Abstract
Three important wine parameters: vineyard, region, and vintage year, were evaluated using fifteen Vitis vinifera L. ‘Pinot noir’ wines derived from the same scion clone (Pinot noir 667). These wines were produced from two vintage years (2015 and 2016) and eight different regions along the Pacific Coast of the United States. We successfully improved the classification of the selected Pinot noir wines by combining an untargeted 1D 1H NMR analysis with a targeted peptide based differential sensing array. NMR spectroscopy was used to evaluate the chemical fingerprint of the wines, whereas the peptide-based sensing array is known to mimic the senses of taste, smell, and palate texture by characterizing the phenolic profile. Multivariate and univariate statistical analyses of the combined NMR and differential sensing array dataset classified the genetically identical Pinot noir wines on the basis of distinctive signatures associated with the region of growth, vineyard, and vintage year.
Keywords: NMR, Differential sensing, chemometrics, wine, Pinot noir
1. Introduction
The growth and cultivation of grapevines for wine production has been a time-honored tradition that dates back thousands of years (McGovern et al., 2017). Over the centuries, winegrowers have developed and refined the tools and techniques of their trade. Many agricultural products are grown in relatively narrow ranges of climatic and soil conditions for optimal yield. Grapevines are frequently cultivated under a broader range of growing conditions to affect flavor profiles and enhance the potential value of the final wine product. The process of wine production consists broadly of three major steps: grape berry growth, fermentation, and wine aging. Subtle differences in each of these steps contribute to the complexity of the products and impart a unique fingerprint to every wine.
The complexity of wine begins with the chemical composition of the fruit. Red wines are known for their complex palate texture, which is commonly attributed to polyphenolic compounds. Phenolics are oligomers of flavonoids and non-flavonoids found in the skin and seeds of grapes (Umali et al., 2011). Some of these compounds such as catechins and epicatechins are found abundantly in red grapes, and have been associated with the bitter taste and antioxidant properties of wines (Gougeon, da Costa, Guyon, & Richard, 2019). Phenolic compounds undergo chemical reactions during the berry development process and have been associated with markers of vintage age in red wine (Gougeon et al., 2019). The chemical profile of the final wine product and the resultant metabolite composition can be differentiated by viticultural practices and environment (Pereira et al., 2005). As with many agricultural products, the maturation of the fruit is a function of the climate during the growing season (Van Leeuwen & Seguin, 2006). Some red wine varieties such as Pinot noir are known to grow in cool regions, and early harvest dates are characteristic of these grape varieties. For this reason, delicate grape berries such as Pinot noir can be difficult to cultivate, especially in the warm California climate (Smith, 2003). Many vineyards have developed viticultural practices to protect the fruit and encourage healthy growth. However, year to year variation in the microclimate ultimately affects the composition of the product beyond the control of any cultivator (Smith, 2003).
Following berry development, the next step in wine production is the vinification process. Fermentation relies heavily on the overall health of the fruit and the presence of sugars and essential amino acids in the grapes (Baiano, Terracone, Longobardi, Ventrella, Agostiano, & Del Nobile, 2012). In addition to sugars, the accumulation of aroma and flavor metabolites or their precursors during fruit maturation enriches the final wine product. During wine fermentation these flavor compounds are released from the berry and undergo various chemical reactions that depend on the temperature and duration of the fermentation process (Gougeon et al., 2019; Lee, Hwang, Berg, Lee, & Hong, 2009). Aging is the final step of wine production. The use of oak wood barrels has also been shown to affect the metabolomic composition of wines (Cassino, Tsolakis, Bonello, Gianotti, & Osella, 2019). A Previous study has shown that wine aging is characterized by a decrease in organic compounds such as lactic acid and succinic acid with an associated increase in esters (Cassino et al., 2019). Barrel aging further amplifies the aromatic flavors that are highly specific to the age and quality of the barrel product (Dumitriu, Peinado, Cotea, & López de Lerma, 2020; Herrera et al., 2020). Overall, the process of winegrowing, from berry growth to wine aging, produces a unique fingerprint that is characteristic of each vineyard’s wine production process, geographic environment, and yearly microclimate.
Due to the economic and cultural value associated with wine, fingerprints of wine have been extensively studied. Both biological and chemical analytical techniques have been used. These techniques have varied from sequencing technologies (Bokulich, Thorngate, Richardson, & Mills, 2014; Gilbert, van der Lelie, & Zarraonaindia, 2014) to nuclear magnetic resonance (NMR) spectroscopy (Amargianitaki & Spyros, 2017; Cassino et al., 2019; Herrera et al., 2020). By suppressing the most abundant solvents (water and ethanol), one-dimensional (1D) 1H NMR affords the ability to characterize and quantify numerous wine components with minimal pretreatment or alteration of the wine sample. Metabolomics studies have used a combination of quantitative untargeted NMR data with chemometrics to distinguish wines based on grape varieties (Gougeon et al., 2019), region of growth (Godelmann et al., 2013; Gougeon et al., 2019), effects of vintage year (Cassino et al., 2019; Lee et al., 2009), and vinification approach (Baiano et al., 2012).
Multivariate statistical techniques such as principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA) are routinely utilized to classify wine by differentiating the samples based on their chemical profile (Godelmann et al., 2013; Gougeon et al., 2019; Grainger, Yeh, Byer, Hjelmeland, Lima, & Runnebaum, 2021). Conversely, univariate techniques utilize a subset of spectral features or metabolites to discriminate samples and classify group membership. Techniques such as random forest (RF) and receiver operating characteristic (ROC) curves have also been utilized to classify wine according to grape varieties by using NMR and mass spectrometry (MS) analytical techniques (Gómez-Meire, Campos, Falqué, Díaz, & Fdez-Riverola, 2014; Mascellani, Hoca, Babisz, Krska, Kloucek, & Havlik, 2021). Univariate techniques such as RF allow high-classification performance while minimizing the risk of over-fitting the data (Mascellani et al., 2021). Please see (Fan, Upadhye, & Worster, 2006; Lo, Rensi, Torng, & Altman, 2018; Worley & Powers, 2013) for a review of multivariate and univariate statistical techniques.
In addition to traditional analytical techniques, the differential sensing (DS) approach (Joydev Hatai, 2020; Patwardhan, Cai, Newson, & Hargrove, 2019) has become a powerful alternative method to detect and distinguish a variety of small molecules (Diehl, Ivy, Rabidoux, Petry, Müller, & Anslyn, 2015; Li, Zamora-Olivares, Diehl, Tian, & Anslyn, 2017) and biomolecules (Zamora-Olivares, Kaoud, Jose, Ellington, Dalby, & Anslyn, 2014; Zamora-Olivares et al., 2020) in complex biological samples. Polymeric materials have been particularly successful in the DS of a variety of beverages (Bender, Bojanowski, Seehafer, & Bunz, 2018; Huang, Seehafer, & Bunz, 2019; Wang et al., 2018). For wine, the DS technique mimics the senses of taste, smell, and palate texture. This technique utilizes a variety of cross-reactive receptors that display different binding affinities for multiple target molecules (Umali & Anslyn, 2010). The DS method has been successfully employed to classify wine varietals (Umali et al., 2011), wine blends (Ghanem et al., 2015), and to differentiate harvest decisions (Umali et al., 2015) on the basis of phenolic composition and distribution. The peptide-based sensors are ensembles of histidine-rich peptides bound to divalent metals and colorimetric indicators (Table S1) containing a catechol moiety. The peptide ensemble variably binds to phenolics, including polyphenolics (i.e., tannins), to create a targeted fingerprint for each wine sample (Nguyen & Anslyn, 2006). The colorimetric indicators are displaced from the peptide ensembles upon phenol binding and the color changes are quantified by UV-Vis spectroscopy. The resulting dataset of color changes are then analyzed as a composite pattern using chemometric routines such as PCA or linear discriminant analysis (LDA) (Stewart, Ivy, & Anslyn, 2014).
The efficient and relatively simple readout of a targeted DS array is distinct and complementary to an untargeted approach offered by 1D 1H NMR spectroscopy. Thus, the combination of untargeted and targeted techniques was expected to improve the analysis and classification of wine and provide results that neither method can efficiently achieve independently (Figure 1). Such analyses are crucial when attempting to capture subtle variations uniquely imparted by growing site and year to year growing conditions. Few studies have evaluated the advantages of combining NMR or mass spectrometry techniques with a targeted DS array. Thus, we hypothesize that the combination of these two analytical techniques will improve the classification of Pinot noir wines grown from fifteen vineyards, from eight distinct American Vitcultural Areas (AVAs) along the United States Pacific Coast, and for two vintage years (2015 and 2016). The Pinot noir wines were classified with a very high accuracy according to vineyard, regions, and vintage by combining 1D 1H-NMR spectroscopy with DS arrays. This was achieved by using multivariate PCA models and univariate statistical modeling based on RF and ROC analyses. Variability attributed to fermentation and aging steps was minimized by using identical fermentation and aging protocols in stainless steel vessels. Quantifying differences that could be attributed to the growing site or conditions is of upmost importance considering the anticipated changes in microclimates and water availability over the upcoming decades due to climate change (Hannah et al., 2013).
Figure 1.
