Abstract
Clear cell renal cell carcinoma (ccRCC) is characterized by accumulation of neutral lipids and adipogenic trans-differentiation. We assessed adipokine expression in ccRCC and found that tumor tissues and patient plasma exhibit obesity-dependent elevations of the adipokine chemerin. Attenuation of chemerin by several approaches led to significant reduction in lipid deposition, and impairment of tumor cell growth in vitro and in vivo. A multi-omics approach revealed that chemerin suppresses fatty acid oxidation, preventing ferroptosis, and maintains fatty acid levels that activate HIF2α expression. The lipid CoQ and mitochondrial complex IV, whose biogenesis are lipid-dependent, were found decreased after chemerin inhibition, contributing to lipid reactive oxygen species production. Monoclonal antibody targeting chemerin led to reduced lipid storage and diminished tumor growth, demonstrating translational potential of chemerin inhibition. Collectively, the results suggest that obesity and tumor cells contribute to ccRCC through the expression of chemerin, which is indispensable in ccRCC biology.
Keywords: RARRES2, chemerin, fatty acid oxidation, ferroptosis, ccRCC
INTRODUCTION
Renal cell carcinoma (RCC) is one of the top ten cancers diagnosed globally, with over 200,000 new cases worldwide each year (1,2). RCC currently results in approximately 74,000 cancer cases and 15,000 mortalities annually in the United States (3,4); and according to the SEER (Surveillance, Epidemiology, and End Results) database, 16% of RCC patients have distant metastatic disease, with a 5-year survival rate of only 11.6% (5).
The most common subtype of renal cancer is clear cell renal cell carcinoma (ccRCC), which accounts for more than 70% of all kidney cancers (6). ccRCC tumors almost uniformly have alterations in the von Hippel-Lindau (VHL) gene, including mutations, deletions or hypermethylation, rendering the VHL protein (pVHL) functionally inactive (7,8). In normal oxygen conditions (normoxia), the hypoxia-inducible factors (HIFs) are hydroxylated by one of three PHD enzymes, leading to recognition and binding to pVHL. pVHL, which functions as an E3 ubiquitin ligase, subsequently targets HIFs for proteasomal degradation (9). In contrast, during hypoxia, or in ccRCC with a dysfunctional pVHL, HIF1α and HIF2α are not degraded and accumulate in cells. The HIFα subunits (predominantly HIF2α in ccRCC) then translocate into nucleus, bind to hypoxia responsive elements (HRE) and drive transcription of a host of hypoxia-responsive adaptation genes (10). Some of the HIF targets include vascular endothelial growth factor (VEGF), glycolysis genes, and CCND1, which contribute to the angiogenesis, Warburg metabolism, and hyperproliferative phenotypes of ccRCC.
The “clear cell” type of RCC is characterized by high lipid and glycogen content (11). Lipid homeostasis in cells is a tightly regulated process to maintain normal cellular functions, ranging from lipid synthesis, uptake, and subcellular distribution or compartmentalization (12). Of all, the storage of fatty acid (FA) in lipid droplets, which is the histologically identifiable phenotype of ccRCC, is an important equilibrating mechanism to prevent lipotoxicity. FAs can be used as building blocks for complex lipids, and can also be used as substrates for beta-oxidation (β-oxidation) in mitochondria to yield energy (13). Dysregulated metabolism is a well-established hallmark of cancer and an enticing target for cancer management. In ccRCC, reports have shown that lipid metabolism contributes to metabolic reprogramming and unbridled cell growth (14). Yet the exact mechanism and the significance of the lipid-storage phenotype remain poorly understood. The potential to exploit a metabolic vulnerability as a therapeutic strategy in lipid-associated cancers also remains largely unexplored.
In the past few decades, the cases of obesity and obesity-related disorders have been increasing steadily in both the developed and developing countries, leading to the declaration by the World Health Organization (WHO) of a global obesity epidemic in 1999 (15). Evidence has also established that tumorigenesis is escalated by adiposity, showing a direct link between metabolism and cancer. In fact, obesity (BMI at least 30 kg/m2) is a known risk factor for ccRCC, and perinephric fat invasion has been associated with poor disease outcome in ccRCC (10,16). Hypertrophic obesity (increase in adipocyte size), hyperplastic obesity (increase in adipocyte number) as well as increased intracellular lipids can contribute to an increase in circulating pro-inflammatory adipokines that act on peripheral tissues and stimulate key cellular biological processes, recently found to include tumorigenesis (17). Realizing the intimate interplay between adiposity and cancer, we hypothesized that adipokines could play detrimental roles in driving tumorigenesis in lipid-laden ccRCC. Notably, the roles and mechanisms of obesity-related factors in increasing cancer risk remain unclear.
In the current study, by combining single-cell resolution transcriptomic data, metabolomic analyses and multiple ccRCC model systems, we discovered that the adipokine chemerin, which is encoded by the retinoic acid receptor responder 2 (RARRES2) gene, is overexpressed in ccRCC due to both an autocrine, tumor-cell-dependent mechanism, as well as obesity-dependent paracrine production, and plays important roles in regulating lipid metabolism and tumorigenesis. We found that chemerin depletion results in dramatic metabolic rewiring in which excessive lipid oxidation leads to mitochondrial dysfunction and ferroptosis. Chemerin controls expression of over 300 genes, most notably HIF1α and HIF2α, through a fatty acid-dependent mechanism. The secretory nature of chemerin makes its elevation amenable to detection in the plasma of ccRCC patients, and further to interruption through the use of a monoclonal antibody. Accordingly, we find targeting chemerin to be effective in controlling orthotopic xenograft tumors in vivo. Together, the data provide a biological link between obesity, suppression of fatty acid oxidation, and tumor growth in lipid-laden ccRCC, suggesting a potential molecular marker and therapeutic target to improve standard therapeutic approaches.
RESULTS
Chemerin is elevated in ccRCC patients through obesity- and tumor-dependent processes
In analysis of bulk tumors, ccRCC cells show evidence of transdifferentiation into adipocytes (18), which are known to secrete a variety of factors that both maintain their own adipocyte functions, and promote adipocyte differentiation of distant cells. To validate transdifferentiation in ccRCC tumor cells, we utilized single cell RNA-sequencing (scRNA-seq) of tumors (n=3) and normal kidney tissues (n=5) (19), and by comparing the tumor cell and normal kidney epithelium clusters (Supplementary Fig. S1, S2A-E, Table S1), we verified that ccRCC tumor cells display a reduced renal epithelial gene signature in favor of an adipogenic signature (Fig. 1A). Tumor cells also demonstrated de-differentiation and epithelial-to-mesenchymal transition, in agreement with the literature (20) (Fig. 1A).
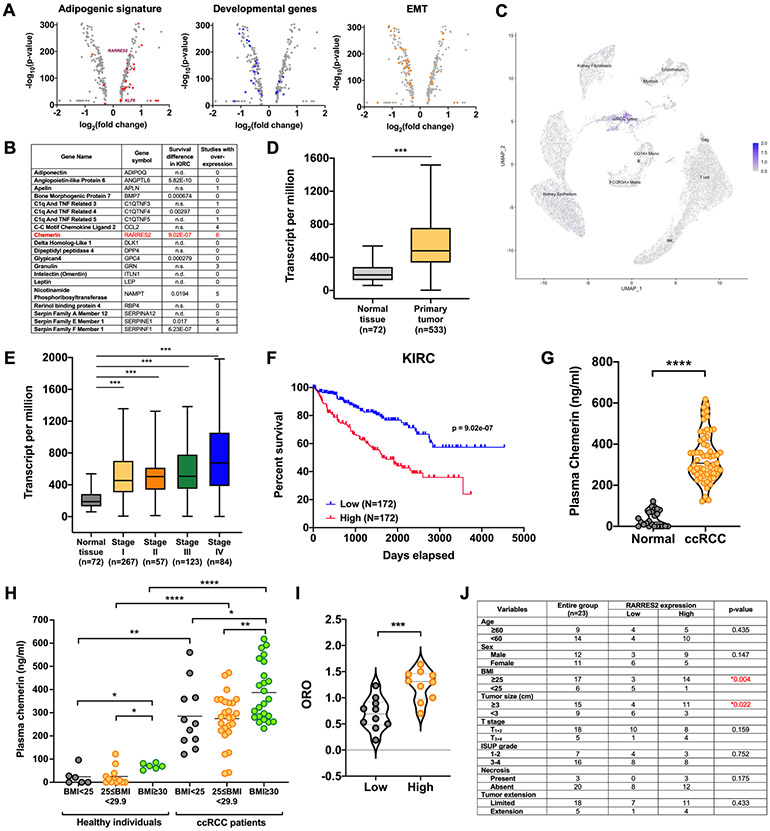
Figure 1. Chemerin is clinically relevant in ccRCC.
