Abstract
The species of the genus Potamolithus inhabiting the southwestern basin of the Andes are difficult to distinguish due to small size and similar shell morphology. Only Potamolithus australis and Potamolithus santiagensis have been traditionally recognized in this region, but the occurrence of several morphologically similar undescribed populations could increase the regional richness. Here we delimit described and potentially undescribed cryptic species of the genus using partial sequences of the mitochondrial cytochrome c oxidase subunit I (COI) gene. Network analysis and diversity indices inferred six highly differentiated haplogroups, many of them sympatric and widespread in the study area. Phylogeographic analyses suggest a scenario of recent diversification and the occurrence of multiple refuges during the successive Pleistocene glaciations. Phylogenetic analysis also recovered six major clades that showed no relationship with physiography. Species delimitation analyses consistently recognized three or four candidate species apart from P. australis and P. santiagensis. Divergence times indicate that speciation of Chilean Potamolithus began at the end of the Pliocene, probably driven by climatic rather than geographic events. Considering the high inter- and intra-basin genetic diversity, conservation efforts should be focused on protecting sympatric taxa in the basins with the highest species richness.
Subject terms: Evolution, Phylogenetics
Introduction
Species identification is increasingly important in biodiversity studies, conservation biology and natural heritage of the countries1–4. The high rate of species extinction as a consequence of habitat loss, pollution and global change, together with the enormous unknown biodiversity in some groups has led to the development of more effective and precise methods of taxonomic discrimination, which has been facilitated by technological advances5. It is now known that a portion of natural diversity is morphologically cryptic, so its identification depends on integrating molecular tools and traditional morphological approaches6.
A significant number of studies has shown that the evolutionary history of the biota in southern South America has been influenced by different geological processes and climatic events such as the Andean orogeny, marine introgressions, drainage reversals and glaciations7–20. The Andean uplift occurred from the Early Miocene generated the drainage divide between Chile and Argentina significantly impacting the distribution of genetic diversity of species, mainly the aquatic fauna10,12,19,21. The Patagonia Region suffered successive glaciations, with more than 10 major cooling events during the Pliocene and Pleistocene22–25. This intermittent melting and cooling of glaciers also had an impact on the underlying biota, leaving a genetic imprint on the phylogeography of several species7,12,19. Some of the strongest glacial events were the Great Patagonian Glaciation (GPG) c. 1 million years ago (Ma), several ancient glaciations collectively called the Pre-GPG occurred in the Lower/Middle Pleistocene (2.1–1.0 Ma) and three other cooling events called the Post-GPG that occurred in the Middle Pleistocene25–28. The boundary between the Piacenzian and Gelasian glaciations marks the boundary between the Pliocene and the Pleistocene, 2.59 Ma29. From the end of the Miocene the land south of 37°S was intermittently covered by ice sheets extending to Antarctica in the successive glaciations and whose greatest extension was reached during the Last Glacial Maximum (LGM) in the late Pleistocene, c. 0.025 Ma24,25,30,31. The tectonics and environmental processes generated expansion and retreat of forests, isolation of species into refugia, habitat fragmentation, extinctions, and the establishment of distributional limits promoted by transverse glacial tongues from the Andes that left open areas of secondary contact after melting10,12,15,19,22.
The superfamily Truncatelloidea Gray, 1840 is one of the most diverse groups of Caenogastropoda, with hundreds of genera32–34. Within this superfamily, the family Tateidae Thiele, 1925 is composed of minute freshwater snails that have a wide geographic distribution throughout the South Pacific and southern South America35–38. Tateid snails were traditionally included in the family Amnicolidae Tryon, 186335, Hydrobiidae Troschel, 185739 or Lithoglyphidae Troschel, 185737,40, revealing inconsistencies in the classification; the monophyly of the Tateidae was recovered by Wilke et al41. Two native genera of this family have been recognized in South America, Potamolithus Pilsbry, 1896, which contains about 47 species42, and the monospecific genus Strobellitatea Cazzaniga, 201743. A third genus is Potamopyrgus Stimpson, 1865, represented in Chile by the invasive species Potamopyrgus antipodarum (Gray, 1843)44. Potamolithus is a diverse genus of thick-shelled operculate snails comprising species that reach between 2.0 and 7.0 mm in length35,45–49. Although there are drawings or illustrations of the soft parts in some taxa, species taxonomy has been based mainly on characters of the external shell morphology39,45,46,48,50. In Chile only two species of the genus have been described based solely on external shell characters and operculum, Potamolithus australis Biese, 1944 and Potamolithus santiagensis (Biese, 1944). Apart from the original description, P. australis, typical of Llanquihue Lake in Chilean Patagonia, appears mentioned in a few later works and lists of species47,51–54 while López Armengol55 considered it as nomen dubium. Potamolithus santiagensis, originally described under the genus Littoridina Souleyet, 1852 from Dehesa Stream36 in central Chile and to which a population of El Yeso Spring was later added56, was subsequently included in the genus Heleobia Stimpson, 1865 but recently transferred to Potamolithus by Collado et al.57. These authors also added two populations from central Chile (Lo Carreño and El Colorado) to the range of P. santiagensis.
Mitochondrial DNA sequences generally constitute a powerful tool for species delimitation, particularly in groups that are difficult to resolve using morphological data58. The advent of species delimitation methods based on DNA sequences has provided a useful method to validate previously described species and identify new evolutionary units in a wide variety of taxa59–64. In the present study, based on the current taxonomic knowledge of the genus Potamolithus, we assessed the hypothesis that only two species exist along its distribution range in Chile. We performed phylogenetic and network analyses in a phylogeographic framework to study the biodiversity of the genus in the southwest basin of the Andes, both within and among species, including potentially cryptic species. We also used molecular species delimitation approaches to define species partitions considering already described species and new lineages in the group.
Methods
Specimen collection
Snails were collected in 2015–2017 from multiple freshwater ecosystems located in central and southern Chile (Fig. 1; Table S1). All animals were collected using a hand sieve and then stored in absolute ethanol. Snail sampling was authorized by the Subsecretaría de Pesca y Acuicultura, Ministerio de Economía, Fomento y Turismo, República de Chile (Resolution No. 3285). The procedure for handling animals was approved by the Bioethics Committee of the Universidad de Valparaíso (Resolution No. 009–2013), institution with which the first author was associated at the beginning of the project. All methods were carried out in accordance with relevant guidelines and regulations proposed. The study was carried out in compliance with the ARRIVE guidelines (http://www.nc3rs.org.uk/page.asp?id=1357).
