Abstract
Transient receptor potential vanilloid member 1 (TRPV1) is a Ca2+-permeable cation channel that serves as the primary heat and capsaicin sensor in humans. Using cryo-EM, we have determined the structures of apo and capsaicin-bound full-length rat TRPV1 reconstituted into lipid nanodiscs over a range of temperatures. This has allowed us to visualize the noxious heat-induced opening of TRPV1 in the presence of capsaicin. Notably, noxious heat-dependent TRPV1 opening comprises stepwise conforma- tional transitions. Global conformational changes across multiple subdomains of TRPV1 are followed by the rearrangement of the outer pore, leading to gate opening. Solvent-accessible surface area analyses and functional studies suggest that a subset of residues form an interaction network that is directly involved in heat sensing. Our study provides a glimpse of the molecular principles underlying noxious physical and chemical stimuli sensing by TRPV1, which can be extended to other thermal sensing ion channels.
Introduction
Noxious heat and capsaicin are two well-known stimuli for nociception. Our ability to sense noxious heat and spiciness is conferred by the transient receptor potential vanilloid member 1 (TRPV1), a calcium permeable ion channel expressed in primary sensory nerve terminals 1. The TRP channel superfamily contains several thermosensitive members2-5, of which TRPV1 is the sensor for noxious stimuli including heat and vanilloid compounds4,6,7. TRPV1, together with the cold sensor TRPM8, have been particularly important model systems to understand the molecular basis of temperature sensing in animals5,8-13. Despite a wealth of studies on TRPV1, our understanding of heat and capsaicin sensing is still limited. It is unclear whether heat sensing in TRPV1 is mediated by a localized/modular heat sensor or global conformational changes. The outer pore, membrane-proximal, and cytosolic regions have all been suggested as the heat sensors in TRPV1, but it remains elusive whether they are directly responsible for heat sensing or are involved in heat-induced downstream conformational changes toward channel opening14-21. More importantly, it is unclear how all of these spatially separate regions are choreographed for heat-dependent gate opening and whether they undergo concerted or sequential conformational changes. A recent model attributes heat capacity change as the origin of the high temperature sensitivity in temperature sensing TRP channels22, which was tested by engineering a non-temperature sensitive K+ channel23. However, whether TRPV1 functions by this principle to physiologically sense heat is yet to be tested.
Capsaicin binds to and activates TRPV1, thereby giving mammals chemically induced heat sensation. Functional studies have shown that heat- and capsaicin-dependent activation pathways are separate yet allosterically linked, as mutations selectively silence one modality over the other14,17,24. How TRPV1 integrates these two signals into channel opening is unclear.
Julius, Cheng, and colleagues elucidated the molecular basis of toxin-dependent opening of TRPV1 by determining cryo-electron microscopy (cryo-EM) structures of the ligand-free closed state and the doubleknot toxin (DkTx) and resiniferatoxin (RTx) bound open state25-27. They also reported an apparent intermediate state of amphipol-reconstituted TRPV1 in the presence of capsaicin, although capsaicin was not visualized25, which has prompted multiple computational studies to model the capsaicin binding site in TRPV128-31. Although these studies are breakthroughs in our understanding of toxin-dependent TRPV1 gating, the mechanisms of heat- and/or capsaicin-dependent TRPV1 gating remain unclear. We have determined six cryo-EM structures of the full-length TRPV1 reconstituted into lipid nanodiscs at various temperatures while in the presence or absence of capsaicin. Our structures visualize the stepwise noxious heat-dependent opening of TRPV1 in the presence of capsaicin, of which the conformation is distinct from the DkTx/RTx-activated TRPV125. Our structural analyses, together with prior and current functional studies, provide the molecular basis for heat- and capsaicin-dependent TRPV1 gating.
Results
Cryo-EM structures of the full-length TRPV1 in nanodiscs
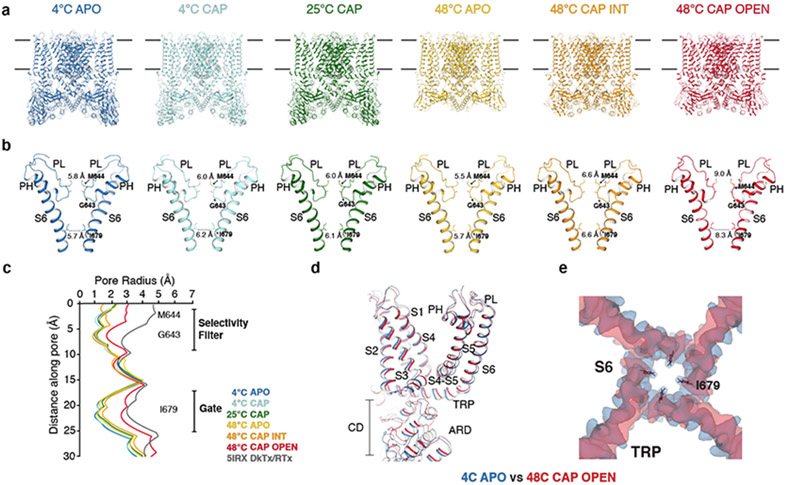
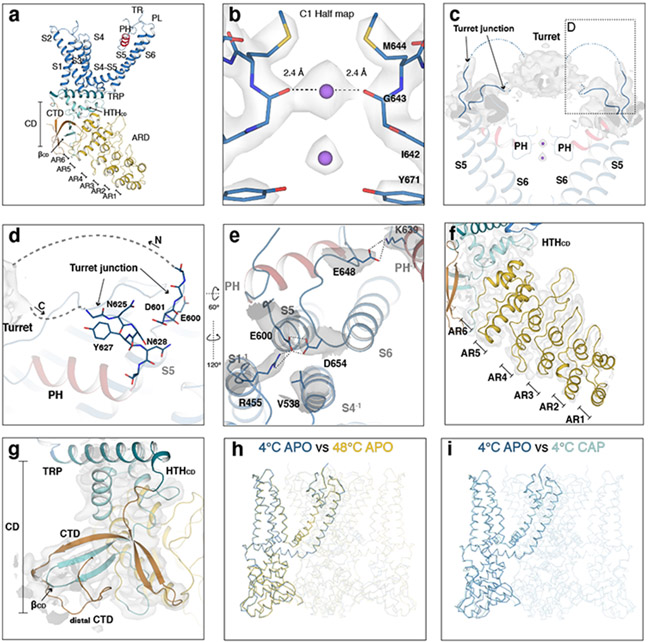
The full-length rat TRPV1 was purified, and reconstituted into lipid nanodiscs at 4 °C then incubated at three different temperatures (4°C, 25°C, and 48°C) for ~30 s before being flash frozen on the cryo-EM grids. For the relevant conditions, capsaicin was administered to the nanodisc-reconstituted TRPV1 prior to heat treatment. We determined six cryo-EM 3D reconstructions (Fig. 1a, b, Extended Data Fig. 1) to overall excellent quality (Extended Data Fig. 2 and Table 1): (i) the 4°C, ligand-free, closed state (TRPV14C,APO) resolved to ~2.63 Å, (ii) the 4°C, capsaicin-bound, closed state (TRPV14C,CAP) to ~3.37 Å, (iii) the 25°C, capsaicin-bound, closed state (TRPV125C,CAP) to ~3.54 Å, (iv) the 48°C, ligand-free, closed state (TRPV148C,APO) to ~3.06 Å, (v) the 48°C, capsaicin-bound, intermediate state (TRPV148C,CAP,INT) to ~3.55 Å, and (vi) the 48°C, capsaicin-bound, open state (TRPV148C,CAP,OPEN) to ~3.72 Å. The TRPV1 channel is homotetrameric with each monomer composed of an N-terminal cytosolic ankyrin repeat domain (ARD) and a transmembrane domain (TMD) comprising six transmembrane helices (S1-S6). The TMD comprises a S1-S4 domain, a pore domain (containing S5, the turret, the pore helix, the selectivity filter, the pore loop, and S6), and the amphipathic TRP helix (Extended Data Fig. 3a). The TMD and ARD are connected via the coupling domain (CD), which includes a helix-loop-helix motif (HLHCD), a β-sheet (βCD), the pre-S1(pre-S1CD) helix, and a C-terminal domain (CTD). HOLE-based pore analysis32 confirmed that TRPV148C,CAP,OPEN adopts an open state, because constriction points at both the selectivity filter and the S6 gate helical bundle crossing are wide enough for cation permeation, where the diagonal distances between the side chains of I679 and M644 are 8.3 Å and 9.0 Å, respectively (Fig. 1b, c). An overlay of the closed and open states (TRPV14C,APO and TRPV148C,CAP,OPEN) reveals global conformational changes across the entire protein (Fig. 1d, e).
Fig. 1. Structures of the full-length TRPV1 at six conditions.
a, Structures of TRPV1 determined at 4°C (TRPV14C,APO, blue), at 4°C with capsaicin (TRPV14C,CAP, sky blue), 25°C with capsaicin (TRPV125C,CAP, green), 48°C (TRPV148C,APO, gold), 48°C with capsaicin in the intermediate state (TRPV148C,CAP,INT, orange), and 48°C with capsaicin in the open state (TRPV148C,CAP,OPEN, red). b, Comparison of the pore domain structures. Only two subunits are shown for clarity with pore loop (PL) and pore helix (PH) as indicated. Diagonal distances at the two narrowest restriction points are shown. c, Pore radii calculated using the HOLE program32 for the TRPV1 structures as color coded in panel a and DkTx/RTx-TRPV1 (PDB 5IRX, gray). d,e, Superposition of the TRPV14C,APO (blue) and the TRPV14C,CAP,OPEN (red) structures (d) and cryo-EM maps (e), highlighting global conformational changes (d) and S6 gate conformation (e), respectively.
Table 1.
