of MyD88 recruits IL-1R-related kinase 4 (IRAK4)261 and activates IRAK1 and IRAK2 through autophosphorylation of its central kinase domain. Then ubiquitin ligase TNF receptor-associated factor 6 (TRAF6) is recruited to form a complex with transforming growth factor (TGF)-β-activated kinase 1 (TAK1) and two TAK-binding proteins (TAB1 and TAB4). TRAF6 is degraded due to its own ubiquitination.262,263 The TAK1–TAB1–TAB4 complex activates the IκB kinase (IKK) complex through phosphorylation. The latter phosphorylates IκB and degrades itself by ubiquitination. NF-κB is released and translocated to the nucleus, thereby regulating the transcription of inflammatory genes.264,265
In the signal pathway mediated by NLRs, when the bacterial component invades the cell, NOD1 and NOD2 recognize the bacterial iE-DAP and MDP, respectively.266,267 And then NOD-like receptors are activated, self-dimerize, and recruit downstream receptor-interacting serine–threonine protein 2 (RIP2) through its CARD.268 Activated RIP2 gathers downstream TAK1, TAK1-binding protein 1, and the NF-κB essential modulator/IKKα/IKKβ complex, and the former activates IKKα/IKKβ,269 thereby activating the transcription of NF-κB and promoting the release of pro-inflammatory factors.
When virus invades cells, RIG-I and MDA5 recognize the corresponding viral RNA through the CTD and undergo conformation changes.270 Activated RIG-I and MDA-5 induce downstream signal transduction by binding with mitochondrial antiviral signaling protein (MAVS). MAVS is an important adaptor protein for downstream signal transduction. The N-terminus contains a CARD-like domain, which binds to RIG-I and MDA-5 through the CARD–CARD interaction.271,272 The proline-enriched domain in MAVS can interact with a series of downstream signaling molecules, such as TRAF3 and 6,273 and activate the protein kinase IKK, which causes phosphorylation of IκB,265 and then IκB is ubiquitinated and degraded by proteases, activating the NF-κB pathway.274
Different from other typical PRR-mediated signaling pathways, spleen tyrosine kinase (Syk) can be activated by associating with the phosphorylated ITAM motif of CLRs.275 In the Dectin-1/Syk pathway, Syk activates protein kinase C-δ, which mediates the phosphorylation of CARD9.276 This allows CARD9 to bind to B cell lymphoma 10277 and para-aspase mucosa-associated lymphoid tissue lymphoma translocation protein 1, forming a three molecular structure that can typically activate NF-κB.278
The mitogen-activated protein kinase (MAPK) signaling
MAPK is a group of serine–threonine protein kinases that can be activated by different extracellular stimuli,279 such as cytokines, neurotransmitters, hormones, cell stress, and cell adhesion. The MAPK pathway is one of the common intersections of signal transduction pathways, such as cell proliferation, stress, inflammation, differentiation, functional synchronization, transformation, and apoptosis.280,281 It is an important transmitter of signals from the cell surface to the inside of the nucleus.
In the MyD88-dependent pathway of TLRs, IRAK-1 is activated by phosphorylation and interacts with TRAF6. In addition to activating the IKK complex, it can also cause the activation of MAPKs (c-Jun N-terminal kinase (JNK), p38 MAPK).282 In addition, when bacterial components invade cells, NLRs are activated, recruiting downstream CARD9, thereby activating p38, JNK, and finally activating the MAPK pathway283 to promote the release of pro-inflammatory factors.
The TBK1–IRF-3 signaling
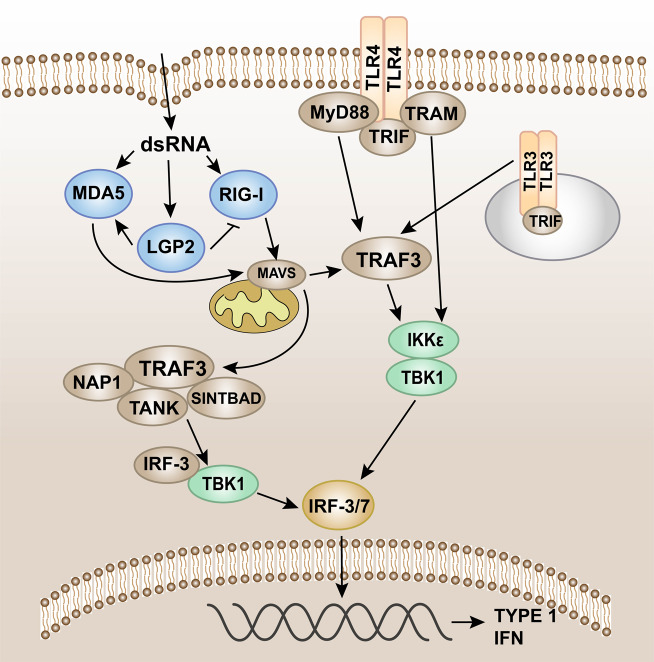
IRF-3 is a key transcription factor that promotes the synthesis of type I IFN and plays an important role in the antiviral innate immune response.284 IRF-3 can be activated through two innate immune antiviral signal pathways, TLR3/TLR4-TIR domain-containing adaptor protein-inducing interferon β (TRIF) and RIG-I-MAVS,285 and then dimerize and merge into the nucleus to work (Fig. 6).286
Fig. 6.
Pattern recognition receptor-mediated TBK1-IRF-3 signaling. Intracellular induction of pathogens is carried out through the detection of foreign molecular components (including cytoplasmic viral and bacterial nucleic acids). Once detected, the innate immune system induces type I interferon (IFN) production through the TANK-binding kinase 1 (TBK1)-interferon regulatory factor-3/7 (IRF-3/7) pathway. IRF-3/7 can be activated through two innate immune antiviral signal pathways, TLR3/TLR4-TIR domain-containing adaptor protein-inducing interferon β (TRIF) and RIG-I-MAVS, and then dimerize and merge into the nucleus to work
The adaptor protein in the MyD88-independent pathway is TRIF. The TRIF axis mainly induces the expression of type I IFNs.287 After the receptor is recognized and combined with the ligand, the pathway is activated by TRIF and TRAF3, leading to the recruitment of IKKε/TANK-binding kinase 1 (TBK1),288 phosphorylation of IRF3, and the activation of type I IFN genes, which promotes the expression of IFN-α and IFN-β, and exerts antiviral effects (Fig. 6).289–291
RLRs such as RIG-I and MDA5 can detect viral nucleic acid. MDA5 and RIG-I will interact with the shared caspase recruitment domain to induce MAVS to dimerize and bind to TRAF3.134,140,292 In turn, TRAF3 recruits the adaptor proteins TANK, NAP1, and SINTBAD. TANK connects upstream RLR signal transduction to TBK1, which induces phosphorylation of IRF-3. IRF-3 phosphorylation and subsequent dimerization induce IRF-3 nuclear translocation, leading to type I IFN gene expression (Fig. 6).192,293,294
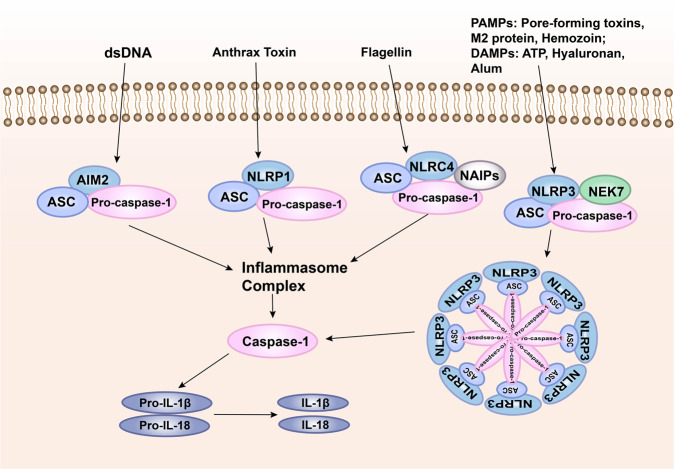
The inflammasome signaling
Inflammasome is the multi-protein complex assembled by PRRs in the cytoplasm and is an important part of the innate immune system.295 The inflammasome can recognize PAMPs or DAMPs and recruit and activate Caspase-1. The activated Caspase-1 spliced proIL-1β/proIL-18 into the corresponding mature cytokine.193,296 There are five main types of inflammasomes that have been discovered, namely, NLRP1 inflammasome,297 NLRP3 inflammasome,298 NLRC4 inflammasome,299,300 IPAF inflammasome, and AIM2 inflammasome.301 Known inflammasomes generally contain ASC, caspase protease, and a protein of the NLR family (e.g., NLRP3) or HIN-200 family protein (e.g., AIM2). Taking NLRP3 as an example,302 the dimerization of NLRP3 under the action of intracellular PAMPs or DAMPs makes the two PYDs to polymerize. With the help of homotype interaction, NLRP3 binds and activates the ASC complex with both PYD and CARD domains, which reactivates the effector complex composed of CARD and caspase-1. In this way, NLRP3 (LRR + NACHT + PYD), ASC (PYD + CARD), and the effector complex (CARD + Caspase-1) together constitute the inflammasome, which produces important pro-inflammatory factors.303–305 After AIM2 recognizes cytoplasmic dsDNA, it also uses inflammasomes to produce IL-1β and IL-18. After AIM2 recognizes cytoplasmic dsDNA, it also produces IL-1β and IL-18 through the inflammasome pathway (Fig. 7).306
Fig. 7.
Pattern recognition receptor-mediated inflammasome signaling. One way for pathogenic microorganisms to induce inflammation is by activating inflammasomes, which are multi-protein complexes assembled by PRRs in the cytoplasm and activate caspase-1 and subsequent activation of pro-inflammatory cytokines IL-1β and IL-18. The inflammasome complex usually contains cytoplasmic PRRs, adaptor protein (ASC), and pro-caspase-1. Many different inflammasome complexes have been detected, each with unique PRRs and activation triggers
Innate immunity not only plays a role in controlling the infection and spread of pathogens in the early stage of infection but also plays an important role in initiating and regulating adaptive immunity.12,307 Innate immune cells produce different types of cytokines through signal transduction initiated after PRRs recognize PAMPs, which directly affect the differentiation of T helper type 1 (Th1), Th2, Th17, and other subgroups in adaptive immunity.15 For example, pathogenic microorganisms activate macrophages to secrete IL-6, TGF-β, IL-23, and other cytokines, which promote Th17 response, leading to excessive immune-inflammatory effects and tissue damage, or activate NK cells to secrete IFN-γ, and then activate macrophages to secrete IL-12, promote the differentiation of Th0 into Th1, promote cellular immune response, and effectively eliminate viral infections.13,308 Therefore, the immune system is a system of mutual influence. Any anti-infection process is completed by mutually activating or inhibiting of different components. These components are as small as each cytokine and as large as the immune system.
PRR-related diseases
PRRs and cancers
The inflammatory microenvironment of tumor constitutes the barrier for tumor growth, which is conducive to tumor formation and development.309 PRRs are widely expressed in a variety of tumor tissues, such as colon cancer, lung cancer, breast cancer, gastric cancer, melanoma, and so on.310,311 The activation of PRRs on the surface of tumor cells can induce the expression of a large number of cytokines, chemokines, hormones, and vascular-promoting factors, which is one of the important factors to induce the formation of tumor inflammatory microenvironment and promote the development of tumor.312,313 At the same time, the activation of PRRs on immune cells can induce antigen-presenting cells including DCs, tumor-associated macrophages, and B cells to activate tumor-specific T cell responses or enhance the antitumor effects of phagocytes. These also indicate that the role of PRRs in immunotherapy against tumors is very important and might represent a new strategy for patients with tumors.314,315
We can see from the data in GEPIA316 (http://gepia.cancer-pku.cn/) that TLR level is significantly increased in tumors, including glioblastoma multiforme, brain lower-grade glioma, kidney renal clear cell carcinoma, acute myeloid leukemia (AML), and pancreatic adenocarcinoma (PAAD). NOD1/2 are highly expressed in AML and PAAD, while RLRs are highly expressed in AML, PAAD, diffuse large B cell lymphoma, head and neck squamous cell carcinoma, and thymoma.
Colorectal cancer (CRC)
CRC, including colon cancer and rectal cancer, is one of the most common gastrointestinal malignancies in clinical practice, and it is also one of the cancers that seriously endanger human health.317,318 Intestinal mucosal epithelial cells and immune cells recognize intestinal microorganisms and their products through TLRs.319–322 TLR2 can recognize peptidoglycans and lipopeptides that infect intestinal epithelial bacteria and produce anti-infection and other immune-protective effects.323 Studies have shown that the expression level of TLR2 protein in colon cancer is significantly upregulated compared with normal epithelial tissues,324 and the use of TLR2 agonists significantly enhances the proliferation, migration, and invasion capabilities of colon cancer cells.325 TLR4 is highly expressed on the surface of colon cancer cells. After stimulation and activation, it can induce a variety of immunosuppressive factors, thus promoting the proliferation and immune escape of colon cancer cells.326 First, TLR4 can produce trophic factors and vascular growth factors through the TLR4/MyD88/NF-κB signaling pathway, thereby promoting tumor cell invasion.327,328 Second, TLR4 can promote tumor proliferation through TLR4/Cyclooxygenase 2 (COX2)/prostaglandin E2 (PGE2). PGE2 is an important cell growth and regulatory factor. After binding to specific receptors, it plays a key role in mediating a series of cell activities, such as cell proliferation, differentiation, and apoptosis, and has immunosuppressive and anti-inflammatory effects. COX2 is the rate-limiting enzyme of prostaglandin synthesis, and it is also highly expressed in inflammation, tumor, and other pathological states.329 Hsu et al.330 found that knocking out the mouse TLR4 gene significantly reduced the expression of COX2 and PGE2 in the intestinal mucosa; after administration of PGE2, the expression of COX2 in the intestinal mucosa increased significantly and promoted the occurrence of intestinal tumors. After administration of PGE2, the expression of COX2 in the intestinal mucosa increased significantly and promoted the occurrence of intestinal tumors. At the same time, it has also been found to promote the expression of amphiregulin and epidermal growth factor receptor (EGFR) in the intestinal mucosa. Finally, the study showed that the abnormal expression of TLR4 in CRC caused by chronic inflammation of the intestine can significantly enhance the expression of PGE2, the upregulation of COX2, and the phosphorylation of EGFR in intestinal mucosal cells, thereby positive feedback promotes the proliferation of tumor cells. TLR5 also plays an important role in tumor immunotherapy.331 In mouse xenograft models of human colon cancer, flagellin around the tumor activates TLR5 to inhibit tumor growth and promote tumor apoptosis.332 In addition, TLR9 is expressed on the surface of the mesentery, which maintains intestinal homeostasis and repairs intestinal damage by generating an immune response.333 TLR9 relies on the MyD88 pathway to induce downstream signals to recruit many inflammatory factors, such as IL-8, TGF-β, PGE2, and other immunosuppressive molecules,334 leading to the continuous development of inflammation, resulting in immune escape, and promoting the unlimited proliferation of tumor cells. After TLR9 recognizes the exogenous ligand, it upregulates the expression of NF-κB signaling factor. Once this pathway is opened, it may induce the secretion of matrix metalloproteinase-13 (MMP-13) and the activation of intercellular adhesion molecule-1, thus promoting the metastasis of tumor cells.335,336 At the same time, the metastatic tumor cells are better adapted and combined with the cell matrix at the metastasis, and the stability of tumor cell metastasis is enhanced. Although TLRs have been shown to enhance colon cancer metastasis, inhibiting these receptors cannot completely hinder tumor progression. Surprisingly, NOD1 is highly expressed in human CRC and its cell lines. After being activated by C12-iE-DAP, it mainly enhances the adhesion, migration, and metastasis of CRC cells through the p38 MAPK pathway.337
Hepatocellular carcinoma (HCC)
HCC is the most common type of primary liver cancer. Among its many influencing factors, inflammation is one of the main reasons that induce liver cancer.338 The expression of TLR2 in liver cancer tissues is significantly higher than that in normal liver tissues, and the expression of TLR2 protein is related to some mutant genes that lead to the occurrence of HCC, such as p53, PIK3CA, and β-catenin.339–342 In addition, Chew et al. revealed that the expression of TLR3 has independent effects on tumor parenchyma and infiltrating NK cells, and the expression of these two parts is related to inhibiting tumor cell proliferation, promoting tumor cell death, and prolonging the survival rate of patients.343 This indicates that TLR3 may directly act on tumor parenchymal cells, promote the recruitment and activation of NK cells, and exert antitumor effects. More and more evidences show that LPS plays a role in the development of HCC. Zhou et al.344 found that LPS activates the TLR4–AKT–SOX2 signaling pathway of liver cancer cell lines to improve the ability of cancer stem cells; Lin et al.345 found that there is a positive feedback loop of COX-2/PGE2/signal transducer and activator of transcription factor 3 (STAT3) activated by LPS in liver cancer cells, which regulates the expression of genes related to tumor proliferation, differentiation, and apoptosis. In the latest research on the treatment of liver cancer, it is found that the antitumor effect of TLR9 agonist combined with anti-PD-1 antibody or anti-PD-L1 is significantly better than single-agent therapy.346 The activation of TLR9 inherent in liver cancer cells regulates the autoarylation and ubiquitination of poly(ADP-ribose) polymerase-1 and the phosphorylation of STAT3, which together upregulate the expression of PD-L1 and eventually induce immune escape. Although TLRs have been reported to be associated with chronic inflammation of the liver, whether they promote the development of HCC remains uncertain. Song et al.347 found that the deficiency of TLR4, TLR9, and their downstream molecule MyD88 in a mouse model characterized by hepatic deletion of TAK1 could block the liver inflammation–fibrosis–cancer axis and reduce liver injury and tumor growth. For TLR3, the downregulation of TLR3 in HCC patients leads to poor prognosis (e.g., defective immune cell recruitment and lack of killing of transformed hepatocytes), leading to protection of transformed hepatocytes from apoptosis, thereby promoting the occurrence of liver cancer.348 Therefore, the expression of TLR3 may become a useful clinical treatment monitoring marker.
In addition to the above reasons, there is now more and more evidence that the imbalance of the gut–hepatic axis may also play a role in the occurrence of HCC.349 Zhou et al.350 discovered that NOD2 acts as a bacterial sensor, linking gut-derived microorganisms to the occurrence of HCC through a known mechanism and a newly discovered mechanism. The known mechanism is that NOD2 activates NF-κB, JAK2/STAT3, and MAPK pathways in a RIP2-dependent manner, leading to liver inflammation.351 It is worth noting that activated NOD2 can also act as the initiator of the nuclear autophagy pathway that does not depend on RIP2, thereby promoting the degradation of the nuclear component lamin A/C, leading to damage to DNA damage repair mechanisms and increased genomic instability, which eventually leads to the occurrence of HCC.350 Meanwhile, the study found that the expression of ALRs was negatively correlated with tumor volume, stage, and metastasis of HCC patients. The researchers proposed that overexpression of ALRs in HCC cells could increase the expression of caspase-1 and IL-1a, and the release of lactate dehydrogenase was also observed,352 which was a marker of the initiation of apoptosis. Thus, ALRs may play an antitumor role by promoting tumor cell apoptosis.
