Abstract
Oncolytic vaccinia viruses have promising efficacy and safety profiles in cancer therapy. Although antitumor activity can be increased by manipulating viral genes, the relative efficacy of individual modifications has been difficult to assess without side-by-side comparisons. This study sought to compare the initial antitumor activity after intravenous administration of five vaccinia virus variants of the same Western Reserve backbone and thymidine kinase gene deletion in RIP-Tag2 transgenic mice with spontaneous pancreatic neuroendocrine tumors. Tumors had focal regions of infection at 5 days after all viruses. Natural killer (NK) cells were restricted to these sites of infection, but CD8+ T cells and tumor cell apoptosis were widespread and varied among the viruses. Antitumor activity of virus VV-A34, bearing amino acid substitution A34K151E to increase viral spreading, and virus VV-IL2v, expressing a mouse IL2 variant (mIL2v) with attenuated IL2 receptor alpha subunit binding, was similar to control virus VV-GFP. However, antitumor activity was significantly greater after virus VV-A34/IL2v, which expressed mIL2v together with A34K151E mutation and viral B18R gene deletion, and virus VV-GMCSF that expressed mouse GM-CSF. Both viruses greatly increased expression of CD8 antigens Cd8a/Cd8b1 and cytotoxicity genes granzyme A, granzyme B, Fas ligand, and perforin-1 in tumors. VV-A34/IL2v led to higher serum IL2 and greater tumor expression of death receptor ligand TRAIL, but VV-GMCSF led to higher serum GM-CSF, greater expression of leukocyte chemokines and adhesion molecules, and more neutrophil recruitment. Together, the results show that antitumor activity is similarly increased by viral expression of GM-CSF or IL2v combined with additional genetic modifications.
Introduction
Vaccinia viruses are widely studied in cancer immunotherapy because they kill tumor cells directly by oncolysis and indirectly by promoting host antitumor immunity and affecting the tumor vasculature (1–4). Vaccinia viruses are promising for treatment of poorly immunogenic tumors because their large genome can accommodate mutations and transgenes, and they target tumors after intravenous administration (1, 3, 5).
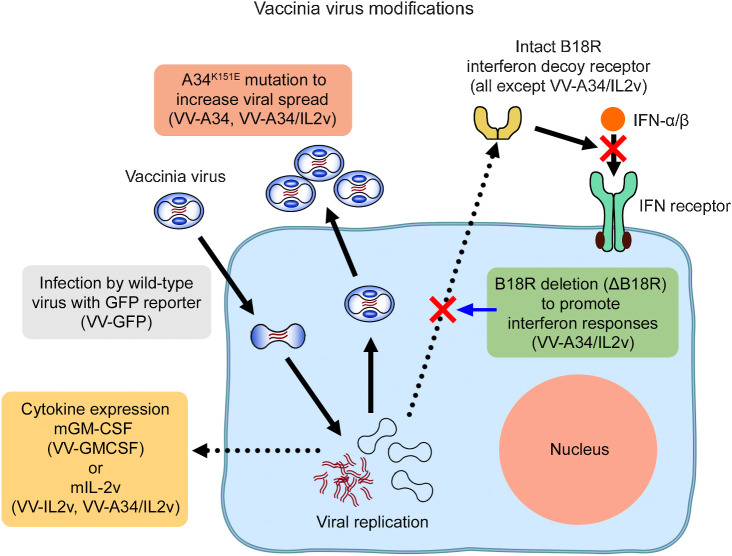
Many vaccinia virus variants have been created to boost antitumor activity. Genetic changes have been made to increase viral replication and spread in tumors and for targeted immunotherapy (6–8). Viral thymidine kinase (TK, J2R gene, VACV-WR94 locus) and vaccinia growth factor have been disrupted by insertional inactivation to favor selective viral replication in tumor cells (7, 9). Substitution of lysine151 to glutamic acid of the viral A34 protein (A34K151E substitution) can increase virus spreading and the formation of extracellular enveloped virus (EEV) in vitro and antitumor activity in vivo (6, 10, 11). Deletion of the vaccinia B18R gene, which encodes a soluble type I IFN decoy receptor that suppresses effects of host IFN (12–14), increases IFNα and IFNβ antitumor activity and specificity (14, 15). In addition to these changes, herpes simplex virus TK (HSV-TK) has been inserted into the virus genome as a selectable reporter and regulator (16, 17).
Tumor immune responses can be amplified by vaccinia virus expression of GM-CSF, IFNα, IFNβ, IL2, IL12, or other cytokines (8, 14, 15, 18–22). GM-CSF–expressing vaccinia viruses that are efficacious in preclinical tumor models are being evaluated in clinical trials (1, 5, 20, 23, 24). Treatment with GM-CSF-expressing vaccinia virus is accompanied by cytotoxic T-cell recruitment to tumors in mice (20, 24), and large increases in blood neutrophils are reported in some patients (25). Vaccinia viruses have also been combined with immune checkpoint inhibitors that block CTLA-4, programmed cell death protein 1 (PD-1), or its ligand PD-L1 (19, 20, 26, 27).
Vaccinia virus expression of IL2, to promote activation of CD8+ T-cells and NK cells and maintenance of immunosuppressive CD4+ Tregs (28, 29), has not been studied as extensively as expression of GM-CSF (18, 19, 30–33). Systemic administration of IL2 as a monotherapy is limited by toxicity (28, 29). IL2 expression by vaccinia virus administered iv has variable toxicity (18, 33). Membrane-bound forms of IL2 have greater antitumor effects and less toxicity in mice, but efficacy can decrease with larger tumor burden (19). A secreted IL2 variant, IL2v, which has attenuated binding to the IL2 receptor alpha subunit, preferentially activates effector immune cells over regulatory T cells (Tregs) with limited toxicity (34).
Few vaccinia virus modifications have been assessed for relative efficacy by side-by-side comparison. Accordingly, our goal was to determine the relative impact of viral modifications in vivo by in-depth side-by-side comparison of the initial antitumor activity of five different mutations and transgene insertions (Table 1, Fig. 1). All vaccinia virus variants were derived from the same parent Western Reserve vaccinia virus strain and had deletion of the viral TK gene. Antitumor activity of the viruses was assessed under standardized conditions on spontaneous pancreatic neuroendocrine tumors in RIP-Tag2 transgenic mice, which have the attribute of developing spontaneous, multifocal tumors that are responsive to oncolytic vaccinia viruses (24). However, long-term survival studies in these mice are difficult because the insulin-secreting tumors are accompanied by hypoglycemia with 50% mortality by 15 weeks of age (24, 35, 36).
Table 1.
Vaccinia virus genetic modifications and cytokine expression.
| Descriptive name | Original name | Genetic modifications | Cytokine expression | Reporter gene | ||
|---|---|---|---|---|---|---|
| VV-GFP | VV03 | TK inactivation | — | — | — | Luc-2A-GFP |
| VV-A34 | VV17 | TK inactivation | A34K151E | — | — | Luc-2A-GFP |
| VV-IL2v | VV39 | TK inactivation | — | — | mIL2v | — |
| VV-A34/IL2v | IGV-100 | TK inactivation | A34K151E | ΔB18R | mIL2v | HSV-TK.007 |
| VV-GMCSF | VV09 | TK inactivation | — | — | mGM-CSF | LacZ |
Genetic modifications and cytokine and reporter gene insertions of five vaccinia virus variants used to compare antitumor activity in RIP-Tag2 mice. All viruses were derived from the same WR backbone. Gene maps for the viruses are shown in Supplementary Fig. S1.
Abbreviations: -, no further change; ΔB18R, vaccinia VACV-WR200 locus deletion; HSV-TK.007, herpes simplex virus-thymidine kinase.007 reporter; LacZ, bacterial beta-galactosidase reporter; Luc-2A-GFP, luciferase 2A-green fluorescent protein dual reporter cassette; mGM-CSF, mouse granulocyte-macrophage colony- stimulating factor; mIL-2v, mouse interleukin-2 variant (34); TK, vaccinia thymidine kinase (J2R gene, VACV-WR94 locus); VV, vaccinia virus.
Figure 1.
Genetic modifications of vaccinia viruses. Diagram illustrating the mutations, deletions, and cytokine gene insertions and intended consequences in the five vaccinia virus variants. Reference virus VV-GFP had viral TK deletion by insertion of genes for GFP and firefly luciferase-2A as reporters. Virus VV-A34 was similar to VV-GFP but also had the A34K151E amino acid substitution to facilitate viral spread by increasing extracellular enveloped virus (6). Virus VV-IL2v had TK deletion by insertion of the gene for the soluble mouse IL2 variant (mIL2v; ref. 34). Virus VV-GMCSF had TK inactivated by insertion of the gene for mouse GM-CSF (mGM-CSF) and an LacZ reporter gene (37, 38). Virus VV-A34/IL2v had a combination of the A34K151E substitution, TK deletion by mIL2v gene insertion, B18R viral gene deletion (ΔB18R) to prevent expression of a decoy receptor that can diminish antitumor effects of IFNα, IFNβ, and other type I IFNs (12, 14), and HSV TK.007 gene insertion as a reporter (not shown).
Intravenous injection of the virus variants was followed by focal vaccinia infection in tumors accompanied by immune cell influx and tumor cell killing at 5 days, but CD8+ T-cell influx and apoptosis in tumors varied widely among the viruses. Virus VV-A34/IL2v, which expressed mouse IL2 variant (mIL2v) together with other modifications, and virus VV-GMCSF, which expressed mouse GM-CSF (mGM-CSF), had significantly greater antitumor activity than the other three viruses. Measurements of serum cytokine levels and tumor gene expression gave insights into why these two viruses with different genetic modifications had equally robust antitumor activity.
