Abstract
Ubiquitin D (UBD) is highly upregulated in many cancers, and plays a pivotal role in the pathophysiological processes of cancers. However, its roles and underlying mechanisms in oral squamous cell carcinoma (OSCC) are still unclear. In the present study, we investigated the role of UBD in patients with OSCC. Quantitative real-time polymerase chain reaction and Western blot were used to measure the expression of UBD in OSCC tissues. Immunohistochemistry assay was used to detect the differential expressions of UBD in 244 OSCC patients and 32 cases of normal oral mucosae. In addition, CCK-8, colony formation, wound healing and Transwell assays were performed to evaluate the effect of UBD on the cell proliferation, migration, and invasion in OSCC. Furthermore, a xenograft tumor model was established to verify the role of UBD on tumor formation in vivo. We found that UBD was upregulated in human OSCC tissues and cell lines and was associated with clinical and pathological features of patients. Moreover, the overexpression of UBD promoted the proliferation, migration and invasion of OSCC cells; however, the knockdown of UBD exerted the opposite effects. In this study, our results also suggested that UBD promoted OSCC progression through NF-κB signaling. Our findings indicated that UBD played a critical role in OSCC and may serve as a prognostic biomarker and potential therapeutic target for OSCC treatment.
Keywords: epithelial-to-mesenchymal transition, NF-kappa B, oral squamous cell carcinoma, tumorigenesis, ubiquitin D
INTRODUCTION
Oral squamous cell carcinoma (OSCC) is a heterogeneous epithelial tumor and the most common malignancy worldwide (Chai et al., 2020). It is reported that 600,000 new cases of OSCC are estimated to be diagnosed worldwide each year (Tampa et al., 2018), and more than half of them have the advanced disease (Terzuoli et al., 2019), with a 5-year survival rate of less than 50% (Petersen, 2009). However, the exact etiology of OSCC is unknown (Ferlay et al., 2010). Alcohol, areca nut, and tobacco exposure are the most common causes of OSCC (Panarese et al., 2019). Despite the recent improvements in therapeutic approaches, the overall survival of OSCC remains unsatisfactory (Zielinska and Katanaev, 2019). Therefore, it is necessary to better elucidate OSCC and the underlying molecular mechanisms of tumorigenesis (Harsha et al., 2020; Ishida et al., 2017).
Ubiquitin D (UBD), also known as Human HLA-F adjacent transcript locus 10 (FAT10), is a protein containing 165 amino acid residues. It belongs to a class of ubiquitin-like proteins and a member of the ubiquitin-like protein family (UBI), whose protein sequence and three-dimensional core structure resemble ubiquitin (UB) (Cappadocia and Lima, 2018; Wang and Zhao, 2019). Previous studies have shown that UBD is involved in various important cellular developmental processes, including immune-mediated inflammation, cell cycle, and proliferation (Aichem and Groettrup, 2016; Wang et al., 2017). In recent years, there have been increasing studies on UBD in malignancies, indicating an association between increased UBD expression and disease progression in various cancers (Aichem and Groettrup, 2016; Qing et al., 2011; Xiang et al., 2020), such as gastric cancer, liver cancer, and glioma (Yan et al., 2018; Yuan et al., 2012; 2014). UBD has been reported to be highly expressed in gastric cancer and a potential marker of patient prognosis (Qing et al., 2011). UBD overexpression in hepatic carcinoma (HCC) tissues plays an important role in cancer development and progression (Luo et al., 2018; Yuan et al., 2014). Furthermore, overexpression of UBD is also strongly associated with poor prognosis in gliomas (Yuan et al., 2012). These findings indicate the role of UBD in the development of tumors. However, the expression pattern of UBD in OSCC and its associated molecular mechanisms have not yet been reported. Besides, the therapeutic potential of UBD as a new option for the treatment of OSCC remains to be elucidated.
The phenomenon of epithelial-to-mesenchymal transition (EMT) is frequently observed in OSCC (Bai et al., 2020). EMT, a critical early time in tumor progression, is a complex cellular mechanism characterized by the loss of epithelial characteristics through epithelial cells and transformation into cells with a mesenchymal phenotype (Yanjia and Xinchun, 2007). The EMT process gives mesenchymal properties to epithelial cells and increases the invasive ability (Siriwardena et al., 2018; Zhou et al., 2015). Besides, NF-κB plays a crucial role in inducing and maintaining EMT and tumor progression. Many studies have shown that UBD can stimulate the activation of the NF-κB pathway (Choi et al., 2014; Gao et al., 2014; Kawamoto et al., 2019). The NF-κB pathway has been reported to promote the proliferation and metastasis of tumor cells, and reduce apoptosis of cancer cells (Mortezaee et al., 2019; Xia et al., 2014). NF-κB is a heterodimeric protein composed of different combinations of members of the Rel family of transcription factors, including NF-κB 1 (p50), NF-κB 2 (p52), RelA (p65), RelB, and c-Rel. They remain in a dormant state in the cytoplasm and can be rapidly activated by stimulation to transfer their position as a nucleus and regulate its target genes (Eluard et al., 2020; Hayden and Ghosh, 2012). RelA, RelB, and c-Rel were synthesized into mature proteins containing the trans-activation domain, while p50 and p52 were synthesized as precursor proteins lacking the domain and dependent on heterodimer to mediate trans-activation. Typically, the activation of the subunit is controlled by the IκB protein of NF-κB isolated in the cytoplasm. This, in turn, is regulated by the IκB kinase (IKK) protein, which phosphorylates IκB and targets it for proteasomal degradation and leads to the release of NF-κB, allowing free transfer to the nucleus and thus affects gene transcription (Harris et al., 2006; Neumann and Naumann, 2007). The initial study found that the nuclear expression of c-Rel plays an important role in human OSCC specimens and cell lines (King et al., 2008). A previous study showed that c-Rel is overexpressed in a variety of solid tumors, including lung, ovary, pancreas, upper digestive tract, thyroid, liver, etc. (Kaltschmidt et al., 2018). Meanwhile, some studies have shown that c-Rel overexpression leads to an increased incidence of breast cancer formation (Sovak et al., 1997; Zamo et al., 2005). The role of the NF-κB signaling pathway in solid tumors has been increasingly recognized. However, there is a lack of direct evidence for cross-links between UBD and the NF-κB pathway in OSCC.
In this study, we analyzed UBD expression in OSCC tissue microarrays by IHC staining and found that UBD expression was significantly correlated with the clinical malignant characteristics of OSCC and patient prognosis. Besides, UBD overexpression in OSCC cell lines confirmed the oncogenic properties of UBD in OSCC. In addition, our findings suggested that UBD activated the NF-κB signaling pathway in OSCC. These characteristics of UBD provided a new perspective to get a deep insight into the progress of OSCC, and may provide a new strategy for the treatment of OSCC.
MATERIALS AND METHODS
OSCC patients and tissue samples
This study was approved by the Ethical Committee of the Stomatological Hospital Affiliated to Nanjing Medical University (No. PJ2018-051-001). Three sets of tissue microarrays including 244 OSCC patients and 32 cases of normal oral mucosae who received surgery at Stomatological Hospital of Jiangsu Province between 2009 and 2014 were included in this study. Additional 50 pairs of OSCC cancer tissues and matched adjacent normal tissue samples from Stomatological Hospital of Jiangsu Province between 2018 and 2019 were used for quantitative real-time reverse transcription (RT)-polymerase chain reaction (PCR) and 10 pairs were used for Western blot analysis. All tissues were stored in liquid nitrogen for further use. Those patients were excluded who accepted chemotherapy, radiotherapy and/or targeted molecular therapy before surgery. All included patients provide written informed consent.
