Abstract
Alternate strategies are needed for B-cell malignancy patients relapsing after CD19-targeted immunotherapy. Here, cell surface proteomics revealed CD72 as an optimal target for poor-prognosis KMT2A/MLL1-rearranged (MLLr) B-ALL, which we further found to be expressed in other B-cell malignancies. Using a recently-described, fully-in vitro system we selected synthetic CD72-specific nanobodies, incorporated them into CARs, and demonstrated robust activity against B-cell malignancy models, including CD19 loss. Taking advantage of CD72’s role in inhibiting B-cell receptor signaling, we found that pharmacologic SHIP1 inhibition increased CD72 surface density. We establish CD72-nanobody CAR T’s as a promising therapy for MLLr B-ALL.
Keywords: Chimeric antigen receptor, CD72, KMT2A, MLLr, acute lymphoblastic leukemia, immunotherapy, lymphoma, nanobody
INTRODUCTION
Surface antigen-targeted immunotherapies have shown great promise for treating B-cell malignancies, particularly CD19 chimeric antigen receptor (CAR) T cells for B-cell acute lymphoblastic leukemia (B-ALL) and diffuse large B-cell lymphoma (DLBCL)(1). However, despite impressive initial responses, many patients relapse, often with loss of CD19 antigen(2). While second-line therapies targeting alternative cell surface proteins, such as CD22, have shown efficacy treating CD19-negative relapses, “antigen escape” has also been observed(3). In this context, several additional B-cell-restricted receptors have emerged as potential targets including CD20(4), CD23(5), CD79b(6), CD37(7), and BAFFR(8). However, it remains unclear which is the optimal target after CD19 failure. Discovery of additional targets may further enable new strategies to overcome resistance as well as engineer novel approaches for cellular therapy.
Thus far, B-ALL patients have shown the most promising responses to CD19 CAR-T therapy(9). However, both pediatric and adult B-ALL patients that harbor translocations of the mixed lineage leukemia (MLL1/KMT2A) gene (MLLr) continue to demonstrate the poorest outcomes, both in the context of standard chemotherapies(10) or, in emerging data, after CAR-T(11-13). Here, our initial goal was discovering new therapeutic targets particularly enriched in MLLr patients, as immunotherapeutic efficacy frequently correlates with surface antigen density(14). While most new immunotherapy targets are found by transcriptome analysis, RNA-based data may not accurately predict surface protein abundance(15). We hypothesized that unbiased quantification of plasma membrane protein density would be most fruitful for uncovering novel targets. Furthermore, while the “surfaceome” comprises ~2% of the total proteome by abundance, it plays key roles in determining cellular fate by mediating external signaling, governing microenvironment interactions, driving cellular migration, and modulating immune surveillance.
Using these methods, we identified CD72, an ITIM-bearing inhibitor of B-cell receptor signaling, as being highly expressed on MLLr B-ALL vs. other B-ALL genotypes. Further investigation also demonstrated consistent CD72 expression in diffuse large B-cell lymphoma. To our knowledge, this antigen has not previously been targeted with cellular therapies. We utilized a recently-described, fully in vitro nanobody yeast display library(16) to develop highly specific binders to CD72. We showed for the first time that CAR T’s developed from these nanobodies, generated without llama immunization, have potent efficacy both in vitro and in vivo. Finally, we demonstrate that modulating B-cell receptor signaling via small molecules can upregulate surface CD72. Based on these results, we propose targeting CD72 as an effective genotype-driven immunotherapy strategy for MLLr B-ALL patients. We further validate in vitro nanobody selection as a versatile platform for cellular therapy development.
RESULTS
Cell surface proteomics of MLLr versus other B-ALL subtypes define distinct surfaceome signatures
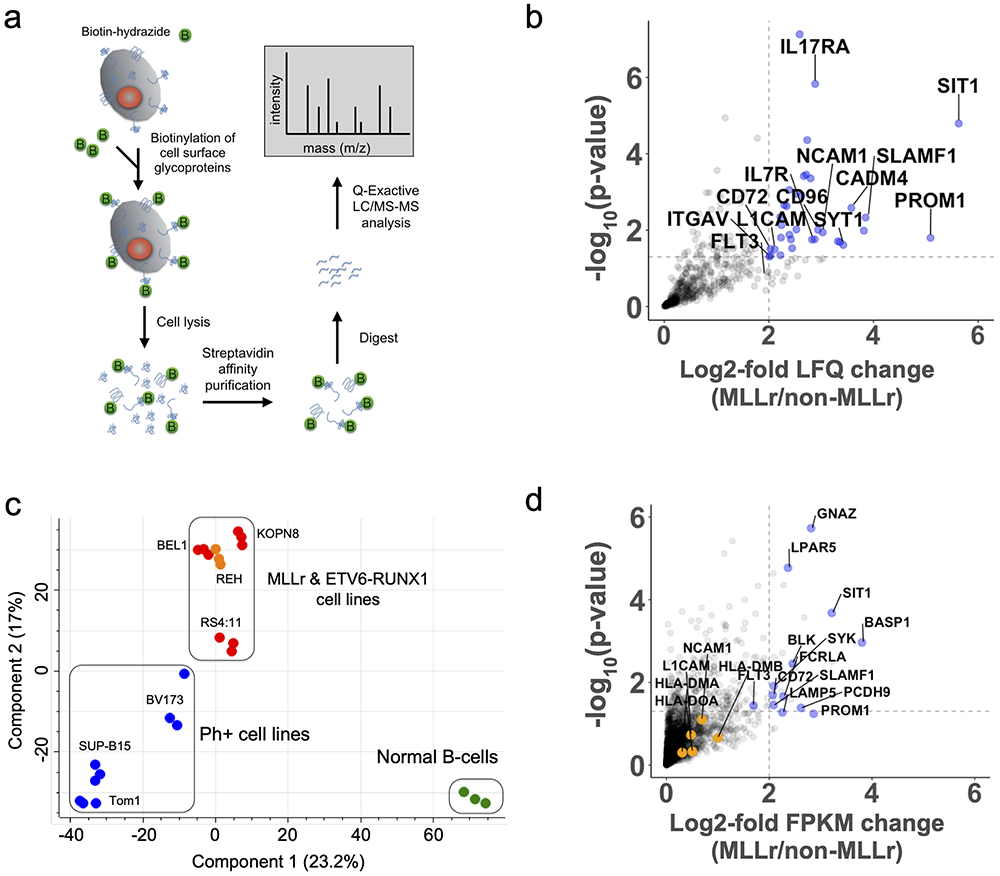
To define the B-ALL cell surfaceome, we enriched N-glycoproteins using a modified version of the Cell Surface Capture method (Fig. 1a and Methods) followed by quantitative mass spectrometry(17). As this method requires sample input of 30-200e6 cells, it is not routinely amenable to primary sample analysis; we therefore performed our analyses on cell lines. We profiled eight B-ALL lines with distinct driver translocations including MLL-AF4 (n=3), MLL-ENL (n=1), BCR-ABL (n=3), and ETV6-RUNX1 (n=1), plus Epstein Barr Virus-immortalized B-cells derived from normal donor umbilical cord blood as a non-malignant comparator (Supplementary Table S1), all performed in biological triplicate. Using label-free quantification (LFQ) in MaxQuant(18) and filtering for Uniprot-annotated membrane or membrane-associated proteins, we quantified 799 membrane proteins (Supplementary Fig. S1a). Biological replicates demonstrated excellent reproducibility (Supplementary Fig. S1b). Using a 2-fold cutoff and p < 0.05 derived from a Welch’s T-test between MLLr vs non-MLLr cell lines, we identified 25 unique membrane proteins specifically enriched on the MLLr surfaceome with 39 downregulated (Fig. 1b, Supplementary Fig. S1c, and Supplementary Data File 1). As positive controls, our analysis identified known hallmarks of MLLr including PROM1 and FLT3 upregulation as well as loss of CD10(19). Principal component analysis showed marked separation of MLLr B-ALL lines from BCR-ABL B-ALL and EBV-immortalized B-cells, implying a distinct cell surfaceome (Fig. 1c). Gene Ontology analysis of differentially expressed membrane proteins indicated enrichment of cell adhesion proteins while interestingly showing a marked decrease of MHC class I and II receptors (Supplementary Fig. S1d).
Figure 1: Multi-omics analysis of the MLLr B-ALL cell surfaceome uncovers unique cell surface signatures and survival dependencies.
(a) Proteomics workflow for quantifying the cell surfaceomes of B-ALL cell lines. (b) Volcano plot displaying MLLr upregulated cell surface proteins. The log2-fold change comparing the label-free quantification values (LFQ) of MLLr versus non-MLLr cell lines is plotted on the x-axis, while the −log10(p-value) is plotted on the y-axis. Proteins with log2-fold change > 2 and −log10(p-value) > 1.3 were considered significantly upregulated and are colored blue, with select proteins labeled. Significance and upregulation cut-offs are shown by dotted lines. Statistical analysis conducted using a two-sided Welch’s T-test. (c) Principal component analysis of the B-ALL cell surfaceome. Cell lines colored as follows: BCR-ABL, blue; MLLr, red; EBV B-cells, green; ETV6-RUNX1, orange. (d) Volcano plot displaying MLLr upregulated transcripts of cell surface proteins. The log2-fold change of the FPKM of different transcripts is shown on the x-axis while the −log10(p-value) is shown on the y-axis. Upregulated transcripts (log2-fold >2 and −log10(p-value) > 1.3) are shown in blue with select genes labeled. Genes identified through proteomics as up or down regulated, but were missed by transcriptome analysis are shown in orange and are labeled. Statistical analysis conducted using a two-sided Welch’s T-test. See also Supplementary Fig. S1 and S2.
To investigate surface protein regulation, we performed parallel RNA-seq. We found a modest correlation between upregulated surface-annotated proteins found by both RNA-seq and surface proteomics, consistent with prior studies(20) (Fig. 1d, Supplementary Fig. S1e-f, and Supplementary Data File 2). We did further note that several adhesion molecules, such as NCAM1, L1CAM, and ITGAV, showed significant upregulation only at the protein level. Similar adhesion proteins have been proposed to play a major role in B-ALL proliferation within the bone marrow niche(21). Furthermore, MHC downregulation is not prominently noted in RNA-seq data. These results reinforce that transcriptome-only analyses may miss biologically- and clinically-relevant findings.
CRISPRi screen of the MLLr cell surfaceome to reveal potential vulnerabilities
We reasoned that cell surface receptors relevant to the growth and survival of MLLr B-ALL may serve as promising immunotherapy targets. We therefore performed a CRISPR-interference functional genomic screen(22) of the MLLr cell line SEM, using an sgRNA library targeting 5973 genes (5 sgRNAs/gene) encoding all Uniprot-annotated membrane-spanning proteins(20) (Supplementary Methods and Supplementary Fig. S2a). We found 60 membrane proteins whose knockdown resulted in significant growth disadvantage after 30 days (>12 doublings) (Supplementary Fig. S2b). However, the only protein that showed specific upregulation on MLLr B-ALL as well as significant genetic dependence was FLT3 (Supplementary Fig. S2c). While FLT3 is being investigated by some groups as a potential immunotherapy target, this protein has marked therapeutic liabilities due to high expression on hematopoietic stem and progenitor cells (HSPCs)(23). Therefore, we explored alternate avenues to find novel targets with more favorable characteristics.
