Abstract
Chelicerate arthropods exhibit dynamic genome evolution, with ancient whole-genome duplication (WGD) events affecting several orders. Yet, genomes remain unavailable for a number of poorly studied orders, such as Opiliones (daddy-long-legs), which has hindered comparative study. We assembled the first harvestman draft genome for the species Phalangium opilio, which bears elongate, prehensile appendages, made possible by numerous distal articles called tarsomeres. Here, we show that the genome of P. opilio exhibits a single Hox cluster and no evidence of WGD. To investigate the developmental genetic basis for the quintessential trait of this group—the elongate legs—we interrogated the function of the Hox genes Deformed (Dfd) and Sex combs reduced (Scr), and a homologue of Epidermal growth factor receptor (Egfr). Knockdown of Dfd incurred homeotic transformation of two pairs of legs into pedipalps, with dramatic shortening of leg segments in the longest leg pair, whereas homeosis in L3 is only achieved upon double Dfd + Scr knockdown. Knockdown of Egfr incurred shortened appendages and the loss of tarsomeres. The similarity of Egfr loss-of-function phenotypic spectra in insects and this arachnid suggest that repeated cooption of EGFR signalling underlies the independent gains of supernumerary tarsomeres across the arthropod tree of life.
Keywords: Hox, Deformed, Sex combs reduced, Egfr, Chelicerata, Arachnida
1. Introduction
The advent of genomic resources has revealed complex dynamics in the evolution of chelicerate genomes. A group of six terrestrial orders (Arachnopulmonata), which includes spiders, scorpions, and pseudoscorpions, exhibit an ancient shared whole-genome duplication (WGD), as evidenced by the architecture of Hox clusters, analyses of synteny, patterns of microRNA enrichment, gene expression patterns and gene tree topologies [1–6] (figure 1a). Separately, genomes of all four living Xiphosura (horseshoe crabs) suggest a lineage-specific twofold genome duplication in this order, with one of these duplications occurring relatively recently [7–9]. While genomes of Acariformes and Parasitiformes (mites and ticks) suggest that these two orders were not included in the genome duplication events, they often deviate from typical arthropod datasets. As examples, the acariform mite Tetranychus urticae exhibits extreme genome compaction (90 Mb), in tandem with the loss of many transcription factors, which has been linked to miniaturization [10]. Similarly, the genome of the parasitiform mite Galendromus occidentalis exhibits an atomized Hox cluster, degradation of synteny and high rates of intron gain and loss [11].
Figure 1.
The significance of Opiliones in evolutionary developmental biology. (a) Consensus phylogeny of Chelicerata (based on [5]) and inferred WGD events in Xiphosura and Arachnopulmonata. (b) Adult male P. opilio climbing on a twig using its prehensile tarsi. (c) Detail of the distal subdivisions (tarsomeres) of the leg 2 tarsus. The distal terminus is to the right. White bars mark tarsomere boundaries. Photographs: Caitlin M. Baker. (Online version in colour.)
One group that may facilitate comparative genomics of Chelicerata is the arachnid order Opiliones (harvestmen) (figure 1b). In phylogenomic datasets, Opiliones exhibit lower evolutionary rates than Parasitiformes or Acariformes, and their placement outside of arachnopulmonates makes this group phylogenetically significant [12,13]. Developmental transcriptomes of the emerging model species Phalangium opilio have suggested that harvestmen do not exhibit systemic genome duplication, as evidenced by the absence of paralogy across the homeobox gene family [2,14] and gene expression patterns of genes with known paralogues in arachnopulmonates [2,6]. As a result, P. opilio has proven useful for the study of chelicerate developmental biology. However, the establishment of a genome for this species is a key prerequisite to validating the assumption of an unduplicated genome in this order, as well as further advancing this model system.
Beyond its use in polarizing developmental traits on phylogenetic trees, Opiliones also exhibit a suite of unique characteristics that are not found in other arthropod models. The most salient of these are the elongate walking legs in some groups (e.g. Phalangioidea, commonly termed ‘daddy-long-legs’). Beyond the hypertrophied growth of certain leg segments, many daddy-long-legs exhibit subdivision of the tarsus into pseudosegments called tarsomeres, with over 100 tarsomeres in some species (figure 1c). Tarsomeres have evolved dynamically across the arthropod tree of life, with gains in tarsomeres across insects [15], scutigeromorph centipedes [16] and several arachnid orders. However, the tarsomeres of daddy-long-legs are sufficiently numerous that they confer prehensility to the distal leg, which is used for climbing, courtship and male–male combat (figure 1b). The largest number of tarsomeres in Phalangioidea typically occurs on the antenniform second leg pair, which serves as a sensory appendage [17] (figure 1c).
These aspects of harvestman biology position them as an opportune group for comparative study, both from the perspective of gene evolution before and after WGD, as well as understanding the genetic basis for morphological convergence (e.g. leg elongation; supernumerary tarsomeres). However, no genomes are available for any Opiliones. Moreover, the developmental genetic basis for arthropod leg elongation and tarsomere patterning is unknown outside of insects. To test the assumption that Opiliones exhibit an unduplicated genome, we generated a draft genome for P. opilio and leveraged this resource to investigate the genetic basis for leg patterning in this iconic arachnid group.
2. Material and methods
For brevity, detailed procedures, protocols and bioinformatic commands for the following operations are provided in the electronic supplementary material.
(a) . Animal husbandry
For genome sequencing, founder population specimens of P. opilio were collected in Madison, WI, USA (43.074628, −89.403904), and a colony was maintained as previously described [14] (electronic supplementary material, methods).
(b) . RNA sequencing
Total RNA was extracted from ca 250 µl of P. opilio embryos spanning an array of stages, reared from females captured in Weston, MA, USA. RNA extraction was performed using TRIzol reagent (ThermoFisher), following the manufacturer's protocol. mRNA purification, library construction and 2 × 150 bp sequencing on an Illumina HiSeq 2500 platform follow our previous procedures [1]. The resulting 79 472 462 paired-end reads (NCBI PRJNA690950) were combined with an older library (16 225 145 paired-end reads sequenced on an Illumina GA II; NBCI PRJNA236471) for annotation of protein-coding regions.