Cell culture, plasmids, and reagents
The human OSCC cell lines HN6 and CAL27 were obtained as previously described. The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (CellMax Cell Technology, China) and 1% penicillin/streptomycin (CellMax Cell Technology) at 37°C in 5% CO2 atmosphere. The full-length coding region of UBD was constructed by GenePharma (China) and the shRNA targeting sequences specific for the human UBD were purchase from GenePharma. The primer sequences were as follows: shUBD-1, 5’-CCUCUCAUCUUAUGGCAUUTT-3’; shUBD-2, 5’-CCUGGCAUGUUAUUGUAUUTT-3’; shUBD-3, 5’-CCAGAUUGUGACUUGCAAUTT-3’; shNC, 5’-TTCTCCGAACGTGTCACGT-3’. Cells were cultured in a 6-well culture with 50%-60% confluency, starved in medium without FBS for 16 h and then transfections of HN6 and CAL27 cells with the related plasmids were performed with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. To induce stable transfection, transfected cells were selected with puromycin (2 μg/ml) (Gibco, USA). NF-κB inhibitor JSH-23 was purchased from Selleck Chemicals (USA).
Real-time RT-PCR
Total RNA was isolated using TRIzol reagent (Vazyme Biotech, China) according to the manufacturer’s instructions. cDNA was synthesized using 5× PrimeScript RT Master Mix (Vazyme Biotech). Quantitative RT-PCR (qRT-PCR) was performed using the SYBR Premix Ex Taq Kit (Vazyme Biotech) and ABI 7900 real-time PCR system (Applied Biosystems, USA) according to the manufacturer’s instructions. GAPDH served as endogenous control and the 2-ΔΔCT method was used for the quantitation of relative gene expression levels. The primers were listed as follows: UBD, F: 5’-AGCCCAGTGATGAGGAGCTG-3’, R: 5’-AGGAGGTGCCTCTTTGCCTC-3’; GAPDH, F: 5’- GAAGGTGAAGGTCGGAGTC -3’, R: 5’-GAGATGGTGATGGGATTTC-3’.
Western blot analysis
Total protein was lysed for 30 min using a lysis buffer (Beyotime, China), and the cells were washed with precooled phosphate-buffered saline (PBS). All proteins were fractionated by SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes (Millipore, USA). The membranes were blocked with 5% BSA and incubated for 2 h at room temperature. The blots were probed using primary antibodies and incubated overnight at 4°C: UBD (ab134077; Abcam, UK), cyclin E1 (#4129; Cell Signaling Technology [CST], USA), cyclin D1 (#55506; CST), CDK2 (#2546; CST), CDK4 (#12790; CST), E-cadherin (#3195; CST), N-cadherin (ab18203; Abcam), Snail (#3879; CST), Slug (#9585; CST), Vimentin (#5741; CST), NF‐κB p65 (#8242; CST), Phospho‐NF‐κB p65 (Ser536) (#3033; CST), IκBα (#4814S; CST), Phospho-IκBα (#5209S; CST), β-actin (AP0733; Bioworld, China), GAPDH (#5174S; CST), and Lamin B1 (#13435S; CST). Then the membrane was washed three times with TBST, and incubated with horseradish peroxidase-conjugated (HRP) secondary antibodies (Proteintech Group, China) for 1 h at room temperature. Finally, immunoreactive bands were detected using an Immobilon Western Chemiluminescent HRP Substrate (Millipore) and visualized using ImageQuantLAS 4000 mini-imaging system (General Electrics, USA). The membranes were stripped and re-probed with a GAPDH (#5174S; CST) or Lamin B1 (#13435S; CST) as a loading control.
Immunohistochemistry
Paraffin-embedded OSCC tissues were dewaxed, rehydrated and antigen-retrieval was performed by heating in EDTA buffer (pH 9) for 15 min in a microwave pressure cooker. After colding at room temperature, 3% hydrogen peroxide solution was used to block endogenous peroxides. Then, the microarrays were washed twice again with PBS and blocked with 5% bovine albumin in PBS for 1 h at room temperature. Tissue microarrays of OSCC tissue were stained with the primary antibody against UBD (ab134077; Abcam) at 4°C overnight. Then, blots were washed 3 times in PBS and incubated for 1 h at room temperature with the second antibody (HRP). The slides were then lightly counterstained with hematoxylin, dehydrated, and mounted. Evaluation of the staining reaction was performed in accordance with the immunoreactive score = staining intensity score × staining extent score. In brief, the intensity was defined as follows: 0, no appreciable staining; 1, weak intensity; 2, moderate intensity; 3, strong intensity; 4, very strong intensity. The percentages of stained-positive cells were divided into four categories: 0 to 1 (0%-25%), 1 to 2 (26%-50%), 2 to 3 (51%-75%), and 3 to 4 (76%-100%). Finally, the above two scores were multiplied for an overall score (range, 1-12). High expression was explained by a staining index score ≥ 4, while low expression was defined as a staining index < 4.
Immunofluorescence staining
Cells (5 × 104) were plated on coverslips to culture for 12 h. Then the cells were fixed with 2.0% formaldehyde for 30 min, washed three times with PBS, and treated with PBS containing 0.5% Triton X-100 for 20 min. Next, cells were stained with primary antibodies against N-cadherin (ab18203; Abcam) or NF‐κB p65 (#8242; CST) overnight at 4°C. Following washing three times with PBS, the cells were incubated with FITC or Cy3-conjugated goat anti-rabbit IgG (1:200; Proteintech Group) for 1 h and then counterstained with 4’,6-di-amidino-2-phenylindole (DAPI; Sigma Chemicals, USA). Plates were taken using a fluorescence microscope (DM4000B; Leica, Germany) and analyzed by ImageJ software (v1.49).
Cell proliferation assay and colony formation assay
Cells transfected with plasmids were plated at a density of 1 × 103 cells per well in 96-well plates. Cell viability was measured using the CCK-8 kit (Dojindo, Japan) to determine the OD value at 450 nm. For colony formation assays, cells were plated in 6-well plates at a concentration of 300 cells/well and incubated for 2 weeks to allow colony formation. After incubation, cells were fixed with formalin and stained with crystal violet solution to visualize plaques. Cell colony formation numbers were counted under the microscope (DM4000B) and analyzed by ImageJ software.
Wound healing and cell invasion assays
Cells transfected with plasmids were cultured in 12-well plates in DMEM until 100% confluence. Monolayer cells were washed with PBS twice and scraped with a plastic 200 µl pipette tip and then incubated with fresh DMEM medium without serum. The wound closure was observed under a microscope at 0 and 12 h and the wound healing rate was calculated and images were captured. Tumor cell invasion was examined using a classical Boyden chamber test (8 μm pore size in a 24-well plate format) with Matrigel-coating (BD Biosciences, USA) according to the manufacturer’s instructions. Cells were seeded into chambers at a density of 1 × 105 cells per well. After 24 h, the cells were fixed with methanol and stained using methylene blue staining. Cell invasive potential was evaluated by counting the stained cells using a light microscope by randomly selecting three fields.
Luciferase activity analysis
NF-κB and UBD reporter plasmids were constructed by GenePharma. One day before transfection, inoculate 1 × 105 cells in a 48-well plate with 350 μl of DMEM complete medium without antibiotics in each well. Place them in a cell culture incubator overnight. Transfection is performed when the confluence is 70% to 80%. After 48 h, the cells were lysed, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Beyotime), according to the manufacturer’s instructions. Each experiment was independently repeated three times.
