Summary
The endoplasmic reticulum (ER)-resident transmembrane protein kinase/RNase Ire1 is a conserved sensor of the cellular unfolded protein response and has been implicated in lipid homeostasis, including lipid synthesis and transport, across species. Here we report a novel catabolic role of Ire1 in regulating lipid mobilization in Drosophila. We found that Ire1 is activated by nutrient deprivation, and, importantly, fat body-specific Ire1 deficiency leads to increased lipid mobilization and sensitizes flies to starvation, whereas fat body Ire1 overexpression results in the opposite phenotypes. Genetic interaction and biochemical analyses revealed that Ire1 regulates lipid mobilization by promoting Xbp1s-associated FoxO degradation and suppressing FoxO-dependent lipolytic programs. Our results demonstrate that Ire1 is a catabolic sensor and acts through the Xbp1s-FoxO axis to hamper the lipolytic response during chronic food deprivation. These findings offer new insights into the conserved Ire1 regulation of lipid homeostasis.
Subject areas: Lipid, Molecular biology, Cell biology
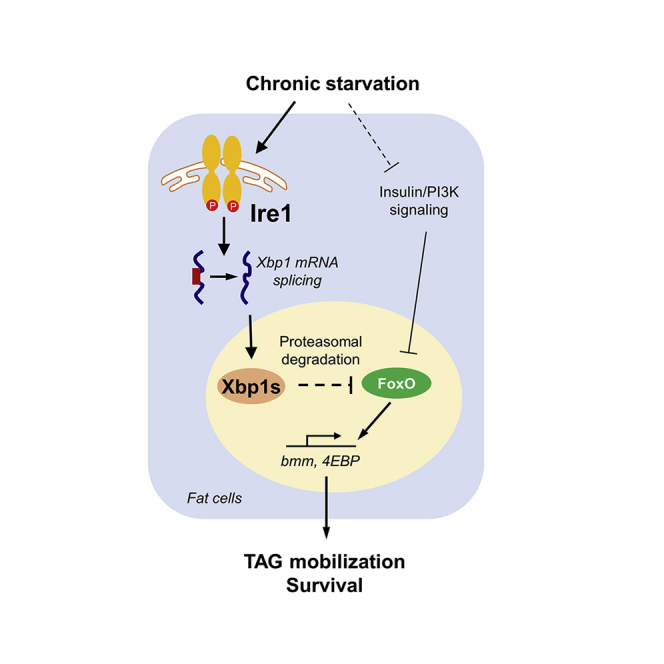
Graphical abstract
Highlights
-
•
Food deprivation systemically activates Ire1 and increases Xbp1 splicing
-
•
Fat body Ire1-Xbp1s axis regulates lipid mobilization and survival during starvation
-
•
Ire1-Xbp1s pathway enhances proteasomal degradation of FoxO
-
•
Fat body Ire1-Xbp1s pathway hampers FoxO-associated lipid mobilization under starvation
Lipid; Molecular biology; Cell biology
Introduction
Providing the energy supply constantly in response to environmental cues is a fundamental feature for both vertebrates and invertebrates. Upon nutrient deprivation, lipid mobilization from body fat storage is an integral component of energy homeostasis, providing energy fuels to meet the body's physiological demand during starvation. Defects in lipid mobilization have been extensively associated with excess fat accumulation and obesity in fly, mice, and humans (Langin et al., 2005).
The endoplasmic reticulum (ER) is the largest organelle in eukaryotic cells and the major site for protein folding and lipid processing in metabolic tissues. Nutrient excess or deprivation alters the state of protein folding within the ER, leading to ER stress and activation of the adaptive unfolded protein responses (UPRER) (Huang et al., 2019). The inositol requiring enzyme 1 (IRE1) is a conserved ER stress sensor from yeast to insects and mammals (Walter and Ron, 2011). Upon activation through trans-autophosphorylation and dimerization or oligomerization during ER stress (Cox et al., 1993; Sidrauski and Walter, 1997; Walter and Ron, 2011), IRE1 splices the X-box-binding protein 1 (XBP1) mRNA to produce XBP1s, the active spliced form of this transcription factor, and induces gene expression involved in protein folding and degradation and ER biogenesis for restoring ER homeostasis (Yoshida et al., 2001). IRE1 also degrades certain mRNAs via a process known as regulated Ire1-dependent decay (RIDD) to alleviate the protein load into the ER (Han et al., 2009; Hollien and Weissman, 2006; Maurel et al., 2014). In addition to the well-recognized role of IRE1 in resolving typical ER stress for cell survival, we and others have also demonstrated that in mammals, IRE1α functions as a metabolic sensor during cellular handling of nutrient stress (Huang et al., 2019). In particular, hepatic IRE1α phosphorylation is coupled to the glucagon/PKA signaling and gluconeogenesis during fasting (Mao et al., 2011), and the hepatic IRE1α-XBP1 pathway modulates the adaptive shift of fuel utilization by enhancing PPARα regulation of fatty acid β-oxidation and ketogenesis following long-term food deprivation (Shao et al., 2014). Hepatic IRE1α was also reported to regulate lipogenesis and lipid secretion by both XBP1-dependent and XBP1-independent mechanisms (Lee et al., 2008; So et al., 2012; Zhang et al., 2011). However, it remains largely unclear whether IRE1 acts to control lipid homeostasis in the peripheral adipose tissues through evolutionarily conserved mechanisms across different species.
The Drosophila has emerged as a powerful genetic model organism for studying the mechanisms of metabolic homeostasis (Baker and Thummel, 2007), including identifying evolutionarily conserved molecules in regulating UPRER and systemic lipid balance (Baumbach et al., 2014; Kuhnlein, 2012; Schlegel and Stainier, 2007). Drosophila Ire1 functions as the homolog of mammalian IRE1α and regulates highly conserved downstream signaling pathways, including Xbp1 splicing, JNK activation, and RIDD (Coelho et al., 2013; Plongthongkum et al., 2007; Yan et al., 2019). Fly Ire1 was reported to control de novo lipogenesis in enterocytes of midgut via Xbp1/Sug signaling to modulate intestinal and systemic lipid homeostasis (Luis et al., 2016), and it was also shown to regulate lipid transport in photoreceptor cells via RIDD degradation of fatty acid transport protein (Fatp) in terms of photoreceptor differentiation (Coelho et al., 2013).
The transcription factor Forkhead box O (FoxO) in Drosophila has been established as a pivotal coordinator in systemic energy balance and nutrient sensing by transcriptionally regulating multiple metabolic pathways involved in food intake control and mobilization of energy stores (Demontis and Perrimon, 2010; Hong et al., 2012; Wang et al., 2011). Particularly, FoxO has been documented to directly promote the expression of Brummer (bmm), which encodes the homolog of human adipose triglyceride lipase (ATGL), and regulate both basal and stimulated lipolysis (Barthel et al., 2005; Kang et al., 2017). Bmm mutant flies have defective fat mobilization with increased TAG storage (Gronke et al., 2005). Therefore, transcriptional activation of lipolysis by FoxO is a critical autonomous determinant of TAG homeostasis in the fat body of Drosophila (Barthel et al., 2005; Kang et al., 2017). Notably, the FoxO-Bmm signaling is tightly controlled through post-translational modifications of FoxO, such as phosphorylation and acetylation, by the insulin and adipokinetic hormone (Akh) pathways, respectively (Kang et al., 2017; Wang et al., 2011), thereby balancing lipid levels in response to nutrient availability and developmental cues.
In this study, we utilized the Drosophila model to characterize the physiological function of Ire1 in lipid homeostasis. We found that nutrient deprivation results in metabolic activation of the Ire1/Xbp1 pathway. Our genetic and biochemical studies provided in vivo evidence suggesting that fat body Ire1 regulates lipid mobilization during starvation response through Xbp1-mediated degradation of FoxO.
