Abstract
Motivation
Ribosome profiling (Ribo-seq) is a powerful approach based on deep sequencing of cDNA libraries generated from ribosome-protected RNA fragments to explore the translatome of a cell, and is especially useful for the detection of small proteins (50–100 amino acids) that are recalcitrant to many standard biochemical and in silico approaches. While pipelines are available to analyze Ribo-seq data, none are designed explicitly for the automatic processing and analysis of data from bacteria, nor are they focused on the discovery of unannotated open reading frames (ORFs).
Results
We present HRIBO (High-throughput annotation by Ribo-seq), a workflow to enable reproducible and high-throughput analysis of bacterial Ribo-seq data. The workflow performs all required pre-processing and quality control steps. Importantly, HRIBO outputs annotation-independent ORF predictions based on two complementary bacteria-focused tools, and integrates them with additional feature information and expression values. This facilitates the rapid and high-confidence discovery of novel ORFs and their prioritization for functional characterization.
Availability and implementation
HRIBO is a free and open source project available under the GPL-3 license at: https://github.com/RickGelhausen/HRIBO.
1 Introduction
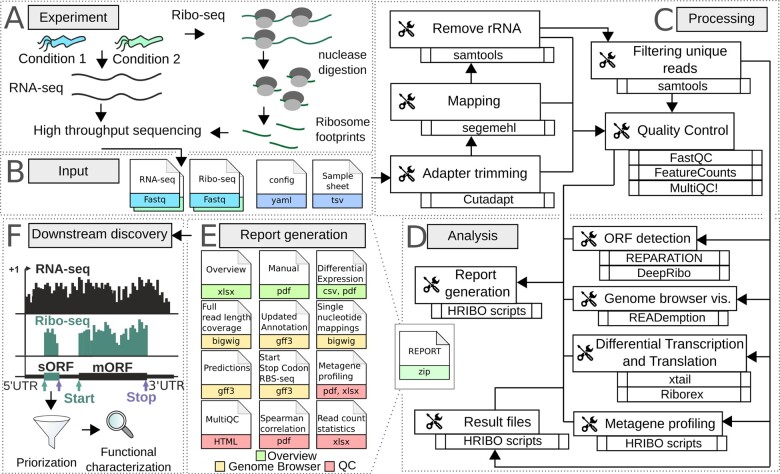
Ribosome profiling (Ribo-seq) (Ingolia et al., 2009) is an RNA-seq based approach that identifies the ribosome-bound fraction of the transcriptome as a proxy for protein expression (Fig. 1A). Because RNase digestion of mRNA regions not protected by translating ribosomes creates so-called ribosome footprints, it also allows definition of open-reading frame (ORF) boundaries. This makes it remarkably suited to detect small proteins, which are currently underrepresented in genome annotations due to length cutoffs imposed during annotation, as well as unique features that preclude their detection by conventional experimental or in silico approaches (Storz et al., 2014). In addition, a parallel whole-transcriptome library allows calculation of translation efficiency (TE, the ratio of footprint library coverage to transcriptome coverage) and identification of ORFs that might be differentially expressed under conditions relevant to the organism studied, such as those encountered during infection. There are existing workflows for the analysis of eukaryotic Ribo-seq data (Chung et al., 2015; Wang et al., 2019), but a dedicated, automatic solution for bacteria is still missing. Here, we present HRIBO (High-throughput annotation by Ribo-seq), a computational pipeline that processes and analyses data from any bacterial Ribo-seq experiment, but also detects translated novel ORFs. The tool is compatible with bacterial annotations and circular chromosomes, uses an optimized mapping approach suitable for small bacterial genomes, integrates machine learning-based ORF prediction tools designed for/trained on bacterial ORF features, and also includes two differential expression tools designed for Ribo-seq data. We implemented HRIBO based on snakemake (Köster et al., 2012), which allows highly reproducible and fully automatic data analysis.
Fig. 1.
