Summary
Background
Methicillin-resistant Staphylococcus aureus (MRSA) is a major public health concern worldwide. Healthcare workers (HCWs) are an important source of transmission of MRSA. We conducted a prospective study to define the frequency of S. aureus nasal colonization with emphasis on the carriage of MRSA in HCWs in relation to the intensity of patient contact.
Methods
Out-of-hospital care emergency medical technicians and students, and HCWs in the emergency department, intensive care unit and a long-term care facility (LTCF) were enrolled to compare the prevalence of MRSA and methicillin-susceptible S. aureus (MSSA) nasal colonization. The MRSA isolates were further identified by their microbiological and molecular characteristics.
Findings
S. aureus was isolated from 63 of 248 HCWs (25.4%). The overall MRSA nasal carriage rate was 15/248, 6%, and the prevalence was higher in the HCWs who had worked for 5–10 years (12.8%), and among female HCWs (10.3%) than male HCWs (0.9%). LTCFs had the highest prevalence (12%). In contrast, the overall carriage of MSSA was 48/248, 19.4%, and most carriers worked for ≥5 years (52.1%). Hospital nurses had the highest rate of MSSA carriage (21.4%). Most of the MRSA isolates were SCCmec IV/ST59 or ST45 (60%), and were resistant to erythromycin and clindamycin (53%).
Conclusions
Hospital nurses have highest S. aureus nasal carriage, whereas HCWs in the LTCFs comprise a significant reservoir of MRSA colonization. The differences in the characteristics of MRSA and MSSA nasal carriage among HCWs highlights the importance on long-term nasal screening of S. aureus in healthcare facilities.
Keywords: MRSA, Long term care facilities, Healthcare worker, Emergency medical technician
Abbreviations: MRSA, Methicillin-resistant Staphylococcus aureus; CA-MRSA, community-associated Methicillin-resistant Staphylococcus aureus; ED, emergency department; EMTs, emergency medical technicians; HA-MRSA, healthcare-associated Methicillin-resistant Staphylococcus aureus; HCWs, Healthcare workers; ICU, intensive care unit; LTCF, long-term care facilities
Introduction
Staphylococcus aureus is an important human pathogen. It has the ability to cause a wide variety of infections ranging from local invasion of skin and soft tissues to life-threatening sepsis. The emergence and spread of methicillin-resistant S. aureus (MRSA) is particularly troublesome because of its association with increased morbidity and mortality [1,2] and the need to select the most appropriate therapy. Biofilm-forming variants are often difficult to treat, even when susceptible to otherwise effective antibiotics.
MRSA infections are usually divided into healthcare-associated (HA-MRSA) or community-associated (CA-MRSA) because of differences in epidemiology, risk factors and choice of drug. HA-MRSA infections are more likely to occur in individuals with underlying diseases, the elderly, recently hospitalised, those undergoin invasive procedures and residents in long term health care facilities [3]. CA-MRSA infections usually occur in otherwise healthy people and with minor trauma. HA-MRSA strains usually possess SCCmec types I, II or III, and tend to be multiple drug resistant. CA-MRSA strains usually possess SCCmec types IV or V and are strongly associated with the Panton-Valentin leucocidin (pvl) gene. CA-MRSA strains are considered to be more virulent, transmittable, and persistent than HA-MRSA [4,5]. CA-MRSA strains can also be transmitted in healthcare facilities and mistaken for HA-MRSA [5].
The ability of S. aureus to colonize the anterior nares and other body sites is a significant predisposing risk factor for infection [5,6]. Elimination of carriage decreases the incidence of S. aureus infection [7,8]. The prevalence of MRSA nasal colonization in worldwide surveys of general populations ranges from 0.7 to 3% [9,10]. The rate is somewhat higher in Taiwan, 3.8% [11]. Hospitalized patients and those in long-term care facilities (LTCF) are at highest risk for MRSA carriage [12,13].
Healthcare workers (HCWs), situated at the interface between hospital and community, are an important reservoir of S. aureus for both HA-MRSA and CA-MRSA [14]. Colonized HCWs can transmit MRSA to patients, their families, and other HCWs and have been implicated as a source of transmission in outbreaks [15,16]. Identification of colonized HCWs combined with hand hygiene and other precautions have been shown to reduce the transmission and control the spread of MRSA [17].
Most studies of the transmission of MRSA have focused on isolates obtained from patients. Less is known about the frequency of carriage in HCWs, their genetic and clonal diversity, virulence gene determinants, and microbiological characteristics. The current study was designed to fill in some of these gaps.
In order to further identify the epidemiologic characteristics of MRSA strains and to clarify the spreading of epidemic clones, we employed molecular methods with sufficient discriminative power for studying clonal distribution. The objectives were to determine the frequency of MRSA compared to methicillin-susceptible S. aureus (MSSA) nasal colonization in different HCWs in relation to the intensity and duration of exposure to patients and characterize the molecular characteristics, antimicrobial resistant profiles, and biofilm-forming abilities of the isolates. The subjects included HCWs in an emergency department (ED), intensive care unit (ICU), and out-of-hospital emergency medical technicians (EMTs), and LTCFs.
Methods
Study design
This one-year prospective study was conducted from January to December 2015 at the National Cheng Kung University Hospital (NCKUH), Tainan, Taiwan, emergency medical service section of the local fire department and long-term care facilities. Healthcare providers who met the study criteria were offered the opportunity to participate in the study. The target population included out-of-hospital and in-hospital healthcare providers. Out-of-hospital providers were EMTs, student EMTs and staffs of LTCF, and in-hospital care providers included nurses and physicians working in the ED and ICU.
