Abstract
Clustered regularly interspaced palindromic repeats (CRISPR)-associated (Cas)9 transactivating CRISPR RNAs (tracrRNAs) form distinct structures essential for target recognition and cleavage and dictate exchangeability between orthologous proteins. As noncoding RNAs that are often apart from the CRISPR array, their identification can be arduous. In this article, a new bioinformatic method for the detection of Cas9 tracrRNAs is presented. The approach utilizes a covariance model based on both sequence homology and predicted secondary structure to locate tracrRNAs. This method predicts a tracrRNA for 98% of CRISPR-Cas9 systems identified by us. To ensure accuracy, we also benchmark our approach against biochemically vetted tracrRNAs finding false-positive and false-negative rates of 5.5% and 7.1%, respectively. Finally, the association between Cas9 amino acid sequence-based phylogeny and tracrRNA secondary structure is evaluated, revealing strong evidence that secondary structure is evolutionarily conserved among Cas9 lineages. Altogether, our findings provide insight into Cas9 tracrRNA evolution and efforts to characterize the tracrRNA of Cas9 systems.
Introduction
Clustered regularly interspaced palindromic repeats (CRISPR) RNA (crRNA) and CRISPR-associated (Cas) proteins cooperate to defend prokaryotic organisms against invading RNA and DNA.1–3 The Cas9 proteins from type II CRISPR-Cas systems are guided to cleave double-strand (ds)DNA targets using two noncoding (nc)RNAs, a crRNA, and a transactivating crRNA (tracrRNA).4,5 The crRNA contains a sequence, termed the spacer, that directly base pairs with the dsDNA target site in the vicinity of a protospacer adjacent motif.6–8 The tracrRNA base pairs with the crRNA and is recognized and bound by Cas9 resulting in the formation of a dual-guide RNA (gRNA) ribonucleoprotein complex.9,10
In recent years, due to its RNA-based programmability, CRISPR-Cas9 has been widely adopted as a genome editing tool for a variety of different genomes, including those from eukaryotic organisms.9,11,12 For these applications, the repair of a Cas9-induced double-strand break has been harnessed to correct disease-causing mutations, introduce beneficial modifications (e.g., plant grain yield), and construct new biosynthetic pathways.13–17 To further simplify its use, the dual-gRNA has been engineered into a single-gRNA (sgRNA) by linking the crRNA and tracrRNA.9 Modifications to the Cas9 protein itself have also been made. By fusing new protein domains to it and impairing its nuclease activity, it has been used as a robust RNA-guided DNA-binding platform. These applications include gene transcriptional activation and repression, epigenomic alteration, base editing, and prime editing.18–26
In prokaryotes, thousands of Cas9s have been identified computationally.27–30 In contrast, the gRNA solution for orthologous Cas9s may not be easily recognizable. This is mainly due to large variation in tracrRNA location, size, and sequence identity.29,31 Consequently, the identification of tracrRNAs represents a bottleneck for the characterization of new Cas9 proteins and their development as genome editing tools. To address this limitation, several approaches have been developed. These include computational methods that locate tracrRNAs by using the CRISPR repeat sequence to search for the sequence in the tracrRNA that has homology to and base pairs with the crRNA (the antirepeat). This is followed by a search for a rho-independent-like termination signal (RTS) in the vicinity of the antirepeat. Other approaches reliant on the sequencing of the small ncRNAs transcribed from the CRISPR-Cas9 locus have also been used.4,29
We and others have developed approaches using sequence and structural covariance models (CMs) to examine the relatedness of Cas9 tracrRNAs.32,33 In this study, we build upon these findings and use CMs as a tool to identify Cas9 tracrRNAs. Using this approach, a tracrRNA was located for >98% of all CRISPR-Cas9 containing assemblies identifiable by us. Comparisons with a diverse collection of experimentally validated tracrRNAs also showed our approach to be accurate. Here, 90.6% of tracrRNAs were identified with false-positive and false-negative rates of 5.5% and 7.1%, respectively. Finally, Bayesian and nonparametric approaches quantifying a phylogenetic signal revealed a strong evolutionary association between the Cas9 phylogeny and the predicted secondary structure of the tracrRNA, confirming previous observations that tracrRNA structures are a main determinant of Cas9-gRNA compatibility.31,34
Materials and Methods
All custom code, scripts, parsers, python objects, covariance models (CMs), and Jupyter notebooks can be found on the primary author's GitHub repository (https://github.com/skDooley/TRACR_RNA).
