Abstract
In mammalian cloning by somatic cell nuclear transfer (SCNT), the treatment of reconstructed embryos with histone deacetylase (HDAC) inhibitors improves efficiency. So far, most of those used for SCNT are hydroxamic acid derivatives—such as trichostatin A—characterized by their broad inhibitory spectrum. Here, we examined whether mouse SCNT efficiency could be improved using chlamydocin analogues, a family of newly designed agents that specifically inhibit class I and IIa HDACs. Development of SCNT-derived embryos in vitro and in vivo revealed that four out of five chlamydocin analogues tested could promote the development of cloned embryos. The highest pup rates (7.1–7.2%) were obtained with Ky-9, similar to those achieved with trichostatin A (7.2–7.3%). Thus, inhibition of class I and/or IIa HDACs in SCNT-derived embryos is enough for significant improvements in full-term development. In mouse SCNT, the exposure of reconstructed oocytes to HDAC inhibitors is limited to 8–10 h because longer inhibition with class I inhibitors causes a two-cell developmental block. Therefore, we used Ky-29, with higher selectivity for class IIa than class I HDACs for longer treatment of SCNT-derived embryos. As expected, 24-h treatment with Ky-29 up to the two-cell stage did not induce a developmental block, but the pup rate was not improved. This suggests that the one-cell stage is a critical period for improving SCNT cloning using HDAC inhibitors. Thus, chlamydocin analogues appear promising for understanding and improving the epigenetic status of mammalian SCNT-derived embryos through their specific inhibitory effects on HDACs.
Keywords: cloned embryo, histone deacetylase, histone deacetylase inhibitor, mouse, somatic cell nuclear transfer
Chlamydocin analogues, a novel family of inhibitors specific for class I and IIb HDACs, significantly improved the ability of mouse SCNT-derived embryos to produce offspring.
Introduction
Cloning by somatic cell nuclear transfer (SCNT) is a technique that produces embryos or animals that are genetically identical to the donor cells. Since the birth of Dolly the sheep derived from an adult mammary gland cell, SCNT has been heralded as a promising assisted reproductive technology with a broad range of applications in regenerative medicine, livestock industries, and pharmaceutical manufacturing [1–5]. Recent advancements in gene-editing technologies, especially the CRISPR/Cas9 system, have further enhanced the use of SCNT in livestock animals including pigs, goats, cattle, and sheep for the production of useful phenotypes such as disease resistance and high meat/milk production [6]. Meanwhile, there have been many attempts to increase the efficiency of SCNT, especially in mice. Cloned mouse embryos, fetuses, and placentas have provided invaluable information on epigenetic aberrations associated with SCNT, and their corrections lead to significant improvements in cloning efficiency [3, 7]. However, many of the effective treatments require genetically modified donor cells or further micromanipulation of reconstructed embryos [8–11]. In this respect, one of the most feasible measures that has been shown to improve mouse cloning is the treatment of SCNT-derived embryos with histone deacetylase (HDAC) inhibitors. HDACs are a family of enzymes involved in the regulation of a number of cellular processes including the control of gene expression through histone acetylation. Therefore, HDAC inhibitors are expected to induce an open chromatin structure of the donor cell-derived genome, leading to efficient transcription of developmentally related genes, especially transcription factors [12, 13]. The first HDAC inhibitor used successfully for mouse SCNT was trichostatin A (TSA), a naturally derived hydroxamate, which has a broad spectrum for inhibiting different HDAC families [14]. Mouse SCNT-derived embryos treated with TSA showed a high potency to develop into blastocysts in vitro and to term offspring in vivo, with pup rates reaching more than 5% per embryos transferred [15]. TSA was also reported to be effective for SCNT cloning in bovines [16, 17], pigs [18, 19], and rabbits [20, 21].
HDACs are grouped into four classes based on structural and functional similarities. The class I isoforms (HDAC1, −2, −3, and −8), class IIa (HDAC4, −5, −7, and −9), and class IIb (HDAC6 and −10) are Zn-dependent enzymes, whereas class III isoforms (SIRT1 to 7) are NAD+-dependent [22]. HDAC inhibitors can also be characterized as pan-HDAC inhibitors or selective HDAC inhibitors according to their selectivity for specific classes. In mouse cloning experiments, it is desirable to use HDAC inhibitors that have selective inhibitory activities for the most efficient and safe SCNT outcomes. However, only hydroxamates—typical pan-HDAC inhibitors including TSA [15, 23], scriptaid [24], suberoylanilide hydroxamic acid (SAHA) [25], oxamflatin [25], m-carboxycinnamic acid bishydroxamide [26], and PXD101 (belinostat) [27]—have been proven to increase mouse cloning efficiencies.
Besides hydroxamates, there are HDAC inhibitors that belong to other molecular families such as short-chain fatty acids, cyclic peptides, and benzamide [28], but none of them have been used effectively for improving mouse SCNT cloning. HDAC inhibitors generally exhibit strong binding to HDACs. For example, TSA is thought to chelate the zinc ion in the active site in a bidentate fashion and binds strongly to HDAC through an aromatic “Cap” structure [29]. Chlamydocin is also a naturally occurring HDAC inhibitor with a cyclic tetrapeptide structure that can bind strongly to HDACs. Based on the structure of chlamydocin, a series of chlamydocin analogues containing various metal-binding groups have been developed as potent HDAC inhibitors [30–32]. These synthetic compounds inhibited class I and IIa HDACs, but not IIb HDACs, at nanomolar concentrations [31] (Figure 1). FK228 (romidepsin), an anti-T-cell lymphoma agent that also has a cyclic tetrapeptide structure (Figure 1), is known to inhibit class I HDACs (HDAC1 and HDAC2) [33]. Here, we examined whether the development of mouse SCNT-derived embryos could be improved by treatment with these newly developed chlamydocin analogue HDAC inhibitors and FK228, which have more selective inhibitory effects than the hydroxamate HDAC inhibitors such as TSA.
Figure 1.
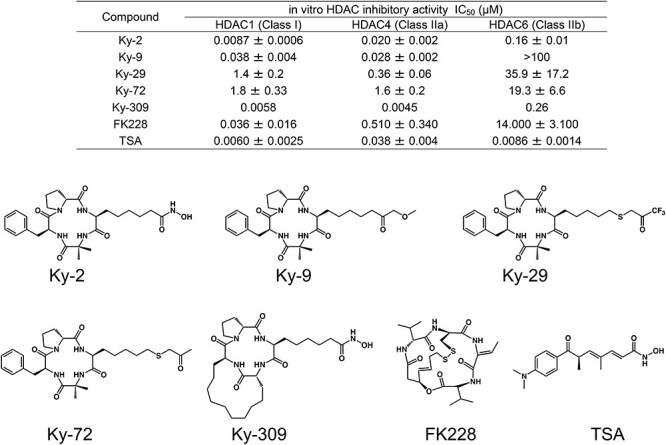
HDAC inhibitory activity and the structure of HDAC inhibitors used in this study. The HDAC inhibitory activities (IC50 values) of HDAC inhibitors were measured as described in the Materials and methods or are quoted from references [30–33, 57].
Materials and methods
Animals
Eight- to 10-week-old B6D2F1 (C57BL/6 × DBA/2 hybrid) female and male mice were purchased from Japan SLC (Shizuoka, Japan) and used for the collection of oocytes and donor cumulus cells. Eight- to 10-week-old C57BL/6N male mice purchased from Japan SLC were used for in vitro fertilization (IVF). ICR strain female mice (CLEA Japan, Inc., Tokyo, Japan), 8–12 weeks old, were used as embryo transfer recipients. The animals were housed under a controlled lighting condition (daily light 07:00–21:00 h) and were maintained under specific pathogen-free conditions. All animal experiments were approved by the Animal Experimentation Committee at the RIKEN Tsukuba Institute and were performed in accordance with the committee’s guiding principles.