Selection of participants
Student EMTs, regarded as representatives of the general population with limited exposure to patients, were enrolled from the annual routine new EMT training program; paramedics and private ambulance EMTs with short term urgent care, transport to hospitals, or transfer of patients between healthcare facilities were selected from two local private ambulance groups. Physicians, nurses and staff who worked in the adult medical ICU (MICU), the medical ED (MED) and a LTCF for more than 6 months of working experience were asked to participate. Participants who had active infections such as fever and known respiratory tract infections, urinary tract infections or other occult illnesses, and who have taken an antibiotic during the prior 21-days were excluded. The protocol and consent forms were approved by the Ethical Review Committee of NCKUH (B-ER-104-029). Written informed consents were obtained from participants prior to taking nasal swabs. Participants were asked to fill out an anonymous questionnaire regarding their place of work (current and previous), wearing adequate personal protective equipment and washing hands before and after patient care.
Microbiologic methods
One sample was taken from each participant. A sterile cotton swab was used to circle the anterior 1 cm of the nasal vestibule of both nares. Swab specimens were inoculated by the streak plate method on Trypticase soy agar with 5% sheep blood plates and incubated overnight at 37°C. All the colonies were sub-cultured on mannitol salt agar and incubated at 37°C for 24h. Mannitol fermenting colonizes that were yellow or golden yellow were selected and subjected to Gram's stain and coagulase test. The presumptive S. aureus isolates were confirmed by coagulase (coa) gene-based PCR [18]. MRSA were identified by the cefoxitin disk-diffusion method according to the recommendations of Clinical and Laboratory Standard Institutes [19].
PFGE
MRSA isolates were identified further by PFGE analysis with chromosomal DNA using the enzyme SmaI. The relatedness of strains was determined by comparison of restriction fragment-length polymorphism in accordance with the guidelines published by Tenover et al. [20] PFGE patterns resulting in 2–3 band differences were considered to be closely related, those with 4–6 band differences were considered to be possibly related, and those with ≥7 band differences were considered to be unrelated.
Susceptibility testing
The antimicrobial susceptibility of MRSA isolates to 10 antibiotics, i.e. oxacillin, trimethoprim/sulfamethoxazole, penicillin, teicoplanin, linezolid, clindamycin, doxycycline, fusidic acid, vancomycin, and erythromycin, was determined in accordance with the guideline of Clinical Laboratory Standards Institute [19].
Biofilm formation assay
Four microliters of a bacterial overnight culture were inoculated into 1 ml of tryptic soy broth containing 0.25% glucose. An aliquot (200 μl) of the sample was poured into each of a 96-well polystyrene microplate (167008, Thermo Fisher Scientific, Waltham, MA, USA), and incubated for 3 days at 37°C. The fluid was removed and the plate was stained with 0.1% safranin solution. The OD490 was measured using a microplate reader (μQuant, BioTek Instruments, Winooski, VT, USA).
Molecular characterization
Genomic DNA was obtained from the MRSA isolates by a Qiamp DNA mini kit protocol (Qiagen, Hilden, Germany) for molecular characterization. The presence of Panton-Valentine leucocidin (pvl) gene and genes for fibronectin binding protein A and B (fnbA, fnbB) were determined by PCR as previous described [21,22]. The S. aureus MLST scheme uses internal fragments of the following seven house-keeping genes: arc, aro, glp, gmk, pta, tpi and yqi. PCR amplification was carried out on chromosomal DNA using an extension time of 30 seconds, and an annealing temperature of 55°C, with Taq polymerase. The PCR products were then sequenced and the data were uploaded to the MLST website (http://www.mlst.net) for further analysis [23]. Typing of the staphylococcal chromosomal cassette mec (SCCmec) was done by PCR with primers and by the methods published previously [24]. PCR for mecA, mupA, and qacA/B were performed by the methods described previously [23,25].
Methods of measurements
Positive nasal swabs for MSSA and MRSA were reported. Further nasal carriage of the MSSA and MRSA prevalence were calculated by descriptive statistics and cross tabulations to determine the frequency distribution of the MRSA nasal carriage among the different groups of healthcare professionals. Pearson's chi-square test, Fisher's exact test Cochran–Mantel–Haenszel test, logistic regression, and generalized linear models were used to compare MRSA colonization between groups. Odd ratios (ORs) were calculated with 95% confidence intervals (CIs). Student t test or Mann–Whitney U test were used to compared continuous variables. SAS software version 9.4 (SAS, Inc., Cary, North Carolina, USA) was applied for data entry, processing and statistical analysis. Positive MRSA samples were enrolled for further pathogenicity by molecular analysis. The drug susceptibility, basic molecular typing, virulence factors such as pvl gene and genes for fibronectin binding protein and the biofilm formation assay were conducted.
