In this study Trakala et al. investigated how chromosome gains and losses, which are a frequent feature of human cancers, can overcome the detrimental effects of aneuploidy. They developed a novel mouse model that enables unprecedented levels of chromosome missegregation in the adult animal and their results show that the initial detrimental effects of random missegregation are outweighed by clonal selection, which is dependent on chromosomal location and the nature of specific genes and is sufficient to drive cancer.
Keywords: aneuploidy, cancer, chromosomal instability, MYC, oncogene, RAD21
Abstract
Chromosome gains and losses are a frequent feature of human cancers. However, how these aberrations can outweigh the detrimental effects of aneuploidy remains unclear. An initial comparison of existing chromosomal instability (CIN) mouse models suggests that aneuploidy accumulates to low levels in these animals. We therefore developed a novel mouse model that enables unprecedented levels of chromosome missegregation in the adult animal. At the earliest stages of T-cell development, cells with random chromosome gains and/or losses are selected against, but CIN eventually results in the expansion of progenitors with clonal chromosomal imbalances. Clonal selection leads to the development of T-cell lymphomas with stereotypic karyotypes in which chromosome 15, containing the Myc oncogene, is gained with high prevalence. Expressing human MYC from chromosome 6 (MYCChr6) is sufficient to change the karyotype of these lymphomas to include universal chromosome 6 gains. Interestingly, while chromosome 15 is still gained in MYCChr6 tumors after genetic ablation of the endogenous Myc locus, this chromosome is not efficiently gained after deletion of one copy of Rad21, suggesting a synergistic effect of both MYC and RAD21 in driving chromosome 15 gains. Our results show that the initial detrimental effects of random missegregation are outbalanced by clonal selection, which is dictated by the chromosomal location and nature of certain genes and is sufficient to drive cancer with high prevalence.
Aneuploidy, or the presence of a number of chromosomes that is not an exact multiple of the haploid number, is a common finding in human cancer. The degree of aneuploidy correlates with poor prognosis and resistance to therapy, although how aberrant chromosome numbers contribute to cancer development is not clear (Weaver and Cleveland 2006; Duijf and Benezra 2013). A number of mouse models of aneuploidy induced by chromosomal instability (CIN) have been generated to study the direct effects of abnormal chromosome numbers at the molecular, cellular, and physiological level. These studies have suggested that aneuploidy can function both as a tumor suppressor or an oncogenic alteration, and whether this outcome is related to aneuploidy levels is unclear due to the different alterations to induce aneuploidy and technical differences in how the degree of aneuploidy was studied (Weaver and Cleveland 2006; Holland and Cleveland 2009; Schvartzman et al. 2010).
Mutations in a surveillance pathway known as the spindle assembly checkpoint (SAC) cause CIN. In wild-type cells, the SAC ensures that anaphase only occurs after chromosomes have properly attached to the metaphase spindle (Musacchio and Salmon 2007). SAC components assemble on unattached kinetochores, where they facilitate the inhibition of a ubiquitin ligase known as the anaphase-promoting complex/cyclosome (APC/C) by sequestering its activator CDC20 throughout the cell. Mutations in CDC20 (CDC20AAA) that prevent binding of CDC20 to the SAC component MAD2 render the ubiquitin ligase refractory to SAC inhibition (Li et al. 2009). As a result, cells enter anaphase before chromosomes are properly bioriented on the mitotic spindle, leading to high levels of chromosome missegregation. When expressed in mice as the sole source of CDC20, the Cdc20AAA allele causes lethality at embryonic day (E) 12.5, indicating that the allele causes levels of chromosome missegregation that are incompatible with embryonic development (Li et al. 2009). Animals heterozygous for the Cdc20AAA allele are viable and develop hepatomas and lymphomas only late in life (Li et al. 2009).
In this work, we performed a systematic analysis of the prevalence of aneuploid cells in previously reported CIN models by using single-cell DNA sequencing technology in freshly isolated cells. To increase the levels of aneuploidy in vivo, we generated an inducible mouse model in which the wild-type Cdc20 allele can be excised at will, generating cells that express CDC20AAA as the sole source of CDC20 (Fig. 1A). Analysis of the dynamics of chromosomal gain and losses in the thymus suggests that random aneuploidies form at initial stages and are eventually substituted by stereotypical karyotypes, including gains in chromosome 15, harboring the Myc gene, leading to high T-cell lymphomas with short latency. Expressing human MYC from chromosome 6 changes tumor karyotypes, leading to universal gains of this chromosome, suggesting that clonal selection of random aneuploidies determines tumor karyotypes and is sufficient to cause malignant cell transformation and cancer. Unexpectedly, the gene encoding the cohesin subunit RAD21 is a major driver of chromosome 15 gains in CIN- and MYC-driven lymphomas.
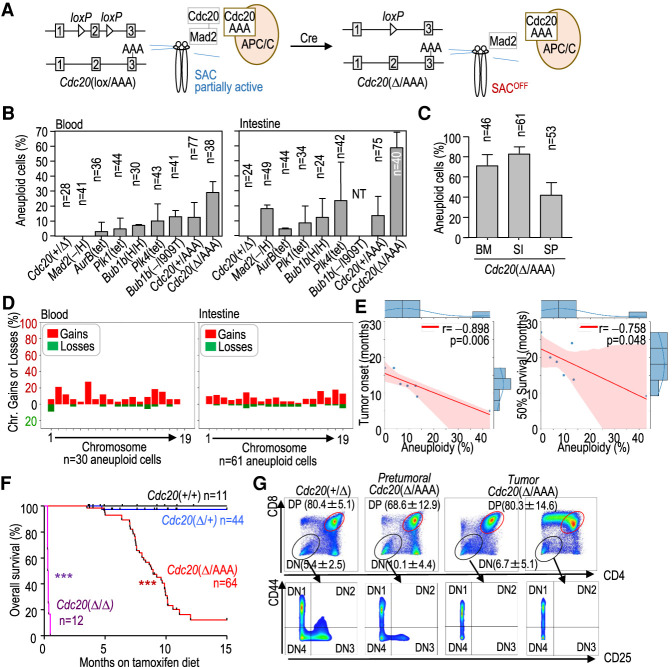
Figure 1.
A novel model with high levels of CIN in adult mouse tissues. (A) Schematic representation of the spindle assembly checkpoint (SAC) and the effect of the new Cdc20Δ/AAA mouse model on checkpoint function, allowing entry into anaphase in the presence of unattached kinetochores. (B) Percentage of blood or intestine aneuploid cells in 3-mo-old mice with the indicated genotypes. (NT) Not tested. (C) Percentage of aneuploid cells in bone marrow (BM), small intestine (SI), and spleen (SP) from three Cdc20Δ/AAA mice 5–9 mo after CRE activation. In B and C, data are mean ± SEM. Data correspond to two mice per genotype (3 mo old; B) or three mice per tissue (5–9 mo old; C). The number of cells analyzed by single-cell DNA sequencing is indicated in every column. (D) Percentage of specific chromosomal gains (red) or losses (green) in the aneuploid (N = 33 blood and 62 intestine) cells from all models represented in B. (E) Correlation between total aneuploidy (taking all blood and intestine cells) and age (in months) at which tumors appear (tumor onset) or at which 50% of the colony is alive in the models indicated in B. Data are taken from Supplemental Table S1. (r) Pearson's correlation coefficient, (p) P-value. Red shading represents the 95% confidence interval. (F) Kaplan-Meier survival curves of mice with the indicated genotypes after treatment with tamoxifen. (***) P < 0.001 (log rank Mantel-Cox test). (G) Representative flow cytometry analysis of the indicated cell populations in thymuses from Cdc20+/Δ and Cdc20Δ/AAA mice or thymic lymphomas in Cdc20Δ/AAA mice. Numeric data indicate mean ± SD (n = 3 mice per group).
Results
A novel model with high levels of chromosomal instability and high prevalence of T-cell lymphomas
We initially assessed the extent of aneuploidy in young (3-mo-old) mice from representative models of CIN with specific genetic modifications in genes encoding mitotic kinases (Plk1, Plk4, and Aurkb) or SAC components (Mad2 and Bub1b, encoding BUBR1) (Supplemental Table S1) using single-cell DNA sequencing (Knouse et al. 2014). A comparative analysis in all these models identified 2%–20% of aneuploid cells in peripheral blood or the intestine (Fig. 1B), in agreement with previous data obtained using a variety of different techniques (see Supplemental Table S1 for details).
To increase levels of aneuploidy in vivo, we generated a new inducible model by combining the Cdc20AAA allele (Li et al. 2009) with a conditional knockout model (Cdc20lox) in which the Cdc20 allele can be excised by a tamoxifen-inducible CRE recombinase ubiquitously expressed from the RNA polymerase II (Polr2a) promoter (RERT2 allele) (Fig. 1A; Manchado et al. 2010). Dietary tamoxifen administration (Supplemental Fig. S1A) resulted in the excision of the functional Cdc20lox allele, thereby generating cells that express CDC20AAA as the sole source of CDC20 (RERT2-Cdc20Δ/AAA mice, referred to here as Cdc20Δ/AAA). Cdc20Δ/AAA splenocytes displayed normal levels of DNA replication but were insensitive to mitotic arrest in the presence of microtubule poisons (Supplemental Fig. S1B), a known effect of the CDC20 AAA mutant protein (Li et al. 2009). As shown in Figure 1B, we did not detect aneuploid cells in the peripheral blood or epithelial intestinal cells from control Cdc20+/Δ mutant mice, and only 10%–12% of cells from Cdc20+/AAA mice were aneuploid. In contrast, aneuploid cells were detected in up to 30% of blood cells or 60% of intestinal cells in young Cdc20Δ/AAA mice (Fig. 1B) and 70% of bone marrow cells or 80% intestine cells in older (5–-9 mo) mice (Fig. 1C).
Analysis of whole-chromosome aberrations in these models showed that most chromosomes were affected, with gains being more prevalent than losses, and that slight preferences of certain aneuploidies might depend on the tissue context (Fig. 1D; Supplemental Fig. S1C). No significant trend was observed in aberrations affecting large versus small chromosomes. To test a possible correlation between aneuploidy and tumor susceptibility, we calculated the aggregated percentage of aneuploidy per model (considering both blood and intestine cells from young mice) and compared it with published data (Supplemental Table S1) or our own data (see below) on tumor onset and median survival for each of the models. Interestingly, the overall ratio of aneuploidy inversely correlated with both tumor onset and age at which 50% of the mouse colony was alive (Fig. 1E), suggesting that the reduced incidence and late latency of tumors in previous models were possibly linked to low levels of aneuploidy in pretumoral tissues.
