Abstract
Multi-omics approaches are novel frameworks that integrate multiple omics datasets generated from the same patients to better understand the molecular and clinical features of cancers. A wide range of emerging omics and multi-view clustering algorithms now provide unprecedented opportunities to further classify cancers into subtypes, improve the survival prediction and therapeutic outcome of these subtypes, and understand key pathophysiological processes through different molecular layers. In this review, we overview the concept and rationale of multi-omics approaches in cancer research. We also introduce recent advances in the development of multi-omics algorithms and integration methods for multiple-layered datasets from cancer patients. Finally, we summarize the latest findings from large-scale multi-omics studies of various cancers and their implications for patient subtyping and drug development.
Keywords: cancer research, genomics, multi-omics approach, proteogenomics, proteomics, systems biology
INTRODUCTION
Living organisms experience millions of signals transferred every second between cells, tissues, organs, and external environmental stimuli. Fine-tuned responses at various degrees and scales within the human body are central to the homeostatic mechanism that copes with potentially harmful environmental perturbations, including pathogens, smoking, and drugs, and interacts with the genetic background arising from spontaneous somatic mutations and numerous germline variants. Thus, a holistic view of homeostatic mechanisms through the study of genomic and epigenetic aberrations is needed to understand the core of cancer biology and the pathophysiological features of cancer during oncogenesis and tumor progression.
A multi-omics study is a data-driven scientific investigation that analyzes a range of high-dimensional datasets at multiple levels and scales to reveal the complexity of cells and their environment. Such type of study can provide novel frameworks to untangle biological phenomena or models to test certain hypotheses using various datasets. In cancer research, a paradigm shift toward multi-omics approaches has been achieved with the recent development of high-throughput technologies in genomics and transcriptomics, increasing effort in large-scale research collaboration, and advancement of computational algorithms (Basu et al., 2013; Berns and Bernards, 2012; Cancer Genome Atlas Network, 2012b; Gentles and Gallahan, 2011; Whitehurst et al., 2007). Together with advances in genomics and transcriptomics, proteomics is emerging as a prominent field to elucidate the dynamics of gene activity. Large-scale proteomic research, such as that promoted by the Clinical Proteomic Tumor Analysis Consortium (CPTAC), has uncovered the ubiquitous link of biomolecules to the environment and disease status (Gillette et al., 2020; Krug et al., 2020; Mertins et al., 2016; Mun et al., 2019; Zhang et al., 2016). Such a transition has extensively deepened our knowledge on the function of driver genes and proteins and has provided a comprehensive understanding of the signaling networks occurring between cells, tissues, organs, and the entire organism. Multi-omics approaches have been applied to numerous clinical studies for better identification of clinical subtypes or drug resistance, prediction of effective combination therapies, and identification of predictive biomarkers to increase the response rate to targeted treatments.
In this review, we introduce the concept of multi-omics approaches in cancer research and provide useful resources for this. We focus on some of the clinical and basic science studies that have benefited from the use of a multi-omics approach to uncover novel concepts and properties. We also discuss some of the challenges connected to multi-omics approaches and how this relatively young field of study can have a positive impact on cancer research.
MULTI-OMICS APPROACHES IN CANCER RESEARCH
Over the past decades, there have been rapid advances in high-throughput technologies, which enable a range of genomic analyses at the cellular and tissue levels. Furthermore, highly developed genome screening technologies, such as whole exome sequencing (WES) and whole genome sequencing (WGS), have enabled comprehensive collection of gene expression data (e.g., RNA sequencing [RNA-seq] and microRNA [miRNA] profiling) and DNA methylation profiles (Cancer Genome Atlas Network, 2012a; 2012b; Cancer Genome Atlas Research Network, 2011, 2013; Cancer Genome Atlas Research Network et al., 2013a; Chin et al., 2006; Hennessy et al., 2010; Neve et al., 2006). Single-cell technologies provide new biological insights for the understanding of gene activity and cytological characteristics at the cellular level (Lee et al., 2021; Stuart et al., 2019; Stuart and Satija, 2019). In addition, large amounts of proteins and metabolites can be detected with high accuracy owing to the maturation of mass spectrometry techniques (Lai et al., 2018; Palmer et al., 2017; Schubert et al., 2017). Proteomics technologies allow to detect almost all human proteins and are advancing toward single-cell resolution (Marx, 2019; Vidova and Spacil, 2017). However, a single platform is insufficient to decipher the complexity underlying cancer genomes or to find a robust association with cancer driver mutations (Bozic et al., 2010; Greenman et al., 2007). Consequently, there is an emerging effort in the development of data-driven mathematical and computational methods to analyze high-dimensional datasets obtained from several novel analysis platforms (Bodenmiller et al., 2012; Hill et al., 2012; Pritchard et al., 2013; Qiu et al., 2011; Sumazin et al., 2011; Tentner et al., 2012; Teves and Won, 2020).
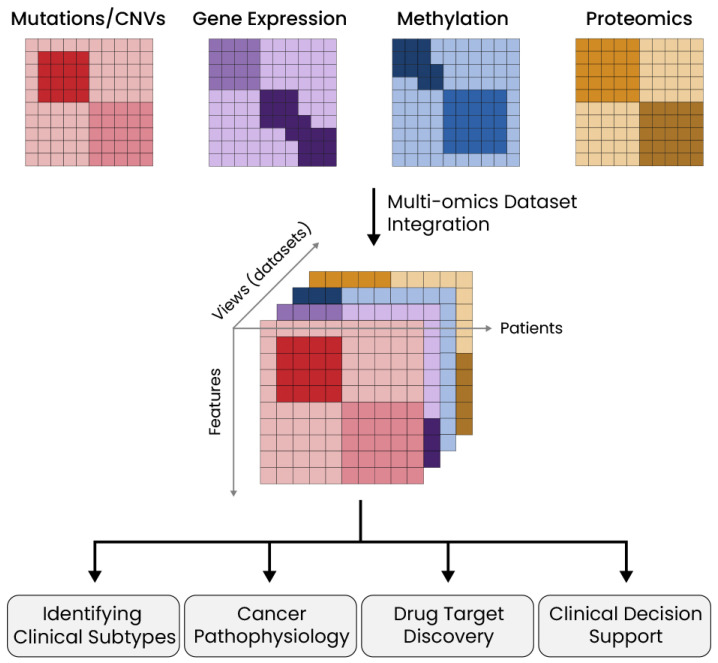
In this regard, multi-omics approaches have been introduced to integrate multiple omics datasets generated from patients and identify coherent and preserved molecular or clinical features across different datasets (Fig. 1). Multi-omics studies aim to identify patient subgroups and biological features underlying cancer pathophysiology; they have been applied to overcome current complexities, due to genetic and phenotypic heterogeneity, that hinder our understanding of cancer genesis and progression, and to design effective predictive models to validate novel therapies and drugs. Within such an integrative framework, there has been an emerging effort to develop computational and mathematical methods that can decipher the complexity of cancer heterogeneity, since genomic and epigenetic instability in tumors can alter intracellular responses to the local environment and affect the individual as a whole through the tumorigenic process.
Fig. 1. Overview of multi-omics approaches in cancer research.