(c) . Genome sequencing, assembly and annotation
Full-sibling inbred lines were established at a colony in Madison, WI, USA. Total genomic DNA was isolated from two specimens (fourth-generation male and sixth-generation female). Long-read sequencing was performed on a PacBio Sequel platform (Pacific Biosciences) using v. 2.1 chemistry. The single-molecule real-time (SMRT) Cells were sequenced on 16 cells with 360 min movie lengths. Short-read sequencing was performed on an Illumina HiSeq 2500 with a 350 bp insert size. Long reads were assembled using Canu v. 1.7 [18]. Contigs were processed using two rounds of scaffolding with SSPACE-LongRead v. 1.1 [19], followed by gap filling with PBJelly v. 15.8 [20] and further polishing with Pilon v. 1.23 [21]. Haplotypic duplicates were identified and removed with Purge_dups v. 1.2.3 [22]. Prior to annotation, a custom repeat library was constructed using RepeatModeler open-1.0.11 [23]. Identified repeats were masked with RepeatMasker open-4.0.6. [23]. For annotation, gene predictions generated with BRAKER2 v. 2.1.5 [24] used previously generated RNA-Seq reads from developmental transcriptomes (NCBI PRJNA236471; PRJNA690950; [12]). A genome browser was generated using MakeHub [25].
(d) . Orthology inference, phylogenetic analysis and discovery of microRNAs
Homologues of Egfr were identified with tBLASTn [26], using query protein sequences of arthropod species for which Egfr expression has been previously studied. Sequence accession data are provided in the electronic supplementary material, table S1 and methods). An initial BLAST search for miRNA families in the genome of P. opilio and seven other chelicerates (electronic supplementary material, table S2) used as queries the miRNAs previously reported from the spider Parasteatoda tepidariorum, the tick Ixodes scapularis and the mite T. urticae [3,5]. To recover unique putative harvestmen miRNAs, we conducted a second search using miRNA families not known in spiders, ticks or mites, but which were shared by at least three mandibulate outgroups.
(e) . Cloning, in situ hybridization and double-stranded RNA microinjection
Cloning of gene fragments, in situ hybridization and embryonic microinjections with dsRNA followed our previous procedures [14,27] (electronic supplementary material, tables S3 and S4). Protocols for fluorescent gene expression assays using hybridization chain reaction (HCR) followed Bruce et al. [28]. For harvestman RNAi experiments, approximately two-thirds of each egg clutch was injected with dsRNA and the remaining third with water (negative control). Details of the phenotype scoring strategy are provided in the electronic supplementary material.
3. Results
(a) . Phalangium opilio draft genome assembly
The draft assembly of the P. opilio genome comprises 580.4 Mbp (37.5% GC content) in 5137 scaffolds (N50: 211 089) and 8349 contigs (N50: 127 429; electronic supplementary material, figure S1 and table S5). The predicted genome repetitiveness is 54.4% and estimated heterozygosity is 1.24%. The number of predicted genes after filtering steps is 20 315, which was further refined with a 98% similarity threshold and manual curation to a final gene set of 18 036. This is comparable to predicted gene sets for the tick I. scapularis (20 486) and the mite T. urticae (18 414) [10,29]. An assessment using the arthropod set of benchmarking universal single-copy orthologues (BUSCOs) [30] indicates 95.1% completeness (electronic supplementary material, figure S1 and table S5). Contamination assessment based on sequence coverage and GC content supports a relatively contamination-free assembly. The detailed description of the genome is provided in the electronic supplementary material, figures S1–S3.
(b) . The genome of Phalangium opilio reveals the absence of Arachnopulmonata-specific whole-genome duplications
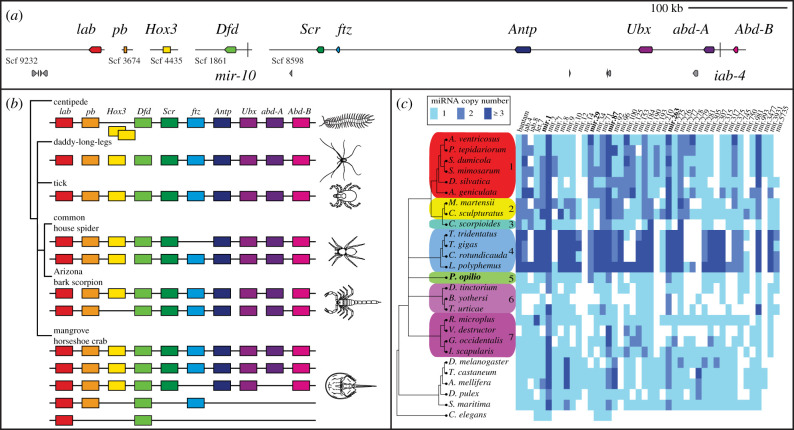
To assess whether P. opilio exhibits ancient WGD, we first examined the architecture of Hox clusters in this species. We discovered one 480 kb scaffold that bore six Hox genes, with the remaining four Hox genes occurring on individual scaffolds (figure 2a,b). In addition to the small size, these scaffolds contained very few or no adjacent genes, suggesting that the position of these four Hox genes outside of the main cluster is an artefact of fragmentary assembly. On the larger 480 kb scaffold, microRNAs mir-10 and iab-4 were located adjacent to Dfd and abdA, respectively, which reflect conserved positions with respect to other arthropods [32] (figure 2a). The complete peptide sequences of all 10 Hox genes corresponded to previous partial sequences predicted from developmental transcriptomes [14].
Figure 2.
Hox genes and microRNAs support an unduplicated genome in the daddy-long-legs P. opilio. (a) Hox gene-containing scaffolds to scale. miRNAs mir-10 and iab-4 are represented by vertical bars. Hox genes are depicted in coloured boxes, and other predicted genes in grey boxes. (b) Hox clusters in selected arthropod genomes (after [5,31]). (c) Comparative analysis of miRNA families and orthologue copy numbers in P. opilio and other chelicerates supports retention of single copies of several families in harvestmen, in contrast to duplication found in arachnopulmonates. Columns correspond to individual miRNA families, with colours representing a different number of paralogues. miRNAs in bold are duplicated in P. opilio and most other chelicerates. 1: Araneae; 2: Scorpiones; 3: Pseudoscorpiones; 4: Xiphosura; 5: Opiliones; 6: Acariformes; 7: Parasitiformes. (Online version in colour.)
We separately examined the genome for evidence of duplicates in genes with known arachnopulmonate-specific paralogues and spatio-temporal subdivisions of expression patterns, focusing on four leg patterning genes (dachshund, homothorax, extradenticle and spineless [4,6,33]) and three retinal determination network genes (sine oculis, Optix and orthodenticle [34–36]). These genes all occurred as single-copy in the harvestman genome (electronic supplementary material, table S6).