Methods
Design and construction of oncolytic vaccinia viruses
Vaccinia viruses with five different genome organizations (Table 1; Supplementary Fig. S1), all carrying heterologous transgenes in place of the viral TK gene (gene name J2R, locus VACV-WR94), were generated from the Western Reserve (WR) strain of vaccinia virus (American Type Culture Collection, Catalog No. VR-2056). The reference virus VV-GFP was constructed by infecting CV-1 African green monkey kidney cells (CV-1 cells) with parental WR virus and transfecting with a homologous recombination plasmid encoding a dual reporter cassette firefly luciferase foot- and -mouth disease virus 2A–enhanced GFP (Luc-2A-GFP) under the control of a synthetic early/late promoter flanked by vaccinia sequences upstream and downstream of VACV-WR94. Virus was isolated by plaque purification of GFP+ viruses, expanded in HeLa cells, and purified by sucrose cushion and gradient ultracentrifugation.
VV-A34 was constructed from VV-GFP, differing only by the presence of a mutation leading to a K151E substitution in the A34 protein (A34K151E). Parental VV-GFP DNA was transfected into Shope fibroma virus (SFV) infected Vero cells along with an A34R targeting homologous recombination DNA fragment carrying the K151E amino acid substitution. Virus was isolated by plaque purification, screened by PCR/sequencing, expanded in HeLa cells, and purified by sucrose cushion and gradient ultracentrifugation.
VV-IL2v encodes mIL2v (F76A, Y79A, and L106G; ref. 34) under the control of a synthetic early/late promoter in place of the vaccinia J2R gene. Parental DNA was transfected into SFV-infected African green monkey kidney cells (BSC40 cells) cells along with a J2R targeting homologous recombination fragment carrying the mIL2v gene. Virus was isolated by GFP(-) plaque purification, screened by PCR/sequencing, expanded in HeLa cells, and purified by sucrose cushion and gradient ultracentrifugation.
VV-GMCSF carries the mGM-CSF gene (Csf2) and the lacZ gene within the J2R region driven off the synthetic early/late promoter and 7.5 promoter, respectively, and as described for JX-594 (37, 38). VV-GMCSF was constructed by infecting BSC40 cells with parental WR virus and transfecting with a J2R targeting homologous recombination plasmid encoding the two transgenes. Virus was isolated by plaque purification of β-galactosidase+ viruses, expanded in HeLa cells, and purified by sucrose cushion and gradient ultracentrifugation.
VV-A34/IL2v carries three genetic modifications of the parental WR virus: (i) replacement of the J2R gene with the mIL2v gene controlled by a synthetic early/late promoter; (ii) mutation in the A34R gene encoding a K151E substitution; and (iii) replacement of the VACV-WR200 gene (WR B18R), designated ΔB18R or B18R viral gene deletion, with a herpes simplex virus thymidine kinase A168H variant gene (TK.007) controlled by a 7.5 promoter as a reporter. Parental VV-IL2v DNA was transfected into SFV infected Vero cells along with two homologous recombination fragments: an A34R viral gene targeting fragment carrying the K151E amino acid substitution, and VACV-WR200 targeting fragment carrying the HSV TK gene. Virus was isolated by plaque purification, screened by PCR/sequencing, expanded in HeLa cells, and purified by sucrose cushion and gradient ultracentrifugation. Influence of the A34K151E mutation (VV-A34, VV-A34/IL2v) on viral spreading in vitro was assessed by comet assay, and viral expression of IL2v (VV-IL2v, VV-A34/IL2v) and GM-CSF (VV-GMCSF) in vitro was assessed by Western blot (Supplementary Fig. S2A and S2B; details in Supplementary Methods).
Mouse tumor model and virus injection
Tumor-bearing RIP-Tag2 transgenic mice (C57BL/6 background) were bred and housed under barrier conditions in the animal care facility at the University of California, San Francisco (UCSF). All experimental procedures were approved by the UCSF Institutional Animal Care and Use Committee. Aliquots of virus were formulated to the concentration of 2 × 107 pfu/100 μL of PBS, pulse vortexed for 30 seconds, withdrawn into 100-μL syringes, and injected via a tail vein of mice at age-13 weeks, which was day 0 of experiments (24). Controls received vehicle (100-μL PBS injected i.v.; details in Supplementary Methods).
Survival of 90% of 42 RIP-Tag2 mice treated at age 13 weeks with vehicle is in scale with historical data for RIP-Tag2 mice, where only 50% live to age 15 to 16 weeks (24, 35). Safety of the viruses is reflected by survival of 100% of 39 mice that received VV-A34/IL2v and 95% of 20 mice that received VV-GMCSF over the 5-day experiment.
Tissue preparation, IHC, and measurements
On day 5 after injection of the virus, mice were anesthetized (ketamine 87 mg/kg and xylazine 13 mg/kg by i.p. injection). Tissues were preserved by perfusion of fixative [1% paraformaldehyde (PFA) in PBS] through the left cardiac ventricle and prepared for IHC staining (24). Cryostat sections 80 μm in thickness of pancreas with tumors were stained by IHC for combinations of viral antigen (vaccinia), tumor vascularity (CD31), apoptosis (activated caspase-3), tumor cells (SV40 T-antigen), CD8+ T cells (CD8 antigen), NK cells (NKp46), and neutrophils (S100A8). Fractional areas (% area density) of staining for vaccinia, CD31, and activated caspase-3 were measured in fluorescence microscopic images, and numerical densities of CD8+ cells, NKp46+ cells, and S100A8+ cells were determined (cells/mm2) in confocal microscopic images by using ImageJ (version 1.52s, http://imagej.nih.gov/ij/; details in Supplementary Methods).
Flow cytometry
Pancreatic islet tumors of RIP-Tag2 mice perfused with PBS after anesthesia and were removed, weighed, and digested in collagenase-II and -IV solution (625U/mL, Gibco) with DNase (60 U/mL, Roche), followed by erythrocyte lysis (24). Dissociated cells were counted and stained for flow cytometric assessment of CD4+, CD8+, Foxp3+, and NK1.1+ cells by FACS (BD LSR Fortessa) and analyzed with FlowJo software (8.8.6 and X; details in Supplementary Methods).
Serum cytokine analysis
IFNα, IFNβ, GM-CSF, and IL2 (including variant IL2v) in serum from blood drawn from RIP-Tag2 mice were measured by U-PLEX multiplex sandwich immunoassays (Meso Scale Discovery, Rockville, MD). IFNγ, TNFα, IL10, IL12p70, and other cytokines were measured by V-PLEX multiplex sandwich immunoassays with the Proinflammatory Panel 1 Mouse Kit (Meso Scale Discovery). Immunoassays were read by the Meso Sector S 600 plate reader (details in Supplementary Methods).
NanoString gene expression analysis
Tumors, spleen, and mesenteric lymph nodes of RIP-Tag2 mice in three treatment groups (vehicle, N = 5 mice; VV-A34/IL2v, N = 5 mice; and VV-GMCSF, N = 4 mice) were removed at 5 days, frozen, and homogenized. RNA was purified and hybridized (25–100 ng RNA). Gene expression was measured with the NanoString Immune Profiling Panel for mice (nCounter PanCancer IO 360) and analyzed using nSolver software 4.0 (NanoString Technologies). Of the 770 genes in the panel, genes in four NanoString Functional Annotation Categories relevant to the IHC readouts were analyzed in detail after removal of duplicates: Apoptosis (39 genes) and Cytotoxicity (42 genes), Cytokine and Chemokine Signaling (94 genes), and Immune Cell Adhesion and Migration subdivided into endothelial cell adhesion (28 genes) and immune cell adhesion (52 genes). Genes in each category were ranked from the greatest to least ratio of VV-A34/IL2v group value to vehicle group value and displayed as rank order plots. Genes in VV-GMCSF group tumors were then ranked in the same order as for VV-A34/IL2v group tumors. Tumor genes were also ranked and displayed from greatest to least ratio of VV-GMCSF group value to vehicle group value. Additional genes related to B cells, T cells, Tregs, NK cells, or neutrophils were also examined (details in Supplementary Methods).
Statistical analysis
Mice of both sexes matched for age were randomly assigned to groups. Group size was determined by power analysis of data from pilot studies to achieve statistical power of 0.8 and P value of 0.05. Sex was tested as a biological variable. Values are expressed as means ± SEM for each group, where the number of mice per group is shown in figure legends. Differences were assessed by Student's t test or ANOVA followed by Tukey test for multiple comparisons (Prism 6, GraphPad). NanoString gene categories were compared using Wilcoxon signed-rank tests, and individual gene differences were assessed by Student's t tests using nSolverTM 4.0 software (NanoString Technologies) or Prism. P values < 0.05 were considered significant.
Results
The experimental design made use of RIP-Tag2 mice with spontaneous tumors that are immunologically “cold” (39) and respond to intravenous administration of vaccinia viruses with peak infection, vascular pruning, CD8+ T-cell infiltration, and apoptosis at 5 days (24). These readouts were compared at this peak after i.v. injection of a standardized dose (107 pfu) of five vaccinia virus variants (Table 1; Fig. 1; Supplementary Fig. S1). The reference vaccinia virus VV-GFP had no modifications other than TK deletion. Virus VV-A34 was similar to VV-GFP but had the A34K151E substitution. Virus VV-IL2v expressed the mIL2v transgene (34). Virus VV-A34/IL2v combined elements of VV-GFP, VV-A34, and VV-IL2v and also had viral B18R gene deletion to enable interferon actions (14, 15) and HSV-TK.007 insertion as a marker gene (16). Virus VV-GMCSF expressed mGM-CSF. After screening all five viruses, the two variants with the greatest antitumor activity were further compared by measuring serum cytokines and tumor gene expression profiles.
