Abstract
GPR43 (also known as FFAR2 or FFA2) is a G-protein-coupled receptor primarily expressed in immune cells, enteroendocrine cells and adipocytes that recognizes short-chain fatty acids, such as acetate, propionate, and butyrate, likely to be implicated in innate immunity and host energy homeostasis. Activated GPR43 suppresses the cAMP level and induces Ca2+ flux via coupling to Gαi and Gαq families, respectively. Additionally, GPR43 is reported to facilitate phosphorylation of ERK through G-protein-dependent pathways and interacts with β-arrestin 2 to inhibit NF-κB signaling. However, other G-protein-dependent and independent signaling pathways involving GPR43 remain to be established. Here, we have demonstrated that GPR43 augments Rho GTPase signaling. Acetate and a synthetic agonist effectively activated RhoA and stabilized YAP/TAZ transcriptional coactivators through interactions of GPR43 with Gαq/11 and Gα12/13. Acetate-induced nuclear accumulation of YAP was blocked by a GPR43-specific inverse agonist. The target genes induced by YAP/TAZ were further regulated by GPR43. Moreover, in THP-1–derived M1-like macrophage cells, the Rho-YAP/TAZ pathway was activated by acetate and a synthetic agonist. Our collective findings suggest that GPR43 acts as a mediator of the Rho-YAP/TAZ pathway.
Keywords: GPR43, RhoA, short-chain fatty acid, TAZ, YAP
INTRODUCTION
The G-protein coupled-receptor (GPCR) GPR43 (also known as FFAR2) is primarily expressed on innate immune cells, such as neutrophils and macrophages (Le Poul et al., 2003; Nilsson et al., 2003). From an immunological perspective, GPR43 is implicated in neutrophil chemotaxis towards its ligands and secretion of inflammatory cytokines (Tedelind et al., 2007; Vinolo et al., 2011). In addition, GPR43 regulates gut hormone secretion in the intestine (Karaki et al., 2006), insulin secretion in pancreatic β cells (Priyadarshini et al., 2015), and lipid metabolism in white adipocytes (Kimura et al., 2013). A recent report showed that GPR43 and GPR109A mediate epithelial protective effects of dietary fiber through inflammasome activation (Macia et al., 2015). Short-chain fatty acids (SCFA), aliphatic compounds with fewer than six carbon atoms derived from gut microbiota fermentation of indigestible carbohydrates, are reported to activate GPR41 and GPR43 receptors (Brown et al., 2003). GPR43 activation by SCFAs triggers inhibition of adenylyl cyclase to reduce cAMP and elevation of Ca2+ levels through Gαi and Gαq families of heterotrimeric G proteins, respectively (Brown et al., 2003; Le Poul et al., 2003). Furthermore, activated GPR43 transmits signals to ERK in a G-protein-dependent manner (Ang et al., 2016) and interacts with β-arrestin 2 to suppress nuclear translocation of NF-κB through a G-protein independent pathway (Lee et al., 2013).
Yes-associated protein (YAP) and its paralog, TAZ, are transcriptional co-activators pivotal in the Hippo signaling pathway (Dong et al., 2007; Lei et al., 2008; Moon and Kim, 2018). Modulation of YAP/TAZ is affected in response to various factors, such as cell-to-cell contact, mechanical tension, metabolism, hypoxia, and ER stress (Dupont et al., 2011; Kim et al., 2011; Ma et al., 2015; Wu et al., 2015). Among the core components of the Hippo pathway, LATS1/2 kinases directly inhibit stabilization of YAP/TAZ via phosphorylation, resulting in cytoplasmic retention by 14-3-3 binding or proteasomal degradation (Hao et al., 2008). Dephosphorylated and stabilized YAP/TAZ translocate to the nucleus and mainly interact with the transcriptional-enhanced associated domain (TEAD) family of transcription factors. The YAP/TAZ-TEAD complex stimulates expression of target genes, such as CTGF and CYR61, in turn, promoting cell proliferation and survival. Thus, YAP/TAZ are known oncogenes implicated in tumor formation (Zhao et al., 2008). Consistently, YAP expression is increased under conditions of tissue damage and shown to be critical in repairing mucosal injury (Taniguchi et al., 2015).
In 2012, Fa-Xing and colleagues reported that both YAP and TAZ are regulated by GPCR signaling (Yu et al., 2012). Their study showed that interactions of lysophosphatidic acid (LPA) and S1P receptors with serum-borne ligands activated RhoA to inhibit LATS1/2, resulting in stimulation of YAP/TAZ activity. Stimulation of protease-activated receptor 1 (PAR1) by thrombin and TRAP6 additionally promoted nuclear translocation of YAP/TAZ via dephosphorylation dependent on the Gα12/13-RhoA pathway (Mo et al., 2012). Furthermore, GPCR coupling to not only Gα12/13, but also Gαq and, to lesser extent, Gαi, regulated YAP/TAZ activity. Another study demonstrated that the β2-adrenergic receptor promotes β-arrestin 2 translocation to the plasma membrane, leading to stimulation of RhoA (Ma et al., 2012). In contrast, GPCR coupling to Gαs activated LATS1/2 kinases to phosphorylate and inhibit YAP/TAZ (Yu et al., 2012). The data collectively suggest that GPCRs capable of modulating RhoA are likely to influence YAP/TAZ activity.
In the current study, we showed induction of Rho GTPase signaling by acetate and a GPR43 specific synthetic agonist, and then YAP/TAZ accumulated in the nucleus in GPR43-expressing HEK293 cells in a manner dependent on Gα12/13 and Gαq/11. Additionally, the genes known to be regulated by YAP/TAZ were specifically altered by GPR43 in stable HEK293 and M1-polarized THP-1 cells. Our findings indicate that GPR43 augments the RhoA signaling pathway to activate YAP/TAZ.
MATERIALS AND METHODS
Reagents and plasmids
The GPR43-selective agonist compound 187 (4-[(2R,6S)-2, 6-dimethylmorpholin-4-yl]-7-(2-fluorobenzenesulfonyl)-2- methyl-5H-pyrrolo[3,2-d]pyrimidin-6-amine), which was invented by Takeda Cambridge Ltd. (https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2015198045), was kindly provided by Dr. Y.S. Kwak (Korea University). The selective inverse agonist (S)-3-(2-(3-chlorophenyl)acetamido)-4-(4-(trifluoromethyl)phenyl)butanoic acid (CATPB), sodium acetate, phorbol 12-myristate 13-acetate (PMA), lipopolysaccharide (LPS), and pertussis toxin (PTX) were purchased from Sigma-Aldrich (USA). Cumate solution (10,000×; PBQM100A-1) was acquired from System Biosciences (USA), YM-254890 from Adipogen (USA) and C3 toxin (C3 transferase from Clostridium botulinum, #CT04) from Cytoskeleton (USA).
The GPR43-myc construct was cloned into PiggyBac cumate switch-inducible vector (PB-CuO, PBQM-812-A1) using the restriction sites NheI and NotI (New England BioLabs, USA).