Results
Drosophila Ire1 is activated by food deprivation and regulates starvation sensitivity
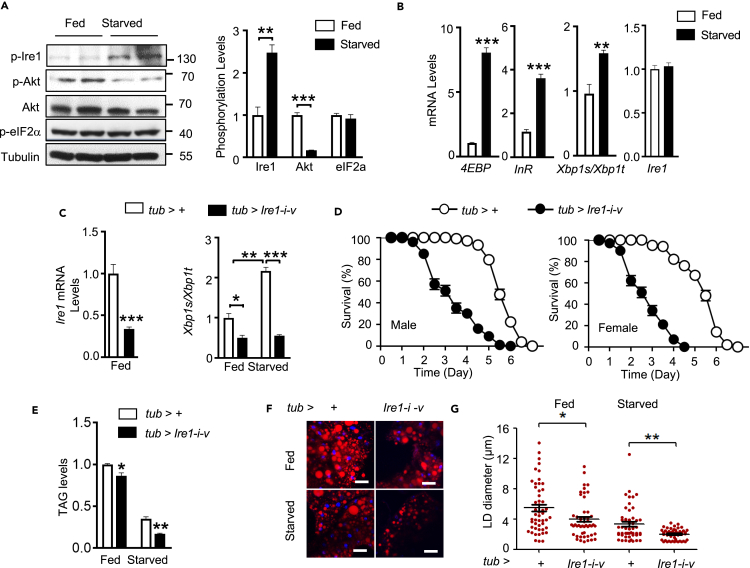
We first examined the expression patterns of Ire1 in w1118 flies. Quantitative RT-PCR (qRT-PCR) analysis revealed that Ire1 is ubiquitously expressed at all developmental stages, with higher expression levels detected in early embryos, pupae, and adults (Figure S1A). We also observed ubiquitous Ire1 mRNA expression in multiple tissues of both larval and adult flies (Figure S1A). To test whether Ire1 is activated by nutrient deprivation, we determined its phosphorylation using a commercial antibody that was able to specifically detect the phosphorylation of fly Ire1 at Ser703 (Figure S1B), a conserved residue corresponding to Ser724 of murine IRE1α located within the kinase activation loop (Korennykh et al., 2009; Song et al., 2017). Indeed, we observed a significant increase of phosphorylated Ire1 in male adult flies following a 48-h starvation (Figure 1A), along with prominently decreased Akt phosphorylation as well as increased expression of 4EBP and InR owing to suppression of insulin signaling (Figures 1A and 1B). Xbp1 mRNA splicing, as detected by either qPCR or a high-gain GFP indicator (Sone et al., 2013), was also elevated upon food deprivation (Figures 1B and Figure S1C). In contrast, we did not observe a strong induction of eIF2α phosphorylation (Figure 1A), another typical ER stress indicator, under starvation (Figures 1A and 1B). These results indicate that the Ire1/Xbp1 pathway is selectively activated in response to starvation in Drosophila.
Figure 1.
Drosophila Ire1 is a crucial sensor of nutrient deprivation.
(A and B) Starvation activates the Ire1-Xbp1 pathway in Drosophila. 3-day-old male adult w1118 flies were ad libitum fed or starved for 48 h. Immunoblot analysis of phosphorylation of Ire1, Akt, and eIF2α in protein extracts of flies using the indicated antibodies (n = 30 flies/group, 15 flies pooled per sample) (A, left). Relative levels of p-Ire1/Tubulin, p-Akt/Akt, and p-eIF2α/Tubulin were quantified (A, right). (B) Quantitative PCR (qPCR) analysis of 4EBP, InR, Xbp1s/Xbp1t, and Ire1 mRNA abundances (n = 40 flies/group, 10 flies pooled per sample). Gene expression levels were normalized to RpL32.
(C) Efficiency of Ire1 knockdown and Xbp1 splicing in male adult tub>Ire1-i-v flies when compared with tub>+ controls (n = 40 flies/group, 10 flies pooled per sample). 3-day old male adult tub>+ and tub>Ire1-i-v flies were fed or starved for 48 h.
(D) Global knockdown of Ire1 expression increases sensitivity to starvation. Survival rate was measured for male and female adult tub>+ and tub>Ire1-i-v flies during food deprivation (3 days of age; n = 160 flies/group). χ2 = 153.7 for males and χ2 = 173.7 for females, P < 0.0001 by log rank test.
(E) Relative whole-body TAG levels under fed or 48-h starved condition were measured and normalized to total protein levels (80 flies/group, 20 flies pooled per sample).
(F and G) Nile red staining of lipid droplets (LDs) in isolated fat bodies from adult flies; scare bars, 50 μm (F) and quantification of LD diameter (G, 12 images/group). Each point represents a single LD. All data are shown as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001 by Student's t test or two-way ANOVA.
Similar to mammals, lipid reserves in flies are primarily stored as triglyceride (TAG) in the lipid droplet of the fat body, which is functionally analogous to mammalian adipose tissue. Under starvation, reserved TAG is mobilized by lipolysis into glycerol and free fatty acids, which can be further delivered to other tissues as energy supplies. We then asked if Ire1 has a physiological role in mediating the starvation response by systemically diminishing Ire1 expression. Because genomic Ire1 disruption led to embryonic lethality (data not shown), we knocked down Ire1 expression in the whole flies by crossing an UAS-Ire1-RNAi line (v39561) to the tub-GAL4 driver line to generate tub-GAL4/UAS-Ire1-RNAi-v39561 (tub>Ire1-i-v) flies. qRT-PCR assessment showed that Ire1 mRNA levels were reduced by ∼60% (Figure 1C). Ire1 deficiency resulted in a significant decrease in Xbp1 mRNA splicing in both feeding and starvation conditions (Figure 1C). Importantly, when compared with survival curves of control flies during starvation, both male and female tub>Ire1-i-v flies lived much shorter and exhibited ∼37% and ∼46% decreases in their median survival rates, respectively (Figure 1D). Moreover, flies with global knockdown of Ire1 expression (tub>Ire1-i-HMC) using another RNAi line (HMC05163) phenocopied this starvation sensitivity (Figure S1D). These results indicate that global Ire1 deficiency sensitizes flies to food deprivation.
Following a 48-h starvation, control male adult flies (tub>+) showed a potent loss of whole-body TAG storages (Figure 1E). Interestingly, the TAG content was significantly decreased by ∼15% under the fed state, and ∼50% after starvation, in tub>Ire1-i-v flies relative to the tub>+ control flies (Figure 1E). Nile Red staining revealed much fewer and smaller lipid droplets in the abdominal fat body of tub>Ire1-i-v flies following starvation (Figures 1F and 1G). Thus, these data demonstrate that Ire1 is a catabolic sensor implicated in maintaining appropriate lipid mobilization in response to food deprivation.
Fat body Ire1 regulates lipid mobilization and survival during starvation
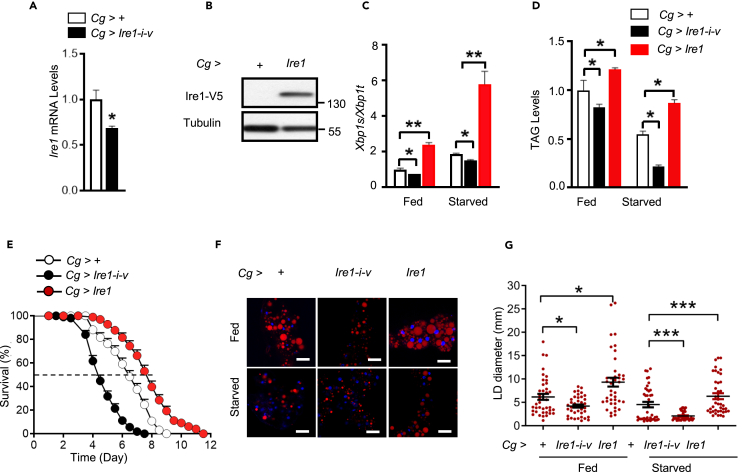
We next asked whether Ire1 regulates lipid mobilization and starvation sensitivity by acting in the fat body, the major metabolic organ sensing nutrient stresses and integrating metabolic regulatory signals. We performed loss- and gain-of-functions studies by knocking down Ire1 expression or overexpressing a wild-type Ire1 protein specifically in the fat body using the Cg-GAL4 driver (Figures 2A and 2B). Cg-GAL4-mediated Ire1 knockdown in the fat body significantly decreased Ire1 mRNA level, as well as Xbp1 splicing, in the abdomen containing large amounts of attached fat bodies, as well as other relatively minor tissues such as the gut, oenocytes, and genitals, in Cg>Ire1-i-v flies (Figures 2A and 2C and Figure S2B). Cg>Ire1-i-v flies exhibited higher sensitivity to starvation stress (an ∼31% decrease in the median survival rate), along with lower TAG storage and smaller lipid droplets in the fed or starved state (Figures 2D–2G). Taking into consideration the potential leaking expression or off-target effects, we also knocked down Ire1 expression using two additional fat body driver lines, R4-GAL4 and Lpp-GAL4 (Figure S2A), as well as another Ire1 RNAi line (HMS03003) with a higher knockdown efficiency (Figure S2B). Consistently, these lines showed similar decreases of TAG levels and sensitivity to starvation (Figure S2C–S2H). Notably, Ire1 knockdown in the oenocytes using the Bo-Gal4 driver had no significant effect on starvation sensitivity (Figure S2I).
Figure 2.
Fat body Ire1 regulates lipid mobilization and starvation resistance
(A) qPCR analysis of Ire1 mRNA abundances in the abdomens of fed male Cg>+ versus Cg>Ire1-i-v flies.
