Summary
Macrophages contribute to host immunity and tissue homeostasis via alternative activation programs. M1-like macrophages control intracellular bacterial pathogens and tumor progression. In contrast, M2-like macrophages shape reparative microenvironments that can be conducive for pathogen survival or tumor growth. An imbalance of these macrophages phenotypes may perpetuate sites of chronic unresolved inflammation, such as infectious granulomas and solid tumors. We have found that plant-derived and synthetic rocaglates sensitize macrophages to low concentrations of the M1-inducing cytokine IFN-gamma and inhibit their responsiveness to IL-4, a prototypical activator of the M2-like phenotype. Treatment of primary macrophages with rocaglates enhanced phagosome-lysosome fusion and control of intracellular mycobacteria. Thus, rocaglates represent a novel class of immunomodulators that can direct macrophage polarization toward the M1-like phenotype in complex microenvironments associated with hypofunction of type 1 and/or hyperactivation of type 2 immunity, e.g., chronic bacterial infections, allergies, and, possibly, certain tumors.
Subject areas: Immunology, Microbiology
Graphical abstract

Highlights
-
•
Rocaglates sensitize MΦ to IFNγ proportional to their translation inhibition activity
-
•
Active rocaglates inhibit MΦ response to IL-4 and CSF1
-
•
Rocaglates upregulate stress response and host defense pathways
-
•
Rocaglates induced autophagy in MΦ and improved control of virulent mycobacteria
Immunology; Microbiology
Introduction
The increasing problem of infectious diseases caused by antibiotic-resistant bacteria has intensified a search for novel therapeutic approaches. Among them are host-directed therapies (HDTs) aimed at either boosting immune-mediated bacterial control or reducing immunopathology, broadly referred to as mechanisms of host resistance or disease tolerance, respectively (Wallis and Hafner, 2015). Many intracellular bacterial pathogens reside in macrophages, the very cells of the innate immune system whose major function is to eliminate invading pathogens. This paradox is enabled by macrophages' plasticity, which successful pathogens exploit to create cellular niches for persistence and replication within inflamed tissue of susceptible hosts (Price and Vance, 2014).
To combat intracellular bacteria, macrophages activate cell autonomous defense mechanisms, such as phagocytosis, production of highly toxic reactive oxygen and nitrogen species, bactericidal peptides, as well as phagosome maturation and autophagy to deliver the ingested pathogens to lysosomes for destruction. This pro-inflammatory type of macrophage activation can be induced by microbial ligands and proinflammatory cytokines. It is broadly referred to as “classical” or M1 type, although this definition encompasses an array of related but non-identical macrophage activation states (Murray et al., 2014). Individuals whose macrophages fail to respond to IFNγ, the prototypical M1 polarizing signal, are extremely susceptible to infections caused by intracellular mycobacterial species, including an avirulent vaccine strain of M. bovis Bacillus Calmette-Guerin (BCG) and normally avirulent environmental non-tuberculous mycobacteria (Fortin et al., 2007). The alternative macrophage activation programs, broadly referred to as M2 type, are mediated by upregulation of anti-inflammatory cytokines. The M2-like macrophages play important homeostatic and adaptive physiological roles in immune regulation, tissue remodeling, fibrosis, and control of parasitic helminth infections (Sica and Mantovani, 2012). However, the activation of an alternative macrophage activation program by IL-4 favors Mtb replication by suppressing autophagy and antigen presentation by the infected macrophages. Overexpression of IL-4 in a mouse model in vivo leads to progression of tuberculosis (TB) and formation of organized necrotic granulomas typical of human TB (Heitmann et al., 2014). A helminth-induced type 2 immune response also antagonizes host protective responses to mycobacteria (Potian et al., 2011). The M2-like differentiation of macrophages is also involved in local immune suppression within tumor microenvironments and tumor promotion (Chanmee et al., 2014).
In vitro, the prototypic M1- and M2-like macrophage polarization states can be artificially induced by treatment with cytokines or microbial ligands and represent polar antagonistic phenotypes. Thus, pretreatment with IL-4 was shown to downregulate the macrophage responsiveness to IFNγ (Salgame et al., 2013; Potian et al., 2011; Harris et al., 2007; Patel et al., 2005; Browne and Holland, 2010), while priming with IFNγ reduces their responses to IL-4(Venkataraman et al., 1999). Within specific in vivo environments, however, macrophages are exposed to multiple and often conflicting polarization signals. In addition to cytokine levels, the macrophage polarization in vivo may be influenced by the growth factors, cell interactions, and environmental stressors. Despite the mutual antagonism of the macrophage polarization programs in vitro, recent studies demonstrated simultaneous presence of M1- and M2-like macrophages within TB lesions, where higher proportion of the M2-like Arginase-1-expressing macrophages was associated with the disease progression (Cadena et al., 2017; Carow et al., 2019; Cronan et al., 2021). Spatial transcriptomic analysis and immunochemistry revealed that the M2-like macrophage markers were more abundant within the inner areas of TB granulomas (Carow et al., 2019; Cadena et al., 2017), whereas the IFNγ-producing T cells were located mostly on the periphery of the organized granulomas (Sakai et al., 2014). Because IFNγ is a labile homodimer, the diffusion of its biologically active dimer within inflammatory lesions is limited. This suggests that the balance of M1/M2 macrophage phenotypes within TB lesions may determine local Mycobacterium tuberculosis (Mtb) control within specific granuloma compartments (Marino et al., 2015).
To identify candidate compounds capable of directing granuloma macrophages toward the M1-like activation and enhancing their anti-mycobacterial activity, we searched for small molecules that enhance M1-like macrophage polarization in synergy with low concentrations of IFNγ. Screening small molecule libraries, we found several synthetic rocaglates that potentiated pleiotropic effects of IFNγ on primary macrophages (Bhattacharya et al., 2016). Rocaglates are a class of bioactive derivatives of natural products isolated from medicinal plants of the Aglaia species that have been used in traditional Chinese medicine for treatment of fever, cough, diarrhea, and inflammation. In modern medicine, the therapeutic potential of synthetic rocaglates has been evaluated for cancer therapy (Santagata et al., 2013; Hwang et al., 2004; Kim et al., 2006, 2007) and viral infections including coronaviruses (Müller et al., 2018). Rocaglates bind to and suppress the RNA helicase activity of the eIF-4A subunit of the eIF4F translation initiation complex and selectively inhibit cap-dependent protein translation (Bordeleau et al., 2008; Santagata et al., 2013). The structurally related rocaglamide A (RocA) has been also shown to inhibit the Raf-MEK-ERK growth factor-activated pathway by targeting prohibitins 1 and 2 (Polier et al., 2012). Although rocaglates are generally known as inhibitors of protein translation, we found that in macrophages, select rocaglates induced the upregulation of IRF1 protein, a master regulator of IFNγ-activated pathways, in the presence of IFNγ at concentrations as low as 0.1 U/ml.
In the present study, we delineate mechanisms of the enhancement the M1 macrophage polarization by active rocaglates. We also demonstrate that rocaglates inhibit macrophage responsiveness to CSF-1 and IL-4 and potentiate control of virulent Mtb by primary macrophages.
Results
Macrophage activation by rocaglates is coupled to partial translational inhibition
To delineate the relationship between translation inhibition and macrophage activation by rocaglates, we screened a library of synthetic rocaglate derivatives. Out of 86 compounds, 23 induced significant Irf1 mRNA upregulation in the presence of 0.2 U/ml of IFNγ. Next, we determined the translation inhibitory concentrations IC50 of these compounds using the 293TR-FLuc translation reporter cell line (Santagata et al., 2013). We observed a tight negative correlation (r = - 0.6832) between the Irf1 mRNA induction and IC50 of those compounds (Figure 1A, Table S1), suggesting a mechanistic link between the translational inhibition and Irf1 co-stimulation properties. To verify this relationship in primary macrophages, we compared translation inhibition activities of two structurally similar rocaglates CMLD010536 and CMLD010850 (Figure 1B) in mouse bone-marrow-derived macrophages (BMDMs) using a puromycin incorporation assay (Rodrigo et al., 2012). CMLD010536 exerted greater translation inhibition than CMLD010850 (Figure 1C), and it was also significantly more potent as an inducer of Irf1 mRNA expression cooperatively with a low dose of IFNγ, as well as of another, IFNγ-independent, rocaglate target gene Txnip (Santagata et al., 2013) (Figure 1D).
Figure 1.
Translation inhibition is required for rocaglate to activate BMDMs.
(A) Twenty three rocaglate analogs were examined (at 0.3 μM for 24 hr) for their translation inhibition activity and induction of Irf1 mRNA expression in the presence of 0.1 U/ml of IFNγ. The correlation coefficient was calculated using GraphPad Prism 8.
(B) Chemical structure of small molecules CMLD010536 and CMLD010850.
(C) Translation inhibition by rocaglates in BMDMs assessed using the puromycin incorporation assay. BMDMs were treated with indicated concentrations of CMLD010536 and CMLD010850 for 2 hr followed by puromycin (5 μg/mL) for 1 hr. Puromycin incorporation was monitored by western blot using puromycin-specific antibody.
(D) BMDMs were treated with 100 nM CMLD010536 (labeled as 536) and CMLD010850 (labeled as 850). The Irf1 (in the presence of IFNγ) and Txnip (without IFNγ) mRNA induction was assessed using quantitative real-time RT-PCR (qRT-PCR). The data are represented as mean ± standard error of the mean (SEM), and p value ≤ 0.05 was considered statistically significant.
(E) BMDMs were treated with 0.1 and 0.3 μM CMLD010536 for 2, 8, and 23 hr followed by a treatment with puromycin (5 μg/mL) for additional 1 hr. Incorporation of puromycin was monitored by western blot using puromycin-specific antibody.
(F) BMDMs were treated with 0.1 μM rocaglate for 6 and 18 hr, and protein levels of Myc, ATF-3, and ATF-4 were monitored by western blot using specific antibody.
Next, we used a puromycin incorporation assay to investigate the dynamics of translation inhibition by CMLD010536 in BMDMs. Protein translation was quantified after 3, 9, and 24 hr of the rocaglate treatment. At each timepoint, puromycin was added for one hour and its incorporation into newly synthesized proteins was determined using western blot with puromycin-specific antibodies. The rocaglate treatment significantly reduced translation at all timepoints in a dose-dependent manner (Figure 1E). However, the rates of residual protein translation were similar at all of the timepoints. These data demonstrate that translation inhibition was partial and that the rocaglate-resistant translation was sustained during the course of the rocaglate treatment.
As reported previously in T cells (Wolfe et al., 2014), translation of c-Myc was sensitive to eIF4A inhibition by rocaglates in BMDMs, as well. After rocaglate treatment the expression of Myc protein was completely abrogated (Figure 1F), while Myc mRNA was upregulated (Figure S1A). Inhibition of the cap-dependent protein translation, however, may trigger a signaling cascade known as the integrated stress response (ISR). It is initiated via cap-independent translation of a constitutively expressed mRNA encoding a stress response transcription factor ATF4, which is followed by the upregulation of its downstream target ATF3. Canonical ISR is induced by several stress-activated kinases via phosphorylation of the translation initiation factor eIF2α. We found that the rocaglate treatment also induced the ATF4 protein followed by the upregulation of the ATF3 mRNA (Figure S1B) and protein (Figure 1F and Figure S1C).
To broadly identify proteins expressed in BMDMs treated with rocaglates for 24 hr, we performed proteomic analysis using mass spectrometry (Table S2). The autophagy adapter, stress response protein Sequestosome 1 (SQSTM1, a.k.a. p62), was found to be the most abundant. SQSTM1/p62 plays an important role in stress resilience by controlling selective autophagy and anti-oxidant defense gene expression (Katsuragi et al., 2015; Moscat and Diaz-Meco, 2011; Knaevelsrud and Simonsen, 2010; Johansen and Lamark, 2011; Taguchi et al., 2011; Jaramillo and Zhang, 2013; Padmanabhan et al., 2006; Fukutomi et al., 2014; Komatsu et al., 2010; Lau et al., 2010).
Taken together, our data demonstrate a reciprocal downregulation of Myc and the upregulation of the ISR TFs ATF4 and ATF3 and p62 in primary macrophages. This response to the rocaglate treatment mimics a coordinated stress response encompassing several adaptive pathways.