Animal experiments
All animal experiments were approved by the institutional guidelines of Nanjing Medical University. To evaluate the role of UBD in tumor progression, HN6 cells (1 × 107 cells/mouse) were injected subcutaneously into the right or left dorsal flank of 6-week-old BALB/c nude mice with control cells and experimental cells. Tumor volume was measured every 3 days and the mice were anesthetized and euthanized after 18 days of injection. The volume of the tumors was calculated using the following formula: Tumor volume = length × width2/2. All tumor tissues were separated, and tumor weight and volume were measured. To detect the role of UBD in lung metastasis, a total of 2 × 106 HN6 cells in 200 μl serum-free DMEM were injected intravenously into the lateral tail vein of the 6-week-old BALB/c nude mice. After 3 weeks, the animals were killed, and the lung tissues were fixed in neutral-buffered formalin for further experiments. Animal experiments were conducted in accordance with institutional and national regulations for the use and welfare of laboratory animals.
Statistical analysis
The Student’s t-test and chi-square test were used for continuous and categorical variables to evaluate the relationship between UBD and clinicopathological parameters, respectively. Data were presented as the mean ± SD from three independent experiments. GraphPad Prism (ver. 7.0; GraphPad Software, USA) was used for statistical analysis. P < 0.05 was considered as statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001).
RESULTS
UBD overexpression was associated with OSCC progression
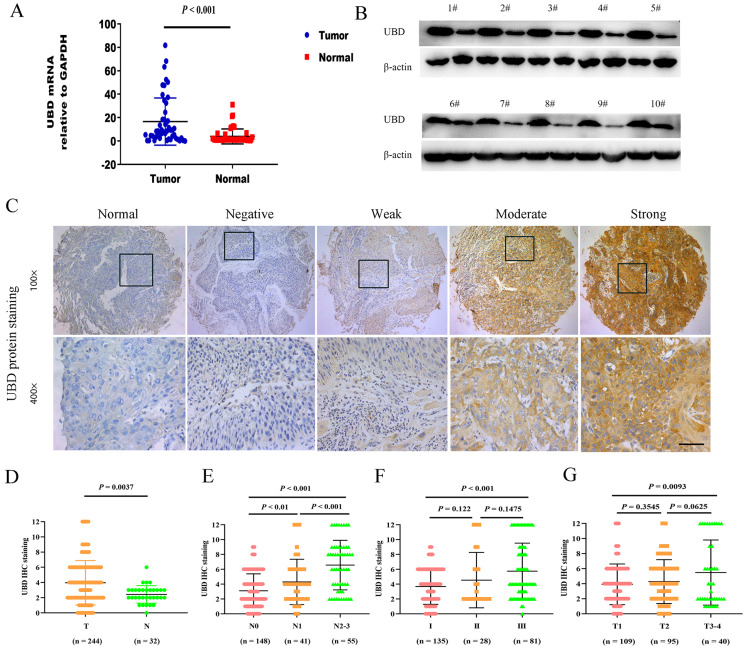
To investigate the role of UBD in OSCC, we first examined the mRNA and protein expression levels of UBD in 50 OSCC tissue samples and the corresponding adjacent normal tissues. Our result showed that the expression level of UBD mRNA was up-regulated in OSCC by qRT-PCR and Western blot analysis was consistent with qRT-PCR results (Figs. 1A and 1B). This was further confirmed by tissue microarray experiments including 244 OSCC tumors and 32 normal tissues (Figs. 1C and 1D). Next, we analyzed the correlation between UBD and clinical characters and pathological features (Table 1). Results showed that the expression of UBD was significantly higher in patients with lymph node metastasis compared to those without metastasis (Fig. 1E) and in patients with higher pathological grade (Fig. 1F). As envisioned, correlation analysis identified a positive correlation between UBD expression level and clinical T stages (Fig. 1G). In summary, our results suggested that the overexpression of UBD might associate with tumor progression and metastasis.
Fig. 1. UBD overexpression was associated with OSCC progression.
(A) Relative expression level of UBD mRNA in 50 OSCC tumor tissues and their matched adjacent non-tumor tissues. (B) Detection of relative protein levels in 10 paired of OSCC tumors and adjacent non-tumor tissues by Western blot. β-actin was used as a loading control. (C and D) Immunohistochemistry (IHC) of UBD expression level on tissue microarrays containing 244 OSCC patient samples and 32 normal oral tissues is shown. Relative negative, weak, moderate and strong stain images compared with normal tissues are shown. (E) Expression levels of UBD in OSCC tissues with different lymph node metastasis status. (F) Expression levels of UBD in OSCC tissues with different pathological grades. (G) Quantification analysis of FAT’10 staining in OSCC with different tumor stages. All error bar values represent the SD. Scale bar = 100 μm.
Table 1.
Association between UBD expression and clinicopathologic parameters
| Pathologic characteristics | n | Overexpression (No. of cases) | Nonoverexpression (No. of cases) | P value |
|---|---|---|---|---|
| Age (y) | ||||
| ≥ 60 | 134 | 58 | 76 | 0.235 |
| < 60 | 110 | 56 | 54 | |
| Sex | ||||
| Male | 148 | 44 | 104 | 0.925 |
| Female | 96 | 28 | 68 | |
| Smoking | ||||
| Yes | 109 | 69 | 40 | < 0.05 |
| No | 135 | 44 | 91 | |
| Drinking | ||||
| Yes | 90 | 63 | 27 | 0.418 |
| No | 154 | 100 | 54 | |
| Location | ||||
| Palate | 23 | 10 | 13 | 0.133 |
| Tongue | 82 | 51 | 31 | |
| Gingiva | 46 | 20 | 26 | |
| Buccal | 68 | 33 | 35 | |
| Mouth floor | 25 | 10 | 15 | |
| Tumor stage | ||||
| T1 | 109 | 44 | 65 | T1 vs T2 = 0.351 |
| T2 | 95 | 40 | 45 | T1 vs T3-T4 < 0.01 |
| T3-T4 | 40 | 28 | 12 | |
| Lymph node status | ||||
| N0 | 148 | 63 | 85 | N0 vs N1 < 0.01 |
| N1 | 41 | 28 | 13 | N0 vs N2-N3 < 0.001 |
| N2-N3 | 55 | 41 | 14 | |
| Pathological grade | ||||
| I | 135 | 58 | 77 | I vs II = 0.736 |
| II | 28 | 13 | 15 | I vs III < 0.001 |
| III | 81 | 55 | 26 |
UBD promoted OSCC cell proliferation in vitro
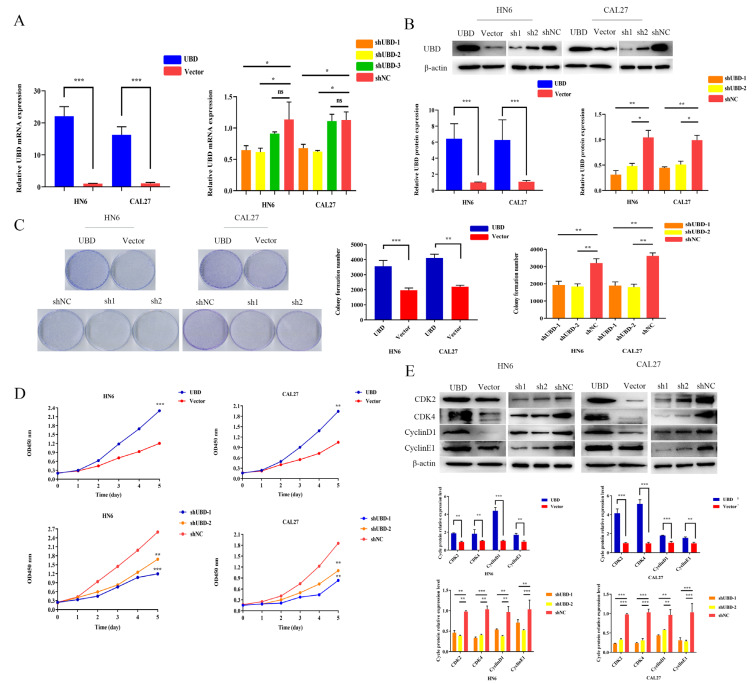
Next, we explored the role of UBD in OSCC cell lines with in vitro experiments. HN6 and CAL27 cells were transfected with either UBD overexpressing plasmid (UBD), or small short hairpin interfering RNA for UBD (shUBD) and measured their proliferation rates. The transfection efficiency was confirmed by qRT-PCR and Western blotting (Figs. 2A and 2B). Next, we tested cell proliferative potential in OSCC cell lines. The colony formation assays revealed that over-expression of UBD promoted cell growth and viability compared with the control group and UBD silencing inhibited colony formation compared with negative control (Fig. 2C), which was further confirmed by CCK-8 assays (Fig. 2D). In addition, we analyzed related cell-cycle proteins by Western blot to determine whether UBD enhanced cell proliferation via alteration of the cell cycle. We found that the major protein associated with cell cycle progression, cyclin-dependent kinases, were activated in cells over expressed UBD (Fig. 2E).