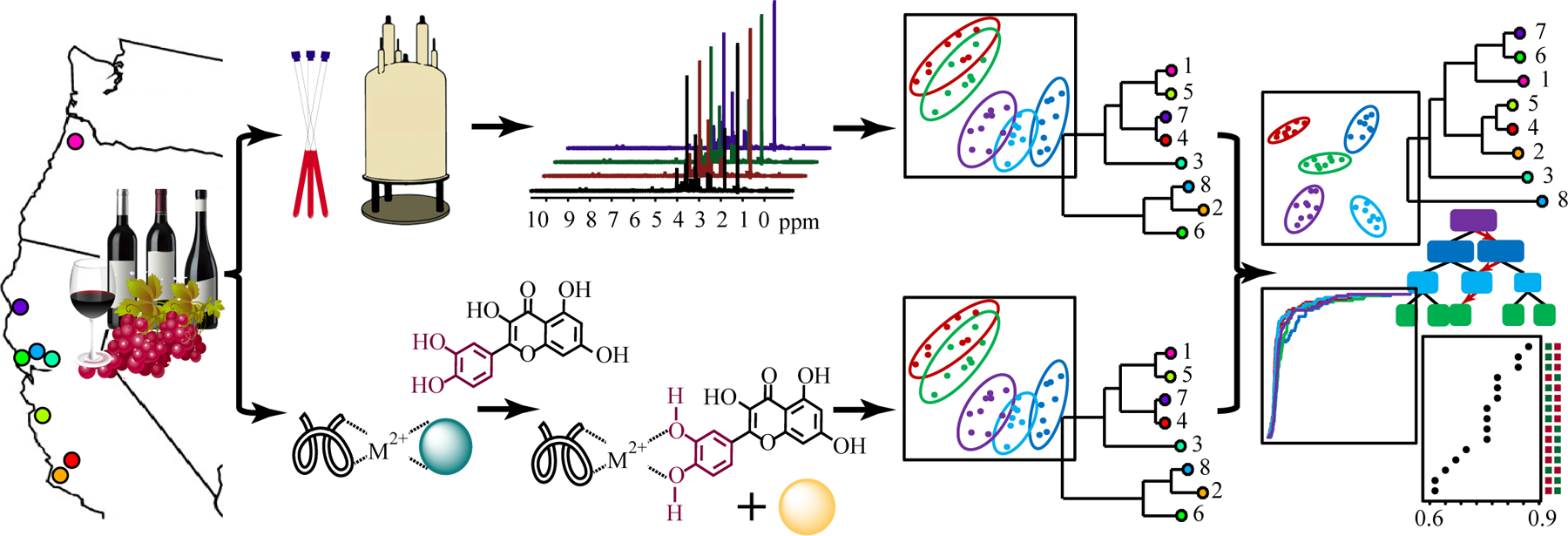
Schematic Representation of Pinot noir Combined Wine Classification. The combined experimental approach utilized NMR spectroscopy and DS array of phenolics (e.g., flavonoids, tannins). The analytical techniques were combined to discriminate fifteen Pinot noir wines from nine AVAs along the coast of California and Oregon. A combination of multivariate and univariate statistical methods was used to produce a classification model that differentiate wines based on vineyard, AVA region or vintage.
2. Materials and Methods
2.1. Vineyard Sites.
Wine grapes (Vitis vinifera L. ‘Pinot noir’ clone 667) from fifteen different vineyard sites along the Pacific Coast of the United States were harvested at a sugar concentration as close as possible to 24 Brix (determined by measuring with a density meter, Anton Paar 35 DMA) between 13 August to 15 September 2015 and between 25 August to 21 September 2016. Eight AVAs, which span a latitudinal distance of approximately 1450 km, are represented in this study: Santa Rita Hills (SRH), Santa Maria Valley (SMV), Arroyo Seco (AS), Carneros (CRN), Sonoma Coast (SNC), Russian River Valley (RRV), Anderson Valley (AV), and Willamette Valley (OR).
2.2. Winemaking.
Grapes were fermented in 200 L stainless steel fermentors at the UC Davis Teaching & Research Winery (Davis, CA). Primary fermentation was initiated by inoculating with Lalvin RC212 (Lallemand) after warming the must to 21°C. The fermentation temperature was held a 21°C for two days after inoculation, and subsequently allowed to rise to 27 °C, where it was held for the remainder of the primary fermentation. Wine was pressed off the red grape skins by using a basket press on the ninth day after grapes were placed into the fermentor. Wines were inoculated with Lalvin VP41 (Lallemand) for malolactic fermentation. Upon completion as measured by conversion of malic acid, the product was stored in stainless steel kegs. Wines were bottled under screw-cap closures approximately six months after harvest. Additional fermentation and winemaking details are available as previously reported (Grainger et al., 2021).
2.3. Differential Sensing Method.
The indicators Chrome Azurol S (CAS) (purity 65%), Bromopyrogagllol Red (BPR), and Pyrocatechol Violet (PCV) (purity 100%) were purchased from Sigma-Aldrich (Saint Louis, MO). Nickel chloride hexahydrate (purity 99.7%), copper (II) sulfate (purity 99.2%), and HEPES buffer were purchased from Fisher Scientific (Hampton, NH). Solid phase peptide synthesis reagents were purchased from P3 BioSystems (Louisville, KY). Peptides were synthesized using standard solid-phase peptide synthesis and a CEM Liberty Blue Automated Microwave Synthesizer (Matthews, NC, USA). Absorbance values were recorded using a Spectra Max Plus 384 plate reader (Molecular Device Inc.).
2.4. Peptide Array and Processing.
A library of nine peptide-based sensors (MM1–MM9) were used for the construction of the DS array. Each sensor was assembled using a histidine peptide, a divalent metal, and a colorimetric indicator. Three different Histidine-containing peptides, WAHEDEFF (TT2), FHFPHHF (SEL1), and WEEHEE (RN8), were used to construct the peptide-metal-indicator ensembles with the corresponding binding ratios shown in Table S1, as previously reported (Umali et al., 2011). Peptides were combined with a metal ion and one of the following indicators: PCV, CAS, and BPR. The imidazole side chain on the peptides chelate the divalent metal ions, and these metals also bind to the colorimetric indicators (Figure 1). Upon addition of the wine to the peptide sensors, the indicators become displaced from the ensembles producing color changes as differential optical responses due to the polyphenols present in the wine, in the manner previously reported in detail (Umali et al., 2011). Arrays were prepared in Fisher Scientific non-treated 96-well plates with flat bottom and clear polystyrene. Final well-plate solutions of peptide ensembles and wine concentration of 1% (v/v) were prepared using 50 mM HEPES in ethanol (1:1 (v/v), pH = 7.4). Absorbance endpoint-values due to the displacement of each indicator by the phenolics were measured at 430 nm, 444 nm, and 560 nm corresponding to the λmax of free CAS, PCV, and BPR, respectively. Eight analytical replicates were used for each of the fifteen wines to ensure reproducibility. Controls consisted of a column of wine alone and a column of the ensemble alone in each plate.
2.5. Sensing Array Batch Correction.
Systematic data variation can arise from known and unknown sources, such as instrument differences, personnel changes, and environmental variation between batches. Such variations are observed in many biological assays (Chakraborty, 2019; Worley & Powers, 2014). Due to the prevalence of batch effects in analytical data, statistical techniques such as PLS have been used as a primary tool to monitor and correct for batch effects when real-time quality measurements are unavailable (Fonville et al., 2010; Nomikos & MacGregor, 1995). In this model, a PLS analysis was used to correct for the variable separation between the two vintage years.
2.6. NMR Sample Preparation.
For each of the fifteen wine samples, eight analytical replicate NMR samples were created. Each sample was prepared by adding 150 μL of wine to 15 μL of 50 mM phosphate buffer prepared in D2O at pH 7.2 (uncorrected). Deuterated sodium-3-trimethylsilypropoinate (TMSP, 50 μM) was used as an internal chemical shift standard.
2.7. NMR Data Collection and Processing.
The NMR experiments were collected on a Bruker AVANCE III 700 MHz spectrometer equipped with a 5 mm quadrupole resonance QCI-P cryoprobe™ (1H, 13C, 15N and 31P) with a 2H lock and decoupling. A SampleJet automated sample changer with Bruker ICON-NMR™ software was used to automate data collection. 1D 1H NOESY experiments with a presaturation pulse were collected for each sample by using a Bruker automation program, Multisupp, to suppress the multiple solvent peaks from water and ethanol. 1D 1H NMR spectra were collected with 65K points, a spectral width of 14705 Hz, 128 scans, 4 dummy scans, and 4s relaxation delay. 1D 1H NMR spectra were batch processed and analyzed using the NMR metabolomics toolbox, MVAPACK (Worley et al., 2014). The 1D 1H NMR spectra were Fourier transformed, auto phased with manual phase adjustment as needed, and TMSP was referenced to 0 ppm. Regions of the spectra containing water and ethanol peaks were removed.
2.8. Multivariate Data Analyses.
The 1D 1H NMR spectra were normalized using probabilistic quotient (PQ) normalization and Pareto scaled. The full resolution NMR spectroscopy dataset was then used to create PCA models and dendrograms based on the Mahalanobis distance between each group (Worley, Halouska, & Powers, 2013). Each dendrogram node is labeled with a p-value indicating the statistical significance of the group separation. DS array data was first batch corrected using a PLSfit correction. The resulting data matrix was Pareto scaled and then used to generate PCA models and dendrograms. The NMR spectroscopy and DS array datasets were also combined with equal contributions of each block (i.e., dataset) to generate multiblock principle component analysis (MB-PCA) models and dendrogram representations (Marshall et al., 2015). PCA models with four components were used to create linear discriminant analysis (PCA-LDA) models (Worley, Halouska, et al., 2013). Models were separately generated for the vineyard data, AVA region data, and vintage years.
2.9. Univariate Analyses.
To carry out univariate analyses, adaptively binned data of the 1D 1H NMR datasets were exported from MVAPACK (Worley & Powers, 2015). DS array data and NMR bins were then combined to create a complete data matrix. The subsequent data analysis was performed in MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/) (Xia, Sinelnikov, Han, & Wishart, 2015). Multivariate ROC curves were obtained for two-group comparisons between each wine and all other groups of wine. The two-group comparison was repeated for the AVA regions. ROC curves were generated using Monte-Carlo cross validation (MCCV) with balanced subsampling. Each MCCV utilized two-thirds of the samples as the training subset, while the remaining one-third was reserved for the testing subset. The dataset was Pareto scaled and the ROC curves were generated using a support vector machine algorithm. The RF algorithm in MetaboAnalyst 4.0, randomForest package (Liaw & Wiener, 2002), was utilized to classify the Pinot noir wines according to vineyard, region, and year. The RF algorithm used an ensemble of 500 decision trees. Each decision tree was grown through random feature selection (maximum of 7 predictors) using a bootstrap sample at each branch. Classification was assigned by majority vote within the ensemble. Two-thirds of the samples was used to construct the training subset, while one-third was reserved for the testing subset.