Remarkably, despite the high levels of aneuploidy and the presence of aberrant mitoses (Supplemental Fig. S1D) in Cdc20Δ/AAA mice, most of these tissues did not display overt macroscopic abnormalities, suggesting that in adult proliferating tissues, aneuploidy is reasonably well tolerated. However, the life span of Cdc20Δ/AAA mice was reduced with a median survival of 8.9 mo (Fig. 1F). While, in agreement with previous work (Manchado et al. 2010), complete lack of CDC20 in Cdc20Δ/Δ mice resulted in early lethality as a consequence of strong mitotic arrest and the severe deterioration of proliferative tissues (Fig. 1F; Supplemental S1D), lethality in Cdc20Δ/AAA mice was due to increased susceptibility to tumor development. When the wild-type Cdc20 allele was acutely eliminated in the presence of the Cdc20AAA allele (Supplemental Fig. S1A), 76% of Cdc20Δ/AAA mice (n = 64) succumbed to thymic lymphomas within the first 10 mo of tamoxifen treatment (Fig. 1F; Supplemental Fig. S1E). These tumors were mostly comprised of proliferating CD8+ or immature CD4+ CD8+ double-positive (DP) T cells (Fig. 1G; Supplemental Fig. S1F). We also detected hyperplasias in other Cdc20AAA tissues such as in squamous epithelia of the forestomach, ovaries, endometrium, and the pancreas (Supplemental Fig. S1G). Carcinomas were rare, likely because animals succumbed to T-cell lymphomas before more aggressive disease could develop. We conclude that high levels of CIN correlate with tumorigenesis and can induce hyperplastic lesions in a variety of tissues, leading to frequent lethal malignant transformation in the T-cell compartment.
T-cell lymphomas harbor characteristic chromosome gains
Bulk DNA sequencing of Cdc20Δ/AAA T-cell lymphomas revealed that these tumors were aneuploid in all cases (Fig. 2A,B). When applying a cutoff in the CNV score that deviates three standard deviations from the euploid value (see the Materials and Methods), all chromosomes with the exception of chromosomes 7 and 19 were found to show numerical alterations in at least some lymphomas (Fig. 2B). Whole-chromosome losses or focal deletions were not a frequent occurrence in Cdc20Δ/AAA T-cell lymphomas. However, we did note the loss of the distal region of chromosome 12 in some tumors (Fig. 2A). This region contains the tumor suppressor Bcl11b, as well as several tumor suppressor microRNAs, and has been previously reported to be lost in T-cell lymphomas in different models (Bueno et al. 2008; De Keersmaecker et al. 2010). However, two chromosome gains stood out. Chromosome 15 was gained in all Cdc20Δ/AAA tumors, whereas chromosome 14 was gained in 89% of tumors (Fig. 2B,C). These chromosome number alterations were also observed using complementary techniques such as Giemsa staining (Fig. 2D) or spectral karyotyping (Fig. 2E) of Cdc20Δ/AAA lymphoma cells. We also confirmed the presence of these alterations using single-cell DNA sequencing from several cells isolated from individual tumors (Fig. 2F). We conclude that T-cell lymphomas harboring recurrent chromosome gains develop with high penetrance in mice missegregating chromosomes at a high frequency.
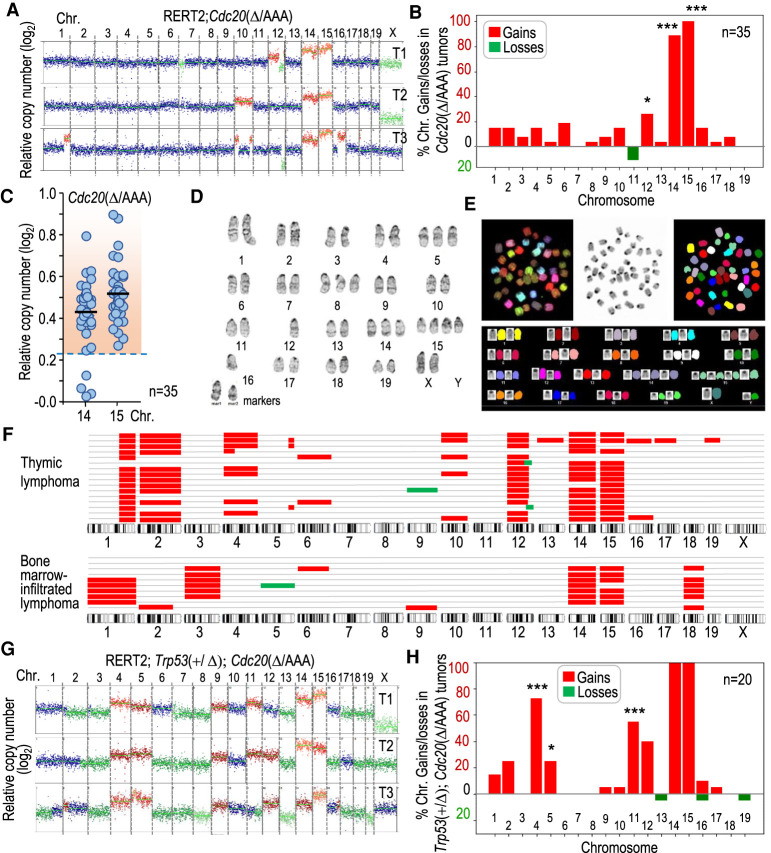
Figure 2.
Karyotype distribution in Cdc20Δ/AAA T-cell lymphomas. (A) Representative segmentation plots of three different T-cell lymphomas from Cdc20Δ/AAA mice, depicting relative copy number per chromosome to a euploid reference on a log2 scale as scored by bulk DNA sequencing. Chromosomal gains are in red and losses in green. (B) Quantification of the percentage of Cdc20Δ/AAA tumors in which each of the individual chromosome gain or loss is present (n = 35 tumors). A chromosome was considered gained when log2 relative copy number > 0.23 and lost when this value is <−0.32 (see the Materials and Methods). Statistics refer to the comparison with lack of gains and losses in 18 Cdc20+/Δ thymic cells analyzed. (C) Relative copy number (log2) of chromosomes 14 and 15 in Cdc20Δ/AAA lymphomas. Each dot indicates an individual tumor. The dashed region indicates samples in which the copy number is above the threshold selected to define chromosomal gains (mean value + 3 SD in euploid cells). (D) Giemsa staining of a representative metaphase spread from a Cdc20Δ/AAA lymphoma. (E) Spectral karyotype of a representative metaphase spread from a Cdc20Δ/AAA lymphoma. (F) Schematic representation of chromosomal gains (red) and losses (green) as detected by single-cell DNA sequencing (scDNA-seq) in primary tumors or bone marrow infiltrations from a tumor. Each row represents a single cell. (G) Representative examples of chromosomal gains (red) or losses (green) in three different T-cell lymphomas from Trp53+/Δ; Cdc20Δ/AAA mice as scored by bulk DNA sequencing. (H) Quantification of the percentage of Trp53+/Δ; Cdc20Δ/AAA tumors with the indicated specific chromosome gains or losses (n = 20 tumors). Statistics refer to the comparison with gains and losses in Cdc20Δ/AAA lymphomas in B. In B and H, (*) P < 0.05, (***) P < 0.001 (Fisher exact test); nonsignificant (P > 0.05) differences not shown.
We next compared the karyotype of Cdc20Δ/AAA tumors with those induced by lack of P53, a well-studied model of T-cell lymphomas. Whereas chromosome 15 was gained in two out of five tumors, Trp53−/− lymphomas displayed a higher karyotypic heterogeneity with frequent gains in chromosome 5 (Supplemental Fig. S2A,B). To analyze whether chromosome 15 gain was dispensable in the presence of a compromised P53 pathway, we crossed the Trp53 conditional knockout mice with RERT2; Cdc20Δ/AAA mice. Unfortunately, both the Trp53 locus and the Polr2a locus, where the Cre-ERT2 cassette is integrated, are located in close proximity on chromosome 11; hence, we could not generate and analyze Trp53Δ/Δ; Cdc20Δ/AAA. However, ablation of one Trp53 allele significantly accelerated tumorigenesis in Trp53+/Δ; Cdc20Δ/AAA mice, reducing the median tumor free survival to 6.13 mo (Supplemental Fig. S2C). These tumors, similarly to Cdc20Δ/AAA, showed universal gain of chromosome 14 and 15 but additionally displayed frequent gains of chromosome 4 and 11 (Fig. 2, cf. G,H and A,B; Supplemental Fig. S2D). Interestingly, the presence of functional Trp53 alleles might prevent the gain of chromosome 11, never observed in Cdc20Δ/AAA mice (Fig. 2B) but present in more than half of Trp53+/Δ; Cdc20Δ/AAA tumors (Fig. 2H). As described previously (Baker et al. 2009), we predicted that Trp53+/Δ; Cdc20Δ/AAA tumors might lose the wild-type Trp53 allele. Indeed, we observed an almost complete absence of P53 at the protein level in Trp53+/Δ; Cdc20Δ/AAA tumors (Supplemental Fig. S2E). In addition, P53 was not detected in these double-mutant T-cell lymphomas after a sublethal dose of γ-irradiation, suggesting loss of the wild-type allele. Interestingly, Cdc20Δ/AAA tumors elicited a robust response of the P53 pathway upon irradiation, indicating that Cdc20Δ/AAA T-cell lymphomas are different from P53-deficient tumors, maintaining at least partially P53 functionality.
