Abstract
Targeted, catalytic degradation of oncoproteins using heterobifunctional small molecules is an attractive modality, particularly for hematologic malignancies, which are often initiated by aberrant transcription factors and are challenging to drug with inhibitors. BRD4, a member of the bromodomain and extraterminal family, is a core transcriptional and epigenetic regulator that recruits the P-TEFb complex, which includes Cdk9 and cyclin T, to RNA polymerase II (pol II). Together, BRD4 and CDK9 phosphorylate serine 2 (pSer2) of heptad repeats in the C-terminal domain of RPB1, the large subunit of pol II, promote transcriptional elongation. Small-molecule degraders of BRD4 have shown encouraging efficacy in preclinical models for several tumor types but less efficacy in other cancers including small-cell lung cancer (SCLC) and pancreatic cancer. Here, we evaluated CFT-2718, a new BRD4-targeting degrader with enhanced catalytic activity and in vivo properties. In vivo, CFT-2718 has significantly greater efficacy than the CDK9 inhibitor dinaciclib in reducing growth of the LX-36 SCLC patient-derived xenograft (PDX) model and performed comparably to dinaciclib in limiting growth of the PNX-001 pancreatic PDX model. In vitro, CFT-2718 reduced cell viability in four SCLC and two pancreatic cancer models. In SCLC models, this activity significantly exceeded that of dinaciclib; furthermore, CFT-2718 selectively increased the expression of cleaved PARP, an indicator of apoptosis. CFT-2718 caused rapid BRD4 degradation and reduced levels of total and pSer2 RPB1 protein. These and other findings suggest that BRD-mediated transcriptional suppression merits further exploration in the setting of SCLC.
Introduction
Epigenetic dysregulation plays a major role during tumorigenesis. The transcriptional and epigenetic regulator BRD4, a member of the bromodomain and extraterminal (BET) family, has emerged as promising therapeutic target (1, 2). BRD4 contains two tandem bromodomains, BD1 and BD2 (3), that allow BRD4 binding to acetylated lysine residues on chromatin-associated proteins, notably histones, to promote transcription (4, 5). During transcriptional initiation and elongation, RPB1, the large subunit of RNA polymerase II (pol II) undergoes multiple posttranslational modifications on a unique regulatory heptad sequence (Tyr1, Ser2, Pro3, Thr4, Ser5, Pro6, Ser7) that is repeated 52 times in the RPB1 carboxy terminal domain (CTD; ref. 6). Initiation requires activity of the cyclin-dependent kinase CDK7, a component of the TFIIH complex, to phosphorylate RPB1 on Ser5 (7). Subsequently, histone-bound BRD4 uses a P-TEFb interaction domain (PID) to recruit P-TEFb, a complex including cyclin T, cyclin K, and CDK9 (8). This interaction both stimulates the P-TEFb kinase activity and targets BRD4- and CDK9-dependent phosphorylation to Ser2 in the RPB1 CTD, promoting transcriptional elongation (8).
Components of the core transcription complexes have attracted considerable interest as drug targets, particularly in cancers driven by oncogenes such as MYC and MYB, which include many fast-growing leukemias. For this reason, drugs have been developed to target CDK7, CDK9, BRD4, and other components of the core transcriptional machinery (9–12). Some of these drugs, such as the CDK9-targeting agent dinaciclib, are classic ATP-competitive kinase inhibitors (13). Others, such as the BRD4-targeting agent JQ1, act by blocking the ability of the bromodomains to bind histones through competing for interaction with acetyl-lysine motifs on histones (14). A particularly promising recent approach has been to use E3 ligase recruiting bifunctional small molecules [also known as PROTACs (proteolysis targeting chimera); ref. 15] to induce degradation of critical transcription factors. In an early application of this approach to BRD4, a BET inhibitor was linked to another small-molecule ligand which binds the cereblon (CRBN) E3 ubiquitin ligase complex, causing significant loss of BRD4 (16). Notably, subsequent comparison of various BRD4-targeted degraders with other classes of BRD4 inhibitors revealed a more profound effect on transcription and cell growth with BRD4 degradation than BRD4 inhibition, and also showed nonequivalent activity of distinct BRD4-targeted degraders (17–20). These results have motivated the development of panels of degraders around targets of interest, to maximize therapeutic flexibility.
Acute loss of BRD4 disproportionally affects genes involved in the core regulatory circuitry resulting in rapid induction of apoptosis and death of transcriptionally addicted cancers such as T-ALL (21). However, BRD4 is critical for global elongation, raising the question as to whether prolonged BRD4 degradation would be tolerated by normal tissues. Data from mathematical modeling to establish the optimal timepoint for drug inhibition of transcription suggests a brief exposure at maximum serum concentration (Cmax) to kill proliferating cells while allowing a recovery time to resting cells (22). Thus, in screening for novel BRD4 degraders, we focused on compounds that were highly potent and displayed pharmacokinetic properties consistent with high systemic exposure, the ability to distribute to peripheral tissues, fast elimination, and a wide therapeutic index. CFT-2718, first described here, is a benzotriazoloazepine-based BRD4 degrader that met all these requirements and enabled us to assess the therapeutic potential for BRD4 degradation in vivo.
Here, we have assessed the activity of CFT-2718 in two models of solid tumors, which are typically more challenging targets than the leukemias and lymphomas in which BRD4-targeting agents have displayed striking activity (20). For this work, we have used models for SCLC, which affects more than 30,000 patients a year and represents approximately 12% to 14% of the total lung cancer population. SCLC has a poor prognosis, with a postdiagnosis median survival of 15 to 20 months if patients are diagnosed with limited-stage (LS) disease and 9 to 12 months for patients with extensive-stage (ES) disease (23). In contrast to other types of lung cancer (24), SCLCs typically do not have kinase-activating mutations and other changes in targetable signaling pathways (25, 26) and are resistant to many therapies targeting signaling proteins.
Similarly, although pancreatic cancer is the twelfth most common cause of cancer, it is the fourth most common cause of cancer-related death. Pancreatic cancer is typically diagnosed at an advanced stage, with 5-year survival rates under 10%; these tumors are also refractory to many treatments targeting signaling proteins. Hence, for both these cancer types, there is considerable interest in evaluating inhibitors with alternative means of action. For SCLC in particular, several preclinical studies have suggested that inhibitors of components of the transcriptional machinery may have value (27–29), motivating us to evaluate CFT-2718 in this tumor type. Overall, the results of this study demonstrate signficant activity of CFT-2718 in causing rapid degradation of BRD4 and MYC, reducing polymerase activity, and in reducing growth of SCLC, both in vitro and in vivo, suggesting the potential benefit of this class of tumor-targeting agent.
Materials and Methods
Cell lines, patient-derived xenograft models, and compounds
MOLT4 CRBN+/+ and CRBN−/− lines were previously described (20). 293T cell lines in which endogenously expressed BRD4 is tagged with a HiBiT Tag (30) were generated using CRISPR-Cas9 System (IDT). The HiBiT coding region was inserted upstream of the N-terminus of endogenous BRD4 coding region using a sgRNA guide from Dharmacon. The template sequence was designed using Benchling. Guide sequence and HR template were ordered from IDT. HiBiT signal was measured via the manufacturer's protocol (catalog No. N3050, Promega) using an EnVision microplate reader (PerkinElmer). The H69 (31), H446 (31), SHP-77 (32), and DMS-114 (33) small-cell lung cancer cell lines (Supplementary Tables S1 and S2) were obtained directly from the ATCC or through the Fox Chase Cancer Center (FCCC) Cell Culture Facility, verified by short tandem repeat (STR) profiling, and cultured in RPMI1640 supplemented with 10% FBS, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. The pancreatic adenocarcinoma cell lines PNX001 and PNX017, derived from patient-derived xenograft (PDX) models were a gift of Igor Astsaturov (Fox Chase Cancer Center, Philadelphia, Pennsylvania; ref. 34). PNX-001 was cultured in RPMI1640 with 15% FBS, 100 U/mL penicillin, 0.1 mg/mL streptomycin, 25 μg/mL insulin, 1× l-Glutamine, 1× NEAA and 1× sodium pyruvate. PNX-017 was cultured in RPMI1640 with 15% FBS, 1× l-Glutamine, 25μg/mL insulin, 1× Transferrin, 1× T3, and 20 ng/mL EGF. All lines were confirmed as Mycoplasma-free, and identity verified by STR profiling by the FCCC Cell Culture Facility. Speckle-type POZ protein (SPOP) wild-type mutational status was reported in cBioPortal for H69 and H446, and directly confirmed by panel testing for PNX-001 and PNX-017.