(A) Volcano plots showing the differentially expressed genes from comparison of the ccRCC tumor cluster and normal kidney epithelium cluster from single-cell RNA-sequencing (scRNA-seq). Adipogenic trans-differentiation of normal kidney epithelium to ccRCC is demonstrated by an enrichment of adipogenic genes (red circles, left), downregulation of developmental genes (blue circles, middle) and epithelial-to-mesenchymal (EMT) transition genes (downregulated epithelial genes and upregulated mesenchymal genes; orange circles, right).
(B) List of adipkines identified by in silico data mining from The Cancer Genome Atlas (TCGA) and Oncomine databases.
(C) scRNA-seq data from human ccRCC specimens presented as a UMAP plot demonstrating overexpression of chemerin in tumor cell cluster. Data derived from Young et al. (19).
(D) mRNA expression transcripts per million (TPM) of chemerin in ccRCC tumor samples (n=533) and the normal adjacent tissues (n=72) from TCGA. Mann-Whitney U-test. Boxplot represents median and 25th and 75th percentiles, whiskers 1.5 times the interquartile range.
(E) mRNA expression TPM of chemerin in Stage I (n=267), Stage II (n=57), Stage III (n=123), Stage IV (n=84) of ccRCC and normal adjacent tissues (n=72) from TCGA. Mann-Whitney U-test. Boxplot represents median and 25th and 75th percentiles, whiskers 1.5 times the interquartile range.
(F) Kaplan-Meier curve showing correlation of higher mRNA expression of RARRES2 with poorer survival of ccRCC patients in the TCGA KIRC dataset. Log rank analysis.
(G) ELISA analysis of plasma chemerin protein level in 24 normal individuals and 59 ccRCC patients. Mann-Whitney U-test.
(H) Plasma chemerin protein level stratified according to BMI status. Normal BMI is defined as value <25kg/m2, overweight as BMI 25kg/m2 ≤BMI<30kg/m2, and obese as BMI≥30kg/m2. Mann-Whitney U-test.
(I) Correlation of 20 plasma chemerin expression with corresponding Oil Red O (ORO) staining of tumor sections. Two-tailed student’s t-test.
(J) Correlation of plasma chemerin expression with patient characteristics and tumor pathology. p-values based on χ2 analysis.
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p<0.05, **p<0.01, ***p<0.001; **** p < 0.0001.
In an effort to identify a potential autocrine adipokine that could be critical for promoting and maintaining ccRCC, we performed in silico data mining using The Cancer Genome Atlas (TCGA) as well as Oncomine databases, and examined association of 100 known adipokines with ccRCC, some of which are shown in Fig. 1B. The adipokines were ranked according to the number of studies showing an overexpression profile in ccRCC, and the correlation with patient survival; the adipokine chemerin, stood out to be a candidate. From scRNA-seq data, we determined that tumor expression of chemerin arises from ccRCC tumor cells rather than other cells in the tumor microenvironment, agreeing with a possible autocrine pathway (Fig. 1C, Supplementary Table S2). In TCGA, chemerin mRNA expression is higher in ccRCC clinical samples (n=533) as compared to adjacent normal kidney tissues (n=72) (Fig. 1D, E); and higher chemerin expression portends a negative prognosis in ccRCC patients (upper third versus lower third) (Fig. 1F). In contrast, chemerin expression is either not associated with outcome in papillary renal cell carcinoma patients (pRCC) (Supplementary Fig. S3A) or chromophobe renal cell carcinoma (Supplementary Fig. S3B) when considering upper and lower thirds, or may even define a better outcome when considering upper and lower fifths (21).
Since chemerin is notably overexpressed in ccRCC tumors, we next measured plasma chemerin levels and investigated the possibility of using chemerin as a biomarker for ccRCC patients. We found that chemerin protein levels are significantly elevated in patient plasma relative to healthy individuals (Fig. 1G). The area under the curve (AUC) was 0.99 at the cut-off value of 121.6 ng/mL, demonstrating a sensitivity of 98.31% and specificity of 95.83% (Supplementary Fig. S3C). Because previous studies have indicated that chemerin is positively correlated with adiposity (22), we considered patient BMI among the patient samples and found that obese patients (BMI ≥30) had significantly higher circulating chemerin levels than overweight patients (25≤BMI<30) or low BMI patients (BMI<25) (Fig. 1H). This confirmed the notion that the paracrine chemerin production in ccRCC is obesity-dependent. Importantly, plasma chemerin remains elevated when comparing low BMI ccRCC patients to low BMI healthy individuals, and high BMI patients to high BMI controls (Fig. 1H). We then assessed chemerin production from tumors, normal adjacent kidney tissue, tumor-adjacent fat, and fat distant to the tumor by performing quantitative reverse transcription-polymerase chain reaction (qRT-PCR) to determine the greatest contributor of chemerin. We again found significantly elevated RARRES2 mRNA in tumor samples versus normal tissue; and that adjacent and distant fat produced equally higher levels than normal kidney, but not as much as the tumor itself (Supplementary Fig. S3D). We validated the expression changes with immunohistochemistry staining of chemerin protein using a tumor microarray (TMA) with 30 ccRCC samples and associated adjacent normal kidney sections (Supplementary Fig. S3E-F). While chemerin was clearly higher in tumors, we noticed somewhat higher than expected signals in the “normal” regions, possibly due to possible diffusion of the soluble protein from nearby tumor. We then stained fresh frozen tumor samples with the lipophilic dye Oil-Red-O (ORO), and compared ORO uptake with plasma chemerin and found a significant positive correlation, suggesting an association between plasma chemerin, regardless of source, and lipid levels in ccRCC tumors (Fig. 1I, Supplementary Fig. S3G). Furthermore, the plasma chemerin levels are also correlated with tumor size independently of BMI (Fig. 1J). Again, we assessed chemerin in a non-clear cell subtype (pRCC), and while circulating levels were elevated over controls, they remained significantly lower than ccRCC (Supplementary Fig. S3H). Together, the data validate that chemerin is a potentially relevant adipokine in ccRCC.
Chemerin sustains ccRCC tumor growth
To functionally dissect a contribution of chemerin in ccRCC, three different shRNAs were used to stably knockdown the chemerin gene RARRES2 in four different ccRCC cell lines: 786-O, 769-P, UOK101 and A-498 (Fig. 2A-D, upper panels; Supplementary Fig. S4A-C). All of these ccRCC cell lines contain detectable amounts of chemerin protein (Supplementary Fig. S4D). All ccRCC isogenic cell lines demonstrated impaired cell proliferation capacity after chemerin knockdown compared to shGFP controls (Fig. 2A-D, lower panels); but not HK-2 cells, which are immortalized proximal tubule epithelial cells (Supplementary Fig. S4E-F). Interestingly, the magnitude of inhibition of proliferation appears to correlate with overall chemerin expression levels (i.e., greatest effect in UOK101 and 769-P). We also observed a reduction in colony forming ability in 786-O cells after chemerin knockdown, another surrogate marker for reduced proliferation ability (Supplementary Fig. 4G). To eliminate potential off-target effects of shRNAs, we also utilized CRISPR-Cas9 technology to knockout chemerin, and the cells showed an even more pronounced impairment of proliferative capacity, where almost all cells died by Day 9 of the assay (Fig. 2E). To investigate whether chemerin is important for ccRCC tumorigenesis in vivo, the 786-O and 769-P cell models infected with either shGFP control or shRARRES2 viruses were transplanted subcutaneously into the flanks of nude mice. Similarly, two unique CRISPR-Cas9 clones of 786-O cells with chemerin knocked-out, and a polyclonal 786-O population with control sgRNA, were also implanted subcutaneously into the flanks of nude mice. Tumors with chemerin inhibition uniformly showed dramatically impaired tumor formation capability in both the knockdown (Fig. 2F, Supplementary Fig. S4H) and knockout systems (Fig. 2G, Supplementary Fig. S4I); as well as reduced Ki67 staining in the knockout system (Supplementary Fig. 4J).
