Highlights
-
•
Investigation of gut microbiome composition and diversity with respect to human personality.
-
•
Analyses targeted bacterial genera linked to behaviour in animal and human psychiatric studies.
-
•
Bacterial genera were modelled (using negative binomial regression) with respect to personality.
-
•
Genera linked to autism are also related to social behaviour in the general population.
-
•
Sociability is associated with higher diversity, and anxiety and stress with reduced diversity.
Keywords: Gut microbiome, Microbiome–gut–brain axis, Metagenomics, Human personality, Social behaviour
Abstract
The gut microbiome has a measurable impact on the brain, influencing stress, anxiety, depressive symptoms and social behaviour. This microbiome–gut–brain axis may be mediated by various mechanisms including neural, immune and endocrine signalling. To date, the majority of research has been conducted in animal models, while the limited number of human studies has focused on psychiatric conditions. Here the composition and diversity of the gut microbiome is investigated with respect to human personality. Using regression models to control for possible confounding factors, the abundances of specific bacterial genera are shown to be significantly predicted by personality traits. Diversity analyses of the gut microbiome reveal that people with larger social networks tend to have a more diverse microbiome, suggesting that social interactions may shape the microbial community of the human gut. In contrast, anxiety and stress are linked to reduced diversity and an altered microbiome composition. Together, these results add a new dimension to our understanding of personality and reveal that the microbiome–gut–brain axis may also be relevant to behavioural variation in the general population as well as to cases of psychiatric disorders.
1. Introduction
Personality shapes our world. It influences our health, our friendships, how we deal with stress, what jobs we succeed in and how we like to spend our time. It is approximately 50% heritable in human populations [1], suggesting that environmental factors also contribute significantly to our personality. In addition to our external environment, the burgeoning field of the microbiome is revealing the many ways that our ‘environment within’ can affect our body’s physiology, including our digestion, immunity, metabolism, development and even our behaviour [2], [3], [4], [5], [6]. In fact, the number of microbial cells in our bodies is estimated to be roughly equal to our own human cells [7], with the majority of these microorganisms inhabiting the gut. Studies in the past decade provide evidence that the gut microbiome interacts with the central nervous system [8] and such findings have the potential to aid the development of new treatments for conditions such as autism [9] and depression [10]. However, research has largely been conducted in animal models and it is unknown how translatable these findings are to humans [11], [12], [13]. While there is some indication that dysbiosis of the gut microbiota may be implicated in neurological and psychiatric disorders [14], an open question remains as to whether variation in the gut microbial community is related to personality, that is behavioural differences between individuals that are consistent over time and different situations and therefore broadly predictable.
Animal studies have demonstrated that the gut microbiome can influence the stress response, anxiety and depressive-like behaviours, as well as social behaviour and communication [6], [15]. Some of the most convincing findings stem from faecal microbiota transplantation whereby behavioural traits can be transferred between mouse strains when their gut microbiota are swapped [16], [17]. For example, when the more anxious and timid Balb/c mice are colonized with the gut microbiota of NIH Swiss mice, their temperament becomes more bold and exploratory like that of the donor NIH Swiss mice, and vice versa [16]. Further support comes from the induction of anxiety and depressive-like behaviours in rodents colonized with the gut microbiota of humans suffering from these symptoms [18], [19], [20]. The transmission of such behaviours via the microbiota therefore suggests that gut microorganisms can contribute causally to behavioural traits. In fact, a recent human study reported improvement in psychiatric symptoms following faecal microbiota transplantation in patients with gastrointestinal disease [21].
There are numerous possible mechanisms that may mediate this interaction between the gut microbial community and the brain, including communication via neural, immune and endocrine pathways [6], [22], [23]. Microorganisms can also produce various neuroactive chemicals [24], [25] and can modulate host neurotransmitter levels [26]. For example, gut bacterial species such as those belonging to the genus Bacteroides have been shown to produce γ-aminobutyric acid (GABA) in large quantities in culture [27]. More recently it has been reported that the relative abundance of Bacteroides is negatively associated with brain signatures of depression [28], suggesting that bacterially derived GABA may play a role in the microbiome–gut–brain axis. Gut dysbiosis might lead to imbalances in neurotransmitters, inflammation or heightened activity of the hypothalamus–pituitary–adrenal axis that regulates the stress response [13]. The observation that psychiatric illnesses are often comorbid with gastrointestinal problems [29], [30] supports the role of the microbiome–gut–brain axis in human biology and psychology. A number of studies has reported associations between the composition of the gut microbiome and conditions such as autism, depression and schizophrenia (Table 1). However, results sometimes differ with regard to changes in the abundances of specific bacterial taxa, likely due to individual variability in gut microbiome composition together with underpowered studies. Furthermore, there is limited understanding of the mechanisms via which each of these bacterial taxa may affect the brain. Indeed, the predominant mechanism likely varies depending on the particular taxon involved, including anti-inflammatory effects, the production of short chain fatty acids, hormonal effects, the release of neuroactive metabolites including neurotransmitters and stimulation of the vagus nerve [23].
Table 1.
Summary of statistically significant associations between bacterial genera in the gut and behavioural or psychiatric traits, as reported in the literature. Table lists findings from research in both animal models and human populations and includes all genera (23 in total) associated with these traits in at least two independent studies.
| Genus | Change in abundance | Behavioural trait/psychiatric condition | Study subject | References |
|---|---|---|---|---|
| Akkermansia | ↓ | Autism | Children | [52] |
| ↑ | Autism | Children | [53] | |
| ↓ | Stress | Mice | [54], [55] | |
| Alistipes | ↓ | Autism | Children | [56] |
| ↑ | Stress | Mice | [57] | |
| ↑ | Depression | Adults | [58], [59] | |
| Bacteroides | ↑ | Autism | Children | [53], [60] |
| ↓ | Stress | Mice | [55], [61] | |
| ↑ | Negative mood | Adults | [62] | |
| ↑ | Psychosis | Adults | [63] | |
| Bifidobacterium | ↓ | Autism | Children | [52], [53], [60], [64], [65], [66], [67] |
| ↑ | Autism | Children | [68] | |
| ↓ | Autism | Mice | [69] | |
| ↓ | Stress | Mice | [70], [71] | |
| ↑ | Stress | Mice | [72] | |
| ↑ | Negative mood | Adults | [62] | |
| ↓ | Depression | Adults | [73] | |
| Blautia | ↓ | Autism | Infants | [74] |
| ↓ | Autism | Children | [75] | |
| ↓ | Autism | Mice | [69] | |
| ↓ | Schizophrenia | Adults | [76] | |
| Clostridium | ↑ | Autism | Children | [53], [56], [67], [75], [77], [78], [79], [80] |
| ↓ | Autism | Children | [60] | |
| ↑ | Stress | Mice | [61], [72], [81] | |
| ↑ | Depression | Adults | [59], [82] | |
| ↑ | Schizophrenia | Adults | [76] | |
| Collinsella | ↑ | Autism | Children | [56] |
| ↓ | Autism | Children | [60] | |
| ↑ | Schizophrenia | Adults | [76] | |
| Corynebacterium | ↑ | Autism | Children | [56] |
| ↓ | Stress | Rats | [83] | |
| Desulfovibrio | ↑ | Autism | Children | [60], [80] |
| Dialister | ↓ | Autism | Children | [56], [60] |
| ↑ | Sociability | Infants | [84] | |
| ↓ | Depression | Adults | [18], [59], [85] | |
| Dorea | ↑ | Autism | Children | [53], [56] |
| ↓ | Autism | Children | [75] | |
| ↓ | Stress | Mice | [61] | |
| Faecalibacterium | ↑ | Autism | Infants | [74] |
| ↓ | Autism | Children | [53], [86] | |
| ↓ | Negative mood | Adults | [87] | |
| ↑ | Negative mood | Adults | [62] | |
| ↓ | Depression | Adults | [59], [85] | |
| ↑ | Depression (prenatal) | Adults | [88] | |
| ↓ | Bipolar disorder | Adults | [89], [90] | |
| Flavonifractor | ↑ | Autism | Children | [75] |
| ↑ | Depression | Adults | [59] | |
| Lactobacillus | ↑ | Autism | Children | [56], [64], [68], [80], [91] |
| ↓ | Autism | Children | [53] | |
| ↓ | Stress | Mice | [61], [72], [92] | |
| ↓ | Stress | Rats | [83] | |
| ↓ | Stress | Macaques | [93] | |
| ↓ | Depression | Adults | [73] | |
| ↑ | Psychosis | Adults | [63] | |
| Lactococcus | ↓ | Autism | Children | [53], [60] |
| Oscillibacter | ↑ | Depression | Adults | [58] |
| ↑ | Stress | Rats | [83] | |
| Oscillospira | ↓ | Autism | Children | [53] |
| ↑ | Sociability | Mice | [94] | |
| ↓ | Stress | Mice | [55], [94], [95] | |
| Parabacteroides | ↓ | Autism | Children | [56] |
| ↑ | Autism | Children | [60] | |
| ↑ | Sociability | Infants | [84] | |
| ↓ | Stress | Mice | [55], [61], [92], [94] | |
| ↑ | Negative mood | Adults | [62] | |
| ↑ | Depression | Adults | [59] | |
| Prevotella | ↑ | Autism | Children | [53] |
| ↓ | Autism | Children | [91] | |
| ↑ | Stress | Mice | [94] | |
| ↓ | Negative mood | Adults | [62] | |
| ↑ | Depression | Adults | [82] | |
| ↓ | Depression | Adults | [18] | |
| Roseburia | ↑ | Autism | Children | [53] |
| ↑ | Stress | Mice | [61] | |
| ↓ | Stress | Mice | [55] | |
| ↓ | Negative mood | Adults | [62] | |
| ↓ | Schizophrenia | Adults | [76] | |
| Streptococcus | ↓ | Autism | Children | [53], [60] |
| ↑ | Depression | Adults | [82] | |
| Sutterella | ↑ | Autism | Children | [52], [65], [96], [97] |
| ↓ | Autism | Children | [75] | |
| ↑ | Stress | Mice | [55] | |
| Turicibacter | ↓ | Autism | Children | [53] |
| ↑ | Sociability | Mice | [94] | |
| ↓ | Stress | Mice | [70], [72], [94] | |
| ↑ | Depression | Adults | [18] |
Intervention studies in humans have started to investigate whether the gut microbiome makes a detectable contribution to the functioning of the central nervous system. Although studies administering probiotics have yielded mixed results in humans, meta-analyses do provide support for their beneficial effect on stress, anxiety and depressive symptoms, including in healthy humans as well as those suffering from psychiatric conditions [31], [32], [33], [34]. In addition, brain imaging studies have revealed that consumption of probiotics in healthy volunteers can affect neural signatures of emotion [35], [36]. For example, probiotic administration over a period of four weeks was found to modulate brain activity, resulting in reduced activity of brain regions involved in emotional processing when participants were presented with negative facial expressions [35]. In studies involving prebiotics, healthy humans show a reduction in salivary cortisol levels after three weeks of taking the supplement [37] and patients with irritable bowel syndrome have reported reduced anxiety following one month of prebiotic supplementation [38].