Results
Prevalence of MRSA and MSSA
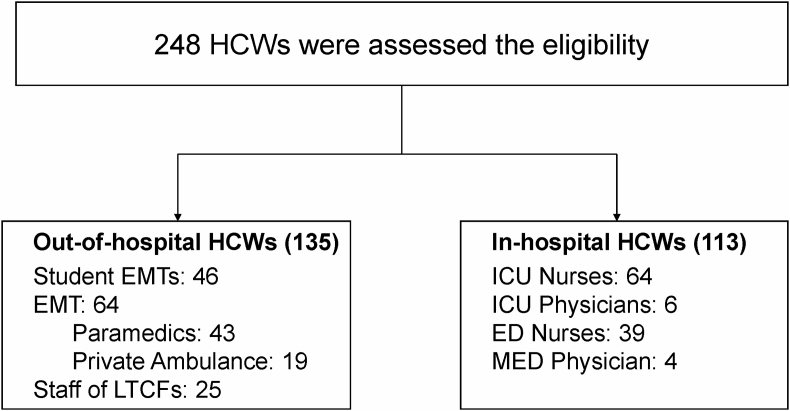
Four hundred and fifteen healthcare providers were invited. Among them, 248 responded and consented to participate in the study. The response rates of EMT, EMT students, in-hospital HCWs and LTCF staffs were 53% (64/120), 92% (46/50), 53% (113/215), and 80% (25/30), respectively. The distribution of the HCWs by site of work is shown in Figure 1. The frequency of isolation of MSSA and MRSA according to the characteristics of the study population is shown in Table 1 and Supplement Table.
Figure 1.
Enrollment of healthcare workers for MRSA and MSSA Nasal Carriage Study. ED, emergency department; EMT, emergency medical technician; ICU, intensive care unit; LTCF, long term care facility; MED, medical emergency department.
Table I.
Demographic characteristics associated with S. aureus, (MSSA and MRSA) colonization
| No. of samples (N=248) (%) | S. aureus (N=63) | MSSA (N=48) | MSSA colonization |
MRSA (N=15) | MRSA colonization |
|||
|---|---|---|---|---|---|---|---|---|
| Prevalence (%) | Or (95% CI) | Prevalence (%) | Or (95% CI) | |||||
| Gender | ||||||||
| Male | 112 (45) | 25 | 24 | 21.4 | 1.27 (0.68, 2.39) | 1 | 0.9 | 0.08 (0.01, 0.61) |
| Female | 136 (55) | 38 | 24 | 17.6 | 14 | 10.3 | ||
| Age (yr) | ||||||||
| Mean ± SD | 31.55±7.52 | 31.10±7.43 | 32.27±7.68 | 31.53±6.76 | ||||
| Working years (yr) | ||||||||
| <3 | 95 (38) | 18 | 15 | 15.8 | Ref | 3 | 3.2 | Ref |
| 3-5 | 31 (13) | 9 | 8 | 25.8 | 1.95 (1.80, 2.11) | 1 | 3.2 | 0.25 (0.20, 0.32) |
| 5-10 | 47 (19) | 14 | 8 | 17 | 0.64 (0.59, 0.70) | 6 | 12.8 | 2.53 (2.26, 2.82) |
| 10-20 | 58 (23) | 19 | 14 | 24.1 | 1.32 (1.22, 1.42) | 5 | 8.6 | 1.33 (1.18, 1.50) |
| >20 | 17 (7) | 3 | 3 | 17.6 | 0.70 (0.62,0.79) | 0 | 0 | - |
| Occupation | ||||||||
| Student EMT | 46 (19) | 7 | 7 | 14.6 | Ref | 0 | 0 | - |
| EMT | 64 (26) | 13 | 12 | 19.4 | 1.39 (1.21, 1.60) | 1 | 1.6 | Ref |
| Staff of LTCFs | 25 (10) | 7 | 4 | 16 | 1.13 (0.97, 1.31) | 3 | 12.0 | 5.52 (4.61, 6.61) |
| Nurses | 103 (41) | 33 | 22 | 21.4 | 1.46 (1.28, 1.67) | 11 | 10.7 | 4.41 (3.74, 5.20) |
| ED | 39 (16) | 12 | 8 | 20.5 | 4 | 10.3 | ||
| ICU | 64 (26) | 21 | 15 | 23.4 | 7 | 10.9 | ||
| Physicians | 10 (4) | 3 | 3 | 30 | 1.82 (1.55, 2.14) | 0 | 0 | - |
S. aureus was isolated from 63 of the 248 HCWs (25.4%). Fifteen (23.8%) of these isolates were MRSA. The overall MRSA nasal carriage rate was 6%. The prevalence of the MRSA nasal carriage was higher in the HCWs who had worked for 5–10 years (12.8%), and among female HCWs (10.3%) than among the male HCWs (0.9%). LTCFs had the highest prevalence (12%, 95% CI 4.61–6.61), followed by hospital nurses in comparison with reference group, the EMTs. In contrast, the overall carriage of MSSA was 48/248, (19.4%) and there was no female preponderance (24/48, 50%). Hospital nurses had the highest rate of MSSA nasal carriage (22/103, 21.4%, 95% CI 1.28–1.67) followed by EMTs and LTCFs in comparison with reference group, the student EMT. None of 10 physicians and student EMTs was colonized by MRSA.
Characterization of MRSA isolates
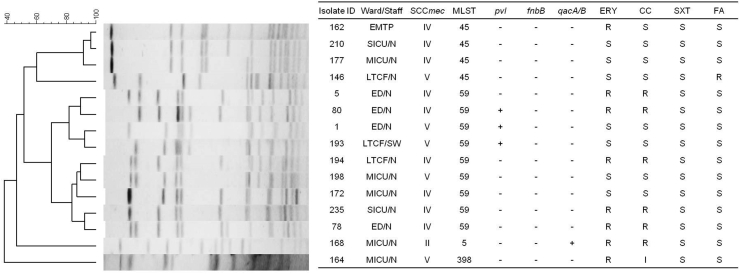
All of the 15 MRSA isolates were positive for mecA by PCR. Most were SCCmec IV (9, 60%) and belonged to two endemic CA-MRSA genotypes, ST59 (6, 67%) and ST45 (3, 33%). All the isolates could be divided into 6 major clones by PFGE pattern analysis (Figure 2). The most predominant pulsotype contained 5 isolates carrying SCCmec IV or V/PVL−/ST59. One isolate, belonging to ST398 and carrying SCCmec V, was isolated from a MICU nurse who had traveled to Europe within 12 months.