The integration of omics datasets is a crucial step in multi-omics studies. Datasets such as somatic mutations, CNV, gene expression, methylation, and proteome datasets are merged using various computational frameworks with distinct methods. The integration enables the comparison of molecular features across multiple viewpoints and the clustering of patients with relevant clinical features. Possible outcomes include enhanced identification of clinical subtypes, understanding of cancer pathophysiology, prediction of potential drug targets, and clinical decision support.
Over the last decade, a range of modeling approaches have been developed to deal with various aspects of cancer. In particular, the integration of large omics datasets has enabled modeling of cellular behaviors at the tissue level to understand cancer pathophysiology or the behavior of cancer cells in response to drugs and angiogenesis (Carro et al., 2010; Hong et al., 2020; Huang et al., 2013; Iadevaia et al., 2010; Pascal et al., 2013; Swanson et al., 2011). Multi-omics studies have opened new avenues for the implementation of targeted therapies for cancer treatment. Integrative approaches with large-scale multi-omics datasets have the potential to delineate the relationship between molecular markers and the response to targeted therapies. A more comprehensive understanding of the molecular characteristics of non-responsive or resistant tumors could enable more precise predictions of therapy outcomes, resulting in an increased therapeutic efficacy or in the ability to bypass drug resistance. In addition, multi-omics approaches might allow to identify subgroups of patients that are most likely to benefit from therapy.
Cancer cells exhibit extreme levels of genetic heterogeneity and genomic instability. Thus, many putative driver aberrations can be observed: some could be bona fide drivers of cancer, but most of them are passenger mutations. Therefore, a major challenge in cancer research is to identify biomarkers or potential targets for cancer treatment (Cancer Genome Atlas Research Network, 2013; Cancer Genome Atlas Research Network et al., 2013a). On the other hand, it remains to be elucidated whether passenger aberrations within cancer genes play a role in cellular functions associated with cancer pathophysiology and response to targeted therapeutics. To evaluate this, a recent study developed a systems-based computational method that can assess low-frequency mutations in impure and heterogeneous samples (Cibulskis et al., 2013). This study successfully reported a range of sub-clonal drivers underpinning tumor progression and treatment resistance. Thus, multi-omics approaches can provide an efficient analytic framework to distinguish drivers from passenger mutations and dissect the genetic heterogeneity of cancer cells.
COMPUTATIONAL FRAMEWORKS FOR MULTI-OMICS STUDIES
Recent advances in high-throughput sequencing technologies have allowed the measurement of a large number of molecular patterns of cancer in a single experiment. High-throughput measurements enable rapid and unbiased profiling of somatic mutations, copy number variations (CNVs), and mRNA, non-coding RNA, and protein expression. Various computational algorithms have been proposed for multi-view clustering, to detect coherent features from heterogeneous inputs. In the biomedical domain, this has facilitated the definition of the clinical subtypes of complex disorders, such as cancers. Clustering methods have been widely developed to identify co-expressed gene modules and subgroups of patients within a certain disease (Langfelder and Horvath, 2008). The integration of multi-omics datasets for the same set of samples has been devised to better understand fine-tuned structures, which are not revealed by examining only a single data type. For instance, cancer subtypes can be classified based on multi-omics datasets, such as gene expression and mutation profiles, from the same patients (Chauvel et al., 2020). Multi-omics clustering can ameliorate potential bias or noise from a single omics dataset as the integration of multiple omics layers can fully represent different cellular aspects from the genomic to the epigenomic level (Nguyen and Wang, 2020; Wang et al., 2014).
To date, various tools have been developed for multi-omics datasets with the following objectives: 1) identify disease subtypes or classify subgroups, 2) identify putative biomarkers for diagnostics and driver genes for diseases, and 3) gain insights into disease biology. Multi-omics frameworks are mostly based on Bayesian statistics (Kirk et al., 2012; Lock and Dunson, 2013; Shen et al., 2009; Vaske et al., 2010; Wu et al., 2015; Yuan et al., 2011), similarity networks (Nguyen et al., 2019; Wang et al., 2014), joint nonnegative matrix factorization (Yang and Michailidis, 2016), and sparse canonical correlation analysis (Witten and Tibshirani, 2009). Several multi-omics tools are highly used in the field or show outperformance for subtype prediction and survival analysis (Table 1). However, most multi-omics tools rely on different mathematical theories and support different ranges of data types. Even when using the same data, their performance varies greatly depending on the biological characteristics of the study objects. Therefore, acquiring biological insights from multi-omics data is a computational and biological challenge, requiring the researcher to select appropriate multi-omics tools.
Table 1.
List of computational frameworks for multi-omics cancer studies
| Study | Findings | Dataset | Principles |
|---|---|---|---|
| iCluster (Curtis et al., 2012; Shen et al., 2009) | Novel subgroups from 2,000 breast tumors | mRNA expressiona
CNVc |
Joint latent variable model-based clustering method |
| iOmicsPASS (Koh et al., 2019) | Novel transcriptional regulatory network from TCGA/CPTAC breast cancer data | mRNA expressiona
CNVd Protein expressione |
Network construction using a modified nearest shrunken centroid algorithm |
| SALMON (Huang et al., 2019) | Improved survival analysis | Mutationh
mRNA/miRNA expression CNVh |
Deep learning based on co-expression modules |
| SNF (Wang et al., 2014) | Subtype classification of clinical relevance | mRNAa/miRNA expressionb
DNA methylationg |
Patient similarity networks using an iterative procedure based on message passing |
| NEMO (Rappoport and Shamir, 2019) | Novel subtypes from even partial AML datasets | mRNAa/miRNA expressionb
DNA methylationg |
Sample clustering from partial datasets using an adjusted Rand index |
| MONET (Rappoport et al., 2020) | Module detection of patient subtypes and improved survival analysis | mRNAa/miRNA expressionb
DNA methylationg |
Detect similar modules commonly present across multi-omics datasets |
| PARADIGM (Vaske et al., 2010) | Detection of pathways affected by cancer with fewer false positives | mRNA expressiona
CNVc |
Pathway recognition algorithm applied to multi-omics datasets |
| LRAcluster (Wu et al., 2015) | Subtype detection in both pan-cancer analysis and single cancer types | Mutationi
mRNA expressiona CNVd DNA methylationg |
Performance of low-rank approximation from probabilistic models |
| BCC (Lock and Dunson, 2013) | Detection of patient subtypes in response to survival rates and driver mutation signatures | mRNAa/miRNA expressionb
DNA methylationg Protein expressionf |
Bayesian framework for estimation of an integrative clustering model |
Gene expression data with normalization (e.g., quantile normalization, fragment per kilobase of transcript per million mapped reads [FPKM]).
Quantification of miRNA expression.
Circular binary segmentation-based copy number segmented means.
Affymetrix 6.0 SNP arrays.
Protein quantification by iTRAQ (isobaric Tags for Relative and Absolute Quantification) protein quantification.
Reverse phase protein array (RPPA).
Illumina Human Methylation arrays.
In the SALMON method, the copy number burden (CNB) is calculated using the total gene length (Kb) from SNP 6 data, and the tumor mutation burden (TMB) is calculated using the total number of mutated genes reported in Mutation Annotation Format (MAF) files.