Since the gut microbiome has been implicated in social development and behaviour [39], [40], [41], [42], recent research has investigated whether it may modulate behavioural symptoms of autism [43]. Autism is not only characterized by impaired social behaviour but is frequently comorbid with gastrointestinal issues [44] and immune dysfunction [45], [46]. Studies comparing autistic individuals with neurotypical controls often report significant differences in gut microbiome composition (Table 1). Interestingly, a trial of faecal microbiota transplantation in children with autism found that it improved not only gastrointestinal symptoms but also social behaviour and communication [47], including two years after completion of the treatment [48]. This suggests that the gut microbial community may play a part in the expression of autistic behavioural traits. Given the evidence that autistic traits are normally distributed across the population [49], [50], [51], gut microbiome composition may also be related to variation in sociability in the general population.
The main aim of this research was to determine whether individual variation in the composition and diversity of the human gut microbiome is related to differences in personality. Previous research has focused on animal models or examined broadscale associations between microbiome composition and psychiatric symptoms. In contrast, here targeted regression analyses were conducted to investigate the relationship between personality traits and the abundances of specific bacterial genera identified in the microbiome–gut–brain axis literature (Table 1). It is hypothesized here that genera previously associated with autism may be related to differences in sociability in the general population, while genera previously linked to depression and stress in animalmodels or psychiatric populations may be differentially abundant in the general population with respect to neurotic traits. As well as assessing genus-level composition, the relationship between personality and measures of microbiome diversity was also investigated, with the hypothesis that variables assessing sociability may be positively related to diversity and those assessing neurotic traits may be negatively related.
2. Materials and methods
2.1. Study population
Adults (over 18 years) were invited to take part in the study (Fig. 1). A faecal sample was received from 671 individuals but sixteen samples were removed (n = 655) since they did not meet the read quality control threshold set at 10,000 reads to ensure detection of low abundance taxa [98]. The research was conducted under a Human Subjects Protocol provided by an institutional review board (Ethical and Independent Review Services, IRB Study #13044-03). All participants provided informed consent and data were analysed anonymously. The sample was 71% female and 29% male, with a mean age of 42 years. Participants resided in twenty different countries and four continents, predominantly North America (83%), with participants from USA accounting for 77% of the sample. 11% of the study participants were from Europe (4% from UK), 5% from Australasia and 2% from Asia.
Fig. 1.
Overview of study design. A faecal sample was provided by each participant using the commercially available uBiome Gut Kit and sample processing and sequencing were carried out by uBiome to generate OTU (operational taxonomic unit) abundances. Participants were asked to complete a comprehensive study questionnaire assessing their behaviour, diet, health, lifestyle and sociodemographics (see Fig. S1 for variables tested within each of these categories). A range of statistical analyses was conducted to determine the relationships between gut microbiome composition and the study variables, with a primary interest in the variables assessing behavioural traits.
2.2. Sample collection, processing and sequencing
The microbiome sequencing company uBiome provided participants with their commercially available Gut Kit. Each participant was supplied with one kit to be used for faecal sample collection at home. Analysis of faecal samples is the standard procedure in microbiome research since they are sufficiently representative of the composition of the gut microbiome [99]. The sampling procedure followed the guidelines of the National Institutes of Health Human Microbiome Project [100]. The sample collection tube contained a lysis and stabilization buffer for DNA preservation during transport at ambient temperatures [98]. The 16S rRNA gene was amplified using universal primers for the V4 hypervariable region and sequencing was performed on the Illumina NextSeq 500 platform, alongside positive and negative controls. Standardized methodology [98] for sample processing and sequencing was conducted by uBiome and has been shown to generate reproducible microbiome profiles [101].
2.3. Questionnaires
An online questionnaire designed for this study was distributed to participants. In total, 44 study variables (continuous and categorical) were analysed relating to behavioural traits, diet, health and lifestyle and also sociodemographics. While the majority of variables were assessed through binary questions or point scales, personality and social behaviour were evaluated with widely used and validated questionnaires from psychological research. The 50-item International Personality Item Pool [102] was used to assess personality traits according to the five-factor model of personality. A large number of studies find that individual differences in human behaviour can broadly be divided into five key domains, known as the ‘Big Five’ [103], [104], comprising extraversion (propensity to seek and enjoy others’ company), agreeableness (trust and cooperation in social interactions), conscientiousness (attention to detail and focus), neuroticism (tendency to feel negative emotions) and openness (creativity, intellectual curiosity and willingness to seek new experiences). These traits are temporally stable within individuals [105] and are found in both sexes, all races and across cultures [106]. In addition, the State–Trait Anxiety Inventory [107], specifically the trait subscale, was used to determine the participants’ general tendency to feel anxious. Participants were also scored for two subscales (social skill and communication) from the Autism Spectrum Quotient, which measures the degree to which any neurotypical adultlies on the continuum between normality and autistic-like behaviours [108]. Social network size was evaluated based on the number of people in their two innermost layers, approximately corresponding to those individuals contacted on a weekly and monthly basis respectively [109].
Detailed dietary habits of the participants were assessed with the EPIC-Norfolk Food Frequency Questionnaire [110]. The associated software, Food Frequency Questionnaire EPIC Tool for Analysis [111], was used to estimate total calorie consumption and intake of food groups, macronutrients, minerals and vitamins (56 variables in total). Food frequency questionnaires with more than ten missing entries were excluded in accordance with the guidelines.
2.4. Multiple regression analyses of bacterial abundances
Microbiome count data are often overdispersed, with a high proportion of zeros [112], [113] and heavily right-skewed [114]. This is because there are relatively few taxa that are shared by the majority of samples, since most taxa are rare and only detected in a small proportion of samples [112]. Given this overdispersion, count data cannot be normalized via transformation and so negative binomial regression was conducted with genus abundance as the response variable (or zero-inflated negative binomial regression in cases where the count data contained an excess of zeros). When modelling microbiome data, it is necessary to account for variation in sequencing read depth between samples. However, rarefying count data or transforming it into proportions can lead to a high rate of false positives and is not recommended for analyses of taxon abundance [115]. Instead, the logarithm of the total read number was included as an offset in the regression models, thereby preserving statistical power by retaining all the count data [112], [116].
As opposed to the majority of microbiome studies which only conduct bivariate correlations between variables and microbial taxon abundances, key variables known to influence the gut microbiome were included in the regression models to avoid potential confounding effects. These variables were sex [117], [118], [119], [120], age [117], [121], body mass index [117], birth delivery mode [120], [122], [123], type of infant feeding method [118], [120], [124], whether participants had been treated with oral antibiotics within the last six months (since studies suggest the gut microbial community largely recovers after this time [125], [126]), whether they suffered from a gut condition [127], [128] and if they took probiotic supplements [36], [129] (though evidence is somewhat lacking as to the degree to which probiotic supplementation can alter faecal microbiota composition [130], [131]). These regression analyses could only be conducted on a subset of the population cohort (n = 261) who supplied the necessary data for all these variables.
The variables of primary interest were those relating to personality traits. Pairwise Kendall’s Tau-b correlation coefficients were computed to assess the correlation structure of all variables in the study, revealing considerable intercorrelation between extraversion, social skill and communication and also between neuroticism, anxiety and stress, as expected from the literature [132], [133]. To avoid multicollinearity in the regression models, two new variables were therefore created by summing the z-scores for the individual variables: sociability (a combined score of an individual’s extraversion, social skill and communication) and neurotic tendencies (a combined score of an individual’s neuroticism, anxiety and stress). These two new variables were used in the regression analyses since social behaviour, anxiety, stress and depressive-like behaviours are the main behavioural traits that have been linked to the gut microbiome [134].
Negative binomial regression analyses were conducted using the R package VGAM to model microbiome count data. All statistical analyses in this study were performed using R 3.2.3 software [135]. An extensive literature review was carried out to identify putative taxa that have been associated with behavioural or psychiatric traits in animal research or human populations. Genera that showed a significant association in at least two independent studies (Table 1) were then targeted in the regression analyses. Genus-level count data were used for the regression models since 16S rRNA sequencing provides an estimated 96% accuracy for genus identification [136] but has limited phylogenetic resolution at the species level [137].
2.5. Analyses of microbiome diversity and community composition
Analyses were also conducted to determine how the overall compositional profile of the microbiome, measured with alpha and beta diversity metrics, was related to the study variables. Alpha diversity is the ecological diversity of a single sample, taking into account the number of different taxa and their relative abundances, while beta diversity measures differences in microbial community composition between individuals [138]. The R package phyloseq was used to determine richness and alpha diversity at the genus level [139]. Since the choice of diversity index can significantly influence results [140], both Shannon’s diversity index and the inverse Simpson’s diversity index were calculated (the inverse of Simpson’s index being more intuitive since its value increases as diversity increases). While these two diversity indices take into account both richness and evenness, Shannon’s diversity index is more sensitive to changes in abundance of rare taxa, whereas the inverse Simpson’s diversity index gives more weight to common taxa [141].
Since the correlation between sequencing depth and diversity was not negligible, microbiome count data were rarefied prior to diversity analyses by randomly sampling counts without replacement such that all samples had an equal number of total counts. Although rarefying the count data was avoided for the regression models, it is still considered a useful normalization technique when conducting diversity analyses as uneven sequence counts across samples can significantly impact diversity estimates [113], [116]. In terms of alpha diversity, associations between each variable and either diversity or richness were measured using Kendall’s Tau-b correlation coefficient. This method is appropriate since it is a non-parametric measure of correlation and also accounts for tied values.
To assess beta diversity, permutational multivariate analysis of variance (PERMANOVA) was conducted to determine which variables significantly affect the overall composition of the gut microbiome. Distance matrices were computed using the distance function in the R package phyloseq [139] to assess the pairwise similarity of gut microbiome composition between individuals. The Bray–Curtis index provides a quantitative measure of dissimilarity between microbial communities, while the Jaccard index is qualitative since it only takes into account presence versus absence of taxa and not their abundances [116]. PERMANOVA was carried out on both Bray–Curtis and Jaccard distance matrices using the adonis function from the R package vegan with 1000 permutations. This function estimates the variance in the distance matrix that is attributable to the variable of interest, that is, the extent to which each variable may explain differences between individuals in microbial community composition. The P values from these analyses were then adjusted for multiple comparisons using the Benjamini–Hochberg method to control for the false discovery rate (FDR), with Padj < 0.1 considered statistically significant.
3. Results
3.1. Summary statistics
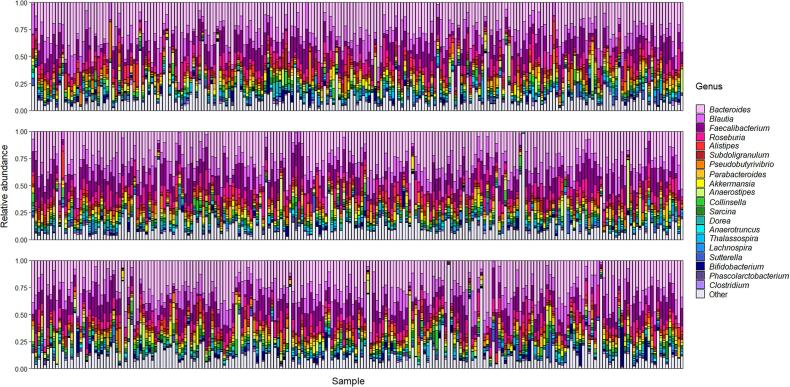
Gut microbiome composition differed markedly between individuals (Fig. 2), as has repeatedly been shown in human populations [142]. Out of all the genera detected across the gut microbiome samples, the top twenty most abundant genera showed close correspondence with previous research [143]. Bacteroides was the most abundant genus, as found in other studies [144], [145]. There was also considerable variation among the study population in the variables assessed in the questionnaire (Tables S1 and S2). The majority of study variables from the questionnaire showed little to modest intercorrelation (Fig. S1), though as expected there was a high degree of correlation between the nutrient variables assessed in the food frequency questionnaire (Fig. S2).
