Abstract
The ongoing coronavirus disease 2019 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), represents a public health crisis of unprecedented proportions. After the emergence of SARS-CoV-1 in 2002, and Middle East respiratory syndrome coronavirus (MERS-CoV) in 2012, this is the third outbreak of a highly pathogenic zoonotic coronavirus (CoV) that the world has witnessed in the last 2 decades. Infection with highly pathogenic human CoVs often results in a severe respiratory disease characterized by a delayed and blunted interferon (IFN) response, accompanied by an excessive production of proinflammatory cytokines. This indicates that CoVs developed effective mechanisms to overcome the host innate immune response and promote viral replication and pathogenesis. In this review, we describe the key innate immune signaling pathways that are activated during infection with SARS-CoV-2 and other well studied pathogenic CoVs. In addition, we summarize the main strategies that these viruses employ to modulate the host immune responses through the antagonism of IFN induction and effector pathways.
Keywords: SARS-CoV-2, coronavirus, innate immunity, interferon, immune evasion
Introduction
In early December of 2019, an outbreak of pneumonia of unknown etiology was reported in Wuhan, China. One month later, viral sequencing identified a novel coronavirus (CoV) as the causative pathogen of this new disease that quickly spread around the globe and has now caused the greatest pandemic the world has seen in a century. As of now, this novel coronavirus disease 2019 (COVID-19) pandemic resulted in over 142 million confirmed infections and 3 million deaths (Coronaviridae Study Group of the International Committee on Taxonomy of Viruses 2020).
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological agent of COVID-19, is an enveloped positive-sense single-stranded RNA (ssRNA) virus that belongs to lineage B of the Betacoronavirus genus, and it is closely related to SARS-CoV-1 that caused the first SARS epidemic in the early 2000s (Lu and others 2020; Wu and others 2020; Zhu and others 2020). In addition to SARS-CoV-1 and SARS-CoV-2, 5 other human coronaviruses (hCoVs) have been reported: Middle East respiratory syndrome coronavirus (MERS-CoV), a highly pathogenic lineage C Betacoronavirus, 4 seasonal hCoVs: 2 Betacoronaviruses (OC43 and HKU1), and 2 Alphacoronaviruses (229E and NL63).
While seasonal CoVs primarily cause upper respiratory tract infection and mild disease (reviewed in Forni and others 2017; Fung and others 2020), infection with high pathogenic CoVs is associated with a wide range of symptoms that can range from mild to severe respiratory illness, including acute respiratory distress syndrome, which requires mechanical ventilation and intensive care (Drosten and others 2003; Fowler and others 2003; Yang and others 2020a). Aberrant activation of the innate immune response by these viruses is believed to contribute significantly to the development of severe symptoms (Faure and others 2014; Channappanavar and others 2016; Blanco-Melo and others 2020). However, there are also great differences between COVID-19, MERS, and SARS. In fact, despite resulting in similar clinical manifestations, MERS and SARS are more likely to be fatal or severe than COVID-19, but their transmissibility is much lower (Liu and others 2020).
Molecular clock analysis indicates that Alphacoronaviruses entered the human population in spillover events from wildlife between 800 and 200 years ago, while the seasonal Betacoronaviruses OC43 and HKU1 emerged more recently (120 and 60 years ago, respectively) (Vijgen and others 2005; Huynh and others 2012). In addition, as mentioned above, in the last 2 decades, the world has seen the emergence of 3 novel highly pathogenic Betacoronaviruses: MERS-CoV, SARS-CoV-1, and more recently SARS-CoV-2.
Genomic analyses have proposed that all hCoVs emerged from bat or rodent reservoirs and could have either directly jumped into human populations or were introduced via intermediate species like cattle, camels, or palm civets (Guan and others 2003; Li and others 2005; Vijgen and others 2006; Corman and others 2015; Lau and others 2015). For instance, while SARS-CoV-2 has been shown to be very closely related to bat CoV isolates, SARS-CoV-2 related viruses were also identified in the Malayan pangolin (Manis javanica), suggesting they may have served as an intermediate host for the transmission to humans (Lam and others 2020; Zhou and others 2020). Interestingly, the current COVID-19 pandemic may not have been the first global outbreak caused by a CoV. In fact, it has been proposed that the seasonal CoV OC43 spilled over from a bovine reservoir in the late 19th century and was the etiological agent of the “Russian flu”-pandemic that caused over 1 million reported deaths worldwide (Vijgen and others 2005).
In this review, we briefly describe our understanding of how CoV infection is sensed by the host innate immune system. In addition, we discuss the main strategies that CoVs employ to modulate and effectively blunt innate immune activation, with an emphasis on the interferon (IFN) induction pathways.
Genome Organization and Life Cycle
All CoVs have a linear genome of 26–30 kb that encode for 16 nonstructural proteins (NSPs), 4 structural proteins, and an array of accessory proteins (reviewed in Fehr and Perlman 2015).
The NSPs of CoVs are encoded by 2 open reading frames (ORFs), ORF1a and ORF1b that are translated into polyproteins pp1a and pp1ab. Autoproteolytic cleavage of these polyproteins generates 15–16 NSPs with different functions during the viral life cycle.
The structural and accessory genes instead are translated from subgenomic RNAs (sgRNAs) that are generated during genome transcription and replication. Among the structural proteins, the spike protein (S) is responsible for attachment to the host receptors and membrane fusion for almost all CoVs (Li 2016). The nucleocapsid protein (N) binds to the RNA genome to form the ribonucleocapsid and facilitate packaging. Membrane (M) and envelope (E) are anchored in the viral envelope to regulate particle maturation and provide structural integrity (Vennema and others 1996). Conversely, the accessory proteins are not essential for virus replication, and their number varies significantly among different CoVs. Notably, they have been shown to play an important role in viral pathogenesis, in some cases by antagonizing the IFN induction and signaling pathways, as discussed more in more detail below (Siu and others 2009; Lu and others 2011; Lui and others 2016; Hu and others 2017; Chang and others 2020).
The CoV life cycle is completely cytoplasmic. Upon binding to the receptor and fusion of the virus with the target membrane, the genomic RNA (gRNA) is released into the cytosol of the infected cells and serves as a transcript to allow translation of the viral polyproteins that are then processed to form functional replication and transcription complexes that are tightly associated with host's cellular membranes. Importantly, the full-length gRNA is replicated via a negative-sense intermediate that serves as a template for the synthesis of new gRNAs. Particle assembly occurs at the ER–Golgi intermediate compartment (ERGIC), and mature virions are released through vesicle fusion with the cell membrane (Romero-Brey and Bartenschlager 2014; Klein and others 2020).
Recognition of Viral Infection by Innate Immune Sensors
Vertebrates have evolved a highly organized defense network against invading pathogens. The innate immune system relies on the recognition of conserved pathogen associated molecular patterns (PAMPs) by a limited number of germline-encoded pattern-recognition receptors (PRRs) (reviewed in Riera Romo and others 2016; Pelka and De Nardo 2018). Type I IFN is an integral component of this defense network and coordinates the expression of antiviral effector genes and the establishment of the adaptive immune response (Le Bon and Tough 2002; Stetson and Medzhitov 2006). In addition, mammalian cells are also equipped with an array of constitutively expressed genes with intrinsic antiviral activity. These proteins, known as restriction factors, inhibit viral replication directly, often before the onset of the IFN response. However, most of these proteins can also be further induced by IFN to amplify their activity (Colomer-Lluch and others 2018).
The importance of these highly coordinated defense mechanisms for the resolution of infection is further emphasized by the fact that pathogenic viruses have developed multiple tools to escape or at least counteract innate immune responses (Garcia-Sastre 2017; Miorin and others 2017). Indeed, while the seasonal CoV 229E has been shown to induce high levels of IFN and IFN stimulated genes (ISGs) (Mesel-Lemoine and others 2012; Loo and others 2020), infection with SARS-CoV-1, MERS-CoV, and SARS-CoV-2 is known to induce only low levels of IFN, likely due to the expression of numerous innate immune antagonists by these highly pathogenic viruses (reviewed in de Wit and others 2016; Xia and Shi 2020).
Virus infections are mainly sensed by PRRs that recognize nucleic acids. These PRRs include retinoic acid associated gene-I (RIG-I)-like receptors (RLRs), the heterogeneous groups of cytosolic DNA sensors (CDSs), and some Toll-like receptors (TLRs) (Baccala and others 2009; Wu and Chen 2014), among others (Fig. 1).
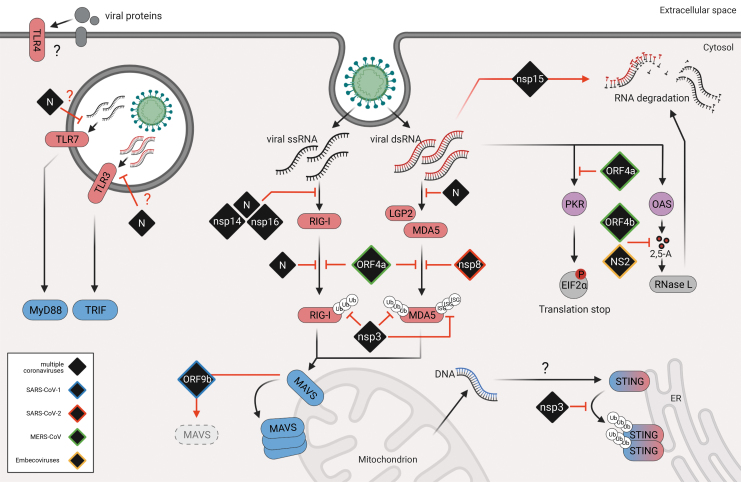
FIG. 1.
Sensing of coronavirus infection and viral antagonism. Upon entry, viral nucleic acids are sensed by PRRs (red). While TLR3 and TLR7 sense endosomal viral RNA and TLR4 recognizes viral glycoproteins in the membrane, the RLRs RIG-I, MDA5, and LGP2 sense cytosolic RNAs. PRRs then activate adaptor proteins MAVS, MyD88, and TRIF (blue) to trigger downstream signaling. Additionally, cytosolic dsRNA also triggers PKR and OASes (purple), which in turn phosphorylates eIF2a to shut down protein translation and activate RNaseL to degrade cytosolic RNA, respectively. Viral infection may also lead to cell damage and subsequent release of DNA into the cytosol, which can trigger cytosolic DNA sensors such as the sensor-adaptor molecule STING (blue-red). Viral proteins (black) that have been described to interfere with interferon induction are depicted in black and have color coded borders to indicate their origin (blue = SARS-CoV-1, red = SARS-CoV-2, green = MERS-CoV, yellow = Embecoviruses, black = multiple coronaviruses). dsRNA, double-stranded RNA; eIF2a, eukaryotic translation initiation factor; LGP2, laboratory of genetics and physiology 2; MAVS, mitochondrial antiviral-signaling protein; MDA5, melanoma differentiation-associated gene 5; MERS-CoV, Middle East respiratory syndrome coronavirus; OASes, 2′,5′-oligoadenylate synthases; PKR, protein kinase R; PRR, pattern-recognition receptor; RIG-I, retinoic acid-associated gene-I; RLR, retinoic acid-associated gene-I (RIG-I)-like receptor; SARS-CoV, severe acute respiratory syndrome coronavirus; TLR, Toll-like receptor.