The rocaglate effect on macrophage activation is mediated by p38 stress kinase
The rocaglate treatment of BMDMs induced de novo p62 protein and mRNA syntheses in an IFNγ-independent manner (Figures 2A and 2B, respectively). Comparing the p62-inducing activities of CMLD010536 with natural rocaglates, silvestrol and rocaglamide A demonstrated that those compounds had similar activities, while CMLD010850 did not induce p62 mRNA expression (Figure 2C). Thus, the upregulation of p62 and Irf1 mRNAs by rocaglates was associated with their ability to selectively inhibit protein translation, although the Irf1 induction required co-stimulation with IFNγ, while the p62 upregulation was IFNγ independent.
Figure 2.
Selective translation inhibition by rocaglate remodels macrophage proteome and upregulates stress response proteins.
(A) BMDMs were treated with CMLD010536 (0, 25, 50, and 100 nM) for 6 and 24 hr. The expression of p62 protein was measured by western blot using p62-specific antibody.
(B) The p62 mRNA induction in BMDMs treated with CMLD010536 (at 25, 50, and 100 nM) for 6 hr. The p62 mRNA levels were determined using qRT-PCR and expressed as a fold induction compared to untreated cells.
(C) Effect of the rocaglate derivatives CMLD010536 (536), sivestrol (sil), rocaglamide A (RocA), and CMLD010850 (850) on p62 mRNA expression. BMDMs were treated with each compound at 100 nM for 24 hr. The p62 mRNA levels were measured by qRT-PCR and expressed as the fold induction compared to untreated cells.
(D) Effect of MAPK inhibitors SB203580 (p38), SB600125 (JNK), and U0126 (ERK) on p62 mRNA expression in BMDMs treated with 100 nM CMLD010536 for 6 hr. Inhibitors were added to a final concentration 10 μM, 30 min prior to the rocaglate treatment. The p62 mRNA levels were determined using qRT-PCR and expressed as fold induction compared to untreated BMDMs.
(E) The kinetics of p38 phosphorylation in BMDMs treated with 0.3 μM CMLD010536 was determined by western blot using phospho-p38 specific antibody.
(F) The effect of the p38 inhibitor SB203580 (10 μM) on IRF1 protein induction by CMLD010536 (300 nM) in synergy with low concentrations of IFNγ (0.05 or 0.1 U/ml). The inhibitor was added to BMDMs 30 min prior to stimulation with the rocaglate and IFNγ. The IRF1 protein levels were determined after 6 hr by western blot using IRF1-specific antibody. The relative densitometric values of IRF1 bands' areas were determined after normalization and are shown as ratios to non-stimulated controls above the blot.
(G) Treatment with 0.1 U/ml alone did not induce the p38 phosphorylation in BMDMs as compared to cells treated with 0.1 U.ml of IFNγ in combination with CMLD010536 (0.3 μM). BMDMs were treated with 0.1 U/ml of IFNγ either in the absence or presence of the rocaglate, and phospho-p38 levels were determined by western blot using phospho-p38-specific antibody.
(H) BMDMs were treated with 0.3 μM rocaglate ,and phosphorylation of STAT1 at Ser727 at indicated timepoints was monitored by western blot using phospho-Ser727-specific antibody.
(I) BMDMs were treated with 0.3 μM rocaglate and 10 μM SB203580 for indicated timepoints and observed STAT1 total level and STAT1 phosphorylation at Ser727 by western blot using specific antibody.
(J) Mass spectrometry measured abundances of proteins which are upregulated by rocaglate (0.1 μM) and IFNγ (0.2 U/mL) in a synergistic way. Bliss independence criterion was used for estimation of synergy effects (see Methods). The data in panels B, C and D are represented as mean ± SEM, and p value ≤ 0.05 was considered statistically significant.
Our previous analysis of rocaglate-mediated macrophage activation using gene expression profiling suggested activation of stress kinase-mediated pathways (Bhattacharya et al., 2016). Therefore, we tested whether p62 induction by rocaglates was mediated by stress-activated MAP kinases using small molecule inhibitors. The p38 inhibitor (SB203580 at 10 uM), added 30 min prior to treatment with 100 nM CMLD010536, completely abrogated p62 mRNA upregulation, while the inhibitors of JNK (SB600125) and ERK (U0126/PD98059) had no effect (Figure 2D). The p38 MAPK inhibitor also abrogated the upregulation of two other rocaglate-induced inflammatory genes, Txnip and Ptgs2 (Figures S2A and S2B, respectively), and also decreased the p62 protein levels (Figure S2C). We confirmed the specificity of p38-mediated inhibition of p62 mRNA using another p38 inhibitor SB212090 (Figure S2D). Of note, both p38 inhibitors were non-toxic during the treatment period, either alone or in combination with rocaglate (Figure S2E).
The p38 activation can be induced by reactive oxygen species (ROS); we tested whether oxidative stress plays a role in the p38-mediated p62 induction by the rocaglate. However, Sqstm1/p62 mRNA induction was unaffected in the presence of ROS scavenger N-acetyl-cysteine (Figure S2F). Notably, the known translation elongation inhibitor cycloheximide also failed to stimulate the expression of p62 (Figure S2G).
Next, we compared the kinetics of p38 phosphorylation (Figure 2E) and p62 mRNA upregulation (Figure S2H) during the course of the rocaglate treatment. Rapid activation of p38, started at 15 min of the treatment, reached a plateau at 30–120 min and declined by 4 hr. Meanwhile, the p62 mRNA continued to increase 4–24 hr of the treatment duration indicating that p38 phosphorylation was necessary to initiate a signaling cascade leading to the p62 upregulation but was dispensable for its maintenance.
Taken together, these data demonstrate that macrophage treatment with rocaglates induces stepwise transcriptome and proteome remodeling. At the early initiation step (0.5–2 hr), the activation of p38 stress kinase drives transcriptional upregulation of p62, Ptgs2, Txnip, and other inflammatory genes. The translation of ISR transcription factors and p62 occurs with a delay at a later stage (16–24 hr) of rocaglate treatment. Interestingly, the p38 inhibitor partially inhibited the p62 protein upregulation by the rocaglate. Perhaps, other signaling pathways, yet to be identified, are involved in the upregulation of p62 protein at the later stage.
The p38 activation mediates synergy of rocaglates with low concentrations of IFNγ
Next, we wanted to determine whether the upregulation of IRF1 by rocaglates in the presence of low concentrations of IFNγ was also mediated by p38. Indeed, CMLD010536 boosted the levels of IRF1 protein in the presence of IFNγ at concentrations as low as 0.05 and 0.1 U/ml, and this boosting effect was abrogated by p38 inhibition (Figure 2F). Importantly, treatment with active rocaglates alone suppressed the basal IRF1 protein expression (Figure 2F). These data demonstrate that synergy of rocaglates with low concentrations of IFNγ in IRF1 upregulation is mediated by p38 MAPK. We observed the p38 phosphorylation in BMDMs treated with rocaglate and low concentration of IFNγ but not with IFNγ alone (Figure 2G). The p38 inhibitor suppressed the Irf1 mRNA co-stimulation by the rocaglate (Figure S3A), but it did not suppress the Irf1 mRNA induction by standard doses of IFNγ alone (Figure S3B). Thus, the synergistic effect of rocaglates with the low concentration of IFNγ does not simply mimic the canonical IFNγ pathway.
The upregulation of the Irf1 gene expression by IFNγ is mediated by the binding of STAT1 homodimers to the gamma-activatable sequence (GAS) in the Irf1 promoter. The STAT1 homodimer formation and promoter binding require STAT1 phosphorylation at the canonical tyrosine-701 by JAK kinases, an event that occurs upon the IFNγ dimer binding to its receptor (Meraz et al., 1996; Decker et al., 1997). However, CMLD010536 did not increase STAT1 phosphorylation at the canonical tyrosine site in our studies. There is emerging evidence that serine (Ser-727) phosphorylation of STAT1 by stress-activated MAP kinases p38 and JNK mediates various stress responses (Dudley et al., 2004) and may account for nearly 80% of IFNγ-induced transcriptional activity by recruiting transcriptional co-activators to the STAT1 dimer. Indeed, we found that the CMLD010536 treatment induced STAT1 Ser727 phosphorylation as early as 15 min (Figure 2H), and this STAT1 Ser727 phosphorylation was inhibited by the p38 MAPK inhibitor (Figure 2I).
To further explore the synergistic effect of rocaglates and low concentration of IFNγ, we compared protein content of BMDMs treated with rocaglate plus IFNγ (0.2 U/ml) vs IFNγ (0.2 U/ml) alone using mass spectrometry. To identify proteins that are upregulated by rocaglate and IFNγ in a synergistic manner, we used the Bliss independence criterion for formal synergy analysis (Greco et al., 1995) (see STAR Methods). Table S3 (“Proteins induced by rocaglates in synergy with IFN-gamma, related to Figure 2J”) presents a list of differentially expressed proteins sorted by the Bliss synergy score in the descending order. Positive values of the synergy score correspond to synergy between rocaglate and IFNγ. A cluster of proteins for which synergistic effect is most pronounced is shown in Figure 2J. This cluster is enriched for known IFN-inducible proteins, including proteins that play important roles in control of intracellular parasites (IFI47, ICAM-1), viruses (IFIT2, SAMHD1, IFI204, Mnda/IFI16), and M1 macrophage polarization (IRF 5). Thus, the rocaglate synergy with IFNγ has a broad effect on macrophage M1-like polarization and activation of diverse mechanisms of innate immunity.
Suppression of the alternative activation of macrophages by rocaglates
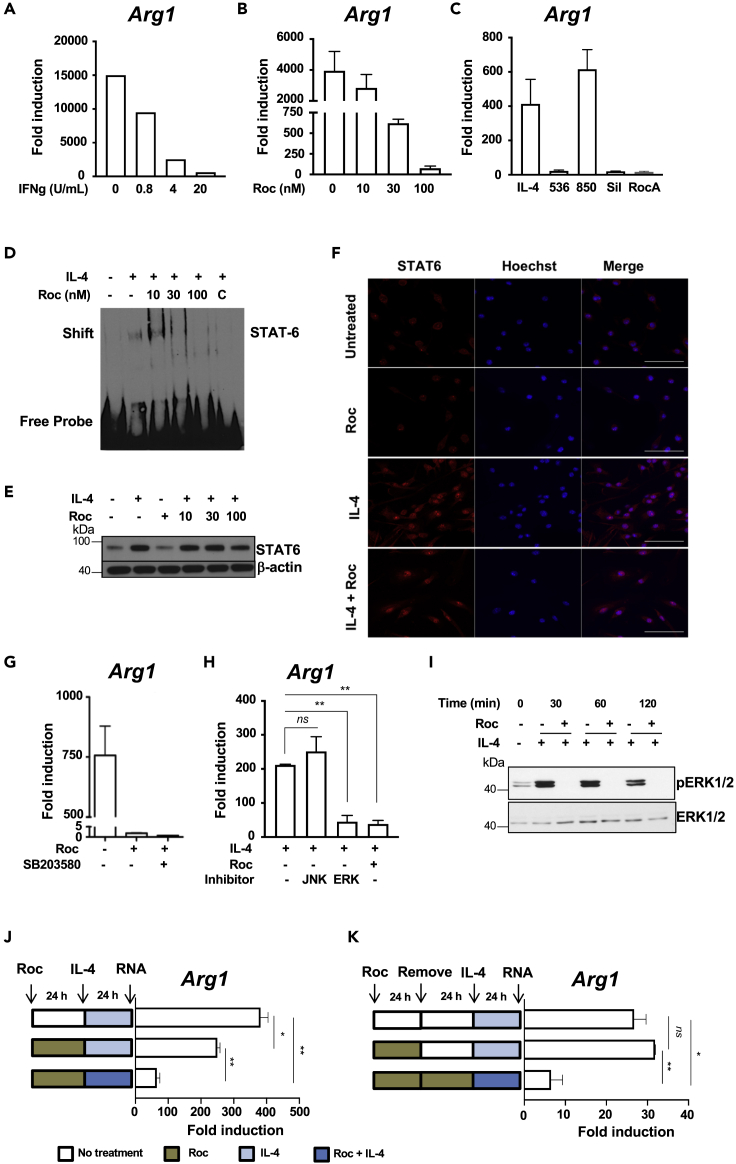
Once we determined that active rocaglates boosted a pro-inflammatory (M1-like) macrophage program, we investigated whether it could concurrently suppress the alternative (M2-like) macrophage activation. In agreement with the known antagonistic relationship of these modes of macrophage activation, pretreatment of macrophages with IFNγ abrogated their response to the M2-polarizing cytokine IL-4, as measured by the Arg1 mRNA expression (Figure 3A). Similarly, the induction of the M2 polarization markers Arg1 and Fizz1 by IL-4 was suppressed by CMLD010536 in a dose-dependent manner (Figure 3B and Figure S4A, respectively). Interestingly, this suppressor effect of the rocaglate does not require the co-stimulation with IFNγ. It does correlate, however, with a rocaglate ability to inhibit mRNA translation: at 0.1 μM concentration, the alternative macrophage activation was suppressed by CMLD010536, silvestrol and rocaglamide A, but not by CMLD010850 (Figure 3C), which is a much less potent translational inhibitor and does not inhibit translation at this concentration (Figure 1C).