2.10. Univariate Analysis of ROC Feature Frequency Selection.
The top features from all vineyard and AVA region ROC curves were cataloged into a frequency map according to the number of times each feature (DS array or NMR) was selected by the ROC curve analysis. A total of 101 unique NMR features (from 210 available features) were selected a total of 571 times. In the same ROC analysis, 27 unique DS array features (from 27 available features) were selected a total of 229 times. NMR chemical shift features were binned using a 0.1 ppm bin width. The most frequently selected NMR features for vineyard (≥ 10) and AVA region (≥ 8) were utilized to putatively assign group-differentiating metabolites. Putative metabolite assignments were based on a set of previously published 1D 1H NMR spectra for 55 known wine metabolites (Gougeon et al., 2019; Hu, Cao, Zhu, Xu, & Wu, 2019; Hu, Gao, Xu, Zhu, Fan, & Zhou, 2020; Mascellani et al., 2021). Assignments were based on consistency with known chemical shifts and coupling patterns. A chemical shift error of 0.1 ppm was used to account for chemical shift variability due to differences in pH, ethanol concentrations, and chemical compositions between the wines and reference 1D 1H NMR spectra. A similar protocol was used to annotate the single ROC curve comparison of the 2015 and 2016 vintage years. A catalog of feature usage was omitted from the ROC curve analysis of vintage since only one comparison was possible.
3. Results and Discussion
3.1. Combining multiple analytical techniques to classify the vintage, vineyard, and the region of Pinot noir wines.
As shown in Figure 1, two complementary techniques were utilized to classify Pinot noir wines derived from the same scion clone (Pinot noir 667). In this regard, an expanded view of the chemical composition of each wine sample could be obtained by combining an untargeted NMR approach with a targeted DS array. The same level of coverage would not be possible with only one of these analytical techniques because they each detect a different set of metabolites. A 1D 1H NMR spectrum was acquired for each wine sample to provide a global chemical profile or metabolic fingerprint. Each peak in the NMR spectrum identifies a particular metabolite, where its relative abundance is indicated by the peak intensity. Thus, each 1D 1H NMR spectrum will vary as the chemical composition of the wine changes. Nonetheless, NMR will only detect the most abundant (> 1 μM) metabolites, so it does not provide a complete picture of the overall chemistry. A DS array was also obtained for each wine sample. In contrast to the NMR data, the DS array was applied to classify only the phenolic composition of each wine sample. An array of nine sensing ensembles (Table S1) was used to identify the unique phenolic profile through the displacement of colorimetric indicators. The NMR spectroscopy and DS array datasets were then subjected to multivariate and univariate statistical analysis. Chemometric analysis was used to identify dataset features that characterized significant differences between the wines (Figure 2) grown in fifteen different vineyard sites, eight distinct AVAs, and two vintage years (2015 and 2016).
Figure 2.
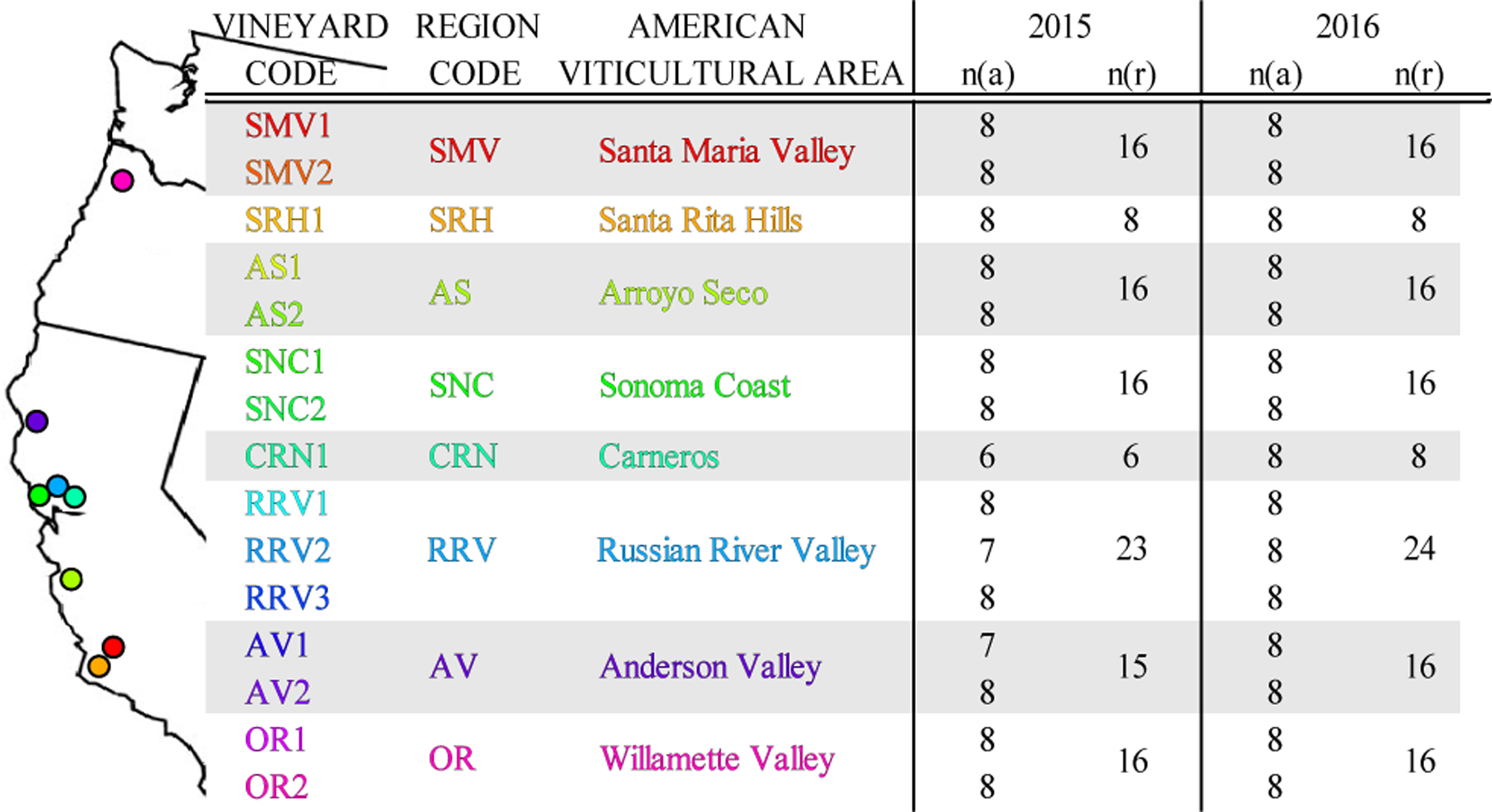
Demographics of the Pinot noir Wine across the Pacific Coast of the United States. Samples displayed by vineyard code, AVA code, and vintage year. The AVA region locations are shown along the coast of California and Oregon. n(a) denotes the number of analytical replicates and n(r) denotes the number of samples from the same AVA region for each vintage year.
3.2. Multivariate analysis highlights unique vineyard and vintage classification.
While all Pinot noir grapes share the same genetic ancestors, the age of the vine and environmental influences lead to vine evolution that plays an important role in the structure of the resulting wine (Smith, 2003). PCA was used to capture maximal group differences in Pinot noir wines from both the NMR and DS array multivariate datasets. For the purpose of analysis, individual PCA models and dendrograms were created for the NMR spectroscopy and DS array data, as well as both the 2015 and 2016 vintage data. The NMR and DS array datasets were then combined with equal contribution to create MB-PCA models, which generated a unified model that captured the maximum variation between the groups and identified the key group-differentiating variables.
Throughout the creation of these PCA and MB-PCA models, specifically with the NMR spectroscopy datasets, it was difficult to display the maximal separation between groups in a two-dimensional (2D) PCA scores plot. The data encompassed more information and complexity than could be explained by two components. Some NMR PCA models required upwards of twelve components. Thus, PCA-LDA models were generated using four components of the respective PCA or MB-PCA models to capture the maximal separation within the model (Figures 3 and S1A, B). This was not a concern for the DS array dataset where a 2D PCA scores plot (Figures S1C, D) was sufficient to display group variation. The resulting PCA-LDA scores plots (Figure 3A and 3B) successfully demonstrated that the classification of Pinot Noir wines produced from an identical Pinot noir clone grown in different geographic regions along the Pacific Coast can be uniquely classified. The corresponding dendrograms display the relative similarity and/or differences between the individual wines with a p-value assigned to each node indicating the statistical significance of these differences.
Figure 3.
