Abstract
Currently, three major circumstances threaten the management of bacterial infections: increasing antimicrobial resistance, expansion of chronic biofilm-associated infections, and lack of an appropriate approach to treat them. To date, the development of accelerated drug susceptibility testing of biofilms and of new antibiofouling systems has not been achieved despite the availability of different methodologies. There is a need for easy-to-use methods of testing the antibiotic susceptibility of bacteria that form biofilms and for screening new possible antibiofilm strategies. Herein, we present a microfluidic platform with an integrated interdigitated sensor (BiofilmChip). This new device allows an irreversible and homogeneous attachment of bacterial cells of clinical origin, even directly from clinical specimens, and the biofilms grown can be monitored by confocal microscopy or electrical impedance spectroscopy. The device proved to be suitable to study polymicrobial communities, as well as to measure the effect of antimicrobials on biofilms without introducing disturbances due to manipulation, thus better mimicking real-life clinical situations. Our results demonstrate that BiofilmChip is a straightforward tool for antimicrobial biofilm susceptibility testing that could be easily implemented in routine clinical laboratories.
Subject terms: Infectious-disease diagnostics, Biofilms, Health care, Biological techniques, Clinical microbiology
Introduction
Biofilms are communities of microorganisms that form on and attach to living and nonliving surfaces. These communities are ubiquitous, as they are found in natural, industrial and medical environments. Biofilms can be beneficial under some conditions, for example, for biodegradation in wastewater treatment, but they are often undesired because of their ability to cause infections, contamination, biofouling, and biocorrosion1.
The attached microorganisms form microcolonies on a surface, where the bacteria are embedded in the extracellular polymeric substances (EPS) that form the biofilm matrix2. The matrix protects the microorganisms and makes biofilms very difficult to eradicate because it increases their resistance to biological, mechanical, physical, and chemical treatments3. Taking into account that approximately 80% of chronic infections in animals and humans are estimated to be biofilm-related4, their formation presents a severe threat in the battle against antimicrobial recalcitrance, and related treatments require several billions of US dollars each year worldwide5,6. Moreover, to date, no antibiotic that can successfully eradicate biofilms has been found, so there is a great need for new strategies to combat biofilms7 while awaiting for the development of effective antibiofilm molecules.
Model systems of in vitro biofilms are essential in research laboratories for testing new antibiofilm compounds, as well as in clinical laboratories for determining the optimal treatment of biofilm-related infections. There is a wide selection of different monitoring techniques for biofilm growth and characterization, varying in the analysis scale, handling time, sensitivity and final detection technique employed8. Standard methods mostly rely on colorimetric measurements and are commercially available, i.e., the microtiter plate method9,10, the MBEC Assay®11,12, the Biofilm Ring Test13,14 and the Lubbock system15. These techniques allow the screening of different antimicrobials in a high-throughput way, but they are generally destructive endpoint diagnostic tools and require removal of the formed biofilm from the growth substrate used. For this reason, these systems cannot be exploited for online monitoring characterization.
Moreover, the majority of these high-throughput screening techniques involve the use of static devices with limited nutrients, which form biofilms that do not resemble all the biofilms’ characteristics found during natural infections. On the other hand, dynamic devices allow bacteria to grow under flow conditions that eliminate the planktonic growth16. Examples of these devices are the Robbins device17, the Drip-Flow reactor18 or the Rotary biofilm reactor19. Although having some limitations (e.g., the prior knowledge of the device flow dynamics, the heterogeneity of the biofilm development, or the low-throughput of these dynamic devices), they can better mimic real in vivo infective conditions20. One of the recurrent problems with such flow conditions is the need for large volumes of media and tubing, which prevent high-throughput screening.
Furthermore, most of the available techniques require advanced microscopy systems to monitor biofilm growth21–24. Recently, Acea Bioscience (San Diego, USA) released xCELLigence based on electrical impedance spectroscopy (EIS) measured on the basis of a defined Cell Index (CI) parameter25. This technique is based on the detection of changes in the diffusion coefficient of a redox solute, which is recorded as an electrochemical reaction on the electrode; thus, this technique provides an excellent nondestructive method for real-time and in situ measurements independent of confocal microscopy. Indeed, EIS was shown to be suitable for online monitoring of biofilm formation, although all the experiments were performed under static conditions and did not resemble all features of natural infections26,27.
Microfluidics represents the next generation of fluidic platforms for biomedical research28. The Bioflux29 and other microfluidic devices30,31 provide dynamic systems with significant control over the flow rate settings, potential for real-time analysis and, particularly for biofilms, greater similarity to the in vivo infective environments by creating the physical conditions encountered in natural environments32.
Even though antibiotics that affect planktonic growth typically become useless when the bacteria form a biofilm, none of the abovementioned techniques have been used in clinical laboratories to find appropriate treatments for a chronic biofilm infection, as they are too complex and require costly and intricate equipment operated by experienced personnel. Instead, antibiotic efficacy and susceptibilities are determined using planktonic bacteria. Furthermore, real biofilm-forming infections are known to be polymicrobial, which modify their antibiotic susceptibility33, but conventional antimicrobial susceptibility test is performed on single isolates obtained from complex samples, thereby missing other important species and their interactions in clinical samples34.
In this work, we developed a microfluidic system to grow and analyze biofilms using samples from different sources (in vitro bacteria cultures, clinical samples, etc.) to be used in clinical or industrial settings. This device can be used to determine a personalized treatment for a patient suffering a chronic infection, even though more studies are necessary to validate this. We proposed an innovative rapid method for studying new antibiofouling strategies, including drug susceptibility testing of different bacterial species using EIS. This microfluidic platform allows homogeneous biofilm growth that can be easily monitored without using a confocal microscope and enables the development and co-culture of polymicrobial biofilms that resemble the real biofilm infections found in complex matrixes such as sputum samples (e.g., cystic fibrosis infections).
Results
Chip fabrication and characterization
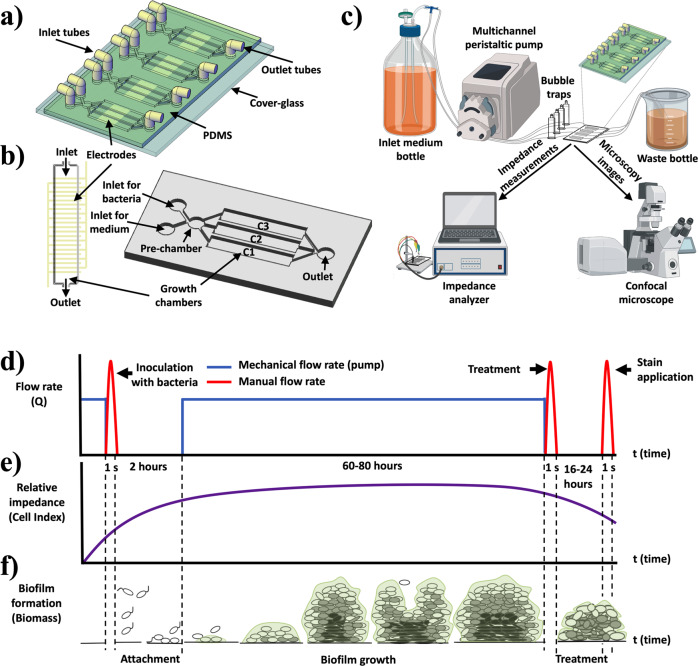
A microfluidic platform with an integrated interdigitated sensor (BiofilmChip) was designed to monitor the growth and treatment of a biofilm in a controlled manner (Fig. 1a, b). The chip was prepared by a combination of standard photolithography and soft lithography techniques. The final system setup is shown in Fig. 1c. Briefly, the medium bottle was connected through tubing to the microfluidic device. Medium was pumped through the system via a high-precision peristaltic pump (see the Material and Methods for details).