Triage of MLLr cell surface proteins identifies CD72 as an optimal immunotherapy target
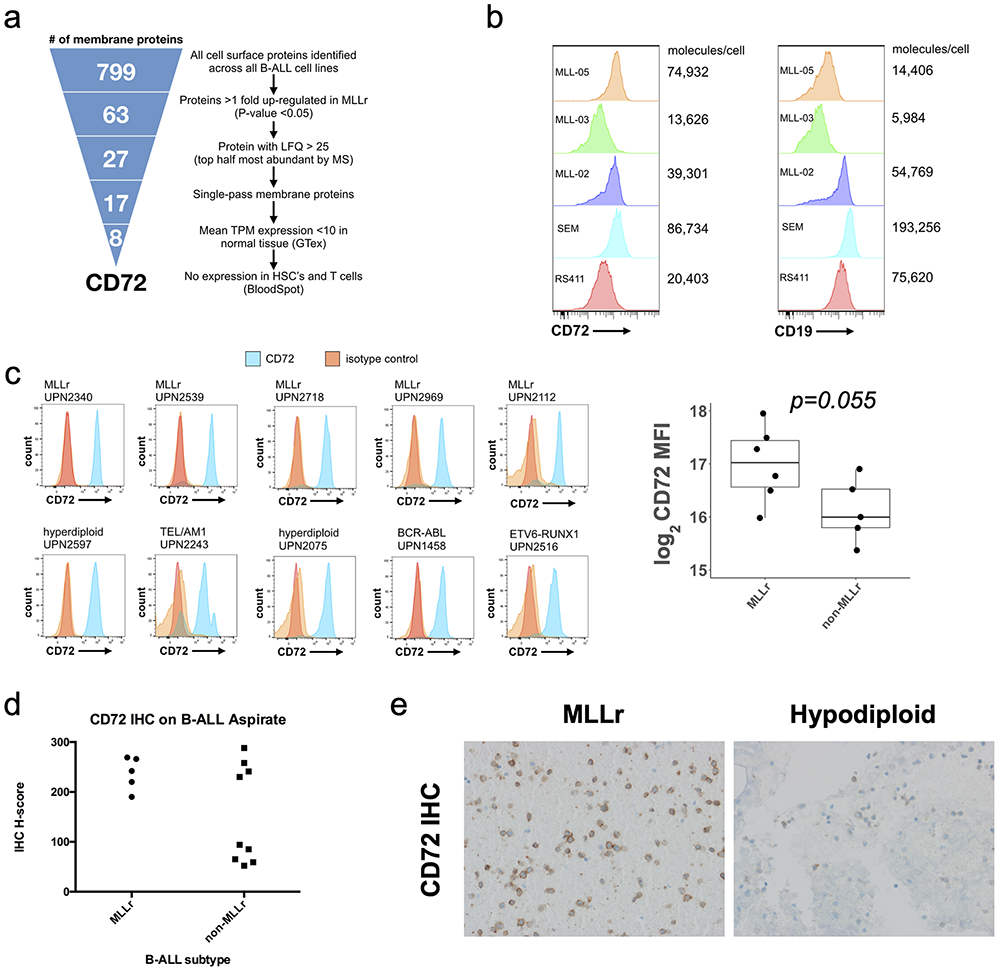
We next bioinformatically triaged our cell surface markers. We sequentially considered up-regulated cell surface markers in MLLr vs. other (63/799 proteins); relatively abundant proteins to find markers with high antigen density (LFQ intensity > 25, 27/799); and single pass membrane proteins to facilitate development of in vitro antibodies (17/799) (Fig. 2a). To avoid “on-target, off-tumor” toxicity, we eliminated protein-encoding genes with a median TPM (transcript per million) > 10 in normal tissue (excluding Spleen) per Genotype-Tissue Expression database (GTex), or any detectable immunohistochemical staining in non-hematopoietic tissues per the Human Protein Atlas(24). This left 8 of 799 proteins (Supplementary Fig. S3a). Finally, to avoid sensitive hematopoietic compartments, we eliminated proteins with any detectable RNA expression in CD34+ stem and progenitor cells (HSPCs) or T-cells in the DMAP resource(25) and Human Blood Atlas (HBA)(26) (Supplementary Fig. S3b-c). After completing this triage, one membrane protein target stood out as best-fulfilling our criteria: CD72.
Figure 2: CD72 is a highly-abundant cell surface marker upregulated on the MLLr cell surface.
(a) Schematic showing triage of cell surface membrane proteins to identify immunotherapy candidates for MLLr B-ALL. (b) Flow cytometry histograms of CD72 and CD19 surface density on MLLr B-ALL patient-derived xenografts and cell lines. Molecules of receptor per cell were calculated using a quantitative flow cytometry assay. (c) Representative flow cytometry histograms of CD72 surface density on viably-frozen, pediatric B-ALL patient samples. Log2 of the Median Fluorescence Intensity (MFI) of CD72 staining is graphed on the y-axis, comparing MLLr to non-MLLr patient samples on the x-axis (total, n =11) (d) Quantification of CD72 abundance by immunohistochemistry (IHC) staining of banked adult B-ALL patient bone marrow aspirates (total, n =15). Each tumor was graded for staining percentage and intensity by two independent pathologists blinded to sample identity, which were used to calculate IHC H-scores (range: 0-300). (e) Representative raw images of CD72 staining intensity by IHC of two different B-ALL subtypes. See also Supplementary Fig. S3, S4, and S5.
Reanalysis of multiple cell line and patient sample transcriptome datasets also confirmed that CD72 is upregulated at the transcript level in MLLr vs. other B-ALL subtypes (Supplementary Fig. S3d-f). In addition, our own RNA-seq data suggested a ~4-fold upregulation of CD72 in MLLr vs. other (Supplementary Fig. S3g), nearly identical to that found in our surface proteomic screen. This integrated transcriptome-proteome analysis therefore suggested that CD72 surface expression is primarily governed by transcriptional control.
CD72, also known as lyb-2 in murine biology, is a single-pass Type-II membrane protein with an extracellular C-type lectin domain and cytoplasmic ITIM motifs. The ITIM motifs on CD72, similar to CD22, serve as scaffolds for inhibitory phosphatases to counteract B-cell receptor (BCR) signaling(27). These proteins, as well as CD19, demonstrate highly similar expression patterns across hematopoietic cell types per the HBA (Supplementary Fig. S4a), beginning expression early in B-cell development but lost on plasmablasts and plasma cells (Supplementary Fig. S4b). Genetic ablation of CD72 in mice demonstrated no lethality but some increase in immune system activation(28). While CD72 knockdown did not affect SEM growth in vitro, the same was true of well-validated targets CD19 and CD22. Taken together, our results suggest CD72 as a promising immunotherapeutic target for MLLr B-ALL.
CD72 is expressed not only in MLLr B-ALL but across B-cell malignancies
A literature search revealed one prior report from 1992 investigating CD72 as a potential biomarker for B-cell malignancies(29). Supporting our results, the authors found positivity by immunohistochemistry (IHC) of CD72 in both MLLr B-ALL patient samples tested. However, they also noted that essentially all B-ALL and B-cell lymphoma samples examined, with the exception of plasmablastic lymphoma, also showed CD72 expression. The Cancer Cell Line Encyclopedia (CCLE) confirmed CD72 to be highly expressed in B-cell leukemia and lymphoma cell lines (Supplementary Fig. S4c). These findings suggested that targeting CD72 may find broader utility beyond MLLr B-ALL.
To independently verify these results, we examined CD72 surface expression on B-ALL patient-derived xenografts (PDX) from the ProXe biobank(30) and viably frozen primary pediatric samples from our institution (Supplementary Tables S2 and S3). By quantitative flow cytometry, we found CD72 to be expressed at several thousand copies per cell in MLLr PDX samples, similar to and sometimes greater than CD19 (Fig. 2b). Primary sample analysis suggested higher CD72 in MLLr cells than non-MLLr, but, notably, revealed CD72 expression even in non-MLLr disease (Fig. 2c). IHC on fixed adult B-ALL bone marrow aspirate found uniformly high CD72 on MLLr B-ALL blasts, compared to variable, but still present, expression in other genomic subtypes (Fig. 2d-e). We also examined peripheral blood samples from patients undergoing autologous stem cell harvest and confirmed no evidence of detectable CD72 expression on CD34+ hematopoieitic stem and progenitor cells (HSPCs) (Supplementary Fig. S4d). Analysis of the Multiple Myeloma Research Foundation CoMMpass dataset (research.themmrf.org) confirmed no detectable CD72 mRNA expression in malignant plasma cells (Supplementary Fig. S4e).
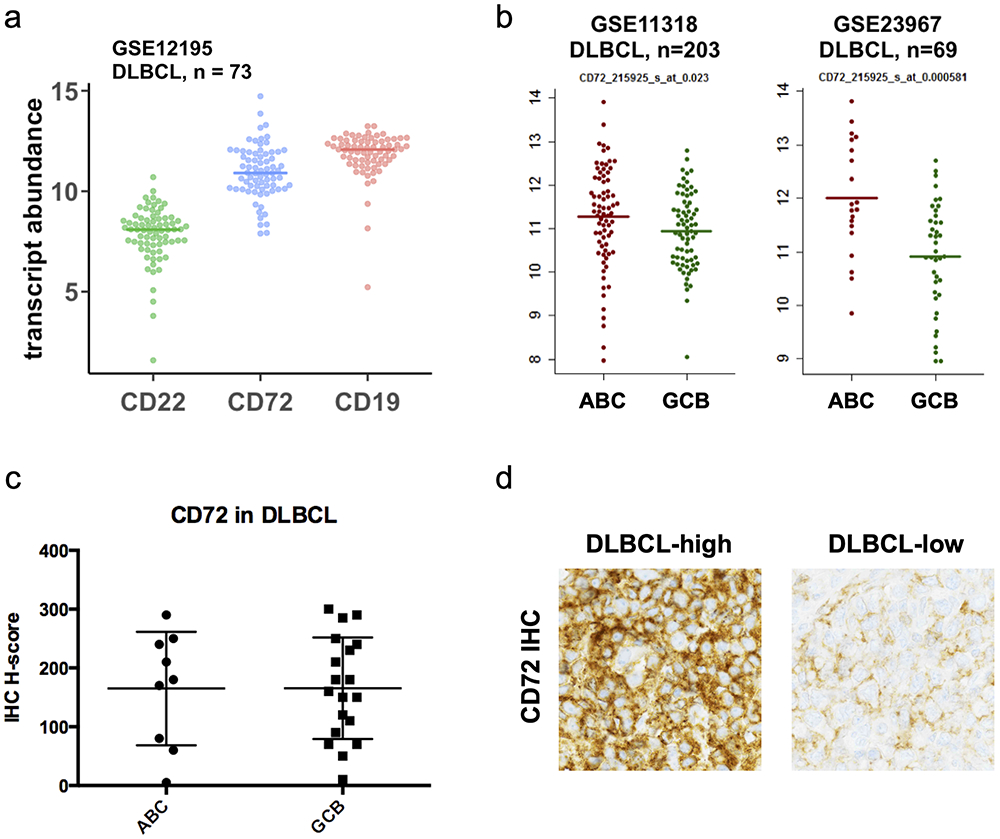
We next validated CD72 expression in DLBCL. We analyzed several publicly-available DLBCL patient transcriptome datasets(31) and found that CD72 mRNA was highly expressed, similar to levels of CD19 and generally higher than CD22 (Fig. 3a). This analysis suggested that poorer-prognosis activated B-cell (ABC) patients may have increased CD72 expression vs. germinal-center B-cell (GCB) (Fig. 3b). While CD72 IHC on an internal cohort of lymph node biopsies did not confirm this hypothesis, we did verify that 93% (26 of 28) showed detectable CD72 expression (Fig. 3c-d).
Figure 3: CD72 is a highly-abundant cell surface marker in Diffuse Large B Cell Lymphoma.
