Oligoastrocytoma has largely vanished from the current classification of infiltrating gliomas following recognition that the majority of cases can be definitively classified as oligodendroglioma or astrocytoma by molecular studies [12]. However, exceptional cases have been reported of “dual-genotype oligoastrocytomas” with distinct oligodendroglioma and astrocytoma cell populations [5, 11].
We identified two individuals with morphologically homogenous infiltrating gliomas, harboring concurrent IDH1/2 mutation (oligodendroglioma/astrocytoma), ATRX and TP53 mutations (astrocytoma), and whole arm loss of 1p/19q (oligodendroglioma), within what we predict to be single tumor clones. Case #1 is a 37-year-old male who underwent resection of a left frontal mass, diagnosed as an anaplastic oligoastrocytoma (WHO grade III) (Fig. 1a). Five years later, a new cystic lesion was found in the previous resection cavity, with histological features in keeping with tumor recurrence (Fig. 1a). Using next-generation sequencing (NGS), both tumors were found to harbor IDH2 (c.515G>A, p.R172K), TP53 (c.569C>G, p.P190R; c.1006G>T, p.E336*), and ATRX (c.3724_3726delinsGG, p.C1242Gfs*36) mutations (Table 1). Chromosome microarray (CMA) analysis of tumor slides (~ 50–60% tumor content) showed whole arm 1p/19q co-deletion and additional genomic alterations (Suppl. Figure 1 and Tables 1, 2).
Fig. 1.
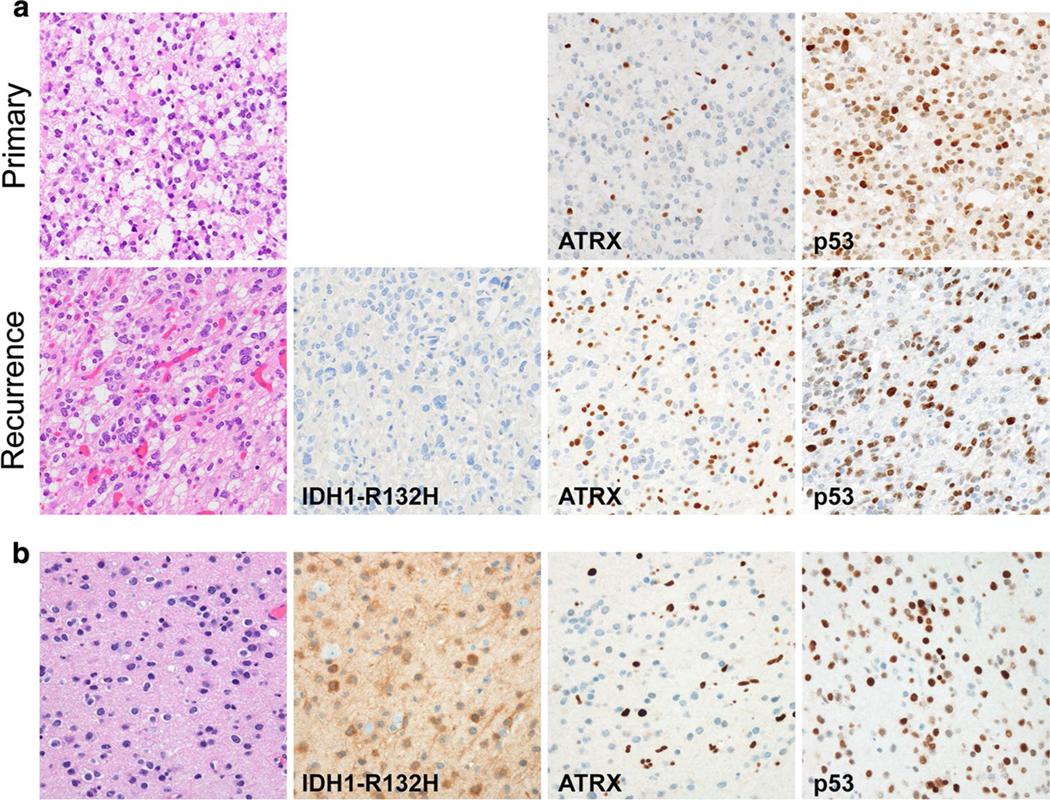
Histological features of dual-genotype gliomas in this study. a Primary moderately cellular infiltrating glioma from Case #1. At tumor recurrence, cells showed greater degrees of atypia and nuclear pleomorphism. Both primary and recurrent tumors were mitotically active without microvascular proliferation or necrosis. IHC was negative for IDH1-R132H, with ATRX loss, and overexpression of p53. b Case #2 demonstrated a low to moderately cellular infiltrating glioma with low mitotic activity. By IHC, the tumor was positive for IDH1-R132H, with loss of ATRX expression, and overexpression of p53
Table 1.