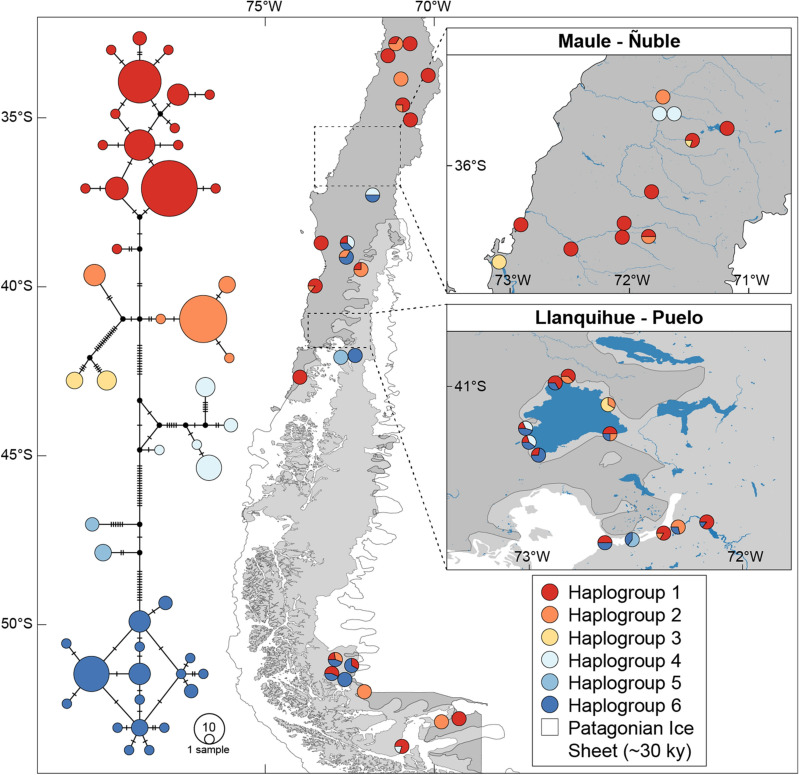
Figure 1.

Sampling localities of Potamolithus in central and southern Chile of the present and other studies44,57,65. The areas of Maule-Ñuble and Llanquihue Lake-Puelo River are enlarged. The locality of El Yeso, historically assigned to Potamolithus santiagensis, is indicated by a white triangle. The type locality of Potamolithus australis, Puerto Chico in Llanquihue Lake, is indicated by a white square. The limits of the ice sheet during the Last Glacial Maximum in Patagonia were generated following different authors24,30,31. The map was created using QGIS Geographic Information System v3.4.9 (http://www.qgis.org). (Map: M.A. Valladares).
Molecular analysis
For DNA extraction, PCR amplifications and sequencing of partial sequences of the mitochondrial cytochrome c oxidase subunit I (COI) gene we followed the methods described by Collado44 and Collado et al.57,65. Forward and reverse strands were corrected for misreads and merged into one sequence file using Sequencher v5.4.6 (Gene Codes Corporation, Ann Arbor, MI, USA). Sequences were aligned with MAFFT v7.47066 using the MAFFT online service for multiple sequence alignment67. To estimate genetic diversity of Potamolithus across the landscape we calculated the number of polymorphic sites (S), number of haplotypes (H), haplotype diversity (Hd) and nucleotide diversity (π) in DnaSP v5.10.168. To visualize the relationships between haplotypes we constructed a median-joining network69 using the program PopART v1.770. Genetic distances were estimated in MEGA X v10.2.671.
Species delimitation in Potamolithus was studied using the Automatic Barcoding Gap Discovery (ABGD) method72 (a non-tree-based method)59, the multi-rate Poisson tree processes (mPTP) analyses73 and a Bayesian general mixed Yule coalescent (bGMYC) approach74. The ABGD method is based on genetic distances and automatically detects the gap between intra and interspecific divergence, which is then used recursively to delimit species hypotheses. The mPTP method delimits species trying to determine the transition from a between‐ to a within‐species process, but also incorporates different levels of intraspecific genetic diversity deriving from differences in either the evolutionary history or sampling of each species73. The GMYC method requires an ultrametric tree (or multiple trees sampled from a posterior distribution in the Bayesian implementation) and attempts to detect the transition between the branching pattern attributed to speciation and intra-species coalescent process. The ABGD method was performed on the online web server (https://bioinfo.mnhn.fr/abi/public/abgd/abgdweb.html) and was run with the default settings (relative gap width of 1.5; intraspecific divergence: between 0.001 and 0.1). For the mPTP and bGMYC analyses, phylogenetic reconstructions were performed previously using the Maximum Likelihood (ML) and Bayesian Inference (BI) algorithms, respectively. Tatea huonensis (Tenison-Woods, 1876) (GenBank accession number: JX97061941) was used as outgroup in both reconstructions. In addition, four species of Potamolithus from Argentina were included in the reconstructions: Potamolithus agapetus Pilsbry, 1911 (GB: KM22091075), Potamolithus buschii (Frauenfeld, 1865) (GB: KM22090975), Potamolithus elenae de Lucía & Gutiérrez Gregoric, 2016 (GB: KX397599, KX39760076) and Potamolithus lapidum supersulcatus Pilsbry, 1896 (GB: KX15884376). The ML reconstruction was performed in RAxML v8.2.1277 using the GTRGAMMAI model of nucleotide substitution and the node support was obtained by performing a bootstrap analysis of 1000 pseudo-replicates. The Bayesian reconstruction for the bGMYC method was performed in BEAST v2.6.378 using the Relaxed Clock Log Normal model and a Coalescent Constant Population tree prior.
We applied two phylogeny‐aware approaches to delimit species in Potamolithus. First, mPTP analyses were performed using the online server (https://mcmc-mptp.h-its.org/mcmc/) and the ML tree from RAxML. For the mPTP method, after removing the outgroup, we performed two independent runs, each of 1,000,000 generations, sampling every 1000 generations and with a 10% burn-in. The second method was a GMYC model implemented in the package bGMYC v1.0.274 in R v3.5.2 software79. For the analyses we used 100 random trees from the BEAST reconstruction (after removing outgroups). Simulations considered 50,000 generations, discarding 40,000 replicates, and setting a thinning every 100th generation. After preliminary analyses with varying parameters on a single tree, the upper and lower bounds of the Yule and coalescent rate change parameters were set to 1.0 and 0.5, respectively, and the upper prior threshold for the number of species was set at 100. Finally, all results were visualized using the package phytools v0.6–2080.
Divergence times were estimated in BEAST using a reduced data set of the haplotypes retrieved by DnaSP. For this analysis, a strict clock model was implemented using a substitution rate of 1.7% with a standard deviation of 0.34%81. For all BEAST analyses the site model was specified using bModelTest v1.2.182, performing three independent analyses with 50 million generations each to infer a Maximum Clade Credibility Tree using a burn-in of 25%. A non-ultrametric Bayesian tree was reconstructed for the complete COI dataset using MrBayes v3.2.783. The analysis used four parallel runs with 20 million generations each using a burn-in period of 25%. ML and BI reconstructions were performed in the CIPRES cluster of the San Diego Supercomputer Center84.