For PDX analysis, the models LX-36 (35), PNX-001 (34), and PNX-017 (34) were used. PDX tumors were obtained from in vivo passaged tumors maintained in C.B17scid mice by Dr. Vladimir Khazak (NexusPharma Inc., Philadelphia, Pennsylvania). Dinaciclib was obtained from MedChemExpress and diluted in HPBCD (2-hydroxypropyl-beta-cyclodextrin). CFT-2718 was obtained from C4 Therapeutics and diluted in D5W (5% dextrose in water); details of chemical synthesis of CFT-2718 are provided in Supplementary Fig. S1. CC-220, CPI-203, bortezomib, and MLN4924 were obtained from Selleckchem.
Cell proliferation assays
Cells were plated in 24- or 96-well plates and cultured for 24 hours. Dinaciclib, CFT-2718, or vehicle were diluted in cell culture medium and added to plates. For some assays, after 72 hours, CellTiter-Blue (Promega) reagent was added to each well. After 2-hour incubation at 37°C, optical density readings were made in the 570- to 600-nm wavelength range, using a Perkin-Elmer ProXpress Visible-UV-fluorescence 16-bit scanner (Perkin-Elmer). In other experiments, a CellTiter-Glo 2.0 luminescence Assay Kit (Promega) was used as an alternative means to assess viability, with data acquired on an EnVision Multilabel Reader (PerkinElmer). In these assays, a time zero plate was incorporated into the assay to define GI50, TGI, and LC50 (36).
In vivo analysis of CFT-2718
All PDX experiments involving mice were performed according to protocols approved by Institutional Animal Care and Use Committees (IACUC) at Fox Chase Cancer Center or WuXi AppTec. Female BALB/C nude mice were used for initial dose-finding experiments to support hematologic cancer xenograft studies. Mice (n = 4/group) were dosed once weekly for 3 weeks with vehicle (D5W) or CFT-2718 administered at 2, 3, or 4 mg/kg in D5W by intravenous injection. Animal body weight was monitored regularly as an indirect measurement of toxicity.
C.B17 scid mice were used for initial dose-finding experiments to support solid tumor xenograft studies. Independent cohorts were injected with vehicle (D5W), or CFT-2718 administered at 1, 1.4, or 1.8 mg/kg in D5W, by retro-orbital injection once a week for 2 weeks. Mice were euthanized 1 to 2 hours after the last injection, and liver tissue was collected for histopathologic and immunoblot assessment.
For RS4;11 xenograft analysis, 6- to 8-week-old C.B17 scid mice were inoculated subcutaneously at the right flank with RS4;11 tumor cells (1 × 107) in 0.2 mL of PBS supplemented with Matrigel (PBS: Matrigel = 1:1). Mice were palpated twice a week after tumor cell implantation to assess tumor onset. Tumor volume was determined by external caliper twice a week, to establish maximum longitudinal diameter (length) and transverse diameter (width). Tumor volume was calculated using the formula, tumor volume = 1/2(length × width2). Animals were grouped for treatment on day 26 when the average tumor volume reached 180 mm3. Body weight also was monitored twice weekly, and mice were regularly monitored for signs of distress.
For solid tumor xenograft analysis, tumors were established in C.B17 scid mice by using a 1-mL syringe and 18G 1½' needle to implant 200 μL of RPMI-Matrigel suspension of PDX tumor fragments subcutaneously in both flanks of a C.B17scid mouse. Mice were palpated twice a week after tumor cells implantation to assess tumor onset. Once tumors volumes were greater than 200 mm3, mice were randomized into cohorts that received vehicle, dinaciclib, and CFT-2718 groups. Mice received intraperitoneal (i.p.) 20% HPBCD, i.p. 20 mg/kg dinaciclib in 20% HPBCD, or retroorbital 1.8 mg/kg CFT-2718 in D5W, once a week for 3 weeks. The dinaciclib dose level was selected based on previous studies demonstrating efficacy (37). Tumor volume, body weight, and distress were determined as for the RS4;11model. Mice were euthanized 1 to 2 hours after a final injection, and tumors excised, divided and prepared for histopathologic and immunoblot assessment, either by formalin fixation or by flash freezing in liquid nitrogen, and stored at −70°C prior to use.
Pharmacokinetic studies were performed in fasted female CD-1 mice at WuXi Apptec. All the procedures related to animal handling, care and the treatment in the study were performed according to the guidelines approved by the IACUC of WuXi AppTec following the guidance of the Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Blood was collected in citrate buffer, and plasma isolated by centrifugation. Protein was precipitated from an aliquot of 8-μL sample with 160-μL internal standard (verapamil). The mixture was mixed by vortexing and centrifuged at 3,220 × g for 15 minutes, 4°C. Supernatant (3 μL) was injected for analysis using a Triple Quad 6500+ LC-MS/MS system. Pharmacokinetic calculations were performed using Phoenix WinNonlin 6.3 using the IV-noncompartmental model 201 (IV bolus input).
Hematoxylin and eosin staining and IHC
Tissues to be analyzed by histopathologic analysis were fixed in 10% phosphate-buffered formaldehyde (formalin) for 24 to 48 hours, embedded in paraffin, sectioned into slides, and prepared for further assessment according to standard protocols. Tumor sections were stained with H&E, and immunostained with antibodies to BRD4 (No. A301–985A100, Bethyl) and with Ki-67 (DAKO) using standard protocols. Immunostained slides and hematoxylin and eosin (H&E)–stained slides were scanned using a Vectra Automated Quantitative Pathology Imaging System (Perkin Elmer). Scanned images then were viewed with Phenochart 1.0.9 software and selected regions of interest were outlined. Expression levels of the proliferative index marker Ki-67 or of BRD4 were quantified using inForm Cell Analysis software (Perkin Elmer) protocols and algorithms. H-score (3 x percentage of strongly staining nuclei + 2 x percentage of moderately staining nuclei + percentage of weakly staining nuclei) was calculated by the system. Percentage of necrotic areas were calculated by ImageJ (NIH, Bethesda, MD) based on the scanned images.
Western blots
For Western blot analysis, cultured cells were disrupted in CelLytic M lysis buffer (Sigma-Aldrich) supplemented with protease and phosphatase inhibitor cocktails (Roche). Tumors were homogenized in T-PER Tissue Protein Extraction Reagent (Thermo Fisher Scientific) supplemented with protease and phosphatase inhibitor cocktails (Roche), using a TerraLyzer Xpedition Sample Processor (No. S6022, ZYMO). The homogenate was centrifuged at 12,000 × g at 4°C for 10 minutes and the supernatant was separated and stored at −20°C.