Clonal evolution of karyotypes in premalignant tissues
To determine whether and how CIN interferes with T-cell maturation and leads to lymphomagenesis, we assessed T-cell development prior to tumor development. Beginning at 3 mo post initiation of tamoxifen treatment, Cdc20Δ/AAA mice presented with a pronounced reduction in total thymocyte number as judged by significant atrophy of the thymus (Fig. 3A), akin to that observed in other T-cell lymphoma models (De Keersmaecker et al. 2010). In these atrophic thymuses, we noted a significant increase in the relative percentage of double-negative (DN) 1 population (CD25low; CD44high) (Supplemental Fig. S3A) that was accompanied by a reduction in DN3, DN4, and CD4+ CD8+ double-positive (DP) cells (Fig. 3B), suggesting an inefficient transition from DN1 to later stages.
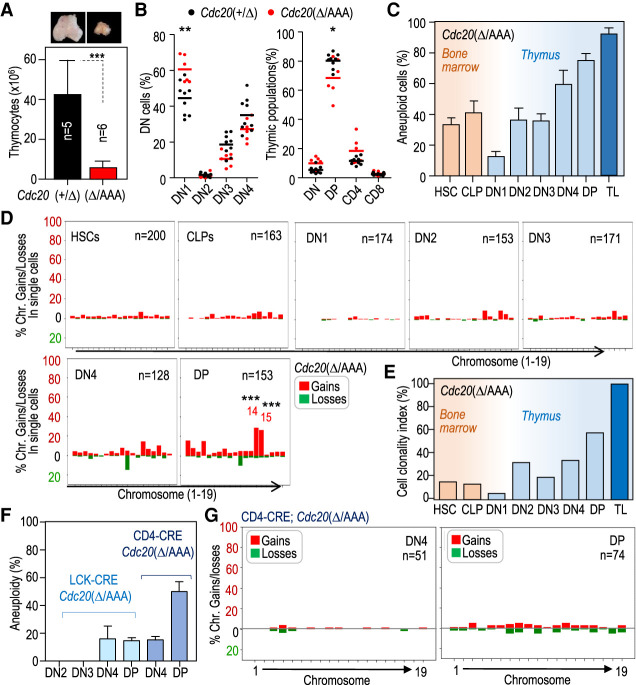
Figure 3.
CIN-induced T-cell lymphomagenesis is preceded by thymic atrophy and selection of specific aneuploidies during T-cell maturation. (A) Representative images of thymuses in 4-mo-old pretumoral Cdc20Δ/AAA mice. The histogram shows the quantification of the number of cells in the thymus from mice with the indicated genotypes. Data are mean ± SD. (***) P = 0.0010 (Student's t-test). (B) Percentage of double-negative (DN) cells (left) of the indicated thymic populations (right) in 4-mo-old pretumoral Cdc20Δ/AAA mice. Horizontal bars indicate mean. (**) P < 0.0032, (*) P = 0.0258 (Student's t-test). (C) Percentage of aneuploid cells in the indicated cell populations as determined by single-cell sequencing (data are mean ± SD). At least 120 single cells were analyzed per cell population obtained from five different mice in each group. (TL) T-cell lymphomas (23 cells from two tumors). No aneuploid cells were found in unsorted thymuses from Cdc20+/Δ mice. (D) Percentage of cells with the indicated chromosome gains and losses as determined by single-cell DNA-seq from the following sorted populations: HSCs and CLPs obtained from the bone marrow and DN1, DN2, DN3, DN4, and DP cells obtained from the thymus from Cdc20Δ/AAA mice 4 mo after tamoxifen addition to the diet. The total number of cells analyzed from five different mice is indicated in the plots. Data from individual mice are shown in Supplemental Fig. S3C. Statistics refer to the comparison with lack of gains and losses in 18 Cdc20+/Δ thymic cells analyzed. (***) P < 0.01 (Fisher exact test); nonsignificant (P > 0.05) differences not shown. (E) Clonality index, defined as the percentage of aneuploid cells carrying clonal imbalances, in the indicated populations of Cdc20Δ/AAA mice. (F) Percentage of aneuploid cells in the indicated thymic populations from LCK-CRE; Cdc20Δ/AAA (N = 2) and CD4-CRE; Cdc20Δ/AAA (N = 2) mice. Data are mean ± SD. (G) Percentage of cells with the indicated chromosome gains and losses as determined by single-cell DNA-seq from DN4 and DP populations obtained from the thymus from CD4-CRE; Cdc20Δ/AAA mice. The total number of cells analyzed from three different mice is indicated in the plots.
We next studied the karyotypic landscape of T-cell precursors in Cdc20Δ/AAA animals prior to disease onset. We analyzed karyotypes at single-cell resolution in sorted hematopoietic stem cells (HSCs) and common lymphoid progenitors (CLPs) in the bone marrow, as well as DN1–4 and DP cells from tumor-free thymuses at 4 mo after the induction of tamoxifen treatment. This analysis revealed that 32% of HSCs were already aneuploid and this number further increased in the CLP population (Fig. 3C; Supplemental Fig. S3B,C). Interestingly, the degree of aneuploidy significantly dropped in the DN1 stage (14% in DN1 vs. 35% in HSCs and 41% in CLPs) (Fig. 3C), suggesting the existence of a bottleneck for aneuploid CLPs during their transit to the thymus and the subsequent development of DN1 progenitors. However, the ratio of aneuploid cells steadily increased in later stages, reaching peak levels of up to 80% in the DP population (Fig. 3C; Supplemental Fig. S3C), in agreement with the known proliferative expansion of T-cell progenitors in the thymus (Supplemental Fig. S1F).
Given that chromosome missegregation during mitosis in SAC-perturbed models happens randomly, we analyzed whether specific chromosome gains or losses were selected. Early progenitors in the bone marrow displayed random gains or losses, affecting most chromosomes in a similar manner without any specific chromosome being gained or lost in >5% of the cells analyzed (200 HSCs and 163 CLPs) (Fig. 3D). Similarly, DN1 cells displayed no enrichment in specific gains or losses, although this analysis was limited due to the low number of aneuploid DN1 cells. Interestingly, the overrepresentation of specific imbalances increased toward the later stages of T-cell maturation (DN2 to DP) (Fig. 3D; Supplemental Fig. S3B,C). Enrichment in specific losses affecting chromosome 11 or gains in chromosomes 14 and 15 was especially obvious and significant, suggesting that cells with these karyotypes are endowed with an advantage. We defined the cell clonality index as the ratio of aneuploid cells carrying identical chromosomal gains or losses (see the Materials and Methods). Interestingly, whereas ∼40% bone marrow cells were aneuploid, most of these alterations were random and <13% of these aneuploid cells were clonal (Fig. 3E). However, the clonality index increased from 28% up to 60% during the DN2–DP transitions. The increase in cell clonality was mostly a consequence of the increase in aneuploid cells with gains of chromosomes 14 and 15 among others (Supplemental Fig. S3D). These results suggest that engraftment into the thymus, which occurs during the transition from the CLP to DN1 stage, favors euploid cells, and proliferative expansion during T-cell maturation is accompanied by clonal selection of cells carrying specific aneuploidies such as chromosomes 14 and 15 gains in Cdc20Δ/AAA animals.
Aneuploid cells require proliferation to undergo clonal selection
To test the specific effect of CIN in later developmental stages during T-cell development, we induced chromosome missegregation in the DN3 state by driving CRE expression from the LCK promoter, or during the DN4–DP transition by driving CRE expression from the CD4 promoter (Supplemental Fig. S3A). We found that animals in which chromosome missegregation was induced during these later stages of T-cell development did not develop T-cell lymphomas during the first 15 mo. Analysis of the degree of aneuploidy in tissues provided a potential explanation for why mice in which aneuploidy was driven by LCK-CRE did not develop tumors. This CRE driver yielded low levels of aneuploidy (Fig. 3F), which is consistent with the known incomplete penetrance of LCK-CRE expression (Lee et al. 2001; Pai et al. 2003). In contrast, CRE expression driven by CD4 led to relatively high levels of aneuploidy in DP cells (50%) (Fig. 3F,G; Supplemental Fig. S3E), similar to levels found in Cdc20Δ/AAA cell populations in which clonal selection has not been taken place (e.g., HSC and CLP in Fig. 3C). Intriguingly, CD4-CRE; Cdc20Δ/AAA DP cells did not accumulate specific aneuploidies (Supplemental Fig. S3D), and gains and losses affected most chromosomes similarly (cell clonality = 0%). The fact the vast majority (>90%) of DP cells do not proliferate (Seitan et al. 2011; Mingueneau et al. 2013) is in agreement with the observation of random losses of chromosomes, which are typically selected against in proliferating cells. It is therefore tempting to speculate that lack of proliferation could preclude clonal selection in this model.
Chromosomal location of MYC determines clonal selection of specific aneuploidies
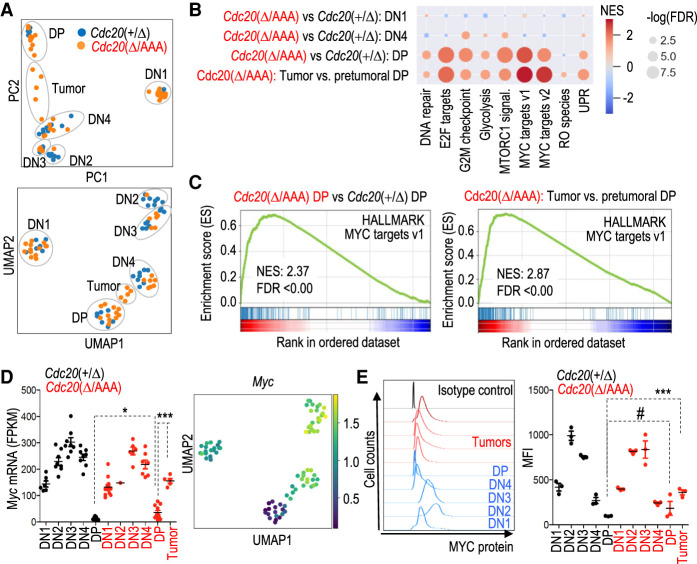
How does CIN drive lymphomagenesis? To address this question, we compared the gene expression profiles of pretumoral T-cell populations and T-cell lymphomas in RERT2; Cdc20Δ/AAA mice. Dimensionality reduction analysis of pretumoral and tumoral samples separated the different T-cell populations and indicated that T-cell lymphomas expressed intermediate levels of CD8 and CD4 (Fig. 4A; Supplemental Fig. S4A). We cannot exclude the possibility that tumor cells clustered between DN4 and DP population as a consequence of increased proliferation, a characteristic feature of DN4 cells opposed to the mainly nonproliferative DP population. The gene expression pattern of pretumoral T-cell precursor populations DN1–4 did not significantly differ between Cdc20Δ/AAA and Cdc20+/Δ animals (Fig. 4A; Supplemental Table S2), but we observed differences in the DP population. Pretumoral DP Cdc20Δ/AAA cells displayed enrichment in pathways related to DNA replication, MTOR, and glycolysis, compared with Cdc20+/Δ DP cells (Fig. 4B; Supplemental Fig. S4B; Supplemental Table S3). Interestingly, the genes significantly deregulated in Cdc20Δ/AAA DP cells were enriched for targets of MYC:MAX (Zeller et al. 2003), a transcription factor of major relevance in many of the biological processes deregulated in these cells (Fig. 4B,C; Supplemental Table S4). Consistent with this observation, Myc RNA levels were significantly up-regulated in pretumoral Cdc20Δ/AAA DP cells when compared with control Cdc20+/Δ DP cells (Fig. 4D), and this tendency was also observed at the protein level (Fig. 4E). Analysis of the gene expression pattern of T-cell lymphomas in Cdc20Δ/AAA animals suggested that Myc expression was further increased in these tumors when compared with pretumoral DP cells from mice of the same genotype (Fig. 4D,E), and up-regulation of MYC target genes was more pronounced (Fig. 4B,C; Supplemental Fig. S4C; Supplemental Tables S5, S6). These results suggested that MYC, a well-known driver of T-cell lymphoma and other hematopoietic malignancies (Gaudet et al. 2003; Meyer and Penn 2008), was also driving tumorigenesis in the Cdc20Δ/AAA CIN model.