Fig. 2. UBD promoted OSCC cell proliferation in vitro.
OSCC cell lines HN6 and CAL27 were transfected with UBD. The efficiency of transfection was validated by qRT-PCR (A) and Western blot (B). (C and D) Colony formation assay and CCK-8 assay showed UBD overexpression increased colony formation efficiency of HN6 and CAL27 cells, and UBD knockdown inhibited the proliferative ability of OSCC cells. OD, optical density. (E) The cell cycle associated-proteins were measured by Western blot, including CDK2, CDK4, cyclin D1, and cyclin E1. Data were presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
UBD promoted OSCC cell migration and invasion
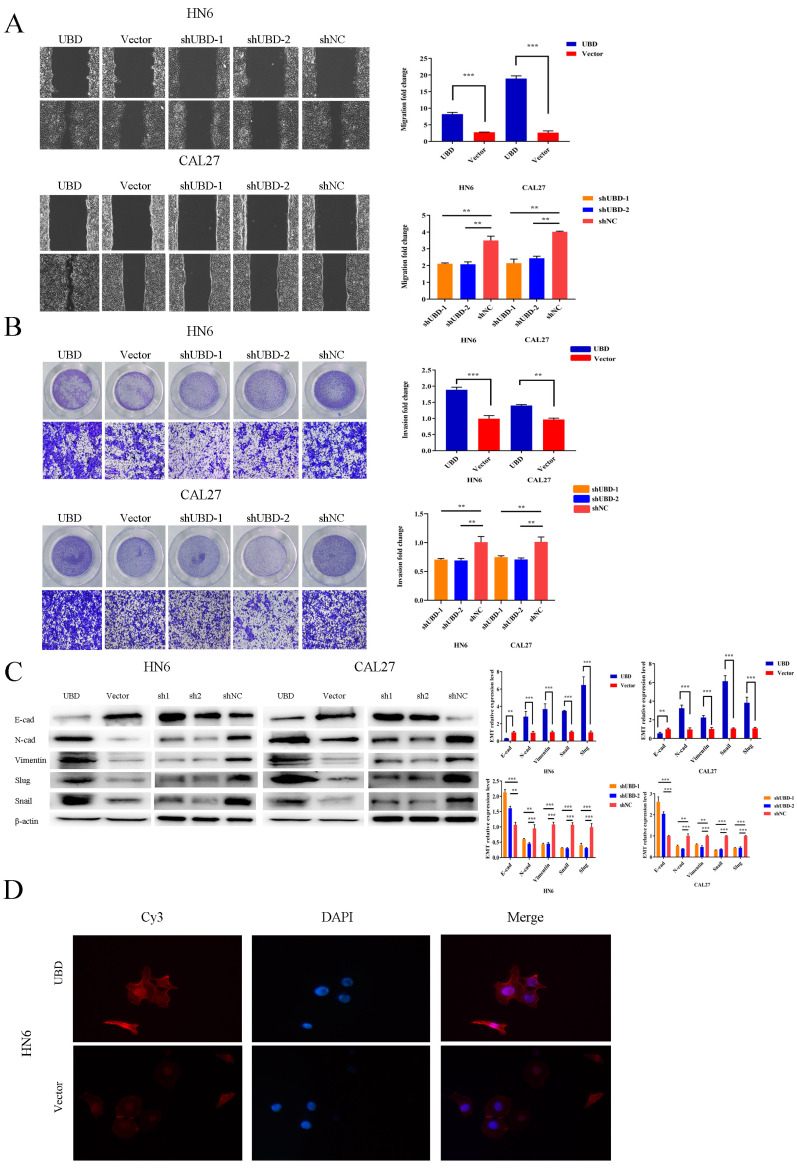
To further examine the role of UBD in OSCC cell lines, we tested cell migration and invasion, which are two critical steps in tumor metastasis (Steinbichler et al., 2020; Yang et al., 2021). We performed wound healing assay with transfected HN6 and CAL27 cells. In wound healing assay, we found that the cell migrative ability was enhanced in the UBD overexpression group (Fig. 3A). The results were further confirmed by transwell migration assay (Fig. 3B). Considering the fact that EMT is a key process in tumor invasion and metastasis, we analyzed the effect of UBD on EMT by measuring the expression levels of EMT markers. Western blot results showed that UBD overexpression increased the expression of N-cadherin, vimentin, Snail family protein and reduced the expression of E-cadherin (Fig. 3C). Downregulation of UBD reversed the EMT process and diminished the metastasis potential of OSCC cells (Fig. 3C). Immunofluorescence staining experiments further confirmed that N-cadherin was increased in UBD-overexpressed cells (Fig. 3D).
Fig. 3. UBD promoted the migration and invasion phenotype of OSCC cells.
(A) Wound healing assays were performed to examine the capacity of cell migration after transfection. The results showed that the cell migration ability was significantly increased in the UBD-overexpression group. Meanwhile, the wound closure was delayed in UBD stable knockdown cells compared with shNC control at both the 12-h time point. (B) Cell invasion growth was analyzed by Transwell assay (×100). (C) Epithelial-mesenchymal-transition (EMT)-related transcription factors and EMT-markers were measured in indicated cells after the same transfection. UBD promoted the expression of mesenchymal markers (N-cadherin [N-cad], Vimentin, Slug, and Snail) and reduced the expression of E-cadherin (E-cad). Downregulation of UBD restrained the EMT process. Data were presented as the mean ± SD. **P < 0.01, ***P < 0.001. (D) The expression of N-cadherin (red) was determined by immunofluorescent staining and co-staining using DAPI (blue) (magnification, ×200).