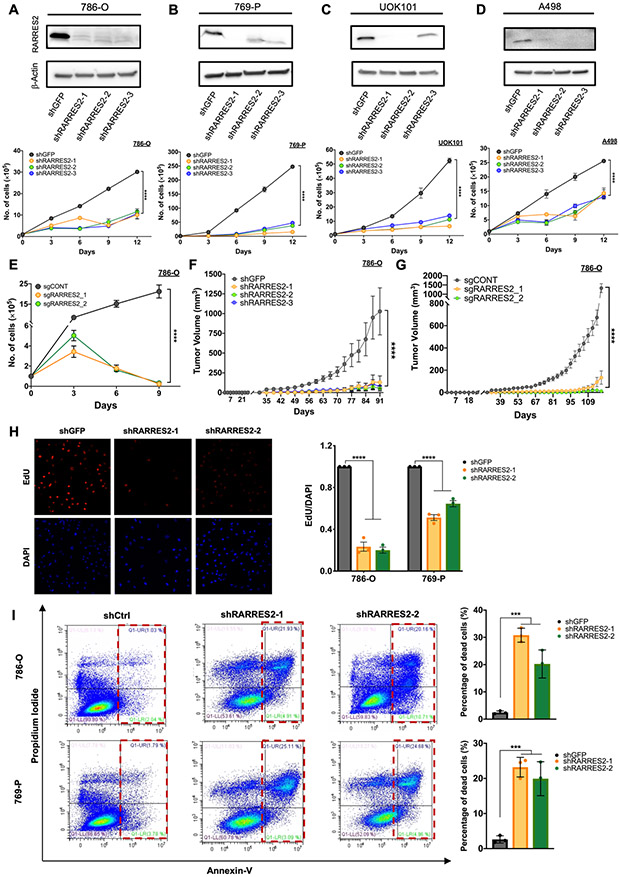
Figure 2. Chemerin regulates ccRCC tumor growth in vitro and in vivo.
(A) Immunoblot of lysate confirming knockdown of chemerin (upper panel) and cell proliferation assay from 786-O cells infected with shGFP control or 3 different shRNAs encoding lentivirus targeting chemerin (shRARRES2-1, shRARRES2-2, shRARRES2-3). Two-way repeated measures ANOVA with Geisser-Greenhouse correction.
(B) Same as above for 769-P cells.
(C) Same as above for UOK101 cells.
(D) Same as above for A-498 cells.
(E) Cell proliferation assay of 786-O cells infected with lentivirus encoding either sgCONT or sgRARRES2 (sgRARRES2_1 and sgRARRES2_2). 2 different clones for sgRARRES2 were selected. Two-way repeated measures ANOVA with Geisser-Greenhouse correction.
(F) Subcutaneous tumor volume measurement in nude mice implanted with 786-O cells which were infected with lentivirus encoding either shGFP (n=7) or 3 different shRARRES2 (n=7 in each arm). Two-way measures ANOVA with Dunnett’s multiple comparison test correction.
(G) Same as (F) from 786-O cells infected with lentivirus encoding either sgCONT (n=5) or 2 different sgRARRES2 (n=5 in each arm). Two-way measures ANOVA with Dunnett’s multiple comparison test correction.
(H) Representative EdU staining and quantification of 786-O and 769-P cells infected with shGFP control or 2 different shRARRES2 (shRARRES2-1, shRARRES2-2). One-way ANOVA.
(I) Flow cytometry for annexin V and propidium iodide staining of 786-O and 769-P cells infected with shGFP control or 2 different shRARRES2 (shRARRES2-1, shRARRES2-2). One-way ANOVA.
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Next, we interrogated the cellular changes that contribute to the functional phenotypes observed by measuring DNA synthesis as a surrogate for proliferation, and by assessing Annexin V/PI staining for programmed cell death. Knockdown of chemerin in both 786-O and 769-P cells reduced DNA synthesis, derived from reduced EdU incorporation (Fig. 2H); and Annexin-V/PI staining showed increased cell death in the cells after chemerin knockdown (Fig. 2I). Thus, chemerin expression in ccRCC tumor cells is functionally critical for sustaining tumor cell survival.
Inhibition of chemerin reprograms lipid metabolism in ccRCC and promotes fatty acid oxidation
To understand whether the chemerin-mediated survival in ccRCC is driven by a change in lipid phenotype, we investigated the effect of chemerin on lipid droplets. To this end, chemerin expression was depleted using shRNAs and the cells were stained with the lipophilic dye BODIPY 493/503 (4,4-Difluoro-1,3,5,7,8-Pentamethyl-4-Bora-3a,4a-Diaza-s-Indacene). Chemerin silencing significantly reduced lipid deposition in the cells; an effect that was partially rescued by incubation of the cells with 50 nM of recombinant chemerin protein for 18 hours (Fig. 3A). The reduction in lipid deposition was also seen in chemerin knockout tumors measured by ORO staining (Fig. 3B). RNA-sequencing of 786-O shGFP control and shRARRES2 cells revealed that chemerin regulates numerous lipid metabolism genes (Fig. 3C-E, Supplementary Fig. S5A). According to genome set enrichment analysis (GSEA) of the transcriptomes, several lipid metabolism pathways were downregulated including the lysophospholipid pathway, steroid hormone biosynthesis and ether lipid metabolism (Fig. 3D, Supplementary Fig. S5B). Some of the genes were then confirmed by qRT-PCR. The genes tested included the fatty acid oxidation (FAO) enzymes CPT1A, ACAD9, DBI and ACOT7 (Fig. 3E), which were upregulated after chemerin silencing in ccRCC and suggested increased FAO. In contrast, the transcription of the lipid uptake protein FABP7 and the lipid droplet component protein PLIN4 were reduced after chemerin knockdown (Fig. 3E). Consistent with the gene expression changes, there was a reduction in fatty acids measured by mass spectrometry (Fig. 3F), especially the saturated fatty acids (SFA) and monounsaturated fatty acids (MUFA). These results confirm dramatic lipid metabolism alterations after chemerin inhibition, highlighting an important role of chemerin in regulating the ccRCC cellular lipid profile.
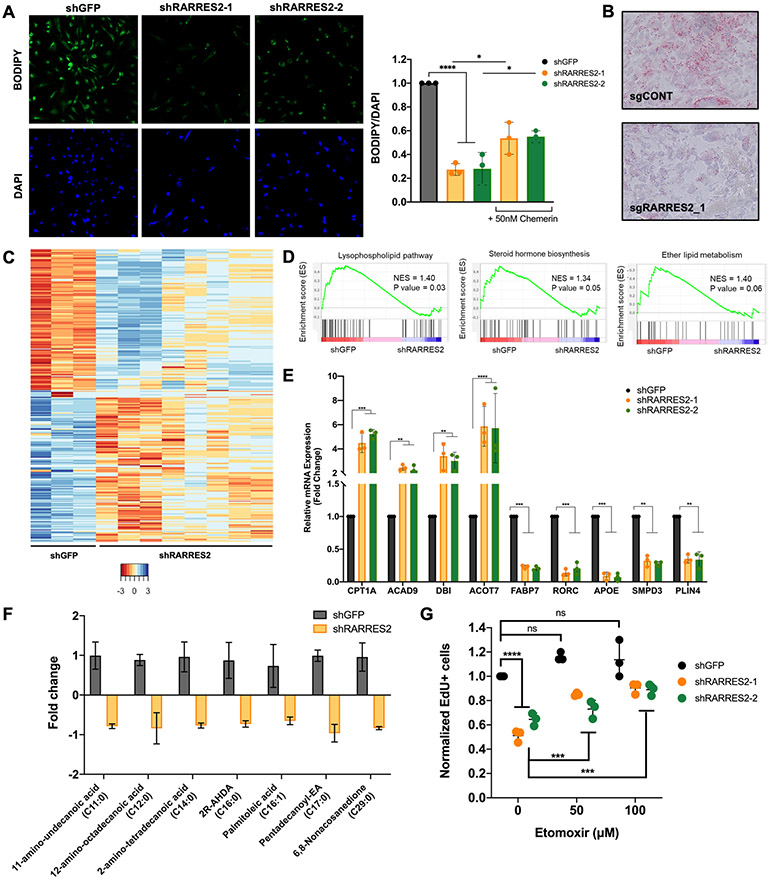
Figure 3. Chemerin suppresses fatty acid oxidation.