Fig. 2.
Stacked bar plots summarizing gut microbiome composition at genus level for the total participant cohort. Microbiome data are plotted showing relative abundance of the top twenty most abundant genera across all samples, with the remaining genera grouped into ‘Other’. The twenty genera in the key are ordered from most to least abundant.
3.2. Regression models of bacterial abundances
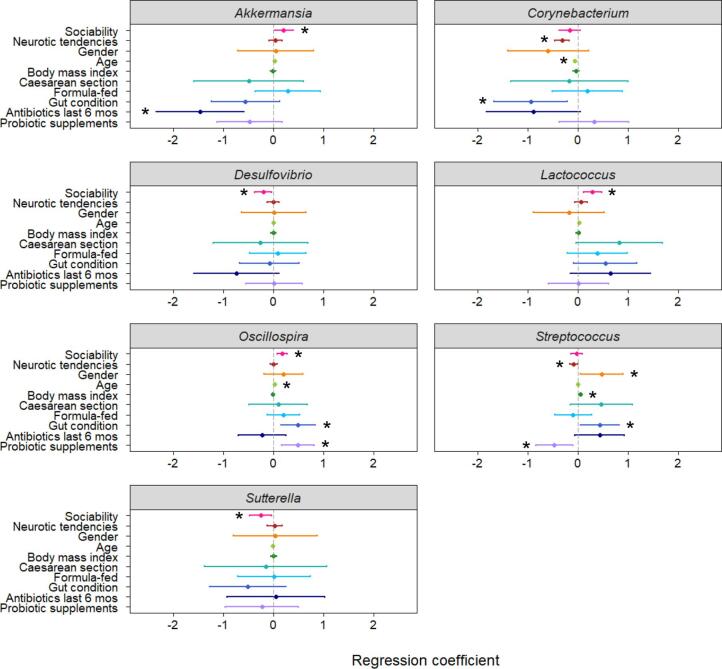
Negative binomial regression analyses revealed that the abundances for seven of the 23 genera were significantly predicted by individual variation in behavioural traits (Fig. 3; Table S3). Sociability (a combined measure of the participant’s extraversion, social skill and communication) was a positive predictor of genus abundance for Akkermansia (P = 0.038), Lactococcus (P = 0.002) and Oscillospira (P < 0.001), and a negative predictor of the abundances of Desulfovibrio (P = 0.019) and Sutterella (P = 0.028). The variable assessing neurotic tendencies (a combined measure of neuroticism, anxiety and stress) negatively predicted genus abundance for Corynebacterium (P < 0.001) and Streptococcus (P = 0.029). Table 2 compares these bacterial genera found to be significantly related to behavioural traits with the results of previous studies on these genera.
Fig. 3.
Coefficient plots from regression models predicting abundances of genera in the human gut microbiome. Asterisks denote significant predictors of genus abundance where P < 0.05 and bars indicate 95% confidence intervals. The key variables of interest for this study were those relating to behavioural traits, namely sociability (extraversion, social skill and communication) and neurotic tendencies (neuroticism, anxiety and stress), while other variables were also included to avoid potential confounding effects. A positive relationship with gender indicates a higher abundance of the genus in females. Plots displayed here depict genera whose abundances were significantly related to differences in behavioural traits (for remaining regression coefficient plots see Fig. S3).
Table 2.
Bacterial genera found to be significantly related to behavioural traits in this study compared with previous research findings extracted from Table 1. For genera significantly related to sociability in this study, it is most relevant to compare these findings to those from studies on sociability or autism (i.e. if genus abundance is higher in sociable individuals, it may be expected to be lower in autism). For genera related to neurotic tendencies it is most relevant to compare these findings to those from studies on stress and depression. Results from previous studies that are clearly in the opposite direction to the findings in this study are marked in brackets.
| Genus | Change in abundance | Behavioural trait/psychiatric condition | Study subject | References |
|---|---|---|---|---|
| Akkermansia | ↓ | Autism | Children | [52] |
| [↑] | Autism | Children | [53] | |
| ↓ | Stress | Mice | [54], [55] | |
| ↑ | Sociability | Adults | This study | |
| Corynebacterium | [↑] | Autism | Children | [56] |
| ↓ | Stress | Rats | [83] | |
| ↓ | Neurotic tendencies | Adults | This study | |
| Desulfovibrio | ↑ | Autism | Children | [60], [80] |
| ↓ | Sociability | Adults | This study | |
| Lactococcus | ↓ | Autism | Children | [53], [60] |
| ↑ | Sociability | Adults | This study | |
| Oscillospira | ↓ | Autism | Children | [53] |
| ↑ | Sociability | Mice | [94] | |
| ↓ | Stress | Mice | [55], [94], [95] | |
| ↑ | Sociability | Adults | This study | |
| Streptococcus | ↓ | Autism | Children | [53], [60] |
| [↑] | Depression | Adults | [82] | |
| ↓ | Neurotic tendencies | Adults | This study | |
| Sutterella | ↑ | Autism | Children | [52], [65], [96], [97] |
| [↓] | Autism | Children | [75] | |
| ↑ | Stress | Mice | [55] | |
| ↓ | Sociability | Adults | This study |
There were also notable results in terms of the possible confounding factors included in the models. Age and body mass index were common predictors of abundance, significantly predicting the abundances of eight and seven genera, respectively. In contrast, birth delivery mode and infant feeding method had little influence on the abundances of most genera. Delivery mode was only a significant predictor of the abundance of one genus, Flavonifractor, with a higher abundance in individuals born by caesarean section. Formula feeding during infancy was a positive predictor of Lactobacillus abundance, in agreement with previous research findings [146]. Surprisingly, abundances of only three of the 23 genera were significantly altered by antibiotic treatment in the past six months, indicative of the differential effects antibiotics can have on bacterial taxa. Three genera, Lactobacillus, Prevotella and Streptococcus, were more abundant in females, which is consistent with previous findings for Lactobacillus [120], [147].
3.3. Microbiome diversity
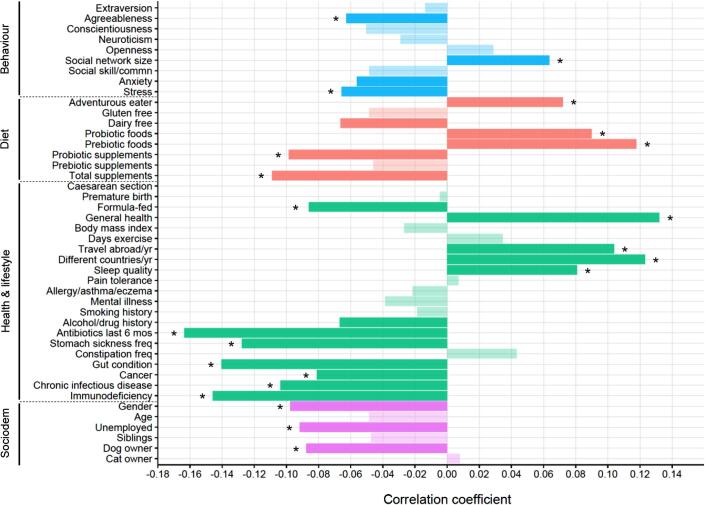
Overall, 25 of the 44 study variables were significantly related to gut microbiome diversity (Padj < 0.1), as measured by Shannon’s diversity index, with the majority of these (22 variables) also retaining significance at FDR < 0.05 (Fig. 4; Table S4). For the primary variables of interest relating to behaviour, both stress (Padj = 0.045) and anxiety (Padj = 0.077) were negatively correlated with Shannon’s diversity index, and agreeableness was also negatively related (Padj = 0.048). There was a positive correlation between Shannon’s diversity index and social network size (Padj = 0.043), such that individuals with a larger social network tended to have a more diverse gut microbiome.
Fig. 4.
Bar plot showing results of Kendall’s Tau-b correlation analysis between gut microbiome diversity and the study variables. Opaque shading indicates a significant correlation at FDR < 0.1 and asterisks denote significance also at FDR < 0.05. The negative relationship with gender reflects a lower diversity in females compared with males. Plot depicts results with Shannon’s diversity index (for results using the inverse Simpson’s diversity index and genus richness see Table S4).
In terms of the other variables in the study, more adventurous eaters and those who ate more foods with naturally occurring probiotics (fermented foods) or prebiotics (non-digestible fibre) tended to have a more diverse gut microbial community. In contrast, consumption of probiotic supplements was significantly correlated with lower diversity. Variables relating to ill health were generally associated with reduced diversity and, not surprisingly, the strongest negative relationship was with antibiotic treatment. Adults who were formula-fed as infants had a significantly less diverse microbiome compared with those that were breast-fed. There was also evidence that females had a lower gut microbiome diversity than males. Diversity was not related to exercise frequency or body mass index, while the strongest positive correlation was with general health. Microbiome diversity was positively correlated with travel, while it was negatively related to dog ownership and also unemployment.
Comparing the correlation results of Shannon’s diversity index and the inverse Simpson’s diversity index revealed good consistencybetween the two indices (Table S4). Most of these relationships were also present when correlating the variables with the observed number of genera (Table S4), though as expected there were some differences between the results since this is a measure of community richness rather than diversity. Notably, there was a significant negative correlation between conscientiousness and genus richness (Padj = 0.014), which was in the same direction as the non-significant trend with diversity.
With respect to nutrient intake estimated from the food frequency questionnaire, the consumption of fruit and cereals, and therefore fibre (and more generally carbohydrates), was positively related to diversity (Table S5). This was in accordance with previous findings that the metabolism of carbohydrates, particularly fibre, promotes microbial diversity [148], [149], [150]. Diversity was also significantly correlated with the dietary abundance of a range of minerals and vitamins (Table S5). There was little relationship between genus richness and nutrient intake with the exception of fish consumption which was positively related.
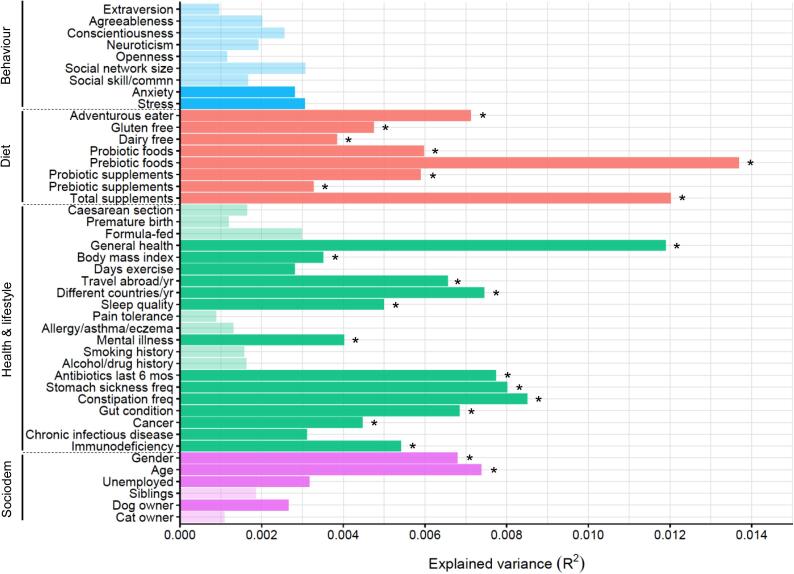
3.4. Microbial community composition
Differences in the composition of the gut microbiome, as quantified by the Bray–Curtis index, were significantly related to 28 of the 44 variables (Padj < 0.1), with most of these (22 variables) retaining significance at FDR < 0.05 (Fig. 5; Table S6). While the P values reported below refer to results with the Bray–Curtis index, the same variables were also significant when using the Jaccard index to measure differences in microbial community composition (Table S6).