Breast cancer
Breast cancer is one of the most common malignant tumors in the female population. It has a strong ability to invade and metastasize.353 It can metastasize to the liver, lung, brain, bone, and other organs, forming complications and increasing the difficulty of treatment. Studies have shown that the promotion of the TLR2 signaling pathway on the metastasis and invasion ability of human breast cancer cells is achieved by upregulating the secretion of inflammatory cytokines.354 LTA, a TLR2 specific ligand, can significantly promote the secretion of tumor metastasis-related factors IL-6, TGF-β, and vascular endothelial growth factor (VEGF) in breast cancer cells,355 thereby promoting the proliferation and metastatic invasion of breast cancer cells, and this promotion is related to the level of TLR2 expression.356,357 In addition, the activation of TLR4 can increase the secretion of IL-6 and IL-10 of cancer cells and induce the production of more MMP-2, MMP-9, and VEGF,358,359 which can significantly enhance the invasion ability of breast cancer. It has been reported that activation of TLR4 on metastatic breast cancer cells can regulate the expression of integrin, which can promote its adhesion and invasion.360
Head and neck squamous cell carcinoma
Squamous cell carcinoma, also known as epidermal carcinoma, is a malignant tumor occurring in the epidermis or adnexal cells.361,362 It is more common in the parts covered by squamous epithelium, such as skin, mouth, lip, esophagus, cervix, vagina, etc.363,364 TLR2, TLR4, and TLR9 are expressed in primary tumors, neck metastases, and recurrent tumors of oral tongue squamous cell carcinoma (OTSCC), and their expression varies from the tumor surface to the invasive front, which may be one of the important factors to promote the invasion of OTSCC.365,366 NOD1 and NOD2 genes are expressed in the human oral squamous cell carcinoma (OSCC) cell line YD-10B, and they may trigger immune responses through the MAPK pathway. Surprisingly, the study revealed that stimulation by the NOD2 agonist MDP can inhibit cell growth by inducing apoptosis. These findings provide the potential value of MDP as a new candidate for OSCC antitumor drugs.367
Respiratory diseases
Aspergillus fumigatus is a fungus widely distributed in nature. It can easily invade the respiratory tract and cause bronchitis and pneumonia in patients.368,369 For the allergic lung inflammation caused by Aspergillus, the recognition of PAMPs by the body’s dendritic cells is mainly negatively regulated through the TLR2-MyD88 pathway. The results showed that PAMPs recognized by TLR2 upregulated IL-10 and decreased the recruitment of pulmonary eosinophils, thus downregulating Th2 response.370,371 In allergic asthma, TLR9–IL-2 affects the Th2 response by regulating the expression of IL-17A, so small molecule inhibitors targeting TLR9 may become a new treatment strategy.372
Nervous diseases
The connection between innate immunity and nervous system is becoming more and more complex and close.373–375 Studies have shown that NOD1/NOD2 may be a new target for the treatment of stress-related gut–brain diseases.376 The gut–brain axis is a biochemical signal of the digestive tract and central nervous system, which affects all events from brain development to the progression of neurological diseases. The hypothalamic–pituitary–adrenal axis (HPA) is one of the main pathways of gut–brain axis signal transmission.377 It has been reported that the immune system plays a key role in brain function and stress response. NLRs are PRRs expressed in the gut and brain. The lack of NOD1 and NOD2 affects the serotonergic signaling of gut and brain, the proliferation of hippocampal cells, and the maturation of neurons, which makes the mice lacking both of them vulnerable to HPA overactivation under stimulation, thus showing anxiety, cognitive impairment, and depression.378–380 TLR4 is very important in the process of neuropathic pain caused by infection and sterile neuronal injury. TLR4 exerts its effects through the activation and nuclear localization of NF-κB and the production of pro-inflammatory cytokines, which can activate pain receptors to cause neuropathic pain.381,382 Relevant studies have revealed for the first time that lysozyme acts as an endogenous ligand for activating TLR4 in sterile nerve injury, thereby promoting neuronal excitement and neuropathic pain.383 The identification of lysozyme as DAMPs has improved our understanding of neuroinflammation and opened up prospects for the treatment of neuropathic pain.
Digestive diseases
Newborns with chronic obstructive jaundice make their livers prone to cholestatic liver disease, and biliary atresia (BA) accounts for half of the cases.384 Viruses have always been considered as the causative pathogen of this disease, and the role of TLRs in the pathogenesis and progression of BA has been determined.385 Subsequent studies have shown that activation of TLR7 can induce type 1 IFN signal transduction, apoptosis, and dysplasia of the neonatal liver and biliary system. This new discovery reveals the pathogenesis of neonatal cholestatic liver disease.386
The expression of TLR5 is closely related to various infectious diseases caused by bacteria.387,388 The lack of TLR5 can cause changes in the intestinal flora and cause colitis.389 The protein–protein interaction between TLR5 and flagellin plays an important role in pathogen defense, immune diseases, and tumors. 4-((4-benzyl-5-(pyridin4yl)-4H-1,2,4-triazol-3-yl)thio)pyrido[3’,2’:4,5]thieno[3,2-d] Pyrimidine (TH1020) is a small molecule inhibitor identified through high-throughput screening, which can disrupt the association between TLR5 and flagellin, and provides a lead compound for new therapies against TLR5.390,391
Alcoholic liver disease is a liver disease caused by long-term heavy drinking. The initial stage usually manifests as fatty liver, which can then develop into alcoholic hepatitis, liver fibrosis, and cirrhosis.392,393 Patients with advanced alcoholic cirrhosis are more susceptible to infection.394 This phenomenon is related to multiple organ failure and immunodeficiency and is usually manifested as insufficient antibacterial activity of neutrophils.395 The neutrophil function to resist microbial infections needs to generate reactive oxygen species through NADPH oxidase 2. Rolas et al. found that, in patients with alcoholic liver cirrhosis, the lack of catalytic core flavocytochrome b558 (gp91phox, p22phox) and p47phox of the NADPH enzyme may be a new factor that patients are susceptible to infection. What is surprising is that the activation of TLR7/8 can reverse the expression and activity of the deficient gp91phox, which provides a direction for restoring the antibacterial response of immunodeficiency patients.396,397
Cardiovascular diseases
Atherosclerosis is a chronic inflammatory disease caused by plaques composed of lipids, cholesterol, calcium, and other substances in blood vessels.398,399 It has been reported that TLRs are extensively and deeply involved in the process of atherosclerosis.400,401 TLR7 has been identified as a good prognosis marker for patients with severe atherosclerosis. TLR7 in the plaque produced in atherosclerotic lesions will secrete IL-10 and TNF-α after being stimulated by ligand. Studies have found that TLR7 may regulate inflammation in atherosclerosis by inhibiting the effects of pro-inflammatory cytokines.402 In addition to the production of plaque, cell-free DNA (cfDNA) is also released in atherosclerotic lesions. TLR9 recognizes cfDNA and plays a key role in the development of vascular inflammation and atherosclerosis by promoting the pro-inflammatory activation of macrophages.403 The formation of initial atherosclerotic plaques is caused by the interaction of macrophages and endothelial cells, macrophage infiltration, and other factors that lead to an increase in neointima.404,405 It is worth noting that studies have shown that TLR5-mediated activation of NADPH oxidase 4 (Nox4) can regulate the migration of SMCs and promote the expression of pro-inflammatory cytokines, which may contribute to the formation of atherosclerotic plaques. This indicates that the flagellin–TLR5–Nox4 cascade is of great significance in atherosclerotic intimal hyperplasia.406,407 In addition, TLR2 activates p38 and extracellular signal-regulated kinase 1/2 signals, thereby upregulating IL-6-mediated receptor activator of NF-κB ligand and downregulating osteoprotegerin. These will make the cartilage formation of vascular SMC transdifferentiate, leading to vascular calcification.408 The expression of TLR3 is reduced in the lung tissue and endothelial cells of patients with pulmonary hypertension, and its deficiency increases the susceptibility to apoptosis and pulmonary hypertension.409 In addition to TLRs, the lack of NOD1 and NOD2 can lead to lipid deposition of atherosclerotic plaques and the reduction of inflammatory cell infiltration, so it has been identified as pre-disease factors.410
Human brain pericytes (HBPs) are an important component of the microvascular wall and contribute to the integrity of the blood–brain barrier (BBB).411 It has been found that TLR4 and NOD1 are expressed in HBP, which also reveals that HBP has the ability to sense systemic infection or blood-borne PAMPs. HBP can perceive Gram-negative bacteria to protect BBB through different pathways mediated by TLR4 and NOD1, or it can trigger paracrine signaling pathways by releasing chemokines and cytokines, leading to the destruction of BBB.412,413 These are all new insights that PRRs are involved in the body’s inflammatory response and may have an impact on the treatment of diseases.
Endocrine diseases
In order to adapt to the constantly changing internal and external environment and maintain the relative stability of the internal environment, the human body must rely on the cooperation and regulation of the nervous, endocrine, and immune systems,414,415 so that the activities of various organs and systems are coordinated, and jointly shoulder all the life phenomena of the body. The immune system is not only regulated by the other two systems but also affects them through chemical information molecules and receptors.416,417
In patients with autoimmune thyroid disease, the expression and activation of TLR2, 3, and 9 are significantly increased.418,419 In addition, it has been reported that TLR9 negatively regulates pancreatic islet development and β cell differentiation, providing new directions for diabetes prevention and treatment strategies.420 In diet-induced obesity, TLR2 and TLR4 inhibit the replication of β cells and affect the nuclear abundance of the cell cycle regulators cyclin D2 and Cdk4. Therefore, targeting TLR2–TLR4 may alleviate the failure of β cells in diabetic patients.421
One of the most common complications of diabetes is diabetic foot ulcers, and the reason why it is not easy to get better is the inflammation of the wound.422,423 Singh et al. believe that changes in the expression level of intracellular TLRs may be one of the reasons for the continuous inflammation of chronic wounds, such as diabetic foot ulcers, and may hinder wound healing in patients with type 2 diabetes mellitus (T2DM).424 The basis of these pro-inflammatory effects is that intracellular TLRs activate dendritic cells and B cells to produce IFN I and III, aggravating the inflammatory response.425–427 The inflammatory phase of the wound healing cascade is the decisive stage of wound development, and studies have found that certain members of innate immunity play an important role in the pathogenesis of chronic wound healing abnormalities.428,429 The signaling pathway of intracellular TLRs is shown to be involved in some chronic inflammatory diseases, such as systemic lupus erythematosus (SLE), multiple sclerosis, hepatitis, and T2DM.430
Skeletal diseases
The expression level of NOD2 in human osteoarthritis (OA) cartilage is significantly higher than that of normal cartilage, and its combined action with TLR2 contributes to the pro-catabolic gene expression induced by 29-kDa amino terminal (matrix degradation product in synovial fluid of patients with OA) in human chondrocytes.431,432 This indicates that the NOD2 and TLR2 cross-regulatory pathway may be a target to prevent the development of arthritis. More and more evidences indicate that the interaction between different signal pathways may be a new way for the occurrence and development of diseases. The latest research combines three biological processes in skeletal muscle during exercise, innate immune response, autophagy protein homeostasis, and adenosine monophosphate-activated protein kinase (AMPK) activation, revealing that TLR9 can regulate the energy metabolism of skeletal muscle during exercise, and TLR9 regulates exercise-induced skeletal muscle AMPK activation by interacting with the core autophagy protein beclin1.433,434
Clinical therapy of PRRs
Based on the important role that PRRs play in innate immunity, they have received extensive attention in the fields of immunology and drug research.310,435 There are many types of PRRs and a wide range of ligands. They can be used as drug targets for tumor, inflammation, autoimmune disease, pathogenic microbial infection, and other diseases, which are important entry points for immunotherapy.314 The activation of PRRs brings double-sided effects: on the one hand, it stimulates innate immunity and adaptive immunity to resist pathogenic microorganisms; on the other hand, it promotes the expression of a large number of cytokines, forms an inflammatory microenvironment, and causes tissue damage.29,436 Therefore, the treatment strategy targeting PRRs is mainly to use ligand analogs to activate PRRs,435 use antagonists to inhibit their activation, or use antibodies and small molecules to inhibit PRR signaling pathways.437 It is reported that most of the agonists and antagonists of TLRs are only in the clinical development stage, which are studied comprehensively in PRRs (Table 2). In addition, microRNA (miRNA), exosomes, and combination therapy also have certain potential in this field.438,439
Table 2.
Clinical trials investigating the use of TLR agonists and antagonists in diseases
| Drug | Phase | Target | Application | Treatment | NCT number | Status |
|---|---|---|---|---|---|---|
| Agonists | ||||||
| MGN1703 | II | TLR9 | Human immunodeficiency virus type 1 (HIV-1) | Monotherapy | NCT02443935 | Completed |
| gp100 | II | TLRs | Melanoma | Combination with resiquimod (R848) | NCT00960752 | Completed |
| MAGE-3 | II | TLRs | Melanoma | Combination with resiquimod (R848) | NCT00960752 | Completed |
| Insulin | II | TLRs | Insulin resistance | Monotherapy | NCT01151605 | Unknown |
| EMD 1201081 | II | TLR9 | Squamous cell carcinoma of the head and neck cancer | Combination with cetuximab | NCT01040832 | Completed |
| Resiquimod | I | TLR7/8 | Influenza vaccination in seniors | Monotherapy | NCT01737580 | Completed |
| CPG 7909 | II | TLR9 | HIV infections | TLR-9 adjuvanted pneumococcal | NCT00562939 | Completed |
| DSP-0509 | II | TLR7 | Advanced solid tumors | Monotherapy and combination with Pembrolizumab | NCT03416335 | Recruiting |
| SD-101 | I | TLR9 | Chronic hepatitis C | Monotherapy and combination with ribavirin | NCT00823862 | Completed |
| Imiquimod | II | TLR7 | Breast cancer (for chest wall recurrences or metastases to the skin), breast neoplasms | Monotherapy | NCT00899574 | Completed |
| Imiquimod | II | TLR7 | Breast cancer, metastatic breast cancer, recurrent breast cancer | Monotherapy and combination with cyclophosphamide (CTX) and radiotherapy (RT) | NCT01421017 | Completed |
| GSK1795091 | I | TLR4 | Cancer | Monotherapy | NCT02798978 | Completed |
| GSK2245035 | II | TLR7 | Mild asthma and allergic rhinitis | Monotherapy | NCT01788813 | Completed |
| SD-101 | I | TLR9 | Metastatic pancreatic adenocarcinoma, refractory pancreatic adenocarcinoma, stage IV pancreatic cancer, AJCC v8 | Combination with nivolumab and radiation therapy | NCT04050085 | Recruiting |
| Motolimod | II | TLR8 | Ovarian cancer | Chemoimmunotherapy with anti-PD-L1 antibody MEDI4736 | NCT02431559 | Active, not recruiting |
| VTX-2337 | I | TLR8 | Locally advanced, recurrent, or metastatic squamous cell cancer of the head and neck (SCCHN) | Combination with cetuximab | NCT01334177 | Completed |
| CPG 7909 | II | TLR9 | Non-Hodgkin lymphoma, mycosis fungoides | Monotherapy | NCT00185965 | Completed |
| PolyICLC | I/II | TLR3 | Melanoma | Adjuvants | NCT04364230 | Recruiting |
| Antagonist | ||||||
| IMO 8400 | II | TLR7, TLR8, TLR9 | Plaque psoriasis | Monotherapy | NCT01899729 | Completed |
| OPN-305 | II | TLR2 | Delayed graft function | Monotherapy | NCT01794663 | Completed |
| Hydroxychloroquine | III | TLR7, TLR9 | Autoimmune diseases, Sjogren’s syndrome, dry eye | Monotherapy | NCT01601028 | Completed |
| Eritoran | II | TLR4 | Insulin sensitivity | Monotherapy | NCT02321111 | Completed |
Limited clinical studies have been carried out investigating PRR agonists and antagonists in related research to date. Most of the agonists and antagonists of TLRs are only in clinical development stage, which are studied comprehensively in PRRs
PRR agonists for therapy
Imiquimod is the first targeted drug for TLRs. It is a specific agonist of the TLR7 receptor. It can induce the production of IFN-α, IL-6, and TNF-α to achieve the purpose of regulating immunity and treating tumors.440 Selgantolimod, a novel TLR8 agonist, can activate TLR8 and elicit cytokine responses in patients with chronic hepatitis B infection.441,442 Further studies with longer durations of selgantolimod treatment are required to evaluate efficacy. The ligand of TLR9 is unmethylated DNA, and CpG oligodinucleotide (CpG ODN) that mimics this structure has good antitumor potential (e.g., IMO-2055, MGN-1703, MGN-1704).443 The research of MGN-1703 on the treatment of advanced CRC is in phase III. MGN-1703 can induce a potent type I IFN response in the intestine, which is related to the subtle changes of intestinal flora.444 Clinically, PRR agonists can be potentially used not only as therapeutic agents to treat but also as adjuvants in conjunction with other immunotherapies.445,446 Due to the instability and drug resistance of existing drugs, some adjuvants targeting PRRs emerge as the times require. A TLR3-specific adjuvant, ARNAX, can enhance DC priming and CTL proliferation without cytokine toxicity.447 And the conjugated STING and TLR1/2 agonist Pam3CSK4-CDGSF can be used as an effective adjuvant for constructing vaccines to enhance antitumor immunotherapy.448 NLR agonists can promote the processing and secretion of IL-1β, which is very important for activating many immune cells.449,450 Therefore, NLR agonists can be served as effective components of vaccines and immune stimulants.
The class II major histocompatibility complex transactivator (CIITA) is a single member of the NLRA subfamily.451 As the main regulator of major histocompatibility complex II, its own modification is very important in the occurrence and development of tumors. Both the deacetylation of histones and the demethylation of DNA will downregulate the transcription of CIITA.452 Decitabine (5-aza-2’-deoxycytidine), Entinostat (MS-275), and Trichostatin A target CIITA to promote its expression recovery in tumors. In addition, andrographolide, arglabin, formononetin, mangiferin, and other botanicals can be used to treat tumors by inhibiting NLRP3 inflammasome.453–457 Representative NLR agonists like Mifamurtide have been approved for clinical trials of non-metastatic sarcoma.