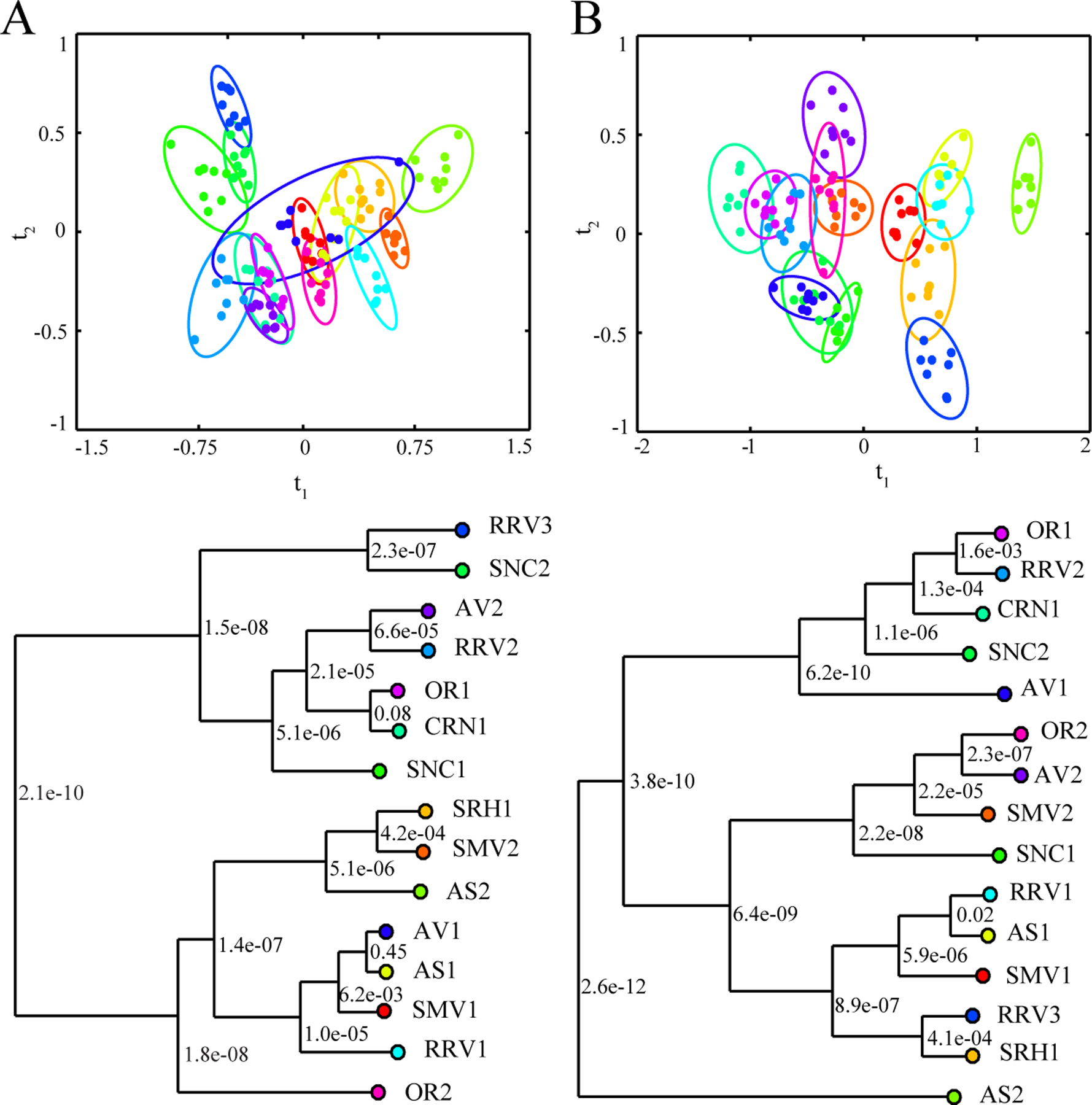
Vineyard Multivariate Scores Plots. LDA-MB-PCA scores plots and associated dendrograms generated from the combined NMR spectroscopy and DS array datasets from the (A) 2015 (R2 0.78, Q2 0.53) and (B) 2016 (R2 0.67, Q2 0.54) wine samples. LDA-MB-PCA scores plots and dendrograms are displayed with color-coded groups labels, symbols and ellipses as defined in Figure 2. Ellipses represent the 95% confidence interval from a normal distribution. Each node of the dendrogram is labeled with a p-value based on Mahalanobis distances between the groups. LDA models were generated from the first four components of the MB-PCA model.
Several conclusions may be drawn from the analysis of the PCA and PCA-LDA scores plots and dendrograms based on the analysis of vineyard classification. First, the PCA-LDA scores plots and dendrograms showed significant differences between the 2015 and 2016 vintage years (Figure 3). The relative clustering in the 2015 and 2016 dendrograms are essentially unique. For example, OR2 is the most unique wine in 2015 (furthest distance from any other wine as shown in Figure 3A), while OR2 clusters close to SRH1 in 2016 (Figure 3B). AS2 showed maximal separation in 2016 while this vineyard clustered close to SMV2 and SRH1 in 2015. Conversely, the wine pairs OR1-CRN1 (p-value 0.08) and AV1-AS1 (p-value 0.45) are not statistically distinct in 2015, but they are distinct in 2016 with p-values of 1.01 × 10−4 and 1.37 × 10−11, respectively (Table S3). Conversely, RRV1-AS1 (p-value 0.02) are not statistically distinct in 2016 but are distinct in 2015 (p-value 4.93 × 10−5 Table S2). A similar level of unique clustering occurs when comparing the NMR LDA-PCA (Figure S1A, B), DS array PCA (Figure S1C, D) and the LDA-MB-PCA (Figure 3) scores plots. These results are consistent with the fact that NMR and DS array capture different chemical features, and these chemical features have different group discriminations. NMR spectroscopy captures a large breadth of information through an untargeted approach and exhibits small within group variation and larger between group variation. Conversely, the DS array specifically targets the phenolic profile, which has limited discrimination and leads to a larger within group variation. As expected, the LDA-MB-PCA is a hybrid of the NMR and DS array models. Now, while the within group variance increases from the NMR LDA-PCA to the LDA-MB-PCA models, both techniques show distinct areas of separation and contribute different features that aid in the overall separation of the wines by vineyard. This is further evident from the univariate analysis shown below.
3.3. Multivariate analysis of regions highlights the complex nature of geographic location and vineyard practices.
In an effort to further demonstrate the applicability of the combined analytical techniques, the classification of eight AVA regions was evaluated by multivariate analysis. While classification of wines according to individual vineyard sites showed distinct clustering in the PCA and PCA-LDA scores plots (Figure 3), wine classification according to AVA region proved to be more nuanced (Figure 4). Vineyard sites such as those within the RRV AVA showed clustering of two wines, RRV1 and RRV3, while the third wine, RRV2, exhibited a distinct signature across both the 2015 and 2016 vintage years. The sub-clustering within this region prevented a multivariate analysis for both the NMR spectroscopy and DS array datasets. Alternatively, valid models with sufficient group separation by region were generated upon the removal of RRV2 from the PCA and PCA-LDA models. The four most significant components of the PCA models were used to generate PCA-LDA models for both the 2015 (Figure 4A) and 2016 (Figure 4B) NMR spectroscopy datasets. PCA models were also generated for the DS array 2015 (Figure S2A) and 2016 datasets (Figure S2B). A valid MB-PCA model could not be generated from the combined 2015 or 2016 region datasets. While the DS array data was a good predictor of vineyard signature, small variations observed in the region DS array datasets hindered our ability to generate valid multiblock models with the combined NMR spectroscopy and DS array datasets. This result suggests that the phenolic profile alone may not be sufficient to differentiate most of the AVA regions.
Figure 4.
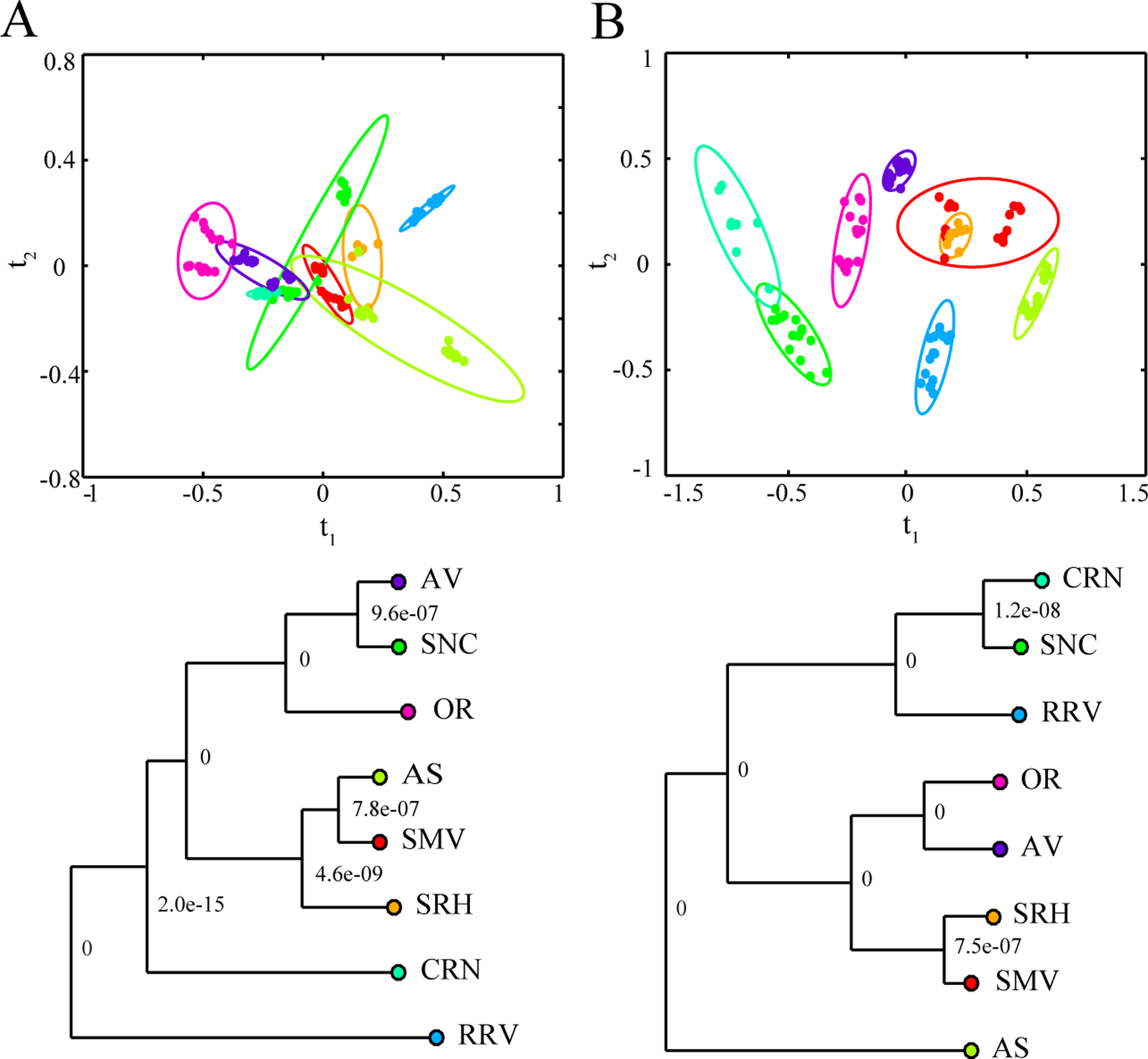
AVA Region Multivariate Scores Plots. LDA-PCA scores plots and associated dendrogram models generated from the NMR spectroscopy region datasets from the (A) 2015 (R2 0.79, Q2 0.74) and (B) 2016 (R2 0.75, Q2 0.70) wine samples. LDA-PCA scores plots and dendrograms are displayed with color-coded groups labels, symbols and ellipses as defined in Figure 2. Ellipses represent the 95% confidence interval from a normal distribution. Each node of the dendrogram is labeled with a p-value based on Mahalanobis distances between the groups. LDA models were generated from the first four components of the PCA model.
The PCA-LDA scores plot and the associated dendrograms for the NMR spectroscopy dataset exhibited good group separation between the eight AVA regions (Figure 4). In fact, several p-values within the dendrogram nodes are zero, which indicates that the region classification is better than the vineyard classification seen in Figure 3. Of course, it is inherently easier to separate eight groups compared to fifteen groups. It is important to note that a closer examination of the individual group clustering indicates some within group sub-clustering. This is particularly noticeable for the SNC and AS regions in 2015 and the SMV region in 2016. This sub-clustering may suggest that group membership within a given AVA region is not uniformly defined by a unique chemical signature.
