Summary
In recent decades, incidence and severity of Clostridioides difficile infection (CDI) has increased dramatically, coinciding with the emergence of hypervirulent strains such as PCR ribotype 027 (RT027). Data on prevalence of distinct C. difficile strains in random CDI cases in Germany are scarce. The aim of this review was to obtain an overview of prevalence and geographical distribution of RT027 among clinical C. difficile isolates from random cases in non-outbreak settings in hospitals in Germany. For this purpose, we performed a literature review on reported cases of C. difficile RT027 in Germany between 2007 and 2019 in three databases (PubMed, Embase and LIVIVO) and conference proceedings. Studies with selection bias for RT027 (e.g. clinical severity, outbreak reports) were excluded. A total of 304 records were screened, from which 21 were included in this analysis. The nationwide prevalence of RT027 in Germany was <1% prior to 2010 but increased continuously thereafter, reaching 21.7% in 2013. The regional prevalence varied markedly between federal states, higher prevalence was reported from North Rhine-Westphalia (37.4%) and Saxony (31.8%) in 2013-2015. However, data on C. difficile RT027 were not available from almost half of the federal states and were scarce at the national level. Our data suggest a remarkable spread of RT027 in Germany during the past decade, which has remained rather unnoticed so far. A national program for molecular surveillance of C. difficile is required to monitor the changing epidemiology of CDI and to adjust the prevention and control measures.
Keywords: Clostridioides difficile, C. difficile infection, PCR ribotype 027, Prevalence, Enhanced surveillance, Germany
Introduction
Clostridioides difficile (C. difficile) is a Gram-positive, anaerobic, spore-forming, toxin-producing bacillus [1]. The bacterium is considered the most frequent cause of antibiotic-associated colitis and healthcare-acquired diarrhoea in developed countries [2]. The disease spectrum is wide and ranges from mild diarrhoea to severe infection [3]. C. difficile pathogenicity is principally mediated by two exotoxins: toxin A (TcdA) and toxin B (TcdB) [4]. C. difficile infection (CDI) primarily affects elderly patients with comorbidities and recent exposure to antibiotics, thereby having major clinical impact on this patient group. Other risk factors for CDI include a compromised immune system, recent stay in a hospital or long-term care facility, and use of acid-suppressive medications [5]. Although CDI is usually associated with hospitalisation, community-acquired CDI have been gaining relevance in recent years [6].
In Germany, CDI was identified as the fourth most commonly diagnosed healthcare-associated infection (HAI) in the national point prevalence survey (PPS) of HAI and antimicrobial use in acute care hospitals in 2016, accounting for 10.0 % of all HAI in the participating hospitals [7]. This rate was twice as high as the average rate (4.8 %) of all countries from the European Union (EU) and European Economic Area (EEA) participating in the EU-wide PPS 2016–2017 organised by the European Centre for Disease Prevention and Control (ECDC) [8]. Furthermore, the prevalence of CDI in German hospitals participating in the PPS in 2016 was significantly higher than the prevalence in 2011 (0.48% vs. 0.34% [7]. As to the burden of HAI in Europe, Cassini et al. estimated a median of 1.7 disability-adjusted life years (DALYs) per case of CDI, which is higher than the burden of other HAI such as urinary tract infection, and almost as high as that of pneumonia in this study [9].
The global increase in incidence and severity of CDI over the last decade is linked to the emergence of certain lineages, including the epidemic PCR ribotype 027 [10]. RT027 has been reported as the most predominant PCR ribotype in Canada, the United States and some countries in Central and South America [[10], [11], [12]]. In Europe, outbreaks of RT027 were reported from different countries mostly in 2005–2007 but also in the following years [[13], [14], [15], [16], [17]]. Studies on C. difficile epidemiology have revealed an inhomogeneous distribution of RT027 between different European regions [[18], [19], [20], [21], [22]]. The proportion of PCR ribotype 027 isolates correlated with the incidence rate in some studies [23]. Rapid emergence and transcontinental spread of RT027 strains occurred through at least two distinct fluoroquinolone-resistant lineages [24,25]. Moreover, it has been postulated that RT027 global dissemination is still ongoing [26].
In Germany, a hospital outbreak of the C. difficile RT027 was reported in 2007 in the south-western state of Rhineland-Palatinate [14]. Mandatory reporting of severe cases of CDI (including detection of RT027 isolates) was introduced as a consequence thereafter [27]. However, since an infection by RT027 does not indicate a severe clinical course per se, and because ribotyping data were not available from most cases of CDI, this criterion was later removed in 2016 [28]. According to the German Infection Protection Act (Infektionsschutzgesetz), severe CDI and CDI outbreaks are subject to mandatory reporting, but strain characterisation is not compulsory. Since 2007 severe cases and/or outbreaks of RT027 have been reported from different geographical regions of Germany [29,30]. However, there is no nationwide enhanced surveillance program in place to analyse the circulating strains and ribotypes. The epidemiology of CDI and the prevalence and distribution of RT027 in particular are therefore not well-understood. The aim of this study was to assess published data on the prevalence, distribution, and temporal evolution of C. difficile RT027 in Germany since 2007.
Methods
Search strategy
We searched electronic databases (PubMed, EMBASE and LIVIVO) using a combination of controlled vocabulary and free text terms (Clostridium difficile, Clostridioides difficile, ribotyping, typing, prevalence, occurrence, Germany) for articles published between 2007 and February 2019 with no language restrictions (see supplementary online material for further details). Conference proceedings were also checked for additional studies.