VAFs detected in Case #1′s gliomas using a NGS panel
| Gene | Transcript | Exon | Genomic | cDNA | Variant classification | Primary tumor variant frequency | Recurrence tumor variant frequency |
|---|---|---|---|---|---|---|---|
| ATRX | NM_000489.3 | Ex9 | chrX:g.76937020TG>T | c.3727delC | Pathogenic | 0.82 | 0.73 |
| IDH2 | NM_002168.3 | Ex4 | chr15:g.90631838C>T | c.515G>A | Pathogenic | 0.44 | 0.33 |
| TP53 | NM_000546.5 | Ex10 | chr17:g.7574021C>A | c.1006G>T | Pathogenic | 0.45 | 0.41 |
| TP53 | NM_000546.5 | Ex6 | chr17:g.7578280G>C | c.569C>G | Pathogenic | 0.47 | 0.33 |
Notice the doubling of the ATRX VAF for this male patient compared to IDH2 and TP53 frequencies, present in autosomes
Table 2.
VAFs detected in Case #2′s glioma using a NGS panel
| Gene | Transcript | Exon | Genomic | cDNA | Variant classification | Variant frequency |
|---|---|---|---|---|---|---|
| ATRX | NM_000489.3 | Ex9 | chrX:g.76939909CA>C | c.838delT | Pathogenic | 0.66 |
| IDH1 | NM_005896.3 | Ex4 | chr2:g.209113112C>T | c.395G>A | Pathogenic | 0.32 |
| TP53 | NM_000546.5 | Ex5 | chr17:g.7578542G>A | c.388C>T | Pathogenic | 0.77 |
Notice the doubling of the ATRX VAF for this male patient compared to IDH1, present in an autosome. TP53 presented with loss-of-heterozygosity (LOH), which is in agreement with the observed mutation frequency being approximately double of IDH1
Case #2 is a 46-year-old male who underwent resection of a right frontal infiltrating glioma (Fig. 1b); the tumor was positive for IDH1-R132H by immunohistochemistry (IHC) and had widespread overexpression of p53 and loss of ATRX. IHC findings were verified by NGS, with detected IDH1 (c.395G>A, p.Arg132His), ATRX (c.838delT), and TP53 (c.388C>T, p.Leu130Phe) mutations (Table 2). CMA analysis of tumor slides (~ 40–50% tumor content) demonstrated whole arm 1p/19q co-deletion present in ~ 50% of the analyzed DNA (Suppl. Figure 1 and Suppl. Table 3).
For both tumors, the variant allele frequency (VAF) of IDH1/2 and TP53 mutations was found to be approximately half of ATRX (± 10% to account for filtered reads with mapping quality < 20) (Tables 1 and 2). Moreover, 1p/19q co-deletion was present in ~ 50–60% of the analyzed DNA, similar to estimated tumor cellularity; FISH experiments demonstrated presence of the co-deletion within the same area in adjacent ATRX IHC slides (Suppl. Figure 2). These observations suggest the 1p/19q co-deletion and the IDH1/2, TP53, and ATRX mutations to be present within a single tumor clone. While partial and isolated deletions within 19q and to a lesser extent 1p have been observed in IDH-mutant astrocytomas (https://www.cancer.gov/tcga), to our knowledge, a tumor harboring concurrent mutations of IDH1/2, ATRX, TP53, and 1p/19q co-deletion has not been previously reported [4]. The biology and classification of these tumors remains uncertain. The absence of a TERT promoter mutation is unusual in an oligodendroglioma, and suggests that the 1p/19q co-deletion may have occurred by chance in tumors with an initial astrocytoma genotype (IDH1/2, ATRX, and TP53 mutations). In addition to the ATRX and TP53 mutations, the relatively more complex microarray copy-number profiles (Suppl. Figure 1) would be more in keeping with astrocytoma, although complex oligodendroglioma copy-number profiles have also been reported [3]. Both cases also lack CIC and FUBP1 mutations, although these alterations are identified in only about half of oligodendrogliomas [2]. An alternative, though less likely explanation to the origin of these tumors is that in which IDH1/2 mutations and 1p/19q co-deletion were the initiating events, followed by TP53 and ATRX mutations. This hypothesis is thought-provoking given that IDH2 mutations are far more frequent in oligondedrogliomas [1]. However, co-occurrence of 1p/19q co-deletion and ATRX mutation is unusual [7]. The observation that the frequencies of 1p/19q co-deletion and IDH1/2, ATRX, and TP53 mutations paralleled the estimated tumor cellularity in the specimens analyzed made us additionally consider the possibility that these gliomas could represent the concomitant (rather than sequential) acquisition of 1p/19q co-deletion and ATRX/TP53 mutations, possibly from an early progenitor neuroglial precursor cell pool in a background of increased risk for IDH1/IDH2 mutant glioma development [6]. Lastly, we considered the possibility of two distinct oligodendroglioma/astrocytoma subclones, as has been previously reported [11]. Nonetheless, the VAF ratios and lack of TERT-promoter mutation argue against this. Overall, we favor that the presence of TP53 and ATRX mutations will lead to biology and behavior more in keeping with an astrocytoma.