(A) BODIPY staining of 786-O cells infected with lentivirus targeting shGFP control or 2 different shRARRES2 (upper panels). DAPI staining of nucleus in lower panels. Quantification of BODIPY staining, normalized to DAPI staining, in 786-O cells infected with lentivirus targeting shGFP control or 2 different shRARRES2. One-way ANOVA. Treatment of chemerin-silenced 786-O cells with 50nM recombinant chemerin protein rescued the lipid deposition defect. Two-tailed student’s t-test.
(B) Oil-Red-O staining of lipid droplets of tumors harvested from mice implanted with 786-O cells infected with lentivirus encoding either sgCONT or sgRARRES2 (4×).
(C) Heatmap of RNA-sequencing for 786-O cells infected with lentivirus encoding either shGFP control (shGFP 1-3) or shRARRES2 (shRARRES2 1-8).
(D) Gene set enrichment analysis of RNA-seq demonstrated downregulation of lipid metabolism pathways, including lysophospholipid pathway, steroid hormone biosynthesis pathway and ether lipid metabolism pathway.
(E) Expression of CPT1A, ACAD9, DBI, ACOT7, FABP7, RORC, APOE, SMPD3, and PLIN4, as measured by qRT-PCR, in the 786-O cells infected with lentivirus targeting shGFP or 2 different shRARRES2. One-way ANOVA.
(F) Fatty acid species that were significantly reduced in metabolite profiling of 786-O cells infected with lentivirus encoding shGFP or 2 different shRARRES2. Two-tailed student’s t-test.
(G) Etomoxir treatment (50 μM and 100μM) rescues tumorigenesis in chemerin-knockdown 786-O cells, measured by EdU flow cytometry. Two-tailed student’s t-test.
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001.
To determine if decreased tumor cell growth (and possibly tumorigenesis) from chemerin suppression is due to the enhanced mitochondrial β-oxidation of lipids, we treated chemerin-inhibited cells with etomoxir, an inhibitor of the fatty acid mitochondrial transporter carnitine-palmitoyltransferase 1 (CPT1). We found that etomoxir treatment rescued the reduction in DNA synthesis (Fig. 3G, Supplementary Fig. S5C) and proliferation defect (Supplementary Fig. S5D) from chemerin silencing. This observation suggests that chemerin-mediated suppression of fatty acid oxidation is necessary to maintain cell proliferation in ccRCC.
Chemerin inhibition increases oxidized lipid and enhances ferroptosis susceptibility
To further characterize the role of chemerin in lipid metabolism, we utilized an untargeted lipidomics approach (Fig. 4A-B, Supplementary Fig. S6A-B). The lipid species in the analysis included phosphatidylcholine (PC), plasmanyl-PC, phosphatidylethanolamine (PE), phosphatidic acid (PA), ceramides, coenzyme Q (CoQ), acetoacetate (AcAc), phosphatidylglycerol (PG), sphingomyelin, sphingosine, oxidized triacylglycerols, oxidized PE, oxidized PC, monogalactosyldiacylglycerol (MGDG), monoglyceride (MG), lysophosphatidylethanolamine (LPE), lysophosphatidylcholine (LPC), oxidized LPC, and diacylglcerols (DG) (Supplementary Fig. S6C-D). Consistent with transcriptomic findings demonstrating increased FAO, we observed an overall increase in oxidized or breakdown lipids and reduction of intact lipids in 786-O cells transfected with shRNAs targeting chemerin compared to shGFP control (Fig. 4C). For instance, during mitochondrial FAO, carnitine is added to fatty acyl-CoA by CPT1 to form acylcarnitines, which are then transported across mitochondrial inner membrane (8). Notably, the fatty acyl-carnitine species which feed the downstream FAO cycle were significantly reduced after knockdown of chemerin, validating an increased in oxidation of acylcarnitine species (Fig. 4D). Pathway analysis of the metabolome also revealed upregulated mitochondrial β-oxidation, taurine and hypotaurine metabolism (which promote lipid metabolism (23)) and amino acid pathways that regulate redox balance (24) in the cells after chemerin knockdown (Fig. 4E).
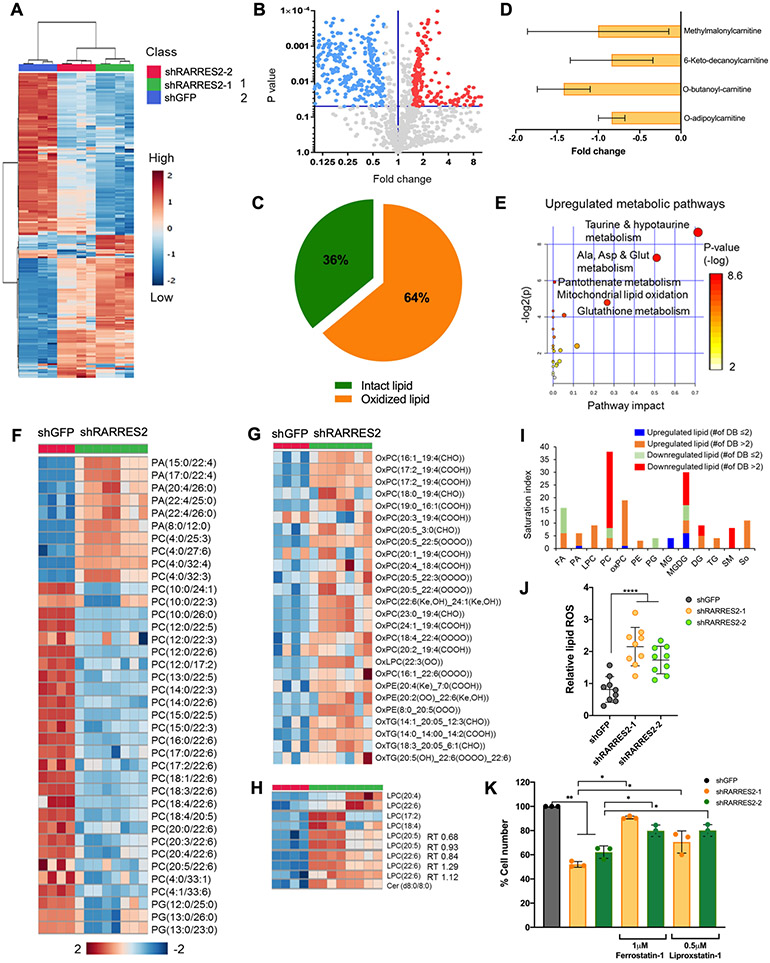
Figure 4. Chemerin-dependent lipid homeostasis pathways affects ferroptosis.
(A) Heatmap from untargeted lipidomics of 786-O infected with lentivirus targeting shGFP control, shRARRES2-1, and shRARRES2-2. Blue, low expression; red, high expression.
(B) Volcano plot showing the differentially expressed lipid metabolites in shGFP in comparison to shRARRES2. Blue and red circles represent lipid species that were significantly downregulated and upregulated, respectively, in RARRES2-targeted cells.