Eligibility criteria
-
-Inclusion criteria:
-
•Publications reporting molecular typing results for C. difficile isolates obtained from patients with the diagnosis of CDI in hospitals or ambulant settings in Germany;
-
•Observational studies, single centre or multicentre, published between 2007 and February 2019.
-
•
-
-Exclusion criteria:
-
•Studies reporting data on outbreaks (either exclusively or mixed);
-
•Studies with overrepresentation of severe disease and/or mandatorily reported cases;
-
•Studies reporting data from asymptomatic individuals.
-
•
Data collection and analysis
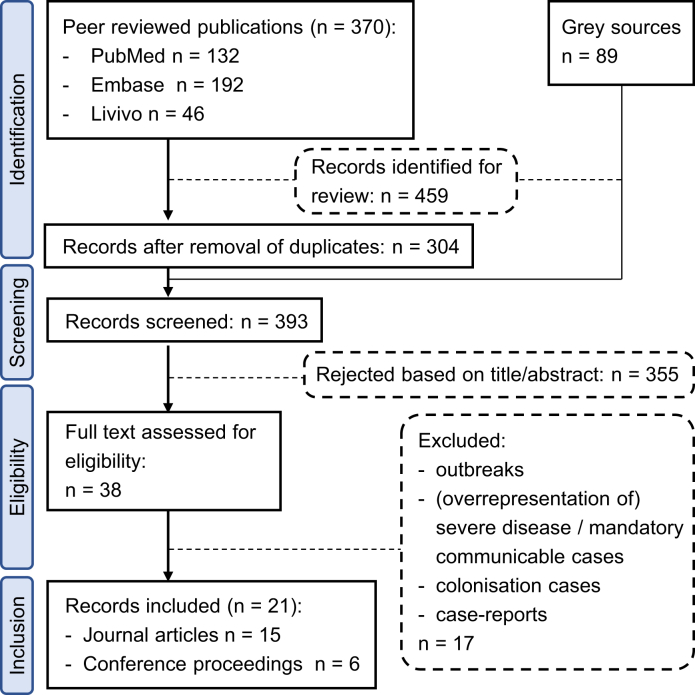
We scanned the titles and abstracts of all initially identified publications. When this was insufficient to rule out eligibility, the full text was obtained. The flow diagram of the study selection process is shown in Figure 1. The following data were retrieved from each study: geographical region, time and duration of the study, total number of isolates with ribotyping results, and total number or percentage of identified RT027 isolates. Study authors were contacted for additional information. When data for the same region and time period were reported in more than one publication, peer-reviewed scientific publications were preferred over conference proceedings.
Figure 1.
Flow diagram of studies included in this analysis.
Statistical analysis
The trend of the RT027 prevalence in Germany between 2005 and 2017 (study time of the included publications) was analysed by a Mann-Kendall test [31] using the Python “mkt” module [32]. The alternative hypothesis of existence of a monotonic upwards trend was tested. For years with RT027 prevalence reported by multiple studies, the mean prevalence of each year, weighted by number of isolates analysed, was used.
Results
Included studies
The literature search identified 459 articles and abstracts from conference proceedings, 21 of which fulfilled the inclusion criteria (15 peer-reviewed journal papers and six abstracts) (Figure 1). From the 21 eligible publications, 15 reported regional or local (single centre) data. Among these, six studies analysed data from 2007 until 2010, whilst nine described data between 2011 and 2017. In addition, we found six publications (corresponding to five studies) that examined national data, mainly in the framework of pan-European investigations such as the ClosER or EUCLID study [[19], [20], [21],33].
Distribution and prevalence of C. difficile RT027 at the regional level
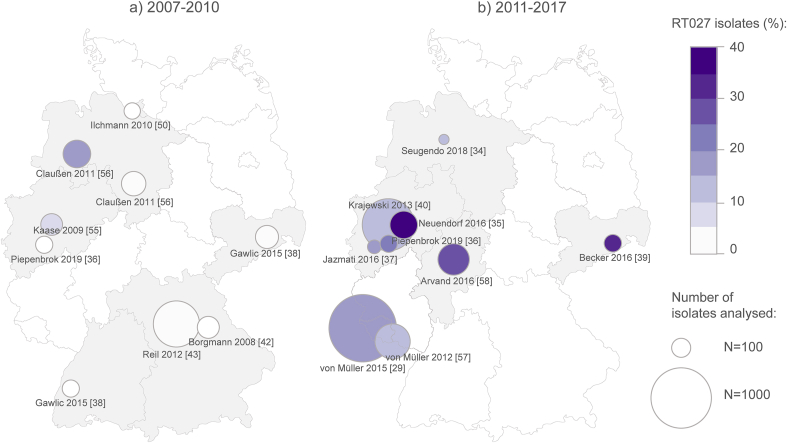
In the regional/local studies, the proportion of RT027 among all C. difficile isolates varied markedly between different regions and different time periods, ranging from zero to 37.4% (Table I). While the prevalence of RT027 was generally low in regional/local studies between 2007 and 2010 (mean = 3.3%), it increased markedly to a mean value of 21.3% in the following years until 2017. Between 2011 and 2017 the prevalence of RT027 ranged between a minimum of 10.3% in Lower Saxony [34] and a maximum of 37.4% in Düsseldorf [35]. A relatively high prevalence was reported mainly from south-west and central Germany, the highest rate being 37.4% reported from Düsseldorf in the State of North Rhine-Westphalia. However, ribotyping data were not available from many regions, particularly from north, north-east and south Germany. Data from different time points were rarely available from the same region or centre. Yet, the few available publications showed a steady increase in prevalence of RT027 over time, i.e. in Cologne from 0% in 2007 to 17.3% in 2014–2015 and 21.3% in 2017 [36,37], in Dresden from 1.4% in 2007–2009 to 31.8% in 2014–2015 [38,39], and in Düsseldorf, from 13.7% in 2010–2012 to 37.4% in 2013–2014 [35,40] (Figure 2).