TLRs are exclusively expressed in lymphocytes and are responsible for detection of a wide range of PAMPs (Baccala and others 2009; Riera Romo and others 2016). TLR3, TLR7, and TLR9 are involved in viral sensing and are all expressed in endosomes or lysosome-related organelles (Kawasaki and Kawai 2014). TLR7 and TLR9 trigger the MyD88 pathway, while TLR3 signals via TRIF to activate an immune response (Kawai and Akira 2007). The group of CDSs comprises proteins like cGAS and IFI-16 that recognize non-self and cellular DNA in the cytoplasm, and signal through the adaptor protein STING, which mainly resides in ER membranes (Ishikawa and Barber 2008; Burdette and Vance 2013; Wu and Chen 2014; Aguirre and Fernandez-Sesma 2017).
The RLR family comprises 2 major sensor proteins RIG-I and melanoma differentiation associated gene 5 (MDA5) and the less characterized regulator laboratory of genetics and physiology 2 (LGP2), and signals through the adaptor protein mitochondrial antiviral-signaling protein (MAVS) (Yoneyama and others 2015).
Upon viral sensing, all the PRRs described above trigger the initiation of harmonized signaling pathways that ultimately lead to the expression of IFN and inflammatory cytokines via transcription factors like NFKB and interferon regulatory transcription factor 3 (IRF3) (reviewed in Wu and Chen 2014; Chen and others 2017).
Secreted IFN then binds to the IFN receptors (IFNAR) in an autocrine and paracrine fashion and activates Janus kinase 1 (JAK1) and Tyrosine kinase 2 (TYK2) (Platanias 2005). These events trigger the phosphorylation of signal transducer and activator of transcription 1 and 2 (STAT1 and STAT2), which then dimerize and interact with IRF9 to form the IGF3 complex (Schindler and others 2007). ISGF3 in turn translocates into the nucleus and binds to the Interferon-Stimulated Response Element (ISRE) promoter to initiate the expression of hundreds of ISGs and the establishment of an antiviral state (reviewed in Schneider and others 2014).
Innate Immune Sensing of CoV Infection
CoVs are RNA viruses that generate double-stranded RNA (dsRNA) intermediates during replication and are therefore subjected to cytosolic RNA recognition by RLRs. While RIG-I senses short dsRNAs carrying a triphosphate or diphosphate moiety at the 5′ terminus, MDA5 has been described to sense primarily long dsRNAs as those generated during picornavirus and CoV replication (Pichlmair and others 2009; Zalinger and others 2015; Yin and others 2021). Interestingly, LGP2, the third member of the family, has the highest affinity to RNA of all RLRs, which may play a key role in its regulatory function (Takahasi and others 2009). However, the physiological RNA agonists of both MDA5 and LGP2 during infection still remain enigmatic.
Both RIG-I and MDA5 have been shown to be involved in the detection of mouse hepatitis virus (MHV) in a cell type-dependent manner (Roth-Cross and others 2008; Li and others 2010). In addition, MDA5-deficient mice failed to induce a protective IFN response and exhibited more severe disease than wild-type mice following MHV infection (Zalinger and others 2015). In line with this observation, we and others have recently shown that MDA5 and its positive regulator LGP2, but not RIG-I, act as the key innate immune sensors in lung epithelial cells during infection with SARS-CoV-2 (Rebendenne and others 2021; Yin and others 2021). However, whether SARS-CoV-2 replication only generates MDA5-dependent agonists, or simply better antagonizes RIG-I activation still remains unclear.
In contrast, RLR signaling appeared to be dispensable for IFN induction during MERS-CoV infection. Indeed, TLR7/MyD88 knockout mice but not MAVS-deficient mice showed more severe disease and reduced IFN induction with respect to control animals after infection with MERS-CoV (Zhao and others 2014; Channappanavar and others 2019).
TLR signaling has also been implicated in the control of other CoV infections. Indeed, several studies have shown that mice deficient in different TLRs or downstream adaptor molecules exhibit increased mortality, weight loss, and viral titers in response to infection. Specifically, TLR3 and TLR7 appear to be of particular importance. Interestingly, both proteins localize at the endosomal membrane, however, while TLR3 is known to sense dsRNA, TLR7 specifically recognizes ssRNA (Alexopoulou and others 2001; Heil and others 2004).
TLR7 has been shown to be critical for the secretion of IFN by plasmacytoid dendritic cells upon infection with both MHV and MERS-CoV (Cervantes-Barragan and others 2006; Scheuplein and others 2015). In addition, mice deficient in MyD88 were shown to be unable to mount a protective immune response and to succumb in response to a SARS-CoV-1 and MERS-CoV challenge (Sheahan and others 2008; Zhao and others 2014).
Interestingly, a similar protective role in the host response to SARS-CoV-1 has also been attributed to TLR3 and the adaptor molecule TRIF. TRIF knockout animals had mortality, weight loss, and viral titers that were comparable to those of the MyD88 knockout mice. However, rather that showing an impaired IFN response, they exhibited elevated levels of IFN and proinflammatory cytokines late in infection that resulted in increased lung pathology This suggest that some additional sensing pathways may compensate for the lack of TLR3 during SARS-CoV-1 infection (Totura and others 2015).
Surprisingly, TLR4, which recognizes bacterial lipopolysaccharides and viral glycoproteins, has also been shown to play an important role in the control of CoV infection. Indeed, TLR4 knockout mice appeared to be more susceptible to MHV and SARS-CoV-1 infection. This may be due to the recognition of the heavily glycosylated spike protein by TLR4 receptors on the surface of innate immune cells (Totura and others 2015). Additionally, the important role of TLR signaling in the establishment of a balanced innate immune response to pathogenic CoV infection has also been recently highlighted by the identification of TLR7 and TLR3 loss-of-function mutations in patients with severe COVID-19 (van der Made and others 2020; Zhang and others 2020).
As mentioned above, virus infected cells can also rapidly trigger expression of different auxiliary RNA-binding proteins with antiviral activity. The interferon-induced proteins with tetratricopeptide repeats (IFITs) belong to this class of antiviral factors, and are among the highest expressed genes during the innate antiviral responses. In addition to antagonizing translation initiation by direct interaction with eukaryotic initiation factor 3 (eIF3), IFIT proteins have been also shown to inhibit infection by sequestering viral RNAs with specific 5′-motifs moieties from the replication sites or by promoting RNA degradation (reviewed in Diamond 2014; Mears and Sweeney 2018).
Specifically, IFIT proteins have been shown to restrict viral replication upon recognition of messenger RNAs (mRNAs) lacking a 2′-O-methylated cap and therefore, CoVs express a 2′-O-methyltransferase to overcome this restriction (Daffis and others 2010). In addition, several studies have indicated that IFIT proteins are important for the immune response against CoV infection. In this respect, knock-down of both IFIT1 and IFIT2 in Vero cells resulted in increased viral titers upon infection with SARS-CoV-1. Moreover, enhanced SARS-CoV-1 replication was also observed in the lungs of IFIT1 deficient mice with respect to control animals (Menachery and others 2014). Similarly, IFIT2 knockout mice also exhibited increased viral replication in the central nervous system and liver in addition to increased morbidity and mortality upon infection with MHV (Butchi and others 2014).
Lastly, increasing evidence suggests that the cGAS-STING pathway, in addition to recognizing dsDNA from DNA viruses, is also playing a role in restricting RNA virus infection by sensing nuclear or mitochondrial DNA that is released in the cytoplasm due to infection-mediated collateral damage (Aguirre and Fernandez-Sesma 2017). In this respect, a recent study suggested that while SARS-CoV-2 infection poorly triggers the activation of the IRF3 and IFN pathways, it quite effectively triggers the activation of NFkB by the cGAS-STING pathway (Neufeldt and others 2020). In addition, the papain-like protease (PLP) of SARS-CoV-1, NL63 and more recently also SARS-CoV-2, which is part of the nsp3 gene, are able to inhibit ubiquitination and activation of STING (Sun and others 2012; Rui and others 2021). However, more studies are needed to explore the impact of the activation of the cGAS-STING pathway in the pathogenesis of hCoV infection.
Mechanisms of Innate Immune Evasion by hCoVs
IFN is the master regulator of the innate antiviral immune response. It can modulate the expression of hundreds of genes and it is extremely effective in restricting viral replication. As a consequence, SARS-CoV-2 and many other CoVs are known to be sensitive to treatment with IFNs (Hensley and others 2004; Li and others 2010; Lei and others 2020; Miorin and others 2020). Therefore, it is not surprising that CoVs developed multiple mechanisms to evade or inhibit IFN induction and signaling pathways during evolution.
Here, we will address the key viral strategies that CoVs exploit to control IFN induction: (1) hiding or modifying viral RNA to avoid recognition by host antiviral sensors; (2) inhibition of PRR activation; and (3) disruption of signaling pathways responsible for IFN induction (Figs. 1 and 2). The existence of these mechanisms explains why IFN induction is significantly attenuated in cells infected with CoVs (Spiegel and others 2005; Kindler and others 2013; Blanco-Melo and others 2020).
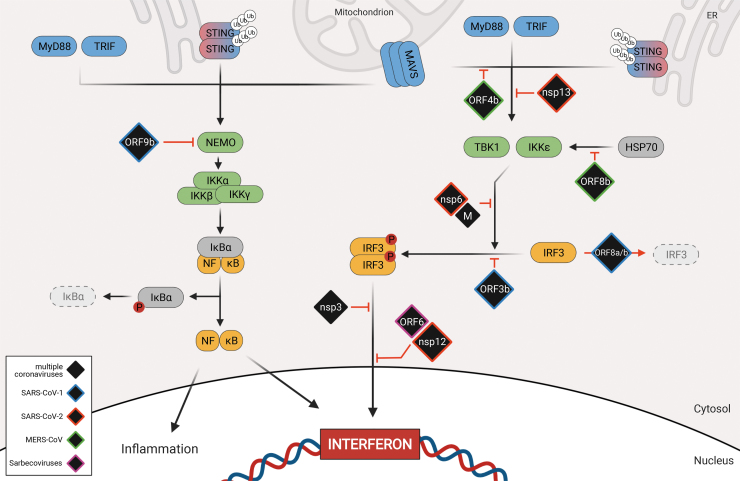
FIG. 2.
Signaling downstream of PRRs activation and viral antagonism. After PRR activation, adaptor molecules initiate the formation of different signaling complexes. TBK1 and IKKe form a complex with other proteins to phosphorylate IRF3, which can then dimerize and translocate into the nucleus. Simultaneously, NEMO triggers the formation of the IKK complex, which then initiates degradation of IKBa and allows for nuclear translocation of NFKB. In the nucleus, the 3 transcription factors IRF3, NFKB, and AP1 together stimulate the production of type I/type III interferon and proinflammatory cytokines. Viral proteins (black) that have been described to interfere with interferon induction are depicted at the step of signaling cascade they antagonize. Viral proteins have color coded borders to indicate their origin (blue = SARS-CoV-1, red = SARS-CoV-2, purple = Sarbecoviruses, green = MERS-CoV, black = multiple coronaviruses). IRF3, interferon regulatory transcription factor 3.