Fig. 5.
Bar plot showing explained variance in gut microbiome composition in relation to the study variables. Opaque shading indicates variables explaining significant inter-individual variation at FDR < 0.1 and asterisks denote significance also at FDR < 0.05. Plot depicts results using the Bray–Curtis distance matrix (for results with the Jaccard distance matrix see Table S6).
Regarding behavioural traits, anxiety (Padj = 0.094) and stress (Padj = 0.052) were significantly related to an altered microbiome composition. Another factor influencing composition was whether the participant suffered from a mental illness (Padj = 0.020). The majority of variables assessing health and lifestyle were also significantly related to composition, whereas there was no effect of early-life events (mode of delivery, premature birth and infant feeding). While exercise frequency did not significantly influence diversity, it did affect overall composition of the gut microbiome, as has been shown in other studies [151]. As was the case with diversity, unemployment was a significant factor influencing microbial community composition. Composition also differed by age and gender, in line with previous research [121].
All the main dietary variables were significantly related to composition (Fig. 5). Both gluten-free and dairy-free diets were associated with an altered microbial community, in accordance with other research findings [152], [153], [154]. The consumption of naturally occurring prebiotic fibre showed the strongest relationship with gut microbiome composition. In terms of the results of the food frequency questionnaire, consumption of fruit, vegetables and cereals, and therefore the total intake of carbohydrates including fibre, were all significantly related to microbiome composition, as well as certain minerals and vitamins (Table S7).
4. Discussion
Investigations of the microbiome–gut–brain axis have predominantly been conducted in animal models or clinical populations, with few studies exploring whether these findings are translatable to the general population. Here both gut microbiome composition and diversity were found to be related to differences in personality. This is the first time that abundances of microbial genera have been modelled with respect to human personality, while also rigorously controlling for the possible confounding effects of key variables known to influence the gut microbiome. The regression results revealed that abundances of particular genera are significantly related to behavioural traits, suggesting that studying the gut microbiome may be relevant to understanding variation in human personality. People with larger social networks were also found to have more diverse microbial communities,indicating that social behaviour may promote diversity of the human gut microbiome. By contrast, lower diversity was associated with increased levels of stress and anxiety, and these traits were also related to differences in overall composition of the microbial community. In addition, the intercorrelation analysis (Fig. S1) revealed that people who ate more foods with naturally occurring probiotics or prebiotics had significantly lower levels of anxiety, stress and neuroticism and were also less likely to suffer from a mental illness, but this relationship was not found in those consuming probiotics or prebiotics in supplement form. Consistent with this, a previous study reported that females consuming more fermented foods (a source of natural probiotics) had fewer symptoms of social anxiety [155] and similarly a traditional Japanese diet rich in fermented soy products has been linked to reduced depressive symptoms [156].
The significant relationships between personality traits and the abundances of particular genera targeted in the regression analyses were largely in the expected direction based on studies in humans with psychiatric conditions and in laboratory animals (Table 2). The genera Akkermansia, Lactococcus and Oscillospira were found to be more abundant in individuals with a higher sociability score. In accordance with these results, previous studies in children have reported a reduction in Lactococcus and Oscillospira in autism, while one study found decreased abundance of Akkermansia (Table 2). Oscillospira has also been positively related to sociability in mice (Table 2). Interestingly, both Akkermansia and Oscillospira are associated with good health; Akkermansia has anti-inflammatory properties and there is some evidence it may be protective against metabolic disorders [157] while lower levels of Oscillospira are linked to inflammatory disease [158]. Two genera, Desulfovibrio and Sutterella, were more abundant in less sociable individuals. This is in line with those studies finding that their abundances are also elevated in people suffering from autism (Table 2) and in fact, it has been hypothesized that Desulfovibrio species may play an important role in the pathophysiology of autism [159]. The variable measuring neurotic tendencies was a significant negative predictor of the abundances of Streptococcus and Corynebacterium. Therefore, individuals that were more neurotic tended to have lower levels of these genera. Since higher neuroticism is a known risk factor for developing depression [160], this result is in agreement with a study in a rat model of stress-induced depression reporting a reduction in Corynebacterium (Table 2). However, in contrast to the findings here, Streptococcus has previously been found to be more prevalent in adults suffering from depression (Table 2). Although Streptococcus species are part of a healthy microbiome, members of this genus can be opportunistic pathogens [161].
The results of this study suggest that some bacterial genera may be more strongly linked to behaviour and, in light of my findings, perhaps future research may benefit from investigating the effects of the genera Akkermansia, Desulfovibrio, Lactococcus, Oscillospira and Sutterella on social behaviour and autistic-like symptoms in animal models, with a view to developing potential new therapies for autism. Notably, not all of the 23 genera targeted in these regression analyses were significantly predicted by behavioural traits (Fig. S3; Table S3), though this was expected given the somewhat inconsistent results reported in the literature (Table 1). In particular, despite extensive findings from animal studies that species belonging to Bifidobacterium and Lactobacillus can alleviate symptoms of anxiety, stress and depression, and also improve social behaviour [42], [162], [163], [164], [165], [166], [167], [168], [169], abundances of these genera were not significantly related to either sociability or neurotic tendencies in this study. This may be because these regression analyses were conducted at the genus level, yet there is some evidence that the behavioural effects of Bifidobacterium and Lactobacillus may be species-specific and even strain-specific [42], [163].
Interpreting results from this study also requires consideration of the bidirectional interactions between the gut microbiome and behaviour. As well as the behavioural effects of gut microorganisms, behaviour can in turn shape the composition of the gut microbiome. For example, the gut microbiome can affect the stress response and stress also disrupts the gut microbiome [6], [15]. In addition, there is growing evidence that how an animal interacts socially can significantly influence its gut microbial community, whose composition is the product of microbial immigration and competition throughout life via transmission of microorganisms, primarily through the faecal–oral route [170]. Research in wild primate populations has revealed that social contact shapes gut microbiome composition [171], [172], [173], [174], [175], [176] and studies in humans find that we share gut microbiota with household members [121], [177], [178], [179], [180], [181]. A key research avenue is untangling the contributions of specific microorganisms to social behaviour versus the influence of social behaviour on microbial colonization. In at least some cases, microbial abundances may differ in relation to autistic traits or sociability because certain taxa are better adapted for transmission between hosts. Notably, the genus Oscillospira, whose abundance was positively predicted by sociability in this study, has also been found to be positively related to host density in a wild animal population [182]. It is important to bear in mind, therefore, that some genera may be efficiently transmitted socially, rather than having a causal effect on social behaviour. Indeed, Oscillospira belongs to the family Ruminococcaceae, many of which are intestinal spore-forming bacteria, thereby facilitating transmission between hosts [183]. In fact, numerous members of the microbiota are well adapted for transmission and it is estimated that at least half the bacterial genera in the gut are able to produce resilient spores adapted for survival and dispersal [183].
The evidence of social transmission of microorganisms is particularly relevant to the finding here that people with larger social networks tend to have a more diverse gut microbiome. Indeed, animal studies have revealed that social interactions are positively associated with microbiome diversity [172], [175], [182], [184]. For example, reduced social contact in a bumblebee colony results in lower gut microbiome diversity [184] and chimpanzees that interact more socially have a more diverse gut microbiome [172]. In fact, horizontal transmission of gut microorganisms through chimpanzee social interactions appears to play a greater role than vertical transmission from the mother in shaping gut microbiome composition [172]. This suggests that although the mother provides the primary inoculum for the newborn’s microbiome, the composition of the microbiome through the individual’s lifetime may be more strongly influenced by social interactions. The relationship between gut microbiome diversity and human social networks has not previously been explored but the positive relationship found here suggests that social interactions may also influence the microbiota of human societies. Interestingly, a study of gut microbiome composition and temperament in infants reported an association between gut microbiome diversity and sociability [84]. By maintaining diversity of the gut microbial community, social transmission of microorganisms may benefit host health in numerous ways [170]. For example,diversity may help to promote stability and resilience of the gut microbiome [142], and is often linked to good health, though this is not always the case [140]. Diversity can provide resistance against infection [185], [186], [187], [188], improve immune function [170] and may reduce the risk of allergies [189]. It is well known that people with more social ties are healthier and live longer, and reduced inflammation is postulated to play a role in this relationship [190], [191]. It is interesting therefore to speculate whether the microbiome may mediate this association between social integration and health, particularly since it is a key regulator of the host immune response [192].
Participants who were more stressed or anxious, and also those reporting poorer sleep quality, tended to have both a less diverse gut microbiome and an altered composition. In line with these results, a recent study found that anxiety status was related to stool consistency, suggesting that anxiety may be associated with differences in gut microbiome composition, perhaps through inducing dysbiosis [193]. Depression has also been associated with differences in microbial community composition [194]. In addition, it has previously been shown in animal studies that stress not only alters the abundances of various microbial taxa but also reduces the diversity of the gut microbial community [61], [195]. In fact, part of the reason why individuals with a larger social network have a more diverse microbiome may be because social support can help buffer the adverse effects of stress on diversity [196]. However, social network size was not significantly correlated with stress in this study (Fig. S1) and the positive relationship between network size and microbiome diversity is consistent with evidence from animal populations that members of the microbiota are transmitted through interactions with the social environment. Further research should attempt to validate this finding and investigate the various microbial transmission routes and their relative importance. Indeed, modern lifestyle choices are geared towards preventing pathogen transmission but maybe we should instead consider ways to promote the spread of beneficial microorganisms [170]. Notably, the negative relationship found here between conscientiousness and genus richness of the gut microbiome may be because conscientious people are more likely to engage in hygienic behaviour [197], which may result in a smaller number of microbial genera inhabiting the gut. The observed relationship between sleep quality and gut microbiome composition and diversity may be due to its intercorrelation with stress, anxiety and neuroticism (Fig. S1) but may also partly reflect the known relationship between the gut microbiome and host circadian rhythms [198].
Although the focus of this research was on personality traits, there are also other novel findings of considerable interest from this study. People who travelled frequently or visited more countries tended to have a more diverse gut microbiome, suggesting that our interaction with the environment does play a considerable role in influencing our gut microbial community. This increased diversity may also partly reflect the different foods people tend to eat when travelling. However, holidays abroad are also positively associated with the prevalence of antibiotic resistance genes in the gut [199]. This suggests that although travelling may increase the taxonomic diversity of the gut, it may also increase the risk of acquiring antibiotic resistance. Another interesting result not previously shown was that more adventurous eaters also had a greater gut microbiome diversity, supporting the idea that microbiome health may be improved through a diverse diet [200]. This finding is particularly pertinent given the increasingly restrictive dietary habits of Western cultures and is also in agreement with a recent study on a wild primate population which reported a positive association between dietary diversity and microbiome richness [175].