Table I.
Regional distribution and prevalence of C. difficile PCR ribotype 027 in Germany, 2006–2017
| First author | Publication year | Geographical scope | Time of study | RT027 (n/N) | RT027 (%) |
|---|---|---|---|---|---|
| Borgmann [42] | 2008 | Bavaria (north) | 2006–2007 | 0/135 | 0.0 |
| Piepenbrock [36] | 2019 | Cologne | 2007 | 0/80 | 0.0 |
| Gawlic [38] | 2015 | Freiburg | 2007–2009 | 0/80 | 0.0 |
| Gawlic [38] | 2015 | Dresden | 2007–2009 | 2/147 | 1.4 |
| Ilchmann [50] | 2010 | Hamburg | 2008 | 0/73 | 0.0 |
| Kaase [55] | 2009 | Bochum | 2008 | 9/130 | 6.9 |
| Reil [43] | 2012 | Bavaria (north) | 2009 | 27/587 | 4.6 |
| Claußen [56] | 2011 | Lower Saxony (southwest) | 2009–2010 | 35/212 | 16.5 |
| Claußen [56] | 2011 | Lower Saxony (south) | 2010 | 1/163 | 0.6 |
| Krajewski [40] | 2013 | Düsseldorf | 2010–2012 | 103/750 | 13.7 |
| von Müller [57] | 2012 | Saarland | 2011–2012 | 50/338 | 14.8 |
| von Müller [29] | 2015 | Saarland | 2008–2013 | 231/1 253 | 18.4 |
| Arvand [58] | 2016 | Hesse | 2011–2014 | 73/270 | 27.0 |
| Seugendo [34] | 2018 | Lower Saxony | 2013–2014 | 3/29 | 10.3 |
| Neuendorf [35] | 2016 | Düsseldorf | 2013–2014 | 79/211 | 37.4 |
| Becker [39] | 2016 | Dresden | 2014–2015 | 27/85 | 31.8 |
| Jazmati [37] | 2016 | Cologne | 2014–2015 | 9/52 | 17.3 |
| Piepenbrock [36] | 2019 | Cologne | 2017 | 17/80 | 21.3 |
n - number of analysed isolates identified as RT027; N - total number of isolates analysed.
Figure 2.
Geographical distribution and prevalence of C. difficile RT027 in Germany in different time periods: a) 2007–2010 and b) 2011–2017 (date of collection of isolates). Marked in grey are federal states from which data is available. The circles illustrate occurrence of RT027. The circle size is proportional to the number of isolates analysed, while the colour reflects the proportion of RT027 among typed isolates. Studies are identified by author name and publication year (see Table I).
Distribution and prevalence of C. difficile RT027 at the national level
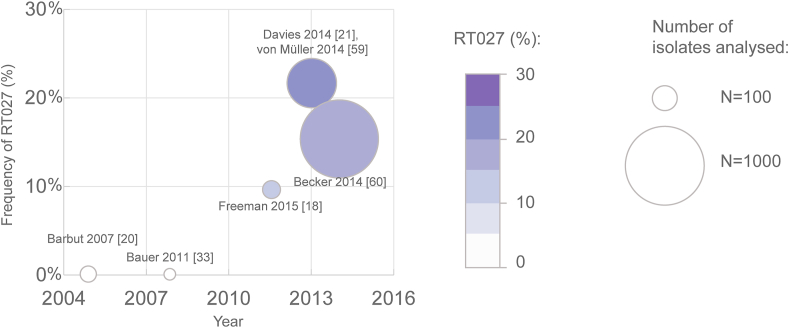
Data were scarce on the national level (Table II). Four out of five national-level studies were performed in the context of pan-European investigations, e.g. the EUCLID study [21]. Whereas no RT027 cases were detected among isolates collected in 2005 and 2008, RT027 accounted for 9.6% of German isolates in 2011–2012, and for 21.7% and 15.8% in studies from 2013 and 2014, respectively (Figure 3).
Table II.
National prevalence of C. difficile PCR ribotype 027 in Germany, 2005–2014
| First author | Publication year | Geographical scope | Time of study | RT027 (n/N) | RT027 (%) |
|---|---|---|---|---|---|
| Barbut [20] | 2007 | Not specified | 2005 | 0/42 | 0.0 |
| Bauer [33] | 2011 | Not specified | 2008 | 0/22 | 0.0 |
| Freeman [18] | 2015 | Not specified (3 sites) | 2011–2012 | 5/52 | 9.6 |
| Davies [21], von Müller [59] | 2014 | All states | 2013 | 86/396 | 21.7 |
| Becker [60] | 2014 | Several states | 2014 | 156/988 | 15.8 |
n –number of analysed isolates identified as RT027; N - total number of isolates analysed.
