Abstract
Prosobranch snails and adult Paramphistomoidea flukes were collected from water bodies and cattle abattoir located in Mpumalanga province of South Africa, respectively. The snails were identified based on morphological characters as well as the ITS-2 and 16S markers as Melanoides sp. and Tarebia granifera, respectively, and the Paramphistomoidea flukes were identified as Calicophoron microbothrium using the ITS-1/5.8S/ITS-2 marker. After confirming identification, the snails were bred to first filial generation (F1) under laboratory conditions. Ninety snails were randomly selected from the laboratory-bred F1 snails and 25 Melanoides sp. and 20 T. granifera were exposed to C. microbothrium miracidia, and the same numbers were maintained as non-exposed controls. Results showed that C. microbothrium successfully established in Melanoides sp. and produced cercariae, and the prepatent period recorded was 21 days. Three snails shed cercariae at day 21 postexposure (PE), and rediae and free cercariae were detected in the soft tissues of one snail on dissection at day 44 PE. The same fluke did not establish in T. granifera. Melanoides sp. started producing offspring at day 7 PE, and T. granifera at day 14 PE. In conclusion, our results showed that Melanoides sp. used in this study is a suitable intermediate host for C. microbothrium under experimental conditions, and given the wide distribution of this snail species, it is important to determine its role in the natural transmission of other Calicophoron species that have been reported in South Africa.
Keywords: Tarebia granifera, Melanoides spp., Melanoides tuberculata, Melanoides victoriae, Thiaridae, Calicophoron microbothrium, experimental infectivity
Background
Amphistomosis is a parasitic infection of domestic and wild ruminants caused by immature amphistomes, also known as stomach flukes and located in the small intestines of susceptible hosts (1, 2). It is the most prevalent snail-borne infection in sub-Saharan Africa (3) caused by digenetic trematodes of the superfamily Paramphistomoidea Fischoeder, 1901 (4). According to Mavenyengwa et al. (2), amphistomosis is characterized by scattered epidemics of acute parasitic gastroenteritis in the definitive hosts and results in loss of production, which is associated with high morbidity and mortality, mainly in young animals.
According to Sey (5), more than 70 amphistomes species have been reported globally; however, differentiating between these species is difficult due to the extreme similarities in their morphological features (6). About 32 of these amphistome species in ruminants belong to the families Paramphistomoidea, Gastrothylacidae, and Stephanopharyngidae, and the majority have been recorded in Africa (7). The superfamily Paramphistomoidea has a cosmopolitan distribution and proven to be the main cause of amphistomosis in eastern and southern Africa (4). However, only 10 amphistome species from the genera Calicophoron, Paramphistomum, Cotylophoron, and Carmyerius are regarded as the most common species infecting domestic ruminants (7, 8). Although various amphistomes species have been documented, often amphistomosis outbreaks are caused by certain species, and out of the many Paramphistomoidea species reported in sub-Sharan Africa (7), Calicophoron microbothrium (Fischoeder, 1901) has the widest distribution in sub-Saharan African countries (4, 8). This species is the main cause of acute amphistomosis in young animals, which results in losses among infected animals (7).
Like other snail-borne parasitic disease, the transmission of amphistomes and prevalence of amphistomosis are largely influenced by the abundance of the definitive host and the availability and efficiency of the snail intermediate hosts in transmitting the parasites (4). A wide range of pulmonate freshwater gastropods belonging to the genera Bulinus Müller (1781), Biomphalaria Preston (1910), and Ceratophallus Brown and Mandahl-Barth (1973), and Galba Müller (1774) from families Planorbidade (Rafinesque, 1815) and Lymnaeidae (Rafinesque, 1815), have been deemed to be the snail intermediate hosts of amphistomes in Africa (4, 9–13). Current literature show that seven freshwater pulmonate snail species—Biomphalaria pfeifferi (Krauss, 1848), Bulinus forskalii (Ehrenberg, 1831), Bulinus globosus (Morelet, 1866), Bulinus nasutus (von Martens, 1879), Bulinus tropicus (Krauss, 1848), Ceratophallus natalensis (Krauss, 1848), and Galba truncatula (Müller, 1774)—have been confirmed as natural intermediate hosts of amphistome species in southern and eastern Africa (4), and species of the genus Bulinus contribute more in the transmission of the various amphistomes species (4).
Prosobranch snails such as Melanoides species and Tarebia granifera (Lamarck, 1816) are tropical and subtropical species native to Southeast Asia, the Mediterranean, Pacific islands, East Africa, and across the Middle East (14–16), and Southeast Asia (17, 18), respectively. In South Africa, two Melanoides species have been reported to date, namely, Melanoides tuberculata (Müller, 1774) and Melanoides victoriae (Dohrn, 1865) (19). According to the authors, M. tuberculata is widely distributed in the present-day Sahara (20), including South Africa, based on the National Freshwater Snail Collection records (19). Melanoides victoriae however appears to be restricted to Southern Africa (14, 21). Of recent, T. granifera has been introduced to Africa and other parts of the world either accidentally through aquarium trades or intentionally for biocontrol of schistosome intermediate hosts (22, 23). According to Appleton et al. (17) and de Kock and Wolmarans (19), these species are known to act as intermediate hosts for several trematodes in their native origin. However, their role in the transmission of snail-borne infections, and for amphistomes in particular, is not known in South Africa, albeit there have been reports of successful experimental infection of M. tuberculata by C. microbothrium in Zimbabwe (24).