Whole-cell lysates and tissue homogenates were used directly for SDS–PAGE and Western blotting, using standard procedures. Primary antibodies included rabbit anti-BRD4 (No. ab128874), β-actin (No. ab49900), anti-polymerase II CTD repeat YSPTSPS (phospho-S5) (No. ab5131), anti-SPOP (ab137537) all from Abcam; anti-phospho-Rpb1 CTD (phospho-S2) (No. 13499S), mouse anti–Rpb1 (No. 2629S), anti-MYC (No. 13987S), anti-CDK9 (No. 2316S), and anti-PARP (No. 9542), all from Cell Signaling Technology; and BRD4 (Sigma, catalog No. PLA-0227, Lot No. 5), actin (Santa Cruz Biotechnology, catalog No. SC-8432, Lot No. A2717), and Ikaros/IKZF1 (Invitrogen, catalog No. PA523728, Lot No. SA2333742A). Secondary anti-mouse and anti-rabbit horseradish peroxidase–conjugated antibodies (GE Healthcare) were used at a dilution of 1:10,000 for visualization of Western blots and blots developed by chemiluminescence using the Immobilon Western Chemiluminescent HRP Substrate (Millipore Sigma). Image analysis was done using ImageJ (NIH, Bethesda, MD), with signal intensity normalized to β-actin or total level of detected proteins. Data were analyzed in Excel by paired t test to determine statistical significance.
Results
Preliminary characterization of CFT-2718
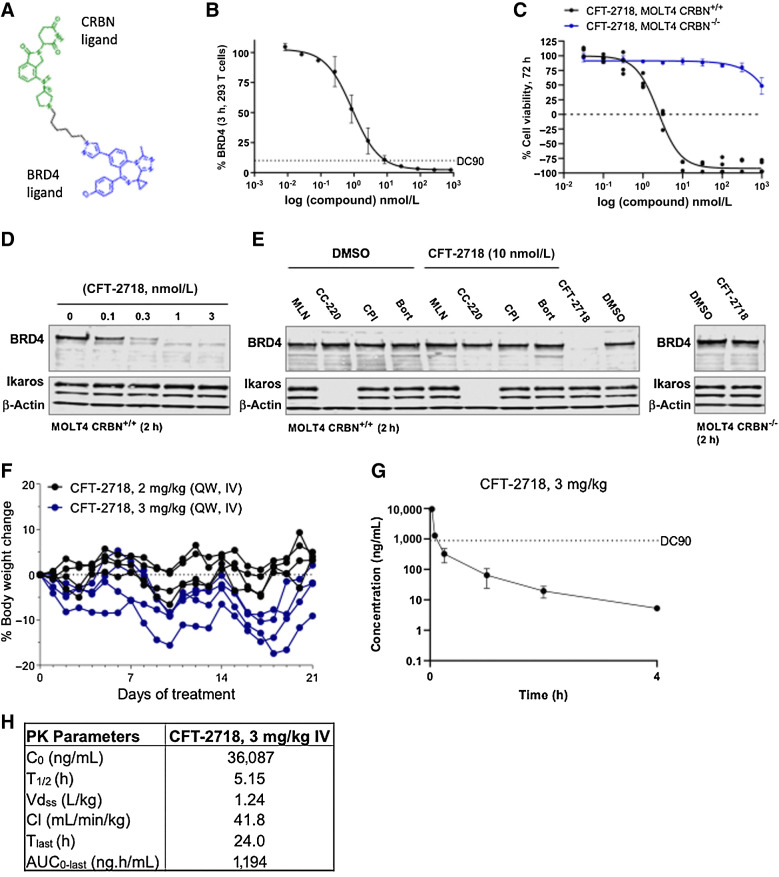
CFT-2718 is composed of a benzotriazoloazepine BET-binding ligand linked via an alkyl chain to an amino-pyrrolidine substituted isoindolone CRBN binding moiety. This compound (Fig. 1A) was selected from a large set of rationally designed BRD4 degraders based on its ability to rapidly and selectively degrade BRD4 in vitro in a CRBN-dependent manner. Degradation of 90% endogenous BRD4 was achieved within 3 hours at a dose of 10 nmol/L in 293T cells (Fig. 1B). Approximately 75% of the acute lymphoblastic leukemia cell line MOLT4 was killed with three days of 10 nmol/L CFT-2718 treatment (MOLT4 CRBN+/+), whereas no effect was observed at this dose in the absence of cereblon (MOLT4 CRBN−/−; Fig. 1C). Degradation was dose-dependent (Fig. 1D), with BRD4 almost completely eliminated at drug concentrations ≤1 nmol/L. Furthermore, CFT-2718 degradation of BRD4 was disrupted by chemical competition with 10 μmol/L of the CRBN-binding ligand CC-220, a BET-binding inhibitor (CPI-203), a proteasome inhibitor (bortezomib), a Nedd8-activating enzyme inhibitor (MLN4924), or in cells lacking cereblon, confirming on-mechanism activity (ref. 16; Fig. 1E). We also determined that while CFT-2718 contains an isoindolinone ring similar to that in lenalidomide, it does not promote Ikaros degradation (ref. 38; Fig. 1D and E).
Figure 1.
Selection of CFT-2718 based on selectivity and rapidity of BRD4 degradation, and rapid turnover pharmacokinetic profile. A, Chemical structure of CFT-2718. B, HiBiT-detection of BRD4 in cell lysate 3 hours after addition of indicated concentrations of CFT-2718 (compound) to 293T cells with endogenously tagged BRD4. Bars represent SD. DC90, 10 nmol/L. C, Viability changes 72 hours after addition of CFT-2718 at indicated concentrations in MOLT4 parental (CRBN+/+) or MOLT4 CRBN−/− cells, assessed by CellTiter Glo. Bars represent SD. D, Western blot indicates dose-dependent degradation of BRD4 by CFT-2718 in MOLT4 cells 2 hours after addition of 10 nmol/L CFT-2718. E, BRD4 degradation 2 hours after addition of 10 nmol/L CFT-2718 (+) alone or with simultaneous addition of 10 μmol/L of the Neddylation inhibitor MLN4924 (ML), the cereblon inhibitor CC-220 (CC), the BET inhibitor CPI-203 (CP), or the proteasome inhibitor bortezomib (BO) to MOLT4 CRBN+/+ or MOLT4 CRBN−/− cells. Actin was used as a loading control for D and E; Ikaros is a control for CFT-2718 specificity. F, Body weight of mice from 3-week tolerability study in naïve female BALB/c nude mice (N = 4/group) dosed intravenously with 2 or 3 mg/kg CFT-2718. Bars represent SEM. G, Plasma concentration of CFT-2718 following a single 3 mg/kg intravenous dose. N = 3; bars represent SD. H, Pharmacokinetic parameters for mice treated as in G, calculated using Phoenix WinNonlin 6.3, IV-Noncompartmental model 201 (i.v. bolus input).
For in vivo testing of antitumor activity, we first tested the tolerability of CFT-2718 by administering the compound at 2 and 3 mg/kg for 3 weeks. When CFT-2718 was administered intravenously at 3 mg/kg once weekly for 3 weeks, four of four mice lost between 9% and 17% of their respective body weights; in contrast, no significant changes were observed at 2 mg/kg (Fig. 1F). From single-dose plasma pharmacokinetic studies at 3 mg/kg, CFT-2718 displayed a high total Cmax of 30.087 ng/mL and moderate clearance of 41.8 mL/min/kg. CFT-2718 was 99.1% bound in mouse plasma. Thus, its time of exposure above the total DC90 (concentration at which 90% of BRD4 has been degraded) of 889 ng/mL was calculated as slightly over 5 minutes at 3 mg/kg and was modeled to be approximately 5 minutes at 1.8 mg/kg (Fig. 1G and H).
