Abstract
Pancreatic ductal adenocarcinoma (PDAC) tumors can originate either from acinar or ductal cells in the adult pancreas. We re-analyze multiple pancreas and PDAC single-cell RNA-seq datasets and find a subset of non-malignant acinar cells, which we refer to as acinar edge (AE) cells, whose transcriptomes highly diverge from a typical acinar cell in each dataset. Genes up-regulated among AE cells are enriched for transcriptomic signatures of pancreatic progenitors, acinar dedifferentiation, and several oncogenic programs. AE-upregulated genes are up-regulated in human PDAC tumors, and consistently, their promoters are hypo-methylated. High expression of these genes is associated with poor patient survival. The fraction of AE-like cells increases with age in healthy pancreatic tissue, which is not explained by clonal mutations, thus pointing to a non-genetic source of variation. The fraction of AE-like cells is also significantly higher in human pancreatitis samples. Finally, we find edge-like states in lung, liver, prostate, and colon tissues, suggesting that sub-populations of healthy cells across tissues can exist in pre-neoplastic states.
Introduction
Pancreatic ductal adenocarcinoma (PDAC) is one of the deadliest cancers with ~8% survival rate at 5 years(1). Pathogenesis of PDAC, and in particular, the cell of origin for PDAC, is not yet fully resolved, thus impeding development of robust therapies. Recent work has demonstrated that in mice, PDAC tumors can be driven from both acinar and ductal cells(2), where an acinar to PDAC transformation is mediated by acinar-ductal metaplasia (ADM)(3).
A classical view of cancer posits that oncogenesis is mediated by a series of somatic mutations in key oncogenes and tumor suppressors, accompanied by clonal selection(4,5). While this clonal genetic model is widely accepted as one of the dominant pathways to oncogenesis, epigenetic alterations also play a key role. Indeed, transcriptional and epigenetic heterogeneity in the progenitor cell population forms the basis for later malignant transformation(6), where such heterogeneity has been shown to be crucial for pre-malignant pancreatic lesions to progress to PDAC(7,8). Furthermore, in a clonal cellular population, pervasive transcriptional fluctuation, in conjunction with complex regulatory networks, can result in a distinct meta-stable cellular states(9–11). For instance, in a clonal population of blood progenitors, high SCA1-expressing outlier cells preferentially commit to the myeloid lineage, whereas cells with low SCA1 expression commit to proerythrocytes(10). Taken together, this suggests a potential non-genetic basis for early stages of tumorigenesis, driven by transcription fluctuation across clonal cells resulting in a distinct cell state primed for malignant transformation in the favorable environment(11). Oncogenic mutations can further amplify this non-genetic heterogeneity, as seen in breast epithelial cell cultures where oncogenic mutations increase the rate of switching between non-stem and stem-like epithelial cells(12). An interplay between genetic and epigenetic alterations is likely to underlie complete malignant transformation(13).
In this work, we investigated the potential role of transcriptional heterogeneity in pancreatic epithelial cells in priming PDAC. We analyzed a published single-cell transcriptomic dataset comprising 57,730 cells from 24 PDAC tumors and 11 pancreas samples from patients having non-PDAC indications(14). We found that non-neoplastic acinar cells contained a sub-population, which we refer to as edge cells (following the terminology in Li et. al(15)), whose transcriptomes diverge from the average acinar cell and show features of pre-malignancy. In particular, genes that are differentially up-regulated among the acinar edge cells are enriched for transcriptomic signatures of pancreatic progenitors and acinar dedifferentiation, as well as several oncogenic programs such as Kras signaling, fatty acid metabolism, and epithelial-mesenchymal transition (EMT). Furthermore, in human PDAC tumors, the genes up-regulated in acinar edge cells are up-regulated and consistently, their promoters are hypo-methylated. Higher expression of these genes also associates with PDAC patient survival. This suggests potential clinical relevance of these early malignancy priming events in acinar cells. Finally, we validate the existence of acinar edge cells in additional independent pancreatic datasets and additionally find that the fraction of edge-like cells increases with age in healthy pancreatic tissue, thus providing a potential mechanism linking the known increase of PDAC incidence with age(1). Intriguingly, we see strong functional similarity between transcriptional drift from non-edge to edge acinar cells and those previously reported in healthy to pre-malignant lung transformation(16), suggesting that our observations in PDAC may possibly be more general. Indeed, we found edge-like cells to be significantly more prevalent in human pancreatitis samples, and furthermore, beyond the pancreas, we found edge-like states among epithelial cells in non-neoplastic lung, liver, prostate and colon tissues.
Overall, our work suggests that transcriptional heterogeneity among non-malignant epithelial cells may be large enough for a fraction to exist in a dedifferentiated, pre-neoplastic state. Since genes up-regulated in this pre-malignant state also increased in expression with age, this may help explain the higher incidence rate of tumors with age in these tissues, in addition to other putative mechanisms associated with the increase in cancer risk with aging(17).
Materials and Methods
The code necessary for reproducing these results are available at https://github.com/hannenhalli-lab/pdac_edge. Details of data downloading and processing procedures are described in Supplementary Methods.
Two-stage statistical test for an edge sub-population.
Our procedure for testing whether a non-malignant cell cluster harbors an edge sub-population consisted of two tests --- the skewness and the proximity tests.
Heterogeneity test:
We selected the 1000 most variable genes (using Seurat’s default FindVariableFeatures function) in the non-malignant cell cluster, z-score normalized their expression, and computed a 50-dimensional PC embedding for each cell; we refer to these PCs as Normal PCs to underscore that they are computed only from the non-malignant cell cluster. We then computed the distance of each cell from the cluster medoid based on Euclidean distance. The 10% of cells that are farthest from the medoid are termed outlier cells. We quantified heterogeneity as the skewness, s, of the distance distribution using the medcouple estimator from the robustbase package in R. To compute the statistical significance of s, we create 100 control cell clusters by shuffling each of the 50 Normal PC coordinates across all cells in the original cluster. For each control cluster, we compute the skewness as above, and based on a Gaussian fit of these 100 control skewness values, we estimated the empirical p-value of s. We used a p-value threshold of 0.01 to consider the cell cluster heterogeneous and proceed to the next test.
Proximity test:
Here we determine whether the outlier cells in the non-malignant cluster are significantly closer to the malignant cell cluster than non-outlier cells. We carry out PCA jointly on both malignant cells and non-malignant cells, using 1000 most highly variable genes across these cells. We refer to these PCs as Pooled PCs. We then define the malignant cell cluster’s medoid using the Euclidean distance metric, and compute the proximity ratio, R, as the ratio between the average distances of outlier cells (in the Pooled PC space) to the malignant cluster medoid to that of the non-outlier cells. A value of R < 1 implies that the outlier cells are closer to malignancy than non-outlier cells. We compute the statistical significance of R by randomly choosing 10% of the non-malignant cells as outlier cells, and re-compute R using these control outlier cells. We repeat this process 100 times, fit a Gaussian to the obtained ratios and estimate the empirical p-value of observing a value less than R. If this p-value is less than 0.01, the outliers are labelled as edge cells.
Modified three-stage statistical test for finding edge heterogeneity.
The three-stage pipeline retains the heterogeneity and proximity tests and incorporates a third collinearity test.
Heterogeneity test:
We computed 5 Normal PCs based on the 1000 most variably expressed genes within the non-malignant cluster of interest. Using each PC individually, as above, we defined the medoid cell, computed the distance of each cell from the medoid, followed by skewness of the distance distribution, s, and its significance based on shuffling the expression separately amongst cells in each sample (this sample-aware shuffling removes any potential bias caused by inter-sample heterogeneity). The p-values of s computed for all 5 PCs are corrected using the Benjamini-Hochberg FDR procedure. Each Normal PC with an FDR < 0.1 is chosen to define outlier cells, i.e., 10% of cells farthest from the cluster medoid.
Proximity test:
5 Pooled PCs are computed based on the 1000 most variably expressed genes across the pooled non-malignant and malignant clusters. For each outlier cell population (defined by a particular Normal PC qualifying the Heterogeneity test), the proximity ratio of the outlier cells, R, and its p-value, is computed separately for each pooled PC as above. The FDR is then computed for each pooled PC, and the set P of all Pooled PCs with an FDR < 0.1 are retained.
Collinearity test:
We compute a 5×5 correlation matrix of Spearman correlation coefficient between every Normal and Pooled PC score pair across all cells in the non-malignant cluster that qualify both heterogeneity and proximity tests. The p-values of each correlation is corrected using the FDR method. For each skewed Normal PC, if there exists at least one collinear Pooled PC with a low proximity ratio (with a correlation FDR < 0.1), then the Normal PC is a direction of edge heterogeneity.
Gene set enrichment comparison to Mascaux et. al.
We divided the acinar cells into three bins based on their distances from the acinar medoid in PC space. We z-scored the normalized expression of each gene across all acinar cells and picked genes that increased in z-score by at least 0.1 between adjacent bins. We carried out a Fisher test for over-representation for 64 gene sets (50 Hallmark gene sets and 14 CancerSEA gene sets), after which we carried out an FDR correction and picked gene sets with a q-value < 0.1 as significant.