Figure 2.
Molecular characterization, antibiogram of nasal carriage isolates and the PFGE dendrogram compares fingerprint patterns of the methicillin-resistant Staphylococcus aureus (MRSA) isolates from 15 healthcare workers. SCCmec and MLST indicate the results for MRSA type. Columns marked “pvl”, “fnbB”, and qacA/B” are the results for genetic tests performed to detect the PVL, fibronectin B and chlorhexidine resistance genes. ERY, erythromycin; CC, clindamycin; SXT, trimethoprim-sufamethoxazole; FA, fucidic acid; S, susceptible; R, resistant. ED, Emergency department; EMTP, Emergency Medical Technician Professional (Paramedic); MICU, Medical ICU; N, nurse; SW, Social Worker; SICU, Surgical ICU; LTCF, Long term care facility.
The MRSA isolates exhibited high rates of resistance to erythromycin (53%) and clindamycin (53%). Only one isolate was resistant to fusidic acid. One isolate from a MICU nurse was detected as qacA/B-positive, conferring resistance to chlorhexidine in S. aureus. The MIC to chlorhexidine was 4mg/L. None of the MRSA isolates were resistant by phenotypic or genetic tests to mupirocin, linezolid or glycopeptide antibiotics (vancomycin or teicoplanin).
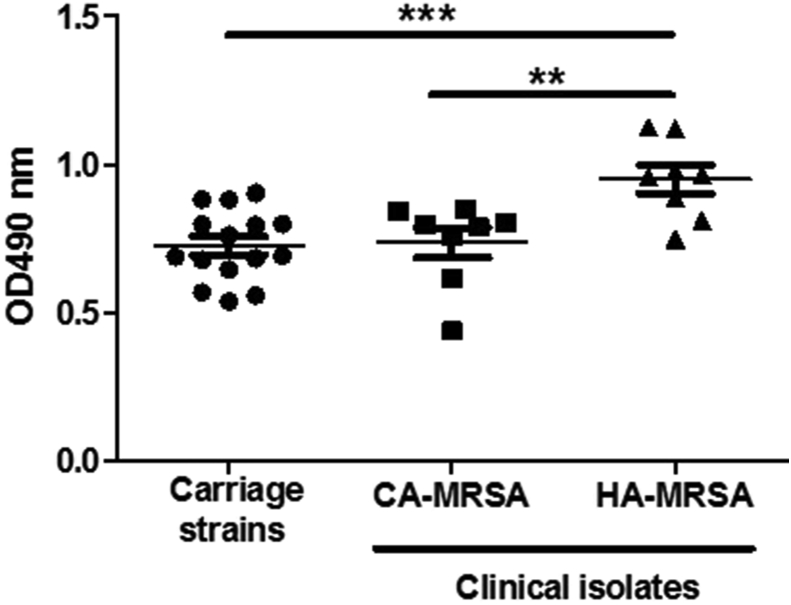
All of the MRSA nasal colonizing isolates, regardless of the SCCmec type, formed low levels of biofilm that were indistinguishable from the CA-MRSA V/PVL+/ST59 clinical isolates (Figure 3). In contrast, the traditional HA-MRSA III/PVL−/ST239 clinical isolates had significantly higher levels of biofilm formation. Additionally, all the MRSA nasal colonizing isolates had fnbA gene but none possessed the fnbB gene.
Figure 3.
Biofilm formation ability. Biofilm formation of the MRSA nasal colonization isolates (carriage), CA-MRSA V/PVL+/ST59 clinical isolates (CA-MRSA), and the HA-MRSA III/PVL−/ST239 (HA-MRSA) onto polystyrene microplates were measured. ∗∗ P<0.01, ∗∗∗ P<0.001 for significant differences based on two-sided unpaired t test.
Discussion
This study aimed to determine the frequency of MRSA compared to MSSA nasal colonization in different HCWs in relation to the intensity and duration of exposure to patients and characterize the molecular characteristics, antimicrobial resistant profiles, and biofilm-forming abilities of the isolates. We found that 25.4% of HCWs were nasal carriers of S. aureus. About a quarter (23.8%) of the HCWs with nasal carriage of S. aureus were colonized with MRSA. The overall frequency of MRSA carriage was 6%. The prevalence of MRSA was highest among LTCF staff, followed by ICU nurses, ED nurses and EMTs. SCCmec typing revealed that the majority of strains were type IV and V. These are the most common types of CA-MRSA in Taiwan [26].
The prevalence of MRSA (6%) in our study is consistent with the frequency of HCW MRSA nasal carriage in previous studies in Taiwan (5.0–7.8%) [27,28], but higher than in other countries [14]. They are also consistent with other studies that found the highest prevalence of MRSA nasal carriage among HCWs with close contact with patients, poor attention to infection control policy, and high work-load [14]. Conditions contributing to MRSA nasal carriage are complex and multifactorial. Direct patient contact is considered to be the main transmission route for MRSA [29]. In the current study the greatest risk for MRSA nasal carriage was in HCWs working for 5–10 years, the most active clinical engagement stage. Many of the HCWs who work in the health facilities for more than 20 years have decreased clinical service but increased the workload of administration, which may be one of the reasons of decreasing the prevalence of MRSA nasal carriage.