Diversity of the gut microbiome was also related to the amount of food people consumed containing natural probiotics and prebiotics. Indeed, fermentation has been practised by human civilization for over 9000 years [201] and is traditionally used in many cultures in the production of foods such as cheese, yogurt, sauerkraut and kimchi. From an evolutionary perspective, fermented foods may represent anadaptation passed on through generations, given their potential beneficial effects on health. However, probiotic supplementation was significantly associated with reduced microbiome diversity. A likely explanation for this finding is that people with reduced microbiome diversity (for example, due to a course of antibiotics or gut dysbiosis) may be more inclined to take probiotic supplements. Indeed, while there was no evidence of a relationship between probiotic consumption and either antibiotic treatment or general health, probiotic supplementation was positively correlated with having a gut condition (Fig. S1).
There was no significant effect of delivery mode, premature birth or infant feeding practice on microbial community composition, though these factors may still influence establishment of the microbiome during infancy, with potential consequences for development of the immune system. However, regression results revealed that one genus, Flavonifractor, was differentially abundant in those born vaginally compared with caesarean section, with a higher abundance in individuals born by caesarean section. Notably, Flavonifractor has also been found to be more abundant in infants with food allergies [202] and birth by caesarean section is often linked to an increased risk of allergies [203]. Although there is some evidence that birth by caesarean section can affect the diversity and community composition of the infant gut microbiome [120], [122], [123], other studies find that delivery mode does not significantly influence development of the microbiome [204], [205], [206] and no lasting differences (except Flavonifractor abundance) were detected here in the adult population. However, individuals that were formula-fed as infants did have a lower gut microbiome diversity. This is the first time the effect of infant diet has been explored with respect to the adult gut microbiome. Most research in infants actually finds that formula feeding is associated with increased diversity compared to breastfeeding [124], but results of a two-year longitudinal study have shown that while formula-fed infants in their first year of life may have higher gut microbiome diversity, breast-fed infants tend to have higher diversity between one and two years of age [123].
Other notable results include females having a significantly lower gut microbiome diversity compared to males. This contrasts a previous human study which found no significant differences in diversity between the sexes [207] but is in the same direction as recent research in a wild baboon population [174]. As expected, antibiotic treatment was associated with reduced diversity of the microbiome and an altered composition, in accordance with other findings [126], [208], [209], [210], [211]. Interestingly, fish consumption was positively associated with genus richness of the gut microbiome, perhaps due to exposure to different bacterial genera inhabiting marine ecosystems. Somewhat surprisingly, dog ownership was linked to lower diversity, which contrasts the finding that infants living with household pets tend to have a more diverse gut microbiome [212]. The reduced microbiome diversity of people in unemployment may partly be a reflection of socioeconomic status which can influence health through lifestyle behaviours, diet, access to medical care and psychosocial factors such as stress [213]. With respect to other comprehensive population studies of the human gut microbiome [204], [194], the results here replicate previous findings that age, gender, body mass index, sleep, constipation, gut conditions, antibiotic treatment and consumption of fruit, vegetables and cereals are related to gut microbiome variation.
As this study was cross-sectional, causation cannot be proved, particularly given the bidirectional nature of the microbiome–gut–brain axis. As discussed, gut bacteria can affect behaviour and behaviour can in turn influence the composition of the gut microbiome. Despite this limitation, the findings here represent an important step in understanding the relationships between the gut microbiome and personality traits, and the potential consequences for mental health. However, since this is one of the first studies linking the gut microbiome to personality, further research would help to confirm the reproducibility of these findings. In addition, it should be borne in mind that there are numerous factors that can affect personality. In particular, genetics account for approximately 50% of variation in personality, while a range of environmental factors contribute to the remaining variation [1]. Since both host genetics and the environment are known to affect microbiome composition [214], it may be that some of the effects of genes or the environment on personality are via their influence on the microbiome.
A key benefit of this study is that rather than examining the expression of extreme traits, as seen in psychiatric disorders, this research focused on behavioural variation in the general population. The findings reported here challenge the results of a recent study concluding a lack of significant relationships between psychiatric measures in healthy humans and microbiome composition and diversity [215]. However, that research was based on a comparatively small sample size and only involved female participants. Given the very exploratory nature of their study, the researchers FDR-corrected their P values for almost 4000 hypotheses which may have led to numerous false negative results. As opposed to their conclusions that the gut microbiome may only be relevant in cases of psychiatric illness, my findings suggest that the microbiome may also be related to personality traits in the healthy population. Indeed, the majority of the results were replicated when anyone suffering from a psychiatric condition was excluded from the dataset, showing that my findings were not driven by those scoring towards the extreme end for the personality traits (Tables S8 to S10). The only notable exception was for the beta diversity analyses, where stress and anxiety were no longer significantly associated with differences in the overall community composition of the gut microbiome. The majority of the regression results were also replicated, although the positive relationship between sociability and Akkermansia abundance was not detected. Additionally, there was evidence in this dataset that Blautia was less abundant in more sociable individuals (P = 0.043) and Prevotella was less abundant in more neurotic individuals (P = 0.002).
In conclusion, differences in gut microbiome composition and diversity are shown to be linked to personality traits in the general population. The results of this study add a new dimension to our understanding of personality and are in line with accumulating evidence that the gut microbiome can influence the central nervous system in humans, with effects on behaviour. Such findings may inform the development of probiotic or prebiotic therapies to help improve mood and treat conditions such as autism, anxiety and depression. Discovering new and effective interventions for mental health conditions is of pressing concern, given the declines in psychological health of our modern society. Finally, it is pertinent to reflect on the ways in which our modern-day living may provide a perfect storm for dysbiosis of the gut. We lead stressful lives with fewer social interactions and less time spent with nature, our diets are typically deficient in fibre, we inhabit oversanitized environments and are dependent on antibiotic treatments. All these factors can influence the gut microbiome and so may be affecting our behaviour and psychological well-being in currently unknown ways.
Acknowledgments
Acknowledgements
Thanks are due to uBiome for their generosity in providing the sampling kits to participants for the study and sequencing the samples. K.V.-A.J. was funded by BBSRC (BB/J014427/1). Thanks to Associate Prof. Philip Burnet and Prof. Robin Dunbar for their support of this project and also to the University of Oxford’s Statistics Department for their discussion of appropriate statistical methods in R.
Author contributions
K.V.-A.J. conceived the idea, designed the study and questionnaire, analysed, interpreted and presented all the data and wrote the manuscript.
Data availability
Data are available from the author upon reasonable request.
Competing interests
The author declares no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.humic.2019.100069.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Power R.A., Pluess M. Heritability estimates of the Big Five personality traits based on common genetic variants. Transl Psychiatry. 2015;5 doi: 10.1038/tp.2015.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Round J.L., Mazmanian S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brestoff J.R., Artis D. Commensal bacteria at the interface of host metabolism and the immune system. Nat Immunol. 2013;14:676–684. doi: 10.1038/ni.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sampson T.R., Mazmanian S.K. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sommer F., Bäckhed F. The gut microbiota – masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 6.Cryan J.F., Dinan T.G. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. 2012;13:701–712. doi: 10.1038/nrn3346. [DOI] [PubMed] [Google Scholar]
- 7.Sender R., Fuchs S., Milo R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell. 2016;164:337–340. doi: 10.1016/j.cell.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Eisenstein M. Microbiome: bacterial broadband. Nature. 2016;533:S104–S106. doi: 10.1038/533S104a. [DOI] [PubMed] [Google Scholar]
- 9.Gilbert J.A., Krajmalnik-Brown R., Porazinska D.L., Weiss S.J., Knight R. Toward effective probiotics for autism and other neurodevelopmental disorders. Cell. 2013;155:1446–1448. doi: 10.1016/j.cell.2013.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dinan T.G., Stanton C., Cryan J.F. Psychobiotics: a novel class of psychotropic. Biol Psychiatry. 2013;74:720–726. doi: 10.1016/j.biopsych.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 11.Foster J.A., McVey Neufeld K.-A. Gut–brain axis: how the microbiome influences anxiety and depression. Trends Neurosci. 2013;36:305–312. doi: 10.1016/j.tins.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 12.Dinan T.G., Cryan J.F. Brain–gut–microbiota axis – mood, metabolism and behaviour. Nat Rev Gastroenterol Hepatol. 2017;14:69–70. doi: 10.1038/nrgastro.2016.200. [DOI] [PubMed] [Google Scholar]
- 13.Bruce-Keller A., Salbaum J.M., Berthoud H.-R. Harnessing gut microbes for mental health: getting from here to there. Biol Psychiatry. 2018;83:214–223. doi: 10.1016/j.biopsych.2017.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rogers G.B. From gut dysbiosis to altered brain function and mental illness: mechanisms and pathways. Mol Psychiatry. 2016;21:738–748. doi: 10.1038/mp.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharon G., Sampson T.R., Geschwind D.H., Mazmanian S.K. The central nervous system and the gut microbiome. Cell. 2016;167:915–932. doi: 10.1016/j.cell.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bercik P. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141:599–609. doi: 10.1053/j.gastro.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 17.Collins S.M., Kassam Z., Bercik P. The adoptive transfer of behavioral phenotype via the intestinal microbiota: experimental evidence and clinical implications. Curr Opin Microbiol. 2013;16:240–245. doi: 10.1016/j.mib.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Kelly J.R. Transferring the blues: depression-associated gut microbiota induces neurobehavioural changes in the rat. J Psychiatr Res. 2016;82:109–118. doi: 10.1016/j.jpsychires.2016.07.019. [DOI] [PubMed] [Google Scholar]
- 19.Zheng P. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21:786–796. doi: 10.1038/mp.2016.44. [DOI] [PubMed] [Google Scholar]
- 20.de Palma G. Transplantation of fecal microbiota from patients with irritable bowel syndrome alters gut function and behavior in recipient mice. Sci Transl Med. 2017;9:eaaf6397. doi: 10.1126/scitranslmed.aaf6397. [DOI] [PubMed] [Google Scholar]
- 21.Kurokawa S. The effect of fecal microbiota transplantation on psychiatric symptoms among patients with irritable bowel syndrome, functional diarrhea and functional constipation: an open-label observational study. J Affect Disord. 2018;235:506–512. doi: 10.1016/j.jad.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 22.Collins S.M., Surette M., Bercik P. The interplay between the intestinal microbiota and the brain. Nat Rev Microbiol. 2012;10:735–742. doi: 10.1038/nrmicro2876. [DOI] [PubMed] [Google Scholar]