The LRAcluster method uses somatic mutation data converted into a binary form.
iCluster
iCluster is an early multi-omics integration method that first integrates multiple inputs and then identifies multi-omics clusters by joint estimation of latent variables and through clustering and expectation–maximization-like algorithms (Shen et al., 2009). It was initially used for large-scale cancer genomic projects, for example for breast and lung cancer, in which gene expression and CNVs were summarized for multiple subgroups of patients. Since the runtime of iCluster increases with the number of features, iCluster+, providing full Bayesian regularization for clustering, has recently been proposed (Mo et al., 2013). iCluster+ identified colorectal cancer subtypes with different cancer progression pathways, one of which was found not to require aggressive drug treatment in addition to surgery.
iOmicsPASS
iOmicsPASS is a network-based algorithm that can merge genome-based networks with multi-omics datasets (Koh et al., 2019). Scores for biological interaction are computed by transformation of omics datasets and used as an input to construct networks, whose edges are defined for phenotypic groups using a modified nearest shrunken centroid algorithm. iOmicsPASS was shown to improve the identification of breast invasive ductal carcinoma (IDC) subtypes by integrating mRNA expression and protein abundance data. Such integrated analysis by iOmicsPASS revealed a new transcriptional regulatory network in a specific breast cancer subtype that could not be found through single-omics analysis.
SALMON (Survival Analysis Learning with Multi-Omics Neural Networks)
SALMON is a deep learning method based on co-expression networks (Huang et al., 2019). It takes multi-omics datasets from cancer patients and computes eigengenes from co-expression modules, and can thus ameliorate the issue of overfitting arising whenever multi-omics approaches are applied to datasets containing many features but few samples are available. For example, by analyzing mRNA and miRNA datasets from 583 female breast invasive carcinoma patients, SALMON provided a good prediction of survival.
SNF (Similarity Network Fusion)
SNF is a novel algorithm for the generation of patient similarity networks that uses an iterative procedure based on message passing (Wang et al., 2014). It calculates similarity networks for individual patients and then merges them to identify disease subtypes and predict phenotypes. In contrast to early integration, SNF takes advantage of individual omics datasets to construct independent single-omics networks and find coherent modules sourced from similar biological features across patients with similar clinical features. SNF iteratively applies a local K-nearest neighbors (KNN) approach to compute a patient similarity matrix for each omics dataset. When merging the global similarity matrices from all omics datasets, SNF conducts averaging of similarity matrices with iterative updating. It has demonstrated high efficiency in identifying clinical subtypes of cancers and other disorders such as autism (Cavalli et al., 2017; Ramaswami et al., 2020).
NEMO (NEighborhood based Multi-Omics clustering)
NEMO is a multi-omics clustering method that can be used for partial datasets without the need for data imputation (Rappoport and Shamir, 2019). NEMO first calculates an inter-patient similarity matrix for each omics dataset and then combines the matrices of different omics datasets into a single matrix. Clusters are identified using an adjusted Rand index to compute the similarity between patients by distance. NEMO was shown to outperform other multi-omics clustering algorithms when tested on multi-omics datasets of 10 cancers, and exhibited enhanced cluster detection from partial datasets.
MONET (Multi Omic clustering by Non-Exhaustive Types)
MONET is a method for detecting similar modules commonly present across multi-omics datasets (Rappoport et al., 2020). MONET utilizes three omics datasets (mRNA expression, DNA methylation, and miRNA expression) to compute an edge-weighted graph per omics dataset, where nodes represent samples and edges represent the similarity between samples. It then detects a disjoint set of modules for patients from multiple omics graphs. MONET was used to conduct benchmarking on 287 patients with ovarian serous cystadenocarcinoma, and revealed four sample modules representing venous invasion status and survival rates.
PARADIGM (PAthway Recognition Algorithm using Data Integration on Genomic Models)
PARADIGM is a method to identify specific biological pathways from a multi-omics dataset (Vaske et al., 2010). It combines multi-omics-scale values derived from an individual sample with gene activities, products, and an overview of the pathway interactions included in the National Cancer Institute (NCI) database, which contains information on protein-protein interactions. PARADIGM utilizes factor graphs derived from variables representing the state of various entities (e.g., a specific mRNA molecule or protein complex), and then creates probabilistic graphical models. Using these, it infers significant and non-significant interactions between pathways involving different entities. This tool proved to be efficient, and revealed four subtypes of glioblastoma leading to significantly different survival outcomes according to the perturbated pathways. This result suggests that the cancer subtype could be used as a basis to support clinical decisions.
LRAcluster (Low Rank Approximation based multi-omics data clustering)
LRAcluster is a multi-omics approach that integrates data on somatic mutations, CNVs, DNA methylation, and gene expression, and performs low-rank approximation from the probabilistic models of various molecular features (Wu et al., 2015). All molecular features from the omics datasets are transformed into variables and arranged in a parameter matrix, which is subject to the low-rank assumption. Next, dimension reduction is conducted, revealing clusters associated with distinct clinical subtypes. LRAcluster outperformed other existing methods in terms of both time and classification accuracy when tested on multi-omics datasets of breast invasive carcinoma, colon adenocarcinoma, and lung adenocarcinoma (LUAD).
BCC (Bayesian Consensus Clustering)
BCC is a data-driven approach that performs consensus clustering across multi-omics datasets (Lock and Dunson, 2013). BCC is based on the finite Dirichlet mixture model to explain not only overall consensus clustering, but also important features inherent to an individual omics dataset. Given that clusters constructed using a single data type are roughly connected, BCC seeks an integrative point for their adherence to an overall cluster. BCC was applied to 384 breast cancer patients from TCGA datasets, including gene expression, DNA methylation, and protein data, and effectively revealed three cancer subtypes associated with specific clinical features.
LATEST FINDINGS AND IMPLICATIONS IN CANCER MULTI-OMICS STUDIES
Cancer research has taken advantage of advances in omics technologies from genomics to transcriptomics and of the wide range of resources of multiple omics datasets originating from the same patients. Multi-omics approaches provide a unique opportunity to identify the molecular and clinical features of cancer patients. In genomics and transcriptomics, there is an unmet need to disentangle incompatibility in related biological processes, such as differences in post-translational modifications or variability in expression profiles due to the role of mRNA transcripts in cancer development (Greenbaum et al., 2003; Hegde et al., 2003; Tyers and Mann, 2003). Recent advances in proteomics through the maturation of several mass spectrometry techniques have enabled the introduction of proteogenomic approaches, which can integrate genomic data with proteomics and information on post-translational modifications (e.g., protein phosphorylation and acetylation). Large-scale proteogenomic research, including that promoted by the CPTAC (Gillette et al., 2020; Krug et al., 2020; Mertins et al., 2016; Mun et al., 2019; Zhang et al., 2016), has been conducted to unravel new biological mechanisms in cancers and provide fundamental information on multi-omics approaches for the development of integration strategies or computational algorithms.