Detection of CRISPR-Cas9 systems
Bacterial and archaeal assemblies were downloaded from PATRIC2, NCBI GenBank, and RefSeq (last downloaded on May 05, 2020). CRISPR arrays were identified using MinCED v0.3.2 and PilerCR v1.06 with relaxed parameter settings (3 or more crRNAs, repeat lengths between 16 and 64 base pairs, and max spacer lengths of 64 base pairs).35,36
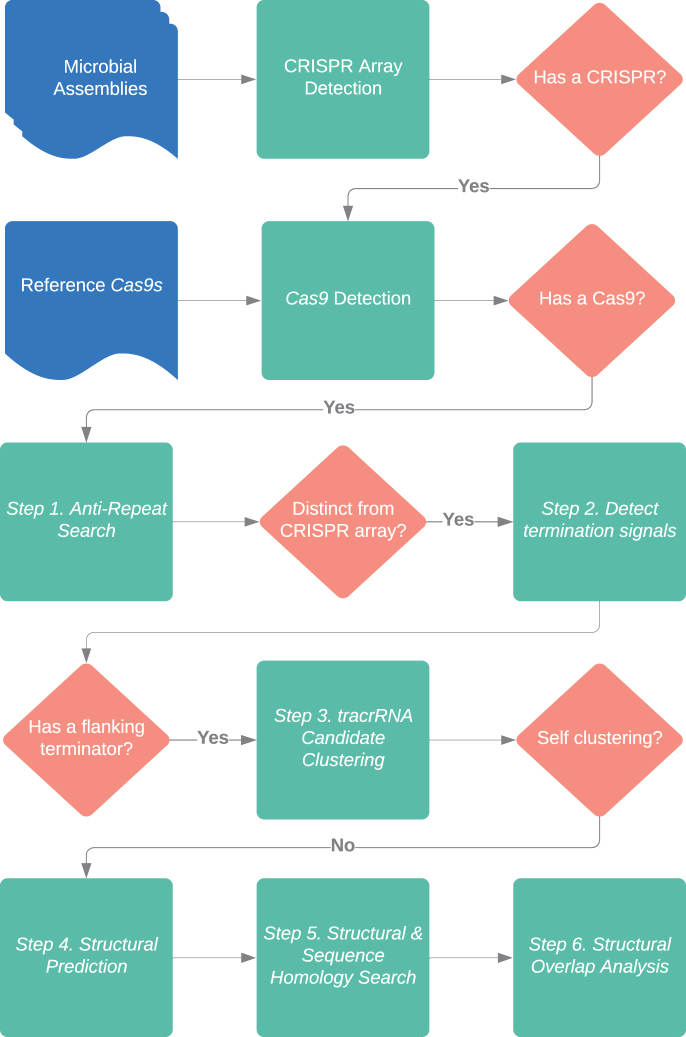
Next, a hidden Markov model (HMM) was generated from 83 previously described diverse Cas9 proteins using HMMER 3.2.1.29,37 The HMM was then used to search for Cas9-like proteins encoded in assemblies containing a CRISPR array. Protein sequences for each assembly were generated by translating open reading frames (ORFs) using Biopython to generate and filter ORFs for sequences between 673 and 2100 amino acids (Fig. 1).38 Next, using the default Python 3.7 hashing function, assemblies duplicated in our collection were removed.
FIG. 1.
Cas9 tracrRNA detection pipeline. Flowchart of the informatic steps and key decisions points used to predict Cas9 tracrRNAs. Cas9-containing CRISPR systems are first identified in microbial DNA assemblies. Assemblies with CRISPR-Cas9 loci are then searched in six steps to predict a tracrRNA. Inputs are shown in blue, informatic activities indicated in green, and key decision points highlighted in orange. Cas, CRISPR-associated; CRISPR, clustered regularly interspaced palindromic repeats; tracrRNA, transactivating CRISPR RNA.
The remaining assemblies and their Cas9 homologues were further examined for the presence of RuvC (protein fold from the E. coli RuvC protein shown to be involved in DNA repair and metabolism) and HNH (protein motif that facilitates DNA cleavage and is characterized by the presence of histidine (H) and asparagine (N) residues) cleavage domains that define a Cas9 nuclease.9,10 This was initially accomplished through the visual inspection of protein alignments performed with multiple sequence comparison by log-expectation (MUSCLE) between 83 diverse Cas9s described earlier for the key catalytic amino acids defining RuvC I, II, and III subdomains and the HNH domain.29,39 Next, the identified regions were extracted and used to generate domain-specific HMMs using HMMER 3.2.1. Each putative Cas9 protein from our collection was then scanned with the cleavage domain-specific HMMs.37 Proteins missing either domain or that had subdomains that were positional outliers were removed. Outlier determination was made by assessing the position of the RuvC I subdomain near the N-terminus and then comparing the relative distance of all other cleavage domains. Anything outside of three standard deviations (distribution of all the search results for the RuvC I subdomain) was removed except for the RuvC III subdomain, where proteins with more than four standard deviations from the mean distance were removed.
For phylogenetic signal analysis, the translated sequences were clustered at 90% sequence homology using CD-HIT v4.7.40 Representative sequences within each cluster were then selected and subsampled for calculating Cas9 and tracrRNA phylogenetic signal.
Identification of Cas9 tracrRNAs
Step1: search for antirepeat signatures
The region of the tracrRNA capable of base pairing with the crRNA, the antirepeat, was identified in Cas9-containing assemblies by searching for sequences with homology to the CRISPR repeat (using BLAST 2.7.3.).41 The parameters used to identify antirepeat signatures include -task “blastn-short,” due to the length of the sequence, as well as disabling the low complexity filter with -dust “no” because CRISPR arrays by definition are low complexity. Sequences that were flanked (distance between hits greater than spacer length) were removed as possible candidates to remove crRNA sequences from consideration (Fig. 1: Step 1). While all assemblies had CRISPR arrays, neither of the two programs (PilerCR or MinCED) accurately detected all of the CRISPR repeats. To correct this and significantly reduce false positives, the coordinates of putative antirepeats in the locus were referenced and used to identify locations that were at least one repeat-spacer unit length away from the CRISPR array.