Fig. 1. Biofilm chip design.
a BiofilmChip 3D view, b 3D view of one chamber with the electrodes and one set of 3 chambers, c experimental setup, d changes in the mechanical flow rate (in blue) and the manual flow rate (in red, only during the inoculation with bacteria and applications of treatment or stain), e expected relative impedance and f biofilm formation over time. Figure created in part with www.biorender.com.
Following the experimental scheme shown in Fig. 1d, f, a biofilm formed and grew inside the chambers via irreversible attachment of bacterial cells to the cover glass. The biofilms could then be visualized under a confocal microscope after a specific staining procedure (Fig. 1c, f) or directly analyzed by impedance measurements (as shown in Fig. 1c, e).
Optimal parameters for uniform biofilm formation
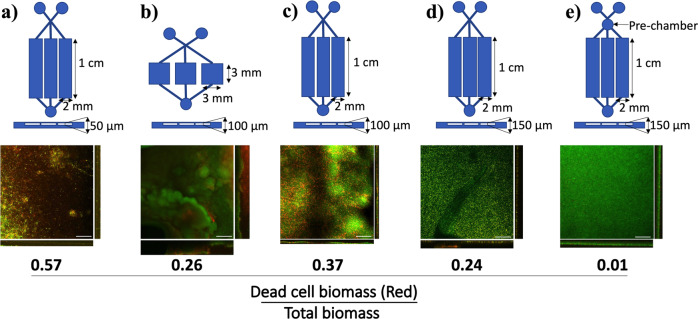
Our primary goal was to design a BiofilmChip with optimized capacity for growing biofilms with maximal uniformity across the different chambers of the chip. First, several prototype designs with chambers that differed in dimension and shape were fabricated and tested by growing a P. aeruginosa PAO1 biofilm. The different prototypes manufactured and tested are shown in Fig. 2, and the formed biofilms were established as described in Fig. 1c, stained with LIVE/DEAD dyes, and visualized under a confocal microscope. As seen in Fig. 2, the chamber geometry and height clearly impacted the biofilm formation: a rectangular chamber morphology was preferred over a square morphology. As observed in prototype b (Fig. 2b), the biofilm formed in the square chamber was nonhomogeneous, while that formed in the rectangular chamber (with a height of 150 μm) remained smooth (Fig. 2d).
Fig. 2. BiofilmChip dimensions, geometry and functional characterization.
schematic representations of the different manufactured chip geometries. The P. aeruginosa PAO1 biofilm structure formed in each chip is shown in the confocal microscopy images (sum and orthogonal views). In all cases, the biofilm was grown for 72 h and stained with the LIVE/DEAD BacLight Bacterial Viability Kit. At the bottom of the images, average proportion of dead cells (stained in red) in the total biofilm biomass from three different experiments is shown. Scale bar represents 50 μm.
P. aeruginosa cell division was impaired in the outer biofilm layers of the 50- and 100-μm height chambers, with evident cell filamentation and increased cell death (stained in red), indicated by a more significant proportion of red biomass (ratios of dead cell biomass to total biomass of 0.57, 0.26 and 0.37) (Fig. 2a–c). On the other hand, bacteria had a morphology characteristic of wild-type Pseudomonas in the chip with 150-μm height chambers and showed uniform green staining covering the entire surface area (a lower proportion of dead cells) (Fig. 2d, e).
Even though the optimal parameters (rectangular shape with 150 μm high) resulted in homogeneous biofilms with a low proportion of dead cells (ratio of 0.24), manual system manipulations (i.e., direct injection during inoculation and staining) caused disturbances in biofilm uniformity (Fig. 2d). There is a tremendous boost of the shear stress all along with the chamber due to the abrupt increase in the flow rate when injecting the bacteria for inoculation. (Fig. 1d). The pressure generated at the inlet was 363 ± 197 psi during manual injection35. At this specific pressure generated during injection, we simulated the influence of fluid velocity and chamber shape on biofilm formation using COMSOL Multiphysics software (COMSOL AB, Sweden). Supplementary Fig. 1 shows that the inclusion of a prechamber ahead of the biofilm growth chambers stabilized the flow and rendered a better distribution of velocity, minimizing the effect of sample loading. The results were confirmed by observing uniform biofilm growth in the chambers, as shown in Fig. 2e. Therefore, the prototype with a rectangular chamber, 150 μm high and with a prechamber (Fig. 2e) was chosen as the optimal design for biofilm formation and used thenceforward.
BiofilmChip robustness
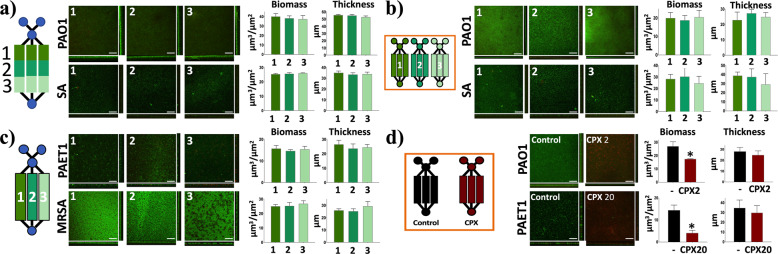
We next evaluated the robustness of the BiofilmChip with the optimized conditions (rectangular chambers 2 mm wide, 10 mm long, and 150 μm high with a 2-mm prechamber diameter) (Fig. 2e) by analyzing some biofilm parameters along with the biofilm growth chamber. For these experiments, we used laboratory strains of Pseudomonas aeruginosa PAO1 and Staphylococcus aureus (ATCC12600). We compared the biofilm biomass and thickness at different locations inside a chamber (close to the inlets, the middle part and the outlet, see Fig. 3a), between sets of chambers (Fig. 3b) and finally among separate chambers of the same set (Fig. 3c). In all cases, we observed robust, homogenous biofilm formation with similar biomass and thickness values among the different locations and chambers, demonstrating the uniformity of the biofilm formed. Note that in Fig. 3c, we used two bacterial strains isolated from chronically infected cystic fibrosis patients (P. aeruginosa PAET1 and methicillin-resistant S. aureus MRSA).
Fig. 3. BiofilmChip robustness evaluation.