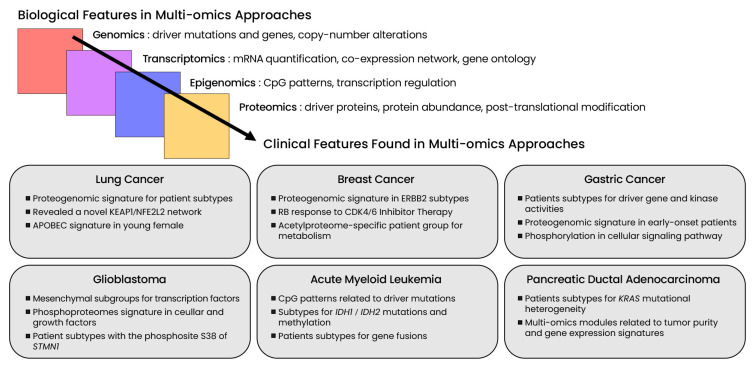
Multi-omics clustering further refined the association between molecular profiles and clinical features among cancer patients (Fig. 2). The identification of coherent subtypes across multiple dataset layers could have major implications for predicting clinical relevance or therapeutic response regardless of the overall tumor mutational load. Moreover, the integration of proteomics datasets enables the identification of a direct connection between mutations and phenotypes, and therefore increases the resolution of clustering patterns across samples. Here, we summarize the latest findings obtained in cancer research using multi-omics approaches.
Fig. 2. Latest findings in cancer multi-omics research.
Multi-omics approaches integrate various high-throughput sequencing datasets across a range of molecular layers. Biological features are subject to multi-view clustering methods and account for distinct subtypes of cancer patients based on relevant clinical features.
Lung cancer
Despite extensive research on its mutation signature and gene expression landscape, LUAD shows a high level of intrinsic or acquired resistance after treatment. Therefore, recent multi-omics-based efforts have been made to integrate genomic, transcriptomic, and proteomic datasets and decipher the molecular features underlying durable treatment responses.
Recently, the CPTAC has conducted a large-scale multi-omics study of LUAD by integrating WES, WGS, RNA-seq, miRNA and DNA methylation profiling, and high-resolution mass spectrometry-based proteomics, phosphoproteomics, and acetylproteomics. Integrative multi-omics clustering revealed four clusters of clinical and molecular features. For example, the patients in Cluster 1 were mostly TP53 positive but STK11 negative, and showed high gene expression in proximal inflammatory structures and high CpG methylation. In contrast, the patients in Cluster 2 were TP53 negative and their transcriptome was enriched in proximal proliferative subcluster genes. This multi-omics approach also enabled to dissect ethnic differences in the cohort, represented by Cluster 3 (Vietnamese patients) and Cluster 4 (Chinese patients), which exhibited distinct mutation signatures (Gillette et al., 2020). Moreover, deep-scale proteogenomic studies revealed a novel KEAP1/NFE2L2 network mechanism based on cis and trans regulation. Driver mutations in KEAP1 did not impact the levels of KEAP1 and NFE2L2 transcripts but were highly correlated with the phosphorylation of NFE2L2 and low protein expression of KEAP1. The KEAP1/NFE2L2 heterocomplex upregulates the antioxidant pathway to protect cancer cells and can be used as a unique biomarker for LUAD.
In another large-scale study, Chen et al. (2020) applied multi-omics approaches for early-stage, non-smoker patients in Taiwan using WES, RNA-seq, and proteomics datasets (Chen et al., 2020). Clustering was performed separately for proteomics, transcriptomics, and phosphoproteomics datasets, and clustering of proteomics data into three subtypes was chosen as the best representative of tumor staging and driver mutation classification. The largest group, Subtype 1, was composed of late-stage tumors (> II) with a high mutation rate, including in TP53. Subtype 2 represented IA- and IB-stage patients that did not carry the EGFR-L858R mutation. Finally, early-stage (IA) patients that lacked the TP53 mutation were classified into Subtype 3. To further decipher the biological features of this cohort, these authors constructed protein-protein interaction network models using STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) (Szklarczyk et al., 2019). The constructed models explained the differential regulation of the three subtypes mentioned above. It was found that extracellular matrix (ECM)-regulated pathways, involving the proteins MMP7, MMP11, and MMP12, were significantly upregulated in Subtype 1 patients. Immunohistochemical staining for these three matrix metalloproteinases (MMPs) revealed that MMP11 was highly associated with patient survival and was a candidate biomarker. This study also showed a clear APOBEC signature in females, associated with upregulation of DNA damage proteins and phosphosites, implicating putative environmental carcinogens in cancer development of non-smoking patients.
Breast cancer
Multi-omics analyses have increased our knowledge of breast cancer biology. In particular, integrative analyses have revealed the recurrence of mutations in the TP53, PIK3CA, and GATA3 genes in breast cancer, but also the presence of specific mutations within subtypes, such as PIK3CA mutations in luminal tumors (Cancer Genome Atlas Network, 2012b). As a result, multi-omics approaches could reveal a new subtype of breast cancer that had not been previously detected from a single dataset. Similarly, integrated analyses revealed the activation of signaling pathways promoting HER2 or epidermal growth factor receptor (EGFR) activity. Given the observed downstream phosphorylation of EGFR, the activation of the HER2 signaling network might reflect the need for a treatment strategy tailored to this subgroup of patients. Endometrial, colon, and rectal cancers have been associated with hypermutation, which might be attributed to microsatellite instability, while a new type of instability driven by mutations of the POLE gene results in ultra-mutated tumors (Cancer Genome Atlas Network, 2012a). Multi-omics analyses have reported MYC-directed activation in aggressive colorectal carcinoma. In clear cell renal cell carcinoma, alterations in cellular oxygen sensing and chromatin remodeling/histone methylation, as well as metabolic shifts in the tricarboxylic acid (TCA) cycle, have been observed, and might be key processes in the pathology of this cancer type (Cancer Genome Atlas Research Network, 2013).
An integrative analysis of gene expression and proteomics has been applied to the survival data of ERBB2-positive patients, and revealed breast tumors with acquired resistance to lapatinib and ability to block EGFR/ERBB2 signaling (Komurov et al., 2012). Nonetheless, an increase in glucose metabolism, unfolded protein response, and endoplasmic reticulum (ER) stress pathways reduced the ability of lapatinib to induce cell death. Arguably, this might imply that targeting both metabolic and signaling networks may improve patient outcomes (Csibi et al., 2013; Komurov et al., 2012).
A recent study on 122 patients integrating data on mutations, mRNA expression, protein expression, and post-translational modifications (phosphorylation and acetylation) has yielded robust profiles to elucidate the biological features of breast cancer (Krug et al., 2020). The resulting subtypes, that is, the basal-inclusive, HER2-inclusive, LumA-inclusive, and LumB-inclusive subtypes, were similar to those generated by the already existing and widely used PAM50 assay but revealed hidden biological structures such as the status of the ERBB2 amplicon, stratified by proteomics assessment; the RB status, which is deeply related to the CDK4/6 inhibitor; and post-translational cross-linkage between proteins involved in cytoplasmic and mitochondrial metabolic pathways. The acetylproteome was found to be useful for distinguishing cancers into luminally and basally enriched subtypes, based on their metabolic activity.
