Abstract
Collagen fibrils are essential for metazoan life. They are the largest, most abundant, and most versatile protein polymers in animals, where they occur in the extracellular matrix to form the structural basis of tissues and organs. Collagen fibrils were first observed at the turn of the 20th century. During the last 40 years, the genes that encode the family of collagens have been identified, the structure of the collagen triple helix has been solved, the many enzymes involved in the post-translational modifications of collagens have been identified, mutations in the genes encoding collagen and collagen-associated proteins have been linked to heritable disorders, and changes in collagen levels have been associated with a wide range of diseases, including cancer. Yet despite extensive research, a full understanding of how cells assemble collagen fibrils remains elusive. Here, we review current models of collagen fibril self-assembly, and how cells might exert control over the self-assembly process to define the number, length and organisation of fibrils in tissues.
Keywords: Collagen, Tendon, Self-assembly, Plasma membrane, Diameter, Polymer length, Mechanical properties
Collagen assembly is a complex multiscale problem
Collagen fibrils appeared at the dawn of multicellular evolution [1], [2], [3], when cells acquired the ability to construct tissues using semi-rigid extracellular polymers to provide long-range mechanical connectivity and sites for cell attachment. These fibrils have evolved to be the primary tensile element in a wide range of tissues, where they are organized into architectural structures ranging from gels, to parallel bundles, and orthogonal lattices.
Collagen fibrils are roughly cylindrical with two tapered tips [4], can be up to 500 nm in diameter, and centimetres in length [5]. The remarkable versatility of collagen to assemble into fibrils of different sizes and pack into a variety of tissue architectures explains how cells were able to construct the skeletons of dinosaurs exceeding 40 m in length, yet use the same fibrils to build a transparent cornea or the stapedius tendon, which is a mere 125 μm in length [6]. The fibrils range in abundance from a few percent in endocrine organs such as pancreas [7] to 15% in lung [8] and 90% of the mass of tissues such as tendon [9], [10]. Overall, collagen fibrils account for 25% of total protein in vertebrates [11].
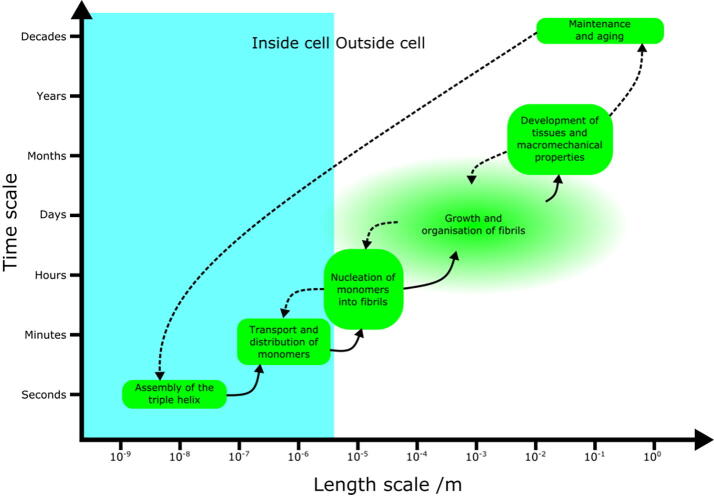
Collagen self-assembly is an inherently multiscale process, in both space and time, that involves the aggregation of monomers to form larger-scale organised structures [Fig. 1]. A multiscale system integrates processes occurring at different length and time scales, making any attempt at understanding the system as a whole challenging. For example, collagen fibrils may be centimetres in length [5], and yet they are formed from monomers with a diameter of only ~1.5 nm; monomers can be produced by cells at a rate of 2x105 per hour [12], and yet crosslinked collagen structures survive for decades. Understanding the assembly and organisation of fibrils thus crosses 8 orders of magnitude in space and 4–5 orders of magnitude in time. Significant challenges exist in bridging this gap in scales. Progress has been made at the macro (large) scale in terms of understanding fibril and tissue mechanics, and at the micro (small) scale in the chemistry of collagen monomers, but the intermediate (mesoscale) regime remains poorly understood. However, the mesoscale regime contains some of the most interesting and fundamental problems in collagen fibril assembly. In this review, we will outline current understanding of collagen fibril assembly. We will first summarise the basic properties of collagen, before describing how in vitro studies, and corresponding mathematical models, have given hints at understanding its self-assembly. Finally, we will outline the limitations of this work, how in vivo collagen assembly differs from self-assembly observed in vitro, and how new approaches could further our understanding of this problem.
Fig. 1.
Diagram to demonstrate how processes involved in the assembly of collagenin vivo cross multiple scales in both space and time. Lines between processes indicate interactions; those lines that are dotted show hypothesised or poorly understood links. The precise length and time scales of the growth and organisation of fibrils in vivo are unknown, as demonstrated by the faded edges of this process bubble, but it is clear that this particular step crosses several scales and is critical for in vivo development.
The multiscale nature of collagen
Collagen is multiscale in space
Collagen monomers are ~300 nm length and ~1.36 nm in diameter in solution [13], with an effective packing diameter of 1.52 nm [14], and assemble in their millions to generate fibrils with a wide variety of diameters. The narrowest collagen fibrils occur in vitreous humor (~10.5–12.0 nm) [15] and cartilage, where “thin” (~16 nm) and thicker (~25–50 nm) fibrils occur [16] alongside hyaluronan and large proteoglycans in a fibre composite gel that resists compression. Relatively thin fibrils (~25 nm) can be found in cornea [17], where they are organized in an exquisite orthogonal lattice [18], [19] that is mechanically strong and yet completely transparent. In contrast, “thick” collagen fibrils occur in tendon, where the ability of the fibrils to transmit high forces is important. “Narrow” (~50 nm) fibrils are deposited by embryonic tenocytes into bundles that are parallel to the tendon long axis. However, soon after birth (in mouse) this unimodal distribution of diameters changes to a multimodal distribution with three peaks at 50 nm, 150 nm, and 250 nm [20].
Much is known about the diameter of fibrils in tissues, but relatively little is known about fibril length. The magnification required to resolve a single fibril in a tissue is such that tracking the fibril from one end to another would require a prohibitively large number of separate images, so direct measurement of fibril lengths in vivo is extremely challenging. Theoretical work based on the frequency of fibril tips in sample images has estimated mean fibril lengths on the order of 10 mm in rat tail [5]. However, the paucity of fibril ends (or tips) introduces large errors in this estimate. Other studies have observed whole fibrils in echinoderms (starfish) and found a maximum length of 600 μm [21].
To further complicate matters, fibrils can fuse by tip-to-tip interactions [22] and by tip-to-shaft interactions to generate Y-shaped branches [23]. Consequently, the question of “How long is a fibril?” is not simple to answer. It most probably varies with developmental stage, anatomical site, and the stage of growth of the fibril. However, if Y-shaped structures are common, then the fibril length is poorly defined. What is clear, however, is that collagen monomers can be assembled into structures several orders of magnitude longer than the monomers themselves.
Collagen is multiscale in time
In vertebrates, collagen fibril assembly begins when the vascular and musculoskeletal systems need to withstand hydrodynamic, compressive, and tensile loads. In mouse, fibril assembly begins at around 14.5 days after fertilisation (embryonic day 14.5 or E14.5) [24], and around embryonic day 10 in chick [25]. By the end of embryonic development, just 9 days later in mice, the fibrils have grown in number and length sufficiently to transmit forces through tendons and ligaments [Fig. 4] for locomotion, and establish the fibrous framework of organs including liver, lung, blood vessels, and skin. This underscores that assembly and organisation of collagen can occur relatively quickly. However, studies in mice have shown that the three-dimensional organization of fibrils that was established during embryonic and early postnatal growth remains unchanged after birth [26]. Thus, the collagen fibril bundles in tendon and ligament, basketweaves in skin and blood vessels, and orthogonal lattices in cornea, form stable, lifelong frameworks, maintained over orders of magnitude longer than the timescales of their original assembly.
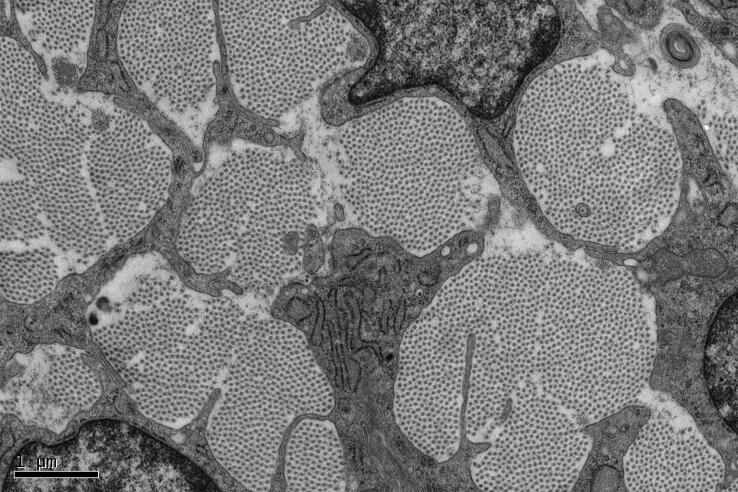
Fig. 4.
Electron micrograph showing a cross section of mouse tail tendon at embryonic 18.5 days. Collagen fibrils appear as bundles of regularly-spaced dark spots, demonstrating the perfect alignment and high degree of spatial order in the long-range organisation of tendon fibrils.
Not only is the collagen matrix effectively permanent, but the constituent fibrils are too. It has been shown that the collagen in fibrils in tendon and cartilage remain throughout life without renewal or turnover [27], [28], [29]. This means that the collagen molecules that are assembled into fibrils during embryonic growth are the same molecules that are in the tissue when the organism dies, perhaps decades later. The high stability of collagen is also apparent in collagens that have been extracted from the remains of extinct animals preserved in permafrost [30], [31], where other proteins and nucleic acids have long since degraded.
The permanency of collagen has advantages: for example, the fibrils provide a stable framework that shields cells against damaging forces. It is estimated that the tendons in a galloping horse experience ~16 kN per kilogram of body mass [32], forces that would readily destroy the resident tenocyte cells if it were not for the stressshielding properties of the collagen fibrils that make up the majority of the mass of the tissue. However, the permanence of fibrils means that if they are damaged or removed by injury or disease, they cannot be replaced in a like-for-like manner. For example, during wound-healing, the new collagen fibrils that are deposited have a different organization and packing compared to the surrounding tissue, leading to the appearance of a scar. These observations demonstrate that collagen crosses orders of magnitude in time, much as it does in space.