Therefore, in order to fully understand the factors influencing the wide distribution of amphistomes species in southern and eastern Africa (4), it is important to accurately identify the snail intermediate hosts and determine their degree of compatibility to C. microbothrium, more especially in alien and/or invasive snail species. This information is critical in designing control measures, particularly where amphistomosis in endemic. Hence, this study aimed at experimentally determining the suitability of two prosobranch snails, Melanoides sp. and T. granifera, as intermediate hosts of C. microbothrium.
Materials and Methods
Snail Collection and Breeding of F1 Generation
Sexually mature Melanoides sp. and T. granifera snails used to breed the first filial generation (F1) snails were collected from various water habitats in Mpumalanga province of South Africa, using metal scoops as described by Coulibaly and Madsen (25). Snails were screened for patent trematode infection through shedding (26). Snails that were not shedding cercariae were bred in 2-L polyvinyl chloride (PVC) containers, with eight snails per container, filled with filtered pond water and allowed to produce an F1 generation. Snails were fed blanched dried lettuce supplemented with commercial tropical fish flakes ad libitum. The water was changed twice a week, and room temperature was maintained at 27 ± 1°C.
Amphistomes Collection, Processing, and Hatching of Eggs
Adult amphistomes were collected from rumen of three naturally infected cattle slaughtered at an abattoir in Mpumalanga province of South Africa, and one portion was preserved in physiological saline and the other in 70% alcohol prior to processing at the laboratory. Amphistomes that were preserved in physiological saline were crushed using pestle and mortar to release eggs. The eggs were recovered by passing the suspension through sieves of descending apertures, and eggs were recovered on a 20-μm sieve. Eggs were washed three times in distilled water and then incubated at 27°C for 14 days. Hatching of the embryonated eggs was stimulated by directly exposing the eggs with artificially illuminated light bulb for an hour (27), and free miracidia were then used for infections. The portion preserved in alcohol were processed and identified according to Eduardo (28).
Snail and Parasite Identification
Thiaridae snails collected from the field were visually and morphologically identified based on the shell images of the South African specimens as described by Brown (14), Miranda et al. (18), and Appleton (29). Soft tissue of up to four individual snails from each species identified based on shell characters were harvested, and DNA was extracted using the Genomic DNA™ Tissue MiniPrep Kit (Zymo Research Corporation, Irvine, CA, USA) according to the manufacturer's instructions. Amplification of the snail DNA was performed based on the 16S rDNA primers and conditions as provided by Palumbi et al. (30) for T. granifera. The 16S primers were not successful for Melanoides sp.; therefore, the ITS-2 primers and cycling conditions described by Malatji et al. (31) were used. A separate PCR amplification was performed on six amphistome specimens from a batch collected from three cattle at an abattoir and preserved in 70% alcohol prior to using the parasite to infect the F1 snails, and this was based on the ITS-1/5.8S/ITS-2 region using primers and thermocycling conditions described by Mucheka et al. (32). Amplicons were sent to Inqaba Biotechnical Industries (Pty) Ltd. in Pretoria, South Africa, for Sanger sequencing.
Sequences were assembled and manually edited using the BioEdit v7.2.5 program (33). Sequences were aligned along with the homologous sequences obtained from the GenBank database using MUSCLE multiple alignment option on MEGA 7 (34), and alignments were trimmed to a common length of 373 nucleotides for amphistomes, and 399 and 481 nucleotides for Melanoides sp. and T. granifera, respectively. jModeltest (35) was used to select the best model test of nucleotide substitution suitable for neighbor-joining, maximum likelihood, and Bayesian inference analyses for the ITS-2, 16S, and ITS-1/5.8S/ITS-2 genes. The following models were selected under the Akaike information criterion: the HKY and HKY+G models (36) for ITS-2 and 16S genes, respectively, and the GTR model for ITS-1/5.8S/ITS-2. Maximum likelihood and neighbor-joining trees were generated using PAUP* 4.0 (37) trees, and the nodal support values were estimated using 1,000 bootstrap replicates. Bayesian inference trees were executed using MrBayes 3.1.2 (38), and the nodal support values were indicated as posterior probabilities.
Experimental Infection
A total of 90 F1 laboratory-bred Melanoides sp. (n = 50) and T. granifera (n = 40) snails, with an average shell height of 11–13 and 6–9 mm, respectively, were randomly selected and used in this study. The snails were divided into two treatment groups per species, 25 infected/25 control for Melanoides sp. and 20 infected/20 control for T. granifera. The selected snails were then allocated to 12 2-L containers, each holding seven to eight snails and were allowed 48 h to acclimatize to the environment.