Figure 3.
Prevalence of C. difficile RT027 in Germany in 2005–2014 (date of collection of isolates). The circles illustrate occurrence of RT027. The circle size is proportional to the number of isolates analysed, while the colour reflects the proportion of RT027 among typed isolates. Studies are identified by author name and publication year (see Table II).
Prevalence of other C. difficile ribotypes
Altogether, 30 different PCR ribotypes were reported between 2005 and 2017 (Tables S1-S6 of the supplementary online material). Among these ribotypes, those with the highest frequency were RT001 (23.0%), RT027 (14.8%), and RT014 (6.9%).
Statistical analysis
Statistical analysis revealed a consistent increase in RT027 prevalence over time (p=0.0002). On the contrary, the prevalence of the other two most frequently detected ribotypes, RT001 and RT014, did not increase during the study period (p=0.759 and p=0.241, respectively).
Discussion
Key findings
Our study provides the first systematic overview of the prevalence and distribution of C. difficile RT027 in Germany in a non-epidemic setting primarily in hospitals in Germany. It identified a remarkable and largely unperceived increase and regional spread of RT027 during the past decade, reaching prevalence rates of >30% in different federal states in 2014–15.
The prevalence was higher in some central regions and in the south-west; however, there was a data gap especially in north, north-east and south Germany. Regionally, ribotyping data were not available from approximately half of the federal states. Among those federal states with available data, the highest prevalence was reported from North Rhine-Westphalia in 2013–2014 and Saxony in 2014–2015, with 37.4% and 31.8%, respectively [35,39]. These findings suggest that RT027 has become endemic in many hospitals in different regions of Germany.
In this work we chose to exclude studies reporting data on outbreaks or severe clinical manifestation in order to avoid selection bias towards a possible overrepresentation of hypervirulent strains. Interestingly, our results focusing on random cases and non-outbreak settings are in line with the results of the National Advisory Laboratory for C. difficile, which showed that RT027 was the second most prevalent ribotype among isolates submitted there between 2011 and 2013 for ribotyping, mainly due to severe clinical disease, recurrence or outbreaks [29].
In our study, we found a remarkable disparity with regard to the prevalence of RT027 between different regions and even within the same region. Data from the same region or centre to different time points were rarely available. However, the few available publications revealed a steadily increasing prevalence of RT027 over time. This finding points towards possible inter- and intra-hospital transmission in some cases. This is in line with previous observations that fluoroquinolone-resistant RT027 strains were imported into Germany at least four times, and that RT027 had been widely disseminated across multiple federal states before the first outbreak was noted in 2007 in south-west Germany [25]. This is in turn in accordance with the data of Eyre et al. suggesting within-country clustering of RT027 in Germany [41]. Furthermore, these authors showed that RT027 clustering also occurred regionally and within-hospitals and found a strong association between clustering and fluoroquinolone resistance [41].
The data included in this review are mainly obtained from hospitalised patients. All studies included either exclusively inpatient samples or a mix of in- and outpatient samples, with the majority being inpatients. However, only two studies provided clear information on this topic [42,43], both of which including > 80% inpatient samples. It is therefore difficult to draw conclusions on the prevalence and circulation of RT027 in the community setting in Germany, yet it would be interesting to assess this in further studies.
Implications
Standardised CDI diagnostics is one of the key components of surveillance. The EUCLID study provided evidence that CDI was underdiagnosed in Europe, probably because of low awareness of the indications and requirements for C. difficile testing among physicians [21]. The high prevalence of CDI in Germany suggests the need for an increased awareness of clinicians of CDI as well as of the spread of epidemic strains in German hospitals. Since testing for C. difficile is not routinely included in the laboratory diagnostic workup for diarrhoeal samples of hospitalized patients in Germany, a test for C. difficile needs to be actively requested when the disease is clinically suspected. In addition, the detection of clusters and outbreaks requires established surveillance, including knowledge of the department-specific frequency of CDI as well as of the prevalence of different ribotypes (e.g. by using the enhanced surveillance protocol). According to the German Infection Protection Act and the German national guidelines on outbreak management and on prevention and control of CDI, a nosocomial outbreak is defined as two or more nosocomial infections for which an epidemiological link is likely or suspected [44]. In case of high CDI incidence or suspicion of nosocomial outbreak, typing of isolates is recommended [45].
Since microbiological culture and characterisation of C. difficile strains are not performed on a regular or systematic basis in Germany, there is currently a lack of information on molecular epidemiology of CDI in the country. At the European level, a CDI surveillance system was in place in 20 countries in 2017 and 21 countries (70% of all EU/EEA countries) participated in ECDC-coordinated CDI surveillance in acute care hospitals in 2016, using a common protocol and thus allowing data comparison [46,47]. The ECDC surveillance protocol allows three options for data collection: ‘minimal’ (aggregated hospital data); ‘light’ (including patient data such as mortality) or ‘enhanced’ (including case-based microbiological data) [48]. Germany did not participate in 2016, but tested the pilot protocol in 2013 [23]. Some countries (e.g. Finland, the Netherlands, and the United Kingdom) performed enhanced surveillance of CDI. Enhanced surveillance supports early recognition of outbreaks due to new C. difficile ribotypes and the distribution of certain ribotypes in specific populations, and also a faster identification of new strains associated with increased morbidity or mortality, facilitating initiation of appropriate control measures [47]. Appropriate microbiological diagnosis and participation in epidemiological surveillance are two pillars of the prevention and control of CDI in healthcare facilities [49]. In conclusion, it would be desirable for Germany to carry out enhanced surveillance in the future.