We next examined the distribution of families of microRNAs (miRNAs), noncoding RNAs with important regulatory roles in animals. miRNAs have been shown to exhibit the signature of genome duplication in both Arachnopulmonata and Xiphosura [3,5]. Thirty conserved miRNA families were identified in the P. opilio genome (figure 2c). Among them, only families mir-2, mir-29, mir-87 and mir-263 had two homologues in Opiliones (figure 2c). These microRNAs, with the exception of mir-29, are also duplicated in most other chelicerates and outgroup arthropods, (electronic supplementary material, table S2), suggesting the origin of paralogues at the arthropod common ancestor (figure 2c). The presence of duplicated mir-29 in harvestmen, horseshoe crabs and a subset of Arachnopulmonata suggests separate independent duplication events in these lineages, although this parsimonious inference is contingent upon the resolution of the position of these groups in arachnid phylogeny. In sum, we found no evidence of miRNA duplications in the harvestman genome that were exclusively shared either with Arachnopulmonata or Xiphosura.
(c) . Deformed and Sex combs reduced are necessary for leg fate specification in Phalangium opilio
Deformed homologues of arachnids are typically expressed in the four leg-bearing segments (L1–L4), whereas Sex combs reduced is expressed from L2 segment onwards (figure 3a; electronic supplementary material, figure S4) [4,14,37]. In the spider P. tepidariorum (a member of Arachnopulmonata), the two Dfd paralogues have divergent expression patterns; only Ptep-DfdA is expressed in the legs and expression levels are uniform across L1–L4 [4]. Knockdown of Ptep-DfdA results in homeotic L1-to-pedipalp transformation [38]. No functional data exist for any single-copy homologue of Dfd in Arachnida, and no functional data exist for Scr in Arachnida altogether. Intriguingly, Po-Dfd is also expressed in L1–L4, but much more strongly in L2 (the longest leg pair) than the other three leg pairs, particularly during leg elongation [14]. Scr is expressed in L3 and L4, but its expression is much stronger in L3 [14].
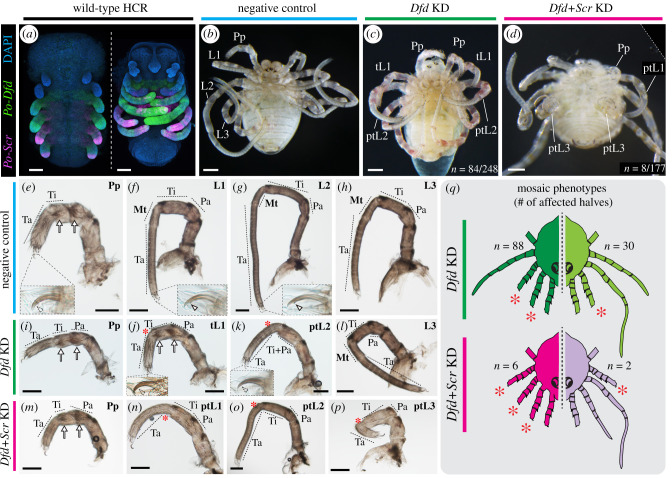
Figure 3.
The single-copy orthologue of Deformed (Dfd) and Sex combs reduced (Scr) in P. opilio are necessary for the appendage identity of three body segments. (a) Wild-type HCR expression patterns of Dfd (green) and Scr (magenta) homologues in stage 11 and 15 embryos. (c) Wild-type (negative control) hatchling of P. opilio. (c) P. opilio hatchling from Po-Dfd RNAi treatment. (d) P. opilio hatchling from Po-Dfd + Scr RNAi treatment. (e–h) Appendage mounts of wild-type P. opilio hatchlings in lateral view. Insets: tarsal claws. (i–l) Appendage mounts of Po-Dfd RNAi hatchlings in lateral view. (m–p) Appendage mounts of Po-Dfd + Scr RNAi hatchlings in lateral view. (q) Schematic depiction of the mosaicism observed in hatchlings of Po-Dfd RNAi (Dfd KD) and Po-Dfd + Scr RNAi (Dfd + Scr KD) and distribution of phenotypic classes. Lighter colours indicate weaker penetrance. Only the two most frequent homeotic conditions are depicted (see electronic supplementary material, figure S5). Asterisk, reduced metatarsus; arrow, setal spurs; arrowhead, claw tooth; L1–L3: leg 1–3; tL1, fully transformed leg 1; ptL1–3: partially transformed legs 1–3; Mt, metatarsus; Pa, patella; Pp, pedipalp; Ta, tarsus; Ti; tibia. Scale bars: 100 µm. (Online version in colour.)
We first investigated the function of Dfd (Po-Dfd) through embryonic RNAi, via microinjection of double-stranded RNA (dsRNA) (electronic supplementary material, figure S5). Upon completion of embryogenesis, 33% (n = 84/248) of RNAi hatchlings exhibited leg-to-pedipalp homeosis affecting L1 and L2 (figure 3b,c; electronic supplementary material, figure S5). Complete leg-to-pedipalp homeotic transformation (figure 3c,j), only observed in L1, was evidenced by the loss of the metatarsus (segment specific to arachnid leg; absent in pedipalp), the presence of pedipalp-specific setal spurs, the loss of the leg-specific tooth of the distal claw (figure 3e,f,i,j) and the undivided tarsus. Partial transformation was evidenced by a defective or absent metatarsus (figure 3g–k). Strongly affected L2 also exhibited a fusion of tibia and patella segments (figure 3k). Importantly, L3 and L4 were unaffected (figure 3h,l). The majority (n = 73/84) of Po-Dfd knockdown phenotypes exhibited mosaicism, with one side of the body more strongly affected (bilateral mosaics), so we further classified homeotic individuals in mosaic classes. This tabulation considered whether a given half presented no homeosis (wild-type), homeosis in both L1 and L2, homeosis in L1 only, or in L2 only (electronic supplementary material, figure S5). This analysis revealed that the most frequent homeotic conditions were having both legs affected (88/168 halves), followed by L1 only (30/168 halves) and L2 only (2/168 halves) (figure 3q; electronic supplementary material, figure S5). In halves with both legs affected, L1 was always more strongly transformed than L2. Notably, knockdown of Po-Dfd resulted in dramatic shortening of the transformed appendages, and particularly for transformed L2 (compare figure 3g,k). Reduced Po-Dfd expression was observed overall in embryos injected with Po-Dfd dsRNA and correlated with the side presenting homeotic transformation in mosaic embryos (electronic supplementary material, figure S6).