Uniformity in the established biofilm biomass and thickness across different chamber locations (a), between sets of chambers (b), and among chambers in the same set (c). Decreases in biomass and thickness after 24 h of treatment of P. aeruginosa PAO1 and PAET1 with ciprofloxacin (d). In all cases, 72-hour-old biofilms were stained and visualized by confocal microscopy (CLSM). Images represent the sum of stacked CLSM images and their corresponding orthogonal views, and bars show the quantified biofilm biomass (μm3/μm2) and average thickness (μm) with error bars of the three different replicates. Scale bar represents 50 μm. The results presented in this figure are representative of the same experiment repeated at least three times, producing similar results every time. PAO1 (P. aeruginosa PAO1 laboratory strain), PAET1 (P. aeruginosa PAET1 clinical isolate strain), SA (S. aureus laboratory strain), MRSA (S. aureus MRSA clinical isolate strain), CPX2 (ciprofloxacin 2 µg/ml), CPX20 (ciprofloxacin 20 µg/ml). *p < 0.05 vs. control in a t-test.
As a proof of concept to evaluate the ability of our BiofilmChip to assess antimicrobial therapy, we determined whether a reduction in the biofilm biomass can be detected with our BiofilmChip, by treating a mature biofilm (72 h old) with the extensively used antibiotic ciprofloxacin (CPX). As shown in Fig. 3d, the treatment of P. aeruginosa laboratory strain PAO1 and clinical P. aeruginosa isolate PAET1 with 2 and 20 μg/ml ciprofloxacin (10× approximately their MIC, See Supplementary Table 1), respectively, reduced the biofilm biomass (35 and 71%) and thickness (12 and 14%) after 24 h, detected by measuring their viability with the LIVE/DEAD staining. Moreover, this reduction of biofilm biomass correlates with an increase of dead cells caused by the ciprofloxacin (see results in Supplementary Table 2). Therefore, BiofilmChip was suitable for use in inhibition experiments to observe changes in biofilm biomass after specific antibiotic treatment.
BiofilmChip is a versatile system for growing biofilms directly from clinical sputum samples
The BiofilmChip prototype has been tested with laboratory and clinical strains of P. aeruginosa and S. aureus (Fig. 3) and directly from patient samples.
Sputum samples from cystic fibrosis patients (Supplementary Table 3) at different stages of P. aeruginosa and/or S. aureus chronic infection were tested. First, the sputum samples were treated with hypotonic media as described in the Material and Methods section and inoculated directly into the BiofilmChip by injecting the sample through the inlet port while the medium flow was stopped. After 2 h, the TSB media was allowed to flow for 72 h. The formed biofilm was stained and visualized under a confocal microscope.
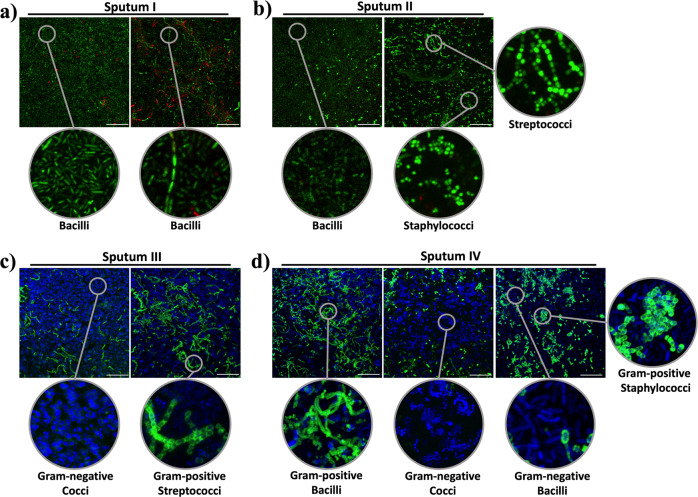
Interestingly, our system allowed the biofilm formation and growth of different bacterial species simultaneously from the sputum samples. Figure 4 displays representative confocal microscopy images of the biofilms formed from four different sputa (I–IV) (Supplementary Table 3). Different bacterial shapes and morphologies can be seen in the enlarged image of biofilms a) and b) (Fig. 4, sputum I-II), stained with the LIVE/DEAD Viability Kit. Furthermore, the biofilms from sputum III and IV (Fig. 4c, d), stained with the Gram Stain Kit, can be distinguished on the basis of the Gram staining of the different bacteria found. Information concerning the identification and antibiotic susceptibility of the bacteria present in each sputum sample is presented in Supplementary Table 3. Sputum II and IV were found to contain P. aeruginosa and S. aureus (Supplementary Table 3), which could correspond to the bacilli and staphylococci found in the biofilm (Fig. 4b, d). However, other bacteria also grew, as observed in the confocal images. On the other hand, when the sputa was grown in agar plates at the Microbiology Service at the Vall d’Hebron Barcelona Hospital, only P. aeruginosa was identified in the cultures of sputum I and III (Fig. 4a, c, Supplementary Table 3), which can correspond to the bacilli found in the biofilm from sputum I, but no signs of Gram-negative bacilli were found in the biofilm from sputum III, demonstrating the existing variability between the planktonic cells used for microbiological identification and those involved in biofilm growth.
Fig. 4. Biofilms formed from four different sputum samples (I–IV) with enlarged images showing the different bacterial shapes found.
Biofilms were stained with a LIVE/DEAD BacLight Bacterial Viability Kit (a and b) and with a Gram Stain Kit (c and d), which stains Gram-negative bacteria blue and Gram-positive bacteria green. Scale bar represents 20 μm. The images shown are representative of one biofilm formed per each sputa, that was repeated at least two times, yielding the same results.
The importance of growing the biofilms directly from sputum samples before analyzing them was corroborated by directly visualizing the sputum by confocal microscopy imaging (Supplementary Fig. 2), showing that it is not possible to directly observe diverse bacteria and their interactions due to the low bacterial concentrations, the presence of eukaryotic cells and the remaining mucus.
Correlation of confocal microscopy and impedance measurements in the BiofilmChip
To validate the use of the BiofilmChip in antibiofilm drug susceptibility tests, 3-day-old biofilms of P. aeruginosa PAO1 (wild-type strain) and P. aeruginosa PAET1 (clinically isolated strain) were treated for 24 h with 2 μg/ml and 20 μg/ml ciprofloxacin, respectively. They were imaged with confocal microscopy to calculate the biofilm biomass. As shown in Fig. 3d, the treated biofilm biomass was significantly decreased compared to that of the control biofilm without treatment. Then, to evaluate whether impedance measurements could be correlated to the established confocal microscopy images for biofilm monitoring, a P. aeruginosa PAO1 strain encoding a green fluorescent protein (GFP) (MK171) was used to grow and easily visualize the biofilm during the time when staining it continuously was not necessary.