Cryo-EM data collection, refinement and validation statistics
| 4 °C APO (EMD- 23473, PDB 7LP9) |
4 °C CAP (EMD- 23474, PDB 7LPA) |
25 °C CAP (EMD- 23475, PDB 7LPB) |
48 °C APO (EMD- 23476, PDB 7LPC) |
48 °C CAP INT (EMD- 23478, PDB 7LPD) |
48 °C CAP OPEN (EMD- 23479, PDB 7LPE) |
48 °C CAP 10 sec (EMD- 23477) |
|
|---|---|---|---|---|---|---|---|
| Data collection and processing | |||||||
| Magnification | 81,000 | 81,000 | 81,000 | 81,000 | 81,000 | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 45 | 60 | 60 | 50 | 45 | 45 | 45 |
| Defocus range (μm) | −1.9 to −0.8 | −2.25 to −1.0 | −2.25 to −1.0 | −1.9 to −0.8 | −1.9 to −0.8 | −1.9 to −0.8 | −1.9 to −0.8 |
| Pixel size (Å) | 1.079 | 1.08 | 1.08 | 1.079 | 1.059 | 1,059 | 1.079 |
| Symmetry imposed | C4 | C4 | C4 | C4 | C4 | C4 | C4 |
| Initial particle images (no.) | 936,518 | 1,167,149 | 622,894 | 1,060,444 | 857,573 | 857,573 | 1,973,776 |
| Final particle images (no.) | 191,327 | 114,269 | 41,958 | 93,835 | 18,991 | 122,813 | 26,461 |
| Map resolution (Å) | 2.63 | 3.37 | 3.54 | 3.07 | 3.55 | 3.72 | 3.7 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Refinement | |||||||
| Initial model used (PDB code) | 5IRZ | 5IRZ | 5IRZ | 5IRZ | 5IRZ | 5IRZ | 5IRZ |
| Model resolution (Å) | 2.63 | 3.37 | 3.54 | 3.07 | 3.55 | 3.72 | |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Model resolution range (Å) | 2.27 - 3.36 | 2.96 – 3.42 | 3.18 - 3.60 | 2.64 – 3.10 | 3.37 – 3.86 | 3.69 – 3.17 | |
| Map sharpening B factor (Å2) | −80 | −80 | −80 | −80 | −80 | −80 | −80 |
| Model composition | |||||||
| Nonhydrogen atoms | 20974 | 18634 | 18412 | 15266 | 15564 | 13844 | |
| Protein residues | 2536 | 2392 | 2392 | 1816 | 1988 | 1944 | |
| Ligands | 46 | 30 | 28 | 42 | 24 | 12 | |
| B factors (Å2) | |||||||
| Protein | 106.26 | 87.49 | 94.91 | 47.73 | 62.98 | 143.91 | |
| Ligand | 90.97 | 43.06 | 45.04 | 23.90 | 36.25 | 129.63 | |
| R.m.s. deviations | |||||||
| Bond lengths (Å) | 0.004 | 0.004 | 0.010 | 0.009 | 0.008 | 0.010 | |
| Bond angles (°) | 0.902 | 0.556 | 1.083 | 0.998 | 1.098 | 1.191 | |
| Validation | |||||||
| MolProbity score | 1.32 | 1.66 | 1.84 | 1.27 | 1.50 | 1.98 | |
| Clashscore | 4.11 | 6.83 | 8.45 | 5.13 | 6.76 | 11.50 | |
| Poor rotamers (%) | 0 | 0 | 0 | 0 | 0 | 0 | |
| Ramachandran plot | |||||||
| Favored (%) | 97.34 | 95.90 | 94.35 | 98.00 | 97.34 | 93.93 | |
| Allowed (%) | 2.66 | 4.10 | 5.65 | 2.00 | 2.66 | 6.07 | |
| Disallowed (%) | 0 | 0 | 0 | 0 | 0 | 0 |
Structure of the full-length apo TRPV1 in nanodiscs
The structure of the full-length TRPV14C,APO in nanodiscs is similar to the previous truncated apo TRPV1 structure25-27, but reveals three notable features not observed in the previous structures. First, we observed two strong cryo-EM density peaks within the selectivity filter that are present in the half-maps of a no symmetry-imposed 3D reconstruction (Extended Data Fig. 3b). These were tentatively attributed to Na+ because of the high concentration (150 mM) used in sample preparation. There were no attributable ion densities within the selectivity filter in the previous apo TRPV1 structure27. Our TRPV14C,APO structure shows that the selectivity filter can bind cations in the apo, closed state. Second, the turret (N604-S626) comprises the loop between S5 and the pore helix, which was truncated in the TRPV1 mutant from the previous studies. Connected to the S5 and the pore helix, the turrets from each subunit assemble a cap like structure33, which we observed as a density on the extracellular side of the four-fold symmetry axis of TRPV1 (Extended Data Fig. 3c). The entry and exit regions of the turret (termed the turret junction) contribute to the interaction network within the outer pore (Extended Data Fig. 3d). The third point highlights the functional role of E600 (turret junction) and E648 (pore loop). E600 is a key allosteric regulatory site, as its mutation leads to elevated basal activity of TRPV1 and altered sensitivity to stimuli including heat, acid, and sodium17,34. Meanwhile, E648 is implicated in proton-dependent TRPV1 activation34. In the TRPV14C,APO structure, the sidechain of E600 is within interaction distance from D654 (pore loop) and R455 (S1), and E648 interacts with K639 (pore helix) from a neighboring subunit (Extended Data Fig. 3e). These interactions stabilize the outer pore interaction network. Last, we are able to resolve the entire ARD as well as the distal CTD, which coils around βCD in an analogous manner to TRPV3 and ground squirrel TRPV1 (Extended Data Fig. 3f-g) 33,35,36.
Neither heat nor capsaicin alone opens TRPV1 in nanodiscs
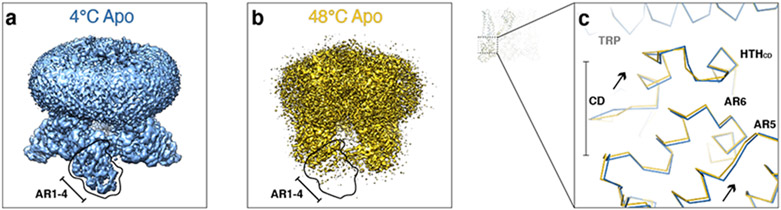
The 3D reconstructions of TRPV148C,APO and TRPV14C,APO are overall very similar, with the intracellular S6 gate closed (Extended Data Fig. 3h). The only recognizable differences are that AR1-AR4 in the ARD are unresolved at 48°C, likely due to their increased flexibility at elevated temperature, and the CD swings closer to the TRP helix (Extended Data Fig. 4).
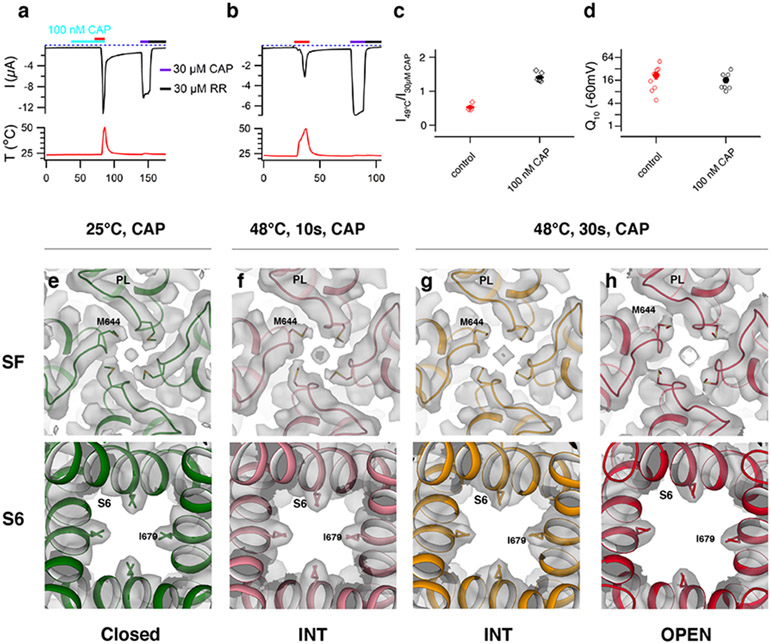
Despite numerous attempts to open the channel using various heat-treatment procedures, we consistently obtained 3D reconstructions similar to TRPV148C,APO. This observation led us to reason that TRPV1 in nanodiscs is more difficult to open than in the cell membrane. It is possible that the intrinsic gating equilibrium of TRPV1 is influenced by the physical and chemical differences between the two environments (e.g. the lack of membrane voltage across nanodiscs system and different lipid compositions and lateral pressure profiles between the two systems). We posit that an additional stimulus is required to capture TRPV1 in the heat-activated open state. We chose to utilize capsaicin because it is well known that heat and capsaicin activate TRPV1 independently, based on mutants that only perturb one activation pathway over the other14,24 and that closed-state TRPV1 with capsaicin bound retains noxious heat sensitivity17. Consistent with prior studies, we found that while 100 nM capsaicin does not elicit observable currents in TRPV1 expressing oocytes, it does sensitize TRPV1 to heat activation with increased efficacy (Fig. 2a-d). Notably, the heat sensitivity (Q10) is not substantially affected by pre-treatment with capsaicin (~16 versus ~22 without capsaicin; Fig. 2d). Since capsaicin becomes enriched in the membrane due to its high partition coefficient (~3x105) 37, we postulate that TRPV1 expressed in oocytes binds capsaicin but that it remains closed at the bulk solution concentration of 100 nM until heat is applied. Our data supports that the capsaicin-bound, closed-state TRPV1 retains sensitivity to noxious heat.
Fig. 2. TRPV1 retains sensitivity to noxious heat after capsaicin sensitization and opens in a stepwise manner.
a, b, Representative time course current traces of TRPV1-expressing oocytes with heat ramp and 100 nM capsaicin (CAP, turquoise) pre-treatment (a) or heat ramp (red) alone (b). Subsequent applications of 30 μM capsaicin (purple) then 30 μM ruthenium red (RR, black) were performed in both cases. The dotted line indicates zero current. The recorded temperature is shown in the bottom panel. c, Ratios of TRPV1 current responses to heat (49°C) relative to saturating capsaicin (30 μM) at room temperature for both protocols. Values for individual oocytes (open diamonds) with mean ±S.E.M. for control (0.54±0.06, n = 4 biological replicates) and 100 nM capsaicin treated (1.40±0.06, n = 6 biological replicates). d, Q10 values for TRPV1 expressing oocytes activated by heat alone or along with 100 nM capsaicin. Values for individual oocytes (open circles) with mean ±S.E.M. for control (21.90±4.50, n = 9 biological replicates) and 100 nM capsaicin treated (16.40±2.76, n = 8 biological replicates). Comparison between pores of TRPV125C,CAP, TRPV148C,CAP,10sec, TRPV148C,CAP,INT and TRPV148C,CAP,OPEN (e-h). Top-down view of the selectivity filter (SF, top) and bottom-up view of the S6 gate (S6, bottom) with cryo-EM density (gray) for: e. TRPV125C,CAP (green), 0.022 thresholding. f. TRPV148C,CAP,10s (pink), 0.045 thresholding. g. TRPV148C,CAP,INT (orange), 0.035 thresholding. h. TRPV148C,CAP,OPEN (red), 0.028 thresholding. Source data for c and d are available online.
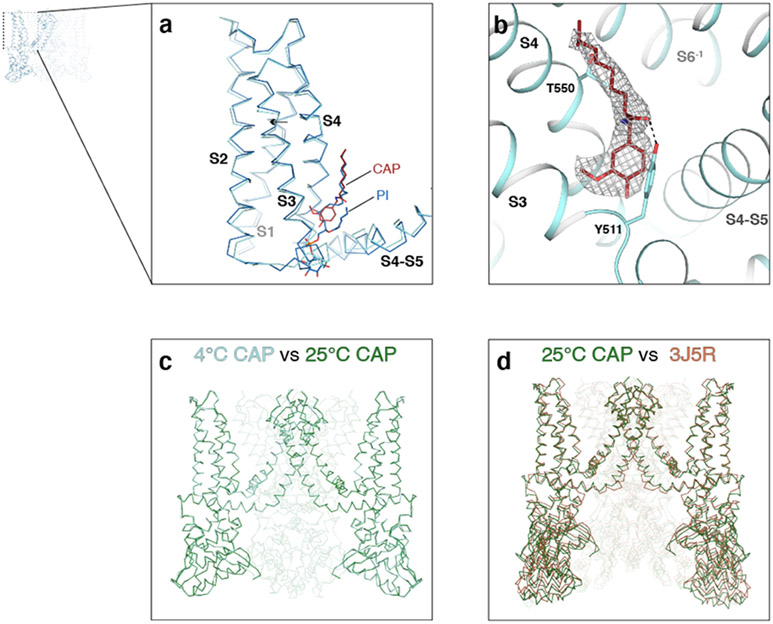
TRPV14C,CAP visualizes the cryo-EM density for capsaicin (Extended Data Fig. 5b) and reveals that capsaicin binding induces a localized conformational change around the capsaicin binding site, such that S2, the S2-S3 loop, the intracellular portion of S3, and the S4-S5 linker move closer to each other (Extended Data Figs. 3i, 5a). There are, however, no noticeable changes in the selectivity filter and the S6 gate (Fig. 1b). We next tested whether increasing temperature from 4°C to 25°C in the presence of capsaicin would lead to channel opening and found that the conformation of TRPV125C,CAP is nearly identical to that of TRPV14C,CAP (Extended Data Fig. 5c), with both structures substantially different from that of the published amphipol-reconstituted TRPV1 in the presence of capsaicin (Extended Data Fig. 5d)25.