RLRs are involved in the recognition of viral infection by the innate immune system, and their agonists include poly I:C.99 Studies have revealed the link between energy metabolism and innate immunity, indicating that lactate may act as a natural inhibitor of RLR signal transduction by targeting the adaptor protein MAVS.458 The latest research shows that activating RIG-I before diffusing alpha-emitting radiation therapy for metastatic and low immunogenic tumors is a more effective combination therapy technique that can inhibit tumor growth and metastasis.459
PRR antagonists for therapy
PRR antagonists belong to immunosuppressants, which are mainly used to inhibit the function of receptors and block the connection between receptors and ligands.460 They can be used to treat diseases with abnormal activation of the immune system. The abnormal activation of TLR7, 8, and 9 contributes to the onset and maintenance of inflammatory diseases.461,462 The inhibitors IMO-3100 and IMO-8400 targeting them can significantly reduce the expression of IL-17A induced by IL-23, which has a potential role in the inflammatory cascade.463,464 In addition, inhibition of TLR7 and TLR9 with IRS-954 or chloroquine may be a new treatment for HCC.465 In addition to the immune inhibitory oligonucleotides that can be used as TLR inhibitors, CPG-52364, a derivative of the small molecular weight chemical compound quinazoline, can also block ligand-induced activation of TLR7, 8, and 9.462 FP7 is a synthetic glycolipid, which can be used as TLR4 inhibitor to treat septic shock.466 Studies have found that inhibitory ODN-containing phosphorothioate modifications of the backbone have potential therapeutic effects for SLE and rheumatoid arthritis. ODN1411 directly binds to the extracellular domain of TLR8 and competitively inhibits its signal transduction.467
In the NLR family, the NLRP3 inflammasome is closely related to the pathophysiology of a variety of inflammatory diseases, so it is usually used as an inhibitor target.298,304,305 MCC950 is a specific NLRP3 antagonist, which can prevent its activation or maintain its activation state.468 β-Carotene has been shown to directly bind to the PYD of NLRP3, thereby blocking the association between NLRP3 and its adaptor protein and ultimately inhibiting the activation of NLRP3 inflammasomes.469 This result further complements the new pharmacological strategy to prevent NLRP3 inflammasome-driven gouty arthritis. The development of small molecule inhibitors for NLRP3 has been in full swing.470,471 Dai et al. reported a series of tetrahydroquinoline inhibitors. In further studies in vitro, it was found that compound 6 directly binds to the NACHT domain of NLRP3 but not to the PYD or LRR domains, which inhibits the assembly and activation of NLRP3 inflammasomes. In vivo, compound 6 can significantly inhibit the expression of IL-1β and alleviate the symptoms of colitis.472 Agarwal et al. identified alkenyl sulfonylurea derivatives as a novel, oral, and biologically effective NLRP3 inhibitor. Compound 7 of alkenyl sulfonylurea derivatives has good pharmacokinetic characteristics and high oral bioavailability.473
Other potential therapies via PRRs
Emerging evidence suggests that the interaction between miRNA and PRR pathway and immunoregulation are intimately interwoven.438 The interaction between the two is particularly significant in the pathogenesis of rheumatoid arthritis (RA).474,475 RA is a common autoimmune disease, which is characterized by synovial hyperplasia and irreversible bone destruction caused by chronic synovitis. Rheumatoid arthritis synovial fibroblast (RASF) plays a central role in this process.476–478 The expression of miR-146a and miR-155 is upregulated in RASF. miR-146a inhibits the expression of TRAF-6 and IRAK-1 in the TLR signaling pathway, thereby inhibiting the production of key adaptor molecules downstream of TLRs in RA.479 And miR-155 negatively regulates the activation of TLRs/IL-1R inflammatory pathway. Both of them will eventually reduce RA inflammation.480 In addition, miR19a/b, miR-20a, and miR-10a are downregulated in RASF. Downregulated miR19a/b directly targets TLR2 to increase its expression481; miR-20a inhibits the expression of ASK1, a key component of the TLR4 pathway482; miR-10a targets IRAK4 and TAK1 to accelerate IκB degradation and NF-κB activation.483 These effects promote the inflammatory response of RA. All these evidences indicate that miRNAs can surpass their conventional functions and act as physiological ligands for TLRs, thereby regulating the expression of cytokines in the inflammatory response. miRNA can also target other PRRs. miR-485 can target RIG-I mRNA for degradation, leading to enhancement of virus replication and inhibition of antiviral response.484 miRNA-155 can aggravate the brain tissue inflammation in neonatal rats with hypoxia and ischemia by regulating the NOD1/NF-κB signaling pathway, thereby causing hypoxia and ischemic brain damage in neonatal rats.485,486
In addition, exosomes are also involved in the complex immune regulatory network of miRNA and PRRs.487 Exosomes are small membrane vesicles containing complex RNA and protein. A variety of cells can secrete exosomes under normal and pathological conditions.488,489 There is evidence that exosomes can regulate innate immunity by combining with TLRs, which provides a new method for the body to resist pathogens and treat diseases.487 Brain exosomes are rich in amyloid-beta (Aβ), the main component of neuritic plaques, which can activate TLR2, TLR4, and TLR9 signaling, thus alleviating the early symptoms of Alzheimer’s disease.490 It was found that the expression of miR-216a-5p in the plasma of patients with syphilis is negatively correlated with the expression of inflammatory cytokines. Exosomes containing miR-216a-5p inhibit the inflammatory response induced by recombinant Tp17 by targeting the TLR4–MyD88 pathway.491 Melphalan, a genotoxic agent, can enhance the ability of multiple myeloma (MM) cells to release exosomes in MM. Hsp70 on the surface of MM-derived exosomes activates TLR2 in NK cells, thereby inducing the production of INFγ.492 This enlightens us that exosomes with significant exposure to DAMPs can link chemotherapy with antitumor innate immune response.
However, whether they are therapeutic agents, adjuvants, or others, it still needs a long time to further improve the research in vitro and in vivo before clinical application in tumor treatment.493
Conclusions and future perspectives
The innate immune system has multiple recognition mechanisms in different cell types with different subcellular structures. Recently, great progress has been made in the study of the interaction between PRRs and ligands. A preliminary understanding of the crystal structure of some PRRs and the commonalities and differences in the structural composition of different types of PRRs has been made. We understand the existing forms of PRRs alone in solution and after binding to ligands, as well as the molecules involved in the process of PRR identification and ligand binding. As the initiation step of the innate immune response and antiviral response, PRR recognition and binding of ligands is critical. PRRs recognize targets through their respective ligand-recognition domains: LRRs, CTDs, CTLDs, and a variety of DNA- or RNA-binding domains. Ligand binding induces activation and transmission of the signal, which is transmitted through cascade amplification by their respective domains—TIR domain, protein interaction domain, CARD, ITAM, and PYD. In some cases, one kind of PRRs can recognize multiple PAMPs, and one kind of PAMPs can also be recognized by different PRRs, which can induce an inflammatory response and adaptive immune response in a synergistic manner.494
PRRs that are difficult to find and identify and complex ligand-recognition mechanisms have hindered the study of the role of the innate immune system in related diseases and even tumors. In previous studies, computational modeling and mutational analysis of human TLR10 showed that the structure of TLR10 was modeled with the explicit inclusion of TLR2 and lipopeptide structures.495 Recently, RRGPredictor, a tool mainly based on text mining and set theory to predict new plant PRRs, makes the identification of PRRs more effective, specific, and sensitive than other available tools.496 And a random forest-based method was proposed to identify PRRs, which is superior to other machine learning methods for PRRs. This method constructs a benchmark database, uses the amino acid composition and the composition transition distribution to formulate the sequences in the dataset, and then uses the maximum relevance maximum distance to select the best features. However, due to the small amount of data and the low sensitivity of the method, the identification results for PRRs are still unsatisfactory.497 Therefore, the ability to develop a vertebrate PRR predictor based on protein domain similarity and homology modeling is becoming an important part of this story. In addition, the future studies on the how membrane PRRs are secreted into body fluids and act as soluble PRRs will undoubtedly further our understandings of their roles in antimicrobial immune defense and autoimmune and autoinflammatory disorders. Therefore, the study of PRR-mediated pattern recognition mechanisms will help to elucidate the signaling pathways and mechanisms of disease, provide new targets and methods for the treatment of diseases, and promote the development of immunology and oncology research and improvements in their theoretical systems.
Acknowledgements
We thank the support from the National Natural Science Foundation of China (82073096), Key Research and Development Plan of Hunan Province (2020SK2053).
Author contributions
M.W. and D.L. conceived the paper, collected the related papers, and finished the manuscript and figures. M.W. provided constructive guidance and made critical revisions. Both authors have read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
References
- 1.Takeda K, Akira S. Toll-like receptors. Curr. Protoc. Immunol. 2015;109:14.12.11–14.12.10. doi: 10.1002/0471142735.im1412s109. [DOI] [PubMed] [Google Scholar]
- 2.Thaiss CA, Zmora N, Levy M, Elinav E. The microbiome and innate immunity. Nature. 2016;535:5–74. doi: 10.1038/nature18847. [DOI] [PubMed] [Google Scholar]
- 3.Riera Romo M, Pérez-Martínez D, Castillo, Ferrer C. Innate immunity in vertebrates: an overview. Immunology. 2016;148:125–139. doi: 10.1111/imm.12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Demaria O, et al. Harnessing innate immunity in cancer therapy. Nature. 2019;574:45–56. doi: 10.1038/s41586-019-1593-5. [DOI] [PubMed] [Google Scholar]
- 5.Kaur BP, Secord E. Innate Immunity. Pediatr. Clin. North Am. 2019;66:905–911. doi: 10.1016/j.pcl.2019.06.011. [DOI] [PubMed] [Google Scholar]
- 6.Taguchi T, Mukai K. Innate immunity signalling and membrane trafficking. Curr. Opin. Cell Biol. 2019;59:1–7. doi: 10.1016/j.ceb.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Ou, C.-L., Sun, Z.-Q. & Li, X.-L. Role of Toll-like receptors in nonresolving inflammation-related cancer. Cancer Cell Res. 1, 76–82 (2014).
- 8.Yan G, Tao C-X. Recent progress in the research on pattern recognition receptors and tumor microenvironment. Chin. J. Cancer Biother. 2015;22:143–150. [Google Scholar]
- 9.Gajewski TF, Schreiber H, Fu Y-X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989;54:1–13. doi: 10.1101/SQB.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- 11.Netea MG, et al. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Černý J, Stříž I. Adaptive innate immunity or innate adaptive immunity? Clin. Sci. 2019;133:1549–1565. doi: 10.1042/CS20180548. [DOI] [PubMed] [Google Scholar]
- 13.Sonnenberg GF, Hepworth MR. Functional interactions between innate lymphoid cells and adaptive immunity. Nat. Rev. Immunol. 2019;19:599–613. doi: 10.1038/s41577-019-0194-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iwasaki A, Medzhitov R. Regulation of adaptive immunity by the innate immune system. Science. 2010;327:291–295. doi: 10.1126/science.1183021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald KA, Kagan JC. Toll-like receptors and the control of immunity. Cell. 2020;180:1044–1066. doi: 10.1016/j.cell.2020.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider DS, Jin Y, Morisato D, Anderson KV. A processed form of the Spätzle protein defines dorsal-ventral polarity in the Drosophila embryo. Development. 1994;120:1243–1250. doi: 10.1242/dev.120.5.1243. [DOI] [PubMed] [Google Scholar]
- 17.Hashimoto C, Hudson KL, Anderson KV. The Toll gene of Drosophila, required for dorsal-ventral embryonic polarity, appears to encode a transmembrane protein. Cell. 1988;52:269–279. doi: 10.1016/0092-8674(88)90516-8. [DOI] [PubMed] [Google Scholar]
- 18.Gay NJ, Keith FJ. Drosophila Toll and IL-1 receptor. Nature. 1991;351:355–356. doi: 10.1038/351355b0. [DOI] [PubMed] [Google Scholar]
- 19.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/S0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 20.Medzhitov R, Preston-Hurlburt P, Janeway CA., Jr. A human homologue of the Drosophila Toll protein signals activation of adaptive immunity. Nature. 1997;388:394–397. doi: 10.1038/41131. [DOI] [PubMed] [Google Scholar]
- 21.He SP, H. J. Research advancement of innate immunity and pattern recognition receptors. Chin. J. Anim. Nutr. 2017;29:3844–3851. [Google Scholar]
- 22.Mortaz E, et al. Pattern recognitions receptors in immunodeficiency disorders. Eur. J. Pharmacol. 2017;808:49–56. doi: 10.1016/j.ejphar.2017.01.014. [DOI] [PubMed] [Google Scholar]
- 23.Park BS, Lee JO. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013;45:e66. doi: 10.1038/emm.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X, Lin G, Han Z, Chai J. Structural biology of NOD-like receptors. Adv. Exp. Med. Biol. 2019;1172:119–141. doi: 10.1007/978-981-13-9367-9_6. [DOI] [PubMed] [Google Scholar]
- 25.Zindel J, Kubes P. DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu. Rev. Pathol. 2020;15:493–518. doi: 10.1146/annurev-pathmechdis-012419-032847. [DOI] [PubMed] [Google Scholar]
- 26.Herwald H, Egesten A. On PAMPs and DAMPs. J. Innate Immun. 2016;8:427–428. doi: 10.1159/000448437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saïd-Sadier N, Ojcius DMAlarmins. inflammasomes and immunity. Biomed. J. 2012;35:437–449. doi: 10.4103/2319-4170.104408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saijo Y, Loo EP, Yasuda S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018;93:592–613. doi: 10.1111/tpj.13808. [DOI] [PubMed] [Google Scholar]
- 29.Brubaker SW, Bonham KS, Zanoni I, Kagan JC. Innate immune pattern recognition: a cell biological perspective. Annu. Rev. Immunol. 2015;33:257–290. doi: 10.1146/annurev-immunol-032414-112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gong T, Liu L, Jiang W, Zhou R. DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 2020;20:95–112. doi: 10.1038/s41577-019-0215-7. [DOI] [PubMed] [Google Scholar]
- 31.Pandolfi F, Altamura S, Frosali S, Conti P. Key role of DAMP in inflammation, cancer, and tissue repair. Clin. Ther. 2016;38:1017–1028. doi: 10.1016/j.clinthera.2016.02.028. [DOI] [PubMed] [Google Scholar]
- 32.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Takeda K, Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 34.Kumar H, Kawai T, Akira S. Toll-like receptors and innate immunity. Biochem. Biophys. Res. Commun. 2009;388:621–625. doi: 10.1016/j.bbrc.2009.08.062. [DOI] [PubMed] [Google Scholar]
- 35.Lacagnina MJ, Watkins LR, Grace PM. Toll-like receptors and their role in persistent pain. Pharmacol. Ther. 2018;184:145–158. doi: 10.1016/j.pharmthera.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Nardo D. Toll-like receptors: activation, signalling and transcriptional modulation. Cytokine. 2015;74:181–189. doi: 10.1016/j.cyto.2015.02.025. [DOI] [PubMed] [Google Scholar]
- 37.Temperley ND, Berlin S, Paton IR, Griffin DK, Burt DW. Evolution of the chicken Toll-like receptor gene family: a story of gene gain and gene loss. BMC Genomics. 2008;9:62. doi: 10.1186/1471-2164-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lauw FN, Caffrey DR, Golenbock DT. Of mice and man: TLR11 (finally) finds profilin. Trends Immunol. 2005;26:509–511. doi: 10.1016/j.it.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 39.Mathur R, Zeng W, Hayden MS, Ghosh S. Mice lacking TLR11 exhibit variable Salmonella typhi susceptibility. Cell. 2016;164:829–830. doi: 10.1016/j.cell.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 40.Andrade WA, et al. Combined action of nucleic acid-sensing Toll-like receptors and TLR11/TLR12 heterodimers imparts resistance to Toxoplasma gondii in mice. Cell Host Microbe. 2013;13:42–53. doi: 10.1016/j.chom.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palti Y. Toll-like receptors in bony fish: from genomics to function. Dev. Comp. Immunol. 2011;35:1263–1272. doi: 10.1016/j.dci.2011.03.006. [DOI] [PubMed] [Google Scholar]
- 42.Chuang T, Ulevitch RJ. Identification of hTLR10: a novel human Toll-like receptor preferentially expressed in immune cells. Biochim. Biophys. Acta. 2001;1518:157–161. doi: 10.1016/S0167-4781(00)00289-X. [DOI] [PubMed] [Google Scholar]
- 43.Chuenchor W, Jin T, Ravilious G, Xiao TS. Structures of pattern recognition receptors reveal molecular mechanisms of autoinhibition, ligand recognition and oligomerization. Curr. Opin. Immunol. 2014;26:14–20. doi: 10.1016/j.coi.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 2010;11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 45.Brennan JJ, Gilmore TD. Evolutionary origins of Toll-like receptor signaling. Mol. Biol. Evol. 2018;35:1576–1587. doi: 10.1093/molbev/msy050. [DOI] [PubMed] [Google Scholar]
- 46.Kim HM, et al. Crystal structure of the TLR4-MD-2complex with bound endotoxin antagonist Eritoran. Cell. 2007;130:906–917. doi: 10.1016/j.cell.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 47.Gay NJ, Gangloff M. Structure and function of Toll receptors andtheir ligands. Annu. Rev. Biochem. 2007;76:141–165. doi: 10.1146/annurev.biochem.76.060305.151318. [DOI] [PubMed] [Google Scholar]
- 48.Armant MA, Fenton MJ. Toll-like receptors: a family of pattern-recognition receptors in mammals. Genome Biol. 2002;3:REVIEWS3011. doi: 10.1186/gb-2002-3-8-reviews3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Botos I, Segal DM, Davies DR. The structural biology of Toll-like receptors. Structure. 2011;19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao D, Li W. Structures and recognition modes of Toll-like receptors. Proteins. 2017;85:3–9. doi: 10.1002/prot.25179. [DOI] [PubMed] [Google Scholar]