Comparison of vaccinia infection in tumors
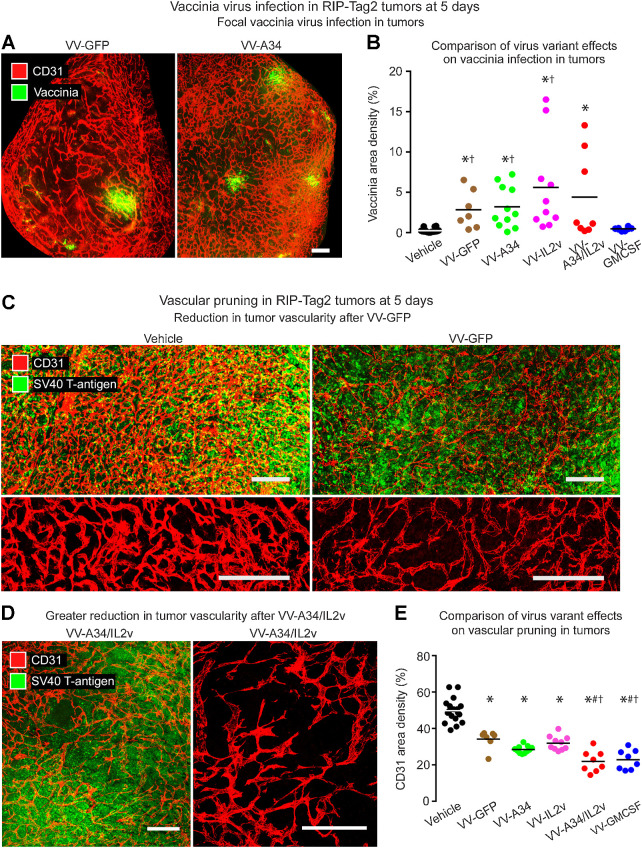
Effects of the A34K151E substitution on infection in tumors were assessed by comparing vaccinia immunoreactivity at 5 days after i.v. injection of vehicle (no virus), VV-GFP (reference virus), or VV-A34 (A34K151E virus). After either virus, focal regions of tumor cells and tumor blood vessels had vaccinia staining (Fig. 2A), which was significantly above the control level (vehicle, area density 0%) but was similar to one another, with area density values of 2.8% for VV-GFP and 3.2% for VV-A34 (Fig. 2B). Accordingly, the A34K151E substitution virus did not have significantly greater spread in tumors than the reference virus under these conditions.
Figure 2.
Staining for vaccinia virus and vasculature in RIP-Tag2 tumors 5 days after i.v. injection of vaccinia virus variants. A, Fluorescence microscopic images of staining for vaccinia (green) and blood vessels (CD31, red) after VV-GFP (left) or VV-A34 (right). B, Mean area density of vaccinia for all tumors in each mouse. Vaccinia staining after reference virus VV-GFP (2.8%) or virus VV-A34 (3.2%) was similar to one another but greater than the control (Vehicle). Values after VV-IL2v (5.6%) and VV-A34/IL2v (4.4%) tended to be greater but were not significantly different from VV-GFP. However, values for VV-GFP, VV-A34, and VV-IL2v were greater than for VV-GMCSF (0.5%). Student's t test: P < 0.05 compared with vehicle* or VV-GMCSF†. C-D, Confocal microscopic images of vasculature (CD31, red) and tumor cells (SV40 T-antigen, green). C, Dense vasculature of control tumor (Vehicle) compared with pruned vasculature after VV-GFP, shown with tumor cells (low magnification, top) and vessels alone (higher magnification, bottom). D, Greater reduction in tumor vascularity after VV-A34/IL2v, where the sparse vasculature is shown with tumor cells (left) and alone (higher magnification, right). E, Measurements revealed reduced vascularity after all viruses and significantly lower values after VV-A34/IL2v or VV-GMCSF. ANOVA: P < 0.05 compared to vehicle*, VV-GFP#, or VV-IL2v†. Mice per group: Vehicle (N = 18), VV-GFP (N = 7), VV-A34 (N = 11), VV-IL2v (N = 10), VV-A34/IL2v (N = 8), VV-GMCSF (N = 8). Scale bar, 200 μm in all images.
We then asked whether expression of mIL2v or mGM-CSF augmented the infection. The amount of vaccinia staining after VV-IL2v (5.6%) or VV-A34/IL2v (4.4%) was not significantly different from the reference virus VV-GFP (2.8%; Fig. 2B). After VV-GMCSF (0.5%), vaccinia staining was highly variable from tumor to tumor, but the low mean values for each mouse were not significantly different from the vehicle controls (Fig. 2B).
To determine whether infection was detectable outside of tumors, vaccinia staining in the exocrine pancreas was compared to staining in adjacent pancreatic islet tumors of the same mice. Mice that received vehicle (no virus) served as negative controls. No vaccinia staining greater than the control level was detected in the exocrine pancreas after any of the viruses tested (Supplementary Fig. S2C).
Comparison of vascular pruning in tumors
Tumors were significantly less vascular at 5 days after the viruses (Fig. 2C–E), consistent with reports of effects on vaccinia viruses delivered by i.v. injection (24, 40). Tumor vessels stained for CD31 (PECAM-1; refs. 41, 42) were similarly reduced after reference virus VV-GFP (38% reduction) or VV-A34 (43% reduction; Fig. 2C and E). The reductions was significantly greater after VV-A34/IL2v (56% reduction) or VV-GMCSF (54% reduction; Fig. 2D and E).
Comparison of CD4, CD8, Treg, and NK cells in tumors
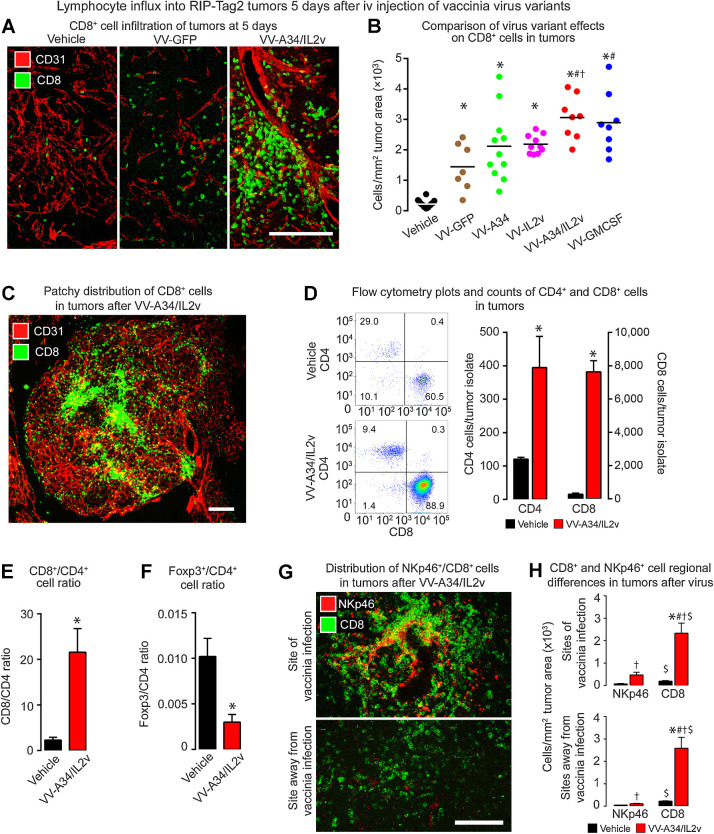
Consistent with previous evidence (24), we found that CD8+ T cells were sparse in tumors of vehicle-treated RIP-Tag2 mice but were widespread after the viruses (Fig. 3A). CD8+ cell influx after VV-A34 or VV-IL2v was about the same as after VV-GFP but was significantly greater after VV-A34/IL2v or VV-GMCSF (Fig. 3B). CD8+ cells were clustered in scattered patches in many tumors after VV-A34/IL2v (Fig. 3C).
Figure 3.
CD8+ T cells in RIP-Tag2 tumors 5 days after vaccinia virus variants. A, Confocal microscopic images show CD8+ cells (green) were sparse in the control (Vehicle), more numerous after reference virus VV-GFP, and even more abundant after virus VV-A34/IL2v. Blood vessels (CD31, red). B, CD8+ cell numerical densities show greater values after all viruses than after vehicle, and greater values after VV-A34/IL2v or VV-GMCSF than other viruses. ANOVA: P < 0.05 compared to vehicle*, VV-GFP#, or VV-IL2v†. Vehicle (N = 18), VV-GFP (N = 7), VV-A34 (N = 11), VV-IL2v (N = 10), VV-A34/IL2v (N = 8), VV-GMCSF (N = 8). C, Fluorescence microscopic image of CD8+ cell clusters (green) and blood vessels (CD31, red) after VV-A34/IL2v. D and E, Flow cytometry data compare CD4+ cells and CD8+ cells identified as the live cell fraction sorted as CD45+/CD19−/NK1.1−/CD11b−/TCRB+ cells from RIP-Tag2 tumors after vehicle or VV-A34/IL2v. D, Flow cytometry dot plots and bar graphs show increases in CD4+ cells and CD8+ cells after the virus, but CD8+ cells predominated. E, CD8+ cell/CD4+ cell ratios reflect the dominance of CD8+ cells after the virus. F, Foxp3+ cell/CD4+ cell ratios show the large decrease in proportion of Foxp3+ cells after the virus. Flow cytometry counts confirmed the small, unchanged number of Foxp3+ cells (1.3 ± 0.5 virus; 1.2 ± 0.2 vehicle) and increase in CD4+ cells (395 ± 93 virus*; 120 ± 6 vehicle) after the virus. Student's t test: P < 0.05 compared with vehicle*. N = 3 mice/group. G and H, CD8+ and NKp46+ cell distribution in tumors after vehicle or VV-A34/IL2v. G, Confocal microscopic images of CD8+ cells (green) and NKp46+ cells (red) in regions of infection (top) and without infection (bottom). H, CD8+ and NKp46+ cell numerical densities in the two regions. ANOVA: P < 0.05 compared with vehicle* (same cell type) and corresponding value for NKp46+ cells#. Student's t test: P < 0.05 compared with vehicle† and corresponding value for NKp46+ cells$. N = 5 mice/group. Scale bars, 200 μm in all images.