Bacterial Ribo-seq data analysis by the HRIBO pipeline. (A) In Ribo-seq, parallel RNA-seq and ribosome footprint libraries are prepared from bacterial cultures, optionally from multiple experimental conditions and replicates, and deep sequenced. (B) The fastq files from each library, config file and a sample sheet defining the experimental setup serve as input for HRIBO, which automatically downloads software dependencies and executes commands for processing (C), which ensures reproducibility. For all the processing steps quality control measures are computed and summarized by MultiQC. (D) First, coverage files for Genome browser visualization are generated, including three variants of single nucleotide mapping, and global metagene profiling of ribosome occupancy near start codons is calculated. Next, two ORF prediction tools (REPARATION and DeepRibo) detect translated annotated and novel ORFs from the ribosome footprint libraries. The detected ORFs are then screened together for differential transcription and translation. Finally, the expression information for each ORF is then aggregated together with additional features (nucleotide sequence, length, etc.) in an overview results table. (E) Files from all analysis steps are bundled together, along with a manual, into a report archive. (F) Overall, HRIBO streamlines the selection of high-confidence, functionally important, translated novel ORFs for further experimental investigation. For more details please refer to the documentation: https://hribo.readthedocs.io/en/latest
2 Approach
HRIBO automatically retrieves tools from bioconda (Grüning et al., 2018) and performs all necessary steps of processing, with pinned and tested tool versions. Detailed documentation, with examples, is available at: https://hribo.readthedocs.io/en/latest/.
HRIBO (Fig. 1B) requires the sequencing data files (paired RNA-seq and Ribo-seq libraries for each sample), the genomic sequence/annotation of the organism and a sample sheet that captures the experimental setup by associating samples/replicates with their experimental condition. As a first step (Fig. 1C), HRIBO performs adapter trimming with Cutadapt (Martin, 2011) and then maps the reads to the genome with segemehl, which has higher sensitivity than other mappers (Otto et al., 2014), but its high computational costs are still acceptable for small genomes. Multimapping/rRNA reads are then removed with samtools (Li et al., 2009) before further processing. FastQC (Andrews et al., 2010) and featurecount (Liao et al., 2014) reports are created after each processing step and aggregated with MultiQC (Ewels et al., 2016), enabling the investigator to identify problems with either the experimental setup (e.g. insufficient rRNA depletion), or the pre-processing (e.g. untrimmed adapters). The resulting mapped reads are then used in subsequent steps (Fig. 1D) to calculate read statistics, detect expressed ORFs (annotated or novel), compute differential expression, generate coverage tracks and perform metagene profiling of global ribosome density near start codons. Notably, we have included two complementary ORF detection tools, both specifically developed for bacterial organisms. REPARATION (Ndah et al., 2017) uses a random forest-based machine-learning approach. DeepRibo (Clauwaert et al., 2019) is based on convolutional and recurrent neural network approaches. The results of the two prediction tools are aggregated by newly developed scripts and compiled into a list of all detected ORFs (both annotated and novel). ORFs that are differentially expressed (both transcription and translation) between samples/conditions are detected using the complementary Ribo-seq-specific tools xtail (Zhang et al., 2017) and Riborex (Li et al., 2017). The list is then enriched with additional computed context and expression information for each ORF, such as length, sequence, normalized read counts, TE, available annotation/novelty, differential expression, and which conditions/replicates the ORF was detected in. Interesting candidates can then be inspected by either full read coverage (Förstner et al., 2014) or single nucleotide mapping (5′/3′ end or center of read) genome browser tracks. Moreover, we developed metagene profiling scripts that globally analyze read density around start codons, including for different read lengths. This allows the inspection of data quality, examination for organism-specific ORF/translatome signatures and potentially the identification of optimal read lengths that could be used to detect three-nucleotide periodicity in ORFs. Finally, the results are collected into a report (Fig. 1E) that can be easily distributed and contains a detailed manual. HRIBO allows priorization and identification of novel ORFs (see Fig. 1F), e.g. sORFs in Salmonella Typhimurium related to virulence (Venturini et al., 2020). An example HRIBO report for a published dataset (Potts et al., 2019), which required 4 h 4 min on 12 cores of an AMD EPYC CPU (@ 1996 MHz) to compute, can be found here: ftp://biftp.informatik.uni-freiburg.de/pub/HRIBO/HRIBO1.4.4_18-09-20.zip.