- 51.Enkhbayar P, Kamiya M, Osaki M, Matsumoto T, Matsushima N. Structural principles of leucine-rich repeat (LRR) proteins. Proteins. 2004;54:394–403. doi: 10.1002/prot.10605. [DOI] [PubMed] [Google Scholar]
- 52.Matsushima N, et al. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics. 2007;8:124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu Y, et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature. 2000;408:111–115. doi: 10.1038/35040600. [DOI] [PubMed] [Google Scholar]
- 54.Netea MG, van Deuren M, Kullberg BJ, Cavaillon JM, Van der Meer JW. Does the shape of lipid A determine the interaction of LPS with Toll-like receptors? Trends Immunol. 2002;23:135–139. doi: 10.1016/S1471-4906(01)02169-X. [DOI] [PubMed] [Google Scholar]
- 55.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 56.Shimizu T. Structural insights into ligand recognition and regulation of nucleic acid-sensing Toll-like receptors. Curr. Opin. Struct. Biol. 2017;47:52–59. doi: 10.1016/j.sbi.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 57.Farhat K, et al. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J. Leukoc. Biol. 2008;83:692–701. doi: 10.1189/jlb.0807586. [DOI] [PubMed] [Google Scholar]
- 58.Cheng K, et al. Specific activation of the TLR1-TLR2 heterodimer by small-molecule agonists. Sci. Adv. 2015;1:e1400139. doi: 10.1126/sciadv.1400139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaur A, et al. TLR2 agonistic small molecules: detailed structure-activity relationship, applications, and future prospects. J. Med. Chem. 2021;64:233–278. doi: 10.1021/acs.jmedchem.0c01627. [DOI] [PubMed] [Google Scholar]
- 60.Omueti KO, Beyer JM, Johnson CM, Lyle EA, Tapping RI. Domain exchange between human toll-like receptors 1 and 6 reveals a region required for lipopeptide discrimination. J. Biol. Chem. 2005;280:36616–36625. doi: 10.1074/jbc.M504320200. [DOI] [PubMed] [Google Scholar]
- 61.Natala SR, et al. Structure based design and synthesis of novel Toll-like Receptor 2 (TLR 2) lipid antagonists. Bioorg. Med. Chem. Lett. 2021;40:127861. doi: 10.1016/j.bmcl.2021.127861. [DOI] [PubMed] [Google Scholar]
- 62.Jin MS, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130:1071–1082. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 63.Kang JY, et al. Recognition of lipopeptide patternsby Toll-like receptor 2-Toll-like receptor 6 heterodimer. Immunity. 2009;31:873–884. doi: 10.1016/j.immuni.2009.09.018. [DOI] [PubMed] [Google Scholar]
- 64.Su L, et al. Structural basis of TLR2/TLR1 activation by the synthetic agonist diprovocim. J. Med. Chem. 2019;62:2938–2949. doi: 10.1021/acs.jmedchem.8b01583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsumoto M, Seya T. TLR3: interferon induction by double-stranded RNA including poly(I:C) Adv. Drug Deliv. Rev. 2008;60:805–812. doi: 10.1016/j.addr.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Vidya MK, et al. Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 2018;37:20–36. doi: 10.1080/08830185.2017.1380200. [DOI] [PubMed] [Google Scholar]
- 68.Qing Z, Lu H, Qiang ZZ. The research progress of Toll-like receptors in innate immune system. J. Biol. 2016;33:83–87. [Google Scholar]
- 69.Anderson KV, Rgens JG, Sslein-Volhard NC. Establish- ment of dorsal-ventral polarity in the Drosophila embryo: genetic studies on the role of the Toll gene product. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- 70.Miyake K. Innate immune sensing of pathogens and danger signals by cell surface Toll-like receptors. Semin Immunol. 2007;19:3–10. doi: 10.1016/j.smim.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Park BS, et al. The structural basis of li- popolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458:1191–1195. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 72.Zhang S, et al. Carnosic acid alleviates bdl-induced liver fibrosis through miR-29b-3p-mediated inhibition of the high-mobility group box 1/Toll-like receptor 4 signaling pathway in rats. Front. Pharmacol. 2018;8:976. doi: 10.3389/fphar.2017.00976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nguyen CT, Kim EH, Luong TT, Pyo S, Rhee DK. TLR4 mediates pneumolysin-induced ATF3 expression through the JNK/p38 pathway in Streptococcus pneumoniae-infected RAW 264.7 cells. Mol. Cells. 2015;38:58–64. doi: 10.14348/molcells.2015.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chiu FF, et al. Domain 4 of pneumolysin from Streptococcus pneumoniae is a multifunctional domain contributing TLR4 activating and hemolytic activity. Biochem. Biophys. Res. Commun. 2019;517:596–602. doi: 10.1016/j.bbrc.2019.07.063. [DOI] [PubMed] [Google Scholar]
- 75.Yoon SI, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335:859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rout AK, et al. Insights into structure and dynamics of extracellular domain of Toll-like receptor 5 in Cirrhinus mrigala (mrigala): a molecular dynamics simulation approach. PLoS ONE. 2021;16:e0245358. doi: 10.1371/journal.pone.0245358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maeda K, Akira S. TLR7 structure: cut in Z-loop. Immunity. 2016;45:705–707. doi: 10.1016/j.immuni.2016.10.003. [DOI] [PubMed] [Google Scholar]
- 78.Song DH, Lee JO. Sensing of microbial molecular patterns by Toll-like receptors. Immunol. Rev. 2012;250:216–229. doi: 10.1111/j.1600-065X.2012.01167.x. [DOI] [PubMed] [Google Scholar]
- 79.Lee SM, et al. Recognition of double-stranded RNA and regulation of interferon pathway by Toll-like receptor 10. Front. Immunol. 2018;9:516. doi: 10.3389/fimmu.2018.00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hedhli D, et al. The antigen-specific response to Toxoplasma gondii profilin, a TLR11/12 ligand, depends on its intrinsic adjuvant properties. Med. Microbiol. Immunol. 2016;205:345–352. doi: 10.1007/s00430-016-0452-3. [DOI] [PubMed] [Google Scholar]
- 81.Raetz M, et al. Cooperation of TLR12 and TLR11 in the IRF8-dependent IL-12 response to Toxoplasma gondii profilin. J. Immunol. 2013;191:4818–4827. doi: 10.4049/jimmunol.1301301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kucera K, et al. Structure-based analysis of Toxoplasma gondii profilin: a parasite-specific motif is required for recognition by Toll-like receptor 11. J. Mol. Biol. 2010;403:616–629. doi: 10.1016/j.jmb.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Motta V, Soares F, Sun T, Philpott DJ. NOD-like receptors: versatile cytosolic sentinels. Physiol. Rev. 2015;95:149–178. doi: 10.1152/physrev.00009.2014. [DOI] [PubMed] [Google Scholar]
- 84.Inohara N, Nuñez G. NODs: intracellular proteins involved in inflammation and apoptosis. Nat. Rev. Immunol. 2003;3:371–382. doi: 10.1038/nri1086. [DOI] [PubMed] [Google Scholar]
- 85.Fritz JH, et al. Nod-like proteins in immunity, inflammation and disease. Nat. Immunol. 2006;7:1250–1257. doi: 10.1038/ni1412. [DOI] [PubMed] [Google Scholar]
- 86.Barbé F, Douglas T, Saleh M. Advances in Nod-like receptors (NLR) biology. Cytokine Growth Factor Rev. 2014;25:681–697. doi: 10.1016/j.cytogfr.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 87.Kim YK, Shin JS, Nahm MH. NOD-like receptors in infection, immunity, and diseases. Yonsei Med. J. 2016;57:5–14. doi: 10.3349/ymj.2016.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meunier E, Broz P. Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. 2017;38:744–757. doi: 10.1016/j.it.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 89.Álvarez CA, et al. Insights into the diversity of NOD-like receptors: identification and expression analysis of NLRC3, NLRC5 and NLRX1 in rainbow trout. Mol. Immunol. 2017;87:102–113. doi: 10.1016/j.molimm.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 90.Pashenkov MV, Dagil YA, Pinegin BV. NOD1 and NOD2: molecular targets in prevention and treatment of infectious diseases. Int. Immunopharmacol. 2018;54:385–400. doi: 10.1016/j.intimp.2017.11.036. [DOI] [PubMed] [Google Scholar]
- 91.Chamaillard M, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat. Immunol. 2003;4:702–707. doi: 10.1038/ni945. [DOI] [PubMed] [Google Scholar]
- 92.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J. Biol. Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 93.Caruso R, Warner N, Inohara N, Núñez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hu Z, et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341:172–175. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 95.Le HT, Harton JA. Pyrin- and CARD-only proteins as regulators of NLR functions. Front. Immunol. 2013;4:275. doi: 10.3389/fimmu.2013.00275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang J, et al. Reconstitution and structure of a plant NLR resistosome conferring immunity. Science. 2019;364:eaav5870. doi: 10.1126/science.aav5870. [DOI] [PubMed] [Google Scholar]
- 97.Yuan M, et al. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature. 2021;592:105–109. doi: 10.1038/s41586-021-03316-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lu Y, et al. Palmitoylation of NOD1 and NOD2 is required for bacterial sensing. Science. 2019;366:460–467. doi: 10.1126/science.aau6391. [DOI] [PubMed] [Google Scholar]
- 99.Rehwinkel J, Gack MU. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat. Rev. Immunol. 2020;20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. Viral RNA detection by RIG-I-like receptors. Curr. Opin. Immunol. 2015;32:48–53. doi: 10.1016/j.coi.2014.12.012. [DOI] [PubMed] [Google Scholar]
- 101.Barral PM, et al. Functions of the cytoplasmic RNA sensors RIG-I and MDA-5: key regulators of innate immunity. Pharmacol. Ther. 2009;124:219–234. doi: 10.1016/j.pharmthera.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 103.Vajjhala PR, Ve T, Bentham A, Stacey KJ, Kobe B. The molecular mechanisms of signaling by cooperative assembly formation in innate immunity pathways. Mol. Immunol. 2017;86:23–37. doi: 10.1016/j.molimm.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 104.Lässig C, Hopfner KP. RIG-I-like receptors: one STrEP forward. Trends Microbiol. 2016;24:517–519. doi: 10.1016/j.tim.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 105.Saito T, et al. Regulation of innate antiviral defenses through a shared repressor domain in RIG-I and LGP2. Proc. Natl Acad. Sci. USA. 2007;104:582–587. doi: 10.1073/pnas.0606699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cui S, et al. The C-terminal regulatory domain is the RNA 5’-triphosphate sensor of RIG-I. Mol. Cell. 2008;29:169–179. doi: 10.1016/j.molcel.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 107.Leung DW, Amarasinghe GK. Structural insights into RNA recognition and activation of RIG-I-like receptors. Curr. Opin. Struct. Biol. 2012;22:297–303. doi: 10.1016/j.sbi.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kowalinski E, et al. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- 109.Gee P, et al. Essential role of the N-terminal domain in the regulation of RIG-I ATPase activity. J. Biol. Chem. 2008;283:9488–9496. doi: 10.1074/jbc.M706777200. [DOI] [PubMed] [Google Scholar]
- 110.Jiang F, et al. Structural basis of RNA recognition and activation by innate immune receptor RIG-I. Nature. 2011;479:423–427. doi: 10.1038/nature10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chang M, et al. Expression and functional characterization of the RIG-I-like receptors MDA5 and LGP2 in Rainbow trout (Oncorhynchus mykiss) J. Virol. 2011;85:8403–8412. doi: 10.1128/JVI.00445-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luo D, et al. Structural insights into RNA recognition by RIG-I. Cell. 2011;147:409–422. doi: 10.1016/j.cell.2011.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Xu XX, et al. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell. 2018;9:246–253. doi: 10.1007/s13238-017-0431-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fan X, Jin T. Structures of RIG-I-like receptors and insights into viral RNA sensing. Adv. Exp. Med. Biol. 2019;1172:157–188. doi: 10.1007/978-981-13-9367-9_8. [DOI] [PubMed] [Google Scholar]
- 115.Chow KT, Gale M, Jr, Loo YM. RIG-I and other RNA sensors in antiviral immunity. Annu. Rev. Immunol. 2018;36:667–694. doi: 10.1146/annurev-immunol-042617-053309. [DOI] [PubMed] [Google Scholar]
- 116.Lässig C, Hopfner KP. Discrimination of cytosolic self and non-self RNA by RIG-I-like receptors. J. Biol. Chem. 2017;292:9000–9009. doi: 10.1074/jbc.R117.788398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Patel JR, et al. ATPase-driven oligomerization of RIG-I on RNA allows optimal activation of type-I interferon. EMBO Rep. 2013;14:780–787. doi: 10.1038/embor.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kang DC, et al. mda-5: An interferon-inducible putative RNA helicase with double-stranded RNA-dependent ATPase activity and melanoma growth-suppressive properties. Proc. Natl Acad. Sci. USA. 2002;99:637–642. doi: 10.1073/pnas.022637199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Brisse M, Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol. 2019;10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Liu G, Gack MU. Distinct and orchestrated functions of RNA sensors in innate immunity. Immunity. 2020;53:26–42. doi: 10.1016/j.immuni.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Crowl JT, Gray EE, Pestal K, Volkman HE, Stetson DB. Intracellular nucleic acid detection in autoimmunity. Annu. Rev. Immunol. 2017;35:313–336. doi: 10.1146/annurev-immunol-051116-052331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hei L, Zhong J. Laboratory of genetics and physiology 2 (LGP2) plays an essential role in hepatitis C virus infection-induced interferon responses. Hepatology. 2017;65:1478–1491. doi: 10.1002/hep.29050. [DOI] [PubMed] [Google Scholar]
- 123.Li Q, Li X, Feng R. Research progress of LGP2 participating in antiviral natural immune response. J. Virol. 2019;35:313–317. [Google Scholar]
- 124.Bruns AM, Leser GP, Lamb RA, Horvath CM. The innate immune sensor LGP2 activates antiviral signaling by regulating MDA5-RNA interaction and filament assembly. Mol. Cell. 2014;55:771–781. doi: 10.1016/j.molcel.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bruns AM, Horvath CM. LGP2 synergy with MDA5 in RLR-mediated RNA recognition and antiviral signaling. Cytokine. 2015;74:198–206. doi: 10.1016/j.cyto.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huo H, Zhao L, Wang D, Chen X, Chen H. LGP2 plays a critical role in MDA5-mediated antiviral activity against duck enteritis virus. Mol. Immunol. 2019;116:160–166. doi: 10.1016/j.molimm.2019.10.006. [DOI] [PubMed] [Google Scholar]
- 127.Duic I, et al. Viral RNA recognition by LGP2 and MDA5, and activation of signaling through step-by-step conformational changes. Nucleic Acids Res. 2020;48:11664–11674. doi: 10.1093/nar/gkaa935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Onomoto K, Onoguchi K, Yoneyama M. Regulation of RIG-I-like receptor-mediated signaling: interaction between host and viral factors. Cell. Mol. Immunol. 2021;18:539–555. doi: 10.1038/s41423-020-00602-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mohanty A, Sadangi S, Paichha M, Samanta M. Molecular characterization and expressional quantification of lgp2, a modulatory co-receptor of RLR-signalling pathway in the Indian major carp Labeo rohita following pathogenic challenges and PAMP stimulations. J. Fish. Biol. 2020;96:1399–1410. doi: 10.1111/jfb.14308. [DOI] [PubMed] [Google Scholar]
- 130.Kato K, et al. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol. Cell. 2021;81:599–613.e8. doi: 10.1016/j.molcel.2020.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 2008;205:1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zou J, et al. Poly IC triggers a cathepsin D- and IPS-1-dependent pathway to enhance cytokine production and mediate dendritic cell necroptosis. Immunity. 2013;38:717–728. doi: 10.1016/j.immuni.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 133.Wang Y, et al. Structural and functional insights into 5’-ppp RNA pattern recognition by the innate immune receptor RIG-I. Nat. Struct. Mol. Biol. 2010;17:781–787. doi: 10.1038/nsmb.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kato H, et al. Differential roles of MDA5 and RIG-1 helicases in the recognition of RNA viruses. Nature. 2006;441:101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 135.Wei W, et al. Screening of antiviral components of Ma Huang Tang and investigation on the ephedra alkaloids efficacy on influenza virus type A. Front. Pharmacol. 2019;10:961. doi: 10.3389/fphar.2019.00961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Tomczyk T, et al. Immune consequences of in vitro infection of human peripheral blood leukocytes with vesicular stomatitis virus. J. Innate Immun. 2018;10:131–144. doi: 10.1159/000485143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yu SP, Ong KC, Perera D, Wong KT. Neuronal transcriptomic responses to Japanese encephalitis virus infection with a special focus on chemokine CXCL11 and pattern recognition receptors RIG-1 and MDA5. Virology. 2019;527:107–115. doi: 10.1016/j.virol.2018.10.015. [DOI] [PubMed] [Google Scholar]
- 138.Uchikawa E, et al. Structural analysis of dsRNA binding to anti-viral pattern recognition receptors LGP2 and MDA5. Mol. Cell. 2016;62:586–602. doi: 10.1016/j.molcel.2016.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walton RW, Brown MC, Sacco MT, Gromeier M. Engineered oncolytic poliovirus PVSRIPO subverts MDA5-dependent innate immune responses in cancer cells. J. Virol. 2018;92:e00879-18. doi: 10.1128/JVI.00879-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brown MC, et al. Viral infection of cells within the tumor microenvironment mediates antitumor immunotherapy via selective TBK1-IRF3 signaling. Nat. Commun. 2021;12:1858. doi: 10.1038/s41467-021-22088-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Peisley A, Wu B, Yao H, Walz T, Hur S. RIG-I forms signaling-competent filaments in an ATP-dependent, ubiquitin-independent manner. Mol. Cell. 2013;51:573–583. doi: 10.1016/j.molcel.2013.07.024. [DOI] [PubMed] [Google Scholar]
- 142.Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat. Rev. Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Dambuza IM, Brown GD. C-type lectins in immunity: recent developments. Curr. Opin. Immunol. 2015;32:21–27. doi: 10.1016/j.coi.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Freeman SA, Grinstein S. Phagocytosis: receptors, signal integration, and the cytoskeleton. Immunol. Rev. 2014;262:193–215. doi: 10.1111/imr.12212. [DOI] [PubMed] [Google Scholar]
- 145.Ebner S, Sharon N, Ben-Tal N. Evolutionary analysis reveals collective properties and specificity in the C-type lectin and lectin-like domain superfamily. Proteins. 2003;53:44–55. doi: 10.1002/prot.10440. [DOI] [PubMed] [Google Scholar]
- 146.Dam TK, Brewer CF. Lectins as pattern recognition molecules: the effects of epitope density in innate immunity. Glycobiology. 2010;20:270–279. doi: 10.1093/glycob/cwp186. [DOI] [PubMed] [Google Scholar]