Evasion of Recognition by Host Antiviral Sensors
Like other positive-sense RNA viruses, CoVs assemble their transcription/replication complexes into membrane-associated compartments that are believe to help concentrating viral and host factors required for efficient replication, while shielding viral nucleic-acids from recognition by host antiviral sensors (Stertz and others 2007; Knoops and others 2008; Paul and Bartenschlager 2013; Snijder and others 2020; Wolff and others 2020). In addition, viral RNA recognition is also prevented by several other mechanisms. For instance, the CoV nucleocapsid protein (N), which has the structural function of encapsidating the viral genome, has also been shown to interfere with IFN induction at different levels.
At the very early steps of infection, N is believed to protect viral RNA from recognition by PRRs through its RNA binding ability (Kopecky-Bromberg and others 2007; Lu and others 2011; Lei and others 2020). In addition, at later steps in the RIG-I activation pathway, both MERS-CoV and SARS-CoV-1 N proteins are able to interact with the E3 ubiquitin ligase TRIM25 to block RIG-I ubiquitination and activation of downstream signaling (Hu and others 2017; Chang and others 2020).
CoV NSP14 and NSP16, which are part of the replicase complex, have also been involved in IFN antagonism. NSP14 has guanine-N7-methyltransferase and is involved in the formation of a 5′ terminal cap structure on the viral RNA that can mimic host mRNA (Chen and others 2009). In addition, NSP16 has 2′-O-MTAse activity that is critical to evade RNA recognition by MDA5 and IFIT proteins. Indeed, MHV mutant viruses that lack 2′-O-MTAse activity trigger and enhanced IFN response and are attenuated in vivo (Daffis and others 2010; Menachery and others 2014, 2017).
Similarly, loss of NSP15 endoribonuclease activity also resulted in robust activation of host dsRNA sensors and MHV attenuation due to an increase in the number of free-dsRNA foci that are not confined within the viral replication compartment (Deng and others 2017; Yuen and others 2020). Finally, the MERS-CoV ORF4a protein has been shown to bind to the RLR cofactor PACT in an RNA-dependent manner to suppress antiviral signaling and promote viral replication (Almazan and others 2013; Yang and others 2013; Siu and others 2014).
Inhibition of PRRs Activation
A rapid and tightly controlled activation of RLR signaling is a crucial step for the initiation of an effective antiviral response. Therefore, the activities of RIG-I, MDA5, and LGP2 are regulated by several types of posttranslational modifications, including ubiquitination, conjugation of ubiquitin-like molecule, and phosphorylation, among others (Cadena and Hur 2019; Kato and others 2021). As a consequence, pathogenic CoVs also evolved strategies to target these critical regulatory mechanisms.
As mentioned above, the N protein of SARS-CoV-1 and MERS-CoV can interact with TRIM25 and impair RIG-I ubiquitination (Hu and others 2017; Chang and others 2020). Similarly, NSP8 of SARS-CoV-2 has recently been described to bind to the CARD domains of MDA5 to suppress its K63-linked polyubiquitination and prevent expression of type I IFN and inflammatory cytokines (Yang and others 2020b). In addition, CoVs also express proteins with deubiquitinase activity (DUBs) that have been shown to target key components of the IFN induction pathway (Devaraj and others 2007; Frieman and others 2009; Bailey-Elkin and others 2014; Shin and others 2020; Liu and others 2021). In this respect, the PLPs of many CoVs have been suggested to remove ubiquitin and the ubiquitin-like molecule ISG15 from cellular proteins (Clementz and others 2010; Klemm and others 2020).
Porcine epidemic diarrhea virus (PEDV) PLP2 has been shown to have DUB activity and interact with RIG-I and STING (Xing and others 2013). Consequently, PLP2 catalytically inactive mutants do not affect ubiquitination of their targets and are less efficient at suppressing IFN induction. MERS-CoV PLP has also been shown to exert DUB and deISGylase activity. However, its substrates still remain to be identified (Mielech and others 2014).
Interestingly, it was recently shown that while SARS-CoV-1 preferentially cleaves Ub from its substrates, SARS-CoV-2 PLP more efficiently reduce the appearance of ISGylated protein substrates (Shin and others 2020). Mechanistically, SARS-CoV-2 PLP was shown to specifically bind to MDA5, but not to RIG-I, and to suppress MDA5 CARD ISGylation to prevent its oligomerization and the activation of downstream signaling events. Notably, this PLP-dependent mechanism of antagonism of MDA5 activation appears to be widely conserved among different CoVs (SARS-CoV-1, MERS, MHV, and NL63), highlighting the key role of MDA5 ISGylation in the IFN induction during CoV infection (Liu and others 2021).
Disruption of Signaling Pathways Responsible for IFN Induction
As mentioned briefly above, upon viral infection, RLR stimulation results in the activation of the adaptor protein MAVS that in turn recruits TNF receptor-associated factors (TRAFs) and NEMO to activate the kinases IKK and TBK1 (Ea and others 2006; Saha and others 2006). While IKK triggers the NFKB pathway through IKBα, TBK1 phosphorylates IRF3, which dimerizes and translocates into the nucleus (Wu and others 2006; Liu and others 2013). IRF3 and NFKB, together with other factors serve as transcription factors for type I IFN induction (Fig. 1, left panel) (Yoneyama and others 2002; Liu and others 2015).
Interestingly, the SARS-CoV-1 accessory protein ORF9b has been shown to localize at the mitochondria and to inhibit RLR signaling by targeting MAVS for ubiquitin-mediated proteasomal degradation (Shi and others 2014). In addition, the ORF9b proteins of both SARS-CoV-1 and SARS-CoV-2 were recently shown to bind to, and colocalize with the mitochondrial outer membrane protein Tom70, which is known to be involved in the activation of MAVS signaling and apoptosis (Lin and others 2010; Gordon and others 2020). However, the functional significance of this interaction remains to be elucidated.
MERS-CoV ORF4b is also an important antagonist of IFN induction. Ectopically expressed ORF4b was shown to interact with IKKe and TBK1 to prevent their interaction with MAVS and inhibit IRF3 phosphorylation (Yang and others 2015). In addition, the MERS-CoV ORF8b protein has been shown to counteract IKKɛ activation by disrupting IKKe-HSP70 interaction, which is known to enhance IRF3 activation. Consistently, infection with a recombinant virus carrying a deletion in ORF8b resulted in an enhanced IFN induction and in the rescue of the colocalization between HSP70 and IKKe (Wong and others 2020).
Moreover, both SARS-CoV-2 NSP6 and NSP13 were shown to bind to TBK1 to prevent IRF3 activation. Interestingly, while binding of NSP13 to TBK1 resulted in an impaired TBK1 phosphorylation, the NSP6-TBK1 interaction resulted in a block of IRF3 phosphorylation (Gordon and others 2020; Xia and others 2020; Yuen and others 2020). More studies will be required to define the detailed molecular mechanism by which these viral proteins exert their antagonistic functions. Interestingly, the M proteins of all 3 highly pathogenic CoVs have also been shown to target TBK1. In fact, M has been proposed to block IRF3 phosphorylation by preventing the formation of a protein complex between TBK1, TRAF3, IKKe, and other factors. However, the specific interactors of M differ between SARS-CoV-1, SARS-CoV-2, and MERS-CoV (Siu and others 2009; Lui and others 2016; Zheng and others 2020).
The key transcription factor IRF3 is also directly targeted by several viral proteins. SARS-CoV-1 ORF8ab and ORF8b have been shown to actively degrade IRF3 in a ubiquitin-dependent fashion (Wong and others 2018). In addition, overexpression of SARS-CoV-1 ORF3b could also inhibit IFN induction by blocking IRF3 phosphorylation and nuclear translocation (Kopecky-Bromberg and others 2007). Interestingly, despite that ORF3b protein of SARS-CoV-2 has relatively low homology with the SARS-CoV-1 protein, due to the presence of a premature stop codon, it appears to exert the same function. Indeed, a recent overexpression study indicates that SARS-CoV-2 ORF3b is able to suppress IFN induction even more efficiently than ORF3b of SARS-CoV-1 (Konno and others 2020). Furthermore, the same study also identified a SARS-CoV-2 ORF3b variant that expresses an elongated protein due to a mutation on the premature stop codon, with even more increased anti-IFN activity in vitro. However, the specific effect of this variant on IFN induction and viral replication during infection, still remains to be addressed.
The PLP proteins of MHV, SARS-CoV-1, and SARS-CoV-2 were also shown to target IRF3. Interestingly, while MHV PLP could interact with IRF3 and block IRF3 ubiquitination and nuclear translocation (Zheng and others 2008), SARS-CoV-1 PLP was shown to inhibit IFN induction at a step downstream of IRF3 nuclear localization and DNA binding (Matthews and others 2014). Conversely, SARS-CoV-2 PLP was recently reported to cleave IRF3 in vitro via its protease activity (Moustaqil and others 2021).
The ORF6 proteins of both SARS-CoV-1 and SARS-CoV-2 have also been proposed to block IRF3 in addition to STAT1 and STAT2 nuclear translocation (Frieman and others 2007; Kopecky-Bromberg and others 2007; Lei and others 2020; Miorin and others 2020; Xia and others 2020; Yuen and others 2020). However, while we could show that IFN-dependent STAT translocation is blocked by direct interaction of ORF6 with the nucleoporin NUP98, it still remains to be clarified whether ORF6 affects IRF3 nuclear import through the same mechanism. Lastly, a recent study also demonstrated that SARS-CoV-2 NSP12 can block nuclear translocation of IRF3 without impairing its phosphorylation. While the polymerase function of NSP12 was dispensable for this phenotype, the exact mode of action needs to be further explored (Wang and others 2021).
Induction and Evasion of the Protein Kinase R and OAS/RNAseL Pathways
dsRNA-dependent protein kinase R (PKR) and 2′,5′-oligoadenylate synthases (OASes) constitute 2 other antiviral defense systems that are activated upon the recognition of cytosolic dsRNAs (reviewed in Sadler and Williams 2008).
PKR is a serine/threonine kinase that is constitutively expressed at basal levels in all tissues and is upregulated in response to IFN (Haines and others 1998). Binding to dsRNA triggers PKR activation and results in its dimerization and autophosphorylation at multiple residues. Active PKR then phosphorylates the alpha subunit of the eukaryotic translation initiation factor (eIF2a) to inhibit viral and host cell translation and trigger stress granule (SGs) formation (Balachandran and others 2000). In addition, PKR can also regulate other signaling pathways to control apoptosis, cell growth, and differentiation through mechanisms that are incompletely understood (Ishii and others 2001; Bonnet and others 2006; Taghavi and Samuel 2012).
Similar to PKR, the OAS family of proteins consists of a homologous set of enzymes that are encoded by ISGs that are also constitutively expressed at low levels at steady state and therefore can act as PRRs for the recognition of cytoplasmic viral RNA. Binding of dsRNA by OASes results in the conversion of ATP into 2′,5′-oligoadenylates (2–5A) that serve as second messengers to activate latent endoribonuclease (RNaseL) (reviewed in Sadler and Williams 2008; Taghavi and Samuel 2012). Activated RNaseL monomers then dimerize and cleave viral and cellular ssRNAs with little specificity to trigger apoptosis of the infected cells (Zhou and others 1997).