Next, to investigate a possible role of Scr in the identity of L3 and L4, we performed RNAi against Po-Scr. Despite verifiable decreased expression (electronic supplementary material, figure S6), we detected no phenotypic effects on dsRNA-injected embryos (five clutches, n = 540 embryos), suggesting that Scr may exhibit functional redundancy in arachnids. We therefore performed a double knockdown of Po-Dfd and Po-Scr. Mortality was high (153/176), but double RNAi resulted in partial leg-to-pedipalp transformation affecting L1, L2 and L3 (n = 8/176) (figure 3d, m–q; electronic supplementary material, figure S5).
Taken together, these results suggest that Po-Dfd is necessary for conferring leg identity of the L1 and L2 segments, and that both Dfd and Scr are necessary for the identity of the L3 segment.
(d) . Epidermal growth factor receptor is necessary for distal leg patterning in the daddy-long-legs
A notable component of leg morphology in daddy-long-legs is the repeated subdivision of the tarsus, the distalmost leg segment. In insects, EGFR signalling is involved in tarsal fate specification [39–41], but it is unknown if this signalling pathway is necessary for leg patterning in chelicerates (figure 4a). Upon surveying the P. opilio genome, we discovered two Egfr paralogues in the harvestman (electronic supplementary material, figure S7). One of these, Po-EgfrB, lacks the transmembrane and intracellular domains seen in other Egfr homologues (electronic supplementary material, figure S8). A 3′ UTR for Po-EgfrB was assembled in both embryonic transcriptomes and corroborated by the genome assembly, disfavouring fragmentary assembly as a possible explanation for missing domains. We therefore focused on Po-EgfrA, the paralogue containing all known Egfr functional domains.
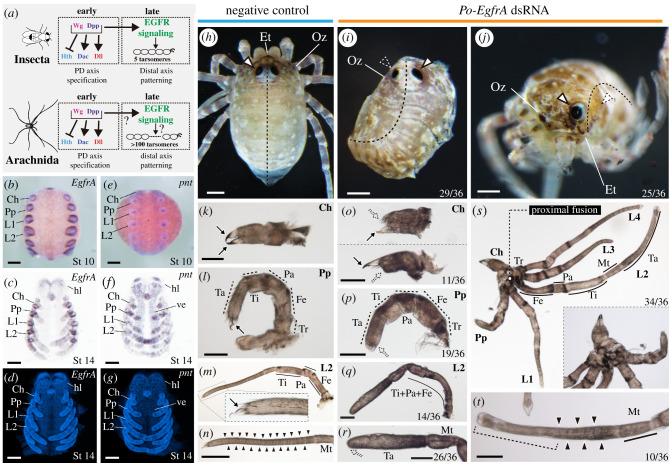
Figure 4.
Po-EgfrA knockdown affects dorsal patterning, eyes and appendage formation. (a) Gene regulatory network specifying PD axis and distal appendage patterning in insects and arachnids. Po-EgfrA (b–d) and Po-pnt (e–g) in situ hybridization wild-type expression. (b,e) Whole-mount stage 10 embryos, merged brightfield and Hoechst nuclear staining, ventral view. (c,f) Flatmounts, stage 14 embryos, brightfield, ventral view. (d,g) Hoechst nuclear counter staining. (h) Negative control hatchling in dorsal view. (h,j) Hatchlings from Po-EgfrA dsRNA-injected treatment (mosaic, left side affected). (b) Hatchling in dorsal view. Note dorsal fusion on the left side of the body (n = 29/36). (c) Hatchling in frontal view, with the left eye absent. A subset of Egfr phenotypes showed eye reduction (25/36) (k–n) Appendage flat mounts of negative control hatchlings, in lateral view. (k) Chelicera. (l) Pedipalp. (m) L2. Inset: detail of the claw. (n) Tarsus of L2. (o–t) Appendage flat mounts of hatchlings of Po-EgfrA dsRNA-injected treatment, in lateral view. (o) Chelicerae with a reduced fixed finger (upper panel), movable finger (lower panel) or both (n = 11/36). (p) Pedipalps lacking claw (n = 19/36). (q) L2, exhibiting podomere fusions proximal to the tarsus (n = 14/36). (r) Distal end of L2, exhibiting claw and tarsomere reduction (n = 26/36). (s) Proximal fusion in adjacent appendages (Ch–L4) (n = 34/36). Inset: Detail of fused coxae. (t) Tarsus of leg 2 shown in (r). Weakly affected legs lacked claws and distal tarsal joints (brackets) but retained proximal joints (n = 10/36). Arrow, claw; outlined white arrowhead, eye; dotted white arrowhead, eye defect; solid black arrowhead, tarsomere joints; Ch, chelicera; Et, egg tooth; Fe, femur; hl, head lobe; L1–L4, legs 1–4; Mt, metatarsus; Oz, ozophore; Pa, patella; Pp, pedipalp; Ta, tarsus; Ti, tibia; Tr, trochanter; ve, ventral ectoderm. Scale bars: 100 µm. (Online version in colour.)
In early limb bud stage, Po-EgfrA is expressed in a strong circular domain around the stomodeum, in a strong ring at the base of each appendage and in a weak stripe along the ventral midline (figure 4b; electronic supplementary material, figure S9). In later stages, additional expression domains occur in the developing eye field, in a strong domain in the medial bridge of the developing brain, and in rings at the boundaries of the segments (podomeres) of the developing appendages (figure 4c,d; electronic supplementary material, figure S9). To investigate EGFR signalling further, we surveyed the expression of a homologue of pointed, an ETS transcription factor that acts as an EGFR signal effector [42]. In early limb bud stages, the single-copy Po-pnt is expressed in the ventral ectoderm and the distal tip of the appendages (figure 4e; electronic supplementary material figure S9). Similar to the beetle T. castaneum [43], in later stages, Po-pnt is also expressed in the head lobes, and groups of cells in the appendages, particularly in the distal region, forming rings (figure 4f,g; electronic supplementary material, figure S9).