- 147.Lepenies B, Lee J, Sonkaria S. Targeting C-type lectin receptors with multivalent carbohydrate ligands. Adv. Drug Deliv. Rev. 2013;65:1271–1281. doi: 10.1016/j.addr.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 148.Lan T, et al. A four-CRD C-type lectin from razor clam Sinonovacula constricta mediates agglutination and phagocytosis. Gene. 2020;728:144287. doi: 10.1016/j.gene.2019.144287. [DOI] [PubMed] [Google Scholar]
- 149.Sancho D, Reis e Sousa C. Signaling by myeloid C-type lectin receptors in immunity and homeostasis. Annu. Rev. Immunol. 2012;30:491–529. doi: 10.1146/annurev-immunol-031210-101352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zelensky AN, Gready JE. Comparative analysis of structural properties of the C-type-lectin-like domain (CTLD) Proteins. 2003;52:466–477. doi: 10.1002/prot.10626. [DOI] [PubMed] [Google Scholar]
- 151.Murugaiah V, Tsolaki AG, Kishore U. Collectins: innate immune pattern recognition molecules. Adv. Exp. Med. Biol. 2020;1204:75–127. doi: 10.1007/978-981-15-1580-4_4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Hou H, et al. C-type lectin receptor: old friend and new player. Med. Chem. 2017;13:536–543. doi: 10.2174/1573406413666170510103030. [DOI] [PubMed] [Google Scholar]
- 153.Mnich ME, van Dalen R, van Sorge NM. C-type lectin receptors in host defense against bacterial pathogens. Front. Cell. Infect. Microbiol. 2020;10:309. doi: 10.3389/fcimb.2020.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Brown GD, Willment JA, Whitehead L. C-type lectins in immunity and homeostasis. Nat. Rev. Immunol. 2018;18:374–389. doi: 10.1038/s41577-018-0004-8. [DOI] [PubMed] [Google Scholar]
- 155.Li K, Underhill DM. C-type lectin receptors in phagocytosis. Curr. Top. Microbiol. Immunol. 2020;429:1–18. doi: 10.1007/82_2020_198. [DOI] [PubMed] [Google Scholar]
- 156.Robinson MJ, Sancho D, Slack EC. Myeloid C-type lectins in innate immunity. Nat. Immunol. 2006;7:1258–1265. doi: 10.1038/ni1417. [DOI] [PubMed] [Google Scholar]
- 157.Martinez-Pomares L. The mannose receptor. J. Leukoc. Biol. 2012;92:1177–1186. doi: 10.1189/jlb.0512231. [DOI] [PubMed] [Google Scholar]
- 158.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J. Exp. Med. 1992;176:287–292. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bravo MF, et al. Synthesis and binding of mannose-specific synthetic carbohydrate receptors. Chemistry. 2020;26:11782–11795. doi: 10.1002/chem.202000481. [DOI] [PubMed] [Google Scholar]
- 160.Jaynes JM, et al. Mannose receptor (CD206) activation in tumor-associated macrophages enhances adaptive and innate antitumor immune responses. Sci. Transl. Med. 2020;12:eaax6337. doi: 10.1126/scitranslmed.aax6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Garcia-Aguilar T, Espinosa-Cueto P, Magallanes-Puebla A, Mancilla R. The mannose receptor is involved in the phagocytosis of mycobacteria-induced apoptotic cells. J. Immunol. Res. 2016;2016:3845247. doi: 10.1155/2016/3845247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lee SJ, et al. Mannose receptor-mediated regulation of serum glycoprotein homeostasis. Science. 2002;295:1898–1901. doi: 10.1126/science.1069540. [DOI] [PubMed] [Google Scholar]
- 163.Kaisar MMM, et al. Dectin-1/2-induced autocrine PGE2 signaling licenses dendritic cells to prime Th2 responses. PLoS Biol. 2018;16:e2005504. doi: 10.1371/journal.pbio.2005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Yamaguchi K, et al. Distinct roles for Dectin-1 and Dectin-2 in skin wound healing and neutrophilic inflammatory responses. J. Invest. Dermatol. 2021;141:164–176.e8. doi: 10.1016/j.jid.2020.04.030. [DOI] [PubMed] [Google Scholar]
- 165.de Quaglia E, et al. Involvement of the Dectin-1 receptor upon the effector mechanisms of human phagocytic cells against Paracoccidioides brasiliensis. J. Immunol. Res. 2019;2019:1529189. doi: 10.1155/2019/1529189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu BY, Li P, Yang GL. The Immunomodulatory Role of C-type Lectin Receptors in Parasitic Infection. Chin. J. Parasitol. Parasit. Dis. 2015;33:228–232. [PubMed] [Google Scholar]
- 167.Saijo S, Iwakura Y. Dectin-1 and Dectin-2 in innate immunity against fungi. Int. Immunol. 2011;23:467–472. doi: 10.1093/intimm/dxr046. [DOI] [PubMed] [Google Scholar]
- 168.Taylor PR, et al. Dectin-1 is required for beta-glucan recognition and control of fungal infection. Nat. Immunol. 2007;8:31–38. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cao M, et al. Dectin-1-induced RIPK1 and RIPK3 activation protects host against Candida albicans infection. Cell Death Differ. 2019;26:2622–2636. doi: 10.1038/s41418-019-0323-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Gow NA, et al. Immune recognition of Candida albicans beta-glucan by dectin-1. J. Infect. Dis. 2007;196:1565–1571. doi: 10.1086/523110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saijo S, et al. Dectin-1 is required for host defense against Pneumocystis carinii but not against Candida albicans. Nat. Immunol. 2007;8:39–46. doi: 10.1038/ni1425. [DOI] [PubMed] [Google Scholar]
- 172.Steele C, et al. Alveolar macrophage-mediated killing of Pneumocystis carinii f. sp. muris involves molecular recognition by the Dectin-1 beta-glucan receptor. J. Exp. Med. 2003;198:1677–1688. doi: 10.1084/jem.20030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ambati S, et al. Dectin-1-targeted antifungal liposomes exhibit enhanced efficacy. mSphere. 2019;4:e00025-19. doi: 10.1128/mSphere.00025-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Walsh NM, Wuthrich M, Wang H, Klein B, Hull CM. Characterization of C-type lectins reveals an unexpectedly limited interaction between Cryptococcus neoformans spores and Dectin-1. PLoS ONE. 2017;12:e0173866. doi: 10.1371/journal.pone.0173866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Yuan K, et al. Dectin-1 is essential for IL-1β production through JNK activation and apoptosis in Aspergillus fumigatus keratitis. Int. Immunopharmacol. 2017;52:168–175. doi: 10.1016/j.intimp.2017.09.008. [DOI] [PubMed] [Google Scholar]
- 176.Fan Y, et al. Perillaldehyde ameliorates Aspergillus fumigatus keratitis by activating the Nrf2/HO-1 signaling pathway and inhibiting Dectin-1-mediated inflammation. Invest. Ophthalmol. Vis. Sci. 2020;61:51. doi: 10.1167/iovs.61.6.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ning YY, Yuan XY, Shan L, Song LX. Research progress on role of C-type lectin receptors in tumor immunotherapy. Chin. J. Immunol. 2019;35:1139–1142. [Google Scholar]
- 178.Zhou MN, et al. N-Carboxyanhydride Polymerization Of Glycopolypeptides That Activate Antigen-presenting Cells Through Dectin-1 and Dectin-2. Angew. Chem. Int. Ed. Engl. 2018;57:3137–3142. doi: 10.1002/anie.201713075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Cadena AP, Cushman TR, Welsh JW. Glycosylation and Antitumor Immunity. Int. Rev. Cell Mol. Biol. 2019;343:111–127. doi: 10.1016/bs.ircmb.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 180.Mereiter S, Balmaña M, Campos D, Gomes J, Reis CA. Glycosylation in the era of cancer-targeted therapy: where are we heading? Cancer Cell. 2019;36:6–16. doi: 10.1016/j.ccell.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 181.Manabe Y, et al. The core fucose on an IgG Antibody Is An Endogenous Ligand of Dectin-1. Angew. Chem. Int. Ed. Engl. 2019;58:18697–18702. doi: 10.1002/anie.201911875. [DOI] [PubMed] [Google Scholar]
- 182.Saijo S, et al. Dectin-2 recognition of alpha-mannans and induction of Th17 cell differentiation is essential for host defense against Candida albicans. Immunity. 2010;32:681–691. doi: 10.1016/j.immuni.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 183.Ambati S, et al. Dectin-2-targeted antifungal liposomes exhibit enhanced efficacy. mSphere. 2019;4:e00715-19. doi: 10.1128/mSphere.00715-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Ritter M, et al. Schistosoma mansoni triggers Dectin-2, which activates the Nlrp3 inflammasome and alters adaptive immune responses. Proc. Natl Acad. Sci. USA. 2010;107:20459–20464. doi: 10.1073/pnas.1010337107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Feinberg H, et al. Mechanism of pathogen recognition by human dectin-2. J. Biol. Chem. 2017;292:13402–13414. doi: 10.1074/jbc.M117.799080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Wittmann A, et al. Dectin-2 recognizes mannosylated O-antigens of human opportunistic pathogens and augments lipopolysaccharide activation of myeloid cells. J. Biol. Chem. 2016;291:17629–17638. doi: 10.1074/jbc.M116.741256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Decout A, et al. Deciphering the molecular basis of mycobacteria and lipoglycan recognition by the C-type lectin Dectin-2. Sci. Rep. 2018;8:16840. doi: 10.1038/s41598-018-35393-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Feng S, et al. Absent in melanoma 2 inflammasome is required for host defence against Streptococcus pneumoniae infection. Innate Immun. 2019;25:412–419. doi: 10.1177/1753425919860252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Zhang Z, Dong X-Q, He S-B. Research advances of absent in melanoma 2 in the development of cancer. J. Postgrad. Med. 2017;30:542–545. [Google Scholar]
- 190.Liu YS, Hu R. Role of absent in melanoma 2 in development of inflammation, immunity and cancer. Pharm. Biotechnol. 2018;25:354–358. [Google Scholar]
- 191.Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur. J. Immunol. 2016;46:269–280. doi: 10.1002/eji.201545839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol. Rev. 2015;265:6–21. doi: 10.1111/imr.12296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Wang B, Tian Y, Yin Q. AIM2 inflammasome assembly and signaling. Adv. Exp. Med. Biol. 2019;1172:143–155. doi: 10.1007/978-981-13-9367-9_7. [DOI] [PubMed] [Google Scholar]
- 195.Gray EE, et al. The AIM2-like receptors are dispensable for the interferon response to intracellular DNA. Immunity. 2016;45:255–266. doi: 10.1016/j.immuni.2016.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Cadena C, Hur S. Filament-like assemblies of intracellular nucleic acid sensors: commonalities and differences. Mol. Cell. 2019;76:243–254. doi: 10.1016/j.molcel.2019.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Caneparo V, Landolfo S, Gariglio M, De Andrea M. The absent in melanoma 2-like receptor IFN-inducible protein 16 as an inflammasome regulator in systemic lupus erythematosus: the dark side of sensing microbes. Front. Immunol. 2018;9:1180. doi: 10.3389/fimmu.2018.01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Morrone SR, et al. Assembly-driven activation of the AIM2 foreign-dsDNA sensor provides a polymerization template for downstream ASC. Nat. Commun. 2015;6:7827. doi: 10.1038/ncomms8827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chen PA, et al. Absent in melanoma 2 regulates tumor cell proliferation in glioblastoma multiforme. J. Neurooncol. 2019;144:265–273. doi: 10.1007/s11060-019-03230-y. [DOI] [PubMed] [Google Scholar]
- 200.Zhang P, Liu X, Cao X. Extracellular pattern recognition molecules in health and diseases. Cell. Mol. Immunol. 2015;12:255–257. doi: 10.1038/cmi.2014.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Presta M, Camozzi M, Salvatori G, Rusnati M. Role of the soluble pattern recognition receptor PTX3 in vascular biology. J. Cell Mol. Med. 2007;11:723–738. doi: 10.1111/j.1582-4934.2007.00061.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Mantovani A, Garlanda C, Doni A, Bottazzi B. Pentraxins in innate immunity: from C-reactive protein to the long pentraxin PTX3. J. Clin. Immunol. 2008;28:1–13. doi: 10.1007/s10875-007-9126-7. [DOI] [PubMed] [Google Scholar]
- 203.Holmskov U, Thiel S, Jensenius JC. Collections and ficolins: humoral lectins of the innate immune defense. Annu. Rev. Immunol. 2003;21:547–578. doi: 10.1146/annurev.immunol.21.120601.140954. [DOI] [PubMed] [Google Scholar]
- 204.Reis ES, Mastellos DC, Hajishengallis G, Lambris JD. New insights into the immune functions of complement. Nat. Rev. Immunol. 2019;19:503–516. doi: 10.1038/s41577-019-0168-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tang Y, et al. Macrophage scavenger receptor 1 contributes to pathogenesis of fulminant hepatitis via neutrophil-mediated complement activation. J. Hepatol. 2018;68:733–743. doi: 10.1016/j.jhep.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Jun JI, Lau LF. CCN1 is an opsonin for bacterial clearance and a direct activator of Toll-like receptor signaling. Nat. Commun. 2020;11:1242. doi: 10.1038/s41467-020-15075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140:805–820. doi: 10.1016/j.cell.2010.01.022. [DOI] [PubMed] [Google Scholar]
- 208.Schlimgen AK, Helms JA, Vogel H, Perin MS. Neuronal pentraxin, a secreted protein with homology to acute phase proteins of the immune system. Neuron. 1995;14:519–526. doi: 10.1016/0896-6273(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 209.Daigo K, et al. Pentraxins in the activation and regulation of innate immunity. Immunol. Rev. 2016;274:202–217. doi: 10.1111/imr.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Muczynski V, et al. Complex formation with pentraxin-2 regulates factor X plasma levels and macrophage interactions. Blood. 2017;129:2443–2454. doi: 10.1182/blood-2016-06-724351. [DOI] [PubMed] [Google Scholar]
- 211.Ma YJ, Garred P. Pentraxins in complement activation and regulation. Front. Immunol. 2018;9:3046. doi: 10.3389/fimmu.2018.03046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Pathak A, Agrawal A. Evolution of C-reactive protein. Front. Immunol. 2019;10:943. doi: 10.3389/fimmu.2019.00943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sproston NR, Ashworth JJ. Role of C-reactive protein at sites of inflammation and infection. Front. Immunol. 2018;9:754. doi: 10.3389/fimmu.2018.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Moutachakkir M, Lamrani Hanchi A, Baraou A, Boukhira A, Chellak S. Immunoanalytical characteristics of C-reactive protein and high sensitivity C-reactive protein. Ann. Biol. Clin. 2017;75:225–229. doi: 10.1684/abc.2017.1232. [DOI] [PubMed] [Google Scholar]
- 215.Pilling D, Gomer RH. The development of serum amyloid P as a possible therapeutic. Front. Immunol. 2018;9:2328. doi: 10.3389/fimmu.2018.02328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Behrens NE, Lipke PN, Pilling D, Gomer RH, Klotz SA. Serum amyloid P component binds fungal surface amyloid and decreases human macrophage phagocytosis and secretion of inflammatory cytokines. mBio. 2019;10:e00218-19. doi: 10.1128/mBio.00218-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ngwa DN, Agrawal A. Structure-function relationships of C-reactive protein in bacterial infection. Front. Immunol. 2019;10:166. doi: 10.3389/fimmu.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Hari-Dass R, Shah C, Meyer DJ, Raynes JG. Serum amyloid A protein binds to outer membrane protein A of Gram-negative bacteria. J. Biol. Chem. 2005;280:18562–18567. doi: 10.1074/jbc.M500490200. [DOI] [PubMed] [Google Scholar]
- 219.Shah C, Hari-Dass R, Raynes JG. Serum amyloid A is an innate immune opsonin for Gram-negative bacteria. Blood. 2006;108:1751–1757. doi: 10.1182/blood-2005-11-011932. [DOI] [PubMed] [Google Scholar]
- 220.Christensen MB, et al. Comparison of serum amyloid A and C-reactive protein as diagnostic markers of systemic inflammation in dogs. Can. Vet. J. 2014;55:161–168. [PMC free article] [PubMed] [Google Scholar]
- 221.Wang W, et al. Diabetic foot disease: grading inflammation by apolipoprotein A-I, C-reactive protein and serum amyloid A. Clin. Lab. 2014;60:1951–1959. doi: 10.7754/clin.lab.2014.140209. [DOI] [PubMed] [Google Scholar]
- 222.Alles VV, et al. Inducible expression of PTX3, a new member of the pentraxin family, in human mononuclear phagocytes. Blood. 1994;84:3483–3493. doi: 10.1182/blood.V84.10.3483.3483. [DOI] [PubMed] [Google Scholar]
- 223.Garlanda C, Bottazzi B, Magrini E, Inforzato A, Mantovani APTX3. PTX3, a humoral pattern recognition molecule, in innate immunity, tissue repair, and cancer. Physiol. Rev. 2018;98:623–639. doi: 10.1152/physrev.00016.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Bottazzi B, et al. The pentraxins PTX3 and SAP in innate immunity, regulation of inflammation and tissue remodelling. J. Hepatol. 2016;64:1416–1427. doi: 10.1016/j.jhep.2016.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Balhara J, et al. Pentraxin 3 deletion aggravates allergic inflammation through a TH17-dominant phenotype and enhanced CD4 T-cell survival. J. Allergy Clin. Immunol. 2017;139:950–963.e9. doi: 10.1016/j.jaci.2016.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Chorny A, et al. The soluble pattern recognition receptor PTX3 links humoral innate and adaptive immune responses by helping marginal zone B cells. J. Exp. Med. 2016;213:2167–2185. doi: 10.1084/jem.20150282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Perea L, et al. Pentraxin-3 modulates lipopolysaccharide-induced inflammatory response and attenuates liver injury. Hepatology. 2017;66:953–968. doi: 10.1002/hep.29215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Brunetta E, et al. Macrophage expression and prognostic significance of the long pentraxin PTX3 in COVID-19. Nat. Immunol. 2021;22:19–24. doi: 10.1038/s41590-020-00832-x. [DOI] [PubMed] [Google Scholar]
- 229.Hickman-Davis JM, et al. Lung surfactant and reactive oxygen-nitrogen species: antimicrobial activity and host-pathogen interactions. Am. J. Physiol. Lung Cell. Mol. Physiol. 2001;281:L517–L523. doi: 10.1152/ajplung.2001.281.3.L517. [DOI] [PubMed] [Google Scholar]
- 230.Vasta GR, et al. Animal lectins as self/non-self recognition molecules. Biochemical and genetic approaches to understanding their biological roles and evolution. Ann. NY Acad. Sci. 1994;712:55–73. doi: 10.1111/j.1749-6632.1994.tb33562.x. [DOI] [PubMed] [Google Scholar]
- 231.Garred P, et al. A journey through the lectin pathway of complement-MBL and beyond. Immunol. Rev. 2016;274:74–97. doi: 10.1111/imr.12468. [DOI] [PubMed] [Google Scholar]
- 232.Singh SS, Cheung RC, Wong JH, Ng TB. Mannose binding lectin: a potential biomarker for many human diseases. Curr. Med. Chem. 2016;23:3847–3860. doi: 10.2174/0929867323666160817162208. [DOI] [PubMed] [Google Scholar]
- 233.Turner MW. The role of mannose-binding lectin in health and disease. Mol. Immunol. 2003;40:423–429. doi: 10.1016/S0161-5890(03)00155-X. [DOI] [PubMed] [Google Scholar]