(a) Plot comparing the log2 transcript abundance of CD22, CD72, and CD19 by microarray analysis of a DLBCL patient cohort (GSE12195, n=73). (b) CD72 transcript abundance by microarray analysis of ABC and GCB subtypes of DLBCL patient samples (GSE11318, n=203 and GSE23967, n=69). (c) Quantification of CD72 abundance by immunohistochemistry (IHC) staining of banked DLBCL patient samples (total, n=28) displayed by ABC or GCB subtype. (d) Representative raw images of CD72 staining intensity by IHC of two different DLBCL patient samples.
As our combined proteomic and RNA-seq results suggested transcription is the primary determinant of CD72 antigen density, we sought to identify potential epigenetic regulators of CD72 by analyzing publicly-available datasets(32,33) (Supplementary Methods). Combining ENCODE ChIP-seq data with ATAC-seq transcription factor motif analysis suggested central B-cell-identity transcription factors, including PAX-5 and EBF-1, as having direct roles as regulators of CD72 (Supplementary Fig. S4f-j and Supplementary Data File 3). Overall, these results support the notion that CD72 is largely restricted to the B-cell compartment, further underscoring its promise as an immunotherapy target.
Development of a CD72-directed CAR-T with nanobody yeast display
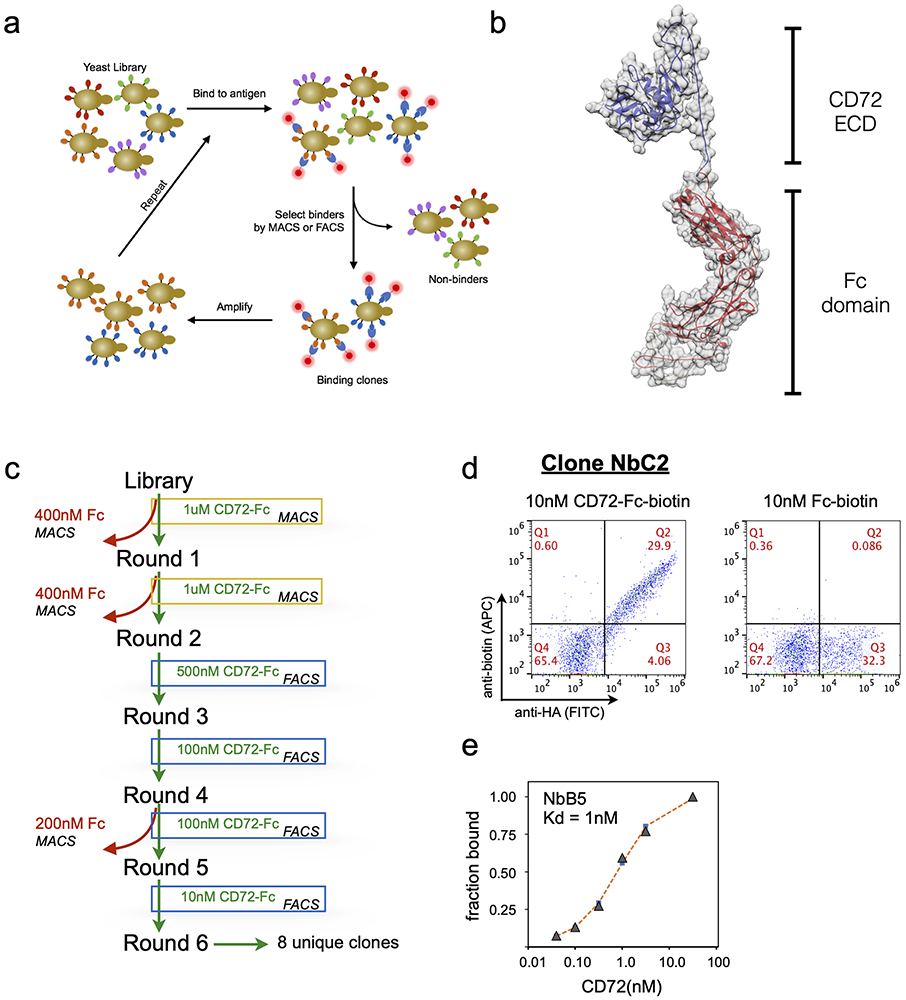
To generate CD72-specific binding reagents for use in a CAR-T cells, we employed a recently developed, fully in vitro nanobody yeast display screening platform(16) (Fig. 4a). Nanobodies are variable heavy chain-only immunoglobulins derived from camelids that, owing to their simple format, small size, and highly modular nature, are finding increasing utility in therapeutic applications. While others have recently shown the potential of nanobodies for CAR-T development(34), to our knowledge all such binders have been generated via llama immunization, a slow, expensive process with limited availability. This fully synthetic yeast display library, which is widely available for academic use, was initially designed for enabling structural biology studies. Here we demonstrate for the first time that this ready-to-use, low cost in vitro approach can also generate binders efficacious in immunotherapy applications.
Figure 4: Isolation of high-affinity CD72 nanobodies with yeast display.
(a) Workflow for in vitro anti-CD72 nanobody selection using yeast display. (b) Structure model of recombinant CD72-Fc fusion used to perform yeast display selections. (c) Schematic displaying the nanobody yeast display selection strategy for each MACS and FACS selection round to enrich for CD72-specific nanobody binders. Two rounds of MACS followed by four rounds of FACS with decreasing concentration of CD72 antigen produced high affinity anti-CD72 nanobodies (d) Flow cytometry plots of yeast clone NbC2 binding to 10nM CD72-ECD-Fc protein (left) or 10nM Fc protein (right). Y-axis displays the anti-biotin-APC signal (corresponding to yeast binding to recombinant protein) and the x-axis displays the anti-HA-FITC signal (corresponding to nanobodies displayed on the yeast surface). (e) Representative plot of on-yeast binding of recombinant CD72-Fc fusion protein for CD72-selected nanobody clone NbB5 expressed on yeast to determine estimated binding affinities. KD determined by curve fitting using non-linear least squares regression.
We expressed in mammalian cells a recombinant fusion protein comprised of the C-terminal extracellular domain of CD72 (aa 117-359) fused to a biotinylated human Fc domain to enable in vitro nanobody panning(35) (Fig. 4b). After six rounds of magnetic bead and flow cytometry-based selection (Fig. 4c and Methods), >50% of the remaining nanobody-expressing yeast specifically bound CD72. Ultimately, we identified eight unique clones that displayed high-affinity for recombinant CD72 and no binding to Fc-domain-only (Fig.4d). CDR3, the major binding determinant for both nanobodies and antibodies(36), possessed a wide range of length and sequence variability. Using on-yeast affinity assays(37), measured clones were estimated to possess KD’s in the low-nM range for recombinant CD72 (Fig. 4e). Taken together, these results indicate that this in vitro-evolved approach can generate nanobody binders to CD72.
In vitro activity of CD72-directed CAR-T against MLLr B-ALL and other B-cell malignancies
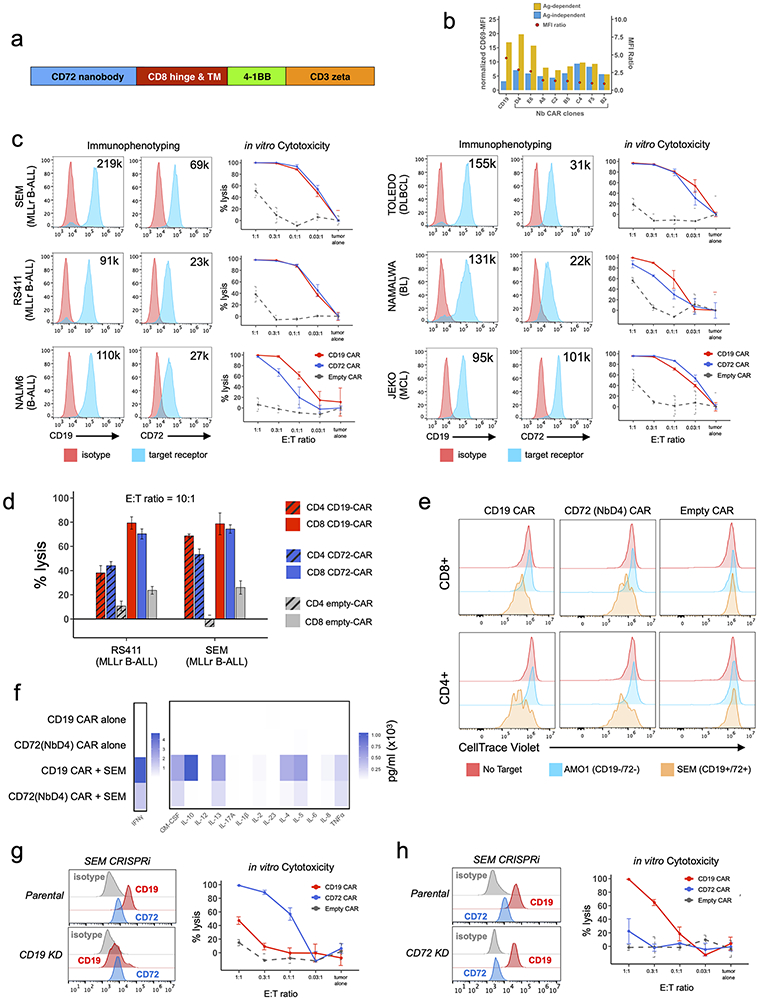
We cloned our eight unique nanobody sequences into a second-generation CAR-T format to screen activity in vitro. Notably, the lentiviral backbone (Fig. 5a) is identical to that used in tisangenlecleucel, an FDA-approved CD19 CAR-T. We first transduced Jurkat cells with our nanobody-based CAR’s to assess their antigen-independent and antigen-dependent activation during co-culture with either a CD72-negative cell line (AMO1, multiple myeloma) or a CD72-positive cell line (RS411, MLLr B-ALL). For all assays we used the tisangenlecleucel single chain variable fragment (scFv) CD19 binder as a positive control. At 1:1 effector:tumor (E:T) ratio, by CD69 staining we found clone NbD4 to possess superior antigen-dependent activation while demonstrating low antigen-independent activation (Fig. 5b). We therefore utilized this clone as our lead binder candidate in all subsequent experiments. CD72(NbD4) CAR’s were generated using normal donor CD4+ and CD8+ T-cells using standard methodology and screened for cytotoxicity against multiple B-cell malignancy cell lines in vitro (see Methods). CD72(NbD4) CAR’s demonstrated potent cytotoxicity against CD72-bearing B-ALL and lymphoma cell lines when co-cultured with tumor cells across varying E:T ratios and incubation times, mirroring efficacy of CD19 CAR-T (Fig. 5c-d). Using the proliferation stain CellTrace Violet, CD72(NbD4) CAR-T’s also demonstrated robust proliferation after 1:1 co-culture for 72-hrs with the SEM cell line, equivalent to CD19 CAR-T (Fig. 5e). We observed robust degranulation of CD72(NbD4) CAR-T’s against these same cell lines during 6-hr co-culture at a 2:1 E:T ratio, as well as against primary B-ALL patient samples, though the degree of degranulation was somewhat less than CD19 CAR-T (Supplementary Fig. S5a-c). Similarly, cytokine profiling of CD72(NbD4) CAR T after 1:1 co-culture for 24-hrs with the SEM B-ALL cell line showed comparable patterns of cytokine release to CD19 CAR-T, though of lower magnitude (Fig. 5f).
Figure 5: Nanobody-based CD72 CAR T demonstrates potent in vitro efficacy against B-cell malignancies.