(B) Immunoblot of Ire1-V5 protein in the abdomens of Cg>+ versus Cg>Ire1-V5 flies.
(C–G) 3-day old male adult Cg>+, Cg>Ire1-i-v, or Cg>Ire1-V5 flies were ad libitum fed or subjected to a starvation for 48 h. (C) qPCR analysis of the body Xbp1 mRNA splicing in flies (40 flies/group, 10 flies pooled per sample). (D) Relative TAG content in the whole body (80 flies/group, 20 flies pooled per sample). (E) Survival rate of male adults flies during starvation (n = 180 flies/group). χ2 = 119.3 for Cg>+ versus Cg>Ire1-i-v, and χ2 = 65.45 for Cg>+ versus Cg>Ire1, P < 0.0001 by log rank test. (F) Nile red staining of LDs in isolated fat bodies from adult flies; scare bars, 50 μm. (G) Quantification of LD diameter on the right. Each point represents a single LD (12 images/group). Results are shown as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001 by Student's t test or two-way ANOVA.
Conversely, overexpression of a wild-type Ire1 in the fat body of Cg>Ire1 flies (Figure 2B), obtained by crossing UAS-Ire1-V5 transgenic flies with the Cg-GAL4 driver, resulted in increased Xbp1 mRNA splicing in the abdomens of adult flies (Figure 2C), significantly elevated TAG levels (Figure 2D), and higher survival rates under starvation (Figure 2E, by ∼23% in median values), along with enlarged lipid droplets in their abdominal fat bodies (Figures 2F and 2G). Moreover, at the larval stage, Ire1 deficiency or overexpression in the fat body also showed similar impacts upon lipid droplets and TAG content (Figure S3). Together, these results reveal that Ire1 can act specifically in the fat body to regulate lipid mobilization and survival during food deprivation.
Fat body Xbp1 mediates Ire1's metabolic effects
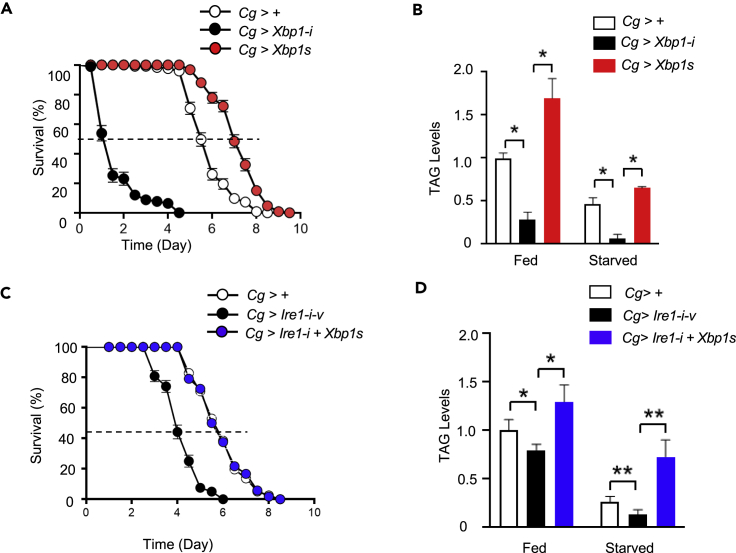
We then determined whether fat body Ire1 exerts its regulatory effects through its downstream effector Xbp1. We generated Cg>Xbp1-i and Cg>Xbp1s flies, in which the expression of Xbp1 was knocked down and the spliced form Xbp1s was overexpressed specifically in the fat body, respectively (Figure S4A and S4B). Knockdown of Xbp1 expression markedly decreased the survival rate (by ∼73% in median values) and whole-body TAG content of starved flies, whereas Xbp1s overexpression resulted in the opposite phenotypes (Figures 3A and 3B). Moreover, fat body overexpression of Xbp1s in the context of Ire1 knockdown was able to rescue the effects of Ire1 deficiency upon starvation sensitivity and lipid mobilization in adult Cg>Ire1-i-v+Xbp1s flies, exhibiting comparable survival rate and TAG content relative to the control Cg>+ flies during starvation (Figures 3C and 3D). These data indicate that fat body Xbp1s mediates, at least in large part, Ire1's regulatory actions in lipid mobilization during the starvation response.
Figure 3.
Fat body Xbp1s contributes to Ire1 regulation of the starvation response
(A and C) Survival rate of starved male adult flies of the indicated genotypes (3 days of age; 160 flies/group). χ2 = 264.4 for Cg>+ versus Cg>Xbp1-i-v, and χ2 = 75.09 for Cg>+ versus Cg>Xbp1s, p < 0.0001 by log rank test. χ2 = 72.80, p < 0.0001 for Cg>+ versus Cg>Ire1-i-v and χ2 = 0.005, p = 0.94 for Cg>+ versus Cg>Ire1-i+Xbp1s by log rank test.
(B and D) Relative TAG content in 3-day old male adult flies of the indicated genotypes following a 48-h starvation (80 flies/group, 20 flies pooled per sample). Results are shown as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.001 by two-way ANOVA.
Fat body Ire1-Xbp1 pathway regulates FoxO-associated gene expression
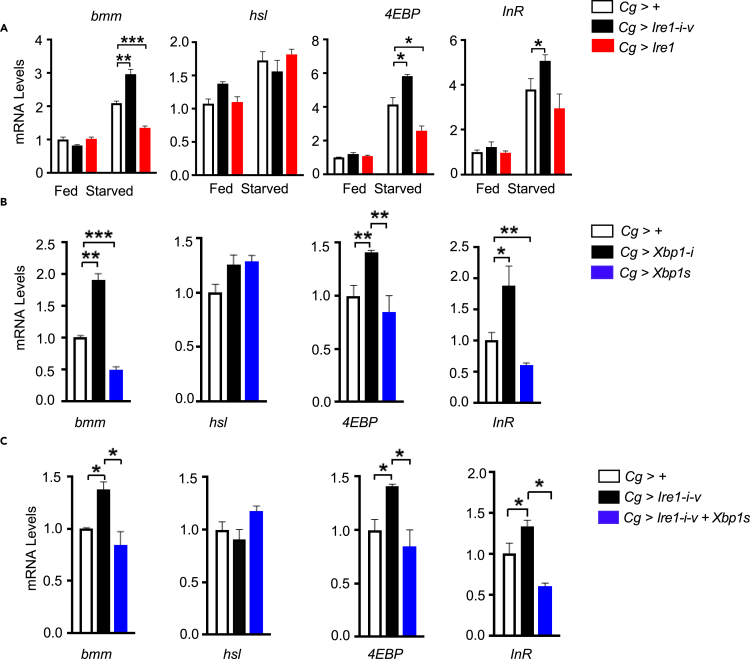
Both Bmm and Hsl are critical lipolytic enzymes in lipid mobilization and TAG storage in the fat body under starvation (Kuhnlein, 2012). To determine if the Ire1/Xbp1 pathway is involved in controlling the expression of these two lipases, we performed qPCR analysis and found that Ire1 deficiency significantly increased starvation-induced upregulation of bmm, but not hsl, in the abdomen of adult Cg>Ire1-i-v flies, whereas Ire1 overexpression effectively reduced bmm expression in Cg>Ire1 flies (Figure 4A). Bmm has been established as the target gene of transcriptional factor FoxO that is activated during starvation response (Kang et al., 2017). Similarly, Ire1 deficiency enhanced, whereas Ire1 overexpression blunted, the induction of two typical FoxO-target genes, 4EBP and InR (Song et al., 2010), in the abdomen of starved adult flies (Figure 4A). Moreover, fat body Xbp1 knockdown and Xbp1s overexpression in adult Cg>Xbp1-i and Cg>Xbp1s flies largely phenocopied, respectively, the effects of Ire1 manipulation upon the expression of these starvation responsive genes following food deprivation (Figure 4B). In accordance with Xbp1s acting as an effector downstream of Ire1, fat body overexpression of Xbp1s was able to sufficiently blunt the augmented expression of bmm, as well as 4EBP and InR, resulting from Ire1 knockdown in starved Cg>Ire1-i-v+Xbp1s flies (Figure 4C). These results suggest that fat body Ire1/Xbp1 pathway may constitute a negative control loop in governing FoxO-directed gene expression program under starvation.
Figure 4.
The Ire1-Xbp1 pathway suppresses starvation-responsive gene expression program
(A) Male adult Cg>+, Cg>Ire1-i-v, and Cg>Ire1 flies were fed or subjected to a 48-h starvation (3 days of age; 40 flies/group, 10 flies pooled per sample). Relative abdomen mRNA abundance of starvation-responsive genes, including bmm, hsl, 4EBP, and InR, was assessed by qPCR. Shown are gene expression levels after normalization to the internal control RpL32.