Figure 4.
MYC is a driver of CIN-induced T-cell lymphomagenesis. (A) Principal component (PC) analysis and uniform manifold approximation and projection (UMAP) of RNA-seq data from the indicated samples. Each dot represents a different tumor. (B) Gene set enrichment analysis (GSEA) of specific HALMARK pathways in the indicated comparisons between pretumoral Cdc20Δ/AAA versus Cdc20+/Δ cells or Cdc20Δ/AAA lymphomas versus pretumoral Cdc20Δ/AAA DP cells. Please refer to Supplemental Fig. S4, B and C, and Supplemental Tables S3–S6 for details. (C) GSEA of MYC targets in pretumoral Cdc20Δ/AAA versus Cdc20+/Δ DP cells (left) or in thymic lymphomas versus pretumoral DP cells from Cdc20Δ/AAA mice (right). (D) Normalized levels of Myc transcripts in the indicated populations from Cdc20Δ/AAA or Cdc20+/Δ samples. Horizontal bars indicate the mean. (*) P = 0.0368, (***) P < 0.001 (Student's t-test). The Myc log2 fold change values for these samples are shown in the UMAP plot. (E) Representative plots of maximum projection (left) and quantification of mean fluorescent intensity (MFI; right) of MYC protein by flow cytometry in DN1, DN2, DN3, DN4, and DP population of control Cdc20+/Δ mice in blue (left) and black (right) and Cdc20Δ/AAA pretumoral and tumoral DP populations in red. Horizontal bars indicate mean; each dot represents a biological replicate. (#) P = 0.3299, (***) P = 0.0008 (Student's t-test).
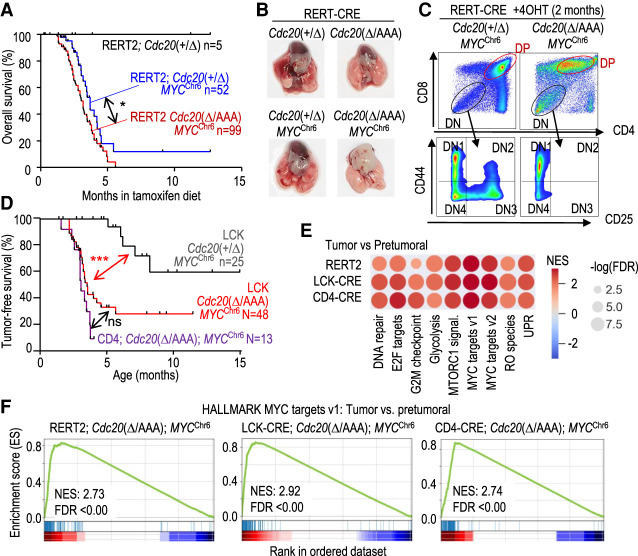
The observation that chromosome 15, where Myc resides, is gained in all Cdc20Δ/AAA-driven T-cell lymphomas (Fig. 2B,C) further raised the interesting possibility that copy number increase of Myc drives this chromosome gain. We directly tested this hypothesis by examining the consequences of expressing MYC from chromosome 6 during karyotype evolution of Cdc20Δ/AAA-driven T-cell lymphomas. To this end, we made use of a MYC transgenic allele (Calado et al. 2012) in which the human MYC cDNA, integrated at the Rosa26 locus, is driven by CAG promoter sequences after CRE-mediated excision of a loxP-STOP-loxP (LSL) cassette (referred to here as MYCChr6 allele) (Supplemental Fig. S5A,B). When we expressed MYCChr6 ubiquitously using the RERT2 allele, all Cdc20Δ/AAA; MYCChr6 mice died before 6 mo of age (Fig. 5A) due to rampant tumor formation in a variety of organs, including spleen, stomach, pancreas, and liver (Supplemental Fig. S5C,D). Despite this early lethality due to these diverse pathologies, we were able to obtain three T-cell lymphomas from these double-mutant mice (Fig. 5B). Similar to tumors in Cdc20Δ/AAA mice, these T-cell lymphomas were formed of DP or CD8+ cells (Fig. 5C; Supplemental Fig. S5E).
Figure 5.
MYCChr6 cooperated with CIN in T-cell lymphomagenesis. (A) Kaplan-Meier overall survival curves of mice with the indicated genotypes after treatment with tamoxifen. Log rank Mantel-Cox test; (*) P = 0.0184. (B) Representative macroscopic images of RERT2-CRE; Cdc20Δ/AAA; MYCChr6 thymic lymphomas. (C) Flow cytometry analysis of thymic lymphomas in RERT2-CRE; Cdc20Δ/AAA; MYCChr6 animals. (D) Tumor-free survival of mice with the indicated genotypes. Log rank Mantel-Cox test; (***) P = 0.0002 (LCK-CRE; Cdc20+/Δ; MYCChr6 vs. LCK-CRE; Cdc20Δ/AAA; MYCChr6) and P < 0.001 (LCK-CRE; Cdc20+/Δ; MYCChr6 vs. CD4-CRE; Cdc20Δ/AAA; MYCChr6). (ns) P = 0.0701. (E) Gene set enrichment analysis (GSEA) of specific HALMARK pathways in tumoral versus pretumoral samples of Cdc20Δ/AAA; MYCChr6 mice driven by the indicated CRE strains. (F) GSEA of MYC targets in the indicated samples from the analysis in E.
The early death of RERT2; Cdc20Δ/AAA; MYCChr6 mice due to tumorigenesis in other tissues precluded an in-depth analysis of T-cell lymphomas. To specifically examine T-cell lymphoma development, we analyzed the consequences of expressing MYCChr6 on tumorigenesis using the LCK- and CD4-CRE recombinase alleles (Supplemental Fig. S3A). The total number of thymic cells or distribution of populations was not altered in these models (Supplemental Fig. S6A,B). Whereas the CRE-inducible MYCChr6 transgene led to a 40% incidence of T-cell lymphomas in a Cdc20+/Δ background, Cdc20Δ/AAA; MYCChr6 developed thymic lymphomas with a median survival of 3.2 and 3 mo, respectively, in mice in which CIN was driven by LCK-CRE and CD4-CRE (Fig. 5D). Transcriptional profiling and differential expression analysis of the different models indicated that Cdc20Δ/AAA; MYCChr6 tumors were also mostly driven by MYC-dependent transcription (Fig. 5E,F).
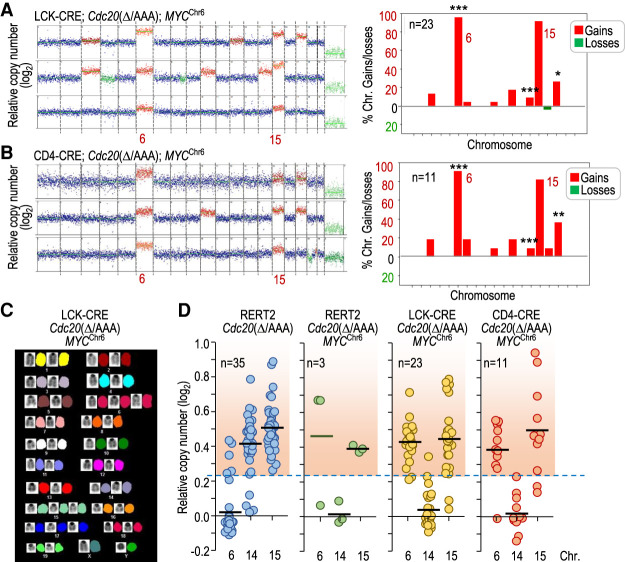
Interestingly, karyotype analysis of LCK-CRE; Cdc20Δ/AAA; MYCChr6- and CD4-CRE; Cdc20Δ/AAA; MYCChr6-driven T-cell lymphomas revealed that, while chromosome 6 was rarely gained in Cdc20Δ/AAA T-cell lymphomas (Fig. 2B), >90% of LCK-CRE; Cdc20Δ/AAA; MYCChr6 and CD4-CRE; Cdc20Δ/AAA; MYCChr6 tumors displayed chromosome 6 gains (Fig. 6A–C). Similarly, chromosome 6 was gained in two out of three tumors in RERT2; Cdc20Δ/AAA; MYCChr6 mice (Fig. 6D). Whereas chromosome 14 gains were not frequent in the presence of the MYCChr6 allele, gains in chromosome 6 did not prevent concomitant gains of chromosome 15 in >80% of these tumors (Fig. 6A,B,D). Partial or complete genetic ablation of Trp53 in CIN- and MYCChr6-driven lymphomas (Supplemental Fig. S6C) similarly resulted in universal gains of chromosome 6. In these tumors, chromosome 6 gains were accompanied of gains of chromosome 15, as well as imbalances observed in P53-null thymic lymphomas such as gains of chromosome 5 (Supplemental Fig. S6D). All together, these data indicate that expressing MYC from a different chromosome is sufficient to lead to almost universal gains of the chromosome where MYC is expressed from, thus changing the stereotypical tumor karyotype.