Limitations
Our study also has some limitations. Firstly, the included studies were heterogeneous with respect to the study design, sampling strategy and sample size. In addition, not all studies aimed to evaluate rates of C. difficile infection and/or classify strains; some were designed to investigate diagnostic tools [35,38] or antimicrobial resistance [18,50]. Nonetheless, these studies were included whenever it was possible to assess the rate of RT027 CDI. Moreover, information on indication for C. difficile testing, definition of diarrhoea or classification of diarrheic stools at the laboratory level was not available in all studies included in this review. With regard to sampling, although C. difficile isolates were collected as part of routine microbiological diagnostics in most studies, a few studies also included samples submitted for typing due to severity of disease. Finally, the sample size was small in most studies, with a median of 135 and a range of 22–1253. Thus, the representativeness of the results is compromised.
Furthermore, different typing methods were used in different studies, which may have had an impact on the ribotyping results [51]. For example, some ribotypes closely related to RT027 (for instance, RT176) may be difficult to distinguish and hence be falsely classified [52,53]. In addition, inter-laboratory standardization is difficult to achieve. Although PCR ribotyping was indeed the most frequent typing method, in line with the European harmonized diagnostic procedures [54], even with this method it is difficult to compare data between laboratories without reference strains [51].
On the other hand, our review has likely identified all available studies on the topic, since we searched three different databases with broad search criteria, complemented by the inclusion of grey literature. We further tried to avoid bias by defining inclusion criteria clearly and thoroughly. Secondly, in the case of studies with partially overlapping investigation periods, only the study with the longer duration (and therefore more samples) was included in order to avoid overrepresentation data.
Conclusions
In summary, this paper provides the first systematic overview of the prevalence and temporal development of C. difficile RT027 among random CDI cases in non-epidemic settings in German hospitals. The results of our study further emphasize the need for a nationwide program for enhanced surveillance of CDI and to monitor the changing epidemiology especially with regard to the nosocomial spread of epidemic strains. These data may help to better adjust the prevention and control strategies in order to reduce the incidence of CDI in Germany.
Author contributions
Writing – Original Draft: V.M.; Writing – Review & Editing: V.M. and M.A.; Conceptualization – M.A.; Methodology – V.M.; Formal Analysis – V.M.; Investigation: V.M.; Data curation – V.M. and M.A.; Visualization – V.M.; Supervision – M.A.
Acknowledgements
The authors are grateful to Dr Esmeralda Valiente for critical review of the manuscript, Dr Melanie Brunke for the idea for the design of the figures, Mandy Krüpfganz for bibliographic assistance, Genevieve Sohl for proofreading and Dr Jordi Casanellas and Angelika Schaffrath Rosario for statistical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.infpip.2020.100102.
Conflict of interest statement
None.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Lawson P.A., Citron D.M., Tyrrell K.L., Finegold S.M. Reclassification of Clostridium difficile as clostridioides difficile (Hall and O'Toole 1935) Prevot 1938. Anaerobe. 2016;40:95–99. doi: 10.1016/j.anaerobe.2016.06.008. [DOI] [PubMed] [Google Scholar]
- 2.Nasiri M.J., Goudarzi M., Hajikhani B., Ghazi M., Goudarzi H., Pouriran R. Clostridioides (Clostridium) difficile infection in hospitalized patients with antibiotic-associated diarrhea: a systematic review and meta-analysis. Anaerobe. 2018;50:32–37. doi: 10.1016/j.anaerobe.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Usacheva E.A., Jin J.P., Peterson L.R. Host response to Clostridium difficile infection: diagnostics and detection. J Glob Antimicrob Resist. 2016;7:93–101. doi: 10.1016/j.jgar.2016.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuehne S.A., Collery M.M., Kelly M.L., Cartman S.T., Cockayne A., Minton N.P. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deneve C., Janoir C., Poilane I., Fantinato C., Collignon A. New trends in Clostridium difficile virulence and pathogenesis. Int J Antimicrob Agents. 2009;33:S24–S28. doi: 10.1016/S0924-8579(09)70012-3. [DOI] [PubMed] [Google Scholar]