(a) CD72-directed nanobody sequences were incorporated into a second-generation CAR backbone design including a CD8 hinge and transmembrane domain (TM), 4-1BB co-stimulatory domain, and CD3 activation domain. (b) Jurkat activation assay measuring antigen-dependent and independent signaling of eight candidate nanobody CAR constructs. Jurkat-CAR’s were incubated overnight (1:1 ratio) with either the CD72-negative cell line AMO1 (antigen-independent, blue bars) or the CD72-positive cell line RS411 (antigen-dependent, yellow bars). Activation measured by CD69 mean fluorescence intensity (MFI) normalized to isotype control MFI (left side y-axis). The ratio of antigen-dependent over antigen-independent MFI is shown with a red circle (right y-axis). (c) Three B-ALL cell lines (SEM, RS411, and Nalm6) and three lymphoma cell lines (TOLEDO-DLBCL, Namalwa-Burkitt’s Lymphoma, and JEKO-Mantle Cell Lymphoma) evaluated for CD19 and CD72 surface expression and susceptibility to either CD19 or CD72-directed CAR T cytotoxicity. FACS histograms display either CD19 or CD72 surface expression along with estimated number of receptors per cell (upper right hand corner of each histogram) measured by quantitative flow cytometry. In vitro cytotoxicity of either CD72 (NbD4, blue), CD19 (red), or empty(grey) CD8 CAR T’s against each cell line at varying effector:target ratios, cocultured for 48 hrs and measured by bioluminesence. (d) In vitro cytotoxicity of either CD72 (NbD4, blue), CD19 (red), or empty(grey) CD8 (solid bars) and CD4 (striped bars) CAR T’s against two B-ALL cell lines (SEM, RS411) at 10:1 effector:target ratio, cocultured for 4 hrs. Cell viability measured via bioluminesence. (e) Proliferation of CD4 and CD8 CAR T cells cultured alone or at 1:1 E:T ratio with either AMO1 tumor cells (multiple myleoma, CD19− and CD72−) or SEM tumor cells (B-ALL, CD19+ and CD72+). CAR T cells were stained with CellTrace Violet (CTV) prior to co-culture, then assayed after 72hrs by flow cytometry. (f) Cytokines released by different CD8 CAR T cells after 1:1 co-culture for 24hr with the SEM B-ALL cell line, cytokines measured by multiplex bead assay using a BioRad Bio-Plex200. (g-h) Cytotoxicity of CD72(NbD4), CD19, or empty CAR T against gene edited SEM cell lines at varying effector:target ratios, cocultured for 48 hrs. (g) Cytotoxicity versus CD19-knockdown CRISPRi-edited SEM cells. (h) Cytotoxicity versus CD72-knockdown CRISPRi-edited SEM cells. FACS histograms demonstrate efficient knockdown of either CD19 or CD72. All target cells stably expressed enhanced-firefly luciferase to enable viability measurements with bioluminescence imaging using D-luciferin. Cytotoxicity experiments were performed in triplicate and signals were normalized to control wells containing only target cells. Data represented by mean +/− SEM. See also Supplementary Fig. S5.
To evaluate the utility of CD72 CAR-T as a therapeutic option after CD19 failure, we suppressed CD19 in SEM cells using CRISPRi to generate a model of CD19 antigen escape. CD72(NbD4) CAR-T was equally efficacious against CD19-negative SEM cells as parental (Fig. 5g), whereas CD19 CAR-T showed greatly diminished activity. Additionally, we knocked down CD72 and showed CD72(NbD4) CAR-T had no detectable activity against these cells, whereas CD19 CAR-T retained robust killing (Fig. 5h). Thus, CD72(NbD4) CAR T therapy is highly- specific and potent against CD72-bearing B-cells, and effective targeting of CD72 is independent of CD19 surface density.
In vivo activity of CD72(NbD4) CAR-T against MLLr B-ALL
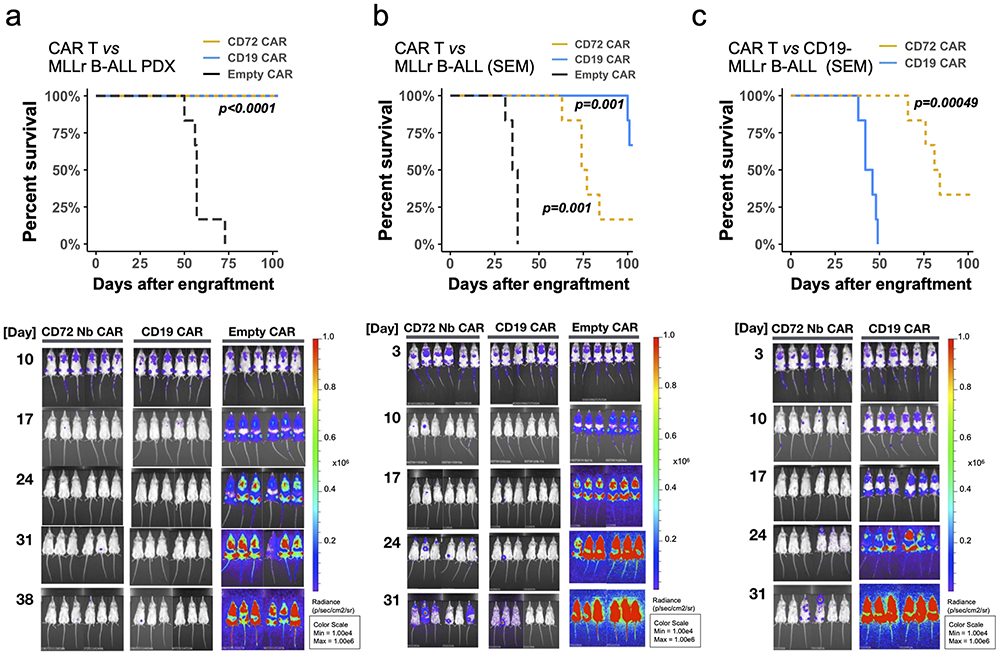
Finally, we examined the in vivo efficacy of our CD72(NbD4) CAR-T against an MLLr B-ALL cell line (SEM) and an MLLr B-ALL PDX in NOD scid gamma (NSG) mice. We engineered both cells to express luciferase for non-invasive bioluminescent imaging (BLI). 1e6 cells were implanted via tail vein injection and engraftment confirmed by BLI at either 3 or 10 days for SEM and PDX, respectively. Each cohort of mice (n=6 per arm) received 5e6 total CAR-T cells (a 1:1 mixture of CD4:CD8 primary T-cells) engineered with either an “empty” CAR backbone, CD72(NbD4) CAR, or CD19 CAR. MLLr PDX-injected mice that received CD72(NbD4) CAR-T showed a strong response and undetectable leukemic burden by BLI, comparable to CD19 CAR-T, and significantly increased survival versus the empty CAR (Fig. 6a). CD72(NbD4) CAR-T performed similarly to CD19 CAR T against wild-type SEM, significantly prolonging survival compared to empty CAR (Fig. 6b). Notably, CD72(NbD4) CAR-T’s robustly controlled CRISPRi CD19-knockdown SEM tumor cell growth in vivo (Fig. 6c). These results underscore the orthogonal nature of these two antigens and verify that targeting CD72 could be effective against CD19-negative relapsed leukemia.
Figure 6: CD72 CAR T eradicates tumors and prolongs survival in cell line and xenograft models of B-ALL.
NSG mice were injected with 1e6 firefly-luciferase labeled tumor cells including an MLLr B-ALL patient-derived xenograft, the parental SEM MLLr B-ALL cell line, and a CD19-knockdown CRISPRi SEM cell line (CD19- MLLr B-ALL). After confirming engraftment, mice were treated with a single dose of 5e6 CAR T cells (1:1 CD8/CD4 mixture) on day 10 (MLLr B-ALL PDX) or day 3 (parental and CD19- SEM MLLr B-ALL). Tumor burden was assessed weekly for five weeks via bioluminescent imaging (BLI), then mice were followed for survival. Survival curves and tumor burden via BLI for mice that received (a) MLLr B-ALL PDX and were treated on day 10 with different CAR T cells (n=6 mice per arm); (b) SEM B-ALL cells, treated on day 3 with different CAR T cells (n=6/arm); (c) CD19-negative SEM B-ALL cells, treated on day 3 with different CAR T cells (n=6/arm). p-values were computed using the log-rank test comparing different CAR constructs to Empty CAR controls, except for 6c where CD72 CAR is compared directly to CD19 CAR. See also Supplementary Fig. S8.
CD72(NbD4) CAR-T displays no toxicity against normal tissues
To evaluate potential off-tumor toxicity in key normal tissue compartments, we performed co-culture experiments combining CD72(NbD4) CAR with normal donor peripheral blood mononuclear cells (PBMCs), as well as other representative normal tissue including human umbilical vein endothelial cells (HUVECs), human bone marrow stromal cells (BMSCs), human induced pluripotent stem cell (iPSC)-derived neural progenitor cells (NPCs), human embryonic stem cell (hESC) derived cardiomyocytes, and macrophages differentiated in vitro from donor monocytes (Supplementary Methods). After 1:1 co-culture for 24hr’s, flow cytometry evaluation of PBMC population subtypes demonstrated B-cell depletion by both CD72(NbD4) CAR-T and CD19 CAR-T, but no effects on T-cells, neutrophils, NK cells, monocytes, or dendritic cells (Supplementary Fig. S6a). In 6-hr degranulation assays, CD72(NbD4) CAR showed strong activation against SEM MLLr B-ALL cells, but no discernable degranulation over empty CAR control in the presence of all normal tissues tested (Supplementary Fig. S6b). Given the recent evidence of CD19 expression in neuronal vascular pericytes, which may explain neurotoxicity associated with CD19-directed CAR-T therapy(38), we additionally analyzed human brain single-cell RNAseq datasets to assess CD72 expression. In contrast to significant CD19 expression in CD248-expressing pericytes, our analysis demonstrated no meaningful CD72 expression in this population or anywhere else in human brain tissue (Supplementary Fig. S7). Taken together, these analyses suggest that targeting CD72 is likely to lead to limited, if any, off-tumor toxicity outside the B-cell compartment.