Somatic cell nuclear transfer
SCNT was carried out as described previously [34, 35]. Briefly, B6D2F1 female mice were superovulated by the injection of 7.5 IU of equine chorionic gonadotropin (Sankyo Yell Yakuhin, Co., Tokyo, Japan) and 7.5 IU of human chorionic gonadotropin (hCG; ASKA Pharmaceutical Co., Ltd., Tokyo, Japan) at a 48-h interval. At 15 h after hCG injection, the mice were euthanized by cervical dislocation and cumulus–oocyte complexes were collected from the oviducts. Cumulus cells were removed by suspending the complexes in potassium-modified simplex optimized medium (KSOM) [36] containing 0.1% bovine testicular hyaluronidase. The oocytes were enucleated in Hepes-buffered KSOM containing 7.5 μg/ml cytochalasin B (CB) using a piezo-driven micropipette (PMM-150FU; Primetech, Ibaraki, Japan). After culture in KSOM under 5% CO2 in air at 37°C for at least 1 h, the enucleated oocytes were injected with donor cumulus cells using a piezo-driven micropipette in Hepes-buffered KSOM without bovine serum albumin (BSA). After culture in KSOM for about 1 h, injected oocytes were incubated in Ca2+-free KSOM containing 2.5 mM SrCl2 and 5 μg/ml CB with or without a HDAC inhibitor (TSA, chlamydocin analogues, or FK228) for 1 h. Next, the oocytes were incubated in KSOM containing 5 μg/ml CB and a HDAC inhibitor for 5 h followed by incubation in KSOM containing a HDAC inhibitor alone (where appropriate) for 2 h. After washing, the SCNT-derived embryos were cultured in KSOM under 5% CO2 in air at 37°C. The embryos reached the two-cell or morula/blastocyst stage after 24 h or 72 h in culture, respectively. The two-cell embryos were transferred into the oviducts of pseudopregnant ICR female mice at 0.5 days postcopulation (dpc; the day following sterile mating with a vasectomized male mouse). The morulae/blastocysts were transferred into the uteri of 2.5 dpc recipients. The pregnant females were euthanized at 19.5 dpc and examined for fetuses and placentas in their uteri. Some live pups were nursed by foster ICR mothers until weaning.
In some experiments, SCNT-derived embryos treated with Ky-9 were additionally treated with 10 μg/ml vitamin C and 0.4% deionized BSA for 8 h [37] (Supplementary Figure S1). The embryos that reached the two-cell stage after 24 h in culture were transferred into the oviducts of recipient females, as described above. The pregnant females were euthanized at 19.5 dpc and examined for fetuses and placentas in their uteri.
In vitro fertilization
IVF-derived embryos were obtained as described previously [38]. Briefly, cumulus–oocyte complexes were collected from the oviducts of B6D2F1 female mice that had been superovulated as described above and were transferred into human tubal fluid (HTF) medium at 37°C under 5% CO2 in humidified air. Spermatozoa were collected from the cauda epididymis of 12-week-old C57BL/6 male mice and preincubated in HTF medium for 1 h at 37°C under 5% CO2 in humidified air. Preactivated spermatozoa were transferred into the oocyte culture medium at concentrations of 200–400 spermatozoa/μl. After 6 h of culture, pronuclear-stage embryos were transferred into KSOM and used for immunostaining as described below.
Intracytoplasmic sperm injection
Intracytoplasmic sperm injection (ICSI)-derived pups were obtained as reported [39]. Briefly, cumulus-free oocytes prepared as described above were each injected with an epididymal sperm head in Hepes-buffered KSOM without BSA. The injected oocytes were cultured in KSOM medium for 24 h at 37°C under 5% CO2 in humidified air. Embryos that reached the two-cell stage were transferred into the oviducts of recipient females. The pregnant females were euthanized at 19.5 dpc and live pups in the uteri were nursed by foster ICR mothers until weaning.
HDAC inhibitors and IC50 measurements
Chlamydocin analogues with different side chains (Ky-2, Ky-9, Ky-29, Ky-72, and Ky-309) were used in this study. The syntheses of Ky-2 (Compound 3 in [31]) and Ky-309 (Compound 3 in [32]) have been reported previously. Ky-9 was synthesized by four steps (epoxidation, ring opening, methylation, and Dess–Martin periodinane reactions) starting from the cyclic tetrapeptide cyclo(-L-Ae9-Aib-L-Phe-D-Pro-), where Ae9 is 2-amino-8-nonenoic acid. The final step gave 270 mg (85%) of white solid after silica gel chromatography. The structure was confirmed by HR-FAB-MS and 500 MHz 1H NMR measurements. Ky-29 and Ky-72 were synthesized by the use of the same starting material, cyclo(−L-Am7-Aib-L-Phe-D-Pro-) [40] where Am7 is 2-amino-7-mercapto-heptanoic acid. In addition to the thiol function for zinc ion coordination, we attempted to add carbonyl function by thioether incorporation. To this cyclic tetrapeptide precursor, we reacted 1,1,1-trifluoro-3-bromoacetone (TCI, Tokyo, Japan) dissolved in N,N-dimethylformamide in the presence of triethylamine. The product, Ky-29, was purified by silica gel chromatography. The structure was confirmed by HR-FAB-MS and 500 MHz 1H NMR measurements. Using the same procedure, bromoacetone was reacted with the thiol moiety of Am7 in the cyclic tetrapeptide to produce Ky-72. Again, the structure was confirmed by HR-FAB-MS and 500 MHz 1H NMR measurements. TSA and FK228 were purchased from Merck KGaA (Darmstadt, Germany) and Abcam Japan Co. (Tokyo, Japan), respectively.
The HDAC inhibitors were dissolved in dimethyl sulfoxide (DMSO; Thermo Fisher Scientific, Waltham, MA) as a stock solution and stored at −40°C before use. They were added to the culture medium on the day of experiments. The DMSO concentration was adjusted to 0.4% in all media used.
Assays for HDAC enzyme activities were performed with fluorescent peptide substrates and recombinant HDAC proteins purified from 293T cells as described [41]. The half-maximal inhibitory concentration (IC50) values of HDAC inhibitors were determined as the mean ± standard deviation (SD) of the concentrations calculated from dose–response curves. The IC50 values and structures of the chlamydocin analogues are shown in Figure 1, together with those of TSA and FK228.
Production of parthenogenetic embryos
Metaphase II (MII) oocytes were collected from B6D2F1 female mice as described above. They were activated by treatment with Ca2+-free CZB medium [42] containing 2.5 mM SrCl2 and 5 μg/ml CB for about 15 min and then transferred into CZB medium containing 5 μg/ml CB with or without an HDAC inhibitor for 6 h followed by incubation in CZB medium containing an HDAC inhibitor alone (where appropriate) for 2 h. The embryos were washed and cultured in CZB medium as described above until analysis.
Immunostaining
Histone acetylation levels of SCNT-derived or IVF-derived embryos treated with HDAC inhibitors were analyzed by immunostaining. The embryos were fixed in 100% ethanol (FUJIFILM Wako Pure Chemical Corporation, Osaka, Japan) at −20°C for 20 min and permeabilized with phosphate-buffered saline (PBS) containing 0.2% Triton X-100 (Merck KGaA) at room temperature (RT) for 1 h. After blocking with 3% BSA in PBS containing 0.2% Triton X-100 for 1 h, they were incubated with primary antibodies (anti-acetyl-Histone H3 antibody; Merck Millipore, Darmstadt, Germany) in PBS containing 30 mg/ml BSA at 4°C overnight. After washing with PBS, the embryos were reacted with secondary antibodies (Anti-rabbit IgG-Alexa 555; Thermo Fisher Scientific) at RT for 1 h. The embryos were mounted on glass slides in Vectashield Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA). The slides were imaged using a CellVoyager CV1000 confocal scanner system (Yokogawa Electronic, Tokyo, Japan) and fluorescence intensity was measured using image processing software (ImageJ; https://imagej.net/Welcome).