Both of these pathways have been shown to be activated during CoV infection. In fact, several studies have shown that infection with multiple CoVs generally triggers the phosphorylation of both PKR and eIF2a, suggesting that dsRNA recognition by PKR is common during CoVs infections (Bechill and others 2008; Wang and others 2009; Cruz and others 2011; Liao and others 2013). Interestingly, infection with SARS-CoV-1 and SARS-CoV-2 was shown to trigger the activation of both PKR and RNAseL in a cell type-dependent manner (Li and others 2021). However, while RNAseL was required to restrict viral replication, eIF2a phosphorylation levels and viral titers were not affected in PKR-deficient cells, indicating that infection with these 2 highly pathogenic CoVs is likely to trigger activation of multiple kinases in the cell stress response (Krahling and others 2009; Li and others 2021).
In contrast to SARS-CoVs, MERS-CoV and MHV appear to be more effective at inhibiting these pathways. In this respect, the accessory protein NS2 of MHV and related betacoronaviruses was shown to have 2′-5′-phosphodiesterase activity that triggers 2–5A cleavage to prevent activation of RNAseL and to block viral RNA degradation. Consistently, a recombinant virus carrying a mutation that abrogates the NS2 enzymatic activity was significantly attenuated in wild-type mice but was highly pathogenic in RNaseL-deficient mice, highlighting the important role of this pathway for viral restriction (Zhao and others 2012; Goldstein and others 2017).
In addition, multiple studies have shown that MERS-CoV ORF4a can bind to dsRNA to block PKR activation and SG formation (Rabouw and others 2016; Comar and others 2019); and ORF4b, similar to NS2, is able to prevent RNAseL activation by degrading 2–5A (Thornbrough and others 2016; Comar and others 2019). However, in this case, recombinant viruses carrying mutations or deletions in these proteins were only modestly attenuated, strongly suggesting that MERS-CoV might have evolved multiple mechanisms for inhibiting the actions of these antiviral host factors (Comar and others 2019; Li and others 2021).
Conclusions
As described above, CoVs have evolved a plethora of mechanisms to control innate immune activation at multiple levels (Table 1). This highlights the importance of bypassing the establishment of a timely and harmonized host response for efficient viral replication. Importantly, the poor and delayed activation of the IFN response combined with excessive production of proinflammatory cytokines constitute the hallmark of the immune dysregulation that is associated with severe illness and poor disease outcome in SARS, MERS, and COVID-19 patients. This model is supported by several studies showing that an appropriate IFN response is necessary to control virus replication and spread in hamsters and mice (Zhao and others 2014; Channappanavar and others 2016, 2019; Boudewijns and others 2020; Hoagland and others 2021).
Table 1.
Interferon Antagonistic Functions of Viral Proteins
| Protein | Target/mechanism | Virus | Citation |
|---|---|---|---|
| Nonstructural | |||
| NSP1 | Inhibition of mRNA export; binding of 40S ribosomal subunit | SARS-CoV-1; SARS-CoV-2; MERS-CoV; hCoV-NL63; hCoV-OC43; MHV | Lei and others (2013); Lokugamage and others (2015); Wang and others (2010); Schubert and others (2020); Zhang and others (2021) |
| NSP3 | Deubiquitylation of RIG-I; deubiquitylation of STING; deubiquitylation and DeISGylation of MDA5 | SARS-CoV-1; SARS-CoV-2; MERS-CoV; HCoV-NL63; MHV, PEDV | Clementz and others (2010); Xing and others (2013); Mielech and others (2014); Klemm and others (2020); Shin and others (2020) |
| NSP6 | Interaction with TBK1 prevents IRF3 phosphorylation | SARS-CoV-2 | Xia and others (2020) |
| NSP8 | Prevention of MDA5 polyubiquitylation | SARS-CoV-2 | Yang and others (2020b) |
| NSP12 | Inhibition of IRF3 nuclear translocation | SARS-CoV-2 | Wang and others (2021) |
| NSP13 | Interaction with TBK1 prevents TBK1 phosphorylation | SARS-CoV-2 | Xia and others (2020); Yuen and others (2020) |
| NSP14 | Capping of viral RNAs | SARS-CoV-2 | Chen and others (2009); Yuen and others (2020) |
| NSP15 | Degradation of viral dsRNA | SARS-CoV-2; MHV | Deng and others (2017); Yuen and others (2020) |
| NSP16 | Capping of viral RNA | SARS-CoV-1; SARS-CoV-2; MERS-CoV; MHV | Daffis and others (2010); Menachery and others (2014); Menachery and others (2017) |
| Structural | |||
| M | Disruption of TBK1-dependent IRF3 phosphorylation | SARS-CoV-1; SARS-CoV-2; MERS-CoV | Siu and others (2009); Lui and others (2016); Zheng and others (2020) |
| N | Shielding of viral RNA; inhibition of TRIM25-dependent RIG-I ubiquitylation | SARS-CoV-1; SARS-CoV-2; MERS-CoV | Kopecky-Bromberg and others (2007); Lu and others (2011); Lei and others (2020) |
| Accessory | |||
| ORF3b | Block of IRF3 phosphorylation | SARS-CoV-1; SARS-CoV-2 | Kopecky-Bromberg and others (2007); Konno and others (2020) |
| ORF4a | RNA-dependent sequestration of PACT (RLR cofactor); sequestration of dsRNA to prevent PKR activation | MERS-CoV | Almazan and others (2013); Yang and others (2013); Siu and others (2014); Rabouw and others (2016); Comar and others (2019) |
| ORF4b | Degradation of 2′,5′-oligoadenylate prevents RNaseL activation; disruption of MAVS interaction with TBK1/IKKe | MERS-CoV | Yang and others (2015); Thornbrough and others (2016); Comar and others (2019); Li and others (2021) |
| ORF6 | Inhibition of IRF3 nuclear translocation | SARS-CoV-1; SARS-CoV-2 | Frieman and others (2007); Kopecky-Bromberg and others (2007); Xia and others (2020); Yuen and others (2020) |
| ORF8(a/b) | Ubiquitin-dependent degradation of IRF3 | SARS-CoV-1 | Wong and others (2018) |
| ORF8b | Inhibition of HSP70-dependent activation of IKKe | MERS-CoV | Wong and others (2020) |
| ORF9b | Ubiquitin-dependent degradation of MAVS | SARS-CoV-1 | Shi and others (2014) |
| NS2 | Degradation of 2′,5′-oligoadenylate prevents RNaseL activation | HCoV-OC43; MHV; ECoV; BEV | Zhao and others (2012); Goldstein and others (2017) |
dsRNA, double-stranded RNA; hCoV, human coronavirus; IRF3, interferon regulatory transcription factor 3; M, membrane; MAVS, mitochondrial antiviral-signaling protein; MDA5, melanoma differentiation-associated gene 5; MERS-CoV, Middle East respiratory syndrome coronavirus; MHV, mouse hepatitis virus; mRNA, messenger RNA; N, nucleocapsid protein; NSP, nonstructural protein; ORF, open reading frame; PEDV, porcine epidemic diarrhea virus; PKR, protein kinase R; RIG-I, retinoic acid-associated gene-I; RLR, retinoic acid-associated gene-I (RIG-I)-like receptor; SARS-CoV, severe acute respiratory syndrome coronavirus.
In addition, autoantibodies against IFN, and monogenic inborn errors of innate immunity have been found in patients with severe COVID-19 (Bastard and others 2020; Zhang and others 2020), further highlighting the key role of the IFN-mediated response in the control of CoV infection and disease progression. Depending on the virus, the efficiency of IFN inhibition may vary significantly, and it is clearly more pronounced during infection with highly pathogenic CoVs. However, there are only a couple of studies that directly compare antagonistic proteins of different CoVs to each other, and even less studies that do it in the context of infection. More comparative studies, especially between low and highly pathogenic viruses are needed to dissect pathogenesis and to identify common and unique mechanisms of CoV-related diseases.
Importantly, differences in the ability of viral proteins to interact with key innate immune factors may also play a critical role in cross-species transmission. For instance, mouse adaptation of both SARS-CoV-1 and SARS-CoV-2 resulted in the acquisition of nonsynonymous mutations in different nonstructural and accessory proteins, some of which are known to antagonize the IFN response (Roberts and others 2007; Leist and others 2020; Rathnasinghe and others 2021). This suggests that additional mutations, which are not related to receptor-binding, are likely required for adaptation to a new host.
Finally, while CoV innate immune antagonism mechanisms have been extensively explored, much less is known about viral RNA sensing during infection. The importance of some PRRs such us TLRs, MDA5, LGP2, and PKR has been demonstrated. However, we still lack a detailed understanding of the molecular mechanisms underlying their activation. More is to be discovered as to where viral PAMPs are being sensed within the cell, and about the kinetics of sensing during the course of infection.
Most importantly, almost nothing is known about the nature of the viral agonists that are detected. Gaining better insight into sensing, viral antagonistic strategies, and innate immune responses to this family of viruses is crucial to develop better antiviral treatments, but also preventive medicines such as vaccines. The attention CoVs have gotten during the last 2 decades, and especially in the last year, must be used to gain a deep understanding of virus–host interactions that result in the activation and evasion of innate immunity, for not only CoVs but also other viral pathogens.
Acknowledgments
We thank Ms. Anastasija Cupic and all members of the A.G.-S. laboratory for helpful feedback. Due to space constraints, we regret our inability to cite all relevant studies. Biorender was used to make all figures.
Author Disclosure Statement
The A.G.-S. laboratory has received research support from Pfizer, Senhwa Biosciences, Kenall Manufacturing, Avimex, Johnson & Johnson, Dynavax, 7Hills Pharma, Pharmamar, ImmunityBio, Accurius, Nanocomposix and Merck. A.G.-S. has consulting agreements for the following companies involving cash and/or stock: Vivaldi Biosciences, Contrafect, 7Hills Pharma, Avimex, Vaxalto, Pagoda, Accurius, Esperovax, Farmak and Pfizer. A.G.-S. is inventor on patents and patent applications on the use of antivirals and vaccines for the treatment and prevention of virus infections, owned by the Icahn School of Medicine at Mount Sinai, New York.
Funding Information
Research on CoV in the A.G.-S. laboratory is partly funded by CRIPT (Center for Research on Influenza Pathogenesis and Transmission), a National Institute of Allergy and Infectious Diseases (NIAID) supported Center of Excellence for Influenza Research and Response (contracts 75N93019R00028 and HHSN272201400008C); by supplements to NIAID grants R35HL135834, U19AI135972, and U19AI142733 and NIAID contract 75N93019C00045; by a National Cancer Institute (NCI) grant U54CA260560; by a supplement to DoD grant W81XWH-20-1-0270; by the Defense Advanced Research Projects Agency (DARPA) grant HR0011-19-20020; by a Fast/Mercatus grant and by the generous support of the JPB Foundation, the Open Philanthropy Project [research grant 2020–215611 (5384)] and anonymous donors to A.G.-S.