CD72 loss alters the B-ALL membrane proteome
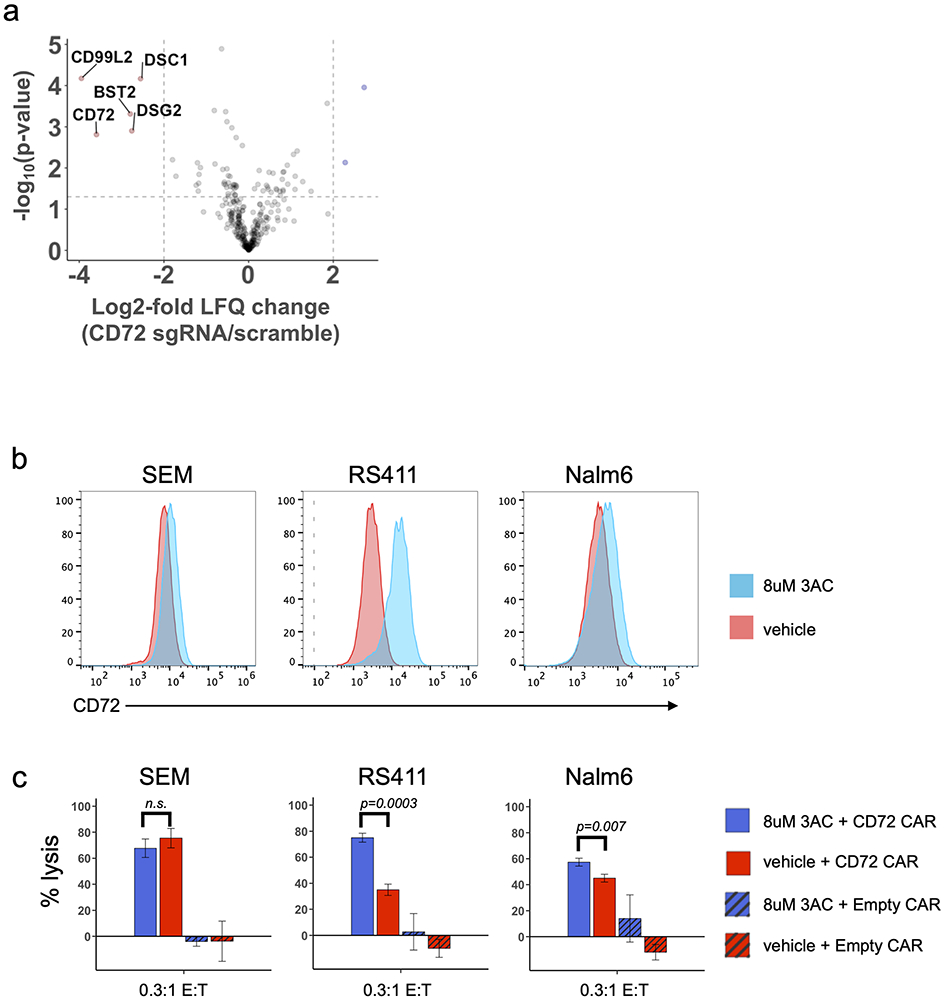
Based on our CRISPRi screen in SEM cells and clinical data with CD19 and CD22 CAR’s(2), we anticipated that CD72 antigen escape is a potential form of resistance to CD72(NbD4) CAR-T. In support of this hypothesis, we noted that bone marrow-isolated MLLr B-ALL PDX cells showed a population with decreased CD72 in NbD4- vs. empty CAR-treated mice (Supplementary Fig. S8a-c). Given CD72’s role in modulating BCR signaling, we hypothesized that other components of this pathway, including other known immunotherapy targets (i.e. CD22, CD79a), may have altered expression levels in the context of CD72 antigen escape. We modeled this scenario by coupling CRISPRi CD72 knockdown in RS411 MLLr B-ALL cells to quantitative cell surface proteomics. Contrary to our hypothesis, we did not find significantly altered regulation of the aforementioned well-known immunotherapeutic targets, nor other BCR signaling components. However, we did find multiple surface proteins downregulated in parallel with CD72, that are primarily involved in cell adhesion, migration, and desmosome junctions (CD99L2, BST2, DSG2) (Fig. 7a). Several of these genes were predictive of poor overall survival when highly expressed (Supplementary Fig. S8d). DSG2 downregulation was confirmed by flow in both SEM and RS411 CD72-knockdown cell lines (Supplementary Fig. S8e). While thoroughly validating this proposal is beyond the scope of our work here, in conjunction with prior studies(39) these findings suggest a hypothesis that loss of CD72 may lead to decreased B-ALL adhesion within the marrow niche, thereby increasing vulnerability to standard chemotherapeutic regimens.
Figure 7: Modulating the B-ALL surfaceome after CD72 antigen escape and increasing CD72 antigen density with SHIP1 inhibition.
(a) Volcano plot displaying differentially regulated cell surface proteins dependent on CD72-knockdown. The log2-fold change comparing the label-free quantification values (LFQ) of RS411-CD72 knockdown versus RS411-scramble cell lines is shown on the x-axis, while the −log10(p-value) is shown on the y-axis. Proteins with log2-fold change < −2 and p < 0.05 were considered significantly downregulated and are colored red. Significance cut-offs are shown by dotted lines. Statistical analysis was conducted using a two-sided Welch’s T-test. (b) Change in CD72 surface density on three B-ALL cell lines after 72 hours of 3AC treatment (8 μM). (c) CAR-T cytotoxicity of three B-ALL cell lines pre-treated with either 3AC (8 μM) or vehicle for 72 hours. Drug was washed out of B-ALL cell lines prior to incubation with either CD72 CAR-T or Empty CAR-T for 24 hours, at 0.3:1 E:T ratio. All target cells stably expressed enhanced-firefly luciferase to enable viability measurements with bioluminescence imaging using D-luciferin. Experiments were performed in triplicate and signals were normalized to control wells containing only target cells. Data is represented as mean +/− SEM. Significance determined using a two-sided t-test. See also Supplementary Fig. S8.
Modulating CD72 antigen density through SHIP1 inhibition
While CD72 is known to be a negative regulator of BCR signaling in normal B-cells, its role in oncogenic BCR signaling remains mostly unexplored. We were inspired by recent work where treatment with ibrutinib, a Bruton’s tyrosine kinase (BTK) inhibitor, led to downregulation of several negative regulators of BCR signaling, including CD22, CD72, and PTPN6, presumably to re-achieve a baseline level of BCR signaling strength(40). We reasoned that the opposite may also be true, whereby pharmacologically agonizing BCR signaling may increase surface CD72. Recent work in BCR-ABL B-ALL has suggested that pharmacologically inhibiting the phosphatase SHIP1 can agonize the BCR pathway, leading to tumor cell death via activation of the autoimmunity checkpoint (AIC) (41). More broadly, upregulating immunotherapy targets using pharmacological intervention has generated great interest as a strategy to mitigate antigen escape(42-44). To test this hypothesis, we treated MLLr cell lines SEM and RS411 with the SHIP1 inhibitor 3AC, finding a strong dose-dependent increase in CD72 surface abundance at 72 h (Supplementary Fig. S8f). In addition, we found that 3AC showed some degree of cytotoxicity to MLLr cell lines, potentially suggestive of AIC-mediated cell death (Supplementary Fig. S8g-h). Using unbiased phospho-proteomics, we notably confirmed increased phosphorylation of Bruton’s tyrosine kinase (BTK) and LYN substrates (among others) in MLLr vs. BCR-ABL B-ALL cell lines, corroborating active BCR signaling pathways (Supplementary Fig. S8i). We next evaluated the ability of 3AC to enhance CD72 CAR-T cytotoxicity. RS411 (MLLr) and NALM6 (non-MLLr) B-ALL cells, with moderate to low baseline CD72 copy number (Fig. 5c), indeed showed significant increases in CD72 CAR-T cytotoxicity after SHIP1 inhibition (Fig. 7b-c). Notably, this increased cytotoxicity was in line with relative increase in CD72 surface antigen density after 3AC (Fig. 7b). These findings are also in agreement with our results in Fig. 5c, where we saw a correlation between CD72 antigen density and CAR-T cytotoxicity across cell lines. In contrast, SEM cells, which express high (Fig. 5c) levels of CD72, did not show any impact on CAR-T cytotoxicity after 3AC. This result suggests a possible “saturation limit” at which CD72 antigen density no longer impacts efficacy, though this conclusion will require more investigation in the future. We do note that relatively high doses of the tool compound 3AC were required to achieve both CD72 upregulation and tumor cell death in these models. However, more potent, ideally clinical-grade inhibitors of SHIP1 are under active investigation. Taken together, our results suggest that combining SHIP1 inhibition with CD72-directed therapy could be a promising rational combination strategy, particularly in the setting of lower baseline CD72 antigen density.
DISCUSSION
Our unbiased assessment of the membrane proteome demonstrates that in B-ALL different oncogenic drivers can generate markedly different cell surface signatures. These results underscore the power of surface proteomic profiling to identify novel immunotherapeutic targets. We found that MLLr B-ALL possesses a unique cell surface signature, marked by increases in receptors related to cell adhesion with significantly depleted MHC I and II. Notably, many of these surface signatures would not be detected by RNA analysis alone. While our results uncover several potential approaches for targeting MLLr biology via the cell surfaceome, and thereby provide a useful resource for the community, here we focused on CD72 as a particularly promising target for MLLr disease. To our knowledge, this poorest-prognosis subtype of B-ALL does not have any previously known preferential immunotherapy targets. Notably, our strategy here may also be applicable to identifying other genotype-enriched surface targets in other malignancies.
Bioinformatic analysis and patient sample examination further indicated that CD72 is highly expressed in other B-cell malignancies, and driven by critical B-cell-related transcription factors, but minimally expressed in other tissues. Given the current clinically-available armamentarium, CD72 is most likely to be placed as a second-line target for patients relapsing after CD19-directed therapy. However , there may be some advantages to targeting CD72 instead of CD19. These include potential for less neurotoxicity, based on our brain scRNA-seq analysis (Supplementary Fig. S7). Therefore, we propose CD72 as an attractive immunotherapeutic target for this poorest-prognosis subtype of B-ALL, while future work may more firmly validate CD72 nanobody CAR-T’s as having broader utility in other B-cell malignancies.
Prior studies have described the development of antibody-drug conjugates (ADC) targeting CD72 for various B-cell cancer indications (45-47). However, to our knowledge no prior work has proposed any specific targeting of CD72 on MLLr B-ALL, as we find here. Furthermore, based on available data this ADC strategy appears limited by modest efficacy, possibly due to poor internalization after antibody binding. In contrast, this property may be beneficial for cellular therapies. We are particularly excited that the fully in vitro nanobody selection platform we employ here can develop robust CAR-T-active binders. The ability to rapidly generate nanobodies for CAR-Ts, without the need for llama immunization and using a library widely available for academic use, promises to greatly expand the utility of this approach. In particular, the small size and strictly monomeric fold of nanobodies makes them advantageous for immunotherapy applications. For example, almost all current CAR-T’s employ scFv binders, which suffer from well-known protein engineering problems including the selection of chain orientation (VH-VL vs VL-VH) and linker length, both of which heavily influence CAR behavior, tonic signaling, and clinical responses(48). The inherent simplicity of nanobodies eliminates these protein engineering difficulties while also potentially facilitating the design of CAR constructs simultaneously targeting multiple antigens.
There are caveats to our approach. First, while our in vitro and bioinformatic analyses suggest low probability of off-tumor toxicity with CD72(NbD4), we cannot fully exclude this possibility on some other hematopoietic cells, particularly if new binders more sensitive for very low levels of CD72 are employed. Encouragingly, based on transcriptional data, CD22 demonstrates roughly similar expression patterns to CD72 (Supplementary Fig. S4a); fortunately, significant off-target activities have not been noted in CD22-directed clinical trials(49). Furthermore, while our preclinical results here are certainly very promising, there is likely additional room for nanobody optimization and CAR engineering. Finally, one concern is the potential for immunogenicity. However, we note that the llama VHH framework residues are >80% homologous to human VH sequences, a vast improvement over murine scFv’s currently used clinically, which possess only ~60% homology yet still have curative potential. Notably, a bi-epitopic, anti-BCMA nanobody CAR-T, generated via llama immunization, is currently in clinical trials for multiple myeloma and thus far has displayed impressive clinical outcomes with minimal impact of binder immunogenicity(34). While this real-world data is highly encouraging, in future work we will continue to evaluate the potential for CD72 nanobody humanization while retaining potent activity.
Importantly, we demonstrate CD72(NbD4) CAR T cells eliminate tumor cells lacking CD19 (Fig. 5g and Fig. 6c), supporting utility as a second-line therapy after CD19 failure, at least in the absence of complete lineage switching(11). However, our results indicate that CD72, like CD19 and CD22, can be lost without any major effect on B-ALL viability. Therefore, we coupled CRISPRi knockdown with cell surface proteomics to define membrane protein alterations in the context of CD72 downregulation. This strategy, which could be considered as “antigen escape profiling”, may prove of broad utility in the study of immunotherapy resistance when applied to other novel targets. In addition, in anticipation of reversing resistance, we investigated the mechanistic hypothesis that agonizing BCR signaling could both increase CD72 expression and directly kill tumor cells. Future pharmacologic approaches to agonize BCR signaling could potentially be combined with CD72-targeted cellular therapy as part of a rational combination strategy in patients. To our knowledge, this is the initial demonstration of CD72 as an attractive cellular therapy target. Our in vitro nanobody engineered T cell platform has potential for immediate clinical translation in MLLr B-ALL and potential future application to an array of tumor types.