Figure 3.
Suppression of the alternative macrophage activation by rocaglates
(A) Pretreatment with IFNγ suppresses macrophage responses to IL-4. BMDMs were pretreated with increasing concentrations of IFNγ for 4h and stimulated with 25 ng/mL IL4 for additional 20 hr. The expression of Arg1 mRNA was measured using qRT-PCR. Fold induction was calculated using the IL4 untreated controls.
(B) BMDMs were pretreated with indicated concentrations of CMLD010536 for 4h, stimulated with IL4 and analyzed for Arg1 mRNA expression as in (A)
(C) BMDMs were pretreated with different rocaglate derivatives (at 100 nM) for 4 hr followed by treatment with 25 ng/mL IL-4 for 20 hr. The Arg1 mRNA induction was determined as in (A)
(D) Effect of CMLD010536 on the IL4-induced STAT6 binding activity in BMDMs was monitored using EMSA. BMDMs were pretreated with the indicated rocaglate concentrations for 2 hr and stimulated with IL4 (25 ng/mL) for additional 4 hr. Nuclear extracts were prepared and STAT6 promoter binding activity was monitored by EMSA using STAT6-specific DNA probe. Competition with unlabeled STAT6 probe (C) demonstrates the specificity of the shifted band.
(E) Rocaglate does not inhibit STAT6 protein upregulation and nuclear translocation by IL4. BMDMs were treated with indicated concentrations of CMLD010536 for 2 hr followed by a treatment with IL4 (25 ng/mL) for additional 4 hr. Total STAT6 protein levels were measured by western blot. The total STAT6 protein levels were not affected by the rocaglate treatments.
(F) BMDMs were treated with 30 nM concentration of rocaglate for 2 hr followed by a treatment with IL4 (25 ng/mL) for additional 4 hr. The BMDMs were stained using STAT6 antibody, nuclear stain Hoechst and observed nuclear translocation of STAT6 using Zeiss LSM 710-Live Duo scan confocal microscopy. Scale bar represents 50 μM.
(G) The induction of the Arg1 mRNA by IL4 (25 ng/mL) was suppressed by CMLD010536 (100 nM), and this suppression was not abrogated by the p38 inhibitor SB203580 (10 μM). The Arg1 mRNA was measured using qRT-PCR as fold induction at 24 hr.
(H) The induction of the Arg1 mRNA by IL4 (25 ng/mL) was not affected by the JNK inhibitor (SB600125) but was suppressed by the ERK inhibitor (U0126) to a degree similar to CMLD010536 (100 nM). The inhibitors were added 30 min prior to IL-4, and the Arg1 mRNA expression was measured using qRT-PCR as fold induction at 24 hr.
(I) The rocaglate treatment abrogates ERK1/2 phosphorylation in IL-4 stimulated macrophages. BMDMs were pre-treated with CMLD010536 (100 nM) for 12 hr and stimulated with IL4 (100 ng/mL). Phospho- and total ERK1/2 proteins were measured using western blot after the indicated periods of the IL-4 stimulation.
(J and K) The suppression of the BMDM response to IL-4 by the rocaglate is reversible. BMDMs were pretreated with rocaglate (100 nM) for 24 hr, washed and stimulated with IL-4 (100 ng/mL) either alone or in the presence of rocaglate (100 nM). The cells not treated with rocaglate and stimulated with IL-4 were used as a positive control. The expression of Arg1 mRNA was measured by using qRT-PCR as above. In (J), BMDMs were pretreated and washed as in (I) but were rested for additional 24 hr before stimulation with IL-4. BMDMs cultured in the presence of the rocaglate for the duration of the experiment served as a control for suppression. TThe data in panels A, B, C, G, H, J and K are represented as mean ± SEM, and p value ≤ 0.05 was considered statistically significant.
The upregulation of the IL-4-inducible genes is mediated by the transcription factor STAT6. Using electrophoretic mobility shift assay (EMSA) with a STAT6 consensus sequence oligonucleotide probe, we determined that pretreatment with 30 and 100 nM of CMLD010536 prior to IL-4 stimulation inhibited STAT6 DNA binding (Figure 3D). This effect was not associated with a decrease in the total STAT6 protein levels (Figure 3E). The rocaglate treatment did not inhibit the IL-4-induced nuclear translocation of STAT-6 (Figure 3F and Figures S4B and S4C). These observations suggested that the suppression of the STAT6 DNA binding activity was a regulatory phenomenon that could not be simply attributed to inhibition of the STAT6 protein translation. Therefore, we sought to determine whether the suppressor activity was mediated by p38. The p38 inhibitor, however, did not restore the expression of Arg1 inhibited by CMLD10536 (Figure 3G). Instead, we observed that the rocaglate treatment mimicked the inhibitory effect of the ERK inhibitor U0126 on IL4-induced Arg1 mRNA expression (Figure 3H). Indeed, the rocaglate treatment completely abrogated the ERK1/2 phosphorylation in IL4-stimulated macrophages without affecting the total ERK1/2 levels (Figures 3I and Figure 4F). This finding is consistent with the previously described role of ERK1/2 in the alternative macrophage activation. Thus, rocaglates inhibit ERK1/2 phosphorylation and Myc translation (Figure 1F), whose activities are necessary for M2-like macrophage polarization (Pello, 2016; Pello et al., 2012).
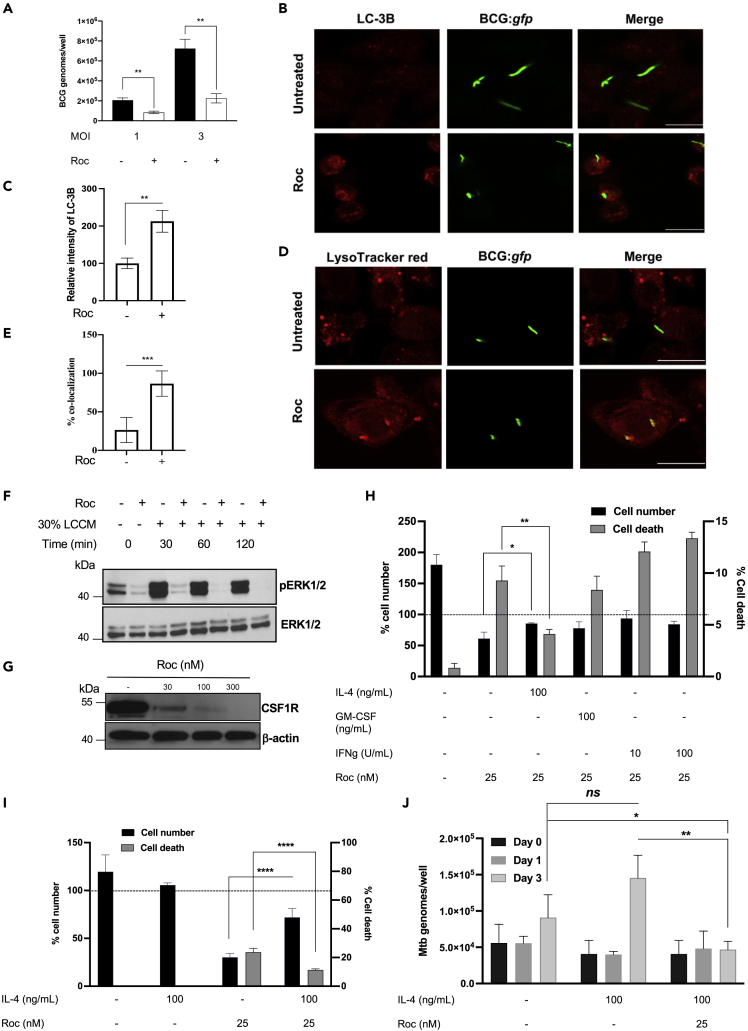
Figure 4.
Rocaglates stimulate antimycobacterial defenses in BMDMs
(A) BMDMs were pretreated with CMLD010536 (50 nM) for 24 hr and subsequently infected with M. bovis BCG at MOI 1 and 3. The intracellular BCG loads were measured 24 hr post infection by determining genome equivalents using qPCR with BCG-specific primers and Taqman probe.
(B and C) BMDMs were infected with BCG:gfp at MOI 1 and rocaglate was added after phagocytosis till 24 hr. The BMDMs were stained using LC-3B antibody and observed fluorescence intensity of LC-3B and its co-localization with BCG:gfp using Zeiss LSM 710-Live Duo scan confocal microscopy. The percent fluorescence intensity was calculated using ImageJ. Scale bar represents 20 μM.
(D and E) Rocaglates promote phagosome-lysosome fusion. BMDMs were pretreated with 25 nM CMLD010536 for 6 hr and infected with M. bovis BCG expressing gfp (BCG:gfp) at MOI 1. At 48 hr p.i., the cells were strained with LysoTracker red for 1 hr, processed for imaging by confocal microscopy and analyzed for co-localization of lysosomes with phagosomes containing BCG:gfp. The percentage of BCG-positive phagosome co-localization with lysosomes and representative images are presented in (D) and (E), respectively. The data in (E) are represented as mean ± SEM (n = 50, phagosomes), p < 0.05. Scale bar represents 20 μM.
(F and G) Rocaglate inhibit ERK phosphorylation induced by CSF1-containing media (LCCM) (F) and the expression of CSF1R (G). BMDMs were pre-treated with 100 nM CMLD010536 for 12 hr and subsequently provided 30% LCCM for the indicated periods of time. (F) Phospho- and total ERK1/2 protein levels were determined by western blot. (G) BMDMs were treated with indicated concentrations of CMLD010536 for 24 hr, and CSF1R protein levels were determined by western blot.
(H) BMDMs were treated with 25 nM CMLD010536 either alone or in combination with IL-4 (100 ng/mL), GM-CSF (100 ng/mL), or IFNγ (10 and 100 U/mL) for 72 hr. The cell numbers and percentage of cell death (right Y axis) were analyzed using LIVE/DEAD staining and automated microscopy. The cell numbers at day 3 were expressed as the percentage of cells at day 0 (left Y axis). The dotted line indicates the cell number at day 0 (100%).
(I). IL-4 improves the survival of the rocaglate-treated and Mtb-infected macrophages. BMDMs were pretreated with IL-4 (100 ng/mL) for 16 hr and infected with Mtb H37Rv at MOI 1. CMLD010536 (25 nM) was added 2 hr p.i. either alone or in combination with IL-4 (100 ng/mL). The percentage of cell numbers and percentage of dead cells at 72 hr p.i. were determined as in (H). The dotted line denotes the cell numbers (100%) immediately after the Mtb infection and washing off the extracellular bacteria (day 0).
(J) The rocaglate treatment reduces the Mtb load in the presence of IL-4. The intracellular bacterial loads were determined after 2 hr phagocytosis (day 0) and at days 1 and 3 p.i. using qPCR and normalized using BCG spike as an internal control. The Mtb loads are presented as fold change compared to Mtb uptake by untreated BMDMs at 0 h. The data in panels A, C, E, H, I and J are represented as mean ± SEM, and p value ≤ 0.05 was considered statistically significant.
Next, we wanted to determine whether the rocaglate treatment induces transient or stable suppression of the M2-like phenotype. As shown in Figure 3J, pre-treatement and removal of the rocaglate prior to the IL-4 stimulation had a substantially weaker inhibitory effect on the Arg1 gene expression, as compared to simultaneous treatment with the rocaglate and IL-4. When we increased the interval between the rocaglate treatment and IL-4 stimulation to 24 hr, the inhibitory effect of the rocaglate pretreatment disappeared completely (Figure 3K). Similar effects were observed when Fizz1 gene expression was used as an M2 polarization marker (Figures S4D and S4E). Thus, the rocaglate inhibition of IL-4 responses is transient and can be completely reversed after their removal. These findings demonstrate that effects of rocaglate on macrophage are regulatory, rather than toxic. No toxicity was observed when macrophages were treated with rocaglate and IL-4 for 72 hr (Figure S4F).
These experiments demonstrated that, unlike the rocaglates' effects on the M1-like polarization, the inhibition of alternative macrophage activation by rocaglates is driven by a separate, p38-independent pathway. Selective translation inhibition by rocaglates, however, is required for both pathways. Perhaps, they represent two modules of a coordinated physiological response of macrophages to attenuated protein translation—a common sign of, and an adaptation to, ongoing stress and microbial pathogenesis (Fontana et al., 2011, 2012).