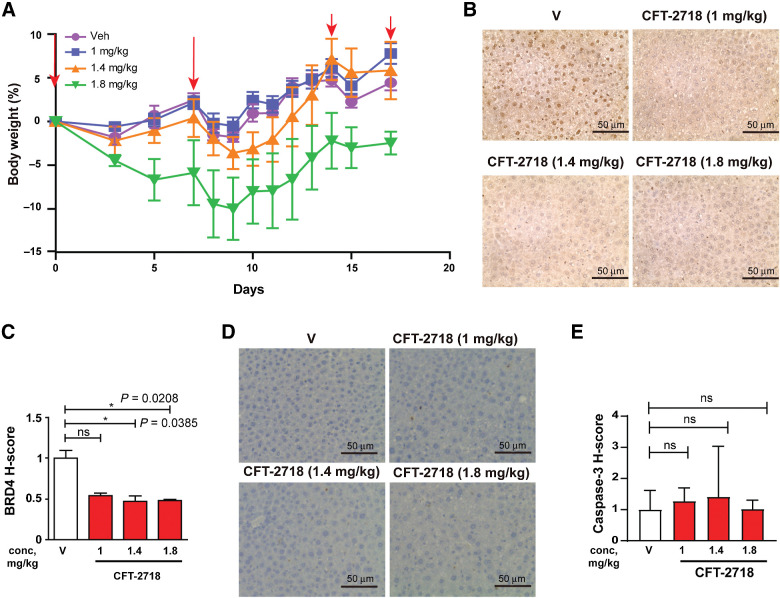
On the basis of these data, we also explored drug tolerability at a lower dose range; C-B17.scid mice were injected with vehicle (D5W) or CFT-2718 administered at doses of 1, 1.4, or 1.8 mg/kg on days 0, 7, and 14; on day 17, mice were euthanized. No effect on mouse body weight was seen with the two lower doses of CFT-2178 over the course of the experiment (Fig. 2A). For the 1.8 mg/kg treatment group, a statistically insignificant and transient drop in body weight was observed following the second dose of the drug, but reversed within a week. No mice manifested signs of distress in the experiment. Observation of liver tissue by IHC at the end of the experiment (Fig. 2B and C) indicated a statistically significant reduction in BRD4 staining. At all dose levels, the expression of the apoptotic indicator cleaved caspase-3 was low, and there was no significant increase in cleaved caspase-3 expression compared with control group even at the highest dose of 1.8 mg/kg (Fig. 2D and E).
Figure 2.
Dose determination for use of CFT-2718 in vivo. A, Change in body weight in mice treated with CFT-2718 on days 0, 7, and 14 with 1, 1.4, 1.8 mg/kg drug, or vehicle. B and C, IHC staining of liver tissue for expression of BRD4; representative images (B) and quantification of H score, normalized to vehicle (C). D and E, Representative image (D) and quantification (E) of cleaved caspase expression determined by IHC in liver tissue from treated mice. For all graphs, *, P < 0.05; **, P < 0.01; and ***, P < 0.001 and ****, P < 0.0001 relative to controls. V represents vehicle.
CFT-2718 is more effective than dinaciclib in control of PDX tumor models in vivo
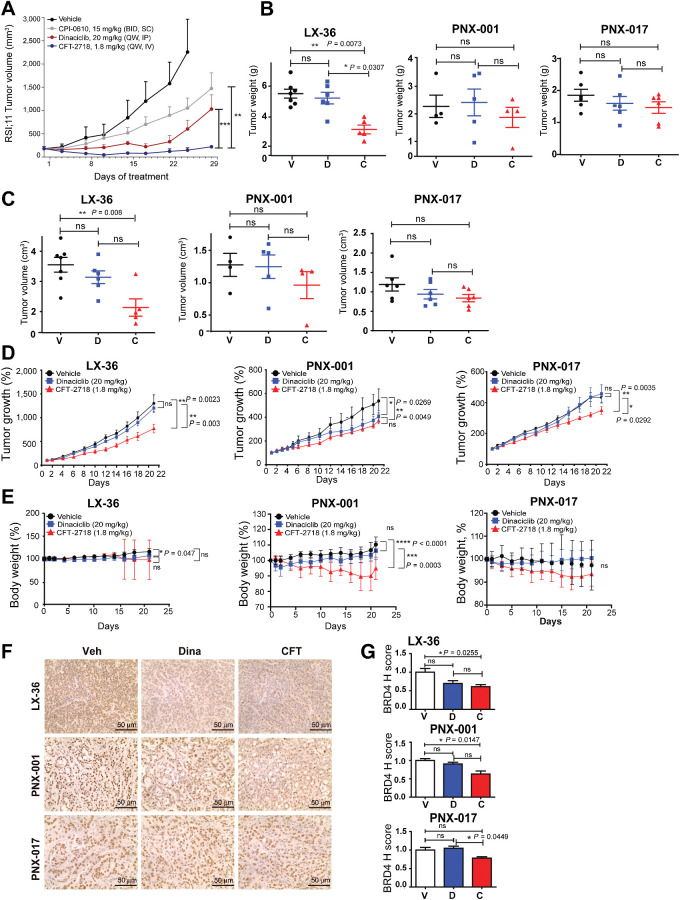
A dose of 1.8 mg/kg was used for subsequent in vivo testing. We first tested whether this dose level would provide sufficient exposure to drive tumor regression in a leukemia xenograft model previously shown to be highly responsive to BRD4 degraders, the human lymphoblastic leukemia cell line RS;411 (39). A low weekly dose of CFT-2718 (1.8 mg/kg QW) was more effective than 20 mg/kg of the CDK9 inhibitor dinaciclib (37) administered weekly, or 15 mg/kg of the BET inhibitor CPI-0610 (40) administered twice a day, at blocking xenograft growth (Fig. 3A).
Figure 3.
Comparison of CFT-2718 and dinaciclib in control of PDX tumor growth in vivo. A, Female CB17 SCID mice bearing RS4;11 human tumor xenografts were treated with vehicle, CIP-0610 (15 mg/kg, twice a day), dinaciclib (20 mg/kg, once weekly), or CFT-2718 (1.8 mg/kg, once weekly), and tumor growth monitored by palpation. Bars represent SEM. **, P <0.01; ***, P < 0.001, based on two-way ANOVA analysis. B–D, Mice bearing established PDX tumors were treated once weekly for 3 weeks (days 0, 7, and 14) with vehicle (V), dinaciclib (D), or CFT-2718 (C). Tumor weight (B), tumor volume (C), and tumor growth (D) are indicated. Tumor growth for each mouse was normalized to day 1 tumor volume. E, Change in body weight in mice treated with CFT-2718 (1.8 mg/kg), dinaciclib (20 mg/kg), or vehicle on days 0, 7, and 14. F and G, IHC staining of excised tumor tissue for expression of BRD4, 3 hours after final dose; representative images (F) and quantification of H score (G). For all graphs, ns, not significant; *, P < 0.05; **, P < 0.01; and ***, P < 0.001 and ****, P < 0.0001 relative to controls.
To assess CFT-2718 control of solid tumor growth, we used three PDX models grown in C.B17 scid; the SCLC model LX-36 (also known as CTG-0199 and BML-4; ref. 35), and the pancreatic cancer models PNX-001 and PNX-017 (34). After implanted tumors became palpable, mice were divided into three groups for treatment with vehicle, 1.8 mg/kg CFT-2718, or 20 mg/kg dinaciclib (37). Drugs were administered weekly on days 0, 7, and 14; mice were euthanized on day 21. Comparison of tumor weight, volume, and growth profile (Fig. 3B–D) indicated that CFT-2718 drug treatment was significantly more effective than dinaciclib in controlling tumor growth of the LX-36 SCLC model, based on in vivo measurement of tumor growth, and weight of excised tumors. In contrast, PDX tumors in dinaciclib-treated mice did not differ significantly from those in mice treated with vehicle.