Most of the MRSA isolates in our study carried either SCCmec type IV or V, suggesting a community origin. Only one MRSA isolate with SCCmec type II, the traditional HA-MRSA type [30], was isolated from an ICU nurse. This finding indicates that the CA- MRSA types (type IV and V) were more common among healthcare providers and the HA-MRSA strain still remained in the hospital. This may be due to strict infection control measures in our hospital. Another possibility is clonal replacement in the hospital setting. In the past decade, CA-MRSA has been increasingly identified as a cause of hospital-onset and healthcare associated infections. A previous report indicated that CA-MRSA has become an increasingly prevalent genetic background among MRSA infections in inpatient and outpatient settings [31]. This suggests that certain clones have the ability to cross the barriers between hospitals and the community [32,33]. Although we did not study S. aureus carriage in patients, detection of CA-MRSA among HCWs in hospital possibly indicates incursion of the community origin strain into hospital setting, and also partly provides insight about the current epidemiology of CA-MRSA in Taiwan.
Several unusual MRSA strains were isolated from the current study population. An isolate from a nurse working in the MICU was SCCmec II ST5. This HA-MRSA strain carried chlorhexidine resistant genes (qacA/B). Chlorhexidine is widely used as antiseptic for central venous catheter care bundle and patient bathing to prevent nosocomial infection in out hospital. Chlorhexidine based soaps and mupirocin ointment are commonly used for cleaning and decontamination of MRSA. Prior investigators have also described MRSA strains carrying chlorhexidine and mupirocin resistant genes [34,35], but these are uncommon in Taiwan.
Another unusual MRSA strain, ST398, was isolated from a MICU nurse in our study. This is an important emerging strain associated with the livestock, mainly in Europe and North America [36,37]. MRSA ST398 is usually associated with pigs and veal calves but can colonize other host species. These include cows, sheep, poultry and farmers who are in frequent contact with MRSA-colonized animals, and can cause infections in humans [38]. The nurse who acquired this strain had traveled to Europe within 12 months and had contact to animals. The role of livestock as a potential source of MRSA infection is a growing public health problem. The risk and impact of HCWs carrying this clone needs to be closely monitored.
Some of the MRSA strains in our study are able to produce biofilm on both mucosal and inanimate surfaces, making them difficult to eradicate [39]. Biofilm formation is considered to have a role in S. aureus colonization [40,41]. Recently fibronectin-binding proteins (Fnb A and Fnb B) have also been reported to play a role in biofilm formation. The association of the expression of Fnb A and Fnb B with increased bacterial aggregation suggests that fibronectin-binding proteins can promote the accumulation phase of biofilms [42]. In the current study, all the MRSA nasal colonization isolates carried the fnbA but not the fnbB gene and only exhibited low levels of biofilm formation. This is consistent with the recent concept that a dispersed mode of growth in the vestibulum nasi is preferable to a biofilm mode during S. aureus nasal colonization [43].
MSSA strains were more abundant than MRSA in nasal carriers (19.4%) and were differently distributed in our study. We believe this is important because all S. aureus have the potential to produce invasive disease and need to be included in surveillance studies and control measures.
This study has several limitations. First, it was conducted in a large metropolitan region in southern Taiwan and the findings may not be generalizable to other localities. We collected nasal swabs from HCWs in different healthcare facilities, however a relatively small number of positive MRSA samples were isolated, and may lead to a higher variability in some groups. Second, single cross-sectional sampling did not allow us to differentiate between transient and persistent carriers of MRSA and MSSA. A longitudinal surveillance study should be conducted to monitor the prevalence change of MRSA and MSSA nasal carriage. Third, samples were collected only from the nares. It has been estimated that 15–50% of MRSA carriers are non-nasal [44]. Therefore, it is likely that we underestimated the overall prevalence of MRSA and MSSA. Fourth, nasal swab samples were not analyzed using pre-enriched culture in the study. Pre-enriched culture was found to be more sensitive than direct culture in the detection of nasal S. aureus [45]. The use of pre-plating enrichment of swabs in TSB to improve nasal S. aureus detection levels is warranted in future studies. Fifth, whole-genome sequencing is more sensitive than molecular analysis to classify MRSA strains as community or hospital-associated. It has the added advantage of establishing genetic relatedness and recent transmission. Finally, we focused on the nasal S. aureus carriage rate in HCWs and did not collect samples from patients or residents in LTCFs. The identification of colonized HCWs allows the appropriate management of these staff, to prevent the spread to others. However, we were unable to determine the source of transmission. A more comprehensive screening of both HCWS and patients is needed to fill the knowledge gap.
The strengths of this study include its prospective design, observation over a full year, adequate sample size of a diverse representative population of HCWs with major differences in their exposure to patients, comprehensive molecular characterization of the MRSA, biofilm-forming ability and antibiotic susceptibility testing.