(B and C) Gene expression levels in (B) male adult Cg>+, Cg>Xbp1-i, and Cg>Xbp1s flies and in (C) male adult Cg>+, Cg>Ire1-i, and Cg>Ire1-i+Xbp1s flies following a 48-h starvation (3 days of age; 40 flies/group, 10 flies pooled per sample). All data are presented as the mean ± SEM (n = 4 independent experiments). ∗p < 0.05, ∗∗p < 0.001, ∗∗∗p < 0.0001 by one-way or two-way ANOVA.
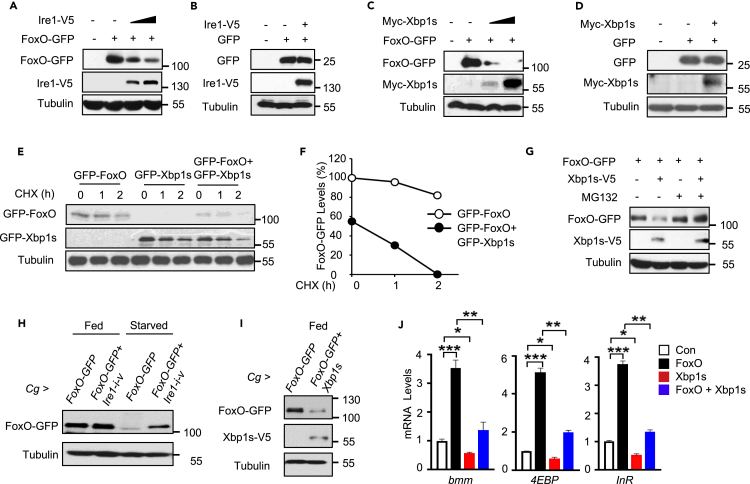
The Ire1/Xbp1 pathway regulates FoxO protein degradation
Upon food deprivation, FoxO shuttles to the nucleus and initiates the transcriptional activation of starvation-responsive genes (Song et al., 2010; Wang et al., 2011). Suppression of 4EBP, InR, and bmm expression (Figure 4) indicates that the Ire1/Xbp1 pathway might decrease FoxO expression and/or activity. We first found that genetic manipulation of Ire1 or Xbp1 in the fat body had no statistically significant effects on foxO mRNA levels in Cg>Ire1-i-v or Cg>Xbp1-i flies (Figures S4C and S4D), excluding the possibility of the transcriptional regulation. We then tested if Ire1 or Xbp1s regulates FoxO via post-transcriptional mechanisms. Indeed, co-expression of FoxO together with Ire1 or Xbp1s in Drosophila S2 cells potently decreased FoxO protein level in a dose-dependent manner (Figures 5A and 5C), whereas co-expression of Ire1 or Xbp1s showed no effect on a neutral GFP protein (Figures 5B and 5D). To further determine if Xbp1s affects the stability of FoxO protein, we blocked protein synthesis in S2 cells with cycloheximide (CHX) and observed a gradual decline of FoxO protein, and co-expression of Xbp1s prominently promoted this process (Figures 5E and 5F). Moreover, addition of MG132, a proteasome inhibitor of protein degradation, largely blocked Xbp1s-mediated decrease of FoxO protein in S2 cells (Figure 5G). Xbp1s overexpression also results in an increase in ubiquitination of FoxO protein (Figure S5A). These data indicate that the Ire1/Xbp1s pathway acts to enhance the proteosomal degradation of FoxO protein. Next, we examined whether Ire1/Xbp1s axis could regulate FoxO protein stability in vivo by specifically overexpressing a GFP-tagged FoxO in adult fat body. Interestingly, chronic starvation (24 h) significantly decreased FoxO-GFP level in the fat body (Figure 5H). Ire1 deficiency potently abolished starvation-induced FoxO decline under starvation hardly affecting FoxO level at fed conditions (Figure 5H). Xbp1s overexpression in the fat body, in contrast, phenocopied starvation-induced FoxO degradation even under fed conditions (Figure 5I). Similar results were observed in R4>FoxO-GFP larvae, as Xbp1s overexpression remarkably reduced FoxO-GFP expression levels in the larval fat body cells (Figure S5B). Note that, Xbp1s does not significantly abolish the nuclear translocation of FoxO under starvation (Van Der Heide et al., 2004) (Figure S5B and S5C). Furthermore, co-expression of Xbp1s in S2 cells, at least in large part, suppressed FoxO-dependent upregulation of bmm, InR, and 4EBP (Figure 5J). These results collectively indicate that Ire1/Xbp1s axis regulates the protein stability of FoxO.
Figure 5.
Ire1/Xbp1 axis regulates proteasome-associated FoxO degradation
(A–D) S2 cells were transiently transfected for 48 h with the plasmids expressing a V5-tagged Ire1 (A, B) or Myc-tagged Xbp1s (C, D), together plasmids expressing a GFP-tagged FoxO (A, C) or neutral GFP (B, D), a negative control. Cell extracts were analyzed by immunoblotting with the GFP or V5 antibody.
(E–G) S2 cells were transiently transfected or co-transfected as indicated for 48 h. Cells were then treated with DMSO (−) or CHX (30 μM) for 1–2 h (E and F) or MG132 (10 μM) for 8 h (G) before immunoblot analysis. (F) Relative FoxO-GFP protein levels were quantified as FoxO-GFP/Tubulin ratios using ImageJ.
(H and I) 5-day-old adult flies with indicated genotypes were fed or starved for 24 h. The whole flies were dissected for immunoblot analysis.
(J) S2 cells were transfected or co-transfected for 48 h as indicated. The mRNA abundance of bmm, 4EBP, and InR was analyzed by qPCR. Values were normalized to the internal control RpL32 (n = 3). Data are shown as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 by one-way ANOVA.
Previous studies have reported that Xbp1-FoxO protein interaction contributes to FoxO degradation in cultured cells (Kishino et al., 2017; Zhou et al., 2011). We wondered whether similar regulation occurs in the fat body by performing immunoprecipitation in Cg>FoxO-GFP+Xbp1s adult flies. However, we failed to observe Xbp1-FoxO binding (data not shown), probably due to insufficient FoxO proteins caused by Xbp1s-induced degradation. On the other hand, in a few fat body cells with remaining FoxO protein expression, we did not observe intranuclear co-localization of FoxO and Xbp1s under starvation as well (Figure S5D). We thus speculate that fat body Xbp1s might regulate FoxO degradation in a manner independent of protein interaction.
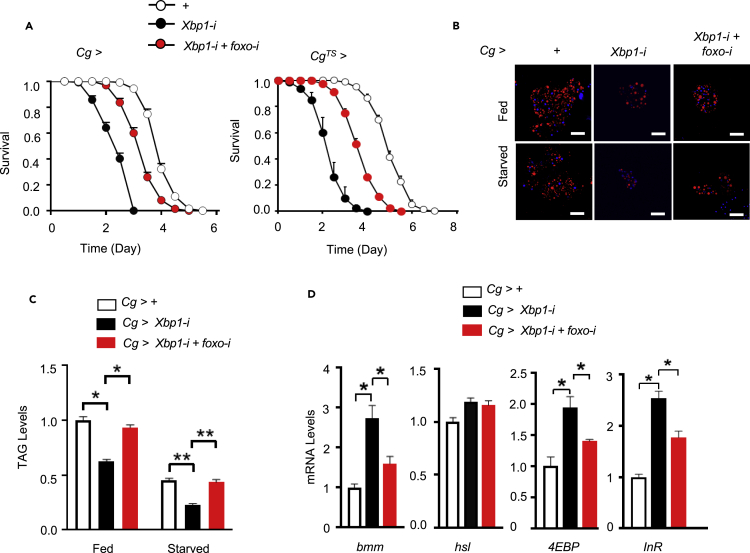
FoxO mediates Xbp1 regulation of lipid metabolism during starvation response
Finally, we investigated the physiological importance of the Xbp1s/FoxO axis. Xbp1 knockdown in the fat body markedly sensitized Cg>Xbp1-i flies to starvation (Figure 3A), whereas fat body foxo knockdown in the context of Xbp1 knockdown (Figure S4A) partially, but significantly, reversed Xbp1 deficiency-associated starvation sensitivity of adult Cg>Xbp1-i+foxo-i flies (Figure 6A, left). Similar results were also obtained when genetic manipulation in the fat body was performed only in adult flies using a Cg-Gal4, tub-Gal80TS (CgTS>) line, the temperature-sensitive driver for the fat body (Figure 6A, right). Moreover, foxo knockdown significantly blunted the decreases, as a result of Xbp1 deficiency, in lipid droplet sizes and TAG levels, as well as the induction of starvation-responsive genes, in Cg>Xbp1-i+foxo-i flies (Figures 6B-D).
Figure 6.