Noxious heat opens TRPV1 in a stepwise manner
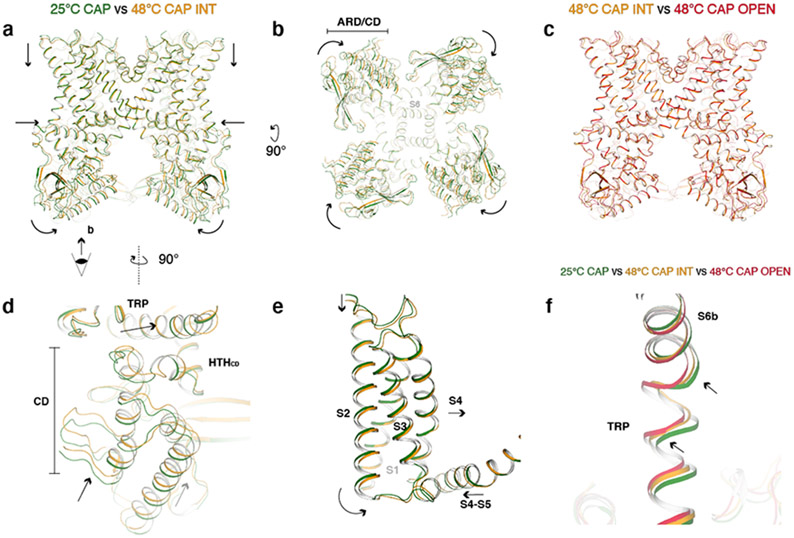
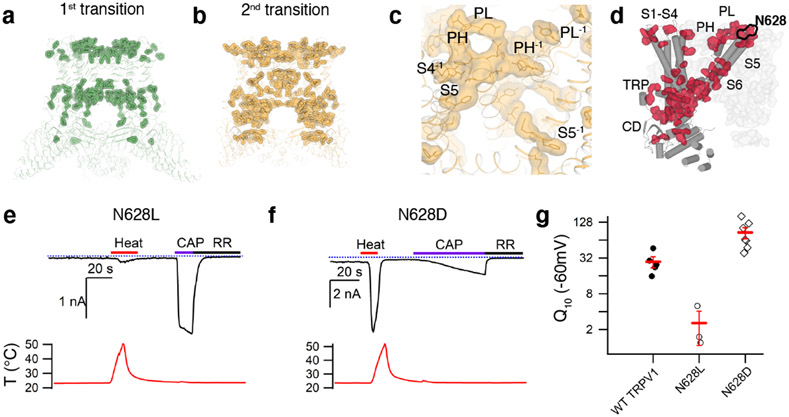
Data processing of TRPV1CAP at 48°C yielded two 3D classes: a major class (~122k particles) and a minor class (~18k particles) (Extended Data Fig. 1). The selectivity filter and the S6 gate of the major class are dilated enough for ion conduction (9.0 and 8.3 Å, respectively), while those of the minor class are dilated to 7.1 and 6.5 Å, respectively, which are wider than those at 25°C but not wide enough for ion permeation (Fig. 1b). Compared to TRPV125C,CAP, both classes at 48°C exhibit similar global conformational changes but differ in their local conformations leading to distinct gating states (Figs. 1b, 2e-g). We suggest that these two classes represent different functional states along the heat-dependent activation pathway for two reasons. First, TRPV1CAP incubated at 48°C for a shorter period (10 s), results in a major 3D class similar to the minor class obtained at 48°C for 30 s (Fig. 2f, g). These observations argue against this minor class as representing a heat desensitized state or an off-pathway state. It is possible that heat-desensitized or partially unfolded states38 might have been discarded during the 3D reconstruction processing. Second, certain regions in the minor class, such as the TRP helix, exhibit conformations between those of TRPV125C,CAP and the major class (Fig. 3f). We therefore attribute the minor class as representing an intermediate state (referred to as TRPV148C,CAP,INT) and the major class as an open state (referred to as TRPV148C,CAP,OPEN) (Figs. 1, 2). We term the conformational changes from TRPV125C,CAP to TRPV148C,CAP,INT as the first transition, and those from TRPV148C,CAP,INT to TRPV148C,CAP,OPEN as the second transition.
Fig. 3. Global conformational changes of the first noxious heat-induced transition.
a, Comparison of TRPV125C,CAP (green) and TRPV148C,CAP,INT (orange) viewed from the membrane. Arrows indicate movements of the ARD, CD, S1-S4 domain, and TRP helix. b, The same as (a), viewed from the intracellular side. ARD/CD movement occurs at an individual protomer level. c, TRPV148C,CAP,INT and TRPV148C,CAP,OPEN adopt similar overall conformations. d, Close-up view of TRPV125C,CAP and TRPV148C,CAP,INT cytoplasmic domains. e, Close-up view of TRPV125C,CAP and TRPV148C,CAP,INT S1-S4 domain. f, Close-up view of the S6b and TRP helix of TRPV125C,CAP, TRPV148C,CAP,INT, and TRPV148C,CAP,OPEN . The arrows indicate TRP helix and S6b movements.
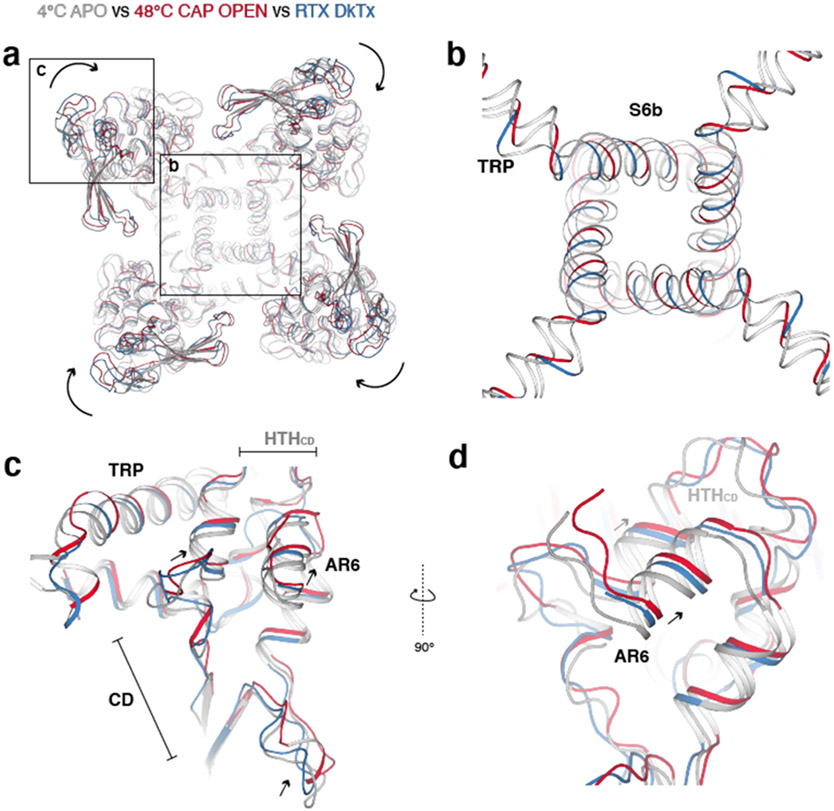
The first transition reveals global conformational changes that occur throughout the channel (Fig. 3a, b, d, e). First, AR1-AR3 become unresolvedly flexible; the CD and ARD (AR4-AR6) undergo a rigid body rotation toward the channel when viewed from the membrane (Fig. 3a). This CD/ARD rotation occurs at the subunit level, rather than rotation of the entire tetrameric ARD ring (Fig. 3b). Second, the TRP helix moves closer to the pore and the CD/ARD movement allows HTHCD to move closer to the TRP helix (Fig. 3d). Third, the S1-S4 domain as well as the C-terminal part of the S4-S5 linker rotate toward the pore when viewed from the membrane (Fig. 3e). Overall, TRPV1 contracts, leading to the slight dilation of the S6 gate during the first transition (Fig. 1b). Previous studies have suggested that the membrane proximal domain (analogous to the CD or HTHCD in our study) as key to the heat sensitivity of TRPV116 and heat-induced ARD motion might play a role in TRPV1 heat sensing39. At the global level, conformational changes in the CD/ARD mediated by 48°C/capsaicin are apparently similar but slightly larger than those mediated by DkTx/RTx, despite the larger gate opening in DkTx/RTx-bound TRPV1 (Extended Data Fig. 6).
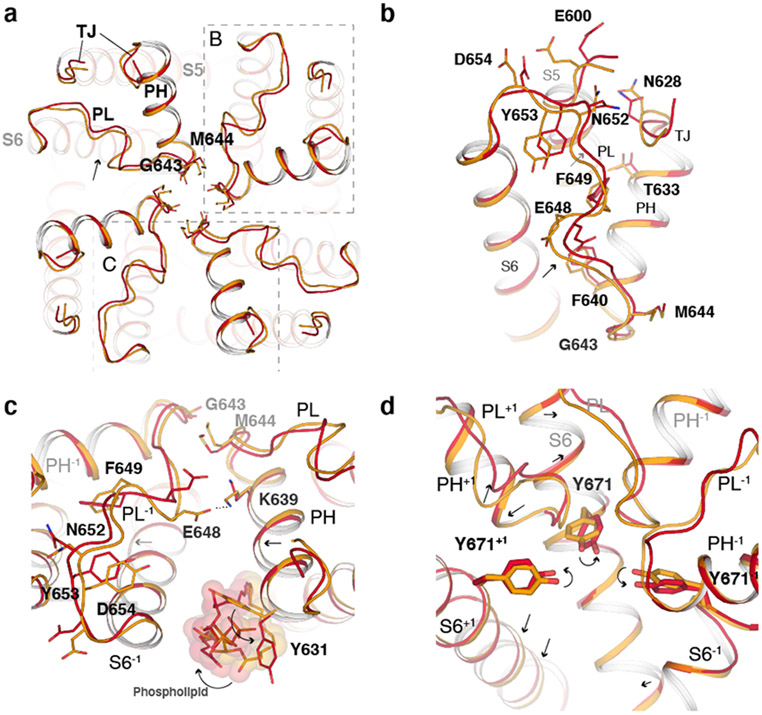
Outer pore rearrangements in the second transition
Following the global conformational changes leading to TRPV148C,CAP,INT (the first transition), progression to the open state TRPV148C,CAP,OPEN (the second transition) is marked by substantial rearrangements in the outer pore. Generally, the outer pore of each subunit appears to tilt and rotate around the membrane with subtle movement of the pore helix and selectivity filter and with more substantial movement of the pore loop and junctional region of the turret (S5-turret and turret-pore helix, termed the turret junction) (Fig. 4a, b). These rearrangements lead to dilation of the selectivity filter, at M644 and G643. This tilting and rotation of the outer pore region within subunits lead to changes at the subunit interfaces in the outer pore. Interestingly, we observed rearrangements of a phospholipid bound at the subunit interface, facilitated by rotamer flipping of Y631 (Fig. 4c). The outer pore rearrangement is apparently coupled to the S6 gate opening. For example, movement of the selectivity filter influences the conformation of Y671 on S6, which is wedged between two selectivity filter loops (Fig. 4d). The outer pore rearrangement seen in TRPV148C,CAP,OPEN is consistent with many previous functional studies. Mutations of N628 (turret junction), T633 (pore helix), N652 (pore loop), and Y653 (pore loop) were shown to selectively reduce heat sensitivity but not sensitivity to capsaicin, with the triple mutant N628K/N652T/Y653T being heat-insensitive 14,15,40. Hydrophobic amino acid residues in the pore loop and pore helix, such as F649, have been previously suggested to be important for heat sensing41. These residues exhibit large heat-induced conformational changes in the TRPV148C,CAP,OPEN structure (Fig. 4b, c). E600 and E648, two residues that are involved in TRPV1 gating and appear to stabilize the outer pore in TRPV14C,APO structure, have lost their interactions with D654 (pore loop-S6) and K639 of the pore helix from the neighboring subunit, respectively (Fig. 4c and Extended Data Fig. 3e). Last, gating-dependent changes in solvent accessibility for the Y671 site has been reported42,43.