Conclusions
This study demonstrates that nasal colonization by S. aureus differs among healthcare professionals in relation to the extent and duration of exposure to different patient groups. Hospital nurses have highest S. aureus nasal carriage rate, whereas HCWs worked in the LTCFs have the highest prevalence of nasal MRSA colonization. Most of the MRSA isolates belong to CA-MRSA strains, exhib high rates of resistance to erythromycin and clindamycin, and produce low level of biofilms. An unusual MRSA strain, ST398, was isolated from an MICU nurse who had exposure to livestock in Europe. Another MICU nurse was colonized a qacA/B-positive strain conferring resistance to chlorhexidine. MRSA represents only the tip of the iceberg of nasal colonization by S. aureus. MSSA strains were 3.2 times more common and much more frequent in hospital nurses and EMTs who had limited exposure to patients. This supports the inclusion of all strains of S. aureus in surveillance and infection control programs both in hospital and out-of-hospital care facilities. Regarding the risk of nosocomial infections, all HCWs should wear mask, gloves, and/or gown all the time during patient care, and be educated and trained periodically about the maintenance of hygiene and infection control to prevent the disease transmission.
Credit Author Statement
Hsin-I Shih: Conceptualization, Writing- Original draft preparation, Formal analysis, Funding acquisition. Chia-Ming Chang: Methodology, Data curation. Fan-Ching Shen: Methodology, Investigation. Yun-Ju Lee: Methodology, Investigation. Chiu Hui Wu: Data curation, Investigation. Hsiang-Chin Hsu: Data curation, Investigation. Chia-Yu Chi: Conceptualization, Resources, Writing- Reviewing and Editing, Supervision.
Acknowledgements
We thank Dr. Calvin M. Kunin for providing invaluable suggestions and critical review of the manuscript.
Footnotes
Previous Presentation: Preliminary results from this study were presented at 30th International Congress of Chemotherapy and Infection.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.infpip.2021.100117.
Conflicts of interest statement
The authors declare that they have no competing interests.
Funding
This study is supported by the Research Centre of National Cheng Kung University Hospital (NCKUH-10406035, NCKUH-10506016, and NCKUH-10605008). The funders played no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cosgrove S.E., Sakoulas G., Perencevich E.N., Schwaber M.J., Karchmer A.W., Carmeli Y. Comparison of mortality associated with methicillin-resistant and methicillin-susceptible Staphylococcus aureus bacteremia: a meta-analysis. Clin Infect Dis. 2003;36:53–59. doi: 10.1086/345476. http://10.1086/345476 [DOI] [PubMed] [Google Scholar]
- 2.Pallin D.J., Egan D.J., Pelletier A.J., Espinola J.A., Hooper D.C., Camargo C.A., Jr. Increased US emergency department visits for skin and soft tissue infections, and changes in antibiotic choices, during the emergence of community-associated methicillin-resistant Staphylococcus aureus. Ann Emerg Med. 2008;51:291–298. doi: 10.1016/j.annemergmed.2007.12.004. http://10.1016/j.annemergmed.2007.12.004 [DOI] [PubMed] [Google Scholar]
- 3.DeLeo F.R., Otto M., Kreiswirth B.N., Chambers H.F. Community-associated meticillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. http://10.1016/S0140-6736(09)61999-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamamoto T., Nishiyama A., Takano T., Yabe S., Higuchi W., Razvina O. Community-acquired methicillin-resistant Staphylococcus aureus: community transmission, pathogenesis, and drug resistance. J Infect Chemother. 2010;16:225–254. doi: 10.1007/s10156-010-0045-9. http://10.1007/s10156-010-0045-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wertheim H.F., Melles D.C., Vos M.C., van Leeuwen W., van Belkum A., Verbrugh H.A. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–762. doi: 10.1016/S1473-3099(05)70295-4. http://10.1016/S1473-3099(05)70295-4 [DOI] [PubMed] [Google Scholar]
- 6.Ellis M.W., Hospenthal D.R., Dooley D.P., Gray P.J., Murray C.K. Natural history of community-acquired methicillin-resistant Staphylococcus aureus colonization and infection in soldiers. Clin Infect Dis. 2004;39:971–979. doi: 10.1086/423965. http://10.1086/423965 [DOI] [PubMed] [Google Scholar]
- 7.Perl T.M., Cullen J.J., Wenzel R.P., Zimmerman M.B., Pfaller M.A., Sheppard D. Intranasal mupirocin to prevent postoperative Staphylococcus aureus infections. N Engl J Med. 2002;346:1871–1877. doi: 10.1056/NEJMoa003069. http://10.1056/NEJMoa003069 [DOI] [PubMed] [Google Scholar]
- 8.van Rijen M.M., Bonten M., Wenzel R.P., Kluytmans J.A. Intranasal mupirocin for reduction of Staphylococcus aureus infections in surgical patients with nasal carriage: a systematic review. J Antimicrob Chemother. 2008;61:254–261. doi: 10.1093/jac/dkm480. http://10.1093/jac/dkm480 [DOI] [PubMed] [Google Scholar]
- 9.Peters C., Dulon M., Kleinmuller O., Nienhaus A., Schablon A. MRSA Prevalence and Risk Factors among Health Personnel and Residents in Nursing Homes in Hamburg, Germany - A Cross-Sectional Study. PLoS One. 2017;12 doi: 10.1371/journal.pone.0169425. http://10.1371/journal.pone.0169425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorwitz R.J., Kruszon-Moran D., McAllister S.K., McQuillan G., McDougal L.K., Fosheim G.E. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis. 2008;197:1226–1234. doi: 10.1086/533494. http://10.1086/533494 [DOI] [PubMed] [Google Scholar]