METHODS
Human cell lines and patient samples
Human cell lines (Supplementary Table S1) were a generous gift from Dr. Markus Müschen, and were authenticated by STR analysis prior to conducting proteomics experiments. Cells were grown in RPMI-1640 supplemented with 20%FBS and 100 U/ml Penicillin-Streptomycin (pen-strep) and passaged for less than 6 months before use. Cell lines were not tested for Mycoplasma. All MLLr B-ALL patient-derived xenografts (Supplementary Table S2) were obtained from the Public Repository for Xenografts (PRoXe) at Dana-Farber Cancer Center except for the ICN3 PDX line (MLLr B-ALL) which was a generous gift from Dr. Markus Müschen, City of Hope. Primary patient samples that had been viably frozen were obtained from the UCSF Helen Diller Cancer Research Center Tissue Bank (Supplementary Table S3). All cells were cultured at 37°C in a humidified incubator with 5% CO2. Modified cell lines were generated using lentivirus as described below.
Cell Surface Protein Labeling
Cell surface proteins were labeled with biotin using the N-linked glycosylation-site biotin labeling method(17). Briefly, 3 × 107 live cells were washed twice and resuspended in 1ml of ice-cold PBS, and treated with 1.6mM sodium metaperiodate (VWR, 13798-22) at 4°C for 20 minutes to oxidize the vicinal diols of sugar residues linked to surface proteins. The cells were then washed twice in PBS in order to remove excess sodium metaperiodate. Cells were resuspended in 1ml of ice-cold PBS and treated with 1mM biocytin hydrazide (Biotium, 90060) and 10mM Aniline (Sigma-Aldrich, 242284) at 4°C for 90 minutes with gentle mixing in order to biotinylate free aldehydes exposed on the sugar residues. After labeling, cells were washed three times with ice-cold PBS to removed excess biotin, frozen in liquid nitrogen, and stored at −80°C until further processing for mass spectrometry. All experiments were performed in biological triplicate with replicates harvested from consecutive passages.
Cell lysis, cell surface protein enrichment, and peptide digestion
Frozen cell pellets were thawed on ice in 1ml of RIPA buffer (Millipore, 20-188) with the addition of 1X HALT protease inhibitors (Pierce, 78442). After incubation on ice for 10 minutes, cells were disrupted by sonication and the lysates were clarified by centrifugation at 17,000 RCF at 4°C for 10 minutes. Clarified lysate was mixed with 500uls of neutravidin agarose resin (Thermo, 29200) and incubated at 4°C for 2 hours with end-over-end mixing. Neutravidin beads with captured biotinylated surface proteins were washed extensively by gravity flow to remove unbound proteins using 50mls of 1X RIPA + 1mM EDTA, followed by 50mls of PBS + 1M NaCl, and finally 50mls of 50mM ABC + 2M Urea buffer. Washed beads were resuspended in digestion buffer (50mM Tris pH 8.5, 10mM TCEP, 20mM 2-Iodoacetamide, 1.6M Urea) with 10ugs of added Trypsin protease (Pierce, 90057) to perform simultaneous disulfide reduction, alkylation, and on-bead peptide digestion at room temperature overnight (16-20 hours). After digestion, the pH was dropped to ~2 with neat trifluoroacetic acid (TFA, Sigma, T6508-10AMP) and the peptide mixture was desalted using a SOLA-HRP column (Thermo, 60109-001) on a vacuum manifold. Desalted peptides were eluted with 50% acetonitrile (ACN, Sigma, 34998-4L) and 50% water with 0.1% TFA and dried down completely in a speedvac. Dried peptides were resuspended in LC/MS grade water (Fisher, W64) with 2% ACN and 0.1% formic acid (FA, Honeywell, 94318-250ML-F). Peptide concentration was measured using 280 nm absorbance on a Nanodrop (Thermo), and the peptide concentration was adjusted to 0.2ug/ul for mass spec runs.
LC-MS and Data Analysis
For each replicate, 1ug of peptide was injected onto a Dionex Ultimate 3000 NanoRSLC instrument with a 15-cm Acclaim PEPMAP C18 (Thermo, 164534) reverse phase column. The samples were separated on a 3.5-hour non-linear gradient using a mixture of Buffer A (0.1% FA) and B (80% ACN/0.1% FA), from 2.4% ACN to 32% ACN. Eluted peptides were analyzed with a Thermo Q-Exactive Plus mass spectrometer. The MS survey scan was performed over a mass range of 350-1500 m/z with a resolution of 70,000, with a maximum injection time (IT) of 100 ms. We performed a data-dependent MS2 acquisition at a resolution of 17,500, AGC of 5e4, and IT of 150 ms. The 15 most intense precursor ions were fragmented in the HCD at a normalized collision energy of 27. Dynamic exclusion was set to 20 seconds to avoid over-sampling of highly abundant species. The raw spectral data files have been deposited at the ProteomeXchange PRIDE repository (PXD016800).
Raw spectral data was analyzed using MaxQuant v1.5.1.2(18) to identify and quantify peptide abundance and searched against the human Swiss-Prot annotated human proteome from Uniprot (downloaded 5/13/18 with 20,303 entries). The “match-between-runs” option was selected to increase peptide identifications while the “fast LFQ” option was selected to calculate label-free quantification values (LFQ) of identified proteins. All other settings were left to the default MaxQuant values. The MaxQuant output data was analyzed using Perseus(50) and the R program (version 3.4.0) in R-Studio. Proteins annotated as “reverse”, “only identified by site”, and “potential contaminant” were filtered out as well as proteins that were quantified in less than 2 out of 3 biological replicates in at least one experimental group. Proteins were further filtered to include only membrane-proteins or membrane-associated proteins using a manually curated list of surfaceome proteins(20). Missing values were imputed based on the normal distribution of the dataset as implemented by Perseus. Volcano plots were generated using output from a two-sample T-test comparing the log-2 transformed LFQ protein abundance values from different cell lines with a false discovery rate (FDR) set to 0.01. All proteomics results figures (as well as all other figures) were produced using the R program package ggplot2(51).
RNA-seq
Total RNA from cell pellets (biological duplicates) were isolated with the QIAgen RNAeasy kit (QIAgen, 74104) and mRNA was further purified from isolated total RNA by poly(A) separation using Oligo (dT)25 magnetic beads (NEB, S1550S) per the manufacturer instructions. Total RNA and mRNA concentration was measured by NanoDrop (Thermo). RNAseq libraries were prepared from purified mRNA using the KAPA Stranded RNAseq kit with riboerase (Kapa Biosystems, KK8483) per the manufacturers protocol and quantified using a 2100 Bioanalyzer with the High Sensitivity DNA Kit (Agilent, 5067-4626). Next generation sequencing was performed on an Illumina NextSeq (High Output) using single end, 75-bp reads at the Chan-Zuckerberg Biohub. Reads were mapped to the reference human (hg19) genome sequence, using Tophat2 aligner(52) with the default parameters. The mRNA expression level for each gene was represented as FPKM, called by Cufflinks(53). The raw sequence files have been deposited at the Gene Expression Omnibus (GEO) repository (GSE142447).
Flow Cytometry
Staining of cells was performed with either 0.5 or 1e6 total cells per sample unless otherwise noted and the manufacturers recommended amount of antibody was used in 100uls total volume of FACS buffer (PBS +2% FBS) for 30-60 minutes prior to washing with excess FACS buffer. Samples were immediately analyzed using either a BD Cytoflex or BD FACS Aria2 flow cytometer. Antibodies used for flow cytometry are listed in Supplementary Table S4.
Immunohistochemistry
Samples were obtained from the UCSF hematopathology archive under an Institutional Review Board-approval protocol. Paraffin-embedded tissue was sectioned at 4 μm onto positively charged glass slides. The slides were incubated at 60°C for 2-12h prior to staining. All immunohistochemistry was performed on a Ventana Medical Systems Discovery Ultra Automated Slide Preparation System. Following deparaffinization, tissue sections were conditioned using an alkaline buffer (CC1, Roche, 950-124) at 97°C for 92 minutes followed by treatment with hydrogen peroxide (Roche, 760-4840) at room temperature for 12 minutes, prior to application of the primary antibody. A rabbit polyclonal antibody to CD72 (Atlas Antibodies, HPA044658, Lot# R40911), applied to the slides at a 1:250 dilution, was incubated for 32 minutes at 36°C. Primary antibody detection used HQ hapten-labelled anti-rabbit antibodies (Roche, 760-4815, incubated for 12 minutes, 37 °C) with the DISCOVERY HQ-HRP Detection system (Roche, 760-4820) and DAB as the chromogen.
Expression of CD72 Fc-fusion antigen
DNA encoding the CD72 extracellular domain (amino acids 117-359) was PCR amplified from a plasmid obtained from the Human ORFeome collection (hORFeome V8.1), and cloned into a mammalian expression vector, fused to the C-terminus of a human constant CH2-CH3 domain (Fc domain), along with a N-terminal AviTag to facilitate site-specific biotinylation during expression. For expression, 30ug of plasmid was transiently transfected into Expi293F cells (Thermo Fisher, A14527, modified to stably express ER-localized BirA) using polyethyleneimine (PEI, Transporter-5, Polysciences, 26008-5) at a 4:1, PEI:DNA mass ratio. Cells were cultured in Expi293 Expression Medium (Thermo Fisher, A1435101) supplemented with 100uM Biotin for 5-7 days to allow for protein expression and biotinylation. To purify recombinant protein, cells were pelleted and the supernatant that contained the protein was recovered, filtered, and pH adjusted with PBS (pH 7.4), prior to loading onto a HiTrap Protein A HP antibody purification column (GE, 29048576) to capture the Fc-fusion protein. The column was washed with PBS and the protein was eluted with 0.1M acetic acid, then buffer exchanged into PBS using a 10K kDa MWCO, Amicon Ultra-4 spin column (EMD Millipore, UFC503008). The concentration of Fc-fusion protein was determined by A280 on a NanoDrop (Thermo) and the molecular weight was confirmed by SDS-PAGE prior to aliquoting, snap freezing in liquid nitrogen, and storage at −80°C until use in yeast display.