Fat body Xbp1s-FoxO axis regulates starvation sensitivity and lipid mobilization
(A) Survival rates of starved male adult flies bearing indicated RNAi lines throughout all developmental stages (left) or only at adult stage (right) (3 days of age; n = 160 flies/group). + versus Xbp1-i (left, χ2 = 241.5; right, χ2 = 170.5), >+ versus Xbp1-i + foxo-i (left, χ2 = 65.55; right, χ2 = 79.9), and Xbp1-i versus Xbp1-i+foxo-i (left, χ2 = 127.2; right, χ2 = 94.47) are all statistically significant. p < 0.0001 by log rank test.
(B–D) Male adult flies at 3 days of age were fed or starved for 24 h. (B) Nile red staining of LDs in isolated fat bodies from adult flies; scare bars, 100 μm. (C) Relative TAG levels in both fed and starved flies (80 flies/group, 20 flies pooled per sample). (D) qPCR analysis of the mRNA abundances of the indicated genes in starved flies with RpL32 used as the internal control for normalization (40 flies/group, 10 flies pooled per sample). Data are shown as the mean ± SEM. ∗p < 0.05 by two-way ANOVA.
To affirm that the Xbp1-FoxO axis cell-autonomously exerts its regulatory actions, we utilized clonal analysis in the fat body to avoid potential systemic feedback effects. We crossed the UAS-Xbp1-RNAi line to the yw, hs-FLP; Act>CD2>Gal4, UAS-GFP line and generated Xbp1-deficient fat body cell clones following heat shock (Song et al., 2019). Interestingly, clones bearing Xbp1 RNAi were much smaller and contained smaller lipid droplets relative to their control clones (Figure S6A), and foxo RNAi resulted in a reversal of Xbp1 RNAi-elicited decreases in fat body cell and lipid droplet sizes (Figure S6A). Moreover, FoxO overexpression also caused smaller fat body cell clones and lipid droplets (Figure S6B and S6C). Consistent with Xbp1s-associated suppression of FoxO, overexpression of Xbp1s significantly restored the sizes of fat body cell and lipid droplet in the context of gain of function of FoxO (Figure S6B and S6C). These results suggest that FoxO is critically involved in mediating Xbp1 regulation of lipid homeostasis and starvation responses. Additionally, FoxO has been reported to regulate fly development besides cell growth and lipid metabolism. In line with this, we observed that FoxO overexpression in the fat body resulted in smaller larval size and delayed pupal development (Figure S6D and S6E). Interestingly, these developmental defects were largely rescued by Xbp1s overexpression (Figure S6D and S6E). Taken together, our data demonstrate that FoxO serves as the downstream effector to mediate the regulatory actions of the Ire1/Xbp1 pathway not only in lipid metabolism but also in growth and development in the fruit fly.
Discussion
Maintaining energy homeostasis is a fundamental aspect of animal physiology, and multiple evolutionarily conserved mechanisms are at work to achieve this highly regulated function. Mobilization of lipid storage during nutrient deprivation constitutes a critical component of catabolism that is under many layers of control mechanisms. Using Drosophila as a model organism with multiple conserved hormonal regulatory networks as well as the ER stress response pathways we identified the Ire1 branch in the fat body that links the UPRER to systemic lipid homeostasis and starvation response. Our findings demonstrate that the Ire1/Xbp1 cascade responds to nutrient availability and regulates starvation-associated lipid mobilization by modulating FoxO protein stability.
In this study, we found that starvation activates the Ire1/Xbp1 pathway, as indicated by Ire1 phosphorylation and Xbp1 mRNA splicing. This reveals Ire1 as a conserved metabolic sensor across species, because similar activation of the IRE1α-XBP1 branch has also been observed in the liver during prolonged starvation of mice (Shao et al., 2014). Although Ire1 activation in metabolic organs has been well documented under nutrient stress conditions including fluctuations of carbohydrates and lipids (Huang et al., 2019), exactly how nutrient deprivation triggers physiological ER stress and adaptive UPRER activation remains largely unclear. It is interesting to note that a recent mouse model study indicated that a low-protein diet could induce IRE1α activation in cancer cells in association with anticancer immune responses (Rubio-Patino et al., 2018), whereas in Drosophila, Ire1 deficiency in the adult intestine was shown to potently abolish lifespan extension caused by a low-protein diet (Luis et al., 2016). This suggests that changes of amino acids or proteins could contribute to Ire1 activation. In addition, metabolic hormones and other endocrine factors could also serve as Ire1 activators. For instance, we have previously shown that Akh, a vital neuroendocrinal hormone in the control of systemic carbolipid metabolism in Drosophila, was able to trigger the cAMP/PKA signaling and result in phosphorylation of Ire1 in the fat body under starvation (Song et al., 2017).
Fat body in the fruit fly functions as a key metabolic organ that modulates the balance between lipid storage and mobilization under nutrient-deprivation conditions. Although our results indicate that Ire1 in the fat body has a critical role in regulating lipid mobilization during starvation, it is worth noting that fat body Ire1 knockdown led to a weaker extent of starvation sensitivity than global Ire1 deficiency, suggesting the existence of Ire1 metabolic actions in other organs. In this regard, we excluded a role for Ire1 in oenocytes, which have been implicated in degrading circulating lipids downstream of the fat body (Gutierrez et al., 2007), since oenocyte Ire1 knockdown did not affect starvation sensitivity. In enterocytes of the gut that are involved in dietary lipid absorption, de novo lipogenesis, and lipid delivery (Song et al., 2014), Ire1 was reported to promote enteric lipid synthesis and suppress systemic lipid turnover (Luis et al., 2016). Therefore, it remains to be unraveled how Ire1 in the fat body as well as in other metabolic tissues, such as the intestine and brain, act together to orchestrate its integrative control on systemic lipid homeostasis.
Given that overexpression of Xbp1s could fully restore the lipid homeostasis and starvation response in Ire1-deficient flies, our results suggest that Ire1 controls lipid mobilization primarily through Xbp1 splicing in the fat body. Furthermore, Ire1-catalyzed Xbp1 splicing generated Xbp1s to destabilize FoxO protein and thus suppress FoxO-dependent lipolytic gene expression program. In accordance, another group also observed a link between Xbp1 deficiency and the induction of FoxO-target gene 4EBP in Drosophila through unclear mechanisms (Huang et al., 2017). Xbp1s appeared to specifically degrade FoxO, but not a neutral GFP, which could be blocked by a proteasome inhibitor MG132. This indicates that Xbp1s employs a proteasome-dependent mechanism for FoxO degradation, the molecular details of which have yet to be fully deciphered. Notably, it was shown that in the state of obesity, disruption of XBP1s-depndent degradation of FoxO1 in the liver contributes to hyperglycemia in diabetic mouse models (Zhou et al., 2011). Thus, Xbp1s-mediated control of FoxO protein stability may represent an evolutionarily conserved mechanism linking the Ire1 branch to ER stress-related pathological processes. However, in contrast to in vitro overexpression assays in cultured cells (Kishino et al., 2017; Zhou et al., 2011), we did not observe Xbp1s-FoxO interaction in adult fat body under chronic starvation. The discrepancy could be explained by the distinct context of fat body tissue. On the other hand, foxo deficiency only partially alleviates starvation sensitivity associated with Xbp1 knockdown, indicating potential FoxO-independent effects downstream of Xbp1s. Previous studies reported that Xbp1s also directly regulates transcriptions of several metabolic genes regarding lipid turnover and synthesis (Luis et al., 2016; Martinez et al., 2020). It would be worthy to investigate the comprehensive signaling networks downstream of Xbp1s in the future.