- 234.Bouwman LH, Roep BO, Roos A. Mannose-binding lectin: clinical implications for infection, transplantation, and autoimmunity. Hum. Immunol. 2006;67:247–256. doi: 10.1016/j.humimm.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 235.Skjoedt MO, et al. Crystal structure and functional characterization of the complement regulator mannose-binding lectin (MBL)/ficolin-associated protein-1 (MAP-1) J. Biol. Chem. 2012;287:32913–32921. doi: 10.1074/jbc.M112.386680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Jack DL, Klein NJ, Turner MW. Mannose-binding lectin: targeting the microbial world for complement attack and opsonophagocytosis. Immunol. Rev. 2001;180:86–99. doi: 10.1034/j.1600-065X.2001.1800108.x. [DOI] [PubMed] [Google Scholar]
- 237.Kilpatrick DC. Mannan-binding lectin and its role in innate immunity. Transfus. Med. 2002;12:335–352. doi: 10.1046/j.1365-3148.2002.00408.x. [DOI] [PubMed] [Google Scholar]
- 238.Neglia L, et al. Mannose-binding lectin has a direct deleterious effect on ischemic brain microvascular endothelial cells. J. Cereb. Blood Flow Metab. 2020;40:1608–1620. doi: 10.1177/0271678X19874509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Dos Santos Silva PM, et al. Insights into anti-pathogenic activities of mannose lectins. Int. J. Biol. Macromol. 2019;140:234–244. doi: 10.1016/j.ijbiomac.2019.08.059. [DOI] [PubMed] [Google Scholar]
- 240.Fortpied J, Vertommen D, Van Schaftingen E. Binding of mannose-binding lectin to fructosamines: a potential link between hyperglycaemia and complement activation in diabetes. Diabetes Metab. Res. Rev. 2010;26:254–260. doi: 10.1002/dmrr.1079. [DOI] [PubMed] [Google Scholar]
- 241.Veldhuizen EJ, van Eijk M, Haagsman HP. The carbohydrate recognition domain of collectins. FEBS J. 2011;278:3930–3941. doi: 10.1111/j.1742-4658.2011.08206.x. [DOI] [PubMed] [Google Scholar]
- 242.Kishore U, et al. Surfactant proteins SP-A and SP-D: structure, function and receptors. Mol. Immunol. 2006;43:1293–1315. doi: 10.1016/j.molimm.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 243.Casals C, García-Fojeda B, Minutti CM. Soluble defense collagens: sweeping up immune threats. Mol. Immunol. 2019;112:291–304. doi: 10.1016/j.molimm.2019.06.007. [DOI] [PubMed] [Google Scholar]
- 244.Watson A, Madsen J, Clark HW. SP-A and SP-D: dual functioning immune molecules with antiviral and immunomodulatory properties. Front. Immunol. 2021;11:622598. doi: 10.3389/fimmu.2020.622598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Carreto-Binaghi LE, Aliouat el M, Taylor ML. Surfactant proteins, SP-A and SP-D, in respiratory fungal infections: their role in the inflammatory response. Respir. Res. 2016;17:66. doi: 10.1186/s12931-016-0385-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Endo Y, Matsushita M, Fujita T. New insights into the role of ficolins in the lectin pathway of innate immunity. Int. Rev. Cell Mol. Biol. 2015;316:49–110. doi: 10.1016/bs.ircmb.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 247.Vassal-Stermann E, et al. Human L-ficolin recognizes phosphocholine moieties of pneumococcal teichoic acid. J. Immunol. 2014;193:5699–5708. doi: 10.4049/jimmunol.1400127. [DOI] [PubMed] [Google Scholar]
- 248.Katayama M, et al. Ficolin-1 is a promising therapeutic target for autoimmune diseases. Int. Immunol. 2019;31:23–32. doi: 10.1093/intimm/dxy056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Ding Q, et al. Ficolin-2 triggers antitumor effect by activating macrophages and CD8+ T cells. Clin. Immunol. 2017;183:145–157. doi: 10.1016/j.clim.2017.08.012. [DOI] [PubMed] [Google Scholar]
- 250.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harb. Perspect. Biol. 2009;1:a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 252.DiDonato JA, Mercurio F, Karin M. NF-κB and the link between inflammation and cancer. Immunol. Rev. 2012;246:379–400. doi: 10.1111/j.1600-065X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 253.Mitchell S, Vargas J, Hoffmann A. Signaling via the NFκB system. Wiley Interdiscip. Rev. Syst. Biol. Med. 2016;8:227–241. doi: 10.1002/wsbm.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Satoh T, Akira S. Toll-like receptor signaling and its inducible proteins. Microbiol. Spectr. 2016;4:10.1128. doi: 10.1128/microbiolspec.MCHD-0040-2016. [DOI] [PubMed] [Google Scholar]
- 255.Anthoney N, Foldi I, Hidalgo A. Toll and Toll-like receptor signalling in development. Development. 2018;145:dev156018. doi: 10.1242/dev.156018. [DOI] [PubMed] [Google Scholar]
- 256.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 2007;7:353–364. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 257.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Huai X, et al. Advances in Toll receptor 4 signaling pathway in colorectal cancer. Bachu Med. J. 2020;3:125–128. [Google Scholar]
- 259.Gay NJ, et al. Assembly and localizationof Toll-like receptor signalling complexes. Nat. Rev. Immunol. 2014;14:546–558. doi: 10.1038/nri3713. [DOI] [PubMed] [Google Scholar]
- 260.Kawai T, Akira S. Toll-like receptors and their crosstalk with other innate receptors in infection and immunity. Immunity. 2011;34:637–650. doi: 10.1016/j.immuni.2011.05.006. [DOI] [PubMed] [Google Scholar]
- 261.De Nardo D, et al. Interleukin-1 receptor-associated kinase 4 (IRAK4) plays a dual role in myddosome formation and Toll-like receptor signaling. J. Biol. Chem. 2018;293:15195–15207. doi: 10.1074/jbc.RA118.003314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Häcker H, et al. Specificity in Toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 263.Verstak B, et al. MyD88 adapter-like (Mal)/TIRAP interaction with TRAF6 is critical for TLR2- and TLR4-mediated NF-kappaB proinflammatory responses. J. Biol. Chem. 2009;284:24192–24203. doi: 10.1074/jbc.M109.023044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat. Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 265.Cohen P, Strickson S. The role of hybrid ubiquitin chains in the MyD88 and other innate immune signalling pathways. Cell Death Differ. 2017;24:1153–1159. doi: 10.1038/cdd.2017.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wei L-Q, Si Z, Qi L-W. Research progress of NOD-like signaling pathways and the relationship between NOD and tumor. Chin. Oncol. 2019;29:223–228. [Google Scholar]
- 267.Platnich JM, Muruve DA. NOD-like receptors and inflammasomes: a review of their canonical and non-canonical signaling pathways. Arch. Biochem. Biophys. 2019;670:4–14. doi: 10.1016/j.abb.2019.02.008. [DOI] [PubMed] [Google Scholar]
- 268.Park JH, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J. Immunol. 2007;178:2380–2386. doi: 10.4049/jimmunol.178.4.2380. [DOI] [PubMed] [Google Scholar]
- 269.Hasegawa M, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–383. doi: 10.1038/sj.emboj.7601962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Zevini A, Olagnier D, Hiscott J. Crosstalk between cytoplasmic RIG-I and STING sensing pathways. Trends Immunol. 2017;38:194–205. doi: 10.1016/j.it.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Hou P, et al. A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection. Cell Res. 2021;31:62–79. doi: 10.1038/s41422-020-0362-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Quicke KM, Diamond MS, Suthar MS. Negative regulators of the RIG-I-like receptor signaling pathway. Eur. J. Immunol. 2017;47:615–628. doi: 10.1002/eji.201646484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Chattopadhyay S, Sen GC. RIG-I-like receptor-induced IRF3 mediated pathway of apoptosis (RIPA): a new antiviral pathway. Protein Cell. 2017;8:165–168. doi: 10.1007/s13238-016-0334-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Eisenächer K, Krug A. Regulation of RLR-mediated innate immune signaling–it is all about keeping the balance. Eur. J. Cell Biol. 2012;91:36–47. doi: 10.1016/j.ejcb.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 275.Rogers NC, et al. Syk-dependent cytokine induction by Dectin-1 reveals a novel pattern recognition pathway for C type lectins. Immunity. 2005;22:507–517. doi: 10.1016/j.immuni.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 276.Leibundgut-Landmann S, Osorio F, Brown GD, Reis e Sousa C. Stimulation of dendritic cells via the dectin-1/Syk pathway allows priming of cytotoxic T-cell responses. Blood. 2008;112:4971–4980. doi: 10.1182/blood-2008-05-158469. [DOI] [PubMed] [Google Scholar]
- 277.Wagener M, Hoving JC, Ndlovu H, Marakalala MJ. Dectin-1-Syk-CARD9 signaling pathway in TB immunity. Front. Immunol. 2018;9:225. doi: 10.3389/fimmu.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Ye XC, et al. Dectin-1/Syk signaling triggers neuroinflammation after ischemic stroke in mice. J. Neuroinflammation. 2020;17:17. doi: 10.1186/s12974-019-1693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science. 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 280.Sakurai K, et al. Cutaneous p38 mitogen-activated protein kinase activation triggers psoriatic dermatitis. J. Allergy Clin. Immunol. 2019;144:1036–1049. doi: 10.1016/j.jaci.2019.06.019. [DOI] [PubMed] [Google Scholar]
- 281.Khorasanizadeh M, Eskian M, Gelfand EW, Rezaei N. Mitogen-activated protein kinases as therapeutic targets for asthma. Pharmacol. Ther. 2017;174:112–126. doi: 10.1016/j.pharmthera.2017.02.024. [DOI] [PubMed] [Google Scholar]
- 282.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413:732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 283.Hsu YM, et al. The adaptor protein CARD9 is required for innate immune responses to intracellular pathogens. Nat. Immunol. 2007;8:198–205. doi: 10.1038/ni1426. [DOI] [PubMed] [Google Scholar]
- 284.Ysebrant de Lendonck L, Martinet V, Goriely S. Interferon regulatory factor 3 in adaptive immune responses. Cell. Mol. Life Sci. 2014;71:3873–3883. doi: 10.1007/s00018-014-1653-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015;347:aaa2630. doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 286.King KR, et al. IRF3 and type I interferons fuel a fatal response to myocardial infarction. Nat. Med. 2017;23:1481–1487. doi: 10.1038/nm.4428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301:640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 288.Samie M, et al. Selective autophagy of the adaptor TRIF regulates innate inflammatory signaling. Nat. Immunol. 2018;19:246–254. doi: 10.1038/s41590-017-0042-6. [DOI] [PubMed] [Google Scholar]
- 289.Wu X, et al. Regulation of TRIF-mediated innate immune response by K27-linked polyubiquitination and deubiquitination. Nat. Commun. 2019;10:4115. doi: 10.1038/s41467-019-12145-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Shalaby KH, et al. The TLR4-TRIF pathway can protect against the development of experimental allergic asthma. Immunology. 2017;152:138–149. doi: 10.1111/imm.12755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Yamamoto M, et al. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 2003;4:1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 292.Banoth B, Cassel SL. Mitochondria in innate immune signaling. Transl. Res. 2018;202:52–68. doi: 10.1016/j.trsl.2018.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Liu XY, Wei B, Shi HX, Shan YF, Wang C. Tom70 mediates activation of interferon regulatory factor 3 on mitochondria. Cell Res. 2010;20:994–1011. doi: 10.1038/cr.2010.103. [DOI] [PubMed] [Google Scholar]
- 294.Zhou R, Zhang Q, Xu PTBK1. a central kinase in innate immune sensing of nucleic acids and beyond. Acta Biochim. Biophys. Sin. 2020;52:757–767. doi: 10.1093/abbs/gmaa051. [DOI] [PubMed] [Google Scholar]
- 295.Rathinam VA, Fitzgerald KA. Inflammasome complexes: emerging mechanisms and effector functions. Cell. 2016;165:792–800. doi: 10.1016/j.cell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–832. doi: 10.1016/j.cell.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 297.Mitchell PS, Sandstrom A, Vance RE. The NLRP1 inflammasome: new mechanistic insights and unresolved mysteries. Curr. Opin. Immunol. 2019;60:37–45. doi: 10.1016/j.coi.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Kelley N, Jeltema D, Duan Y, He Y. The NLRP3 inflammasome: an overview of mechanisms of activation and regulation. Int. J. Mol. Sci. 2019;20:3328. doi: 10.3390/ijms20133328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Duncan JA, Canna SW. The NLRC4 inflammasome. Immunol. Rev. 2018;281:115–123. doi: 10.1111/imr.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Karki R, et al. IRF8 regulates transcription of Naips for NLRC4 inflammasome activation. Cell. 2018;173:920–933. doi: 10.1016/j.cell.2018.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lugrin J, Martinon F. The AIM2 inflammasome: sensor of pathogens and cellular perturbations. Immunol. Rev. 2018;281:99–114. doi: 10.1111/imr.12618. [DOI] [PubMed] [Google Scholar]
- 302.Malik A, Kanneganti TD. Inflammasome activation and assembly at a glance. J. Cell Sci. 2017;130:3955–3963. doi: 10.1242/jcs.207365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Elliott EI, Sutterwala FS. Initiation and perpetuation of NLRP3 inflammasome activation and assembly. Immunol. Rev. 2015;265:35–52. doi: 10.1111/imr.12286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Yang Y, Wang H, Kouadir M, Song H, Shi F. Recent advances in the mechanisms of NLRP3 inflammasome activation and its inhibitors. Cell Death Dis. 2019;10:128. doi: 10.1038/s41419-019-1413-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Zhao C, Zhao W. NLRP3 inflammasome-a key player in antiviral responses. Front. Immunol. 2020;11:211. doi: 10.3389/fimmu.2020.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Wang B, Yin Q. AIM2 inflammasome activation and regulation: a structural perspective. J. Struct. Biol. 2017;200:279–282. doi: 10.1016/j.jsb.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Vesely MD, Kershaw MH, Schreiber RD, Smyth MJ. Natural innate and adaptive immunity to cancer. Annu. Rev. Immunol. 2011;29:235–271. doi: 10.1146/annurev-immunol-031210-101324. [DOI] [PubMed] [Google Scholar]
- 308.Netea MG, Schlitzer A, Placek K, Joosten LAB, Schultze JL. Innate and adaptive immune memory: an evolutionary continuum in the host’s response to pathogens. Cell Host Microbe. 2019;25:13–26. doi: 10.1016/j.chom.2018.12.006. [DOI] [PubMed] [Google Scholar]
- 309.Greten FR, Grivennikov SI. Inflammation and cancer: triggers, mechanisms, and consequences. Immunity. 2019;51:27–41. doi: 10.1016/j.immuni.2019.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Żeromski J, et al. Significance and role of pattern recognition receptors in malignancy. Arch. Immunol. Ther. Exp. 2019;67:133–141. doi: 10.1007/s00005-019-00540-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Hopcraft SE, Damania B. Tumour viruses and innate immunity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017;372:20160267. doi: 10.1098/rstb.2016.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Burn OK, Prasit KK, Hermans IF. Modulating the tumour microenvironment by intratumoural injection of pattern recognition receptor agonists. Cancers. 2020;12:3824. doi: 10.3390/cancers12123824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Hangai, S., Kimura, Y., Taniguchi, T. & Yanai, H. Signal-transducing innate receptors in tumor immunity. Cancer Sci. 10.1111/cas.14848 (2021). [DOI] [PMC free article] [PubMed]
- 314.Shekarian T, et al. Pattern recognition receptors: immune targets to enhance cancer immunotherapy. Ann. Oncol. 2017;28:1756–1766. doi: 10.1093/annonc/mdx179. [DOI] [PubMed] [Google Scholar]
- 315.Rameshbabu S, Labadie BW, Argulian A, Patnaik A. Targeting innate immunity in cancer therapy. Vaccines. 2021;9:138. doi: 10.3390/vaccines9020138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Tang Z, et al. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Weitz J, et al. Colorectal cancer. Lancet. 2005;365:153–165. doi: 10.1016/S0140-6736(05)17706-X. [DOI] [PubMed] [Google Scholar]
- 318.Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients. 2019;11:164. doi: 10.3390/nu11010164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Janney A, Powrie F, Mann EH. Host-microbiota maladaptation in colorectal cancer. Nature. 2020;585:509–517. doi: 10.1038/s41586-020-2729-3. [DOI] [PubMed] [Google Scholar]
- 320.Tilg H, Adolph TE, Gerner RR, Moschen AR. The intestinal microbiota in colorectal cancer. Cancer Cell. 2018;33:954–964. doi: 10.1016/j.ccell.2018.03.004. [DOI] [PubMed] [Google Scholar]
- 321.Hennessy C, O’Connell S, Egan LJ, McKernan DP. Inhibition of anti-viral responses in intestinal epithelial cells by epigenetic modifying drugs is mediated by a reduction in viral pattern recognition receptor expression and activity. Immunopharmacol. Immunotoxicol. 2019;41:527–537. doi: 10.1080/08923973.2019.1661430. [DOI] [PubMed] [Google Scholar]
- 322.Magalhães D, Soares-da-Silva P, Magro F. The effect of PRR ligands on the membrane potential of intestinal epithelial cells. Pharmacol. Rep. 2017;69:978–984. doi: 10.1016/j.pharep.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 323.Abreu MT. Toll-like receptor signalling in the intestinal epithelium: how bacterial recognition shapes intestinal function. Nat. Rev. Immunol. 2010;10:131–144. doi: 10.1038/nri2707. [DOI] [PubMed] [Google Scholar]
- 324.Kuugbee ED, et al. Structural change in microbiota by a probiotic cocktail enhances the gut barrier and reduces cancer via TLR2 signaling in a rat model of colon cancer. Dig. Dis. Sci. 2016;61:2908–2920. doi: 10.1007/s10620-016-4238-7. [DOI] [PubMed] [Google Scholar]
- 325.Meng S, et al. Effect of TLR2 on the proliferation of inflammation-related colorectal cancer and sporadic colorectal cancer. Cancer Cell Int. 2020;20:95. doi: 10.1186/s12935-020-01184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Fukata M, et al. Toll-like receptor-4 promotes the development of colitis-associated colorectal tumors. Gastroenterology. 2007;133:1869–1881. doi: 10.1053/j.gastro.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Cario E. The human TLR4 variant D299G mediates inflammation-associated cancer progression in the intestinal epithelium. Oncoimmunology. 2013;2:e24890. doi: 10.4161/onci.24890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Yesudhas D, Gosu V, Anwar MA, Choi S. Multiple roles of Toll-like receptor 4 in colorectal cancer. Front. Immunol. 2014;5:334. doi: 10.3389/fimmu.2014.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Liu S, et al. FOXP3 inhibits cancer stem cell self-renewal via transcriptional repression of COX2 in colorectal cancer cells. Oncotarget. 2017;8:44694–44704. doi: 10.18632/oncotarget.17974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Hsu HH, et al. Prostaglandin E2-induced COX-2 expressions via EP2 and EP4 signaling pathways in human LoVo colon cancer cells. Int. J. Mol. Sci. 2017;18:1132. doi: 10.3390/ijms18061132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Brackett CM, et al. Toll-like receptor-5 agonist, entolimod, suppresses metastasis and induces immunity by stimulating an NK-dendritic-CD8+ T-cell axis. Proc. Natl Acad. Sci. USA. 2016;113:E874–E883. doi: 10.1073/pnas.1521359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Moradi-Marjaneh R, et al. Toll like receptor signaling pathway as a potential therapeutic target in colorectal cancer. J. Cell. Physiol. 2018;233:5613–5622. doi: 10.1002/jcp.26273. [DOI] [PubMed] [Google Scholar]