- 23.Johnson K.V.-A., Foster K.R. Why does the microbiome affect behaviour? Nat Rev Microbiol. 2018;16:647–655. doi: 10.1038/s41579-018-0014-3. [DOI] [PubMed] [Google Scholar]
- 24.Strandwitz P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018;1693:128–133. doi: 10.1016/j.brainres.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wall R. Bacterial neuroactive compounds produced by psychobiotics. In: Lyte M., Cryan J.F., editors. Microbial endocrinology: the microbiota–gut–brain axis in health and disease. Springer; New York: 2014. pp. 221–239. [Google Scholar]
- 26.Yano J.M. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell. 2015;161:264–276. doi: 10.1016/j.cell.2015.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett E., Ross R.P., O’Toole P.W., Fitzgerald G.F., Stanton C. γ-Aminobutyric acid production by culturable bacteria from the human intestine. J Appl Microbiol. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 28.Strandwitz P. GABA-modulating bacteria of the human gut microbiota. Nat Microbiol. 2019;4:396–403. doi: 10.1038/s41564-018-0307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mussell M. Gastrointestinal symptoms in primary care: prevalence and association with depression and anxiety. J Psychosom Res. 2008;64:605–612. doi: 10.1016/j.jpsychores.2008.02.019. [DOI] [PubMed] [Google Scholar]
- 30.Folks D.G. The interface of psychiatry and irritable bowel syndrome. Curr. Psychiatry Rep. 2004;6:210–215. doi: 10.1007/s11920-004-0066-0. [DOI] [PubMed] [Google Scholar]
- 31.McKean J., Naug H., Nikbakht E., Amiet B., Colson N. Probiotics and subclinical psychological symptoms in healthy participants: a systematic review and meta-analysis. J Altern Complement Med. 2017;23:249–258. doi: 10.1089/acm.2016.0023. [DOI] [PubMed] [Google Scholar]
- 32.Wallace C.J.K., Milev R. The effects of probiotics on depressive symptoms in humans: a systematic review. Ann. Gen. Psychiatry. 2017;16:14. doi: 10.1186/s12991-017-0138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang H., Lee I.-S., Braun C., Enck P. Effect of probiotics on central nervous system functions in animals and humans: a systematic review. J Neurogastroenterol Motil. 2016;22:589–605. doi: 10.5056/jnm16018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pirbaglou M. Probiotic supplementation can positively affect anxiety and depressive symptoms: a systematic review of randomized controlled trials. Nutr Res. 2016;36:889–898. doi: 10.1016/j.nutres.2016.06.009. [DOI] [PubMed] [Google Scholar]
- 35.Tillisch K. Consumption of fermented milk product with probiotic modulates brain activity. Gastroenterology. 2013;144:1394–1401. doi: 10.1053/j.gastro.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bagga D. Probiotics drive gut microbiome triggering emotional brain signatures. Gut Microbes. 2018;9:486–496. doi: 10.1080/19490976.2018.1460015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmidt K. Prebiotic intake reduces the waking cortisol response and alters emotional bias in healthy volunteers. Psychopharmacology. 2015;232:1793–1801. doi: 10.1007/s00213-014-3810-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Azpiroz F. Effects of scFOS on the composition of fecal microbiota and anxiety in patients with irritable bowel syndrome: a randomized, double blind, placebo controlled study. Neurogastroenterol Motil. 2017;29 doi: 10.1111/nmo.12911. [DOI] [PubMed] [Google Scholar]
- 39.Hsiao E.Y. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155:1451–1463. doi: 10.1016/j.cell.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desbonnet L., Clarke G., Shanahan F., Dinan T.G., Cryan J.F. Microbiota is essential for social development in the mouse. Mol Psychiatry. 2014;19:146–148. doi: 10.1038/mp.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Theije C.G.M. Altered gut microbiota and activity in a murine model of autism spectrum disorders. Brain Behav Immun. 2014;37:197–206. doi: 10.1016/j.bbi.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 42.Buffington S.A. Microbial reconstitution reverses maternal diet-induced social and synaptic deficits in offspring. Cell. 2016;165:1762–1775. doi: 10.1016/j.cell.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krajmalnik-Brown R., Lozupone C., Kang D., Adams J.B. Gut bacteria in children with autism spectrum disorders: challenges and promise of studying how a complex community influences a complex disease. Microb. Ecol. Health Dis. 2015;26:26914. doi: 10.3402/mehd.v26.26914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hsiao E.Y. Gastrointestinal issues in autism spectrum disorder. Harv Rev Psychiatry. 2014;22:104–111. doi: 10.1097/HRP.0000000000000029. [DOI] [PubMed] [Google Scholar]
- 45.Meltzer A., van de Water J. The role of the immune system in autism spectrum disorder. Neuropsychopharmacology. 2017;42:284–298. doi: 10.1038/npp.2016.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsiao E.Y. Immune dysregulation in autism spectrum disorder. Int Rev Neurobiol. 2013;113:269–302. doi: 10.1016/B978-0-12-418700-9.00009-5. [DOI] [PubMed] [Google Scholar]
- 47.Kang D.W. Microbiota Transfer Therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome. 2017;5:10. doi: 10.1186/s40168-016-0225-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kang D. Long-term benefit of Microbiota Transfer Therapy on autism symptoms and gut microbiota. Sci Rep. 2019;9:5821. doi: 10.1038/s41598-019-42183-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wing L. The autistic continuum. In: Wing L., editor. Aspects of Autism: Biological Research. The Royal College of Psychiatrists; London: 1988. pp. 5–8. [Google Scholar]
- 50.Constantino J.N., Todd R.D. Autistic traits in the general population: a twin study. Arch Gen Psychiatry. 2003;60:524–530. doi: 10.1001/archpsyc.60.5.524. [DOI] [PubMed] [Google Scholar]
- 51.Ruzich E. Measuring autistic traits in the general population: a systematic review of the Autism-Spectrum Quotient (AQ) in a nonclinical population sample of 6,900 typical adult males and females. Mol. Autism. 2015;6:2. doi: 10.1186/2040-2392-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang L. Low relative abundances of the mucolytic bacterium Akkermansia muciniphila and Bifidobacterium spp. in feces of children with autism. Appl Environ Microbiol. 2011;77:6718–6721. doi: 10.1128/AEM.05212-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Angelis M. Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS One. 2013;8 doi: 10.1371/journal.pone.0076993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bharwani A. Structural & functional consequences of chronic psychosocial stress on the microbiome & host. Psychoneuroendocrinology. 2016;63:217–227. doi: 10.1016/j.psyneuen.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 55.Maltz R. Social stressor exposure in C57BL/6 mice impacts inflammation, microbiota, short chain fatty acids in a murine model of colitis. Inflamm Bowel Dis. 2017;23:S103–S104. [Google Scholar]
- 56.Strati F. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome. 2017;5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bangsgaard Bendtsen K.M. Gut microbiota composition is correlated to grid floor induced stress and behavior in the BALB/c mouse. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naseribafrouei A. Correlation between the human fecal microbiota and depression. Neurogastroenterol Motil. 2014;26:1155–1162. doi: 10.1111/nmo.12378. [DOI] [PubMed] [Google Scholar]
- 59.Jiang H. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav Immun. 2015;48:186–194. doi: 10.1016/j.bbi.2015.03.016. [DOI] [PubMed] [Google Scholar]
- 60.Finegold S.M. Pyrosequencing study of fecal microflora of autistic and control children. Anaerobe. 2010;16:444–453. doi: 10.1016/j.anaerobe.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 61.Bailey M.T. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain Behav Immun. 2011;25:397–407. doi: 10.1016/j.bbi.2010.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li L. Gut microbes in correlation with mood: case study in a closed experimental human life support system. Neurogastroenterol Motil. 2016;28:1233–1240. doi: 10.1111/nmo.12822. [DOI] [PubMed] [Google Scholar]
- 63.Schwarz E. Analysis of microbiota in first episode psychosis identifies preliminary associations with symptom severity and treatment response. Schizophr Res. 2017;192:398–403. doi: 10.1016/j.schres.2017.04.017. [DOI] [PubMed] [Google Scholar]
- 64.Adams J.B., Johansen L.J., Powell L.D., Quig D., Rubin R.A. Gastrointestinal flora and gastrointestinal status in children with autism – comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011;11:22. doi: 10.1186/1471-230X-11-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang L. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol Autism. 2013;4:42. doi: 10.1186/2040-2392-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Son J.S. Comparison of fecal microbiota in children with autism spectrum disorders and neurotypical siblings in the Simons Simplex Collection. PLoS One. 2015;10 doi: 10.1371/journal.pone.0137725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grimaldi R. In vitro fermentation of B-GOS: impact on faecal bacterial populations and metabolic activity in autistic and non-autistic children. FEMS Microbiol Ecol. 2017;93:fiw23. doi: 10.1093/femsec/fiw233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pulikkan J. Gut microbial dysbiosis in Indian children with autism spectrum disorders. Microb Ecol. 2018;76:1102–1114. doi: 10.1007/s00248-018-1176-2. [DOI] [PubMed] [Google Scholar]
- 69.Golubeva A.V. Microbiota-related changes in bile acid & tryptophan metabolism are associated with gastrointestinal dysfunction in a mouse model of autism. EBioMedicine. 2017;24:166–178. doi: 10.1016/j.ebiom.2017.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong M.-L. Inflammasome signaling affects anxiety- and depressive-like behavior and gut microbiome composition. Mol Psychiatry. 2016;21:797–805. doi: 10.1038/mp.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang C. Bifidobacterium in the gut microbiota confer resilience to chronic social defeat stress in mice. Sci Rep. 2017;7:45942. doi: 10.1038/srep45942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marin I.A. Microbiota alteration is associated with the development of stress-induced despair behavior. Sci Rep. 2017;7:43859. doi: 10.1038/srep43859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Aizawa E. Possible association of Bifidobacterium and Lactobacillus in the gut microbiota of patients with major depressive disorder. J Affect Disord. 2016;202:254–257. doi: 10.1016/j.jad.2016.05.038. [DOI] [PubMed] [Google Scholar]
- 74.Inoue R. A preliminary investigation on the relationship between gut microbiota and gene expressions in peripheral mononuclear cells of infants with autism spectrum disorders. Biosci Biotechnol Biochem. 2016;80:2450–2458. doi: 10.1080/09168451.2016.1222267. [DOI] [PubMed] [Google Scholar]
- 75.Luna R.A. Distinct microbiome–neuroimmune signatures correlate with functional abdominal pain in children with autism spectrum disorder. Cell Mol Gastroenterol Hepatol. 2017;3:218–230. doi: 10.1016/j.jcmgh.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shen Y. Analysis of gut microbiota diversity and auxiliary diagnosis as a biomarker in patients with schizophrenia: a cross-sectional study. Schizophr Res. 2018;197:470–477. doi: 10.1016/j.schres.2018.01.002. [DOI] [PubMed] [Google Scholar]
- 77.Finegold S.M. Gastrointestinal microflora studies in late-onset autism. Clin Infect Dis. 2002;35(Suppl. 1):S6–S16. doi: 10.1086/341914. [DOI] [PubMed] [Google Scholar]
- 78.Song Y., Liu C., Finegold S.M. Real-time PCR quantitation of Clostridia in feces of autistic children. Appl Environ Microbiol. 2004;70:6459–6465. doi: 10.1128/AEM.70.11.6459-6465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Parracho H.M.R.T., Bingham M.O., Gibson G.R., McCartney A.L. Differences between the gut microflora of children with autistic spectrum disorders and that of healthy children. J Med Microbiol. 2005;54:987–991. doi: 10.1099/jmm.0.46101-0. [DOI] [PubMed] [Google Scholar]
- 80.Tomova A. Gastrointestinal microbiota in children with autism in Slovakia. Physiol Behav. 2015;138:179–187. doi: 10.1016/j.physbeh.2014.10.033. [DOI] [PubMed] [Google Scholar]
- 81.Arase S., Watanabe Y., Setoyama H., Nagaoka N., Kawai M. Disturbance in the mucosa-associated commensal bacteria is associated with the exacerbation of chronic colitis by repeated psychological stress; is that the new target of probiotics? PLoS One. 2016;11 doi: 10.1371/journal.pone.0160736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lin P. Prevotella and Klebsiella proportions in fecal microbial communities are potential characteristic parameters for patients with major depressive disorder. J Affect Disord. 2017;207:300–304. doi: 10.1016/j.jad.2016.09.051. [DOI] [PubMed] [Google Scholar]