Flow cytometric analysis of cells dissociated from tumors confirmed that CD4+ cells and CD8+ cells, expressed as proportions of CD45+/CD19−/NK1.1−/CD11b−/TCRB+ cells, were significantly more abundant at 5 days after VV-A34/IL2v than after vehicle (Fig. 3D). However, the ratio of CD8+ cells to CD4+ cells was significantly greater after the virus (Fig. 3E). The ratio of Foxp3+ cells to CD4+ cells was much lower because Foxp3+ cells did not increase after the virus (Fig. 3F). Consistent with these data, expression of Cd4, Cd8a, and Cd8b1 increased in tumors after VV-A34/IL2v or VV-GMCSF but Foxp3 did not (Supplementary Table S1).
As oncolytic vaccinia viruses induce the influx of NK cells into tumors (29), we assessed the number and distribution of NK cells stained for NKp46 immunoreactivity. NKp46+ cells were sparse in tumors after vehicle but were numerous only in focal sites of vaccinia infection in tumors at 5 days after VV-A34/IL2v (Fig. 3G and H). This distinctive distribution of NKp46+ cells contrasted with CD8+ T cells, which were widely distributed and much more abundant in the same tumors (Fig. 3G and H). Corresponding flow cytometry data confirmed that the number of NK1.1+ cells in tumors increased after VV-A34/IL2v, but the increase was significantly less than for CD8+ cells (Supplementary Fig. S2D). The data are also consistent with the lack of significant change in tumor expression of Ncr1 (Nkp46) or Klrb1 (Nkrp1g) genes after the virus (Supplementary Table S1).
Comparison of tumor cell killing
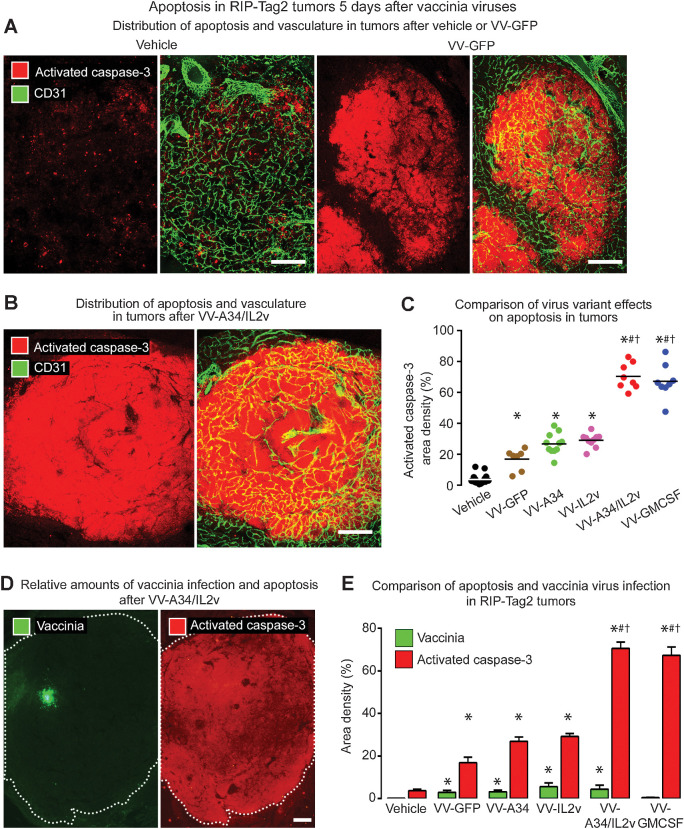
Building on evidence from CD8+ T-cell depletion studies for involvement of cytotoxic T cells in tumor cell killing (24), we found that CD8+ T-cell influx was positively correlated with apoptosis reflected by activated caspase-3 staining across all treatment groups (Supplementary Fig. S2E). Unlike the scattered apoptotic cells in vehicle-treated tumors (area density 4%), broad regions of activated caspase-3 staining were found after all viral variants (Fig. 4A–C). However, the size and extent of the regions varied with the variants. Activated caspase-3 staining after VV-GFP (area density 18%) was nearly fivefold the control value (Fig. 4C). Apoptosis in tumors after VV-A34 (area density 26%) or VV-IL2v (area density 27%) tended to be greater than the reference virus, but the values overlapped and were not significantly different (Fig. 4C). Apoptosis was significantly more widespread after VV-A34/IL2v (area density 70%) or VV-GMCSF (area density 67%; Fig. 4B and C).
Figure 4.
Apoptosis in RIP-Tag2 tumors 5 days after vaccinia virus variants. A and B, Staining for apoptosis (activated caspase-3, red) and blood vessels (CD31, green). A, Confocal microscopic images show little apoptosis after vehicle, widespread apoptosis after reference virus VV-GFP, and even more extensive apoptosis after combination virus VV-A34/IL2v. B, Widespread apoptosis after VV-A34/IL2v. C, Activated caspase-3 in tumors of each mouse after virus or vehicle. Values after VV-GFP or A34K151E substitution virus VV-A34 were similar to one another but significantly greater than after vehicle. By comparison, values after VV-A34/IL2v or VV-GMCSF were significantly greater than after the other viruses. D, Fluorescence microscopic images of tumor after VV-A34/IL2v compare focal staining for vaccinia (green) to widespread staining for activated caspase-3 (red) in adjacent sections (white dotted line outlines tumor border). E, Staining for vaccinia (green bars) and activated caspase-3 (red bars) after vehicle and five virus variants show consistently larger amounts of apoptosis than vaccinia infection, where the ratios ranged from 6 for VV-GFP, to 16 for VV-A34/IL2v, to 143 for VV-GMCSF. ANOVA: P < 0.05 compared with vehicle*, VV-GFP#, or VV-IL2v†. Vehicle (N = 18), VV-GFP (N = 7), VV-A34 (N = 11), VV-IL2v (N = 10), VV-A34/IL2v (N = 8), VV-GMCSF (N = 8). Scale bar, 200 μm in all images.
Apoptosis in tumors after all viruses was much more widespread than vaccinia staining (Fig. 4D and E). Ratios of apoptosis to vaccinia staining ranged from 6 for VV-GFP (N = 7 mice), to 16 for VV-A34/IL2v (N = 8 mice), to 143 for VV-GMCSF (N = 8 mice; Fig. 4E). The amount of apoptosis after VV-GMCSF was similar to that found after VV-A34/IL2v, despite the exceptionally high ratio due to low values for vaccinia staining (Fig. 4E), providing further evidence that the amount of vaccinia staining was not predictive of antitumor activity.
Viral properties accompanying robust antitumor activity
We sought to identify properties that could explain similarly robust antitumor activities despite different genetic modifications of viruses VV-A34/IL2v and VV-GMCSF by comparing changes in serum cytokines and tumor gene expression related to tumor cell killing and immune activation.
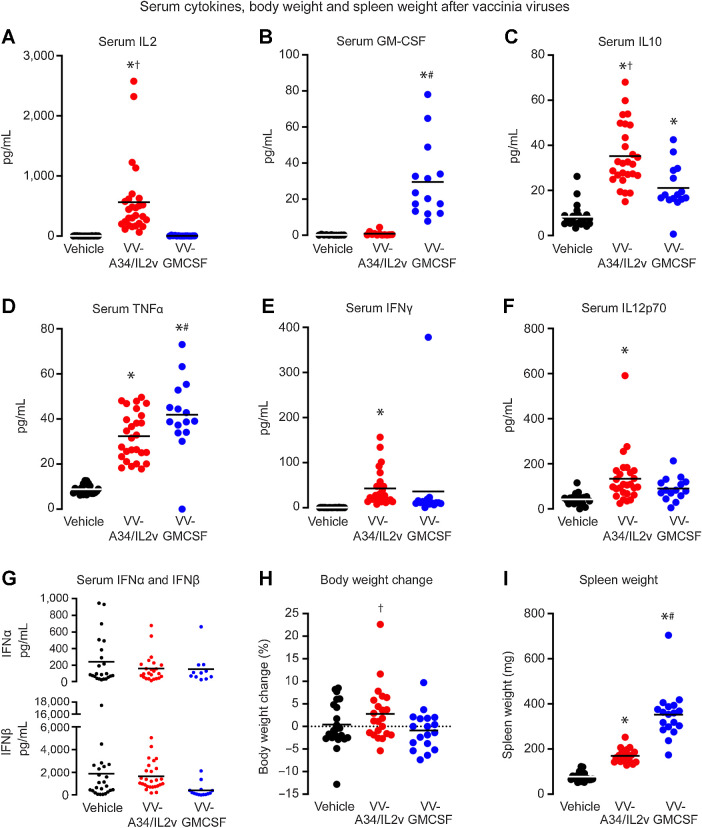
Serum cytokines
Serum IL2 levels were much higher after VV-A34/IL2v, and serum GM-CSF was much higher after VV-GMCSF (Fig. 5A and B), as expected for these cytokine-expressing viruses. Serum anti-inflammatory IL10 and proinflammatory/antitumor TNF-α were increased after both viruses, but IL10 was greater after VV-A34/IL2v, and TNF-α was greater after VV-GMCSF (Fig. 5C and D). Serum IFNγ and IL12 heterodimer IL12p70 were also increased after both viruses, but the increases were significant only after VV-A34/IL2v (Fig. 5E and F). IL-1β, IL4, and IL5 were significantly increased after VV-A34/IL2v, and IL6 was increased after VV-GMCSF (Supplementary Table S2). Serum levels of type I interferons IFNα and IFNβ after VV-A34/IL2v or VV-GMCSF were not significantly above baseline at 5 days (Fig. 5G, Supplementary Table S2), consistent with other reports at this time point (43). Similarly, expression of IFNα gene (Ifna1) and its receptor (Ifnar1) was not increased in tumors after VV-A34/IL2v or VV-GMCSF at 5 days (Supplementary Table S1).
Figure 5.