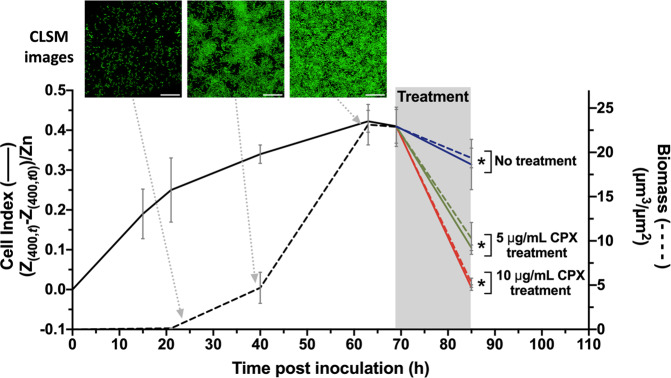
We next used BiofilmChip devices with integrated interdigitated sensors (see Fig. 1a, b and the Material and Methods). Initially, impedance was measured at frequency values between 40 Hz and 400 kHz every 12 h during the entire biofilm growth period (from before inoculation until after treatment). Measures were plotted in a Bode diagram (shown in Supplementary Fig. 3). Ward et al. proposed different mechanisms on how biofilm growth can interfere in electrode-electrolyte impedance value (i.e., production of redox compounds; biofilm material deposition onto the electrode surface; the presence of microbial cells close to electrode surface; among others)36. They suggested that measurements at low frequencies are dominated by charge transfer and mass diffusion of electroactive compounds. In concordance with their results, impedance value at 400 Hz frequency showed substantial differences in impedance at the different steps of the experiment. Accordingly, impedance measurements at 400 Hz and confocal microscopy images were performed at the same time to calculate the cell index (CI) and the biofilm biomass, respectively. CI is a normalized parameter where the impedance value of the blank sample is subtracted from the value measured at a specific time point of the biofilm growth (see the Materials and Methods). In the early stages of biofilm formation, the cells can be imaged under a confocal microscope, but the calculated biomass is nearly zero. The CI, however, varied during the initial biofilm formation stages and thus, can better assess the cell attachment to the substrate as seen in Fig. 5. Thereafter, 69 h old biofilms were treated with ciprofloxacin at 5 μg/ml and 10 μg/ml for 16 h, and the impedance and biomass were measured. Both the CI and biomass presented the same response to the antibiotic at the two concentrations (green and red lines, Fig. 5) or the lack of treatment (blue lines, Fig. 5).
Fig. 5. Correlation in real time the biofilm formation and removal using confocal microscopy and impedance measurement.
Monitored P. aeruginosa MK171 (expressing eGFP) biofilm growth and effect of antibiotic treatment evaluated on the basis of electrical impedance (represented as the CI, left y-axis, solid line) and biomass calculated from confocal microscopy images (defined as μm3/μm2, right y-axis, dashed line). In the shaded part, corresponding to the treatment duration, green and red lines represent 5 μg/ml and 10 μg/ml ciprofloxacin treatment, respectively, and the blue line illustrates the control without treatment. The values shown are the average of three independent experiments, and the error bars indicate the standard deviation between the experiments. Representative confocal images from three different time points are shown. Scale bar represents 20 μm. *p < 0.05 in a Pearson’s correlation test.
Considering the impedance changes on the basis of antibiotic treatment seen in Fig. 5, the applicability of these impedance measurements to the antibiotic sensitivity of real patient samples was tested (Supplementary Table 3, sputum V–VII). Biofilms were grown from different sputum samples having a pathogen with a known susceptibility profile, and they were treated with different antibiotics. Biofilm impedance diminished in the presence of an effective antimicrobial treatment, as proven by the difference in CI values calculated before and after antibiotic exposure of biofilms formed from sputa V–VII (Supplementary Table 3 and Supplementary Fig. 4). This CI response agrees with the response found from Fig. 5, validating the use of BiofilmChip for biofilm susceptibility testing directly from clinical samples.
Discussion
Biofilm-related infections are currently a serious threat around the world. The overuse and misuse of antibiotics, along with the increase of biofilm-associated chronic infections is leading to an increasing prevalence of multi-drug resistant bacteria, resulting in a rise in the morbidity and mortality of these infections, which are expected to cause the death of ten million people by 205037. The susceptibility of bacteria embedded in a biofilm to antimicrobial treatment can significantly differ from that of the same free-living bacteria, as it is known that bacteria growing in biofilms have an intrinsically higher antibiotic resistance than their planktonic counterparts due to different mechanisms38. Nevertheless, currently, the treatment decisions for biofilm infections are based on susceptibility tests performed in planktonic bacteria immediately after individual bacterial isolation. This makes the treatment choice inappropriate for the effective management of biofilm-associated infections39. Moreover, standard antimicrobial susceptibility tests are performed on individual isolates considered relevant by the microbiologist among the full range of growing microorganisms, so in case more than one pathogen is isolated, multiple tests have to be carried out. Taken together, part of the antimicrobial misuse could be attributed to the lack of tools and readily implemented technologies applied in typical microbiology laboratories to easily determine the proper treatment for biofilm infections.
The tool we developed consists of a microfluidic chamber aligned to an interdigitated electrode on which the biofilm grows. It was designed to evaluate biomass formation during the establishment of a biofilm and test antimicrobial drugs and treatments using EIS. Due to the integration of EIS into this BiofilmChip, it is unnecessary to use sophisticated technology such as advanced confocal microscopy, making it suitable for typical microbiology laboratories or industry.
The fabrication cost of the BiofilmChip once you have a specific mask fabrication is cheap (less than one US dollar) and the impedance measurement device can be fabricated easily by an electronic enterprise with cost less than four hundred US dollars. For sure extended manufacturing of all the different BiofilmChip components will reduce the price considerably.
BiofilmChip has several advantages. For example, the microfluidics better mimic growth and resemble a naturally occurring biofilm and reduce the amount of medium and reagents needed to run these continuous biofilm experiments (~16 ml of media per each set of three chambers per day, which is 90% of media reduction compared to the flow cell system). We have optimized this chip’s development by producing different variants of the growth chamber geometry and dimensions to enable optimal biofilm development. It has been documented that the flow cell geometry and the shear stress in the chambers affect biofilm growth40. In agreement with that, we observed that both the chamber shape and height were key factors to be considered when reproducing a homogeneous biofilm. It was found that a rectangular shape and 150 μm height were optimal for this purpose, as shown in Fig. 2.
Moreover, an important feature of our device is the inclusion of a prechamber in the design (Fig. 1a), which prevents biofilm disturbances caused by manual inoculations that suddenly increase the flow rate (and consequently the shear stress) inside the chambers. The simulations of the media flow velocity inside the chambers (Supplementary Fig. 1) demonstrated that the design with a prechamber enables a more homogeneous media distribution among the three chambers of the same set, leading to a more uniform biofilm and minimizing the impact of manual injection. Although biofilms in nature are rather heterogeneous41, homogeneity is a key factor for further reliable detection of susceptibility under different treatments. As Fig. 3d shows, significant decreases in P. aeruginosa PAO1 and PAET1 biofilm biomass were detected after treatment with 2 μg/ml and 20 μg/ml ciprofloxacin, respectively. To our knowledge, the prechamber is an innovative component that has never been published before; therefore, this new component represents a breakthrough in microfluidic devices, where flow rate control is critical.