Similar to the vineyards, the AVA region analysis showed a unique clustering pattern between the 2015 and 2016 vintages. Likewise, the dendrograms presented a distinct set of nearest neighbors. For example, RRV was the most unique AVA region in 2015, but RRV clustered near SNC and CRN in 2016. Conversely, AS was the most unique AVA region in 2016, but it clustered closer to SMV in 2015. Overall, the Pinot noir wine clustering using the combined NMR spectroscopy and DS array dataset contained a stronger metabolic signature for the individual vineyard than for the geographic location of growth. Nevertheless, a classification-based AVA region was still achieved.
3.4. Univariate analyses demonstrate an improved wine classification through a combination of analytical techniques.
In addition to multivariate analysis, univariate analyses were used to determine specific features from the NMR spectroscopy and DS array datasets that could be used to distinguish Pinot noir wines by vineyard or region. Univariate analysis was carried out with 1D 1H-NMR binned data and raw DS array data. Specifically, NMR features corresponded to a given ppm bin, whereas DS array features attributed the absorbance (λmax 430 nm, 444 nm, or 560 nm) of a particular peptide sensor (MM1–MM9, Table S1). RF analysis was used to establish the classification accuracy of the Pinot noir wines across vineyard and region. As shown in Table 1, the vineyard classification accuracy for the NMR spectroscopy or DS array datasets was high with an average value of 0.94 ± 0.10 and 0.88 ± 0.13, respectively. The classification accuracy ranges from 0 to 1, where a value of 1 indicates a perfect classification. The vineyard classification accuracy for the combined NMR spectroscopy and DS array dataset exceeded the individual results and reached an average of 0.98 ± 0.05 with p-values < 0.05 when compared to the results from the individual techniques. There were no notable differences in vineyard classification accuracy between the 2015 and 2016 vintages. Interestingly, the performance of the individual analytical techniques varied between the two vintages. The NMR spectroscopy dataset had a higher vineyard classification accuracy for 2016 (0.98 ± 0.05) compared to 2015 (0.90 ± 0.12). The opposite was observed for the DS array dataset where the 2015 dataset (0.92 ± 0.08) was better than the 2016 dataset (0.80 ± 0.15).
Table 1.
Summary of Pinot noir Wines Univariate Analysis
| Sample Namea | Random Forest Classificationb | NMR+DS Array ROC Curvec | |||||
|---|---|---|---|---|---|---|---|
| NMR | DS Array | NMR+DS Array | AUC | NMR Ratiod (% NMR) | DS Array Ratioe (% DS) | ||
| 2015 Vineyard | |||||||
| SMV1 | Santa Maria Valley | 0.88 | 1.00 | 1.00 | 0.95 | 0.93 (0.07) | 0.07 (0.04) |
| SMV2 | Satna Maria Valley | 1.00 | 0.88 | 1.00 | 0.98 | 0.73 (0.05) | 0.27 (0.15) |
| SRH1 | Santa Rita Hills | 0.75 | 0.88 | 0.88 | 0.91 | 0.4 (0.05) | 0.6 (0.56) |
| AS1 | Arroyo Seco | 0.63 | 1.00 | 0.88 | 0.81 | 0.88 (0.1) | 0.12 (0.11) |
| AS2 | Arroyo Seco | 0.75 | 0.88 | 1.00 | 0.98 | 1.00 (0.07) | 0.00 (0.00) |
| SNC1 | Sonoma Coast | 1.00 | 0.88 | 1.00 | 0.98 | 0.68 (0.08) | 0.32 (0.3) |
| SNC2 | Sonoma Coast | 0.75 | 1.00 | 1.00 | 0.99 | 0.2 (0.01) | 0.8 (0.44) |
| CRN1 | Carneros | 1.00 | 1.00 | 1.00 | 0.92 | 0.64 (0.08) | 0.36 (0.33) |
| RRV1 | Russian River Valley | 1.00 | 0.75 | 1.00 | 0.99 | 0.93 (0.07) | 0.07 (0.04) |
| RRV2 | Russian River Valley | 1.00 | 0.86 | 1.00 | 0.98 | 0.52 (0.06) | 0.48 (0.44) |
| RRV3 | Russian River Valley | 1.00 | 1.00 | 1.00 | 0.99 | 0.67 (0.05) | 0.33 (0.19) |
| AV1 | Anderson Valley | 0.86 | 0.86 | 0.86 | 0.94 | 0.92 (0.11) | 0.08 (0.07) |
| AV2 | Anderson Valley | 0.88 | 1.00 | 1.00 | 0.94 | 0.64 (0.08) | 0.36 (0.33) |
| OR1 | Willamette Valley | 1.00 | 1.00 | 1.00 | 0.99 | 0.80 (0.06) | 0.20 (0.11) |
| OR2 | Willamette Valley | 1.00 | 0.88 | 1.00 | 0.95 | 0.32 (0.04) | 0.68 (0.63) |
| Averagef | 0.90±0.12 | 0.92±0.08 | 0.97±0.05 | 0.95±0.05 | |||
| p-valueg (individual vs combination) | 0.045 | 0.05 | |||||
| 2016 Vineyard | |||||||
| SMV1 | Santa Maria Valley | 1.00 | 0.75 | 1.00 | 0.97 | 0.87 (0.06) | 0.13 (0.07) |
| SMV2 | Satna Maria Valley | 0.88 | 0.88 | 1.00 | 0.96 | 0.72 (0.09) | 0.28 (0.26) |
| SRH1 | Santa Rita Hills | 1.00 | 1.00 | 1.00 | 0.95 | 0.64 (0.08) | 0.36 (0.33) |
| AS1 | Arroyo Seco | 1.00 | 0.88 | 1.00 | 0.95 | 0.47 (0.03) | 0.53 (0.3) |
| AS2 | Arroyo Seco | 0.88 | 0.88 | 0.88 | 0.95 | 0.40 (0.03) | 0.60 (0.33) |
| SNC1 | Sonoma Coast | 1.00 | 1.00 | 1.00 | 0.97 | 0.67 (0.05) | 0.33 (0.19) |
| SNC2 | Sonoma Coast | 1.00 | 0.75 | 1.00 | 0.96 | 0.80 (0.10) | 0.20 (0.19) |
| CRN1 | Carneros | 0.88 | 0.50 | 0.88 | 0.99 | 0.73 (0.05) | 0.27 (0.15) |
| RRV1 | Russian River Valley | 1.00 | 0.75 | 1.00 | 1.00 | 1.00 (0.07) | 0.00 (0.00) |
| RRV2 | Russian River Valley | 1.00 | 0.75 | 1.00 | 0.96 | 0.88 (0.10) | 0.12 (0.11) |
| RRV3 | Russian River Valley | 1.00 | 1.00 | 1.00 | 0.99 | 0.40 (0.02) | 0.60 (0.22) |
| AV1 | Anderson Valley | 1.00 | 0.75 | 1.00 | 1.00 | 0.87 (0.06) | 0.13 (0.07) |
| AV2 | Anderson Valley | 1.00 | 1.00 | 1.00 | 0.97 | 0.72 (0.09) | 0.28 (0.26) |
| OR1 | Willamette Valley | 1.00 | 1.00 | 1.00 | 0.99 | 0.80 (0.04) | 0.20 (0.07) |
| OR2 | Willamette Valley | 1.00 | 0.63 | 1.00 | 0.99 | 1.00 (0.07) | 0.00 (0.00) |
| Average | 0.975±0.05 | 0.83±0.15 | 0.98±0.04 | 0.97±0.02 | |||
| p-value (individual vs combination) | 0.64 | 0.001 | |||||
| Vineyard Totals | |||||||
| Average | 0.94±0.10 | 0.88±0.13 | 0.98±0.05 | 0.96±0.04 | |||
| p-value (vineyard vs combination) | 0.050 | 0.0002 | |||||
| 2015 Region | |||||||
| Santa Maria Valley | SMV1, SMV2 | 1.00 | 0.94 | 1.00 | 0.97 | 0.93 (0.07) | 0.07 (0.04) |
| Santa Rita Hills | SRH1 | 0.63 | 0.63 | 0.88 | 0.91 | 0.36 (0.04) | 0.64 (0.59) |
| Arroyo Seco | AS1, AS2 | 0.94 | 1.00 | 0.94 | 0.93 | 0.72 (0.09) | 0.28 (0.26) |
| Sonoma Coast | SNC1, SNC2 | 0.81 | 0.81 | 1.00 | 0.96 | 0.60 (0.03) | 0.40 (0.15) |
| Carneros | CRN1 | 0.83 | 0.83 | 1.00 | 0.87 | 0.53 (0.04) | 0.40 (0.22) |
| Russian River Valley | RRV1, RRV2, RRV3 | 1.00 | 0.78 | 1.00 | 0.99 | 0.87 (0.06) | 0.13 (0.07) |
| Anderson Valley | AV1, AV2 | 0.93 | 0.87 | 0.93 | 0.96 | 0.73 (0.05) | 0.27 (0.15) |
| Willamette Valley | OR1, OR2 | 1.00 | 0.94 | 1.00 | 0.99 | 0.80 (0.04) | 0.20 (0.07) |
| Average | 0.089±0.12 | 0.85±0.11 | 0.97±0.05 | 0.948±0.039 | |||
| p-value (individual vs combination) | 0.147 | 0.018 | |||||
| 2016 Region | |||||||
| Santa Maria Valley | SMV1, SMV2 | 0.94 | 1.00 | 1.00 | 0.99 | 0.80 (0.04) | 0.20 (0.07) |
| Santa Rita Hills | SRH1 | 1.00 | 0.75 | 1.00 | 0.93 | 0.80 (0.06) | 0.20 (0.11) |
| Arroyo Seco | AS1, AS2 | 1.00 | 0.94 | 1.00 | 0.98 | 0.90 (0.04) | 0.10 (0.04) |
| Sonoma Coast | SNC1, SNC2 | 1.00 | 0.88 | 1.00 | 0.99 | 0.67 (0.05) | 0.33 (0.19) |
| Carneros | CRN1 | 0.88 | 0.50 | 0.88 | 0.99 | 0.80 (0.06) | 0.20 (0.11) |
| Russian River Valley | RRV1, RRV2, RRV3 | 1.00 | 0.83 | 1.00 | 0.98 | 1.00 (0.05) | 0.00 (0.00) |
| Anderson Valley | AV1, AV2 | 1.00 | 0.75 | 1.00 | 0.99 | 1.00 (0.05) | 0.00 (0.00) |
| Willamette Valley | OR1, OR2 | 1.00 | 0.69 | 1.00 | 0.98 | 0.73 (0.05) | 0.27 (0.15) |
| Average | 0.977±0.043 | 0.792±0.147 | 0.984±0.041 | 0.980±0.020 | |||
| p-value (individual vs combination) | 0.736 | 0.005 | |||||
| Region Totals | |||||||
| Average | 0.935±0.101 | 0.821±0.132 | 0.976±0.044 | 0.964±0.035 | |||
| p-value (region vs combination) | 0.1529 | 0.0002 | |||||
list of the fifteen vineyard IDs and the associated regions.