Fig. 4. Conformational changes in the outer pore during the second transition.
a, Extracellular view comparing the pore region in TRPV148C,CAP,INT (orange) and TRPV148C,CAP,OPEN (red). b, Close-up view of the outer pore. c, Close-up view of the pore loop and pore helix interface of adjacent subunits. Phospholipids are shown as spheres and sticks. d, Rearrangement of the interface between S6 and the outer pore, where +1 and −1 indicate features from neighboring subunits. Indicated residues are shown as sticks. Key regions abbreviated as turret junction (TJ), pore helix (PH) and pore loop (PL).
This outer pore rearrangement observed in TRPV148C,CAP,OPEN is drastically different from that of the DkTx/RTx-bound TRPV1 structure (Extended Data Fig. 7a) 25,26. In the DkTx/RTx-TRPV1 structure, the interaction network between the pore loop and pore helix is disrupted leading to displacement from each other. However, in TRPV148C,CAP,OPEN the pore loop, turret junction and the pore helix move together in the same direction (clockwise when viewed from the extracellular side).
Noxious heat opens the S6 gate in the second transition
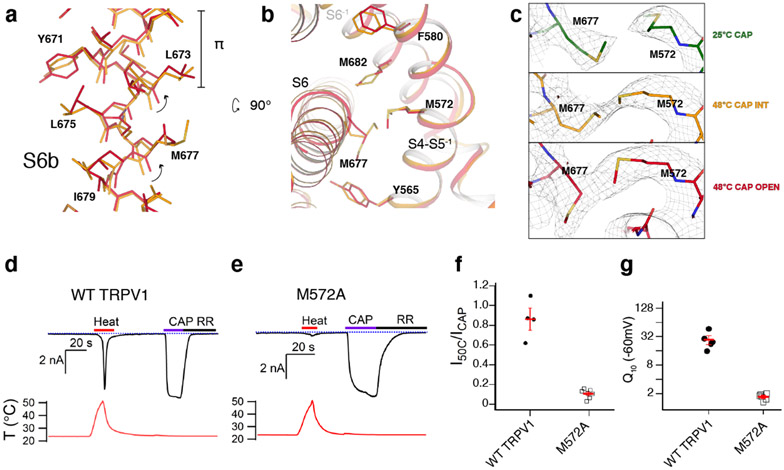
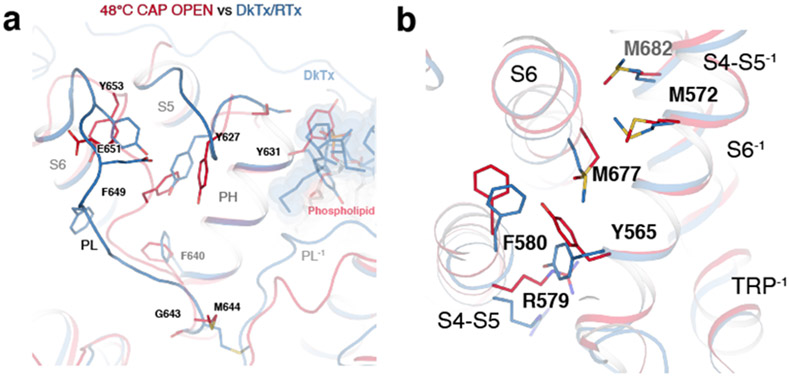
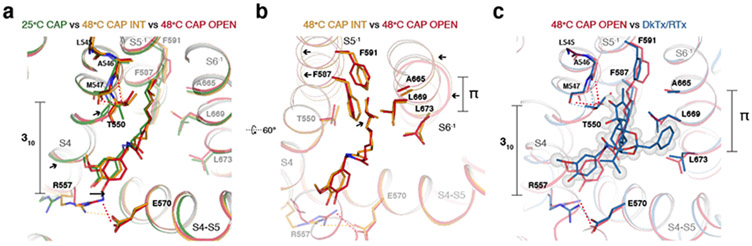
Two key rearrangements of S6 and its surrounding helices during the second transition appear to open the S6 gate. First, in all the resting and intermediate states, I668-L673 within S6 form a π helix-like turn44 (Fig. 5a). During the second transition, S6b (I668-V686) undergoes conformational changes, which include rearrangement of the π helix-like turn leading to rotation of S6b as well as Y671 ring rotation (Fig. 5a). Second, this rotation of S6b during the second transition appears either to be stabilized or induced by the concerted rotamer changes between M677 (S6) and M572 (the S4-S5 linker) (Fig. 5a,b). These concerted rotamer changes are associated with conformational changes of the surrounding residues such as Y565 (the S4-S5 linker), F580 (the neighboring S5), and M682 (the neighboring S6) (Fig. 5b). Notably, inspection of this interface reveals that there are stepwise, concerted rotamer changes involving M572 and M677 during the first and the second transitions (Fig. 5c). In the TRPV148C,CAP,INT structure, M572 rotamerizes to be closer to M677. In the TRPV148C,CAP,OPEN structure, the side chain of M677 moves away from the S4-S5 linker, together with S6b rotation. To test the importance of the concerted movements of M572 and M677 in heat dependent TRPV1 gating, we tested M572A and found that heat-dependent opening of M572A is nearly abolished (Q10 ~1.7) with little effect on capsaicin gating (Fig. 5d-g). This strongly suggests that heat does contribute substantially to the conformational changes we observe. Previous functional studies also suggest that heat contributes appreciably to the observed conformational changes. For instance, alanine substitutions of Y671, I672, and M677 more impact heat-dependent TRPV1 gating than capsaicin-dependent gating45. Large heat-dependent accessibility changes were also observed for I668 and Y67142. It is noteworthy that there are differences between the DkTx/RTx-bound TRPV1 open state and TRPV148C,CAP,OPEN around S6 and its interaction network (Extended Data Fig. 7b).
Fig. 5. Conformational changes in the S6 gate during the second transition.
a, Rearrangement of the S6 π bulge (π) between TRPV148C,CAP,INT (orange) and TRPV148C,CAP,OPEN (red). b, Side chain rearrangements between S6 and the S4-S5 linker of the neighboring subunit. c, Temperature-dependent conformational change of M572 and M677: 25°C (green); 48°C, intermediate (orange); 48°C, open (red). Cryo-EM density thresholdings are 0.022, 0.019, and 0.018 respectively. d, e, Representative time-course whole cell current traces for wild type (WT) TRPV1 (d) and TRPV1 M572A (e). Currents elicited by heat (red) at −60 mV followed by application of 10 μM capsaicin (CAP, purple) then 50μM ruthenium red (black). The dotted blue line indicates zero current. The measured temperature is shown in the lower panel in red. f, Summary of current responses to heat (50°C) relative to saturating capsaicin (10 μM) at room temperature. Values for individual cells with mean ±S.E.M. (red lines) for WT TRPV1 (closed circles, 0.86±0.11, n = 4 biological replicates) and TRPV1 M572A (open squares, 0.11±0.02, n = 7 biological replicates). g, Q10 values obtained from WT TRPV1 and TRPV1 M572A expressing cells activated by heat. Values for individual cells are shown with mean ± S.E.M. for wild type (close circles, Q10 = 28.2 ±7.5, n = 4 biological replicates) and M572A (open squares, Q10 = 1.7±0.1, n = 6 biological replicates). Source data for f and g are available online.