Cell culture and THP-1 polarization
HEK293 cells expressing GPR43 induced by cumate (GPR43-HEK293) were cultured in Dulbecco’s modified Eagle’s medium (DMEM; WelGene, Korea) supplemented with 10% fetal bovine serum (FBS, #16000-044; Thermo Fisher Scientific, USA), 1× GlutaMAX, 1% Pen-Strep, and 2 μg/ml puromycin. To induce GPR43 expression, GPR43-HEK293 cells were incubated with 1× cumate-containing (30 μg/ml cumate) medium at least for 48 h. The human acute monocytic leukemia cell line, THP-1 (ATCC TIB-202; ATCC, USA), was cultured in ATCC-modified RPMI 1640 (Life Technologies, USA) supplemented with 10% FBS, 1% Pen-Strep, and 50 μM β-mercaptoethanol (Sigma-Aldrich). Both cell lines were maintained in a humidified atmosphere of 5% CO2 at 37°C.
To induce polarization of THP-1, 1.0 × 106 cells/ml suspended cells were stimulated with 20 ng/ml PMA for 24 h to establish Mφ macrophage-like cells. Adherent Mφ-like cells were washed once with fresh RPMI 1640 (WelGene) to eliminate residual traces of PMA. To generate M1-like macrophages, adherent cells were exposed to 100 ng/ml LPS and 20 ng/ml interferon-γ (IFN-γ; PeproTech, USA) for 48 h. To generate M2-like macrophages, cells were exposed to 20 ng/ml interleukin-4 (IL-4; PeproTech) and 20 ng/ml interleukin-13 (IL-13; PeproTech) for a further 48 h, followed by incubation in fresh medium for 12 h that was replaced with RPMI 1640 including 2 mM glucose and 0.5% FBS for an additional 12 h.
Luciferase assay
The serum response factor-response element (SRF-RE) luciferase reporter plasmid was purchased from Promega (USA). The reporter plasmid was transiently co-transfected with CMV promoter-renilla luciferase (pRL-CMV) into HEK293 cells using FuGene 6 (Promega), followed by incubation in 5% CO2 at 37°C for 24 h. On day 1, cells were treated with 1× cumate for an additional 24 h. On day 2, cells were replaced with serum-starved medium including 0.1% FBS and cumate overnight. Prior to stimulation, cells were deprived of serum and glucose for 1 h. Reporter activities were measured utilizing the Dual-Luciferase Reporter Assay System (Promega).
RNA interference
Specific siRNAs against GNAQ (L-008562-00), GNA11 (L-010860-00), GNA12 (L-008435-00), and GNA13 (L-009948-00) were obtained from Dharmacon (USA) and siRNA against β-arrestin 2 (No. 409) from Bioneer (Korea). Cells were transfected with the indicated siRNAs using RNAiMAX transfection reagent (Invitrogen, USA) according to the manufacturer’s protocol and incubated for 72 h.
Antibodies and immunoblotting
Antibodies against YAP, phospho-YAP Ser127, TAZ, LATS1, phospho-LATS1 Thr1079, ERK1/2, and phospho-ERK1/2 were purchased from Cell Signaling Technology (USA). Anti-β-actin was obtained from Abfrontier (Korea). To detect the active form of RhoA, the Active Rho Detection Kit (#8820S) was utilized (Cell Signaling Technology).
Immunoblotting was performed as follows: cells were harvested with lysis buffer (0.1% SDS, 50 mM Tris-HCl pH7.5, 150 mM NaCl, 1% NP-40, 0.25% sodium deoxycholate) containing 1× protease inhibitor cocktail (P3100-001; GenDEPOT, USA) and 1× phosphatase inhibitor cocktail (P3200-001; GenDEPOT). Lysates were centrifuged at 18,000g for 20 min prior to boiling with Laemmli SDS sample buffer for 10 min, subjected to SDS-PAGE and transferred to PVDF membranes, which were subsequently blocked for 30 min in 2% bovine serum albumin (BSA) in Tris-buffered saline containing 0.1% Tween 20 and probed with the relevant primary antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibody for detection of protein bands.
Immunocytochemistry
Cells were cultured in 8-well μ-slides (#80826; Ibidi, Germany) prior to treatment with 1× cumate. After stimulation, samples were washed with cold phosphate-buffered saline (PBS) and fixed in 4% paraformaldehyde for 20 min. Cells were permeabilized with 0.5% Triton X-100 in PBS for 5 min after quenching of residual paraformaldehyde with three drops of 1 M glycine, pH 6.8. Samples were incubated with blocking solution (2% BSA, 1% normal goat serum in 0.1% Triton X-100 in PBS) for 20 min and probed with primary antibodies at 37°C for 1 h, followed by secondary Alexa Fluor-488 conjugated IgG (Life Technologies) for 45 min at room temperature away from light. Cells were mounted using Fluoroshield Mounting Medium with DAPI (Abcam, UK) and subjected to laser-scanning LSM 880 confocal microscopy (Zeiss, Germany).
Real-time polymerase chain reaction
Total RNA was isolated using the RNeasy Plus mini kit (Qiagen, Germany) according to manufacturer’s instructions and cDNA synthesized using the RevertAid H Minus first-strand cDNA synthesis kit (Thermo Fisher Scientific). Real-time polymerase chain reaction (PCR) was performed using SolgTM 2× Real-Time PCR Smart mix (SolGent, Korea) and the indicated primers: β-actin, forward 5’-CATGTACGTTGCTATCCAGGC-3’, reverse 5’-CTCCTTAATGTCACGCACGAT-3’, as well as primer pairs corresponding to FFAR2, CTGF, CYR61, YAP, TAZ, IL-1β, IL-6, TNF, CD163, IL-10, CCL22, and TP53 obtained from Bioneer. To analyze relative gene expression, cycle thresholds (Ct) were normalized to that of β-actin as the reference gene and expressed as fold change using the 2-ΔΔCt method (Schmittgen and Livak, 2008).
RNA sequencing
The RNA sequencing library was prepared using TruSeq RNA Sample Prep Kit (Illumina, USA) and the sequencing was performed using Illumina HiSeq2000 platform to generate 100 bp paired-end reads. The reference genome of human was obtained from the NCBI genome (https://www.ncbi.nlm.nih.gov/genome), and genome indexing was performed using STAR (v.2.5.1) (Dobin et al., 2013). The sequenced reads were mapped to the human genome (hg19) STAR, and the gene expression levels were quantified with count module in the STAR. The edgeR (v.3.12.1) (McCarthy et al., 2012) package was used to select differentially expressed genes from the RNA-seq count data between conditions (fold change > 1.5, false discovery rate [FDR] < 0.05). Meanwhile, the TMM (the trimmed mean of M-values normalization) normalized CPM (counts pre million) value of each gene was added to 1, and log2-transformed for further analysis. The heapmap was generated using Microsoft Excel 2016 program (Microsoft, USA).
Data access
NGS (next-generation sequencing) data was deposited in the NCBI Gene Expression Omnibus under accession number GSE171873. The raw sequence tags were deposited in the NCBI Short Read Archive (SRA) under accession number SRP314432.
Statistical analysis
Results were obtained from at least two or three independent experiments and presented as means ± SEM. Statistical significance was determined using Student’s t-test and data were considered significant at P values < 0.05.