RF classification accuracy ranges from 0 to 1, where 1 is perfect classification. RF classification accuracy using just the NMR or DS array data alone or using the combined dataset.
ROC - receiver operating characteristic curve, AUC-area under the ROC curve. AUC ranges from 0 to 1, where 1 indicates perfect classification. ROC and AUC were calculated using the combined NMR spectroscopy and DS array datasets.
NMR ratio identifies the percentage of the total features used in the ROC curve that are from the 1D 1H NMR spectrum. %NMR identifies the percentage of the total number of NMR features used in the ROC curve.
DS array ratio identifies the percentage of the total features used in the ROC curve that are from the DS array data. %DS identifies the percentage of the total number of DS array features used in the ROC curve
column averages are presented as average ± standard deviation
p-values are calculated from a Student’s t-test
A similar level of classification success was achieved using the AVA regions (Table 1). The AVA region classification accuracy for the NMR spectroscopy and DS array datasets was similarly high with an average value of 0.94 ± 0.10 and 0.82 ± 0.13, respectively. The AVA region classification accuracy for the combined NMR spectroscopy and DS array datasets equaled or exceeded the individual results. The AVA region classification accuracy reached an average of 0.98 ± 0.04. While the improvement was statistically significant relative to the DS array dataset (p-value 0.0002), it was not significant when compared to the NMR spectroscopy dataset (p-value 0.15). This is in part due to the limited AVA region variance (attributed to the DS array data described earlier). Overall, the NMR spectroscopy dataset showed a greater ability to distinguish the nuances of AVA regions. Nevertheless, there were specific situations where the classification accuracy was low when only the NMR spectroscopy or the DS array dataset was used independently, but the accuracy improved significantly for the combined dataset. For example, in the case of the 2015 SRH region, both the NMR spectroscopy and DS array datasets alone were only capable of accurately classifying 63% of the wine samples. Specifically, only five of the eight wine samples were correctly classified as SRH. Conversely, the combined NMR spectroscopy and DS array data improved the classification accuracy to 88%, in which seven out of the eight wine samples were correctly classified as SRH. Overall, the combination of analytical techniques improved the classification accuracy for both vineyards and AVA regions.
ROC curves were also generated to advance our understanding of the metabolic fingerprint that defined Pinot noir wines derived from grapes grown along the Pacific Coast. ROC curves were utilized to further evaluate the performance of the NMR spectroscopy and DS array datasets, and to identify unique features that distinguished vineyards and AVA regions. ROC curves compare the true positive rate (1-specificity) with the false positive rate (sensitivity) where the AUC provides a measure of model performance and accuracy (Fan et al., 2006). AUC typically ranges from 0.5 to 1, where 1 indicates perfection and 0.5 identifies a random outcome. For each model, the ROC curve with the fewest number of features and the highest AUC was chosen. Representative ROC curves for vineyard SNC2 and the associated feature list are shown in Figure S3. ROC curves were only generated from the combined NMR spectroscopy and DS array dataset. Table 1 summarizes the AUC for each ROC curve for the vineyards, AVA regions and vintages. An average AUC of 0.96 ± 0.04 was observed for both the vineyard and AVA regions. Likewise, the ROC curves indicate that a high classification accuracy was obtained by combining the NMR spectroscopy and DS array datasets.
The ratio of NMR and DS array features used to generate the ROC curves was also evaluated. Table 1 lists the NMR spectroscopy and DS array percent contribution to both the model and to the total number of available features. Notably, a variable amount of NMR and DS array features was used to define each individual ROC curve. For example, vineyards SNC2 2015 showed an AUC of 0.99 with a 20:80 ratio of NMR to DS array features. Interestingly, the ROC curve for the 2016 vintage exhibited a similar AUC of 0.96, but the relative feature contributions changed to an 80:20 ratio of NMR spectroscopy to DS array features (Figure S3). The consistently high AUC values for both vineyard and region indicated that the combined NMR spectroscopy and DS array data contained distinct features for all fifteen wines and eight regions. These features may be used to separate each vineyard and region from the entire collection of Pinot noir wines. There were a few circumstances where all features were selected from a single dataset (i.e., NMR). These instances corresponded to vineyards AS2 2015, RRV1 2016, and OR2 2016; and AVA regions RRV 2016 and AV 2016. Notably, the lower contribution of DS array features to the 2015 and 2016 AVA regions was consistent with the multivariate analysis as described above.
ROC curves were also used to identify DS array and NMR spectroscopy features that were frequently utilized to distinguish Pinot noir wines by vineyard or AVA region (Figure 5). The top features selected from the ROC curve analysis of the combined NMR spectroscopy and DS array dataset for the 2015 SNC2 vineyard are shown in Figure 5A. The ROC feature selection plot in Figure 5A identifies the specific NMR bins (i.e., ppm) and peptide sensors (i.e., MM1–MM9) that differentiated the 2015 SNC2 vineyard from the remaining vineyards. The plot also identified the relative directional change in the features and how often each feature was selected to differentiated between the vineyards. A similar analysis was completed for each AVA region, where a representative plot of the top features is shown in Figure 5B for the 2015 SNC AVA region.
Figure 5.
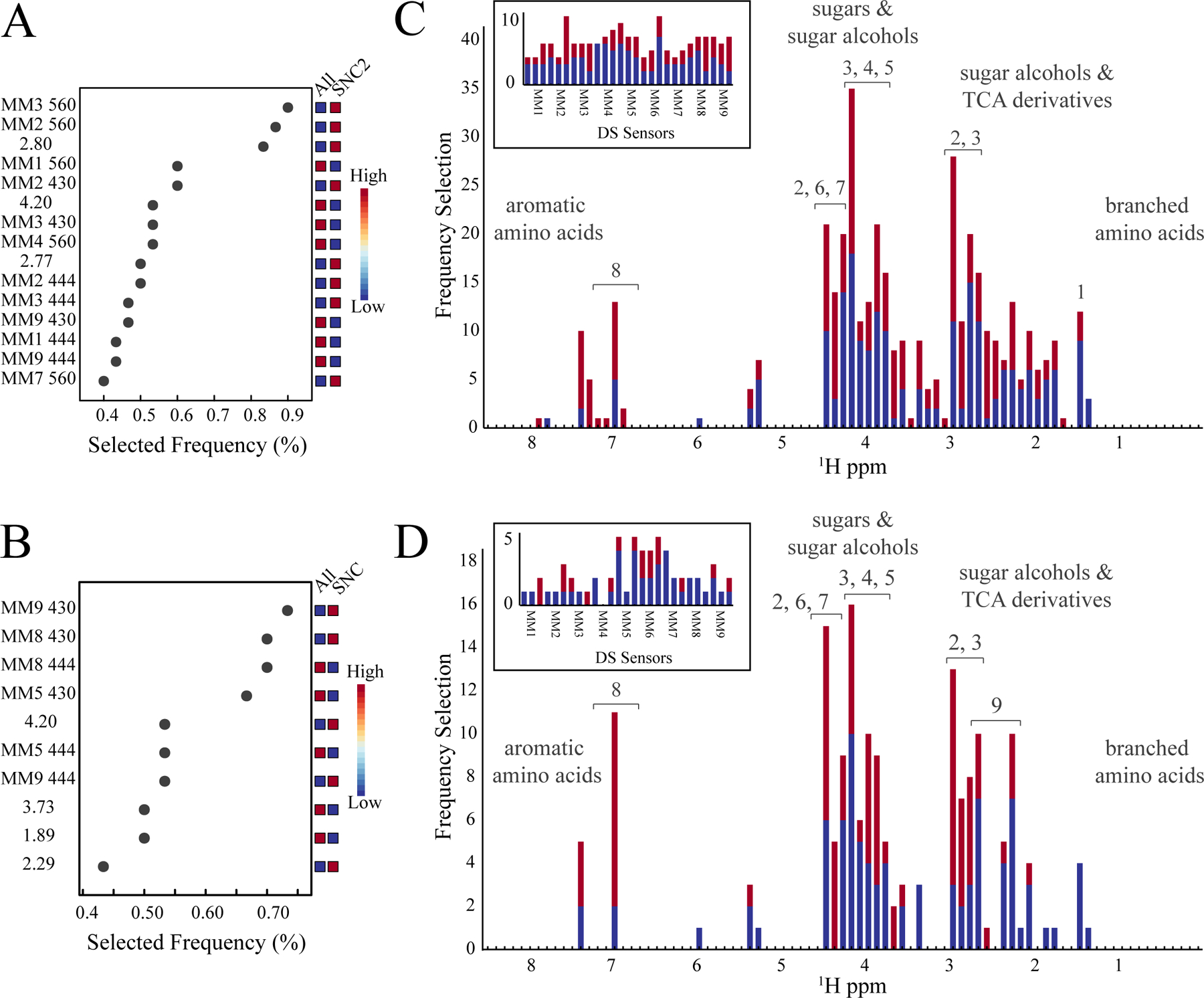
ROC Curve Analysis and Feature Selection Frequency. Representative ROC curve feature selection charts from the combined NMR spectroscopy and DS array dataset for the (A) 2105 SNC2 vineyard analysis and (B)2015 SNC region analysis. The ROC curves were generated with MetaboAnalyst 4.0 (https://www.metaboanalyst.ca/) (Xia et al., 2015). NMR feature usage from all of the ROC curves is plotted using an NMR bin (ppm) size of 0.1 ppm for (C) vineyard and (D) region analysis. 2015 data are colored blue, and the 2016 data is colored red. A plot of the DS array feature (MM1 to MM9) usage from the same ROC curve analyses are displayed as an insert. Putative metabolite assignments correspond to 1, isobutanol; 2, malic acid; 3, phenethyl alcohol; 4, mannitol; 5, fructose; 6, ethyl acetate; 7, ethyl lactate; 8, tyrosine; and 9, citric acid.