It is well known that continuous biofilms produced in vitro are more reliable for antibiotic susceptibility studies42, as they are more reproducible and similar to the in vivo infection than are in vitro static biofilms performed in microtiter plates. However, the analysis of these continuous biofilms by confocal microscopy is expensive, requires further staining and trained personnel and is extraordinarily time-consuming. We demonstrate an excellent correlation of the biofilm biomass measured via a standardized methodology involving confocal microscopy with the impedance results, which clearly validates the use of EIS to evaluate biofilm growth. Besides, the use of an EIS analyzer is quicker, simpler, and does not require any training. One interesting observation from the use of impedance measurements at 400 Hz is that such measurements can detect the attachment of bacterial cells to the substrate in the early stages of biofilm growth, when usually there aren’t enough cells to be detected as a biofilm by the COMSTAT software analysis of confocal microscopy images (Fig. 5). Once a biofilm is mature, the measures under confocal microscopy are comparable to those under EIS.
Moreover, the impedimetric variations can be used to measure the response of biofilms to antibiotic exposure. In the biofilms in Fig. 5, the CI has the same pattern as the quantified biomass. For the biofilms formed from sputum samples described in Supplementary Table 3 (V–VII), the impedimetric response of the biofilms to the antibiotic was on the same wavelength as the sensitivity reported from the sputum (Supplementary Fig. 4), demonstrating the applicability of the BiofilmChip for antibiofilm testing and choosing the right personalized therapy in the future.
Interestingly, the BiofilmChip was designed to be used for growing biofilms with different characteristics. As far, it has been tested with biofilms composed of different bacterial species (from laboratory use or clinical isolates) and isolated directly from sputum samples obtained from patients suffering Cystic Fibrosis, but the applicability of the BiofilmChip can be extended to different patient samples (sputum from patients suffering from bronchiectasis, urinary infections, etc.) or hypothetically from contaminated surfaces such as medical devices (catheters, cardiac valves, infected joints, etc.) or samples from the food industry (surfaces, etc.), among others.
Under natural conditions, most of these biofilms are formed from a polymicrobial community of bacteria43 which interact and affect the other’s pathogenicity and antibiotic susceptibility44. Specifically, a clear example is found in chronic infections in the cystic fibrosis lung, which is known as a polymicrobial consortium formed from different bacterial species45,46. Under these conditions, the antibiotic concentration needed to remove the formed biofilm is different from the needed to treat isolated planktonic bacteria or even to treat bacteria grown in a monomicrobial biofilm.
Our BiofilmChip enables the growth of polymicrobial biofilms, as it makes the growth of a biofilm directly from a patient sputum sample possible and ensures that no species are excluded, and helps determine the best treatment needed for personalized therapy. For instance, the sputum samples used to form the biofilm shown in Fig. 4b, d) are reportedly from a CF patient suffering from a lung infection by P. aeruginosa and S. aureus (sputum II and IV in Supplementary Table 3). However, when these sputum samples were directly grown in the BiofilmChip, a diverse population of other microorganisms was found, and the polymicrobial nature of these infections was demonstrated. The combination of confocal microscopy with the impedance measurements in the BiofilmChip would allow to determine which species from the polymicrobial biofilm is affected by the antibiotic, rendering a better treatment, as already described by Müsken et al.22, where the evolution of the polymicrobial biofilms in response to different antibiotic treatments was analyzed. However, it has to be taken into account that more species would be able to grow if using different mediums and/or other growing conditions. The antibiotic treatment prescribed for these patients has probably been determined from the individual susceptibility of P. aeruginosa and S. aureus grown in isolation and planktonically, but as we have previously reported33, antibiotic resistance can significantly change in polymicrobial biofilms47,48. The importance of individualized diagnostic of biofilm resistance in chronically infected patients was previously highlighted to increase the effectiveness of the treatment49. Ongoing work will reveal whether the BiofilmChip allows personalized treatment, ensuring a more accurate diagnosis and a better clinical outcome.
Methods
Laboratory and clinically isolated bacterial strains and growth conditions
Wild-type Pseudomonas aeruginosa PAO1 strain CECT 4122 (ATCC 15692) and Staphylococcus aureus CECT 86 (ATCC 12600) were obtained from the Spanish Type Culture Collection (CECT). The P. aeruginosa PAET1 strain isolated from a cystic fibrosis patient suffering from persistant infection and S. aureus MRSA were from our laboratory stock50,51. Pseudomonas aeruginosa PAO1::eGFP (MK171) was kindly provided by Prof. Tim Tolker-Nielsen52. To obtain inocula for examination, the strains were cultured overnight at 37 °C in Luria Bertani (LB) liquid medium (Scharlab, Spain) for P. aeruginosa and tryptic soy broth (TSB) medium (Scharlab, Spain) for S. aureus. Bacterial growth was measured by reading the absorbance at 550 nm (A550).
Obtaining and processing clinical sputum samples
Excess sputum samples (Supplementary Table 3) from cystic fibrosis patients were collected at the Hospital Universitari Vall d’Hebron Microbiology Department. Approval regarding human sample collection and manipulation was obtained from the Clinical Research Ethics Committee (Comitè Ètic d’Investigació Clínica, CEIC) under the number PR(AG)275/2019. Sputum samples were diluted in 10 mM Tris-HCl hypotonic buffer at pH 7.5 (Fisher Scientific, Spain) in a 1:1 ratio and incubated at 4 °C for 5 min before inoculation.
Microfluidic device design and fabrication: chip fabrication and electrode fabrication
The fabrication of the BiofilmChip (Fig. 1a) involved a multistep procedure using photolithographic and soft lithographic techniques carried out in the MicroFabSpace and Microscopy Characterization Facility, Unit 7 of ICTS “NANBIOSIS” from CIBER-BBN at IBEC. All solvents and chemicals were obtained from Sigma Aldrich unless otherwise specified. The SU-8 photoresist and developer were from MicroChem (Newton, MA). The Ordyl photoresist and developer were from Elga (Italy).
Microfluidic chambers were designed using CAD software (Autodesk, USA), and the master was built over a glass substrate (TED PELLA, Inc., USA) (75 mm × 25 mm). Briefly, the fabrication process started with three consecutive solvent baths (acetone, isopropanol, and ethanol) applied to the glass slide. Following this cleaning protocol, up to three layers of Ordyl SY 550 were laminated on top of the support material using a hot/cold laminator to obtain a smooth attached film surface (150 μm high). Then, the slide was exposed through an acetate photomask to UV light (5 s, 24 mW cm2, 345 nm) in the mask aligner and subsequently placed on a hot plate at 65 °C for 3 min.