Emerging evidence from research in our group as well as many others has uncovered Ire1 as an evolutionarily conserved regulator linking UPRER to multiple physiological processes in both fly and mammals. Both fly Ire1 and mammalian IRE1α have been connected to glucagon/cAMP/PKA signaling, thereby modulating carbohydrate production following a short-term starvation (Mao et al., 2011; Song et al., 2017). Excessive activation of fly Ire1/mouse IRE1α was also documented to decrease neuronal survival and contribute to neurodegeneration (Ni et al., 2018; Yan et al., 2019). Mammalian IRE1α has been well established as an important regulator in hepatic lipid metabolism, including fatty acid synthesis, β-oxidation, ketogenesis, as well as lipid secretion (Huang et al., 2019), whereas whether IRE1α is involved in modulation of lipid homeostasis in peripheral adipose tissues remains much less understood. Although the IRE1α-XBP1s signaling branch has been implicated in adipocyte differentiation in vitro from cultured pre-adipocytes (Sha et al., 2009), adipocyte XBP1 abrogation in mice was reported to have no apparent impact on lipid metabolism in adipose tissue except during lactation (Gregor et al., 2013). Interestingly, adipose XBP1s overexpression was shown to significantly decrease adiposity by enhancing thermogenesis in both normal and obese mice (Deng et al., 2018). In this scenario, it remains unclear whether Ire1-directed suppression via the Xbp1s-FoxO axis of lipid mobilization may operate in distinct types of adipocytes in mammalian adipose tissues. Nonetheless, our findings herein highlight Ire1 as an ancient catabolic sensor that contributes to the delicate homeostatic control of lipid metabolism.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-p-Ire1α | Novus Biologicals | Cat: NB100-2323, RRID: AB_10145203 |
| Rabbit anti-p-Akt | Cell Signaling | Cat: 4056S, RRID: AB_331163 |
| Rabbit anti-Akt | Cell Signaling | Cat: 9272S, RRID: AB_329827 |
| Rabbit anti-p-eIF2α | Cell Signaling | Cat: 9721S, RRID: AB_330951 |
| Mouse anti-Tubulin | Sigma-Aldrich | Cat: T6199, RRID: AB_477583 |
| Mouse anti-V5 | Invitrogen | Cat: R96025, RRID: AB_159313 |
| Mouse anti-GFP | Abclonal | Cat: AE012 |
| Mouse anti-poly-ubiquitination | Enzo Life Sciences | Cat: BM-PW8805, RRID: AB_10541434 |
| Mouse anti Myc | Cell Signaling | Cat: 2276S, RRID: AB_331783 |
| Experimental Models: Cell Lines | ||
| Drosophila S2R+ cell | Gift from Norbert Perrimon | NA |
| Experimental Models: Organisms/Strains | ||
| Drosophila, w1118 | Bloomington | 3,605 |
| Drosophila, tub-GAL4 | Bloomington | 5,138 |
| Drosophila, Cg-GAL4 | Bloomington | 7,011 |
| Drosophila, R4-GAL4 | Bloomington | 33,832 |
| Drosophila, Lpp-GAL4 | Song et al., 2019 | NA |
| Drosophila, Tub-GAL80 | Song et al., 2019 | NA |
| Drosophila, UAS-Ire1-i-HMC05163 | Bloomington | 62,156 |
| Drosophila, UAS-Ire1-i-HMS03003 | Bloomington | 36,743 |
| Drosophila, UAS-Xbp1.HG | Bloomington | 60,730 |
| Drosophila, UAS-Ire1-i | VDRC | v39561 |
| Drosophila, UAS-Xbp1-i | VDRC | v109312 |
| Drosophila, UAS-foxo-i | VDRC | v107786 |
| yw,hs-Flp;Act>CD2>GAL4 | Song et al., 2019 | NA |
| Drosophila, UAS-GFP | Song et al. (2010) | NA |
| Drosophila, UAS-Xbp1s | This Paper | NA |
| Drosophila, UAS-Ire1 | Yan et al. (2019) | NA |
| Drosophila, UAS-foxo | Gift from Lei Xue lab | NA |
| Drosophila, UAS-foxo-GFP | Gift from Lei Xue lab | NA |
| Drosophila, Bo-Gal4 | Gift from Alex P. Gould lab | NA |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TRizol reagent | Sigma-Aldrich | Cat: T9424 |
| BSA | Sigma-Aldrich | Cat: A7030-1KG |
| iScript Reverse Transcription mix | Vazyme | Cat: R333-01 |
| iQ SYBR Green Supermix | Vazyme | Cat: Q111-02/03 |
| Nile Red | Santa Cruz | Cat: 7385-67-3 |
| Protease inhibitor cocktail | Sigma | Cat: P8340 |
| Phosphatase inhibitor cocktail | Sigma | Cat: P2850 |
| MG132 | Sigma-Aldrich | Cat: M7449 |
| Cycloheximide | Sigma-Aldrich | Cat: 5087390001 |
| Triglyceride reagent | Sigma-Aldrich | Cat: T2449 |
| Glycerol standard | Sigma-Aldrich | Cat: G7793-5ML |
| Free glycerol reagent | Sigma-Aldrich | Cat: F6428 |
| Bacterial and Virus Strains | ||
| DH5a | TIANGEN | Cat: CB101 |
| Software and Algorithms | ||
| Photoshop | Adobe | NA |
| Excel | Microsoft | NA |
| ImageJ | NIH | NA |
| GraphPad Prism 6 | GraphPad | NA |
| Others | ||
| OLYMPUS-FV1200 | OLYMPUS microsystems | NA |
| Carl Zeiss | Zeiss microsystems | NA |
| Oligonucleotides | ||
| Primers for Drosophila Ire1 qPCR F: CTGAAGCGACAGGCCAACA R: CCGATACAACGAGCTGGAGG | ||
| Primers for Drosophila Xbp1s qPCR F: TGGATCTGCCGCAGGGTAT, R: GCGCTTGACGTCGAACTCTT | ||
| Primers for Drosophila Xbp1t qPCR F: AGAAGGCACGCATGGAGG R: AGCAGTGACTCGTTGATGGC | ||
| Primers for Drosophila foxo qPCR F: CCACCGACGAGTTGGACAGTA, R: TGCGACGCGATGAGTTCTT | ||
| Primers for Drosophila bmm qPCR F: CAGGGTGGTGAACGAAGCTC R: CCGCTTGTGAGCATCGTCT | ||
| Primers for Drosophila 4EBP qPCR F: CTCCTGGAGGCACCAAACTTATC R: TTCCCCTCAGCAAGCAACTG | ||
| Primers for Drosophila InR qPCR F: ACAAAATGTAAAACCTTGCAAATCC R: GCAGGAAGCCCTCGATGA | ||
| Primers for Drosophila hsl qPCR F: AGTGATGAGTGGCTTTCCCAAC R: AAATTTAGGAATCCGTGCGGC | ||
| Primers for Drosophila RpL32 qPCR F: GCTAAGCTGTCGCACAAATG R: GTTCGATCCGTAACCGATGT | ||
Resource availability
Lead contact
All requests for reagent and resources should be directed to the Lead Contact, Dr. Wei Song (songw@whu.edu.cn).
Materials availability statement
Available through lead contact.
Experimental model and subject details
Fly strains and culture
Flies were raised on yeast-cornmeal-agar standard food medium (20 g yeast powder Instant Dry Yeast, Yichang, China), 60 g corn flour, 10 g agar, 100 g sucrose, and 15 mL 10% Tegosept (Apex, USA) in ethanol per liter) at 25°C with 50% humidity under 12-hr light-dark cycles, and 5-day old male adult flies for these experiments. The w1118 (#3605), tub-GAL4 (#5138), Cg-GAL4 (#7011), R4-GAL4 (#33832), Ire1-RNAi-HMC05163 (#62156), Ire1-RNAi-HMS03003 (#36743), UAS-Xbp1.HG (#60730) lines were obtained from the Bloomington Stock Centre. The Ire1-RNAi (v39561), Xbp1-RNAi (v109312) and foxo-RNAi (v107786) lines were obtained from the Vienna Drosophila RNAi Center. The transgenic lines, generated by the Core Facility of Drosophila Resource and Technique, SIBCB, CAS, were backcrossed for >5 generations into the w1118 background. The yw, hs-Flp; Act>CD2>GAL4, UAS-GFP, Lpp-GAL4, tub-GAL80TS and UAS-Ire1 were described previously (Song et al., 2010, Song et al., 2019; Yan et al., 2019). UAS-foxo and UAS-foxo-GFP line were kind gifts from Dr. Lei Xue from Tongji University. Bo-Gal4 was a kind gift from Dr. Alex P Gould.
Method details
Generation of Xbp1s transgenic flies
To generate the transgenic UAS-Xbp1s lines, the Xbp1s cDNA was amplified via the RT-PCR-based strategy using total RNA extracts from w1118 flies. The oligonucleotide primers for the RT-PCR were as follows.
Sense: 5′-CGGGGTACCATGGCACCCACAGCAAACAC-3’
Antisense: 5′-CTAGTCTAGATCAGATCAAACTGGGAAACA-3′
The PCR fragment for Xbp1s was first inserted into pAc5.1/V5-His-B plasmid (V4110-20, Invitrogen) before subcloning into the pUAST plasmid for the expression of V5-tagged Xbp1s protein. Following the standard germ-line transformation procedures, three independent UAS-Xbp1s transgenic lines were generated using the pUAS-Xbp1s plasmid.
Starvation resistance measurement
20 adult flies were cultured on 1% agar, and were transferred every two days to avoid fungal contamination. The number of dead flies was recorded every 12 hr. Each measurement was performed with 5–6 replicates.