Motif enrichment and network analysis.
To find a list of motifs enriched near acinar-expressed genes, we used the SPRY-SARUS motif scanner(18) to scan the central 100 bp region of ATAC-seq peaks for matches to motifs in the JASPAR 2020 vertebrate motif collection(19). Out of 746 motifs, we restricted our scans to 589 motifs that involved a TF that was expressed in at least 10% of all acinar cells. We split the ATAC-seq peak regions into foreground or background sets depending on whether or not the peaks were at most 10kb upstream of a gene expressed in at least one acinar cell. We scanned both sets of regions for motif matches (p-value < 10−4) and carried out a Fisher test of over-representation among the foreground sequences for each motif. We then computed q-values for each TF and retained TFs with a q-value < 0.1.
For each retained enriched TF, we created gene sets that consisted of its putative gene targets in the foreground set. We scored each gene set’s activity in each acinar cell using AUCell and used AUCell’s internal Global_k1 threshold to declare a gene set as active or inactive in each acinar cell. We then computed the fraction of acinar cells in each of the 3 bins with an active gene set, with the same cell - bin assignment that was computed in Fig. 2C.
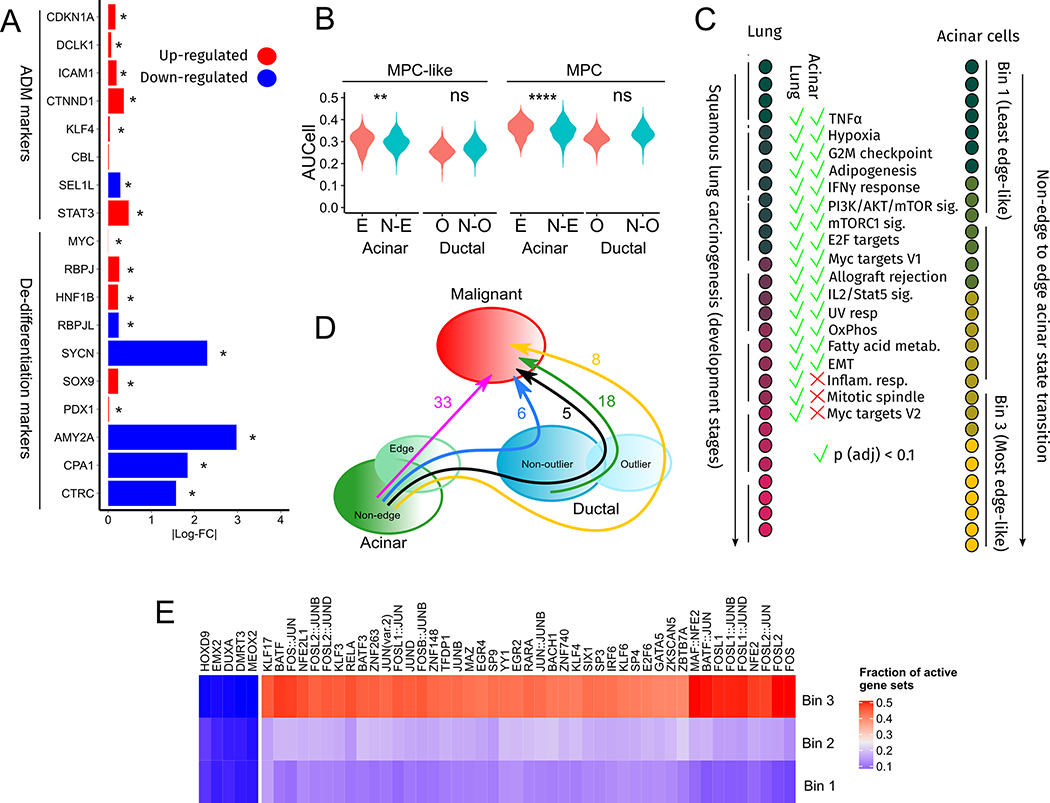
Fig. 2. Functional analysis of acinar edge cells.
(A) Bars indicate log-fold changes between edge and non-edge acinar cells. (B) The Y-axis is the gene set activity, computed by AUCell, of multi-potent-cell (MPC) and multipotent-cell-like (MPC-like) gene sets across cells in acinar and ductal cell sub-populations shown on the X-axis. **** indicates a p-value less than 10−4. (C) Gene sets enriched among genes increasing monotonically in expression during lung cancer progression (Mascaux et. al, left) and from Bin 1 to Bin 3 of the non-edge to edge acinar transition (right). (D) The number of oncogenic gene sets enriched among genes increasing in expression along the cell state transitions indicated by arrows. (E) The fraction of acinar cells in each bin that have an active regulon of the TF indicated along the columns. These are the 50 most variably activated regulons across the three bins.
Variant calling.
We called variants in acinar cells from the raw sequencing reads in GSE81547 and GSE85241 datasets using the GATK best practices workflow. We then removed variants that were (a) shared across donors, (b) were annotated in dbSNP v138, ( c ) had fewer than 5 reads aligning to the locus or had fewer than 3 reads supporting the alternate allele.
Comparing healthy tissue donors and cancer patient donors in Tosti et. al, 2021 :
Three of the samples --- TUM-13, TUM-C1 and TUM-25 --- were derived from histologically normal pancreas locations in cancer patients (Neuroendocrine tumor, PDAC, and Mixed Mullerian Tumor, respectively). To verify that samples from cancer patient did not possess higher edge gene set activity than healthy donors, we modelled edge gene set activity as a Gaussian linear mixed model with donor (random effect), sample type (healthy or tumor-adjacent, fixed effect) and cell type (random effect) as regressors. Since the effect of sample type on the edge gene set activity (coefficient = 0.0014, standard error = 0.016) was not statistically significant (t-value= 0.089), we considered cells from both cancer patients and healthy donors as normal.
Adaptive AUCell threshold computation.
When running AUCell on the datasets analyzed in Figs. 6 and 7, we found that the AUCell Global_k1 threshold, which was computed after pooling all cells in a given study, was affected by variations in library sizes between cells collected from different donors. We thus developed an adaptive thresholding strategy where an activity threshold for the edge gene set was computed separately for each donor.
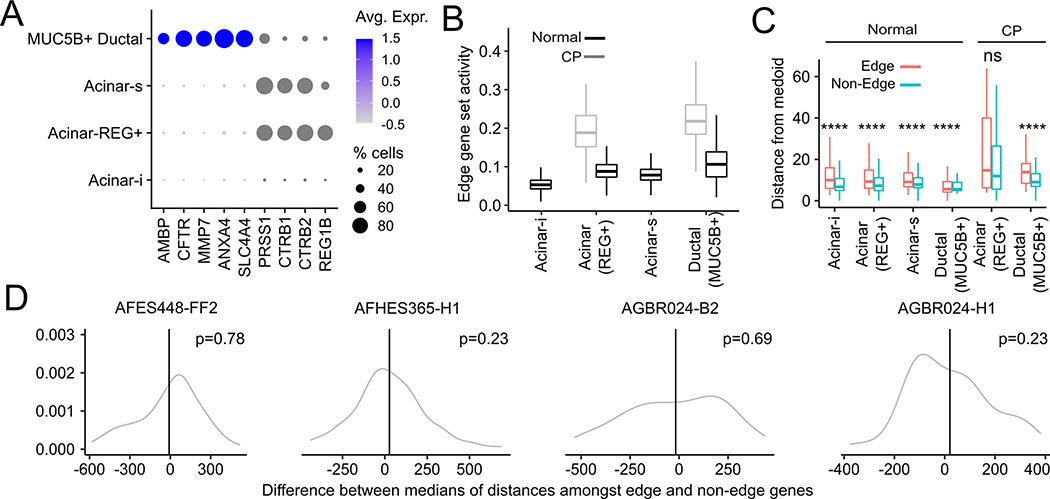
Fig. 6. Analysis of single-nucleus RNA-seq and spatial transcriptomic data from healthy and chronic pancreatitis samples.
(A) Expression of ductal marker genes (AMBP, CFTR, MMP7, ANXA4) and acinar marker genes (PRSS1, CTRB1, CTRB2, REG1B) in acinar sub-types and MUC5B+ ductal cells. (B) Activity of the edge gene set across acinar sub-types and MUC5B+ ductal cells in normal and chronic pancreatitis (CP) biopsies. There were no Acinar-i and Acinar-s cells in the CP biopsy. (C) Euclidean distance of edge and non-edge cells, identified using an independently ascertained signature, from the medoid of pooled acinar and MUC5B+ ductal cells in PCA space. (D) Distributions (gray) of the difference between the median inter-pixel distances among randomly chosen edge genes on the one hand and non-edge genes on the other in different healthy donors (donor ID shown in panel titles). Vertical black lines are the corresponding difference between median inter-pixel distances amongst actual edge and non-edge genes. One-sided p-values are computed from a Gaussian approximation to the distribution shown in gray.