Macrophage activation by rocaglates improves bacterial control
The ability of rocaglates to synergize with low concentrations of IFNγ inhibits the alternative macrophage activation and activates protective stress responses, making this class of compounds an attractive candidate for HDT of infections caused by intracellular bacterial pathogens. The SQSTM1/p62 protein, which is induced by rocaglates in a p38-dependent but IFNγ-independent manner, coordinates activation of central stress response pathways: antioxidative defense and autophagy (Katsuragi et al., 2015; Moscat and Diaz-Meco, 2011; Rogov et al., 2014; Komatsu et al., 2010; Taguchi et al., 2011; Jaramillo and Zhang, 2013; Padmanabhan et al., 2006; Fukutomi et al., 2014; Lau et al., 2010). It is a major adapter protein that targets intracellular bacteria to LC3-positive autophagosomes (Zheng et al., 2009). Previously, we have reported that the rocaglate treatment stimulates autophagic flux and increases the lipidated form of LC3 associated with autophagosome maturation (Bhattacharya et al., 2016). To test whether rocaglate treatment could enhance macrophage resistance to intracellular bacteria, we treated BMDMs with CMLD010536 (50 nM), infected them with Mycobacterium bovis BCG, and determined their bacterial loads and viability at 24 hr post infection (p.i.). The rocaglate treatment significantly reduced the bacterial loads, as determined by quantitative PCR of the bacterial genomes (Figure 4A), and viability, as determined by colony forming units plating (Figure S5A). The rocaglate treatment increased co-localization of M. bovis BCG (BCG:gfp) with autophagosome marker LC-3B (Figures 4B and 4C) and the delivery of the intracellular bacteria to lysosomes (Figures 4D and 4E, and Figure S5B and S5C). In agreement with the upregulation of p62 mRNA and protein (Figures 2A–2C), we observed abundant p62 puncta in the cytoplasm of the rocaglate-treated BMDMs and the co-localization of BCG:gfp with p62 (Figure S5D). These data demonstrate that rocaglate-mediated macrophage activation significantly increased autophagy and clearance of vacuolar intracellular bacteria (BCG). Compared to BCG vaccine, virulent Mycobacterium tuberculosis (Mtb) is more resilient to macrophage attack. In naive macrophages, the bacteria block phagosome-lysosome fusion, then escape from phagosomes to the cytoplasm, grow, and eventually destroy the host cells. Because of a slow replication rate of the bacteria, this cycle takes several days, unless a high multiplicity of infection (MOI) is used. To test whether rocaglates would improve the macrophage ability to control virulent Mtb in vitro, we used low MOI and extended the time p.i. to 72 hr. Infection with virulent Mtb dramatically decreased the viability of BMDMs treated with CMLD10536 at 25–50 nM for 3 days. Even in the absence of infection, the rocaglate treatment for 48 hr decreased cell survival (Figure S5E).
The highly homogeneous BMDM population prepared in our laboratory according to (Chitu et al., 2011) is critically dependent on CSF1 for survival. Increasing the CSF1-containing conditioned media 3-fold did not improve cell survival in the presence of rocaglates. The ERK1/2 phosphorylation, which is downstream of CSF1 receptor (CSF1R) signaling, also could not be restored in the rocaglate-treated cells by increasing CSF1 (Figure 4F). Finally, we found that rocaglates suppressed the expression of the CSF1 receptor protein (Figure 4G and Figure S5F) and, thus, compromised a major survival pathway. To identify alternative survival factors, we compared the survival of the rocaglate-treated BMDMs in the presence of GM-CSF, IFNγ, and IL-4 (Figure 4H). Surprisingly, IL-4 significantly improved the rocaglate-treated macrophage survival within 72 hr, as compared to GM-CSF and IFNγ. The pro-survival effect of IL-4 on rocaglate-treated BMDMs was not mediated by either STAT6 or ERK1/2 since both were inhibited by rocaglate in the presence of IL-4 (Figures 3D and 3I). Possibly, this effect is mediated by activation of an alternative survival pathway downstream of the IL-4 receptor, such as Akt (Vergadi et al., 2017).
The pro-survival effect of IL-4 was also observed in Mtb-infected BMDM cultures at 72 hr p.i. (Figure 4I). In contrast, GM-CSF failed to improve the macrophage survival in these settings (Figure S5F). Therefore, we tested the effects of rocaglates on Mtb replication in macrophages in the presence of IL-4. The bacterial loads were estimated using an optimized quantitative PCR method developed in our laboratory (Yabaji et al., submitted for publication). We observed an approximately 2-fold increase between 0 and 72 hr p.i. in naive macrophages (Figure 4J). Pretreatment with IL-4 had no effect on the bacterial uptake. In the presence of IL-4, the bacterial loads increased by 72 hr p.i., as compared to naive macrophages. In contrast, no bacterial growth was observed in BMDMs treated with rocaglate and IL-4, and the bacterial loads at 72 hr p.i. were significantly lower as compared to naive and IL-4-treated macrophages. Thus, rocaglate treatment improves macrophage ability to control Mtb growth even in the presence of an M2-polarizing cytokine. Notably, ARG1-positive (M2-polarized) macrophages have been detected in advanced TB granulomas in zebrafish, mice, non-human primates, and humans (Mattila et al., 2013). Therefore, this rocaglate activity may be beneficial in the context of active TB disease.
To begin testing effects of rocaglates in vivo, we have chosen a model of acute respiratory challenge with Streptococcus pneumoniae (serotype 3) (Yang et al., 2014). Alveolar macrophages (AMs) play an important role at the early phase of this infection (Dockrell et al., 2003; Gordon et al., 2000). It is also worth noting that in naive hosts, the AM displays an M2-like phenotype. We wondered whether rocaglate pretreatment could prime macrophages for more efficient control of the acute bacterial infection. Outbred CD-1 IGS male mice were treated with CMLD010536 (25 g/kg, i.p.) at 48, 24, and 0 hr before the respiratory challenge with Strep. pneumoniae. At 24 hr p.i., bronchoalveolar lavage (BAL) was performed to harvest 2 mL of BAL fluid. We observed significant reduction of live bacteria (as determined by BAL colony forming units counts, Figure S6 left panel) and BAL cell numbers (Figure S6, right panel) in rocaglate pretreated mice as compared to vehicle-treated controls. These results show that rocaglate pretreatment increased bacterial control and reduced inflammatory cell recruitment to the lungs at the early stage of respiratory infection with Strep. pneumoniae.
Discussion
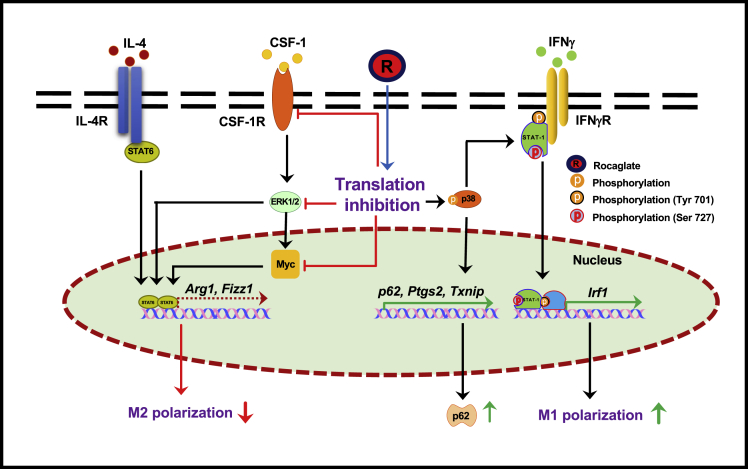
Taken together, our data demonstrate that rocaglates enhance pleiotropic effects of IFNγ on primary macrophages. These effects are associated with their selective translation inhibition activity and cannot be recapitulated by general inhibitors of translation such as cycloheximide and puromycin. Using metabolic labeling with puromycin, we have determined that despite overall translational repression in cells exposed to rocaglates, a fraction of translational activities persisted. Proteins involved in stress response such as ATF4, ATF3, and SQSTM1/p62 were directly upregulated by the translationally active rocaglates. In addition, we identified a set of proteins whose upregulation with rocaglates required co-stimulation with low concentrations of IFNγ. This category encompasses interferon-inducible proteins that govern M1-like macrophage differentiation and activation, such as transcription factors IRF1, IRF5, as well as anti-proliferative and anti-viral proteins (Figure 2J). Thus, exposure of primary macrophage to rocaglates induced stress responses and restricted their differentiation, channeling it toward the M1-like phenotype, as depicted in Figure 5.
Figure 5.
Pleiotropic effects of rocaglates' on primary macrophages
Active rocaglate (R) induces p38-dependent and independent effects. The p38-dependent effects include synergy with low concentrations of IFNγ (via STAT1 phosphorylation at Ser727 and upregulation of IRF1), as well as an IFNγ-independent induction of several inflammatory and stress response genes including SQSTM1/p62 protein. The p38-independent effects include inhibition of anabolic pathways mediated by the CSF1 receptor, the IL-4-STAT6 axis, ERK1/2 MAPK, and Myc. In toto, macrophage response to selective protein translation inhibition amounts to adaptive proteome remodeling, inhibition of cell growth, and increased expression of stress response proteins. This adaptive response increases macrophage resistance to intracellular mycobacteria.
The macrophage-activating effects of the rocaglates, either IFN-dependent or independent, partially relied on activation of a stress-activated MAP kinase p38. Thus, the p62, Txnip, and Ptgs2 mRNAs could be induced in naive BMDMs by the treatment with rocaglates alone in a p38-dependent manner. Our data demonstrate that the p38 MAPK pathway activation also plays a central role in rocaglates' synergy with IFNγ in macrophage activation via p38-mediated phosphorylation of STAT1 at a non-canonical site. Canonically, the upregulation of the Irf1 gene expression by IFNγ is mediated by the binding of STAT1 homodimers to the GAS in the Irf1 promoter. The STAT1 homodimer formation and promoter binding require STAT1 phosphorylation following the IFNγ binding to its receptor (Meraz et al., 1996; Decker et al., 1997). However, rocaglates did not increase STAT1 phosphorylation at the canonical tyrosine site in our studies. There is an emerging evidence that serine (Ser-727) phosphorylation of STAT1 by stress-activated MAP kinases p38 and JNK mediates various stress responses (Dudley et al., 2004) and may account for nearly 80% of IFNγ-induced transcriptional activity. Importantly, this STAT1 modification does not increase DNA binding or nuclear translocation of STAT1 dimers (Wen et al., 1995; Kovarik et al., 1998; Goh et al., 1999). Therefore, we tested whether the synergistic effect of the rocaglate on IRF1 induction was mediated via the alternative mechanism(s) involving MAP kinase activation. We found (i) that rocaglates induced STAT1 Ser-727 phosphorylation in macrophages and (ii) p38 inhibitors abrogated Ser-727 phosphorylation of STAT-1 and the synergistic effect on the IRF1 expression. We propose that low doses of IFNγ are necessary to initiate the STAT1 dimer formation and binding to the IRF1 promoter, while the p38 activation by rocaglates and STAT1 Ser-727 phosphorylation boosts and sustains the Irf1 transcription, most likely, by facilitating interactions of STAT1 with transcriptional co-activators. This hypothesis is further supported by the fact that p38 is known to translocate to the nucleus and phosphorylate chromatin-associated substrates (Maik-Rachline et al., 2020).
The synergistic effect of rocaglates does not simply mimic the canonical IFNγ pathway. Indeed, treatment with active rocaglates alone induced no Irf1 mRNA upregulation (Bhattacharya et al., 2016) and even suppressed the basal IRF1 protein expression (Figure 2G). Furthermore, the p38 inhibition did not suppress the Irf1 mRNA induction by standard doses of IFNγ alone (Figure S3B), demonstrating that p38 does not participate in the IFNγ receptor-mediated Irf1 induction in vitro. However in vivo, p38 may act as a stress-regulated controller of macrophage responses to activating stimuli.
Similar to mTORC1 inhibitors that also inhibit protein translation, rocaglate treatment of macrophages activated autophagic pathway. Previously, we have demonstrated the upregulation of autophagic flux in rocaglate-treated macrophages. Our current study demonstrates that the treatment of BMDMs with rocaglates results into the upregulation of p62 mRNA in a p38-dependent manner as early as at 6 hr. The p62 protein levels were significantly increased at 16–24 hr, as demonstrated by western blot and the macrophage staining with p62-specific antibodies. After infection, mycobacteria co-localized with p62, LC-3B, and lysosomes. These observations are consistent with previous studies demonstrating that p62 may serve as an autophagy adapter that directs mycobacteria toward lysosomes. Although the p62 protein accumulation inside the cells may be caused by blocking autophagic flux, the rocaglate-treated macrophages upregulated p62 at transcriptional and translational levels during prolonged rocaglate treatment. Similarly, prolonged starvation of mouse embryonic fibroblasts induced p62 protein via transcriptional upregulation to overcome starvation (Sahani et al., 2014). This suggests that rocaglate exposure might mimic starvation-induced autophagy.