Immunoblotting and quantitative RT-PCR
For immunoblot analysis, protein extracts were prepared from flies (at 3–5 days of ages; 20 flies per group) by homogenization in radioimmunoprecipitation assay (RIPA) buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl) using a tissue homogenizer (Tissernlyser-24, JingXin company, Shanghai, China). Protein concentration was determined using Bradford (Bio-Rad, Hercules, CA). The extracts were analyzed by SDS-PAGE before immunoblotting using the desired antibody. The blots were subsequently developed with an Amersham Biosciences ECL detection system. For quantitative RT-PCR analysis, total RNA was extracted from flies with TRIzol reagent (Invitrogen, Carlsbad, CA, USA). After DNase treatment, total RNA (2 μg) was converted to cDNA using Moloney Murine Leukemia Virus (MMLV) reverse transcriptase and random hexamer primers (Invitrogen). qPCR was then performed with an ABI 7500 fast real-time PCR system (Applied Biosystems, Foster City, CA) using SYBR green PCR master mix (Applied Biosystems). Ribosomal protein L32 (RpL32) was used as the internal control.
Triglyceride content measurement
For measurement of the whole-body triglyceride (TAG) content, 20 flies per group were homogenized in 0.5 mL PBS. After sufficient mixing of 0.4-mL homogenates with 1.6 mL of CHCl3:CH3OH (2:1, vol/vol), the suspension was centrifuged at 2,500 rpm. for 10 min at room temperature. The lower organic phase was transferred and air-driedovernight in a chemical hood. The residual liquid was re-suspended in 0.5 mL of 1% Triton X-100 in absolute ethanol, and the concentration of TAG was determined using the serum triglyceride determination kit (triglyceride reagent T2449 and free glycerol reagent F6428, Sigma-Aldrich, MO, USA).
Nile Red staining
Fat body were dissected from 3 rd instar larvae or male adults in PBS (Phosphate Buffered Salts, TaKaRa, Japan) and fixed in 4% paraformaldehyde (Sigma-Aldrich)/PBS for 15 min. Tissues were rinsed three times with PBST (PBS with 0.2% Triton), and then incubated for 30 min with Nile Red reagents (1–5 μg/mL in 75% glycerol, Santa Cruz) and DAPI (1:10,000 diluted by PBS, Sigmal, D5964). After rinsing three times with PBST, tissues were soaked in 60% glycerol (Sigma-Aldrich) for confocal microscopy analysis on Olympus (BX61).
Mosaic analysis
The hs-Flp; Act>CD2>Gal4/UAS system was used to generate clones in larval fat body cells. At 6 hr after egg deposition, transgenes were induced at 37°C for 30 min. Fat bodies were dissected from 3 rd instar larvae and fixed with 4% formaldehyde (Sigma-Aldrich)/PBS for 15 min. For clonal analysis of cell sizes, larval fat body cells were stained with Alexa Fluor 568 phalloidin (1:1,000, Invitrogen) or Nile Red after blocking with 5% normal Donkey serum for 2 hr to visualize cell boundaries. For clonal analysis of lipid droplets size, tissues were incubated with Nile red (0.5 μg/mL, Sigma) for 30 min. Samples were mounted using Vectashield mounting medium with DAPI (Vector Laboratories, Burlingame, CA, USA) and imaged by laser confocal microscopy (Olympus, BX61).
Cell culture and transfection
Drosophila S2 cells were cultured at 26°C in Schneider's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin and 100 μg/mL streptomycin. Transfection was performed using Effectene (Qiagen), according to the manufacturer's instructions. Transfected cells were treated with 10 μM MG132 or 30 μM CHX (Sigma-Aldrich) for the indicated time intervals. Cell protein lysates or total RNA extracts were then prepared for further analysis.
Antibodies and chemicals
Antibodies are listed as follows: p-Akt (1:1,000, Cell Signaling, 4056), Akt (1:1,000, Cell Signaling, 9272), p-eIF2α (1:1,000, Cell Signaling, 9721), p-IRE1α -S724 (1:1,000, Novus Biologicals, NB100-2323), GFP (1:10,000, Abclonal, AE012), poly-ubiquitination (1:1,000, FK1 clone, Enzo, BML-PW8805), V5 (1:5,000, Invitrogen, R960-25), and α-Tubulin (1:8,000, Sigma, T6199). The secondary antibodies used are as follows: goat anti-rabbit IgG-HRP (1:3,000, Santa Cruz) and goat anti-mouse IgG-HRP (1:3,000, Santa Cruz).
Quantification and statistical analysis
Statistical analysis was performed using unpaired two-tailed Student's t-test, one-way or two-way analysis of variance (ANOVA) followed by Bonferroni's posttest using GraphPad Prism 5.0. Log rank test was used to analyze starvation-survival curves. Data are presented as the mean ± SEM. P < 0.05 was considered as statistically significant.
Acknowledgments
We thank the Core Facility of Drosophila Resource and Technology, SIBCB, CAS, for generating the desired fly lines. This study was supported by grants from the National Natural Science Foundation of China (No. 31690102, 91857204, and 32021003 to Y.L.; 31300971 and 31471130 to P.H. and J.L.; 91957118, 31971079, and 31800999 to W.S.), the Fundamental Research Funds for the Central Universities, and from the Ministry of Science and Technology (National Key R&D Program of China 2018YFA0800700 and 2016YFA0500100 to Y.L.).
Author contributions
P.Z., P.H., T.X., X.X., Y.S., J.L., C.Y., L.W., J.G., S.C., X.W., and L.Z. performed the experiments. H.S. and J.L. analyzed the data. Y.L. and W.S. conceived and designed the experiments and wrote the manuscript.
Declaration of interests
The authors declare no conflict of interest.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102819.
Contributor Information
Wei Song, Email: songw@whu.edu.cn.
Yong Liu, Email: liuyong31279@whu.edu.cn.
Supplemental information
References
- Baker K.D., Thummel C.S. Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 2007;6:257–266. doi: 10.1016/j.cmet.2007.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel A., Schmoll D., Unterman T.G. FoxO proteins in insulin action and metabolism. Trends Endocrinol. Metab. 2005;16:183–189. doi: 10.1016/j.tem.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Baumbach J., Hummel P., Bickmeyer I., Kowalczyk K.M., Frank M., Knorr K., Hildebrandt A., Riedel D., Jackle H., Kuhnlein R.P. A Drosophila in vivo screen identifies store-operated calcium entry as a key regulator of adiposity. Cell Metab. 2014;19:331–343. doi: 10.1016/j.cmet.2013.12.004. [DOI] [PubMed] [Google Scholar]
- Coelho D.S., Cairrao F., Zeng X., Pires E., Coelho A.V., Ron D., Ryoo H.D., Domingos P.M. Xbp1-independent Ire1 signaling is required for photoreceptor differentiation and rhabdomere morphogenesis in Drosophila. Cell Rep. 2013;5:791–801. doi: 10.1016/j.celrep.2013.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J.S., Shamu C.E., Walter P. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell. 1993;73:1197–1206. doi: 10.1016/0092-8674(93)90648-a. [DOI] [PubMed] [Google Scholar]
- Demontis F., Perrimon N. FOXO/4E-BP signaling in Drosophila muscles regulates organism-wide proteostasis during aging. Cell. 2010;143:813–825. doi: 10.1016/j.cell.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Wang Z.V., Gordillo R., Zhu Y., Ali A., Zhang C., Wang X., Shao M., Zhang Z., Iyengar P. Adipocyte Xbp1s overexpression drives uridine production and reduces obesity. Mol. Metab. 2018;11:1–17. doi: 10.1016/j.molmet.2018.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor M.F., Misch E.S., Yang L., Hummasti S., Inouye K.E., Lee A.H., Bierie B., Hotamisligil G.S. The role of adipocyte XBP1 in metabolic regulation during lactation. Cell Rep. 2013;3:1430–1439. doi: 10.1016/j.celrep.2013.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronke S., Mildner A., Fellert S., Tennagels N., Petry S., Muller G., Jackle H., Kuhnlein R.P. Brummer lipase is an evolutionary conserved fat storage regulator in Drosophila. Cell Metab. 2005;1:323–330. doi: 10.1016/j.cmet.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Gutierrez E., Wiggins D., Fielding B., Gould A.P. Specialized hepatocyte-like cells regulate Drosophila lipid metabolism. Nature. 2007;445:275–280. doi: 10.1038/nature05382. [DOI] [PubMed] [Google Scholar]