Molecular structure and spatial organisation of collagen
Collagen nomenclature
The criteria for classifying a protein as a collagen have not been established precisely. All collagens comprise three polypeptide chains (called α-chains) wound into a supercoiled triple helix in which each α-chain is formed from repeating Gly-X-Y triplets [33], a residue of glycine, frequently combined with imino acids proline and hydroxyproline [34], [35]. As with all proteins, these polypeptide chains have an N- and C- terminal end; in collagens, the triple helix is formed by association of the C-termini and zippering of the helix in a C to N direction [36]. However, not all proteins that contain a triple helix are considered collagens [37]. It is generally agreed that there are 28 distinct “types” of collagen in vertebrates [37], [38], but almost 200 in C. elegans [39].
Collagens can be homotrimers comprising α-chains encoded by a single gene or heterotrimers comprising two or three different types of α-chain encoded by different genes. Each α-chain is identified by an Arabic number and a Roman numeral in parentheses. For example, type II collagen has three identical α1(II) chains and type III collagen has three identical α1(III) chains, encoded by genes Col2a1 and Col3a1, respectively. In the gene nomenclature, the first Arabic number refers to the collagen “type” and the second Arabic number refers to the α-chain. Some collagens are heterotrimers of two or three different α-chains. For example, type I collagen is a heterotrimer of α1(I)2α2(I) encoded by two genes, Col1a1 and Col1a2. Type V collagen is an example of a heterotrimeric collagen that can exist in two forms: α1(V)2α2(V) and α1(V)α2(V)α3(V).
Collagen molecules are synthesized as procollagens
The classical fibril-forming collagens are types I, II, III, V, and XI, with collagens XXIV and XXVII included on the basis of structural similarities. Analysis of protein-coding variations in the human genome showed that genes encoding collagens I, II, III, V, XI and XXVII are intolerant to protein-truncating variants [40], which definitively illustrates the requirement of collagen fibrils for life.
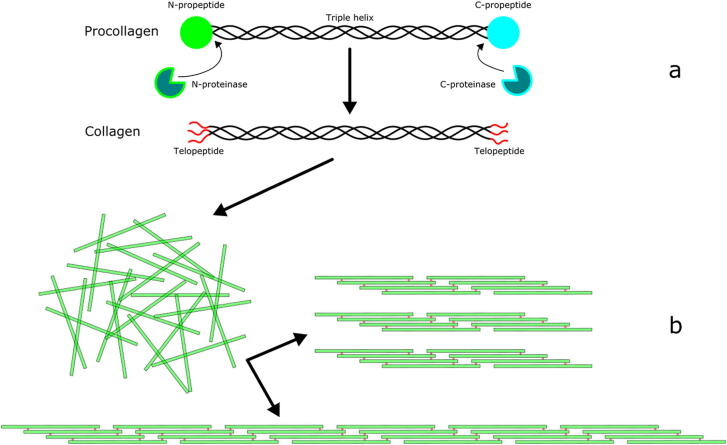
An important feature of fibrillar collagens is that they are synthesized as procollagens, containing an uninterrupted triple helix with globular domains at each end. These globular domains are the N- and C-propeptides, which are removed by procollagen N-proteinases and C-proteinases, respectively [Fig. 2a]. There are three N-proteinases: ADAMTS-2, −3, and −14 [41]. It has been shown that the N-proteinases exhibit maximal activity when procollagen is properly folded into a triple helix [42]. In fact, mutations in collagen genes that change the structure of the triple helix (even hundreds of residues C-terminal to the N-proteinase cleavage sites) can stop cleavage of the N-propeptides [43], [44] and result in abnormal fibril assembly [45]. It was postulated in these studies that as the procollagen triple helix folds in a C to N direction, changes in chain alignment caused by amino acid substitutions or deletions would be propagated to the N-terminus and change the conformation of the procollagen molecule where N-proteinase cleaves. The reason why N-proteinases require a triple helical conformation of procollagen molecule is unclear. Electron microscope evidence suggests that the N-propeptides of type I procollagen are in a “bent back” conformation [46], which might present the scissile bonds to each of the three α-chains to N-proteinase. Whether or not the N-propeptides of type II and III procollagen are also ’bent back’ needs to be confirmed. These observations point to the N-propeptides having more than simply a propeptide function to help keep procollagen soluble, but might instead suggest an important role in controlling fibril assembly. In contrast, the C-proteinases (which are members of the BMP1/mTld family) have a broad substrate specificity not restricted to procollagens [47].
Fig. 2.
a, Diagram of procollagen cleavage to collagen by removal of the N- and C-propeptides from a monomer. b, Diagram showing two possible distribution outcomes for 24 collagen monomers.
Following the removal of propeptides, each end of the collagen triple helix terminates at a set of short extra-helical peptides, known as telopeptides [Fig. 2a]. These non–helical strands are critical for normal fibril assembly, and are involved in collagen cross-linking, mediated by the lysyl-oxidase enzyme (LOX), which permanently binds adjacent monomers together. Proteolytic removal of telopeptides has been shown to inhibit collagen I fibril self-assembly [48], and adding synthetic telopeptide-like peptides to a self-assembly system inhibits fibril assembly by blocking binding between telopeptides and neighbouring monomers [49].
The presence of a propeptide (or proprotein) domain that renders a protein inactive is typical of secreted proteinases, growth factors and hormones. The presence of two propeptides, as in procollagen, is unusual. Given the impact of propeptide cleavage on assembly, it is possible that having two propeptides, and different families of proteinases to remove them, provides spatial and temporal control of fibril assembly. Removal of the C-propeptides, to generate pNcollagen, dramatically decreases the solubility of procollagen and is essential for fibril formation [50]. Removal of the N-propeptides is not essential for fibril formation. In the case of type I procollagen, retention of the N-propeptides can modulate the shape of fibrils in vitro [51] and in vivo [46]. Retained N-propeptides of type I and III collagen have been detected in human skin [52].
Collagen fibrils are “molecular alloys”
Collagen fibrils are heterotypic polymers comprising 3 main components: a “major” fibrillar collagen, specifically type I or II collagen; a “minor” fibrillar collagen such as type V in cornea [53] or type XI in cartilage; and a FACIT (Fibril Associated Collagen with Interrupted Triple helices) collagen such as type IX, XII, or XIV, which can also bind a wide range of soluble and membrane-associated molecules [54], [55], [56]. Absence of type I [57] or II [58] collagen leads to major skeletal deficiencies and is incompatible with life; mutations in genes encoding these collagens result in severe skeletal disorders, notably (but not exclusively) osteogenesis imperfecta and achondrogenesis, respectively. Mutations in genes encoding type V and XI collagens cause the Ehlers-Danlos and Stickler/Marshall syndromes, in which affected individuals can have a range of skeletal and occular problems [59]. Studies in developing mouse have shown that the core fibrillar network in cartilage is a cross-linked copolymer of collagens II, IX, and XI [60]. Of special interest, autosomal recessive chondrodysplasia (cho) in mice lacking collagen XI affects cartilage in limbs, ribs, mandibles, and trachea, and is accompanied by the absence of thin fibrils and the appearance of thick fibrils [61]. The thin fibrils have an exquisite 10 + 4 microfibril structure with the inner core and outer shell of microfibrils tilted by ~3° to the fibril axis [62]. The absence of thin fibrils in the cho/cho mouse suggests that collagen XI is either required to initiate the assembly of the thin fibrils, or has a major role in limiting the lateral growth of fibrils.
In fibrous, mineralized and vascular tissues, the fibrils are predominately type I collagen with smaller amounts of type III and V collagen, and can have either type XII or XIV at their surfaces [63]. Studies have shown that collagen-I containing fibrils do not form in the absence of collagen V in vivo [64], which illustrates the importance of type V collagen in the assembly of type I collagen-containing fibrils. Collagen XII- and XIV-null mice demonstrate delayed endothelial maturation [65], and mutations in Col12a1 cause myopathic Ehlers-Danlos Syndrome with a clinical phenotype involving both joints and tendons [66]. The fibrils can also bind small proteoglycans including decorin [67], [68], fibromodulin [69], and lumican [70], with important roles in controlling fibrillar alignment and size [71].
These studies have shown that fibrils formed in vivo are “molecular alloys” (the term first coined by [72]) comprising a major, minor, and FACIT collagen. The ability of type I collagen to co-polymerise with types III and V, and for type II collagen to co-polymerise with collagens types XI and III [73], and for these fibrils to bind a variety of molecules at their surfaces, helps to explain the versatility of fibrils as the primary scaffolding component of tissues. However, precisely how the multitude of combinations are controlled in vivo remains unclear.
Long-range organization of fibrils
The three-dimensional organization of the fibrils within a tissue plays a crucial role in determining its mechanical properties, and this organisation varies radically from tissue to tissue. The orientation, degree of alignment, cross-link density, volume fraction, and relative length distribution of the fibrils all contribute to the differing mechanical needs of each tissue. The orientation of the fibrils determines what kinds of loads the tissue can resist effectively. In tendons, the fibrils are strongly aligned with the tendon’s longitudinal axis to withstand uniaxial tensile loading [74]; in arteries, fibrils are arranged helically in lamellar units with alternating chirality [75] to resist radial deformations; in skin, fibrils exhibit a high degree of dispersion, which provides resistance to deformations in multiple directions [76]. In some ways, these tissues behave, mechanically, like modern, man-made, fibre-reinforced composite materials, which are commonly produced using glass, carbon, or aramid fibres for engineering applications. In both cases, the fibres are the main load-bearing components as they have a much higher tangent modulus than the matrix in which they are embedded. Uniaxial composites (like tendons) display transversely isotropic material behaviour, whereas laminated materials (like arteries) are orthotropic. As a result, many of the mathematical methods that are used to model these materials [77] can be borrowed and adapted to model soft tissues. In contrast to these engineering composites, in which the fibres are often embedded in a relatively stiff, solid matrix such as cured epoxy, however, collagen fibres are embedded in a proteoglycan-rich, viscous matrix, which enables the fibres to slide relative to each other [78]. To mimic this structure, biomaterials scientists have created soft composites, consisting of electrospun [79], solution blow spun [80], or three-dimensionally printed [81] polymer microfibres embedded in a hydrogel matrix. The fibres provide mechanical reinforcement, whilst the hydrogels can be loaded with biological agents to improve biocompatibility and allow fibre sliding.
The relative length distribution of the fibrils directly determines the shape of a tissue’s stress–strain curve [76], [82], [83]. Within a fixed length of tissue at rest, the fibrils all have an arc length that is longer than the section of tissue, with this extra length being accommodated by fibril tortuosity, or crimp. Each fibril has a slightly different arc length to the others, producing a distribution. As a tissue is stretched, its fibrils straighten one-by-one and contribute to the stiffness of the tissue only once taut. This process is often called collagen recruitment [84], [85], and gives rise to a J-shaped stress–strain curve that is characteristic of all soft tissues. By manufacturing solution blow spun biomaterials with a distribution of wavy fibres, it has recently become possible to reproduce this J-shaped stress–strain behaviour [80].