Gastric cancer
Multi-omics research on gastric cancers revealed four subtypes: 1) an Epstein–Barr virus subtype with recurrent PIK3CA mutations, 2) a microsatellite-unstable subtype with a high mutation rate, 3) a genomically stable type enriched in a diffuse histological variant, and 4) a chromosomally unstable type with aneuploidy and focal amplification of receptor tyrosine kinases (Cancer Genome Atlas Research Network, 2014). A recent proteogenomic study of early-onset gastric cancer revealed four subtypes through integrated analysis; moreover, phosphorylation data supported the classification into four subtypes and provided information about active signaling pathways (Mun et al., 2019). The authors of this study applied a network propagation method to mutation and phosphorylation data and calculated two types of network-smoothed scores. Two functionally related cellular processes, affiliated with gastric cancer pathogenesis, were identified using network-smoothed scores for pairs of mutated genes and phosphorylated proteins. The first cellular process was represented by Notch and caspase signaling with mutated genes and phosphorylated proteins. The second cellular process was associated with MAPK, AMPK, FOXO, mTOR, and T-cell receptor signaling. Therefore, multi-omics approaches enable the discovery of various subtypes of gastric cancer, thereby allowing a comprehensive understanding of patient stratification and suggesting novel possibilities for personalized targeted therapy.
Glioblastoma
In highly characterized samples of glioblastoma patients, a multi-omics approach has delineated core transcriptional factors (CEBP and STAT3) that widely regulate mesenchymal transformation in glioblastoma (Carro et al., 2010). Integrative analyses of gene expression and phosphoproteomes have identified several cellular features that respond to stress and growth factors (Hill et al., 2012; Huang et al., 2013), are key regulators of the EGFR signaling pathway, and are associated with patient survival outcomes (Amit et al., 2007). Similarly, combining proteomic and metabolomic profiles also revealed a unique regulatory function in a cellular network of stress and growth factors (Bordbar et al., 2012). Dekker et al. (2020) conducted an integrative multi-omics analysis of gene and protein expression, as well as phosphoproteomic profiles, using paired primary recurrent tissue samples from eight glioblastoma patients (Dekker et al., 2020). Half of the patients showed a marked difference in the phosphorylation of STMN1 (S38), a component of the ERBB4 signaling pathway.
Acute myeloid leukemia
Integrating methylation profiles with genomic and transcriptomic datasets can substantiate the utility of studying acute myeloid leukemia (AML). A multi-omics analysis of 200 adult patients with AML showed distinct gene expression and methylation patterns across samples (Cancer Genome Atlas Research Network et al., 2013b). In particular, CpG-sparse regions showed a marked difference in methylation due to gene mutations. AML cells with IDH1 and IDH2 mutations exhibited more extensive methylation than normal CD34+CD38- cells, whereas AML cells with MLL fusions or co-occurring NPM1, DNMT3A, and FLT3 mutations were related to loss of DNA methylation.
Pancreatic ductal adenocarcinoma
A multi-omics approach has also been applied to pancreatic ductal adenocarcinoma (PDAC) by integrating omics profiling of 150 patients for mutations, gene expression (mRNA, miRNA, and long non-coding RNA [lncRNA]), DNA methylation, and protein expression (Cancer Genome Atlas Research Network, 2017). KRAS mutational heterogeneity and signatures of individual pancreatic cancers have been identified, indicating the existence of distinct molecular subtypes of pancreatic cancer. For multi-omics clustering, the SNF method was applied to mRNA, miRNA, and DNA methylation data, and allowed to identify three clusters, which are mostly associated with tumor purity and gene expression signatures. This provides insights into the importance of considering neoplastic cellularity for further analysis of PDAC and the need for molecular characterization platforms to further stratify samples.
ADVANCES IN DRUG TARGET DISCOVERY USING CANCER MULTI-OMICS
Drug target discovery is a critical step in the development of cancer drugs and personalized therapeutics. In traditional drug target discovery, biomolecules with a confirmed mechanism of action are selected through a series of studies, which require enormous manpower (Lindsay, 2003; Paananen and Fortino, 2020). Over the last decade, putative drug targets have been identified through the latest high-throughput genomic approaches in combination with experimental validation, including overexpression or knockdown by RNAi and the use of transgenic animals and model organisms (Benson et al., 2006). Multi-omics is an interdisciplinary approach to study biological characteristics, and can comprehensively yield many drug target candidates in a cost-effective manner. The analysis of 14 cancer subtypes from TCGA multi-omics datasets revealed 40 driver genes associated with the Wnt, Notch, Hedgehog, JAK/STAT, NK-KB, and MAPK signaling pathways (Chen et al., 2014). Among them, well-known driver genes such as EGFR, ERBB2, PIK3CA, and KRAS were confirmed to be upregulated in several cancers, and DCUN1D1 and NSD3 were identified as new diver genes. Along with the success of trastuzumab (an agent targeting HER2), the use of multi-omics approaches for the discovery of new druggable targets in breast cancer has emerged. A recent proteomic analysis of 105 breast cancer patients has elucidated the association of this cancer type with CDK12, PAK1, PTK2, RIPK2, and TLK2 amplicons, and highlighted the overexpression of EGFR following the loss of CETN3 and SKP1 (Mertins et al., 2016). Progress has also been made with regard to tumor metabolites. Jain et al. (2012) detected consumption and release (CORE) profiles of 219 metabolites from NCI-60 cell lines. After the integrated analysis of CORE profiles with gene expression data, these authors demonstrated that glycine consumption and upregulation of the mitochondrial glycine biosynthetic pathway were highly correlated with the proliferation of cancer cells.
Multi-omics approaches may allow systematic assessment of drug discovery for personalized cancer therapy and improve the efficacy of chemotherapy (Aguirre et al., 2018; Li et al., 2013; Pauli et al., 2017). Refining molecular-defined subsets of patients can provide information on drug response and resistance, which vary among patients. Cui et al. (2020) integrated the expression of lncRNA, miRNA, mRNA, methylation, and the profile of somatic mutations with the expression of drug response-related lncRNAs. These authors found that lncRNAs respond to diverse chemotherapeutic drugs and characterized some key lncRNAs, such as HOXA-AS2, which mediate resistance to the drug adriamycin in BRCA patients (Cui et al., 2020). Another proteogenomic study of breast cancer found that triple-negative BRCA (TNBC) tumors with RB1 mutations or deletions are resistant to the CDK4/6 inhibitor palbociclib, unlike wild-type TNBC. However, most of the TNBC samples showed a small level of RB protein expression along with that of the wild-type RB1 gene. Based on previous findings, the Genomics of Drug Sensitivity in Cancer (GDSC) data analysis showed that the response to palbociclib was correlated with the total amount of RB protein, regardless of the RB1 genotype. An exception to this is that the I388S, P515L, and N480 (in-frame) mutations of the RB1 gene led to poor palbociclib response (Krug et al., 2020). Collectively, these studies indicate that multi-omics analysis can unravel new biological characteristics and enable to discover drug targets that cannot be pinpointed based on single-omics data.