Amino acid clusters exhibit changes in solvent exposure
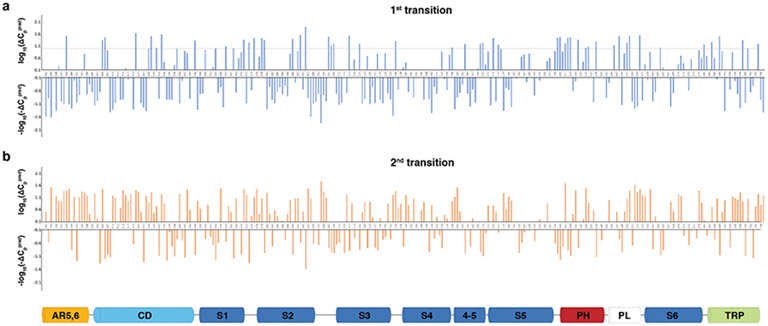
Miller and Clapham have proposed that heat capacity change plays an important role in thermal sensing by the temperature sensitive TRP channels22. Heat capacity can be increased by greater solvent exposure of hydrophobic amino acids and by decreased solvent exposure of hydrophilic amino acids. We calculated the solvent accessible surface areas (SASA) per amino acid residue in three structures of TRPV1CAP, TRPV125C,CAP, TRPV148C,CAP,INT, and TRPV148C,CAP,OPEN. Using the calculated SASA changes and the equation describing thermal protein denaturation46, we calculated predicted heat capacity change (ΔCppred) per residue for the first transition (from TRPV125C,CAP to TRPV148C,CAP,INT) and for the second transition (from TRPV148C,CAP,INT to TRPV148C,CAP,OPEN ). We excluded amino acids for which side chains were not built in all three models. Due to various technical limitations (e.g. modest resolution of our structures, the uncertainty of the ΔCppred values for events involving small SASA changes [as opposed to protein unfolding], the different water properties of bulk water versus bound water47,48, and the uncertainty of solvent exposure at the protein-lipid interface), our aim was to utilize ΔCppred to qualitatively identify the localized region(s), rather than to identify key residues, that potentially contribute to heat sensing. Notably, although conformational changes are larger in the first transition, markedly more amino acid residues exhibit relatively large ΔCppred (from overall larger SASA changes) in the second transition (Fig. 6a,b). The total ΔCppred of the second transition is ~2.1 kJ mol −1 K −1 while that of the first transition is negligibly small. The ΔCppred value of the second transition is about four times smaller than that predicted by Miller and Clapham (8-20 kJ mol −1 K −1) for ΔCp to be the dominant contributer to heat sensing. However, in addition to the uncertainty of ΔCppred due to the above-described limitations, we caution against interpretation of our absolute ΔCppred value as a large fraction of the channel was not included in our calculation of total ΔCppred. Qualitatively, our analysis suggests that heat capacity change contributes mainly to the second heat-dependent transition of TRPV1 (Fig. 6a, b, Extended Data Fig. 8). Residues exhibiting large ΔCppred values in both transitions are mainly clustered in two regions: the outer pore and the CD/ARD (AR6)/TRP helix regions (Fig. 6). The outer pore residues with relatively large ΔCppred are clustered at the interfaces between neighboring subunits comprising the pore loop, the pore helix, S4, S5, and S6 (termed the outer pore cluster, Fig. 6c). Interestingly, many sites that were previously identified as important for heat-dependent TRPV1 opening (e.g. N628, N652, Y671) 14,42 also exhibit large ΔCppred values. The CD/ARD(AR6)/TRP helix interaction network extends into the S4-S5 linker and the S6 gate (termed the CD cluster). These two clusters converge at the S6 gate, as if these spatially separate subdomains are thermally coupled (Fig. 6d). Among these sites, to test the role of heat capacity change in TRPV1 heat-dependent gating, we chose to probe N628 that is exposed to extracellular side in TRPV125C,CAP but buried in TRPV148C,CAP,OPEN (Figs. 4b, 6d) and introduced isosteric mutations to affect only the polarity of the site: leucine (hydrophobic, heat capacity decrease) or aspartate (hydrophilic, heat capacity increase). Surprisingly, increased polarity of the site (N628D) exhibits greater heat sensitivity (Q10 ~86), while reduced polarity (N628L) decreases Q10 (~2.6) (Fig. 6e-g). The point mutation functional effects correlate well with the direction of heat capacity change, but the dramatic effect on heat sensitivity by such a small solvent accessibility change cannot be solely explained by Miller and Clapham’s heat capacity model 22. It is possible that for conformational changes of a folded protein, the heat capacity change due to changes in localized solvent exposure is position specific within the protein structure and likely deviates from the equation describing thermal protein denaturation. To the best of our knowledge, this is the first observation that a point mutation can substantially increase or decrease the heat sensitivity of TRPV1, implicating this site on the outer pore cluster as being directly involved in TRPV1 heat sensing.
Fig. 6. Structure mapping of SASA-based heat capacity changes.
a, Amino acid residues with relatively large heat capacity change (ΔCPpred > 15 J mol−1 K−1) during the first transition are highlighted as sticks and surfaces on the TRPV14C,APO structure. b, High ΔCPpred residues for the second transition mapped on the TRPV148C,CAP,INT structure. c, Zoomed in view of the outer pore region in b. High ΔCPpred residues for the second transition form a contiguous network encompassing interfaces between neighboring subunits comprising the pore loop (PL), pore helix(PH), S4, S5, and S6. d, The interaction network formed by high ΔCPpred residues for the first and second transitions are shown as surface representations in red (one subunit) and gray (the rest of the channel). e, f, Representative time-course of TRPV1-mediated currents for N628L (e) and N628D (f). Currents elicited by heat (red) at −60 mV followed by application of 10μM capsaicin (purple) and 50μM ruthenium red (black). The dotted blue line indicates zero current. The recorded temperature is shown in the lower panels in red. g, Q10 values obtained from WT, N628L and N628D TRPV1-expressing cells activated by heat. Data for individual cells are shown with mean ± S.E.M. (red lines) for wild type (solid circles, Q10 = 28.2 ±7.5, n=4 biological replicates N628L (open circles, Q10 = 2.6 ±1.5, n = 3 biological replicates) and N628D (open diamonds, Q10=86.0±18.2, n = 7 biological replicates). Source data for g are available online.
A capsaicin site salt bridge primes TRPV1 opening
Because heat-dependent opening of TRPV1CAP was sensitized by capsaicin, we cannot exclude the contribution of capsaicin in our observed conformational changes. Inspection of the capsaicin binding site provides us clues to understand the mechanistic link that integrates heat and capsaicin gating in TRPV1. Our high-quality 3D reconstruction allows us to unambiguously assign the configuration of capsaicin in the TRPV1 structure (Extended Data Figs. 5, 9), which differs from previous models 28,29,31,33. The vanillyl methoxy group of capsaicin points toward the crevice between S3 and S4, while the amide oxygen points away from the crevice and within the hydrogen bonding distance to the hydroxyl of Y511 on S3. The major structural change during the first transition involves the movement of R557 on S4 to form a hydrogen bond with E570 on the S4-S5 linker, leading to the S4-S5 linker swivel (Extended Data Fig. 9a). This salt bridge formation is made possible by the motion of the 310 helical part of S4 (S4b, N551-G558), which is stabilized by the hydrogen bonding of the T550 sidechain with the backbone carbonyls of A546 and M547. Therefore, contrary to the previous predictions28,30,31, T550 does not bind capsaicin but propogates conformational changes. Formation of the R557-E570 salt bridge was proposed to be key to TRPV1 opening in the DkTx/RTx activated TRPV1 structure26. This salt bridge, however, is formed in the intermediate state of TRPV148C,CAP. Capsaicin binding differs between TRPV148C,CAP,INT and TRPV148C,CAP,OPEN due to the heat-induced conformational change of S5 and S6 (particularly the π helical region) of the neighboring subunit, which abut the capsaicin lipidic tail. (Extended Data Fig. 9b). In the DkTx/RTx-activated TRPV1 structure the bulky tricyclic daphnane ring interacts directly with the π bulge of S6 and neighboring S5, which explains the high potency and efficacy of RTx (Extended Data Fig. 9c). Taken together, we propose that formation of the R557-E570 salt bridge primes the channel for opening, but additional conformational changes on S5 and S6 are required for TRPV1 to open.
Discussion
Our study offers explanations for several aspects of TRPV1 function. First, it was reported that DkTx-bound TRPV1 cannot be further activated by heat17. The critical involvement of the pore loop in heat-dependent TRPV1 opening means that the conformational locking of the loop by DkTx likely blocks heat-dependent activation pathways (Extended Data Fig. 7a). Notably, DkTx binds at the outer pore interface and competes off the phospholipid whose position changes in a heat-dependent manner (Fig. 4c and Extended Data Fig. 7a). Second, our TRPV148C,CAP,OPEN structure is distinct from that of the DkTx/RTx open TRPV1 structure, suggesting stimulus-specific activation pathways for TRPV1. This is in contrast to the recent mouse TRPV3 study where nearly identical heat and ligand-open structures were observed36. However, a gain-of-function mutation was introduced for the mTRPV3 structural studies which essentially eliminates heat sensitivity (Q10 ~1.2), so it is possible that the observed conformational changes reflect the intrinsic channel gating, rather than heat- or ligand-dependent gating36.
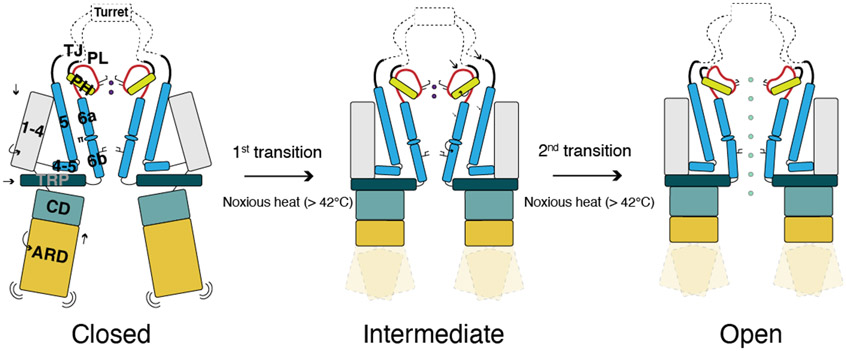
We have captured two noxious heat-dependent conformational transitions that likely take place in a stepwise manner (Fig. 7). The first transition involves global domain rearrangements and primes the channel for opening, while the second transition involves the coupled rearrangement of the outer pore and the interface between S6 and the S4-S5 linker leading to channel opening. Two lines of evidence point to the ARD/CD initiating the first conformational transition: 1) in our apo, heat-treated TRPV148C,APO structure ARD/CD movement is similar, but attenuated, compared to the first transition in the presence of capsaicin. 2) At elevated temperature a large portion of the ARD is not resolved due to its increased flexibility. We speculate that the heat induced flexibility in the N-terminal ARD (AR1-AR4) weakens the interactions between adjacent ARDs within the ARD ring, leading to the movement of the CD and C-terminal ARD (AR5-AR6). The contraction of all the subdomains (CD/ARD, the TRP helix, the S1-S4 domain) during the first transition apparently enables the allosteric communication between the subdomains during the second transition, which we propose is the role of this first transition in heat sensing. Our SASA analysis suggests that heat capacity change contributes more to the second heat transition of TRPV1.
Fig 7. Working model of TRPV1 heat-activation.
In the 1st transition, all subdomains (ARD/CD, S1-S4, and TRP) become contracted. In the 2nd transition, local conformational changes of the outer pore and S6 result in dilation of the selectivity filter and S6 gate, opening the channel.
Structure-based analyses and functional studies provide important insights into the heat-dependent gating of TRPV1. First, although there is no dedicated temperature sensor domain in TRPV1, a subset of residues around the outer pore (outer pore cluster) and CD (CD cluster) that are important for temperature sensing appear to form a coupled network throughout the channel. We show that point mutation of a site in the outer pore cluster (N628) controls TRPV1 heat sensitivity, suggesting that this cluster is directly involved in heat sensing. Heat-driven, choreographed movements within this network could be the basis of heat sensitivity. We speculate that the heat capacity changes contribute to this choreographed movement. However, other factors may contribute to the heat sensitivity of TRPV1, for instance changes in M572 and M677 conformation do not involve any changes in solvent exposure (Fig. 5c and Extended Data Fig. 8) and ΔH and ΔS values for TRPV1 heat-dependent opening appear to be relatively constant over the physiological temperature range49. It is possible that coupled, heat-dependent motions of the poorly resolved ARD and cytoplasmic domain contribute substantially to large ΔS and ΔH values associated with TRPV1 heat gating. Second, large conformational changes are not a requisite for high heat sensitivity of TRPV1. Previous and current studies demonstrate that changes to even a single amino acid may give rise to large effects on heat sensing14,23,50. We posit that the exquisitely fine tuned interaction network renders TRPV1 functionally sensitive to small conformational changes. Last, because we utilized capsaicin to sensitize TRPV1 in our study, the fully heat-dependent open state of TRPV1 remains to be revealed. We believe our findings on TRPV1 may be generally applied to understand the polymodal sensing mechanisms of other thermal sensing TRP channels.