Dinaciclib and CFT-2718 had less marked activity in the two pancreatic cancer models. Tumor growth was inhibited at endpoint stage by dinaciclib and CFT-2718 in PNX-001, and by CFT-2718 in PNX-017 in comparison with the vehicle group (Fig. 3D); however, the weight of excised tumors did not differ significantly from that of dinaciclib-treated tumors, or vehicle-treated controls. For the LX-36 model, mouse weight was stable for all treatment groups throughout the experiment, but, for the two pancreatic PDX models, there was a statistically significant reduction in weight with CFT-2718 versus the other two groups, although this did not exceed 10% at any point (Fig. 3E).
Mice were euthanized 21 days after the beginning of treatment (three hours after the final dose of drug). At this time point, based on IHC assessment, expression of BRD4 was significantly reduced by CFT-2718 in the LX36 tumor model (P = 0.026), the PNX-001 model (P = 0.0147), and the PNX-017 model (P = 0.0449; Fig. 3F and G). In contrast, dinaciclib did not significantly reduce BRD4 expression in any cell model. Parallel measurement of Ki-67 levels by IHC indicated neither CFT-2718 nor dinaciclib significantly reduce dKi-67 expression in the LX-36, PNX-001, and PNX-017 models (Supplementary Fig. S2A and S2B). Together, these data indicate that CFT-2718 is effective in gaining access to BRD4 in these PDX tumors and that the subsequent removal of BRD4 is durable. Furthermore, these pharmacodynamic effects translate to variable response in tumor growth reductions across models, with the most significant reductions observed in the LX-36 SCLC model.
Comparison of CFT-2718 and dinaciclib activity in control of cancer cell growth in vitro
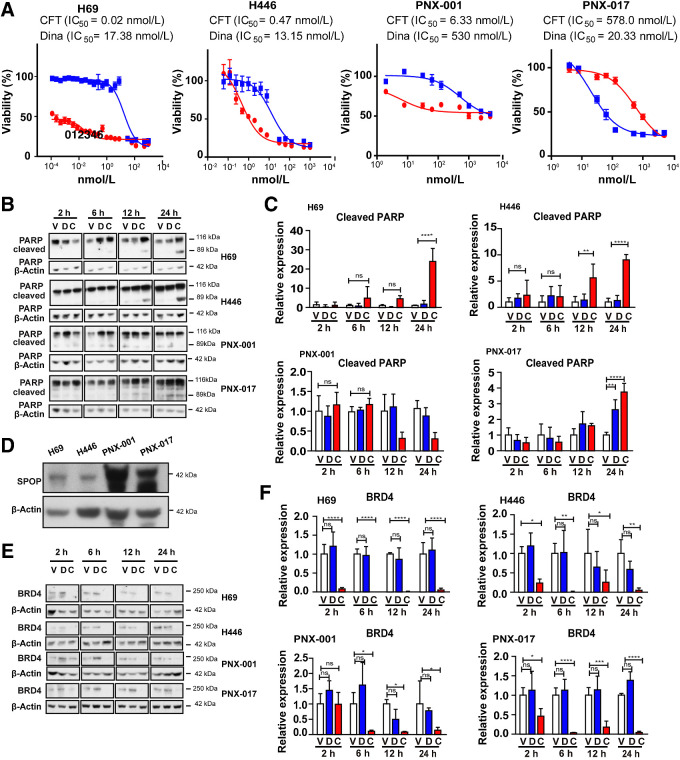
To further analyze the activity in CFT-2718 in reference to dinaciclib, we used two SCLC cell lines (H69 and H446; ref. 31) and two pancreatic cell lines derived from the PNX-001 and PNX-017 PDX models (34). CFT-2718 greatly reduced viability of both SCLC cell models (Fig. 4A); in both cases, IC50 values were lower than 1 nmol/L, in contrast to IC50 values of 13 to 17 nmol/L for dinaciclib, reflecting a 20- to 100-fold difference. Increasing doses of each drug reduced viability of each cell line by >90%. In contrast, the two pancreatic models were much less responsive to both drugs. Although CFT-2718 was more active than dinaciclib in the PNX-001 model (IC50 6.3 versus 530 nmol/L), dinaciclib was more active than CFT-2718 in the PNX-017 model (20 versus 578 nmol/L).
Figure 4.
CFT-2718 is more effective than dinaciclib in controlling cell growth and efficiently reduces BRD4 expression in vitro. A, Cell viability determination by CTB assay, 72 hours after addition of CFT-2718 (CFT, red line) or dinaciclib (blue line). IC50 values are indicated. B and C are representative images (B) and quantification (C) of PARP cleavage in cells treated at time points indicated. D, Expression of SPOP in cell lines indicated. For all graphs, ns, not significant; *, P < 0.05; **, P < 0.01; and ***, P < 0.001 and ****, P < 0.0001 relative to controls. E and F, BRD4 expression assessed by Western blot following treatment of cell lines with 10 nmol/L CFT-2718 (C), or dinaciclib (D), or vehicle for times indicated. Representative data (E) and quantification (F) are shown. At each time point, BRD4 expression in drug -treated groups was normalized to vehicle. All Western blots were performed at least three times. For all graphs, ns, not significant; *, P < 0.05; **, P < 0.01; and ***, P < 0.001 and ****, P < 0.0001 relative to controls.
To further benchmark drug activity, we compared induction of apoptosis up to 24 hours following treatment of cells with 10 nmol/L CFT-2718, 10 nmol/L dinaciclib, or vehicle (Fig. 4B and C). Measurement of PARP cleavage indicated significantly greater activity of CFT-2718 than dinaciclib in inducing apoptosis in the two SCLC models. In contrast, in the pancreatic cancer models, CFT-2718 and dinaciclib comparably induced apoptosis in PNX-017 by 24 hours after exposure (a modest effect compared with the SCLC models), while neither drug was effective in inducing apoptosis in the PNX-001 cell line.
Mechanistically, cellular sensitivity to BRD4 inhibitors has been shown to be influenced by mutations in the ubiquitin ligase adaptor SPOP in some cancer types, with mutated SPOP associated with greater response to BRD4-targeting drugs in endometrial cancer, but resistance in prostate cancer (41, 42). Analysis of the SPOP gene in all four cell models confirmed all four lines were wild-type; however, Western blot analysis indicated considerably higher intrinsic expression of SPOP in the two pancreatic cell lines versus the SCLC cell lines (Fig. 4D). SPOP levels were not affected by treatment with dinaciclib or CFT-2718 (Supplementary Fig. S3).
Comparison of CFT-2718 and dinaciclib activity in control of BRD4-related signaling
Western blot analysis of the expression of BRD4 and functionally interacting proteins was performed over a similar time course. CFT-2718 caused rapid and profound reduction in the expression of BRD4 expression in each of the four cell models (Fig. 4E and F). This was observed within 2 hours of drug addition, in the H69 and H446 cell models; a more delayed effect characterized both pancreatic cell lines, with reduction in BRD4 becoming marked only 6 hours after treatment. In all models, BRD4 levels remained depressed at 24 hours after drug addition. In contrast, treatment with dinaciclib did not significantly affect expression of BRD4 at any time point (Fig. 4E and F).