The suppression of alternative macrophage activation occurred via inhibition of STAT6 transcriptional activity in a p38-independent manner. The inhibitory rocaglate activity also targeted anabolic pathways via coordinated downregulation of the growth factor CSF1 receptor and Myc proteins and inhibition of the phosphorylation of ERK1/2 (Figures 1F, 3I, 4F and 4G). We found that rocaglates potently inhibit ERK phosphorylation, a key activation step in the growth factor signal transduction relay. Phosphorylated ERK promotes cell proliferation and provides anabolic and pro-survival signals primarily via phosphorylation of transcription factors Myc and CREB. Because Myc is known to be critically important for the induction and maintenance of the “trophic” M2-like macrophage state (Pello, 2016; Pello et al., 2012), ERK inhibition is likely to downregulate the Myc-dependent pathways involved in M2 polarization. According to the literature, ERK can also directly phosphorylate STAT6 and increase its DNA binding and transcriptional activities (So et al., 2007). We also observed that rocaglates inhibited IL-4-induced STAT6 DNA binding. Thus, profound inhibition of ERK phosphorylation by rocaglates may explain, or significantly contribute to, the inhibition of both the STAT6- and Myc-dependent pathways of the M2 polarization and, thus, suppress both the induction and maintenance of the M2-like macrophage phenotype.
It appears that the rocaglate treatement modifies the balance of MAP kinase-regulated physiological modules: decreasing the ERK1/2-mediated anabolic activities and increasing the p38-mediated stress and IFNγ responses. This coordinated change may suggest an existence of a common upstream regulatory mechanism. We documented the upregulation of ATF4 protein at an early stage of the rocaglate treatment (Figure 1F). This transcription factor is activated via a so called upstream open reading frame mechanism from pre-existing mRNA template in response to inhibition of cap-mediated protein translation by several stress-activated eIF2α kinases (Medenbach et al., 2011). These effects are consistent with the known ability of rocaglates to inhibit eIF4A helicase, a component of the cap-dependent translation initiation complex. The ensuing activation of the ISR involves activation of a downstream TF ATF3 at both mRNA and protein levels. Depending on stress intensity, duration, and cellular context, the ISR activation can be transient and adaptive promoting cell survival or induce apoptosis. Our studies demonstrate that at low concentrations, rocaglates do not induce apoptosis but activate p38-mediated macrophage activation and M1-like polarization. Thus, in contrast to stress-induced inhibition of cap-dependent protein translation, the effect of rocaglates is partial and more selective, allowing for the upregulation of proteins that activate stress adaptation pathways. We hypothesize that by mimicking ISR, rocaglates shift the balance of the MAP kinase modules in macrophages de-prioritizing responses to the growth factors and sensitizing to the activator(s) of antimicrobial defenses. This implies an existence of an extended ISR network integrating the ISR proper and MAP kinase modules.
Mechanistically, synthetic and natural rocaglamide derivatives have been shown to inhibit translation by binding to a component of the eIF4F translation initiation complex—the RNA helicase eIF4A—that prevents its incorporation into the eIF4F complex (Cencic et al., 2009). However, not all protein translation is equally sensitive to the inhibition of eIF4A helicase activity. Translation of mRNA species containing structural barriers, such as stem-loop structures or protein-polypurine RNA complexes, within their mRNA 5’ UTR is most sensitive to eIF4A inhibition (Iwasaki et al., 2016, 2019). Among the most eIF4A-dependent and rocaglate-sensitive transcripts in acute lymphoblastic leukemia cells were oncogene-encoding transcripts and superenhancer-associated transcription factors, such as Myc (Wolfe et al., 2014). Because various stressors converge on a common pathway known as ISR leading to inhibition of cap-dependent protein translation, we hypothesize that the eIF4A-mediated selective downregulation of protein translation enables rapid adjustment of the anabolic pathways at a translational level to match environmental inputs. Therefore, small molecules that downregulate the eIF4A RNA helicase activity may mimic those stress signals causing adaptive and reversible proteome remodeling.
The canonical eIF4A-dependent translation inhibition by rocaglates may provide a stress-like “conservation of resources” signal that leads to the inhibition of ERK phosphorylation and, thus, to the coordinated downregulation of several anabolic pathways. Yet another mechanism may be associated with a recently described alternative target of rocaglates—prohibitin. Rocaglates have been shown to bind prohibitins directly and prevent their interaction with c-Raf, thereby inhibiting Raf activation and Raf-MEK-ERK signaling in Jurkat tumor cell line (Polier et al., 2012). This pathway may lead to translation inhibition by rocaglates via an eIF4A-independent but ERK-dependent pathway since MEK-ERK signaling has been shown to promote cap-dependent translation via phosphorylation of eIF4E, the key translation initiation factor (Polier et al., 2012). Whether the rocaglate binding to prohibitin mediates ERK inhibition and/or translation inhibition in macrophages remains to be established (Chu et al., 2016). Our data suggest that an alternative pathway of ERK downregulation in macrophages can occur via downregulation of CSF1R, a receptor for a major macrophage growth factor.
Taken together, our new data demonstrate that biological effects of rocaglates on primary macrophage activation are mechanistically linked to p38 activation and translation inhibition and are not limited to IRF1 induction. They significantly overlap with pleiotropic effects of IFNγ on macrophages, including translation inhibition (Su et al., 2015), p38-mediated induction of autophagy (Matsuzawa et al., 2012), and the enhancement of phagosome maturation and mycobacterial control (Gutierrez et al., 2004; MacMicking et al., 2003; Shenoy et al., 2007; Singh et al., 2006), as well as suppression of anabolic pathways mediated by IL-4, CSF1R, and ERK(Dickensheets and Donnelly, 1999; Naka et al., 2001; Yu et al., 2004) (Table S4). This coordinated macrophage reprogramming resembles stress preconditioning. The exposure to low, subthreshold, doses of stress prior to encountering more potent stressors was shown to broadly increase stress resilience and prevent subsequent damage by higher doses of stressors in many biological systems. This phenomenon has been termed “hormesis” (Calabrese, 2008). In this conceptual framework, the effect of rocaglates may be viewed as anticipatory cross-protection against various stressors, including intracellular bacteria.
The above data suggest that rocaglates may represent a novel class of HDTs with potential for broad applications in vivo. The dissection of signal transduction pathways activated by rocaglates demonstrate that most of their biological effects are achieved via regulatory pathways distinct from those activated by standard concentrations of IFNγ and can be IFNγ receptor independent. Therefore, rocaglates may be particularly useful for directing macrophage polarization when the receptor signaling is disrupted by mutations, microbial factors, or negative biological regulators. In mycobacterial infections, these small molecules may penetrate mycobacterial granulomas better than acid-labile IFNγ dimers and compensate for its decreased bioavailability, especially in HIV-TB co-infections. Helminth co-infections inducing the M2 macrophage polarization increase susceptibility to mycobacterial infections (Salgame et al., 2013). In these settings, the ability of rocaglates to guide monocyte/macrophages toward M1-like phenotype and inhibit triggers of the alternative, M2-like, activation may be particularly useful.
Rocaglates are active against protozoan infections, such as malaria (Langlais et al., 2018) and visceral leishmaniasis (Chaparro et al., 2020). Silvestrol has been shown to display a broad spectrum of antiviral activities against Corona-, Ebola-, Zika-, Picorna-, Hepatis E and Chikungunya viruses (Blum et al., 2020; Müller et al., 2018). Recent analysis of a SARS-CoV-2 protein interaction map predicted that the rocaglate target eIF4A could be the antiviral drug target (Gordon et al., 2020). Our data suggest that rocaglates may also be useful for boosting macrophage-mediated immunity and preventing bacterial pneumonias following viral infections. Recently, it has been suggested that Coronavirus Disease 2019 (COVID-19) pandemics may lead to reactivation of latent TB infection and an upsurge of TB in endemic areas (Nordling, 2020). In these settings, treatment of patients withTB infected with COVID-19 with a rocaglate may also reduce risks of TB progression.
The rocaglates' ability to direct M1-like macrophage polarization also warrants their evaluation in chronic non-infectious pathologies, where macrophages are exposed to complex, often conflicting environmental signals. Tumor-associated macrophages stimulate tumor cell proliferation, angiogenesis, metastasis, and resistance to therapy. Those properties have been associated with M2-like differentiation of monocytes guided by cytokines, CSF-1 among them, within tumor microenvironments (Schmid and Varner, 2010; Murray, 2018; Grivennikov et al., 2010). Thus, in solid tumors, rocaglates may target both the tumor cells decreasing their proliferation and myeloid cells that comprise the immune and inflammatory milieu. In asthma, allergic reactions may be associated with chronic bacterial infections. Currently, asthma therapy relies on immune suppression by corticosteroids. However, prolonged treatment with steroids has numerous undesirable side effects, including chronic pulmonary infections caused by opportunistic mycobacteria and fungi that further exacerbate pulmonary pathology (Fritscher et al., 2011). The important theoretical advantage of rocaglates as compared to corticosteroids is that rocaglates would shift the balance of macrophage activation toward M1-like phenotype without suppressing mechanisms of macrophage resistance to bacterial and viral infections. Thus, rocaglates may represent a novel class of HDTs broadly applicable to diseases associated with hypofunction of type 1 and/or hyperactivation of type 2 immunity, e.g., chronic bacterial infections, allergies, and, possibly, certain tumors.
Limitations of the study
One limitation of our study is that we did not determine whether the pleiotropic effects of rocaglates on macrophages can be solely explained by their inhibition of translation (by binding to RNA helicase eIF4A). Further mechanistic dissection, however, may reveal additional, macrophage-specific targets and mechanisms. For example, expanded analyses of mRNA translation using Ribo-seq could provide more precise information about the rocaglate-mediated regulation of protein biosynthesis in activated macrophages. Although we have found that activating effects of rocaglates in macrophages were mediated by stress kinase p38, precise mechanisms linking translation inhibition with p38 activation, as well as macrophage-specific downstream cascades, e.g. IRF1 upregulation, remain to be elucidated. Mechanisms of the downregulation of STAT6 DNA binding by rocaglates in macrophages also require further studies. In addition, cell type-specific effects of rocaglates on immune and stromal cells need to be delineated in vitro and in vivo to more comprehensively evaluate their therapeutic potential.