CONCLUDING REMARKS
In this review, we introduce computational methods for multi-omics studies and report the latest findings in cancer research based on them. Multi-omics approaches can fully characterize the intersection between different layers of quantitative information, systematically summarizing biological interactions from an individual cell or tissue to an individual patient with a primary tumor and possible metastases. In addition, such integration can reflect the molecular characteristics of tumors at various levels, from genes to proteins, and different cancer stages through multidisciplinary analysis.
Multi-omics approaches may hold the potential to study different cancer types with a high level of similarity, in terms of molecular characteristics, to basal-like breast cancer, high-grade serous ovarian cancer, and serous endometrial cancer (Cancer Genome Atlas Research Network et al., 2013a). A systems approach integrating multi-omics data is key to understanding cancer biology and investigating the molecular pathogenesis of cancer. Multi-omics data analysis across tumor types can identify molecular characteristics commonly underlying a range of cancer types and further detail patient subgroups as well as the molecular classification of cancer subtypes.
Therefore, multiple data layers, including genomics, transcriptomics, epigenomics, and proteomics datasets, are required to fully represent the molecular and clinical structures of cancer patients. The generation of high-quality and unbiased datasets is a critical part of multi-omics approaches. In addition, further studies should consider proper integration methods and computational algorithms for robust and systematic assessment to obtain solid findings and predictive models.
ACKNOWLEDGMENTS
This work was supported by the Korean NRF Grant 2019M3E5D3073568 (to J.Y.A.) and a Korea University Grant.
Footnotes
AUTHOR CONTRIBUTIONS
Y.J.H. and J.Y.A. wrote the original draft. Y.J.H., C.H., G.H.L., J.M.P., and J.Y.A. reviewed and edited the manuscript. Y.J.H., C.H., and J.Y.A. provided a figure and table.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Aguirre A.J., Nowak J.A., Camarda N.D., Moffitt R.A., Ghazani A.A., Hazar-Rethinam M., Raghavan S., Kim J., Brais L.K., Ragon D., et al. Real-time genomic characterization of advanced pancreatic cancer to enable precision medicine. Cancer Discov. 2018;8:1096–1111. doi: 10.1158/2159-8290.CD-18-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amit I., Citri A., Shay T., Lu Y., Katz M., Zhang F., Tarcic G., Siwak D., Lahad J., Jacob-Hirsch J., et al. A module of negative feedback regulators defines growth factor signaling. Nat. Genet. 2007;39:503–512. doi: 10.1038/ng1987. [DOI] [PubMed] [Google Scholar]
- Basu A., Bodycombe N.E., Cheah J.H., Price E.V., Liu K., Schaefer G.I., Ebright R.Y., Stewart M.L., Ito D., Wang S., et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154:1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson J.D., Chen Y.N., Cornell-Kennon S.A., Dorsch M., Kim S., Leszczyniecka M., Sellers W.R., Lengauer C. Validating cancer drug targets. Nature. 2006;441:451–456. doi: 10.1038/nature04873. [DOI] [PubMed] [Google Scholar]
- Berns K., Bernards R. Understanding resistance to targeted cancer drugs through loss of function genetic screens. Drug Resist. Updat. 2012;15:268–275. doi: 10.1016/j.drup.2012.10.002. [DOI] [PubMed] [Google Scholar]
- Bodenmiller B., Zunder E.R., Finck R., Chen T.J., Savig E.S., Bruggner R.V., Simonds E.F., Bendall S.C., Sachs K., Krutzik P.O., et al. Multiplexed mass cytometry profiling of cellular states perturbed by small-molecule regulators. Nat. Biotechnol. 2012;30:858–867. doi: 10.1038/nbt.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordbar A., Mo M.L., Nakayasu E.S., Schrimpe-Rutledge A.C., Kim Y.M., Metz T.O., Jones M.B., Frank B.C., Smith R.D., Peterson S.N., et al. Model-driven multi-omic data analysis elucidates metabolic immunomodulators of macrophage activation. Mol. Syst. Biol. 2012;8:558. doi: 10.1038/msb.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozic I., Antal T., Ohtsuki H., Carter H., Kim D., Chen S., Karchin R., Kinzler K.W., Vogelstein B., Nowak M.A. Accumulation of driver and passenger mutations during tumor progression. Proc. Natl. Acad. Sci. U. S. A. 2010;107:18545–18550. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network, author. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012a;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network, author. Comprehensive molecular portraits of human breast tumours. Nature. 2012b;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, author. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, author. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, author. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–209. doi: 10.1038/nature13480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandoth C., Schultz N., Cherniack A.D., Akbani R., Liu Y., Shen H., Robertson A.G., Pashtan I., Shen R., et al. Cancer Genome Atlas Research Network. Integrated genomic characterization of endometrial carcinoma. Nature. 2013a;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T.J., Miller C., Ding L., Raphael B.J., Mungall A.J., Robertson A., Hoadley K., Triche T.J., Jr., Laird P.W., Jr., et al. Cancer Genome Atlas Research Network, author. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med. 2013b;368:2059–2074. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, author. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 2017;32:185–203.e13. doi: 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carro M.S., Lim W.K., Alvarez M.J., Bollo R.J., Zhao X., Snyder E.Y., Sulman E.P., Anne S.L., Doetsch F., Colman H., et al. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli F.M.G., Remke M., Rampasek L., Peacock J., Shih D.J.H., Luu B., Garzia L., Torchia J., Nor C., Morrissy A.S., et al. Intertumoral heterogeneity within medulloblastoma subgroups. Cancer Cell. 2017;31:737–754.e6. doi: 10.1016/j.ccell.2017.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvel C., Novoloaca A., Veyre P., Reynier F., Becker J. Evaluation of integrative clustering methods for the analysis of multi-omics data. Brief. Bioinform. 2020;21:541–552. doi: 10.1093/bib/bbz015. [DOI] [PubMed] [Google Scholar]
- Chen Y., McGee J., Chen X., Doman T.N., Gong X., Zhang Y., Hamm N., Ma X., Higgs R.E., Bhagwat S.V., et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS One. 2014;9:e98293. doi: 10.1371/journal.pone.0098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.J., Roumeliotis T.I., Chang Y.H., Chen C.T., Han C.L., Lin M.H., Chen H.W., Chang G.C., Chang Y.L., Wu C.T., et al. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–244.e17. doi: 10.1016/j.cell.2020.06.012. [DOI] [PubMed] [Google Scholar]