References
- Aguirre S, Fernandez-Sesma A. 2017. Collateral damage during dengue virus infection: making sense of DNA by cGAS. J Virol 91(14):e01081-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. 2001. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413(6857):732–738 [DOI] [PubMed] [Google Scholar]
- Almazan F, DeDiego ML, Sola I, Zuniga S, Nieto-Torres JL, Marquez-Jurado S, Andres G, Enjuanes L. 2013. Engineering a replication-competent, propagation-defective Middle East respiratory syndrome coronavirus as a vaccine candidate. mBio 4(5):e00650-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccala R, Gonzalez-Quintial R, Lawson BR, Stern ME, Kono DH, Beutler B, Theofilopoulos AN. 2009. Sensors of the innate immune system: their mode of action. Nat Rev Rheumatol 5(8):448–456 [DOI] [PubMed] [Google Scholar]
- Bailey-Elkin BA, Knaap RC, Johnson GG, Dalebout TJ, Ninaber DK, van Kasteren PB, Bredenbeek PJ, Snijder EJ, Kikkert M, Mark BL. 2014. Crystal structure of the Middle East respiratory syndrome coronavirus (MERS-CoV) papain-like protease bound to ubiquitin facilitates targeted disruption of deubiquitinating activity to demonstrate its role in innate immune suppression. J Biol Chem 289(50):34667–34682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran S, Roberts PC, Brown LE, Truong H, Pattnaik AK, Archer DR, Barber GN. 2000. Essential role for the dsRNA-dependent protein kinase PKR in innate immunity to viral infection. Immunity 13(1):129–141 [DOI] [PubMed] [Google Scholar]
- Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, Dorgham K, Philippot Q, Rosain J, Beziat V, Manry J, Shaw E, Haljasmagi L, Peterson P, Lorenzo L, Bizien L, Trouillet-Assant S, Dobbs K, de Jesus AA, Belot A, Kallaste A, Catherinot E, Tandjaoui-Lambiotte Y, Le Pen J, Kerner G, Bigio B, Seeleuthner Y, Yang R, Bolze A, Spaan AN, Delmonte OM, Abers MS, Aiuti A, Casari G, Lampasona V, Piemonti L, Ciceri F, Bilguvar K, Lifton RP, Vasse M, Smadja DM, Migaud M, Hadjadj J, Terrier B, Duffy D, Quintana-Murci L, van de Beek D, Roussel L, Vinh DC, Tangye SG, Haerynck F, Dalmau D, Martinez-Picado J, Brodin P, Nussenzweig MC, Boisson-Dupuis S, Rodriguez-Gallego C, Vogt G, Mogensen TH, Oler AJ, Gu J, Burbelo PD, Cohen JI, Biondi A, Bettini LR, D'Angio M, Bonfanti P, Rossignol P, Mayaux J, Rieux-Laucat F, Husebye ES, Fusco F, Ursini MV, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Castagnoli R, Montagna D, Licari A, Marseglia GL, Duval X, Ghosn J; HGID Lab; NIAID-USUHS Immune Response to COVID Group; COVID Clinicians; COVID-STORM Clinicians; Imagine COVID Group; French COVID Cohort Study Group; Milieu Interieur Consortium; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; Tsang JS, Goldbach-Mansky R, Kisand K, Lionakis MS, Puel A, Zhang SY, Holland SM, Gorochov G, Jouanguy E, Rice CM, Cobat A, Notarangelo LD, Abel L, Su HC, Casanova JL. 2020. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science 370(6515):eabd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechill J, Chen Z, Brewer JW, Baker SC. 2008. Coronavirus infection modulates the unfolded protein response and mediates sustained translational repression. J Virol 82(9):4492–4501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, Nilsson-Payant BE, Liu WC, Uhl S, Hoagland D, Moller R, Jordan TX, Oishi K, Panis M, Sachs D, Wang TT, Schwartz RE, Lim JK, Albrecht RA, tenOever BR. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5):1036..e9–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnet MC, Daurat C, Ottone C, Meurs EF. 2006. The N-terminus of PKR is responsible for the activation of the NF-kappaB signaling pathway by interacting with the IKK complex. Cell Signal 18(11):1865–1875 [DOI] [PubMed] [Google Scholar]
- Boudewijns R, Thibaut HJ, Kaptein SJF, Li R, Vergote V, Seldeslachts L, Van Weyenbergh J, De Keyzer C, Bervoets L, Sharma S, Liesenborghs L, Ma J, Jansen S, Van Looveren D, Vercruysse T, Wang X, Jochmans D, Martens E, Roose K, De Vlieger D, Schepens B, Van Buyten T, Jacobs S, Liu Y, Marti-Carreras J, Vanmechelen B, Wawina-Bokalanga T, Delang L, Rocha-Pereira J, Coelmont L, Chiu W, Leyssen P, Heylen E, Schols D, Wang L, Close L, Matthijnssens J, Van Ranst M, Compernolle V, Schramm G, Van Laere K, Saelens X, Callewaert N, Opdenakker G, Maes P, Weynand B, Cawthorne C, Vande Velde G, Wang Z, Neyts J, Dallmeier K. 2020. STAT2 signaling restricts viral dissemination but drives severe pneumonia in SARS-CoV-2 infected hamsters. Nat Commun 11(1):5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burdette DL, Vance RE. 2013. STING and the innate immune response to nucleic acids in the cytosol. Nat Immunol 14(1):19–26 [DOI] [PubMed] [Google Scholar]
- Butchi NB, Hinton DR, Stohlman SA, Kapil P, Fensterl V, Sen GC, Bergmann CC. 2014. Ifit2 deficiency results in uncontrolled neurotropic coronavirus replication and enhanced encephalitis via impaired alpha/beta interferon induction in macrophages. J Virol 88(2):1051–1064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena C, Hur S. 2019. Filament-like assemblies of intracellular nucleic acid sensors: commonalities and differences. Mol Cell 76(2):243–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Züst R, Weber F, Spiegel M, Lang KS, Akira S, Thiel V, Ludewig B. 2006. Control of coronavirus infection through plasmacytoid dendritic-cell–derived type I interferon. Blood 109(3):1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CY, Liu HM, Chang MF, Chang SC. 2020. Middle East respiratory syndrome coronavirus nucleocapsid protein suppresses type I and type III interferon induction by targeting RIG-I signaling. J Virol 94(13):e00099-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, Perlman S. 2016. Dysregulated type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe 19(2):181–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Channappanavar R, Fehr AR, Zheng J, Wohlford-Lenane C, Abrahante JE, Mack M, Sompallae R, McCray PB Jr., Meyerholz DK, Perlman S.. 2019. IFN-I response timing relative to virus replication determines MERS coronavirus infection outcomes. J Clin Invest 129(9):3625–3639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Xia P, Li S, Zhang T, Wang TT, Zhu J. 2017. RNA sensors of the innate immune system and their detection of pathogens. IUBMB Life 69(5):297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Cai H, Pan J, Xiang N, Tien P, Ahola T, Guo D. 2009. Functional screen reveals SARS coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc Natl Acad Sci U S A 106(9):3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J Virol 84(9):4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colomer-Lluch M, Ruiz A, Moris A, Prado JG. 2018. Restriction factors: from intrinsic viral restriction to shaping cellular immunity against HIV-1. Front Immunol 9:2876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comar CE, Goldstein SA, Li Y, Yount B, Baric RS, Weiss SR. 2019. Antagonism of dsRNA-induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. mBio 10(2):e00319-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Baldwin HJ, Tateno AF, Zerbinati RM, Annan A, Owusu M, Nkrumah EE, Maganga GD, Oppong S, Adu-Sarkodie Y, Vallo P, da Silva Filho LVRF, Leroy EM, Thiel V, van der Hoek L, Poon LLM, Tschapka M, Drosten C, Drexler JF, Schultz-Cherry S. 2015. Evidence for an ancestral association of human coronavirus 229E with bats. J Virol 89(23):11858–11870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses. 2020. The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5(4):536–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz JL, Sola I, Becares M, Alberca B, Plana J, Enjuanes L, Zuniga S. 2011. Coronavirus gene 7 counteracts host defenses and modulates virus virulence. PLoS Pathog 7(6):e1002090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daffis S, Szretter KJ, Schriewer J, Li J, Youn S, Errett J, Lin TY, Schneller S, Zust R, Dong H, Thiel V, Sen GC, Fensterl V, Klimstra WB, Pierson TC, Buller RM, Gale M Jr., Shi PY, Diamond MS.. 2010. 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468(7322):452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, van Doremalen N, Falzarano D, Munster VJ. 2016. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol 14(8):523–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Hackbart M, Mettelman RC, O'Brien A, Mielech AM, Yi G, Kao CC, Baker SC. 2017. Coronavirus nonstructural protein 15 mediates evasion of dsRNA sensors and limits apoptosis in macrophages. Proc Natl Acad Sci U S A 114(21):E4251–E4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devaraj SG, Wang N, Chen Z, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J Biol Chem 282(44):32208–32221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS. 2014. IFIT1: a dual sensor and effector molecule that detects non-2′-O methylated viral RNA and inhibits its translation. Cytokine Growth Factor Rev 25(5):543–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosten C, Gunther S, Preiser W, van der Werf S, Brodt HR, Becker S, Rabenau H, Panning M, Kolesnikova L, Fouchier RA, Berger A, Burguiere AM, Cinatl J, Eickmann M, Escriou N, Grywna K, Kramme S, Manuguerra JC, Muller S, Rickerts V, Sturmer M, Vieth S, Klenk HD, Osterhaus AD, Schmitz H, Doerr HW. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med 348(20):1967–1976 [DOI] [PubMed] [Google Scholar]
- Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. 2006. Activation of IKK by TNFalpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell 22(2):245–257 [DOI] [PubMed] [Google Scholar]
- Faure E, Poissy J, Goffard A, Fournier C, Kipnis E, Titecat M, Bortolotti P, Martinez L, Dubucquoi S, Dessein R, Gosset P, Mathieu D, Guery B. 2014. Distinct immune response in two MERS-CoV-infected patients: can we go from bench to bedside? PLoS One 9(2):e88716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. 2015. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol 1282:1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forni D, Cagliani R, Clerici M, Sironi M. 2017. Molecular evolution of human coronavirus genomes. Trends Microbiol 25(1):35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler RA, Lapinsky SE, Hallett D, Detsky AS, Sibbald WJ, Slutsky AS, Stewart TE; Toronto SARS Critical Care Group. 2003. Critically ill patients with severe acute respiratory syndrome. JAMA 290(3):367–373 [DOI] [PubMed] [Google Scholar]