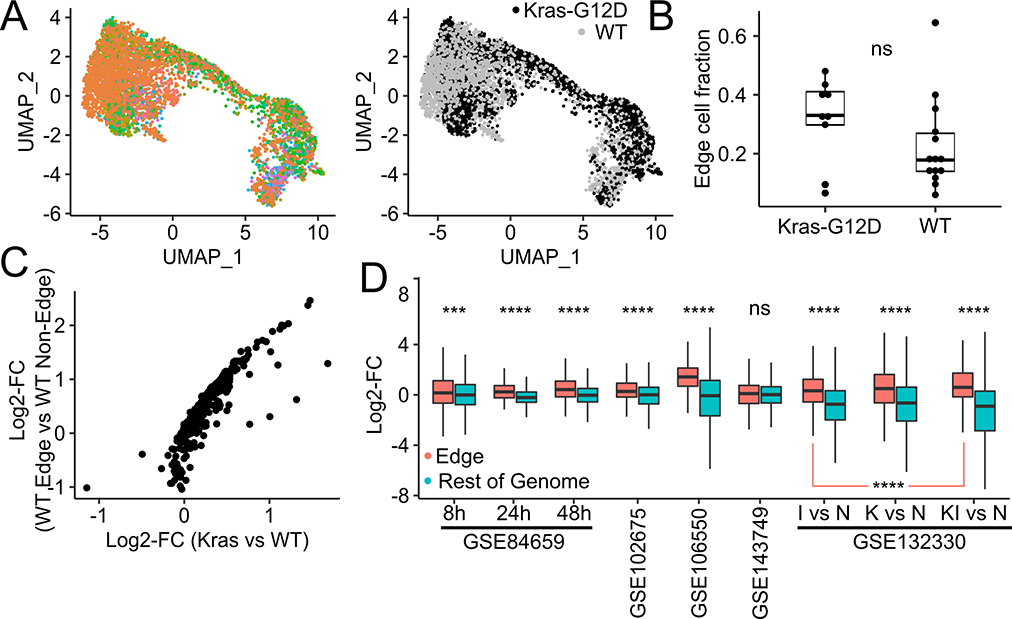
Fig. 7. Analysis of single-cell RNA-seq and bulk RNA-seq from healthy and Kras-G12D-bearing mice.
(A) UMAP plots of acinar cells from batch-integrated KrasWT and KrasG12D mice. Left : Cells are colored according to batch. Right : Cells are colored by Kras mutation status. (B) Fraction of edge cells in KrasG12D mice and KrasWT mice. (C) Average gene expression log-fold differences between acinar cells from KrasG12D mice and KrasWT mice (x-axis) and between edge and non-edge cells in KrasWT mice (y-axis). (D) Log-fold changes of expression of edge genes (red) and non-edge genes (blue) observed in published studies where RNA-seq is carried out before and after pancreatitis is induced by caerulein treatment.
For a given donor, we generated a collection of expression-controlled gene sets containing the same number of genes as the edge gene set. We first divided all expressed genes in a donor into 10 bins based on their mean normalized expression and assigned each edge gene to a bin based on its normalized expression level. For each edge gene, we then picked a gene at random from the same expression bin. The activity of the resulting control gene set was then scored using AUCell, where the 95th percentile of the AUCell scores was stored as a putative activity threshold. This process was repeated 100 times, with the largest putative activity threshold chosen as the final edge gene set activity threshold. Any cell with a AUCell score higher than this threshold was considered to be an edge cell.
Survival analysis of TCGA cancer patients.
For each cancer type investigated here, we obtained the mRNA expression (in TPM units) and clinical data for TCGA cancer patients from UCSC-xena browser (https://xena.ucsc.edu/). We used Cox regression to model the overall survival of patients by using the median expression of each signature gene set (y-axis of Fig. 3C) as an explanatory variable. Additionally, we used the age of patients as covariate and stratified the model based on their gender to control for these potential confounders. The resulting p-values were corrected for multiple comparisons using the FDR method and hazard ratios were plotted on log scale.
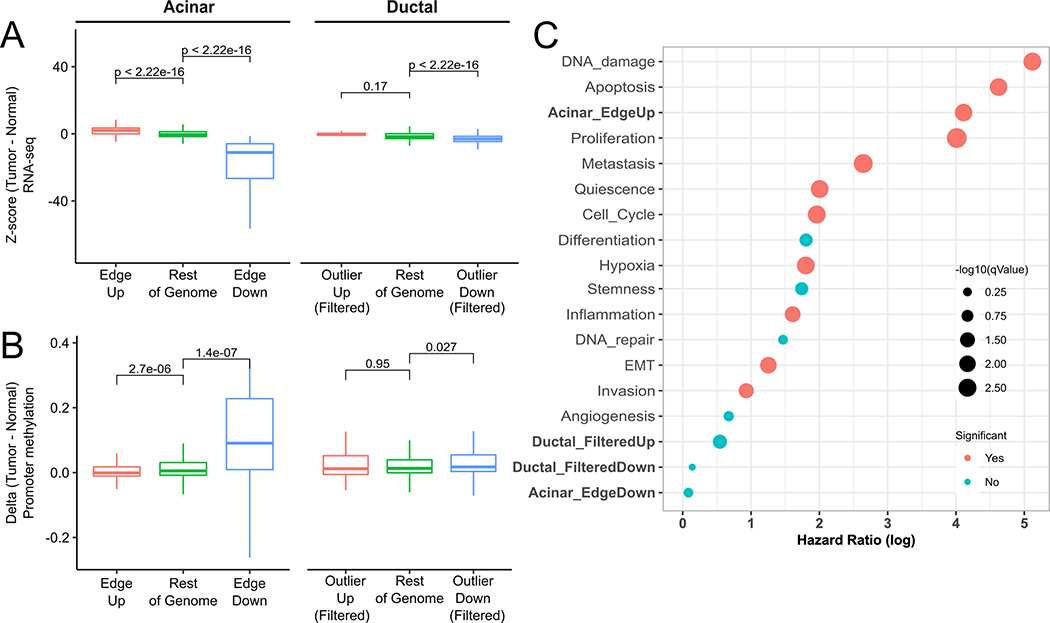
Fig. 3. Acinar Edge and Ductal Outlier genes in TCGA PDAC.
(A) RNA-seq expression z-scores in PDAC samples (using GTEx pancreas RNA-seq as a reference) of up-regulated (red), down-regulated (blue) and remaining (green) genes in edge-acinar cells and outlier-ductal cells. Genes in the Outlier-Up and Outlier-Down datasets are filtered to remove overlapping Edge-Up and Edge-Down genes. (B) Methylation z-scores among PDAC using methylation samples from healthy samples as a reference (see Methods) of gene promoters in A, (C) Log of Hazard ratios obtained from Cox regression of gene sets in TCGA PDAC samples.
Expression analysis in bulk tumor data.
For each cancer type investigated, we z-scored the expression of each gene in TCGA cancer patients based on its mean and standard deviation in normal samples of corresponding tissue from GTEx and used the averaged z-scores to compare different gene sets. Prior to z-scoring, we performed quantile normalization in order to make the two datasets comparable.
DNA methylation analysis in bulk tumor data.
We used 450k DNA methylation data of cancer and normal samples from array-expression for pancreas(20) and from GEO database for lung (GSE66836) and liver (GSE54503) samples. The coordinates of 450k methylation array probes were obtained using the COHCAP library in R and were mapped to the 5kb upstream promoter region of each gene using bedtools. We used the mean and standard deviation of aggregated methylation of each promoter in normal samples to compute the z-scores of the same in the cancer samples and plotted the averaged z-scores to compare different gene sets.
Results
Normal acinar cells include a transcriptionally divergent Edge subpopulation shifted toward a malignant state
We obtained processed gene-wise read counts from RNA-seq profiling of 57,730 pre-annotated cells across 24 PDAC and 11 non-PDAC samples(14). The non-PDAC samples were taken from the normal pancreatic sites (Table S1 in Peng et. al(14) ) of patients with other conditions: neuroendocrine tumors (n=3), solid pseudopapillary tumors (n=3), serous cystic neoplasia (n=1), mucinous cystic neoplasia (n=2), duodenal intraepithelial neoplasia (n=1) and small intestine papillary adenocarcinoma (n=1). We processed the data using Seurat v3.0(21), following which we used doubletFinder(22) to discard 2,877 potentially doublet cells, leaving us with 54,853 cells. These cells comprised 10 annotated types -- T cells, B cells, Macrophages, Stellate cells, Fibroblasts, Endothelial cells, Acinar cells, Ductal cells (Type 1 and Type 2) and Endocrine cells. A UMAP plot of the data shows that the annotated cell types are well-separated (Fig. S1). In the original annotations of the data, Ductal cell type 2 refers to malignant ductal cells, to contrast them with non-malignant ductal cells (type 1).