Nanobody selections using yeast display
Nanobody yeast selections were performed as previously described (16) with some modifications. Briefly, the first MACS selection round utilized 5e9 induced yeast from the naïve library with an initial pre-clear depletion step prior to specific enrichment. Depletion was performed by washing and resuspending induced yeast in selection buffer (20mM HEPES pH 7.5, 150mM NaCl, 0.5% BSA, 20mM Maltose) with 400nM recombinant Fc-biotin protein plus 500uls of anti-biotin microbeads (Miltenyi,130-090-485) and incubating for 40 minutes at 4°C. The sample was passed over a MACS LD column (Miltenyi, 130-042-901) to capture nanobody-yeast with affinity for the magnetic beads or the Fc protein domain. For enrichment, yeast recovered in the flow-through were pelleted, then resuspended in selection buffer with 1uM recombinant CD72-Fc-biotin and anti-biotin microbeads and incubated for 1 hour at 4°C. The sample was passed over a MACS LS column (Miltenyi, 130-042-401), washed extensively, then eluted with selection buffer off-magnet to recover nanobody-yeast specific for the CD72 ECD. Recovered yeast were grown overnight in glucose containing Trp-dropout media (United States Biological, D9530), then induced with galactose containing Trp-dropout media to begin the next round. Two rounds of MACS selection were performed with equivalent amounts of reagents used in each round. For subsequent FACS selection rounds, induced yeast from previous rounds were washed, resuspended in selection buffer, and incubated with recombinant CD72-Fc-biotin protein for 1 hour at 4°C. To stain for FACS sorting, yeast were washed, then resuspended in fresh selection buffer and incubated with anti-HA-FITC (Miltenyi, 130-099-389) and anti-biotin-APC (Miltenyi, 130-113-288) antibodies for 30 minutes at 4°C in the dark, then washed and resuspended in cold selection buffer prior to FACS sorting. Yeast were sorted on a BD FACS Aria2 by gating on the double positive FITC/APC population of yeast (yeast expressing HA-tagged nanobodies on their surface and bound by recombinant CD72-Fc-biotin protein, respectively). To isolate nanobody binders with increased affinity for CD72 ECD, four rounds of FACS were conducted using decreasing amounts of recombinant protein (500nM, 100nM, 100nM, 10nM). To isolate individual nanobody-yeast clones after the final selection round, yeast were sparsely plated on YPD-agar plates, and individual yeast colonies were picked, grown, and induced in 96-well deep well microplates. Nanobody-yeast clones were screened in 96-well plate format for CD72-ECD specific binding by incubating with 10nM CD72-Fc-biotin protein, or in a separate plate to rule out Fc binders, 10nM Fc-biotin protein. Clones were stained with antibodies and analyzed with a BD Cytoflex flow cytometer. Nanobody-yeast clones with both positive binding to CD72-Fc-biotin and negative binding to Fc-biotin were subsequently selected for plasmid isolation and sanger sequencing.
Nanobody binding affinity determination by on-yeast FACS
Estimated nanobody binding affinities for recombinant CD72-ECD were determined on-yeast using established methods as previously described(37). Briefly, nanobody-expressing yeast clones were incubated in selection buffer with multiple, decreasing concentrations of recombinant CD72-Fc-biotin protein for several hours at 4°C to allow for complete binding equilibrium to occur. Yeast samples were washed, then stained with anti-HA-FITC and anti-biotin-APC for 30 minutes at 4°C in the dark. Yeast were washed and resuspended in selection buffer, then analyzed by flow cytometry on a BD Cytoflex flow cytometer. To determine binding constants, spectra was gated on double-positive APC/FITC populations, and the median fluorescent intensity of the APC signal was plotted against the concentration of CD72-ECD recombinant protein to construct binding curves. Dissociation constants (KD) were determined by curve fitting the data using non-linear least squares regression.
CAR constructs
All CAR expression plasmids utilized identical components aside from the variable extracellular binding domains (CD72-directed nanobodies or CD19-directed scFv). The signaling components including the CD8 hinge and transmembrane domain, 4-1BB co-stimulatory domain, and CD3ζ signaling domain are identical to that utilized in the clinically approved CD19-directed CAR construct tisangenlecleucel. Nanobody binders were cloned into the CAR backbone plasmid using Gibson Assembly. CAR expression vectors utilized a GFP marker for identification of CAR+ cells.
Jurkat-CAR activation testing
Jurkat T-cells were transduced with experimental CAR constructs including CD72-directed nanobody-based CAR’s, empty CAR, and CD19-directed CAR T as previously described. Jurkat CAR-expressing cells were incubated with target cells at a 1:1 ratio overnight (50,000 cell each), then assessed for activation by examining CD69 cell surface upregulation by flow cytometry (anti-CD69-APC, BD, 560967) on a BD Cytoflex flow cytometer. Antigen-independent activation was assessed by incubation of Jurkat-CAR cells alone or with the CD72 & CD19 negative, multiple myeloma cell line AMO1. Antigen-dependent activation was assessed by incubation of Jurkat-CAR cells with the CD72/CD19 positive B-ALL cell line SEM.
Transduction and expansion of human T cells
Primary human T cells were purified from the leukapheresis products of anonymous healthy blood donors from the Blood Centers of the Pacific (now Vitalant) under an IRB-exempt protocol in accordance with the U.S. Common Rule (Category 4). CD8+ and CD4+ T cell populations were isolated separately using RosetteSep Human T cell Enrichment kits (STEMCELL Technologies, 15023 and 15022). T-cells were cultured in X-VIVO 15 T cell media (Lonza, c04-418Q) supplemented with 10% human AB serum (Valley Medical, HP1022), 10mM neutralized N-acetyl cystine (Sigma-Aldrich, A9165-5G) and 55uM 2-mercaptoethanol (Thermo Fisher, 21985023), and were passaged every two days. For expansion, T-cells were stimulated with CD3/CD28 Dynabeads (Life Technologies, 11131D) according to the manufacturer’s instructions (25uls of beads per 1 million T cells) for five days and grown in the presence of 30U/ml recombinant IL-2 (ProSpec, CYT-209). Transduction with CAR lentiviruses was performed one day after the start of bead stimulation without spinfection or the use of polybrene supplements. After the removal of activation beads, transduction efficiency was assessed by flow cytometry (GFP+) and T cells were optionally MACS enriched, then expanded for another four days. CAR T cells used in all experiments were normalized for CAR expression.
CAR T in vitro cytotoxicity assays
Cytotoxicity assays were conducted by mixing target cells with CAR T cells for either 4, 24, or 48 hours, at ratios as described for each experiment. For measuring cytotoxicity by bioluminescence with target cell lines stably expressing effLuc, 150ug/ml of D-luciferin (Gold Biotechnology, LUCK-1G) was added to each sample, incubated for ten minutes at room temperature, then read using a GloMax Explorer luminescence plate reader (Promega). Percent viable cells were normalized to bioluminescence of target cells incubated alone (100% viable) and experiments were performed with technical triplicates.
CAR T proliferation assays
CAR-T cells were stained with CellTrace Violet (CTV) according to the manufacturers protocol, then co-cultured with or without target cells (1:1 E:T ratio) for 72 hours. CAR-T proliferation and cell doublings were measured via flow cytometry by observing dilution of the CTV signal on a BD Cytoflex flow cytometer (BD Biosciences).
CAR T cytokine release assays
CAR-T Cytokine release was assessed after 24-hrs of CAR-T’s in culture alone or in co-culture with target cells at a 1:1 effector-to-target ratio. Cytokine concentrations in collected supernatant was measured by Eve Technologies using a multiplex bead assay with a BioRad Bio-Plex200 and a Milliplex Human High Sensitivity T-Cell panel (Millipore) according to their protocol.
Murine experiments
NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were obtained from in-house breeding stocks at the UCSF Preclinical Therapeutics Core (PTC) facility or from The Jackson Laboratory. A mixture of male and female, 6-8 weeks old NSG mice were transplanted via tail-vein injection with 1e6 B-ALL tumor cells including either the SEM cell line, SEM-CRISPRi knockdown lines, or the ICN3 B-ALL xenograft, all previously modified to stably express luciferase. Tumor burden was assessed either three or ten days post-transplantation (for cell lines or xenografts, respectively) through non-invasive bioluminescent imaging at the UCSF PTC on a Xenogen In Vivo Imaging System. Mice were distributed to different experimental arms such that each arm had equal initial tumor burden. One day after distribution, mice received 5e6 CAR T cells with different targeting domains described for each experimental arm (1:1 mixture of CD4/CD8) via tail vein injection. Tumor burden was subsequently monitored weekly for one month. Mice were followed for survival which is denoted by the time to develop overt leukemia symptoms at which point mice were sacrificed and their bone marrow was harvested and viably frozen for later analysis. All studies were approved by the UCSF Institutional Animal Care and Usage Committee.
Inhibitors
The INNP5D/SHIP1 inhibitor 3AC (3-alpha-aminocholestane) was obtained from Calbiochem (Millipore Sigma, 565835). Concentrated stocks were made at 10mM in sterile DMSO and stored at −20°C until used.
Cell viability assay
Cells were aliquoted into 96-well plates in 100uls of complete media at 1,000 - 10,000 cells per well prior to addition of 3AC diluted into complete media at varying concentrations. After two days cell viability was measured using the CellTiter-Glo luminescence cell viability assay (Promega, G7573) according to the manufacturer’s instructions. Luminescence was measured on a GloMax Explorer luminescence plate reader (Promega). Percent viable cells at each dose were normalized to untreated wells and all experiments were performed with technical triplicates at each dose.
CD19 and CD72 surface density measured by flow cytometry
Quantitative measurement of CD19 and CD72 surface density was determined by flow cytometry using the Bangs Laboratory Quantum MESF Kit (Bangs Labs, Quantum R-PE MESF, 827) and was performed according to the manufacturers recommended procedure. Flow cytometry was performed on a BD Cytoflex flow cytometer (BD Biosciences).
Statistical analysis
All data is presented as mean +/− standard deviation. Statistical significance in proteomics and transcriptomics analysis was determined by Welch’s t-test; two-sample t-test with null hypothesis that the different in log2-transformation of proteomic LFQ’s, transcriptome FPKM’s for RNA-seq, or transcriptome microarray intensity is equal to 0. A p < 0.05 is considered statistically significant. For Kaplan-Meier survival analysis we used the log-ranked test to determine statistical significance.
Supplementary Material
STATEMENT OF SIGNIFIGANCE.
KMT2A/MLL1-rearranged (MLLr) B-ALL patients have poor prognoses despite recent immunotherapy advances. Here surface proteomics identifies CD72 as enriched on MLLr B-ALL but also widely expressed across B-cell cancers. We show a recently-described, fully-in vitro nanobody platform generates binders highly active in CAR-Ts, and demonstrate its broad applicability for immunotherapy development.
Acknowledgements:
We thank Dr. Markus Müschen for providing B-ALL cell lines and insightful comments on the manuscript, Drs. Alex Martinko and Max Horlbeck for providing CRISPRi sgRNA libraries and advice on data analysis, Drs. Axel Hyrenius-Wittstein, Kole Roybal, and Justin Eyquem for providing CAR-T backbone construct, protocols for generating CAR-T cells, and advice on murine studies, Dr. Andrew Leavitt for providing access to stem cell harvest samples, and the staff of the UCSF Helen Diller Family Comprehensive Cancer Center (HDFCCC) Tissue Core for performing immunohistochemistry. We also thank the staff of the UCSF Antibiome Center for advice and assistance with recombinant protein expression as well as the Chan-Zuckerberg Biohub staff for assistance with sequencing. Finally, we would also like to acknowledge the family of a patient who experienced relapse due to CD19-escape for raising funds to support this project. This work was supported by the UCSF Dept. of Laboratory Medicine, a lymphoma pilot grant from the UCSF HDFCCC, NIH DP2 OD022552, and NIH K08 CA184116 (to A.P.W.); NIH K08 CA230188, a Career Award for Medical Sciences from the Burroughs Wellcome Fund, and Technology Impact Award from the Cancer Research Institute (to A.T.S.); and NIH P30 CA082103 (to Preclinical Therapeutics Core managed by B.C.H., the HDFCCC Pediatric Malignancies Tissue Bank, and HDFCCC Tissue Core).
Footnotes
Author’s Disclosures
M.A.N. and A.P.W. have filed a provisional patent based on technology described here. A.P.W. is an equity holder and scientific advisory board member of Indapta Therapeutics and Protocol Intelligence. A.T.S. is a founder of Imunnai and Cartography Biosciences and has received research funding from Arsenal Biosciences and 10x Genomics. The other authors have no relevant conflicts of interest to declare.