- 11.Wang J.T., Liao C.H., Fang C.T., Chie W.C., Lai M.S., Lauderdale T.L. Prevalence of and risk factors for colonization by methicillin-resistant Staphylococcus aureus among adults in community settings in Taiwan. J Clin Microbiol. 2009;47:2957–2963. doi: 10.1128/JCM.00853-09. http://10.1128/JCM.00853-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mody L., Kauffman C.A., Donabedian S., Zervos M., Bradley S.F. Epidemiology of Staphylococcus aureus colonization in nursing home residents. Clin Infect Dis. 2008;46:1368–1373. doi: 10.1086/586751. http://10.1086/586751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hidron A.I., Kourbatova E.V., Halvosa J.S., Terrell B.J., McDougal L.K., Tenover F.C. Risk factors for colonization with methicillin-resistant Staphylococcus aureus (MRSA) in patients admitted to an urban hospital: emergence of community-associated MRSA nasal carriage. Clin Infect Dis. 2005;41:159–166. doi: 10.1086/430910. http://10.1086/430910 [DOI] [PubMed] [Google Scholar]
- 14.Albrich W.C., Harbarth S. Health-care workers: source, vector, or victim of MRSA? Lancet Infect Dis. 2008;8:289–301. doi: 10.1016/S1473-3099(08)70097-5. http://10.1016/S1473-3099(08)70097-5 [DOI] [PubMed] [Google Scholar]
- 15.Eveillard M., Martin Y., Hidri N., Boussougant Y., Joly-Guillou M.L. Carriage of methicillin-resistant Staphylococcus aureus among hospital employees: prevalence, duration, and transmission to households. Infect Control Hosp Epidemiol. 2004;25:114–120. doi: 10.1086/502360. http://10.1086/502360 [DOI] [PubMed] [Google Scholar]
- 16.Harris S.R., Cartwright E.J., Torok M.E., Holden M.T., Brown N.M., Ogilvy-Stuart A.L. Whole-genome sequencing for analysis of an outbreak of meticillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis. 2013;13:130–136. doi: 10.1016/S1473-3099(12)70268-2. http://10.1016/S1473-3099(12)70268-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mo Health . Ministry of Health; 2002. Guidelines for the control of methicillin-resistant Staphylococcus aureus in New Zealand. [Google Scholar]
- 18.Hookey J.V., Richardson J.F., Cookson B.D. Molecular typing of Staphylococcus aureus based on PCR restriction fragment length polymorphism and DNA sequence analysis of the coagulase gene. J Clin Microbiol. 1998;36:1083–1089. doi: 10.1128/jcm.36.4.1083-1089.1998. http://10.1128/JCM.36.4.1083-1089.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CaLS Institute., editor. 27th Edition edn. Clinical and Laboratory Standards Institute; 2017. M100 Performance Standards for Antimicrobial Susceptibility Testing. [Google Scholar]
- 20.Tenover F.C., Arbeit R.D., Goering R.V., Mickelsen P.A., Murray B.E., Persing D.H. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. http://10.1128/JCM.33.9.2233-2239.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lina G., Piemont Y., Godail-Gamot F., Bes M., Peter M.O., Gauduchon V. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin Infect Dis. 1999;29:1128–1132. doi: 10.1086/313461. http://10.1086/313461 [DOI] [PubMed] [Google Scholar]
- 22.Mehrotra M., Wang G., Johnson W.M. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol. 2000;38:1032–1035. doi: 10.1128/jcm.38.3.1032-1035.2000. http://10.1128/JCM.38.3.1032-1035.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enright M.C., Day N.P., Davies C.E., Peacock S.J., Spratt B.G. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–1015. doi: 10.1128/jcm.38.3.1008-1015.2000. http://10.1128/JCM.38.3.1008-1015.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira D.C., de Lencastre H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 2002;46:2155–2161. doi: 10.1128/AAC.46.7.2155-2161.2002. http://10.1128/aac.46.7.2155-2161.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mc Gann P., Milillo M., Kwak Y.I., Quintero R., Waterman P.E., Lesho E. Rapid and simultaneous detection of the chlorhexidine and mupirocin resistance genes qacA/B and mupA in clinical isolates of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis. 2013;77:270–272. doi: 10.1016/j.diagmicrobio.2013.06.006. http://10.1016/j.diagmicrobio.2013.06.006 [DOI] [PubMed] [Google Scholar]
- 26.Huang Y.C., Chen C.J. Community-associated meticillin-resistant Staphylococcus aureus in children in Taiwan, 2000s. Int J Antimicrob Agents. 2011;38:2–8. doi: 10.1016/j.ijantimicag.2011.01.011. http://10.1016/j.ijantimicag.2011.01.011 [DOI] [PubMed] [Google Scholar]
- 27.Huang Y.C., Su L.H., Lin T.Y. Nasal carriage of methicillin-resistant Staphylococcus aureus in contacts of an adolescent with community-acquired disseminated disease. Pediatr Infect Dis J. 2004;23:919–922. doi: 10.1097/01.inf.0000141745.12941.ef. http://10.1097/01.inf.0000141745.12941.ef [DOI] [PubMed] [Google Scholar]