Serum cytokines (pg/mL) and spleen and body weights of RIP-Tag2 mice at 5 days after vehicle, VV-A34/IL2v, or VV-GMCSF. A and B, Elevated levels of IL2 after VV-A34/IL2v (A) and GM-CSF after VV-GMCSF (B) fit with expression of these cytokines by the respective viruses. C–G, Serum IL10 (C), TNFα (D), IFNγ (E), and IL12p/70 (F) were increased after one or both viruses, but serum type I IFNs, IFNα (G, top) and IFNβ (G, bottom) were similar to vehicle after both viruses (values for these and other cytokines are in Supplementary Table S2). ANOVA: P < 0.05 compared with vehicle* (N = 11–24), VV-GMCSF† (N = 14–15), or VV-A34/IL2v# (N = 12–27). H, Percent body weight gain (mean ± SEM) over 5-day experiment was greater for mice after VV-A34/IL2v (N = 22) than after VV-GMCSF (N = 18), but did not differ from mice that received vehicle (N = 22). Student's t test: P < 0.05 compared with VV-GMCSF†. I, Spleen weight after VV-A34/IL2v (N = 20) was greater than after vehicle (N = 20) but less than after VV-GMCSF (N = 18). ANOVA: P < 0.05 compared with vehicle* or VV-A34/IL2v#.
Other readouts
Mice treated with vehicle had a mean body weight gain of 0.4% during the 5-day experiment (Fig. 5H). Mice that received VV-A34/IL2v had a 2.8% increase in body weight, but mice given VV-GMCSF had a 0.9% weight loss, which was small but significantly different from the VV-A34/IL2v group (Fig. 5H). Increased spleen weight at the end of the experiment, reflecting splenomegaly that accompanies vaccinia virus infection (44), was found after both viruses and was greater after VV-GMCSF (Fig. 5I).
Gene expression in tumors
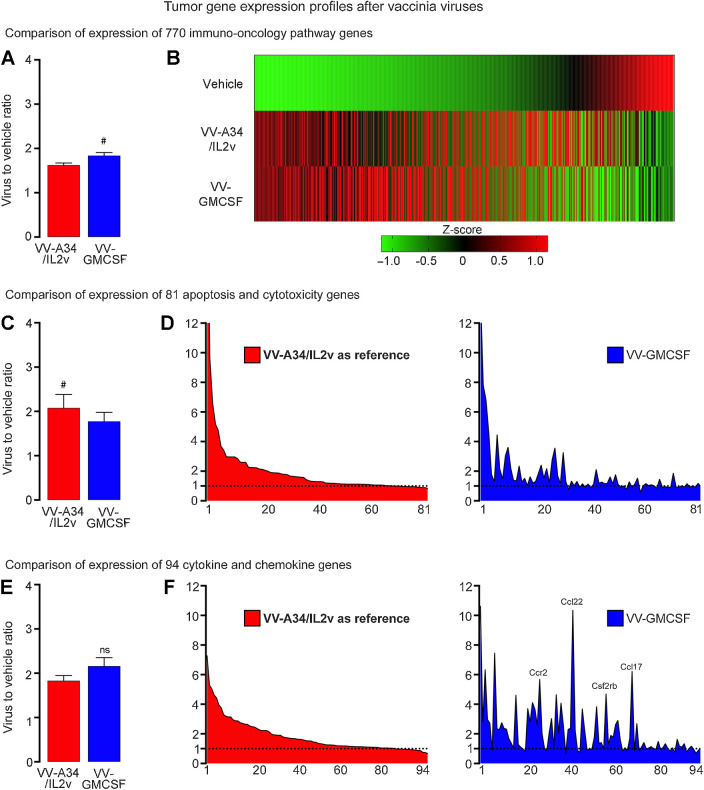
Many of the 770 immuno-oncology genes assessed by the NanoString Mouse PanCancer IO 360 Panel had greater expression in tumors at 5 days after VV-A34/IL2v or VV-GMCSF compared with vehicle. The overall expression was significantly higher after VV-GMCSF than after VV-A34/IL2v (Fig. 6A and B). However, distinctive similarities and differences between the two viruses were revealed by analysis of gene subcategories encoding proteins related to apoptosis and cytotoxicity, chemokines, cytokines, endothelial cell adhesion, and immune cell adhesion (Fig. 6C–F; Supplementary Figs. S3 and S4A-S4E; Supplementary Tables S1 and S3-S6).
Figure 6.
Gene expression profiles in RIP-Tag2 tumors 5 days after VV-A34/IL2v (red) or VV-GMCSF (blue) relative to vehicle. A and B, Expression of 770 immuno-oncology pathway genes after the two viruses compared as groups (A) and as heatmaps that show mean expression of genes converted into Z-scores (B). C and D, Expression of 81 apoptosis and cytotoxicity genes (Supplementary Table S3) compared as groups (C) and ranked from greatest to least ratio of VV-A34/IL2v to vehicle (D, left), with the same rank order used for the genes after VV-GMCSF (D, right). Value for granzyme A (Gzma) in D after VV-A34/IL2v (23.33) or VV-GMCSF (13.91) exceeded the y-axis maximum of 12 and is truncated. E and F, Expression of 94 cytokine, chemokine, and related genes (Supplementary Table S4) compared as groups (E) and ranked (F) as in C and D. High values for Ccr2, Ccl22, Csf2rb, and Ccl17 after VV-GMCSF are labeled (F, right). Wilcoxon signed-rank test: #, P < 0.05 for genes compared as groups. NS, not significant. Error bars show SEM. Vehicle (N = 5), VV-A34/IL2v (N = 5), 4 VV-GMCSF (N = 4). Horizontal dotted line marks value for vehicle group normalized to 1.0. Expression analyzed by NanoString Mouse PanCancer IO 360 Panel.
For the 81 apoptosis and cytotoxicity genes, expression was significantly greater for 26 after VV-A34/IL2v, 20 after VV-GMCSF, 15 after both viruses, and overall was greater after VV-A34/IL2v than after VV-GMCSF (Fig. 6C; Supplementary Table S3). However, the most highly expressed genes in this category, including granzyme A (Gzma), granzyme B (Gzmb), Fas ligand (Fasl), perforin 1 (Prf1), and killer cell lectin-like receptor genes Klrd1 and Klrk1, underwent similarly large increases after both viruses (Fig. 6D; Supplementary Figs. S3E and S4C). Expression of 7 genes, including TRAIL (Tnfsf10), was significantly greater after VV-A34/IL2v, and expression of caspase 8 (Casp8), toll-like receptor 4 (Tlr4), and receptor-interacting serine/threonine kinase 3 (Ripk3), a marker of necroptosis (45), was greater after VV-GMCSF (Supplementary Table S3).
Overall expression of 94 genes encoding cytokines, chemokines, and related proteins was similar after both viruses, but this proved deceptive (Fig. 6E; Supplementary Table S4). Only 15 genes in this category were significantly upregulated after both viruses, but 23 were greater after VV-A34/IL2v and 33 were greater after VV-GMCSF. Genes upregulated after both viruses included IL2 receptor alpha, beta, and gamma chains (Il2ra, Il2rb, Il2rg) and chemokines Ccl5, Ccr5, and Cxcr3 related to T-cell trafficking (Supplementary Fig. S4D; ref. 46). Lymphotoxin B (Ltb) expression was also increased after both viruses (Supplementary Table S1). Genes that had greater expression after VV-A34/IL2v included lymphocyte trafficking chemokines Ccl1, Cxcl9, and Cxcl10 (Supplementary Fig. S4D; ref. 46). Those with greater expression after VV-GMCSF included GM-CSF receptor beta subunit (Csf2rb) and leukocyte recruitment chemokines Ccr2, Ccl22, Csf2rb, and Ccl17 that formed tall peaks in rank-order plots (Fig. 6F; Supplementary Fig. S4D; refs. 47, 48).
The 28 genes encoding endothelial cell adhesion proteins gave insight into endothelial cell-immune cell interactions involved in antitumor activity (47, 49). Although VV-A34/IL2v and VV-GMCSF promoted equivalent CD8+ T-cell influx into tumors, overall expression of genes in this category was greater after VV-GMCSF (Supplementary Figs. S3A and S3B and S4A). Expression of eight genes increased significantly after VV-GMCSF, four after VV-A34/IL2v, and four after both viruses (Supplementary Table S5). Intercellular adhesion molecule 1 (Icam1) and integrin subunits Itgal, Itga4, and Itgb2 were upregulated after both viruses, but the increase in Itga4 subunit of integrin α4β1 (VLA-4, very late antigen-4) was greater after VV-GMCSF, and Vcam1 and increases in integrin alpha chains Itgax (CD11c) and Itgae (CD103) were significant only after VV-GMCSF (Supplementary Fig. S4E).
Expression of 52 genes encoding immune cell adhesion proteins was significantly greater overall after VV-GMCSF (Supplementary Figs. S3C and S3D, S4B). Expression of 30 genes in this category increased significantly after VV-GMCSF, 22 after VV-A34/IL2v, and 20 after both viruses (Supplementary Table S6). Consistent with the robust recruitment of CD8+ T cells to tumors, CD8 antigen alpha and beta chains (Cd8a, Cd8b1) had greater than eightfold increases after both viruses (Supplementary Tables S1 and S6).
VV-GMCSF also induced strikingly higher expression in tumors of multiple neutrophil-associated genes, including calgranulin A/B (S100a8/a9) and granulocyte stimulating factor receptor (Csf3r; Supplementary Fig. S4F). Counts of neutrophils identified by S100A8+ staining confirmed that neutrophils were significantly more abundant in tumor after VV-GMCSF (Supplementary Fig. S4G and S4H).