Forty-five snails, 25 Melanoides sp. and 20 T. granifera, were then individually exposed to three C. microbothrium miracidia as described by Chingwena et al. (24). After exposure, snails were then returned into their respective 2-L containers and fed blanched dried lettuce supplemented with commercial tropical fish flakes three times a week ad libitum throughout the duration of the experiment and kept at the same temperature of 27°C.
Measurements of Parameters
Snail mortality was recorded daily for the experimental groups for 44 days postexposure (PE) to miracidia. Snails were deemed dead if they did not respond to mechanical stimuli, and those that showed movements were returned back into their respective experimental treatments (39). Since female Melanoides sp. and T. granifera are ovoviviparous, to evaluate reproduction rate, the number of offspring(s) produced were counted every 2 days, and the offspring were removed from the experimental containers. Numbers of offspring were combined to compose weekly collections, and this was done over the 5-week duration of the experiments.
At 3 weeks (21 days) PE, all infected snails were individually exposed to artificial light for 1 h to stimulate cercariae shedding. Shedding was stimulated weekly until the end of the experiments. On day 44 postinfection, shells of surviving snails in the infected group were cracked using a blunt forceps, and the soft tissue were squashed between two glass slides and observed under a stereo microscope (x40) to determine the presence or absence of larval stages of C. microbothrium. Snails were deemed infected with C. microbothrium if they contained actively crawling elongate rediae with a muscular pharynx or pigmented cercariae in their tissues as described by Chingwena et al. (24). Kolmogorov–Smirnov test on SPSS 24 was used to check if the data sets were normally distributed, while Mann–Whitney U-test was used to test if there was a difference in the mean number of offspring produced between the control and exposed groups of both Thiaridae snails. Bar graphs were plotted using mean and standard deviations to show the trend in the average weekly offspring production using Microsoft Excel 2016.
Results
Molecular Identification
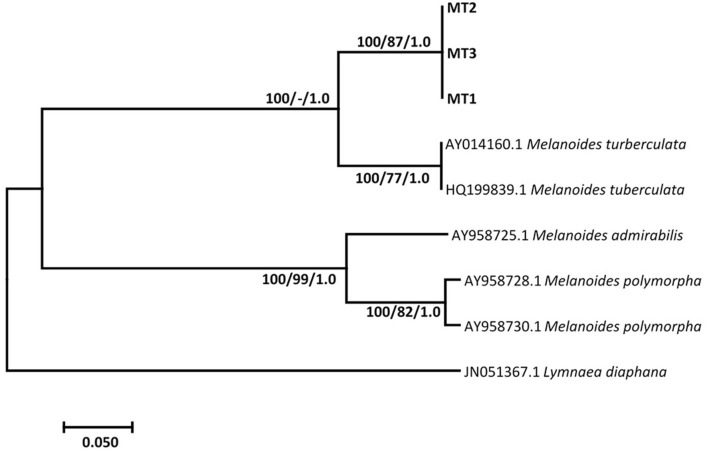
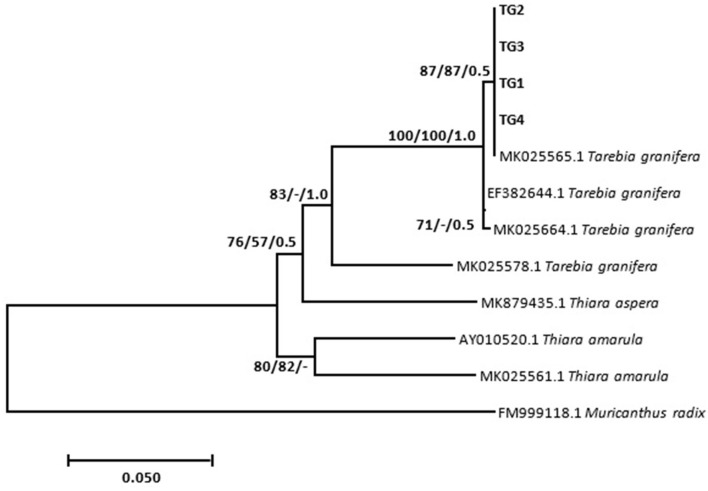
Results show that specimens presumed morphologically to be M. tuberculata had homogeneous sequences, which gave a homology of 83.45% with M. tuberculata isolate (HQ199839.1). These sequences were deposited in GenBank under the accession numbers MZ229766–MZ229768. Phylogenetic analysis showed that these isolates formed a strong-supported monophyletic sister clade with the clade consisting of GenBank M. tuberculata (AY014160.1 and HQ199839.1) (Figure 1). Results also showed that all selected Tarebia specimens were confirmed as T. granifera. BLAST analysis showed that the specimens gave a 100% homology with T. granifera isolate (MK025565.1), and further formed a moderately supported with this GenBank isolate (Figure 2). These specimen's sequences were deposited in GenBank under the accession numbers MZ312250–MZ312253.
Figure 1.