The role of cross-link density in collagen fibrils is clear: the higher the cross-link density, the stiffer the tissue [86]. Fibril volume fraction is also thought to affect tissue mechanics, but the relationship is less clear. Theoretically, since collagen is generally much stiffer than other extracellular matrix components, the larger the ratio of collagen to non-collagen, the stiffer the tissue should be [87]. Mathematical models often assume that the amount of strain energy stored by the collagenous and non-collagenous parts of the extracellular matrix is linearly proportional to their respective volume fractions [88], [89]. It is extremely difficult to test experimentally whether this theory holds in soft tissues, however, as this would require the production of tissue samples that differ only in their collagen fibril volume fraction, without also affecting other factors such as the relative length distribution.
A large and continually developing body of work has begun to disentangle the relationships between collagen architecture and tissue mechanical function, but the mechanisms by which the cells produce such complex three-dimensional structures remain largely unknown. However, it is reasonable to suppose that the lengths, diameters, and organisation of fibrils play a crucial role in determining tissue mechanical properties, and therefore cells must exert careful control of these factors during development.
Collagen self-assembly
Self-assembly in vitro
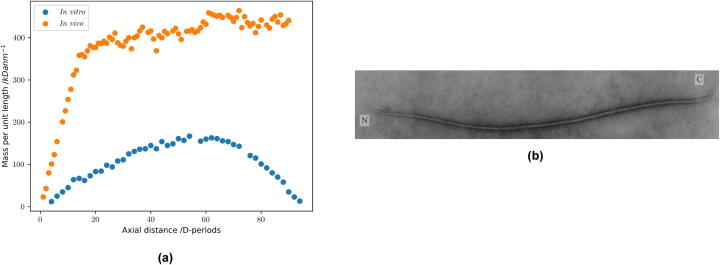
The nucleation and growth of collagen fibrils can occur in a minimal system of purified collagen molecules in a warm, neutral buffer [90], [91], and indicates an intrinsic tendency to self-assemble. Warming a cold acetic acid solution of collagen, followed by neutralisation, results in an accumulation of early collagen fibrils with smoothly tapered tips when less than 1% of the collagen has assembled [90]. These fibrils are unipolar, meaning the N-to-C orientations of all monomers are aligned [Fig. 3b]. They are typically ~20 nm in diameter and ~5 μm in length. A diameter limitation is evident at this early stage of growth, as the tips show a gradual flattening of the mass profile [Fig. 3a], but lack the abrupt diameter limitation observed in the tips of collagen fibrils formed in embryonic tissues [92]. These early fibrils can readily fuse as their concentration increases and this process contributes to an increase in fibril size leading to the broad range of diameters in the final gel, typically 20–80 nm [90]. Growing short reconstituted fibril seeds in a dilute collagen solution avoids inter-fibrillar fusion, allowing individual fibrils to grow in length at uniform diameter [93]. The fibril assembly pathway is critically dependent both on the intactness of the telopeptides [94] and also on the order of warming and neutralisation of the initial cold, acid solution of collagen: neutralisation followed by warming leads to an accumulation of thin filaments in the early stages of fibril assembly, rather than short fibrils with tapered ends [90].
Fig. 3.
a, Comparison of fibril mass profiles in vivo and in vitro. b, Image of a single collagen fibril formed in vitro. The N and C ends of the fibril are deduced by analysis of the D-periodic staining pattern. This fibril is unipolar with a length of 6 μm.
An alternative fibril assembly system, to match the steps occurring in tissue, is based on a starting solution of pCcollagen and C-proteinase [95]. Fibrils assemble along with the enzymatic cleavage of the pCcollagen to form the collagen monomer. These fibrils are found to have two polarised tips with C-termini of the molecules in both tips pointing to a point of maximum diameter in the fibril where molecules are in anti-parallel register [96]. Fibrils of this polarity type, with two N-terminal tips and a central region of polarity reversal, have subsequently been identified in tissue and are described as N,N bipolar fibrils [97], [98]. Despite the symmetry of molecular polarity in the two tips, the fibrils generated in this cell-free system have a shape polarity with a relatively coarse α-tip and fine β-tip. The tips show a near uniform increase in mass per unit length with distance from the end [4]. The gradients of mass per unit length are found to have a dependence on the C-proteinase to pCcollagen ratio used in the assembly system, with higher enzyme:substrate ratios giving rise to greater mass slopes at the tips [99].
Evidence of surface nucleation in collagen fibril growth was obtained from a simple seeding experiment where fibril length fragments from chick embryonic tendon were used as seeds in a solution of type I collagen [100]. Almost all fibril fragments initially had square broken ends after the mechanical extraction procedure, but ~95% of these formed spurs of growth in the solution of collagen monomer, which slowly elongated over several hours to form smoothly tapered tips.
Self-assembly in silico
Theoretical and computational models provide important tools in understanding the mechanisms of complex systems, with the ultimate aim being to formulate a complete multiscale model of a given system. For collagen assembly, the inherent diversity of length and time scales in the hierarchical self-assembly process makes a full multiscale theoretical understanding particularly challenging [Fig. 1]. This problem motivates a variety of theoretical approaches; each approach typically addresses a specific scale in length and time.
An organising principle acting across all scales is the idea of free energy minimisation. An energy that reflects the intermolecular interactions, which could be electrostatic, covalent or otherwise, plus any external constraints, such as spatial confinement, can be associated with any configuration of monomers. Self-assembly involves changes in molecular configurations to lower free energy states, against a viscous drag from the environment, generally in the presence of noise associated with Brownian motion. There are likely to be multiple minima in the free energy landscape, the majority representing disorganised configurations, and a fundamental question is how the self-assembly process can be driven towards those few states with high spatial organisation.
Existing work on macromolecular assembly has typically involved explicit, discrete Brownian dynamics, Monte-Carlo, or molecular dynamics models [101], [102]. Collagen can be modelled as a semi-flexible diffusive rod using a discrete worm-like-chain model [103] with Brownian motion. However, the narrow diameter of a collagen triple helix relative to the scale of a fibril ensures that any explicit simulation of assembly will impose a large computational cost; very small spatial perturbations could result in very large changes in force for neighbouring monomers, requiring very small time steps in system integration. Such small time steps make simulating the diffusion of a monomer over the lengthscale of a fibril challenging. This computational cost limits the scale of such simulations to small systems in space, or short simulations in time. For example, explicit modelling of collagen triple helix folding has been performed using the OpenMM molecular dynamics library [104], but such systems require only 3 individual worm-like-chains [105]. Modelling larger systems comes with a penalty in simulation length. This issue is exemplified by the work of McCluskey et al. [106], who use the LAMMPS molecular dynamics library [107] to model the in vitro self-assembly of collagen. Each collagen monomer is treated as a flexible chain of 200 1.5 nm beads. This chain is divided into nine regions, with even-numbered segments being slightly longer (37.5 nm) than the odd-numbered ones (30 nm), and attractive interactions between even segments in separate monomers, attractive interactions between odd segments, and repulsive interactions between even and odd segments. Monomers are predicted to aggregate first into dimers and trimers, then into long, narrow filaments a few monomers in cross-section. Thus they predict that axial growth precedes lateral growth, and that monomers aggregate into initially disordered fibrils, which slowly cystrallize into an ordered state with a clear D-band pattern. Accompanying experiments reveal the timescale of the process, showing a rise in turbidity over 20–40 min as monomers in solution assemble and crystallize into fibrils, and the model shows some success in matching this behaviour. However, this model is subject to the computational limits discussed previously, and is only able to model relatively small assembly systems for a short period of time. Furthermore, it gives no consideration to cross-linking, despite experiments in the paper demonstrating that telopeptides are required for assembly.
These computations [106] represent the current state of the art in simulating collagen self-assembly. Similar work, also using LAMMPS, has studied assembly of “collagen-mimetic” rods [108], but neglects specific details of collagen itself to achieve simpler dynamics. Other recent studies using coarse-grained molecular dynamics have explored the influence of axial stretch on the organisation of the assembled fibril [109], but without simulating the origin of this organisation. Additional work has studied the self-assembly of rod-like particles [110], and of collagen specifically [111], [112], by diffusion limited aggregation, but it is notable that such principles lead to branching structures that lack the hallmarks of collagen fibril self-assembly.
It is worth comparing modelling work on collagen assembly to the broad literature of theoretical work on actin assembly [113], microtubule assembly [114], [115], and amyloid plaque formation [116]. These systems appear similar: all involve the formation of filamentous structures from smaller subunits. However, collagen is again distinguished by its multiple lengthscales. Actin, microtubules, and amyloid plaques form much shorter fibres from globular subunits that lack the enormous aspect ratio of a collagen monomer. These differences make such systems more tractable in traditional particle-based modelling.
Collagen assembly in vivo
Assembly in vitro does not explain in vivo observations
We have seen how past work has begun to elucidate the mechanisms of in vitro collagen assembly. However, complicating matters further, fibrils formed by self-assembly in vitro differ from those observed in vivo. For example, in vivo fibrils show a distinct radius limitation that is not observed in vitro, as was discussed previously and shown in Fig. 3a. Additionally, collagen self-assembled in vitro forms a randomly oriented gel, in which there is no control of fibril length, number, or orientation. A given number of collagen monomers could form any distribution of fibril number, lengths, and diameter [Fig. 2b]; we have previously discussed how these properties are critical in determining tissue mechanics [76], and that in vivo fibrils are very precisely distributed, for example into parallel bundles in tendon [Fig. 4]. We are thus left with a number of unanswered questions: How is fibril diameter controlled in vivo? What determines the number of fibrils in vivo? What determines the lengths of these fibrils? How are fibrils oriented correctly for a given purpose? Given the additional differences between in vivo and in vitro assembly, novel approaches will be required to answer these questions.
Additional insights into the molecular regulation of collagen fibril assembly in vivo have come from studies of the Ehlers-Danlos syndrome (EDS), which is a heterogeneous group of connective tissue disorders featuring hyperextensible, fragile, easy bruising skin, joint hypermobility, and abnormal wound-healing. The majority of the genes linked to EDS encode the fibrillar collagens types I, III and V, and enzymes that modify procollagen including ADAMTS2 [117]. These studies showed a direct cause and effect of structural changes in type I-containing collagen fibrils and the mechanical properties of tissues. Tenascin-X is an extracellular matrix glycoprotein found in skin, muscle, tendons and blood vessels, and was the first non-collagen gene shown to cause classical-like EDS [118]. Mao and colleagues showed that the skin of tenascin-X deficient mice had 40% reduction in fibril numbers. Solving the conundrum of how tenascin-X helps to regulate collagen fibril number will be an important step forward in understanding the complexity of cellular control of collagen fibril formation in vivo.