Figure 6.
MYC drives specific chromosome gains in Cdc20Δ/AAA tumors. (A) Representative karyotype of three tumors from LCK-CRE; Cdc20Δ/AAA; MYCChr6 mice as determined by DNA sequencing. The plot to the right shows the distribution of chromosomal gains and losses in these animals (n = 23). (B) Representative karyotype of three tumors from CD4-CRE; Cdc20Δ/AAA; MYCChr6 mice as determined by DNA sequencing. The plot to the right shows the distribution of chromosomal gains and losses in these animals (n = 11). In A and B, statistics refer to the comparison versus gains and losses in Cdc20Δ/AAA thymic lymphomas (Fig. 2B). (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 (Fisher exact test); nonsignificant (P > 0.05) differences not shown. (C) Representative SKY analysis of a T-cell lymphoma in LCK-CRE; Cdc20Δ/AAA; MYCChr6 mice showing specific gains of chromosomes 6 and 15, among others. (D) Relative copy number of chromosomes 6, 14, and 15 in T-cell lymphomas from mice with the indicated genotypes. The red-shaded region indicates copy number values higher than the threshold selected for chromosome gains (the mean + 3 SD of the euploid value). Data for chromosomes 14 and 15 in RERT2; Cdc20Δ/AAA are taken from Figure 2C for comparison. Each dot indicates an individual tumor.
Genes in addition to MYC drive chromosome 15 gain in CIN-induced T-cell lymphoma
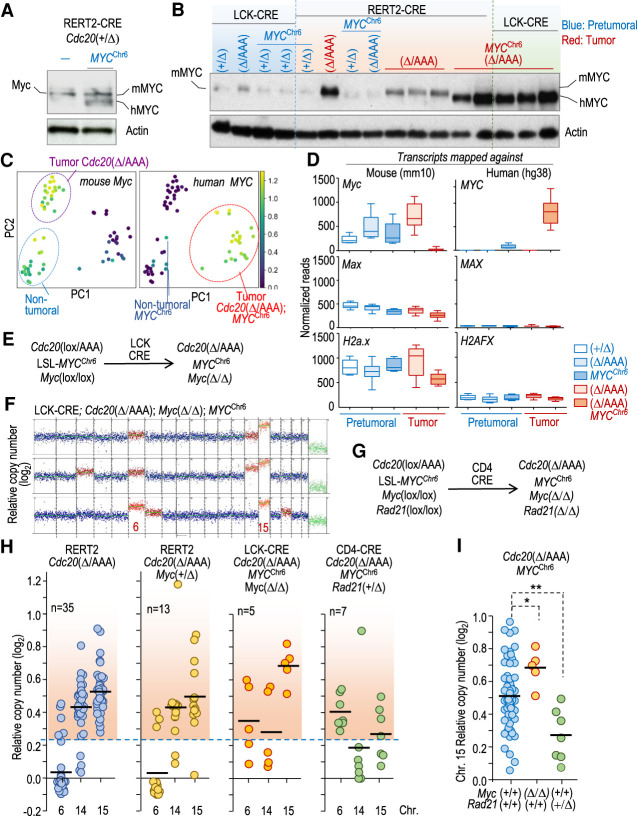
Although chromosome 15 gain was slightly less prevalent in T-cell lymphomas from Cdc20Δ/AAA; MYCChr6 samples when compared with RERT2-CRE; Cdc20Δ/AAA -driven T-cell malignancies, it was nevertheless a frequent occurrence (Fig. 6D). This result raised the possibility that, even in the presence of exogenous MYC, the endogenous Myc encoded on chromosome 15 was still contributing to T-cell lymphomagenesis. To test this possibility, we first examined the expression of transgenic human (h) and endogenous murine (m) MYC. We were able to distinguish these two forms of MYC because hMYC migrates slightly faster on SDS-PAGE than mMYC. hMYC was expressed at similar levels as mMYC in pretumoral T cells of RERT2-Cre; Cdc20+/Δ; MYCChr6 mice (Fig. 7A). Surprisingly, Cdc20Δ/AAA; MYCChr6 tumors, driven by both the RERT2-CRE or LCK-CRE allele, primarily expressed hMYC and not mMYC (Fig. 7B). This loss of mMYC expression was due to down-regulation of transcription. Human MYC transcripts were low in pretumoral MYCChr6 samples and significantly increased in tumoral MYCChr6 samples (Fig. 7C,D) in agreement to what observed at the protein level (Fig. 7B). Murine Myc transcripts, on the other hand, were detected in pretumoral T cells and Cdc20Δ/AAA tumors but not in T-cell lymphomas of Cdc20Δ/AAA; MYCChr6 mice (Fig. 7C,D). This down-regulation is likely a consequence of negative feedback loops described previously (Cole 2014), which limit expression of the endogenous Myc gene in presence of abundant MYC signaling. Importantly, these data indicated that increasing mMyc levels is unlikely to be the only reason for why chromosome 15 is frequently gained in Cdc20Δ/AAA; MYCChr6-driven T-cell lymphomas. Instead, the lack of Myc expression indicates that other genes located on chromosome 15 may contribute to T-cell lymphomagenesis in Cdc20Δ/AAA; MYCChr6 mice.
Figure 7.
Endogenous Myc is dispensable for MYCChr6-driven T-cell lymphomas. (A) Immunodetection of mouse (mMYC) and human (hMYC) MYC in protein lysates from pretumoral RERT2-CRE; Cdc20+/Δ thymuses in the absence and presence of the MYCChr6 allele. β-Actin was used as a loading control. (B) Immunodetection of the indicated proteins in pretumoral (blue) and tumoral (red) thymic samples from mice with the indicated genotypes. β-Actin was used as a loading control. (C) PCA plot showing the levels of murine Myc or human MYC transcripts in the same samples as in Figure 5E. (D) Relative normalized counts for mouse Myc and human MYC transcripts from pretumoral (blue) and tumoral (red) samples with the indicated genotypes. Reads from these samples were mapped against the mouse (mm10) or human (hg38) reference genomes to discriminate between murine Myc and human MYC transcripts. Whiskers: 10–90 percentile. Mouse and human Max/MAX and H2a.x/H2AFX transcripts are shown for comparison. (E) Schematic representation of the simultaneous editing of Cdc20, Myc, and MYCChr6 alleles by CRE recombinase. (F) Representative karyograms of three thymic lymphomas from LCK-CRE; Cdc20Δ/AAA; MycΔ/Δ; MYCChr6 mice. (G) Schematic representation of the simultaneous editing of Cdc20, MYCChr6, Myc, and Rad21 mutant alleles by CRE recombinase. (H) Relative copy number of chromosomes 6, 14, and 15 in T-cell lymphomas from mice with the indicated genotypes. Each dot indicates an individual tumor. The red-shaded region indicates copy number values higher than the threshold selected for chromosome gains (mean + 3 SD of the euploid value). (I) Quantification of chromosome 15 gains in multiple CIN- and MYCChr6-driven T-cell lymphomas either in a Rad21+/+ and Myc+/+ background (n = 69) versus MycΔ/Δ (n = 5) or Rad21+/Δ (n = 7) malignancies. (*) P = 0.0284, (**) P = 0.0052 (two-tailed Student's t-test).
To directly test the importance of endogenous Myc in MYCChr6 -driven T-cell lymphomagenesis, we deleted the endogenous Myc gene encoded by chromosome 15 using an additional Myc(lox) allele in a Cdc20Δ/AAA; MYCChr6 background. CRE activity in this model results in the concomitant activation of MYC on chromosome 6 and ablation of Cdc20 and Myc floxed alleles (Fig. 7E). Unexpectedly, karyotype analysis showed that chromosome 15 was invariably gained in T-cell lymphomas obtained from this model despite the complete lack of endogenous Myc in this model (Fig. 7F). This result, along with the silencing of the endogenous Myc in Cdc20Δ/AAA; MYCChr6 tumors, indicated that genes in addition to Myc may contribute to chromosome 15 gains in T-cell lymphomas in these models.
Rad21 contributes to chromosome 15 gains in CIN-induced T-cell lymphoma
MYC is commonly amplified with RAD21 in multiple human tumors (8p24 amplicon), and both genes map close together in human chromosome 8 or mouse chromosome 15. RAD21 expression, which is transcriptionally regulated by MYC, prevents excessive replicative stress caused by this oncogene, thus contributing to MYC-induced tumorigenesis (Rohban and Campaner 2015; Rohban et al. 2017). We therefore directly tested the possibility that increased copies of Rad21 may contribute to chromosome 15 gains by using a Rad21lox conditional allele driven by CD4-CRE expression (Fig. 7G; Seitan et al. 2011). Homozygous CD4-CRE-mediated deletion of Rad21 results in lethality when thymocytes are forced to proliferate (Seitan et al. 2011), preventing further tumorigenesis studies in vivo. However, ablation of a single copy of Rad21 in a CIN- and MYCChr6-driven background (CD4-CRE; Cdc20Δ/AAA; MYCChr6; Rad21+/Δ) resulted in the development of T-cell lymphomas with slightly increased dynamics compared with Rad21+/+ models (Supplemental Fig. S7A). This observation is in agreement with a haploinsufficient role for RAD21 in vivo and the tumorigenic effect of decreasing RAD21 expression levels in the hematopoietic system (Mullenders et al. 2015). Interestingly, whereas gains of chromosome 6 or chromosome 14 were similar to mice with wild-type Rad21, loss of a single copy of Rad21 resulted in decreased number of tumors (four out of seven) with 15 gains compared with control models (Fig. 7H; Supplemental Fig. S7B). Unexpectedly, MycΔ/Δ lymphomas displayed a higher CNV score for chromosome 15 gains, possibly suggesting more than three copies of this chromosome in Cdc20Δ/AAA- and MYCChr6-driven tumors. The CNV score for chromosome 15, on the other hand, was significantly lower in Rad21+/Δ T-cell lymphomas when compared with Rad21+/+ tumors (Fig. 7I). Based on these observations, we speculate that, in the presence of exogenous MYC on chromosome 6 and a CIN background, chromosome 15 gains are driven by Rad21.