- 6.Kuntz J.L., Chrischilles E.A., Pendergast J.F., Herwaldt L.A., Polgreen P.M. Incidence of and risk factors for community-associated Clostridium difficile infection: a nested case-control study. BMC Infect Dis. 2011;11:194. doi: 10.1186/1471-2334-11-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nationales Referenzzentrum (NRZ) für Surveillance von nosokomialen Infektionen . Abschlussbericht; 2017. Deutsche nationale Punkt-Prävalenzerhebung zu nosokomialen Infektionen und Antibiotika-Anwendung 2016.http://www.nrz-hygiene.de/fileadmin/nrz/download/pps2016/PPS_2016_Abschlussbericht_20.07.2017.pdf Available at: [Google Scholar]
- 8.Suetens C., Latour K., Kärki T., Ricchizzi E., Kinross P., Moro M.L. Prevalence of healthcare-associated infections, estimated incidence and composite antimicrobial resistance index in acute care hospitals and long-term care facilities: results from two European point prevalence surveys, 2016 to 2017. Euro Surveill. 2018;23:1800516. doi: 10.2807/1560-7917.ES.2018.23.46.1800516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cassini A., Plachouras D., Eckmanns T., Abu Sin M., Blank H.P., Ducomble T. Burden of Six Healthcare-Associated Infections on European Population Health: Estimating Incidence-Based Disability-Adjusted Life Years through a Population Prevalence-Based Modelling Study. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freeman J., Bauer M.P., Baines S.D., Corver J., Fawley W.N., Goorhuis B. The Changing Epidemiology of Clostridium difficile Infections. Clin Microbiol Rev. 2010;23:529–549. doi: 10.1128/cmr.00082-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tamez-Torres K.M., Torres-Gonzalez P., Leal-Vega F., García-Alderete A., López García N.I., Mendoza-Aguilar R. Impact of Clostridium difficile infection caused by the NAP1/RT027 strain on severity and recurrence during an outbreak and transition to endemicity in a Mexican tertiary care center. Int J Infect Dis. 2017;65:44–49. doi: 10.1016/j.ijid.2017.09.022. [DOI] [PubMed] [Google Scholar]
- 12.Aguayo C., Flores R., Levesque S., Araya P., Ulloa S., Lagos J. Rapid spread of Clostridium difficile NAP1/027/ST1 in Chile confirms the emergence of the epidemic strain in Latin America. Epidemiol Infect. 2015;143:3069–3073. doi: 10.1017/s0950268815000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuijper E.J., Coignard B., Tull P. Emergence of Clostridium difficile-associated disease in North America and Europe. Clin Microbiol Infect. 2006;12:2–18. doi: 10.1111/j.1469-0691.2006.01580.x. [DOI] [PubMed] [Google Scholar]
- 14.Jansen A., Kleinkauf N., Weiss B., Zaiss N.H., Witte W., Bornhofen B. [Emergence of clostridium difficile ribotype 027 in Germany: epidemiological and clinical characteristics] Z Gastroenterol. 2010;48:1120–1125. doi: 10.1055/s-0029-1245269. [DOI] [PubMed] [Google Scholar]
- 15.Birgand G., Blanckaert K., Carbonne A., Coignard B., Barbut F., Eckert C. Investigation of a large outbreak of Clostridium difficile PCR-ribotype 027 infections in northern France, 2006-2007 and associated clusters in 2008-2009. Euro Surveill. 2010;15:19597. doi: 10.2807/ese.15.25.19597-en. [DOI] [PubMed] [Google Scholar]
- 16.Indra A., Huhulescu S., Fiedler A., Kernbichler S., Blaschitz M., Allerberger F. Outbreak of Clostridium difficile 027 infection in Vienna, Austria 2008-2009. Euro Surveill. 2009;14:19186. doi: 10.2807/ese.14.17.19186-en. [DOI] [PubMed] [Google Scholar]
- 17.Oleastro M., Coelho M., Gião M., Coutinho S., Mota S., Santos A. Outbreak of Clostridium difficile PCR ribotype 027 - the recent experience of a regional hospital. BMC Infect Dis. 2014;14:209. doi: 10.1186/1471-2334-14-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freeman J., Vernon J., Morris K., Nicholson S., Todhunter S., Longshaw C. Pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin Microbiol Infect. 2015;21 doi: 10.1016/j.cmi.2014.09.017. 248.e9-16. [DOI] [PubMed] [Google Scholar]
- 19.Freeman J., Vernon J., Pilling S., Morris K., Nicholson S., Shearman S. The ClosER study: results from a three-year pan-European longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes, 2011-2014. Clin Microbiol Infect. 2018;24:724–731. doi: 10.1016/j.cmi.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 20.Barbut F., Mastrantonio P., Delmée M., Brazier J., Kuijper E., Poxton I. Prospective study of Clostridium difficile infections in Europe with phenotypic and genotypic characterisation of the isolates. Clin Microbiol Infect. 2007;13:1048–1057. doi: 10.1111/j.1469-0691.2007.01824.x. [DOI] [PubMed] [Google Scholar]
- 21.Davies K.A., Longshaw C.M., Davis G.L., Bouza E., Barbut F., Barna Z. Underdiagnosis of Clostridium difficile across Europe: the European, multicentre, prospective, biannual, point-prevalence study of Clostridium difficile infection in hospitalised patients with diarrhoea (EUCLID) Lancet Infect Dis. 2014;14:1208–1219. doi: 10.1016/S1473-3099(14)70991-0. [DOI] [PubMed] [Google Scholar]
- 22.Kuijper E.J., Barbut F., Brazier J.S., Kleinkauf N., Eckmanns T., Lambert M.L. Update of Clostridium difficile infection due to PCR ribotype 027 in Europe, 2008. Euro Surveill. 2008;13:18942. doi: 10.2807/ese.13.31.18942-en. [DOI] [PubMed] [Google Scholar]