RNAi against EgfrA resulted in 39.6% (n = 36/91) of hatchlings exhibiting segmentation and appendage defects (figure 4h–t; electronic supplementary material, figure S10); all affected embryos were bilateral mosaics (figure 4i,j) and defects correlated with reduced Po-EgfrA expression in embryos (electronic supplementary material, figure S11). Po-EgfrA dsRNA-injected hatchlings showed defects of antero-posterior (body) segmentation, with dorsal tissue fusion on the side of the body affected (n = 29/36), correlating with a characteristic curved shape of the body of mosaic individuals (figure 4i). Defects of the eyes (n = 25/36) ranged from a small reduction in size to complete absence (figure 4j).
The appendages exhibited an array of defects in terminal structures (figure 4k–t). The chelicera showed the reduction of the fixed finger, movable finger or both (n = 11/36) (figure 4o); the pedipalp showed a reduced claw (n = 19/36) (figure 4p); and, in the case of all legs, the claw and tarsomeres were reduced (n = 26/36) (figure 4q,r). Most notably, a subset of weakly affected individuals (n = 10) showed a condition in which the distalmost tarsomeres were fused and the claw was missing, or just the claw was missing (figure 4t). Segmental fusions in limbs occurred in a subset of hatchlings (n = 14/36) (figure 4q). The proximal segment of the appendages (coxa) showed defects ranging from reduction in size to complete proximal fusion of adjacent appendage coxae (n = 34/36) (figure 4s), which correlated with the strong ring of expression at the base of all appendages.
These results are broadly consistent with expression and functional data available for Egfr homologues in insect models [43–46]. The loss-of-function phenotypic spectrum in P. opilio suggests that EgfrA may underpin both leg elongation and tarsomere morphogenesis in daddy-long-legs.
4. Discussion
(a) . Genomics and Hox-logic in a phylogenetically significant arachnid model
Spatio-temporal subdivision of expression domains of duplicated developmental patterning genes are systemic in Arachnopulmonata, as established by gene expression surveys in model spiders, scorpions and whip spiders [1,2,4,47]. This phenomenon makes arachnopulmonates an ideal taxon for investigating the role of gene duplication in generating body plan disparity. Identifying subfunctionalization or neofunctionalization of duplicates requires a clear inference of the ancestral single-copy homologue's expression pattern and function. In this regard, P. opilio has played a central role in polarizing developmental phenomena, given its phylogenetic position, low evolutionary rate and tractability in the laboratory [2,6,48].
Nevertheless, the assumed unduplicated condition of the Opiliones genome has not been rigorously tested. The efficiency of developmental transcriptomes in discovering paralogy is a function of sequencing depth and sampling strategy, and thus transcriptomes frequently fail to capture paralogues [1,2,5]. As a first step in validating the use of P. opilio as an effective outgroup to Arachnopulmonata, we assembled and interrogated the genome of P. opilio, which revealed no evidence of systemic genome duplication events previously reported for arachnopulmonates or horseshoe crabs. Together with the genome architecture of model systems like I. scapularis and T. urticae, the condition of the harvestman genome strongly supports the inference that an unduplicated genome is the ancestral condition for arachnids.
Having established that Hox genes of P. opilio are bona fide single-copy, we targeted the first functional datapoints for single-copy Hox genes in any arachnid species, focusing on genes that pattern walking leg identity. We were able to show that the knockdown of Po-Dfd affects the identity of legs 1 and 2. This discovery is significant for two reasons. First, two copies of Dfd occur in spiders and these exhibit subdivision of expression pattern, with Ptep-DfdA being expressed throughout leg tissues and ventral ectoderm, and Ptep-DfdB mostly restricted to the ventral ectoderm [4]. It has been shown that the spider paralogue Ptep-DfdA is necessary for repressing pedipalp identity on a single-body segment (L1); there is no effect on L2 [38]. Compared to the Po-Dfd RNAi phenotype, these data are congruent with the hypothesis that arachnopulmonate Dfd paralogues have undergone subfunctionalization. Further functional studies of DfdB paralogues in arachnopulmonates are necessary to test this scenario, specifically targeting DfdB (alone and also through double knockdown with DfdA). Second, segments affected by Dfd knockdown in harvestman (L1 and L2) are positionally homologous to those affected by Dfd knockdown in pancrustaceans (mandibular and maxillary segments [49–51]). These results bring further support for the notion that the establishment of some Hox anterior boundaries predates the evolution of tagmata, with further substantiation from Hox anterior boundaries in Onychophora [52].
Furthermore, we found that the knockdown of Po-Scr alone has no discernible phenotype, whereas the double knockdown of Po-Dfd and Po-Scr resulted in homeotic transformation of L1–L3 into pedipalps. These data suggest functional redundancy of Scr in L3, paralleling the dynamics of Ubx and abdA in insects; the knockdown of abdA alone has a limited effect on homeotic abdominal-to-thoracic segment transformations, whereas the double knockdown of both these genes results in more complete transformations of abdominal segments into thoracic identities [53]. Similar functional redundancy has been shown for Antp-1 and Ubx-1 in the spider P. tepidariorum [54].
As these experiments show, P. opilio has the potential to serve as an informative outgroup to Arachnopulmonata for the study of paralogue divergence after duplication. In addition to newly generated genomic resources, the effectiveness of single and double RNAi in this system makes P. opilio an opportune point of comparison for future investigations of arachnid body plan evolution.
(b) . A conserved role for epidermal growth factor receptor signalling in appendage patterning across Arthropoda
In the fruit fly Drosophila melanogaster, EGFR-Ras signalling is responsible for patterning the legs in two phases. First, EGFR signalling begins with a distal expression (central in the leg disc) of the EGFR ligands in the leg disc [40,41]. The reduction of EGFR signalling results in progressively greater deletion of more distal leg structures, which is consistent with a distal-to-proximal requirement gradient of EGFR signalling by downstream tarsal patterning genes, and with a distal source of EGFR signalling [15,40,41]. A distal source of EGFR signalling is also in accordance with the distal-to-proximal requirement for EGFR signalling in the regenerating leg of the cricket G. bimaculatus [45]. In P. opilio, 72% (n = 26/36) of the hatchlings resulting from RNAi against Egfr exhibited defects in the tarsus. These defects ranged from the absence of claws and distal tarsomere fusions, to complete failure to form all tarsal subdivisions. The spectrum of increasingly severe defects from distal-to-proximal observed upon Egfr knockdown in P. opilio suggests that daddy-long-leg tarsomeres are patterned by a gradient of EGFR signalling similar to D. melanogaster and G. bimaculatus. This conclusion is further supported by the expression of the EGFR signalling effector Po-pnt, which we showed to occur on the tip of the developing appendages at the limb bud stage.