STAR★Methods
Key resources table
| REAGENTS OR RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Puromycin Antibody, clone 12D10 | Millipore | Cat# MABE343; RRID: AB_2566826 |
| Anti-IRF-1 antibody | Cell Signaling Technology | Cat# 8478; RRID: AB_10949108 |
| Anti-SQSTM1/p62 antibody | Cell Signaling Technology | Cat# 5114; RRID: AB_10624872 |
| Monoclonal Anti-β-Actin antibody | Millipore Sigma | Cat# A2228; RRID: AB_476697 |
| Anti-ATF-3 (44C3a) antibody | Santa Cruz Biotechnology | Cat# SC-81189: RRID: AB_2058591 |
| Anti-ATF-4 antibody | Abcam | Cat# ab23760; RRID: AB_725569 |
| Anti-STAT1 antibody [SM1] | Cell Signaling Technology | Cat# 9172S; RRID: AB_2198300 |
| Anti-STAT1 (phospho S727) antibody | Cell Signaling Technology | Cat# 9177S; RRID: AB_2197983 |
| Anti-p38 MAPK antibody | Cell Signaling Technology | Cat# 9212S; RRID: AB_330713 |
| Anti-Phospho-p38 MAPK antibody | Cell Signaling Technology | Cat# 9211S; RRID: AB_331641 |
| Anti-STAT-6 antibody | Abcam | Cat# 44718; RRID: AB_778114 |
| Anti-STAT6 (phospho Y641) antibody | Abcam | Cat# ab54461; RRID: AB_882721 |
| p44/42 MAPK (Erk1/2) (L34F12) Mouse mAb | Cell Signaling Technology | Cat# 4696S; RRID: AB_390780 |
| Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204) Antibody | Cell Signaling Technology | Cat#9101; RRID: AB_331646 |
| M-CSF receptor antibody | Cell Signaling Technology | Cat# 3152; RRID: AB_2085233 |
| Anti-c-Myc/N-Myc antibody | Cell Signaling Technology | Cat# 13987S; RRID: AB_2631168 |
| Anti-Histone H3 antibody | Cell Signaling Technology | Cat# 9715S; RRID: AB_331563 |
| Anti-Alpha tubulin antibody | Sigma | Cat# T9026; RRID: AB_477593 |
| Anti-LC-3B antibody | Cell Signaling Technology | Cat# 2775S; RRID: AB_915950 |
| Anti-mouse IgG HRP linked antibody | Cell Signaling Technology | Cat#7076S; RRID: AB_330924 |
| Anti-rabbit IgG HRP linked antibody | Cell Signaling Technology | Cat#7074S; RRID: AB_2099233 |
| F(ab')2-Goat anti-Rabbit IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Invitrogen | Cat# A-11072; RRID: AB_142057 |
| Chemicals | ||
| Rocaglamide A | MedChemExpress | Cat# HY-19356 |
| SB203580 | Calbiochem | Cat# 559389 |
| SB600125 | Calbiochem | Cat# 129566 |
| U0126 | Calbiochem | Cat# 662005 |
| SP202190 | MedChemExpress | Cat# HY-10295 |
| Recombinant murine IFN-γ | Peprotech | Cat# 315-05 |
| Recombinant murine Interleukin -3 | Peprotech | Cat# 213-13 |
| Recombinant murine Interleukin-4 | Peprotech | Cat# 214-14 |
| Recombinant murine TNF-α | Peprotech | Cat# 315-01A |
| Recombinant murine GM-CSF | Peprotech | Cat# 315-03 |
| Puromycin dihydrochloride from Streptomyces alboniger | Millipore Sigma | Cat# P8833-10MG |
| Characterized Fetal Bovine Serum | GE Healthcare | Cat# SH30396 |
| Promega GoTaq™ qPCR Master Mix | Fisher Scientific | Cat# PRA6002 |
| Protease inhibitor cocktail | sigma | P8340-5mL |
| Phosphatase inhibitor cocktail II | sigma | P0044-5mL |
| Phosphatase inhibitor cocktail III | sigma | P5726-5mL |
| Corning™ Costar™ Ultra-Low Attachment Microplates | Thermo Fisher | Cat# 3471 |
| Corning™ Falcon™ Tissue Culture Dish with Grid | Thermo Fisher | Cat# 353025 |
| Qiagen RNeasy Plus Mini Kit (250) | Qiagen | Cat# 74136 |
| Live-or-Dye™ 594/614 Fixable Viability Staining Kits | Biotium | Cat# 32006 |
| EMSA kits | Signosis Inc. | Cat# GS-0047 |
| LysoTracker™ Red DND-99 | Invitrogen | Cat# L7528 |
| nuclear extraction kits | Signosis Inc. | Cat# SK-0001 |
| Hoechst 33342 | Fisher Scientific | Cat# H3570 |
| Paraformaldehyde Solution 4% in PBS | Fisher Scientific | Cat# J19943-K2 |
| TaqMan™ Environmental Master Mix 2.0 | Fisher Scientific | Cat#4396838-5mL |
| DMEM/Ham's F-12 50/50 Mix [+] L-glutamine | Corning® | Cat# 45000-344 |
| Glutamine | Corning® | Cat# 25-005-CI |
| Penicillin Streptomycin solution | Corning® | Cat# 30-002-CI |
| HEPES buffer | Corning® | Cat# 25-060-CI |
| Middlebrook 7H9 Broth | BD Biosciences | Cat# 271310 |
| Middlebrook 7H10 Agar | BD Biosciences | Cat# 262710 |
| Experimental models: Organisms/Strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock No.: 000664 |
| Mouse: B6J.C3-Sst1C3HeB/FejKrmn | Mouse Mutant Resource and Research Center | MMRRC Stock No: 043908-UNC (Pichugin et al., 2009) |
| Mycobacterium tuberculosis H37Rv | ATCC | Cat# 27294 |
| Mycobacterium bovis BCG | ATCC | Cat# 35737 |
| Mycobacterium bovis BCG:gfp | This paper | N/A |
| Streptococcus pneumoniae | ATCC | Cat# 33400 |
| Sequence-Based reagents | ||
| qPCR primers: Irf-1 Forward: CAGAGGAAAGAGAGAAAGTCC Reverse: CACACGGTGACAGTGCTGG |
This paper | N/A |
| qPCR primers: Txnip Forward: TATGTACGCCCCTGAGTTCC Reverse: GCTCACTGCACGTTGTTGTT |
This paper | N/A |
| qPCR primers: p62 Forward: AGCTGCCCTCAGCCCTCT Reverse: GGCTTCTCTTCCCTCCATGTT |
This paper | N/A |
| qPCR primers: Arginase 1 Forward: AAGAAAAGGCCGATTCACCT Reverse: CATGATATCTAGTCCTGAAAGG |
This paper | N/A |
| qPCR primers: Ptgs2 Forward: TCTCCAACCTCTCCTACTAC Reverse: ACTCTCTCCGTAGAAGAACC |
This paper | N/A |
| qPCR primers: Fizz1 Forward: GGTCCCAGTGCATATGGATGAGACCATAGA Reverse: CACCTCTTCACTCGAGGGACAGTTGGCAGC |
This paper | N/A |
| qPCR primers: Csf1R Forward: CATGGCGAGGGTTCATTATC Reverse: GCTTGCTAGGCTCCAATTT |
This paper | N/A |
| qPCR primers: Myc Forward: GTGCTGCATGAGGAGACACC Reverse: GACCTCTTGGCAGGGGTTTG |
This paper | N/A |
| qPCR primers: 18S Forward: TCAAGAACGAAAGTCGGAGGT Reverse: CGGGTCATGGGAATAACG |
This paper | N/A |
| qPCR primers: Beta actin Forward: ATCACCCAGATCATGTTTGA Reverse: TACGACCAGAGGCATACA |
This paper | N/A |
| qPCR primers: Mtb specific primers Forward: GGAAATGTCACGTCCATTCATTC Reverse: CGTTGTTCAGCTCGGTA Probe: 56-FAM/AGCTTGGTCAGGGACTGCTTCC/36-TAMSp/ |
This paper | N/A |
| qPCR primers: BCG specific primers Forward: GTG GTG GAG CGG ATT TGA Reverse: CAA CCG GAC GGT GAT CC Probe: /5Cy5/TTCTGGTCG/TAO/ACGATTGGCACATCC/3IAbRQSp/ |
This paper | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should contact to Dr.Igor Kramnik (ikramnik@bu.edu).
Materials availability
This study did not generate new unique reagents.
Data and code availability
This study did not generate datasets/code and any additional information will be available from the lead contact upon request.
Experimental model and subject details
C57BL/6 J mice were obtained from the Jackson Laboratory (USA). The B6J.C3-Sst1C3HeB/FejKrmn mice were developed in our laboratory (Pichugin et al., 2009) (available from MMRRC stock # 043908-UNC). Adult mice (6–12 weeks old) of both sexes were used in experiments. No sex-dependent variation was observed. All experiments were performed with the full knowledge and approval of the Standing Committee on Animals at Boston University (IACUC protocol number PROTO201800218).
Method details
Bacterial strains
The wild type M. tuberculosis H37Rv, M. bovis BCG Pasteur and recombinant M. bovis BCG expressing gfp strains were grown at 37°C in Middlebrook 7H9 broth (BD Biosciences) or on 7H10 agar plates (BD Biosciences), respectively. The both solid and liquid media contained contained glycerol (0.5% v/v) and Tween 80 (0.05%). The MB7H9 broth was enriched using 10% ADC and MB7H10 agar was enriched using with 10% OADC. The M. bovis BCG strain expressing gfp were grown with 50 μg/mL Hygromycin B.
BMDM culture
Isolation of mouse bone marrow and culture of BMDMs were carried out as previously described (Pan et al., 2008). In brief, bone marrow cells were isolated from femurs and cultured in DMEM/F12 with 10% heat-inactivated endotoxin-free fetal calf serum (Hyclone) in the presence of IL-3 (10 ng/ml) and L929 conditioned medium as a source of CSF-1.
Bliss independence criterion
Synergy effects between Rocaglate and IFNγ were assessed using the Bliss independence criterion (Greco et al., 1995). According to this criterion, if drug A applied separately results in fold-change of the level of protein i, and drug B applied separately results in fold-change of the level of protein i, drugs A and B applied together affect protein i independently, i.e. without effects of antagonism or synergy, when this combination results in fold change of the protein level. If the effect of the drug combination is less than , drugs antagonize for protein i, and if the effect of the drug combination is more than , drugs synergize. Importantly, the Bliss independence criterion assumes that drugs are given in the same doses, both separately and in combination.
To assess the strength of synergy effects, for each differentially expressed protein i we have calculated the synergy score Si by taking the difference between the observed effect of a combination of rocaglate and IFNγ , and the effect estimated from Bliss independence:
If Si > 0 rocaglate and IFNγ synergize in inducing expression of the protein i, and if Si < 0 rocaglate and IFNγ antagonize in inducing expression of the protein i. We assumed that if , the combination of rocaglate and IFNγ gives approximately independent effect. The file “Bliss_synergy_analysis.xlsx” presents a list of differentially expressed proteins sorted by Bliss synergy score Si in descending order.
Gel shift assay
The nuclear extracts were prepared by nuclear extraction kits (Signosis Inc). Gel shift assays were done with EMSA kits (Signosis Inc). 5 μg nuclear extracts were incubated with 1× binding buffer and biotin-labeled probe for 30 min at room temperature. The samples were electrophoresed on a 6 % polyacrylamide gel in 0.5 % TBE at 120 V for 45 min and transferred onto a nylon membrane in 0.5 % TBE at 300 mA for 1 h. After transfer and UV cross-linking, the membrane was detected with Streptavidin-HRP.
Immunoblotting
Equal amounts of protein extracts were separated by SDS-PAGE and transferred to PVDF membrane (Millipore). Bands were detected with enhanced chemiluminescence (ECL) kit (Perkin Elmer).
RNA isolation and quantitative PCR
Total RNA was isolated using the RNeasy Plus mini kit (Qiagen). cDNA synthesis was performed using the SuperScript II (Invitrogen). Quantitative real-time RT-PCR (qRT-PCR) was performed with the GoTaq qPCR Mastermix (Promega) using the CFX-90 real-time PCR System (Bio-Rad). For calculating fold induction the cycle threshold (Ct) of the test gene was normalized to the Ct of the internal control (18S) gene.
Hoechst/PI staining method for cell cytotoxicity
For cell viability assays BMDMs were plated in 96 well tissue culture plates. The supernatant was aspirated and to each well 100 μl PBS containing Hoechst (Invitrogen, 10 μM) and PI (Calbiochem, 2 μM) were added. The % of total and dead cells was calculated by automated cell cytometer.
Reporter assay for measuring translation inhibition
293TR-Fluc cells, a gift of Dr. Whitesell (Santagata et al., 2013) were grown to confluence. Compounds were added at different dilutions and kept for 18 h. 100 ul of Nanolight Firefly Luc Assay reagent was added to the wells and the luminescence was measured using a Tecan-plate reader after 2 mins. Cells treated with DMSO served as negative controls and with 100 μg/mL cycloheximide served as positive controls. Percentage of translation inhibition for each compound was calculated in triplicates from two independent experiments.
Puromycin incorporation assay
Puromycin labeling for measuring the intensity of translation was performed as described (Schmidt et al., 2009). In brief, 5 μg/ml puromycin (Sigma) was added in the culture medium and incubated for 1 h at 37 °C and 5% CO2. Cells were lysed and the cell lysate was separated on 12% SDS-PAGE gels, and transferred to PVDF membranes (Millipore). Immunoblotting was performed with the anti-puromycin antibody clone 12D10 (1:10000) (Millipore).
Immunofluorescence microscopy
BMDMs from C57BL/6J mice were grown on coverslips and infected with BCG:gfp followed by treatment with rocaglate for 24 h. Cells were fixed with chilled 100% methanol for 5 min at room temperature and then blocked for 60 min with 1% BSA containing 22.52 mg/mL glycine in PBST (PBS+ 0.1% Tween 20). Cells were incubated with primary antibodies purchased from cell signaling technology [p62(1:200) and LC3B(1:200)] overnight at 4 °C in 1% BSA, and incubated with Alexa Fluor 594-conjugated Goat anti-Rabbit IgG (H+L) secondary Antibody (Invitrogen) in 1% BSA in dark for 1 h. The cells were mounted using ProlongTM Gold antifade reagent (Thermo Fisher Scientific) and Images were acquired using Zeiss LSM 710-Live Duo scan confocal microscope. All images were processed using ImageJ software.