Discussion
This study compared the initial antitumor activity of five vaccinia virus variants constructed from the same Western Reserve vaccinia backbone with different genetic modifications (Fig. 1). To enable side-by-side comparisons, the viruses were examined under standardized conditions in a mouse tumor model known to be immunologically “cold” at baseline and to be responsive to oncolytic vaccinia viruses. After intravenous administration of the viruses, vaccinia infection was limited to focal patches of tumors and was absent in normal tissue. In contrast, antitumor activity, evidenced by tumor vascular pruning, influx of CD8+ T cells and NK cells, and tumor cell apoptosis, was much more widespread, but the magnitude differed among the five viruses. Effects of viruses expressing mIL2v alone (VV-IL2v) or bearing the A34K151E substitution (VV-A34) were largely similar to the reference virus (VV-GFP). However, tumor cell killing was significantly greater after viruses that expressed mGM-CSF (VV-GMCSF) or mIL2v together with A34K151E substitution, B18R viral gene deletion, and other changes (VV-A34/IL2v). Analysis of gene expression and serum cytokines indicated similarities consistent with equivalent antitumor activities, but accompanying differences revealed mechanistic dissimilarities of the two viruses.
Approach for comparing antitumor activities
Experiments were designed to determine the relative contributions of single and multiple genetic changes in vaccinia viruses to antitumor activity. Viral variants were derived from the same Western Reserve backbone, administered in the same dose, intravenous route, and model of spontaneous tumors, and assessed by the same panel of readouts. The RIP-Tag2 mouse model of pancreatic neuroendocrine tumors has been shown to be predictive of clinical success of therapeutics (39, 50, 51). Measurements of initial antitumor activity were made at 5 days, when vaccinia infection, CD8+ T-cell influx, and tumor cell killing peaked (24). Although these initial responses are not necessarily indicative of long-term outcome, the approach revealed clear differences in the contributions of the genetic variants to antitumor activity. Informative next steps will be to build on these findings by using similar approaches to assess responses in other tumor models and at other time points and to extend the mechanistic understanding by using additional readouts.
Widespread tumor cell killing despite focal tumor infection
Vaccinia viral infection in tumors, assessed by IHC staining, was patchy, differed little among the viruses, and was consistently less extensive than tumor cell killing. Infection after vaccinia virus VV-A34 bearing the A34K151E substitution, which increases EEV production after infection of cultured cells (6, 52), was similar to the reference virus (VV-GFP) that lacked this modification. The lack of greater spreading of infection by vaccinia virus with the A34K151E mutation contrasts with the behavior of such viruses reported for cultured cells (6) and other tumor models (10, 11). More widespread infection has been reported in implanted tumors at 1 day after intravenous injection of a vaccinia virus strain that bears the A34K151E substitution (10), raising the question of timing of peak infection. Efficacy in a model of peritoneal carcinomatosis after intraperitoneal injection of virus (11) may not be predictive of spontaneous tumors after by intravenous administration. The survival advantage in mice with implanted tumors attributed to viruses with the A34K151E substitution (10, 11) deserves further study in spontaneous tumor models after intravenous delivery. Nonetheless, importantly, our studies revealed that the extent of vaccinia infection was not predictive of overall antitumor activity at 5 days, which instead was more closely linked to CD8+ T-cell recruitment.
The finding of greater antitumor activity after viruses expressing mGM-CSF or mIL2v in combination with other modifications differs from a previous report of equivalent effects at 5 days after vaccinia viruses that expressed human, mouse, or no GM-CSF in the RIP-Tag2 model (24). This apparent discrepancy could reflect changes in the latter viruses during mouse adaptation or differences in the Western Reserve viral backbone or viral constructs. The differences highlight the rationale and importance of deriving all five viral variants from the same viral backbone to enable meaningful side-by-side comparisons.
Vascular pruning, CD8+ T-cell influx, and tumor cell killing
Pruning of the tumor vasculature, a well-documented effect of oncolytic vaccinia viruses (20, 24, 40), occurred after all five viruses but was greatest after VV-A34/IL2v and VV-GMCSF. Reduced vascularity has been linked to necrosis in some tumors (20, 40) but cannot fully explain the robust antitumor of effects of vaccinia viruses (24).
Dependency of widespread tumor cell killing by vaccinia viruses on CD8+ T-cell recruitment was shown by CD8+ T-cell depletion, which resulted in narrowing of apoptosis to regions of vaccinia virus infection in tumors (24).
CD8+ T cells are sparse in RIP-Tag2 tumors at baseline (24) but were recruited to tumors by all five viruses. CD8+ T-cell recruitment was similar after the reference virus (VV-GFP) and viruses with the single A34K151E substitution (VV-A34) or mIL2v expression (VV-IL2v), but was greater after viruses VV-GMCSF and VV-A34/IL2v. Although other immune cells were recruited with CD8+ cells into tumors, influx of CD8+ cells was much greater than CD4+ cells or NK cells, and Foxp3+ Tregs did not change.
Consistent with the essential contribution of CD8+ T cells to widespread tumor cell killing after vaccina virus administered intravenously (24), the amount of CD8+ T-cell influx had a significant positive correlation with the amount of activated caspase-3 staining in tumors. NK cells, which were largely restricted to sites of vaccinia infection at 5 days, appear not to have a major contribution to the widespread tumor cell killing.
IL2v expression (VV-IL2v) as a single modification did not amplify tumor cell killing, but together with other modifications (VV-A34/IL2v) resulted in significantly greater antitumor activity. Expression of mGM-CSF (VV-GMCSF) led to similarly robust antitumor activity.
Similarities of the most efficacious viruses
Tumor gene expression profiling and serum cytokine measurements provided insights into mechanisms that contributed to the robust antitumor activities of viruses VV-A34/IL2v and VV-GMCSF. Both viruses increased expression of genes associated with CD8+ T-cell–mediated tumor cell killing, including granzyme A, granzyme B, perforin1, and Fas ligand. Granzymes from CD8+ T cells have lethal effects after entering cells through pores formed by perforin1 (53, 54). Granzyme B triggers apoptosis through activation of caspase-3 cascades, whereas granzyme A acts in a caspase-independent manner (53–55).
Both viruses also increased expression of vascular adhesion molecules involved in leukocyte trafficking into tumors. Icam1 upregulation is particularly important because of the large influx of CD8+ T cells into RIP-Tag2 tumors. Endothelial cell Icam1 expression is required for entry of activated antigen-specific T cells into these tumors (56). Both viruses were also accompanied by upregulation of alpha-L and beta-2 integrin subunits (Itgal and Itgb2) of LFA-1 (leukocyte function associated antigen-1), the receptor for ICAM-1-mediated leukocyte transmigration (47, 57).
Differences between the most efficacious viruses
Similarities in gene expression were accompanied by striking differences between the two viruses. VV-A34/IL2v was followed by higher expression of death receptor ligand TRAIL (Tnfsf10) and CXCR3 ligands CXCL9 and CXCL10 in tumors and by higher serum levels of IL2, IFNγ, IL10, and heterodimeric IL12 (IL12p70), which promote CD8+ T-cell activation, recruitment, and cytotoxicity (58–61). However, serum IFNα and expression of Ifna1 and Ifnar1 genes in tumors after VV-A34/IL2v were similar to VV-GMCSF or vehicle. Serum IFNα is reported to be elevated at 7 days after intratumoral injection of vaccinia virus, but the level at 5 days is not significantly greater than baseline (43). These findings bring into question the contribution of host IFNα activity amplification by viral B18R gene deletion (14, 15) to the robust antitumor action of VV-A34/IL2v at 5 days in this model.
VV-GMCSF led to higher tumor expression of Vcam1, toll-like receptor 4 Tlr4, necroptosis inducer Ripk3 (45, 54), and many genes for cytokine/chemokine ligands and receptors involved in leukocyte trafficking (IL16, Ccl9, Ccl17, Cxcl16, Ccr2, Csf1r, Csf2rb, Csf3r). VV-GMCSF also not only increased serum GM-CSF but also serum TNFα, which kills cancer cells directly and increases Vcam1 expression and immune cell influx (62, 63). Another distinctive feature of VV-GMCSF was greater neutrophil recruitment, a known effect of GM-CSF (64). Neutrophils contribute to the antitumor action of GM-CSF expressing measles virus (65) but have uncertain effects after GM-CSF–expressing vaccinia virus. The contribution of mechanistic differences in CD8+ T-cell recruitment to robust tumor cell killing by VV-A34/IL2v and VV-GMCSF deserves further study by selective inhibition or deletion of leukocyte recruitment factors, use of tumor models with genetically altered immune responses, and single-cell analysis of gene expression.
Tolerability
All vaccinia virus variants studied were well tolerated over 5 days after intravenous administration. This finding is important because wild-type IL2 has dose-limiting toxicity. In one report, 60% of mice that received vaccinia virus expressing wild-type IL2 by intravenous injection lost more than 20% body weight and required euthanasia by day 5 (33). Similarly, mice that received vaccinia virus expressing a soluble form of IL2 had serum IL2 levels of 22,000 pg/mL and 80% mortality by 5 days (19). These toxicities are attributed to binding of wild-type IL2 to the alpha subunit of IL2 receptor. By comparison, mice that received intravenous injection of VV-A34/IL2v, which expressed IL2v with attenuated binding to the alpha subunit (34), had serum IL2 levels averaging 564 pg/mL, gained weight, and had 100% survival. Serum IL2 levels in these mice evidently reflected viral cytokine expression, because IL2 levels in mice that received VV-GMCSF, which did not express IL2, were similar to vehicle-treated mice.