Transcriptional activity assay
For detecting nascent RNA synthesis during zygotic gene activation (ZGA) at the two-cell stage, diploid parthenogenetic embryos were generated as above and treated with or without an HDAC inhibitor (Ky-9, Ky-309, TSA, or FK228). Transcription was detected by Click-iT RNA Alexa Fluor 594 Imaging Kits (Thermo Fisher Scientific) according to the manufacturer’s instructions. Briefly, parthenogenetic embryos at 5 h after activation were cultured in CZB medium containing 2 mM 5-ethynyl uridine (EU) until 24 h after activation (19 h exposure). They were fixed with 4% paraformaldehyde (FUJIFILM Wako Pure Chemical Corporation) and permeabilized with 0.2% Triton X-100 in PBS containing 30 mg/ml BSA. The parthenogenetic embryos were then incubated in the Click-iT reaction cocktail at RT for 30 min and washed with the Click-iT reaction rinse buffer. Fluorescence images of EU were detected using a CellVoyager CV1000 confocal microscope scanner system with consistent laser strength and fluorescence intensity was measured using ImageJ.
Statistical analysis
The developmental rates of embryos (Tables 1,2, and4; Supplementary Table S1 and Figure S1) were analyzed using Fisher exact probability test. Immunofluorescence (Figure 2) and body and placental weights (Figures 2 and 3; Table 3) were analyzed using Turkey test for multiple comparisons between groups. P values <0.05 were considered statistically significant.
Table 1.
Development in vitro and in vivo (morula/blastocyst transfer) of SCNT-derived embryos treated with HDAC inhibitors
| HDAC inhibitor | Inhibitor concentration (nM) | No. of embryos cultured | No. of two-cell embryos at 24 h (%) | No. of four-cell embryos at 48 h (% per 2 cells) | No. of morula/blast at 72 h (% per 2 cells) | No. of embryos transferred (no. of recipients) | No. of implantations (%) | No. of cloned offspring (%) | No. of placentas only (%) |
|---|---|---|---|---|---|---|---|---|---|
| Control | – | 530 | 484 (91.3) | 382 (78.9) | 271 (56.0) | 210 (14) | 83 (39.5) | 0 (0) | 3 (1.4) |
| TSA | 5 | 64 | 49 (76.5)* | 45 (91.8)* | 32 (65.3) | 15 (1) | 6 (40.0) | 1 (6.7) | 0 (0) |
| Ky-2 | 100 | 63 | 52 (82.5)* | 45 (86.5) | 31 (59.6) | 31 (2) | 7 (22.6)* | 1 (3.2) | 1 (3.2) |
| 10 | 67 | 62 (92.5) | 57 (91.9)* | 38 (61.3) | 38 (2) | 18 (47.4) | 1 (2.6) | 0 (0) | |
| Ky-9 | 1600 | 69 | 57 (82.6)* | 44 (77.2) | 28 (49.1) | 28 (2) | 19 (67.9)* | 2 (7.1)* | 3 (10.7)* |
| 32 | 66 | 60 (90.9) | 50 (83.3) | 35 (58.3) | 25 (2) | 10 (40.0) | 0 (0) | 1 (4.0) | |
| Ky-29 | 2000 | 61 | 54 (88.5) | 51 (94.4)* | 37 (68.5) | 37 (2) | 21 (56.8)* | 1 (2.7) | 4 (10.8)* |
| 400 | 164 | 155 (94.5) | 132 (85.2) | 109 (70.3)* | 88 (5) | 29 (33.0) | 4 (4.5)* | 1 (1.1) | |
| Ky-72 | 1600 | 57 | 47 (82.5)* | 38 (80.9) | 28 (59.6) | 28 (2) | 18 (64.3)* | 2 (7.1)* | 3 (10.7)* |
| Ky-309 | 100 | 69 | 58 (84.0)* | 27 (46.6)* | 6 (10.3)* | 6 (2) | 4 (66.7)* | 1 (16.7)* | 1 (16.7) |
| 5 | 113 | 106 (93.8) | 89 (84.0) | 61 (57.5) | 21 (1) | 4 (19.0)* | 0 (0) | 0 (0) | |
| FK228 | 20 | 67 | 63 (94.0) | 2 (3.2)* | 0 (0)* | N.D. | N.D. | N.D. | N.D. |
* P < 0.05 (vs. control, Fisher exact probability test).
Table 2.
Development in vitro and in vivo (two-cell transfer) of SCNT-derived embryos treated with HDAC inhibitors
| HDAC inhibitor | Concentration of inhibitor (nM) | No. of embryos cultured | No. (%) of two-cell embryos at 24 h | No. of embryos transferred (no. of recipients) | No. of implantations (%) | No. of offspring (%) | No. of placentas only (%) |
|---|---|---|---|---|---|---|---|
| Control | – | 487 | 416 (85.4) | 213 (8) | 93 (43.7) | 6 (2.8) | 6 (2.8) |
| TSA | 5 | 153 | 133 (86.9) | 109 (6) | 69 (63.3)* | 8 (7.3) | 3 (2.8) |
| Ky-2 | 100 | 153 | 142 (92.8)* | 137 (6) | 55 (40.1) | 3 (2.2) | 3 (2.2) |
| 10 | 74 | 68 (91.9) | 73 (4) | 40 (54.8) | 1 (1.4) | 3 (4.1) | |
| Ky-9 | 3200 | 232 | 203 (87.5) | 135 (6) | 78 (57.8)* | 9 (6.7) | 2 (1.5) |
| 1600 | 196 | 179 (91.3)* | 139 (7) | 74 (53.2)* | 10 (7.2)* | 4 (2.9) | |
| 320 | NR | NR | 194 (6) | 103 (53.1)* | 9 (4.6) | 3 (1.5) | |
| 32 | 71 | 60 (84.5) | 69 (4) | 39 (56.5)* | 2 (2.9) | 3 (4.3) | |
| Ky-29 | 2000 | 335 | 292 (87.2) | 217 (9) | 85 (39.2) | 3 (1.4) | 3 (1.4) |
| 400 | 228 | 203 (89.0) | 181 (9) | 116 (64.1)* | 6 (3.3) | 2 (1.1) | |
| Ky-72 | 1600 | 113 | 105 (92.9)* | 85 (3) | 36 (42.4) | 2 (2.4) | 1 (1.2) |
| Ky-309 | 5 | 73 | 66 (90.4) | 66 (4) | 33 (50.0) | 1 (1.5) | 2 (3.0) |
| 1 | 247 | 190 (76.9)* | 68 (4) | 21 (30.9)* | 0 (0) | 0 (0) | |
| 0.5 | 288 | 251 (87.2) | 84 (3) | 32 (38.1) | 1 (1.2) | 0 (0) |
NR, not recorded.
* P < 0.05 (vs. control, Fisher exact probability test).
Table 4.