Results
The amplification of the COI gene in the Chilean Potamolithus (176 individuals) produced a fragment of 510 nucleotides in length. The mtDNA COI sequence alignment recovered a total of 86 polymorphic and 69 parsimony-informative sites, defining 47 haplotypes from 196 individuals considering our original sequences and those obtained from GenBank. The nucleotide composition in the complete dataset (excluding outgroup) was 38.7% A, 19.1% C, 16.1% G and 26.1% T. The best-fitting model of nucleotide substitution, as determined by bModelTest, was the HKY + I + G. The sequences showed a pattern of high haplotype diversity (Hd = 0.928) and nucleotide diversity (π = 0.0382). The diversity indices showed the co-occurrence of several genetic entities in the same locality (considering sites with N > 2). This was reflected by a large number of mutational steps and high nucleotide diversity within localities (Table S1). The median-joining network recovered six well-differentiated haplogroups without any evident latitudinal segregation (Fig. 2). Haplogroup 1 was composed of 16 haplotypes (89 individuals) detected in 33 out of the 48 localities analyzed, representing the largest latitudinal distribution ranging from Central Chile to the southernmost locality sampled in Patagonia (32° to 53°S). Haplogroup 2 was composed of five haplotypes (33 individuals) distributed in 17 localities from central Chile to Tierra del Fuego (32° to 52°S). Haplogroup 3 contained two haplotypes (7 individuals) presents in three localities in the southcentral area of Chile (35° to 41°S). Haplogroup 4 recovered five haplotypes (15 individuals) presents in seven localities in the south-central area (35° to 53°S). Haplogroup 5 was composed of two haplotypes (5 individuals) presents in two neighboring localities from the south of Chile (41°S), representing the narrowest distribution among all haplogroups. Haplogroup 6 was composed of 17 haplotypes (42 individuals) presents in 16 localities in the southcentral area of Chile (37° to 51°S). All haplogroups showed high degrees of geographic overlap with no clear physiographic separation between or within groups. Only haplogroups 1 and 6 showed a star-like structure, however, this can be attributed to the fact that these groups contain most sampled individuals, and that the rest of the haplogroups could have been underrepresented.
Figure 2.
The median-joining haplotype networks of Potamolithus populations obtained in the present study. The areas of Maule-Ñuble and Llanquihue Lake and Puelo River are enlarged. The number of mutational steps among haplogroups is indicated by small bars. The limits of the ice sheet during the Last Glacial Maximum in Patagonia were generated following different authors24,30,31. The map was created using QGIS Geographic Information System v3.4.9 (http://www.qgis.org). (Map: M.A. Valladares).
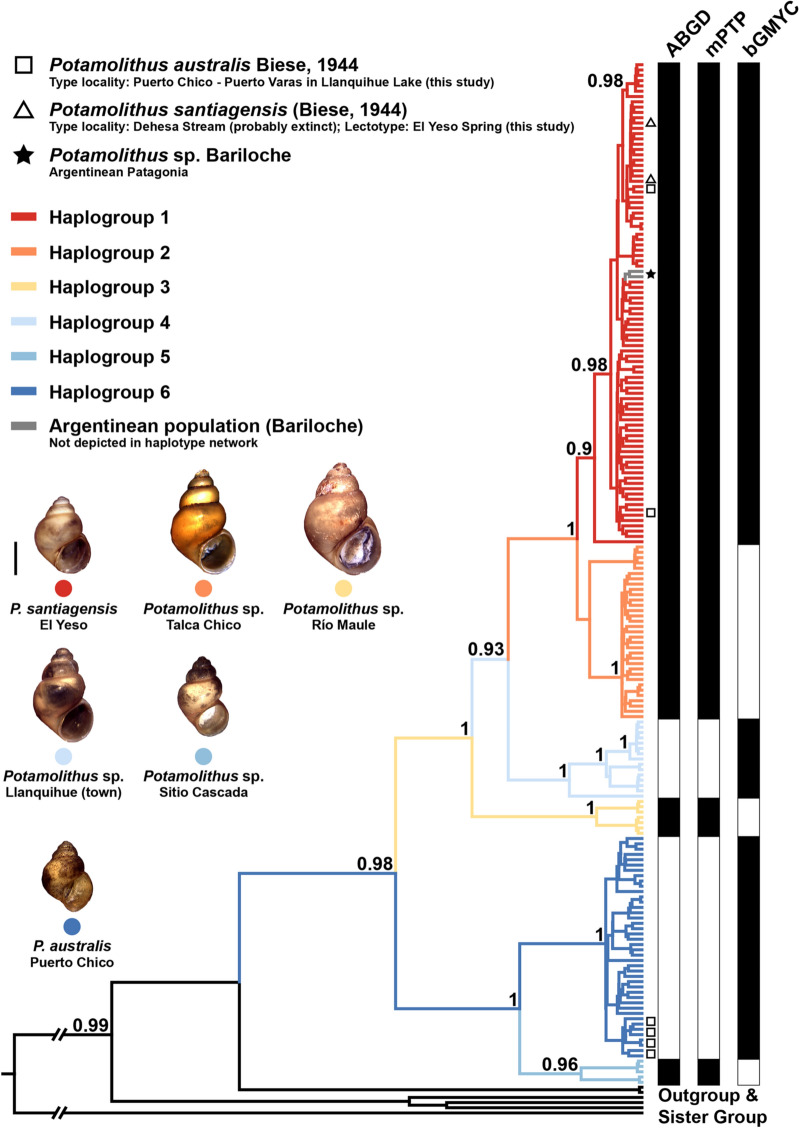
Both ML and BI methods recovered congruent tree topologies inferring six clades with high values of node support (Fig. S1), which correspond to the six haplogroups described by the haplotype network. These clades conform two major clades, one including haplogroups 1 to 4 and the other haplogroups 5 and 6. The species P. elenae (Estero Valcheta, Argentinean Patagonia) was recovered as the sister lineage of all Chilean Potamolithus populations. Potamolithus lapidum supersulcatus (Uruguay River, Argentina), P. agapetus and P. buschii (Buenos Aires, Argentina) formed a more distant lineage from the Chilean species. In addition to the Chilean populations and GenBank sequences, the phylogenetic reconstructions included two individuals from Bariloche, Argentinean Patagonia, which were recovered in Haplogroup 1 (star in Figs. 3 and S1). Genetic distances among Potamolithus species or haplogroups ranged from 3.1 to 11% (Table S2).
Figure 3.
Species delimitation of Chilean Potamolithus populations using the COI gene. Delimitation of molecular clusters was performed using ABGD, mPTP and bGMYC. Posterior probability (> 0.9) values obtained in the Bayesian COI tree are shown at the nodes (BEAST reconstruction). Scale bar next to the shells represents 1 mm.