The plasma membrane is the limit of direct cellular control
Uncontrolled self-assembly of collagen fibrils is a combination of chemistry and physics. The results of uncontrolled self-assembly are observed in vitro; the particular issue of in vivo assembly now becomes a question of how cells can influence the environment around them to change the physical and chemical landscape such that ordered fibril development will occur.
Whilst the procollagen and collagen molecules are transiting through the secretory pathway the cell has full control of pH and other solution conditions that can influence protein assembly and aggregation. Therefore, even though the procollagen can be processed to pNcollagen, pCcollagen and collagen during transport [24], [119], [120], the cell can exert active negative control of collagen fibrillogenesis [121]. How the cell achieves this is unknown. What is clear, however, is that the plasma membrane is the limit of the cell’s ability to directly control the collagen it produces, and is the final point from which it can influence its environment and must therefore exert control over collagen fibril assembly, fibril number, and matrix organization.
The suggestion that the plasma membrane is the site of collagen fibril formation is not new. In the 1940s, fibres were observed at the cell surface [122], and in the 1970s, collagen fibrils were seen in plasma membrane invaginations of embryonic fibroblasts [123]. With improvements in image handling software [124], the commercialization of serial block face-scanning electron microscopy (SBF-SEM) [125], and the development of suitable preparation protocols [126], it has been possible to obtain volumetric three-dimensional reconstructions of embryonic tendon [127] and cornea [18] showing collagen fibrils in plasma membrane invaginations, known as fibripositors. Fibripositors exhibit a range of morphologies, from simple invaginations of the plasma membrane (recessed fibripositors) to finger-like projections (protruding fibripositors) [127]. They are actin-dependent structures and usually contain just one fibril tip.
Although transmission EM and SBF-SEM provide high resolution images of fibripositors, the samples are chemically fixed, dried and embedded in resin prior to imaging. Consequently, it is challenging to perform time-resolved studies to obtain a detailed insight into the role, or roles, of fibripositors. For example, we do not know their rate of formation, if they are static or dynamic structures, or if they are exclusively involved in fibril assembly at the exclusion of fibril turnover. More recently, using CRISPR-Cas9 to tag the proα2(I) chain of type I collagen with photoswitchable Dendra2, it has been possible to image, live, collagen fibril synthesis by fibroblasts in culture [128]. All the fibrils synthesized by the cells were attached to the plasma membrane. Also, cells migrating over collagen fibrils pull and align the fibrils to facilitate end-to-end fusion, which had been suggested to be a mode of rapid growth in length of fibrils [129].
In addition to directly interacting with fibrils at the plasma membrane via fibripositors, cells are able to exert indirect influence over longer distances. For example, it has been shown that cells can remodel existing collagen fibril networks around them to transmit forces and communicate with other cells simply by exerting stress on nearby fibrils [130], [131], [132]. Such stresses are propagated through a fibril network by steric interactions with other fibrils.
These results give hints as to how cells control in vivo fibril assembly, but we still lack a comprehensive understanding that integrates all factors and scales.
New approaches to unanswered questions
We have previously discussed how collagen assembly is an inherently multiscale process for which a single theoretical framework may never be sufficient to capture all aspects of the system. This is especially true in vivo. We saw how, in past work, explicitly modelling collagen monomers in Brownian dynamics and Monte-Carlo simulations helped to elucidate the behaviour of collagen self-assembly in vitro. We also recognized the limitations of such systems to capture the length scales of both a single monomer and a large fibril due to their computational cost. This cost poses a barrier to using such techniques to study assembly for an in vivo system, in which effects on the scale of cells and tissue cannot be neglected.
Just as one need not track every molecule in the air to understand the weather, considering larger lengthscales allows us to avoid explicitly modelling the behaviour of individual collagen monomers. Instead, we can describe properties of the system as functions that vary continuously in space and time. For example, the work of Rutenberg and colleagues [133], [134], [135], [136], [137] has made extensive use of mesoscopic continuum models, inspired by liquid crystals, to study the organisation of monomers within a fibril. Instead of resolving individual monomers, their properties, such as orientation, are described with functions that vary smoothly with position in a fibril. For example, using a so-called “director field” [133], a function that captures molecular orientations from the core of a fibril towards its periphery. Such continuum models require assumptions such as cylindrical symmetry. Balancing energetic penalties for distortions of the molecular array with a surface free energy allows fibril radius to be related to the twist angle of monomers arranged helically relative to the fibril axis [134]. The same group [133] also used a phase field crystal model to represent D band density modulation, the axially periodic striations of fibrillar collagen, demonstrating the coexistence of fibrils of different sizes and distinct structures, indicating an underlying phase transition. A further refinement of this approach incorporates enzymatic cross-linking that is confined to the fibril surface, assuming the enzyme LOX is too large to penetrate beneath, resulting in a core where the director field is insensitive to the D-band surrounded by a shell that is strongly coupled to the D band, with fibril growth being controlled by the availability of collagen outside the fibril [137]. While the energies underpinning these models remain an empirical representation of the underlying intermolecular interactions, these models provide promising mechanistic explanations for internal fibril structure and radius control. However, these studies remain agnostic about the precise mechanisms of assembly, and do not answer the question of how multiple fibrils assemble and organise in vivo to form tendons and other tissues.
By moving up in lengthscale again, we can consider macroscopic models that address the organisation and properties of bundles of fibrils in the extracellular space. The organisation of fibrils in the extracellular space on lengthscales comparable with the size of individual cells are described by a variety of macroscale models, ranging from organised bundles of fibrils in tendon to more disordered matrix structures seen in in vitro systems. In the former category, the diffusion of pCcollagen from the periphery of a fibre bundle towards its centre was addressed by Rutenberg et al. [138], who showed that diffusion must be sufficiently rapid for fibril growth to be homogeneous across the bundle. They identified a threshold monomer concentration, determined in terms of uptake rate and diffusion coefficient, necessary for homogeneous fibril expansion to take place.
The origin of organization within disordered matrices has been described with both discrete and continuum approaches. For example, discrete approaches have been used to predict the reorganisation of a random fibre network in response to cellular stresses using a computational representation of individual filaments and calculating their interactions explicitly [132]. Work such as this has shown how random fibre networks can be remodelled to produce dense, parallel bundles in response to stress. Continuum methods can approach these problems by describing a disordered matrix as a continuous function that varies in space and time. For example, Grekas and colleagues [139] used a continuum approach to demonstrate that the strongly nonlinear relation between stress and strain for individual fibrils translates at the network level to a mechanical phase transition, with tension applied by cells forming bundles of laterally compressed fibres within the matrix. A network of fibrils can thereby reorganise into “tethers” that radiate out from a contracting cell, allowing the cell to remodel its environment. Continuum and discrete network models can also be combined, as in recent work by Ban and colleagues [140], showing how remodelling into dense fibril tethers may be mediated by crosslinks. These are examples of macroscopic self-assembly, orchestrated by fibroblasts but exploiting mechanical and biochemical interactions, which can extend over appreciable distances. This work on tethers raises the questions: where does a fibril end and a tether begin? Could similar processes be at work in fibril assembly in vivo? The answers to such questions remain unclear, but novel approaches hold great promise in providing new answers to old problems.
Competing interests
The authors declare no competing interests.
Acknowledgements
The research in the authors’ laboratories is funded by BBSRC (BB/T001984/1) and Wellcome (110126/Z/15/Z and 203128/Z/16/Z). The authors thank Helena Raymond-Hayling for careful reading of the manuscript.