(C) Proportion of oxidized or breakdown versus intact lipid levels in shRARRES2 cells compared to shGFP.
(D) Fold change of acylcarnitine species in RARRES2-targeted 786-O cells versus shGFP.
(E) Metabolic Pathway Analysis using KEGG human metabolome database that shows the significantly upregulated metabolic pathways according to untargeted lipidomics and metabolomics data after chemerin knockdown.
(F) Heatmap from untargeted lipidomics of 786-O infected with lentivirus targeting shGFP (n=4) or shRARRES2 (including shRARRES2-1 and shRARRES2-2, n=8) showing lower glycerophospholipid species in chemerin deficient cells. Blue, low expression; red, high expression. PA, phosphatidic acid; PC, phosphatidylcholine; PG, phosphatidylglycerol.
(G) Heatmap from untargeted lipidomics showing higher oxidized glycerophospholipid species in chemerin deficient cells. Blue, low expression; red, high expression. OxPC, oxidized phosphatidylcholine; OxLPC, oxidized lysophosphatidylcholine; OxPE, oxidized phosphatidylethanolamine; OxTG, oxidized triacylglycerols.
(H) Heatmap from untargeted lipidomics showing higher lysophosphatidylcholine, the breakdown product of phosphatidylcholine, in chemerin deficient cells. Blue, low expression; red, high expression. LPC, lysophosphatidylcholine; Cer, ceramides.
(I) Saturation index of different lipid species after chemerin knockdown. Stacked bar chart shows different saturation abundances of significant lipid classes in chemerin knockdown cells compared to controls. DB, double bond.
(J) Lipid reactive oxygen species (ROS) measurement by BODIPY 581/591 C-11 in 786-O infected with lentivirus encoding either shGFP or shRARRES2. One-way ANOVA.
(K) 786-O cells infected with lentivirus encoding either shGFP or shRARRES2, were treated with 2 different ferroptosis inhibitors (1 μM Ferrostatin-1 or 0.5 μM Liproxstatin-1). Two-tailed student’s t-test.
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001.
The significant reduction in glycerophospholipid (GPL) species including PA, PC and PG (Fig. 4F) observed in chemerin-deficient cells is accompanied with a complementary increase in corresponding oxidized GPL products (Fig. 4G). Concurrently, the LPC, which are produced after removal of a single acyl group from standard phospholipids with two tails, was increased (Fig. 4H, Supplementary Fig. S6E). This suggests that chemerin-knockdown cells scavenge fatty acid tails from most of the GPL species as substrates for β-oxidation. In addition to oxidation in GPL, other lipid classes were also observed to have undergone oxidation or breakdown. In sphingolipids, intact sphingomyelin was reduced after chemerin knockdown, and the breakdown product sphingosine was significantly increased (Supplementary Fig. S6F). Another product in sphingolipid metabolism, ceramide, also known to induce cell death, was increased after chemerin knockdown (Supplementary Fig. S6F). A similar trend was observed in glycerolipids (Supplementary Fig. S6G).
As accumulation of oxidized lipid products are known to induce ferroptosis in cells (25), we next sought to investigate if the chemerin knockdown-induced cell death could be explained by ferroptosis. We found that polyunsaturated fatty acid (PUFA) species, potent ferroptosis inducers, increased after chemerin inhibition (Fig. 4I, orange bars). The increase in PUFA was observed in peroxidized glycerophospholipids, particularly oxidized PC that contain PUFA chains including arachidonic acid (C20:4) and docosahexaenoic acid (C22:6). On the other hand, monounsaturated fatty acid (MUFA) which reduces PUFA incorporation into phospholipid and decreases ferroptosis, was decreased (Fig. 3F). In addition, the increase in LPC is a lipidomic signature of ferroptosis when oxidized PUFA is cleaved from the sn2 position of phospholipid (Fig. 4H, Supplementary S6E). PUFAs have been reported to be tumoricidal by enhancing free radical generation (26). This was also evidenced in cells with chemerin knockdown where we observed an increase in the production of lipid reactive oxygen species (ROS) measured by BODIPY C-11 581/591 (Fig. 4J). As a proof-of-concept, the treatment of chemerin-deficient cells with ferroptosis inhibitors, 1 μM Ferrostatin-1 and 0.5 μM Liproxstatin-1, rescued the proliferation defect (Fig. 4K). Collectively, the results suggest that ferroptosis is induced in ccRCC after chemerin inhibition by peroxidized lipid species with increased PUFAs and elevated lipid ROS.
Reduction of OXPHOS complex IV after chemerin inhibition enhances ROS production
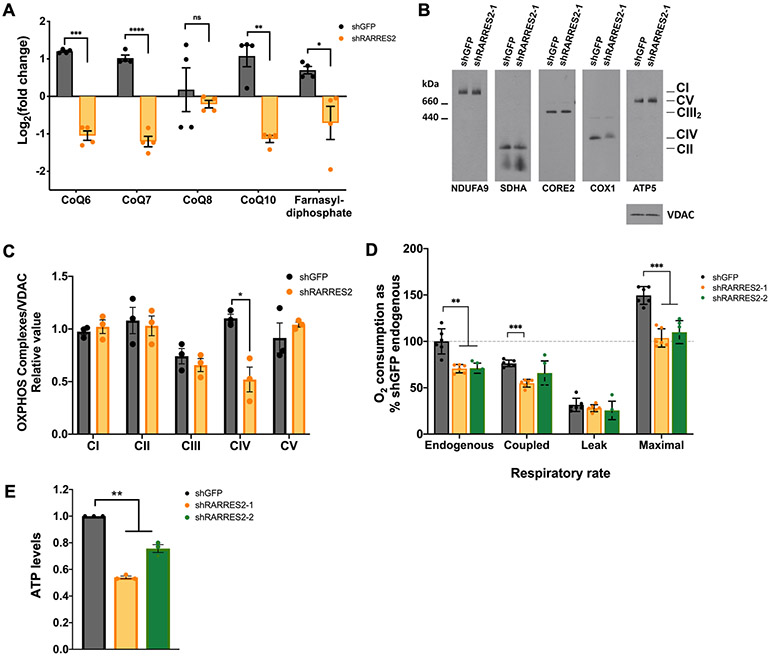
As part of the lipid reprogramming in ccRCC cells after chemerin inhibition, we found that CoQ10, the major form of CoQ found in humans, was also decreased (Fig. 5A). Since CoQ is an electron carrier of the mitochondrial respiratory chain (MRC), we hypothesized that chemerin silencing results in mitochondrial dysfunction that could promote ferroptosis. We then analyzed the other components of the MRC and oxidative phosphorylation system (OXPHOS) upon chemerin knockdown. We observed that the level of complex IV (CIV), the fourth and last enzyme of the MRC, was also decreased (Fig. 5B, 5C, and Supplementary Fig. S7A). It is relevant to note that CIV contains heme A as a cofactor (27). Heme A is essential for CIV enzymatic activity and it is also required for CIV biogenesis, since defects in heme A synthesis severely compromise CIV holoenzyme formation (28). In cells, heme A is present exclusively in CIV and differs from heme B (the more common type of heme in the cell) as it has a farnesyl group at the C2 position of the porphyrin ring. Lipid profiling confirmed that farnesyl-diphosphate, which is the farnesyl group donor for heme A biosynthesis, was reduced after chemerin silencing (Fig. 5A), suggesting a possible defect in heme A and, consequently, CIV levels. Furthermore, several phospholipids were shown to intimately interact with CIV and were proposed to play a role in its function and stability (29). The reduction of phospholipid after chemerin inhibition (Fig. 4F) could also contribute to the observed decrease in CIV.
Figure 5. Genetic deletion of chemerin in ccRCC cells affects mitochondrial OXPHOS.
(A) Metabolic profiling of CoQ (CoQ6, CoQ7, CoQ8, and CoQ10) and farnesyl-diphosphate from mass spectrometry analysis of 786-O cells infected with lentivirus targeting shGFP control or shRARRES2. Four technical replicates were performed.