These “dual-genotype” infiltrating gliomas pose a diagnostic dilemma as they do not fit neatly within the current understanding of glioma genetics and classification. Based on WHO 2016 criteria, the tumors reported here would be consistent with the diagnosis of oligodendroglioma, IDH-mutant and 1p/19q co-deleted [9]. However, a common diagnostic approach, which utilizes immunohistochemistry for IDH1-R132H, ATRX, and p53, would lead to classification of such cases as IDH-mutant astrocytomas [8]. The cIMPACT-NOW update 2 allows for tumors positive for IDH-R132H with loss of ATRX and p53 overexpression (as in Case #2) to be diagnosed as IDH-mutant astrocytomas without 1p/19q co-deletion testing [8]. These results suggest that expansion of recommendations for 1p/19q co-deletion testing may be warranted. However, it remains uncertain which alterations will dictate the tumor phenotype and prognosis, the 1p/19q co-deletion or ATRX/TP53 mutations. As such, these tumors do not fit neatly within the diagnosis of oligodendroglioma. Thus, we suggest classification of such tumors as oligoastrocytoma, IDH-mutant and 1p/19q-codeleted, not elsewhere classified (NEC) or, alternatively, as diffuse glioma, IDH-mutant and 1p/19q-codeleted, NEC. In keeping with the cIMPACT-NOW recommendations, this diagnosis conveys the unusual molecular features and uncertainty with respect to tumor classification [10]. As molecular testing is more widely implemented in the characterization of infiltrating gliomas, it is clear that there are unusual tumors which run counter to our understanding of glioma biology. Larger case series and single-cell sequencing analyses of tumor heterogeneity are needed to clearly define the prognostic implications of these dual-genotype gliomas.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this article (https://doi.org/10.1007/s00401-020-02141-x) contains supplementary material, which is available to authorized users.
References
- 1.Appay R, Tabouret E, Macagno N, Touat M, Carpentier C, Colin C et al. (2018) IDH2 mutations are commonly associated with 1p/19q co-deletion in diffuse adult gliomas. Neuro Oncol 20:716–718. 10.1093/neuonc/noy014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brat DJ, Verhaak RG, Aldape KD, Yung WK, Salama SR, Cooper LA et al. (2015) Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med 372:2481–2498. 10.1056/NEJMoa1402121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dahlback HS, Gorunova L, Brandal P, Scheie D, Helseth E, Meling TR et al. (2011) Genomic aberrations in diffuse low-grade gliomas. Genes Chromosomes Cancer 50:409–420. 10.1002/gcc.20866 [DOI] [PubMed] [Google Scholar]
- 4.Eckel-Passow JE, Lachance DH, Molinaro AM, Walsh KM, Decker PA, Sicotte H et al. (2015) Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med 372:2499–2508. 10.1056/NEJMoa1407279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huse JT, Diamond EL, Wang L, Rosenblum MK (2015) Mixed glioma with molecular features of composite oligodendroglioma and astrocytoma: a true “oligoastrocytoma”? Acta Neuropathol 129:151–153. 10.1007/s00401-014-1359-y [DOI] [PubMed] [Google Scholar]
- 6.Jenkins RB, Xiao Y, Sicotte H, Decker PA, Kollmeyer TM, Hansen HM et al. (2012) A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet 44:1122–1125. 10.1038/ng.2388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leeper HE, Caron AA, Decker PA, Jenkins RB, Lachance DH, Giannini C (2015) IDH mutation, 1p19q co-deletion and ATRX loss in WHO grade II gliomas. Oncotarget 6:30295–30305. 10.18632/oncotarget.4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Louis DN, Giannini C, Capper D, Paulus W, Figarella-Branger D, Lopes MB et al. (2018) cIMPACT-NOW update 2: diagnostic clarifications for diffuse midline glioma, H3 K27M-mutant and diffuse astrocytoma/anaplastic astrocytoma, IDH-mutant. Acta Neuropathol 135:639–642. 10.1007/s00401-018-1826-y [DOI] [PubMed] [Google Scholar]
- 9.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK et al. (2016) The 2016 World Health Organization Classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 10.Louis DN, Wesseling P, Paulus W, Giannini C, Batchelor TT, Cairncross JG et al. (2018) cIMPACT-NOW update 1: not otherwise specified (NOS) and not elsewhere classified (NEC). Acta Neuropathol 135:481–484. 10.1007/s00401-018-1808-0 [DOI] [PubMed] [Google Scholar]
- 11.Qu M, Olofsson T, Sigurdardottir S, You C, Kalimo H, Nister M et al. (2007) Genetically distinct astrocytic and oligodendroglial components in oligoastrocytomas. Acta Neuropathol 113:129–136. 10.1007/s00401-006-0142-0 [DOI] [PubMed] [Google Scholar]
- 12.Sahm F, Reuss D, Koelsche C, Capper D, Schittenhelm J, Heim S et al. (2014) Farewell to oligoastrocytoma: in situ molecular genetics favor classification as either oligodendroglioma or astrocytoma. Acta Neuropathol 128:551–559. 10.1007/s00401-014-1326-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