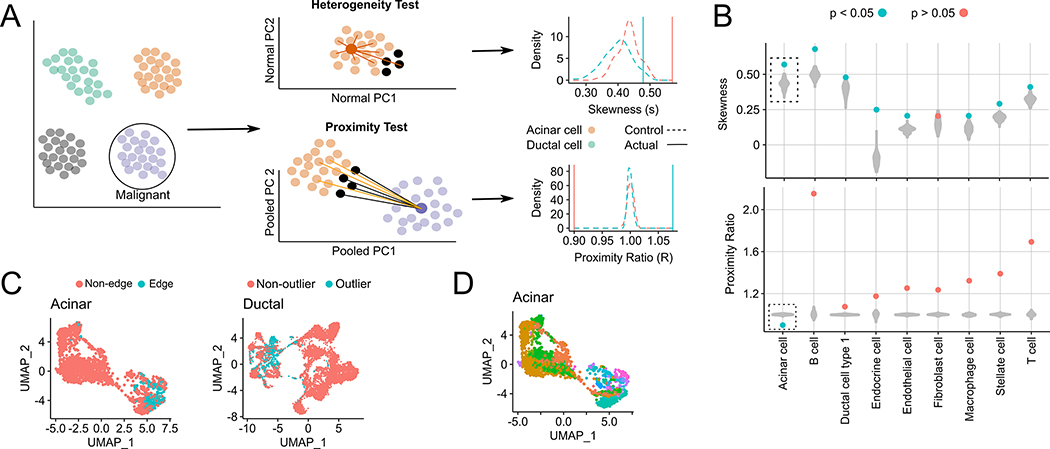
If a given non-malignant cell cluster, say X, passes the two statistical filters below, we state that X contains edge cells (Fig. 1A). The first filter – heterogeneity test – checks if a subset of cell in X have significantly diverged from X’s medoid in Principal Component (PC) space. These PCs, which we call Normal PCs, are computed based on transcriptomes only in X to capture gene expression variation within X. If X passes the filter, we consider the 10% of cells farthest from X’s medoid as candidate edge cells. The second filter – proximity test – checks if the candidate edge cells are significantly closer to the malignant cluster than the remaining cells in X. The proximity test is based on PC coordinates computed from cells in both X and the malignant cluster, which we call Pooled PCs. Technically, the heterogeneity test can also be carried out in Pooled PC space. However, since Pooled PCs also capture gene expression differences between X and the malignant cluster, they do not provide an unbiased measure of heterogeneity within X.
Fig. 1. Testing the presence of an edge sub-population among non-malignant cells in scRNA-seq data.
(A) Within each non-malignant cluster, every cell’s distance from the cluster medoid (in Normal PC space) is calculated, and the resulting distance distribution is tested for positive skewness. In the proximity test, we test if the 10% of non-malignant cells farthest from their own medoid (black, termed outlier cells) are significantly closer, in the Pooled PC space, to the malignant cluster medoid (dark purple) than the remaining 90% of cells (orange). If both test conditions hold, the outlier cells are called edge cells. For both tests, examples of the distributions of skewness and the proximity ratio are shown for acinar and ductal cells, as well as their respective control populations (B) Violin plots of medoid distance distribution skewness values (top) and malignant proximity ratio (bottom) after shuffling is performed 100 times for each indicated cluster. Filled circles indicate skewness and proximity ratio values of actual cells, where blue and red indicate a significant (< 0.05) or insignificant p-value for each test. (C) UMAP plots of edge and non-edge acinar cells (left) and non-outlier and outlier ductal cells (right). (D) UMAP plots of acinar cells colored by their sample of origin (34 samples in total, as acinar cells from one sample were discarded as they were likely doublets).
We assessed all 9 non-malignant cell types and found that only acinar cells harbored edge cells, having uniquely passed both heterogeneity and proximity tests (Fig. 1B). The existence of edge cells in the acinar population is not due to copy number alterations (CNA) as the acinar cells were shown not to harbor CNAs, in contrast to malignant ductal cells (14). The non-malignant ductal cells passed the heterogeneity test but not the proximity test, suggesting that ductal cells are highly heterogeneous but that the candidate ductal edge cells (Fig. 1C) do not significantly drift towards malignancy. For clarity, we henceforth refer to the candidate edge ductal cells as outlier ductal cells.
We performed several controls (Supplementary Fig. 2A–E and Supplementary Section S1) to ensure that the acinar edge population (Fig. 1C,D) did not arise from common artefacts related to single cell sequencing such as cell cycle, inter-donor/batch variation, the presence of tumor-adjacent cells, and library size differences. Since our computational approach bears similarities to trajectory analysis, we also assessed an analogous trajectory-based pipeline based on Monocle3(23) for detecting edge cells, where pseudotime values of cells were used for the heterogeneity and proximity tests. This alternative strategy (Supplementary Section S2), however, failed to detect edge cells.
Overall, these results reveal an edge subpopulation uniquely in non-neoplastic acinar cells that have transcriptionally drifted away from the acinar medoid and toward malignant ductal cells. In contrast, ductal cells possess an outlier ductal sub-population that drift away from the ductal medoid but do not drift towards a malignant state.
Edge acinar cells diverge from a normal acinar phenotype and represent a pre-neoplastic state
Edge acinar cells expressed PRSS1, a marker of acinar cells, at much lower levels than non-edge acinar cells (Fig. S2B, p < 10−62). To further check if edge acinar cells differentially expressed markers of dedifferentiation, we assessed the expression of genes curated by Baldan et. al(24) that are up- and down-regulated during acinar dedifferentiation. Four genes (RBPJ,HNF1B,SOX9, MYC) that are up-regulated during dedifferentiation are also up-regulated in edge acinar cells, while five genes (AMY2A,RBPJL,SYCN,CPA1,CTRC) that are down-regulated during dedifferentiation are also down-regulated in edge acinar cells (Fig. 2A). Acinar dedifferentiation precedes acinar-ductal metaplasia -- the conversion of acinar to ductal cells during pancreatic injury -- which in turn is potentially a precursor to PDAC(25). We checked expression changes of the genes STAT3, SEL1L, CBL, KLF4, CTNND1, ICAM1, DCLK1 and CDKN1A, which are known to increase in expression during acinar-to-ductal metaplasia(3). With the exception of SEL1L, all other genes were up-regulated in edge-acinar cells (Fig. 2A).
The acinar cell response during pancreatic injury has been suggested to represent a reversion to a multipotent embryonic pancreatic progenitor state(26), which, in mice embryos are marked by expression of Sox9, Ptf1a, Pdx1 and Nkx6–1(27). We found that SOX9 and PDX1 were up-regulated in edge acinar cells (Fig. 2A), although NKX6–1 showed a negligible up-regulation and PTF1A was down-regulated. Nonetheless, we checked if other genes active in pancreatic progenitors were also expressed in edge acinar cells by processing (see Supplementary Section S4 and Supplementary Fig. S2A–B) a single-cell RNA-seq dataset of human fetal (15.4 weeks gestational age) pancreatic tissue(28). We used AUCell (29) to score acinar and ductal cells for the activity of genes that were up-regulated in SOX9+PDX1+ multipotent cell (MPC)-like and SOX9+PTF1A+NKX6–1+PDX1+ MPC cells. Both gene sets were significantly more active in edge acinar cells than non-edge acinar cells (Fig. 2B), but not in outlier ductal cells when compared to non-outlier cells.
Since edge acinar cells are transcriptionally closer to malignant ductal cells than non-edge cells, we checked if the non-edge to edge transition involved known pathways of tumorigenesis. To interpolate intermediate states between non-edge and edge states, we divided acinar cells into three equal-sized bins based on their distance from the acinar cluster medoid. We tested genes monotonically increasing expression across these bins for enrichment of genes from 50 Hallmark gene sets and 14 gene sets from the CancerSEA(30) database. Out of 19,276 genes expressed in acinar cells, 3,273 genes exhibited a monotonic increase in expression from the first to the third bin and were enriched for 43 of the 64 gene sets (q-value < 0.1, Supplementary Table 1). 15 of these gene sets overlapped with gene sets enriched among genes increasing in expression across early stages of lung malignant transformation documented in Mascaux et. al (16) (Fig 2C), including genes related to Myc targets, mTOR signaling, IL2 STAT5 signaling, TNF-alpha signaling via NFKB, response to IFN-gamma, EMT, and UV response.
Next, we investigated four potential paths between non-outlier acinar to malignant cell states (Fig 2D). We identified the genes monotonically increasing in expression along each of these paths and identified enriched oncogenic pathways (Fig. S3C) among these genes. We observed most oncogenic changes (33 pathways enriched) along the path “non-edge acinar -> edge acinar -> malignant” (Fig. 2D). We contrasted this with two other paths, namely, “non-edge -> edge -> outlier ductal -> malignant” and “non-edge -> edge -> all ductal cells -> malignant”, where respectively only 8 and 5 pathways were enriched. This contrast suggests that ductal cells may not always be an intermediate transition state between edge acinar and malignant ductal cells.
Next, leveraging transcription factor (TF) motifs and acinar-specific ATAC-seq data (31), we analyzed TF activity in each of the three acinar cell bins to understand the transcriptional networks potentially driving the edge acinar state (see Supplementary Section S4). We focused on the 50 TFs whose putative target gene sets showed the most variable activity among all bins (Fig. 2E, see Supplementary Table 2 for a complete table of all 230 TFs). The RBPJ gene set showed high activity in Bin 3, which, along with the increase in RBPJ expression in edge acinar cells, provides a putative mechanistic link to the re-activation of embryonic progenitor genes in edge acinar cells(32). The activity of several KLF factors increased in Bin 3, including KLF5, whose knock-out is known to reduce proliferation in low-grade PanIN cell lines(33). HES1 activity, which maintains acinar plasticity(34), also increased from Bin 1 to Bin 3.