(B) Steady-state levels of mitochondrial individual OXPHOS complexes were evaluated by BN-PAGE analysis of whole cell extracts prepared in the presence of 1% lauryl maltoside (LM) and probed with antibodies against complex I subunit NDUFA9, complex II subunit SDHA, complex III subunit CORE2, complex IV subunit COX1 and complex V subunit ATP5.
(C) Signals from (B) were quantified and normalized by VDAC using the histogram function of Adobe Photoshop on digitalized images. Error bars represent the mean ± SD of three independent experiments with two technical replicates. Unpaired T-test with Welch’s correction.
(D) NADH substrate-driven cellular respiratory rates expressed as percentile of shGFP control endogenous O2 consumption. Error bars represent the mean ± SD of three independent experiments with two technical replicates. Unpaired t-test with Welch’s correction.
(E) ATP measurement following chemerin knockdown in 786-O cells. One-way ANOVA.
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001; ns, not significant.
Due to the alteration in the MRC components CoQ and CIV, we next sought to investigate whether the mitochondrial respiratory function was affected by chemerin inhibition by polarographic analysis of cellular respiration. Consistently, we observed a defect in respiratory capacities, including endogenous, coupled and maximal respiration (Fig. 5D), which were similarly observed in Seahorse Mito Stress Test assay (Supplementary Fig. S7B). This was associated with a reduction in ATP production (Fig. 5E). In order for β-oxidation to support ATP synthesis, cells need a functional MRC so that the electrons from lipid oxidation can be transferred to MRC complexes by reducing equivalents NADH and FADH2. However, chemerin knockdown cells have high FA β-oxidation yet low respiratory capacity. This is likely to create an unbalance between the high electron influx from the β-oxidation pathway and the downstream slow electron flux in the MRC. A large input of electrons in a CIV-compromised MRC represents a favorable condition for the premature electron escape from MRC CI and CIII to molecular oxygen, generating superoxide radicals. The increase in ROS species could in turn contribute to promote ferroptosis (Fig. 4J), consistent with previous observations (30). It is important also to note that there was no structural change in the filamentous mitochondrial network nor the number of mitochondria after silencing of chemerin (Supplementary Fig. S7C).
Chemerin expression is VHL dependent and regulates HIF in ccRCC
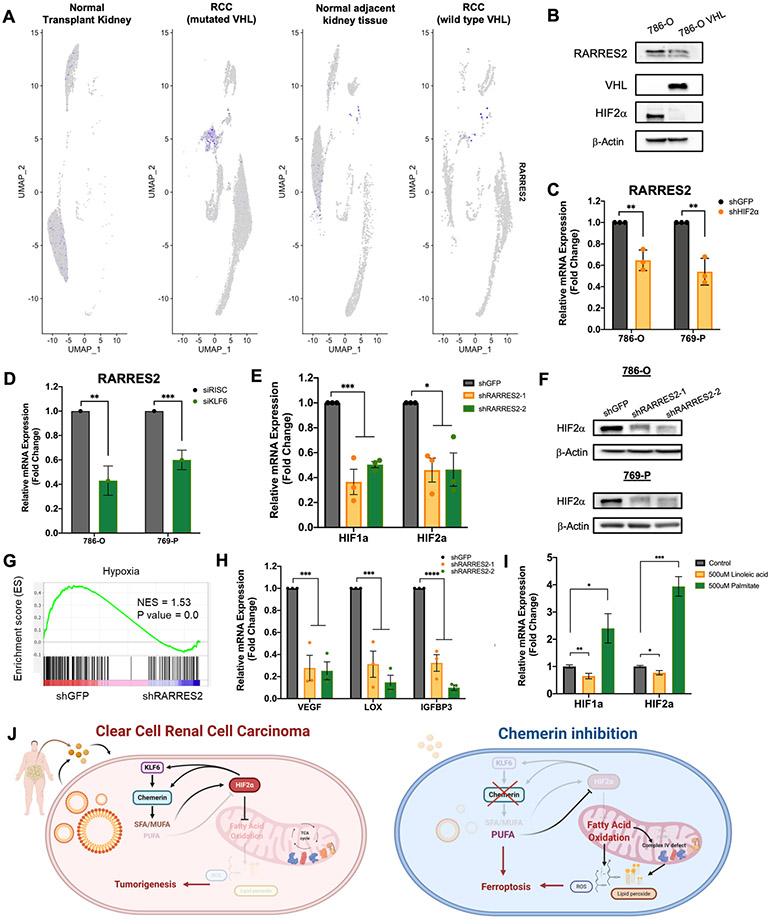
We next sought to investigate underlying signaling mechanism of how chemerin regulates fatty acid oxidation and mitochondrial functions. Since most of the ccRCC tumors are VHL null, we first examined whether chemerin expression is VHL dependent. By interrogating the scRNA-seq data, we found that chemerin was detected at higher expression only in ccRCC with VHL mutation, but generally not in rare ccRCC that retain wild-type VHL, or in normal kidney epithelium (Fig. 6A, Supplementary Fig. S8A). To validate this observation, 786-O cells with VHL mutation, were transduced with an adenovirus overexpressing VHL protein (Fig. 6B) to restore VHL function of HIF regulation. When VHL was expressed, we observed destabilization of HIF2α, and a subsequent reduction in both the chemerin protein as well as mRNA level (Fig. 6B, Supplementary Fig. S8B). HIF2α silencing also transcriptionally reduced chemerin expression (Fig. 6C, Supplementary Fig. S8C). Thus, chemerin expression in ccRCC is VHL dependent. In the case of wild type VHL and HIF2α dependent, hypoxia could therefore lead to elevated expression as well.
Figure 6. Chemerin expression is VHL dependent and regulates HIF expression.
(A) scRNA-seq data from human normal transplant kidney, normal kidney tissue adjacent to tumor, ccRCC specimens with wild-type VHL and ccRCC specimens with VHL mutation, presented as UMAP plots.
(B) Immunoblots of lysates from 786-O transfected with VHL plasmids.
(C) Relative mRNA expression of chemerin in 786-O and 760-P cells infected with lentivirus encoding shGFP or shHIF2α. Two-tailed student’s t-test.
(D) Relative mRNA expression of chemerin in 786-O and 769-P cells transfected with control siRISC or siKLF6. Two-tailed student’s t-test.
(E) Expression of HIF1α and HIF2α after chemerin knockdown in 786-O cells, measured by qRT-PCR. One-way ANOVA.
(F) Immunoblots of lysates from 786-O and 769-P for HIF2α after infection with lentivirus targeting shGFP or 2 different shRARRES2.
(G) Gene set enrichment analysis of RNA-sequencing of 786-O infected with lentivirus encoding either shGFP or shRARRES2, demonstrating downregulation of hypoxia associated pathways. Blue, low expression; red, high expression.
(H) Relative mRNA expression of HIF target genes, VEGF, LOX and IGFBP3 after chemerin knockdown in 786-O cells, measured by qRT-PCR. One-way ANOVA.
(I) Relative mRNA expression of HIF1α and HIF2α in 786-O after incubation with 500μM linoleic acid or 500μM palmitic acid for 24 hours, measured by qRT-PCR. One-way ANOVA.
(J) Proposed mechanism of chemerin regulation of ccRCC tumorigenesis. HIF2α, hypoxia inducible factor 2 alpha; MUFA, monounsaturated fatty acid; PUFA, polyunsaturated fatty acid; ROS, reactive oxygen species; SFA, saturated fatty acid; TCA cycle, tricarboxylic acid cycle.
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001.
We noted from the literature the recent description of the zinc finger DNA-binding transcription factor KLF6 as a target of the VHL-HIF2 axis in ccRCC, and after interrogating the list of KLF6 responsive genes, we found chemerin (Supplementary Fig. S8D) (31). Accordingly, knockdown of KLF6 leads to reduced chemerin expression (Fig. 6D, Supplementary Fig. S8E-F). Furthermore, RNA-sequencing data of multiple ccRCC cell lines from the Cancer Dependency Map (www.depmap.org), demonstrate a positive correlation between KLF6 and RARRES2 expression (R2 = 0.8344, p-value = 0.0302). (Supplementary Figure S8G). Together, the data suggest that VHL alterations, through deregulation of HIF2α and KLF6, mediates tumor-derived chemerin expression in ccRCC.