Step 2: detect rho-independent termination signals
Next, tracrRNA boundaries and directionality were assessed by identifying RTS using ERPIN v5.5 (parameters -add 1 4 1 and -cutoff 100%) and an RTS database (Fig. 1, Step 2).42 Assuming a prototypical Cas9 tracrRNA structure (i.e., antirepeat followed by additional tracrRNA sequence and finally an RTS), the up- and downstream regions adjacent to the antirepeat were scanned for the presence of an RTS. Initially, each antirepeat candidate with its respective RTS was considered a viable tracrRNA candidate. In addition, if an antirepeat had a termination signal on both sides, the pair was considered as potential tracrRNAs. Next, all tracrRNA candidates were conservatively filtered by removing sequences whose combined length was >300 base-pairs. This cutoff value was based on the longest characterized Cas9 tracrRNA length plus a generous buffer.27
Step3: clustering tracrRNA candidates
Putative tracrRNAs were next clustered at 95% sequence identity with a 90% sequence coverage cutoff using cd-hit-est v.4.7.40 Sequence clusters that did not map back to at least five different assemblies were removed from initial clustering under the assumption that rare sequences (found in fewer than five genomes) may be false positives (Fig. 1, Step 3). The resulting sequences and their respective clusters then formed the basis for structural predictions.
Steps 4 and 5: tracrRNA structural predictions and searches for orthologous sequences
To generate a consensus secondary structure for each sequence-based tracrRNA cluster, sequences from each cluster (both 5′ antirepeat and 3′ hairpin-like encoding sequences) were first aligned using MAFFT (—maxiterate 1000—globalpair) and then fed into RNAalifold 2.4.5 (Fig. 1, Step 4).43,44 The resulting consensus folds and sequences were then used as CMs within INFERNAL 1.1.2 to find RNA orthologs within the Cas9-associated DNA assemblies identified earlier (Fig. 1, Step 5).45 To ensure all identified tracrRNAs contained an antirepeat, CM search results were next filtered to remove any hits whose corresponding nucleotide sequence had <55% pairing with either the consensus repeat (from the specific type II system being evaluated) or the reverse complement of it.
Step 6: analysis of CM overlap
Following tracrRNA identification, a final analysis was performed to examine the overlap between CMs. For this, CMs from Steps 4 and 5 were used with INFERNAL 1.1.2 to identify similarities between each putative tracrRNA sequence cataloged in Steps 1–5.45 Results were next visualized by creating an undirected graph. In the graph, CMs were represented as vertices and a line was added between the two vertices if the CMs identified the same putative tracrRNA sequence. Connecting line widths were scaled by the percentage of shared sequences (percent similarity = [no. of shared sequences]/[min (no. found with model 1, no. found with model 2)]). Each network was then pruned for lines separating weakly connected vertices to isolate highly similar clusters. For phylogenetic analyses, all clusters not associated with the top 10 most common structures were removed to make statistical calculations computationally feasible.
Calculating phylogenetic signal
To estimate the degree to which tracrRNA secondary structure associations are evolutionarily conserved among Cas9 lineages, the phylogenetic signal was quantified using both Bayesian and nonparametric approaches. For the Bayesian approach, ancestral states of tracrRNA secondary structures along the Cas9 phylogeny were estimated using maximum likelihood under an All-Rates-Differ model in the R package diversitree.46 Achieving convergence with the full data set was unattainable due to the computational complexity of estimating transition rates with more than 10 discrete tracrRNA states. Thus, the original data set was pruned to include only the 10 most common tracrRNA secondary structures (as described above). Subsequently, this data set was subsampled to represent 25% of the pruned data (512 lineages) while preserving 62 verified tracrRNA sequences.33
With the ancestral state estimates, phylogenetic delta was calculated using time-continuous discrete-trait Markov chain models (2 chains, 100,000 iterations each, thinned every 10 iterations, 100 iterations deleted as burn-in, see Borges et al. for more details).47 Values of phylogenetic delta above 1 indicate a close correspondence of the trait with the phylogeny, with increasing values representing increasing correspondence (i.e., strong phylogenetic signal), whereas values near 0 indicate a weak phylogenetic signal. The results presented below were generated from a subsampling procedure that successfully converged. The Cas9 lineages involved in this calculation can be found in the Supplementary Table S1. To ensure the results were not biased by subsampling, 10 iterations were performed, and the resulting delta values for each round of subsampling can be found in the Supplementary Table S2.
The second approach for quantifying the phylogenetic signal was a modified two-block partial least-squares test.48 This procedure utilized the pruned data set described above before the resampling procedures (2050 lineages included) and quantified the correlation coefficient of the Cas9 phylogeny (converted to a phylogenetic covariance matrix) with the trait matrix. Multivariate effect size and significance were calculated using residual randomization via permutation procedures (1000 iterations, R package geomorph).49,50
Results
Identification of type II CRISPR-Cas9 systems
A total of 41,999 putative type II CRISPR-Cas9 systems from over 1 million microbial nucleotide sequences were identified (Supplementary Table S3). Cas9 length ranged from 700 to 1800 amino acids and exhibited a bimodal length distribution centered around 1100 and 1400 amino acids (Supplementary Fig. S1). To calculate the phylogenetic relationship between tracrRNA and Cas9, 2724 diverse and representative systems were also selected (Supplementary Table S3) and subsampled (the Methods section and Supplementary Table S1). In addition, as a control for our methods, 79 type II CRISPR-Cas9 systems with an experimentally validated tracrRNA were also included in our analysis (Supplementary Table S4).33 Of these, a Cas9 encoding ORF was only detected for 73 using our methods.