In a second phase, EGFR ligands are expressed as rings at the boundaries of embryonic tarsomeres [39]. The reduction or upregulation of EGFR signalling at this later stage in D. melanogaster results in defects in medial tarsomeres and in failure to correctly pattern the tarsal joints [40,41]. However, in short germ insect models (e.g. beetle, water strider and cricket), Egfr is expressed as rings at the boundaries of all leg segments proximal to the tarsus, in contrast to rings restricted to the tarsus in the fruit fly [43–45]. RNAi-mediated knockdown in two insect models resulted in leg segment truncations proximal to the tarsus [43,44]. Our data in the arachnid P. opilio largely accord with the expression and functional results in short germ insects: 38% (n = 14/36) of Po-EgfrA RNAi phenotypes also exhibited leg segment fusions proximal to the tarsus. The signal effector Po-pnt is also expressed in a distal domain forming rings in later stages of appendage development, which accords with the second phase of expression in model insects [39,43]. Together, these results suggest that the second role of EGFR signalling in leg segmentation may also be conserved in P. opilio.
Disentangling the effects of early versus late EGFR signalling phases in the phenotypes observed in Po-EgfrA RNAi could be further explored by disrupting EGFR signalling in later development, to surpass the early function in PD axis development. We anticipate that the genome of P. opilio will facilitate the development of more sophisticated tools for functional genetics, toward refining the understanding of how daddy-long-legs make their long legs.
Supplementary Material
Acknowledgements
RNA sequencing was performed at the Center for Systems Biology, Harvard University. Genomic laboratory work was conducted at the Smithsonian Laboratories of Analytical Biology (LAB). Specimen vouchering and long-term cryogenic curation was provided by the Smithsonian National Museum of Natural History. HCR was made possible by the Patel lab and the 2021 MBL embryology course. Microscopy was performed at the Newcomb Imaging Center, Department of Botany, University of Wisconsin-Madison. Computing was conducted by the Smithsonian Institution High Performance Cluster (https://doi.org/10.25572/SIHPC). Audrey R. Crawford and Calvin So assisted with harvestman rearing and RNAi experiments.
Contributor Information
Guilherme Gainett, Email: guilherme.gainett@wisc.edu.
Vanessa L. González, Email: gonzalezv@si.edu.
Data accessibility
The data underlying this article are available in the article and in its online supplementary material. The genome assembly, EGFR tree and alignment, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.ht76hdrds [55]. Sequencing data are deposited in NCBI under accession numbers NCBI PRJNA690950 (RNA-seq), NCBI PRJNA647749 (genome), NCBI SRR12286133 (long reads), NCBI SRR12286133 (short reads). The genome browser is available at: http://phalangium-opilio.sigenomehub.org/.
The data are provided in electronic supplementary material [56].
Authors' contributions
G.G.: conceptualization, data curation, formal analysis, investigation, methodology, validation, visualization, writing-original draft, writing-review and editing; V.L.G.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, software, visualization, writing-original draft, writing-review and editing; J.B.: data curation, formal analysis, methodology, software, writing-review and editing; E.V.W.S.: data curation, methodology, writing-review and editing; C.M.B.: resources, writing-review and editing; L.B.G.: data curation, investigation, writing-review and editing; C.E.S.-L.: data curation, formal analysis, investigation, methodology, writing-review and editing; J.A.C.: funding acquisition, resources, supervision, writing-review and editing; P.S.: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, resources, supervision, validation, writing-original draft, writing-review and editing
All authors gave final approval for publication and agreed to be held accountable for the work performed therein.
Competing interests
We declare we have no competing interests.
Funding
This material is based on work supported by the Food and Drug Administration (J.A.C.), the Global Genome Initiative grant no. GGI-Exploratory-2016-047 (V.L.G.), and National Science Foundation grant nos. IOS-1552610 and IOS-2019141 (P.P.S.). G.G. was supported by a Wisconsin Alumni Research Foundation Fall Research Competition award.
References
- 1.Sharma PP, Schwager EE, Extavour CG, Wheeler WC. 2014. Hox gene duplications correlate with posterior heteronomy in scorpions. Proc. R. Soc. B 281, 20140661. ( 10.1098/rspb.2014.0661) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leite DJ, et al. 2018. Homeobox gene duplication and divergence in arachnids. Mol. Biol. Evol. 35, 2240-2253. ( 10.1093/molbev/msy125) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leite DJ, Ninova M, Hilbrant M, Arif S, Griffiths-Jones S, Ronshaugen M, McGregor AP. 2016. Pervasive microRNA duplication in chelicerates: insights from the embryonic microRNA repertoire of the spider Parasteatoda tepidariorum. Genome Biol. Evol. 8, 2133-2144. ( 10.1093/gbe/evw143) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwager EE, et al. 2017. The house spider genome reveals an ancient whole-genome duplication during arachnid evolution. BMC Biol. 15, 62. ( 10.1186/s12915-017-0399-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ontano AZ, et al. 2021. Taxonomic sampling and rare genomic changes overcome long-branch attraction in the phylogenetic placement of pseudoscorpions. Mol. Biol. Evol. 38, 2446-2467. ( 10.1093/molbev/msab038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan ED, Santibáñez López CE, Sharma PP. 2020. Developmental gene expression as a phylogenetic data class: support for the monophyly of Arachnopulmonata. Dev. Genes Evol. 230, 137-153. ( 10.1007/s00427-019-00644-6) [DOI] [PubMed] [Google Scholar]