- Chin K., DeVries S., Fridlyand J., Spellman P.T., Roydasgupta R., Kuo W.L., Lapuk A., Neve R.M., Qian Z., Ryder T., et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csibi A., Fendt S.M., Li C., Poulogiannis G., Choo A.Y., Chapski D.J., Jeong S.M., Dempsey J.M., Parkhitko A., Morrison T., et al. The mTORC1 pathway stimulates glutamine metabolism and cell proliferation by repressing SIRT4. Cell. 2013;153:840–854. doi: 10.1016/j.cell.2013.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H., Kong H., Peng F., Wang C., Zhang D., Tian J., Zhang L. Inferences of individual drug response-related long non-coding RNAs based on integrating multi-omics data in breast cancer. Mol. Ther. Nucleic Acids. 2020;20:128–139. doi: 10.1016/j.omtn.2020.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C., Shah S.P., Chin S.F., Turashvili G., Rueda O.M., Dunning M.J., Speed D., Lynch A.G., Samarajiwa S., Yuan Y., et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker L.J.M., Kannegieter N.M., Haerkens F., Toth E., Kros J.M., Steenhoff Hov D.A., Fillebeen J., Verschuren L., Leenstra S., Ressa A., et al. Multiomics profiling of paired primary and recurrent glioblastoma patient tissues. Neurooncol. Adv. 2020;2:vdaa083. doi: 10.1093/noajnl/vdaa083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentles A.J., Gallahan D. Systems biology: confronting the complexity of cancer. Cancer Res. 2011;71:5961–5964. doi: 10.1158/0008-5472.CAN-11-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette M.A., Satpathy S., Cao S., Dhanasekaran S.M., Vasaikar S.V., Krug K., Petralia F., Li Y., Liang W.W., Reva B., et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell. 2020;182:200–225.e35. doi: 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum D., Colangelo C., Williams K., Gerstein M. Comparing protein abundance and mRNA expression levels on a genomic scale. Genome Biol. 2003;4:117. doi: 10.1186/gb-2003-4-9-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde P.S., White I.R., Debouck C. Interplay of transcriptomics and proteomics. Curr. Opin. Biotechnol. 2003;14:647–651. doi: 10.1016/j.copbio.2003.10.006. [DOI] [PubMed] [Google Scholar]
- Hennessy B.T., Lu Y., Gonzalez-Angulo A.M., Carey M.S., Myhre S., Ju Z., Davies M.A., Liu W., Coombes K., Meric-Bernstam F., et al. A technical assessment of the utility of reverse phase protein arrays for the study of the functional proteome in non-microdissected human breast cancers. Clin. Proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S.M., Lu Y., Molina J., Heiser L.M., Spellman P.T., Speed T.P., Gray J.W., Mills G.B., Mukherjee S. Bayesian inference of signaling network topology in a cancer cell line. Bioinformatics. 2012;28:2804–2810. doi: 10.1093/bioinformatics/bts514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Choi S., Kim R., Koh J. Mechanisms of macromolecular interactions mediated by protein intrinsic disorder. Mol. Cells. 2020;43:899–908. doi: 10.14348/molcells.2020.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S.S., Clarke D.C., Gosline S.J., Labadorf A., Chouinard C.R., Gordon W., Lauffenburger D.A., Fraenkel E. Linking proteomic and transcriptional data through the interactome and epigenome reveals a map of oncogene-induced signaling. PLoS Comput. Biol. 2013;9:e1002887. doi: 10.1371/journal.pcbi.1002887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z., Zhan X., Xiang S., Johnson T.S., Helm B., Yu C.Y., Zhang J., Salama P., Rizkalla M., Han Z., et al. SALMON: Survival Analysis Learning With Multi-Omics Neural Networks on breast cancer. Front. Genet. 2019;10:166. doi: 10.3389/fgene.2019.00166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iadevaia S., Lu Y., Morales F.C., Mills G.B., Ram P.T. Identification of optimal drug combinations targeting cellular networks: integrating phospho-proteomics and computational network analysis. Cancer Res. 2010;70:6704–6714. doi: 10.1158/0008-5472.CAN-10-0460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M., Nilsson R., Sharma S., Madhusudhan N., Kitami T., Souza A.L., Kafri R., Kirschner M.W., Clish C.B., Mootha V.K. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science. 2012;336:1040–1044. doi: 10.1126/science.1218595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirk P., Griffin J.E., Savage R.S., Ghahramani Z., Wild D.L. Bayesian correlated clustering to integrate multiple datasets. Bioinformatics. 2012;28:3290–3297. doi: 10.1093/bioinformatics/bts595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh H.W.L., Fermin D., Vogel C., Choi K.P., Ewing R.M., Choi H. iOmicsPASS: network-based integration of multiomics data for predictive subnetwork discovery. NPJ Syst. Biol. Appl. 2019;5:22. doi: 10.1038/s41540-019-0099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komurov K., Tseng J.T., Muller M., Seviour E.G., Moss T.J., Yang L., Nagrath D., Ram P.T. The glucose-deprivation network counteracts lapatinib-induced toxicity in resistant ErbB2-positive breast cancer cells. Mol. Syst. Biol. 2012;8:596. doi: 10.1038/msb.2012.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krug K., Jaehnig E.J., Satpathy S., Blumenberg L., Karpova A., Anurag M., Miles G., Mertins P., Geffen Y., Tang L.C., et al. Proteogenomic landscape of breast cancer tumorigenesis and targeted therapy. Cell. 2020;183:1436–1456.e31. doi: 10.1016/j.cell.2020.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Tsugawa H., Wohlgemuth G., Mehta S., Mueller M., Zheng Y., Ogiwara A., Meissen J., Showalter M., Takeuchi K., et al. Identifying metabolites by integrating metabolome databases with mass spectrometry cheminformatics. Nat. Methods. 2018;15:53–56. doi: 10.1038/nmeth.4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P., Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kim J., Park J.E. Single-cell toolkits opening a new era for cell engineering. Mol. Cells. 2021;44:127–135. doi: 10.14348/molcells.2021.0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Kung H.J., Mack P.C., Gandara D.R. Genotyping and genomic profiling of non-small-cell lung cancer: implications for current and future therapies. J. Clin. Oncol. 2013;31:1039–1049. doi: 10.1200/JCO.2012.45.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M.A. Target discovery. Nat. Rev. Drug Discov. 2003;2:831–838. doi: 10.1038/s41573-019-0046-z. [DOI] [PubMed] [Google Scholar]
- Lock E.F., Dunson D.B. Bayesian consensus clustering. Bioinformatics. 2013;29:2610–2616. doi: 10.1093/bioinformatics/btt425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx V. A dream of single-cell proteomics. Nat. Methods. 2019;16:809–812. doi: 10.1038/s41592-019-0540-6. [DOI] [PubMed] [Google Scholar]
- Mertins P., Mani D.R., Ruggles K.V., Gillette M.A., Clauser K.R., Wang P., Wang X., Qiao J.W., Cao S., Petralia F., et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. 2016;534:55–62. doi: 10.1038/nature18003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Q., Wang S., Seshan V.E., Olshen A.B., Schultz N., Sander C., Powers R.S., Ladanyi M., Shen R. Pattern discovery and cancer gene identification in integrated cancer genomic data. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4245–4250. doi: 10.1073/pnas.1208949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun D.G., Bhin J., Kim S., Kim H., Jung J.H., Jung Y., Jang Y.E., Park J.M., Kim H., Jung Y., et al. Proteogenomic characterization of human early-onset gastric cancer. Cancer Cell. 2019;35:111–124.e10. doi: 10.1016/j.ccell.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Neve R.M., Chin K., Fridlyand J., Yeh J., Baehner F.L., Fevr T., Clark L., Bayani N., Coppe J.P., Tong F., et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–527. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H., Shrestha S., Draghici S., Nguyen T. PINSPlus: a tool for tumor subtype discovery in integrated genomic data. Bioinformatics. 2019;35:2843–2846. doi: 10.1093/bioinformatics/bty1049. [DOI] [PubMed] [Google Scholar]