Strikingly, several FAO genes in our analysis that were regulated by chemerin have been reported as HIF target genes (14,32), and chemerin inhibition reduced transcription of both HIF1α and HIF2α in cells (Fig. 6E). HIF2α protein level was also significantly decreased by chemerin silencing in both 786-O and 769-P cells (Fig. 6F); where HIF1α protein is not detectable. GSEA analysis of the RNA-seq data confirmed that in addition to the lipid pathways noted in Fig. 3D, the hypoxia pathway was one of the most downregulated pathways after chemerin knockdown (Fig. 6G, Supplementary Fig. S8H). We validated reduced expression of the HIF target genes VEGF, LOX and IGFBP3 by qRT-PCR (Fig. 6H). Interestingly, as a potential link between lipid metabolism and HIFα mRNA expression, Yoo and colleagues demonstrated previously that the HepG2 cells induce HIF1α expression as a protective measure against fatty acid-induced toxicity (33). Exogenous addition of palmitate, for example, led to a robust induction of HIF1α mRNA. We thus tested whether levels of free FA in ccRCC cells could promote HIF1α or HIF2α expression. We found that palmitate (an SFA), opposite pattern seen in linoleic acid (a PUFA), increases mRNA (Fig. 6I) and protein levels of HIF2α, and subsequently chemerin in ccRCC cells (Supplementary Fig. S8I), and thus we propose that through maintenance of low FA metabolism, chemerin participates in a positive feedback loop to maintain high levels of HIF (Fig. 6J).
Monoclonal antibody against chemerin reduces ccRCC tumor growth
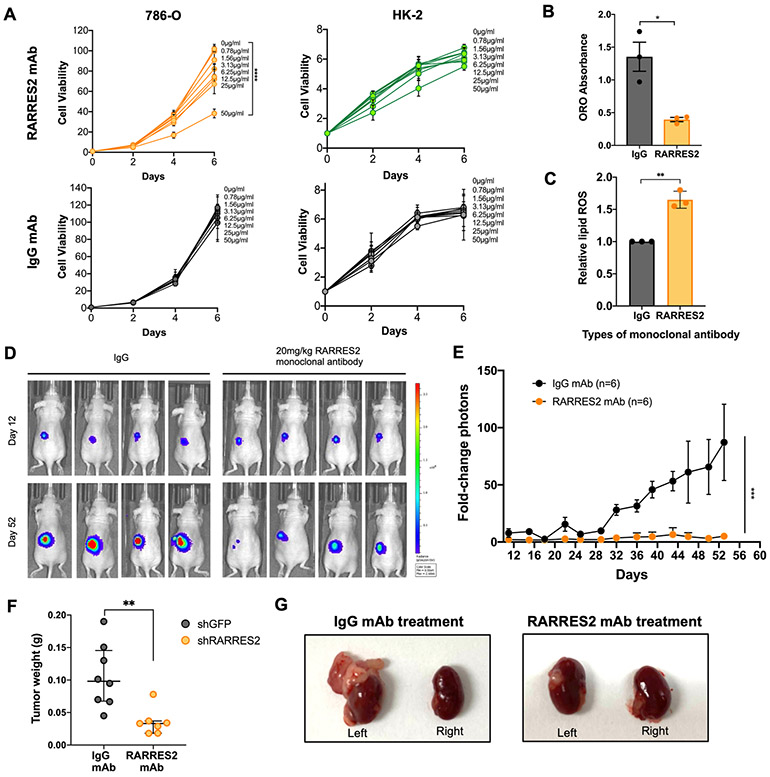
Given the significant clinical correlates of chemerin in ccRCC, we investigated the potential translational impact of inhibiting chemerin with a monoclonal antibody (mAb). Treatment of ccRCC 786-O cells showed a dose-dependent reduction in cell viability with increasing concentrations of chemerin mAb (Fig. 7A, left upper panel). In contrast, cell viability was not affected when HK-2 cells were treated with a similar dose range of the antibody (Fig. 7A, upper right panel). Likewise, neither cell line was affected by control IgG mAb treatment at the same doses (Fig. 7A, lower panels). Importantly, 786-O cells treated with chemerin mAb also showed a decrease in lipid droplets (Fig. 7B) and an increase in lipid ROS (Fig. 7C) as compared to treatment with IgG.
Figure 7. Monoclonal antibody (mAb) targeting chemerin reduces ccRCC tumor burden.
(A) Cell viability assay of 786-O and HK-2 cells treated with RARRES2 mAb (upper panels) and IgG mAb control (lower panels). Two-way repeated measures ANOVA was used for statistical analysis with Tukey correction.
(B) ORO staining quantification in 786-O cells treated with either IgG mAb or RARRES2 mAb. Two tailed student’s t-test.
(C) Relative lipid ROS measured using BODIPY 581/591 C11 assay in 786-O cells treated with either IgG mAb or RARRES2 mAb. Two tailed student’s t-test.
(D) Representative bioluminescence imaging of before (Day 0) and Day 28 post-treatment in mice receiving 20 mg/kg of either mAb (n=6 each arm), after 786-O was implanted orthotopically under the left kidney capsule of nude mice.
(E) Quantification of bioluminescence imaging of before (day 0) and day 52 post-treatment in mice receiving 20 mg/kg of either mAb (n=6 each arm), after 786-O cells were implanted orthotopically under the left kidney capsule of nude mice. One-tailed student’s t-test.
(F) Tumor weight measurement (n=6 each group) at the end of the assay. Student’s t-test.
(G) Representative kidney tumor from (E).
Error bars represent SEM of three independent experiments and three technical replica per experiment. *p < 0.05; **p < 0.01; ***p < 0.001; **** p < 0.0001.
Next, we examined the potential for therapeutic targeting of chemerin using the antibody in an orthotopic ccRCC model. 786-O cells that were engineered to express firefly luciferase were transplanted under the left kidney capsules in nude mice, and tumor growth was monitored using bioluminescent imaging (BLI). The mice were randomly assigned into two different treatment arms: one receiving 20 mg/kg chemerin mAb twice weekly, and the other receiving IgG control mAb of the same dose. The doses of antibody used in this study did not result in significant changes in animal weight, suggesting minimal toxicity (Supplementary Fig. S9A). Consistent with the cell culture results, in vivo 786-O tumor growth was significantly reduced by treatment of antibody targeting chemerin (Fig. 7D-E). The mice treated with chemerin mAb also had lower tumor weights than those receiving IgG mAb upon sacrifice (Fig. 7F-G). Collectively, the results suggest that targeting chemerin therapeutically in ccRCC represents a promising avenue that could have translational potential.
DISCUSSION
In the current report, we utilized a multidisciplinary approach including scRNA-seq, functional genomics, untargeted lipidomics and metabolomics to provide new insights into the role of the adipokine chemerin in lipid metabolism and tumorigenesis in ccRCC. The identification of HIF-dependent chemerin expression to prevent FAO and escape from ferroptosis, highlights a critical metabolic dependency of ccRCC on suppression of lipid metabolism. The findings have several direct implications, including the linkage of an obesity-driven systemic factor with tumor promotion, as well as a novel regulatory mechanism of HIF signaling. The soluble nature of chemerin suggests that both biomarker and therapeutic opportunities may be useful in devising new strategies to treat kidney cancer.