RESULTS
RhoA activation is induced by GPR43
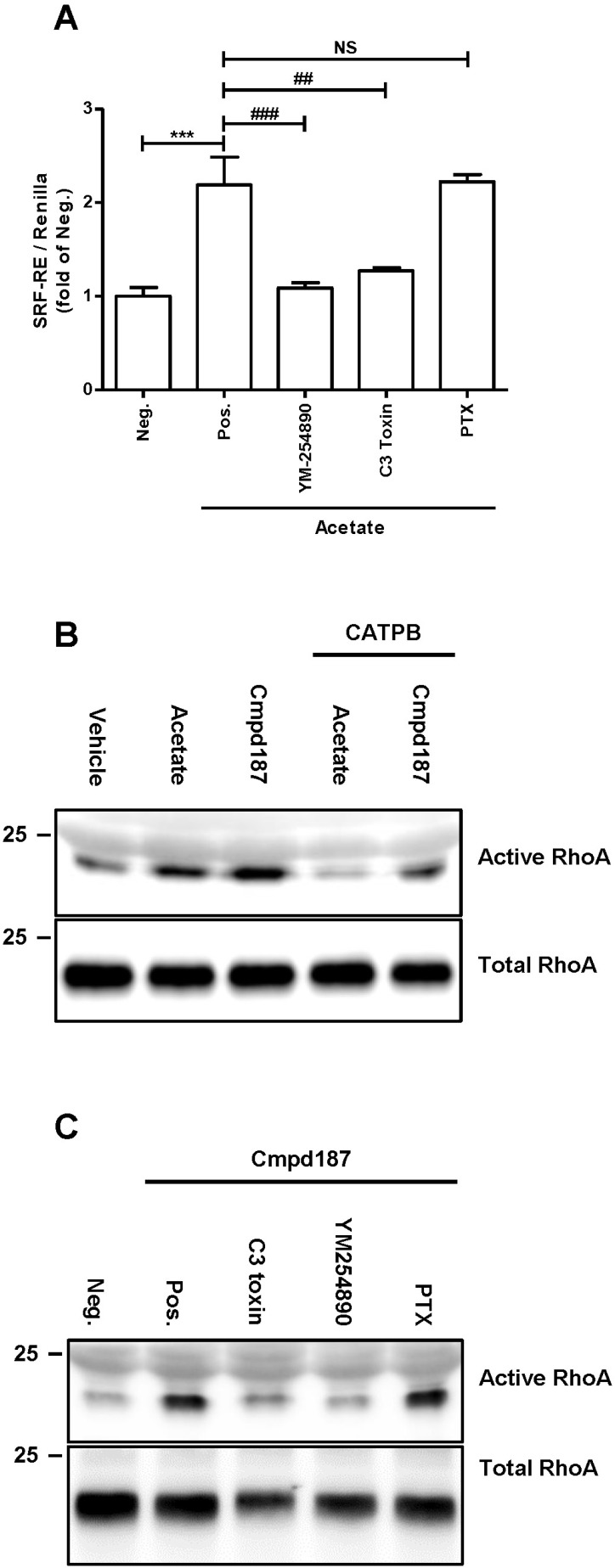
Binding of ligands to GPR43 stimulates activation of Gαi and Gαq to initiate downstream signaling pathways, followed by interactions with β-arrestin 2, resulting in desensitization on the membrane and inhibition of NF-κB signaling (Lee et al., 2013). To ascertain whether activated GPR43 stimulates RhoA activity, we performed the SRF-RE dual luciferase assay capable of verifying correlations of GPCRs with RhoA (Cheng et al., 2010). Augmentation of luminescence with acetate treatment was significantly suppressed by YM-254890 and botulinum C3 toxin, which are Gαq family and Rho GTPase inhibitors, respectively, but not PTX, a Gαi family inhibitor (Fig. 1A). SCFAs serve as ligands for both GPR43 and GPR41 and inhibit histone deacetylase activity (Waldecker et al., 2008). To exclude the possibility that acetate exerts off-target effects in the SRF-RE assay, we employed compound 187, a specific synthetic agonist for GPR43. Both acetate and compound 187 induced an increase in active RhoA (Fig. 1B), with compound 187 displaying more efficacious activity than acetate. Conversely, CATPB, an inverse agonist for GPR43, reduced the ligand-induced activation of RhoA (Fig. 1B). In addition, active RhoA induced by compound 187 was reduced by C3 toxin and YM-254890, but not PTX (Fig. 1C), further supporting the theory that GPR43 is a potential activator of RhoA signaling.
Fig. 1. RhoA activation is induced by GPR43.
(A) Human GPR43/HEK293 cells were treated with acetate and the indicated reagents for 6 h to evaluate SRF-RE activity. Data from at least two independent experiments are presented as mean ± SEM. The P values were calculated by Student’s t-test. ## P < 0.01, ### P < 0.005, ***P < 0.005. Neg., negative; Pos., positive. (B) GPR43/HEK293 cells were stimulated with 10 mM acetate, 10 μM agonist and 10 μM CATPB for 5 min. (C) HEK293 cells were pre-treated with 0.5 μg/ml C3 toxin and 0.2 μg/ml PTX for 16 h. The next day, cells were pretreated with YM-254980 for 15 min and stimulated with agonist for 5 min, followed by immunoblotting with anti-RhoA.
YAP/TAZ is dephosphorylated and activated by GPR43 via RhoA
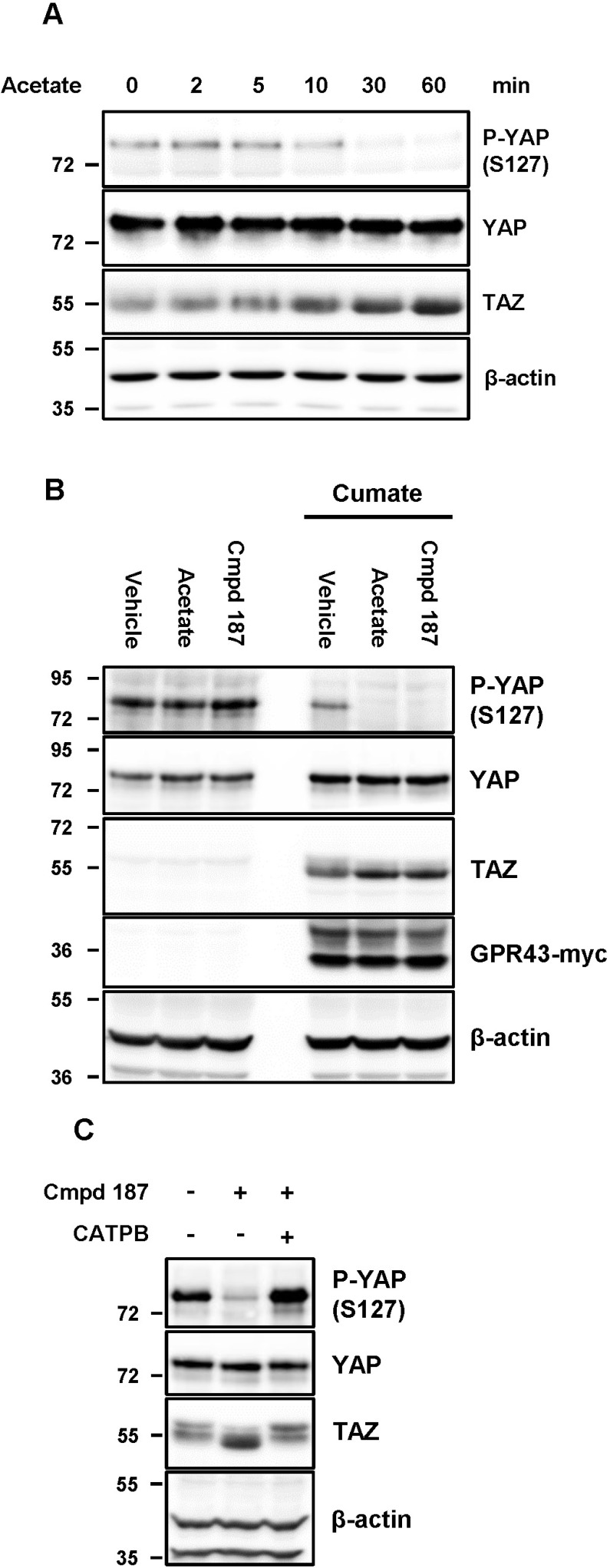
In view of the above finding that GPR43 significantly influences RhoA activity (Fig. 1), we examined the effect of GPR43 on YAP/TAZ, one of the downstream effectors of RhoA. Activated GPR43 by SCFAs such as acetate, propionate, and butyrate, significantly reduced phosphorylation of YAP at Ser127 in a time-dependent manner and increased the amount of TAZ protein with faster migration on SDS-PAGE (Fig. 2A, Supplementary Fig. S1). We identified that the stability of YAP/TAZ is specifically dependent on the GPR43 expression and activity (Figs. 2B and 2C).