In summary, this side-by-side comparison revealed marked differences among five vaccinia viruses engineered with different genetic modifications reported to increase cell killing. Standardized comparison by a panel of readouts identified two viruses with especially robust antitumor activity after intavenous delivery; one expressed GM-CSF (VV-GMCSF) and the other expressed an IL2 variant (VV-A34/IL2v) together with the A34K151E substitution and B18R viral gene deletion. The viruses promoted widespread influx of CD8+ T cells accompanied by strong upregulation of cytotoxicity genes and extensive apoptosis in tumors, but NK cell recruitment was limited to focal sites of vaccinia infection in tumors, and Treg recruitment was suppressed. Despite the similarities, VV-A34/IL2v resulted in greater tumor expression of death receptor ligand TRAIL and higher serum IL2 and IFNγ, whereas VV-GMCSF led to greater expression of leukocyte recruitment cytokines and vascular adhesion molecules, higher serum GM-CSF and TNFα, and more neutrophil recruitment. Together, this standardized comparison of vaccinia virus variants enabled identification of genetic modifications that amplify tumor cell killing. Antitumor activity of oncolytic viruses can be further advanced by extending this approach to other modifications of the virus, treatment regimens, and tumor models.
Authors' Disclosures
P. Wall is a scientist at Ignite Immunotherapy and an employee of Pfizer. A. Wang reports a patent for WO2020/148612 pending and a patent for PCT/IB2020/051025 pending. M.R. Dermyer reports a patent for WO2020/148612 pending and a patent for PCT/IB2020/051025 pending. M.R. Dermyer is an employee and stockholder in Pfizer. H. Laklai reports a patent for WO2020/148612 pending and a patent for PCT/IB2020/051025 pending. H. Laklai is a scientist at Pfizer. J.J. Binder reports a patent for WO2020/148612 pending and a patent for PCT/IB2020/051025 pending. J.J. Binder is an employee and stockholder of Pfizer. C. Lees reports a patent for WO2020/148612 pending and a patent for PCT/IB2020/051025 pending. C. Lees is an employee and stockholder in Pfizer. R. Hollingsworth reports patents WO2020/148612 and PCT/IB2020/051025 pending to Pfizer. R. Hollingsworth is an employee and shareholder of Pfizer. L. Maruri-Avidal reports personal fees from Pfizer outside the submitted work. L. Maruri-Avidal is an employee and stockholder of Ignite Immunotherapy. D.H. Kirn reports personal fees and other support from Ignite Immunotherapeutics; personal fees from Pfizer during the conduct of the study; and personal fees and other support from 4D Molecular Therapeutics outside the submitted work. In addition, D.H. Kirn has a patent for oncolytic vaccinia pending to Pfizer. D.H. Kirn is an employee and stockholder of Pfizer. D.M. McDonald reports grants from the NIH; grants and nonfinancial support from Ignite Immunotherapy; and nonfinancial support from Pfizer during the conduct of the study. No disclosures were reported by the other authors.
Acknowledgments
This work was supported in part by National Heart, Lung, and Blood Institute grants R01 HL143896, R01 HL059157, and R01 HL127402 from the NIH and grant CA-0121573_20180427 from Ignite Immunotherapy to D.M. McDonald. We thank Darin Abbadessa at Ignite Immunotherapy for preparing the viruses for injection into mice at UCSF and Jon Osborne for assistance with flow cytometry. We also thank David Murray for NanoString measurements of gene expression in tumors, Talia Nguyen for NanoString and serum cytokine analyses, and Elena Galitovskaya for performing comet assays and Western blots, all at Pfizer San Diego.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
T. Inoue: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. T. Byrne: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. M. Inoue: data curation, formal analysis, investigation, visualization, methodology, writing–review and editing. M.E. Tait: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. P. Wall: Data curation, formal analysis, validation, investigation, visualization, methodology, writing–review and editing. A. Wang: Conceptualization, resources, data curation, formal analysis, validation, methodology, writing–review and editing. M.R. Dermyer: Conceptualization, data curation, formal analysis, supervision, validation, methodology. H. Laklai: Data curation, formal analysis, validation, methodology, writing–review and editing. J.J. Binder: Conceptualization, resources, data curation, supervision, validation. C. Lees: Conceptualization, data curation, validation, methodology, writing–review and editing. R. Hollingsworth: Conceptualization, resources, data curation, formal analysis, supervision, validation, methodology. L. Maruri-Avidal: Conceptualization, resources, data curation, formal analysis, validation, methodology, writing–review and editing. D.H. Kirn: Conceptualization, data curation, formal analysis, supervision, validation, methodology, writing–review and editing. D.M. McDonald: Conceptualization, resources, data curation, formal analysis, supervision, funding acquisition, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Breitbach CJ, Burke J, Jonker D, Stephenson J, Haas AR, Chow LQ, et al. Intravenous delivery of a multi-mechanistic cancer-targeted oncolytic poxvirus in humans. Nature 2011;477:99–102. [DOI] [PubMed] [Google Scholar]
- 2. Lawler SE, Speranza MC, Cho CF, Chiocca EA. Oncolytic viruses in cancer treatment: a review. JAMA Oncol 2017;3:841–49. [DOI] [PubMed] [Google Scholar]
- 3. Guo ZS, Lu B, Guo Z, Giehl E, Feist M, Dai E, et al. Vaccinia virus-mediated cancer immunotherapy: cancer vaccines and oncolytics. J Immunother Cancer 2019;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Thorne SH. Immunotherapeutic potential of oncolytic vaccinia virus. Front Oncol 2014;4:155–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Breitbach CJ, Bell JC, Hwang TH, Kirn DH, Burke J. The emerging therapeutic potential of the oncolytic immunotherapeutic Pexa-Vec (JX-594). Oncolytic Virother 2015;4:25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blasco R, Sisler JR, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J Virol 1993;67:3319–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McCart JA, Ward JM, Lee J, Hu Y, Alexander HR, Libutti SK, et al. Systemic cancer therapy with a tumor-selective vaccinia virus mutant lacking thymidine kinase and vaccinia growth factor genes. Cancer Res 2001;61:8751–7. [PubMed] [Google Scholar]
- 8. Cho E, Islam S, Jiang F, Park JE, Lee B, Kim ND, et al. Characterization of oncolytic vaccinia virus harboring the human IFNB1 and CES2 transgenes. Cancer Res Treat 2020;52:309–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Puhlmann M, Brown CK, Gnant M, Huang J, Libutti SK, Alexander HR, et al. Vaccinia as a vector for tumor-directed gene therapy: biodistribution of a thymidine kinase-deleted mutant. Cancer Gene Ther 2000;7:66–73. [DOI] [PubMed] [Google Scholar]
- 10. Kirn DH, Wang Y, Liang W, Contag CH, Thorne SH. Enhancing poxvirus oncolytic effects through increased spread and immune evasion. Cancer Res 2008;68:2071–5. [DOI] [PubMed] [Google Scholar]
- 11. Thirunavukarasu P, Sathaiah M, Gorry MC, O'Malley ME, Ravindranathan R, Austin F, et al. A rationally designed A34R mutant oncolytic poxvirus: improved efficacy in peritoneal carcinomatosis. Mol Ther 2013;21:1024–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Symons JA, Alcami A, Smith GL. Vaccinia virus encodes a soluble type I interferon receptor of novel structure and broad species specificity. Cell 1995;81:551–60. [DOI] [PubMed] [Google Scholar]
- 13. Alcami A, Symons JA, Smith GL. The vaccinia virus soluble alpha/beta interferon (IFN) receptor binds to the cell surface and protects cells from the antiviral effects of IFN. J Virol 2000;74:11230–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kirn DH, Wang Y, Le Boeuf F, Bell J, Thorne SH. Targeting of interferon-beta to produce a specific, multi-mechanistic oncolytic vaccinia virus. PLoS Med 2007;4:e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ahmed CM, Johnson HM. Type I interferon mimetics bypass vaccinia virus decoy receptor virulence factor for protection of mice against lethal infection. Clin Vaccine Immunol 2014;21:1178–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coupar BE, Andrew ME, Boyle DB. A general method for the construction of recombinant vaccinia viruses expressing multiple foreign genes. Gene 1988;68:1–10. [DOI] [PubMed] [Google Scholar]
- 17. Islam S, Lee B, Jiang F, Kim EK, Ahn SC, Hwang TH. Engineering and characterization of oncolytic vaccinia virus expressing truncated herpes simplex virus thymidine kinase. Cancers (Basel) 2020;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaufman HL, Flanagan K, Lee CS, Perretta DJ, Horig H. Insertion of interleukin-2 (IL2) and interleukin-12 (IL-12) genes into vaccinia virus results in effective anti-tumor responses without toxicity. Vaccine 2002;20:1862–9. [DOI] [PubMed] [Google Scholar]
- 19. Liu Z, Ge Y, Wang H, Ma C, Feist M, Ju S, et al. Modifying the cancer-immune set point using vaccinia virus expressing re-designed interleukin-2. Nat Commun 2018;9:4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chon HJ, Lee WS, Yang H, Kong SJ, Lee NK, Moon ES, et al. Tumor microenvironment remodeling by intratumoral oncolytic vaccinia virus enhances the efficacy of immune-checkpoint blockade. Clin Cancer Res 2019;25:1612–23. [DOI] [PubMed] [Google Scholar]
- 21. Zhang Z, Zhang J, Zhang Y, Xing J, Yu Z. Vaccinia virus expressing IL-37 promotes antitumor immune responses in hepatocellular carcinoma. Cell Biochem Funct 2019;37:618–24. [DOI] [PubMed] [Google Scholar]