The three different species delimitation approaches were partially congruent detecting five (ABGD and mPTP) or six (bGMYC) species partitions (Fig. 3). Potamolithus snails from El Yeso, originally assigned to P. santiagensis by Biese56 (triangles in Figs. 1, 3 and S1), were consistently identified as a partition with the three methods of species delimitation employed here, although the ABGD and mPTP analyses included haplogroups 1 and 2 whereas the bGMYC only Haplogroup 1. Samples from Puerto Chico (Llanquihue Lake), type locality of P. australis, were recovered in haplogroups 1 and 6 (square in Fig. 1, 3 and S1); most samples sequenced from this locality were haplogroup 6. Following Biese36, individuals of Haplogroup 1 were assigned to P. santiagensis and Haplogroup 6 to P. australis. The ABGD and mPTP analyses also identified haplogroups 3 to 5 as separate partitions, while the bGMYC method haplogroups 2 to 5. Divergence times estimated in BEAST suggest that the splitting of a Potamolithus ancestor from T. huonensis, a closely related tateid, occurred about 12.4 (7.4–18.1) Ma (Fig. S2). The splitting of Chilean Potamolithus species occurred between 2.7 and 0.68 Ma. Potamolithus santiagensis, circumscribed to the distribution area covered by Haplogroup 1, split from Haplogroup 2 approximately 0.68 (0.39–0.98) Ma whereas P. australis from Haplogroup 5 approximately 1.5 (0.89–2.2) Ma (Fig. S2).
Discussion
Using different molecular analyses, in the present study we showed that the biodiversity of the genus Potamolithus in Chile is underestimated, with more species than the two previously recognized. Our results suggest that there is cryptic diversity in the group and at least three species partitions would require validation and formal description. However, besides relying on single locus analyses, an integrative approach is recommended considering multiple loci and combined with other characters such as morphology, internal anatomy, radula, larval development and ecology, among others59. The phylogenetic analysis inferred at least six well-supported monophyletic groups, depicting several phylogenetic species, which is congruent with the haplogroups recovered by the network analysis.
Two of the three species delimitation methods were congruent in the number and conformation of the species partitions recovered. Five partitions were delimited using ABGD and mPTP and six with bGMYC. Previous studies have shown that the Generalized Mixed Yule Coalescent model is useful in detecting incipient speciation events85, when the threshold of intra- and interspecific distances is low, but on certain occasions it can overestimate the number of species due to pronounced intraspecific genetic variability86,87. The tree recovered in our analysis shows large branch lengths between the main clades, but short within each lineage, which is also noticeable in the haplotype network. These results suggest that the evolutionary process of the species is consistent with a recent diversification scenario (see below). Considering this, it is likely that the model implemented in the mPTP method is more appropriate to the Potamolithus pattern, mainly since it considers different levels of intraspecific diversity within a species as a result of their evolutionary history or uneven sampling73. Using a conservative approach88, the species delimitation analyses were consistent with the definition of five species partitions in Potamolithus in Chile, three of them previously undetected.
Potamolithus australis and P. santiagensis were described in the same article36, and partly on the same sheet, using characters from the external shell morphology and operculum, which are not entirely clear, despite they were described in different genera. Although these species have not been evaluated until now with independent morphological evidence, recent phylogenetic analyses based on COI sequences performed with samples of P. australis from Puerto Chico and P. santiagensis from El Yeso showed that they are different species42,57,65. Three methods of species delimitation in the present study identified these morphologically cryptic taxa as separate units, with P. santiagensis covering from Valparaíso Region to Tierra del Fuego in southern Patagonia, including Chiloé Island. Potamolithus australis has a narrower range, covering from Trupán Lake in the Bío-Bío Region to Sofía Lake in the southern section of western Patagonia in continental territory. Apart from these two species, the ABGD and mPTP methods identified three additional partitions in Potamolithus whereas the bGMYC method four. One of these lineages (Haplogroup 2) has a similar range to that of P. santiagensis, although it was not found on Chiloé Island. A second lineage (Haplogroup 4) ranges from the Maule Region to the Strait of Magellan in southern Patagonia, covering continental territory. A third lineage (Haplogroup 3) is distributed from Putagán in the Maule Region to Llanquihue Lake in Los Lagos Region, while a fourth lineage (Haplogroup 5), found inhabiting a small waterfall whose waters flow into the Puelo River in Los Lagos Region, is restricted to that single location.
The COI phylogenetic tree and network analysis of Potamolithus populations showed a deep genetic divergence but weak phylogenetic structure regarding the landscape physiography. Haplogroup 1 (P. santiagensis), Haplogroup 6 (P. australis), Haplogroup 2, and to a lesser degree haplogroups 3 and 4, have a wide geographic distribution in the country so the hypotheses of vicariance and speciation by basins do not explain the diversity pattern observed in the group. Moreover, we did not find evidence that phylogenetic structure corresponds to the faunistic units proposed by Stuardo & Vega89 for the land mollusks in continental Chile, i.e., a “northern fauna” in the north of the country and a “forest fauna” from around 38° S to Cape Horn together with a transitional zone of overlap between the units from 31 to 38° S. Furthermore, we did not find evidence of any congruence between the haplotype distribution and the biogeographic regions proposed by Dyer90 for Chilean fishes. Similar studies performed in different faunal groups have shown contradictory results14, but in all cases population differentiation was due to a mixture of evolutionary processes. For instance, in an area similar to the one studied here, a rather weak influence of geographic barriers in the differentiation of several species of the crustaceans and catfishes has been reported 7,12,19,91, although vicariance has also contributed to divergence in several fish species10,12,21.
The splitting of Chilean Potamolithus populations in the two major clades 2.73 (1.8–3.69) Ma is congruent with the Pliocene–Pleistocene limit about 2.59 Ma (ICS, 2008). Subsequent pulses of speciation are also congruent with the Pre-GPG in the Lower/Middle Pleistocene 2.1–1.0 Ma and the GPG in the Middle Pleistocene. The origin of P. australis is congruent with both dates whereas P. santiagensis with the last one; the coldest Pleistocene glaciation occurred 0.7 Ma23. We did not find association between contemporary levels of genetic diversity of Potamolithus species and the LGM. The haplotypes distributed in the north of the studied area are also found in southern Patagonia, and five of the haplogroups occur in Llanquihue Lake, a large waterbody that was completely glaciated during the early Pleistocene23. The presence of P. santiagensis, P. australis and several candidate species in southern Patagonia, an area that was covered by ice during the last ice age, may be due to the presence of refuge areas that remained in northern and/or southern Chile from which the species could have recolonized the different waterbodies once the ice melted. Colonization signals were detected in several species of sigmodontine rodents from lower latitudes in southern Patagonia14, with local differentiation also contributing to species diversity. Divergence times in Potamolithus also suggest persistence through the Pleistocene glacial cycles, similar to the pattern inferred in the sigmodontine rodents of the area14.