- Han D., Lerner A.G., Vande Walle L., Upton J.P., Xu W., Hagen A., Backes B.J., Oakes S.A., Papa F.R. IRE1alpha kinase activation modes control alternate endoribonuclease outputs to determine divergent cell fates. Cell. 2009;138:562–575. doi: 10.1016/j.cell.2009.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J., Weissman J.S. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science (New York, NY) 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- Hong S.H., Lee K.S., Kwak S.J., Kim A.K., Bai H., Jung M.S., Kwon O.Y., Song W.J., Tatar M., Yu K. Minibrain/Dyrk1a regulates food intake through the Sir2-FOXO-sNPF/NPY pathway in Drosophila and mammals. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H.W., Zeng X., Rhim T., Ron D., Ryoo H.D. The requirement of IRE1 and XBP1 in resolving physiological stress during Drosophila development. J. Cell Sci. 2017;130:3040–3049. doi: 10.1242/jcs.203612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Xing Y., Liu Y. Emerging roles for the ER stress sensor IRE1alpha in metabolic regulation and disease. J. Biol. Chem. 2019;294:18726–18741. doi: 10.1074/jbc.REV119.007036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P., Chang K., Liu Y., Bouska M., Birnbaum A., Karashchuk G., Thakore R., Zheng W., Post S., Brent C.S. Drosophila Kruppel homolog 1 represses lipolysis through interaction with dFOXO. Sci. Rep. 2017;7:16369. doi: 10.1038/s41598-017-16638-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kishino A., Hayashi K., Hidai C., Masuda T., Nomura Y., Oshima T. XBP1-FoxO1 interaction regulates ER stress-induced autophagy in auditory cells. Sci. Rep. 2017;7:4442. doi: 10.1038/s41598-017-02960-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korennykh A.V., Egea P.F., Korostelev A.A., Finer-Moore J., Zhang C., Shokat K.M., Stroud R.M., Walter P. The unfolded protein response signals through high-order assembly of Ire1. Nature. 2009;457:687–693. doi: 10.1038/nature07661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhnlein R.P. Thematic review series: lipid droplet synthesis and metabolism: from yeast to man. Lipid droplet-based storage fat metabolism in Drosophila. J. Lipid Res. 2012;53:1430–1436. doi: 10.1194/jlr.R024299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langin D., Dicker A., Tavernier G., Hoffstedt J., Mairal A., Ryden M., Arner E., Sicard A., Jenkins C.M., Viguerie N. Adipocyte lipases and defect of lipolysis in human obesity. Diabetes. 2005;54:3190–3197. doi: 10.2337/diabetes.54.11.3190. [DOI] [PubMed] [Google Scholar]
- Lee A.H., Scapa E.F., Cohen D.E., Glimcher L.H. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis N.M., Wang L., Ortega M., Deng H., Katewa S.D., Li P.W., Karpac J., Jasper H., Kapahi P. Intestinal IRE1 is required for increased triglyceride metabolism and longer lifespan under dietary restriction. Cell Rep. 2016;17:1207–1216. doi: 10.1016/j.celrep.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao T., Shao M., Qiu Y., Huang J., Zhang Y., Song B., Wang Q., Jiang L., Liu Y., Han J.D. PKA phosphorylation couples hepatic inositol-requiring enzyme 1alpha to glucagon signaling in glucose metabolism. Proc. Natl. Acad. Sci. U S A. 2011;108:15852–15857. doi: 10.1073/pnas.1107394108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez B.A., Hoyle R.G., Yeudall S., Granade M.E., Harris T.E., Castle J.D., Leitinger N., Bland M.L. Innate immune signaling in Drosophila shifts anabolic lipid metabolism from triglyceride storage to phospholipid synthesis to support immune function. PLoS Genet. 2020;16 doi: 10.1371/journal.pgen.1009192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel M., Chevet E., Tavernier J., Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem. Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Ni H., Rui Q., Li D., Gao R., Chen G. The role of IRE1 signaling in the central nervous system diseases. Curr. Neuropharmacol. 2018;16:1340–1347. doi: 10.2174/1570159X16666180416094646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plongthongkum N., Kullawong N., Panyim S., Tirasophon W. Ire1 regulated XBP1 mRNA splicing is essential for the unfolded protein response (UPR) in Drosophila melanogaster. Biochem. Biophys. Res. Commun. 2007;354:789–794. doi: 10.1016/j.bbrc.2007.01.056. [DOI] [PubMed] [Google Scholar]
- Rubio-Patino C., Bossowski J.P., De Donatis G.M., Mondragon L., Villa E., Aira L.E., Chiche J., Mhaidly R., Lebeaupin C., Marchetti S. Low-protein diet induces IRE1alpha-dependent anticancer immunosurveillance. Cell Metab. 2018;27:828–842.e827. doi: 10.1016/j.cmet.2018.02.009. [DOI] [PubMed] [Google Scholar]
- Schlegel A., Stainier D.Y. Lessons from "lower" organisms: what worms, flies, and zebrafish can teach us about human energy metabolism. PLoS Genet. 2007;3:e199. doi: 10.1371/journal.pgen.0030199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha H., He Y., Chen H., Wang C., Zenno A., Shi H., Yang X., Zhang X., Qi L. The IRE1alpha-XBP1 pathway of the unfolded protein response is required for adipogenesis. Cell Metab. 2009;9:556–564. doi: 10.1016/j.cmet.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M., Shan B., Liu Y., Deng Y., Yan C., Wu Y., Mao T., Qiu Y., Zhou Y., Jiang S. Hepatic IRE1alpha regulates fasting-induced metabolic adaptive programs through the XBP1s-PPARalpha axis signalling. Nat. Commun. 2014;5:3528. doi: 10.1038/ncomms4528. [DOI] [PubMed] [Google Scholar]
- Sidrauski C., Walter P. The transmembrane kinase Ire1p is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–1039. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- So J.S., Hur K.Y., Tarrio M., Ruda V., Frank-Kamenetsky M., Fitzgerald K., Koteliansky V., Lichtman A.H., Iwawaki T., Glimcher L.H. Silencing of lipid metabolism genes through IRE1alpha-mediated mRNA decay lowers plasma lipids in mice. Cell Metab. 2012;16:487–499. doi: 10.1016/j.cmet.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sone M., Zeng X., Larese J., Ryoo H.D. A modified UPR stress sensing system reveals a novel tissue distribution of IRE1/XBP1 activity during normal Drosophila development. Cell Stress Chaperones. 2013;18:307–319. doi: 10.1007/s12192-012-0383-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Cheng D., Hong S., Sappe B., Hu Y., Wei N., Zhu C., O'Connor M.B., Pissios P., Perrimon N. Midgut-derived activin regulates glucagon-like action in the fat body and glycemic control. Cell Metab. 2017;25:386–399. doi: 10.1016/j.cmet.2017.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Kir S., Hong S., Hu Y., Wang X., Binari R., Tang H.W., Chung V., Banks A.S., Spiegelman B. Tumor-derived ligands trigger tumor growth and host wasting via differential MEK activation. Dev. Cell. 2019;48:277–286. doi: 10.1016/j.devcel.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Ren D., Li W., Jiang L., Cho K.W., Huang P., Fan C., Song Y., Liu Y., Rui L. SH2B regulation of growth, metabolism, and longevity in both insects and mammals. Cell Metab. 2010;11:427–437. doi: 10.1016/j.cmet.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Veenstra J.A., Perrimon N. Control of lipid metabolism by tachykinin in Drosophila. Cell Rep. 2014;9:40–47. doi: 10.1016/j.celrep.2014.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Heide L.P., Hoekman M.F., Smidt M.P. The ins and outs of FoxO shuttling: mechanisms of FoxO translocation and transcriptional regulation. Biochem. J. 2004;380:297–309. doi: 10.1042/BJ20040167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P., Ron D. The unfolded protein response: from stress pathway to homeostatic regulation. Science (New York, NY) 2011;334:1081–1086. doi: 10.1126/science.1209038. [DOI] [PubMed] [Google Scholar]
- Wang B., Moya N., Niessen S., Hoover H., Mihaylova M.M., Shaw R.J., Yates J.R., 3rd, Fischer W.H., Thomas J.B., Montminy M. A hormone-dependent module regulating energy balance. Cell. 2011;145:596–606. doi: 10.1016/j.cell.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Liu J., Gao J., Sun Y., Zhang L., Song H., Xue L., Zhan L., Gao G., Ke Z. IRE1 promotes neurodegeneration through autophagy-dependent neuron death in the Drosophila model of Parkinson's disease. Cell Death Dis. 2019;10:800. doi: 10.1038/s41419-019-2039-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H., Matsui T., Yamamoto A., Okada T., Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- Zhang K., Wang S., Malhotra J., Hassler J.R., Back S.H., Wang G., Chang L., Xu W., Miao H., Leonardi R. The unfolded protein response transducer IRE1alpha prevents ER stress-induced hepatic steatosis. EMBO J. 2011;30:1357–1375. doi: 10.1038/emboj.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Lee J., Reno C.M., Sun C., Park S.W., Chung J., Fisher S.J., White M.F., Biddinger S.B., Ozcan U. Regulation of glucose homeostasis through a XBP-1-FoxO1 interaction. Nat. Med. 2011;17:356–365. doi: 10.1038/nm.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.