- 23.van Dorp S.M., Kinross P., Gastmeier P., Behnke M., Kola A., Delmée M. Standardised surveillance of Clostridium difficile infection in European acute care hospitals: a pilot study, 2013. Euro Surveill. 2016;21:30293. doi: 10.2807/1560-7917.ES.2016.21.29.30293. [DOI] [PubMed] [Google Scholar]
- 24.He M., Miyajima F., Roberts P., Ellison L., Pickard D.J., Martin M.J. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet. 2013;45:109–113. doi: 10.1038/ng.2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Steglich M., Nitsche A., von Muller L., Herrmann M., Kohl T.A., Niemann S. Tracing the Spread of Clostridium difficile Ribotype 027 in Germany Based on Bacterial Genome Sequences. PLoS One. 2015;10 doi: 10.1371/journal.pone.0139811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valiente E., Cairns M.D., Wren B.W. The Clostridium difficile PCR ribotype 027 lineage: a pathogen on the move. Clin Microbiol Infect. 2014;20:396–404. doi: 10.1111/1469-0691.12619. [DOI] [PubMed] [Google Scholar]
- 27.Robert Koch-Institut (RKI) Schwer verlaufende Clostridium-difficile-Infektionen: IfSG-Surveillancedaten von 2013. Epid Bull. 2014;27:233–237. [Google Scholar]
- 28.Robert Koch-Institut (RKI) IfSG-Meldepflicht-Anpassungsverordnung: Zur Umsetzung der neuen Meldepflichten. Epid Bull. 2016;16:135–136. doi: 10.17886/EpiBull-2016-026. [DOI] [Google Scholar]
- 29.von Müller L., Mock M., Halfmann A., Stahlmann J., Simon A., Herrmann M. Epidemiology of Clostridium difficile in Germany based on a single center long-term surveillance and German-wide genotyping of recent isolates provided to the advisory laboratory for diagnostic reasons. Int J Med Microbiol. 2015;305:807–813. doi: 10.1016/j.ijmm.2015.08.035. [DOI] [PubMed] [Google Scholar]
- 30.Berger F. Ausbruchsuntersuchungen bei Clostridium (Clostridioides) difficile. Epid Bull. 2018;14:137–139. doi: 10.17886/EpiBull-2018-017. [DOI] [Google Scholar]
- 31.Mann H.B. Nonparametric tests against trend. Econometrica. 1945;13:245–259. [Google Scholar]
- 32.Goswami B. 2017. Mann-Kendall Test (mkt)https://up-rs-esp.github.io/mkt/ [Google Scholar]
- 33.Bauer M.P., Notermans D.W., van Benthem B.H.B., Brazier J.S., Wilcox M.H., Rupnik M. Clostridium difficile infection in Europe: a hospital-based survey. Lancet. 2011;377:63–73. doi: 10.1016/S0140-6736(10)61266-4. [DOI] [PubMed] [Google Scholar]
- 34.Seugendo M., Janssen I., Lang V., Hasibuan I., Bohne W., Cooper P. Prevalence and Strain Characterization of Clostridioides (Clostridium) difficile in Representative Regions of Germany, Ghana, Tanzania and Indonesia - A Comparative Multi-Center Cross-Sectional Study. Front Microbiol. 2018;9:1843. doi: 10.3389/fmicb.2018.01843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neuendorf M., Guadarrama-Gonzalez R., Lamik B., MacKenzie C.R. A prospective study of two isothermal amplification assays compared with real-time PCR, CCNA and toxigenic culture for the diagnosis of Clostridium difficile infection. BMC Microbiology. 2016;16:19. doi: 10.1186/s12866-016-0635-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Piepenbrock E., Stelzer Y., Berger F., Jazmati N. Changes in Clostridium (Clostridioides) difficile PCR-Ribotype Distribution and Antimicrobial Resistance in a German Tertiary Care Hospital Over the Last 10 Years. Curr Microbiol. 2019;76:520–526. doi: 10.1007/s00284-019-01654-3. [DOI] [PubMed] [Google Scholar]
- 37.Jazmati N., Hain O., Kaasch A., Plum G. Würzburg; Germany: 2016. Clinical significance of presumptive identification of Clostridium difficile ribotype 027 in clinical stool samples by real-time PCR. eP-146. 13er Kongress für Infektionskrankheiten und Tropenmedizin (KIT) [Google Scholar]
- 38.Gawlik D., Slickers P., Engelmann I., Müller E., Lück C., Friedrichs A. DNA-Microarray-based Genotyping of Clostridium difficile. BMC Microbiol. 2015;15:158. doi: 10.1186/s12866-015-0489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Becker M., Schon S., Ruppelt A., Jatzwauk L., Ehricht R., Monecke S. 2016. Typisierung von Clostridium difficile mittels Microarray-Hybridisierung. 13er Kongress für Krankenhaushygiene DGKH, Berlin, Germany. [Google Scholar]
- 40.Krajewski C., Miller V., Guadarrama R., Lamik-Wolters B., Mackenzie C.R. 2013. A retrospective and prospective analysis of the slpA-type, toxome and clinical characteristics of 750 Clostridium difficile isolates collected over a three year period. P2493. 23rd ECCMID, Berlin, Germany. [Google Scholar]
- 41.Eyre D.W., Davies K.A., Davis G., Fawley W.N., Dingle K.E., De Maio N. Two Distinct Patterns of Clostridium difficile Diversity Across Europe Indicating Contrasting Routes of Spread. Clin Infect Dis. 2018;67:1035–1044. doi: 10.1093/cid/ciy252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borgmann S., Kist M., Jakobiak T., Reil M., Scholz E., von Eichel-Streiber C. Increased number of Clostridium difficile infections and prevalence of Clostridium difficile PCR ribotype 001 in southern Germany. Euro Surveill. 2008;13:19057. doi: 10.2807/ese.13.49.19057-en. [DOI] [PubMed] [Google Scholar]