REFERENCES
- 1.Park JH, Geyer MB, Brentjens RJ. CD19-targeted CAR T-cell therapeutics for hematologic malignancies: interpreting clinical outcomes to date. Blood. American Society of Hematology; 2016;127:3312–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majzner RG, Mackall CL. Tumor Antigen Escape from CAR T-cell Therapy. Cancer Discovery. American Association for Cancer Research; 2018;8:1219–26. [DOI] [PubMed] [Google Scholar]
- 3.Fry TJ, Shah NN, Orentas RJ, Stetler-Stevenson M, Yuan CM, Ramakrishna S, et al. CD22-targeted CAR T cells induce remission in B-ALL that is naive or resistant to CD19-targeted CAR immunotherapy. Nature Medicine. Nature Publishing Group; 2018;24:20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang W-Y, Wang Y, Guo Y-L, Dai H-R, Yang Q-M, Zhang Y-J, et al. Treatment of CD20-directed Chimeric Antigen Receptor-modified T cells in patients with relapsed or refractory B-cell non-Hodgkin lymphoma: an early phase IIa trial report. Signal Transduct Target Ther. Nature Publishing Group; 2016;1:16002–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordano Attianese GMP, Marin V, Hoyos V, Savoldo B, Pizzitola I, Tettamanti S, et al. In vitro and in vivo model of a novel immunotherapy approach for chronic lymphocytic leukemia by anti-CD23 chimeric antigen receptor. Blood. 2011;117:4736–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ormhøj M, Scarfò I, Cabral ML, Bailey SR, Lorrey SJ, Bouffard AA, et al. Chimeric antigen receptor T cells targeting CD79b show efficacy in lymphoma with or without co-targeting CD19. Clinical Cancer Research. American Association for Cancer Research; 2019;:clincanres.1337.2019. [Google Scholar]
- 7.Köksal H, Dillard P, Josefsson SE, Maggadottir SM, Pollmann S, Fåne A, et al. Preclinical development of CD37CAR T-cell therapy for treatment of B-cell lymphoma. Blood Adv. 2019;3:1230–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin H, Dong Z, Wang X, Cheng WA, Wen F, Xue W, et al. CAR T cells targeting BAFF-R can overcome CD19 antigen loss in B cell malignancies. Sci Transl Med. American Association for the Advancement of Science; 2019;11:eaaw9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacoby E, Shahani SA, Shah NN. Updates on CAR T-cell therapy in B-cell malignancies. Immunological Reviews. 2019;290:39–59. [DOI] [PubMed] [Google Scholar]
- 10.Sun Y-N, Hu Y-X, Gao L, Xiao P-F, Lu J, Wu S-Y, et al. The therapeutic efficacy of pediatric ALL patients with MLL gene rearrangement treated with CCLG-ALL2008 protocol. Eur Rev Med Pharmacol Sci. 2018;22:6020–9. [DOI] [PubMed] [Google Scholar]
- 11.Gardner R, Wu D, Cherian S, Fang M, Hanafi L-A, Finney O, et al. Acquisition of a CD19-negative myeloid phenotype allows immune escape of MLL-rearranged B-ALL from CD19 CAR-T-cell therapy. Blood. 2016;127:2406–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanjuan-Pla A, Bueno C, Prieto C, Acha P, Stam RW, Marschalek R, et al. Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia. Blood. 2015;126:2676–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiland J, Pal D, Case M, Irving J, Ponthan F, Koschmieder S, et al. BCP-ALL blasts are not dependent on CD19 expression for leukaemic maintenance. Leukemia. Nature Publishing Group; 2016;30:1920–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shah NN, Stevenson MS, Yuan CM, Richards K, Delbrook C, Kreitman RJ, et al. Characterization of CD22 expression in acute lymphoblastic leukemia. Pediatr Blood Cancer. John Wiley & Sons, Ltd; 2015;62:964–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–42. [DOI] [PubMed] [Google Scholar]
- 16.McMahon C, Baier AS, Pascolutti R, Wegrecki M, Zheng S, Ong JX, et al. Yeast surface display platform for rapid discovery of conformationally selective nanobodies. Nat Struct Mol Biol. Springer US; 2018;:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wollscheid B, Bausch-Fluck D, Henderson C, O’Brien R, Bibel M, Schiess R, et al. Mass-spectrometric identification and relative quantification of N-linked cell surface glycoproteins. Nature Biotechnology. 2009;27:378–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. Nature Research; 2016;11:2301–19. [DOI] [PubMed] [Google Scholar]
- 19.Geng H, Brennan S, Milne TA, Chen W-Y, Li Y, Hurtz C, et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discovery. American Association for Cancer Research; 2012;2:1004–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinko AJ, Truillet C, Julien O, Diaz JE, Horlbeck MA, Whiteley G, et al. Targeting RAS-driven human cancer cells with antibodies to upregulated and essential cell-surface proteins. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Klamer S, Voermans C. The role of novel and known extracellular matrix and adhesion molecules in the homeostatic and regenerative bone marrow microenvironment. Cell Adhesion & Migration. 2014;8:563–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. CELL. 2014;159:647–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kikushige Y, Yoshimoto G, Miyamoto T, Iino T, Mori Y, Iwasaki H, et al. Human Flt3 is expressed at the hematopoietic stem cell and the granulocyte/macrophage progenitor stages to maintain cell survival. The Journal of Immunology. American Association of Immunologists; 2008;180:7358–67. [DOI] [PubMed] [Google Scholar]
- 24.Uhlén M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. American Association for the Advancement of Science; 2017;357:eaan2507. [DOI] [PubMed] [Google Scholar]
- 25.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER, et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature. Nature Publishing Group; 2019;569:503–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Novershtern N, Subramanian A, Lawton LN, Mak RH, Haining WN, McConkey ME, et al. Densely interconnected transcriptional circuits control cell states in human hematopoiesis. CELL. 2011;144:296–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsubata T Inhibitory B cell co-receptors and autoimmune diseases. Immunol Med. Taylor & Francis; 2019;75:1–9. [DOI] [PubMed] [Google Scholar]
- 28.Xu M, Hou R, Sato-Hayashizaki A, Man R, Zhu C, Wakabayashi C, et al. Cd72(c) is a modifier gene that regulates Fas(lpr)-induced autoimmune disease. J Immunol. American Association of Immunologists; 2013;190:5436–45. [DOI] [PubMed] [Google Scholar]
- 29.Schwarting R, Castello R, Moldenhauer G, Pezzutto A, Hoegen von I, Ludwig WD, et al. Human Lyb-2 homolog CD72 is a marker for progenitor B-cell leukemias. Am J Hematol. 1992;41:151–8. [DOI] [PubMed] [Google Scholar]
- 30.Townsend EC, Murakami MA, Christodoulou A, Christie AL, Köster J, DeSouza TA, et al. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell. Elsevier Inc; 2016;29:574–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, et al. Mutations of multiple genes cause deregulation of NF-κB in diffuse large B-cell lymphoma. Nature. Nature Publishing Group; 2009;459:717–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Godfrey L, Crump NT, Thorne R, Lau I-J, Repapi E, Dimou D, et al. DOT1L inhibition reveals a distinct subset of enhancers dependent on H3K79 methylation. Nature Communications. Nature Publishing Group; 2019;10:–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerry J, Godfrey L, Repapi E, Tapia M, Blackledge NP, Ma H, et al. MLL-AF4 Spreading Identifies Binding Sites that Are Distinct from Super-Enhancers and that Govern Sensitivity to DOT1L Inhibition in Leukemia. CellReports. 2017;18:482–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu J, Chen L-J, Yang S-S, Sun Y, Wu W, Liu Y-F, et al. Exploratory trial of a biepitopic CAR T-targeting B cell maturation antigen in relapsed/refractory multiple myeloma. Proc Natl Acad Sci USA. 2019;116:9543–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Czajkowsky DM, Hu J, Shao Z, Pleass RJ. Fc-fusion proteins: new developments and future perspectives. EMBO Mol Med. John Wiley & Sons, Ltd; 2012;4:1015–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zemlin M, Klinger M, Link J, Zemlin C, Bauer K, Engler JA, et al. Expressed murine and human CDR-H3 intervals of equal length exhibit distinct repertoires that differ in their amino acid composition and predicted range of structures. Journal of Molecular Biology. 2003;334:733–49. [DOI] [PubMed] [Google Scholar]
- 37.Hunter SA, Cochran JR. Cell-Binding Assays for Determining the Affinity of Protein-Protein Interactions: Technologies and Considerations. Meth Enzymol. Elsevier; 2016;580:21–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Parker KR, Migliorini D, Perkey E, Yost KE, Bhaduri A, Bagga P, et al. Single-Cell Analyses Identify Brain Mural Cells Expressing CD19 as Potential Off-Tumor Targets for CAR-T Immunotherapies. CELL. 2020;183:126–142.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsieh Y-T, Gang EJ, Geng H, Park E, Huantes S, Chudziak D, et al. Integrin alpha4 blockade sensitizes drug resistant pre-B acute lymphoblastic leukemia to chemotherapy. Blood. 2013;121:1814–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim E, Hurtz C, Koehrer S, Wang Z, Balasubramanian S, Chang BY, et al. Ibrutinib inhibits pre-BCR+ B-cell acute lymphoblastic leukemia progression by targeting BTK and BLK. Blood. 2017;129:1155–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müschen M Autoimmunity checkpoints as therapeutic targets in B cell malignancies. Nature Publishing Group. Nature Publishing Group; 2018;18:103–16. [DOI] [PubMed] [Google Scholar]
- 42.Nijhof IS, Groen RWJ, Lokhorst HM, van Kessel B, Bloem AC, van Velzen J, et al. Upregulation of CD38 expression on multiple myeloma cells by all-trans retinoic acid improves the efficacy of daratumumab. Leukemia. Nature Publishing Group; 2015;29:2039–49. [DOI] [PubMed] [Google Scholar]
- 43.Ramakrishna S, Highfill SL, Walsh Z, Nguyen SM, Lei H, Shern JF, et al. Modulation of Target Antigen Density Improves CAR T-cell Functionality and Persistence. Clinical Cancer Research. American Association for Cancer Research; 2019;25:5329–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pont MJ, Hill T, Cole GO, Abbott JJ, Kelliher J, Salter AI, et al. γ-Secretase inhibition increases efficacy of BCMA-specific chimeric antigen receptor T cells in multiple myeloma. Blood. 2019;134:1585–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myers DE, Uckun FM. An anti-CD72 immunotoxin against therapy-refractory B-lineage acute lymphoblastic leukemia. Leuk Lymphoma. Taylor & Francis; 1995;18:119–22. [DOI] [PubMed] [Google Scholar]
- 46.Polson AG, Calemine-Fenaux J, Chan P, Chang W, Christensen E, Clark S, et al. Antibody-drug conjugates for the treatment of non-Hodgkin’s lymphoma: target and linker-drug selection. Cancer Research. American Association for Cancer Research; 2009;69:2358–64. [DOI] [PubMed] [Google Scholar]
- 47.Euw EV, Taschereau E, Ou DM, Mead M, Larson S, Slamon D. Abstract 3041: CD72 as a potential new therapeutic target in diffuse large B-cell lymphoma (DLBCL) and mantle cell lymphoma (MCL). Cancer Res. 2019;79:3041. [Google Scholar]
- 48.Guedan S, Calderon H, Posey AD, Maus MV. Engineering and Design of Chimeric Antigen Receptors. Mol Ther Methods Clin Dev. 2019;12:145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pan J, Niu Q, Deng B, Liu S, Wu T, Gao Z, et al. CD22 CAR T-cell therapy in refractory or relapsed B acute lymphoblastic leukemia. Leukemia. 2019;33:2854–66. [DOI] [PubMed] [Google Scholar]
- 50.Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Meth. 2016;:1–10. [DOI] [PubMed] [Google Scholar]
- 51.Wickham H. ggplot2. Springer Science & Business Media; 2009. [Google Scholar]
- 52.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology. 2010;28:511–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.