Thus, edge acinar cells differentially up-regulate markers of acinar dedifferentiation and acinar-ductal metaplasia, and reactivate genes expressed in embryonic pancreas progenitor cells. This is concomitant with the activation of several oncogenic processes, driven by key TFs, during transition from a non-edge to edge acinar cell state. More surprisingly, there is a substantial commonality between the processes up-regulated in transition from a non-edge to edge acinar cell state and those up-regulated during lung pre-malignant progression.
Genes up-regulated in edge acinar cells are predictive of PDAC survival
We created gene sets consisting of genes significantly up-regulated and down-regulated in edge acinar cells (Edge-Up and Edge-Down, Supplementary Table 3) and outlier ductal cells (Outlier-Up and Outlier-Down), compared to their respective non-edge and non-outlier counterparts, and analyzed their RNA-seq expression and promoter methylation in both healthy pancreatic tissues and human PDAC tumor samples. We found that Edge-Up genes were up-regulated, while Edge-Down genes were down-regulated in PDAC tumors from the TCGA database, compared to healthy pancreatic tissues from the GTEx database (Fig. 3A). Consistent with gene expression, we found significant hypomethylation at promoters of Edge-Up and hypermethylation of Edge-Down gene promoters in PDAC samples (Fig. 3B). This suggests that gene expression and methylation changes in acinar edge cells foreshadow changes in PDAC tumors in a consistent manner.
When we repeat these analyses for ductal Outlier-Up and Outlier-Down gene sets, counterintuitively (since outlier ductal cells do not exhibit a drift towards malignancy), we found a similar trend as for ductal cells, where Outlier-Up genes were up-regulated while Outlier-Down genes were down-regulated in PDAC tumors (Fig. S3E), and Outlier-Up gene promoters were hypomethylated (Fig. 3B), though Outlier-Down gene promoters were not hypermethylated. We scrutinized these counter-intuitive observations and found that this is likely because over half the Outlier-Up genes were also Edge-Up genes, with only 8 Outlier-Up (and 177 Outlier-Down genes) being ductal-specific in their expression pattern. Removal of these overlapping genes eliminates these trends in RNA-seq and methylation patterns (Fig. 3A,B).
We further assessed, using a Cox proportional-hazards model, whether the four gene sets’ activity in PDAC tumors are associated with patient survival. As shown in Fig. 3C and Fig. S3E, both Edge-Up and Outlier-Up gene sets have a significant hazard ratio (q-value < 0.1), but Edge-Up gene set has a higher hazard ratio than Outlier-Up genes. Notably, neither Edge-Down nor Outlier-Down gene sets are significantly associated with survival. As above, repeating the survival analysis based on ductal-specific Outlier-Up genes does not show significant association with survival. We also performed Cox regression for oncogenic gene sets in CancerSEA and found that a majority of these sets were predictive of survival, albeit with a lower hazard ratio than the Edge-Up gene set.
These results suggest that the genes increasing in expression in the edge-acinar state were key to tumor progression and are in line with our findings (Fig. 2C) that several oncogenic processes are enriched only among genes increasing in expression during the non-edge to edge transformation.
Acinar edge cells are found in independent healthy pancreas samples
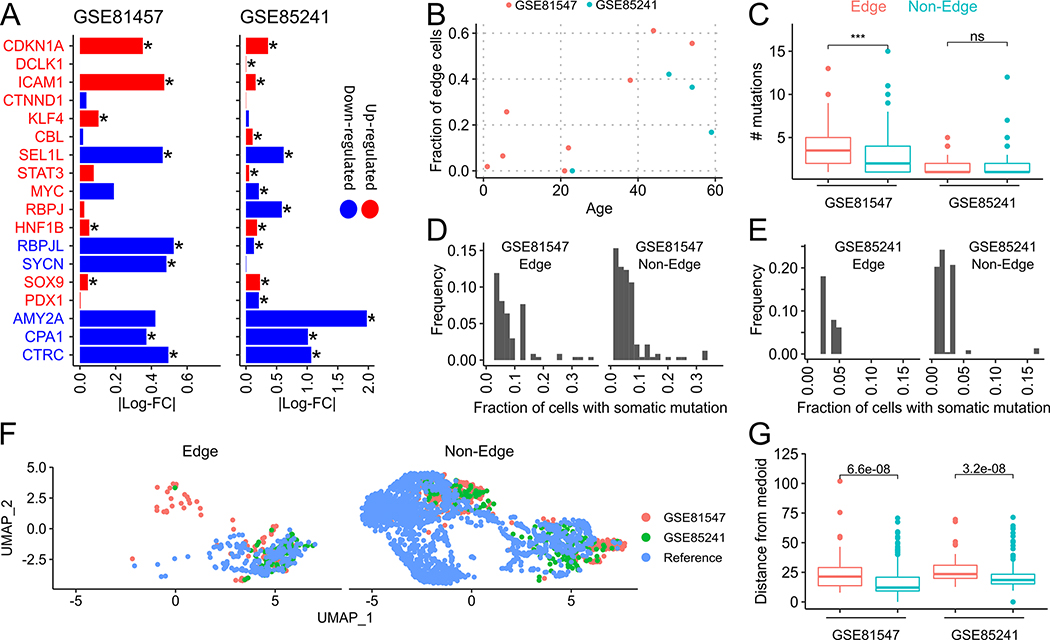
We checked if edge states can be found among acinar cells in other published single-cell datasets of human pancreatic tissues. We re-analyzed published SMART-seq(27) (GSE81547) and CEL-seq(35) (GSE85241) single-cell RNA-seq datasets of healthy human pancreas samples from donors spanning four decades of age. We removed genes that showed an age-associated increase in expression from our edge signature (see Supplementary Section S4) and used AUCell to score edge gene set activity in both datasets separately. Cells were declared as edge or non-edge based on the Global_k1 activity threshold computed by AUCell. First, similar to Fig. 2A, we compared the log-fold changes of acinar-ductal metaplasia and acinar dedifferentiation markers between edge and non-edge acinar cells (Fig. 4A). In GSE81547, all 9 dedifferentiation markers, and 6 out of 9 ADM markers, showed consistent fold-changes with edge acinar cells. In GSE85241, 6 out of 9 dedifferentiation markers, and 4 out of 9 ADM markers, showed consistent fold-changes with edge acinar cells.
Fig. 4. Acinar edge cells in independent datasets and links with aging.
(A) Bars indicate log-fold changes between edge and non-edge acinar cells in GSE81547 and GSE85241. Markers in red and blue fonts are known to be up-regulated and down-regulated, respectively, during ADM (CDKN1A to STAT3) and dedifferentiation (MYC to CTRC). Matching color of the marker text and the bar indicates that the observed log-fold change matches the expected gene expression change of the marker. (B) Scatter plot of fraction of edge-acinar cells with tissue donor age. (C) Number of mutations in edge and non-edge acinar cells (D,E) Histogram of the fraction of edge and non-edge cells that contain a somatic mutation. (F) UMAP of acinar cells from GSE81547, GSE85241 and the reference dataset (Peng et. al) (G) Distance of edge (red) and non-edge (cyan) cells from the medoid acinar cell in PCA space computed separately for each dataset.
Consistent with PDAC risk increases with age, we found an age-dependent increase in the fraction of edge cells (R2 = 0.66, p = 0.02) across both datasets (Fig. 4B). As tissues accumulate somatic mutations during aging, we assessed whether edge cells possessed somatic, especially oncogenic, mutations, using the GATK pipeline (see Methods). The number of somatic mutations in these cells agreed with estimates of somatic mutations rates in pancreas tissue in GTEx data(36). We found that edge acinar cells had more somatic mutations than non-edge acinar cells in GSE81547 but not in GSE85241 (Fig. 4C). The differences between both datasets likely stem from differences in their library sizes, with GSE81547 being sequenced to a much higher depth(37). Nonetheless, in both datasets, all these mutations were rare, and were present, on average, in 2.18% and 3.47% of non-edge and edge cells in GSE85241, and in 6.34% and 8.53% of edge cells, respectively in GSE81547 (Figures 4D,E), which does not support a clonal origin for edge cells. This modest difference in mutation frequency between edge and non-edge cells was not significant based on sampling that preserves the number of edge and non-edge cells in each sample. Further, none of the mutations in edge and non-edge cells were classified as oncogenic driver mutations in the COSMIC cancer gene census (v92)(38).
We compared the edge cells from these two datasets with the edge cells found in our reference dataset. After batch-correction, edge and non-edge cells overlapped each other in UMAP space (Fig. 4F), and edge cells in GSE81547 and GSE85241 were significantly farther from their medoid than non-edge cells (Fig. 4G). Thus, the edge states in each of these datasets are similar and represent a transcriptional drift away from the normal acinar state in each of them.