Detection of Cas9 tracrRNAs using CMs
The detection of Cas9 tracrRNAs was automated using a multistep approach that combines both homology and structural searches (Fig. 1). First, building upon previous methods, the identification of the tracrRNA antirepeat and rho-independent termination-like signal was automated similar to that described in Chyou et al. (Fig. 1, Steps 1 and 2).29,32,51,52 Next, based on functional associations between the gRNA secondary structure and orthogonality, we reasoned that conserved tracrRNA structural features could be used to complement homology-dependent methods in the identification of a tracrRNA.34
To accomplish this, sequences of tracrRNAs predicted in Steps 1 and 2 (Fig. 1) were first aligned and clustered based on sequence similarity (Fig. 1, Step 3). Next, sequences (including both 5′ antirepeat and 3′ hairpin-like structures) within each cluster were used to predict a consensus secondary structure (Fig. 1, Step 4). CMs were next generated from each cluster based on sequence and structural homology and used to search for related tracrRNAs (Fig. 1, Step 5).
Finally, to examine the relationship between tracrRNAs in our collection, a last clustering step based on CM similarity was applied (Fig. 1, Step 6). For some systems, multiple solutions were observed within the CRISPR-Cas9 locus after Step 6 (Fig. 1) (Supplementary Fig. S2A–E). In these cases, additional filtering was applied to permit the selection of a single tracrRNA. For this, we initially experimented with several different metrics (most thermodynamically stable structure, network cluster with highest connectivity between CMs, and largest cluster network), but proximity to the cas9 gene produced the best fit. In situations where two or more tracrRNAs in the region closest to the cas9 gene had overlapping locations, the tracrRNA with the most stable secondary structure (based on minimum free energy calculations [Supplementary Table S3]) was chosen.
Next, our pipeline was used to predict tracrRNA solutions for the 41,999 CRISPR-Cas9 systems identified earlier. To establish false-positive and false-negative rates of our approach, its ability to accurately predict the tracrRNA from a curated set of 73 experimentally validated Cas9 tracrRNAs was also evaluated.33 For this, the loci containing the curated tracrRNAs were identified and flagged in our collection. Altogether, our algorithm predicted a tracrRNA for 98% (41,741 out of 41,999) of the Cas9 systems searched (Supplementary Table S3). For the curated set of tracrRNAs proven to support Cas9 functionality, our approach correctly identified 90.4% (66 out of 73) resulting in false-positive and false-negative rates of 5.5% (5 out of 69) and 2.7% (2 out of 73), respectively. Of the five systems where a different tracrRNA was identified, four (Cco, Kki, Lsp1, and Nsa) were predicted to have a tracrRNA that was transcribed in the opposite direction from the antirepeat than described earlier. For the fifth system, the tracrRNA was predicted to be in a different location (Ghy3) (Supplementary Fig. S2A–E).33
To compare our method with previous ones, we also calculated the accuracy of two other tracrRNA prediction pipelines, CRISPROne and TracrPredictor, using the diverse collection of 73 experimentally determined Cas9 tracrRNAs used to benchmark our approach.32,33,51 For this, the genomes encoding the Cas9 systems from the validated collection were used as input in CRISPROne and TracrPredictor and the location and orientation of the identified tracrRNA used to assess each approach. TracrPredictor successfully predicted 50.7% (37 out of 73) of these with false-positive and false-negative rates of 9.5% (4 out of 42) and 42.5% (31 out of 73), respectively (Supplementary Table S5). For CRISPROne, 17.8% of the tracrRNAs (identified as an antirepeat) were successfully located with a false-positive rate of 76.4% (42 out of 55) and a false-negative rate of 24.7% (18 out of 73) (Supplementary Table S5).