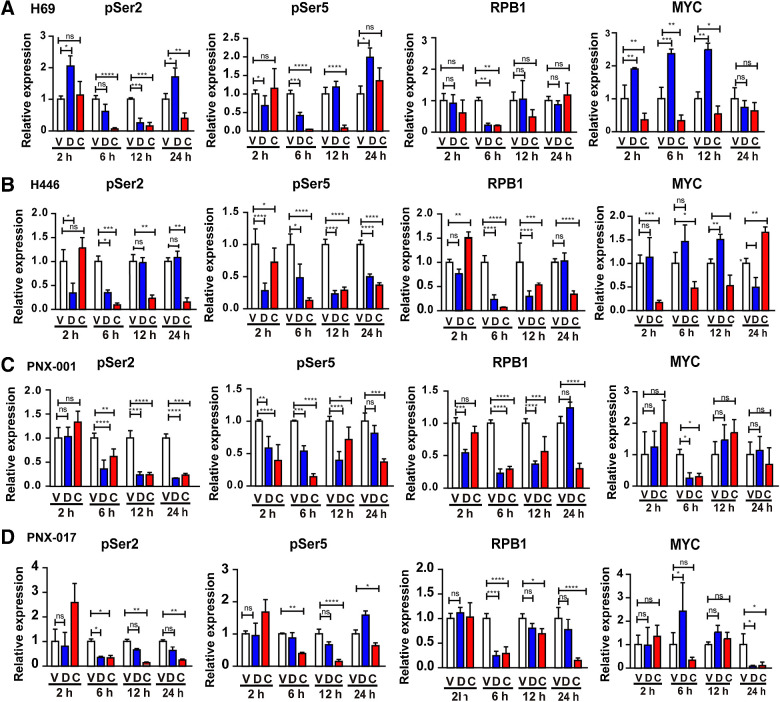
Phosphorylation of serine 2 (Ser2) and serine 5 (Ser5) on the carboxy-terminal domain (CTD) of the large subunit (RPB1) of RNA polymerase 2 (pol II) is required for transcriptional progression (43). Ser5 is phosphorylated by the TFIIH-associated kinase CDK7 near transcription start sites. During elongation, BRD4 binds to the CTD of RPB1 and directly phosphorylates Ser2, but not Ser5; in addition, BRD4 promotes the recruitment of CDK9, which also phosphorylates Ser2 (44). In all cells treated with CFT-2718, Western blot analysis indicates that CFT-2718 significantly reduces levels of both phosphorylated Ser2 and Ser5 (pSer2, pSer5), as well as total RPB1, within 6 hours of drug addition (Fig. 5; Supplementary Fig. S4). Levels of pSer2 remained strongly depressed for 24 hours following drug addition; in some of the cell models, levels of pSer5 and RPB1 also remained depressed later time points. In comparison, treatment with dinaciclib reduced pSer2 strongly at 6-, 12-, and 24-hour time points in the pancreatic models, but only transiently reduced pSer2 at 6 to 12 hours after drug addition in the two SCLC lines, to a lesser degree than observed with CFT-2718. Dinaciclib also reduced pSer5 and RPB1 expression, with maximal effect observed at the 6-hour time point; for pSer5, magnitude of inhibition was also less than observed with CFT-2718.
Figure 5.
Comparison of CFT-2718 and dinaciclib inhibition of RNA polymerase II. The H69, H446, PNX-001, and PNX-017 cell lines were treated with10 nmol/L CFT-2718 (C), 10 nmol/L dinaciclib (D), or vehicle (V), for times indicated. A–D are quantification of phosphorylated Ser2 (pSer2), phosphorylated Ser 5 (pSer5), RPB1, and Myc expression in H69 (A), H446 (B), PNX-001 (C), and PNX-017 (D) cells. Expression of each protein is normalized to vehicle-treated lanes. All experiments were performed >3 times; representative Western blots are shown in Supplementary Fig. S4. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ****, P < 0.0001 relative to vehicle.
The fact that inhibition of BRD4 by small molecules causes a rapid reduction in transcription (within 60 minutes) also leads to a decrease in the protein expression of MYC (45). CFT-2718 treatment reduced MYC expression in all cell models, with maximal loss observed by 2 hours in the SCLC cell lines, and by 6 hours in the pancreatic cancer models (Fig. 5; Supplementary Fig. S4). Although levels remained depressed relative to vehicle-treated cells in the SCLC models, they rebounded in the pancreatic cancer models. Notably, dinaciclib had little effect on MYC expression in any of the models.
Finally, to extend our analysis of CFT-2718 action in additional SCLC cell models, we treated two additional cell lines, DMS-114 and SHP-77 with CFT-2718, dinaciclib, or vehicle, and analyzed effect on signaling. As for the other SCLC cell lines, this analysis demonstrated a greater effect of CFT-2718 than dinaciclib on induction of PARP, associated with degradation of BRD4 and rapid degradation of MYC, and inhibition of cell growth at significantly lower concentrations than with use of dinaciclib (IC50s for CFT-2718 vs. dinaciclib: 12.5 nmol/L vs. 202 nmol/L in SHP-77 cells, and 1.5 nmol/L vs. 159 for DMS-114 cells; Supplementary Fig. S5).
Discussion
For the first time, in this study, we characterized the activity of CFT-2718 in two types of treatment-refractory solid tumors, SCLC and pancreatic cancer. CFT-2718 was selected for evaluation based on evidence of rapid and specific degradation of BRD4 in a CRBN-dependent manner, and its favorable pharmacokinetic profile in vivo. Both in vitro and in vivo, this compound is active in reducing the growth of a PDX model and cell lines for SCLC; in direct benchmarking, the IC50 of CFT-2718 is significantly lower than that of dinaciclib in multiple SCLC cell line models and the compound is much more effective than dinaciclib at inducing apoptosis when the two compounds are administered at similar doses. CFT-2718 rapidly, efficiently, and durably eliminated expression of BRD4, pSer2, and MYC in multiple SCLC models, confirming on-target activity. We also found that CFT-2718 effectively targeted BRD4 and pSer2 in pancreatic cancer models, but IC50 for this drug was three orders of magnitude higher than observed in SCLC cell lines in vitro, indicating the likely presence of compensatory signaling processes in the pancreatic cell lines.
BRD4 emerged early as a target of interest for small molecule inhibition and compounds such as JQ1 displayed promising early preclinical results in control of cancers with known dysfunction of BRD4, such as NUT midline carcinomas (ref. 14; reviewed in ref. 46). These types of catalytic inhibitors have shown value in some tumor types and a number are presently in clinical trials (e.g., GSK525762/molibresib, NCT01587703; BMS-986158, NCT03936465; AZD5153, NCT03205176). Although these types of agents are useful in suppressing BRD4 interaction with the targets of its catalytic function, part of BRD4 activity involves activity as an adaptor/scaffold protein for an additional component of the transcriptional machinery and is noncatalytic, suggesting that targeted protein degradation is a more effective strategy.
A number of distinct BRD4 degraders have been developed, based on differences in degradation targeting moiety, BRD4-binding moiety, and variation of linker domains (discussed in refs. 46, 47). The JQ1-based degrader, dBET6, was highly efficacious in two human systemic leukemia models; however, this agent required daily i.p. dosing at 7.5 mg/kg (20). ARV-825 and ARV-771 were built on the BRD4 inhibitor OTX015 scaffold and differed in their ligase-targeting ligands (CRBN and VHL, respectively). ARV-825, dosed daily oral at 25 mg/kg, significantly inhibited growth of a thyroid cancer xenograft model (48). ARV-771 dosed daily subcutaneously at 30 mg/kg, promoted tumor regression in a prostate cancer xenograft model (18). Advances in potency of BRD4 degraders led to improvements in potency and efficacy of BET degraders, which enabled lower drug exposures and less frequent dosing. Notably, QCA570 (39) and compound 23 (49) achieved complete and durable tumor regression in the RS4;11 human leukemia tumor xenograft model when dosed three times weekly at 5 mg/kg intravenously, with minimal changes in animal body weight. QCA570 also enabled disease control when dosed at 5 mg/kg once weekly, but tumors ultimately progressed after 3 weeks of treatment (39). In this study, we found similar effects with CFT-2718 in the same tumor model but with a reduced schedule and dose level (once weekly at 1.8 mg/kg); regressions were observed in the first 2 weeks of dosing, followed by progression beginning in the third week of treatment. Furthermore, all three BRD4 degraders (QCA570, compound 23, CFT-2718) were rapidly eliminated from plasma, which may be a contributing factor to their positive therapeutic index. On the basis of these observations, CFT-2718 is equal to, or slightly better, than the most potent BET degraders published to date.