Neighbor-joining tree based on the 399-nucleotide sequence of the ITS-2, confirming the identity of Melanoides sp. (MT1-3) (in bold) collected from Mpumalanga province of South Africa and their relationship with other Melanoides species obtained from the NCBI GenBank database. The support value indicated on the nodes follows the order neighbor-joining bootstrap value/maximum likelihood bootstrap value/Bayesian inference posterior probability.
Figure 2.
Neighbor-joining tree based on the 481-nucleotide sequence of the 16S rDNA, confirming the identity of Tarebia granifera (TG1–4) (in bold) collected from Mpumalanga province of South Africa and their relationship with other Thiaridae species obtained from the NCBI GenBank database. The support value indicated on the nodes follows the order neighbor-joining bootstrap value/maximum likelihood bootstrap value/Bayesian inference posterior probability.
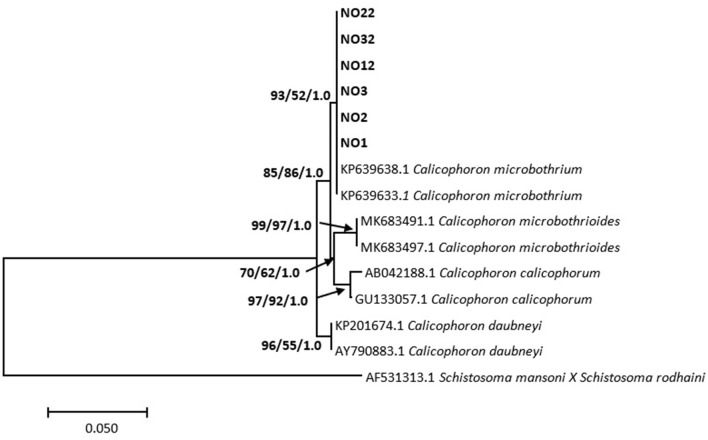
Phylogenetic analysis showed the relationship between the Calicophoron species as presented in Figure 3. Molecular analysis confirmed the identification of the six analyzed specimens (GenBank accession numbers MZ229627–MZ229632) as C. microbothrium, with a homology ranging from 99.25 to 99.50% (C. microbothrium, KP639638.1). These specimens formed a novel well-supported clade by neighbor joining with the GenBank C. microbothrium isolates (KP639633.1 and KP639638.1) (Figure 3).
Figure 3.
Neighbor-joining tree based on the 373-nucleotide sequence of the ITS-1/5.8S/ITS-2 marker, confirming the identity of amphistomes from Mpumalanga province (in bold) as Calicophoron microbothrium and their relationship with Calicophoron species obtained from the NCBI GenBank database. The support value indicated on the nodes follows the order neighbor-joining bootstrap value/maximum likelihood bootstrap value/Bayesian inference posterior probability.
Experimental Infectivity
Experimental infection results showed that C. microbothrium established in 4 of 25 (16%) exposed Melanoides sp. (Table 1). However, only three of these snails were shedding cercariae as from day 21 PE until day 44 (PE), and one snail was found with rediae when the experiment was terminated. None of the T. granifera snails were shedding cercariae during the experiment, and no larval stages were found in the tissues at day 44 PE. There was no mortality recorded from the experimental groups (Table 1).
Table 1.
Summary of infection and mortality rates of Melanoides sp. and Tarebia granifera exposed to Calicophoron microbothrium miracidia.
| Snail species | No. exposed | Miracidia per snail | NS (Day 44 PE) | Infectivity | ||||
|---|---|---|---|---|---|---|---|---|
| NSC (Day 21 PE) | NWL (44 days PE) | Total infected | % Infectivity | Mortality (%) | ||||
| Melanoides spp. —exposed group | 25 | 3–5 | 25 | 3 | 1 | 4 | 16 | 0 |
| Melanoides spp. —Control | 25 | N/A | 25 | N/A | N/A | N/A | N/A | 0 |
| T. granifera—exposed group | 20 | 3–5 | 20 | 0 | 0 | 0 | 0 | 0 |
| T. granifera-control | 20 | N/A | 20 | N/A | N/A | N/A | N/A | 0 |
NS, Number of snails surviving at day 44 post-exposure to miracidia; NSC, Number of snails shedding cercariae at day 21 post-exposure to miracidia; NWL, Number of snails with larvae at day 44 post-exposure to miracidia; N/A, Not applicable.
Table 2 shows the average number of offspring produced by both the exposed and non-exposed thiarid snails for a period of 44 days. The nonexposed groups (Melanoides sp. = 150, T. granifera = 30) generally produced more offspring for both thiarid species over the 44 days of the experiments as compared to the exposed groups (Melanoides sp. = 121, T. granifera = 8); however, the differences were only statistically significant (p < 0.05) at week 2 for the Melanoides sp. groups, and at weeks 4 and 5 for the T. granifera groups (p < 0.05) (Table 2).
Table 2.