Methods
TRPV1 Protein expression and purification
The Rattus norvegicus full-length TRPV1 was cloned into the pEG BacMam vector51, in-frame with a FLAG-tag and 10× His-tag at C-terminus. All structural studies were performed using the wild-type (WT) rat TRPV1 construct. Baculovirus was generated according to manufacturer’s protocol (Bac-to-Bac, Invitrogen). For rat TRPV1 protein expression, HEK293S GnTI− cells (ATCC) was cultured in FreeStyle 293 media (Life Technologies) supplemented with 2% (v/v) FBS (Gibco) at 8% (w/v) CO2. Cultures at 3×106 mL−1 cell density were infected with 6% (v/v) P3 baculovirus. After 20-22 hrs of shaking incubation at 37°C, 10 mM sodium butyrate (Sigma-Aldrich) was added and the temperature was lowered to 30°C to boost protein expression. After 40-44 hrs, the cells were harvested by centrifugation at 550× g and were subsequently resuspended in lysis buffer (20 mM Tris pH 8, 150 mM NaCl, 12 μg mL−1 leupeptin, 12 μg mL−1 pepstatin, 12 μg mL−1 aprotinin, 2 μg mL−1 DNase I, 1 mM phenylmethylsulphonyl fluoride (PMSF), and 1% (w/v) digitonin). Membrane protein extraction was performed at 4°C for 1 hr, followed by centrifugation at 13000× g for 30 min to remove insoluble material. The supernatant was subsequently incubated with anti-FLAG M2 resin (Sigma-Aldrich) at 4°C for 1 hr to allow protein binding. The resin was then packed onto a gravity-flow column (BioRad), and washed with 10 column volumes of wash buffer (20 mM Tris pH 8, 150 mM NaCl, 0.07% digitonin). The TRPV1 protein was then eluted with 5 column volumes of elution buffer (20 mM Tris pH 8, 150 mM NaCl, 0.07% digitonin, 100 μg mL−1 FLAG peptide (GenScript)). The eluted protein was collected and subjected for nanodiscs reconstitution.
Nanodiscs Reconstitution
MSP2N2 was purified according to the previously published protocol52. FLAG-purified TRPV1 was concentrated to 1-1.5 mg mL−1, and mixed with purified MSP2N2 and lipid mix [1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1'-rac-glycerol) (POPG), Avanti Polar Lipids; POPC:POPE:POPG=3:1:1] at 1:3:200 molar ratio, where TRPV1 was treated as tetramer in molar ratio calculations. The mixture was incubated at 4°C for 30 min with constant rocking. Subsequently, 100 mg mL−1 Bio-Beads SM2 (Bio-Rad) was added to the mixture to initiate the reconstitution reaction. The Bio-Beads were exchanged with a fresh batch after two hours (100 mg mL−1), and the mixture was incubated with constant rocking at 4°C for 12-15 hrs. The sample was then subjected to size exclusion chromatography on a Superose 6 Increase 10/300 GL column (Cytiva) pre-equilibrated with buffer containing 20 mM HEPES pH 7.5, 150 mM NaCl.
Cryo-EM sample preparation and data collection
Peak fractions containing nanodisc-reconstituted TRPV1 from the size exclusion chromatography were concentrated to ~0.8 mg mL−1. All cryo-EM samples in this study were prepared on freshly glow-discharged UltrAuFoil R1.2/1.3 300 mesh grids (Quantifoil), using a Leica EM GP2 plunge freezer. (i) For TRPV14C,APO sample, 3 μL of TRPV1-nanodiscs sample was applied to the grid, blotted for 2 s in the chamber set at 4°C and 95% humidity, followed by plunge-freezing in liquid-ethane cooled by liquid nitrogen. (ii) For TRPV148C,APO sample, 10 μl of the TRPV1 sample was incubated in a heat block set at 48°C for 30 s, 3 μL of which was then applied to the grid, blotted for 2 s in the chamber set at 48°C and 80% humidity, followed by plunge-freezing in liquid-ethane cooled by liquid nitrogen. (iii) For TRPV14C,CAP sample, TRPV1 sample was mixed with 30 μM capsaicin (Sigma) for 30 min before applying to the grid, blotted for 2 s in the chamber set at 4°C and 95% humidity, then plunge-frozen in liquid-ethane cooled by liquid nitrogen. (iv) For TRPV125C,CAP sample, TRPV1 sample was mixed with 30 μM capsaicin for 30 min before applying to the grid, blotted for 2 s in the chamber set at 25°C and 95% humidity, then plunge-froze in liquid-ethane cooled by liquid nitrogen. (v) For TRPV148C,CAP sample, TRPV1 sample was mixed with 30 μM capsaicin for 30 min before applying to the grid, incubated in a heat block set at 48°C for 30 s, blotted for 2 s in the chamber set at 48°C and 80% humidity, then plunge-froze in liquid-ethane cooled by liquid nitrogen. For 25°C and 48°C freezing, all the tools (grid, tube, forceps) were equilibrated to the repective temperatures before use.
TRPV14C,CAP and TRPV125C,CAP datasets were collected with a Titan Krios microscope (Thermo Fisher) operating at 300 kV equipped with a K3 detector (Gatan) in counting mode, using the Latitude-S automated data acquisition program. Movie datasets were collected at a nominal magnification of 81,000 × with a pixel size of 1.08 Å/pix at specimen level. Each movie contains 60 frames over a 4.6 s exposure time, using a dose rate of about 15 e−/Å2/s, resulting in the total accumulated dose of ~60 e−/Å2. The nominal defocus range was set from −1 to −2.25 μm.
TRPV14C,APO and TRPV148C,APO datasets were collected with a Titan Krios microscope (Thermo Fisher) operating at 300 kV equipped with a K3 detector (Gatan) in counting mode with GIF BioQuantum energy filter (slit width 20 eV), using the Serial-EM automated data acquisition program. Movie datasets were collected at a nominal magnification of 81,000 × with a pixel size of 0.5395 Å/pix in super-resolution mode. Each movie contains 74 frames over a 3.5 s exposure time, using a dose rate of about 15 e−/Å2/s, resulting at the total accumulated dose of ~50 e−/Å2. The nominal defocus range was set from −0.75 to −2.0 μm.
TRPV148C,CAP dataset was collected with a Titan Krios microscope (Thermo Fisher) operating at 300 kV equipped with a K3 detector (Gatan) in counting mode with GIF BioQuantum energy filter (slit width 20 eV), using the Serial-EM automated data acquisition program. Movie datasets were collected at a nominal magnification of 81,000 × with a pixel size of 0.5295 Å/pix in super-resolution mode. Each movie contains 50 frames over a 2.4 s exposure time, using a dose rate of about 18.5 e−/Å2/s, resulting in the total accumulated dose of ~45 e−/Å2. The nominal defocus range was set from −0.8 to −1.9 μm.
Cryo-EM data processing
All datasets were processed using a similar procedure. Beam-induced motion correction and dose-weighing were performed using MotionCor253. For TRPV14C,APO, TRPV148C,APO and TRPV148C,CAP datasets, the movies are 2× fourier-binned to 1.079 Å per pixel (TRPV14C,APO and TRPV148C,APO) and 1.059 Å per pixel (TRPV148C,CAP), respectively. The motion corrected micrographs were then subjected to contast transfer function (CTF)estimation using Gctf54. Micrographs were subsequently selected based on CTF fit quality and CTF estimated resolution. An initial set of particles were manually picked and subjected to a reference-free 2D classification (k=10, T=2), from which the best 3-5 classes were selected as reference for automated particle picking in RELION 3.055. Particles were extracted by 4x4 Fourier binning with 4.32 Å/pix pixel size and 64 pixel box size. Reference-free 2D classification (k=50, T=2) was performed in RELION and classes showing clear secondary structure features of TRPV1 were selected. These particles were subsequently subjected to 3D auto-refinement in RELION, using a previously published TRPV1 map (EMD-8118, low-passed filtered to 30 Å) as reference without masking. Refined particles were re-extracted, re-centered and un-binned and subjected to another round of 3D refinement, using the result of the previous 3D refinement as a reference. Several rounds of 3D classification without image alignment were then performed using the output from the un-binned 3D refinement, with a soft solvent mask covering the best resolved region of the channel. A single class showing the clearest and the best resolved features was selected and subjected to 3D auto-refinement. If needed, another round of 3D classification was performed followed by 3D refinement in cryoSPARC56. Several rounds of CTF refinement and Bayesian polishing were performed, which improved resolution and map quality. Local resolution was calculated using RELION 3.1 or cryoSPARC56. Pixel size calibration was performed with the final maps.
Model building, refinement, and alignment
The model-building process is similar for all structures reported in this paper. A previously published TRPV1 structure (PDB 5IRZ) was used as a reference. During model building the register assignment was guided by the presence of large aromatic side chains. The placement of individual structural elements was performed by rigid body fitting and the structures were manually refined using real space refinement in Coot with ideal geometry restrains57. The restraints for lipids and ligands, including POPC, POPE, POPG, and capsaicin, were calculated in Elbow (as implemented in Phenix58) from isomeric SMILES strings and optimized using the REEL QM2 method (as implemented in the Phenix suite58). These were then inspected and adjusted manually to ensure correct stereochemistry before being fitted into the cryo-EM maps in Coot. The MolProbity59 server (http://molprobity.biochem.duke.edu) was utilized to identify problematic regions in the models, which were then manually adjusted in Coot. The final refinement was performed using the phenix-real_space_refine function with global minimization and secondary structure restrains as implemented in the Phenix suite58. The Fourier shell correlation of the half- and full-maps against the model, calculated in Phenix, were in good agreement, indicating that the models were not over-refined. Structural analyses and illustrations were performed using PYMOL (Schrödinger)60 and UCSF Chimera61. Structure alignments and cryo-EM density map alignments was performed by Fit In Map in UCSF Chimera. Based on the aligned map, each structural model was aligned to its corresponding map by Fit In Map.
Two-electrode voltage clamp electrophysiology in X. laevis oocytes
The WT rat TRPV1 DNA was subcloned into the pGEM-HE vector, and the construct was linearized wih SphI, and complementary RNA (cRNA) was synthesized by in vitro transcription using T7 RNA polymerase (Thermo Fisher). All defolliculated oocytes were ordered from Ecocyte (Austin, Texas). Rat TRPV1 cRNA was injected to Xenopus laevis oocytes and incubated at 17°C for 3-4 days in a solution containing (in mM) 96 NaCl, 2 KCl, 1 MgCl2, 1.8 CaCl2, 5 HEPES, pH 7.6 (with NaOH). For the two-electrode voltage clamp (TEVC) recording, oocyte membrane voltage was controlled using an OC-725C oocyte clamp (Warner Instruments, Hamden, CT). Data were filtered at 1–3 kHz and digitized at 20 kHz using pClamp software (Molecular Devices, Sunnyvale, CA) and a Digidata 1440A digitizer (Axon Instruments). Microelectrode resistances were 0.1–1 MΩ when filled with 3 M KCl. The external recording solution contained 100 mM KCl, 2 mM MgCl2, 10 mM HEPES, pH 7.6 (with KOH) and 0.01 mM 2-[(4-methoxy)-2-naphthalenyl)amino]-5-nitro-benzoic acid (Tocris), and 0.01 mM flufenamic acid were added to the recording solution to minimize calcium-activated chloride currents. Capsaicin and Ruthenium Red (RR) were applied using a gravity-fed perfusion system. Voltage was initially held at −60 mV and ramped to +60 mV for 300 ms every 0.5 s. Heat stimuli were achieved by passing the external recording solution through glass capillary coils immersed in a water bath maintained at about 70-80°C, and recordings were performed during constant perfusion with temperature measured using a thermistor (TA-29, Warner Instruments). The thermistor was connected to the digitizer via a temperature controller (TC-324B, Warner Instruments). All data analysis was carried out using Igor Pro 6.3 (Wavemetrics, Portland, OR). Q10 values were calculated using the equation
where SArrhe is the slope of linear fit to Arrhenius plotted data between absolute temperatures T1 and T262.