An additional potential advantage of CFT-2718 derives from its rapid kinetics. Because of the essential nature of BRD4 in normal as well as transformed cells, chronic in vivo use of BRD4 inhibitors has been associated with some side effects, including reactivation of latent HIV infection, immunosuppression, and some effects in the central nervous system (46). Infrequent, low-dose administration of CFT-2718 exhibits rapid impacts on tumor growth viability of BRD4-dependent tumors, while minimizing effects on nontumor tissue.
In contrast to many other forms of cancer, SCLC outcomes have shown limited improvement with the advent of targeted and immunotherapies (50). Of the 25% of patients with SCLC diagnosed with LS-SCLC (with the cancer limited to one side of the thorax and proximal lymph nodes), concurrent treatment with radiotherapy and etoposide/platin chemotherapy has long been standard of care (51). The 75% of patients diagnosed with ES-SCLC typically are treated with chemotherapy which benefits most patients for only a short period of time (52). Immune checkpoint inhibitors (ICIs) have been evaluated in SCLC, with the PD-L1–targeting antibody atezolizumab recently given FDA approval, for use in first-line treatment of ES-SCLC in combination with chemotherapy (53, 54). Although the increase in overall survival is so far limited, this may be a promising approach; unfortunately, less than 29% of ES-SCLC express PD-L1 (8), emphasizing the need for more broadly active agents. Three previous studies have investigated BRD4 inhibition in SCLC, identifying the BET inhibitor JQ1 as active in SCLC (27–29). This study confirms efficacy of targeting BRD4 in multiple cell culture models of SCLC and, for the first time, additionally demonstrates efficacy of BRD degradation in an SCLC PDX model in vivo. Together with our current findings, these data suggest that transcriptional suppression, and degradation of BRD4 in particular, warrants further exploration in the setting of SCLC.
Authors' Disclosures
M.E. Fitzgerald is an employee and stockholder of C4 Therapeutics. R.E. Michael is an employee and stockholder of C4 Therapeutics. S.L. Fisher reports other support from C4 Therapeutics outside the submitted work. C.G. Nasveschuk is an employee and stockholder of C4 Therapeutics. D.A. Proia reports other support from C4 Therapeutics outside the submitted work and is a stockholder of C4 Therapeutics. E.A. Golemis reports nonfinancial support from C4 Therapeutics and grants from C4 Therapeutics during the conduct of the study. No disclosures were reported by the other authors.
Acknowledgments
E.A. Golemis and A.S. Nikonova were supported by NCI Core Grant P30 CA006927 (to Fox Chase Cancer Center), by NIH P50 DE030707, and the William Wikoff Smith Charitable Trust (to E.A. Golemis). We would like to thank Anna Kiseleva for help in the preparation of figures, Margret Einarson of the Fox Chase Cancer Center High Throughput Screening Facility. We thank Roy Pollock for helpful critique of the manuscript.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
D. Sun: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, investigation, visualization, writing–original draft, project administration, writing–review and editing. A.S. Nikonova: Data curation, formal analysis, supervision, validation, investigation, visualization, methodology. P. Zhang: Data curation, formal analysis, supervision, validation, investigation, visualization, methodology. A.Y. Deneka: Data curation, formal analysis, investigation, visualization, writing–review and editing. M.E. Fitzgerald: Conceptualization, resources, data curation, formal analysis, validation, investigation, methodology. R.E. Michael: Data curation, formal analysis, validation, investigation, visualization, methodology. L. Lee: Data curation, formal analysis, validation, investigation, visualization, methodology. A.C. Lilly: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. S.L. Fisher: Conceptualization, formal analysis, validation, investigation, visualization, methodology. A.J. Phillips: Data curation, formal analysis, validation, investigation, visualization, methodology. C.G. Nasveschuk: Conceptualization, data curation, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. D.A. Proia: Conceptualization, data curation, formal analysis, supervision, writing–original draft, project administration, writing–review and editing. Z. Tu: Formal analysis, supervision, visualization, writing–review and editing. E.A. Golemis: Conceptualization, resources, formal analysis, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Shi J, Vakoc CR. The mechanisms behind the therapeutic activity of BET bromodomain inhibition. Mol Cell 2014;54:728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer 2018;17:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature 1999;399:491–6. [DOI] [PubMed] [Google Scholar]
- 4. Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell 2012;149:214–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature 2009;461:664–8. [DOI] [PubMed] [Google Scholar]
- 6. Brookes E, Pombo A. Modifications of RNA polymerase II are pivotal in regulating gene expression states. EMBO Rep 2009;10:1213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Trigon S, Serizawa H, Conaway JW, Conaway RC, Jackson SP, Morange M. Characterization of the residues phosphorylated in vitro by different C-terminal domain kinases. J Biol Chem 1998;273:6769–75. [DOI] [PubMed] [Google Scholar]
- 8. Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 2006;23:297–305. [DOI] [PubMed] [Google Scholar]
- 9. Prinjha R, Tarakhovsky A. Chromatin targeting drugs in cancer and immunity. Genes Dev 2013;27:1731–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle 2016;15:519–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Perez-Salvia M, Esteller M. Bromodomain inhibitors and cancer therapy: from structures to applications. Epigenetics 2017;12:323–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. White ME, Fenger JM, Carson WE III. Emerging roles of and therapeutic strategies targeting BRD4 in cancer. Cell Immunol 2019;337:48–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parry D, Guzi T, Shanahan F, Davis N, Prabhavalkar D, Wiswell D, et al. Dinaciclib (SCH 727965), a novel and potent cyclin-dependent kinase inhibitor. Mol Cancer Ther 2010;9:2344–53. [DOI] [PubMed] [Google Scholar]
- 14. Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sakamoto KM, Kim KB, Kumagai A, Mercurio F, Crews CM, Deshaies RJ. Protacs: chimeric molecules that target proteins to the Skp1-Cullin-F box complex for ubiquitination and degradation. Proc Natl Acad Sci U S A 2001;98:8554–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winter GE, Buckley DL, Paulk J, Roberts JM, Souza A, Dhe-Paganon S, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015;348:1376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saenz DT, Fiskus W, Qian Y, Manshouri T, Rajapakshe K, Raina K, et al. Novel BET protein proteolysis-targeting chimera exerts superior lethal activity than bromodomain inhibitor (BETi) against post-myeloproliferative neoplasm secondary (s) AML cells. Leukemia 2017;31:1951–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Raina K, Lu J, Qian Y, Altieri M, Gordon D, Rossi AM, et al. PROTAC-induced BET protein degradation as a therapy for castration-resistant prostate cancer. Proc Natl Acad Sci U S A 2016;113:7124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohoka N, Okuhira K, Ito M, Nagai K, Shibata N, Hattori T, et al. In vivo knockdown of pathogenic proteins via specific and nongenetic inhibitor of apoptosis protein (IAP)-dependent protein erasers (SNIPERs). J Biol Chem 2017;292:4556–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Winter GE, Mayer A, Buckley DL, Erb MA, Roderick JE, Vittori S, et al. BET bromodomain proteins function as master transcription elongation factors independent of CDK9 recruitment. Mol Cell 2017;67:5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knoechel B, Roderick JE, Williamson KE, Zhu J, Lohr JG, Cotton MJ, et al. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat Genet 2014;46:364–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chen Y, Wen H, Wu CI. A mathematical theory of the transcription repression (TR) therapy of cancer - whether and how it may work. Oncotarget 2017;8:38642–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer 2015;121:664–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell 2012;150:1121–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peifer M, Fernandez-Cuesta L, Sos ML, George J, Seidel D, Kasper LH, et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat Genet 2012;44:1104–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. George J, Lim JS, Jang SJ, Cun Y, Ozretic L, Kong G, et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015;524:47–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lenhart R, Kirov S, Desilva H, Cao J, Lei M, Johnston K, et al. Sensitivity of small cell lung cancer to BET inhibition is mediated by regulation of ASCL1 gene expression. Mol Cancer Ther 2015;14:2167–74. [DOI] [PubMed] [Google Scholar]
- 28. Kato F, Fiorentino FP, Alibes A, Perucho M, Sanchez-Cespedes M, Kohno T, et al. MYCL is a target of a BET bromodomain inhibitor, JQ1, on growth suppression efficacy in small cell lung cancer cells. Oncotarget 2016;7:77378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kaur G, Reinhart RA, Monks A, Evans D, Morris J, Polley E, et al. Bromodomain and hedgehog pathway targets in small cell lung cancer. Cancer Lett 2016;371:225–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Schwinn MK, Machleidt T, Zimmerman K, Eggers CT, Dixon AS, Hurst R, et al. CRISPR-mediated tagging of endogenous proteins with a luminescent peptide. ACS Chem Biol 2018;13:467–74. [DOI] [PubMed] [Google Scholar]
- 31. Little CD, Nau MM, Carney DN, Gazdar AF, Minna JD. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature 1983;306:194–6. [DOI] [PubMed] [Google Scholar]
- 32. Fisher ER, Paulson JD. A new in vitro cell line established from human large cell variant of oat cell lung cancer. Cancer Res 1978;38(11 Pt 1):3830–5. [PubMed] [Google Scholar]
- 33. Pettengill OS, Sorenson GD, Wurster-Hill DH, Curphey TJ, Noll WW, Cate CC, et al. Isolation and growth characteristics of continuous cell lines from small-cell carcinoma of the lung. Cancer 1980;45:906–18. [DOI] [PubMed] [Google Scholar]
- 34. Bobrov E, Skobeleva N, Restifo D, Beglyarova N, Cai KQ, Handorf E, et al. Targeted delivery of chemotherapy using HSP90 inhibitor drug conjugates is highly active against pancreatic cancer models. Oncotarget 2017;8:4399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gaponova AV, Nikonova AS, Deneka AY, Kopp MC, Kudinov AE, Skobeleva N, et al. A novel HSP90 inhibitor-drug conjugate to SN38 is highly effective in small cell lung cancer. Clin Cancer Res 2016;22:5120–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. NCI. Available from: https://dtp.cancer.gov/databases_tools/docs/compare/compare_methodology.htm#:~:text=SpecialConcentraDonParametersGI50CTGI,50(3C9).
- 37. Baker A, Gregory GP, Verbrugge I, Kats L, Hilton JJ, Vidacs E, et al. The CDK9 inhibitor dinaciclib exerts potent apoptotic and antitumor effects in preclinical models of MLL-rearranged acute myeloid leukemia. Cancer Res 2016;76:1158–69. [DOI] [PubMed] [Google Scholar]
- 38. Chamberlain PP, Lopez-Girona A, Miller K, Carmel G, Pagarigan B, Chie-Leon B, et al. Structure of the human Cereblon-DDB1-lenalidomide complex reveals basis for responsiveness to thalidomide analogs. Nat Struct Mol Biol 2014;21:803–9. [DOI] [PubMed] [Google Scholar]
- 39. Qin C, Hu Y, Zhou B, Fernandez-Salas E, Yang CY, Liu L, et al. Discovery of QCA570 as an exceptionally potent and efficacious proteolysis targeting chimera (PROTAC) degrader of the bromodomain and extra-terminal (BET) proteins capable of inducing complete and durable tumor regression. J Med Chem 2018;61:6685–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Albrecht BK, Gehling VS, Hewitt MC, Vaswani RG, Cote A, Leblanc Y, et al. Identification of a benzoisoxazoloazepine inhibitor (CPI-0610) of the bromodomain and extra-terminal (BET) family as a candidate for human clinical trials. J Med Chem 2016;59:1330–9. [DOI] [PubMed] [Google Scholar]
- 41. Janouskova H, El Tekle G, Bellini E, Udeshi ND, Rinaldi A, Ulbricht A, et al. Opposing effects of cancer-type-specific SPOP mutants on BET protein degradation and sensitivity to BET inhibitors. Nat Med 2017;23:1046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang P, Wang D, Zhao Y, Ren S, Gao K, Ye Z, et al. Intrinsic BET inhibitor resistance in SPOP-mutated prostate cancer is mediated by BET protein stabilization and AKT-mTORC1 activation. Nat Med 2017;23:1055–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol Cell 2009;36:541–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Devaiah BN, Lewis BA, Cherman N, Hewitt MC, Albrecht BK, Robey PG, et al. BRD4 is an atypical kinase that phosphorylates serine2 of the RNA polymerase II carboxy-terminal domain. Proc Natl Acad Sci U S A 2012;109:6927–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, et al. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 2011;478:524–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu Y, Vakoc CR. Targeting cancer cells with BET bromodomain inhibitors. cold spring harb perspect med 2017;7:a026674 doi 10.1101/cshperspect.a026674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yang CY, Qin C, Bai L, Wang S. Small-molecule PROTAC degraders of the bromodomain and extra terminal (BET) proteins - a review. Drug Discov Today Technol 2019;31:43–51. [DOI] [PubMed] [Google Scholar]
- 48. He L, Chen C, Gao G, Xu K, Ma Z. ARV-825-induced BRD4 protein degradation as a therapy for thyroid carcinoma. Aging (Albany NY) 2020;12:4547–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhou B, Hu J, Xu F, Chen Z, Bai L, Fernandez-Salas E, et al. Discovery of a small-molecule degrader of bromodomain and extra-terminal (BET) proteins with picomolar cellular potencies and capable of achieving tumor regression. J Med Chem 2018;61:462–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Deneka AY, Boumber Y, Beck T, Golemis EA. Tumor-targeted drug conjugates as an emerging novel therapeutic approach in small cell lung cancer (SCLC). Cancers 2019;11:1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Parsons HM, Harlan LC, Stevens JL, Ullmann CD. Treatment of small cell lung cancer in academic and community settings: factors associated with receiving standard therapy and survival. Cancer journal 2014;20:97–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kalemkerian GP, Loo BW, Akerley W, Attia A, Bassetti M, Boumber Y, et al. NCCN guidelines insights: small cell lung cancer, version 2.2018. J Natl Compr Canc Netw 2018;16:1171–82. [DOI] [PubMed] [Google Scholar]
- 53. Horn L, Mansfield AS, Szczesna A, Havel L, Krzakowski M, Hochmair MJ, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med 2018;379:2220–9. [DOI] [PubMed] [Google Scholar]
- 54. Armstrong SA, Liu SV. Dashing decades of defeat: long anticipated advances in the first-line treatment of extensive-stage small cell lung cancer. Curr Oncol Rep 2020;22:20. [DOI] [PubMed] [Google Scholar]