Fig. 2. YAP and TAZ are stabilized and activated by GPR43.
(A) GPR43/HEK293 cells were stimulated with 10 mM acetate for the indicated time-points. (B) Cells were incubated with or without 1× cumate for 48 h. And then, the cells were subjected to serum-deprivation for 16 h prior to being stimulated with the indicated reagents for 1 h. (C) GPR43/HEK293 cells were stimulated with Cmpd 187 and CATPB for 1 h.
Phosphorylation of YAP and TAZ serves as an important step for nuclear export and dephosphorylation, along with other modifications, may be required for nuclear import, although the precise mechanisms remain elusive (Shreberk-Shaked and Oren, 2019). Since translocation to the nucleus is a prerequisite for the transcriptional activities of YAP and TAZ, we examined the cellular localization of these proteins following treatment with GPR43 modulators. YAP and TAZ are usually distributed throughout the cytoplasm and nucleus at high cell density (Figs. 3A [upper panels] and 3B). Empty spaces between the cells had been occupied by the cells and they were washed away during the course of immunocytochemistry. Even at nearly 100% confluence, nuclear localization of YAP and TAZ was markedly increased upon acetate treatment in GPR43-expressing HEK293 cells (Figs. 3A [middle panels] and 3B), which was completely abrogated by the GPR43 antagonist CATPB (Figs. 3A [bottom panels] and 3B).
Fig. 3. Active GPR43 induces translocation to the nucleus.
Cells were treated with agonist for 1 h. Cells were treated with DMSO, acetate and CATPB before fixation. (A) YAP and TAZ localization in cells were determined via immunocytochemical staining for endogenous YAP/TAZ (green) or DAPI (blue) for nuclei. Scale bars = 20 μm. (B) Quantification of the number of YAP/TAZ in nucleus.
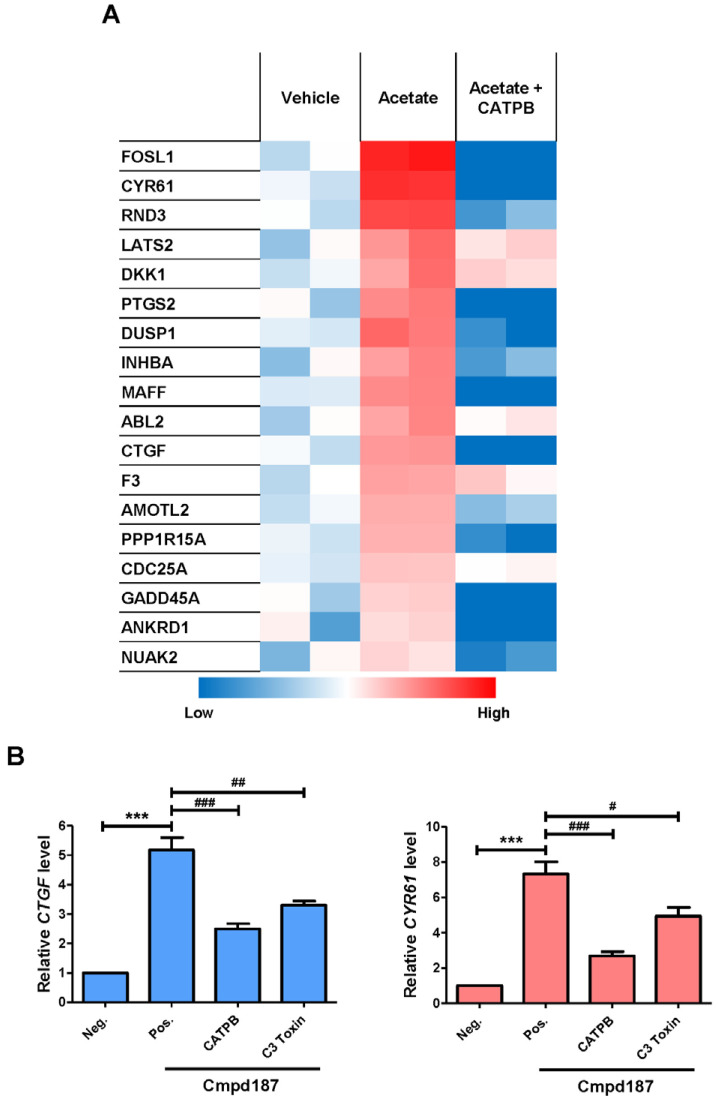
Next, changes in transcription induced by activation of GPR43 were examined. To analyze the transcriptome of GPR43-expressing HEK293 cells, RNA-seq experiments were conducted, as described in Materials and Methods. Among the differentially expressed genes, target genes of YAP/TAZ transcription factors were induced by acetate and downregulated by CATPB (Fig. 4A, Table 1). Expression of two of the genes, CTGF and CYR61, was re-confirmed via qRT-PCR by treating with compound 187 instead of acetate (Fig. 4B). Compound 187-induced expression of these genes was reduced following treating with CATPB and C3 toxin (Fig. 4B).
Fig. 4. YAP/TAZ target genes are directed by GPR43.
(A) Established target genes of YAP/TAZ from RNAseq data. (B) GPR43-expressing cells were stimulated with agonist and CATPB for 90 min and pre-treatment with C3 toxin was conducted for 16 h. Data from three independent experiments are presented as mean ± SEM (Student’s t-test). ***P < 0.005, # P < 0.05, ## P < 0.01, ### P < 0.005. Neg., negative; Pos., positive.
Table 1.