Discussion
Previous computational studies suggested that cancer aneuploidies are driven by copy number changes in oncogenes and tumor suppressor genes (Davoli et al. 2013). However, whole-chromosome gains and losses are generally detrimental, causing multiple cellular stresses that are accompanied by a p53-dependent antiproliferative response (Thompson and Compton 2010; Santaguida and Amon 2015). In fact, single-chromosome gains can be robust inhibitors of tumor growth (Sheltzer et al. 2017). Early work investigating the role of CIN in driving LOH found that chromosome missegregation initially promotes loss of the chromosome carrying the wild-type Trp53 allele in Trp53+/− mice, thus resulting in P53-null cells. However, aneuploidy was quickly reversed as tumor cells gained a second copy of the chromosome carrying the mutant Trp53(−) allele (Baker et al. 2009). The fact that Trp53+/− cells require two chromosome missegregation events to develop into tumors (and restore euploidy) thus indicates that losing a copy of the whole chromosome containing Trp53 antagonizes tumorigenesis, presumably due to haploinsufficiency of genes located on the monosomic chromosome. Given the significant fitness penalties associated with whole-chromosome aneuploidies, it thus was not clear whether gains or losses of specific oncogenes or tumor suppressor genes could outweigh these adverse effects to drive tumorigenesis.
Our comparative analysis of a number of CIN models, using single-cell DNA sequencing as a standardized protocol, suggests that previous murine CIN models display low or moderate levels of aneuploidy. A novel model in which high levels of CIN are induced by expressing CDC20 AAA as the only cellular source of CDC20 in cells results in high prevalence tumorigenesis, and the aggregated data from all these models suggest that levels of aneuploidy in healthy tissues positively correlate with tumor susceptibility later in life.
A detailed analysis of the dynamics of aneuploidy during T-cell maturation suggests that CIN initially results in random accumulation of chromosomal gains and losses in early progenitors. However, specific karyotypes are selected during the proliferative phases of T-cell maturation, leading to the emergence of clones further selected during malignant transformation, giving rise to T-cell lymphomas. Gain of chromosome 15, where the Myc-encoding gene resides, is an obligatory alteration that probably prevents DP cells from down-regulating pathways that stimulate proliferation. MYC is a potent oncogene involved in 70% of human cancers and is a major driver of T-cell acute lymphoblastic leukemia (T-ALL) and T-cell lymphomas, where it functions as a central node downstream from major oncogenic pathways, including NOTCH or TAL1/ARID5B (Weng et al. 2006; Palomero et al. 2007; Leong et al. 2017). In murine thymic lymphomas, MYC is typically overexpressed (McMorrow et al. 1988; Gaudet et al. 2003; Yoshida et al. 2007; Bueno et al. 2008; Timakhov et al. 2009; De Keersmaecker et al. 2010). Furthermore, gain of chromosome 15 is a frequent occurrence in T-cell lymphomas induced by carcinogens and in genetically modified mice, including LCK-TLX1 transgenics (McMorrow et al. 1988; Yoshida et al. 2007; Bueno et al. 2008; De Keersmaecker et al. 2010; Bakker et al. 2016). P53 loss, which is typically associated with increased chromosomal instability, is also accompanied by chromosome 15 gains (Venkatachalam et al. 1998; Haines et al. 2006; Bakker et al. 2016). Interestingly, p53 seems to be at least partially functional in Cdc20Δ/AAA lymphomas, suggesting that high levels of CIN may overcome the need of eliminating p53 function. This is an interesting observation that would require further research in the future.
How does MYC drive thymic lymphomagenesis in Cdc20Δ/AAA animals? DN3 thymocytes depend on ligand-induced Notch signaling to maintain MYC expression (Weng et al. 2006). Upon pre-T-cell receptor (pre-TCR) expression, they lose their dependence on Notch and cytokine signaling and differentiate into CD4+ CD8+ DP cells, in which MYC is typically repressed. We propose that in aneuploid Cdc20Δ/AAA DP cells, this repression is lost as a consequence of frequent chromosome 15 gain, which then contributes to the development of lymphomas. Interestingly, while MYC expression is higher in Cdc20Δ/AAA thymic lymphomas compared with pretumoral DP cells, it is still lower than in normal DN2, DN3, and DN4 cells. Thus, it appears that the amount of MYC needed to drive T-cell lymphomagenesis is not greater than what is observed in highly proliferative lymphocyte populations. In fact, Cdc20Δ/AAA; MYCChr6; MycΔ/Δ mice display higher CNV score for chromosome 15 gains than the corresponding Myc(+/+) controls (Fig. 7). One possible explanation for this observation arises from the fact that MYC overexpression induces replicative stress and a DNA damage response that acts as a barrier to tumor development (Halazonetis et al. 2008; Rohban and Campaner 2015). This replication stress can be managed in tumor cells through the expression of RAD21, a component of the cohesin complex involved in transcription, repair of DNA double-strand breaks, as well as in chromatid cohesion during mitosis (Losada 2014). MYC and cohesin cooperate in triggering transcription from specific sites in the genome (Yan et al. 2013), and RAD21 reinforces cohesion-mediated DNA synthesis, thus limiting the negative effect of MYC-induced replicative stress (Rohban et al. 2017). Human RAD21 has recently been reported to drive chromosome 8 gains in Ewing sarcoma, a tumor with high levels of replicative stress (Su et al. 2021). MYC and RAD21 are frequently coamplified in human cancer (Rohban et al. 2017) and, importantly, loss of RAD21 is synthetic lethal with MYC overexpression (Toyoshima et al. 2012), suggesting the possibility of inhibiting cohesion complexes to restrain MYC-driven proliferation in cancer (Toyoshima et al. 2012; Rohban and Campaner 2015).
CIN-induced T-cell lymphomas are characterized by almost universal gains of chromosome 14 and 15. On the other hand, T-cell lymphomas generated in a Trp53-null background typically accumulate gains in chromosomes 5 and 15 among others. When MYC is expressed from chromosome 6, chromosome 6 gain becomes essential either in CIN- or p53-loss-induced tumors, indicating that a single oncogene can determine gains of a specific chromosome in cancer. Whereas chromosome 14 is frequently gained in Cdc20Δ/AAA thymic lymphomas, this chromosome is rarely gained in the presence of MYCChr6. An interesting possibility for these observations is that chromosome 14 gain may contribute to the deregulation of the endogenous Myc locus, and that this event is totally dispensable when MYC is provided exogenously. Overall, these data support the hypothesis that random aneuploidies in premalignant tissues are subjected to clonal selection and that the ultimate aneuploidy spectrum observed in tumors is likely dictated by the chromosomal position of specific driver oncogenes (MYC) or genes modulating tumorigenesis (RAD21) in these malignancies. It has not escaped our attention that in this model, both the adverse (defective proliferation of random aneuploidies) (Ben-David and Amon 2020) and the beneficial (gain of oncogenes and “helper” genes in selected aneuploidies) effects of aneuploidy would contribute to tumorigenesis by favoring clonal selection at early stages of tumor development.
Materials and methods
Mouse models
The Cdc20AAA (Li et al. 2009), Cdc20lox (Manchado et al. 2010), Myclox (de Alboran et al. 2001), Rad21lox (Seitan et al. 2011), and Rosa26-LSL-CAG-Myc-hCD2 (MYCChr6)(Calado et al. 2012) alleles were reported previously. For excision of loxP-flanked sequences, we used a tamoxifen-inducible CRE recombinase ubiquitously expressed under the control of RNAPII sequences (Manchado et al. 2010). To activate Cre during specific stages of T-cell development, we used constitutive LCK-CRE or CD4-CRE (Lee et al. 2001). Tamoxifen was administered in the diet (Envigo TD.130860), alternating tamoxifen diet with normal minimal phytoestrogen diet (Teklad global 16% protein, Envigo). Other mouse models used in this work are listed in Supplemental Table S1 (Baker et al. 2004; Malureanu et al. 2010; González-Loyola et al. 2015; Levine et al. 2017; de Cárcer et al. 2018). Mice were in a mixed C57/BL6J x 129/J mixed background. All animal procedures were performed following the institutional guidelines and protocols approved by the Massachusetts Institute of Technology Committee for Animal Care and the corresponding national legislation.
For histological analyses, dissected organs were fixed in 10% buffered formalin (Sigma) and embedded in paraffin wax. Sections of 3- or 5-μm thickness were stained with hematoxylin and eosin (H&E). Additional immunohistochemical examinations of the tissues and pathologies were performed using specific antibodies against KI67 (Abcam ab16667), phospho-histone H3 S10 (Millipore 06-570), and active caspase 3 (Cell Signaling 9661).
Cell isolation from tissues and tumors and flow cytometry
Bone marrow, spleenocytes, thymocytes, and intestinal crypt cells were isolated as described previously (Pfau et al. 2016). Briefly, after obtaining a single-cell suspension, cells were incubated for 3 min in ACK buffer for red blood cell lysis and washed in IMDM containing 0.5% BSA. Cells were counted with a Coulter counter analyzer, and 3 million cells were stained with antibodies for CD8a, CD4, CD25, and CD44 (BD Biosciences) for thymic profile analysis. For the isolation of HSCs and CLPs, 50 million to 80 million bone marrow cells were incubated with a mouse lineage antibody cocktail labeled with biotin obtained from Miltenyi Biotech. Cells were incubated with antibiotin microbeads, and subsequently, lineage-positive cells were depleted by retention of biotin-positive cells on a MACS column (Miltenyi Biotech). Lineage-depleted cells were stained for CD48, CD150, CD117, and Sca-1 (BD Biosciences and BioLegend) and sorted on a FACSARIA4 cell sorter (Becton Dickinson). For splenocyte stimulation, cells were treated with 50 ng/mL phorbol 12-myristate 13-acetate (PMA) + 50 ng/mL ionomycin and treated with 10 μM EdU for 2 h before analysis by flow cytometry. In all cases, the fixable Near-IR live/dead dye (Invitrogen) was used to exclude dead cells from the analysis. FlowJo software was used for flow cytometry analysis.