In vitro development of SCNT-derived embryos following extended treatment with HDAC inhibitors for 24 h
| Group | Time (h) of inhibitor treatment | No. of embryos cultured | No. (%) of two-cell embryos at 24 h (%) | No. (%) of four-cell embryos at 48 h (per two cells) | No. of embryos at 96 h (% per two cells) | |
|---|---|---|---|---|---|---|
| Morulae | Blastocysts | |||||
| Control | None | 20 | 20 (100) | 16 (80.0) | 0 (0) | 8 (40.0) |
| TSA | 8 | 22 | 22 (100) | 19 (86.4) | 0 (0) | 15 (68.2) |
| Ky-29 | 8 | 23 | 23 (100) | 20 (87.0) | 0 (0) | 15 (65.2) |
| TSA + TSA | 24 | 20 | 20 (100) | 3 (15.0)* | 3 (15.0) | 0 (0)* |
| TSA + Ky-29 | 24 | 21 | 19 (90.5) | 9 (47.4)* | 0 (0) | 13 (68.4) |
| Ky-29 + Ky-29 | 24 | 20 | 20 (100) | 17 (85.0) | 0 (0) | 11 (55.0) |
* P < 0.05 (vs. control, Fisher exact probability test).
Figure 2.
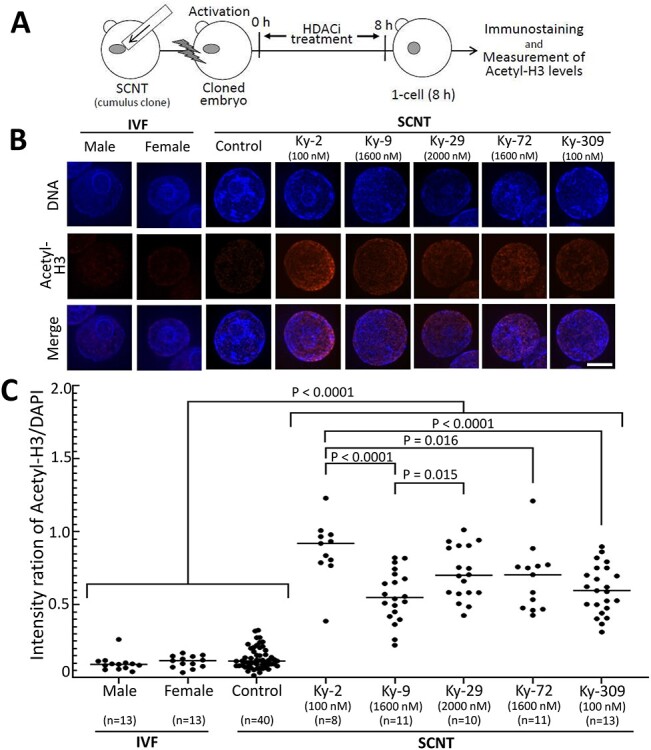
Acetyl-H3 levels of nuclei of one-cell SCNT-derived embryos treated with chlamydocin analogue HDAC inhibitors. (A) Scheme of the experiment. (B) Representative images of acetyl-H3 immunofluorescence and DNA (DAPI) in the nuclei of one-cell SCNT-derived embryos with or without (control) HDAC inhibitor treatment and IVF-derived embryos. The male and female pronuclei of IVF-derived embryos were analyzed separately. Bar = 5 μm. (C) Relative fluorescent intensity of acetyl-H3 measured. The DAPI density was set to 1. Each dot represents one embryo. The fluorescence levels of all HDAC inhibitor treatment groups were significantly higher than that of the control embryos (P < 0.0001, by Tukey test).
Figure 3.
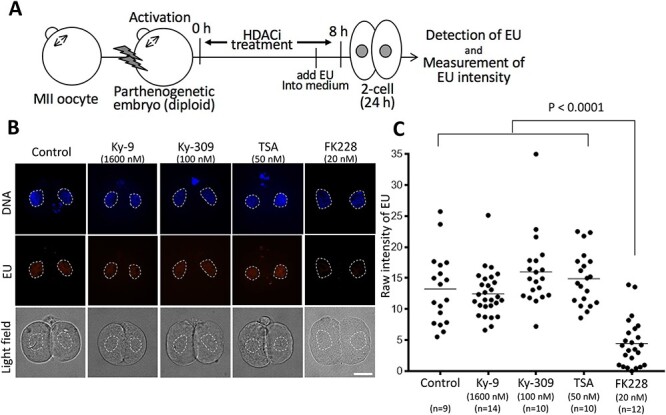
Transcriptional activity assay at the two-cell stage in parthenogenetic embryos treated with FK228 and other HDAC inhibitors. (A) Scheme of the transcriptional activity assay using parthenogenetic embryos. 5-EU was added to the medium at 5 h after activation and the embryos were cultured until 24 h. Nascent RNA transcription during the major zygote gene activation (ZGA) phase at the two-cell stage was measured by incorporation of EU detected by specific fluorescence. (B) Representative images of fluorescence specific for incorporated EU in control and HDAC inhibitor-treated parthenogenetic embryos. Bar = 20 μm. (C) Relative fluorescent intensity for incorporated EU (see Materials and methods). Each dot represents one nucleus (two nuclei per embryo). The fluorescence level of the FK228-treated embryos was significantly lower that of the embryos treated with other HDAC inhibitors (P < 0.0001 by Tukey test).
Table 3.
Body and placental weights of pups derived from HDAC inhibitor-treated SCNT-derived embryos
| Control | Ky-2 | Ky-9 | Ky-29 | Ky-72 | Ky-309 | TSA | |
|---|---|---|---|---|---|---|---|
| No. of offspring and placentas | 4 | 5 | 25 | 13 | 3 | 2 | 7 |
| Body weight at term (g ± SD) | 1.49 ± 0.18 | 1.64 ± 0.19 | 1.39 ± 0.29 | 1.41 ± 0.27 | 1.78 ± 0.16 | 1.61 ± 0.21 | 1.57 ± 0.15 |
| Placental weight at term (g ± SD) | 0.36 ± 0.09 | 0.41 ± 0.09 | 0.30 ± 0.11 | 0.36 ± 0.06 | 0.40 ± 0.04 | 0.31 ± 0.05 | 0.28 ± 0.07 |
There were no significant differences between the HDAC inhibitor groups and the control group for each parameter (Tukey multiple comparisons test).
Results
Histone acetylation levels in SCNT-derived embryos after treatment with chlamydocin analogues
We first sought to identify which of the chlamydocin analogues had the potential to increase the histone acetylation levels of one-cell mouse embryos. The concentrations of the analogues in the culture medium were determined based on their inhibitory activities (IC50 values) for HDAC1 and HDAC4 (Figure 1). The histone acetylation level was measured from the fluorescent intensity of the nuclei in SCNT-derived embryos at 8 h of HDAC inhibitor treatment using an anti-acetyl-Histone H3 antibody (Figure 2A). All the analogues tested significantly increased the histone acetylation levels in SCNT-derived embryos compared with those of nontreated SCNT-derived control embryos (Figure 2B and C). The histone acetylation levels of nontreated SCNT-derived control embryos were similar to those of IVF-derived embryos (Figure 2B and C), consistent with a previous report [43]. These results indicate that these analogues might induce a loosened chromatin structure, reflecting a more reprogrammable genomic state following SCNT.
Developmental ability of SCNT-derived embryos treated with chlamydocin analogues
Next, we examined whether these analogues could improve the in vitro developmental ability of SCNT-derived embryos. In addition to the concentrations above, lower concentrations were also tested for some analogues. Compared with untreated SCNT-derived embryos (controls), only Ky-29 at the higher concentration (2000 nM) significantly increased both four-cell and morula/blastocyst formation rates (P < 0.05; Table 1). By contrast, Ky-309 at the higher concentration (100 nM) significantly decreased both four-cell and morula/blastocyst formation rates (P < 0.05; Table 1). FK228 induced a strong developmental arrest of SCNT-derived embryos at the two-cell stage and no morulae were obtained (Table 1). In other experimental groups including controls, about 80 and 50% of embryos (per two-cell stage) developed into four-cell and morulae/blastocyst stages, respectively (Table 1). After transfer of these morulae/blastocysts into recipient female mice, at least one cloned offspring was born at term, except for the control group, and in the lower concentration Ky-9 and Ky-309 groups. Of these, the higher concentration Ky-9, lower concentration Ky-29, and higher concentration Ky-72 and Ky-309 treatment groups resulted in significantly higher pup rates than in the control group. Except for Ky-309, which seemed to have embryonic toxicity, Ky-9 gave the best pup rate (7.1%), similar to that of TSA (6.7%).