3 Conclusion
HRIBO is a reproducible and standardized pipeline that includes all tools required to process Ribo-seq datasets from bacterial organisms, from pre-processing and quality control to ORF prediction and differential expression analysis. Read information, differential transcription/translation and additional computed features for both annotated and predicted ORFs are summarized in a single table that can be easily inspected together with the generated genome browser tracks. This streamlines the selection of high confidence, functionally important, translated novel ORFs for further experimental investigation. Existing pipelines are, other than the exceptions discussed below (Choe et al., 2020; Fremin et al., 2020; Weaver et al., 2019), not specifically developed for bacterial organisms. Most existing pipelines do not cover the complete analysis workflow and lack the initial processing steps, including mapping (see Fig. 1 of Ribominer) (Li et al., 2020). Moreover, the complete workflows frequently use mapping tools with a lower runtime, at the cost of lower sensitivity, to allow processing of large eukaryotic genomes in an acceptable time-scale (Michel et al., 2016), or they support only a small set of bacterial organisms (Verbruggen et al., 2019). Furthermore, none of the pipelines feature ORF discovery tools specialized for bacteria. This is not only of importance due to decreased sensitivity for bacterial translation signatures, but also because some tools are not compatible with circular bacterial genomes, meaning that ORFs that span the origin might have negative coordinates. In addition, the different architecture of bacterial annotations, which include operons instead of introns and exons, also preclude their use with some eukaryotic tools (Calviello et al., 2016). The three pipelines and protocols also developed for bacterial organisms are either built for different objectives, or they offer other functionality than HRIBO. The pipeline used to process Ribo-Seq and translation initiation site profiling data in bacteria (Weaver et al., 2019), as well as in archaea (Gelsinger et al., 2020) consists of a collection of semi-automatic ipython notebooks which perform the pre-processing of the data. In addition, while the pipeline performs some post-processing calculations (such as pause score and gene density analysis), it does not include bacterial ORF prediction tools that work solely on Ribo-seq (rather than initiation site profiling) coverage files. Moreover, it does not perform the additional analysis steps on predicted ORFs performed by HRIBO such as calculation of translation efficiency, differential expression analysis and computation of additional ORF features. MetaRiboSeq (Fremin et al., 2020) is an experimental and computational protocol to investigate the transitional landscape of a metagenomic sample. However, it considers only proteins that satisfy a specific RPKM threshold and are also considered homologous to proteins in a database as translated, while HRIBO, which has been designed for use with a single organism, allows the use of more sophisticated and computationally more costly tools. Furthermore, the MetaRiboSeq computational approach is only described in the manuscript and not implemented as a script or tool, therefore it does not offer any automation. STATR (Choe et al., 2020), in contrast to HRIBO, is a semi-automatic analysis protocol, requiring the manual installation of all software dependencies and execution of tool commands and does not, unlike HRIBO, offer quantification and removal of rRNA and multi-mapping reads, sequencing quality control reports, ORF detection/prediction, differential transcription and translation output and customized genome browser tracks. Recently, translation initiation site profiling, where ribosomes are enriched at start codons by treatment with antibiotics that specifically target initiating ribosomes (Gelsinger et al., 2020; Meydan et al., 2019; Weaver et al., 2019), has now been adapted for bacteria and archaea. Therefore, as an update to HRIBO, we plan to add a module allowing the reproducible analysis of translation initiation site profiling data.
Acknowledgements
The authors thank the anonymous referees for their helpful comments.
Funding
This work was supported by the DFG SCHM 2663/3; High Performance and Cloud Computing Group, University Tübingen via bwHPC; DFG INST 37/935-1 FUGG. R.G.; DFG grant BA 2168/21-1 SPP 2002; DFG grant 322977937/GRK2344 2017; BMBF ‘RNAProNet—031L0164B’, DFG grants SH580/7-1: Sh580/8-1 within the DFG SPP2002 to C.M.S and DFG, Germany's Excellence Strategy—EXC-2189—Project ID: 390939984.
Conflict of Interest: none declared.
Data availability
The publically available third-party data used in our ReadTheDocs Tutorial can be accessed via the GEO accession number GSE107834 [https://doi.org/10.1371/journal.pone.0211430]. All source code created for this publication is available on our GitHub page: https://github.com/RickGelhausen/HRIBO.
Contributor Information
Rick Gelhausen, Bioinformatics Group, Department of Computer Science, University of Freiburg, 79110 Freiburg, Germany.
Sarah L Svensson, Chair of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, 97080 Würzburg, Germany.
Kathrin Froschauer, Chair of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, 97080 Würzburg, Germany.
Florian Heyl, Bioinformatics Group, Department of Computer Science, University of Freiburg, 79110 Freiburg, Germany.
Lydia Hadjeras, Chair of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, 97080 Würzburg, Germany.
Cynthia M Sharma, Chair of Molecular Infection Biology II, Institute of Molecular Infection Biology (IMIB), University of Würzburg, 97080 Würzburg, Germany.
Florian Eggenhofer, Bioinformatics Group, Department of Computer Science, University of Freiburg, 79110 Freiburg, Germany.