The 3D master mold fabrication was finished by developing the Ordyl film using the developer solvent for 3 min. To replicate this mold, a poly(dimethylsiloxane) (PDMS) prepolymer mixture (a curing agent-to-PDMS ratio of 1:10, Sylgard®184, Dow Corning) was placed in a desiccator with a vacuum applied to remove bubbles. The mixture was then poured on top of the Ordyl master to fabricate a PDMS replica and heated overnight at 65 °C. The casted PDMS was peeled off carefully, and inlet and outlet holes were made using a Harris Uni-Core 0.5 mm puncher. A general view of the PDMS BiofilmChip used for biofilm growth and analysis is shown in Fig. 1a. The BiofilmChip includes several sets of independent chambers, each connected to two inlets (one for medium and one for bacterial inoculation), one prechamber, three different growth chambers (C1, C2, C3) and one outlet (Fig. 1b). Each growth chamber was 2 mm wide, 10 mm long and 150 μm high. The BiofilmChip unit was fabricated as three connected chambers (C1, C2 and C3) and considered representative with coherent replicates for straightforward analysis of a given sample.
Gold interdigitated sensors (Fig. 1b) were fabricated onto a glass coverslip to facilitate inspection under a confocal microscope and correlation to impedance measurements. The dimensions and characteristics of the interdigitated electrode were 16 pairs of gold fingers with 75 μm width and 75 μm gap between them. The design of these fingers was selected following the microfluidic chamber size (Fig. 1b). AZ5214 was spread using a spinner to create a uniform layer over the glass substrate. After that, the resin was exposed to UV light in the desired pattern. Then, it was reversed, followed by UV flood light exposure. Next, the glass coverslip was dipped into a solution of AZ Developer 400 K. Then, 20 nm chromium and 80 nm gold were thermally evaporated on glass. After this step, the glass slip was dipped into an AZ Remover 100 solution to remove the photoresist, leaving only the desired pattern’s gold/chromium layer. For bonding to the PDMS microfluidic chamber, an O2 plasma cleaner was used to activate both surfaces. PDMS slabs were carefully placed in contact with the glass slide and left on a hot plate to create permanent bonding. The PDMS microfluidic chamber chip was ready to use. Before every assay, the microfluidic device was exposed to UV/ozone for 15 min to make the surface hydrophilic and sterile. Then, the device was filled with sterile Milli-Q water.
Experimental setup
The BiofilmChip system operated in the three different biofilm formation stages (inoculation, growth and measurement). The final setup is shown in Fig. 1c. LB or TSB +0.2% glucose was stored in a 100 ml bottle connected through Tygon E-3603 tubing (1.5 mm diameter) (DD BioLab) to the microfluidic device. Tubes were attached to the chip by insertion into the inlet and outlet access holes. The medium was pumped through the system with a high-precision Ismatec IPC16 ISM933 multichannel peristaltic pump (Ismatec). Bubble traps (DTU Systems Biology, Technical University of Denmark) were added to the system to avoid the formation of bubbles inside the growth chambers. A constant flow rate of 11.4 μl/min for each set of three chambers was established. The system was filled with media through one inlet, and then, at the inoculation stage, a solution of bacteria (A550 ≈ 0.1) or diluted hypotonic buffer sputum sample was pumped into the chip chamber through the other inlet.
At the start of the growth stage, the peristaltic pump was stopped for two hours to allow cell adherence to the glass surface. Then, the medium flowed through the chambers for 65–72 h. The system was previously sterilized with 0.5% v/v sodium hypochlorite in water as previously described53, and experiments were carried out at room temperature.
Confocal laser scanning microscopy and image analysis
Pseudomonas aeruginosa and Staphylococcus aureus nonfluorescent biofilms were stained with the LIVE/DEAD BacLight Bacterial Viability Kit (Thermo Scientific), consisting of SYTO9 and propidium iodide. Staining of the biofilms formed from sputum was performed with the Bacterial Viability and Gram Stain Kit (Biotium), which differentiates Gram-positive and Gram-negative bacteria. This kit comprises wheat germ agglutinin (WGA) coupled to a CFTM-488 fluorophore that stains the walls of Gram-positive bacteria and the dye DAPI, which stains both Gram-negative and Gram-positive bacteria. Biofilms formed with Pseudomonas aeruginosa MK171 (expressing eGFP) were visualized under the microscope directly without staining.
All biofilms were visualized under a Zeiss LSM 800 confocal scanning laser microscope (CSLM) using 20X/0.80 air or 63X/1.4 oil objectives. Simulated fluorescence projections and orthogonal sections were generated using ImageJ software, and COMSTAT 2 software was used to quantify the biomass and thickness of the biofilms54.
Biofilm treatment
Biofilms between 65 and 72 h old were treated for 16 or 24 h with different antibiotic concentrations while the medium flow was stopped. After the treatment, the flow was set at 11.4 μl/min for 30 min to wash and exclude detached bacteria affected by the antibiotic from the biofilm.
Impedance measurements and cell index calculation
We based our monitoring on the cell index (CI) parameter55, as we wanted to use an independent, standardized parameter to compare measurements among different samples. The measurements were performed at 400 Hz as we detected bigger changes in bold diagram (see Supplementary Fig. 3).
| 1 |
Where Zn is the corresponding impedance value at 400 Hz of medium with no cells, Z(400,t) is the measured impedance at 400 Hz at time point t of biofilm growth and Z(400,t0) is the background impedance 2 h after bacteria inoculation, before the biofilm started to grow.
The device used to record EIS values was an Agilent 4294 A 40 Hz–110 MHz precision impedance analyzer. This equipment allowed measuring the impedance along a range of frequencies with an oscilloscope level of 0.5 V. The impedance was measured in the module and phase format and plotted in Bode diagrams. Once the microfluidic device was connected, two impedance measurements were recorded each day, one during the morning and the second one during the afternoon.
Statistical analysis
Significant differences in biofilm biomass of the ciprofloxacin-treated biofilms vs. non-treated biofilms were calculated using an unpaired t-test with the GraphPad Prism 9.0 software. Correlation between Biomass and Cell Index parameters in the biofilm treatment was calculated using Pearson’s correlation with the GraphPad Prism 9.0 software.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank Prof. Tim Tolker-Nielsen from Costerton Biofilm Center, University of Copenhagen, for providing the P. aeruginosa MK171 strain. This work was supported in part through grants to ET from the Ministerio de Ciencia, Innovación y Universidades (BIO2015-63557-R and RTI2018-098573-B-100) (MINECO/FEDER), the Generalitat de Catalunya (2017 SGR1079 and CERCA program), and the Spanish Cystic Fibrosis Foundation and La Caixa Foundation. The authors want to acknowledge MicroFabSpace and the Microscopy Characterization Facility, Unit 7 of ICTS “NANBIOSIS” from CIBER-BBN at IBEC. This research was supported by the Networking Biomedical Research Center (CIBER), Spain. CIBER is an initiative funded by the VI National R&D&i Plan 2008–2011, Iniciativa Ingenio 2010, Consolider Program, CIBER Actions, and the Instituto de Salud Carlos III (RD16/0006/0012), with the support of the European Regional Development Fund. This work was funded by the CERCA Programme and by the Commission for Universities and Research of the Department of Innovation, Universities, and Enterprise of the Generalitat de Catalunya (2017 SGR 1079). This work was developed in the context of AdvanceCat and Base3D with the support of ACCIÓ (Catalonia Trade and Investment; Generalitat de Catalunya) under the Catalonian ERDF operational program (European Regional Development Fund) 2014–2020. This work was funded by the Spanish Ministry of Economy and Competitiveness (MINECO) through the projects MINDS (Proyectos I+D Excelencia + FEDER): TEC2015-70104-P, CTQ2016-75870-P.