Our results indicate that these novel chlamydocin analogues can have beneficial effects on the in vivo development of SCNT-derived embryos. To further examine this possibility, we undertook embryo transfer experiments using two-cell embryos. As expected, Ky-9 at 1600 nM gave the highest pup rate (7.2%), which was significantly higher than in control (2.8%, P < 0.05; Table 2). We then tested a higher concentration of Ky-9 (3200 nM). This treatment did not further increase the pup rate of SCNT-derived pups, but it was still high (6.7%, P = 0.075 vs. control; Table 2). This result indicates that the embryotoxicity of Ky-9 was low even at a concentration higher than the IC50 (see Figure 1). The pup rates obtained with Ky-9 were similar to that produced with TSA (7.3%, P = 0.059 vs. control), although TSA promoted the development of SCNT-derived embryos at a much lower concentration (5 nM) than Ky-9. We also tested an intermediate concentration of Ky-9 (320 nM) and found that the effect was modest (4.6% pup rate; Table 2).
Cloned pups obtained following HDAC inhibitor treatment commenced respiration soon after Caesarian section and showed no obvious abnormalities in appearance. The HDAC inhibitor treatment did not affect the mean body or placental weights at birth when compared with those of the control group (Table 3). We also observed the growth of pups obtained from SCNT embryos treated with Ky-9 or TSA and found that they were weaned at 3 weeks of age at rates similar to those of ICSI-derived pups (Supplementary Table S1). They showed no discernible abnormalities in behavior and appearance. All the Ky-9-derived clones tested for fertility (n = 4) produced offspring after mating with male mice (Supplementary Figure S2).
Developmental block induced by FK228 was associated with the inhibition of ZGA
As mentioned above, FK228 induced a strong two-cell developmental block following treatment for SCNT-derived embryos at the one-cell stage (Table 1). It is known that inhibition of the major ZGA phase that occurs at the two-cell stage in mouse embryos causes a developmental block [44]. To evaluate this possibility, we performed EU incorporation assays using parthenogenetic embryos treated with FK228 or other HDAC inhibitors. As shown in Figure 3, the EU fluorescence intensity was significantly lower in embryos treated with FK228 than that in control embryos, indicating that FK228 strongly inhibited ZGA at the two-cell stage. By contrast, other HDAC inhibitors (e.g., Ky-9, Ky-309, and TSA) had no adverse effects on ZGA (Figure 3), although Ky-309 induced a moderate two-cell developmental block (Table 1).
Ky-29 permitted 24-h treatment for SCNT-derived embryos but did not improve cloning efficiency
In all mouse SCNT experiments so far reported, HDAC inhibitor treatment for the resulting embryos has been limited to 8–10 h following oocyte activation. This is because HDAC1 expressed at the zygote to two-cell transition is essential for mouse embryos to develop further. Therefore, we surmised that, if longer treatments with HDAC inhibitors (e.g., 24 h) do not inhibit HDAC1, they might further improve the developmental efficiency of SCNT-derived embryos. According to the HDAC selectivity of chlamydocin analogues based on their IC50 values, Ky-29 seemed to be appropriate for the selective inhibition of HDACs other than HDAC1 (Figure 1). We then tested the effects of extended inhibitor treatments for 24 h on embryo development by an additional 16-h treatment with Ky-29 (500 nM; Figure 4A). As expected, this additional treatment following Ky-29 or TSA treatment for 8 h did not inhibit the development of SCNT-derived embryos and supported their development until blastocysts at rates of 72 and 55%, respectively (Table 4; Figure 4). As expected, no SCNT-derived embryos developed to blastocysts when they were treated with TSA for 24 h (Table 4, Figure 4). We then examined the in vivo developmental ability of SCNT-derived embryos treated sequentially with TSA then with Ky-29 by embryo transfer. However, only 1.1% of the embryos transferred reached term (Table 5).
Figure 4.
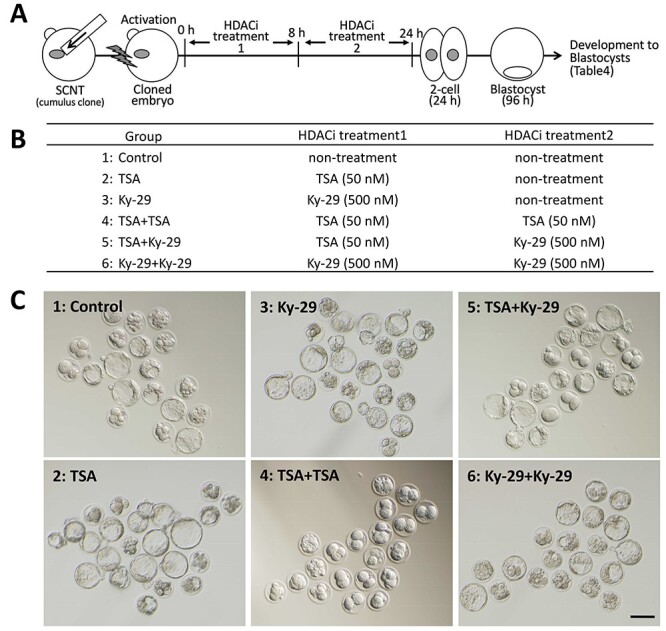
Analysis of the developmental ability of SCNT-derived embryos treated with HDAC inhibitor (s) for 24 h. (A) Scheme of the experiment. SCNT-derived embryos treated with an inhibitor for 8 h were further treated with the same or different inhibitor until 24 h. (B) The experimental groups including three groups of 24-h HDAC inhibitor treatment. (C) Bright-field images of SCNT-derived embryos treated with HDAC inhibitors for 24 h. The images were taken at 72 h after oocyte activation. For the details of the results, see Table 4.
Table 5.
Full-term development of SCNT-derived embryos following sequential treatment with TSA and Ky-29 for 24 h
| Group | No. of embryos cultured | No. of embryos at 24 h (%) | No. of embryos transferred (no. of recipients) | No. of implantations (%) | No. of offspring (%) | No. of placentas only (%) |
|---|---|---|---|---|---|---|
| TSA + Ky-29 | 184 | 179 (97.3) | 179 (6) | 62 (34.6) | 2 (1.1) | 6 (3.3) |
Synergistic effect of vitamin C and deionized BSA on the development of embryos derived from SCNT-derived embryos treated with Ky-9
It has been reported that an additional treatment with vitamin C and deionized BSA to TSA-treated SCNT embryos further improved their developmental efficiency [37]. To test whether this was also true for Ky-9-treated SCNT-derived embryos, we cultured them in the presence of vitamin C and deionized BSA for 8 h following treatment with Ky-9. As expected, the cloning efficiency was significantly increased by additional treatment with vitamin C and deionized BSA (Supplementary Figure S1).