- 333.Gao C, et al. TLR9 signaling activation at different stages in colorectal cancer and NF-kappaB expression. Onco Targets Ther. 2018;11:5963–5971. doi: 10.2147/OTT.S174274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Alvarez-Arellano L, et al. TLR9 and NF-κB are partially involved in activation of human neutrophils by Helicobacter pylori and its purified DNA. PLoS ONE. 2014;9:e101342. doi: 10.1371/journal.pone.0101342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Merrell MA, et al. Toll-like receptor 9 agonists promote cellular invasion by increasing matrix metalloproteinase activity. Mol. Cancer Res. 2006;4:437–447. doi: 10.1158/1541-7786.MCR-06-0007. [DOI] [PubMed] [Google Scholar]
- 336.Rath T, et al. Matrix metalloproteinase-13 is regulated by toll-like receptor-9 in colorectal cancer cells and mediates cellular migration. Oncol. Lett. 2011;2:483–488. doi: 10.3892/ol.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Jiang HY, et al. Activation of the pattern recognition receptor NOD1 augments colon cancer metastasis. Protein Cell. 2020;11:187–201. doi: 10.1007/s13238-019-00687-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Tarao K, et al. Inflammation in background cirrhosis evokes malignant progression in HCC development from HCV-associated liver cirrhosis. Scand. J. Gastroenterol. 2013;48:729–735. doi: 10.3109/00365521.2013.782064. [DOI] [PubMed] [Google Scholar]
- 339.Li H, et al. Expressions of TLR2 and TLR4 protein and their relationship tomutations in genes in hepatocellular carcinoma. Chin J. Cancer Prev. Treat. 2014;21:401–406. [Google Scholar]
- 340.Junjie X, et al. The association between Toll-like receptor 2 single-nucleotide polymorphisms and hepatocellular carcinoma susceptibility. BMC Cancer. 2012;12:57. doi: 10.1186/1471-2407-12-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Nischalke HD, et al. The Toll-like receptor 2 (TLR2) -196 to -174 del/ins polymorphism affects viral loads and susceptibility to hepatocellular carcinoma in chronic hepatitis C. Int. J. Cancer. 2012;130:1470–1475. doi: 10.1002/ijc.26143. [DOI] [PubMed] [Google Scholar]
- 342.Kairaluoma V, Kemi N, Huhta H, Pohjanen VM, Helminen O. Prognostic role of TLR4 and TLR2 in hepatocellular carcinoma. Acta Oncol. 2021;60:554–558. doi: 10.1080/0284186X.2021.1877346. [DOI] [PubMed] [Google Scholar]
- 343.Chew V, et al. Toll-like receptor 3 expressing tumor parenchyma and infiltrating natural killer cells in hepatocellular carcinoma patients. J. Natl Cancer Inst. 2012;104:1796–1807. doi: 10.1093/jnci/djs436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Zhou S, et al. TLR4 increases the stemness and is highly expressed in relapsed human hepatocellular carcinoma. Cancer Med. 2019;8:2325–2337. doi: 10.1002/cam4.2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Lin A, et al. TLR4 signaling promotes a COX-2/PGE2/STAT3 positive feedback loop in hepatocellular carcinoma (HCC) cells. Oncoimmunology. 2015;5:e1074376. doi: 10.1080/2162402X.2015.1074376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Zhou B, et al. Hepatoma cell-intrinsic TLR9 activation induces immune escape through PD-L1 upregulation in hepatocellular carcinoma. Theranostics. 2020;10:6530–6543. doi: 10.7150/thno.44417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Song IJ, et al. The contribution of toll-like receptor signaling to the development of liver fibrosis and cancer in hepatocyte-specific TAK1-deleted mice. Int. J. Cancer. 2018;142:81–91. doi: 10.1002/ijc.31029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Bonnin, M. et al. Toll-like receptor 3 downregulation is an escape mechanism from apoptosis during hepatocarcinogenesis J. Hepatol.71, 763–772 (2019); corrigendum 72, 594 (2020). [DOI] [PubMed]
- 349.Yu LX, Schwabe RF. The gut microbiome and liver cancer: mechanisms and clinical translation. Nat. Rev. Gastroenterol. Hepatol. 2017;14:527–539. doi: 10.1038/nrgastro.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Zhou Y, et al. Hepatic NOD2 promotes hepatocarcinogenesis via a RIP2-mediated proinflammatory response and a novel nuclear autophagy-mediated DNA damage mechanism. J. Hematol. Oncol. 2021;14:9. doi: 10.1186/s13045-020-01028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Caruso R, Warner N, Inohara N, Núñez G. NOD1 and NOD2: signaling, host defense, and inflammatory disease. Immunity. 2014;41:898–908. doi: 10.1016/j.immuni.2014.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Faloppi L, et al. Lactate dehydrogenase in hepatocellular carcinoma: something old, something new. Biomed. Res. Int. 2016;2016:7196280. doi: 10.1155/2016/7196280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Woolston C. Breast cancer. Nature. 2015;527:S101. doi: 10.1038/527S101a. [DOI] [PubMed] [Google Scholar]
- 354.Bhatelia K, Singh K, Singh R. TLRs: linking inflammation and breast cancer. Cell. Signal. 2014;26:2350–2357. doi: 10.1016/j.cellsig.2014.07.035. [DOI] [PubMed] [Google Scholar]
- 355.Hummell DS, Winkelstein JA. Bacterial lipoteichoic acid sensitizes host cells for destruction by autologous complement. J. Clin. Investig. 1986;77:1533–1538. doi: 10.1172/JCI112468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Schröder NW, et al. Lipoteichoic acid (LTA) of Streptococcus pneumoniae and Staphylococcus aureus activates immune cells via Toll-like receptor (TLR)-2, lipopolysaccharide-binding protein (LBP), and CD14, whereas TLR-4 and MD-2 are not involved. J. Biol. Chem. 2003;278:15587–15594. doi: 10.1074/jbc.M212829200. [DOI] [PubMed] [Google Scholar]
- 357.Schafer ZT, Brugge JS. IL-6 involvement in epithelial cancers. J. Clin. Investig. 2007;117:3660–3663. doi: 10.1172/JCI34237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Sansone P, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J. Clin. Investig. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Di Domenico M, Ricciardi C, Fusco A, Pierantoni GM. Anti-VEGF therapy in breast and lung mouse models of cancers. J. Biomed. Biotechnol. 2011;2011:947928. doi: 10.1155/2011/947928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Liao SJ, et al. Triggering of Toll-like receptor 4 on metastatic breast cancer cells promotes αvβ3-mediated adhesion and invasive migration. Breast Cancer Res. Treat. 2012;133:853–863. doi: 10.1007/s10549-011-1844-0. [DOI] [PubMed] [Google Scholar]
- 361.Moskovitz J, Moy J, Ferris RL. Immunotherapy for head and neck squamous cell carcinoma. Curr. Oncol. Rep. 2018;20:22. doi: 10.1007/s11912-018-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Solomon B, Young RJ, Rischin D. Head and neck squamous cell carcinoma: Genomics and emerging biomarkers for immunomodulatory cancer treatments. Semin. Cancer Biol. 2018;52:228–240. doi: 10.1016/j.semcancer.2018.01.008. [DOI] [PubMed] [Google Scholar]
- 363.Oliva M, et al. Immune biomarkers of response to immune-checkpoint inhibitors in head and neck squamous cell carcinoma. Ann. Oncol. 2019;30:57–67. doi: 10.1093/annonc/mdy507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Waldman A, Schmults C. Cutaneous squamous cell carcinoma. Hematol. Oncol. Clin. North Am. 2019;33:1–12. doi: 10.1016/j.hoc.2018.08.001. [DOI] [PubMed] [Google Scholar]
- 365.Mäkinen LK, et al. Toll-like receptors 2, 4, and 9 in primary, metastasized, and recurrent oral tongue squamous cell carcinomas. J. Oral. Pathol. Med. 2016;45:338–345. doi: 10.1111/jop.12373. [DOI] [PubMed] [Google Scholar]
- 366.Mäkinen LK, et al. Predictive role of Toll-like receptors 2, 4, and 9 in oral tongue squamous cell carcinoma. Oral. Oncol. 2015;51:96–102. doi: 10.1016/j.oraloncology.2014.08.017. [DOI] [PubMed] [Google Scholar]
- 367.Yoon HE, et al. Nucleotide-binding oligomerization domain 2 (NOD2) activation induces apoptosis of human oral squamous cell carcinoma cells. J. Oral. Pathol. Med. 2016;45:262–267. doi: 10.1111/jop.12354. [DOI] [PubMed] [Google Scholar]
- 368.van de Veerdonk FL, Gresnigt MS, Romani L, Netea MG, Latgé JP. Aspergillus fumigatus morphology and dynamic host interactions. Nat. Rev. Microbiol. 2017;15:661–674. doi: 10.1038/nrmicro.2017.90. [DOI] [PubMed] [Google Scholar]
- 369.Latgé JP, Chamilos G. Aspergillus fumigatus and Aspergillosis in 2019. Clin. Microbiol. Rev. 2019;33:e00140–18. doi: 10.1128/CMR.00140-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Zhang PP, et al. Toll-like receptor 2 and dectin-1 function as promising biomarker for Aspergillus fumigatus infection. Exp. Ther. Med. 2017;14:3836–3840. doi: 10.3892/etm.2017.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Percier P, et al. Aspergillus fumigatus recognition by dendritic cells negatively regulates allergic lung inflammation through a TLR2/MyD88 pathway. Am. J. Respir. Cell Mol. Biol. 2021;64:39–49. doi: 10.1165/rcmb.2020-0083OC. [DOI] [PubMed] [Google Scholar]
- 372.Murakami Y, et al. TLR9-IL-2 axis exacerbates allergic asthma by preventing IL-17A hyperproduction. Sci. Rep. 2020;10:18110. doi: 10.1038/s41598-020-75153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Ransohoff RM, Brown MA. Innate immunity in the central nervous system. J. Clin. Investig. 2012;122:1164–1171. doi: 10.1172/JCI58644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Klein RS, Garber C, Howard N. Infectious immunity in the central nervous system and brain function. Nat. Immunol. 2017;18:132–141. doi: 10.1038/ni.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Kipnis J. Multifaceted interactions between adaptive immunity and the central nervous system. Science. 2016;353:766–771. doi: 10.1126/science.aag2638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 376.Keogh CE, Rude KM, Gareau MG. Role of pattern recognition receptors and the microbiota in neurological disorders. J. Physiol. 2021;599:1379–1389. doi: 10.1113/JP279771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Dick A, Provencal N. Central neuroepigenetic regulation of the hypothalamic-pituitary-adrenal axis. Prog. Mol. Biol. Transl. Sci. 2018;158:105–127. doi: 10.1016/bs.pmbts.2018.04.006. [DOI] [PubMed] [Google Scholar]
- 378.Pusceddu MM, et al. Nod-like receptors are critical for gut-brain axis signalling in mice. J. Physiol. 2019;597:5777–5797. doi: 10.1113/JP278640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Pariante CM. Neuroscience, mental health and the immune system: overcoming the brain-mind-body trichotomy. Epidemiol. Psychiatr. Sci. 2016;25:101–105. doi: 10.1017/S204579601500089X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Hill AS, Sahay A, Hen R. Increasing adult hippocampal neurogenesis is sufficient to reduce anxiety and depression-like behaviors. Neuropsychopharmacology. 2015;40:2368–2378. doi: 10.1038/npp.2015.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Yadav S, Surolia A. Lysozyme elicits pain during nerve injury by neuronal Toll-like receptor 4 activation and has therapeutic potential in neuropathic pain. Sci. Transl. Med. 2019;11:eaav4176. doi: 10.1126/scitranslmed.aav4176. [DOI] [PubMed] [Google Scholar]
- 382.Tanga FY, Nutile-McMenemy N, DeLeo JA. The CNS role of Toll-like receptor 4 in innate neuroimmunity and painful neuropathy. Proc. Natl Acad. Sci. USA. 2005;102:5856–5861. doi: 10.1073/pnas.0501634102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Guo LH, Schluesener HJ. The innate immunity of the central nervous system in chronic pain: the role of Toll-like receptors. Cell. Mol. Life Sci. 2007;64:1128–1136. doi: 10.1007/s00018-007-6494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Poddar U, et al. Neonatal cholestasis: differentiation of biliary atresia from neonatal hepatitis in a developing country. Acta Paediatr. 2009;98:1260–1264. doi: 10.1111/j.1651-2227.2009.01338.x. [DOI] [PubMed] [Google Scholar]
- 385.Huang YH, et al. Expression of toll-like receptors and type 1 interferon specific protein MxA in biliary atresia. Lab. Invest. 2007;87:66–74. doi: 10.1038/labinvest.3700490. [DOI] [PubMed] [Google Scholar]
- 386.Huang YH, et al. Toll-like receptor 7 agonist induces hypoplasia of the biliary system in a neonatal mouse model. J. Microbiol. Immunol. Infect. 2018;51:166–173. doi: 10.1016/j.jmii.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 387.Price AE, et al. A map of toll-like receptor expression in the intestinal epithelium reveals distinct spatial, cell type-specific, and temporal patterns. Immunity. 2018;49:560–575. doi: 10.1016/j.immuni.2018.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Zhang B, et al. Viral infection. Prevention and cure of rotavirus infection via TLR5/NLRC4-mediated production of IL-22 and IL-18. Science. 2014;346:861–865. doi: 10.1126/science.1256999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Vijay-Kumar M, et al. Deletion of TLR5 results in spontaneous colitis in mice. J. Clin. Investig. 2007;117:3909–3921. doi: 10.1172/JCI33084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Yan L, et al. Pyrimidine triazole thioether derivatives as toll-like receptor 5 (TLR5)/flagellin complex inhibitors. ChemMedChem. 2016;11:822–826. doi: 10.1002/cmdc.201500471. [DOI] [PubMed] [Google Scholar]
- 391.Zhou Z, et al. Exogenous activation of Toll-like receptor 5 signaling mitigates acetaminophen-induced hepatotoxicity in mice. Toxicol. Lett. 2021;342:58–72. doi: 10.1016/j.toxlet.2021.01.022. [DOI] [PubMed] [Google Scholar]
- 392.Jalan R, et al. Acute-on chronic liver failure. J. Hepatol. 2012;57:1336–1348. doi: 10.1016/j.jhep.2012.06.026. [DOI] [PubMed] [Google Scholar]
- 393.Gustot T, et al. Clinical course of acute-on-chronic liver failure syndrome and effects on prognosis. Hepatology. 2015;62:243–252. doi: 10.1002/hep.27849. [DOI] [PubMed] [Google Scholar]
- 394.Singal AK, Bataller R, Ahn J, Kamath PS, Shah VH. ACG clinical guideline: alcoholic liver disease. Am. J. Gastroenterol. 2018;113:175–194. doi: 10.1038/ajg.2017.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Boussif A, et al. Impaired intracellular signaling, myeloperoxidase release and bactericidal activity of neutrophils from patients with alcoholic cirrhosis. J. Hepatol. 2016;64:1041–1048. doi: 10.1016/j.jhep.2015.12.005. [DOI] [PubMed] [Google Scholar]
- 396.Rolas L, et al. NADPH oxidase depletion in neutrophils from patients with cirrhosis and restoration via toll-like receptor 7/8 activation. Gut. 2018;67:1505–1516. doi: 10.1136/gutjnl-2016-313443. [DOI] [PubMed] [Google Scholar]
- 397.Bajaj JS. Alcohol, liver disease and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2019;16:235–246. doi: 10.1038/s41575-018-0099-1. [DOI] [PubMed] [Google Scholar]
- 398.Lin J, Kakkar V, Lu X. Essential roles of Toll-like receptors in atherosclerosis. Curr. Med. Chem. 2016;23:431–454. doi: 10.2174/0929867323666151207111408. [DOI] [PubMed] [Google Scholar]
- 399.Falck-Hansen M, Kassiteridi C, Monaco C. Toll-like receptors in atherosclerosis. Int. J. Mol. Sci. 2013;14:14008–14023. doi: 10.3390/ijms140714008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Li B, Xia Y, Hu B. Infection and atherosclerosis: TLR-dependent pathways. Cell. Mol. Life Sci. 2020;77:2751–2769. doi: 10.1007/s00018-020-03453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Wolf D, Ley K. Immunity and inflammation in atherosclerosis. Circ. Res. 2019;124:315–327. doi: 10.1161/CIRCRESAHA.118.313591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Karadimou G, et al. Low TLR7 gene expression in atherosclerotic plaques is associated with major adverse cardio- and cerebrovascular events. Cardiovasc. Res. 2017;113:30–39. doi: 10.1093/cvr/cvw231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Fukuda D, et al. Toll-like receptor 9 plays a pivotal role in angiotensin II-induced atherosclerosis. J. Am. Heart Assoc. 2019;8:e010860. doi: 10.1161/JAHA.118.010860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Libby P, Ridker PM, Hansson GK, Ather LTN. Inflammation in atherosclerosis from pathophysiology to practice. J. Am. Coll. Cardiol. 2009;54:2129–2138. doi: 10.1016/j.jacc.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Gisterå A, Hansson GK. The immunology of atherosclerosis. Nat. Rev. Nephrol. 2017;13:368–380. doi: 10.1038/nrneph.2017.51. [DOI] [PubMed] [Google Scholar]
- 406.Kim J, Seo M, Kim SK, Bae YS. Flagellin-induced NADPH oxidase 4 activation is involved in atherosclerosis. Sci. Rep. 2016;6:25437. doi: 10.1038/srep25437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Kim J, et al. The flagellin-TLR5-Nox4 axis promotes the migration of smooth muscle cells in atherosclerosis. Exp. Mol. Med. 2019;51:1–13. doi: 10.1038/s12276-019-0275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Lee GL, et al. TLR2 promotes vascular smooth muscle cell chondrogenic differentiation and consequent calcification via the concerted actions of osteoprotegerin suppression and IL-6-mediated RANKL induction. Arterioscler. Thromb. Vasc. Biol. 2019;39:432–445. doi: 10.1161/ATVBAHA.118.311874. [DOI] [PubMed] [Google Scholar]
- 409.Farkas D, et al. Toll-like receptor 3 is a therapeutic target for pulmonary hypertension. Am. J. Respir. Crit. Care Med. 2019;199:199–210. doi: 10.1164/rccm.201707-1370OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Vlacil AK, et al. Deficiency of nucleotide-binding oligomerization domain-containing proteins (NOD) 1 and 2 reduces atherosclerosis. Basic Res. Cardiol. 2020;115:47. doi: 10.1007/s00395-020-0806-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Armulik A, Genové G, Betsholtz C. Pericytes: developmental, physiological, and pathological perspectives, problems, and promises. Dev. Cell. 2011;21:193–215. doi: 10.1016/j.devcel.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 412.Nyúl-Tóth Á, et al. Expression of pattern recognition receptors and activation of the non-canonical inflammasome pathway in brain pericytes. Brain Behav. Immun. 2017;64:220–231. doi: 10.1016/j.bbi.2017.04.010. [DOI] [PubMed] [Google Scholar]
- 413.Navarro R, et al. Role of nucleotide-binding oligomerization domain 1 (NOD1) in pericyte-mediated vascular inflammation. J. Cell Mol. Med. 2016;20:980–986. doi: 10.1111/jcmm.12804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Mravec B, Gidron Y, Hulin I. Neurobiology of cancer: interactions between nervous, endocrine and immune systems as a base for monitoring and modulating the tumorigenesis by the brain. Semin. Cancer Biol. 2008;18:150–163. doi: 10.1016/j.semcancer.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 415.Del Rey A, Besedovsky HO. Immune-neuro-endocrine reflexes, circuits, and networks: physiologic and evolutionary implications. Front. Horm. Res. 2017;48:1–18. doi: 10.1159/000452902. [DOI] [PubMed] [Google Scholar]