Bulk and single-cell DNA sequencing
Single-cell DNA sequencing was performed as previously described (Knouse et al. 2014, 2016). Briefly, single cells were isolated either by microaspiration or by 96-well plate sorting on a FACSAria4 cell sorter, and genomic DNA was amplified with the GenomePlex single-cell whole-genome amplification kit (Sigma 25-457-8). Amplified DNA was purified, barcoded, pooled, and sequenced on an Illumina HiSeq2000 at the Massachusetts Institute of Technology Bio-Micro Center. For bulk DNA sequencing analysis, 50 ng of purified DNA was prepared and barcoded using Nextera reagents (Illumina). Libraries were quantified using an AATI fragment analyzer before pooling and were sequenced on an Illumina HiSeq2000. Sequencing reads were trimmed to 40 nucleotides and aligned to mouse (mm9) genome reference using the BWA (0.7.17) backtrack algorithm with default options. HMMcopy (0.1.1) was used to detect copy number variations (CNVs) by estimating DNA copy number in 500-kb bins, controlling for mappability and GC content (calculated by HMMcopy gcCounter). Variability scores (VSs) were calculated as previously described (Knouse et al. 2014). Cells with high variability in copy number across bins (VS > 0.4) were excluded from the analysis. We used the tail values of the kernel density estimation to define a chromosome as being lost when the CNV score log2 < −0.32 and gained when this value was log2 > 0.23, corresponding to the mean value ± 3 standard deviations of the copy number value of chromosome scores in euploid cels.
To define the clonality of a cell population, we scored chromosomes that were gained or lost with a frequency higher or lower than the average ± 2 SD of the overall gains and losses in that cell population after single-cell DNA sequencing analysis. These chromosomes were defined as clonally gained or lost, and the percentage of cells carrying these clonal imbalances was taken as the clonality index.
RNA sequencing
Total RNA was isolated from primary cells or tumors, using the RNeasy plus mini kit (Qiagen) and sequenced on an Illumina HiSeq2000. Read quality was monitored by the MIT BMC/IGB in house pipeline, including sequencing error rate estimation, sequencing reads complexity estimation, fastqc report, sample contamination estimation, intergenic reads percentile calculation, intronic reads percentile calculation, UTR reads percentile calculation, gene coding reads percentile calculation, rRNA contamination estimation, and sense to antisense read ratio indicating strand specificity. Alignments were carried out using STAR/2.5.3a, and counting was performed by RSEM/1.3.0 based on r/3.3.1. Sample hierarchical clustering and heat map creation were performed by TIBCO Spotfire build version 7.11.1.0.12 based on log2(FPKM + 1) values calculated by RSEM/1.3.0. To avoid potential noise caused by noncoding genes, as well as nonexpressed genes, clustering was computed by protein coding genes with expression (sample-wise average expression ≥1 FPKM). Normalization and differential expression were carried out by DESeq2 1.10.1 using r/3.2.3. Up-regulated genes were defined as baseMean > 10, log2 fold change > 0.5, and multitesting adjusted P-values < 0.05; down-regulated genes were baseMean > 10, log2 fold change < −0.5, and multitesting adjusted P-values < 0.05. For the analysis of human MYC expression, sequencing reads were mapped to the human genome (hg38) using bowtie2, and alignments with mismatches were filtered out. Gene counts were quantified with featureCounts and data normalized using DESeq2. Gene set enrichment analysis was performed using GSEA version 4.0.3 obtained from the Broad Institute and GSEApy (https://gseapy.readthedocs.io/en/latest/index.html) using custom gene sets for MYC targets (Zeller et al. 2003) or canonical pathway gene sets available at MsigDB (https://www.gsea-msigdb.org/gsea/index.jsp). Enrichr (https://amp.pharm.mssm.edu/Enrichr) and DAVID (https://david.ncifcrf.gov) were used for functional annotation of gene signatures.
Cell culture, metaphase spreads, and spectral karyotyping
Single-cell suspensions were obtained from primary tumors after ACK red blood cell lysis and were cultured in the presence of the CDK1 inhibitor RO3306 (7.5 μM) for 6 h to enrich for cells in G2. The CDK1 inhibitor was then washed out, and cells were cultured for an additional hour to enter mitosis. Subsequently, samples were treated with (0.075M KCl) hypotonic solution and were fixed in methanol:acetic acid (3:1 volume ratio). Metaphases were prepared in a controlled humidity chamber (Thermotron). Spectral karyotyping was performed and analyzed as previously described (Liyanage et al. 1996), using a combination of five different fluorochromes. A minimum of 20 metaphases were imaged and karyotyped using the hiSKY 7.2.7 software (ASI) on a Leica DMRXA microscope.
Protein analysis
For immunodetection in protein lysates, proteins were separated on SDS-PAGE, transferred to nitrocellulose membranes (Bio-Rad), and probed using specific primary antibodies against MYC (Cell Signaling), cyclin B1 (Cell Signaling), P53 (Cell Signaling), P21 (Santa Cruz Biotechnology), and phosphorylated H2AX (γ-H2AX; Millipore 05-636). Actin (1:10,000; Sigma) was used as a loading control. Ki67 (Abcam ab16667), phospho-histone H3 S10 (Millipore 06-570), and active caspase 3 (Cell Signaling 9661) were used for detection of the corresponding antigens in paraffin sections.
Statistical analysis
Statistical analyses were performed using GraphPad Prism version 8 and Python v. 3.8. All statistical tests of comparative data were performed using two-sided, unpaired Student's t-test, Fisher exact tests, or one-way or two-way ANOVA for differential comparison between two or more groups, respectively. Data with P < 0.05 were considered statistically significant (P < 0.05 [*], P < 0.01 [**], and P < 0.001 [***]). Specific statistical parameters for each assay are indicated in the corresponding figure legends.
Data availability
Sequencing data are available at the Gene Expression Omnibus (ID GSE174012) and the Sequence Read Archive (SRA) under the master accession number PRJNA728729.
Supplementary Material
Acknowledgments
We thank past and present members of the Amon laboratory, particularly John Replogle, Hilla Weidberg, Gabriel Neurohr, Sarah Pfau, Daniel Corbi, Summer Morrill, Rebecca Silberman, and Timothy Mullen, for helpful discussions, reading of the manuscript, and technical advice. We thank Shannon MacMann for assistance with mouse colony management. We thank Dr. Roderick Bronson for the histopathological analysis. We thank the Histology, Flow Cytometry, and Genomics Core Facilities at the Swanson Biotechnology Center for technical assistance. This work was supported by National Institutes of Health grant CA206157 to A.A., who is an investigator of the Howard Hughes Medical Institute, the Paul F. Glenn Center for the Biology of Aging Research at Massachusetts Institute of Technology, and the Ludwig Center at Massachusetts Institute of Technology, and by a synergy grant from the European Research Council (ERC-2019-SyG-855158). M.T. was supported by a long-term EMBO fellowship and funding from the Ludwig Center at Massachusetts Institute of Technology.
Author contributions: M.T. and A.A. designed and directed the study. M.T. performed most assays with the help of M.A., C.S., A.C., A.H., and P.-h.H. L.Z. contributed to the single-cell sequencing of CIN models. X.A.S. helped in the analysis of Rad21 models. D.W. performed the SKY analysis. G.P. assisted with flow cytometry. D.M. conducted bioinformatic analysis. C.S. and J.Z. generated the Bub1b−/I909T and Mad2−/H models, respectively. T.R., A.H., and J.V.D. provided conceptual advice. M.T. wrote the manuscript with comments from all authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348341.121.
Competing interest statement
The authors declare no competing interests.