Sequence and structural homology of Cas9 gRNAs
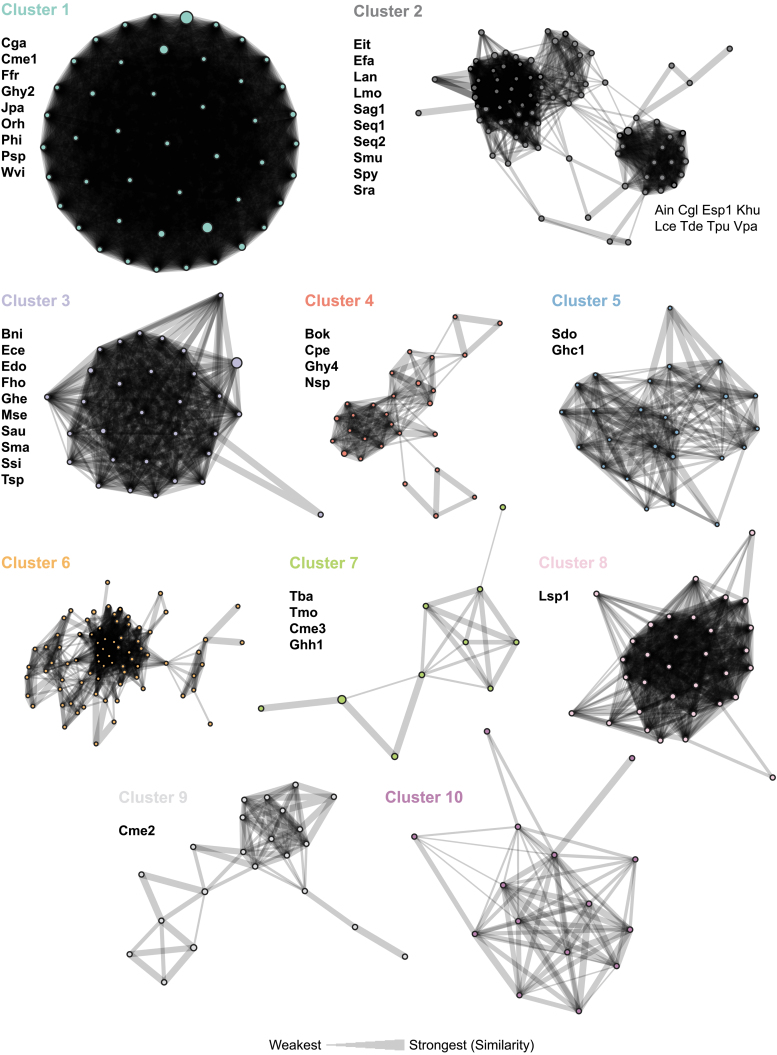
Based on sequence and structural overlap, 94.7% (39,527 out of 41,741) of the identified tracrRNAs could be categorized into 10 clusters (Fig. 2). One thousand three hundred and eighty-eight of the 2214 remaining tracrRNAs were classified into 31 additional CM-based similarity groups and 826 of the remainders represented as singletons in the data set (Supplementary Table S3). The majority of previously characterized tracrRNAs could be found in the 10 most abundant clusters (Fig. 2, clusters 1–7 and 10).
FIG. 2.
Top 10 covariance models and clustering of Cas9 tracrRNAs. Undirected graphs of the top 10 Cas9 tracrRNA clusters based on similarity between sequence and predicted secondary structure CMs. Vertices represent a CM and are colored according to the designated cluster. The width of the connecting lines indicates the percentage of similarity or relatedness among CMs. The number of circles in each cluster indicates the degree of sequence diversity. Previously characterized tracrRNAs associated with each cluster are indicated. CM, covariance model.
The relatedness among CMs within each cluster was also examined. Here, CMs representing tracrRNA sequences that clustered at 90% sequence identity were used as vertices in undirected graphs (Fig. 2). In addition, shared sequence and structural homology between CMs were connected with a line scaled to reflect the percentage of tracrRNAs that fit both models (Fig. 2). In this way, the relationship and diversity of CMs within each cluster could be visualized (Fig. 2). Cluster 1 was the most diverse as it contained the most nodes, however, all CMs were highly related (Fig. 2). In contrast, clusters 2, 4, 6, 7, and 9 were smaller and yielded subgroups that were weakly connected by three of fewer CMs (Fig. 2). Clusters 3, 5, 8, and 10 showed higher rates of connectivity among CMs than clusters 2, 4, 6, 7, and 9, with a few more distantly related tracrRNA groups on the periphery (Fig. 2).
To visualize tracrRNA structural features in the context of the gRNA used by Cas9, an sgRNA was generated by linking the 3′ end of the full-length CRISPR repeat with a self-folding tetraloop (5′-GAAA-3′) to the 5′ end of the antirepeat in the tracrRNA as described previously (Supplementary Fig. S3).9 This was done once for each of the top 10 clusters using the most abundant tracrRNA sequence and respective CRISPR repeat.
As observed previously, most sgRNA structures comprised varying degrees of complementation between the repeat and antirepeat followed by two or more hairpin-like structures in the tracrRNA (Supplementary Fig. S3).27,31,33,34 Likewise, a repeat:antirepeat mismatch resulting in a bulge was detected in some but not all instances (Supplementary Fig. S3). In most cases, the nexus fold, a functionally important and conserved hairpin structure hypothesized to orient the spacer away from the rest of the dual gRNA, was detected almost immediately (within 2 or 3 nts) after the repeat:antirepeat duplex (Supplementary Fig. S3, clusters 1, 2, 4–6, 7, and 9).27,34 For clusters 3, 8, and 10, it was located ∼9 nts after the repeat:antirepeat (Supplementary Fig. S3). The nexus-like fold itself varied in length from 10 to 80 nts with an average length of 24 nts and ranged from simple 3 nts stem loop structures (Supplementary Fig. S3, clusters 1, 4, and 6) to more complex structures with additional bulges and stems (Supplementary Fig. S3, clusters 3 and 9).