Rolf Backofen, Bioinformatics Group, Department of Computer Science, University of Freiburg, 79110 Freiburg, Germany; Signalling Research Centres BIOSS and CIBSS, University of Freiburg, 79104 Freiburg, Germany.
References
- Andrews S. et al. (2010) Fastqc: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Calviello L. et al. (2016) Detecting actively translated open reading frames in ribosome profiling data. Nat. Methods, 13, 165–170. [DOI] [PubMed] [Google Scholar]
- Choe D. et al. (2020) STATR: a simple analysis pipeline of Ribo-Seq in bacteria. J. Microbiol., 58, 217–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.Y. et al. (2015) The use of duplex-specific nuclease in ribosome profiling and a user-friendly software package for Ribo-Seq data analysis. RNA, 21, 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clauwaert J. et al. (2019) DeepRibo: a neural network for precise gene annotation of prokaryotes by combining ribosome profiling signal and binding site patterns. Nucleic Acids Res., 47, e36–e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewels P. et al. (2016) MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics, 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förstner K.U. et al. (2014) READemptiona tool for the computational analysis of deep-sequencing–based transcriptome data. Bioinformatics, 30, 3421–3423. [DOI] [PubMed] [Google Scholar]
- Fremin B.J. et al. (2020) MetaRibo-Seq measures translation in microbiomes. Nat. Commun., 11, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelsinger D.R. et al. (2020) Ribosome profiling in archaea reveals leaderless translation, novel translational initiation sites, and ribosome pausing at single codon resolution. Nucleic Acids Res., 48, 5201–5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüning B. et al. ; The Bioconda Team. (2018) Bioconda: sustainable and comprehensive software distribution for the life sciences. Nat. Methods, 15, 475–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia N.T. et al. (2009) Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science, 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster J. et al. (2012) Snakemake—a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Li F. et al. (2020) RiboMiner: a toolset for mining multi-dimensional features of the translatome with ribosome profiling data. BMC Bioinformatics, 21, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. et al. 1000 Genome Project Data Processing Subgroup. (2009) The sequence alignment/map format and SAMtools. Bioinformatics, 25, 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W. et al. (2017) Riborex: fast and flexible identification of differential translation from Ribo-seq data. Bioinformatics, 33, 1735–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y. et al. (2014) featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Martin M. (2011) CUTADAPT removes adapter sequences from high-throughput sequencing reads. EMBnet. J., 17, 10–12. [Google Scholar]
- Meydan S. et al. (2019) Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Mol. Cell, 74, 481–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel A.M. et al. (2016) RiboGalaxy: a browser based platform for the alignment, analysis and visualization of ribosome profiling data. RNA Biol., 13, 316–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndah E. et al. (2017) Reparation: ribosome profiling assisted (re-)annotation of bacterial genomes. Nucleic Acids Res., 45, e168–e168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto C. et al. (2014) Lacking alignments? The next-generation sequencing mapper segemehl revisited. Bioinformatics, 30, 1837–1843. [DOI] [PubMed] [Google Scholar]
- Potts A.H. et al. (2019) Role of CsrA in stress responses and metabolism important for salmonella virulence revealed by integrated transcriptomics. PLoS One, 14, e0211430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G. et al. (2014) Small proteins can no longer be ignored. AnnuRevBiochem, 83, 753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venturini E. et al. (2020) A global data-driven census of salmonella small proteins and their potential functions in bacterial virulence. microLife, uqaa002, doi: 10.1093/femsml/uqaa002. [DOI] [PMC free article] [PubMed]
- Verbruggen S. et al. (2019) PROTEOFORMER 2.0: further developments in the ribosome profiling-assisted proteogenomic hunt for new proteoforms. Mol. Cell. Proteomics, 18, S126–S140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H. et al. (2019) Computational resources for ribosome profiling: from database to web server and software. Brief. Bioinf., 20, 144–155. [DOI] [PubMed] [Google Scholar]
- Weaver J. et al. (2019) Identifying small proteins by ribosome profiling with stalled initiation complexes. mBio, 10, e02819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P. et al. (2017) Genome-wide identification and differential analysis of translational initiation. Nat. Commun., 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The publically available third-party data used in our ReadTheDocs Tutorial can be accessed via the GEO accession number GSE107834 [https://doi.org/10.1371/journal.pone.0211430]. All source code created for this publication is available on our GitHub page: https://github.com/RickGelhausen/HRIBO.