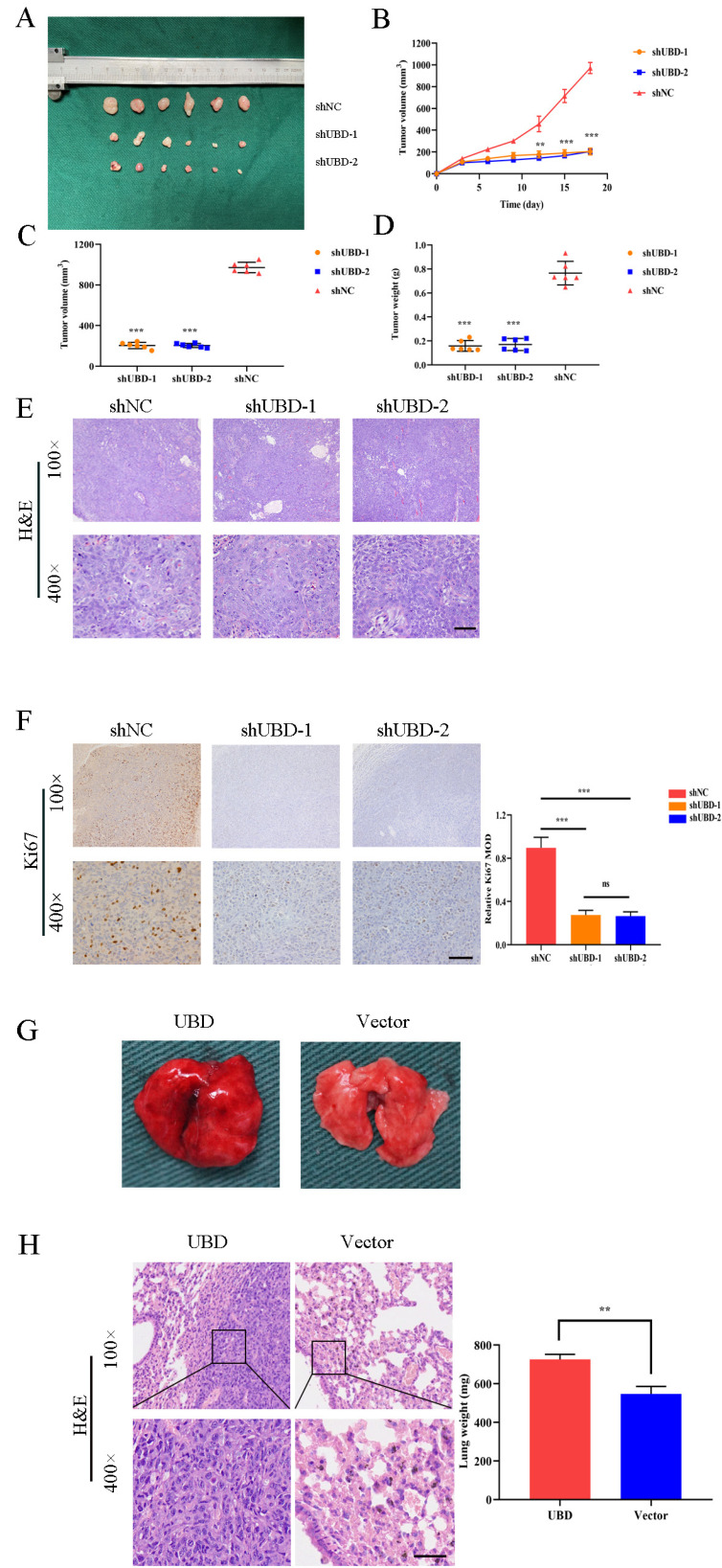
Knockdown of UBD diminished tumorigenicity of OSCC cells in vivo
The contribution of UBD in tumorigenicity was further explored in the xenograft cancer model established with BALB/c nude mice. 1 × 106 HN6 cells transfected with shUBD or shNC were subcutaneously injected into the left or right flank of the mice respectively. The tumor growth was measured until the mice were sacrificed 20 days later (Fig. 4A). The results demonstrated that knockdown of UBD significantly decreased the tumor volume and weight compared to controls (Figs. 4B-4D). Furthermore, there were no morphological difference between HN6 negative control group and HN6‐shUBD group in xenografted tumors (Fig. 4E). Immunohistochemistry assays showed that UBD knockdown reduced the expression of Ki67 in xenograft tumors (Fig. 4F). To further determine the effect of UBD overexpression on OSCC cell metastasis in vivo, we performed the distal dissemination assays using a nude mouse model. A total of 2 × 106 HN6 cells were injected with UBD or Vector transfected cells via the tail vein (group 2, n = 3) and nude mice were killed at 2 weeks. Then the lungs were isolated and pictures were taken (Fig. 4G). H&E staining revealed that the metastatic colony numbersin lungs at 2 weeks were significantly higher compared to control groups, and lung weight were also significantly elevated at 2 weeks (P < 0.01) (Fig. 4H). These data suggest that UBD promote OSCC cell metastasis in vivo.
Fig. 4. Knockdown of UBD diminished tumorigenicity of OSCC cells in vivo.
(A) Knockdown of UBD inhibited tumor growth in vivo. Images showed representative tumors that dissected from our model. The tumors dissected from mice were presented below. Each group contains six nude mice. (B-D) Growth curves of xenograft tumors (B), tumor volumes (C), and tumor weight (D). (E) Representative H&E staining of tumor tissues and (F) IHC staining of Ki67. MOD, mean optical density. (G) Whole mount pictures of the lung tissues are shown. (H) H&E staining of lung tissue sections at 2 weeks are shown. Scale bars = 50 μm (E, F, and H). Data were presented as the mean ± SD. ns, not significant. **P < 0.01, ***P < 0.001, based on Student’s t-test.
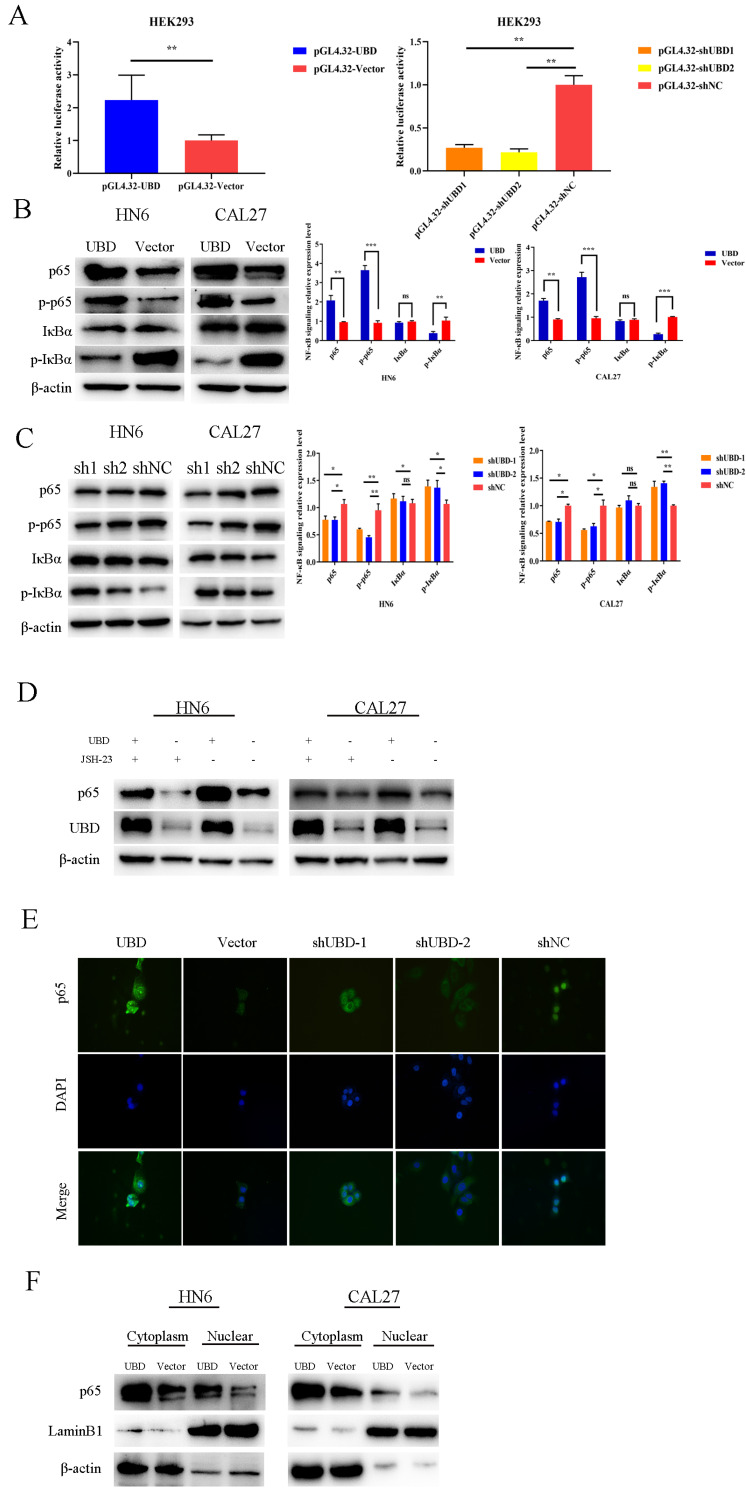
UBD activated NF-κB signaling pathway in OSCC cells
Recent studies indicated that overexpression of UBD enhanced tumor growth through NF-κB signaling pathway. Therefore, we firstly performed a luciferase reporter gene assays, and a luciferase plasmid with the top 2000 nt of the promoter domain of p65 (pGL4.32) was generated. Luciferase reporter assays demonstrated that the overexpression of UBD increased the luciferase activity of pGL4.32 in HEK293 (Fig. 5A). The opposite results were obtained after UBD knockdown (Fig. 5A). To explore the association between UBD and NF-κB signaling pathway in OSCC cells, the protein expressions of phosphorylated NF-κB p65, NF-κB p65, phosphorylated IκBα, and IκBα were examined by Western blot (Figs. 5B and 5C). Notably, we found that UBD increased the S536 phosphorylation levels of p65 (Fig. 5B). In contrast, knockdown of UBD decreased the phosphorylation levels of p65 (Fig. 5C). Moreover, we used the NF-κB transcription inhibitor JSH-23. We found that the expression of p65 in cells overexpressing UBD was higher than in untreated cells (Fig. 5D, Supplementary Fig. S1). Furthermore, immunofluorescence and cellular fractionation experiments showed that nuclear translocation of NF-κB p65 was enhanced in OSCC cells overexpress UBD. Compared with the control group, when UBD was overexpressed, p65 of HN6 entered into the nucleus, but p65 of HN6 shUBD cells was still in the cytoplasm (Figs. 5E and 5F). Taken together, our findings suggested that overexpression of UBD contributed to disease progression through activating NF-κB signaling pathway.