The top selected features from all ROC curve analyses were cataloged and the usage-frequency between vineyards and AVA regions are plotted in Figures 5C and 5D, respectively. Notably, the DS feature selection rate was relatively consistent across the nine sensors, which suggests an equal contribution of phenols to the group classification. Conversely, the selection rate of the NMR features was highly variable, which suggests certain metabolites were preferred discriminators of the Pinot noir wines. Thus, the high-usage NMR features were assigned to potential metabolites or chemical classes using a set of reference 1D 1H NMR spectra of known wine metabolites. Specifically, 55 metabolites were previously identified from four prior NMR metabolomics studies of similar wines (Gougeon et al., 2019; Hu et al., 2019; Hu et al., 2020; Mascellani et al., 2021). Figures 5C and 5D depicts several metabolite classes and putative metabolite assignments that may preferentially classify Pinot noir wines according to vineyard or AVA region, respectively. Our analysis suggests that metabolites such as branched-chain amino acids, sugar alcohols including ethyl alcohols and phenyl alcohols, derivatives of the tricarboxylic acid cycle (TCA) such as malic acid and citric acid, sugars including fructose, and aromatic amino acids may play an important role in classifying Pinot noir wines across the Pacific Coast of the United States. It is important to note that the primary purpose of our study was to demonstrate the value of combining NMR and DS array features to classify and differentiate Pinot noir wines based on vineyard, region, and vintage. Assigning a metabolite to each of the group-defining features was not our original intent. As a result, the accuracy of the subsequent metabolite identification was greatly hindered by solely relying on 1D 1H NMR spectra and, importantly, by the limited availability of data resources consisting of reference NMR spectra specific to the wines and geographic locations used in this study. Accordingly, only the 55 known wine metabolites were used to assign the NMR frequency plots in Figures 5C and 5D. The remaining unassigned NMR bins correspond to currently unknown or unverified wine metabolites.
The ROC curves further supported the observation that the 2015 and 2016 vintage years exhibited distinct features that were independent of vineyard and AVA region. This was evident by the variable number of NMR spectroscopy and DS array features contributing to each pair of 2015/2016 vineyard or AVA region model (Figure S3 and Table 1). The univariate analyses (RF and ROC) clearly highlighted the value and importance of combining multiple analytical techniques to identify distinct regions of the metabolic fingerprint. In addition to the improved accuracy of predicting vineyard or AVA region membership, the univariate analyses corroborate that different combinations of features were required to accurately classify each vineyard or AVA region.
3.5. Vintage year analysis highlights the effects of microclimate on both vineyard and region classification.
Along with vineyard and AVA region classification, vintage year was also evaluated for a metabolic fingerprint that contributed to distinguishing the various Pinot noir wines. As our results indicate, we have found that the AVA region, and more importantly, the specific vineyard site, can significantly impact the metabolic fingerprint of the Pinot noir wines. It is evident from the PCA (and PCA-LDA) models (Figure 3 and Figure 4) that significant differences are present between the 2015 and 2016 vintages. The dendrograms show no consistent clustering patterns between the two vintage datasets. All attempts to create a unified multivariate model with the combined datasets proved ineffective. A detailed RF analysis of the entire 2015 and 2016 wine dataset further illustrated the uniqueness of the two vintages. Specifically, the 116 replicates (4 outliers were excluded) from 2015 and 120 replicates from 2016 were combined for a single RF analysis. The resulting RF model resulted in 97.4% and 100% classification accuracy for the 2015 and 2016 vintages, respectively. Again, this outcome suggests that the wine datasets can be readily classified according to vintage year alone. In a similar manner, a ROC curve was created from the entire 2015 and 2016 wine dataset. A resulting ROC curve consisting of 25 NMR spectroscopy and DS array features yielded an AUC of 0.88 for differentiating between the 2015 and 2016 vintage years (Figure S4). A putative annotation of the top ROC curve features revealed that almost all of the identified metabolites such as sugars, sugar alcohols, and TCA derivatives were decreased in 2015 compared to 2016 (Figure S4). Only one NMR feature was increased in 2015. Together, the entirety of the univariate and multivariate statistical analyses described above demonstrates that Pinot noir wines along the Pacific Coast of the United States can be distinguished by vintage year with a high level of accuracy.
4. Conclusions
Pinot noir wines from fifteen vineyards in eight wine-producing regions along the Pacific Coast of the United States were evaluated for metabolic features that classified the wines according to vineyard, region, and vintage year. A variety of biological or analytical techniques have previously been employed to distinguish between various types of wines. NMR, gas chromatography-MS, and various sensors have been used to identify unique fingerprints of European and American wines. The combination of such techniques has also been utilized to discriminate wine varietals from unique geographic regions (Duley et al., 2021; Kioroglou, Mas, & Portillo, 2020; Wu et al., 2019). Nevertheless, little attention has been paid to combining multiple analytical techniques to improve the classification accuracy of identical Pinot noir clones grown across distinct geographic locations. Toward this end, we report that the combination of untargeted metabolomics fingerprinting using 1D 1H NMR spectroscopy with a targeted analysis of phenolic profiles using a colorimetric DS peptide-based array has proven to be a highly effective approach to distinguish wines produced from genetically identical grapevines across vineyard location, geographic region, and vintage year. Our analysis highlights that targeted and untargeted techniques can be combined to successfully classify wine varietals solely based on geographic location and vintage year. In this study, Pinot noir wines were classified according to vineyard and AVA region with an accuracy of 0.96 ± 0.04. We have demonstrated through multivariate and univariate statistical techniques that the combination of NMR spectroscopy and DS array showed a marked improvement in distinguishing vineyards and regions that were at times difficult to verify by these techniques individually (Table 1). Together this data demonstrated that the combined analysis of both untargeted and targeted analytical techniques provides an improved and efficient method of wine variety verification by vineyard, region, and vintage year.
Supplementary Material
Acknowledgments
We would like to thank Dr. Martha Morton for her assistance with the NMR spectroscopy data collection of the wine samples and Dr. Thomas Payne for his contribution to the batch correction. This work was supported in part by funding from the Redox Biology Center (P30 GM103335, NIGMS), and the Nebraska Center for Integrated Biomolecular Communication (P20 GM113126, NIGMS). The research was performed in facilities renovated with support from the National Institutes of Health (RR015468-01). We also acknowledge the Freshman Research Initiative at UT Austin HHMI 52008124 (DZO), NSF 1212971 (EVA), and the Welch Reagents Chair F-0046 (EVA). The wine grapes and funding for winemaking at the UC Davis Teaching & Research Winery were provided by Jackson Family Wines (RCR). Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
The authors declare no competing interest.
References
- Amargianitaki M, & Spyros A (2017). NMR-based metabolomics in wine quality control and authentication. Chem. Biol. Technol. Agric, 4(1), 9. 10.1186/s40538-017-0092-x. [DOI] [Google Scholar]
- Baiano A, Terracone C, Longobardi F, Ventrella A, Agostiano A, & Del Nobile MA (2012). Effects of different vinification technologies on physical and chemical characteristics of Sauvignon blanc wines. Food Chem, 135(4), 2694–2701. 10.1016/j.foodchem.2012.07.075. [DOI] [PubMed] [Google Scholar]
- Bender M, Bojanowski NM, Seehafer K, & Bunz UHF (2018). Immobilized Poly(aryleneethynylene) pH Strips Discriminate Different Brands of Cola. Chemistry – A European Journal, 24(50), 13102–13105. 10.1002/chem.201803103. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Thorngate JH, Richardson PM, & Mills DA (2014). Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proceedings of the National Academy of Sciences, 111(1), E139. 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassino C, Tsolakis C, Bonello F, Gianotti V, & Osella D (2019). Wine evolution during bottle aging, studied by 1H NMR spectroscopy and multivariate statistical analysis. Food Res. Int, 116, 566–577. 10.1016/j.foodres.2018.08.075. [DOI] [PubMed] [Google Scholar]
- Chakraborty S (2019). Use of Partial Least Squares improves the efficacy of removing unwanted variability in differential expression analyses based on RNA-Seq data. Genomics, 111(4), 893–898. 10.1016/j.ygeno.2018.05.018. [DOI] [PubMed] [Google Scholar]
- Diehl KL, Ivy MA, Rabidoux S, Petry SM, Müller G, & Anslyn EV (2015). Differential sensing for the regio- and stereoselective identification and quantitation of glycerides. PNAS, 112(30), E3977–E3986. 10.1073/pnas.1508848112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duley G, Dujourdy L, Klein S, Werwein A, Spartz C, Gougeon RD, & Taylor DK (2021). Regionality in Australian Pinot noir wines: A study on the use of NMR and ICP-MS on commercial wines. Food Chemistry, 340, 127906. 10.1016/j.foodchem.2020.127906. [DOI] [PubMed] [Google Scholar]