Patch clamp electrophysiology
Whole-cell patch clamp recordings from transiently transfected HEK293T cells were performed at room temperature (22–24°C) and with heat (up to ~50°C) followed. Current responses were low-pass filtered at 2 kHz (Axopatch 200B), digitally sampled at 5–10 kHz (Digidata 1440A), converted to digital files in Clampex10.7 (Molecular Devices) and stored on an external hard drive for offline analyses (Clampfit10.7, Molecular Devices; Excel 2010, Microsoft Office; Igor Pro 6.34A, Wavemetrics). Pipettes were pulled from borosilicate glass and heat-polished to final resistances between 3 and 7 MΩ. Electrodes were filled with an intracellular solution containing (in mM) 140 NaCl, 5 MgCl2, 10 HEPES, 5 EGTA, and adjusted to pH 7.4 (NaOH). MgCl2 was included to both increase the quality of the seals and to block endogenous HEK293T channels. The extracellular solution consisted of (in mM): 140 NaCl, 10 HEPES, 5 EDTA, pH 7.4 (NaOH). For heat activation, we used the same method as described in oocyte temperature recordings above.
Calculation of Solvent Accessible Area
Apolar and polar solvent-accessible surface area (SASA) per residue for the TRPV125C,CAP, TRPV148C,CAP,INT, and TRPV148C,CAP,OPEN structures were calculated using the GETAREA server (http://curie.utmb.edu/getarea.html)63. Calculations were not done on residues whose side chains could not be built. Heat capacity change, ΔCP, was calculated by the Makhatadze and Privalov’s equation below46, using the change in solvent-accessible surface area for non-polar and polar residues (ΔASAnpol and ΔASApol, respectively).
Extended Data
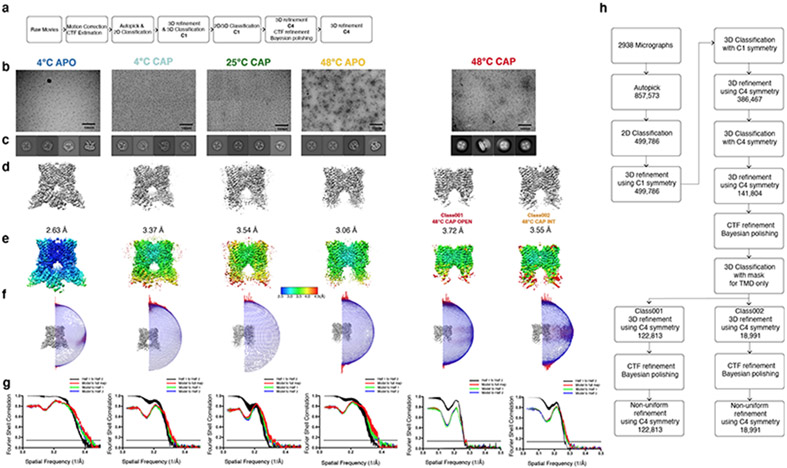
Extended Data Fig. 1. TRPV1 data collection and processing.
Data processing procedures, a, Data processing flow chart for TRPV14C,APO, TRPV14C,CAP, TRPV125C,CAP, TRPV148C,APO. b, representative micrographs, see Table 1 for details. c, 2D classification images, d, 3D reconstructions, e, local resolution estimation, f, the Euler distribution plot, g, FSC curves for TRPV14C,APO, TRPV14C,CAP, TRPV125C,CAP, TRPV148C,APO, TRPV148C,CAP,OPEN and TRPV148C,CAP,INT, respectively. h, Data processing flow chart for TRPV148C,CAP,INT and TRPV148C,CAP,OPEN.
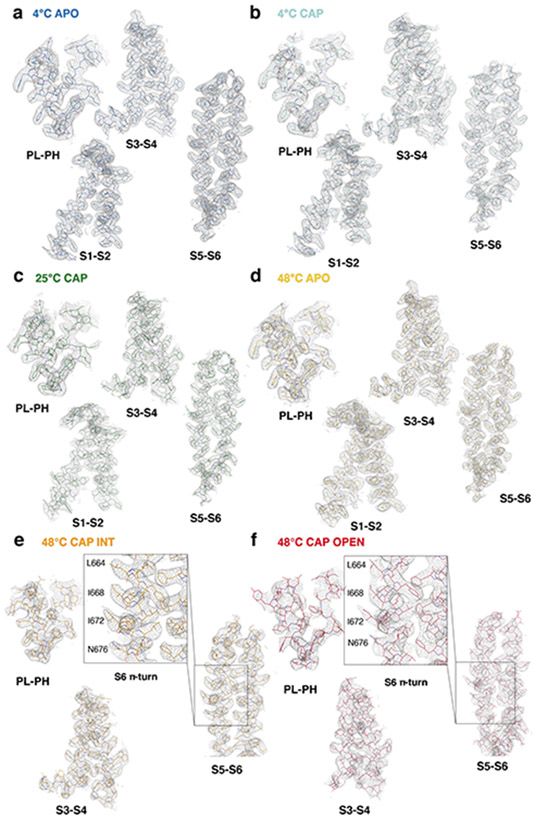
Extended Data Fig. 2. Representative Cryo-EM density of the TRPV1 structures.
a-f, cryo-EM density for subdomains in TRPV14C,APO (a, thresholding 0.014), TRPV14C,CAP (b, thresholding 0.014), TRPV125C,CAP (c, thresholding 0.025), TRPV148C,APO (d, thresholding 0.019), TRPV148C,CAP,INT (e, thresholding 0.28), TRPV148C,CAP,OPEN (f, thresholding 0.3). Structural elements are shown as sticks and EM density as gray mesh.
Extended Data Fig. 3. Structural features of the full-length TRPV1.
a, Architecture of the TRPV1 protomer with subdomains indicated: ankyrin repeat domain (ARD), coupling domain (CD), transmembrane helices S1-S6, TRP helix, and C-terminal domain (CTD). b, Cryo-EM density (half-map without symmetry) for the selectivity filter of TRPV14C,APO corresponding to putative sodium ions at 0.04 thresholding. c, Cryo-EM density of the turret and turret junction (0.012 thresholding). d, Close-up view of the outer pore and turret junction (0.012 thresholding). e, Interaction networks spanning the outer pore region and the S1-S4 domain (0.03 thresholding). Key residues interacting with E600 and E648 are shown as sticks with surrounding cryo-EM density. f, g, Cryo-EM density of the CD, TRP, ARD (f), and CTD (g). The ARD is colored in gold, the CD and its individual elements (HTHCD, βCD,) in sky blue, the TRP domain in dark green, and the CTD in orange. The cryo-EM density (gray) is shown at 0.012 thresholding. h, Superposition of a single protomer from TRPV14C,APO (blue) and TRPV148C,APO (gold). i, Superposition of a single protomer from TRPV14C,APO (blue) and TRPV14C,CAP (cyan).
Extended Data Fig. 4. Comparison of TRPV14C,APO and TRPV148C,APO.
a, b, Cryo-EM 3D reconstructions of TRPV14C,APO (a, blue) and TRPV148C,APO (b, gold), respectively. Outlines indicate AR1-AR4. c, Close-up comparison of the cytoplasmic domains between TRPV14C,APO (blue) and TRPV148C,APO (gold).
Extended Data Fig. 5. Comparison of TRPV14C,APO, TRPV14C,CAP, TRPV125C,CAP and the published structure of TRPV1 in the presence of capsaicin.
a, Close-up view of the S1-S4 domain of TRPV14C,APO (blue) and TRPV14C,CAP (cyan). Capsaicin (red) and phosphatidyl inositol (blue) molecules are shown as sticks. b, Close-up view of capsaicin in the vanilloid pocket of TRPV14C,CAP. The cryo-EM density is shown at 0.025 thresholding. c, Side view comparison of TRPV14C,CAP (cyan) and TRPV125C,CAP (green). d, Side view comparison of TRPV14C,CAP (green) and the published TRPV1 structure in the presence of capsaicin (PDB ID: 3J5R, brown).
Extended Data Fig. 6. Comparison between the overall structures of TRPV14C,APO, TRPV148C,CAP,OPEN and DkTx/RTx-bound TRPV1.
a, Comparison of TRPV14C,APO (silver), TRPV148C,CAP,OPEN (red), and DkTx/RTx-TRPV1 (blue) viewed from the intracellular side. ARD/CD movement occurs at an individual protomer level. b, Comparison of the S6b and TRP domain of TRPV14C,APO, TRPV148C,CAP,OPEN, and DkTx/RTx-TRPV1. c, Close-up view of TRPV14C,APO, TRPV148C,CAP,OPEN, and DkTx/RTx-TRPV1 in the cytoplasmic domains. d, Alternate angle and close-up view of TRPV14C,APO, TRPV148C,CAP,OPEN, and DkTx/ RTx-TRPV1 in the cytoplasmic domains.
Extended Data Fig. 7. Comparison of TRPV148C,CAP,OPEN and DkTx/RTx-bound TRPV1 structures.
a, The overlapping locations of phospholipid (TRPV148C,CAP,OPEN, red) and DkTx (DkTx/RTx-TRPV1, blue), shown as sticks and spheres, between the pore loop and pore helix. Several side chains are shown as sticks to illustrate the differences in the outer pore of the two structures. b, Structural differences between TRPV148C,CAP,OPEN and DkTx/RTx-TRPV1 at S6, the S4-S5 linker, and the TRP helix.
Extended Data Fig. 8. Solvent accessible surface area-based heat capacity change plots for the first and the second transitions.
a, b, ΔCPpred plots for the first (a) and second (b) transitions. For each transition, residues exhibiting positive ΔCPpred are plotted in the upper graph using log10(ΔCPpred), and residues exhibiting negative ΔCPpred are plotted in the lower graph using −log10(−ΔCPpred). The dotted line denotes the 15 J mol−1 K−1 threshold. ΔCPpred was calculated as described in the Methods. Residues for which the side chains were not resolved were not included in the calculation.
Extended Data Fig. 9. Rearrangement in the vanilloid pocket during the heat-dependent transitions.
a, Close-up view of the vanilloid binding sitein TRPV125C,CAP (green), TRPV148C,CAP,INT (orange), and TRPV148C,CAP,OPEN (red). Several key residues in capsaicin are shown as sticks. Dotted lines denote either H-bond or salt bridge interactions. The 310 helical region of S4 is indicated as 310. b, Close-up view of S5 and S6 in TRPV148C,CAP,INT (orange), and TRPV148C,CAP,OPEN (red). The π helical turn in S6 is denoted by π. c, Comparison of TRPV148C,CAP,OPEN (red) and DkTx/RTx-bound TRPV1 (PDB ID: 5IRX, blue). DkTx is shown as sticks and gray spheres; capsaicin is depicted as sticks only.