Phagosome-lysosome fusion assay
The cells were pretreated with CMLD010536 (25 nM) for 6 h and subsequently infected with M. bovis BCG expressing gfp (BCG:gfp) for 1 h at MOI 1. To remove the extracellular bacteria, the cells were treated with 200 ug/ml of amikacin for 1 h. After 1 h, 24 h and 48 h post-infection, cells were stained with 200 nM of LysoTracker red dye for 1 h at 37 °C and analyzed using Zeiss LSM 710-Live Duo scan confocal microscope. The images were processed using FIJI software and the percentage of co-localization of BCG:gfp containing phagosomes with LysoTracker red dye was calculated by dividing the number of co-localized phagosomes by the total number of phagosomes. At least 50 cells are measured at each condition and the mean ± SEM (p value was ≤ 0.05) was calculated using GraphPad Prism 8.
Macrophage infection and determination of intracellular bacterial loads
BMDM from B6J.C3-Sst1C3HeB/FejKrmn mice were infected with the mycobacteria in 96 well plate at preferred multiplicity of infection (MOI) by suspending bacteria in BMDM specific medium. The infected cells were centrifuged for 5 min at 200xg and incubated at 37°C with 5% CO2 for 1 h and extracellular bacteria were killed off by incubating the cells in medium containing 200 μg/ml amikacin for 1 h. Further BMDM were washed 3 times with 1% PBS containing 2% FBS and incubated at 37°C with 5% CO2 until further analysis. The intracellular bacterial load was determined using quantitative real time PCR and standard CFU methods. BMDM were lysed using 0.05% Tween 80 in 1X PBS for 5 min at room temperature. The lysates were serially diluted and used for plating on Middlebrook 7H10 agar. DNA for quantitative PCR method was isolated using modified Mtb lysis buffer and magnetic bead purification, as described in detail in Yabaji et al., submitted for publication. Briefly, the infected macrophages were lysed directly in wells using lysis buffer (25 mM NaOH and 0.2 mM EDTA), and total DNA was isolated using magnetic beads. The intracellular bacterial load was determined by quantitative real time PCR (qPCR) using specific set of M.tb and M. bovis-BCG primer/probes.
Quantification and statistical analysis
The densitometric analyses were performed using ImageJ 2.0 software and all the graphs were plotted using Graphpad Prism 8 (GraphPad, San Diego, CA, USA). For multiple treatment experiments, Students’s unpaired t-test and one-way ANOVA analysis was performed in Graphpad Prism 8. The p values of ≤ 0.05 were considered statistically significant.
Acknowledgments
This work was supported by the National Institutes of Health, award numbers R33 AI105944–04 and R01 HL133190-01 (IK), 1R01GM120272 (ARI), and R01CA218500 (ARI). The authors are grateful to Dr. John Connor for helpful discussions.
Author contributions
Conceptualization, I.K. and J.A.P.; methodology, S.C., S.M.Y., O.S.R., B.B., B.N.K., L.E.B., A.B.B., A.R.I., and L.K.; investigation S.C., S.M.Y., B.B., E.W., and N.V.; formal analysis, S.M.Y., O.S.R., and S.R.; writing – original draft I.K., S.C., and S.M.Y.; writing – review and editing, L.K., J.A.P., and A.R.I.; resources, L.E.B. and A.B.B; data curation, S.R. and O.S.R.; funding acquisition I.K., J.A.P., L.K., and A.R.I.; supervision, I.K., J.A.P., and L.K.
Declaration of interests
The authors declare no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102845.
Supplemental information
References
- Bhattacharya B., Chatterjee S., Devine W.G., Kobzik L., Beeler A.B., Porco J.A., Jr., Kramnik I. Fine-tuning of macrophage activation using synthetic rocaglate derivatives. Sci. Rep. 2016;6:24409. doi: 10.1038/srep24409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum L., Geisslinger G., Parnham M.J., Grünweller A., Schiffmann S. Natural antiviral compound silvestrol modulates human monocyte-derived macrophages and dendritic cells. J. Cell Mol. Med. 2020;24:6988–6999. doi: 10.1111/jcmm.15360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau M.E., Robert F., Gerard B., Lindqvist L., Chen S.M., Wendel H.G., Brem B., Greger H., Lowe S.W., Porco J.A., Jr., Pelletier J. Therapeutic suppression of translation initiation modulates chemosensitivity in a mouse lymphoma model. J. Clin. Invest. 2008;118:2651–2660. doi: 10.1172/JCI34753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne S.K., Holland S.M. Anticytokine autoantibodies in infectious diseases: pathogenesis and mechanisms. Lancet Infect Dis. 2010;10:875–885. doi: 10.1016/S1473-3099(10)70196-1. [DOI] [PubMed] [Google Scholar]
- Cadena A.M., Fortune S.M., Flynn J.L. Heterogeneity in tuberculosis. Nat. Rev. Immunol. 2017;17:691–702. doi: 10.1038/nri.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E.J. Hormesis: why it is important to toxicology and toxicologists. Environ. Toxicol. Chem. 2008;27:1451–1474. doi: 10.1897/07-541. [DOI] [PubMed] [Google Scholar]
- Carow B., Hauling T., Qian X., Kramnik I., Nilsson M., Rottenberg M.E. Spatial and temporal localization of immune transcripts defines hallmarks and diversity in the tuberculosis granuloma. Nat. Commun. 2019;10:1823. doi: 10.1038/s41467-019-09816-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cencic R., Carrier M., Galicia-Vazquez G., Bordeleau M.E., Sukarieh R., Bourdeau A., Brem B., Teodoro J.G., Greger H., Tremblay M.L. Antitumor activity and mechanism of action of the cyclopenta[b]benzofuran, silvestrol. PLoS One. 2009;4:e5223. doi: 10.1371/journal.pone.0005223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanmee T., Ontong P., Konno K., Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers (Basel) 2014;6:1670–1690. doi: 10.3390/cancers6031670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro V., Leroux L.-P., Masvidal L., Lorent J., Graber T.E., Zimmermann A., Arango Duque G., Descoteaux A., Alain T., Larsson O., Jaramillo M. Translational profiling of macrophages infected with Leishmania donovani identifies mTOR- and eIF4A-sensitive immune-related transcripts. PLoS Pathog. 2020;16:e1008291. doi: 10.1371/journal.ppat.1008291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitu V., Yeung Y.G., Yu W., Nandi S., Stanley E.R. Measurement of macrophage growth and differentiation. Curr. Protoc. Immunol. 2011;92:14–20. doi: 10.1002/0471142735.im1420s92. 1-14.20.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J., Galicia-Vázquez G., Cencic R., Mills J.R., Katigbak A., Porco J.A., Pelletier J. CRISPR-mediated drug-target validation reveals selective pharmacological inhibition of the RNA helicase, eIF4A. Cell Rep. 2016;15:2340–2347. doi: 10.1016/j.celrep.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan M.R., Hughes E.J., Brewer W.J., Viswanathan G., Hunt E.G., Singh B., Mehra S., Oehlers S.H., Gregory S.G., Kaushal D., Tobin D.M. A non-canonical type 2 immune response coordinates tuberculous granuloma formation and epithelialization. Cell. 2021;184:1757–1774.e14. doi: 10.1016/j.cell.2021.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T., Kovarik P., Meinke A. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J. Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- Dickensheets H.L., Donnelly R.P. Inhibition of IL-4-inducible gene expression in human monocytes by type I and type II interferons. J. Leukoc. Biol. 1999;65:307–312. doi: 10.1002/jlb.65.3.307. [DOI] [PubMed] [Google Scholar]
- Dockrell D.H., Marriott H.M., Prince L.R., Ridger V.C., Ince P.G., Hellewell P.G., Whyte M.K. Alveolar macrophage apoptosis contributes to pneumococcal clearance in a resolving model of pulmonary infection. J. Immunol. 2003;171:5380–5388. doi: 10.4049/jimmunol.171.10.5380. [DOI] [PubMed] [Google Scholar]
- Dudley A.C., Thomas D., Best J., Jenkins A. The STATs in cell stress-type responses. Cell Commun. Signal. 2004;2:8. doi: 10.1186/1478-811X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M.F., Banga S., Barry K.C., Shen X., Tan Y., Luo Z.-Q., Vance R.E. Secreted bacterial effectors that inhibit host protein synthesis are critical for induction of the innate immune response to virulent Legionella pneumophila. PLoS Pathog. 2011;7:e1001289. doi: 10.1371/journal.ppat.1001289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana M.F., Shin S., Vance R.E. Activation of host mitogen-activated protein kinases by secreted Legionella pneumophila effectors that inhibit host protein translation. Infect. Immun. 2012;80:3570–3575. doi: 10.1128/IAI.00557-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin A., Abel L., Casanova J.L., Gros P. Host genetics of mycobacterial diseases in mice and men: forward genetic studies of BCG-osis and tuberculosis. Annu. Rev. Genomics Hum. Genet. 2007;8:163–192. doi: 10.1146/annurev.genom.8.080706.092315. [DOI] [PubMed] [Google Scholar]
- Fritscher L.G., Marras T.K., Bradi A.C., Fritscher C.C., Balter M.S., Chapman K.R. Nontuberculous mycobacterial infection as a cause of difficult-to-control asthma: a case-control study. Chest. 2011;139:23–27. doi: 10.1378/chest.10-0186. [DOI] [PubMed] [Google Scholar]
- Fukutomi T., Takagi K., Mizushima T., Ohuchi N., Yamamoto M. Kinetic, thermodynamic, and structural characterizations of the association between Nrf2-DLGex degron and Keap1. Mol. Cell. Biol. 2014;34:832–846. doi: 10.1128/MCB.01191-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goh K.C., Haque S.J., Williams B.R. p38 MAP kinase is required for STAT1 serine phosphorylation and transcriptional activation induced by interferons. EMBO J. 1999;18:5601–5608. doi: 10.1093/emboj/18.20.5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O'Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon S.B., Irving G.R., Lawson R.A., Lee M.E., Read R.C. Intracellular trafficking and killing of Streptococcus pneumoniae by human alveolar macrophages are influenced by opsonins. Infect Immun. 2000;68:2286–2293. doi: 10.1128/iai.68.4.2286-2293.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco W.R., Bravo G., Parsons J.C. The search for synergy: a critical review from a response surface perspective. Pharmacol. Rev. 1995;47:331–385. [PubMed] [Google Scholar]
- Grivennikov S.I., Greten F.R., Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Harris J., De Haro S.A., Master S.S., Keane J., Roberts E.A., Delgado M., Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Heitmann L., Abad Dar M., Schreiber T., Erdmann H., Behrends J., Mckenzie A.N., Brombacher F., Ehlers S., Holscher C. The IL-13/IL-4Ralpha axis is involved in tuberculosis-associated pathology. J. Pathol. 2014;234:338–350. doi: 10.1002/path.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang B.Y., Su B.N., Chai H., Mi Q., Kardono L.B., Afriastini J.J., Riswan S., Santarsiero B.D., Mesecar A.D., Wild R. Silvestrol and episilvestrol, potential anticancer rocaglate derivatives from Aglaia silvestris. J. Org. Chem. 2004;69:3350–3358. doi: 10.1021/jo040120f. [DOI] [PubMed] [Google Scholar]
- Iwasaki S., Floor S.N., Ingolia N.T. Rocaglates convert DEAD-box protein eIF4A into a sequence-selective translational repressor. Nature. 2016;534:558–561. doi: 10.1038/nature17978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki S., Iwasaki W., Takahashi M., Sakamoto A., Watanabe C., Shichino Y., Floor S.N., Fujiwara K., Mito M., Dodo K. The translation inhibitor rocaglamide targets a bimolecular cavity between eIF4A and polypurine RNA. Mol. Cell. 2019;73:738–748.e9. doi: 10.1016/j.molcel.2018.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo M.C., Zhang D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:279–296. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi Y., Ichimura Y., Komatsu M. p62/SQSTM1 functions as a signaling hub and an autophagy adaptor. FEBS J. 2015;282:4672–4678. doi: 10.1111/febs.13540. [DOI] [PubMed] [Google Scholar]
- Kim S., Hwang B.Y., Su B.N., Chai H., Mi Q., Kinghorn A.D., Wild R., Swanson S.M. Silvestrol, a potential anticancer rocaglate derivative from Aglaia foveolata, induces apoptosis in LNCaP cells through the mitochondrial/apoptosome pathway without activation of executioner caspase-3 or -7. Anticancer Res. 2007;27:2175–2183. [PMC free article] [PubMed] [Google Scholar]