These findings validate the existence of edge-like acinar subpopulation cells in additional datasets, where they consistently exhibit expression profiles of ADM and dedifferentiation markers as in the PDAC dataset. Furthermore, we observe a strong correlation between frequency of edge cells and age.
Edge-like variation in other tissues
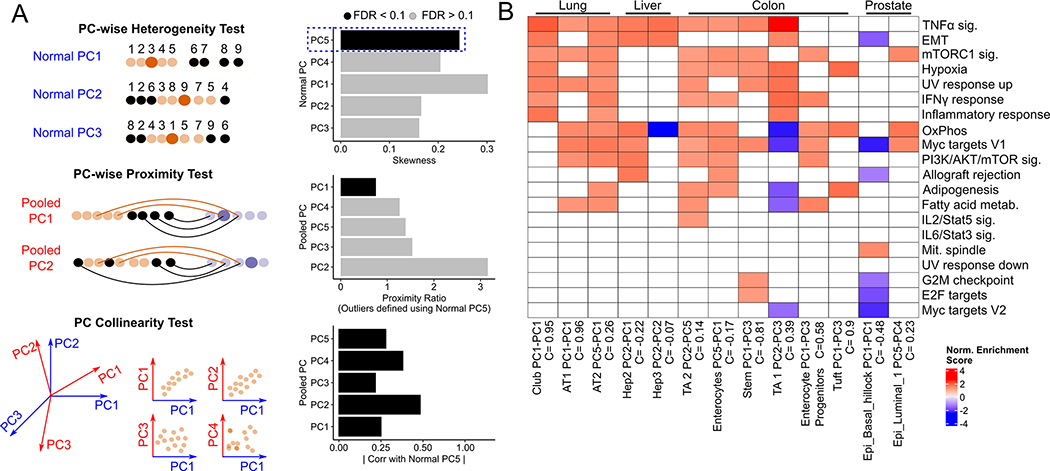
Alveolar type 2 (AT2) cells are believed to be the cell-of-origin(39) of lung adenocarcinoma (LUAD) tumors. However, application of our original pipeline on scRNA-seq data from non-malignant (AT2) and LUAD samples(40) did not detect an edge sub-population among AT2 cells, or any other non-malignant lung epithelial cluster. We then modified our original pipeline to check if any individual principal components reflected significant gene expression heterogeneity and a drift towards malignancy. Here, heterogeneity and the proximity tests are done for individual Normal and Pooled PCs respectively, and an additional test of collinearity between the qualifying Normal PC and the qualifying Pooled PCs (Fig. 5A, Methods). We note that multiple Normal PCs can show heterogeneity and drifts towards malignancy, reflecting the activation and inhibition of different gene sets in a subset of non-malignant cells. With this refined pipeline, we found that Normal PC5 of AT2 cells defined an edge population that showed a drift towards a malignant cell cluster (Tumor State 2) along Pooled PC 1, which is collinear with Normal PC5 (Correlation coefficient = 0.82, q-value < 10−9). Additionally, Normal PC1 of Club cells, and Normal PCs 1 and 2 of AT1 cells, also represented drifts towards malignancy.
Fig. 5. Edge heterogeneity in epithelial cells of lung, liver, prostate and intestine.
(A) Schematic of three-stage pipeline to detect directions of edge heterogeneity. Each Normal PC is tested for positive skewness, and each PC that passes the test is used to define an outlier cell population (in black). For each outlier population, each pooled PC is then used to compute distances between non-neoplastic and malignant cells and carry out the proximity test, with all PCs tested for collinearity with the Normal PC used to define the outlier cells. Collinearity is defined as the Spearman correlation between the Normal and Pooled PC scores of all non-neoplastic cells in the cluster. Those skewed Normal PCs that are collinear (FDR < 0.1) with a Pooled PC that passes the proximity test represent directions of edge heterogeneity within the non-neoplastic cluster. The bar plots shown are from running the three-stage test on Alveolar Type 2 (AT2) cells, where Normal PC 5 is used to define outlier cells (B) Normalized enrichment scores of gene sets (those that are active during lung cancer progression in Mascaux et. al) enriched in indicated edge-like populations. The Normal and Pooled PC pair that pass heterogeneity and proximity ratio tests, along with their collinearity scores (which have q-value < 0.1) are shown.
We then tested our pipeline on non-malignant liver(41,42)(caveats with this dataset discussed in Supplementary Section S3), colon(43,44), and prostate tissues(45) to find edge sub-populations that showed a drift towards liver hepatocellular carcinoma (LIHC), colorectal cancer (CRC), and prostate adenocarcinoma (PAAD), respectively. We found multiple clusters in each dataset that showed a drift towards malignancy in each of these tissues, including two hepatocyte clusters (Hep2 and Hep3) in the liver, transit-amplifying cells (TA1 and TA2), enterocytes, enterocyte progenitors, and intestinal stem cell clusters in the colon, and basal and luminal cells in the prostate. Genes up-regulated in the edge-like populations in these tissues were enriched for several oncogenic gene sets (Supplementary Fig. S3D). Edge-like AT2 and AT1 cells in the lung, and TA1 (Transit Amplifying) cells in the colon, were enriched (Fisher test, p = 0.042, p=0.05, and p=0.027, respectively) for gene sets active in lung cancer progression in the Mascaux et. al study (Fig 5B).
Overall, while we did not find a global transcriptomic shift toward malignancy in lung, liver, prostate and colon, our results suggest significant heterogeneity in specific oncogenic programs in multiple epithelial clusters in these tissues.
The edge acinar state is activated during chronic pancreatitis
Pancreatitis is associated with an increased PDAC risk(46). To investigate whether the increased risk is associated with the presence of edge cells, we analyzed a single-nucleus RNA-seq dataset of 120,000 cells in pancreas samples from healthy donors, chronic pancreatitis (CP) patients, and histologically normal tissues adjacent to pancreatic tumors(47). In addition to reporting three acinar cell states (Acinar-i, Acinar-s and Acinar-REG+), the study had reported a novel MUC5B+ ductal population that also expressed acinar cell markers, where 45% of these cells expressed PRSS1 (Fig. 6A). We used AUCell to compute edge gene set activity among the acinar and MUC5B+ ductal cells, and labelled cells as “edge” based on a more stringent and adaptive activity threshold (see Methods) than AUCell’s Global_k1 threshold. The detected edge population was enriched for cells from the MUC5B+ ductal cell population (Odds ratio = 4.962, p < 10−116 ), Acinar-REG+ (Odds ratio = 4.45, p < 10−170) and Acinar-s populations (Odds ratio = 5.07, p < 10−170), but not the Acinar-i population (the odds ratio was relative to the expectation based on all acinar cells). Further, relative to normal samples, acinar cells from chronic pancreatitis biopsies were over-represented in the edge population (Odds ratio = 3.83, p < 10−66), consistent with higher expression of the edge gene set in Acinar-REG+ and MUC5B+ ductal cells in chronic pancreatitis biopsies (Fig. 6B). Additionally, when we performed PCA separately for each cell type and donor type, we found that, by and large, edge cells were farther away from the medoid cell than non-edge cells (Fig. 6C).
Next, we checked if any of the edge genes were detectable in the normal pancreas in histopathology data. MMP7 was one of the highest up-regulated genes in our edge gene set (Average Log-FC = 1.87), and is known to contribute to PDAC initiation and progression(48). While MMP7 expression occurs in PDAC cells(49), multiple studies reported that a small fraction of normal pancreatic samples showed low-antibody staining for MMP7 (49–51). This observation is consistent with a small fraction of acinar cells in the healthy pancreas being in an edge state in some of the normal samples.
Finally, we evaluated the spatial localization of edge cells among spatial transcriptomic datasets assayed from a subset of healthy donors. This data was collected using the Cartana in situ sequencing platform, where pixel-wise locations of expression of each of 98 chosen genes were measured. We used the locations of ten genes --- nine from the edge gene set (LCN2,CALD1, B2M, HLA-DRA, CD74, CD3D, KRT19, REG3G), and MUC5B --- as an indicator of the location of edge cells. We reasoned that, if edge cells exist in a single cluster, the distance between a given pair of pixels expressing an edge gene would, on average, be significantly shorter than that of a non-edge gene. We checked if the median inter-pixel distance of each of the 10 edge genes was shorter than that of a random chosen set of 10 genes and found that this was not the case in any of the tissue slices (Figure 6D). However, this does not preclude the possibility that the edge cells could exist as clusters at multiple foci distributed across the pancreas.