Weekly average production of offspring by exposed to Calicophoron microbothrium miracidia and non-exposed groups of Melanoides sp. and Tarebia granifera.
| Melanoides spp. | T. granifera | |||||
|---|---|---|---|---|---|---|
| Weeks | Non-exposed (± SD) | Exposed (± SD) | p-value | Non-exposed (± SD) | Exposed (± SD) | P-value |
| 1 | 2.50 ± 2.11 | 1.33 ± 1.57 | 0.418 | 0 | 0 | – |
| 2 | 1.75 ± 0.87 | 1.58 ± 1.08 | 0.001* | 0.25 ± 0.70 | 0.13 ± 0.35 | 0.262 |
| 3 | 2.58 ± 1.17 | 2.33 ± 1.07 | 0.420 | 0.38 ± 0.74 | 0.13 ± 0.35 | 0.405 |
| 4 | 2.75 ± 1.06 | 2.41 ± 1.72 | 0.425 | 1.75 ± 1.28 | 0.38 ± 0.52 | 0.011* |
| 5 | 2.98 ± 1.24 | 2.41 ± 0.79 | 0.123 | 2.00 ± 1.72 | 1.18 ± 1.02 | 0.014* |
Significant difference at P < 0.05.
Discussion
Molecular and BLAST analyses based on the ITS-1/5.8S/ITS-2 characterized all six isolates of amphistomes collected from the abattoir for this study as C. microbothrium, with a percentage similarity of more than 99% with an isolate from Zimbabwe (KP639638.1). The isolates formed a strongly supported clade with the GenBank C. microbothrium sequences and formed a sister clade to the C. microbothrioides clade by a bootstrap value of 92% by neighbor joining. Furthermore, there was no sequence divergence and diversity found within the C. microbothrium populations from Mpumalanga (South Africa) with GenBank sequences from Zimbabwe (KP639638.1) and South Africa (KP639633.1). Based on the literature (4, 8), C. microbothrium is widely distributed in southern Africa, and this species has been implicated in several amphistomosis cases in young ruminants in eastern and southern Africa (4).
Comparing the sequences of Thiaridae species through BLAST, the sequences from this study confirmed the identification of our specimens as Melanoides sp. and T. granifera with percentage similarities of 83.45 % and 100 % to Melanoides tuberculata and T. granifera, respectively. The Melanoides isolates showed a mean genetic distance of 10.4% from isolates obtained from the GenBank database. This divergence was further illustrated on the phylogenetic tree, where three of the Melanoides sp. species formed a strong supported sister clade to the Iranian (HQ199839.1) and United Kingdom (AY014160.1) isolates. M. tuberculata and M. victoriae are the only two species from the genus that have been reported in South Africa. The absence of M. victoriae in the GenBank database meant that we could not have a complete comparison of the sequences we got from our isolates considering that two Melanoides species have been documented in South Africa. However, on comparison of shell characters of our experimental specimens with those documented for M. victoriae and M. tuberculata (14), our specimens were more compatible with M. tuberculata rather than M. victoriae. As a result, the species we used in our experiments were designated as Melanoides sp., and the genetic difference observed could be due existence of clones as some populations have been reported to be entirely female and propagate parthenogentically (14), with a possibility that it could be a mixture of two clones.
The T. granifera isolates from this study showed a genetic variation ranging from 0.0 to 0.08% from the reference sequences obtained from GenBank. However, no genetic variation was observed within sequences of our study population. These species formed a strongly supported clade by neighbor joining and Bayesian inference, supporting the identification of our isolates as T. granifera. Although M. tuberculata is regarded indigenous to South Africa (28, 40), both Thiaridae species are considered invasive in their allochthonous regions (15, 16, 18, 22, 23), and their role in the transmission of snail-borne parasites has not yet been intensively explored in South Africa. Experimental studies have shown that parasites tend to be more infective and increase in virulence and transmission against the native or local hosts (sympatric) as compared to the foreign host (allopatric) population (41). This was observed in this study, where results showed that of the two Thiaridae snail species studied, only Melanoides sp. snails were susceptible to C. microbothrium. This is not surprising as two Melanoides species have been recorded in South Africa to date (19), and of these two species, M. tuberculata is not only considered endemic and widespread in South Africa (15, 29) but also proven to be a compatible intermediate host of various snail-borne parasites (19, 27); this includes successful experimental infections of this amphistome species in Zimbabwe (24). The unsuccessful infection in T. granifera recorded in this study may possibly be due to failure of C. microbothrium to adapt to this invasive freshwater snail (42).
Chingwena et al. (24) reported an infection rate of 5.9% in M. tuberculata snails infected with C. microbothrium, with a mortality rate of 29.2%. This infection rate is lower than the observed 16% infection recorded in Melanoides sp. from this study, with no mortalities. Although the author further concluded that this snail species could potentially act as an intermediate host of C. microbothrium, since the infected snails did not shed the cercariae, infection was only confirmed through the observation of the rediae with cercariae upon dissection (24). However, in this study, 12% of the infected Melanoides sp. snails started shedding cercariae at day 21 PE until day 44 PE, with only one snail (4%) found with rediae and free cercariae on dissection. The findings from this study and that of Chingwena et al. (24) show that not only can Melanoides sp. sustain full development of C. microbothrium, but this also demonstrates that this snail species is able to shed cercariae and hence be able to transmit the parasite to the definitive host. However, the magnitude in which Melanoides sp. contributes to the transmission of C. microbothrium to ruminants under natural conditions still needs to be assessed (24).