- Kim S., Salim A.A., Swanson S.M., Kinghorn A.D. Potential of cyclopenta[b]benzofurans from Aglaia species in cancer chemotherapy. Anticancer Agents Med. Chem. 2006;6:319–345. doi: 10.2174/187152006777698123. [DOI] [PubMed] [Google Scholar]
- Knaevelsrud H., Simonsen A. Fighting disease by selective autophagy of aggregate-prone proteins. FEBS Lett. 2010;584:2635–2645. doi: 10.1016/j.febslet.2010.04.041. [DOI] [PubMed] [Google Scholar]
- Komatsu M., Kurokawa H., Waguri S., Taguchi K., Kobayashi A., Ichimura Y., Sou Y.S., Ueno I., Sakamoto A., Tong K.I. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nat. Cell Biol. 2010;12:213–223. doi: 10.1038/ncb2021. [DOI] [PubMed] [Google Scholar]
- Kovarik P., Stoiber D., Novy M., Decker T. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. EMBO J. 1998;17:3660–3668. doi: 10.1093/emboj/17.13.3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langlais D., Cencic R., Moradin N., Kennedy J.M., Ayi K., Brown L.E., Crandall I., Tarry M.J., Schmeing M., Kain K.C. Rocaglates as dual-targeting agents for experimental cerebral malaria. Proc. Natl. Acad. Sci. U. S. A. 2018;8:201713000. doi: 10.1073/pnas.1713000115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau A., Wang X.J., Zhao F., Villeneuve N.F., Wu T., Jiang T., Sun Z., White E., Zhang D.D. A noncanonical mechanism of Nrf2 activation by autophagy deficiency: direct interaction between Keap1 and p62. Mol. Cell Biol. 2010;30:3275–3285. doi: 10.1128/MCB.00248-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacMicking J.D., Taylor G.A., Mckinney J.D. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science. 2003;302:654–659. doi: 10.1126/science.1088063. [DOI] [PubMed] [Google Scholar]
- Maik-Rachline G., Lifshits L., Seger R. Nuclear P38: roles in physiological and pathological processes and regulation of nuclear translocation. Int. J. Mol. Sci. 2020;21:6102. doi: 10.3390/ijms21176102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino S., Cilfone N.A., Mattila J.T., Linderman J.J., Flynn J.L., Kirschner D.E. Macrophage polarization drives granuloma outcome during Mycobacterium tuberculosis infection. Infect Immun. 2015;83:324–338. doi: 10.1128/IAI.02494-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzawa T., Kim B.H., Shenoy A.R., Kamitani S., Miyake M., Macmicking J.D. IFN-gamma elicits macrophage autophagy via the p38 MAPK signaling pathway. J. Immunol. 2012;189:813–818. doi: 10.4049/jimmunol.1102041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila J.T., Ojo O.O., Kepka-Lenhart D., Marino S., Kim J.H., Eum S.Y., Via L.E., Barry C.E., Klein E., Kirschner D.E. Microenvironments in tuberculous granulomas are delineated by distinct populations of macrophage subsets and expression of nitric oxide synthase and arginase isoforms. J. Immunol. 2013;191:773–784. doi: 10.4049/jimmunol.1300113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J., Seiler M., Hentze M.W. Translational control via protein-regulated upstream open reading frames. Cell. 2011;145:902–913. doi: 10.1016/j.cell.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Meraz M.A., White J.M., Sheehan K.C., Bach E.A., Rodig S.J., Dighe A.S., Kaplan D.H., Riley J.K., Greenlund A.C., Campbell D. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T. Feedback on fat: p62-mTORC1-autophagy connections. Cell. 2011;147:724–727. doi: 10.1016/j.cell.2011.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller C., Schulte F.W., Lange-Grünweller K., Obermann W., Madhugiri R., Pleschka S., Ziebuhr J., Hartmann R.K., Grünweller A. Broad-spectrum antiviral activity of the eIF4A inhibitor silvestrol against corona- and picornaviruses. Antiviral Res. 2018;150:123–129. doi: 10.1016/j.antiviral.2017.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P.J. Nonresolving macrophage-mediated inflammation in malignancy. FEBS J. 2018;285:641–653. doi: 10.1111/febs.14210. [DOI] [PubMed] [Google Scholar]
- Murray P.J., Allen J.E., Biswas S.K., Fisher E.A., Gilroy D.W., Goerdt S., Gordon S., Hamilton J.A., Ivashkiv L.B., Lawrence T. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41:14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naka T., Tsutsui H., Fujimoto M., Kawazoe Y., Kohzaki H., Morita Y., Nakagawa R., Narazaki M., Adachi K., Yoshimoto T. SOCS-1/SSI-1-deficient NKT cells participate in severe hepatitis through dysregulated cross-talk inhibition of IFN-gamma and IL-4 signaling in vivo. Immunity. 2001;14:535–545. doi: 10.1016/s1074-7613(01)00132-7. [DOI] [PubMed] [Google Scholar]
- Nordling L. Science; 2020. HIV and TB Increase Death Risk from COVID-19, Study Finds—But Not by Much. [Google Scholar]
- Padmanabhan B., Tong K.I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M.I., Kobayashi A., Yokoyama S., Yamamoto M. Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell. 2006;21:689–700. doi: 10.1016/j.molcel.2006.01.013. [DOI] [PubMed] [Google Scholar]
- Pan H., Mostoslavsky G., Eruslanov E., Kotton D.N., Kramnik I. Dual-promoter lentiviral system allows inducible expression of noxious proteins in macrophages. J. Immunol. Methods. 2008;329:31–44. doi: 10.1016/j.jim.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S.Y., Ding L., Brown M.R., Lantz L., Gay T., Cohen S., Martyak L.A., Kubak B., Holland S.M. Anti-IFN-gamma autoantibodies in disseminated nontuberculous mycobacterial infections. J. Immunol. 2005;175:4769–4776. doi: 10.4049/jimmunol.175.7.4769. [DOI] [PubMed] [Google Scholar]
- Pello O.M. Macrophages and c-Myc cross paths. Oncoimmunology. 2016;5:e1151991. doi: 10.1080/2162402X.2016.1151991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pello O.M., De Pizzol M., Mirolo M., Soucek L., Zammataro L., Amabile A., Doni A., Nebuloni M., Swigart L.B., Evan G.I. Role of c-MYC in alternative activation of human macrophages and tumor-associated macrophage biology. Blood. 2012;119:411–421. doi: 10.1182/blood-2011-02-339911. [DOI] [PubMed] [Google Scholar]
- Pichugin A.V., Yan B.S., Sloutsky A., Kobzik L., Kramnik I. Dominant role of the sst1 locus in pathogenesis of necrotizing lung granulomas during chronic tuberculosis infection and reactivation in genetically resistant hosts. Am. J. Pathol. 2009;174:2190–2201. doi: 10.2353/ajpath.2009.081075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier G., Neumann J., Thuaud F., Ribeiro N., Gelhaus C., Schmidt H., Giaisi M., Kohler R., Muller W.W., Proksch P. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem. Biol. 2012;19:1093–1104. doi: 10.1016/j.chembiol.2012.07.012. [DOI] [PubMed] [Google Scholar]
- Potian J.A., Rafi W., Bhatt K., Mcbride A., Gause W.C., Salgame P. Preexisting helminth infection induces inhibition of innate pulmonary anti-tuberculosis defense by engaging the IL-4 receptor pathway. J. Exp. Med. 2011;208:1863–1874. doi: 10.1084/jem.20091473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.V., Vance R.E. The macrophage paradox. Immunity. 2014;41:685–693. doi: 10.1016/j.immuni.2014.10.015. [DOI] [PubMed] [Google Scholar]
- Rodrigo C.M., Cencic R., Roche S.P., Pelletier J., Porco J.A., Jr. Synthesis of rocaglamide hydroxamates and related compounds as eukaryotic translation inhibitors: synthetic and biological studies. J. Med. Chem. 2012;55:558–562. doi: 10.1021/jm201263k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dotsch V., Johansen T., Kirkin V. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 2014;53:167–178. doi: 10.1016/j.molcel.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Sahani M.H., Itakura E., Mizushima N. Expression of the autophagy substrate SQSTM1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy. 2014;10:431–441. doi: 10.4161/auto.27344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S., Mayer-Barber K.D., Barber D.L. Defining features of protective CD4 T cell responses to Mycobacterium tuberculosis. Curr. Opin. Immunol. 2014;29:137–142. doi: 10.1016/j.coi.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgame P., Yap G.S., Gause W.C. Effect of helminth-induced immunity on infections with microbial pathogens. Nat. Immunol. 2013;14:1118–1126. doi: 10.1038/ni.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagata S., Mendillo M.L., Tang Y.C., Subramanian A., Perley C.C., Roche S.P., Wong B., Narayan R., Kwon H., Koeva M. Tight coordination of protein translation and HSF1 activation supports the anabolic malignant state. Science. 2013;341:1238303. doi: 10.1126/science.1238303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid M.C., Varner J.A. Myeloid cells in the tumor microenvironment: modulation of tumor angiogenesis and tumor inflammation. J. Oncol. 2010;2010:201026. doi: 10.1155/2010/201026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E.K., Clavarino G., Ceppi M., Pierre P. SUnSET, a nonradioactive method to monitor protein synthesis. Nat. Methods. 2009;6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]
- Shenoy A.R., Kim B.H., Choi H.P., Matsuzawa T., Tiwari S., Macmicking J.D. Emerging themes in IFN-gamma-induced macrophage immunity by the p47 and p65 GTPase families. Immunobiology. 2007;212:771–784. doi: 10.1016/j.imbio.2007.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sica A., Mantovani A. Macrophage plasticity and polarization: in vivo veritas. J. Clin. Invest. 2012;122:787–795. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S.B., Davis A.S., Taylor G.A., Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- So E.Y., Oh J., Jang J.Y., Kim J.H., Lee C.E. Ras/Erk pathway positively regulates Jak1/STAT6 activity and IL-4 gene expression in Jurkat T cells. Mol. Immunol. 2007;44:3416–3426. doi: 10.1016/j.molimm.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Su X., Yu Y., Zhong Y., Giannopoulou E.G., Hu X., Liu H., Cross J.R., Ratsch G., Rice C.M., Ivashkiv L.B. Interferon-gamma regulates cellular metabolism and mRNA translation to potentiate macrophage activation. Nat. Immunol. 2015;16:838–849. doi: 10.1038/ni.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi K., Motohashi H., Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Venkataraman C., Leung S., Salvekar A., Mano H., Schindler U. Repression of IL-4-induced gene expression by IFN-gamma requires Stat1 activation. J. Immunol. 1999;162:4053–4061. [PubMed] [Google Scholar]
- Vergadi E., Ieronymaki E., Lyroni K., Vaporidi K., Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J. Immunol. 2017;198:1006–1014. doi: 10.4049/jimmunol.1601515. [DOI] [PubMed] [Google Scholar]
- Wallis R.S., Hafner R. Advancing host-directed therapy for tuberculosis. Nat. Rev. Immunol. 2015;15:255–263. doi: 10.1038/nri3813. [DOI] [PubMed] [Google Scholar]
- Wen Z., Zhong Z., Darnell J.E., Jr. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82:241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- Wolfe A.L., Singh K., Zhong Y., Drewe P., Rajasekhar V.K., Sanghvi V.R., Mavrakis K.J., Jiang M., Roderick J.E., Van Der Meulen J. RNA G-quadruplexes cause eIF4A-dependent oncogene translation in cancer. Nature. 2014;513:65–70. doi: 10.1038/nature13485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Huang Y.-C.T., Koziel H., De Crom R., Ruetten H., Wohlfart P., Thomsen R.W., Kahlert J.A., Sørensen H.T., Jozefowski S. Female resistance to pneumonia identifies lung macrophage nitric oxide synthase-3 as a therapeutic target. eLife. 2014;3:265. doi: 10.7554/eLife.03711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.R., Mahdi R.M., Ebong S., Vistica B.P., Chen J., Guo Y., Gery I., Egwuagu C.E. Cell proliferation and STAT6 pathways are negatively regulated in T cells by STAT1 and suppressors of cytokine signaling. J. Immunol. 2004;173:737–746. doi: 10.4049/jimmunol.173.2.737. [DOI] [PubMed] [Google Scholar]
- Zheng Y.T., Shahnazari S., Brech A., Lamark T., Johansen T., Brumell J.H. The adaptor protein p62/SQSTM1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009;183:5909–5916. doi: 10.4049/jimmunol.0900441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate datasets/code and any additional information will be available from the lead contact upon request.