List of genes influenced by YAP/TAZ
| Name | Description | Reference |
|---|---|---|
| FOSL1 | FOS-like 1, AP-1 transcription factor subunit | (Zanconato et al., 2015) |
| CYR61 | Cysteine-rich angiogenic inducer 61, CCN family member 1 (CCN1) |
(Zhao et al., 2008), GSEAa |
| RND3 | Rho family GTPase 3 | (Ito et al., 2016) |
| LATS2 | Large tumor suppressor kinase 2 |
(Moroishi et al., 2015; Wang et al., 2018) |
| DKK1 | Dickkopf WNT signaling pathway inhibitor 1 |
(Park et al., 2015) |
| PTGS2 | Prostaglandin-endoperoxide synthase 2 | (Corley et al., 2018) |
| DUSP1 | Dual specificity phosphatase 1 | GSEAa |
| INHBA | Inhibin subunit β A | (Mo et al., 2012) |
| MAFF | MAF BZIP transcription factor F | (Ito et al., 2016) |
| ABL2 | ABL proto-oncogene 2, Non-receptor tyrosine kinase |
(Hoj et al., 2019) |
| CTGF | Connective tissue growth factor CCN family member 2 (CCN2) |
(Zhao et al., 2008), GSEAa |
| F3 | Coagulation Factor III | (Wang et al., 2018) |
| AMOTL2 | Angiomotin like 2 | (Wang et al., 2018; Zanconato et al., 2015), GSEAa |
| PPP1R15A | Protein phosphatase 1 regulatory subunit 15A |
(Ito et al., 2016) |
| CDC25A | Cell division cycle 25A | (Zanconato et al., 2015) |
| GADD45A | Growth arrest and DNA damage inducible α |
(Wang et al., 2018) |
| ANKRD1 | Ankyrin repeat domain 1 | (Yu et al., 2012), GSEAa |
| NUAK2 | NUAK family kinase 2, Omphalocele kinase 2 |
(Wang et al., 2018) |
GSEA, gene set enrichment analysis.
GSEA – CORDENONSI_YAP_CONSERVED_SIGNATURE.
YAP/TAZ is stabilized and activated through GPR43-Gαq/12/13-RhoA axis
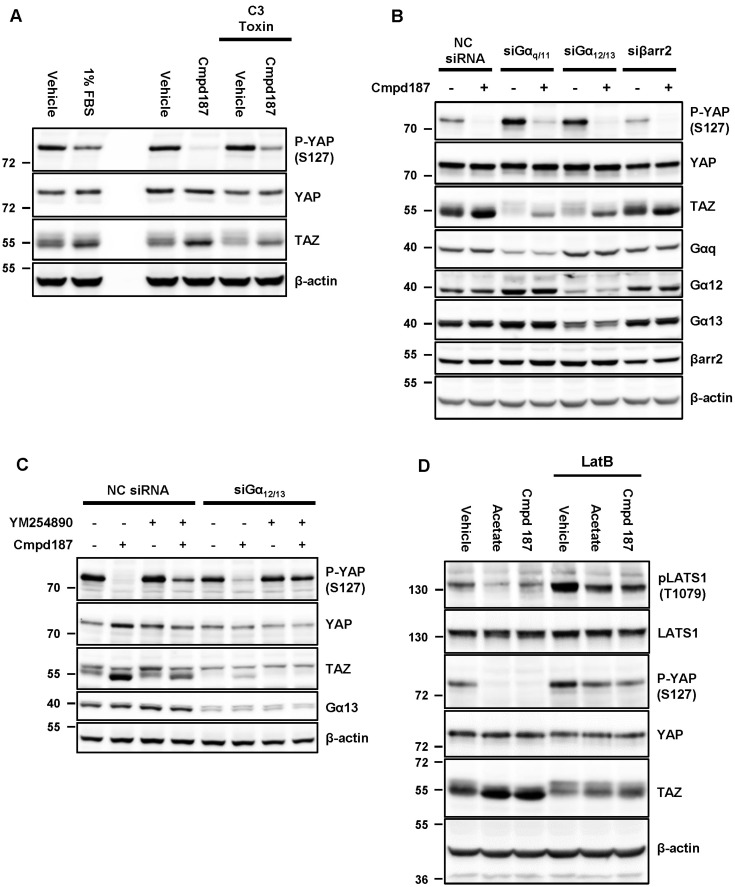
Next, we tested whether C3 toxin could affect the stability of YAP and TAZ. Compound 187 strongly suppressed phosphorylation of YAP and elevated TAZ to a greater extent than 1% FBS, a known RhoA activator (Fig. 5A, left lanes). C3 toxin counteracted the effects of compound 187, indicating that RhoA mediates GPR43 signaling to YAP/TAZ (Fig. 5A, right lanes). While GPR43 is widely known to be coupled to Gαi and Gαq, this receptor has also been shown to interact with Gα12 and Gα13 in the yeast pheromone system (Brown et al., 2003). To establish GPR43-mediated signaling pathway underlying YAP/TAZ regulation, GPR43-expressing HEK293 cells were transfected with siRNAs for G-proteins and β-arrestin 2. Knockdown of Gαq/11 and Gα12/13 significantly augmented phosphorylation of YAP and the slow migrating bands of TAZ while knockdown of β-arrestin 2 exerted no such effects and compound 187 still induced dephosphorylation of YAP and TAZ (Fig. 5B), suggesting that multiple G proteins may be involved in the regulation of YAP/TAZ. Upon simultaneous treatment with YM-254890 and siRNA of Gα12/13, the activity of compound 187 was completely abrogated (Fig. 5C), supporting the requirement for both Gαq/11 and Gα12/13 for YAP/TAZ activation through GPR43. Moreover, acetate and compound 187 reduced the phosphorylation of Thr 1079 in LATS1, an upstream inhibitory kinase of YAP/TAZ and latruculin B, a F-actin assembly blocker, restored the phosphorylation (Fig. 5D), suggesting that RhoA-mediated F-actin assembly modulated LATS1-YAP/TAZ axis. Taken together, our results suggest that GPR43 induces the activation of YAP/TAZ via Gαq/11- and Gα12/13-RhoA pathways.
Fig. 5. YAP and TAZ are stabilized and activated through αq/12/13-RhoA axis.
GPR43-G(A) Cells were serum-deprivation and were simultaneously treated with 0.5 μg/ml C3 toxin for overnight prior to being treated with the indicated reagents for 1 h. (B) GPR43/HEK293 cells were transfected with the indicated siRNAs for 72 h. (C) The cells were transfected with non-targeted and Gα12/13 targeted siRNAs for 72 h prior to treating with the indicated reagents after pre-incubating with YM-254890 for 15 min. (D) GPR43/HEK293 cells were pretreated with 1 μg/ml latrunculin B for 30 min and then treated with acetate and Cmpd 187 for 1 h.