Author contributions
E.T. and J.S. conceived the idea; N.B-C., M.J.L-M., B.V.A-J. J.S. and E.T. conceived and designed the experimental studies; N.B-C., M.J.L-M., B.V.A-J. conducted the experiments; M.T.M-G supplied the clinical isolates; N.B-C., M.J.L-M., J.S. and E.T wrote the manuscript with inputs from B.V.A-J. and M.T.M-G. All authors read and approved the manuscript.
Data availability
The authors declare that all relevant data supporting the findings of the study are available within the article and the Supplementary information file. Additional details are also available upon reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Núria Blanco-Cabra, Maria José López-Martínez.
Supplementary information
The online version contains supplementary material available at 10.1038/s41522-021-00236-1.
References
- 1.Karunakaran E, Mukherjee J, Ramalingam B, Biggs CA. “Biofilmology”: a multidisciplinary review of the study of microbial biofilms. Appl. Microbiol. Biotechnol. 2011;90:1869–1881. doi: 10.1007/s00253-011-3293-4. [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Lebeaux D, Chauhan A, Rendueles O, Beloin C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens. 2013;2:288–356. doi: 10.3390/pathogens2020288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control. 2019;8:76. doi: 10.1186/s13756-019-0533-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yebra DM, Kiil S, Dam-Johansen K. Antifouling technology—past, present and future steps towards efficient and environmentally friendly antifouling coatings. Prog. Org. Coat. 2004;50:75–104. doi: 10.1016/j.porgcoat.2003.06.001. [DOI] [Google Scholar]
- 6.Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol. Mol. Biol. Rev. 2014;78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wolfmeier H, Pletzer D, Mansour SC, Hancock REW. New perspectives in biofilm eradication. ACS Infect. Dis. 2018;4:93–106. doi: 10.1021/acsinfecdis.7b00170. [DOI] [PubMed] [Google Scholar]
- 8.Haney, E. F., Trimble, M. J., Cheng, J. T., Valle, Q. & Hancock, R. E. W. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules8, 10.3390/biom8020029 (2018). [DOI] [PMC free article] [PubMed]
- 9.Christensen GD, et al. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: a quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985;22:996–1006. doi: 10.1128/jcm.22.6.996-1006.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O’Toole, G. A. Microtiter dish biofilm formation assay. J. Vis. Exp.10.3791/2437 (2011). [DOI] [PMC free article] [PubMed]
- 11.Harrison JJ, et al. Microtiter susceptibility testing of microbes growing on peg lids: a miniaturized biofilm model for high-throughput screening. Nat. Protoc. 2010;5:1236–1254. doi: 10.1038/nprot.2010.71. [DOI] [PubMed] [Google Scholar]
- 12.Ceri H, et al. The Calgary Biofilm Device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J. Clin. Microbiol. 1999;37:1771–1776. doi: 10.1128/JCM.37.6.1771-1776.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Olivares E, et al. The BioFilm ring test: a rapid method for routine analysis of Pseudomonas aeruginosa Biofilm formation kinetics. J. Clin. Microbiol. 2016;54:657–661. doi: 10.1128/JCM.02938-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chavant P, Gaillard-Martinie B, Talon R, Hébraud M, Bernardi T. A new device for rapid evaluation of biofilm formation potential by bacteria. J. Microbiol. Methods. 2007;68:605–612. doi: 10.1016/j.mimet.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Sun Y, Dowd SE, Smith E, Rhoads DD, Wolcott RD. In vitro multispecies Lubbock chronic wound biofilm model. Wound Repair Regen. 2008;16:805–813. doi: 10.1111/j.1524-475X.2008.00434.x. [DOI] [PubMed] [Google Scholar]
- 16.Macia MD, Rojo-Molinero E, Oliver A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014;20:981–990. doi: 10.1111/1469-0691.12651. [DOI] [PubMed] [Google Scholar]
- 17.Nickel JC, Ruseska I, Wright JB, Costerton JW. Tobramycin resistance of Pseudomonas aeruginosa cells growing as a biofilm on urinary catheter material. Antimicrob. Agents Chemother. 1985;27:619–624. doi: 10.1128/AAC.27.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goeres DM, et al. A method for growing a biofilm under low shear at the air-liquid interface using the drip flow biofilm reactor. Nat. Protoc. 2009;4:783–788. doi: 10.1038/nprot.2009.59. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence JR, Swerhone GD, Neu TR. A simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods. 2000;42:215–224. doi: 10.1016/S0167-7012(00)00195-0. [DOI] [PubMed] [Google Scholar]
- 20.Azeredo J, et al. Critical review on biofilm methods. Crit. Rev. Microbiol. 2017;43:313–351. doi: 10.1080/1040841X.2016.1208146. [DOI] [PubMed] [Google Scholar]
- 21.Tolker-Nielsen T, Sternberg C. Methods for studying biofilm formation: flow cells and confocal laser scanning microscopy. Methods Mol. Biol. 2014;1149:615–629. doi: 10.1007/978-1-4939-0473-0_47. [DOI] [PubMed] [Google Scholar]
- 22.Musken M, Di Fiore S, Romling U, Haussler S. A 96-well-plate-based optical method for the quantitative and qualitative evaluation of Pseudomonas aeruginosa biofilm formation and its application to susceptibility testing. Nat. Protoc. 2010;5:1460–1469. doi: 10.1038/nprot.2010.110. [DOI] [PubMed] [Google Scholar]
- 23.Rath H, Stumpp SN, Stiesch M. Development of a flow chamber system for the reproducible in vitro analysis of biofilm formation on implant materials. PLoS One. 2017;12:e0172095. doi: 10.1371/journal.pone.0172095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Musken, M. et al. Breaking the vicious cycle of antibiotic killing and regrowth of biofilm-residing Pseudomonas aeruginosa. Antimicrob. Agents Chemother., 10.1128/AAC.01635-18 (2018). [DOI] [PMC free article] [PubMed]
- 25.van Duuren J, et al. Use of single-frequency impedance spectroscopy to characterize the growth dynamics of biofilm formation in Pseudomonas aeruginosa. Sci. Rep. 2017;7:5223. doi: 10.1038/s41598-017-05273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pires L, et al. Online monitoring of biofilm growth and activity using a combined multi-channel impedimetric and amperometric sensor. Biosens. Bioelectron. 2013;47:157–163. doi: 10.1016/j.bios.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 27.Paredes J, Becerro S, Arana S. Comparison of real time impedance monitoring of bacterial biofilm cultures in different experimental setups mimicking real field environments. Sens. Actuators B: Chem. 2014;195:667–676. doi: 10.1016/j.snb.2014.01.098. [DOI] [Google Scholar]