- 416.Glaser R, Kiecolt-Glaser JK. Stress-induced immune dysfunction: implications for health. Nat. Rev. Immunol. 2005;5:243–251. doi: 10.1038/nri1571. [DOI] [PubMed] [Google Scholar]
- 417.Renner U, Sapochnik M, Lucia K, Stalla GK, Arzt E. Intrahypophyseal immune-endocrine interactions: endocrine integration of the inflammatory inputs. Front. Horm. Res. 2017;48:37–47. doi: 10.1159/000452904. [DOI] [PubMed] [Google Scholar]
- 418.Peng, S. et al. Increased Toll-like receptors activity and TLR ligands in patients with autoimmune thyroid diseases. Front. Immunol. 7, 578 (2016); correction 8, 551 (2017). [DOI] [PMC free article] [PubMed]
- 419.Dvornikova KA, Bystrova EY, Platonova ON, Churilov LP. Polymorphism of Toll-like receptor genes and autoimmune endocrine diseases. Autoimmun. Rev. 2020;19:102496. doi: 10.1016/j.autrev.2020.102496. [DOI] [PubMed] [Google Scholar]
- 420.Liu M, et al. Toll-like receptor 9 negatively regulates pancreatic islet beta cell growth and function in a mouse model of type 1 diabetes. Diabetologia. 2018;61:2333–2343. doi: 10.1007/s00125-018-4705-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Ji Y, et al. Toll-like receptors TLR2 and TLR4 block the replication of pancreatic β cells in diet-induced obesity. Nat. Immunol. 2019;20:677–686. doi: 10.1038/s41590-019-0396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 422.Grennan D. Diabetic foot ulcers. JAMA. 2019;321:114. doi: 10.1001/jama.2018.18323. [DOI] [PubMed] [Google Scholar]
- 423.Armstrong DG, Boulton AJM, Bus SA. Diabetic foot ulcers and their recurrence. N. Engl. J. Med. 2017;376:2367–2375. doi: 10.1056/NEJMra1615439. [DOI] [PubMed] [Google Scholar]
- 424.Singh K, et al. Increased expression of endosomal members of toll-like receptor family abrogates wound healing in patients with type 2 diabetes mellitus. Int. Wound J. 2016;13:927–935. doi: 10.1111/iwj.12411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Guiducci C, et al. Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9. J. Exp. Med. 2010;207:2931–2942. doi: 10.1084/jem.20101048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 426.McKelvey KJ, Highton J, Hessian PA. Cell-specific expression of TLR9 isoforms in inflammation. J. Autoimmun. 2011;36:76–86. doi: 10.1016/j.jaut.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 427.Marshak-Rothstein A, Rifkin IR. Immunologically active autoantigens: the role of toll-like receptors in the development of chronic inflammatory disease. Annu. Rev. Immunol. 2007;25:419–441. doi: 10.1146/annurev.immunol.22.012703.104514. [DOI] [PubMed] [Google Scholar]
- 428.Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 2014;14:181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 429.Han G, Ceilley R. Chronic wound healing: a review of current management and treatments. Adv. Ther. 2017;34:599–610. doi: 10.1007/s12325-017-0478-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Cavalcante P, et al. Increased expression of Toll-like receptors 7 and 9 in myasthenia gravis thymus characterized by active Epstein-Barr virus infection. Immunobiology. 2016;221:516–527. doi: 10.1016/j.imbio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 431.Hwang HS, Lee MH, Choi MH, Kim HA. NOD2 signaling pathway is involved in fibronectin fragment-induced pro-catabolic factor expressions in human articular chondrocytes. BMB Rep. 2019;52:373–378. doi: 10.5483/BMBRep.2019.52.6.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 432.Hwang HS, Park SJ, Cheon EJ, Lee MH, Kim HA. Fibronectin fragment-induced expression of matrix metalloproteinases is mediated by MyD88-dependent TLR-2 signaling pathway in human chondrocytes. Arthritis Res. Ther. 2015;17:320. doi: 10.1186/s13075-015-0833-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 433.Hoffman NJ, et al. Global phosphoproteomic analysis of human skeletal muscle reveals a network of exercise-regulated kinases and AMPK substrates. Cell Metab. 2015;22:922–935. doi: 10.1016/j.cmet.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Liu Y, et al. TLR9 and beclin 1 crosstalk regulates muscle AMPK activation in exercise. Nature. 2020;578:605–609. doi: 10.1038/s41586-020-1992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 435.Aleynick M, Svensson-Arvelund J, Flowers CR, Marabelle A, Brody JD. Pathogen molecular pattern receptor agonists: treating cancer by mimicking infection. Clin. Cancer Res. 2019;25:6283–6294. doi: 10.1158/1078-0432.CCR-18-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 436.Cao X. Self-regulation and cross-regulation of pattern-recognition receptor signalling in health and disease. Nat. Rev. Immunol. 2016;16:35–50. doi: 10.1038/nri.2015.8. [DOI] [PubMed] [Google Scholar]
- 437.Pizzolla A, Smith JM, Brooks AG, Reading PC. Pattern recognition receptor immunomodulation of innate immunity as a strategy to limit the impact of influenza virus. J. Leukoc. Biol. 2017;101:851–861. doi: 10.1189/jlb.4MR0716-290R. [DOI] [PubMed] [Google Scholar]
- 438.Fabbri M. TLRs as miRNA receptors. Cancer Res. 2012;72:6333–6337. doi: 10.1158/0008-5472.CAN-12-3229. [DOI] [PubMed] [Google Scholar]
- 439.Nabet BY, et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell. 2017;170:352.e13–366.e13. doi: 10.1016/j.cell.2017.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Hemmi H, et al. Small anti-viral compounds activate immune cells via the TLR7 MyD88-dependent signaling pathway. Nat. Immunol. 2002;3:196–200. doi: 10.1038/ni758. [DOI] [PubMed] [Google Scholar]
- 441.Gane, E. J. et al. Safety, pharmacokinetics, and pharmacodynamics of the oral TLR8 agonist selgantolimod in chronic hepatitis B. Hepatology10.1002/hep.31795 (2021). [DOI] [PubMed]
- 442.Amin, O. E. et al. Therapeutic potential of TLR8 agonist GS-9688 (selgantolimod) in chronic hepatitis B: re-modelling of antiviral and regulatory mediators. Hepatology10.1002/hep.31695 (2020). [DOI] [PMC free article] [PubMed]
- 443.Smith DA, et al. Antitumor activity and safety of combination therapy with the Toll-like receptor 9 agonist IMO-2055, erlotinib, and bevacizumab in advanced or metastatic non-small cell lung cancer patients who have progressed following chemotherapy. Cancer Immunol. Immunother. 2014;63:787–796. doi: 10.1007/s00262-014-1547-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 444.Krarup AR, et al. The TLR9 agonist MGN1703 triggers a potent type I interferon response in the sigmoid colon. Mucosal Immunol. 2018;11:449–461. doi: 10.1038/mi.2017.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Owen AM, Fults JB, Patil NK, Hernandez A, Bohannon JK. TLR agonists as mediators of trained immunity: mechanistic insight and immunotherapeutic potential to combat infection. Front. Immunol. 2021;11:622614. doi: 10.3389/fimmu.2020.622614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Cui L, Wang X, Zhang D. TLRs as a promise target along with immune checkpoint against gastric cancer. Front. Cell Dev. Biol. 2021;8:611444. doi: 10.3389/fcell.2020.611444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 447.Takeda Y, et al. A TLR3-specific adjuvant relieves innate resistance to PD-L1 blockade without cytokine toxicity in tumor vaccine immunotherapy. Cell Rep. 2017;19:1874–1887. doi: 10.1016/j.celrep.2017.05.015. [DOI] [PubMed] [Google Scholar]
- 448.Hu HG, Wu JJ, Zhang BD, Li WH, Li YM. Pam3CSK4-CDGSF augments antitumor immunotherapy by synergistically activating TLR1/2 and STING. Bioconjug. Chem. 2020;31:2499–2503. doi: 10.1021/acs.bioconjchem.0c00522. [DOI] [PubMed] [Google Scholar]
- 449.He Y, Hara H, Núñez G. Mechanism and regulation of NLRP3 inflammasome activation. Trends Biochem. Sci. 2016;41:1012–1021. doi: 10.1016/j.tibs.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 450.Liu P, et al. NOD-like receptor signaling in inflammation-associated cancers: from functions to targeted therapies. Phytomedicine. 2019;64:152925. doi: 10.1016/j.phymed.2019.152925. [DOI] [PubMed] [Google Scholar]
- 451.Londhe P, Zhu B, Abraham J, Keller C, Davie J. CIITA is silenced by epigenetic mechanisms that prevent the recruitment of transactivating factors in rhabdomyosarcoma cells. Int. J. Cancer. 2012;131:E437–E448. doi: 10.1002/ijc.26478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Cycon KA, Mulvaney K, Rimsza LM, Persky D, Murphy SP. Histone deacetylase inhibitors activate CIITA and MHC class II antigen expression in diffuse large B-cell lymphoma. Immunology. 2013;140:259–272. doi: 10.1111/imm.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Lee J, et al. Sulforaphane attenuates activation of NLRP3 and NLRC4 inflammasomes but not AIM2 inflammasome. Cell Immunol. 2016;306-307:53–60. doi: 10.1016/j.cellimm.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 454.Guo W, et al. Small molecule-driven mitophagy-mediated NLRP3 inflammasome inhibition is responsible for the prevention of colitis-associated cancer. Autophagy. 2014;10:972–985. doi: 10.4161/auto.28374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Zhao Y, et al. Small molecule GL-V9 protects against colitis-associated colorectal cancer by limiting NLRP3 inflammasome through autophagy. Oncoimmunology. 2017;7:e1375640. doi: 10.1080/2162402X.2017.1375640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 456.Abderrazak A, et al. Anti-inflammatory and antiatherogenic effects of the NLRP3 inflammasome inhibitor arglabin in ApoE2.Ki mice fed a high-fat diet. Circulation. 2015;131:1061–1070. doi: 10.1161/CIRCULATIONAHA.114.013730. [DOI] [PubMed] [Google Scholar]
- 457.Wu D, et al. Formononetin administration ameliorates dextran sulfate sodium-induced acute colitis by inhibiting NLRP3 inflammasome signaling pathway. Mediators Inflamm. 2018;2018:3048532. doi: 10.1155/2018/3048532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Zhang W, et al. Lactate is a natural suppressor of RLR signaling by targeting MAVS. Cell. 2019;178:176–189.e15. doi: 10.1016/j.cell.2019.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Domankevich V, et al. RIG-1-like receptor activation synergizes with intratumoral alpha radiation to induce pancreatic tumor rejection, triple-negative breast metastases clearance, and antitumor immune memory in mice. Front. Oncol. 2020;10:990. doi: 10.3389/fonc.2020.00990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Patra MC, Choi S. Recent progress in the development of Toll-like receptor (TLR) antagonists. Expert Opin. Ther. Pat. 2016;26:719–730. doi: 10.1080/13543776.2016.1185415. [DOI] [PubMed] [Google Scholar]
- 461.Balak DM, et al. IMO-8400, a toll-like receptor 7, 8, and 9 antagonist, demonstrates clinical activity in a phase 2a, randomized, placebo-controlled trial in patients with moderate-to-severe plaque psoriasis. Clin. Immunol. 2017;174:63–72. doi: 10.1016/j.clim.2016.09.015. [DOI] [PubMed] [Google Scholar]
- 462.Lai CY, Su YW, Lin KI, Hsu LC, Chuang TH. Natural modulators of endosomal Toll-like receptor-mediated psoriatic skin inflammation. J. Immunol. Res. 2017;2017:7807313. doi: 10.1155/2017/7807313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Calore F, et al. The TLR7/8/9 antagonist IMO-8503 inhibits cancer-induced cachexia. Cancer Res. 2018;78:6680–6690. doi: 10.1158/0008-5472.CAN-17-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Suárez-Fariñas M, et al. Suppression of molecular inflammatory pathways by Toll-like receptor 7, 8, and 9 antagonists in a model of IL-23-induced skin inflammation. PLoS ONE. 2013;8:e84634. doi: 10.1371/journal.pone.0084634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 465.Mohamed FE, et al. Effect of toll-like receptor 7 and 9 targeted therapy to prevent the development of hepatocellular carcinoma. Liver Int. 2015;35:1063–1076. doi: 10.1111/liv.12626. [DOI] [PubMed] [Google Scholar]
- 466.Li, C., Wang, J., Zhao, M., Zhang, S. & Zhang, Y. Toll-like receptor 4 antagonist FP7 alleviates lipopolysaccharide-induced septic shock via NF-kB signaling pathway. Chem. Biol. Drug Des.10.1111/cbdd.13837 (2021). [DOI] [PubMed]
- 467.Sacre S, et al. Oligodeoxynucleotide inhibition of Toll-like receptors 3, 7, 8, and 9 suppresses cytokine production in a human rheumatoid arthritis model. Eur. J. Immunol. 2016;46:772–781. doi: 10.1002/eji.201546123. [DOI] [PubMed] [Google Scholar]
- 468.Yaw ACK, Chan EWL, Yap JKY, Mai CW. The effects of NLRP3 inflammasome inhibition by MCC950 on LPS-induced pancreatic adenocarcinoma inflammation. J. Cancer Res. Clin. Oncol. 2020;146:2219–2229. doi: 10.1007/s00432-020-03274-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 469.Yang G, et al. Direct binding to NLRP3 pyrin domain as a novel strategy to prevent NLRP3-driven inflammation and gouty arthritis. Arthritis Rheumatol. 2020;72:1192–1202. doi: 10.1002/art.41245. [DOI] [PubMed] [Google Scholar]
- 470.Zhang X, et al. Development of small molecule inhibitors targeting NLRP3 inflammasome pathway for inflammatory diseases. Eur. J. Med. Chem. 2020;185:111822. doi: 10.1016/j.ejmech.2019.111822. [DOI] [PubMed] [Google Scholar]
- 471.Coll RC, et al. A small-molecule inhibitor of the NLRP3 inflammasome for the treatment of inflammatory diseases. Nat. Med. 2015;21:248–255. doi: 10.1038/nm.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 472.Dai Z, et al. Development of novel tetrahydroquinoline inhibitors of NLRP3 inflammasome for potential treatment of DSS-induced mouse colitis. J. Med. Chem. 2021;64:871–889. doi: 10.1021/acs.jmedchem.0c01924. [DOI] [PubMed] [Google Scholar]
- 473.Agarwal S, et al. Identification of a novel orally bioavailable NLRP3 inflammasome inhibitor. Bioorg. Med. Chem. Lett. 2020;30:127571. doi: 10.1016/j.bmcl.2020.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 474.Sparks JA. Rheumatoid arthritis. Ann. Intern. Med. 2019;170:ITC1–ITC16. doi: 10.7326/AITC201901010. [DOI] [PubMed] [Google Scholar]
- 475.Burmester GR, Pope JE. Novel treatment strategies in rheumatoid arthritis. Lancet. 2017;389:2338–2348. doi: 10.1016/S0140-6736(17)31491-5. [DOI] [PubMed] [Google Scholar]
- 476.Nygaard G, Firestein GS. Restoring synovial homeostasis in rheumatoid arthritis by targeting fibroblast-like synoviocytes. Nat. Rev. Rheumatol. 2020;16:316–333. doi: 10.1038/s41584-020-0413-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Zhang F, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat. Immunol. 2019;20:928–942. doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 478.Croft AP, et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature. 2019;570:246–251. doi: 10.1038/s41586-019-1263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 479.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc. Natl Acad. Sci. USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 480.Ceppi M, et al. MicroRNA-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl Acad. Sci. USA. 2009;106:2735–2740. doi: 10.1073/pnas.0811073106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 481.Li Z, Cai J, Cao X. MiR-19 suppresses fibroblast-like synoviocytes cytokine release by targeting toll like receptor 2 in rheumatoid arthritis. Am. J. Transl. Res. 2016;8:5512–5518. [PMC free article] [PubMed] [Google Scholar]
- 482.Philippe L, et al. MiR-20a regulates ASK1 expression and TLR4-dependent cytokine release in rheumatoid fibroblast-like synoviocytes. Ann. Rheum. Dis. 2013;72:1071–1079. doi: 10.1136/annrheumdis-2012-201654. [DOI] [PubMed] [Google Scholar]
- 483.Mu N, et al. A novel NF-κB/YY1/microRNA-10a regulatory circuit in fibroblast-like synoviocytes regulates inflammation in rheumatoid arthritis. Sci. Rep. 2016;6:20059. doi: 10.1038/srep20059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Ingle H, et al. The microRNA miR-485 targets host and influenza virus transcripts to regulate antiviral immunity and restrict viral replication. Sci. Signal. 2015;8:ra126. doi: 10.1126/scisignal.aab3183. [DOI] [PubMed] [Google Scholar]
- 485.Lukiw WJ, Pogue AI. Vesicular transport of encapsulated microRNA between glial and neuronal cells. Int. J. Mol. Sci. 2020;21:5078. doi: 10.3390/ijms21145078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Alexandrov P, Zhai Y, Li W, Lukiw W. Lipopolysaccharide-stimulated, NF-kB-, miRNA-146a- and miRNA-155-mediated molecular-genetic communication between the human gastrointestinal tract microbiome and the brain. Folia Neuropathol. 2019;57:211–219. doi: 10.5114/fn.2019.88449. [DOI] [PubMed] [Google Scholar]
- 487.Guo HY, Cheng AC, Wang MS, Yin ZQ, Jia RY. Exosomes: potential therapies for disease via regulating TLRs. Mediators Inflamm. 2020;2020:2319616. doi: 10.1155/2020/2319616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 488.Pegtel DM, Gould SJ. Exosomes. Annu. Rev. Biochem. 2019;88:487–514. doi: 10.1146/annurev-biochem-013118-111902. [DOI] [PubMed] [Google Scholar]
- 489.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977. doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 490.Yuyama K, et al. Decreased amyloid-β pathologies by intracerebral loading of glycosphingolipid-enriched exosomes in Alzheimer model mice. J. Biol. Chem. 2014;289:24488–24498. doi: 10.1074/jbc.M114.577213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 491.Peng RR, Shang SX, Zhao LS, Long FQ. MiR-216a-5p-containing exosomes suppress rTp17-induced inflammatory response by targeting TLR4. Biosci. Rep. 2019;39:BSR20190686. doi: 10.1042/BSR20190686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 492.Vulpis E, et al. Genotoxic stress modulates the release of exosomes from multiple myeloma cells capable of activating NK cell cytokine production: role of HSP70/TLR2/NF-kB axis. Oncoimmunology. 2017;6:e1279372. doi: 10.1080/2162402X.2017.1279372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 493.Banstola A, Jeong JH, Yook S. Immunoadjuvants for cancer immunotherapy: a review of recent developments. Acta Biomater. 2020;114:16–30. doi: 10.1016/j.actbio.2020.07.063. [DOI] [PubMed] [Google Scholar]
- 494.Tartey S, Takeuchi O. Pathogen recognition and Toll-like receptor targeted therapeutics in innate immune cells. Int. Rev. Immunol. 2017;36:57–73. doi: 10.1080/08830185.2016.1261318. [DOI] [PubMed] [Google Scholar]
- 495.Guan Y, et al. Human TLRs 10 and 1 share common mechanisms of innate immune sensing but not signaling. J. Immunol. 2010;184:5094–5103. doi: 10.4049/jimmunol.0901888. [DOI] [PubMed] [Google Scholar]
- 496.Santana Silva RJ, Micheli F. RRGPredictor, a set-theory-based tool for predicting pathogen-associated molecular pattern receptors (PRRs) and resistance (R) proteins from plants. Genomics. 2020;112:2666–2676. doi: 10.1016/j.ygeno.2020.03.001. [DOI] [PubMed] [Google Scholar]
- 497.Feng P, Feng L. Sequence based prediction of pattern recognition receptors by using feature selection technique. Int. J. Biol. Macromol. 2020;162:931–934. doi: 10.1016/j.ijbiomac.2020.06.234. [DOI] [PubMed] [Google Scholar]