According to Brown (14) and Veeravechsukij et al. (43), both T. granifera and M. tuberculata are ovoviviparous and reproduce parthenogenetically (14, 43), and this might be one of the key characteristic of T. granifera's success as an invader. Results from the present study showed that Melanoides sp. experimental groups started producing offspring from the first week of the experiments. However, the number of offspring produced by the control group (non-exposed) significantly declined at week 2 (p < 0.05), followed by a gradual increase from week 3 to termination, and this might be due to multiple biological factors. However, T. granifera snails only started on week 2 of the experiments, and the number of offspring produced gradually increased up to week 5. The number of offspring produced per week was significantly different (p < 0.05) on weeks 4 and 5 for both exposed and control groups. This delay in reproduction is surprising since previous field and experimental studies in Florida and Puerto Rico showed that an estimate sexual maturing of T. granifera species takes place when the snail shell height is 5.5–8.0 mm (44) and 6.0–7.0 mm (45). Furthermore, Appleton et al. (17) reported the presence of blasted stage embryos in the brood pouches of snails as small as 8.0 mm. Despite the delay in reproduction observed from this study, sexual maturity started when the shell heights were about 8 mm for the infected and 10 mm for the control, which is in line with other studies (17, 44, 46).
Conclusion
Results from this study have shown the ability of Melanoides sp. to sustain infection with C. microbothrium and to shed cercariae as early as day 21 PE. In this study, the snail can act as an invertebrate host C. microbothrium under laboratory conditions, but the extent to which it is contributing to natural transmission of this parasite in South Africa and whether the snail is susceptible to other amphistome species from the genus Calicophoron is unknown. Furthermore, this study has also confirmed that the strain of T. granifera used in this study is refractory to infection with C. microbothrium under laboratory conditions. Future research efforts should focus on collecting other invasive freshwater snail species and assessing trematode infections using combined techniques such as shedding of cercariae, detection of presence of larval stages in fresh snail tissues, and use of molecular techniques (PCR) to confirm infection as done in this study.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: GenBank under the accession numbers MZ229766–MZ229768.
Ethics Statement
The study was reviewed and approved by the Animal Research Ethics Committee (AREC/041/017M) of the University of KwaZulu-Natal, South Africa.
Author Contributions
SM and MM conceptualized the study and collected samples. MM and NM performed laboratory work and analyzed the results. MM wrote the first draft of the manuscript. All authors read, edited, and agreed on the final version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Louis La Grange for assisting with snail collection and amphistomes collection from an abattoir in Mpumalanga province.
Footnotes
Funding. Financial assistance was from SM's Research Productivity funds (UKZN) and the National Research Foundation of South Africa provided bursaries for MM and NM.
References
- 1.Gupta NK. Amphistomes, Systematics and Biology. New Dehli: Oxford; IBH Publishing Co. (1993). [Google Scholar]
- 2.Mavenyengwa M, Mukaratirwa S, Obwolo M, Monrad J. Observations on mass production of Calicophoron microbothrium metacercariae from experimentally and naturally infected Bulinus tropicus. Onderstepoort J Vet Res. (2006) 73:95–100. 10.4102/ojvr.v73i2.153 [DOI] [PubMed] [Google Scholar]
- 3.Howell A. Snail-borne diseases in bovids at high and low altitude in Eastern Uganda: integrated parasitological and malacological mapping (MSc thesis), Liverpool School of Tropical Medicine, Liverpool, United Kingdom: (2011). [Google Scholar]
- 4.Pfukenyi DM, Mukaratirwa S. Amphistome infections in domestic and wild ruminants in East and Southern Africa: a review. Onderstepoort J Vet Res. (2018) 85:1–13. 10.4102/ojvr.v85i1.1584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sey O. CRC Handbook of the Zoology of Amphistomes. Boca Raton, FL: Taylor and Francis Group; (2019). [Google Scholar]
- 6.Shylla JA, Ghatani S, Chatterjee A, Tandon V. Secondary structure analysis of ITS2 in the rDNA of three Indian paramphistomid species found in local livestock. Parasitol Res. (2011) 108:1027–32. 10.1007/s00436-010-2148-8 [DOI] [PubMed] [Google Scholar]