- 43.Reil M., Hensgens M.P., Kuijper E.J., Jakobiak T., Gruber H., Kist M. Seasonality of Clostridium difficile infections in Southern Germany. Epidemiol Infect. 2012;140:1787–1793. doi: 10.1017/s0950268811002627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO) Ausbruchsmanagement und struktuiertes Vorgehen bei gehäuftem Auftreten nosokomialer Infektionen. Bundesgesundheitsbl. 2002;45:180–186. doi: 10.1007/s00103-012-1549-5. [DOI] [Google Scholar]
- 45.Kommission für Krankenhaushygiene und Infektionsprävention (KRINKO) Hygienemaßnahmen bei Clostridioides difficile-Infektion (CDI) Bundesgesundheitsbl. 2019;62:906–923. doi: 10.1007/s00103-019-02959-1. [DOI] [PubMed] [Google Scholar]
- 46.European Centre for Disease Prevention and Control (ECDC) Stockholm ECDC; 2018. Healthcare-associated infections: Clostridium difficile infections - annual epidemiological report for 2016.https://www.ecdc.europa.eu/en/publications-data/healthcare-associated-infections-clostridium-difficile-infections-annual Available at: [Google Scholar]
- 47.Krutova M., Kinross P., Barbut F., Hajdu A., Wilcox M.H., Kuijper E.J. How to: Surveillance of Clostridium difficile infections. Clin Microbiol Infect. 2018;24:469–475. doi: 10.1016/j.cmi.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 48.European Centre for Disease Prevention and Control (ECDC) ECDC; Stockholm: 2019. European surveillance of Clostridioides (Clostridium) difficile infections - surveillance protocol version 2.4 (ECDC Technical Document)https://www.ecdc.europa.eu/en/publications-data/european-surveillance-clostridium-difficile-infections-surveillance-protocol-2 Available at: [Google Scholar]
- 49.Tschudin-Sutter S., Kuijper E.J., Durovic A., Vehreschild M.J.G.T., Barbut F., Eckert C. Guidance document for prevention of Clostridium difficile infection in acute healthcare settings. Clin Microbiol Infect. 2018;24:1051–1054. doi: 10.1016/j.cmi.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 50.Ilchmann C., Zaiss N.H., Speicher A., Christner M., Ackermann G., Rohde H. Comparison of resistance against erythromycin and moxifloxacin, presence of binary toxin gene and PCR ribotypes in Clostridium difficile isolates from 1990 and 2008. Eur J Clin Microbiol Infect Dis. 2010;29:1571–1573. doi: 10.1007/s10096-010-1017-9. [DOI] [PubMed] [Google Scholar]
- 51.Elliott B., Androga G.O., Knight D.R., Riley T.V. Clostridium difficile infection: Evolution, phylogeny and molecular epidemiology. Infect Genet Evol. 2017;49:1–11. doi: 10.1016/j.meegid.2016.12.018. [DOI] [PubMed] [Google Scholar]
- 52.Berger F. Auftreten von Clostridium difficile Ribotyp 176 in Deutschland. Epid Bull. 2017;10:93–95. doi: 10.17886/EpiBull-2017-010.3. [DOI] [Google Scholar]
- 53.Valiente E., Dawson L.F., Cairns M.D., Stabler R.A., Wren B.W. Emergence of new PCR ribotypes from the hypervirulent Clostridium difficile 027 lineage. J Med Microbiol. 2012;61:49–56. doi: 10.1099/jmm.0.036194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Crobach M.J., Planche T., Eckert C., Barbut F., Terveer E.M., Dekkers O.M. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect. 2016;22:S63–S81. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 55.Kaase M., Szabados F., Tix H., Anders A., Gatermann S. First detection of Clostridium difficile ribotype 027 in Bochum, Germany, confirmed by slpA sequencing. (KMP07) Int J Med Microbiol. 2009;299:39. [Google Scholar]
- 56.Claußen K., Scharlach M., Pulz M. Zum Vorkommen von Clostridium difficile in zwei Regionen Niedersachsens. Epid Bull. 2011;40:363–366. doi: 10.25646/4526. [DOI] [Google Scholar]
- 57.von Müller L., Halfmann A., Herrmann M. Aktuelle Daten und Trends zur Antibiotikaresistenzentwicklung von Clostridium difficile. Bundesgesundheitsbl. 2012;55:1410–1417. doi: 10.1007/s00103-012-1556-6. [DOI] [PubMed] [Google Scholar]
- 58.Arvand M., Bettge-Weller G. Clostridium difficile ribotype 027 is not evenly distributed in Hesse, Germany. Anaerobe. 2016;40:1–4. doi: 10.1016/j.anaerobe.2016.04.006. https://doi.org/j.anaerobe.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 59.von Müller L., Zevallos D., Nimmesgern A., Herrmann M., EUCLID study group . 2014. Analysis of actual Clostridium difficile epidemiology in Germany based on a multicenter bi-annual point prevalance study in european countries (EUCLID). O010. 24th ECCMID, Barcelona, Spain. [Google Scholar]
- 60.Becker J., Braeu T., Schellberger M. 2014. Surveillance of Clostridium difficile binary toxin and ribotype 027 using laboratory data in Germany. P0763. 24th ECCMID, Barcelona, Spain. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.