Several species of Potamolithus from Argentina, Uruguay and Brazil have been proclaimed with some degree of threat, mainly due to their restricted range, habitat loss, water pollution, tourism and overcollection37,49,50,92,93, although only seven appear classified in the IUCN Red List of Threatened Species as Data Deficient or Least Concern (e.g. Pastorino & Darrigran94,95). The conservation status of P. santiagensis and P. australis has not been evaluated until now. Although the wide range that both species occupy would mean classifying them in a lower conservation category, the presence of invasive species constitutes a serious threat, considering that in the type locality of P. santiagensis the species is probably extinct, possibly linked to the presence of the invasive P. antipodarum44,57,65. The occurrence of Physa acuta Draparnaud, 1805 in several freshwater ecosystems in Chile96 also constitutes a potential threat to these species, in addition to those already mentioned for other South American congeners.
Supplementary Information
Acknowledgements
This paper was supported by Proyecto DIUBB 193309 3/R. MAV acknowledges a Postdoctoral Fellowship (VRIP UBB Nº 352/6786/2020 and 352/1941/2021). GAC and CT-D acknowledges the Grupo de Investigación en Biodiversidad y Cambio Global (GI 170509/EF) of the UBB. We also thank Nicolás Villalobos and Francis Miño for their assistance during samplings and Darío Farías for molecular amplifications.
Author contributions
The three authors designed the study. G.A.C. collected samples. M.A.V. carried out molecular analyses and prepared figures. G.A.C. and M.A.V. wrote the paper with input from C.T-D. All authors reviewed and discussed the manuscript.
Data availability
Original mitochondrial sequences obtained in the present study were submitted to GenBank (accession numbers MW916963–MW917138) (Table S1). Voucher specimens are housed at the Laboratorio de Malacología y Sistemática Molecular, Universidad del Bío-Bío, Chillán, Chile.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94900-3.
References
- 1.Agapow PM, Bininda-Emonds OR, Crandall KA, Gittleman JL, Mace GM, Marshall JC, Purvis A. The impact of species concept on biodiversity studies. Q. Rev. Biol. 2004;79:161–179. doi: 10.1086/383542. [DOI] [PubMed] [Google Scholar]
- 2.Sites JW, Marshall JC. Operational criteria for delimiting species. Annu. Rev. Ecol. Evol. Syst. 2004;35:199–227. doi: 10.1146/annurev.ecolsys.35.112202.130128. [DOI] [Google Scholar]
- 3.Hooper DU, Chapin FS, Ewel JJ, Hector A, Inchausti P, Lavorel S, Lawton JH, Lodge DM, Loreau M, Naeem S, Schmid B, Setälä H, Symstad AJ, Vandermeer J, Wardle DA. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005;75:3–35. doi: 10.1890/04-0922. [DOI] [Google Scholar]
- 4.Bickford D, Lohman DJ, Sohdi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I. Cryptic species as a window on diversity and conservation. Trends Ecol. Evol. 2007;22:148–155. doi: 10.1016/j.tree.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Wiens JJ. Species delimitation: New approaches for discovering diversity. Syst. Biol. 2007;56:875–878. doi: 10.1080/10635150701748506. [DOI] [PubMed] [Google Scholar]
- 6.Ladner JT, Palumbi SR. Extensive sympatry, cryptic diversity and introgression throughout the geographic distribution of two coral species complexes. Mol. Ecol. 2012;21:2224–2238. doi: 10.1111/j.1365-294X.2012.05528.x. [DOI] [PubMed] [Google Scholar]
- 7.Pérez-Losada M, Jara CG, Bond-Buckup G, Crandall KA. Phylogenetic relationships among the species of Aegla (Anomura: Aeglidae) freshwater crabs from Chile. J. Crust. Biol. 2002;22:304–313. doi: 10.1163/20021975-99990237. [DOI] [Google Scholar]
- 8.Pérez-Losada M, Bond-Buckup G, Jara CG, Crandall KA. Molecular systematics and biogeography of the southern South American freshwater "crabs" Aegla (Decapoda: Anomura: Aeglidae) using multiple heuristic tree search approaches. Syst. Biol. 2004;53:767–780. doi: 10.1080/10635150490522331. [DOI] [PubMed] [Google Scholar]
- 9.Hernández R, Jordan TE, Dalenz Farjat A, Echavarria L, Idleman B, Reynolds JH. Age, distribution, tectonics, and eustatic controls of the Paranense and Caribbean marine transgressions in southern Bolivia and Argentina. J. South. Am. Earth. Sci. 2005;19:495–512. doi: 10.1016/j.jsames.2005.06.007. [DOI] [Google Scholar]
- 10.Ruzzante DE, Walde SJ, Cussac VE, Dalebout ML, Seibert J, Ortubay S, Habit E. Phylogeography of the Percichthyidae (Pisces) in Patagonia: Roles of orogeny, glaciation, and volcanism. Mol. Ecol. 2006;15:2949–2968. doi: 10.1111/j.1365-294X.2006.03010.x. [DOI] [PubMed] [Google Scholar]
- 11.Zemlak TS, Habit EM, Walde SJ, Battini MA, Adams EDM, Ruzzante DE. Across the southern Andes on fin: Glacial refugia, drainage reversals and a secondary contact zone revealed by the phylogeographical signal of Galaxias platei in Patagonia. Mol. Ecol. 2008;17:5049–5061. doi: 10.1111/j.1365-294X.2008.03987.x. [DOI] [PubMed] [Google Scholar]
- 12.Unmack PJ, Bennin AP, Habit EM, Victoriano PF, Johnson JB. Impact of ocean barriers, topography, and glaciation on the phylogeography of the catfish Trichomycterus areolatus (Teleostei: Trichomycteridae) in Chile. Biol. J. Linn. Soc. 2009;97:876–892. doi: 10.1111/j.1095-8312.2009.01224.x. [DOI] [Google Scholar]