Fig. 5. UBD activated NF-κB signaling pathway in OSCC cells.
(A) Luciferase reporter assays showed that overexpression of UBD in HEK293 could upregulate the luciferase activities, while UBD knockdown in HEK293 will correspondingly reduce the luciferase activities. (B and C) Effects of overexpression or knockdown of UBD on the activation of NF-κB signaling pathway were detected by Western blot analysis. Data were presented as the mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant. (D) Overexpression of UBD rescued the expression of p65 suppressed by JSH-23. (E) Immunofluorescent staining of the p65 in indicated cells. (F) Cytoplasmic fractions and nuclear fractions were analyzed by immunoblotting with p65. LaminB1 was used as a nuclear protein marker.
DISCUSSION
UBD is a member of the ubiquitin-like protein family and is encoded in the HLA-F locus at the telomeric end of class I MHC (Groettrup et al., 2008). Accumulating evidence indicates that UBD is highly expressed in multiple cancers, while overexpressed UBD promotes the malignant progression of tumors. Yuan et al. (2014) found that UBD promoted tumor invasion and metastasis by modifying the degradation of β-catenin in liver cancer. Xue et al. (2016) found that UBD promoted tumor progression and led to drug-resistance development in non-small cell lung cancer. Herein, we demonstrate for the first time the role of UBD in the progression of OSCC and its potential value as a new therapeutic target for human cancer. As an ubiquitin, UBD positively affects both the catalytic and noncatalytic activities of OTUB1 in reducing the polyubiquitylation of its substrate protein TRAF3 (TNF receptor-associated factor 3) and the formation of ubiquitin conjugates in cellulo (Bialas et al., 2019). Deng et al. (2020) showed that UBD is directly bound to epidermal growth factor receptor (EGFR) in osteosarcoma. Furthermore, UBD knockdown significantly promoted EGFR ubiquitination, while overexpression of UBD reduced EGFR ubiquitination. Another study has shown that UBD stabilizes the expression of EGFR by regulating its degradation and ubiquitination effects, and positively regulates HK2 via the EGFR/AKT pathway to promote the development of bladder cancer (Zou et al., 2021). UBD also stabilizes the expression of Nav1.5 by antagonizing the ubiquitination and degradation of Nav1.5, thereby preventing the degradation of Nav1.5 protein. Loss of UBD in cardiomyocytes can lead to increased arrhythmia and mortality after myocardial infarction. Therefore, UBD prevents ischemic arrhythmia by binding to Nav1.5 and preventing its degradation (Liu et al., 2021). Through these studies, we found that UBD, as a member of the ubiquitin-like family, can exert its ubiquitination effect by stabilizing or degrading the expression of other proteins, whether in tumors or other diseases.
To investigate the correlation between UBD expression and the progression of OSCC, we first analyzed the expression of UBD in tissue microarrays. The clinical results indicated that the expression of UBD in OSCC patients was positively correlated with clinical progression and histological tumor grade. Therefore, UBD may become a potential therapeutic target for OSCC. Our gain- and loss-of-function studies showed that UBD induced OSCC cells to enhance invasiveness and migration; furthermore, the proliferation was also enhanced both in vivo and in vitro. This suggests that UBD is related to the carcinogenicity of OSCC. It has been previously shown that there is a molecular link between UBD and the NF-κB pathway; therefore, we investigated whether UBD exerts its function through the NF-κB pathway in OSCC. Our previous studies showed a significant correlation between NF-κB activity and invasion and proliferation of OSCC (Chu et al., 2014; Wang et al., 2016). The inhibition of NF-κB activity inhibited the migration and proliferation of tumor cells. Our immunofluorescence results showed that knockdown of UBD also decreased the nuclear fraction of the NF-κB key transcription factor p65 protein, while after the overexpression of UBD, the NF-κB pathway was significantly activated in the nucleus. Besides, this study also showed that overexpression of UBD rescued the activation of NF-κB transcription due to JSH-23 inhibition. The results of Western blot also showed that IkBα, a repressor of NF-κB, was downregulated in cells overexpressing UBD. Therefore, we conclude that UBD promotes OSCC tumorigenesis and development through the NF-κB signaling pathway, which defines a key mechanism for UBD-mediated activation of NF-κB signaling and identifies a novel molecule with therapeutic potential for the treatment of OSCC. The NF-κB pathway is an oncogenic signaling pathway that is closely related to tumor development, including cell cycle, invasion, migration, and proliferation (Awasthee et al., 2019; Yang et al., 2020). It has been confirmed that the NF-κB signaling pathway is activated in OSCC, demonstrating that the NF-κB signaling pathway plays a crucial role in the OSCC progression (Derakhshan et al., 2017; Wilken et al., 2011).
The EMT refers to the biological process by which epithelial cells transform into cells with mesenchymal phenotype through a specific program and is considered to be the main mechanism of cancer cell migration and invasion (Patel et al., 2015). The EMT process is characterized by the loss of epithelial cell adhesion molecules and their substitution by such mesenchymal markers as vimentin, fibronectin, collagen, and matrix metalloproteinases. In the present study, we found that overexpression of UBD significantly promoted cell invasion and migration. We observed that overexpression of UBD attenuated E-cadherin expression and increased the protein levels of N-cadherin and vimentin in HN6 and CAL27 cells, while after knocking down UBD, the results were reversed. We believe that the study of UBD for EMT and tumor metastasis will certainly open new horizons for the treatment of OSCC.
In conclusion, we demonstrated that UBD has a pro-malignant function in OSCC and exerts an oncogenic role by activating the NF-κB signaling pathway. Further elucidation of the mechanism of UBD in OSCC development would be helpful to provide new approaches for the treatment of OSCC and new anticancer strategies.
Supplemental Materials
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (81772887). Jiangsu Provincial Medical Innovation Team (CXTDA2017036), the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2018-87), Jiangsu Provincial Medical Youth Talent (QNRC2016854), and Natural Science Foundation of Jiangsu Province of China (BK20171488).