References
- Baker DJ, Jeganathan KB, Cameron JD, Thompson M, Juneja S, Kopecka A, Kumar R, Jenkins RB, de Groen PC, Roche P, et al. 2004. Bubr1 insufficiency causes early onset of aging-associated phenotypes and infertility in mice. Nat Genet 36: 744–749. 10.1038/ng1382 [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. 2009. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell 16: 475–486. 10.1016/j.ccr.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DC, de Jong TV, Halsema N, Kazemier HG, Hoekstra-Wakker K, Bradley A, et al. 2016. Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol 17: 115. 10.1186/s13059-016-0971-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David U, Amon A. 2020. Context is everything: aneuploidy in cancer. Nat Rev Genet 21: 44–62. 10.1038/s41576-019-0171-x [DOI] [PubMed] [Google Scholar]
- Bueno MJ, Pérez de Castro I, Gómez de Cedron M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. 2008. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell 13: 496–506. 10.1016/j.ccr.2008.04.018 [DOI] [PubMed] [Google Scholar]
- Calado DP, Sasaki Y, Godinho SA, Pellerin A, Köchert K, Sleckman BP, de Alboran IM, Janz M, Rodig S, Rajewsky K. 2012. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol 13: 1092–1100. 10.1038/ni.2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MD. 2014. MYC association with cancer risk and a new model of MYC-mediated repression. Cold Spring Harb Perspect Med 4: a014316. 10.1101/cshperspect.a014316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Xu AW, Mengwasser KE, Sack LM, Yoon JC, Park PJ, Elledge SJ. 2013. Cumulative haploinsufficiency and triplosensitivity drive aneuploidy patterns and shape the cancer genome. Cell 155: 948–962. 10.1016/j.cell.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Alboran IM, O'Hagan RC, Gärtner F, Malynn B, Davidson L, Rickert R, Rajewsky K, DePinho RA, Alt FW. 2001. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14: 45–55. 10.1016/S1074-7613(01)00088-7 [DOI] [PubMed] [Google Scholar]
- de Cárcer G, Venkateswaran SV, Salgueiro L, El Bakkali A, Somogyi K, Rowald K, Montañés P, Sanclemente M, Escobar B, de Martino A, et al. 2018. Plk1 overexpression induces chromosomal instability and suppresses tumor development. Nat Commun 9: 3012. 10.1038/s41467-018-05429-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Keersmaecker K, Real PJ, Gatta GD, Palomero T, Sulis ML, Tosello V, Van Vlierberghe P, Barnes K, Castillo M, Sole X, et al. 2010. The TLX1 oncogene drives aneuploidy in T cell transformation. Nat Med 16: 1321–1327. 10.1038/nm.2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duijf PH, Benezra R. 2013. The cancer biology of whole-chromosome instability. Oncogene 32: 4727–4736. 10.1038/onc.2012.616 [DOI] [PubMed] [Google Scholar]
- Gaudet F, Hodgson JG, Eden A, Jackson-Grusby L, Dausman J, Gray JW, Leonhardt H, Jaenisch R. 2003. Induction of tumors in mice by genomic hypomethylation. Science 300: 489–492. 10.1126/science.1083558 [DOI] [PubMed] [Google Scholar]
- González-Loyola A, Fernández-Miranda G, Trakala M, Partida D, Samejima K, Ogawa H, Canamero M, de Martino A, Martinez-Ramirez A, de Cárcer G, et al. 2015. Aurora B overexpression causes aneuploidy and p21Cip1 repression during tumor development. Mol Cell Biol 35: 3566–3578. 10.1128/MCB.01286-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines BB, Ryu CJ, Chang S, Protopopov A, Luch A, Kang YH, Draganov DD, Fragoso MF, Paik SG, Hong HJ, et al. 2006. Block of T cell development in P53-deficient mice accelerates development of lymphomas with characteristic RAG-dependent cytogenetic alterations. Cancer Cell 9: 109–120. 10.1016/j.ccr.2006.01.004 [DOI] [PubMed] [Google Scholar]
- Halazonetis TD, Gorgoulis VG, Bartek J. 2008. An oncogene-induced DNA damage model for cancer development. Science 319: 1352–1355. 10.1126/science.1140735 [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. 2009. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol 10: 478–487. 10.1038/nrm2718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Whittaker CA, Amon A. 2014. Single cell sequencing reveals low levels of aneuploidy across mammalian tissues. Proc Natl Acad Sci 111: 13409–13414. 10.1073/pnas.1415287111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knouse KA, Wu J, Amon A. 2016. Assessment of megabase-scale somatic copy number variation using single-cell sequencing. Genome Res 26: 376–384. 10.1101/gr.198937.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PP, Fitzpatrick DR, Beard C, Jessup HK, Lehar S, Makar KW, Pérez-Melgosa M, Sweetser MT, Schlissel MS, Nguyen S, et al. 2001. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 15: 763–774. 10.1016/S1074-7613(01)00227-8 [DOI] [PubMed] [Google Scholar]
- Leong WZ, Tan SH, Ngoc PCT, Amanda S, Yam AWY, Liau WS, Gong Z, Lawton LN, Tenen DG, Sanda T. 2017. ARID5B as a critical downstream target of the TAL1 complex that activates the oncogenic transcriptional program and promotes T-cell leukemogenesis. Genes Dev 31: 2343–2360. 10.1101/gad.302646.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MS, Bakker B, Boeckx B, Moyett J, Lu J, Vitre B, Spierings DC, Lansdorp PM, Cleveland DW, Lambrechts D, et al. 2017. Centrosome amplification is sufficient to promote spontaneous tumorigenesis in mammals. Dev Cell 40: 313–322 e315. 10.1016/j.devcel.2016.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Fang X, Wei Z, York JP, Zhang P. 2009. Loss of spindle assembly checkpoint-mediated inhibition of Cdc20 promotes tumorigenesis in mice. J Cell Biol 185: 983–994. 10.1083/jcb.200904020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liyanage M, Coleman A, du Manoir S, Veldman T, McCormack S, Dickson RB, Barlow C, Wynshaw-Boris A, Janz S, Wienberg J, et al. 1996. Multicolour spectral karyotyping of mouse chromosomes. Nat Genet 14: 312–315. 10.1038/ng1196-312 [DOI] [PubMed] [Google Scholar]
- Losada A. 2014. Cohesin in cancer: chromosome segregation and beyond. Nat Rev Cancer 14: 389–393. 10.1038/nrc3743 [DOI] [PubMed] [Google Scholar]
- Malureanu L, Jeganathan KB, Jin F, Baker DJ, van Ree JH, Gullon O, Chen Z, Henley JR, van Deursen JM. 2010. Cdc20 hypomorphic mice fail to counteract de novo synthesis of cyclin B1 in mitosis. J Cell Biol 191: 313–329. 10.1083/jcb.201003090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E, Guillamot M, de Cárcer G, Eguren M, Trickey M, Garcia-Higuera I, Moreno S, Yamano H, Canamero M, Malumbres M. 2010. Targeting mitotic exit leads to tumor regression in vivo: modulation by Cdk1, Mastl, and the PP2A/B55α,δ phosphatase. Cancer Cell 18: 641–654. 10.1016/j.ccr.2010.10.028 [DOI] [PubMed] [Google Scholar]
- McMorrow LE, Newcomb EW, Pellicer A. 1988. Identification of a specific marker chromosome early in tumor development in γ-irradiated C57BL/6J mice. Leukemia 2: 115–119. [PubMed] [Google Scholar]
- Meyer N, Penn LZ. 2008. Reflecting on 25 years with MYC. Nat Rev Cancer 8: 976–990. 10.1038/nrc2231 [DOI] [PubMed] [Google Scholar]
- Mingueneau M, Kreslavsky T, Gray D, Heng T, Cruse R, Ericson J, Bendall S, Spitzer MH, Nolan GP, Kobayashi K, et al. 2013. The transcriptional landscape of αβ T cell differentiation. Nat Immunol 14: 619–632. 10.1038/ni.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, Keller M, Pae J, Wang K, Kayembe C, Rocha PP, Raviram R, Gong Y, et al. 2015. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med 212: 1833–1850. 10.1084/jem.20151323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. 2007. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Pai SY, Truitt ML, Ting CN, Leiden JM, Glimcher LH, Ho IC. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity 19: 863–875. 10.1016/S1074-7613(03)00328-5 [DOI] [PubMed] [Google Scholar]
- Palomero T, Sulis ML, Cortina M, Real PJ, Barnes K, Ciofani M, Caparros E, Buteau J, Brown K, Perkins SL, et al. 2007. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med 13: 1203–1210. 10.1038/nm1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfau SJ, Silberman RE, Knouse KA, Amon A. 2016. Aneuploidy impairs hematopoietic stem cell fitness and is selected against in regenerating tissues in vivo. Genes Dev 30: 1395–1408. 10.1101/gad.278820.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohban S, Campaner S. 2015. Myc induced replicative stress response: how to cope with it and exploit it. Biochim Biophys Acta 1849: 517–524. 10.1016/j.bbagrm.2014.04.008 [DOI] [PubMed] [Google Scholar]
- Rohban S, Cerutti A, Morelli MJ, di Fagagna FDA, Campaner S. 2017. The cohesin complex prevents Myc-induced replication stress. Cell Death Dis 8: e2956. 10.1038/cddis.2017.345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santaguida S, Amon A. 2015. Short- and long-term effects of chromosome mis-segregation and aneuploidy. Nat Rev Mol Cell Biol 16: 473–485. 10.1038/nrm4025 [DOI] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R. 2010. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer 10: 102–115. 10.1038/nrc2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, et al. 2011. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature 476: 467–471. 10.1038/nature10312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Ko JH, Replogle JM, Habibe Burgos NC, Chung ES, Meehl CM, Sayles NM, Passerini V, Storchova Z, Amon A. 2017. Single-chromosome gains commonly function as tumor suppressors. Cancer Cell 31: 240–255. 10.1016/j.ccell.2016.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su XA, Ma D, Parsons JV, Replogle JM, Amatruda JF, Whittaker CA, Stegmaier K, Amon A. 2021. RAD21 is a driver of chromosome 8 gain in Ewing sarcoma to mitigate replication stress. Genes Dev 35: 556–572. 10.1101/gad.345454.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SL, Compton DA. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J Cell Biol 188: 369–381. 10.1083/jcb.200905057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timakhov RA, Tan Y, Rao M, Liu Z, Altomare DA, Pei J, Wiest DL, Favorova OO, Knepper JE, Testa JR. 2009. Recurrent chromosomal rearrangements implicate oncogenes contributing to T-cell lymphomagenesis in Lck-MyrAkt2 transgenic mice. Genes Chromosomes Cancer 48: 786–794. 10.1002/gcc.20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoshima M, Howie HL, Imakura M, Walsh RM, Annis JE, Chang AN, Frazier J, Chau BN, Loboda A, Linsley PS, et al. 2012. Functional genomics identifies therapeutic targets for MYC-driven cancer. Proc Natl Acad Sci 109: 9545–9550. 10.1073/pnas.1121119109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachalam S, Shi YP, Jones SN, Vogel H, Bradley A, Pinkel D, Donehower LA. 1998. Retention of wild-type p53 in tumors from p53 heterozygous mice: reduction of p53 dosage can promote cancer formation. EMBO J 17: 4657–4667. 10.1093/emboj/17.16.4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver BA, Cleveland DW. 2006. Does aneuploidy cause cancer? Curr Opin Cell Biol 18: 658–667. 10.1016/j.ceb.2006.10.002 [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. 2006. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev 20: 2096–2109. 10.1101/gad.1450406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Enge M, Whitington T, Dave K, Liu J, Sur I, Schmierer B, Jolma A, Kivioja T, Taipale M, et al. 2013. Transcription factor binding in human cells occurs in dense clusters formed around cohesin anchor sites. Cell 154: 801–813. 10.1016/j.cell.2013.07.034 [DOI] [PubMed] [Google Scholar]
- Yoshida MA, Nakata A, Akiyama M, Kakinuma S, Sado T, Nishimura M, Shimada Y. 2007. Distinct structural abnormalities of chromosomes 11 and 12 associated with loss of heterozygosity in X-ray-induced mouse thymic lymphomas. Cancer Genet Cytogenet 179: 1–10. 10.1016/j.cancergencyto.2007.06.016 [DOI] [PubMed] [Google Scholar]
- Zeller KI, Jegga AG, Aronow BJ, O'Donnell KA, Dang CV. 2003. An integrated database of genes responsive to the Myc oncogenic transcription factor: identification of direct genomic targets. Genome Biol 4: R69. 10.1186/gb-2003-4-10-r69 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data are available at the Gene Expression Omnibus (ID GSE174012) and the Sequence Read Archive (SRA) under the master accession number PRJNA728729.