- 13.Xu J, Perez-Losada M, Jara C, Crandall KA. Pleistocene glaciation leaves deep signature on the freshwater crab Aegla alacalufi in Chilean Patagonia. Mol. Ecol. 2009;18:904–918. doi: 10.1111/j.1365-294X.2008.04070.x. [DOI] [PubMed] [Google Scholar]
- 14.Lessa EP, D’Elía G, Pardiñas UFJ. Genetic footprints of late Quaternary climate change in the diversity of Patagonian-Fueguian rodents. Mol. Ecol. 2010;19:3031–3037. doi: 10.1111/j.1365-294X.2010.04734.x. [DOI] [PubMed] [Google Scholar]
- 15.Zemlak TS, Habit EM, Walde SJ, Carrea C, Ruzzante DE. Surviving historical Patagonian landscapes and climate: Molecular insights from Galaxias maculatus. BMC Evol. Biol. 2010;10:67. doi: 10.1186/1471-2148-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuñez JJ, Wood NK, Rabanal FE, Fontanella FM, Sites JW., Jr Amphibian phylogeography in the Antipodes: Refugia and postglacial colonization explain mitochondrial haplotype distribution in the Patagonian frog Eupsophus calcaratus (Cycloramphidae) Mol. Phylogenet. Evol. 2011;58:343–352. doi: 10.1016/j.ympev.2010.11.026. [DOI] [PubMed] [Google Scholar]
- 17.Ruzzante DE, Walde SJ, Macchi PJ, Alonso M, Barriga JP. Phylogeography and phenotypic diversification in the Patagonian fish Percichthys trucha: The roles of quaternary glacial cycles and natural selection. Biol. J. Linn. Soc. 2011;103:514–529. doi: 10.1111/j.1095-8312.2011.01682.x. [DOI] [Google Scholar]
- 18.Sérsic AN, Cosacov A, Cocucci AA, Johnson LA, Pozner R, Avila LJ, Sites JW, Jr, Morando M. Emerging phylogeographic patterns of plants and terrestrial vertebrates from Patagonia. Biol. J. Linn. Soc. 2011;103:475–494. doi: 10.1111/j.1095-8312.2011.01656.x. [DOI] [Google Scholar]
- 19.Unmack PJ, Barriga JP, Battini MA, Habit EM, Johnson JB. Phylogeography of the catfish Hatcheria macraei reveals a negligible role of drainage divides in structuring populations. Mol. Ecol. 2012;21:942–959. doi: 10.1111/j.1365-294X.2011.05408.x. [DOI] [PubMed] [Google Scholar]
- 20.Teta P, D’Elia G. Taxonomical notes on the long-clawed mole mice of the genus Geoxus (Cricetidae), with the description of a new species from an oceanic island of southern Chile. Hystrix. 2017;27:1–10. [Google Scholar]
- 21.Muñoz-Ramírez CP, Unmack PJ, Habit E, Johnson JB, Cussac VE, Victoriano P. Phylogeography of the ancient catfish family Diplomystidae: Biogeographic, systematic, and conservation implications. Mol. Phylogenet. Evol. 2014;73:146–160. doi: 10.1016/j.ympev.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Simpson-Vuilleumier B. Pleistocene changes in the fauna and flora of South America. Science. 1971;173:771–780. doi: 10.1126/science.173.3999.771. [DOI] [PubMed] [Google Scholar]
- 23.Rabassa J, Clapperton CH. Quaternary glaciations of the Southern Andes. Quaternary Sci. Rev. 1990;9:153–174. doi: 10.1016/0277-3791(90)90016-4. [DOI] [Google Scholar]
- 24.Clapperton CM. Quaternary Geology and Geomorphology of South America. New York: Elsevier; 1993. pp. 1–769. [Google Scholar]
- 25.Rabassa J, Coronato A, Martínez O. Late Cenozoic glaciations in Patagonia and Tierra del Fuego: An updated review. Biol. J. Linn. Soc. 2011;103:316–335. doi: 10.1111/j.1095-8312.2011.01681.x. [DOI] [Google Scholar]
- 26.Malagnino, E. The discovery of the oldest extra-Andean glaciation in the lago Buenos Aires basin, Argentina. in Quaternary of South America and Antarctic Peninsula (ed. Rabassa, J. & Salemme, M.) 9, 69–83. A.A. (Balkema Publishers, 1995).
- 27.Coronato A, Martínez O, Rabassa J. 2004. Pleistocene glaciations in Argentine Patagonia, South America. In: Ehlers J, Gibbard PL, editors. Quaternary Glaciations—Extent and Chronology (Part III) New York: Elsevier; 2004. pp. 49–67. [Google Scholar]
- 28.Coronato A, Meglioli A, Rabassa J. Glaciations in the Magellan Straits and Tierra del Fuego, southernmost South America. In: Ehlers J, Gibbard PL, editors. Quaternary Glaciations—Extent and Chronology (Part III) New York: Elsevier; 2004. pp. 45–48. [Google Scholar]
- 29.ICS (International Commission on Stratigraphy). https://stratigraphy.org/ (2008).
- 30.McCulloch RD, Bentley MJ, Purves RS. Climatic inferences from glacial and palaeoecological evidence at the last glacial termination, southern South America. J. Quaternary Sci. 2000;15:409–417. doi: 10.1002/1099-1417(200005)15:4<409::AID-JQS539>3.0.CO;2-#. [DOI] [Google Scholar]
- 31.Sugden DE, Hulton NRJ, Purves RS. Modelling the inception of the Patagonian ice-sheet. Quaternary Int. 2002;95–96:55–64. doi: 10.1016/S1040-6182(02)00027-7. [DOI] [Google Scholar]
- 32.Hershler R, Ponder WF. A review of morphological characters of hydrobioid snails. Smithson. Contr. Zool. 1998;600:1–55. doi: 10.5479/si.00810282.600. [DOI] [Google Scholar]
- 33.Bouchet P, Rocroi J-P, Frýda J, Hausdorf B, Ponder W, Valdés Á, Warén A. Classification and nomenclator of gastropod families. Malacologia. 2005;47:1–397. [Google Scholar]
- 34.Criscione F, Ponder WF. A phylogenetic analysis of rissooidean and cingulopsoidean families (Gastropoda: Caenogastropoda) Mol. Phylogenet. Evol. 2013;66:1075–1082. doi: 10.1016/j.ympev.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 35.Pilsbry, H. A. Non–marine Mollusca of Patagonia. Reports of the Princeton University Expedition to Patagonia 1896–1899 (ed. Scott, W.B) 513–633 (1911).