Footnotes
AUTHOR CONTRIBUTIONS
A.S. and Y.W. conceived the study design and drafted the article together. F.J. and E.Y. proofread the manuscript. J.Z. and J.Y. performed the experiments and analyzed the data. H.Z., X.D., G.L., and Y.W. performed some experiments and secured funding. Y.Z. and X.S. sought approval from the ethics committee, and provided funding.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Aichem A., Groettrup M. The ubiquitin-like modifier FAT10 in cancer development. Int. J. Biochem. Cell Biol. 2016;79:451–461. doi: 10.1016/j.biocel.2016.07.001. [DOI] [PubMed] [Google Scholar]
- Awasthee N., Rai V., Chava S., Nallasamy P., Kunnumakkara A.B., Bishayee A., Chauhan S.C., Challagundla K.B., Gupta S.C. Targeting IkappaappaB kinases for cancer therapy. Semin. Cancer Biol. 2019;56:12–24. doi: 10.1016/j.semcancer.2018.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai Y., Sha J., Kanno T. The role of carcinogenesis-related biomarkers in the Wnt pathway and their effects on epithelial-mesenchymal transition (EMT) in oral squamous cell carcinoma. Cancers (Basel) 2020;12:555. doi: 10.3390/cancers12030555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bialas J., Boehm A.N., Catone N., Aichem A., Groettrup M. The ubiquitin-like modifier FAT10 stimulates the activity of deubiquitylating enzyme OTUB1. J. Biol. Chem. 2019;294:4315–4330. doi: 10.1074/jbc.RA118.005406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappadocia L., Lima C.D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 2018;118:889–918. doi: 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai A.W.Y., Lim K.P., Cheong S.C. Translational genomics and recent advances in oral squamous cell carcinoma. Semin. Cancer Biol. 2020;61:71–83. doi: 10.1016/j.semcancer.2019.09.011. [DOI] [PubMed] [Google Scholar]
- Choi Y., Kim J.K., Yoo J.Y. NFkappaB and STAT3 synergistically activate the expression of FAT10, a gene counteracting the tumor suppressor p53. Mol. Oncol. 2014;8:642–655. doi: 10.1016/j.molonc.2014.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W., Song X., Yang X., Ma L., Zhu J., He M., Wang Z., Wu Y. Neuropilin-1 promotes epithelial-to-mesenchymal transition by stimulating nuclear factor-kappa B and is associated with poor prognosis in human oral squamous cell carcinoma. PLoS One. 2014;9:e101931. doi: 10.1371/journal.pone.0101931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X., Deng J., Yi X., Zou Y., Liu H., Li C., Deng B., Fan H., Hao L. Ubiquitin-like protein FAT10 promotes osteosarcoma glycolysis and growth by upregulating PFKFB3 via stabilization of EGFR. Am. J. Cancer Res. 2020;10:2066–2082. [PMC free article] [PubMed] [Google Scholar]
- Derakhshan A., Chen Z., Van Waes C. Therapeutic small molecules target inhibitor of apoptosis proteins in cancers with deregulation of extrinsic and intrinsic cell death pathways. Clin. Cancer Res. 2017;23:1379–1387. doi: 10.1158/1078-0432.CCR-16-2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eluard B., Thieblemont C., Baud V. NF-kappaB in the new era of cancer therapy. Trends Cancer. 2020;6:677–687. doi: 10.1016/j.trecan.2020.04.003. [DOI] [PubMed] [Google Scholar]
- Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer. 2010;127:2893–2917. doi: 10.1002/ijc.25516,. [DOI] [PubMed] [Google Scholar]
- Gao Y., Theng S.S., Zhuo J., Teo W.B., Ren J., Lee C.G. FAT10, an ubiquitin-like protein, confers malignant properties in non-tumorigenic and tumorigenic cells. Carcinogenesis. 2014;35:923–934. doi: 10.1093/carcin/bgt407. [DOI] [PubMed] [Google Scholar]
- Groettrup M., Pelzer C., Schmidtke G., Hofmann K. Activating the ubiquitin family: UBA6 challenges the field. Trends Biochem. Sci. 2008;33:230–237. doi: 10.1016/j.tibs.2008.01.005. [DOI] [PubMed] [Google Scholar]
- Harris J., Oliere S., Sharma S., Sun Q., Lin R., Hiscott J., Grandvaux N. Nuclear accumulation of cRel following C-terminal phosphorylation by TBK1/IKK epsilon. J. Immunol. 2006;177:2527–2535. doi: 10.4049/jimmunol.177.4.2527. [DOI] [PubMed] [Google Scholar]
- Harsha C., Banik K., Ang H.L., Girisa S., Vikkurthi R., Parama D., Rana V., Shabnam B., Khatoon E., Kumar A.P., et al. Targeting AKT/mTOR in oral cancer: mechanisms and advances in clinical trials. Int. J. Mol. Sci. 2020;21:3285. doi: 10.3390/ijms21093285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayden M.S., Ghosh S. NF-kappaB, the first quarter-century: remarkable progress and outstanding questions. Genes Dev. 2012;26:203–234. doi: 10.1101/gad.183434.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida K., Tomita H., Nakashima T., Hirata A., Tanaka T., Shibata T., Hara A. Current mouse models of oral squamous cell carcinoma: genetic and chemically induced models. Oral Oncol. 2017;73:16–20. doi: 10.1016/j.oraloncology.2017.07.028. [DOI] [PubMed] [Google Scholar]
- Kaltschmidt B., Greiner J.F.W., Kadhim H.M., Kaltschmidt C. Subunit-specific role of NF-kappaB in cancer. Biomedicines. 2018;6:44. doi: 10.3390/biomedicines6020044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto A., Nagata S., Anzai S., Takahashi J., Kawai M., Hama M., Nogawa D., Yamamoto K., Kuno R., Suzuki K., et al. Ubiquitin D is upregulated by synergy of Notch signalling and TNF-alpha in the inflamed intestinal epithelia of IBD patients. J. Crohns Colitis. 2019;13:495–509. doi: 10.1093/ecco-jcc/jjy180. [DOI] [PubMed] [Google Scholar]
- King K.E., Ponnamperuma R.M., Allen C., Lu H., Duggal P., Chen Z., Van Waes C., Weinberg W.C. The p53 homologue DeltaNp63alpha interacts with the nuclear factor-kappaB pathway to modulate epithelial cell growth. Cancer Res. 2008;68:5122–5131. doi: 10.1158/0008-5472.CAN-07-6123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Ge J., Chen C., Shen Y., Xie J., Zhu X., Liu M., Hu J., Chen L., Guo L., et al. FAT10 protects against ischemia-induced ventricular arrhythmia by decreasing Nedd4-2/Nav1.5 complex formation. Cell Death Dis. 2021;12:25. doi: 10.1038/s41419-020-03290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Xiong H., Chen L., Liu X., Zou S., Guan J., Wang K. GRP78 promotes hepatocellular carcinoma proliferation by increasing FAT10 expression through the NF-kappaB pathway. Exp. Cell Res. 2018;365:1–11. doi: 10.1016/j.yexcr.2018.02.007. [DOI] [PubMed] [Google Scholar]
- Mortezaee K., Najafi M., Farhood B., Ahmadi A., Shabeeb D., Musa A.E. NF-kappaB targeting for overcoming tumor resistance and normal tissues toxicity. J. Cell. Physiol. 2019;234:17187–17204. doi: 10.1002/jcp.28504. [DOI] [PubMed] [Google Scholar]
- Neumann M., Naumann M. Beyond IkappaBs: alternative regulation of NF-kappaB activity. FASEB J. 2007;21:2642–2654. doi: 10.1096/fj.06-7615rev. [DOI] [PubMed] [Google Scholar]