Acknowledgments:
Cryo-EM data were collected at the Duke University Shared Materials Instrumentation Facility (SMIF) and at the Pacific Northwest Center for Cryo-EM (PNCC) at OHSU. We thank Janette Myers at PNCC for assistance in data collection, Nilakshee Bhattacharya at SMIF for assistance with the microscope operation, Lucas B. Dillard at NIEHS for assistance in sample screening, and Alberto Bartesaghi at Duke for a pre-processing interface for data collection. We thank Justin Fedor and Ying Yin for critical reading of the manuscript and discussion, William Borschel for training Feng Zhang on the patch clamp recording, Jorg Grandl for discussion and Wonpil Im for advice on the solvent accessibility analysis. This research was supported by NIH grant (R35NS097241 to S.-Y.L.) and by the National Institute of Health Intramural Research Program; US National Institutes of Environmental Health Sciences (ZIC ES103326 to M.J.B). A portion of this research was supported by NIH grant U24GM129547 and performed at the PNCC at OHSU and accessed through EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. DUKE SMIF is affiliated with the North Carolina Research Triangle Nanotechnology Network, which is in part supported by the NSF (ECCS-2025064).
Footnotes
Competing interests: The authors declare no competing interests.
Data Availability:
The coordinates are deposited in the Protein Data Bank with the PDB IDs 7LP9 (TRPV14C,APO), 7LPA (TRPV14C,CAP), 7LPB (TRPV125C,CAP), 7LPC (TRPV148C,APO), 7LPD (TRPV148C,CAP,INT) and 7LPE (TRPV148C,CAP,OPEN), respectively. The cryo-EM maps are deposited in the Electron Microscopy Data Bank with the IDs EMD-23473 (TRPV14C,APO), EMD-23474 (TRPV14C,CAP), EMD-23475 (TRPV125C,CAP), EMD-23476 (TRPV148C,APO), EMD-23477 (TRPV148C,CAP,10sec), EMD-23478 (TRPV148C,CAP,INT) and EMD-23479 (TRPV148C,CAP,OPEN), respectively. Source data are provided with this paper.
References
- 1.Caterina MJ & Julius D The vanilloid receptor: a molecular gateway to the pain pathway. Annu Rev Neurosci 24, 487–517 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Ramsey IS, Delling M & Clapham DE An introduction to TRP channels. Annu Rev Physiol 68, 619–47 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Vriens J & Voets T Heat sensing involves a TRiPlet of ion channels. Br J Pharmacol 176, 3893–3898 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vay L, Gu C & McNaughton PA The thermo-TRP ion channel family: properties and therapeutic implications. Br J Pharmacol 165, 787–801 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandell M, Macpherson LJ & Patapoutian A From chills to chilis: mechanisms for thermosensation and chemesthesis via thermoTRPs. Curr Opin Neurobiol 17, 490–7 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caterina MJ et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature 389, 816–24 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Szallasi A et al. Resiniferatoxin-type phorboid vanilloids display capsaicin-like selectivity at native vanilloid receptors on rat DRG neurons and at the cloned vanilloid receptor VR1. Br J Pharmacol 128, 428–34 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Voets T et al. The principle of temperature-dependent gating in cold- and heat-sensitive TRP channels. Nature 430, 748–54 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Brauchi S, Orio P & Latorre R Clues to understanding cold sensation: thermodynamics and electrophysiological analysis of the cold receptor TRPM8. Proc Natl Acad Sci U S A 101, 15494–9 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin Y et al. Structure of the cold- and menthol-sensing ion channel TRPM8. Science 359, 237–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang S et al. A paradigm of thermal adaptation in penguins and elephants by tuning cold activation in TRPM8. Proc Natl Acad Sci U S A 117, 8633–8638 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paricio-Montesinos R et al. The Sensory Coding of Warm Perception. Neuron 106, 830–841 e3 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao J, Liu B & Qin F Kinetic and energetic analysis of thermally activated TRPV1 channels. Biophys J 99, 1743–53 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grandl J et al. Temperature-induced opening of TRPV1 ion channel is stabilized by the pore domain. Nat Neurosci 13, 708–14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SE, Patapoutian A & Grandl J Single residues in the outer pore of TRPV1 and TRPV3 have temperature-dependent conformations. PLoS One 8, e59593 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao J, Liu B & Qin F Modular thermal sensors in temperature-gated transient receptor potential (TRP) channels. Proc Natl Acad Sci U S A 108, 11109–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jara-Oseguera A, Bae C & Swartz KJ An external sodium ion binding site controls allosteric gating in TRPV1 channels. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang F et al. Heat activation is intrinsic to the pore domain of TRPV1. Proc Natl Acad Sci U S A 115, E317–E324 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vlachova V et al. Functional role of C-terminal cytoplasmic tail of rat vanilloid receptor 1. J Neurosci 23, 1340–50 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F, Cui Y, Wang K & Zheng J Thermosensitive TRP channel pore turret is part of the temperature activation pathway. Proc Natl Acad Sci U S A 107, 7083–8 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim M et al. Evidence that the TRPV1 S1-S4 membrane domain contributes to thermosensing. Nat Commun 11, 4169 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clapham DE & Miller C A thermodynamic framework for understanding temperature sensing by transient receptor potential (TRP) channels. Proc Natl Acad Sci U S A 108, 19492–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chowdhury S, Jarecki BW & Chanda B A molecular framework for temperature-dependent gating of ion channels. Cell 158, 1148–1158 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordt SE & Julius D Molecular basis for species-specific sensitivity to "hot" chili peppers. Cell 108, 421–30 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Cao E, Liao M, Cheng Y & Julius D TRPV1 structures in distinct conformations reveal activation mechanisms. Nature 504, 113–8 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao Y, Cao E, Julius D & Cheng Y TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liao M, Cao E, Julius D & Cheng Y Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature 504, 107–12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F et al. Structural mechanism underlying capsaicin binding and activation of the TRPV1 ion channel. Nat Chem Biol 11, 518–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elokely K et al. Understanding TRPV1 activation by ligands: Insights from the binding modes of capsaicin and resiniferatoxin. Proc Natl Acad Sci U S A 113, E137–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darre L & Domene C Binding of Capsaicin to the TRPV1 Ion Channel. Mol Pharm 12, 4454–65 (2015). [DOI] [PubMed] [Google Scholar]
- 31.Hanson SM, Newstead S, Swartz KJ & Sansom MSP Capsaicin interaction with TRPV1 channels in a lipid bilayer: molecular dynamics simulation. Biophys J 108, 1425–1434 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smart OS, Neduvelil JG, Wang X, Wallace BA & Sansom MS HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Graph 14, 354–60, 376 (1996). [DOI] [PubMed] [Google Scholar]
- 33.Nadezhdin KD et al. Extracellular cap domain is an essential component of the TRPV1 gating mechanism. Nat Commun 12, 2154 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jordt SE, Tominaga M & Julius D Acid potentiation of the capsaicin receptor determined by a key extracellular site. Proc Natl Acad Sci U S A 97, 8134–9 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zubcevic L, Borschel WF, Hsu AL, Borgnia MJ & Lee SY Regulatory switch at the cytoplasmic interface controls TRPV channel gating. Elife 8(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh AK et al. Structural basis of temperature sensation by the TRP channel TRPV3. Nat Struct Mol Biol 26, 994–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swain J & Kumar Mishra A Location, Partitioning Behavior, and Interaction of Capsaicin with Lipid Bilayer Membrane: Study Using Its Intrinsic Fluorescence. J Phys Chem B 119, 12086–93 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Sanchez-Moreno A et al. Irreversible temperature gating in trpv1 sheds light on channel activation. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ladron-de-Guevara E et al. The Contribution of the Ankyrin Repeat Domain of TRPV1 as a Thermal Module. Biophys J 118, 836–845 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryu S, Liu B, Yao J, Fu Q & Qin F Uncoupling proton activation of vanilloid receptor TRPV1. J Neurosci 27, 12797–807 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bae C et al. Structural insights into the mechanism of activation of the TRPV1 channel by a membrane-bound tarantula toxin. Elife 5(2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salazar H et al. Structural determinants of gating in the TRPV1 channel. Nat Struct Mol Biol 16, 704–10 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Steinberg X et al. Conformational dynamics in TRPV1 channels reported by an encoded coumarin amino acid. Elife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zubcevic L & Lee SY The role of pi-helices in TRP channel gating. Curr Opin Struct Biol 58, 314–323 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Susankova K, Ettrich R, Vyklicky L, Teisinger J & Vlachova V Contribution of the putative inner-pore region to the gating of the transient receptor potential vanilloid subtype 1 channel (TRPV1). J Neurosci 27, 7578–85 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Makhatadze GI & Privalov PL Energetics of protein structure. Adv Protein Chem 47, 307–425 (1995). [DOI] [PubMed] [Google Scholar]
- 47.Henriques DA, Ladbury JE & Jackson RM Comparison of binding energies of SrcSH2-phosphotyrosyl peptides with structure-based prediction using surface area based empirical parameterization. Protein Sci 9, 1975–85 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Franzese G & Rubi M Aspects of Physical Biology: Biological Water, Protein Solutions, Transport and Replication, (Springer Berlin Heidelberg, 2008). [Google Scholar]
- 49.Voets T Quantifying and modeling the temperature-dependent gating of TRP channels. Rev Physiol Biochem Pharmacol 162, 91–119 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Chen H, Deng J, Cui Q, Chanda B & Henzler-Wildman K Mapping temperature-dependent conformational change in the voltage-sensing domain of an engineered heat-activated K(+) channel. Proc Natl Acad Sci U S A 118(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 51.Goehring A et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–85 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritchie TK et al. Chapter 11 - Reconstitution of membrane proteins in phospholipid bilayer nanodiscs. Methods Enzymol 464, 211–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang K Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Punjani A, Rubinstein JL, Fleet DJ & Brubaker MA cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods 14, 290–296 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Emsley P & Cowtan K Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–32 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Adams PD et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen VB et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Delano WL The PyMol Molecular Graphics System, (DeLano Scientific, Palo Alto, CA, USA, 2002). [Google Scholar]
- 61.Goddard TD, Huang CC & Ferrin TE Visualizing density maps with UCSF Chimera. J Struct Biol 157, 281–7 (2007). [DOI] [PubMed] [Google Scholar]
- 62.Kang K et al. Modulation of TRPA1 thermal sensitivity enables sensory discrimination in Drosophila. Nature 481, 76–80 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fraczkiewicz R & Braun W Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. Journal of Computational Chemistry 19, 319–333 (1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The coordinates are deposited in the Protein Data Bank with the PDB IDs 7LP9 (TRPV14C,APO), 7LPA (TRPV14C,CAP), 7LPB (TRPV125C,CAP), 7LPC (TRPV148C,APO), 7LPD (TRPV148C,CAP,INT) and 7LPE (TRPV148C,CAP,OPEN), respectively. The cryo-EM maps are deposited in the Electron Microscopy Data Bank with the IDs EMD-23473 (TRPV14C,APO), EMD-23474 (TRPV14C,CAP), EMD-23475 (TRPV125C,CAP), EMD-23476 (TRPV148C,APO), EMD-23477 (TRPV148C,CAP,10sec), EMD-23478 (TRPV148C,CAP,INT) and EMD-23479 (TRPV148C,CAP,OPEN), respectively. Source data are provided with this paper.