Cas9 and tracrRNA evolutionary association
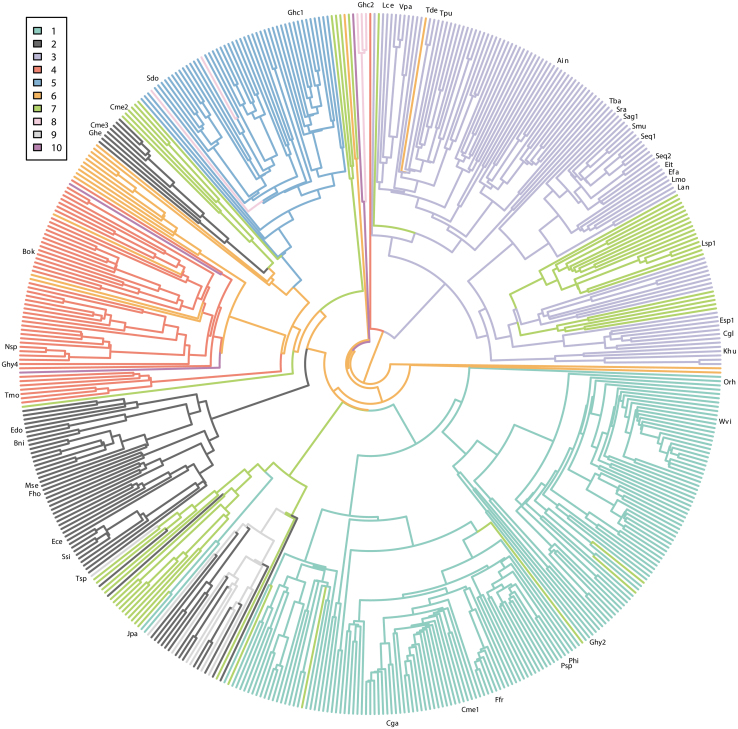
Cas9 phylogeny and predicted gRNA secondary structures have been linked to exchangeability between orthologous Cas9s.31,34 This suggests a tight evolutionary association between Cas9 and its gRNA structural features. To further test this observation, we examined the phylogenetic association between tracrRNA secondary structure and Cas9 protein. For this, two statistical methods, Bayesian estimation of the phylogenetic delta statistic and a nonparametric-modified two-block partial least-squares model, were used to evaluate the phylogenetic relationship between the 10 primary tracrRNA secondary structures (encompassing 83.3% of all representative tracrRNAs identified) and our representative collection of Cas9 proteins. First, a diverse and representative collection of Cas9s were subsampled and a phylogenetic tree was constructed. Next, tracrRNA structures were mapped to it (Fig. 3). Ancestral states were then estimated, from which the delta statistic was calculated. In these scenarios, a significant phylogenetic signal was detected using both approaches (delta = 244.107 and r-PLS (partial least squares) = 0.912, effect size = 28.952, p = 1e-04) as can be visualized by the strong clustering of tracrRNA secondary structures across Cas9 phylogeny (Fig. 3).47,49 Rare exceptions to this were observed as the occurrence of the same tracrRNA structure in distantly related orthologs (Fig. 3).
FIG. 3.
Cas9 phylogeny and tracrRNA secondary structure. Predicted tracrRNA secondary structures associated with Cas9 phylogeny. Each color represents tracrRNA secondary structure associated with clusters 1–10 (Fig. 2). Cas9 proteins characterized previously are indicated (Supplementary Table S4).
Discussion
We provide a framework for the global identification of CRISPR-Cas9 tracrRNAs. Our method builds upon previous approaches that have sought to identify the key components that define a tracrRNA, the antirepeat, and 3′ hairpin-like secondary structures, and adds to them by utilizing CMs to identify sequence and structural homologues (Fig. 1, Steps 1–5).29,32,51,52 In total, we predicted a tracrRNA solution from 98% of the identified Cas9 systems.
In comparison with a diverse collection of experimentally determined tracrRNAs, we also showed that our approach in most cases (66 out of 73 [90.4%]) could accurately identify a Cas9 tracrRNA.33 This can be contrasted with other tracrRNA detection methods, CRISPROne and TracrPredictor, that had difficulty identifying the majority of tracrRNAs in our diverse benchmarking set. In the five instances where a different tracrRNA was detected (Cco, Ghy3, Kki, Lsp1, and Nsa) with our pipeline, it is also a possibility that a second tracrRNA may have evolved in the CRISPR-Cas9 locus as described previously.27 This is supported, in part, by the identification of alternative tracrRNAs in these loci that exhibit CM homology to tracrRNAs known to support Cas9 functionality (Fig. 2 and Supplementary Figs. S2A–E). In addition, the location of the alternate tracrRNA within the CRISPR-Cas9 locus is consistent with other characterized systems. These locations include regions near the end of the CRISPR array or directly adjacent to the cas9 gene (Supplementary Figs. S2A–E).