- 28.Sackmann EK, Fulton AL, Beebe DJ. The present and future role of microfluidics in biomedical research. Nature. 2014;507:181–189. doi: 10.1038/nature13118. [DOI] [PubMed] [Google Scholar]
- 29.Benoit MR, Conant CG, Ionescu-Zanetti C, Schwartz M, Matin A. New device for high-throughput viability screening of flow biofilms. Appl. Environ. Microbiol. 2010;76:4136–4142. doi: 10.1128/AEM.03065-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang, X. Y., Sun, K., Abulimiti, A., Xu, P. P. & Li, Z. Y. Microfluidic system for observation of bacterial culture and effects on Biofilm formation at microscale. Micromachines10, 10.3390/mi10090606 (2019). [DOI] [PMC free article] [PubMed]
- 31.Straub H, et al. A microfluidic platform for in situ investigation of biofilm formation and its treatment under controlled conditions. J. Nanobiotechnol. 2020;18:166. doi: 10.1186/s12951-020-00724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Janakiraman V, Englert D, Jayaraman A, Baskaran H. Modeling growth and quorum sensing in biofilms grown in microfluidic chambers. Ann. Biomed. Eng. 2009;37:1206–1216. doi: 10.1007/s10439-009-9671-8. [DOI] [PubMed] [Google Scholar]
- 33.Cendra MDM, Blanco-Cabra N, Pedraz L, Torrents E. Optimal environmental and culture conditions allow the in vitro coexistence of Pseudomonas aeruginosa and Staphylococcus aureus in stable biofilms. Sci. Rep. 2019;9:16284. doi: 10.1038/s41598-019-52726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hall-Stoodley L, Stoodley P. Evolving concepts in biofilm infections. Cell Microbiol. 2009;11:1034–1043. doi: 10.1111/j.1462-5822.2009.01323.x. [DOI] [PubMed] [Google Scholar]
- 35.Hayward WA, et al. Pressure generated by syringes: implications for hydrodissection and injection of dense connective tissue lesions. Scand. J. Rheumatol. 2011;40:379–382. doi: 10.3109/03009742.2011.560892. [DOI] [PubMed] [Google Scholar]
- 36.Ward AC, Connolly P, Tucker NP. Pseudomonas aeruginosa can be detected in a polymicrobial competition model using impedance spectroscopy with a novel biosensor. PLoS One. 2014;9:e91732. doi: 10.1371/journal.pone.0091732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Inoue H. Strategic approach for combating antimicrobial resistance (AMR) Glob. Health Med. 2019;1:61–64. doi: 10.35772/ghm.2019.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol. Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 39.Coenye T, Goeres D, Van Bambeke F, Bjarnsholt T. Should standardized susceptibility testing for microbial biofilms be introduced in clinical practice? Clin. Microbiol. Infect. 2018;24:570–572. doi: 10.1016/j.cmi.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Salek MM, Jones SM, Martinuzzi RJ. The influence of flow cell geometry related shear stresses on the distribution, structure and susceptibility of Pseudomonas aeruginosa 01 biofilms. Biofouling. 2009;25:711–725. doi: 10.1080/08927010903114603. [DOI] [PubMed] [Google Scholar]
- 41.Wimpenny J, Manz W, Szewzyk U. Heterogeneity in biofilms. FEMS Microbiol. Rev. 2000;24:661–671. doi: 10.1111/j.1574-6976.2000.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 42.Reichhardt C, Parsek MR. Confocal laser scanning microscopy for analysis of Pseudomonas aeruginosa Biofilm architecture and matrix localization. Front. Microbiol. 2019;10:677. doi: 10.3389/fmicb.2019.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cendra MDM, Torrents E. Pseudomonas aeruginosa biofilms and their partners in crime. Biotechnol. Adv. 2021;49:107734. doi: 10.1016/j.biotechadv.2021.107734. [DOI] [PubMed] [Google Scholar]
- 45.Kidd, T. J. et al. Defining antimicrobial resistance in cystic fibrosis. J. Cyst. Fibros.10.1016/j.jcf.2018.08.014 (2018). [DOI] [PubMed]
- 46.Filkins LM, O’Toole GA. Cystic fibrosis lung infections: polymicrobial, complex, and hard to treat. PLoS Pathog. 2015;11:e1005258. doi: 10.1371/journal.ppat.1005258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Orazi, G. & O’Toole, G. A. Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a Biofilm model of cystic fibrosis infection. mBio8, 10.1128/mBio.00873-17 (2017). [DOI] [PMC free article] [PubMed]
- 48.Orazi, G., Ruoff, K. L. & O’Toole, G. A. Pseudomonas aeruginosa increases the sensitivity of Biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. MBio,10, 10.1128/mBio.01501-19 (2019). [DOI] [PMC free article] [PubMed]
- 49.Musken M, et al. Towards individualized diagnostics of biofilm-associated infections: a case study. NPJ Biofilms Microbiomes. 2017;3:22. doi: 10.1038/s41522-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Crespo A, Gavalda J, Julian E, Torrents E. A single point mutation in class III ribonucleotide reductase promoter renders Pseudomonas aeruginosa PAO1 inefficient for anaerobic growth and infection. Sci. Rep. 2017;7:13350. doi: 10.1038/s41598-017-14051-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Blanco-Cabra, N. et al. Novel Oleanolic and Maslinic Acids derivatives as a promising treatment against bacterial biofilm in nosocomial infections: an in Vitro and in Vivo study. ACS Infect. Dis.10.1021/acsinfecdis.9b00125 (2019). [DOI] [PubMed]
- 52.Klausen M, et al. Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol. Microbiol. 2003;48:1511–1524. doi: 10.1046/j.1365-2958.2003.03525.x. [DOI] [PubMed] [Google Scholar]
- 53.Crespo A, Blanco-Cabra N, Torrents E. Aerobic vitamin B12 biosynthesis is essential for Pseudomonas aeruginosa class ii ribonucleotide reductase activity during planktonic and Biofilm growth. Front. Microbiol. 2018;9:986. doi: 10.3389/fmicb.2018.00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weiss Nielsen, M., Sternberg, C., Molin, S. & Regenberg, B. Pseudomonas aeruginosa and Saccharomyces cerevisiae biofilm in flow cells. J. Vis. Exp.10.3791/2383 (2011). [DOI] [PMC free article] [PubMed]
- 55.Gutiérrez D, Hidalgo-Cantabrana C, Rodríguez A, García P, Ruas-Madiedo P. Monitoring in real time the formation and removal of Biofilms from clinical related pathogens using an impedance-based technology. PLoS One. 2016;11:e0163966. doi: 10.1371/journal.pone.0163966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors declare that all relevant data supporting the findings of the study are available within the article and the Supplementary information file. Additional details are also available upon reasonable request.