References
- 1.Aouacheria Abdel, Cluzel Caroline, Lethias Claire, Gouy Manolo, Garrone Robert, Exposito Jean-Yves. Invertebrate data predict an early emergence of vertebrate fibrillar collagen clades and an anti-incest model*. J. Biol. Chem. 2004;279(46):47711–47719. doi: 10.1074/jbc.M408950200. [DOI] [PubMed] [Google Scholar]
- 2.Boot-Handford Raymond P., Tuckwell Danny S. Fibrillar collagen: the key to vertebrate evolution? A tale of molecular incest. BioEssays. 2003;25(2):142–151. doi: 10.1002/bies.10230. [DOI] [PubMed] [Google Scholar]
- 3.Huxley-Jones Julie, Robertson David L., Boot-Handford Raymond P. On the origins of the extracellular matrix in vertebrates. Matrix Biol.: J. Int. Soc. Matrix Biol. January 2007;26(1):2–11. doi: 10.1016/j.matbio.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 4.Holmes D.F., Chapman J.A., Prockop D.J., Kadler K.E. Growing tips of type I collagen fibrils formed in vitro are near-paraboloidal in shape, implying a reciprocal relationship between accretion and diameter. Proc. Natl. Acad. Sci. U.S.A. October 1992;89(20):9855–9859. doi: 10.1073/pnas.89.20.9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig A.S., Birtles M.J., Conway J.F., Parry D.A. An estimate of the mean length of collagen fibrils in rat tail-tendon as a function of age. Connect. Tissue Res. 1989;19(1):51–62. doi: 10.3109/03008208909016814. [DOI] [PubMed] [Google Scholar]
- 6.Svensson Rene B., Herchenhan Andreas, Starborg Tobias, Larsen Michael, Kadler Karl E., Qvortrup Klaus, Peter Magnusson S. Evidence of structurally continuous collagen fibrils in tendons. Acta Biomater. 2017;50:293–301. doi: 10.1016/j.actbio.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 7.van Suylichem P.T., van Deijnen J.E., Wolters G.H., van Schilfgaarde R. Amount and distribution of collagen in pancreatic tissue of different species in the perspective of islet isolation procedures. Cell Transplant. 1995;4(6):609–614. doi: 10.1177/096368979500400610. [DOI] [PubMed] [Google Scholar]
- 8.Laurent G.J. Lung collagen: More than scaffolding. Thorax. June 1986;41(6):418–428. doi: 10.1136/thx.41.6.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckley Mark R, Evans Elisabeth, Satchel Lauren N, Matuszewski Paul E, Chen Yi-Ling, Elliott Dawn M, Soslowsky Louis J, Dodge George R. Distributions of Types I, II and III Collagen by Region in the Human Supraspinatus Tendon. Connective Tissue Res. 2013;54(6):374–379. doi: 10.3109/03008207.2013.847096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kastelic J., Galeski A., Baer E. The multicomposite structure of tendon. Connect. Tissue Res. 1978;6(1):11–23. doi: 10.3109/03008207809152283. [DOI] [PubMed] [Google Scholar]
- 11.Smejkal Gary B, Fitzgerald Cody. Revised Estimate of Total Collagen in the Human Body. Int. J. Proteom. Bioinform. 2017;2(1):001–002. [Google Scholar]
- 12.Calverley Ben C., Kadler Karl E., Pickard Adam. Dynamic High-Sensitivity Quantitation of Procollagen-I by Endogenous CRISPR-Cas9 NanoLuciferase Tagging. Cells. September 2020;9(9):2070. doi: 10.3390/cells9092070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boedtker Helga, Doty Paul. The Native and Denatured States of Soluble Collagen. J. Am. Chem. Soc. September 1956;78(17):4267–4280. [Google Scholar]
- 14.Chapman John A. The regulation of size and form in the assembly of collagen fibrils in vivo. Biopolymers. August 1989;28(8):1367–1382. doi: 10.1002/bip.360280803. [DOI] [PubMed] [Google Scholar]
- 15.Snowden John M, Swann David A. Vitreous structure: V. The morphology and thermal stability of vitreous collagen fibers and comparison to articular cartilage (type II) collagen. Investigat. Ophthalmol. Vis. Sci. 19(6):9, 1980. [PubMed] [Google Scholar]
- 16.Bos Kees Jan, Holmes David F., Kadler Karl E., McLeod David, Morris Nicholas P., Bishop Paul N. Axial structure of the heterotypic collagen fibrils of vitreous humour and cartilage1 1Edited by W. Baumeister. J. Mol. Biol. 2001;306(5):1011–1022. doi: 10.1006/jmbi.2000.4429. [DOI] [PubMed] [Google Scholar]
- 17.Birk D.E., Fitch J.M., Linsenmayer T.F. Organization of collagen types I and V in the embryonic chicken cornea. Investigat. Ophthalmol. Vis. Sci. October 1986;27(10):1470–1477. [PubMed] [Google Scholar]
- 18.Young Robert D., Knupp Carlo, Pinali Christian, Png Kenneth M.Y., Ralphs James R., Bushby Andrew J., Starborg Tobias, Kadler Karl E., Quantock Andrew J. Three-dimensional aspects of matrix assembly by cells in the developing cornea. Proc. Natl. Acad. Sci. 2014;111(2):687–692. doi: 10.1073/pnas.1313561110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes D.F., Gilpin C.J., Baldock C., Ziese U., Koster A.J., Kadler K.E. Corneal collagen fibril structure in three dimensions: Structural insights into fibril assembly, mechanical properties, and tissue organization. Proc. Natl. Acad. Sci. 2001;98(13):7307–7312. doi: 10.1073/pnas.111150598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang Joan, Garva Richa, Pickard Adam, Yeung Ching-Yan Chloé, Mallikarjun Venkatesh, Swift Joe, Holmes David F., Calverley Ben, Lu Yinhui, Adamson Antony, Raymond-Hayling Helena, Jensen Oliver, Shearer Tom, Meng Qing Jun, Kadler Karl E. Circadian control of the secretory pathway maintains collagen homeostasis. Nat. Cell Biol. 2020;22(1):74–86. doi: 10.1038/s41556-019-0441-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trotter John A., Koob Thomas J. Collagen and proteoglycan in a sea urchin ligament with mutable mechanical properties. Cell Tissue Res. 1989;258(3) doi: 10.1007/BF00218864. [DOI] [PubMed] [Google Scholar]
- 22.Kadler Karl E., Holmes David F., Graham Helen, Starborg Tobias. Tip-mediated fusion involving unipolar collagen fibrils accounts for rapid fibril elongation, the occurrence of fibrillar branched networks in skin and the paucity of collagen fibril ends in vertebrates. Matrix Biol. August 2000;19(4):359–365. doi: 10.1016/s0945-053x(00)00082-2. [DOI] [PubMed] [Google Scholar]
- 23.Starborg T., Lu Y., Huffman A., Holmes D.F., Kadler K.E. Electron microscope 3D reconstruction of branched collagen fibrils in vivo. Scand. J. Med. Sci. Sports. 2009;19(4):547–552. doi: 10.1111/j.1600-0838.2009.00907.x. [DOI] [PubMed] [Google Scholar]
- 24.Canty Elizabeth G., Yinhui Lu., Meadows Roger S., Shaw Michael K., Holmes David F., Kadler Karl E. Coalignment of plasma membrane channels and protrusions (fibripositors) specifies the parallelism of tendon. J. Cell Biol. May 2004;165(4):553–563. doi: 10.1083/jcb.200312071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Birk D.E., Trelstad R.L. Extracellular compartments in tendon morphogenesis: Collagen fibril, bundle, and macroaggregate formation. J. Cell Biol. July 1986;103(1):231–240. doi: 10.1083/jcb.103.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kalson Nicholas S., Lu Yinhui, Taylor Susan H., Starborg Tobias, Holmes David F., Kadler Karl E. A structure-based extracellular matrix expansion mechanism of fibrous tissue growth. eLife. 2015;4:e05958. doi: 10.7554/eLife.05958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katja Maria Heinemeier, Peter Schjerling, Jan Heinemeier, Stig Peter Magnusson, and Michael Kjaer, Lack of tissue renewal in human adult Achilles tendon is revealed by nuclear bomb 14C. FASEB J. 27(5) (2013), 2074–2079. [DOI] [PMC free article] [PubMed]
- 28.Katja M. Heinemeier, Peter Schjerling, Jan Heinemeier, Mathias B. Møller, Michael R. Krogsgaard, Tomas Grum-Schwensen, Michael M. Petersen, and Michael Kjaer, Radiocarbon dating reveals minimal collagen turnover in both healthy and osteoarthritic human cartilage. Sci. Transl. Med. 8(346) (July 2016), 346ra90–346ra90. [DOI] [PubMed]
- 29.Adam E.M. Jørgensen, Peter Schjerling, Michael R. Krogsgaard, Michael M. Petersen, Jesper Olsen, Michael Kjær, and Katja M. Heinemeier, Collagen Growth Pattern in Human Articular Cartilage of the Knee. CARTILAGE, page 1947603520971016, November 2020. [DOI] [PMC free article] [PubMed]
- 30.Michael Buckley, Craig Lawless, and Natalia Rybczynski, Collagen sequence analysis of fossil camels, Camelops and c.f. Paracamelus, from the Arctic and sub-Arctic of Plio-Pleistocene North America. J. Proteom., 194 (March 2019), 218–225. [DOI] [PubMed]
- 31.Shunji Hattori, Tomomi Kiriyama-Tanaka, Masashi Kusubata, Yuki Taga, Testuya Ebihara, Katsuyuki Imai, Mitsutaka Miura, Yoshihiro Mezaki, Alexei Tikhonov, and Haruki Senoo, Preservation of collagen in the soft tissues of frozen mammoths. bioRxiv, page 2021.04.12.439423, April 2021. [DOI] [PMC free article] [PubMed]
- 32.Harrison Simon M., Chris Whitton R., Kawcak Chris E., Stover Susan M., Pandy Marcus G. Relationship between muscle forces, joint loading and utilization of elastic strain energy in equine locomotion. J. Exp. Biol. December 2010;213(23):3998–4009. doi: 10.1242/jeb.044545. [DOI] [PubMed] [Google Scholar]
- 33.Ramshaw John A.M., Shah Naina K, Brodsky Barbara. Gly-X-Y Tripeptide Frequencies in Collagen: A Context for Host-Guest Triple-Helical Peptides. J. Struct. Biol. 1998;122(1–2):86–91. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- 34.Bella Jordi. Collagen structure: New tricks from a very old dog. Biochem. J. April 2016;473(8):1001–1025. doi: 10.1042/BJ20151169. [DOI] [PubMed] [Google Scholar]
- 35.Shoulders Matthew D., Raines Ronald T. Collagen structure and stability. Annu. Rev. Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hans Peter Bächinger, Peter Bruckner, Rupert Timpl, Darwin J. Prockop, and Jürgen Engel, Folding Mechanism of the Triple Helix in Type-III Collagen and Type-III pN–Collagen. Eur. J. Biochem. 106(2) (May 1980), 619–632. [DOI] [PubMed]
- 37.Ricard-Blum Sylvie. The Collagen Family. Cold Spring Harbor Perspect. Biol. January 2011;3(1) doi: 10.1101/cshperspect.a004978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kadler Karl E., Baldock Clair, Bella Jordi, Boot-Handford Raymond P. Collagens at a glance. J. Cell Sci. 2007;120(12):1955–1958. doi: 10.1242/jcs.03453. [DOI] [PubMed] [Google Scholar]