- Nguyen N.D., Wang D. Multiview learning for understanding functional multiomics. PLoS Comput. Biol. 2020;16:e1007677. doi: 10.1371/journal.pcbi.1007677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paananen J., Fortino V. An omics perspective on drug target discovery platforms. Brief. Bioinform. 2020;21:1937–1953. doi: 10.1093/bib/bbz122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer A., Phapale P., Chernyavsky I., Lavigne R., Fay D., Tarasov A., Kovalev V., Fuchser J., Nikolenko S., Pineau C., et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat. Methods. 2017;14:57–60. doi: 10.1038/nmeth.4072. [DOI] [PubMed] [Google Scholar]
- Pascal J., Bearer E.L., Wang Z., Koay E.J., Curley S.A., Cristini V. Mechanistic patient-specific predictive correlation of tumor drug response with microenvironment and perfusion measurements. Proc. Natl. Acad. Sci. U. S. A. 2013;110:14266–14271. doi: 10.1073/pnas.1300619110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli C., Hopkins B.D., Prandi D., Shaw R., Fedrizzi T., Sboner A., Sailer V., Augello M., Puca L., Rosati R., et al. Personalized in vitro and in vivo cancer models to guide precision medicine. Cancer Discov. 2017;7:462–477. doi: 10.1158/2159-8290.CD-16-1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.R., Bruno P.M., Gilbert L.A., Capron K.L., Lauffenburger D.A., Hemann M.T. Defining principles of combination drug mechanisms of action. Proc. Natl. Acad. Sci. U. S. A. 2013;110:E170–E179. doi: 10.1073/pnas.1210419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu P., Simonds E.F., Bendall S.C., Gibbs K.D., Jr., Bruggner R.V., Jr., Linderman M.D., Jr., Sachs K., Jr., Nolan G.P., Jr., Plevritis S.K., Jr. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat. Biotechnol. 2011;29:886–891. doi: 10.1038/nbt.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G., Won H., Gandal M.J., Haney J., Wang J.C., Wong C.C.Y., Sun W., Prabhakar S., Mill J., Geschwind D.H. Integrative genomics identifies a convergent molecular subtype that links epigenomic with transcriptomic differences in autism. Nat. Commun. 2020;11:4873. doi: 10.1038/s41467-020-18526-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport N., Safra R., Shamir R. MONET: multi-omic module discovery by omic selection. PLoS Comput. Biol. 2020;16:e1008182. doi: 10.1371/journal.pcbi.1008182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappoport N., Shamir R. NEMO: cancer subtyping by integration of partial multi-omic data. Bioinformatics. 2019;35:3348–3356. doi: 10.1093/bioinformatics/btz058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert O.T., Rost H.L., Collins B.C., Rosenberger G., Aebersold R. Quantitative proteomics: challenges and opportunities in basic and applied research. Nat. Protoc. 2017;12:1289–1294. doi: 10.1038/nprot.2017.040. [DOI] [PubMed] [Google Scholar]
- Shen R., Olshen A.B., Ladanyi M. Integrative clustering of multiple genomic data types using a joint latent variable model with application to breast and lung cancer subtype analysis. Bioinformatics. 2009;25:2906–2912. doi: 10.1093/bioinformatics/btp543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., 3rd, Stoeckius M., 3rd, Smibert P., 3rd, Satija R., 3rd Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Satija R. Integrative single-cell analysis. Nat. Rev. Genet. 2019;20:257–272. doi: 10.1038/s41576-019-0093-7. [DOI] [PubMed] [Google Scholar]
- Sumazin P., Yang X., Chiu H.S., Chung W.J., Iyer A., Llobet-Navas D., Rajbhandari P., Bansal M., Guarnieri P., Silva J., et al. An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell. 2011;147:370–381. doi: 10.1016/j.cell.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K.R., Rockne R.C., Claridge J., Chaplain M.A., Alvord E.C., Jr., Anderson A.R., Jr. Quantifying the role of angiogenesis in malignant progression of gliomas: in silico modeling integrates imaging and histology. Cancer Res. 2011;71:7366–7375. doi: 10.1158/0008-5472.CAN-11-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., Simonovic M., Doncheva N.T., Morris J.H., Bork P., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tentner A.R., Lee M.J., Ostheimer G.J., Samson L.D., Lauffenburger D.A., Yaffe M.B. Combined experimental and computational analysis of DNA damage signaling reveals context-dependent roles for Erk in apoptosis and G1/S arrest after genotoxic stress. Mol. Syst. Biol. 2012;8:568. doi: 10.1038/msb.2012.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teves J.M., Won K.J. Mapping cellular coordinates through advances in spatial transcriptomics technology. Mol. Cells. 2020;43:591–599. doi: 10.14348/molcells.2020.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyers M., Mann M. From genomics to proteomics. Nature. 2003;422:193–197. doi: 10.1038/nature01510. [DOI] [PubMed] [Google Scholar]
- Vaske C.J., Benz S.C., Sanborn J.Z., Earl D., Szeto C., Zhu J., Haussler D., Stuart J.M. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–i245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidova V., Spacil Z. A review on mass spectrometry-based quantitative proteomics: targeted and data independent acquisition. Anal. Chim. Acta. 2017;964:7–23. doi: 10.1016/j.aca.2017.01.059. [DOI] [PubMed] [Google Scholar]
- Wang B., Mezlini A.M., Demir F., Fiume M., Tu Z., Brudno M., Haibe-Kains B., Goldenberg A. Similarity network fusion for aggregating data types on a genomic scale. Nat. Methods. 2014;11:333–337. doi: 10.1038/nmeth.2810. [DOI] [PubMed] [Google Scholar]
- Whitehurst A.W., Bodemann B.O., Cardenas J., Ferguson D., Girard L., Peyton M., Minna J.D., Michnoff C., Hao W., Roth M.G., et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815–819. doi: 10.1038/nature05697. [DOI] [PubMed] [Google Scholar]
- Witten D.M., Tibshirani R.J. Extensions of sparse canonical correlation analysis with applications to genomic data. Stat. Appl. Genet. Mol. Biol. 2009;8:Article28. doi: 10.2202/1544-6115.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., Wang D., Zhang M.Q., Gu J. Fast dimension reduction and integrative clustering of multi-omics data using low-rank approximation: application to cancer molecular classification. BMC Genomics. 2015;16:1022. doi: 10.1186/s12864-015-2223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Michailidis G. A non-negative matrix factorization method for detecting modules in heterogeneous omics multi-modal data. Bioinformatics. 2016;32:1–8. doi: 10.1093/bioinformatics/btv544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y., Savage R.S., Markowetz F. Patient-specific data fusion defines prognostic cancer subtypes. PLoS Comput. Biol. 2011;7:e1002227. doi: 10.1371/journal.pcbi.1002227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Liu T., Zhang Z., Payne S.H., Zhang B., McDermott J.E., Zhou J.Y., Petyuk V.A., Chen L., Ray D., et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. 2016;166:755–765. doi: 10.1016/j.cell.2016.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]