Activation of GPR43 in M1-like THP-1 promotes YAP/TAZ stabilization
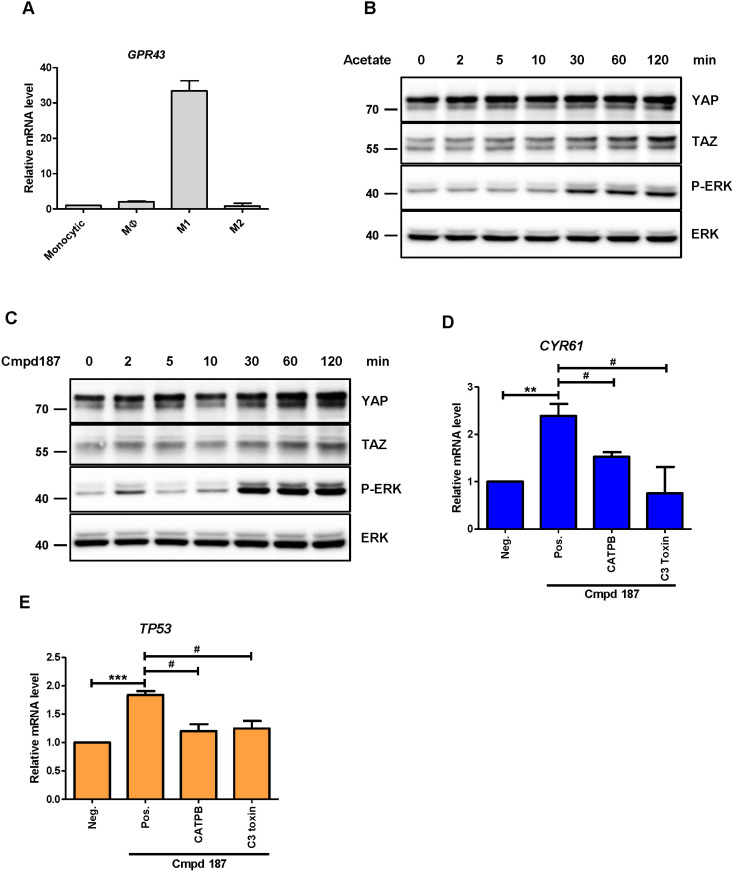
According to the human protein atlas (http://www.proteinatlas.org/), GPR43 is highly expressed in myeloid lineages. THP-1 is a human monocyte-like cancer cell line capable of differentiating into polarized macrophages, such as M1-like or M2-like types (Chanput et al., 2013). Therefore, we further examined whether expression of GPR43 differs depending on the macrophage type. Our experiment disclosed higher expression of GPR43 in M1-like polarized THP-1 compared to other cells (Fig. 6A). We also identified that YAP and TAZ were highly expressed in M1-like cells, as already reported (Zhou et al., 2019) (Supplementary Fig. S2). GPR43-overexpressing THP-1 cells were examined with some markers for M1 and M2 polarization, but no statistically significant change was observed (Supplementary Fig. S3). It was interesting that mRNAs of IL-1β and IL-6, markers of M1 polarized macrophages, were augmented by acetate in Mφ-differentiated THP-1 cells (Supplementary Fig. S4B), while mRNA of GPR43 itself was not altered (Supplementary Fig. S4A). Although compound 187 elicited a very small change in these markers and M2 markers were not significantly changed (Supplementary Fig. S4C), these data suggested that GPR43 may facilitate or mediate M1 polarization. After differentiation of THP-1 cells into M1 macrophage-like cells, stimulation with acetate (Fig. 6B) and compound 187 (Fig. 6C) promoted YAP and TAZ stabilization over time, although phosphorylation of YAP was not detected (data not shown). Compound 187-induced CYR61 gene expression was reduced by CATPB and C3 toxin (Fig. 6D). In addition, p53, which was identified as a target gene for YAP in macrophages (Zhou et al., 2019), was positively regulated by GPR43-RhoA (Fig. 6E). Collectively, these results support GPR43-mediated activation of YAP and TAZ via Gαq/11- and Gα12/13-RhoA pathways in macrophages.
Fig. 6. Activation of GPR43 induces YAP/TAZ stabilization in M1-like THP-1.
(A) RT-PCR analysis of mRNA levels of human GPR43 in monocytic THP-1 and polarized cell lines, such as Mφ, M1, and M2-like. THP-1 cells were stimulated with (B) 10 mM acetate and (C) 10 μM Cmpd187 for the indicated times. RT-PCR analysis of (D) CYR61 and (E) TP53 in M1-like THP-1 cells. Cells were pre-treated with CATPB for 15 min and C3 toxin overnight, followed by agonist stimulation for 4 h. Data from three independent experiments are presented as mean ± SEM (Student’s t-test). **P < 0.01, ***P < 0.005, # P < 0.05.
DISCUSSION
The main objective of this investigation was to determine whether GPR43 acts as a bona fide modulator of the RhoA-YAP/TAZ axis. Our results suggest that GPR43 exerts regulatory activity through Gαq/11 and Gα12/13 signaling. Earlier, Vaque et al. (2013) reported that Gαq/11-mediated activation of RhoA is achieved through TRIO-RhoGEF. The group showed that RhoA is stimulated through Gαq-TRIO signaling by the synthetic Gαq-coupled receptor, Sy-Rq, and a cognate agonist (Vaque et al., 2013). Therefore, both Gα12/13 and Gαq/11 signaling could be critically implicated in YAP/TAZ stabilization upon activation of RhoA by GPR43. However, not all GPCRs use both Gα12/13 and Gαq/11 to promote RhoA activity. For example, with regard to the RhoA-YAP/TAZ axis, PAR1 couples with both Gαq/11 and Gα12/13 but is reported to depend solely on the Gα12/13 pathway to activate RhoA (Gadepalli et al., 2013; Mo et al., 2012). On the other hand, CXCR4 induces a slight increase rather than decrease in phosphorylation of YAP/TAZ, albeit being capable of coupling with Gαq/11, Gα12/13 as well as Gαi/o (Yu et al., 2012). Thus, it is likely that activation of RhoA-YAP/TAZ axis is dependent on which GPCR is involved and cellular context. Upon treatment with the GPR43 agonist for 1 h, YAP/TAZ activity reached peak levels in HEK293 cells while peak activity was detected after 2 h in M1-like THP-1 macrophages (Figs. 6B and 6C). The GPR43-YAP/TAZ signaling pathway requires further investigation in distinct cellular contexts.
It has been known that YAP/TAZ helps to recover and regenerate colonic tissue by promoting the intestinal stem cell (ISC) in dextran sodium sulfate (DSS)-induced colitis model (Mahoney et al., 2014; Taniguchi et al., 2015). Activated GPR43 by SCFAs has also been revealed to promote gut epithelial integrity (Macia et al., 2015). From these previous reports, it is possible to deduce that activated YAP/TAZ by GPR43 is implicated in damaged tissue repair.
A recent report demonstrated that YAP proteins increased in M1-type and decreased in M2-type macrophages (Zhou et al., 2019). In line with this article, our data showed the increased YAP/TAZ expression levels in M1-polarized THP-1 (Supplementary Fig. S2) and the elevated gene expression of TP53 as well as CYR61 through GPR43-RhoA axis (Figs. 6D and 6E). Since increased p53 by YAP blocks the M2 polarization and M1 macrophages have a pro-inflammatory function (He et al., 2015; Zhou et al., 2019), SCFA-GPR43-RhoA-YAP axis in M1 macrophages may aggravate inflammatory bowel disease in a certain circumstance. This hypothesis is contrary to the fact that SCFAs are well known anti-inflammatory agents and GPR43 knockout mice exhibited deteriorated colitis phenotype (Agus et al., 2016; Macia et al., 2015; Maslowski et al., 2009; Masui et al., 2013). However, this discrepancy may explain the contradictory report that colitis was mitigated in GPR43-deficient mice (Kim et al., 2013; Sina et al., 2009). In other words, GPR43 may exert an anti-inflammatory effect through neutrophils and a pro-inflammatory effect through M1 macrophages. Further investigation should address this complex mechanism of GPR43 in terms of the regulation of innate immune system.