- 39.I.L. Johnstone, Cuticle collagen genes. Expression in Caenorhabditis elegans. Trends in genetics: TIG, 16(1) (January 2000), 21–27. [DOI] [PubMed]
- 40.Monkol Lek, Konrad J. Karczewski, Eric V. Minikel, Kaitlin E. Samocha, Eric Banks, Timothy Fennell, Anne H. O’Donnell-Luria, James S. Ware, Andrew J. Hill, Beryl B. Cummings, Taru Tukiainen, Daniel P. Birnbaum, Jack A. Kosmicki, Laramie E. Duncan, Karol Estrada, Fengmei Zhao, James Zou, Emma Pierce-Hoffman, Joanne Berghout, David N. Cooper, Nicole Deflaux, Mark DePristo, Ron Do, Jason Flannick, Menachem Fromer, Laura Gauthier, Jackie Goldstein, Namrata Gupta, Daniel Howrigan, Adam Kiezun, Mitja I. Kurki, Ami Levy Moonshine, Pradeep Natarajan, Lorena Orozco, Gina M. Peloso, Ryan Poplin, Manuel A. Rivas, Valentin Ruano-Rubio, Samuel A. Rose, Douglas M. Ruderfer, Khalid Shakir, Peter D. Stenson, Christine Stevens, Brett P. Thomas, Grace Tiao, Maria T. Tusie-Luna, Ben Weisburd, Hong-Hee Won, Dongmei Yu, David M. Altshuler, Diego Ardissino, Michael Boehnke, John Danesh, Stacey Donnelly, Roberto Elosua, Jose C. Florez, Stacey B. Gabriel, Gad Getz, Stephen J. Glatt, Christina M. Hultman, Sekar Kathiresan, Markku Laakso, Steven McCarroll, Mark I. McCarthy, Dermot McGovern, Ruth McPherson, Benjamin M. Neale, Aarno Palotie, Shaun M. Purcell, Danish Saleheen, Jeremiah M. Scharf, Pamela Sklar, Patrick F. Sullivan, Jaakko Tuomilehto, Ming T. Tsuang, Hugh C. Watkins, James G. Wilson, Mark J. Daly, Daniel G. MacArthur, and Exome Aggregation Consortium, Analysis of protein-coding genetic variation in 60,706 humans. Nature 536(7616) (August 2016), 285–291. [DOI] [PMC free article] [PubMed]
- 41.Bekhouche Mourad, Colige Alain. The procollagen N-proteinases ADAMTS2, 3 and 14 in pathophysiology. Matrix Biol. May 2015;44–46:46–53. doi: 10.1016/j.matbio.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 42.K. Tanzawa, J. Berger, and D.J. Prockop,Type I procollagen N-proteinase from whole chick embryos. Cleavage of a homotrimer of pro-alpha 1(I) chains and the requirement for procollagen with a triple-helical conformation. J. Biolog. Chem. 260(2) (January 1985), 1120–1126. [PubMed]
- 43.Dombrowski Kenneth E., Vogel Bruce E., Prockop Darwin J. Mutations that alter the primary structure of type I procollagen have long-range effects on its cleavage by procollagen N-proteinase. Biochemistry. August 1989;28(17):7107–7112. doi: 10.1021/bi00443a048. [DOI] [PubMed] [Google Scholar]
- 44.Vogel B.E., Doelz R., Kadler K.E., Hojima Y., Engel J., Prockop D.J. A substitution of cysteine for glycine 748 of the alpha 1 chain produces a kink at this site in the procollagen I molecule and an altered N-proteinase cleavage site over 225 nm away. J. Biol. Chem. December 1988;263(35):19249–19255. [PubMed] [Google Scholar]
- 45.Culbert A.A., Lowe M.P., Atkinson M., Byers P.H., Wallis G.A., Kadler K.E. Substitutions of aspartic acid for glycine-220 and of arginine for glycine-664 in the triple helix of the pro alpha 1(I) chain of type I procollagen produce lethal osteogenesis imperfecta and disrupt the ability of collagen fibrils to incorporate crystalline hydroxyapatite. Biochem. J. November 1995;311(Pt 3):815–820. doi: 10.1042/bj3110815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.D.F. Holmes, R.B. Watson, B. Steinmann, and K.E. Kadler, Ehlers-Danlos syndrome type VIIB. Morphology of type I collagen fibrils formed in vivo and in vitro is determined by the conformation of the retained N-propeptide. J. Biol. Chem. 268(21) (July 1993), 15758–15765. [PubMed]
- 47.Sandrine Vadon-Le Goff, David J.S. Hulmes, and Catherine Moali, BMP-1/tolloid-like proteinases synchronize matrix assembly with growth factor activation to promote morphogenesis and tissue remodeling. Matrix Biol. 44–46 (May 2015),14–23. [DOI] [PubMed]
- 48.Leibvich S.J., Weiss J.B. Evidence for the role of terminal regions of tropocollagen in fibrillogenesis: Specific enzymic degradation studies. Biochem. J. April 1970;117(2):22P. doi: 10.1042/bj1170022pa. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prockop Darwin J., Fertala Andrzej. Inhibition of the Self-assembly of Collagen I into Fibrils with Synthetic Peptides: Demonstration that assembly is driven by specific binding sites on the monomers *. J. Biol. Chem. June 1998;273(25):15598–15604. doi: 10.1074/jbc.273.25.15598. [DOI] [PubMed] [Google Scholar]
- 50.Kadler K.E., Hojima Y., Prockop D.J. Assembly of collagen fibrils de novo by cleavage of the type I pC-collagen with procollagen C-proteinase. Assay of critical concentration demonstrates that collagen self-assembly is a classical example of an entropy-driven process. J. Biol. Chem. November 1987;262(32):15696–15701. [PubMed] [Google Scholar]
- 51.Hulmes David J.S., Kadler Karl E., Paul Mould A., Hojima Yoshio, Holmes David F., Cummings Christine, Chapman John A., Prockop Darwin J. Pleomorphism in type I collagen fibrils produced by persistence of the procollagen N-propeptide. J. Mol. Biol. November 1989;210(2):337–345. doi: 10.1016/0022-2836(89)90335-5. [DOI] [PubMed] [Google Scholar]
- 52.Fleischmajer R., Timpl R., Tuderman L., Raisher L., Wiestner M., Perlish J.S., Graves P.N. Ultrastructural identification of extension aminopropeptides of type I and III collagens in human skin. Proc. Natl. Acad. Sci. U.S.A. December 1981;78(12):7360–7364. doi: 10.1073/pnas.78.12.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Birk D.E., Fitch J.M., Babiarz J.P., Linsenmayer T.F. Collagen type I and type V are present in the same fibril in the avian corneal stroma. J. Cell Biol. March 1988;106(3):999–1008. doi: 10.1083/jcb.106.3.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Farndale Richard W. Collagen-binding proteins: Insights from the Collagen Toolkits. Essays Biochem. September 2019;63(3):337–348. doi: 10.1042/EBC20180070. [DOI] [PubMed] [Google Scholar]
- 55.Svensson L., Oldberg Å., Heinegård D. Collagen binding proteins. Osteoarth. Cartilage. August 2001;9:S23–S28. doi: 10.1053/joca.2001.0440. [DOI] [PubMed] [Google Scholar]
- 56.Shawn M. Sweeney, Joseph P. Orgel, Andrzej Fertala, Jon D. McAuliffe, Kevin R. Turner, Gloria A. Di Lullo, Steven Chen, Olga Antipova, Shiamalee Perumal, Leena Ala-Kokko, Antonella Forlino, Wayne A. Cabral, Aileen M. Barnes, Joan C. Marini, and James D. San Antonio, Candidate Cell and Matrix Interaction Domains on the Collagen Fibril, the Predominant Protein of Vertebrates. J. Biol. Chem. 283(30) (July 2008), 21187–21197. [DOI] [PMC free article] [PubMed]
- 57.K Harbers, M Kuehn, H Delius, and R Jaenisch, Insertion of retrovirus into the first intron of alpha 1(I) collagen gene to embryonic lethal mutation in mice. Proc. Natl. Acad. Sci. U.S.A. 81(5) (March 1984), 1504–1508. [DOI] [PMC free article] [PubMed]
- 58.Li S.W., Prockop D.J., Helminen H., Fässler R., Lapveteläinen T., Kiraly K., Peltarri A., Arokoski J., Lui H., Arita M. Transgenic mice with targeted inactivation of the Col2 alpha 1 gene for collagen II develop a skeleton with membranous and periosteal bone but no endochondral bone. Genes Dev. November 1995;9(22):2821–2830. doi: 10.1101/gad.9.22.2821. [DOI] [PubMed] [Google Scholar]
- 59.Shireen R. Lamandé and John F. Bateman, Genetic Disorders of the Extracellular Matrix. Anatomical Record (Hoboken, N.J.: 2007), 303(6) (June 2020), 1527–1542. [DOI] [PMC free article] [PubMed]
- 60.Eyre David. Collagen of articular cartilage. Arthritis Res. 2002;4(1):30–35. doi: 10.1186/ar380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li Y., Lacerda D.A., Warman M.L., Beier D.R., Yoshioka H., Ninomiya Y., Oxford J.T., Morris N.P., Andrikopoulos K., Ramirez F. A fibrillar collagen gene, Col11a1, is essential for skeletal morphogenesis. Cell. February 1995;80(3):423–430. doi: 10.1016/0092-8674(95)90492-1. [DOI] [PubMed] [Google Scholar]
- 62.Holmes D.F., Kadler K.E. The 10+4 microfibril structure of thin cartilage fibrils. Proc. Nat. Acad. Sci. November 2006;103(46):17249–17254. doi: 10.1073/pnas.0608417103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Keene D.R., Lunstrum G.P., Morris N.P., Stoddard D.W., Burgeson R.E. Two type XII-like collagens localize to the surface of banded collagen fibrils. J. Cell Biol. May 1991;113(4):971–978. doi: 10.1083/jcb.113.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wenstrup Richard J., Florer Jane B., Brunskill Eric W., Bell Sheila M., Chervoneva Inna, Birk David E. Type V collagen controls the initiation of collagen fibril assembly. J. Biol. Chem. December 2004;279(51):53331–53337. doi: 10.1074/jbc.M409622200. [DOI] [PubMed] [Google Scholar]
- 65.Agarwal Pallavi, Zwolanek Daniela, Keene Douglas R., Schulz Jan-Niklas, Blumbach Katrin, Heinegård Dick, Zaucke Frank, Paulsson Mats, Krieg Thomas, Koch Manuel, Eckes Beate. Collagen XII and XIV, New Partners of Cartilage Oligomeric Matrix Protein in the Skin Extracellular Matrix Suprastructure. J. Biol. Chem. June 2012;287(27):22549–22559. doi: 10.1074/jbc.M111.335935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fransiska Malfait, Clair Francomano, Peter Byers, John Belmont, Britta Berglund, James Black, Lara Bloom, Jessica M. Bowen, Angela F. Brady, Nigel P. Burrows, Marco Castori, Helen Cohen, Marina Colombi, Serwet Demirdas, Julie De Backer, Anne De Paepe, Sylvie Fournel-Gigleux, Michael Frank, Neeti Ghali, Cecilia Giunta, Rodney Grahame, Alan Hakim, Xavier Jeunemaitre, Diana Johnson, Birgit Juul-Kristensen, Ines Kapferer-Seebacher, Hanadi Kazkaz, Tomoki Kosho, Mark E. Lavallee, Howard Levy, Roberto Mendoza-Londono, Melanie Pepin, F. Michael Pope, Eyal Reinstein, Leema Robert, Marianne Rohrbach, Lynn Sanders, Glenda J. Sobey, Tim Van Damme, Anthony Vandersteen, Caroline van Mourik, Nicol Voermans, Nigel Wheeldon, Johannes Zschocke, and Brad Tinkle, The 2017 international classification of the Ehlers-Danlos syndromes. Amer. J. Med. Genet. Part C: Seminars Med. Genet. 175(1) (2017), 8–26.