Discussion
Since the first reports on the beneficial effect of TSA treatment on the development of mouse SCNT-derived embryos [15, 23], this method has become one of the standard cloning approaches. The same idea prompted cloning researchers to search for other HDAC inhibitors that might promote the development of SCNT-derived embryos. Those include scriptaid [24], SAHA [25], oxamflatin [25], and CBHA [26], which were found to significantly improve cloning efficiency in the mouse. All these HDAC inhibitors are hydroxamic acid derivatives and have a broad inhibitory spectrum against class I, IIa, and IIb HDACs. Here, we tested the activity of chlamydocin analogues, which include nonhydroxamic acid-type HDAC inhibitors and preferentially inhibit class I and IIa HDACs, and demonstrated that at least some of them—such as Ky-9, which possesses a methoxymethyl ketone moiety as the zinc-binding group—strongly promoted the development of SCNT-derived embryos to a level similar to that of TSA. Therefore, our results appear to have narrowed down the target HDACs for successful mouse SCNT to these two classes. By contrast, Ono et al. [25] considered class IIb HDAC to be the key for improving mouse SCNT. This was because valproic acid (VPA), an inhibitor for class I and IIa HDACs, had no effect on mouse SCNT. As our results using FK228 infer, HDAC inhibitors can have unexpected adverse effects on embryonic development. Indeed, the effective dose of VPA is high (in the mM range) and might not have exerted its beneficial effect on SCNT-derived embryos within its safe range. Indeed, VPA even at the minimal concentration that enhanced nuclear acetylation levels showed significant cell toxicity in vitro [45].
We expected that the use of FK228, an inhibitor of class I HDACs, would provide important clues to understanding how such inhibitors might affect mouse SCNT. Unexpectedly, FK228 treatment of SCNT-derived embryos resulted in a strong developmental block at the two-cell stage. Our EU incorporation assay revealed that this was a typical two-cell block caused by failure of major ZGA. Although it is not known how the inhibition of HDAC by FK228 explains this adverse effect at a molecular level, it is known that the sensitivity of cells to FK228 is increased with the intracellular level of glutathione [33]. As oocytes and early preimplantation embryos contain high amounts of glutathione [46], they might be particularly sensitive to FK228. Although this HDAC inhibitor is approved for the treatment of cutaneous and peripheral T-cell lymphomas [47], it is reported to be associated with a risk of cardiac toxicities [48]. As FK228 is a natural product, its analogues have been developed by refining its HDAC inhibitor activity and isoform selectivity [49]. It would be interesting to know whether the use of these FK228 analogues might help avoid the FK228-specific developmental arrest of mouse embryos. Intriguingly, Ky-309 (at 100 nM), a bicyclic analogue of chlamydocin, seemed to induce a two-cell developmental block by some mechanism (s) other than inhibition of ZGA because embryos treated with this agent showed normal de novo RNA synthesis (Figure 3). Although we do not know the exact cause of the developmental block induced by Ky-309, it might be related to the unique structure of Ky-309. This molecule is characterized by a bicyclic tetrapeptide structure with a CH2 (n = 10) loop (Figure 1). This long CH2 loop brings a bulky, hydrophobic region situated out of the active pocket of HDACs [50, 51]. This likely alters the structure and/or behavior of HDAC itself or HDAC-interacting proteins, which might compromise the metabolism of developing embryos.
In mouse SCNT, the duration of HDAC inhibitor treatment is limited to 10 h at the maximum because extended exposure to reconstructed embryos readily induces developmental arrest at the two-cell stage. This is because HDAC1 activation is essential for major ZGA, as demonstrated by an HDAC1 gene knockdown study [52]. Therefore, we expected that the developmental block would not occur following longer inhibition of HDACs other than HDAC1. To test this, we used Ky-29, which has a higher selectivity for HDAC4 than for HDAC1. As expected, prolonged treatment with Ky-29 did not cause a developmental block in SCNT-derived embryos and most of them developed into blastocysts. However, the subsequent birth rate of clones was not improved by this longer treatment. It was reported that TSA treatment for SCNT-derived embryos increased the expression of major ZGA genes [12]. Some of these are also upregulated by Kdm4d treatment, which removes excessive histone methylation (H3K9me3) from SCNT-derived embryo genomes [9]. The birth rates of clones following Kdm4d treatment was 7.6–8.4% [9, 53], similar to those following TSA treatment in our experiments (6.7–7.3%) using the same oocytes and donor cells (B6D2F1 oocytes and cumulus cells). Therefore, we assume that inhibition of HDAC (s) during the first 10 h after oocyte activation might help in maximizing such effects on the development of SCNT-derived embryos.
It would be interesting to identify which HDAC (s) are key target (s) for improving the development of SCNT-derived embryos. Our findings here suggest that these target HDACs are class I and/or class IIa. Class I HDACs comprise HDAC1, −2, −3, and −8. All these are expressed in MII oocytes or embryos, but HDAC3 and −8 play essential roles in controlling meiotic protein complexes, such as cohesin, not by regulating the histone acetylation levels [54, 55]. Class IIa HDACs comprise HDAC4, −5, −7, and −9; among these, HDAC4 is known to be expressed in mouse oocytes [56]. However, it is also assumed to stabilize the oocyte’s meiotic genome [56]. Thus, it is important to accurately identify those HDACs whose primary function is histone deacetylation in oocytes and embryos, which would enable us to improve mammalian SCNT procedures by targeting them using HDAC inhibitors with high selectivity and low toxicity.
Author contributions
KI, NN, TW, MY, and AO conceived the project and designed the experiments. SK, AI, NN, MY, and AO wrote the manuscript. SK, EM, JM-K, KI, HI, NO, KM, SI, NI, AI, NN, and AO performed the experiments.
Supplementary Material
Acknowledgments
We thank S. Maeda and A. Nakata, Seed Compounds Exploratory Unit for Drug Discovery Platform, RIKEN Center for Sustainable Resource Science, for technical help with HDAC enzyme assays.
† Grant Support: This study was supported by KAKENHI Grant Numbers JP25112009 (AO), JP19H05640 (MY) and JP19H05758 (AO) and Epigenome Project of the All-RIKEN Projects (AI, MY, and AO).
Contributor Information
Satoshi Kamimura, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan; Faculty of Life and Environmental Sciences, University of Yamanashi, Kofu, Yamanashi, Japan; Department of Basic Medical Sciences for Radiation Damages, National Institute of Radiological Sciences, National Institutes for Quantum and Radiological Science and Technology, Chiba, Japan.
Kimiko Inoue, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan; Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Ibaraki, Japan.
Eiji Mizutani, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan; Faculty of Life and Environmental Sciences, University of Yamanashi, Kofu, Yamanashi, Japan; Laboratory of Stem Cell Therapy, Faculty of Medicine, University of Tsukuba, Ibaraki, Japan; Division of Stem Cell Therapy, Institute of Medical Science, University of Tokyo, Tokyo, Japan.
Jin-Moon Kim, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan.
Hiroki Inoue, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan.
Narumi Ogonuki, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan.
Kei Miyamoto, Faculty of Biology-Oriented Science and Technology, Kindai University, Kinokawa-shi, Wakayama-ken, Japan.
Shunya Ihashi, Faculty of Biology-Oriented Science and Technology, Kindai University, Kinokawa-shi, Wakayama-ken, Japan.
Nobuhiko Itami, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan.
Teruhiko Wakayama, Faculty of Life and Environmental Sciences, University of Yamanashi, Kofu, Yamanashi, Japan.
Akihiro Ito, School of Life Sciences, Tokyo University of Pharmacy and Life Sciences, Hachioji, Tokyo, Japan; RIKEN Center for Sustainable Resource Science, Wako, Saitama, Japan.
Norikazu Nishino, RIKEN Center for Sustainable Resource Science, Wako, Saitama, Japan; Graduate School of Life Science and Systems Engineering, Kyushu Institute of Technology, Kitakyushu, Japan.
Minoru Yoshida, RIKEN Center for Sustainable Resource Science, Wako, Saitama, Japan; Department of Biotechnology, The University of Tokyo, Bunkyo-ku, Tokyo, Japan.