- Frieman M, Ratia K, Johnston RE, Mesecar AD, Baric RS. 2009. Severe acute respiratory syndrome coronavirus papain-like protease ubiquitin-like domain and catalytic domain regulate antagonism of IRF3 and NF-kappaB signaling. J Virol 83(13):6689–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frieman M, Yount B, Heise M, Kopecky-Bromberg SA, Palese P, Baric RS. 2007. Severe acute respiratory syndrome coronavirus ORF6 antagonizes STAT1 function by sequestering nuclear import factors on the rough endoplasmic reticulum/Golgi membrane. J Virol 81(18):9812–9824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung SY, Yuen KS, Ye ZW, Chan CP, Jin DY. 2020. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg Microbes Infect 9(1):558–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Sastre A. 2017. Ten strategies of interferon evasion by viruses. Cell Host Microbe 22(2):176–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein SA, Thornbrough JM, Zhang R, Jha BK, Li Y, Elliott R, Quiroz-Figueroa K, Chen AI, Silverman RH, Weiss SR. 2017. Lineage A betacoronavirus NS2 proteins and the homologous Torovirus Berne pp1a carboxy-terminal domain are phosphodiesterases that antagonize activation of RNase L. J Virol 91(5):e02201-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon DE, Hiatt J, Bouhaddou M, Rezelj VV, Ulferts S, Braberg H, Jureka AS, Obernier K, Guo JZ, Batra J, Kaake RM, Weckstein AR, Owens TW, Gupta M, Pourmal S, Titus EW, Cakir M, Soucheray M, McGregor M, Cakir Z, Jang G, O'Meara MJ, Tummino TA, Zhang Z, Foussard H, Rojc A, Zhou Y, Kuchenov D, Huttenhain R, Xu J, Eckhardt M, Swaney DL, Fabius JM, Ummadi M, Tutuncuoglu B, Rathore U, Modak M, Haas P, Haas KM, Naing ZZC, Pulido EH, Shi Y, Barrio-Hernandez I, Memon D, Petsalaki E, Dunham A, Marrero MC, Burke D, Koh C, Vallet T, Silvas JA, Azumaya CM, Billesbolle C, Brilot AF, Campbell MG, Diallo A, Dickinson MS, Diwanji D, Herrera N, Hoppe N, Kratochvil HT, Liu Y, Merz GE, Moritz M, Nguyen HC, Nowotny C, Puchades C, Rizo AN, Schulze-Gahmen U, Smith AM, Sun M, Young ID, Zhao J, Asarnow D, Biel J, Bowen A, Braxton JR, Chen J, Chio CM, Chio US, Deshpande I, Doan L, Faust B, Flores S, Jin M, Kim K, Lam VL, Li F, Li J, Li YL, Li Y, Liu X, Lo M, Lopez KE, Melo AA, Moss FR, 3rd, Nguyen P, Paulino J, Pawar KI, Peters JK, Pospiech TH Jr., Safari M, Sangwan S, Schaefer K, Thomas PV, Thwin AC, Trenker R, Tse E, Tsui TKM, Wang F, Whitis N, Yu Z, Zhang K, Zhang Y, Zhou F, Saltzberg D, QCRG Structural Biology Consortium; Hodder AJ, Shun-Shion AS, Williams DM, White KM, Rosales R, Kehrer T, Miorin L, Moreno E, Patel AH, Rihn S, Khalid MM, Vallejo-Gracia A, Fozouni P, Simoneau CR, Roth TL, Wu D, Karim MA, Ghoussaini M, Dunham I, Berardi F, Weigang S, Chazal M, Park J, Logue J, McGrath M, Weston S, Haupt R, Hastie CJ, Elliott M, Brown F, Burness KA, Reid E, Dorward M, Johnson C, Wilkinson SG, Geyer A, Giesel DM, Baillie C, Raggett S, Leech H, Toth R, Goodman N, Keough KC, Lind AL, Zoonomia C, Klesh RJ, Hemphill KR, Carlson-Stevermer J, Oki J, Holden K, Maures T, Pollard KS, Sali A, Agard DA, Cheng Y, Fraser JS, Frost A, Jura N, Kortemme T, Manglik A, Southworth DR, Stroud RM, Alessi DR, Davies P, Frieman MB, Ideker T, Abate C, Jouvenet N, Kochs G, Shoichet B, Ott M, Palmarini M, Shokat KM, Garcia-Sastre A, Rassen JA, Grosse R, Rosenberg OS, Verba KA, Basler CF, Vignuzzi M, Peden AA, Beltrao P, Krogan NJ.. 2020. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science 370(6521):eabe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, Luo SW, Li PH, Zhang LJ, Guan YJ, Butt KM, Wong KL, Chan KW, Lim W, Shortridge KF, Yuen KY, Peiris JS, Poon LL. 2003. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 302(5643):276–278 [DOI] [PubMed] [Google Scholar]
- Haines GK, 3rd, Panos RJ, Bak PM, Brown T, Zielinski M, Leyland J, Radosevich JA. 1998. Interferon-responsive protein kinase (p68) and proliferating cell nuclear antigen are inversely distributed in head and neck squamous cell carcinoma. Tumour Biol 19(1):52–59 [DOI] [PubMed] [Google Scholar]
- Heil F, Hemmi H, Hochrein H, Ampenberger F, Kirschning C, Akira S, Lipford G, Wagner H, Bauer S. 2004. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 303(5663):1526–1529 [DOI] [PubMed] [Google Scholar]
- Hensley LE, Fritz LE, Jahrling PB, Karp CL, Huggins JW, Geisbert TW. 2004. Interferon-beta 1a and SARS coronavirus replication. Emerg Infect Dis 10(2):317–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DA, Moller R, Uhl SA, Oishi K, Frere J, Golynker I, Horiuchi S, Panis M, Blanco-Melo D, Sachs D, Arkun K, Lim JK, tenOever BR. 2021. Leveraging the antiviral type I interferon system as a first line of defense against SARS-CoV-2 pathogenicity. Immunity 54(3):557..e5–570.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Li W, Gao T, Cui Y, Jin YW, Li P, Ma QJ, Liu X, Cao C. 2017. The severe acute respiratory syndrome coronavirus nucleocapsid inhibits type I interferon production by interfering with TRIM25-mediated RIG-I ubiquitination. J Virol 91(8):e02143-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram S, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86(23):12816–12825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Kwon H, Hiscott J, Mosialos G, Koromilas AE. 2001. Activation of the I kappa B alpha kinase (IKK) complex by double-stranded RNA-binding defective and catalytic inactive mutants of the interferon-inducible protein kinase PKR. Oncogene 20(15):1900–1912 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Barber GN. 2008. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature 455(7213):674–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Ahmad S, Zhu Z, Young JM, Mu X, Park S, Malik HS, Hur S. 2021. Structural analysis of RIG-I-like receptors reveals ancient rules of engagement between diverse RNA helicases and TRIM ubiquitin ligases. Mol Cell 81(3):599..e8–613.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Akira S. 2007. TLR signaling. Semin Immunol 19(1):24–32 [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Kawai T. 2014. Toll-like receptor signaling pathways. Front Immunol 5:461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindler E, Jonsdottir HR, Muth D, Hamming OJ, Hartmann R, Rodriguez R, Geffers R, Fouchier RA, Drosten C, Muller MA, Dijkman R, Thiel V. 2013. Efficient replication of the novel human betacoronavirus EMC on primary human epithelium highlights its zoonotic potential. mBio 4(1):e00611-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P. 2020. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun 11(1):5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemm T, Ebert G, Calleja DJ, Allison CC, Richardson LW, Bernardini JP, Lu BG, Kuchel NW, Grohmann C, Shibata Y, Gan ZY, Cooney JP, Doerflinger M, Au AE, Blackmore TR, van der Heden van Noort GJ, Geurink PP, Ovaa H, Newman J, Riboldi-Tunnicliffe A, Czabotar PE, Mitchell JP, Feltham R, Lechtenberg BC, Lowes KN, Dewson G, Pellegrini M, Lessene G, Komander D. 2020. Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2. EMBO J 39(18):e106275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. 2008. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6(9):e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konno Y, Kimura I, Uriu K, Fukushi M, Irie T, Koyanagi Y, Sauter D, Gifford RJ, Consortium U-C, Nakagawa S, Sato K. 2020. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep 32(12):108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecky-Bromberg SA, Martinez-Sobrido L, Frieman M, Baric RA, Palese P. 2007. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. J Virol 81(2):548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahling V, Stein DA, Spiegel M, Weber F, Muhlberger E. 2009. Severe acute respiratory syndrome coronavirus triggers apoptosis via protein kinase R but is resistant to its antiviral activity. J Virol 83(5):2298–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT, Jia N, Zhang YW, Shum MH, Jiang JF, Zhu HC, Tong YG, Shi YX, Ni XB, Liao YS, Li WJ, Jiang BG, Wei W, Yuan TT, Zheng K, Cui XM, Li J, Pei GQ, Qiang X, Cheung WY, Li LF, Sun FF, Qin S, Huang JC, Leung GM, Holmes EC, Hu YL, Guan Y, Cao WC. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285 [DOI] [PubMed] [Google Scholar]
- Lau SKP, Woo PCY, Li KSM, Tsang AKL, Fan RYY, Luk HKH, Cai J-P, Chan K-H, Zheng B-J, Wang M, Yuen K-Y, Sandri-Goldin RM. 2015. Discovery of a novel coronavirus, China rattus coronavirus HKU24, from Norway rats supports the murine origin of betacoronavirus 1 and has implications for the ancestor of betacoronavirus lineage A. J Virol 89(6):3076–3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bon A, Tough DF. 2002. Links between innate and adaptive immunity via type I interferon. Curr Opin Immunol 14(4):432–436 [DOI] [PubMed] [Google Scholar]
- Lei L, Ying S, Baojun L, Yi Y, Xiang H, Wenli S, Zounan S, Deyin G, Qingyu Z, Jingmei L, Guohui C. 2013. Attenuation of mouse hepatitis virus by deletion of the LLRKxGxKG region of Nsp1. PLoS One 8(4):e61166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei X, Dong X, Ma R, Wang W, Xiao X, Tian Z, Wang C, Wang Y, Li L, Ren L, Guo F, Zhao Z, Zhou Z, Xiang Z, Wang J. 2020. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat Commun 11(1):3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist SR, Dinnon KH, 3rd, Schafer A, Tse LV, Okuda K, Hou YJ, West A, Edwards CE, Sanders W, Fritch EJ, Gully KL, Scobey T, Brown AJ, Sheahan TP, Moorman NJ, Boucher RC, Gralinski LE, Montgomery SA, Baric RS. 2020. A mouse-adapted SARS-CoV-2 induces acute lung injury and mortality in standard laboratory mice. Cell 183(4):1070..e12–1085.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. 2016. Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3(1):237–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Liu Y, Zhang X. 2010. Murine coronavirus induces type I interferon in oligodendrocytes through recognition by RIG-I and MDA5. J Virol 84(13):6472–6482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. 2005. Bats are natural reservoirs of SARS-like coronaviruses. Science 310(5748):676–679 [DOI] [PubMed] [Google Scholar]
- Li Y, Renner DM, Comar CE, Whelan JN, Reyes HM, Cardenas-Diaz FL, Truitt R, Tan LH, Dong B, Alysandratos KD, Huang J, Palmer JN, Adappa ND, Kohanski MA, Kotton DN, Silverman RH, Yang W, Morrisey EE, Cohen NA, Weiss SR. 2021. SARS-CoV-2 induces double-stranded RNA-mediated innate immune responses in respiratory epithelial-derived cells and cardiomyocytes. Proc Natl Acad Sci U S A 118(16):eabe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Fung TS, Huang M, Fang SG, Zhong Y, Liu DX. 2013. Upregulation of CHOP/GADD153 during coronavirus infectious bronchitis virus infection modulates apoptosis by restricting activation of the extracellular signal-regulated kinase pathway. J Virol 87(14):8124–8134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Paz S, Hiscott J. 2010. Tom70 imports antiviral immunity to the mitochondria. Cell Res 20(9):971–973 [DOI] [PubMed] [Google Scholar]
- Liu G, Lee JH, Parker ZM, Acharya D, Chiang JJ, van Gent M, Riedl W, Davis-Gardner ME, Wies E, Chiang C, Gack MU. 2021. ISG15-dependent activation of the sensor MDA5 is antagonized by the SARS-CoV-2 papain-like protease to evade host innate immunity. Nat Microbiol 6(4):467–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Xie W, Wang Y, Xiong Y, Chen S, Han J, Wu Q. 2020. A comparative overview of COVID-19, MERS and SARS: review article. Int J Surg 81:1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Cai X, Wu J, Cong Q, Chen X, Li T, Du F, Ren J, Wu YT, Grishin NV, Chen ZJ. 2015. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science 347(6227):aaa2630. [DOI] [PubMed] [Google Scholar]