- 28.Huang Y.C., Su L.H., Lin T.Y. Nasal carriage of methicillin-resistant Staphylococcus aureus among pediatricians in Taiwan. PLoS One. 2013;8 doi: 10.1371/journal.pone.0082472. http://10.1371/journal.pone.0082472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weigelt J.A., Lipsky B.A., Tabak Y.P., Derby K.G., Kim M., Gupta V. Surgical site infections: Causative pathogens and associated outcomes. Am J Infect Control. 2010;38:112–120. doi: 10.1016/j.ajic.2009.06.010. http://10.1016/j.ajic.2009.06.010 [DOI] [PubMed] [Google Scholar]
- 30.Huang Y.C., Su L.H., Wu T.L., Liu C.E., Young T.G., Chen P.Y. Molecular epidemiology of clinical isolates of methicillin-resistant Staphylococcus aureus in Taiwan. J Clin Microbiol. 2004;42:307–310. doi: 10.1128/JCM.42.1.307-310.2004. http://10.1128/jcm.42.1.307-310.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.David M.Z., Cadilla A., Boyle-Vavra S., Daum R.S. Replacement of HA-MRSA by CA-MRSA infections at an academic medical center in the midwestern United States, 2004-5 to 2008. PLoS One. 2014;9 doi: 10.1371/journal.pone.0092760. http://10.1371/journal.pone.0092760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O'Hara F.P., Amrine-Madsen H., Mera R.M., Brown M.L., Close N.M., Suaya J.A. Molecular characterization of Staphylococcus aureus in the United States 2004-2008 reveals the rapid expansion of USA300 among inpatients and outpatients. Microb Drug Resist. 2012;18:555–561. doi: 10.1089/mdr.2012.0056. http://10.1089/mdr.2012.0056 [DOI] [PubMed] [Google Scholar]
- 33.Chen C.J., Huang Y.C., Su L.H., Wu T.L., Huang S.H., Chien C.C. Molecular epidemiology and antimicrobial resistance of methicillin-resistant Staphylococcus aureus bloodstream isolates in Taiwan, 2010. PLoS One. 2014;9 doi: 10.1371/journal.pone.0101184. http://10.1371/journal.pone.0101184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheng W.H., Wang J.T., Lauderdale T.L., Weng C.M., Chen D., Chang S.C. Epidemiology and susceptibilities of methicillin-resistant Staphylococcus aureus in Taiwan: emphasis on chlorhexidine susceptibility. Diagn Microbiol Infect Dis. 2009;63:309–313. doi: 10.1016/j.diagmicrobio.2008.11.014. http://10.1016/j.diagmicrobio.2008.11.014 [DOI] [PubMed] [Google Scholar]
- 35.Lee H., Lim H., Bae I.K., Yong D., Jeong S.H., Lee K. Coexistence of mupirocin and antiseptic resistance in methicillin-resistant Staphylococcus aureus isolates from Korea. Diagn Microbiol Infect Dis. 2013;75:308–312. doi: 10.1016/j.diagmicrobio.2012.11.025. http://10.1016/j.diagmicrobio.2012.11.025 [DOI] [PubMed] [Google Scholar]
- 36.Smith T.C., Male M.J., Harper A.L., Kroeger J.S., Tinkler G.P., Moritz E.D. Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One. 2009;4:e4258. doi: 10.1371/journal.pone.0004258. http://10.1371/journal.pone.0004258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wassenberg M.W., Bootsma M.C., Troelstra A., Kluytmans J.A., Bonten M.J. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin Microbiol Infect. 2011;17:316–319. doi: 10.1111/j.1469-0691.2010.03260.x. http://10.1111/j.1469-0691.2010.03260.x [DOI] [PubMed] [Google Scholar]
- 38.Garcia-Alvarez L., Holden M.T., Lindsay H., Webb C.R., Brown D.F., Curran M.D. Meticillin-resistant Staphylococcus aureus with a novel mecA homologue in human and bovine populations in the UK and Denmark: a descriptive study. Lancet Infect Dis. 2011;11:595–603. doi: 10.1016/S1473-3099(11)70126-8. http://10.1016/S1473-3099(11)70126-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Donlan R.M., Costerton J.W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. http://10.1128/cmr.15.2.167-193.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwase T., Uehara Y., Shinji H., Tajima A., Seo H., Takada K. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature. 2010;465:346–349. doi: 10.1038/nature09074. http://10.1038/nature09074 [DOI] [PubMed] [Google Scholar]
- 41.Quinn G.A., Tarwater P.M., Cole A.M. Subversion of interleukin-1-mediated host defence by a nasal carrier strain of Staphylococcus aureus. Immunology. 2009;128:e222–e229. doi: 10.1111/j.1365-2567.2008.02952.x. http://10.1111/j.1365-2567.2008.02952.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCourt J., O'Halloran D.P., McCarthy H., O'Gara J.P., Geoghegan J.A. Fibronectin-binding proteins are required for biofilm formation by community-associated methicillin-resistant Staphylococcus aureus strain LAC. FEMS Microbiol Lett. 2014;353:157–164. doi: 10.1111/1574-6968.12424. http://10.1111/1574-6968.12424 [DOI] [PubMed] [Google Scholar]
- 43.Krismer B., Peschel A. Does Staphylococcus aureus nasal colonization involve biofilm formation? Future Microbiol. 2011;6:489–493. doi: 10.2217/fmb.11.37. http://10.2217/fmb.11.37 [DOI] [PubMed] [Google Scholar]
- 44.Lautenbach E., Nachamkin I., Hu B., No Fishman, Tolomeo P., Prasad P. Surveillance cultures for detection of methicillin-resistant Staphylococcus aureus: diagnostic yield of anatomic sites and comparison of provider- and patient-collected samples. Infect Control Hosp Epidemiol. 2009;30:380–382. doi: 10.1086/596045. http://10.1086/596045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nadimpalli M., Heaney C., Stewart J.R. Identification of Staphylococcus aureus from enriched nasal swabs within 24 h is improved with use of multiple culture media. J Med Microbiol. 2013;62:1365–1367. doi: 10.1099/jmm.0.058248-0. http://10.1099/jmm.0.058248-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.