- 22. Ahmed J, Chard LS, Yuan M, Wang J, Howells A, Li Y, et al. A new oncolytic V accinia virus augments antitumor immune responses to prevent tumor recurrence and metastasis after surgery. J Immunother Cancer 2020;8:e000415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thorne SH, Hwang TH, O'Gorman WE, Bartlett DL, Sei S, Kanji F, et al. Rational strain selection and engineering creates a broad-spectrum, systemically effective oncolytic poxvirus, JX-963. J Clin Invest 2007;117:3350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim M, Nitschke M, Sennino B, Murer P, Schriver BJ, Bell A, et al. Amplification of oncolytic vaccinia virus widespread tumor cell killing by sunitinib through multiple mechanisms. Cancer Res 2018;78:922–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Park BH, Hwang T, Liu TC, Sze DY, Kim JS, Kwon HC, et al. Use of a targeted oncolytic poxvirus, JX-594, in patients with refractory primary or metastatic liver cancer: a phase I trial. Lancet Oncol 2008;9:533–42. [DOI] [PubMed] [Google Scholar]
- 26. Rojas JJ, Sampath P, Hou W, Thorne SH. Defining effective combinations of immune checkpoint blockade and oncolytic virotherapy. Clin Cancer Res 2015;21:5543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Z, Ravindranathan R, Kalinski P, Guo ZS, Bartlett DL. Rational combination of oncolytic vaccinia virus and PD-L1 blockade works synergistically to enhance therapeutic efficacy. Nat Commun 2017;8:14754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Amin A, White RL. Interleukin-2 in renal cell carcinoma: a has-been or a still-viable option? J Kidney Cancer VHL 2014;1:74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jiang T, Zhou C, Ren S. Role of IL2 in cancer immunotherapy. Oncoimmunology 2016;5:e1163462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hugin AW, Flexner C, Moss B. Clearance of recombinant vaccinia virus expressing IL2: role of local host immune responses. Cell Immunol 1993;152:499–509. [DOI] [PubMed] [Google Scholar]
- 31. Coupar BE, Oke PG, Andrew ME. Insertion sites for recombinant vaccinia virus construction: effects on expression of a foreign protein. J Gen Virol 2000;81:431–9. [DOI] [PubMed] [Google Scholar]
- 32. Dasgupta S, Tripathi PK, Bhattacharya-Chatterjee M, O'Malley B Jr., Chatterjee SK. Recombinant vaccinia virus expressing IL2 generates effective anti-tumor responses in an orthotopic murine model of head and neck carcinoma. Mol Ther 2003;8:238–48. [DOI] [PubMed] [Google Scholar]
- 33. Chen H, Sampath P, Hou W, Thorne SH. Regulating cytokine function enhances safety and activity of genetic cancer therapies. Mol Ther 2013;21:167–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Klein C, Waldhauer I, Nicolini VG, Freimoser-Grundschober A, Nayak T, Vugts DJ, et al. Cergutuzumab amunaleukin (CEA-IL2v), a CEA-targeted IL2 variant-based immunocytokine for combination cancer immunotherapy: Overcoming limitations of aldesleukin and conventional IL2-based immunocytokines. Oncoimmunology 2017;6:e1277306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sennino B, Ishiguro-Oonuma T, Wei Y, Naylor RM, Williamson CW, Bhagwandin V, et al. Suppression of tumor invasion and metastasis by concurrent inhibition of c-Met and VEGF signaling in pancreatic neuroendocrine tumors. Cancer Discov 2012;2:270–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kobayashi S, Contractor T, Vosburgh E, Du YN, Tang LH, Clausen R, et al. Alleles of Insm1 determine whether RIP1-Tag2 mice produce insulinomas or nonfunctioning pancreatic neuroendocrine tumors. Oncogenesis 2019;8:16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mastrangelo MJ, Maguire HC Jr, Eisenlohr LC, Laughlin CE, Monken CE, McCue PA, et al. Intratumoral recombinant GM-CSF-encoding virus as gene therapy in patients with cutaneous melanoma. Cancer Gene Ther 1999;6:409–22. [DOI] [PubMed] [Google Scholar]
- 38. Kim JH, Oh JY, Park BH, Lee DE, Kim JS, Park HE, et al. Systemic armed oncolytic and immunologic therapy for cancer with JX-594, a targeted poxvirus expressing GM-CSF. Mol Ther 2006;14:361–70. [DOI] [PubMed] [Google Scholar]
- 39. Sadanandam A, Wullschleger S, Lyssiotis CA, Grotzinger C, Barbi S, Bersani S, et al. A cross-species analysis in pancreatic neuroendocrine tumors reveals molecular subtypes with distinctive clinical, metastatic, developmental, and metabolic characteristics. Cancer Discov 2015;5:1296–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Breitbach CJ, Arulanandam R, De Silva N, Thorne SH, Patt R, Daneshmand M, et al. Oncolytic vaccinia virus disrupts tumor-associated vasculature in humans. Cancer Res 2013;73:1265–75. [DOI] [PubMed] [Google Scholar]
- 41. Inai T, Mancuso M, Hashizume H, Baffert F, Haskell A, Baluk P, et al. Inhibition of vascular endothelial growth factor (VEGF) signaling in cancer causes loss of endothelial fenestrations, regression of tumor vessels, and appearance of basement membrane ghosts. Am J Pathol 2004;165:35–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mancuso MR, Davis R, Norberg SM, O'Brien S, Sennino B, Nakahara T, et al. Rapid vascular regrowth in tumors after reversal of VEGF inhibition. J Clin Invest 2006;116:2610–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fend L, Yamazaki T, Remy C, Fahrner C, Gantzer M, Nourtier V, et al. Immune checkpoint blockade, immunogenic chemotherapy or IFN-alpha blockade boost the local and abscopal effects of oncolytic virotherapy. Cancer Res 2017;77:4146–57. [DOI] [PubMed] [Google Scholar]
- 44. Mathurin KS, Martens GW, Kornfeld H, Welsh RM. CD4 T-cell-mediated heterologous immunity between mycobacteria and poxviruses. J Virol 2009;83:3528–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G. Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 2010;11:700–14. [DOI] [PubMed] [Google Scholar]
- 46. Mikucki ME, Fisher DT, Matsuzaki J, Skitzki JJ, Gaulin NB, Muhitch JB, et al. Non-redundant requirement for CXCR3 signalling during tumoricidal T-cell trafficking across tumour vascular checkpoints. Nat Commun 2015;6:7458–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peske JD, Woods AB, Engelhard VH. Control of CD8 T-cell infiltration into tumors by vasculature and microenvironment. Adv Cancer Res 2015;128:263–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bakos E, Thaiss CA, Kramer MP, Cohen S, Radomir L, Orr I, et al. CCR2 Regulates the immune response by modulating the interconversion and function of effector and regulatory T cells. J Immunol 2017;198:4659–71. [DOI] [PubMed] [Google Scholar]
- 49. Erdag G, Schaefer JT, Smolkin ME, Deacon DH, Shea SM, Dengel LT, et al. Immunotype and immunohistologic characteristics of tumor-infiltrating immune cells are associated with clinical outcome in metastatic melanoma. Cancer Res 2012;72:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hanahan D. Heritable formation of pancreatic beta-cell tumours in transgenic mice expressing recombinant insulin/simian virus 40 oncogenes. Nature 1985;315:115–22. [DOI] [PubMed] [Google Scholar]
- 51. Raymond E, Dahan L, Raoul JL, Bang YJ, Borbath I, Lombard-Bohas C, et al. Sunitinib malate for the treatment of pancreatic neuroendocrine tumors. N Engl J Med 2011;364:501–13. [DOI] [PubMed] [Google Scholar]
- 52. Perdiguero B, Lorenzo MM, Blasco R. Vaccinia virus A34 glycoprotein determines the protein composition of the extracellular virus envelope. J Virol 2008;82:2150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Metkar SS, Wang B, Ebbs ML, Kim JH, Lee YJ, Raja SM, et al. Granzyme B activates procaspase-3 which signals a mitochondrial amplification loop for maximal apoptosis. J Cell Biol 2003;160:875–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Martinez-Lostao L, Anel A, Pardo J. How do cytotoxic lymphocytes kill cancer cells? Clin Cancer Res 2015;21:5047–56. [DOI] [PubMed] [Google Scholar]
- 55. Lieberman J, Fan Z. Nuclear war: the granzyme A-bomb. Curr Opin Immunol 2003;15:553–9. [DOI] [PubMed] [Google Scholar]
- 56. Garbi N, Arnold B, Gordon S, Hammerling GJ, Ganss R. CpG motifs as proinflammatory factors render autochthonous tumors permissive for infiltration and destruction. J Immunol 2004;172:5861–9. [DOI] [PubMed] [Google Scholar]
- 57. Smith DJ, Vainio PJ. Targeting vascular adhesion protein-1 to treat autoimmune and inflammatory diseases. Ann N Y Acad Sci 2007;1110:382–8. [DOI] [PubMed] [Google Scholar]
- 58. Henry CJ, Ornelles DA, Mitchell LM, Brzoza-Lewis KL, Hiltbold EM. IL-12 produced by dendritic cells augments CD8+ T cell activation through the production of the chemokines CCL1 and CCL17. J Immunol 2008;181:8576–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Groom JR, Luster AD. CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 2011;89:207–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mumm JB, Emmerich J, Zhang X, Chan I, Wu L, Mauze S, et al. IL-10 elicits IFNgamma-dependent tumor immune surveillance. Cancer Cell 2011;20:781–96. [DOI] [PubMed] [Google Scholar]
- 61. Bhat P, Leggatt G, Waterhouse N, Frazer IH. Interferon-gamma derived from cytotoxic lymphocytes directly enhances their motility and cytotoxicity. Cell Death Dis 2017;8:e2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhou Z, Connell MC, MacEwan DJ. TNFR1-induced NF-kappaB, but not ERK, p38MAPK or JNK activation, mediates TNF-induced ICAM-1 and VCAM-1 expression on endothelial cells. Cell Signal 2007;19:1238–48. [DOI] [PubMed] [Google Scholar]
- 63. Josephs SF, Ichim TE, Prince SM, Kesari S, Marincola FM, Escobedo AR, et al. Unleashing endogenous TNF-alpha as a cancer immunotherapeutic. J Transl Med 2018;16:242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Becher B, Tugues S, Greter M. GM-CSF: from growth factor to central mediator of tissue inflammation. Immunity 2016;45:963–73. [DOI] [PubMed] [Google Scholar]
- 65. Grote D, Cattaneo R, Fielding AK. Neutrophils contribute to the measles virus-induced antitumor effect: enhancement by granulocyte macrophage colony-stimulating factor expression. Cancer Res 2003;63:6463–8. [PubMed] [Google Scholar]