- Panarese I., Aquino G., Ronchi A., Longo F., Montella M., Cozzolino I., Roccuzzo G., Colella G., Caraglia M., Franco R. Oral and oropharyngeal squamous cell carcinoma: prognostic and predictive parameters in the etiopathogenetic route. Expert Rev. Anticancer Ther. 2019;19:105–119. doi: 10.1080/14737140.2019.1561288. [DOI] [PubMed] [Google Scholar]
- Patel S., Shah K., Mirza S., Daga A., Rawal R. Epigenetic regulators governing cancer stem cells and epithelial-mesenchymal transition in oral squamous cell carcinoma. Curr. Stem Cell Res. Ther. 2015;10:140–152. doi: 10.2174/1574888x09666141020163700. [DOI] [PubMed] [Google Scholar]
- Petersen P.E. Oral cancer prevention and control--the approach of the World Health Organization. Oral Oncol. 2009;45:454–460. doi: 10.1016/j.oraloncology.2008.05.023. [DOI] [PubMed] [Google Scholar]
- Qing X., French B.A., Oliva J., French S.W. Increased expression of FAT10 in colon benign, premalignant and malignant epithelial neoplasms. Exp. Mol. Pathol. 2011;90:51–54. doi: 10.1016/j.yexmp.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siriwardena S., Tsunematsu T., Qi G., Ishimaru N., Kudo Y. Invasion-related factors as potential diagnostic and therapeutic targets in oral squamous cell carcinoma-a review. Int. J. Mol. Sci. 2018;19:1462. doi: 10.3390/ijms19051462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sovak M.A., Bellas R.E., Kim D.W., Zanieski G.J., Rogers A.E., Traish A.M., Sonenshein G.E. Aberrant nuclear factor-kappaB/Rel expression and the pathogenesis of breast cancer. J. Clin. Invest. 1997;100:2952–2960. doi: 10.1172/JCI119848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbichler T.B., Savic D., Dudas J., Kvitsaridze I., Skvortsov S., Riechelmann H., Skvortsova I.I. Cancer stem cells and their unique role in metastatic spread. Semin. Cancer Biol. 2020;60:148–156. doi: 10.1016/j.semcancer.2019.09.007. [DOI] [PubMed] [Google Scholar]
- Tampa M., Mitran M.I., Mitran C.I., Sarbu M.I., Matei C., Nicolae I., Caruntu A., Tocut S.M., Popa M.I., Caruntu C., et al. Mediators of inflammation - a potential source of biomarkers in oral squamous cell carcinoma. J. Immunol. Res. 2018;2018:1061780. doi: 10.1155/2018/1061780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzuoli E., Bellan C., Aversa S., Ciccone V., Morbidelli L., Giachetti A., Donnini S., Ziche M. ALDH3A1 overexpression in melanoma and lung tumors drives cancer stem cell expansion, impairing immune surveillance through enhanced PD-L1 output. Cancers (Basel) 2019;11:1963. doi: 10.3390/cancers11121963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Zhao B. UBA6 and its bispecific pathways for ubiquitin and FAT10. Int. J. Mol. Sci. 2019;20:2250. doi: 10.3390/ijms20092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Chen J., Zhang W., Zheng Y., Wang Z., Liu L., Wu H., Ye J., Zhang W., Qi B., et al. Axon guidance molecule semaphorin3A is a novel tumor suppressor in head and neck squamous cell carcinoma. Oncotarget. 2016;7:6048–6062. doi: 10.18632/oncotarget.6831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Zhu W.G., Xu X. Ubiquitin-like modifications in the DNA damage response. Mutat. Res. 2017;803-805:56–75. doi: 10.1016/j.mrfmmm.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Wilken R., Veena M.S., Wang M.B., Srivatsan E.S. Curcumin: a review of anti-cancer properties and therapeutic activity in head and neck squamous cell carcinoma. Mol. Cancer. 2011;10:12. doi: 10.1186/1476-4598-10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Y., Shen S., Verma I.M. NF-kappaB, an active player in human cancers. Cancer Immunol. Res. 2014;2:823–830. doi: 10.1158/2326-6066.CIR-14-0112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang S., Shao X., Cao J., Yang B., He Q., Ying M. FAT10: function and relationship with cancer. Curr. Mol. Pharmacol. 2020;13:182–191. doi: 10.2174/1874467212666191113130312. [DOI] [PubMed] [Google Scholar]
- Xue F., Zhu L., Meng Q.W., Wang L., Chen X.S., Zhao Y.B., Xing Y., Wang X.Y., Cai L. FAT10 is associated with the malignancy and drug resistance of non-small-cell lung cancer. Onco Targets Ther. 2016;9:4397–4409. doi: 10.2147/OTT.S98410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Lei J., Chen L., Deng H., Dong D., Jin T., Liu X., Yuan R., Qiu Y., Ge J., et al. Human leukocyte antigen F locus adjacent transcript 10 overexpression disturbs WISP1 protein and mRNA expression to promote hepatocellular carcinoma progression. Hepatology. 2018;68:2268–2284. doi: 10.1002/hep.30105. [DOI] [PubMed] [Google Scholar]
- Yang H., Kuo Y.H., Smith Z.I., Spangler J. Targeting cancer metastasis with antibody therapeutics. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2021 doi: 10.1002/wnan.1698. 2021 Jan 18 [Epub]. https://doi.org/10.1002/wnan.1698. [DOI] [PubMed] [Google Scholar]
- Yang L., Shi P., Zhao G., Xu J., Peng W., Zhang J., Zhang G., Wang X., Dong Z., Chen F., et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct. Target. Ther. 2020;5:8. doi: 10.1038/s41392-020-0110-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanjia H., Xinchun J. The role of epithelial-mesenchymal transition in oral squamous cell carcinoma and oral submucous fibrosis. Clin. Chim. Acta. 2007;383:51–56. doi: 10.1016/j.cca.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Yuan J., Tu Y., Mao X., He S., Wang L., Fu G., Zong J., Zhang Y. Increased expression of FAT10 is correlated with progression and prognosis of human glioma. Pathol. Oncol. Res. 2012;18:833–839. doi: 10.1007/s12253-012-9511-2. [DOI] [PubMed] [Google Scholar]
- Yuan R., Wang K., Hu J., Yan C., Li M., Yu X., Liu X., Lei J., Guo W., Wu L., et al. Ubiquitin-like protein FAT10 promotes the invasion and metastasis of hepatocellular carcinoma by modifying beta-catenin degradation. Cancer Res. 2014;74:5287–5300. doi: 10.1158/0008-5472.CAN-14-0284. [DOI] [PubMed] [Google Scholar]
- Zamo A., Malpeli G., Scarpa A., Doglioni C., Chilosi M., Menestrina F. Expression of TP73L is a helpful diagnostic marker of primary mediastinal large B-cell lymphomas. Mod. Pathol. 2005;18:1448–1453. doi: 10.1038/modpathol.3800440. [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu S., Cai G., Kong L., Zhang T., Ren Y., Wu Y., Mei M., Zhang L., Wang X. Long non coding RNA MALAT1 promotes tumor growth and metastasis by inducing epithelial-mesenchymal transition in oral squamous cell carcinoma. Sci. Rep. 2015;5:15972. doi: 10.1038/srep15972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zielinska K.A., Katanaev V.L. Information theory: new look at oncogenic signaling pathways. Trends Cell Biol. 2019;29:862–875. doi: 10.1016/j.tcb.2019.08.005. [DOI] [PubMed] [Google Scholar]
- Zou Y., Du Y., Cheng C., Deng X., Shi Z., Lu X., Hu H., Qiu J., Jiang W. FAT10 promotes the progression of bladder cancer by upregulating HK2 through the EGFR/AKT pathway. Exp. Cell Res. 2021;398:112401. doi: 10.1016/j.yexcr.2020.112401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.