- 7.Pfukenyi DM, Monrad J, Mukaratirwa S. Epidemiology and control of trematode infections in cattle in Zimbabwe: a review. J S Afr Vet Assoc. (2005) 76:9–17. 10.4102/jsava.v76i1.387 [DOI] [PubMed] [Google Scholar]
- 8.Dinnik JA. Paramphistomum sukumum sp. nov. and other stomach-flukes from cattle in the Sukumaland area of the Lake Region, Tanganyika. Parasitology. (1964) 54:201–9. 10.1017/S0031182000067858 [DOI] [PubMed] [Google Scholar]
- 9.Dinnik JA, Dinnik NN. The life cycle of Paramphistomum microbothrium Fischoeder, 1901 (Trematoda: Paramphistomidae). Parasitology. (1954) 44:285–99. 10.1017/S0031182000018916 [DOI] [PubMed] [Google Scholar]
- 10.Dinnik JA. Paramphistomum phillerouxi sp. nov. (Trematoda: Paramphistomidae) and its development in Bulinus forskalii. J Helminthol. (1961) 35:69–90. 10.1017/S0022149X00024792 [DOI] [PubMed] [Google Scholar]
- 11.Dinnik JA. The snail hosts of certain Paramphistomidae and Gastrothylacidae (Trematode) discovered by the late Dr. P. L. LeRoux in Africa. J Helminthol. (1965) 39:141–50. 10.1017/S0022149X00020551 [DOI] [PubMed] [Google Scholar]
- 12.Wright CA, Southgate VR, Howard GW. A note on the life cycles of some amphistomes in Zambia. J Helminthol. (1979) 53:251–2. 10.1017/S0022149X00006039 [DOI] [Google Scholar]
- 13.Southgate VR, Brown DS, Warlow A, Knowles RJ, Jones A. The influence of Calicophoron microbothrium on the susceptibility of Bulinus tropicus to Schistosoma bovis. Parasitol Res. (1989) 75:381–91. 10.1007/BF00931134 [DOI] [PubMed] [Google Scholar]
- 14.Brown DS. Freshwater Snails of Africa and Their Medical Importance. 2nd ed. London: Taylor & Francis Ltd. (1994). [Google Scholar]
- 15.Facon B, Pointier JP, Glaubrecht M, Poux C, Jarne P, David P. A molecular phylogeography approach to biological invasions of the New World by parthenogenetic thiarid snails. Mol Ecol. (2003) 12:3027–39. 10.1046/j.1365-294X.2003.01972.x [DOI] [PubMed] [Google Scholar]
- 16.Raw JL, Perissinotto R, Miranda NAF, Peer N. Feeding dynamics of Melanoides tuberculata (Müller, 1774). J Molluscan Stud. (2016) 82:328–35. 10.1093/mollus/eyv070 [DOI] [Google Scholar]
- 17.Appleton CC, Forbes AT, Demetriades NT. The occurrence, bionomics and potential impacts of the invasive freshwater snail Tarebia granifera (Lamarck, 1822) (Gastropoda: Thiaridae) in South Africa. Zool Meded. (2009) 83:525–36. [Google Scholar]
- 18.Miranda NAF, Measey GJ, Peer N, Raw JL, Perissinotto R, Appleton CC. Shell crushing resistance of alien and native thiarid gastropods to prey crabs in South Africa. Aquat Invasions. (2016) 3:303–11. 10.3391/ai.2016.11.3.08 [DOI] [Google Scholar]
- 19.De Kock KN, Wolmarans CT. Distribution and habitats of Melanoides tuberculata (Müller, 1774) and M. victoriae (Dohrn, 1865) (Mollusca: Prosobranchia: Thiaridae) in South Africa. Water SA. (2009) 35:713. 10.4314/wsa.v35i5.49197 [DOI] [Google Scholar]
- 20.Van Damme D. The freshwater Mollusca of northern Africa: distribution, biogeography and palaeoecology. Dev Hydrol. (1984) 25. [Google Scholar]
- 21.Appleton CC. Mollusca. In: De Moor IJ, Day JA. editors. Guides to the Freshwater Invertebrates of Southern Africa (Volume 6) Chapter 3: Arachnida & Mollusca, Araneae, Water Mites & Mollusca. WRC Report No. 182/02. Pretoria: Water SA; (2002). p. 42–125. [Google Scholar]
- 22.Derraik JGB. The potential significance to human health associated with the establishment of the snail Melanoides tuberculata in New Zealand. N Z Med J. (2008) 121:25–32. [PubMed] [Google Scholar]
- 23.Work K, Mills C. Rapid population growth countered high mortality in a demographic study of the invasive snail, Melanoides tuberculata (Müller, 1774), in Florida. Aquat Invasions. (2013) 8:417–25. 10.3391/ai.2013.8.4.05 [DOI] [Google Scholar]
- 24.Chingwena G, Mukaratirwa S, Kristensen TK, Chimbari M. Susceptibility of freshwater snails to the amphistome Calicophoron microbothrium and the influence of the species on susceptibility of Bulinus tropicus to Schistosoma haematobium and Schistosoma mattheei infections. J Parasitol. (2002) 88:880–3. 10.1645/0022-3395(2002)088[0880:SOFSTT]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 25.Coulibaly G, Madsen H. Seasonal density fluctuations of intermediate hosts of schistosomes in two streams in Bamako, Mali. J Afr Zool. (1990) 104:201–12. [Google Scholar]