The data from this study demonstrate that GPR43 and YAP/TAZ are closely associated through RhoA and provide further insights into the physiological roles and mechanisms of action of GPR43.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This work was supported by a grant (NRF-2019M3E5D 4069882) from the National Research Foundation, Ministry of Science and ICT and Future Planning, and a grant from the KRIBB Initiative Program.
Footnotes
AUTHOR CONTRIBUTIONS
B.O.P. conceived and performed experiments, and wrote the manuscript. S.H.K. performed experiments. J.H.K. and S.Y.K. analyzed RNA-seq data. B.C.P. and S.B.H. provided expertise and feedback. S.G.P., J.H.K., and S.K. conceived experiments, wrote the manuscript, and secured funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Agus A., Denizot J., Thevenot J., Martinez-Medina M., Massier S., Sauvanet P., Bernalier-Donadille A., Denis S., Hofman P., Bonnet R., et al. Western diet induces a shift in microbiota composition enhancing susceptibility to Adherent-Invasive E. coli infection and intestinal inflammation. Sci. Rep. 2016;6:19032. doi: 10.1038/srep19032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang Z., Er J.Z., Tan N.S., Lu J., Liou Y.C., Grosse J., Ding J.L. Human and mouse monocytes display distinct signalling and cytokine profiles upon stimulation with FFAR2/FFAR3 short-chain fatty acid receptor agonists. Sci. Rep. 2016;6:34145. doi: 10.1038/srep34145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker G., Davenport R., Downham R., Farnaby W., Goldby A., Hannah D., Harrison D., Willems H. 3-SUBSTITUTED 2-AMINO-INDOLE DERIVATIVES. WIPO WO 2015/198045 A1 2015 [Google Scholar]
- Brown A.J., Goldsworthy S.M., Barnes A.A., Eilert M.M., Tcheang L., Daniels D., Muir A.I., Wigglesworth M.J., Kinghorn I., Fraser N.J., et al. The Orphan G protein-coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J. Biol. Chem. 2003;278:11312–11319. doi: 10.1074/jbc.M211609200. [DOI] [PubMed] [Google Scholar]
- Chanput W., Mes J.J., Savelkoul H.F., Wichers H.J. Characterization of polarized THP-1 macrophages and polarizing ability of LPS and food compounds. Food Funct. 2013;4:266–276. doi: 10.1039/c2fo30156c. [DOI] [PubMed] [Google Scholar]
- Cheng Z., Garvin D., Paguio A., Stecha P., Wood K., Fan F. Luciferase reporter assay system for deciphering GPCR pathways. Curr. Chem. Genomics. 2010;4:84–91. doi: 10.2174/1875397301004010084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley S.M., Mendoza-Reinoso V., Giles N., Singer E.S., Common J.E., Wilkins M.R., Beverdam A. Plau and Tgfbr3 are YAP-regulated genes that promote keratinocyte proliferation. Cell Death Dis. 2018;9:1106. doi: 10.1038/s41419-018-1141-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S.A., Gayyed M.F., Anders R.A., Maitra A., Pan D. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S., et al. Role of YAP/TAZ in mechanotransduction. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Gadepalli R., Kotla S., Heckle M.R., Verma S.K., Singh N.K., Rao G.N. Novel role for p21-activated kinase 2 in thrombin-induced monocyte migration. J. Biol. Chem. 2013;288:30815–30831. doi: 10.1074/jbc.M113.463414. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hao Y., Chun A., Cheung K., Rashidi B., Yang X. Tumor suppressor LATS1 is a negative regulator of oncogene YAP. J. Biol. Chem. 2008;283:5496–5509. doi: 10.1074/jbc.M709037200. [DOI] [PubMed] [Google Scholar]
- He X.Y., Xiang C., Zhang C.X., Xie Y.Y., Chen L., Zhang G.X., Lu Y., Liu G. p53 in the myeloid lineage modulates an inflammatory microenvironment limiting initiation and invasion of intestinal tumors. Cell Rep. 2015;13:888–897. doi: 10.1016/j.celrep.2015.09.045. [DOI] [PubMed] [Google Scholar]
- Hoj J.P., Mayro B., Pendergast A.M. A TAZ-AXL-ABL2 feed-forward signaling axis promotes lung adenocarcinoma brain metastasis. Cell Rep. 2019;29:3421–3434.e8. doi: 10.1016/j.celrep.2019.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Matsubara D., Tanaka I., Makiya K., Tanei Z.I., Kumagai Y., Shiu S.J., Nakaoka H.J., Ishikawa S., Isagawa T., et al. Loss of YAP1 defines neuroendocrine differentiation of lung tumors. Cancer Sci. 2016;107:1527–1538. doi: 10.1111/cas.13013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaki S., Mitsui R., Hayashi H., Kato I., Sugiya H., Iwanaga T., Furness J.B., Kuwahara A. Short-chain fatty acid receptor, GPR43, is expressed by enteroendocrine cells and mucosal mast cells in rat intestine. Cell Tissue Res. 2006;324:353–360. doi: 10.1007/s00441-005-0140-x. [DOI] [PubMed] [Google Scholar]
- Kim M.H., Kang S.G., Park J.H., Yanagisawa M., Kim C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology. 2013;145:396–406.e10. doi: 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- Kim N.G., Koh E., Chen X., Gumbiner B.M. E-cadherin mediates contact inhibition of proliferation through Hippo signaling-pathway components. Proc. Natl. Acad. Sci. U. S. A. 2011;108:11930–11935. doi: 10.1073/pnas.1103345108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura I., Ozawa K., Inoue D., Imamura T., Kimura K., Maeda T., Terasawa K., Kashihara D., Hirano K., Tani T., et al. The gut microbiota suppresses insulin-mediated fat accumulation via the short-chain fatty acid receptor GPR43. Nat. Commun. 2013;4:1829. doi: 10.1038/ncomms2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Poul E., Loison C., Struyf S., Springael J.Y., Lannoy V., Decobecq M.E., Brezillon S., Dupriez V., Vassart G., Van Damme J., et al. Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J. Biol. Chem. 2003;278:25481–25489. doi: 10.1074/jbc.M301403200. [DOI] [PubMed] [Google Scholar]
- Lee S.U., In H.J., Kwon M.S., Park B.O., Jo M., Kim M.O., Cho S., Lee S., Lee H.J., Kwak Y.S., et al. beta-Arrestin 2 mediates G protein-coupled receptor 43 signals to nuclear factor-kappaB. Biol. Pharm. Bull. 2013;36:1754–1759. doi: 10.1248/bpb.b13-00312. [DOI] [PubMed] [Google Scholar]
- Lei Q.Y., Zhang H., Zhao B., Zha Z.Y., Bai F., Pei X.H., Zhao S., Xiong Y., Guan K.L. TAZ promotes cell proliferation and epithelial-mesenchymal transition and is inhibited by the hippo pathway. Mol. Cell. Biol. 2008;28:2426–2436. doi: 10.1128/MCB.01874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B., Chen Y., Chen L., Cheng H., Mu C., Li J., Gao R., Zhou C., Cao L., Liu J., et al. Hypoxia regulates Hippo signalling through the SIAH2 ubiquitin E3 ligase. Nat. Cell Biol. 2015;17:95–103. doi: 10.1038/ncb3073. [DOI] [PubMed] [Google Scholar]