Atsuo Ogura, RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan; Graduate School of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Ibaraki, Japan; RIKEN Cluster for Pioneering Research, Wako, Saitama, Japan.
Conflict of interest
The authors have declared that no conflict of interest exists.
Data availability
All data are incorporated into the article and its online supplementary material.
References
- 1. Ogura A, Inoue K, Wakayama T. Recent advancements in cloning by somatic cell nuclear transfer. Philos Trans R Soc Lond B Biol Sci 2013; 368:20110329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Niemann H, Lucas-Hahn A. Somatic cell nuclear transfer cloning: practical applications and current legislation. Reprod Domest Anim 2012; 47:2–10. [DOI] [PubMed] [Google Scholar]
- 3. Matoba S, Zhang Y. Somatic cell nuclear transfer reprogramming: mechanisms and applications. Cell Stem Cell 2018; 23:471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nagashima H, Matsunari H. Growing human organs in pigs-a dream or reality? Theriogenology 2016; 86:422–426. [DOI] [PubMed] [Google Scholar]
- 5. Lee SH, Oh HJ, Kim MJ, Kim GA, Setyawan EMN, Ra K, Abdillah DA, Lee BC. Dog cloning-no longer science fiction. Reprod Domest Anim 2018; 53:133–138. [DOI] [PubMed] [Google Scholar]
- 6. Perisse IV, Fan Z, Singina GN, White KL, Polejaeva IA. Improvements in gene editing technology boost its applications in livestock. Front Genet 2021; 11:614688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Loi P, Iuso D, Czernik M, Ogura A. A new, dynamic era for somatic cell nuclear transfer? Trends Biotechnol 2016; 34:791–797. [DOI] [PubMed] [Google Scholar]
- 8. Inoue K, Kohda T, Sugimoto M, Sado T, Ogonuki N, Matoba S, Shiura H, Ikeda R, Mochida K, Fujii T, Sawai K, Otte AP et al. Impeding Xist expression from the active X chromosome improves mouse somatic cell nuclear transfer. Science 2010; 330:496–499. [DOI] [PubMed] [Google Scholar]
- 9. Matoba S, Liu Y, Lu F, Iwabuchi KA, Shen L, Inoue A, Zhang Y. Embryonic development following somatic cell nuclear transfer impeded by persisting histone methylation. Cell 2014; 159:884–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang LY, Li ZK, Wang LB, Liu C, Sun XH, Feng GH, Wang JQ, Li YF, Qiao LY, Nie H, Jiang LY, Sun H et al. Overcoming intrinsic H3K27me3 imprinting barriers improves post-implantation development after somatic cell nuclear transfer. Cell Stem Cell 2020; 27:315, e315–325. [DOI] [PubMed] [Google Scholar]
- 11. Inoue K, Ogonuki N, Kamimura S, Inoue H, Matoba S, Hirose M, Honda A, Miura K, Hada M, Hasegawa A, Watanabe N, Dodo Y et al. Loss of H3K27me3 imprinting in the Sfmbt 2 miRNA cluster causes enlargement of cloned mouse placentas. Nat Commun 2020; 11:2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Inoue K, Oikawa M, Kamimura S, Ogonuki N, Nakamura T, Nakano T, Abe K, Ogura A. Trichostatin A specifically improves the aberrant expression of transcription factor genes in embryos produced by somatic cell nuclear transfer. Sci Rep 2015; 5:10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang G, Zhang L, Liu W, Qiao Z, Shen S, Zhu Q, Gao R, Wang M, Wang M, Li C, Liu M, Sun J et al. Dux-mediated corrections of aberrant H3K9ac during 2-cell genome activation optimize efficiency of somatic cell nuclear transfer. Cell Stem Cell 2021; 28:150–163. [DOI] [PubMed] [Google Scholar]
- 14. Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 1990; 265:17174–17179. [PubMed] [Google Scholar]
- 15. Kishigami S, Mizutani E, Ohta H, Hikichi T, Thuan NV, Wakayama S, Bui HT, Wakayama T. Significant improvement of mouse cloning technique by treatment with trichostatin A after somatic nuclear transfer. Biochem Biophys Res Commun 2006; 340:183–189. [DOI] [PubMed] [Google Scholar]
- 16. Iager AE, Ragina NP, Ross PJ, Beyhan Z, Cunniff K, Rodriguez RM, Cibelli JB. Trichostatin A improves histone acetylation in bovine somatic cell nuclear transfer early embryos. Cloning Stem Cells 2008; 10:371–379. [DOI] [PubMed] [Google Scholar]
- 17. Wu X, Li Y, Li GP, Yang D, Yue Y, Wang L, Li K, Xin P, Bou S, Yu H. Trichostatin A improved epigenetic modifications of transfected cells but did not improve subsequent cloned embryo development. Anim Biotechnol 2008; 19:211–224. [DOI] [PubMed] [Google Scholar]
- 18. Li J, Svarcova O, Villemoes K, Kragh PM, Schmidt M, Bogh IB, Zhang Y, Du Y, Lin L, Purup S, Xue Q, Bolund L et al. High in vitro development after somatic cell nuclear transfer and trichostatin A treatment of reconstructed porcine embryos. Theriogenology 2008; 70:800–808. [DOI] [PubMed] [Google Scholar]
- 19. Yamanaka K, Sugimura S, Wakai T, Kawahara M, Sato E. Acetylation level of histone H3 in early embryonic stages affects subsequent development of miniature pig somatic cell nuclear transfer embryos. J Reprod Dev 2009; 55:638–644. [DOI] [PubMed] [Google Scholar]
- 20. Shi LH, Miao YL, Ouyang YC, Huang JC, Lei ZL, Yang JW, Han ZM, Song XF, Sun QY, Chen DY. Trichostatin A (TSA) improves the development of rabbit-rabbit intraspecies cloned embryos, but not rabbit-human interspecies cloned embryos. Dev Dyn 2008; 237:640–648. [DOI] [PubMed] [Google Scholar]
- 21. Meng Q, Polgar Z, Liu J, Dinnyes A. Live birth of somatic cell-cloned rabbits following trichostatin A treatment and cotransfer of parthenogenetic embryos. Cloning Stem Cells 2009; 11:203–208. [DOI] [PubMed] [Google Scholar]
- 22. Haberland M, Montgomery RL, Olson EN. The many roles of histone deacetylases in development and physiology: implications for disease and therapy. Nat Rev Genet 2009; 10:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Rybouchkin A, Kato Y, Tsunoda Y. Role of histone acetylation in reprogramming of somatic nuclei following nuclear transfer. Biol Reprod 2006; 74:1083–1089. [DOI] [PubMed] [Google Scholar]
- 24. Van Thuan N, Bui HT, Kim JH, Hikichi T, Wakayama S, Kishigami S, Mizutani E, Wakayama T. The histone deacetylase inhibitor scriptaid enhances nascent mRNA production and rescues full-term development in cloned inbred mice. Reproduction 2009; 138:309–317. [DOI] [PubMed] [Google Scholar]
- 25. Ono T, Li C, Mizutani E, Terashita Y, Yamagata K, Wakayama T. Inhibition of class IIb histone deacetylase significantly improves cloning efficiency in mice. Biol Reprod 2010; 83:929–937. [DOI] [PubMed] [Google Scholar]
- 26. Dai X, Hao J, Hou XJ, Hai T, Fan Y, Yu Y, Jouneau A, Wang L, Zhou Q. Somatic nucleus reprogramming is significantly improved by m-carboxycinnamic acid bishydroxamide, a histone deacetylase inhibitor. J Biol Chem 2010; 285:31002–31010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Qiu X, Li N, Xiao X, Zhang L, You H, Li Y. Effects of embryo aggregation and PXD101 on the in vitro development of mouse somatic cell nuclear transfer embryos. Cell Reprogram 2017; 19:337–343. [DOI] [PubMed] [Google Scholar]