- 83.Yu M. Variations in gut microbiota and fecal metabolic phenotype associated with depression by 16S rRNA gene sequencing and LC/MS-based metabolomics. J Pharm Biomed Anal. 2017;138:231–239. doi: 10.1016/j.jpba.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 84.Christian L.M. Gut microbiome composition is associated with temperament during early childhood. Brain Behav Immun. 2015;45:118–127. doi: 10.1016/j.bbi.2014.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valles-Colomer M. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol. 2019;4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 86.Kang D.-W. Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe. 2018;49:121–131. doi: 10.1016/j.anaerobe.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 87.Turroni S. Temporal dynamics of the gut microbiota in people sharing a confined environment, a 520-day ground-based space simulation, MARS500. Microbiome. 2017;5:39. doi: 10.1186/s40168-017-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Togher K. Depression-associated alterations in the maternal microbiome during pregnancy: priming for adverse infant outcomes? Gut. 2017;66(Suppl. 1):A15–A16. [Google Scholar]
- 89.Evans S.J. The gut microbiome composition associates with bipolar disorder and illness severity. J Psychiatr Res. 2017;87:23–29. doi: 10.1016/j.jpsychires.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Painold A. A step ahead: exploring the gut microbiota in inpatients with bipolar disorder during a depressive episode. Bipolar Disord. 2019;21:40–49. doi: 10.1111/bdi.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kang D. Reduced incidence of Prevotella and other fermenters in intestinal microflora of autistic children. PLoS One. 2013;8 doi: 10.1371/journal.pone.0068322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Galley J.D. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC Microbiol. 2014;14:189. doi: 10.1186/1471-2180-14-189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bailey M.T., Coe C.L. Maternal separation disrupts the integrity of the intestinal microflora in infant rhesus monkeys. Dev Psychobiol. 1999;35:146–155. [PubMed] [Google Scholar]
- 94.Szyszkowicz J.K., Wong A., Anisman H., Merali Z., Audet M.-C. Implications of the gut microbiota in vulnerability to the social avoidance effects of chronic social defeat in male mice. Brain Behav Immun. 2017;66:45–55. doi: 10.1016/j.bbi.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 95.Zhang J. Blockade of interleukin-6 receptor in the periphery promotes rapid and sustained antidepressant actions: a possible role of gut–microbiota–brain axis. Transl Psychiatry. 2017;7 doi: 10.1038/tp.2017.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Williams B.L. Impaired carbohydrate digestion and transport and mucosal dysbiosis in the intestines of children with autism and gastrointestinal disturbances. PLoS One. 2011;6 doi: 10.1371/journal.pone.0024585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Williams B.L., Hornig M., Parekh T., Lipkin W.I. Application of novel PCR-based methods for detection, quantitation, and phylogenetic characterization of Sutterella species in intestinal biopsy samples from children with autism and gastrointestinal disturbances. MBio. 2012;3:e00261–11. doi: 10.1128/mBio.00261-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Almonacid D.E. 16S rRNA gene sequencing and healthy reference ranges for 28 clinically relevant microbial taxa from the human gut microbiome. PLoS One. 2017;12 doi: 10.1371/journal.pone.0176555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Eckburg P.B. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.McInnes, P. & Cutting, M. Manual of Procedures for Human Microbiome Project: Core Microbiome Sampling, Protocol A, HMP Protocol no. 07-001, Version 11 (2010).
- 101.Vera-Wolf P. Measures of reproducibility in sampling and laboratory processing methods in high-throughput microbiome analysis. bioRxiv. 2018 doi: 10.1101/322677. [DOI] [Google Scholar]
- 102.Goldberg, L. R. International Personality Item Pool. (2001). Available at: http://ipip.ori.org.
- 103.Digman J.M. Personality structure: emergence of the five-factor model. Annu Rev Psychol. 1990;41:417–440. [Google Scholar]
- 104.McCrae R.R., John O.P. An introduction to the five-factor model and its applications. J Pers. 1992;60:175–215. doi: 10.1111/j.1467-6494.1992.tb00970.x. [DOI] [PubMed] [Google Scholar]
- 105.McCrae R., Costa P., Ostendorf F., Angleitner A. Age differences in personality across the adult lifespan. Dev Psychol. 1999;35:466–477. doi: 10.1037//0012-1649.35.2.466. [DOI] [PubMed] [Google Scholar]
- 106.McCrae R.R., Costa P.T. Personality trait structure as a human universal. Am Psychol. 1997;52:509–516. doi: 10.1037//0003-066x.52.5.509. [DOI] [PubMed] [Google Scholar]
- 107.Spielberger C.D., Gorsuch R.L., Lushene R., Vagg P.R., Jacobs G.A. Consulting Psychologists Press; California: 1983. Manual for the state-trait anxiety inventory. [Google Scholar]
- 108.Baron-Cohen S., Wheelwright S., Skinner R., Martin J., Clubley E. The autism-spectrum quotient (AQ): evidence from Asperger syndrome/high-functioning autism, males and females, scientists and mathematicians. J Autism Dev Disord. 2001;31:5–17. doi: 10.1023/a:1005653411471. [DOI] [PubMed] [Google Scholar]
- 109.Roberts S.G.B., Wilson R., Fedurek P., Dunbar R.I.M. Individual differences and personal social network size and structure. Pers Individ Dif. 2008;44:954–964. [Google Scholar]
- 110.Bingham S.A. Nutritional methods in the European prospective investigation of cancer in Norfolk. Public Health Nutr. 2001;4:847–858. doi: 10.1079/phn2000102. [DOI] [PubMed] [Google Scholar]
- 111.Mulligan A.A. A new tool for converting food frequency questionnaire data into nutrient and food group values: FETA research methods and availability. BMJ Open. 2014;4 doi: 10.1136/bmjopen-2013-004503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xu L., Paterson A.D., Turpin W., Xu W. Assessment and selection of competing models for zero-inflated microbiome data. PLoS One. 2015;10 doi: 10.1371/journal.pone.0129606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Weiss S. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xia Y., Sun J. Hypothesis testing and statistical analysis of microbiome. Genes Dis. 2017;4:138–148. doi: 10.1016/j.gendis.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McMurdie P.J., Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Goodrich J.K. Conducting a microbiome study. Cell. 2014;158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Arumugam M. Enterotypes of the human gut microbiome. Nature. 2011;473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ding T., Schloss P.D. Dynamics and associations of microbial community types across the human body. Nature. 2014;509:357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Jašarević E., Morrison K.E., Bale T.L. Sex differences in the gut microbiome–brain axis across the lifespan. Philos Trans R Soc B Biol Sci. 2016;371:20150122. doi: 10.1098/rstb.2015.0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Martin R. Early-life events, including mode of delivery and type of feeding, siblings and gender, shape the developing gut microbiota. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yatsunenko T. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dominguez-Bello M.G. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. PNAS. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Bokulich N.A., Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. 2016;8:343ra82. doi: 10.1126/scitranslmed.aad7121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Praveen P., Jordan F., Priami C., Morine M.J. The role of breast-feeding in infant immune system: a systems perspective on the intestinal microbiome. Microbiome. 2015;3:41. doi: 10.1186/s40168-015-0104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.de la Cochetière M.F. Resilience of the dominant human fecal microbiota upon short-course antibiotic challenge. J Clin Microbiol. 2005;43:5588–5592. doi: 10.1128/JCM.43.11.5588-5592.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dethlefsen L., Huse S.M., Sogin M.L., Relman D.A. The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bhattarai Y., Muniz Pedrogo D.A., Kashyap P.C. Irritable bowel syndrome: a gut microbiota-related disorder? Am J Physiol Gastrointest Liver Physiol. 2017;312:G52–G62. doi: 10.1152/ajpgi.00338.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Halfvarson J. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol. 2017;2:17004. doi: 10.1038/nmicrobiol.2017.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hemarajata P., Versalovic J. Effects of probiotics on gut microbiota: mechanisms of intestinal immunomodulation and neuromodulation. Therap. Adv. Gastroenterol. 2013;6:39–51. doi: 10.1177/1756283X12459294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kristensen N.B. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8:52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zmora N. Personalized gut mucosal colonization resistance to empiric probiotics is associated with unique host and microbiome features. Cell. 2018;174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 132.Austin E.J. Personality correlates of the broader autism phenotype as assessed by the Autism Spectrum Quotient (AQ) Pers. Individ. Dif. 2005;38:451–460. [Google Scholar]
- 133.Uliaszek A.A. The role of neuroticism and extraversion in the stress–anxiety and stress–depression relationships. Anxiety Stress Coping. 2010;23:363–381. doi: 10.1080/10615800903377264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Vuong H.E., Yano J.M., Fung T.C., Hsiao E.Y. The microbiome and host behavior. Annu Rev Neurosci. 2017;40:21–49. doi: 10.1146/annurev-neuro-072116-031347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.R Development Core Team . R Foundation for Statistical Computing; Vienna: 2015. R: A Language and Environment for Statistical Computing. Available at: http://www.R-project.org. [Google Scholar]
- 136.Srinivasan R. Use of 16S rRNA gene for identification of a broad range of clinically relevant bacterial pathogens. PLoS One. 2015;10 doi: 10.1371/journal.pone.0117617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Janda J.M., Abbott S.L. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. 2007;45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Finotello F., Mastrorilli E., Di Camillo B. Measuring the diversity of the human microbiota with targeted next-generation sequencing. Brief Bioinform. 2016;19:679–692. doi: 10.1093/bib/bbw119. [DOI] [PubMed] [Google Scholar]
- 139.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8 doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Johnson K.V.-A., Burnet P.W.J. Microbiome: should we diversify from diversity? Gut Microbes. 2016;7:455–458. doi: 10.1080/19490976.2016.1241933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hill T.C.J., Walsh K.A., Harris J.A., Moffett B.F. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol. 2003;43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 142.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Riedel C.U., Schwiertz A., Egert M. The stomach and small and large intestinal microbiomes. In: Marchesi J.R., editor. The human microbiota and microbiome. CABI; Wallingford: 2013. pp. 1–19. [Google Scholar]
- 144.The Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Faith J.J. The long-term stability of the human gut microbiota. Science. 2013;341:1237439. doi: 10.1126/science.1237439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Penders J. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. 2006;118:511–521. doi: 10.1542/peds.2005-2824. [DOI] [PubMed] [Google Scholar]
- 147.Fransen F. The impact of gut microbiota on gender-specific differences in immunity. Front Immunol. 2017;8:754. doi: 10.3389/fimmu.2017.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Koropatkin N.M., Cameron E.A., Martens E.C. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sonnenburg E.D., Sonnenburg J.L. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20:779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sonnenburg E.D. Diet-induced extinctions in the gut microbiota compound over generations. Nature. 2016;529:212–215. doi: 10.1038/nature16504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mach N., Fuster-Botella D. Endurance exercise and gut microbiota: a review. J Sport Heal Sci. 2017;6:179–197. doi: 10.1016/j.jshs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.David L.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Zhu Y. Meat, dairy and plant proteins alter bacterial composition of rat gut bacteria. Sci Rep. 2015;5:15220. doi: 10.1038/srep15220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Bonder M.J. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016;8:45. doi: 10.1186/s13073-016-0295-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hilimire M.R., DeVylder J.E., Forestell C.A. Fermented foods, neuroticism, and social anxiety: an interaction model. Psychiatry Res. 2015;228:203–208. doi: 10.1016/j.psychres.2015.04.023. [DOI] [PubMed] [Google Scholar]