- Ma X., Zhao Y., Daaka Y., Nie Z. Acute activation of beta2-adrenergic receptor regulates focal adhesions through betaArrestin2- and p115RhoGEF protein-mediated activation of RhoA. J. Biol. Chem. 2012;287:18925–18936. doi: 10.1074/jbc.M112.352260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macia L., Tan J., Vieira A.T., Leach K., Stanley D., Luong S., Maruya M., Ian McKenzie C., Hijikata A., Wong C., et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nat. Commun. 2015;6:6734. doi: 10.1038/ncomms7734. [DOI] [PubMed] [Google Scholar]
- Mahoney J.E., Mori M., Szymaniak A.D., Varelas X., Cardoso W.V. The hippo pathway effector Yap controls patterning and differentiation of airway epithelial progenitors. Dev. Cell. 2014;30:137–150. doi: 10.1016/j.devcel.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maslowski K.M., Vieira A.T., Ng A., Kranich J., Sierro F., Yu D., Schilter H.C., Rolph M.S., Mackay F., Artis D., et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masui R., Sasaki M., Funaki Y., Ogasawara N., Mizuno M., Iida A., Izawa S., Kondo Y., Ito Y., Tamura Y., et al. G protein-coupled receptor 43 moderates gut inflammation through cytokine regulation from mononuclear cells. Inflamm. Bowel Dis. 2013;19:2848–2856. doi: 10.1097/01.MIB.0000435444.14860.ea. [DOI] [PubMed] [Google Scholar]
- McCarthy D.J., Chen Y., Smyth G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo J.S., Yu F.X., Gong R., Brown J.H., Guan K.L. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon K.H., Kim J.W. Hippo signaling circuit and divergent tissue growth in mammalian eye. Mol. Cells. 2018;41:257–263. doi: 10.14348/molcells.2018.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroishi T., Park H.W., Qin B., Chen Q., Meng Z., Plouffe S.W., Taniguchi K., Yu F.X., Karin M., Pan D., et al. A YAP/TAZ-induced feedback mechanism regulates Hippo pathway homeostasis. Genes Dev. 2015;29:1271–1284. doi: 10.1101/gad.262816.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson N.E., Kotarsky K., Owman C., Olde B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short-chain fatty acids. Biochem. Biophys. Res. Commun. 2003;303:1047–1052. doi: 10.1016/s0006-291x(03)00488-1. [DOI] [PubMed] [Google Scholar]
- Park H.W., Kim Y.C., Yu B., Moroishi T., Mo J.S., Plouffe S.W., Meng Z., Lin K.C., Yu F.X., Alexander C.M., et al. Alternative Wnt signaling activates YAP/TAZ. Cell. 2015;162:780–794. doi: 10.1016/j.cell.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini M., Villa S.R., Fuller M., Wicksteed B., Mackay C.R., Alquier T., Poitout V., Mancebo H., Mirmira R.G., Gilchrist A., et al. An acetate-specific GPCR, FFAR2, regulates insulin secretion. Mol. Endocrinol. 2015;29:1055–1066. doi: 10.1210/me.2015-1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shreberk-Shaked M., Oren M. New insights into YAP/TAZ nucleo-cytoplasmic shuttling: new cancer therapeutic opportunities? Mol. Oncol. 2019;13:1335–1341. doi: 10.1002/1878-0261.12498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sina C., Gavrilova O., Forster M., Till A., Derer S., Hildebrand F., Raabe B., Chalaris A., Scheller J., Rehmann A., et al. G protein-coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J. Immunol. 2009;183:7514–7522. doi: 10.4049/jimmunol.0900063. [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Wu L.W., Grivennikov S.I., de Jong P.R., Lian I., Yu F.X., Wang K., Ho S.B., Boland B.S., Chang J.T., et al. A gp130-Src-YAP module links inflammation to epithelial regeneration. Nature. 2015;519:57–62. doi: 10.1038/nature14228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedelind S., Westberg F., Kjerrulf M., Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: a study with relevance to inflammatory bowel disease. World J. Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaque J.P., Dorsam R.T., Feng X., Iglesias-Bartolome R., Forsthoefel D.J., Chen Q., Debant A., Seeger M.A., Ksander B.R., Teramoto H., et al. A genome-wide RNAi screen reveals a Trio-regulated Rho GTPase circuitry transducing mitogenic signals initiated by G protein-coupled receptors. Mol. Cell. 2013;49:94–108. doi: 10.1016/j.molcel.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinolo M.A., Ferguson G.J., Kulkarni S., Damoulakis G., Anderson K., Bohlooly Y.M., Stephens L., Hawkins P.T., Curi R. SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One. 2011;6:e21205. doi: 10.1371/journal.pone.0021205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldecker M., Kautenburger T., Daumann H., Busch C., Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J. Nutr. Biochem. 2008;19:587–593. doi: 10.1016/j.jnutbio.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Wang Y., Xu X., Maglic D., Dill M.T., Mojumdar K., Ng P.K., Jeong K.J., Tsang Y.H., Moreno D., Bhavana V.H., et al. Comprehensive molecular characterization of the Hippo signaling pathway in cancer. Cell Rep. 2018;25:1304–1317.e5. doi: 10.1016/j.celrep.2018.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Wei L., Fan F., Ji S., Zhang S., Geng J., Hong L., Fan X., Chen Q., Tian J., et al. Integration of Hippo signalling and the unfolded protein response to restrain liver overgrowth and tumorigenesis. Nat. Commun. 2015;6:6239. doi: 10.1038/ncomms7239. [DOI] [PubMed] [Google Scholar]
- Yu F.X., Zhao B., Panupinthu N., Jewell J.L., Lian I., Wang L.H., Zhao J., Yuan H., Tumaneng K., Li H., et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanconato F., Forcato M., Battilana G., Azzolin L., Quaranta E., Bodega B., Rosato A., Bicciato S., Cordenonsi M., Piccolo S. Genome-wide association between YAP/TAZ/TEAD and AP-1 at enhancers drives oncogenic growth. Nat. Cell Biol. 2015;17:1218–1227. doi: 10.1038/ncb3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ye X., Yu J., Li L., Li W., Li S., Yu J., Lin J.D., Wang C.Y., Chinnaiyan A.M., et al. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev. 2008;22:1962–1971. doi: 10.1101/gad.1664408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Li W., Wang S., Zhang P., Wang Q., Xiao J., Zhang C., Zheng X., Xu X., Xue S., et al. YAP aggravates inflammatory bowel disease by regulating M1/M2 macrophage polarization and gut microbial homeostasis. Cell Rep. 2019;27:1176–1189.e5. doi: 10.1016/j.celrep.2019.03.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.