- Liu S, Chen J, Cai X, Wu J, Chen X, Wu YT, Sun L, Chen ZJ. 2013. MAVS recruits multiple ubiquitin E3 ligases to activate antiviral signaling cascades. Elife 2:e00785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokugamage KG, Narayanan K, Nakagawa K, Terasaki K, Ramirez SI, Tseng CT, Makino S. 2015. Middle East respiratory syndrome coronavirus nsp1 inhibits host gene expression by selectively targeting mRNAs transcribed in the nucleus while sparing mRNAs of cytoplasmic origin. J Virol 89(21):10970–10981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo SL, Wark PAB, Esneau C, Nichol KS, Hsu AC, Bartlett NW. 2020. Human coronaviruses 229E and OC43 replicate and induce distinct antiviral responses in differentiated primary human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol 319(6):L926–L931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet 395(10224):565–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Pan J, Tao J, Guo D. 2011. SARS-CoV nucleocapsid protein antagonizes IFN-beta response by targeting initial step of IFN-beta induction pathway, and its C-terminal region is critical for the antagonism. Virus Genes 42(1):37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui PY, Wong LY, Fung CL, Siu KL, Yeung ML, Yuen KS, Chan CP, Woo PC, Yuen KY, Jin DY. 2016. Middle East respiratory syndrome coronavirus M protein suppresses type I interferon expression through the inhibition of TBK1-dependent phosphorylation of IRF3. Emerg Microbes Infect 5:e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K, Schafer A, Pham A, Frieman M. 2014. The SARS coronavirus papain like protease can inhibit IRF3 at a post activation step that requires deubiquitination activity. Virol J 11:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mears HV, Sweeney TR. 2018. Better together: the role of IFIT protein-protein interactions in the antiviral response. J Gen Virol 99(11):1463–1477 [DOI] [PubMed] [Google Scholar]
- Menachery VD, Gralinski LE, Mitchell HD, Dinnon KH, 3rd, Leist SR, Yount BL Jr., Graham RL, McAnarney ET, Stratton KG, Cockrell AS, Debbink K, Sims AC, Waters KM, Baric RS.. 2017. Middle East respiratory syndrome coronavirus nonstructural protein 16 is necessary for interferon resistance and viral pathogenesis. mSphere 2(6):e00346-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menachery VD, Yount BL Jr., Josset L, Gralinski LE, Scobey T, Agnihothram S, Katze MG, Baric RS.. 2014. Attenuation and restoration of severe acute respiratory syndrome coronavirus mutant lacking 2′-o-methyltransferase activity. J Virol 88(8):4251–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesel-Lemoine M, Millet J, Vidalain PO, Law H, Vabret A, Lorin V, Escriou N, Albert ML, Nal B, Tangy F. 2012. A human coronavirus responsible for the common cold massively kills dendritic cells but not monocytes. J Virol 86(14):7577–7587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mielech AM, Kilianski A, Baez-Santos YM, Mesecar AD, Baker SC. 2014. MERS-CoV papain-like protease has deISGylating and deubiquitinating activities. Virology 450–451:64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L, Kehrer T, Sanchez-Aparicio MT, Zhang K, Cohen P, Patel RS, Cupic A, Makio T, Mei M, Moreno E, Danziger O, White KM, Rathnasinghe R, Uccellini M, Gao S, Aydillo T, Mena I, Yin X, Martin-Sancho L, Krogan NJ, Chanda SK, Schotsaert M, Wozniak RW, Ren Y, Rosenberg BR, Fontoura BMA, García-Sastre A. 2020. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc Natl Acad Sci U S A 117(45):28344–28354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L, Maestre AM, Fernandez-Sesma A, Garcia-Sastre A. 2017. Antagonism of type I interferon by flaviviruses. Biochem Biophys Res Commun 492(4):587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustaqil M, Ollivier E, Chiu HP, Van Tol S, Rudolffi-Soto P, Stevens C, Bhumkar A, Hunter DJB, Freiberg AN, Jacques D, Lee B, Sierecki E, Gambin Y. 2021. SARS-CoV-2 proteases PLpro and 3CLpro cleave IRF3 and critical modulators of inflammatory pathways (NLRP12 and TAB1): implications for disease presentation across species. Emerg Microbes Infect 10(1):178–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Cerikan B, Cortese M, et al. 2020. SARS-CoV-2 infection induces a pro-inflammatory cytokine response through cGAS-STING and NF-κB. bioRxiv. [Epub ahead of print]; DOI: 10.1101/2020.07.21.212639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Bartenschlager R. 2013. Architecture and biogenesis of plus-strand RNA virus replication factories. World J Virol 2(2):32–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelka K, De Nardo D. 2018. Emerging concepts in innate immunity. Methods Mol Biol 1714:1–18 [DOI] [PubMed] [Google Scholar]
- Pichlmair A, Schulz O, Tan CP, Rehwinkel J, Kato H, Takeuchi O, Akira S, Way M, Schiavo G, Reis e Sousa C. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J Virol 83(20):10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5(5):375–386 [DOI] [PubMed] [Google Scholar]
- Rabouw HH, Langereis MA, Knaap RC, Dalebout TJ, Canton J, Sola I, Enjuanes L, Bredenbeek PJ, Kikkert M, de Groot RJ, van Kuppeveld FJ. 2016. Middle East respiratory coronavirus accessory protein 4a inhibits PKR-mediated antiviral stress responses. PLoS Pathog 12(10):e1005982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathnasinghe R, Jangra S, Cupic A, Martinez-Romero C, Mulder LCF, Kehrer T, Yildiz S, Choi A, Mena I, De Vrieze J, Aslam S, Stadlbauer D, Meekins DA, McDowell CD, Balaraman V, Richt JA, De Geest BG, Miorin L, Krammer F, Simon V, Garcia-Sastre A, Schotsaert M. 2021. The N501Y mutation in SARS-CoV-2 spike leads to morbidity in obese and aged mice and is neutralized by convalescent and post-vaccination human sera. medRxiv. [Epub ahead of print]; DOI: 10.1101/2021.01.19.21249592 [DOI] [Google Scholar]
- Rebendenne A, Valadao ALC, Tauziet M, Maarifi G, Bonaventure B, McKellar J, Planes R, Nisole S, Arnaud-Arnould M, Moncorge O, Goujon C. 2021. SARS-CoV-2 triggers an MDA-5-dependent interferon response which is unable to control replication in lung epithelial cells. J Virol 95(8):e02415-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera Romo M, Perez-Martinez D, Castillo Ferrer C. 2016. Innate immunity in vertebrates: an overview. Immunology 148(2):125–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Deming D, Paddock CD, Cheng A, Yount B, Vogel L, Herman BD, Sheahan T, Heise M, Genrich GL, Zaki SR, Baric R, Subbarao K. 2007. A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3(1):e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, Bartenschlager R. 2014. Membranous replication factories induced by plus-strand RNA viruses. Viruses 6(7):2826–2857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth-Cross JK, Bender SJ, Weiss SR. 2008. Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J Virol 82(20):9829–9838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Su J, Shen S, Hu Y, Huang D, Zheng W, Lou M, Shi Y, Wang M, Chen S, Zhao N, Dong Q, Cai Y, Xu R, Zheng S, Yu XF. 2021. Unique and complementary suppression of cGAS-STING and RNA sensing- triggered innate immune responses by SARS-CoV-2 proteins. Signal Transduct Target Ther 6(1):123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler AJ, Williams BR. 2008. Interferon-inducible antiviral effectors. Nat Rev Immunol 8(7):559–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha SK, Pietras EM, He JQ, Kang JR, Liu SY, Oganesyan G, Shahangian A, Zarnegar B, Shiba TL, Wang Y, Cheng G. 2006. Regulation of antiviral responses by a direct and specific interaction between TRAF3 and Cardif. EMBO J 25(14):3257–3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuplein VA, Seifried J, Malczyk AH, Miller L, Hocker L, Vergara-Alert J, Dolnik O, Zielecki F, Becker B, Spreitzer I, Konig R, Becker S, Waibler Z, Muhlebach MD. 2015. High secretion of interferons by human plasmacytoid dendritic cells upon recognition of Middle East respiratory syndrome coronavirus. J Virol 89(7):3859–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Levy DE, Decker T. 2007. JAK-STAT signaling: from interferons to cytokines. J Biol Chem 282(28):20059–20063 [DOI] [PubMed] [Google Scholar]
- Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert K, Karousis ED, Jomaa A, Scaiola A, Echeverria B, Gurzeler LA, Leibundgut M, Thiel V, Muhlemann O, Ban N. 2020. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 27(10):959–966 [DOI] [PubMed] [Google Scholar]
- Sheahan T, Morrison TE, Funkhouser W, Uematsu S, Akira S, Baric RS, Heise MT. 2008. MyD88 is required for protection from lethal infection with a mouse-adapted SARS-CoV. PLoS Pathog 4(12):e1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi CS, Qi HY, Boularan C, Huang NN, Abu-Asab M, Shelhamer JH, Kehrl JH. 2014. SARS-coronavirus open reading frame-9b suppresses innate immunity by targeting mitochondria and the MAVS/TRAF3/TRAF6 signalosome. J Immunol 193(6):3080–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin D, Mukherjee R, Grewe D, Bojkova D, Baek K, Bhattacharya A, Schulz L, Widera M, Mehdipour AR, Tascher G, Geurink PP, Wilhelm A, van der Heden van Noort GJ, Ovaa H, Muller S, Knobeloch KP, Rajalingam K, Schulman BA, Cinatl J, Hummer G, Ciesek S, Dikic I. 2020. Papain-like protease regulates SARS-CoV-2 viral spread and innate immunity. Nature 587(7835):657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, Kok KH, Ng MH, Poon VK, Yuen KY, Zheng BJ, Jin DY. 2009. Severe acute respiratory syndrome coronavirus M protein inhibits type I interferon production by impeding the formation of TRAF3.TANK.TBK1/IKKepsilon complex. J Biol Chem 284(24):16202–16209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu KL, Yeung ML, Kok KH, Yuen KS, Kew C, Lui PY, Chan CP, Tse H, Woo PCY, Yuen KY, Jin DY. 2014. Middle East respiratory syndrome coronavirus 4a protein is a double-stranded RNA-binding protein that suppresses PACT-induced activation of RIG-I and MDA5 in the innate antiviral response. J Virol 88(9):4866–4876 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Snijder EJ, Limpens R, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas F, Koster AJ, Barcena M. 2020. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol 18(6):e3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegel M, Pichlmair A, Martinez-Sobrido L, Cros J, Garcia-Sastre A, Haller O, Weber F. 2005. Inhibition of Beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J Virol 79(4):2079–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S, Reichelt M, Spiegel M, Kuri T, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Kochs G. 2007. The intracellular sites of early replication and budding of SARS-coronavirus. Virology 361(2):304–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson DB, Medzhitov R. 2006. Type I interferons in host defense. Immunity 25(3):373–381 [DOI] [PubMed] [Google Scholar]
- Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, Chen Z. 2012. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One 7(2):e30802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi N, Samuel CE. 2012. Protein kinase PKR catalytic activity is required for the PKR-dependent activation of mitogen-activated protein kinases and amplification of interferon beta induction following virus infection. Virology 427(2):208–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahasi K, Kumeta H, Tsuduki N, Narita R, Shigemoto T, Hirai R, Yoneyama M, Horiuchi M, Ogura K, Fujita T, Inagaki F. 2009. Solution structures of cytosolic RNA sensor MDA5 and LGP2 C-terminal domains: identification of the RNA recognition loop in RIG-I-like receptors. J Biol Chem 284(26):17465–17474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornbrough JM, Jha BK, Yount B, Goldstein SA, Li Y, Elliott R, Sims AC, Baric RS, Silverman RH, Weiss SR. 2016. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. mBio 7(2):e00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totura AL, Whitmore A, Agnihothram S, Schafer A, Katze MG, Heise MT, Baric RS. 2015. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection. mBio 6(3):e00638-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Made CI, Simons A, Schuurs-Hoeijmakers J, van den Heuvel G, Mantere T, Kersten S, van Deuren RC, Steehouwer M, van Reijmersdal SV, Jaeger M, Hofste T, Astuti G, Corominas Galbany J, van der Schoot V, van der Hoeven H, Hagmolen Of Ten Have W, Klijn E, van den Meer C, Fiddelaers J, de Mast Q, Bleeker-Rovers CP, Joosten LAB, Yntema HG, Gilissen C, Nelen M, van der Meer JWM, Brunner HG, Netea MG, van de Veerdonk FL, Hoischen A. 2020. Presence of genetic variants among young men with severe COVID-19. JAMA 324(7):663–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vennema H, Godeke GJ, Rossen JW, Voorhout WF, Horzinek MC, Opstelten DJ, Rottier PJ. 1996. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J 15(8):2020–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Lemey P, Maes P, Van Reeth K, Nauwynck H, Pensaert M, Van Ranst M. 2006. Evolutionary history of the closely related group 2 coronaviruses: porcine hemagglutinating encephalomyelitis virus, bovine coronavirus, and human coronavirus OC43. J Virol 80(14):7270–7274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijgen L, Keyaerts E, Moes E, Thoelen I, Wollants E, Lemey P, Vandamme AM, Van Ranst M. 2005. Complete genomic sequence of human coronavirus OC43: molecular clock analysis suggests a relatively recent zoonotic coronavirus transmission event. J Virol 79(3):1595–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhou Z, Xiao X, Tian Z, Dong X, Wang C, Li L, Ren L, Lei X, Xiang Z, Wang J. 2021. SARS-CoV-2 nsp12 attenuates type I interferon production by inhibiting IRF3 nuclear translocation. Cell Mol Immunol 18(4):945–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liao Y, Yap PL, Png KJ, Tam JP, Liu DX. 2009. Inhibition of protein kinase R activation and upregulation of GADD34 expression play a synergistic role in facilitating coronavirus replication by maintaining de novo protein synthesis in virus-infected cells. J Virol 83(23):12462–12472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shi H, Rigolet P, Wu N, Zhu L, Xi XG, Vabret A, Wang X, Wang T. 2010. Nsp1 proteins of group I and SARS coronaviruses share structural and functional similarities. Infect Genet Evol 10(7):919–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Melia CE, Snijder EJ, Barcena M. 2020. Double-membrane vesicles as platforms for viral replication. Trends Microbiol 28(12):1022–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong HH, Fung TS, Fang S, Huang M, Le MT, Liu DX. 2018. Accessory proteins 8b and 8ab of severe acute respiratory syndrome coronavirus suppress the interferon signaling pathway by mediating ubiquitin-dependent rapid degradation of interferon regulatory factor 3. Virology 515:165–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LR, Ye ZW, Lui PY, Zheng X, Yuan S, Zhu L, Fung SY, Yuen KS, Siu KL, Yeung ML, Cai Z, Woo PC, Yuen KY, Chan CP, Jin DY. 2020. Middle East respiratory syndrome coronavirus ORF8b accessory protein suppresses type I IFN expression by impeding HSP70-dependent activation of IRF3 kinase IKKepsilon. J Immunol 205(6):1564–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. 2006. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-kappaB activation [corrected]. Nat Cell Biol 8(4):398–406 [DOI] [PubMed] [Google Scholar]
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Chen ZJ. 2014. Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488 [DOI] [PubMed] [Google Scholar]
- Xia H, Cao Z, Xie X, Zhang X, Chen JY, Wang H, Menachery VD, Rajsbaum R, Shi PY. 2020. Evasion of type I interferon by SARS-CoV-2. Cell Rep 33(1):108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Shi PY. 2020. Antagonism of type I interferon by severe acute respiratory syndrome coronavirus 2. J Interferon Cytokine Res 40(12):543–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Chen J, Tu J, Zhang B, Chen X, Shi H, Baker SC, Feng L, Chen Z. 2013. The papain-like protease of porcine epidemic diarrhea virus negatively regulates type I interferon pathway by acting as a viral deubiquitinase. J Gen Virol 94(Pt 7):1554–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu Y, Xu J, Shu H, Xia Ja, Liu H, Wu Y, Zhang L, Yu Z, Fang M, Yu T, Wang Y, Pan S, Zou X, Yuan S, Shang Y. 2020a. Clinical course and outcomes of critically ill patients with SARS-CoV-2 pneumonia in Wuhan, China: a single-centered, retrospective, observational study. Lancet Respir Med 8(5):475–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Ye F, Zhu N, Wang WL, Deng Y, Zhao ZD, Tan WJ. 2015. Middle East respiratory syndrome coronavirus ORF4b protein inhibits type I interferon production through both cytoplasmic and nuclear targets. Sci Rep 5:17554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Zhang L, Geng H, Deng Y, Huang B, Guo Y, Zhao Z, Tan W. 2013. The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein Cell 4(12):951–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhang X, Wang F, Wang P, Kuang E, Li X. 2020b. Suppression of MDA5-mediated antiviral immune responses by NSP8 of SARS-CoV-2. bioRxiv. [Epub ahead of print]; DOI: 10.1101/2020.08.12.247767 [DOI] [Google Scholar]
- Yin X, Riva L, Pu Y, Martin-Sancho L, Kanamune J, Yamamoto Y, Sakai K, Gotoh S, Miorin L, De Jesus PD, Yang CC, Herbert KM, Yoh S, Hultquist JF, Garcia-Sastre A, Chanda SK. 2021. MDA5 governs the innate immune response to SARS-CoV-2 in lung epithelial cells. Cell Rep 34(2):108628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneyama M, Onomoto K, Jogi M, Akaboshi T, Fujita T. 2015. Viral RNA detection by RIG-I-like receptors. Curr Opin Immunol 32:48–53 [DOI] [PubMed] [Google Scholar]
- Yoneyama M, Suhara W, Fujita T. 2002. Control of IRF-3 activation by phosphorylation. J Interferon Cytokine Res 22(1):73–76 [DOI] [PubMed] [Google Scholar]
- Yuen CK, Lam JY, Wong WM, Mak LF, Wang X, Chu H, Cai JP, Jin DY, To KK, Chan JF, Yuen KY, Kok KH. 2020. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect 9(1):1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalinger ZB, Elliott R, Rose KM, Weiss SR. 2015. MDA5 is critical to host defense during infection with murine coronavirus. J Virol 89(24):12330–12340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Miorin L, Makio T, Dehghan I, Gao S, Xie Y, Zhong H, Esparza M, Kehrer T, Kumar A, Hobman TC, Ptak C, Gao B, Minna JD, Chen Z, Garcia-Sastre A, Ren Y, Wozniak RW, Fontoura BMA. 2021. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci Adv 7(6):eabe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, Ogishi M, Sabli IKD, Hodeib S, Korol C, Rosain J, Bilguvar K, Ye J, Bolze A, Bigio B, Yang R, Arias AA, Zhou Q, Zhang Y, Onodi F, Korniotis S, Karpf L, Philippot Q, Chbihi M, Bonnet-Madin L, Dorgham K, Smith N, Schneider WM, Razooky BS, Hoffmann HH, Michailidis E, Moens L, Han JE, Lorenzo L, Bizien L, Meade P, Neehus AL, Ugurbil AC, Corneau A, Kerner G, Zhang P, Rapaport F, Seeleuthner Y, Manry J, Masson C, Schmitt Y, Schluter A, Le Voyer T, Khan T, Li J, Fellay J, Roussel L, Shahrooei M, Alosaimi MF, Mansouri D, Al-Saud H, Al-Mulla F, Almourfi F, Al-Muhsen SZ, Alsohime F, Al Turki S, Hasanato R, van de Beek D, Biondi A, Bettini LR, D'Angio M, Bonfanti P, Imberti L, Sottini A, Paghera S, Quiros-Roldan E, Rossi C, Oler AJ, Tompkins MF, Alba C, Vandernoot I, Goffard JC, Smits G, Migeotte I, Haerynck F, Soler-Palacin P, Martin-Nalda A, Colobran R, Morange PE, Keles S, Colkesen F, Ozcelik T, Yasar KK, Senoglu S, Karabela SN, Rodriguez-Gallego C, Novelli G, Hraiech S, Tandjaoui-Lambiotte Y, Duval X, Laouenan C, COVID-STORM Clinicians; COVID Clinicians; Imagine COVID Group; French COVID Cohort Study Group; CoV-Contact Cohort; Amsterdam UMC Covid-19 Biobank; COVID Human Genetic Effort; NIAID-USUHS/TAGC COVID Immunity Group; Snow AL, Dalgard CL, Milner JD, Vinh DC, Mogensen TH, Marr N, Spaan AN, Boisson B, Boisson-Dupuis S, Bustamante J, Puel A, Ciancanelli MJ, Meyts I, Maniatis T, Soumelis V, Amara A, Nussenzweig M, Garcia-Sastre A, Krammer F, Pujol A, Duffy D, Lifton RP, Zhang SY, Gorochov G, Beziat V, Jouanguy E, Sancho-Shimizu V, Rice CM, Abel L, Notarangelo LD, Cobat A, Su HC, Casanova JL. 2020. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science 370(6515):eabd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ Jr., Baric RS, Enjuanes L, Gallagher T, McCray PB Jr., Perlman S.. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 111(13):4970–4975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Jha BK, Wu A, Elliott R, Ziebuhr J, Gorbalenya AE, Silverman RH, Weiss SR. 2012. Antagonism of the interferon-induced OAS-RNase L pathway by murine coronavirus ns2 protein is required for virus replication and liver pathology. Cell Host Microbe 11(6):607–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Chen G, Guo B, Cheng G, Tang H. 2008. PLP2, a potent deubiquitinase from murine hepatitis virus, strongly inhibits cellular type I interferon production. Cell Res 18(11):1105–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Zhuang MW, Han L, Zhang J, Nan ML, Zhan P, Kang D, Liu X, Gao C, Wang PH. 2020. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) membrane (M) protein inhibits type I and III interferon production by targeting RIG-I/MDA-5 signaling. Signal Transduct Target Ther 5(1):299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou A, Paranjape J, Brown TL, Nie H, Naik S, Dong B, Chang A, Trapp B, Fairchild R, Colmenares C, Silverman RH. 1997. Interferon action and apoptosis are defective in mice devoid of 2′,5′-oligoadenylate-dependent RNase L. EMBO J 16(21):6355–6363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W; China Novel Coronavirus Investigating and Research Team. 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382(8):727–733 [DOI] [PMC free article] [PubMed] [Google Scholar]