- 26.Sharif M, Daryani A, Karimi SA. A faunistic survey of cercariae isolated from lymnaeid snails in central areas of Mazandaran, Iran. Pak J Biol Sci. (2010) 1:158–63. 10.3923/pjbs.2010.158.163 [DOI] [PubMed] [Google Scholar]
- 27.Mukaratirwa S, Munjere IF, Takawira M, Chingwena G. Susceptibility of 7 freshwater gastropod species in Zimbabwe to infection with Gastrodiscus aegyptiacus (Cobbold, 1876) Looss, 1896. J S Afr Vet Assoc. (2004) 75:186–8. 10.4102/jsava.v75i4.481 [DOI] [PubMed] [Google Scholar]
- 28.Eduardo SL. The taxonomy of the family Paramphistomidae Fischoeder, 1901 with special reference to the morphology of species occurring in ruminants. III. Revision of the genus Calicophoron Näsmark, 1937. Syst Parasitol. (1983) 5:25–79. 10.1007/BF00010983 [DOI] [PubMed] [Google Scholar]
- 29.Appleton CC. Alien and invasive fresh water Gastropoda in South Africa. Afr J Aquat Sci. (2003) 28:69–81. 10.2989/16085914.2003.9626602 [DOI] [Google Scholar]
- 30.Palumbi S, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. The Simple Fool's Guide to PCR, Version 2.0. Honolulu, HL: Department of Zoology and Kewalo Marine Laboratory, University of Hawaii; (1991). [Google Scholar]
- 31.Malatji MP, Lamb J, Mukaratirwa S. Molecular characterization of liver fluke intermediate host lymnaeids (Gastropoda: Pulmonata) snails from selected regions of Okavango Delta of Botswana, KwaZulu-Natal and Mpumalanga provinces of South Africa. Vet Parasitol Regl Stud Rep. (2019) 17:100318. 10.1016/j.vprsr.2019.100318 [DOI] [PubMed] [Google Scholar]
- 32.Mucheka VT, Lamb JM, Pfukenyi DM, Mukaratirwa S. DNA sequence analyses reveal co-occurrence of novel haplotypes of Fasciola gigantica with F. hepatica in South Africa and Zimbabwe. Vet Parasitol. (2015) 214:144–51. 10.1016/j.vetpar.2015.09.024 [DOI] [PubMed] [Google Scholar]
- 33.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. (1999) 41:95–8. [Google Scholar]
- 34.Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. (2016) 33:1870–4. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. (2008) 25:1253–6. 10.1093/molbev/msn083 [DOI] [PubMed] [Google Scholar]
- 36.Hasegawa M, Kishino H, Yano T. Dating the human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. (1985) 22:160–74. 10.1007/BF02101694 [DOI] [PubMed] [Google Scholar]
- 37.Swofford D. Phylogenetic Analysis Using Parsimony (paup), Version 4. Sunderland, MA: Sinauer Associates; (2002). [Google Scholar]
- 38.Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. (2001) 17:754–5. 10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- 39.Kalinda C, Chimbari MJ, Malatji MP, Mukaratirwa S. Influence of desiccation on the survival of Bulinus globosus under laboratory conditions. J Freshw Ecol. (2018) 33:461–73. 10.1080/02705060.2018.1520157 [DOI] [Google Scholar]
- 40.Appleton CC, Miranda NAF. Two Asian freshwater snails newly introduced into South Africa and an analysis of alien species reported to date. Afr Invertebr. (2015) 56:1–17. 10.5733/afin.056.0102 [DOI] [Google Scholar]
- 41.Hurtrez-Boussès S, Meunier C, Durand P, Renaud F. Dynamics of host–parasite interactions: the example of population biology of the liver fluke (Fasciola hepatica). Microb Infect. (2001) 3:841–9. 10.1016/S1286-4579(01)01442-3 [DOI] [PubMed] [Google Scholar]
- 42.Blackburn TM, Pyšek P, Bacher S, Carlton JT, Duncan RP, Jarošik V, et al. A proposed unified framework for biological invasions. Trends Ecol Evol. (2011) 26:333–9. 10.1016/j.tree.2011.03.023 [DOI] [PubMed] [Google Scholar]
- 43.Veeravechsukij N, Krailas D, Namchote S, Wiggering B, Neiber MT, Glaubrecht M. Molecular phylogeography and reproductive biology of the freshwater snail Tarebia granifera in Thailand and Timor (Cerithioidea, Thiaridae): morphological disparity versus genetic diversity. Zoosyst Evol. (2018) 94:461. 10.3897/zse.94.28981 [DOI] [Google Scholar]
- 44.Tucker Abbott R. A study of an intermediate snail host (Thiara granifera) of the oriental lung fluke (Paragonimus). Proc US Natl Mus. (1952) 102:71–116. 10.5479/si.00963801.102-3292.71 [DOI] [Google Scholar]
- 45.Chaniotis BN, Butler JM, Ferguson FF, Jobin WR. Bionomics of Tarebia granifera (Gastropoda: Thiaridae) in Puerto Rico, an Asian vector of paragonimiasis westermani. Caribb J Sci. (1980) 16:81–9. [Google Scholar]
- 46.Appleton CC, Nadasan DS. First report of Tarebia granifera (Lamarck, 1816) (Gastropoda: Thiaridae) from Africa. J Molluscan Stud. (2002) 68:399–402. 10.1093/mollus/68.4.399 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found at: GenBank under the accession numbers MZ229766–MZ229768.