In examining the sequence and structural overlap of the identified tracrRNAs using CMs (Fig. 1, Step 6), we found that they could be classified mainly into 10 groups (Fig. 2). In general, when observing the distribution for previously determined tracrRNAs, it seems that our structural classifications also correlated with Cas9-gRNA compatibility (Fig. 2).31,33,53 Exceptions to this were observed in cluster 2, where Cas9 and gRNAs (Spy and Tde) previously shown to be incompatible were grouped together (Fig. 2).53 This finding also matches what was observed in our clustering analysis and indicates that our methods could be improved by removing edges with low connectivity between CMs. Altogether, our findings suggest that the number of noncross reactive gRNA groupings may be extended from 7 to 10 or more pending further experimentation.33,34,53
Both approaches for calculating the phylogenetic signal showed that the tracrRNA structure is an evolutionarily conserved trait among Cas9 lineages. This matches previous observations that Cas9-gRNA exchangeability is associated with the gRNA secondary structure and Cas9 phylogeny.31,34 Interestingly, exceptions to this were noted in our analysis. In those instances, distantly related Cas9 orthologs were associated with the same tracrRNA structural classification. This observation may provide evidence of more recent evolutionary events resulting from the resetting of the tracrRNA or recombination between different CRISPR-Cas9 systems.27
Conclusion
Using CMs based on both sequence homology and predicted structure, an informatic approach, enabling the identification of Cas9 tracrRNAs, was developed. This method permitted the global identification of more than 41K tracrRNAs and the development of sgRNA solutions for nearly all CRISPR-Cas9 systems detected by us. Structural predictions revealed strong homology among the tracrRNA secondary structures that tightly correlated with Cas9 phylogeny. Altogether, the results presented here will aid in the characterization and development of new Cas9s as genome editing tools and may be extended to other CRISPR systems that utilize a tracrRNA.23,53–57
Supplementary Material
Authors' Contributions
S.K.D., E.K.B., W.N.M., A.H., and J.K.Y. designed the research; S.K.D. performed the research; S.K.D., E.K.B., and J.K.Y. analyzed the data. S.K.D., E.K.B., A.H., and J.K.Y. wrote the article. All authors read and approved the final article and it has not been published, in press, or submitted elsewhere.
Acknowledgments
We thank Dr. Dean Adams (Iowa State University) for his advice and expertise on how to calculate the phylogenetic signal and for his quick replies to bugs in the phylogenetic analysis. In addition, we thank Kevin Hayes for supporting the early stages of this analysis and Corteva for funding the initial stages of this research. We also thank the ISU legal team and Lynne Mumm for her work in facilitating the collaboration agreements between Corteva and Iowa State. Finally, we thank Dr. James Reece and the Office of the Vice President for Research at Iowa State University for funding me (S.K.D.).
Author Disclosure Statement
S.K.D., E.K.B., W.N.M., and A.H. have no competing financial interests. J.K.Y. is an employee of Corteva Agriscience™.
Funding Information
This research was funded by Iowa State University.
Supplementary Material
References
- 1. Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. 10.1099/mic.0.27437-0 [DOI] [PubMed] [Google Scholar]
- 2. Bolotin A, Quinquis B, Sorokin A, et al. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology (Reading). 2005;151:2551–2561. 10.1099/mic.0.28048-0 [DOI] [PubMed] [Google Scholar]
- 3. Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. 10.1007/s00239-004-0046-3 [DOI] [PubMed] [Google Scholar]
- 4. Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. 10.1038/nature09886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Garneau JE, Dupuis ME, Villion M, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. 10.1038/nature09523 [DOI] [PubMed] [Google Scholar]
- 6. Horvath P, Romero DA, Coute-Monvoisin AC, et al. Diversity, activity, and evolution of CRISPR loci in Streptococcus thermophilus. J Bacteriol. 2008;190:1401–1412. 10.1128/JB.01415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mojica FJM, Díez-Villaseñor C, García-Martínez J, et al. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155(Pt 3):733–740. DOI: 10.1099/mic.0.023960-0 [DOI] [PubMed] [Google Scholar]
- 8. Deveau H, Barrangou R, Garneau JE, et al. Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol. 2008;190:1390–1400. 10.1128/JB.01412-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jinek M, Chylinski K, Fonfara I, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gasiunas G, Barrangou R, Horvath P, et al. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci U S A. 2012;109:E2579–E2586. 10.1073/pnas.1208507109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. 10.1126/science.1232033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schwank G, Koo BK, Sasselli V, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. 10.1016/j.stem.2013.11.002 [DOI] [PubMed] [Google Scholar]
- 14. Wu Y, Liang D, Wang Y, et al. Correction of a genetic disease in mouse via use of CRISPR-Cas9. Cell Stem Cell. 2013;13:659–662. 10.1016/J.STEM.2013.10.016 [DOI] [PubMed] [Google Scholar]
- 15. Shi J, Gao H, Wang H, et al. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15:207–216. 10.1111/pbi.12603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wang Z, Wang Y, Wang S, et al. CRISPR-Cas9 HDR system enhances AQP1 gene expression. Oncotarget. 2017;8:111683–111696. 10.18632/oncotarget.22901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jakociunas T, Bonde I, Herrgard M, et al. Multiplex metabolic pathway engineering using CRISPR/Cas9 in Saccharomyces cerevisiae. Metab Eng. 2015;28:213–222. 10.1016/j.ymben.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 18. Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. 10.1016/j.cell.2013.06.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31:833–838. 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Perez-Pinera P, Kocak DD, Vockley CM, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Methods. 2013;10:973–976. 