Kras mutations induce an edge-like transcriptional state in acinar cells in mice
KRAS is the most frequently mutated oncogene in human PDAC, and is mutated in nearly all PDAC samples in TCGA(52), with the KRASG12D mutation believed to drive PDAC initiation. We checked if KrasG12D mutation-bearing acinar cells in mice are more likely to be in an edge-like state by detecting them among pooled single-cell acinar transcriptomes (Fig. 7A) from KrasWT mice (from the Tabula Muris(53) and Tabula Muris Senis(54) projects) and from neoplastic PDAC lesions in KrasG12D bearing mice in the KIC model (KrasLSL−G12D/+Ink4afl/flPtf1aCre/+, GSE125588(8)) and the PRT mouse model (Ptf1a-CreER, LSL-KrasG12D, LSL-tdTomato, GSE141017(55)). We used AUCell (with an adaptive threshold) and mouse orthologs of the human edge gene set to detect edge cells and found that KrasG12D mice contained a larger fraction of edge acinar cells than KrasWT mice, although not at a statistically significant level (W = 86, Wilcoxon one-sided test, p = 0.16, Fig. 7B). Interestingly, we found a high correlation (Spearman rho = 0.88, p < 2.2 × 10−16, Fig. 7C) between log-fold changes in gene expression between edge and non-edge cells in KrasWT mice on the one hand with log-fold changes in gene expression between KrasG12D acinar cells and KrasWT acinar cells on the other. There is thus a large concordance between gene expression programs activated by the KrasG12D mutation and those activated during a non-edge to edge transition. Finally, we analyzed published bulk RNA-seq profiles of mouse pancreatic samples before and after pancreatitis induction(56–60). With the exception of GSE143749(59), where log-fold changes were compared between pancreatic tuft and non-tuft cells, the edge gene set was more strongly up-regulated than non-edge genes in the remaining pancreatitis samples (Fig. 7D). In particular, in GSE132330(60), edge genes were more strongly up-regulated after pancreatic injury in KrasG12D mice (KI vs N) than in KrasWT mice (I vs N, p=1.07 × 10−5).
There is thus a clear concordance between gene expression changes during the non-edge to edge transition and those produced by KrasG12D induction and pancreatitis in mice.
Discussion
Here we show the existence of a subset of non-malignant acinar cells that we refer to as edge cells(15), that are transcriptionally distinct from a typical acinar cell, and significantly closer to malignant PDAC cells. This phenomenon is observed broadly across individuals and in multiple datasets. Although edge cells do not seem to be driven by clonal somatic mutations, interestingly, we see evidence of increased prevalence of edge cells with age, and consistently, an enrichment of edge-up-regulated genes among genes increasing in expression with age. Our analysis of spatial transcriptomic data suggests that edge cells likely do not exist as a single cluster within the healthy pancreas, and are potentially distributed across the pancreas, either as isolated cells or in multiple clusters.
One way to interpret the observed global transcriptional drift in acinar edge cells toward malignancy is that there are overlapping oncogenic programs that individually show heterogeneity in the non-malignant cell population and are broadly concordant with each other. Ultimately, an increased transcriptional activity along multiple oncogenic programs in a subset of cells is revealed as the edge cells by our approach. Importantly, gene expression changes during the non-edge to edge transition are similar to those induced by the KrasG12D mutation and during pancreatitis. This suggests that the edge acinar state, which can be found even in histologically normal pancreas samples, is associated with, and possibly contributes to, both pancreatitis and PDAC. Furthermore, since the gene expression differences between edge and non-edge acinar cells in mice are similar to those induced by the KrasG12D mutation, it is possible that edge cells “pre-activate” a KrasG12D induced program that leads rapidly to oncogenesis upon mutation. Our results also reveal significant heterogeneity involving several oncogenic programs in non-malignant epithelial cells of lung, liver, prostate and colon.
There is a significant overlap between pathways activated in the non-edge to edge transitions in acinar cells on the one hand, and those activated during pre-malignant progression in the lung on the other. In acinar edge cells, we see an up-regulation of the targets of transcription factors RBPJ, HES1, and KLF5 targets, which are known to mediate acinar cell plasticity and a reversion to a multi-potent pancreatic progenitor state(32,34). This suggests a role for known transcriptional networks playing a role in the transition to an edge state. In the context of regulatory networks, transcriptional fluctuations can lead to non-genetic phenotypic heterogeneity (9,10,61), which, in malignant cells, can lead to drug-resistance (62) in a manner that can be perturbed by targeting key transcription factors(63).
The duration for which a cell remains in an edge state may involve epigenetic mechanisms like DNA methylation and histone modifications (64,65). Coupled single cell transcriptomics and DNA methylation data from the same acinar cell, which is needed to precisely assess the role of DNA methylation in sustaining the edge cell population, is currently not available. However, we found that the promoters of genes that are up-regulated in the edge acinar cells relative to non-edge cells, were hypomethylated in PDAC tumors, and the converse was true for genes down-regulated in edge acinar cells, suggesting a potential role of epigenetics in maintaining the edge cell state.
A potential role of the tissue environment, and DNA methylation, in giving rise to edge cells is further supported by our observed link between age and the fraction of edge cells in healthy acinar cells. Aging is the greatest risk factor for most cancers(66). While clonal expansion of somatic mutations does occur with age in certain tissues such as skin and oesophagus(67), we found no evidence of clonal expansion in the edge acinar cells. Beyond the role of mutations, epigenetic changes from age-related hypomethylation(68) likely contribute to the stability and rate of switching to an edge state with age. Though we do not find mutations underlying the edge cells in the pancreas, the edge state may represent a state primed for malignant transformation by oncogenic mutation (13) or other age-associated transcriptomic changes (17,69,70)
Overall, our results support the notion of an edge transcriptomic state in healthy tissues that is pre-malignant. Pancreatic acinar cells likely switch between edge and non-edge states, although the time spent by cells in either state is unclear. Establishing the stability of these states would require the tracing of lineages of acinar cells to infer the regulatory changes underlying the switching process.
Supplementary Material
Statement of Significance.
We find ‘edge’ epithelial cell states with oncogenic transcriptional activity in human organs without oncogenic mutations. In the pancreas, the fraction of acinar cells increases with age.
Acknowledgments
This work utilized the computational resources of the NIH HPC Biowulf cluster and was supported by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, NIH. We would like to thank Curtis Harris, Xin Wang, Shouhui Yang and Cenk Sahinalp for discussions and feedback. We especially thank Argiris Efstratiadis for his feedback and comments on our re-analysis of fetal pancreatic single-cell RNA-seq data. We thank Arati Rajeevan for help with illustration.
Footnotes
The authors declare no potential conflicts of interest.
References
- 1.Kleeff J, Korc M, Apte M, La Vecchia C, Johnson CD, Biankin A V., et al. Pancreatic cancer. Nat Rev Dis Prim 2016; [DOI] [PubMed] [Google Scholar]
- 2.Xu Y, Liu J, Nipper M, Wang P. Ductal vs. acinar? Recent insights into identifying cell lineage of pancreatic ductal adenocarcinoma. Ann Pancreat Cancer. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storz P Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011. page 646–74. [DOI] [PubMed] [Google Scholar]
- 5.Nowell PC. The clonal evolution of tumor cell populations. Science (80- ). 1976; [DOI] [PubMed] [Google Scholar]
- 6.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet 2006. [DOI] [PubMed] [Google Scholar]
- 7.Bernard V, Semaan A, Huang J, Lucas FAS, Mulu F, Stephens B, et al. Single Cell Transcriptomics of Pancreatic Cancer Precursors Demonstrates Epithelial and Microenvironmental Heterogeneity as an Early Event in Neoplastic Progression. bioRxiv. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hosein AN, Huang H, Wang Z, Parmar K, Du W, Huang J, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thattai M, Van Oudenaarden A. Intrinsic noise in gene regulatory networks. Proc Natl Acad Sci U S A. 2001; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Transcriptome-wide noise controls lineage choice in mammalian progenitor cells. Nature. 2008; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brock A, Chang H, Huang S. Non-genetic heterogeneity a mutation-independent driving force for the somatic evolution of tumours. Nat. Rev. Genet 2009. [DOI] [PubMed] [Google Scholar]
- 12.Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci U S A 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaz M, Hwang SY, Kagiampakis I, Phallen J, Patil A, O’Hagan HM, et al. Chronic Cigarette Smoke-Induced Epigenomic Changes Precede Sensitization of Bronchial Epithelial Cells to Single-Step Transformation by KRAS Mutations. Cancer Cell. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peng J, Sun BF, Chen CY, Zhou JY, Chen YS, Chen H, et al. Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q, Wennborg A, Aurell E, Dekel E, Zou JZ, Xu Y, et al. Dynamics inside the cancer cell attractor reveal cell heterogeneity, limits of stability, and escape. Proc Natl Acad Sci U S A 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mascaux C, Angelova M, Vasaturo A, Beane J, Hijazi K, Anthoine G, et al. Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature. Nature Publishing Group; 2019;571:570–5. [DOI] [PubMed] [Google Scholar]