- 67.Danielson Keith G., Baribault Helene, Holmes David F., Graham Helen, Kadler Karl E., Iozzo Renato V. Targeted Disruption of Decorin Leads to Abnormal Collagen Fibril Morphology and Skin Fragility. J. Cell Biol. February 1997;136(3):729–743. doi: 10.1083/jcb.136.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Raul Fleischmajer, Larry W. Fisher, E. Douglas MacDonald, Lloydstone Jacobs, Jerome S. Perlish, and John D. Termine, Decorin interacts with fibrillar collagen of embryonic and adult human skin. J. Struct. Biol. 106(1) (February 1991), 82–90. [DOI] [PubMed]
- 69.Hedbom E., Heinegård D. Binding of fibromodulin and decorin to separate sites on fibrillar collagens. J. Biol. Chem. December 1993;268(36):27307–27312. [PubMed] [Google Scholar]
- 70.Chakravarti Shukti, Magnuson Terry, Lass Jonathan H., Jepsen Karl J., LaMantia Christian, Carroll Heidi. Lumican Regulates Collagen Fibril Assembly: Skin Fragility and Corneal Opacity in the Absence of Lumican. J. Cell Biol. June 1998;141(5):1277–1286. doi: 10.1083/jcb.141.5.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Robinson Kelsey A., Sun Mei, Barnum Carrie E., Weiss Stephanie N., Huegel Julianne, Shetye Snehal S., Lin Linda, Saez Daniel, Adams Sheila M., Iozzo Renato V., Soslowsky Louis J., Birk David E. Decorin and biglycan are necessary for maintaining collagen fibril structure, fiber realignment, and mechanical properties of mature tendons. Matrix Biol.: J. Int. Soc. Matrix Biol. December 2017;64:81–93. doi: 10.1016/j.matbio.2017.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hansen Uwe, Bruckner Peter. Macromolecular Specificity of Collagen Fibrillogenesis. J. Biol. Chem. September 2003;278(39):37352–37359. doi: 10.1074/jbc.M304325200. [DOI] [PubMed] [Google Scholar]
- 73.Young Robert D., Lawrence Paul A., Duance Victor C., Aigner Thomas, Monaghan Paul. Immunolocalization of Collagen Types II and III in Single Fibrils of Human Articular Cartilage. J. Histochem. Cytochem. March 2000;48(3):423–432. doi: 10.1177/002215540004800312. [DOI] [PubMed] [Google Scholar]
- 74.Chavaunne T. Thorpe and Hazel R.C. Screen, Tendon structure and composition. Metabolic influences on risk for tendon disorders, pages 3–10, 2016. [DOI] [PubMed]
- 75.Holzapfel Gerhard A. Collagen. Springer; 2008. Collagen in arterial walls: biomechanical aspects; pp. 285–324. [Google Scholar]
- 76.Cormac Flynn, Fiber-matrix models of the dermis. Computational Biophysics of the Skin, page 133, 2014.
- 77.Bhagwan D. Agarwal, Lawrence J. Broutman, and K. Chandrashekhara, Analysis and performance of fiber composites. John Wiley & Sons, 2017.
- 78.Screen H.R.C., Bader D.L., Lee D.A., Shelton J.C. Local strain measurement within tendon. Strain. 2004;40(4):157–163. [Google Scholar]
- 79.Bosworth Lucy A, Turner Lesley-Anne, Cartmell Sarah H. State of the art composites comprising electrospun fibres coupled with hydrogels: a review. Nanomed. Nanotechnol. Biol. Med. 2013;9(3):322–335. doi: 10.1016/j.nano.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 80.Tamara L Akentjew, Claudia Terraza, Cristian Suazo, Jekaterina Maksimcuka, Camila A Wilkens, Francisco Vargas, Gabriela Zavala, Macarena Ocaña, Javier Enrione, Claudio M García-Herrera, et al. Rapid fabrication of reinforced and cell-laden vascular grafts structurally inspired by human coronary arteries. Nat. Commun. 10(1) (2019), 1–15. [DOI] [PMC free article] [PubMed]
- 81.Jetze Visser, Ferry P.W. Melchels, June E. Jeon, Erik M. Van Bussel, Laura S. Kimpton, Helen M. Byrne, Wouter J.A. Dhert, Paul D. Dalton, Dietmar W. Hutmacher, and Jos Malda, Reinforcement of hydrogels using three-dimensionally printed microfibres. Nat. Commun. 6(1) (2015), 1–10. [DOI] [PubMed]
- 82.Kastelic J., Palley I., Baer E. A structural mechanical model for tendon crimping. J. Biomech. 1980;13(10):887–893. doi: 10.1016/0021-9290(80)90177-3. [DOI] [PubMed] [Google Scholar]
- 83.Tim Bevan, Nadege Merabet, Jack Hornsby, Paul N. Watton, and Mark S. Thompson, A biomechanical model for fibril recruitment: evaluation in tendons and arteries. J. Biomech. 74 (2018), 192–196. [DOI] [PubMed]
- 84.Raz Einat, Lanir Yoram. Recruitment viscoelasticity of the tendon. J. Biomech. Eng. 2009;131(11) doi: 10.1115/1.3212107. [DOI] [PubMed] [Google Scholar]
- 85.Tom Shearer, William J. Parnell, Barbara Lynch, Hazel R.C. Screen, and I. David Abrahams, A recruitment model of tendon viscoelasticity that incorporates fibril creep and explains strain-dependent relaxation. J. Biomech. Eng. 142(7), 2020. [DOI] [PubMed]
- 86.Avery N.C., Bailey A.J. Collagen. Springer; 2008. Restraining cross-links responsible for the mechanical properties of collagen fibers: natural and artificial; pp. 81–110. [Google Scholar]
- 87.Stapleton Scott E, Moreira Ricardo, Jockenhoevel Stefan, Mela Petra, Reese Stefanie. Effect of reinforcement volume fraction and orientation on a hybrid tissue engineered aortic heart valve with a tubular leaflet design. Adv. Model. Simul. Eng. Sci. 2015;2(1):1–17. [Google Scholar]
- 88.Shearer Tom. A new strain energy function for the hyperelastic modelling of ligaments and tendons based on fascicle microstructure. J. Biomech. 2015;48(2):290–297. doi: 10.1016/j.jbiomech.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 89.Chanda Arnab, Chatterjee Subhodip, Gupta Vivek. Soft composite based hyperelastic model for anisotropic tissue characterization. J. Compos. Mater. 2020;54(28):4525–4534. [Google Scholar]
- 90.Holmes D.F., Capaldi M.J., Chapman J.A. Reconstitution of collagen fibrils in vitro; the assembly process depends on the initiating procedure. Int. J. Biol. Macromol. 1986;8(3):161–166. [Google Scholar]
- 91.Holmes David F., Yinhui Lu., Starborg Tobias, Kadler Karl E. Collagen Fibril Assembly and Function. Curr. Top. Dev. Biol. 2018;130:107–142. doi: 10.1016/bs.ctdb.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 92.Holmes David F, Graham Helen K, Kadler Karl E. Collagen fibrils forming in developing tendon show an early and abrupt limitation in diameter at the growing tips11Edited by M. F. Moody. J. Mol. Biol. November 1998;283(5):1049–1058. doi: 10.1006/jmbi.1998.2153. [DOI] [PubMed] [Google Scholar]
- 93.Haworth R.A., Chapman J.A. A study of the growth of normal and iodinated collagen fibrils in vitro using electron microscope autoradiography. Biopolymers. September 1977;16(9):1895–1906. doi: 10.1002/bip.1977.360160906. [DOI] [PubMed] [Google Scholar]
- 94.Capaldi M.J., Chapman J.A. The C-terminal extrahelical peptide of type I collagen and its role in fibrillogenesis in vitro. Biopolymers. November 1982;21(11):2291–2313. doi: 10.1002/bip.360211115. [DOI] [PubMed] [Google Scholar]
- 95.Kadler K.E., Hulmes D.J., Hojima Y., Prockop D.J. Assembly of type I collagen fibrils de novo by the specific enzymic cleavage of pC collagen. The fibrils formed at about 37 degrees C are similar in diameter, roundness, and apparent flexibility to the collagen fibrils seen in connective tissue. Ann. N. Y. Acad. Sci. 1990;580:214–224. doi: 10.1111/j.1749-6632.1990.tb17930.x. [DOI] [PubMed] [Google Scholar]
- 96.Kadler K.E., Hojima Y., Prockop D.J. Collagen fibrils in vitro grow from pointed tips in the C- to N-terminal direction. Biochem. J. June 1990;268(2):339–343. doi: 10.1042/bj2680339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Thurmond F.A., Trotter J.A. Native collagen fibrils from echinoderms are molecularly bipolar. J. Mol. Biol. January 1994;235(1):73–79. doi: 10.1016/s0022-2836(05)80015-4. [DOI] [PubMed] [Google Scholar]
- 98.Holmes D.F., Lowe M.P., Chapman J.A. Vertebrate (chick) collagen fibrils formed in vivo can exhibit a reversal in molecular polarity. J. Mol. Biol. January 1994;235(1):80–83. doi: 10.1016/s0022-2836(05)80016-6. [DOI] [PubMed] [Google Scholar]
- 99.Holmes D.F., Watson R.B., Chapman J.A., Kadler K.E. Enzymic control of collagen fibril shape. J. Mol. Biol. August 1996;261(2):93–97. doi: 10.1006/jmbi.1996.0443. [DOI] [PubMed] [Google Scholar]
- 100.Holmes David F., Tait Alexander, Hodson Nigel W., Sherratt Michael J., Kadler Karl E. Growth of collagen fibril seeds from embryonic tendon: Fractured fibril ends nucleate new tip growth. J. Mol. Biol. May 2010;399(1):9–16. doi: 10.1016/j.jmb.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hafner Anne E, Krausser Johannes, Šarić Andela. Minimal coarse-grained models for molecular self-organisation in biology. Curr. Opin. Struct. Biol. October 2019;58:43–52. doi: 10.1016/j.sbi.2019.05.018. [DOI] [PubMed] [Google Scholar]
- 102.Thomas Marcus, Schwartz Russell. Quantitative computational models of molecular self-assembly in systems biology. Phys. Biol. May 2017;14(3) doi: 10.1088/1478-3975/aa6cdc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Masao Doi, Samuel Frederick Edwards, and Samuel Frederick Edwards, The theory of polymer dynamics, volume 73. oxford University Press, 1988.