10.1038/nmeth.2600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hilton IB, D'Ippolito AM, Vockley CM, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat Biotechnol. 2015;33:510–517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gaudelli NM, Komor AC, Rees HA, et al. Programmable base editing of A*T to G*C in genomic DNA without DNA cleavage. Nature. 2017;551:464–471. 10.1038/nature24644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Morgan SL, Mariano NC, Bermudez A, et al. Manipulation of nuclear architecture through CRISPR-mediated chromosomal looping. Nat Commun. 2017;8:15993. 10.1038/ncomms15993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhou Y, Wang P, Tian F, et al. Painting a specific chromosome with CRISPR/Cas9 for live-cell imaging. Cell Res. 2017;27:298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576:149–157. 10.1038/s41586-019-1711-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yang L, Yang B, Chen J. One prime for all editing. Cell. 2019;179:1448–1450 [DOI] [PubMed] [Google Scholar]
- 27. Faure G, Shmakov SA, Makarova KS, et al. Comparative genomics and evolution of trans-activating RNAs in Class 2 CRISPR-Cas systems. RNA Biol. 2019;16:435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mohanraju P, Makarova KS, Zetsche B, et al. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353:aad5147. [DOI] [PubMed] [Google Scholar]
- 29. Chylinski K, Le Rhun A, Charpentier E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013;10:726–737. 10.4161/rna.24321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shmakov S, Smargon A, Scott D, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15:169–182. 10.1038/nrmicro.2016.184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fonfara I, Le Rhun A, Chylinski K, et al. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014;42:2577–2590. 10.1093/nar/gkt1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chyou TY, Brown CM. Prediction and diversity of tracrRNAs from type II CRISPR-Cas systems. RNA Biol. 2019;16:423–434. 10.1080/15476286.2018.1498281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gasiunas G, Young JK, Karvelis T, et al. A catalogue of biochemically diverse CRISPR-Cas9 orthologs. Nat Commun. 2020;11:5512. 10.1038/s41467-020-19344-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Briner AE, Donohoue PD, Gomaa AA, et al. Guide RNA functional modules direct Cas9 activity and orthogonality. Mol Cell. 2014;56:333–339. 10.1016/j.molcel.2014.09.019 [DOI] [PubMed] [Google Scholar]
- 35. Edgar RC. PILER-CR: Fast and accurate identification of CRISPR repeats. BMC Bioinform. 2007;8:18. 10.1186/1471-2105-8-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bland C, Ramsey TL, Sabree F, et al. CRISPR recognition tool (CRT): A tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinform. 2007;8:209. 10.1186/1471-2105-8-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. 10.1371/journal.pcbi.1002195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cock PJ, Antao T, Chang JT, et al. Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics. 2009;25:1422–1423. 10.1093/bioinformatics/btp163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Edgar RC. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004;5:113. 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fu L, Niu B, Zhu Z, et al. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Altschul SF, Gish W, Miller W, et al. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 42. Gautheret D, Lambert A. Direct RNA motif definition and identification from multiple sequence alignments using secondary structure profiles. J Mol Biol. 2001;313:1003–1011. 10.1006/jmbi.2001.5102 [DOI] [PubMed] [Google Scholar]
- 43. Katoh K, Misawa K, Kuma K, et al. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lorenz R, Bernhart SH, Höner zu Siederdissen C, et al. ViennaRNA package 2.0. Algorithms Mol Biol. 2011;6:26. 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. 10.1093/bioinformatics/btt509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. FitzJohn RG. Diversitree: Comparative phylogenetic analyses of diversification in R. Methods Ecol Evol. 2012;3:1084–1092. 10.1111/j.2041-210X.2012.00234.x [DOI] [Google Scholar]
- 47. Borges R, Machado JP, Gomes C, et al. Measuring phylogenetic signal between categorical traits and phylogenies. Bioinformatics. 2019;35:1862–1869. 10.1093/bioinformatics/bty800 [DOI] [PubMed] [Google Scholar]
- 48. Rohlf FJ, Corti M. Use of two-block partial least-squares to study covariation in shape. Syst Biol. 2000;49:740–753. 10.1080/106351500750049806 [DOI] [PubMed] [Google Scholar]
- 49. Collyer ML, Adams DC, Freckleton R. RRPP: An R package for fitting linear models to high-dimensional data using residual randomization. Methods Ecol Evol. 2018;9:1772–1779. 10.1111/2041-210x.13029 [DOI] [Google Scholar]
- 50. Adams DC, Collyer ML. Phylogenetic ANOVA: Group-clade aggregation, biological challenges, and a refined permutation procedure. Evolution. 2018;72:1204–1215. 10.1111/evo.13492 [DOI] [PubMed] [Google Scholar]
- 51. Zhang Q, Ye Y. Not all predicted CRISPR-Cas systems are equal: Isolated cas genes and classes of CRISPR like elements. BMC Bioinform. 2017;18:92. 10.1186/s12859-017-1512-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Karvelis T, Gasiunas G, Young J, et al. Rapid characterization of CRISPR-Cas9 protospacer adjacent motif sequence elements. Genome Biol. 2015;16:253. 10.1186/s13059-015-0818-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Esvelt KM, Mali P, Braff JL, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. 10.1038/nmeth.2681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zetsche B, Gootenberg JS, Abudayyeh OO, et al. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163:759–771. 10.1016/j.cell.2015.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Burstein D, Harrington LB, Strutt SC, et al. New CRISPR-Cas systems from uncultivated microbes. Nature. 2017;542:237–241. 10.1038/nature21059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Harrington LB, Burstein D, Chen JS, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362:839–842. 10.1126/science.aav4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yan WX, Hunnewell P, Alfonse LE, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363:88–91. 10.1126/science.aav7271 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.