- Dumitriu G-D, Peinado RA, Cotea VV, & López de Lerma N (2020). Volatilome fingerprint of red wines aged with chips or staves: Influence of the aging time and toasting degree. Food Chem, 310, 125801. 10.1016/j.foodchem.2019.125801. [DOI] [PubMed] [Google Scholar]
- Fan J, Upadhye S, & Worster A (2006). Understanding receiver operating characteristic (ROC) curves. J. Emerg. Med, 8(1), 19–20. 10.1017/S1481803500013336. [DOI] [PubMed] [Google Scholar]
- Fonville JM, Richards SE, Barton RH, Boulange CL, Ebbels TMD, Nicholson JK, … Dumas M-E (2010). The evolution of partial least squares models and related chemometric approaches in metabonomics and metabolic phenotyping. J. Chemom, 24(11‐12), 636–649. 10.1002/cem.1359. [DOI] [Google Scholar]
- Ghanem E, Hopfer H, Navarro A, Ritzer MS, Mahmood L, Fredell M, … Anslyn EV (2015). Predicting the composition of red wine blends using an array of multicomponent Peptide-based sensors. Molecules, 20(5), 9170–9182. 10.3390/molecules20059170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert JA, van der Lelie D, & Zarraonaindia I (2014). Microbial terroir for wine grapes. Proceedings of the National Academy of Sciences, 111(1), 5. 10.1073/pnas.1320471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godelmann R, Fang F, Humpfer E, Schütz B, Bansbach M, Schäfer H, & Spraul M (2013). Targeted and Nontargeted Wine Analysis by 1H NMR Spectroscopy Combined with Multivariate Statistical Analysis. Differentiation of Important Parameters: Grape Variety, Geographical Origin, Year of Vintage. J. Agric. Food Chem, 61(23), 5610–5619. 10.1021/jf400800d. [DOI] [PubMed] [Google Scholar]
- Gómez-Meire S, Campos C, Falqué E, Díaz F, & Fdez-Riverola F (2014). Assuring the authenticity of northwest Spain white wine varieties using machine learning techniques. Food Research International, 60, 230–240. 10.1016/j.foodres.2013.09.032. [DOI] [Google Scholar]
- Gougeon L, da Costa G, Guyon F, & Richard T (2019). 1H NMR metabolomics applied to Bordeaux red wines. Food Chem, 301, 125257. 10.1016/j.foodchem.2019.125257. [DOI] [PubMed] [Google Scholar]
- Grainger C, Yeh A, Byer S, Hjelmeland A, Lima MMM, & Runnebaum RC (2021). Vineyard site impact on the elemental composition of Pinot noir wines. Food Chemistry, 334, 127386. 10.1016/j.foodchem.2020.127386. [DOI] [PubMed] [Google Scholar]
- Hannah L, Roehrdanz PR, Ikegami M, Shepard AV, Shaw MR, Tabor G, … Hijmans RJ (2013). Climate change, wine, and conservation. Proceedings of the National Academy of Sciences, 110(17), 6907. 10.1073/pnas.1210127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera P, Durán-Guerrero E, Sánchez-Guillén MM, García-Moreno MV, Guillén DA, Barroso CG, & Castro R (2020). Effect of the type of wood used for ageing on the volatile composition of Pedro Ximénez sweet wine. J. Sci. Food Agric, 100(6), 2512–2521. 10.1002/jsfa.10276. [DOI] [PubMed] [Google Scholar]
- Hu B, Cao Y, Zhu J, Xu W, & Wu W (2019). Analysis of metabolites in chardonnay dry white wine with various inactive yeasts by (1)H NMR spectroscopy combined with pattern recognition analysis. AMB Express, 9(1), 140. 10.1186/s13568-019-0861-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Gao J, Xu S, Zhu J, Fan X, & Zhou X (2020). Quality evaluation of different varieties of dry red wine based on nuclear magnetic resonance metabolomics. Applied Biological Chemistry, 63(1), 24. 10.1186/s13765-020-00509-x. [DOI] [Google Scholar]
- Huang W, Seehafer K, & Bunz UHF (2019). Discrimination of Flavonoids by a Hypothesis Free Sensor Array. ACS Applied Polymer Materials, 1(6), 1301–1307. 10.1021/acsapm.9b00116. [DOI] [Google Scholar]
- Joydev Hatai CS (2020). Artificial Peptide and Protein Receptors. In Supramolecular Chemistry in Water (pp. 79–113). [Google Scholar]
- Kioroglou D, Mas A, & Portillo MC (2020). Qualitative Factor-Based Comparison of NMR, Targeted and Untargeted GC-MS and LC-MS on the Metabolomic Profiles of Rioja and Priorat Red Wines. Foods, 9(10). 10.3390/foods9101381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Hwang G-S, Berg F, Lee C-H, & Hong Y-S (2009). Evidence of vintage effects on grape wines using H-1 NMR-based metabolomic study. Anal. Chim. Acta 648, 71–76. 10.1016/j.aca.2009.06.039. [DOI] [PubMed] [Google Scholar]
- Li X, Zamora-Olivares D, Diehl KL, Tian W, & Anslyn EV (2017). Differential sensing of oils by conjugates of serum albumins and 9,10-distyrylanthracene probes: a cautionary tale. Supramolecular Chemistry, 29(4), 308–314. 10.1080/10610278.2016.1228934. [DOI] [Google Scholar]
- Liaw A, & Wiener M (2002). Classification and Regression by randomForest. R News, 2, 18–22. [Google Scholar]
- Lo Y-C, Rensi SE, Torng W, & Altman RB (2018). Machine learning in chemoinformatics and drug discovery. Drug Discovery Today, 23(8), 1538–1546. 10.1016/j.drudis.2018.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall DD, Lei S, Worley B, Huang Y, Garcia-Garcia A, Franco R, … Powers R (2015). Combining DI-ESI-MS and NMR datasets for metabolic profiling. Metabolomics, 11(2), 391–402. 10.1007/s11306-014-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascellani A, Hoca G, Babisz M, Krska P, Kloucek P, & Havlik J (2021). 1H NMR chemometric models for classification of Czech wine type and variety. Food Chemistry, 339, 127852. 10.1016/j.foodchem.2020.127852. [DOI] [PubMed] [Google Scholar]
- McGovern P, Jalabadze M, Batiuk S, Callahan MP, Smith KE, Hall GR, … Lordkipanidze D (2017). Early Neolithic wine of Georgia in the South Caucasus. Proceedings of the National Academy of Sciences, 114(48), E10309. 10.1073/pnas.1714728114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen BT, & Anslyn EV (2006). Indicator Displacement Assays. Coord. Chem. Rev, 250(23–24), 3118–3127. 10.1016/j.ccr.2006.04.009. [DOI] [Google Scholar]
- Nomikos P, & MacGregor JF (1995). Multi-way partial least squares in monitoring batch processes. Chemometrics and Intelligent Laboratory Systems, 30(1), 97–108. 10.1016/0169-7439(95)00043-7. [DOI] [Google Scholar]
- Patwardhan NN, Cai Z, Newson CN, & Hargrove AE (2019). Fluorescent peptide displacement as a general assay for screening small molecule libraries against RNA. Org. Biomol. Chem, 17(7), 1778–1786. 10.1039/C8OB02467G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira GE, Gaudillere J-P, Van Leeuwen C, Hilbert G, Lavialle O, Maucourt M, … Rolin D (2005). 1H NMR and Chemometrics To Characterize Mature Grape Berries in Four Wine-Growing Areas in Bordeaux, France. J. Agric. Food Chem, 53(16), 6382–6389. 10.1021/jf058058q. [DOI] [PubMed] [Google Scholar]
- Smith RJ (2003). Wine Grape Varieties in California. (1st ed.). University of California Agriculture and Natural Resources. [Google Scholar]
- Stewart S, Ivy MA, & Anslyn EV (2014). The use of principal component analysis and discriminant analysis in differential sensing routines. Chem. Soc. Rev, 43(1), 70–84. 10.1039/c3cs60183h. [DOI] [PubMed] [Google Scholar]
- Umali AP, & Anslyn EV (2010). A General Approach to Differential Sensing Using Synthetic Molecular Receptors. Curr. Opin. Chem. Biol, 14(6), 685–692. 10.1016/j.cbpa.2010.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umali AP, Ghanem E, Hopfer H, Hussain A, Kao Y, Zabanal LG, … Anslyn EV (2015). Grape and wine sensory attributes correlate with pattern-based discrimination of Cabernet Sauvignon wines by a peptidic sensor array. Tetrahedron, 71(20), 3095–3099. 10.1016/j.tet.2014.09.062. [DOI] [Google Scholar]
- Umali AP, LeBoeuf SE, Newberry RW, Kim S, Tran L, Rome WA, … Anslyn EV (2011). Discrimination of flavonoids and red wine varietals by arrays of differential peptidic sensors. Chemical Science, 2(3), 439–445. 10.1039/c0sc00487a. [DOI] [Google Scholar]
- Van Leeuwen C, & Seguin G (2006). The concept of terroir in viticulture. Journal of Wine Research, 17(1), 1–10. 10.1080/09571260600633135. [DOI] [Google Scholar]
- Wang B, Han J, Zhang H, Bender M, Biella A, Seehafer K, & Bunz UHF (2018). Detecting Counterfeit Brandies. Chemistry – A European Journal, 24(65), 17361–17366. 10.1002/chem.201804607. [DOI] [PubMed] [Google Scholar]
- Worley B, Halouska S, & Powers R (2013). Utilities for quantifying separation in PCA/PLS-DA scores plots. Anal Biochem, 433(2), 102–104. 10.1016/j.ab.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley B, & Powers R (2013). Multivariate Analysis in Metabolomics. Curr Metabolomics, 1(1), 92–107. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley B, & Powers R (2014). MVAPACK: A Complete Data Handling Package for NMR Metabolomics. ACS Chem. Biol, 9(5), 1138–1144. 10.1021/cb4008937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worley B, & Powers R (2015). Generalized adaptive intelligent binning of multiway data. Chemom. Intell. Lab. Syst, 146, 42–46. 10.1016/j.chemolab.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Tian L, Chen B, Jin B, Tian B, Xie L, … Lin G (2019). Verification of imported red wine origin into China using multi isotope and elemental analyses. Food Chemistry, 301, 125137. 10.1016/j.foodchem.2019.125137. [DOI] [PubMed] [Google Scholar]
- Xia J, Sinelnikov IV, Han B, & Wishart DS (2015). MetaboAnalyst 3.0--making metabolomics more meaningful. Nucleic Acids Res, 43(W1), W251–257. 10.1093/nar/gkv380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamora-Olivares D, Kaoud TS, Jose J, Ellington A, Dalby KN, & Anslyn EV (2014). Differential Sensing of MAP Kinases Using SOX-Peptides. Angew. Chem. Int. Ed, 53(51), 14064–14068. 10.1002/anie.201408256. [DOI] [PubMed] [Google Scholar]
- Zamora-Olivares D, Kaoud TS, Zeng L, Pridgen JR, Zhuang DL, Ekpo YE, … Dalby KN. (2020). Quantification of ERK Kinase Activity in Biological Samples Using Differential Sensing. ACS Chem. Biol, 15(1), 83–92. 10.1021/acschembio.9b00580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