- 7.Kenny NJ, et al. 2015. Ancestral whole-genome duplication in the marine chelicerate horseshoe crabs. Heredity 116, 190-199. ( 10.1038/hdy.2015.89) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shingate P, Ravi V, Prasad A, Tay BH, Garg KM, Chattopadhyay B, Yap LM, Rheindt FE, Venkatesh B. 2020. Chromosome-level assembly of the horseshoe crab genome provides insights into its genome evolution. Nat. Commun. 11, 1-13. ( 10.1038/s41467-020-16180-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shingate P, Ravi V, Prasad A, Tay BH, Venkatesh B. 2020. Chromosome-level genome assembly of the coastal horseshoe crab (Tachypleus gigas). Mol. Ecol. Resour. 20, 1748-1760. ( 10.1111/1755-0998.13233) [DOI] [PubMed] [Google Scholar]
- 10.Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487-492. ( 10.1038/nature10640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoy MA, et al. 2016. Genome sequencing of the phytoseiid predatory mite Metaseiulus occidentalis reveals completely atomized Hox genes and superdynamic intron evolution. Genome Biol. Evol. 8, 1762-1775. ( 10.1093/gbe/evw048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballesteros JA, Sharma PP. 2019. A critical appraisal of the placement of Xiphosura (Chelicerata) with account of known sources of phylogenetic error. Syst. Biol. 68, 896-917. ( 10.1093/sysbio/syz011) [DOI] [PubMed] [Google Scholar]
- 13.Lozano-Fernandez J, Tanner AR, Giacomelli M, Carton R, Vinther J, Edgecombe GD, Pisani D. 2019. Increasing species sampling in chelicerate genomic-scale datasets provides support for monophyly of Acari and Arachnida. Nat. Commun. 10, 2295-2298. ( 10.1038/s41467-019-10244-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma PP, Schwager EE, Extavour CG, Giribet G. 2012. Hox gene expression in the harvestman Phalangium opilio reveals divergent patterning of the chelicerate opisthosoma. Evol. Dev. 14, 450-463. ( 10.1111/j.1525-142X.2012.00565.x) [DOI] [PubMed] [Google Scholar]
- 15.Kojima T. 2017. Developmental mechanism of the tarsus in insect legs. Curr. Opin. Insect. Sci. 19, 36-42. ( 10.1016/j.cois.2016.11.002) [DOI] [PubMed] [Google Scholar]
- 16.Kenning M, Müller CHG, Sombke A. 2017. The ultimate legs of Chilopoda (Myriapoda): a review on their morphological disparity and functional variability. PeerJ 5, e4023-e4036. ( 10.7717/peerj.4023) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willemart RH, Farine JP, Gnaspini P. 2009. Sensory biology of Phalangida harvestmen (Arachnida, Opiliones): a review, with new morphological data on 18 species. Acta Zool. 90, 209-227. ( 10.1111/j.1463-6395.2008.00341.x) [DOI] [Google Scholar]
- 18.Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. 2017. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722-736. ( 10.1101/gr.215087.116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boetzer M, Pirovano W. 2014. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinf. 15, 211. ( 10.1186/1471-2105-15-211) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.English AC, et al. 2012. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS ONE 7, e47768. ( 10.1371/journal.pone.0047768) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963. ( 10.1371/journal.pone.0112963) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guan D, McCarthy SA, Wood J, Howe K, Wang Y, Durbin R. 2020. Identifying and removing haplotypic duplication in primary genome assemblies. Bioinformatics 36, 2896-2898. ( 10.1093/bioinformatics/btaa025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smit A, Hubley R, Green P. 2015. RepeatMasker Open-4.0. 2013–2015.
- 24.Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. 2019. Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962, 65-95. ( 10.1007/978-1-4939-9173-0_5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoff KJ. 2019. MakeHub: fully automated generation of UCSC genome browser assembly hubs. Genom. Proteom. Bioinform. 17, 546-549. ( 10.1016/j.gpb.2019.05.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403-410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 27.Sharma PP, Schwager EE, Giribet G, Jockusch EL, Extavour CG. 2013. Distal-less and dachshund pattern both plesiomorphic and apomorphic structures in chelicerates: RNA interference in the harvestman Phalangium opilio (Opiliones). Evol. Dev. 15, 228-242. ( 10.1111/ede.12029) [DOI] [PubMed] [Google Scholar]
- 28.Bruce HS, Jerz G, Kelly S, McCarthy J, Pomerantz A, Senevirathne G, Sherrard A, Sun DA, Wolff C, Patel NH. 2021. Hybridization chain reaction (HCR) in situ protocol. ( 10.17504/protocols.io.bunznvf6) [DOI] [Google Scholar]
- 29.Gulia-Nuss M, et al. 2016. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun. 7, 10507. ( 10.1038/ncomms10507) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, Zdobnov EM. 2017. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543-548. ( 10.1093/molbev/msx319) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chipman AD, et al. 2014. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLOS Biol. 12, e1002005. ( 10.1371/journal.pbio.1002005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pace RM, Grbić M, Nagy LM. 2016. Composition and genomic organization of arthropod Hox clusters. EvoDevo 7, 1-11. ( 10.1186/s13227-016-0048-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Setton EVW, March LE, Nolan ED, Jones TE, Cho H, Wheeler WC, Extavour CG, Sharma PP. 2017. Expression and function of spineless orthologs correlate with distal deutocerebral appendage morphology across Arthropoda. Dev. Biol. 430, 224-236. ( 10.1016/j.ydbio.2017.07.016) [DOI] [PubMed] [Google Scholar]
- 34.Schomburg C, Turetzek N, Schacht MI, Schneider J, Kirfel P, Prpic NM, Posnien N. 2015. Molecular characterization and embryonic origin of the eyes in the common house spider Parasteatoda tepidariorum. EvoDevo 6, 15. ( 10.1186/s13227-015-0011-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Samadi L, Schmid A, Eriksson BJ. 2015. Differential expression of retinal determination genes in the principal and secondary eyes of Cupiennius salei Keyserling (1877). EvoDevo 6, 16. ( 10.1186/s13227-015-0010-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gainett G, Ballesteros JA, Kanzler CR, Zehms JT, Zern JM, Aharon S, Gavish-Regev E, Sharma PP. 2020. Systemic paralogy and function of retinal determination network homologs in arachnids. BMC Genom. 21, 811-817. ( 10.1186/s12864-020-07149-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Telford MJ, Thomas RH. 1998. Expression of homeobox genes shows chelicerate arthropods retain their deutocerebral segment. Proc. Natl Acad. Sci. USA 95, 10 671-10 675. ( 10.1073/pnas.95.18.10671) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pechmann M, Schwager EE, Turetzek N, Prpic NM. 2015. Regressive evolution of the arthropod tritocerebral segment linked to functional divergence of the Hox gene labial. Proc. R. Soc. B 282, 20151162. ( 10.1098/rspb.2015.1162) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galindo MI, Bishop SA, Couso JP. 2005. Dynamic EGFR-Ras signalling in Drosophila leg development. Dev. Dyn. 233, 1496-1508. ( 10.1002/dvdy.20452) [DOI] [PubMed] [Google Scholar]