- 156.Selhub E.M., Logan A.C., Bested A.C. Fermented foods, microbiota, and mental health: ancient practice meets nutritional psychiatry. J Physiol Anthropol. 2014;33:2. doi: 10.1186/1880-6805-33-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cani P.D., de Vos W.M. Next-generation beneficial microbes: the case of Akkermansia muciniphila. Front Microbiol. 2017;8:1765. doi: 10.3389/fmicb.2017.01765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Konikoff T., Gophna U. Oscillospira: a central, enigmatic component of the human gut microbiota. Trends Microbiol. 2016;24:523–524. doi: 10.1016/j.tim.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 159.Finegold S.M. Desulfovibrio species are potentially important in regressive autism. Med Hypotheses. 2011;77:270–274. doi: 10.1016/j.mehy.2011.04.032. [DOI] [PubMed] [Google Scholar]
- 160.Klein D.N., Durbin C.E., Shankman S.A. Guilford Press; New York: 2009. Personality and mood disorders; pp. 93–112. [Google Scholar]
- 161.Henriques-Normark B., Normark S. Commensal pathogens, with a focus on Streptococcus pneumoniae, and interactions with the human host. Exp Cell Res. 2010;316:1408–1414. doi: 10.1016/j.yexcr.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 162.Bharwani A., Mian M.F., Surette M.G., Bienenstock J., Forsythe P. Oral treatment with Lactobacillus rhamnosus attenuates behavioural deficits and immune changes in chronic social stress. BMC Med. 2017;15:7. doi: 10.1186/s12916-016-0771-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Savignac H.M., Kiely B., Dinan T.G., Cryan J.F. Bifidobacteria exert strain-specific effects on stress-related behavior and physiology in BALB/c mice. Neurogastroenterol Motil. 2014;26:1615–1627. doi: 10.1111/nmo.12427. [DOI] [PubMed] [Google Scholar]
- 164.Davis D.J. Lactobacillus plantarum attenuates anxiety-related behavior and protects against stress-induced dysbiosis in adult zebrafish. Sci Rep. 2016;6:33726. doi: 10.1038/srep33726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Messaoudi M. Assessment of psychotropic-like properties of a probiotic formulation (Lactobacillus helveticus R0052 and Bifidobacterium longum R0175) in rats and human subjects. Br J Nutr. 2011;105:755–764. doi: 10.1017/S0007114510004319. [DOI] [PubMed] [Google Scholar]
- 166.Bravo J.A. Ingestion of Lactobacillus strain regulates emotional behavior and central GABA receptor expression in a mouse via the vagus nerve. PNAS. 2011;108:16050–16055. doi: 10.1073/pnas.1102999108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Bercik P. The anxiolytic effect of Bifidobacterium longum NCC3001 involves vagal pathways for gut–brain communication. Neurogastroenterol Motil. 2011;23:1132–1139. doi: 10.1111/j.1365-2982.2011.01796.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Desbonnet L. Effects of the probiotic Bifidobacterium infantis in the maternal separation model of depression. Neuroscience. 2010;170:1179–1188. doi: 10.1016/j.neuroscience.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 169.Jang H.-M., Lee H., Jang S.-E., Han M.J., Kim D.-H. Evidence for interplay among antibacterial-induced gut microbiota disturbance, neuro-inflammation, and anxiety in mice. Mucosal Immunol. 2018;11:1386–1397. doi: 10.1038/s41385-018-0042-3. [DOI] [PubMed] [Google Scholar]
- 170.Browne H.P., Neville B.A., Forster S.C., Lawley T.D. Transmission of the gut microbiota: spreading of health. Nat Rev Microbiol. 2017;15:531–543. doi: 10.1038/nrmicro.2017.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Tung J. Social networks predict gut microbiome composition in wild baboons. Elife. 2015;4 doi: 10.7554/eLife.05224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Moeller A. Social behavior shapes the chimpanzee pan-microbiome. Sci Adv. 2016;2 doi: 10.1126/sciadv.1500997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Amato K.R. Patterns in gut microbiota similarity associated with degree of sociality among sex classes of a neotropical primate. Microb Ecol. 2017;74:250–258. doi: 10.1007/s00248-017-0938-6. [DOI] [PubMed] [Google Scholar]
- 174.Grieneisen L.E., Livermore J., Alberts S., Tung J., Archie E.A. Group living and male dispersal predict the core gut microbiome in wild baboons. Integr Comp Biol. 2017;57:770–785. doi: 10.1093/icb/icx046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Perofsky A.C., Lewis R.J., Abondano L.A., Di Fiore A., Meyers L.A. Hierarchical social networks shape gut microbial composition in wild Verreaux’s sifaka. Proc R Soc B Biol Sci. 2017;284:20172274. doi: 10.1098/rspb.2017.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Raulo A. Social behavior and gut microbiota in red-bellied lemurs (Eulemur rubriventer): in search of the role of immunity in the evolution of sociality. J Anim Ecol. 2017;87:388–399. doi: 10.1111/1365-2656.12781. [DOI] [PubMed] [Google Scholar]
- 177.Song S.J. Cohabiting family members share microbiota with one another and with their dogs. Elife. 2013;2 doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lax S. Longitudinal analysis of microbial interaction between humans and the indoor environment. Science. 2014;345:1048–1052. doi: 10.1126/science.1254529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Schloss P.D., Iverson K.D., Petrosino J.F., Schloss S.J. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome. 2014;2:25. doi: 10.1186/2049-2618-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Odamaki T. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci Rep. 2018;8:85. doi: 10.1038/s41598-017-18391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Brito I.L. Transmission of human-associated microbiota along family and social networks. Nat Microbiol. 2019;4:964–971. doi: 10.1038/s41564-019-0409-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Li H. Pika population density is associated with the composition and diversity of gut microbiota. Front Microbiol. 2016;7:758. doi: 10.3389/fmicb.2016.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Browne H.P. Culturing of ‘unculturable’ human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533:543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Billiet A. Colony contact contributes to the diversity of gut bacteria in bumblebees (Bombus terrestris) Insect Sci. 2017;24:270–277. doi: 10.1111/1744-7917.12284. [DOI] [PubMed] [Google Scholar]
- 185.Dillon R.J., Vennard C.T., Buckling A., Charnley A.K. Diversity of locust gut bacteria protects against pathogen invasion. Ecol Lett. 2005;8:1291–1298. [Google Scholar]
- 186.Lawley T.D. Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Abt M.C., Pamer E.G. Commensal bacteria mediated defenses against pathogens. Curr Opin Immunol. 2014;29:16–22. doi: 10.1016/j.coi.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Koch H., Schmid-Hempel P. Socially transmitted gut microbiota protect bumble bees against an intestinal parasite. PNAS. 2011;108:19288–19292. doi: 10.1073/pnas.1110474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Bloomfield S.F. Time to abandon the hygiene hypothesis: new perspectives on allergic disease, the human microbiome, infectious disease prevention and the role of targeted hygiene. Perspect. Public Health. 2016;136:213–224. doi: 10.1177/1757913916650225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Holt-Lunstad J., Smith T.B., Layton J.B. Social relationships and mortality risk: a meta-analytic review. PLoS Med. 2010;7 doi: 10.1371/journal.pmed.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Yang Y.C. Social relationships and physiological determinants of longevity across the human life span. PNAS. 2016;113:578–583. doi: 10.1073/pnas.1511085112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Rooks M.G., Garrett W.S. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016;16:341–352. doi: 10.1038/nri.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Shiro Y., Arai Y.-C., Ikemoto T., Hayashi K. Stool consistency is significantly associated with pain perception. PLoS One. 2017;12 doi: 10.1371/journal.pone.0182859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Zhernakova A. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352:565–569. doi: 10.1126/science.aad3369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Partrick K.A. Acute and repeated exposure to social stress reduces gut microbiota diversity in Syrian hamsters. Behav Brain Res. 2018;345:39–48. doi: 10.1016/j.bbr.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Johnson K.V.-A., Dunbar R.I.M. Pain tolerance predicts human social network size. Sci Rep. 2016;6:25267. doi: 10.1038/srep25267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Aunger R. The determinants of reported personal and household hygiene behaviour: a multi-country study. PLoS One. 2016;11 doi: 10.1371/journal.pone.0159551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Paschos G.K., FitzGerald G.A. Circadian clocks and metabolism: implications for microbiome and aging. Trends Genet. 2017;33:760–769. doi: 10.1016/j.tig.2017.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.von Wintersdorff C.J.H. High rates of antimicrobial drug resistance gene acquisition after international travel, the Netherlands. Emerg Infect Dis. 2014;20:649–657. doi: 10.3201/eid2004.131718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Heiman M.L., Greenway F.L. A healthy gastrointestinal microbiome is dependent on dietary diversity. Mol Metab. 2016;5:317–320. doi: 10.1016/j.molmet.2016.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.McGovern P.E. Fermented beverages of pre- and proto-historic China. PNAS. 2004;101:17593–17598. doi: 10.1073/pnas.0407921102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Ling Z. Altered fecal microbiota composition associated with food allergy in infants. Appl Environ Microbiol. 2014;80:2546–2554. doi: 10.1128/AEM.00003-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Papathoma E., Triga M., Fouzas S., Dimitriou G. Cesarean section delivery and development of food allergy and atopic dermatitis in early childhood. Pediatr Allergy Immunol. 2016;27:419–424. doi: 10.1111/pai.12552. [DOI] [PubMed] [Google Scholar]
- 204.Falcony G. Population-level analysis of gut microbiome variation. Science. 2016;352:560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 205.Chu D.M. Maturation of the infant microbiome community structure and function across multiple body sites and in relation to mode of delivery. Nat Med. 2017;23:314–326. doi: 10.1038/nm.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Stewart C.J. Cesarean or vaginal birth does not impact the longitudinal development of the gut microbiome in a cohort of exclusively preterm infants. Front Microbiol. 2017;8:1008. doi: 10.3389/fmicb.2017.01008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Haro C. Intestinal microbiota is influenced by gender and body mass index. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Jakobsson H.E. Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Dethlefsen L., Relman D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. PNAS. 2010;108:4554–4561. doi: 10.1073/pnas.1000087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Jernberg C., Löfmark S., Edlund C., Jansson J.K. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 211.Dominguez-Bello M.G. Same exposure but two radically different responses to antibiotics: resilience of the salivary microbiome versus long-term microbial shifts in feces. MBio. 2015;6:e01693–15. doi: 10.1128/mBio.01693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Azad M.B. Infant gut microbiota and the hygiene hypothesis of allergic disease: impact of household pets and siblings on microbiota composition and diversity. Allergy Asthma Clin Immunol. 2013;9:15. doi: 10.1186/1710-1492-9-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Findley K., Williams D.R., Grice E.A., Bonham V.L. Health disparities and the microbiome. Trends Microbiol. 2016;24:847–850. doi: 10.1016/j.tim.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Rothschild D. Environmental factors dominate over host genetics in shaping human gut microbiota composition. Nature. 2018;555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 215.Kleiman S.C. The gut–brain axis in healthy females: lack of significant association between microbial composition and diversity with psychiatric measures. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170208. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from the author upon reasonable request.