- 36.Biese WA. Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte I, Familia Amnicolidae. Bol. Mus. Nac. Hist. Nat. 1944;22:169–190. [Google Scholar]
- 37.Rumi A, Gutiérrez Gregoric DE, Núñez V, Cesar II, Roche MA, Tassara MP, Martín SM, López Armengol MF. Freshwater Gastropoda from Argentina: Species richness, distribution patterns, and an evaluation of endangered species. Malacologia. 2006;49:189–208. doi: 10.4002/1543-8120-49.1.189. [DOI] [Google Scholar]
- 38.Zielske S, Haase M. Molecular phylogeny and a modified approach of character-based barcoding refining the taxonomy of New Caledonian freshwater gastropods (Caenogastropoda, Truncatelloidea, Tateidae) Mol. Phylogenet. Evol. 2015;89:171–181. doi: 10.1016/j.ympev.2015.04.020. [DOI] [PubMed] [Google Scholar]
- 39.Davis GM, Pons da Silva MC. Potamolithus: Morphology, convergence, and relationships among hydrobioid snails. Malacologia. 1984;25:73–108. [Google Scholar]
- 40.Scarabino F. Lista sistemática de los Gastropoda dulciacuícolas vivientes de Uruguay. Comun. Soc. Malacol. Urug. 2004;8:347–356. [Google Scholar]
- 41.Wilke T, Haase M, Hershler R, Liu H-P, Misof B, Ponder W. Pushing short DNA fragments to the limit: Phylogenetic relationships of “hydrobioid” gastropods (Caenogastropoda: Rissooidea) Mol. Phylogenet. Evol. 2013;66:715–736. doi: 10.1016/j.ympev.2012.10.025. [DOI] [PubMed] [Google Scholar]
- 42.de Lucía M, Gonçalves ICB, Barbosa dos Santos S, Collado GA, Gutiérrez Gregoric DE. Phylogenetic and morphological study of the genus Potamolithus (Truncatelloidea: Tateidae) in hotspots of diversity at the Paranaense Forest, Argentina, with the addition of six new species. Zool. Anz. 2021;292:92–110. doi: 10.1016/j.jcz.2021.03.003. [DOI] [Google Scholar]
- 43.Cazzaniga NJ. Strobelitatea, a replacement name for Strobeliella Cazzaniga, 1981 (Gastropoda: Tateidae), non Nordsieck, 1977 (Gastropoda: Clausiliidae) Zootaxa. 2017;4216:188–189. doi: 10.11646/zootaxa.4216.2.4. [DOI] [PubMed] [Google Scholar]
- 44.Collado GA. Out of New Zealand: Molecular identification of the highly invasive freshwater mollusk Potamopyrgus antipodarum (Gray, 1843) in South America. Zool. Stud. 2014;53:70. doi: 10.1186/s40555-014-0070-y. [DOI] [Google Scholar]
- 45.Simone LRL, Moracchioli N. Hydrobiidae (Gastropoda: Hydrobioidea) from the Ribeira valley, SE Brazil, with descriptions of two new cavernicolous species. J. Mollus. Stud. 1994;60:445–459. doi: 10.1093/mollus/60.4.445. [DOI] [Google Scholar]
- 46.López Armengol MF. Taxonomic revision of Potamolithus agapetus Pilsbry, 1911, and Potamolithus buschii (Frauenfeld, 1865) (Gastropoda: Hydrobiidae) Malacologia. 1996;38:1–17. [Google Scholar]
- 47.López Armengol MF, Darrigran G. Distribución del género neotropical Potamolithus Pilsbry y Rush, 1896 (Gastropoda: Hydrobiidae) en el estuario del Río de la Plata. Iberus. 1998;12:67–74. [Google Scholar]
- 48.Pons Da Silva MC, Veitenheimer-Mendes IL. Redescrição de Potamolithus catharinae com base em topotipos (Gastropoda, Hydrobiidae), rio Hercílio, Santa Catarina, Brasil. Iheringia Ser. Zool. 2004;94:83–88. [Google Scholar]
- 49.Bichuette ME, Trajano E. Diversity of Potamolithus (Littorinimorpha, Truncatelloidea) in a high-diversity spot for troglobites in southeastern Brazil: Role of habitat fragmentation in the origin of subterranean fauna, and conservation status. Subterr. Biol. 2018;25:61–88. doi: 10.3897/subtbiol.25.23778. [DOI] [Google Scholar]
- 50.Núñez V. Redescription of Potamolithus paranensis (Pilsbry, 1911) and Potamolithus simplex (Pilsbry, 1911) (Gastropoda: Tateidae) Moll. Res. 2017;37:17–30. doi: 10.1080/13235818.2016.1201038. [DOI] [Google Scholar]
- 51.Stuardo J. Contribución a un catálogo de los moluscos chilenos de agua dulce. Gayana. 1961;1:7–32. [Google Scholar]
- 52.Valdovinos C. Biodiversidad de moluscos chilenos: Base de datos taxonómica y distribucional. Gayana. 1999;63:111–164. [Google Scholar]
- 53.Sielfeld, W. Phylum Mollusca. Guías de Identificación. Universidad Arturo Prat, Iquique, Chile, 1–15 (2001).
- 54.Valdovinos C. Estado de conocimiento de los gastrópodos dulceacuícolas de chile. Gayana. 2006;70:88–95. [Google Scholar]
- 55.López Armengol M. F. Estudio sistemático y bioecológico del género Potamolithus (Hydrobiidae) utilizando técnicas de taxonomía numérica. Tesis Doctoral N° 455, Facultad Ciencias Naturales y Museo, Universidad Nacional de La Plata, La Plata, Argentina (1985).
- 56.Biese WA. Revisión de los moluscos terrestres y de agua dulce provistos de concha de Chile. Parte II, Familia Amnicolidae (continuación) Bol. Mus. Nac. Hist. Nat. 1947;23:63–77. [Google Scholar]
- 57.Collado GA, Vidal MA, Aguayo KP, Méndez MA, Valladares MA, Cabrera FJ, Pastenes L, Gutiérrez Gregoric DE, Puillandre N. Morphological and molecular analysis of cryptic native and invasive freshwater snails in Chile. Sci. Rep. 2019;9:7846. doi: 10.1038/s41598-019-41279-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiens J, Penkrot T. Delimiting species using DNA and morphological variation and discordant species limits in spiny lizards (Sceloporus) Syst. Biol. 2002;51:69–91. doi: 10.1080/106351502753475880. [DOI] [PubMed] [Google Scholar]
- 59.Puillandre N, Modica MV, Zhang Y, Sirovich L, Boisselier M-C, Cruaud C, Holford M, Samadi S. Large-scale species delimitation method for hyperdiverse groups. Mol. Ecol. 2012;21:2671–2691. doi: 10.1111/j.1365-294X.2012.05559.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang JJ, Kapli P, Pavlidis P, Stamatakis A. A general species delimitation method with applications to phylogenetic placements. Bioinformatics. 2013;29:2869–2876. doi: 10.1093/bioinformatics/btt499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Larson ER, Castelin M, Williams BW, Olden JD, Abbott CL. Phylogenetic species delimitation for crayfishes of the genus Pacifastacus. PeerJ. 2016;4:e1915. doi: 10.7717/peerj.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chroni A, Djan M, Vidaković D, Petanidou T, Vujić A. Molecular species delimitation in the genus Eumerus (Diptera: Syrphidae) Bull. Entomol. Res. 2017;107:126–138. doi: 10.1017/S0007485316000729. [DOI] [PubMed] [Google Scholar]
- 63.Nantarat N, Sutjarit J, Tongkerd P, Wade C, Naggs F, Panha S. Phylogenetics and species delimitations of the operculated land snail Cyclophorus volvulus (Gastropoda: Cyclophoridae) reveal cryptic diversity and new species in Thailand. Sci. Rep. 2019;9:7041. doi: 10.1038/s41598-019-43382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Leria L, Vila-Farré M, Álvarez-Presas M, Sánchez-Gracia A, Rozas J, Sluys R, Riutort M. Cryptic species delineation in freshwater planarians of the genus Dugesia (Platyhelminthes, Tricladida): Extreme intraindividual genetic diversity, morphological stasis, and karyological variability. Mol. Phylogenet. Evol. 2020;143:106496. doi: 10.1016/j.ympev.2019.05.010. [DOI] [PubMed] [Google Scholar]