- 17.Aunan JR, Cho WC, Søreide K. The biology of aging and cancer: A brief overview of shared and divergent molecular hallmarks. Aging Dis. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kulakovskiy I, Levitsky V, Oshchepkov D, Bryzgalov L, Vorontsov I, Makeev V. From binding motifs in chip-seq data to improved models of transcription factor binding sites. J Bioinform Comput Biol 2013. [DOI] [PubMed] [Google Scholar]
- 19.Fornes O, Castro-Mondragon JA, Khan A, Van Der Lee R, Zhang X, Richmond PA, et al. JASPAR 2020: Update of the open-Access database of transcription factor binding profiles. Nucleic Acids Res 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer AS, Nazarov PV., Giese NA, Beghelli S, Heller A, Greenhalf W, et al. Transcriptional variations in the wider peritumoral tissue environment of pancreatic cancer. Int J Cancer. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, et al. Comprehensive Integration of Single-Cell Data. Cell. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGinnis CS, Murrow LM, Gartner ZJ. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Syst. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, et al. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Baldan J, Houbracken I, Rooman I, Bouwens L. Adult human pancreatic acinar cells dedifferentiate into an embryonic progenitor-like state in 3D suspension culture. Sci Rep. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Puri S, Folias AE, Hebrok M. Plasticity and dedifferentiation within the pancreas: Development, homeostasis, and disease. Cell Stem Cell. 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan FC, Bankaitis ED, Boyer D, Xu X, Van de Casteele M, Magnuson MA, et al. Spatiotemporal patterns of multipotentiality in Ptf1aexpressing cells during pancreas organogenesis and injuryinduced facultative restoration. Dev. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Enge M, Arda HE, Mignardi M, Beausang J, Bottino R, Kim SK, et al. Single-Cell Analysis of Human Pancreas Reveals Transcriptional Signatures of Aging and Somatic Mutation Patterns. Cell. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villani V, Thornton ME, Zook HN, Crook CJ, Grubbs BH, Orlando G, et al. SOX9+/PTF1A+ Cells Define the Tip Progenitor Cells of the Human Fetal Pancreas of the Second Trimester. Stem Cells Transl Med. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan H, Yan M, Zhang G, Liu W, Deng C, Liao G, et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arda HE, Tsai J, Rosli YR, Giresi P, Bottino R, Greenleaf WJ, et al. A Chromatin Basis for Cell Lineage and Disease Risk in the Human Pancreas. Cell Syst. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arda HE, Benitez CM, Kim SK. Gene regulatory networks governing pancreas development. Dev. Cell 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He P, Yang JW, Yang VW, Bialkowska AB. Krüppel-like Factor 5, Increased in Pancreatic Ductal Adenocarcinoma, Promotes Proliferation, Acinar-to-Ductal Metaplasia, Pancreatic Intraepithelial Neoplasia, and Tumor Growth in Mice. Gastroenterology. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hidalgo-Sastre A, Brodylo RL, Lubeseder-Martellato C, Sipos B, Steiger K, Lee M, et al. Hes1 Controls Exocrine Cell Plasticity and Restricts Development of Pancreatic Ductal Adenocarcinoma in a Mouse Model. Am J Pathol. Elsevier Inc.; 2016;186:2934–44. [DOI] [PubMed] [Google Scholar]
- 35.Muraro MJ, Dharmadhikari G, Grün D, Groen N, Dielen T, Jansen E, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yizhak K, Aguet F, Kim J, Hess JM, Kübler K, Grimsby J, et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science (80- ). 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu F, Zhang Y, Zhang L, Li Z, Fang Q, Gao R, et al. Systematic comparative analysis of single-nucleotide variant detection methods from single-cell RNA sequencing data. Genome Biol. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sondka Z, Bamford S, Cole CG, Ward SA, Dunham I, Forbes SA. The COSMIC Cancer Gene Census: describing genetic dysfunction across all human cancers. Nat. Rev. Cancer. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sutherland KD, Song JY, Kwon MC, Proost N, Zevenhoven J, Berns A. Multiple cells-of-origin of mutant K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci U S A. 2014; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim N, Kim HK, Lee K, Hong Y, Cho JH, Choi JW, et al. Single-cell RNA sequencing demonstrates the molecular and cellular reprogramming of metastatic lung adenocarcinoma. Nat Commun. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma L, Hernandez MO, Zhao Y, Mehta M, Tran B, Kelly M, et al. Tumor Cell Biodiversity Drives Microenvironmental Reprogramming in Liver Cancer. Cancer Cell. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smillie CS, Biton M, Ordovas-Montanes J, Sullivan KM, Burgin G, Graham DB, et al. Intra- and Inter-cellular Rewiring of the Human Colon during Ulcerative Colitis. Cell. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qian J, Olbrecht S, Boeckx B, Vos H, Laoui D, Etlioglu E, et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Karthaus WR, Hofree M, Choi D, Linton EL, Turkekul M, Bejnood A, et al. Regenerative potential of prostate luminal cells revealed by single-cell analysis. Science (80- ). 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tosti L, Hang Y, Debnath O, Tiesmeyer S, Trefzer T, Steiger K, et al. Single-Nucleus and In Situ RNA–Sequencing Reveal Cell Topographies in the Human Pancreas. Gastroenterology. 2021; [DOI] [PubMed] [Google Scholar]
- 48.Fukuda A, Wang SC, Morris JP, Folias AE, Liou A, Kim GE, et al. Stat3 and MMP7 Contribute to Pancreatic Ductal Adenocarcinoma Initiation and Progression. Cancer Cell. 2011; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jakubowska K, Pryczynicz A, Januszewska J, Sidorkiewicz I, Kemona A, Niewiński A, et al. Expressions of matrix metalloproteinases 2, 7, and 9 in carcinogenesis of pancreatic ductal adenocarcinoma. Dis Markers. 2016; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuhlmann KFD, Van Till JWO, Boermeester MA, De Reuver PR, Tzvetanova ID, Offerhaus GJA, et al. Evaluation of matrix metalloproteinase 7 in plasma and pancreatic juice as a biomarker for pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2007; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones LE, Humphreys MJ, Campbell F, Neoptolemos JP, Boyd MT. Comprehensive Analysis of Matrix Metalloproteinase and Tissue Inhibitor Expression in Pancreatic Cancer: Increased Expression of Matrix Metalloproteinase-7 Predicts Poor Survival. Clin Cancer Res. 2004; [DOI] [PubMed] [Google Scholar]
- 52.Waters AM, Der CJ. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb Perspect Med. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schaum N, Karkanias J, Neff NF, May AP, Quake SR, Wyss-Coray T, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Almanzar N, Antony J, Baghel AS, Bakerman I, Bansal I, Barres BA, et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schlesinger Y, Yosefov-Levi O, Kolodkin-Gal D, Granit RZ, Peters L, Kalifa R, et al. Single-cell transcriptomes of pancreatic preinvasive lesions and cancer reveal acinar metaplastic cells’ heterogeneity. Nat Commun. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cobo I, Martinelli P, Flández M, Bakiri L, Zhang M, Carrillo-De-Santa-Pau E, et al. Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature. 2018; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fazio EN, Young CC, Toma J, Levy M, Berger KR, Johnson CL, et al. Activating transcription factor 3 promotes loss of the acinar cell phenotype in response to cerulein-induced pancreatitis in mice. Bronner M, editor. Mol Biol Cell [Internet]. American Society for Cell Biology; 2017. [cited 2020 May 7];28:2347–59. Available from: 10.1091/mbc.e17-04-0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li X, Lin Z, Wang L, Liu Q, Cao Z, Huang Z, et al. RNA-Seq analyses of the role of miR-21 in acute pancreatitis. Cell Physiol Biochem. 2018; [DOI] [PubMed] [Google Scholar]
- 59.DelGiorno KE, Naeem RF, Fang L, Chung CY, Ramos C, Luhtala N, et al. Tuft Cell Formation Reflects Epithelial Plasticity in Pancreatic Injury: Implications for Modeling Human Pancreatitis. Front Physiol. 2020; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alonso-Curbelo D, Ho YJ, Burdziak C, Maag JLV, Morris JP, Chandwani R, et al. A gene–environment-induced epigenetic program initiates tumorigenesis. Nature. 2021; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Huang S Non-genetic heterogeneity of cells in development: More than just noise. Development. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Emert BL, Coté C, Torre EA, Dardani IP, Jiang CL, Jain N, et al. Variability within rare cell states enables multiple paths towards drug resistance. bioRxiv [Internet]. 2020;2020.03.18.996660. Available from: http://biorxiv.org/content/early/2020/05/12/2020.03.18.996660.abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bird A DNA methylation patterns and epigenetic memory. Genes Dev. 2002. [DOI] [PubMed] [Google Scholar]
- 65.Dodd IB, Micheelsen MA, Sneppen K, Thon G. Theoretical Analysis of Epigenetic Cell Memory by Nucleosome Modification. Cell. 2007; [DOI] [PubMed] [Google Scholar]
- 66.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020; [DOI] [PubMed] [Google Scholar]
- 67.Martincorena I Somatic mutation and clonal expansions in human tissues. Genome Med. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Horvath S DNA methylation age of human tissues and cell types. Genome Biol. 2013; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hinkal G, Parikh N, Donehower LA. Timed somatic deletion of p53 in mice reveals age-associated differences in tumor progression. PLoS One. 2009; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tao Y, Kang B, Petkovich DA, Bhandari YR, In J, Stein-O’Brien G, et al. Aging-like Spontaneous Epigenetic Silencing Facilitates Wnt Activation, Stemness, and Braf V600E -Induced Tumorigenesis. Cancer Cell. 2019; [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.