- 40.Campbell G. 2002. Distalization of the Drosophila leg by graded EGF-receptor activity. Nature 418, 781-785. ( 10.1038/nature00971) [DOI] [PubMed] [Google Scholar]
- 41.Galindo MI, Bishop SA, Greig S, Couso JP. 2002. Leg patterning driven by proximal–distal interactions and EGFR signaling. Science 297, 256-259. ( 10.1126/science.1072311) [DOI] [PubMed] [Google Scholar]
- 42.Brunner D, Dücker K, Oellers N, Hafen E, Scholzi H, Klämbt C. 1994. The ETS domain protein Pointed-P2 is a target of MAP kinase in the Sevenless signal transduction pathway. Nature 370, 386-389. ( 10.1038/370386a0) [DOI] [PubMed] [Google Scholar]
- 43.Grossmann D, Prpic NM. 2012. Egfr signaling regulates distal as well as medial fate in the embryonic leg of Tribolium castaneum. Dev. Biol. 370, 264-272. ( 10.1016/j.ydbio.2012.08.005) [DOI] [PubMed] [Google Scholar]
- 44.Refki PN, Khila A. 2015. Key patterning genes contribute to leg elongation in water striders. EvoDevo 6, 14. ( 10.1186/s13227-015-0015-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakamura T, Mito T, Miyawaki K, Ohuchi H, Noji S. 2008. EGFR signaling is required for re-establishing the proximodistal axis during distal leg regeneration in the cricket Gryllus bimaculatus nymph. Dev. Biol. 319, 46-55. ( 10.1016/j.ydbio.2008.04.002) [DOI] [PubMed] [Google Scholar]
- 46.Halfar K, Rommel C, Stocker H, Hafen E. 2001. Ras controls growth, survival and differentiation in the Drosophila eye by different thresholds of MAP kinase activity. Development 128, 1687-1696. ( 10.5167/uzh-625) [DOI] [PubMed] [Google Scholar]
- 47.Gainett G, Sharma PP. 2020. Genomic resources and toolkits for developmental study of whip spiders (Amblypygi) provide insights into arachnid genome evolution and antenniform leg patterning. EvoDevo 11, 18-18. ( 10.1186/s13227-020-00163-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Baudouin-Gonzalez L, et al. 2021. The evolution of Sox gene repertoires and regulation of segmentation in arachnids. Mol. Biol. Evol. msab088. ( 10.1093/molbev/msab088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brown S, DeCamillis M, Gonzalez-Charneco K, Denell M, Beeman R, Nie W, Denell R. 2000. Implications of the Tribolium Deformed mutant phenotype for the evolution of Hox gene function. Proc. Natl Acad. Sci. USA 97, 4510-4514. ( 10.1073/pnas.97.9.4510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hughes CL, Kaufman TC. 2000. RNAi analysis of Deformed, proboscipedia and Sex combs reduced in the milkweed bug Oncopeltus fasciatus: novel roles for Hox genes in the hemipteran head. Development 127, 3683-3694. ( 10.1242/dev.127.17.3683) [DOI] [PubMed] [Google Scholar]
- 51.Martin A, Serano JM, Jarvis E, Bruce HS, Wang J, Ray S, Barker CA, O'Connell LC, Patel NH. 2016. CRISPR/Cas9 mutagenesis reveals versatile roles of Hox genes in crustacean limb specification and evolution. Curr. Biol. 26, 14-26. ( 10.1016/j.cub.2015.11.021) [DOI] [PubMed] [Google Scholar]
- 52.Janssen R, Eriksson BJ, Tait NN, Budd GE. 2014. Onychophoran Hox genes and the evolution of arthropod Hox gene expression. Front. Zool. 11, 22. ( 10.1186/1742-9994-11-22) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Angelini DR, Liu PZ, Hughes CL, Kaufman TC. 2005. Hox gene function and interaction in the milkweed bug Oncopeltus fasciatus (Hemiptera). Dev. Biol. 287, 440-455. ( 10.1016/j.ydbio.2005.08.010) [DOI] [PubMed] [Google Scholar]
- 54.Khadjeh S, Turetzek N, Pechmann M, Schwager EE, Wimmer EA, Damen WGM, Prpic NM. 2012. Divergent role of the Hox gene Antennapedia in spiders is responsible for the convergent evolution of abdominal limb repression. Proc. Natl Acad. Sci. USA 109, 4921-4926. ( 10.1073/pnas.1116421109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gainett G, González VL, Ballesteros JA, Setton EVW, Baker CM, Barolo Gargiulo L, Santibáñez-López CE, Coddington JA, Sharma PP. 2021. Data from: The genome of a daddy-long-legs (Opiliones) illuminates the evolution of arachnid appendages. Dryad Digital Repository. ( 10.5061/dryad.ht76hdrds) [DOI] [PMC free article] [PubMed]
- 56.Gainett G, González VL, Ballesteros JA, Setton EVW, Baker CM, Barolo Gargiulo L, Santibáñez-López CE, Coddington JA, Sharma PP. 2021. Data from: The genome of a daddy-long-legs (Opiliones) illuminates the evolution of arachnid appendages. Figshare. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Gainett G, González VL, Ballesteros JA, Setton EVW, Baker CM, Barolo Gargiulo L, Santibáñez-López CE, Coddington JA, Sharma PP. 2021. Data from: The genome of a daddy-long-legs (Opiliones) illuminates the evolution of arachnid appendages. Dryad Digital Repository. ( 10.5061/dryad.ht76hdrds) [DOI] [PMC free article] [PubMed]
- Gainett G, González VL, Ballesteros JA, Setton EVW, Baker CM, Barolo Gargiulo L, Santibáñez-López CE, Coddington JA, Sharma PP. 2021. Data from: The genome of a daddy-long-legs (Opiliones) illuminates the evolution of arachnid appendages. Figshare. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material. The genome assembly, EGFR tree and alignment, are available from the Dryad Digital Repository: https://doi.org/10.5061/dryad.ht76hdrds [55]. Sequencing data are deposited in NCBI under accession numbers NCBI PRJNA690950 (RNA-seq), NCBI PRJNA647749 (genome), NCBI SRR12286133 (long reads), NCBI SRR12286133 (short reads). The genome browser is available at: http://phalangium-opilio.sigenomehub.org/.
The data are provided in electronic supplementary material [56].