- 65.Collado GA, Aguayo KP, Cazzaniga NJ, Gutiérrez Gregoric DE, de Lucía M, Haase M. Systematic evaluation of cryptic freshwater snails from central Chile, including the enigmatic Littoridina santiagensis (Gastropoda, Truncatelloidea) Eur. J. Taxon. 2019;524:1–15. [Google Scholar]
- 66.Katoh K, Standley DM. MAFFT Multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 69.Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 70.Leigh JW, Bryant D. POPART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015;6:1110–1116. doi: 10.1111/2041-210X.12410. [DOI] [Google Scholar]
- 71.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Puillandre N, Lambert A, Brouillet S, Achaz G. ABGD, automatic barcode gap discovery for primary species delimitation. Mol. Ecol. 2011;21:1864–1877. doi: 10.1111/j.1365-294X.2011.05239.x. [DOI] [PubMed] [Google Scholar]
- 73.Kapli P, Lutteropp S, Zhang J, Kobert K, Pavlidis P, Stamatakis A, Flouri T. Multi-rate Poisson Tree Processes for single-locus species delimitation under Maximum Likelihood and Markov Chain Monte Carlo. Bioinformatics. 2017;33:1630–1638. doi: 10.1093/bioinformatics/btx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reid NM, Carstens BC. Phylogenetic estimation error can decrease the accuracy of species delimitation: A Bayesian implementation of the general mixed Yule-coalescent model. BMC Evol. Biol. 2012;12:196. doi: 10.1186/1471-2148-12-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Koch E, Martin SM, Ciocco NF. A molecular contribution to the controversial taxonomical status of some freshwater snails (Caenogastropoda: Rissooidea, Cochliopidae) from the Central Andes desert to Patagonia. Iheringia Ser. Zool. 2015;105:69–75. doi: 10.1590/1678-4766201510516975. [DOI] [Google Scholar]
- 76.de Lucía M, Gutiérrez Gregoric DE. The genus Potamolithus Pilsbry, 1896 (Gastropoda: Tateidae) on the Somuncurá Plateau, Patagonia. Argentina. Moll. Res. 2017;37:202–211. doi: 10.1080/13235818.2017.1279476. [DOI] [Google Scholar]
- 77.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bouckaert R, Vaughan TG, Barido-Sottani J, Duchêne S, Fourment M, Gavryushkina A, Heled J, Jones G, Kühnert D, De Maio N, Matschiner M, Mendes FK, Müller NF, Ogilvie HA, du Plessis L, Popinga A, Rambaut A, Rasmussen D, Siveroni I, Suchard MA, Wu C-H, Xie D, Zhang C, Stadler T, Drummond AJ. BEAST 2.5: An advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:e1006650. doi: 10.1371/journal.pcbi.1006650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.R Core Team. R: A Language and Environment for Statistical Computing, Vienna, Austria. R Foundation for Statistical Computing (2018). https://www.R-project.org (2019).
- 80.Revell LJ. Phytools: An R package for phylogenetic comparative biology (and other things) Methods Ecol. Evol. 2012;3:217–223. doi: 10.1111/j.2041-210X.2011.00169.x. [DOI] [Google Scholar]
- 81.Wilke T, Schultheiß R, Albrecht C. As time Goes by: A simple fool’s guide to molecular clock approaches in invertebrates. Am. Malacol. Bull. 2009;27:25–45. doi: 10.4003/006.027.0203. [DOI] [Google Scholar]
- 82.Bouckaert RR, Drummond AJ. bModelTest: Bayesian phylogenetic site model averaging and model comparison. BMC Evol. Biol. 2017;17:42. doi: 10.1186/s12862-017-0890-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES science gateway for inference of large phylogenetic trees. Proc. Gatew. Comput. Environ. Workshop (GCE) 2010;1:1–8. [Google Scholar]
- 85.García-Melo JE, Oliveira C, Da Costa Silva GJ, Ochoa-Orrego LE, Garcia Pereira LH, Maldonado-Ocampo JA. Species delimitation of neotropical characins (Stevardiinae): Implications for taxonomy of complex groups. PLoS ONE. 2019;14:1–22. doi: 10.1371/journal.pone.0216786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Talavera G, Dincă V, Vila R. Factors affecting species delimitations with the GMYC model: Insights from a butterfly survey. Methods Ecol. Evol. 2013;4:1101–1110. doi: 10.1111/2041-210X.12107. [DOI] [Google Scholar]
- 87.Jurado-Rivera JA, Zapelloni F, Pons J, Juan C, Jaume D. Morphological and molecular species boundaries in the Hyalella species flock of Lake Titicaca (Crustacea: Amphipoda) Contrib. Zool. 2020;89:353–372. doi: 10.1163/18759866-bja10004. [DOI] [Google Scholar]
- 88.Carstens BC, Pelletier TA, Reid NM, Satler JD. How to fail at species delimitation. Mol. Ecol. 2013;22:4369–4383. doi: 10.1111/mec.12413. [DOI] [PubMed] [Google Scholar]
- 89.Stuardo J, Vega R. Synopsis of the Land Mollusca of Chile. With remarks on distribution. Stud. Neotrop. Fauna Environ. 1985;20:125–146. doi: 10.1080/01650528509360682. [DOI] [Google Scholar]
- 90.Dyer BS. Systematic review and biogeography of the freshwater fishes of Chile. Estud. Oceanol. 2000;19:77–98. [Google Scholar]
- 91.Muñoz-Ramírez CP, Victoriano PF, Habit E. Inter-basin dispersal through irrigation canals explains low genetic structure in Diplomystes cf. chilensis, an endangered freshwater catfish from Central Chile. Limnologica. 2015;53:10–16. doi: 10.1016/j.limno.2015.04.004. [DOI] [Google Scholar]
- 92.Gutiérrez Gregoric DE, Núñez V, Rumi A, Roche MA. Freshwater gastropods from del Plata Basin, Argentina. Checklist and new locality records. Comun. Soc. Malacol. Urug. 2006;9:51–60. [Google Scholar]
- 93.Soutullo, A. et al. Especies prioritarias para la conservación en Uruguay 2009. Serie de Informes N° 16. Proyecto Fortalecimiento del Proceso de Implementación del Sistema Nacional de Áreas Protegidas. http://www.ruguayambiental.com/documentos/EspeciesPrioritariasConservacion.pdf (2009).
- 94.Pastorino, G. & Darrigan, G. Potamolithus doeringi. The IUCN Red List of Threatened Species 2011: e.T189312A84458605. https://www.iucnredlist.org/species/189312/84458605 (2011).
- 95.Pastorino, G. & Darrigan, G. Potamolithus simplex. The IUCN Red List of Threatened Species 2011: e.T189074A84458205. https://www.iucnredlist.org/species/189074/84458205 (2011).
- 96.Collado GA. Unraveling cryptic invasion of a freshwater snail in Chile based on molecular and morphological data. Biodivers. Conserv. 2017;26:567–578. doi: 10.1007/s10531-016-1255-y. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original mitochondrial sequences obtained in the present study were submitted to GenBank (accession numbers MW916963–MW917138) (Table S1). Voucher specimens are housed at the Laboratorio de Malacología y Sistemática Molecular, Universidad del Bío-Bío, Chillán, Chile.