- 28. Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem 2005; 96:293–304. [DOI] [PubMed] [Google Scholar]
- 29. Gallinari P, Di Marco S, Jones P, Pallaoro M, Steinkuhler C. HDACs, histone deacetylation and gene transcription: from molecular biology to cancer therapeutics. Cell Res 2007; 17:195–211. [DOI] [PubMed] [Google Scholar]
- 30. Furumai R, Komatsu Y, Nishino N, Khochbin S, Yoshida M, Horinouchi S. Potent histone deacetylase inhibitors built from trichostatin A and cyclic tetrapeptide antibiotics including trapoxin. Proc Natl Acad Sci U S A 2001; 98:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nishino N, Jose B, Shinta R, Kato T, Komatsu Y, Yoshida M. Chlamydocin-hydroxamic acid analogues as histone deacetylase inhibitors. Bioorg Med Chem 2004; 12:5777–5784. [DOI] [PubMed] [Google Scholar]
- 32. Islam NM, Kato T, Nishino N, Kim HJ, Ito A, Yoshida M. Bicyclic peptides as potent inhibitors of histone deacetylases: optimization of alkyl loop length. Bioorg Med Chem Lett 2010; 20:997–999. [DOI] [PubMed] [Google Scholar]
- 33. Furumai R, Matsuyama A, Kobashi N, Lee KH, Nishiyama M, Nakajima H, Tanaka A, Komatsu Y, Nishino N, Yoshida M, Horinouchi S. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res 2002; 62:4916–4921. [PubMed] [Google Scholar]
- 34. Wakayama T, Perry AC, Zuccotti M, Johnson KR, Yanagimachi R. Full-term development of mice from enucleated oocytes injected with cumulus cell nuclei. Nature 1998; 394:369–374. [DOI] [PubMed] [Google Scholar]
- 35. Ogura. Cloning mice. Cold Spring Harb Protoc 2017; 2017:pdb prot094425. [DOI] [PubMed] [Google Scholar]
- 36. Lawitts JA, Biggers JD. Culture of preimplantation embryos. Methods Enzymol 1993; 225:153–164. [DOI] [PubMed] [Google Scholar]
- 37. Miyamoto K, Tajima Y, Yoshida K, Oikawa M, Azuma R, Allen GE, Tsujikawa T, Tsukaguchi T, Bradshaw CR, Jullien J, Yamagata K, Matsumoto K et al. Reprogramming towards totipotency is greatly facilitated by synergistic effects of small molecules. Biol Open 2017; 6:415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mochida K, Hasegawa A, Ogonuki N, Inoue K, Ogura A. Early production of offspring by in vitro fertilization using first-wave spermatozoa from prepubertal male mice. J Reprod Dev 2019; 65:467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ogonuki N, Mori M, Shinmen A, Inoue K, Mochida K, Ohta A, Ogura A. The effect on intracytoplasmic sperm injection outcome of genotype, male germ cell stage and freeze-thawing in mice. PLoS One 2010; 5:e11062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishino N, Jose B, Okamura S, Ebisusaki S, Kato T, Sumida Y, Yoshida M. Cyclic tetrapeptides bearing a sulfhydryl group potently inhibit histone deacetylases. Org Lett 2003; 5:5079–5082. [DOI] [PubMed] [Google Scholar]
- 41. Shivashimpi GM, Amagai S, Kato T, Nishino N, Maeda S, Nishino TG, Yoshida M. Molecular design of histone deacetylase inhibitors by aromatic ring shifting in chlamydocin framework. Bioorg Med Chem 2007; 15:7830–7839. [DOI] [PubMed] [Google Scholar]
- 42. Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 1989; 86:679–688. [DOI] [PubMed] [Google Scholar]
- 43. Tanabe Y, Kuwayama H, Wakayama S, Nagatomo H, Ooga M, Kamimura S, Kishigami S, Wakayama T. Production of cloned mice using oocytes derived from ICR-outbred strain. Reproduction 2017; 154:859–866. [DOI] [PubMed] [Google Scholar]
- 44. Warner CM, Versteegh LR. In vivo and in vitro effect of alpha-amanitin on preimplantation mouse embryo RNA polymerase. Nature 1974; 248:678–680. [DOI] [PubMed] [Google Scholar]
- 45. Li X, Ao X, Bai L, Li D, Liu X, Wei Z, Bou S, Li G. VPA selectively regulates pluripotency gene expression on donor cell and improve SCNT embryo development. In Vitro Cell Dev Biol Anim 2018; 54:496–504. [DOI] [PubMed] [Google Scholar]
- 46. Nasr-Esfahani MH, Johnson MH. Quantitative analysis of cellular glutathione in early preimplantation mouse embryos developing in vivo and in vitro. Hum Reprod 1992; 7:1281–1290. [DOI] [PubMed] [Google Scholar]
- 47. Ceccacci E, Minucci S. Inhibition of histone deacetylases in cancer therapy: lessons from leukaemia. Br J Cancer 2016; 114:605–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rivers ZT, Oostra DR, Westholder JS, Vercellotti GM. Romidepsin-associated cardiac toxicity and ECG changes: a case report and review of the literature. J Oncol Pharm Pract 2018; 24:56–62. [DOI] [PubMed] [Google Scholar]
- 49. Narita K, Matsuhara K, Itoh J, Akiyama Y, Dan S, Yamori T, Ito A, Yoshida M, Katoh T. Synthesis and biological evaluation of novel FK228 analogues as potential isoform selective HDAC inhibitors. Eur J Med Chem 2016; 121:592–609. [DOI] [PubMed] [Google Scholar]
- 50. Yoshida M, Matsuyama A, Komatsu Y, Nishino N. From discovery to the coming generation of histone deacetylase inhibitors. Curr Med Chem 2003; 10:2351–2358. [DOI] [PubMed] [Google Scholar]
- 51. Yoshida M, Kudo N, Kosono S, Ito A. Chemical and structural biology of protein lysine deacetylases. Proc Jpn Acad Ser B Phys Biol Sci 2017; 93:297–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol 2008; 319:110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Matoba S, Wang H, Jiang L, Lu F, Iwabuchi KA, Wu X, Inoue K, Yang L, Press W, Lee JT, Ogura A, Shen L et al. Loss of H3K27me3 imprinting in somatic cell nuclear transfer embryos disrupts post-implantation development. Cell Stem Cell 2018; 23:343, e345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. He Y, Li X, Gao M, Liu H, Gu L. Loss of HDAC3 contributes to meiotic defects in aged oocytes. Aging Cell 2019; 18:e13036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Singh VP, Yueh WT, Gerton JL, Duncan FE. Oocyte-specific deletion of Hdac8 in mice reveals stage-specific effects on fertility. Reproduction 2019; 157:305–316. [DOI] [PubMed] [Google Scholar]
- 56. Kageyama S, Liu H, Nagata M, Aoki F. Stage specific expression of histone deacetylase 4 (HDAC4) during oogenesis and early preimplantation development in mice. J Reprod Dev 2006; 52:99–106. [DOI] [PubMed] [Google Scholar]
- 57. Nguyen HM, Sako K, Matsui A, Ueda M, Tanaka M, Ito A, Nishino N, Yoshida M, Seki M. Transcriptomic analysis of Arabidopsis thaliana plants treated with the Ky-9 and Ky-72 histone deacetylase inhibitors. Plant Signal Behav 2018; 13:e1448333. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are incorporated into the article and its online supplementary material.