- 104.Eastman Peter, Swails Jason, Chodera John D., McGibbon Robert T., Zhao Yutong, Beauchamp Kyle A., Wang Lee-Ping, Simmonett Andrew C., Harrigan Matthew P., Stern Chaya D., Wiewiora Rafal P., Brooks Bernard R., Pande Vijay S. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLOS Computat. Biol. July 2017;13(7) doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Martin J. Falk, Amy Duwel, Lucy J. Colwell, and Michael P Brenner, Collagen-inspired self-assembly of twisted filaments. Phys. Rev. Lett. 123(23) (2019), 238102. [DOI] [PubMed]
- 106.McCluskey Andrew R., Hung Kennes S.W., Marzec Bartosz, Sindt Julien O., Sommerdijk Nico A.J.M., Camp Philip J., Nudelman Fabio. Disordered Filaments Mediate the Fibrillogenesis of Type I Collagen in Solution. Biomacromolecules. September 2020;21(9):3631–3643. doi: 10.1021/acs.biomac.0c00667. [DOI] [PubMed] [Google Scholar]
- 107.Plimpton Steve. Fast parallel algorithms for short-range molecular dynamics. J. Comput. Phys. 1995;117(1):1–19. [Google Scholar]
- 108.Hafner Anne E., Gyori Noemi G., Bench Ciaran A., Davis Luke K., Šarić Andela. Modeling Fibrillogenesis of Collagen-Mimetic Molecules. Biophys. J . November 2020;119(9):1791–1799. doi: 10.1016/j.bpj.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Baptiste Depalle, Zhao Qin, Sandra J. Shefelbine, and Markus J. Buehler, Influence of cross-link structure, density and mechanical properties in the mesoscale deformation mechanisms of collagen fibrils. J. Mech. Behav. Biomed. Mater. 52 (2015), 1–13. [DOI] [PMC free article] [PubMed]
- 110.Song Hongyan, Parkinson John. Modelling the Self-Assembly of Elastomeric Proteins Provides Insights into the Evolution of Their Domain Architectures. PLoS Comput. Biol. March 2012;8(3) doi: 10.1371/journal.pcbi.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Parkinson John, Kadler Karl E., Brass Andy. Self-assembly of rodlike particles in two dimensions: A simple model for collagen fibrillogenesis. Phys. Rev. E. October 1994;50(4):2963–2966. doi: 10.1103/physreve.50.2963. [DOI] [PubMed] [Google Scholar]
- 112.Parkinson John, Kadler Karl E., Brass Andy. Simple physical model of collagen fibrillogenesis based on diffusion limited aggregation. J. Mol. Biol. April 1995;247(4):823–831. doi: 10.1006/jmbi.1994.0182. [DOI] [PubMed] [Google Scholar]
- 113.Lee Kun-Chun, Liu Andrea J. Force-Velocity Relation for Actin-Polymerization-Driven Motility from Brownian Dynamics Simulations. Biophys. J . September 2009;97(5):1295–1304. doi: 10.1016/j.bpj.2009.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gardner Melissa K., Charlebois Blake D., Jánosi Imre M., Howard Jonathon, Hunt Alan J., Odde David J. Rapid Microtubule Self-Assembly Kinetics. Cell. August 2011;146(4):582–592. doi: 10.1016/j.cell.2011.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Castle Brian T., Odde David J. Brownian Dynamics of Subunit Addition-Loss Kinetics and Thermodynamics in Linear Polymer Self-Assembly. Biophys. J . December 2013;105(11):2528–2540. doi: 10.1016/j.bpj.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Andela Šarić, Yassmine C. Chebaro, Tuomas P.J. Knowles, and Daan Frenkel, Crucial role of nonspecific interactions in amyloid nucleation. Proc. Natl. Acad. Sci. 111(50) (December 2014), 17869–17874. [DOI] [PMC free article] [PubMed]
- 117.Malfait Fransiska, Castori Marco, Francomano Clair A., Giunta Cecilia, Kosho Tomoki, Byers Peter H. The Ehlers-Danlos syndromes. Nat. Rev. Disease Primers. July 2020;6(1):64. doi: 10.1038/s41572-020-0194-9. [DOI] [PubMed] [Google Scholar]
- 118.Jau Ren Mao, Glen Taylor, Willow B. Dean, Diane R. Wagner, Veena Afzal, Jeffrey C. Lotz, Edward M. Rubin, and James Bristow, Tenascin-X deficiency mimics Ehlers-Danlos syndrome in mice through alteration of collagen deposition. Nat. Genet. 30(4) (April 2002), 421–425. [DOI] [PubMed]
- 119.Canty-Laird Elizabeth G., Yinhui Lu., Kadler Karl E. Stepwise proteolytic activation of type I procollagen to collagen within the secretory pathway of tendon fibroblasts in situ. Biochem. J. January 2012;441(2):707–717. doi: 10.1042/BJ20111379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nicola L. Stevenson, Dylan J.M. Bergen, Yinhui Lu, M. Esther Prada-Sanchez, Karl E. Kadler, Chrissy L. Hammond, and David J. Stephens, Giantin is required for intracellular N-terminal processing of type I procollagen. J. Cell Biol. 220(6) (June 2021), e202005166. [DOI] [PMC free article] [PubMed]
- 121.Humphries Sally M., Yinhui Lu., Canty Elizabeth G., Kadler Karl E. Active Negative Control of Collagen Fibrillogenesis in Vivo. J. Biol. Chem. May 2008;283(18):12129–12135. doi: 10.1074/jbc.M708198200. [DOI] [PubMed] [Google Scholar]
- 122.Stearns Mary L. Studies on the development of connective tissue in transparent chambers in the rabbit’s ear. II. Am. J. Anatom. 1940;67(1):55–97. [Google Scholar]
- 123.Trelstad Robert L., Hayashi Kimiko. Tendon collagen fibrillogenesis: Intracellular subassemblies and cell surface changes associated with fibril growth. Dev. Biol. September 1979;71(2):228–242. doi: 10.1016/0012-1606(79)90166-0. [DOI] [PubMed] [Google Scholar]
- 124.James R. Kremer, David N. Mastronarde, and J. Richard McIntosh, Computer Visualization of Three-Dimensional Image Data Using IMOD. J. Struct. Biol. 116(1) (January 1996), 71–76. [DOI] [PubMed]
- 125.Denk Winfried, Horstmann Heinz. Serial Block-Face Scanning Electron Microscopy to Reconstruct Three-Dimensional Tissue Nanostructure. PLOS Biol. October 2004;2(11) doi: 10.1371/journal.pbio.0020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Starborg Tobias, Kalson Nicholas S., Yinhui Lu., Mironov Aleksandr, Cootes Timothy F., Holmes David F., Kadler Karl E. Using transmission electron microscopy and 3View to determine collagen fibril size and three-dimensional organization. Nat. Protoc. July 2013;8(7):1433–1448. doi: 10.1038/nprot.2013.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kalson N.S., Starborg T., Lu Y., Mironov A., Humphries S.M., Holmes D.F., Kadler K.E. Nonmuscle myosin II powered transport of newly formed collagen fibrils at the plasma membrane. Proc. Nat. Acad. Sci. December 2013;110(49):E4743–E4752. doi: 10.1073/pnas.1314348110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Adam Pickard, Antony Adamson, Yinhui Lu, Joan Chang, Richa Garva, Nigel Hodson, and Karl E. Kadler, Collagen assembly and turnover imaged with a CRISPR-Cas9 engineered Dendra2 tag. bioRxiv, page 331496, June 2018.
- 129.H.K. Graham, D.F. Holmes, R.B. Watson, and K.E. Kadler, Identification of collagen fibril fusion during vertebrate tendon morphogenesis. The process relies on unipolar fibrils and is regulated by collagen-proteoglycan interaction. J. Mol. Biol. 295(4) (January 2000), 891–902. [DOI] [PubMed]
- 130.Ma Xiaoyue, Schickel Maureen E., Stevenson Mark D., Sarang-Sieminski Alisha L., Gooch Keith J., Ghadiali Samir N., Hart Richard T. Fibers in the Extracellular Matrix Enable Long-Range Stress Transmission between Cells. Biophys. J . April 2013;104(7):1410–1418. doi: 10.1016/j.bpj.2013.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Abhilash A.S., Baker Brendon M., Trappmann Britta, Chen Christopher S., Shenoy Vivek B. Remodeling of Fibrous Extracellular Matrices by Contractile Cells: Predictions from Discrete Fiber Network Simulations. Biophys. J . October 2014;107(8):1829–1840. doi: 10.1016/j.bpj.2014.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hailong Wang, A.S. Abhilash, Christopher S. Chen, Rebecca G. Wells, and Vivek B. Shenoy, Long-range force transmission in fibrous matrices enabled by tension-driven alignment of fibers. Biophys. J. 107(11) (2014), 2592–2603. [DOI] [PMC free article] [PubMed]
- 133.Cameron Samuel, Kreplak Laurent, Rutenberg Andrew D. Phase-field collagen fibrils: Coupling chirality and density modulations. Phys. Rev. Res. March 2020;2(1) [Google Scholar]
- 134.Brown Aidan I., Kreplak Laurent, Rutenberg Andrew D. An equilibrium double-twist model for the radial structure of collagen fibrils. Soft Matter. October 2014;10(42):8500–8511. doi: 10.1039/c4sm01359j. [DOI] [PubMed] [Google Scholar]
- 135.Cameron Samuel, Kreplak Laurent, Rutenberg Andrew D. Polymorphism of stable collagen fibrils. Soft Matter. June 2018;14(23):4772–4783. doi: 10.1039/c8sm00377g. [DOI] [PubMed] [Google Scholar]
- 136.Matthew P. Leighton, Laurent Kreplak, and Andrew D. Rutenberg, Chiral phase-coexistence in compressed double-twist elastomers. arXiv:2102.02242 [cond-mat, physics:physics], February 2021. [DOI] [PubMed]
- 137.Leighton Matthew P., Kreplak Laurent, Rutenberg Andrew D. Non-equilibrium growth and twist of cross-linked collagen fibrils. Soft Matter. 2021;17(5):1415–1427. doi: 10.1039/d0sm01830a. [DOI] [PubMed] [Google Scholar]
- 138.Andrew D. Rutenberg, Aidan I. Brown, and Laurent Kreplak, Uniform spatial distribution of collagen fibril radii within tendon implies local activation of pC-collagen at individual fibrils. Phys. Biol. 13(4) (August 2016), 046008. [DOI] [PubMed]
- 139.Grekas Georgios, Proestaki Maria, Rosakis Phoebus, Notbohm Jacob, Makridakis Charalambos, Ravichandran Guruswami. Cells exploit a phase transition to mechanically remodel the fibrous extracellular matrix. J. R. Soc. Interface. February 2021;18(175):20200823. doi: 10.1098/rsif.2020.0823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ehsan Ban, J. Matthew Franklin, Sungmin Nam, Lucas R. Smith, Hailong Wang, Rebecca G. Wells, Ovijit Chaudhuri, Jan T. Liphardt, and Vivek B. Shenoy, Mechanisms of Plastic Deformation in Collagen Networks Induced by Cellular Forces. Biophys. J. 114(2) (January 2018), 450–461. [DOI] [PMC free article] [PubMed]