Obesity has been recognized as a global public health issue, and it correlates with higher incidences of cancer. With more than half of the US population currently overweight (BMI more than 25), it is thought that increased body mass is contributing to the rise in kidney cancer incidence (34). The hypertrophied adipose tissues in obesity can trigger inflammation, hypoxia, and angiogenesis, which subsequently may promote tumor formation (35). While efforts are underway to elucidate mechanisms explaining the link between obesity and cancer, the dysregulated secretion of signaling molecules by adipose tissue (adipokines), is a likely culprit (36). Over the last decade, many reports have demonstrated that adipokines play roles in cancer development. For instance, some adipokines, including leptin, have proinflammatory roles and can stimulate cancer stem cells to become more tumorigenic (35). Chemerin, a recently discovered multifunctional adipokine, has been shown to regulate adipogenesis, inflammation, angiogenesis and energy metabolism (17). Chemerin expression increases when preadipocytes differentiate into adipocytes and circulating chemerin has been correlated with BMI of patients (17). However, the underlying mechanisms of adipokines on cancer pathogenesis and lipid metabolism remain enigmatic. Because normal kidney epithelium undergoes adipogenic trans-differentiation when becoming ccRCC, we explored the dependency of autocrine and paracrine derived chemerin in lipid metabolism and tumor survival.
While we found consistently that chemerin expression is elevated at both the mRNA (from database analyses) and at the protein level (from the TMA) in ccRCC as compared to normal adjacent tumors, it is interesting to note that the magnitude of change tended to be less in the protein staining. Considering the extracellular role of chemerin, it is not unreasonable to hypothesize that tumor-derived protein diffuses into the adjacent tissue, as has been shown for VEGF (37), much like chemerin production from systemic or adjacent fat can diffuse into the tumor or a premalignant lesion and contribute to ccRCC biology. The nature of a secreted factor thus holds promise of both a detectable biomarker and an actionable target pathway. One significant question is relative contributions of systemic chemerin (potentially related to obesity) versus tumor-derived chemerin to ccRCC. Our data support a model where obesity elevates chemerin in a supporting manner, that is superseded by tumor production to maintain tumor growth. Considering the levels in our cohort of patients, it appears that approximately 25% of the circulating chemerin could be attributable to obesity, while 75% could be associated with the presence of a tumor. One could also argue that chemerin is likely not driving tumorigenesis as an oncogene, but rather promoting expansion and survival of an incipient VHL-deficient cell as it rewires its metabolism via constitutive HIF activation. An obesity environment may therefore be tumor promoting.
Critically, although obesity is a recognized as risk factor of ccRCC, it is commonly accepted that higher BMI is associated with improved survival (38-40). It remains unclear as to whether this phenomenon, known as the “obesity paradox,” is due to the nature of tumors in obese patients, reverse causation of cancer cachexia, or detection bias. Nevertheless, how chemerin would fit into a model as an obesity-dependent driver of ccRCC and yet be prognostic of worse outcome could be explained by several possibilities. First, it is unlikely that chemerin is the only adipokine playing a role in ccRCC. Indeed, in our initial informatics screening process, we identified other potential proteins that associate with poor outcome and are also overexpressed in tumors. We did not look for potentially counteracting molecules that could explain better outcomes, but it is possible that they exist. Secondly, our studies have not attempted as of yet to determine how chemerin impacts responses to standard of care approaches such as tyrosine kinase inhibitors. It is possible that obesity contributes to tumor formation, but also results in better drug distribution or responses. In fact, there are reports that obesity associates with more highly angiogenic tumors; and chemerin itself has noted roles in recruiting endothelial progenitor cells through a proangiogenic role (41,42). Thus, further work will be required to delineate all of the implications of obesity in ccRCC, and specifically the functions of chemerin beyond metabolic rewiring.
Accumulation of lipids in lipid droplets is a well-established phenomenon in ccRCC, but the significance of lipid storage is still ambiguous. Some have suggested that the significance is to prevent lipotoxicity, while others have proposed a role in suppressing ER stress (32,43). The observation that chemerin prevents ferroptosis by suppressing FAO aligns more with the evasion of lipotoxicity theory; but the fact that reduced FAO also supports elevated HIF expression adds a new layer of benefit to ccRCC tumors. Ferroptosis is a recently described form of regulated cell death that results from iron-dependent lipid peroxidation, and is a promising strategy to induce cancer cell death (44). It was reported by Yang et al. that RCC cells are more prone to ferroptosis if they are grown at low cell density (<50% confluency) as compared to high cell density (45). This could be due to the inadequate production of a secreted factor, such as chemerin, to support cell survival. Interestingly, we also noted in a prior work that induction of lipid droplets in vitro is density dependent (14). Thus it is intriguing to think that the link between ferroptosis evasion and lipid droplet production is the common secretion of chemerin.
While our current work demonstrated an important role of chemerin in ccRCC tumor biology, the question remains if a receptor is involved for the action of this circulating adipokine. In the literature, chemerin acts as a natural ligand for chemokine-like receptor 1 (CMKLR1, also known as ChemR23), G protein-coupled receptor 1 (GPR1) and C-C chemokine receptor-like 2 (CCRL2). Among these, CMKLR1 is the most well-studied in cancers and chemerin/CMKLR1 has been shown to be involved in gastric cancer (46), hepatocellular carcinoma metastasis (47) and immune cell trafficking (48). In our case, after knocking out CMKLR1 in 769-P (Supplementary Fig. S10A-B), we observed a decrease in cell proliferation ability (Supplementary Fig. S10C) and lipid deposition (Supplementary Fig. S10D), similar to the effect after chemerin silencing. Some of the lipid metabolism genes regulated by chemerin (CPT1A and PLIN4) were also downregulated by CMKLR1 knockout (Supplementary Fig. S10E). This suggests the involvement of CMKLR1 receptor in lipid biology of chemerin in ccRCC. Yet more work needs to be done to fully elucidate the functional role of CMKLR1 in the ferroptosis pathway. In addition, chemerin binds to GPR1 with similar affinity as CMKLR1 and GPR1 has been demonstrated to be involved in glucose homeostasis in obesity (49), breast cancer (50) and adipogenesis (50). On the other hand, CCRL2 is not involved in signal transduction and its role in human biology is still poorly understood. Hence, it is of high interest to clearly delineate the roles of these receptors in ccRCC biology as they may also be useful therapeutic targets.
Chemerin has a physiological role as an inflammatory mediator in homeostasis as well as recently described pathological inflammatory roles in multiple cancers (51). Thus, the effect of chemerin on tumor growth could also be driven by its function on inflammatory cells. Chemerin has been reported to recruit immune effector cells and increase T-cell mediated cytotoxicity in breast cancer (52), prostate cancer (53) and sarcoma models (54). However, these cancers, in contrast to ccRCC, have lower chemerin expression in tumor tissues than healthy individuals, and higher chemerin expression is correlated with better survival outcome (52). It was demonstrated in multiple reports that chemerin increases NK cell and CD8+ T cell infiltration in orthotopic syngeneic models, thus these tumors downregulate chemerin expression as a means of immune escape (52). On the other hand, chemerin has also been shown to promote Treg differentiation, and promotes polarization of M2 macrophages in pRCC, both of which are tumor-promoting (55). Interestingly, it was also noted that chemerin may play a role in increasing lipid deposition in macrophages (52), showing the interplay of chemerin in immune cell modulation, lipid deposition and tumor progression. In the study of pRCC, elevated chemerin did associate with worse survival (21). Our studies in ccRCC thus far have only considered chemerin in an immunodeficient setting, thereby excluding potential additional roles of inflammatory cell modulation. Future experiments are planned to delineate the broader roles of chemerin in ccRCC in an immunocompetent model, and also to address a potential interaction with efficacy of immunotherapy.
The classic triad symptoms of RCC are hematuria, abdominal pain and a palpable mass, yet most patients are asymptomatic until incidental detection by radiographic imaging (34). Most of the cases therefore have advanced to higher stages or even metastasized when diagnosed (34). Hence, there is a need to have a circulating biomarker that could be useful for early detection. Several adipokines have been described as possible biomarker candidates. For instance, a large case-control study in 2013 reported reduced serum adiponectin in RCC patients, but one of the biggest limitations of the study is that only males were included (56). Moreover, the correlations of adipokines (including adiponectin and leptin) and RCC risks are inconsistent in multiple reports, further weakening their potential as biomarkers (57,58). In a recent report by Lu et al., serum chemerin is higher in oral squamous cell carcinoma patients than healthy individuals (59). In the current report, we detected an elevated chemerin level in ccRCC patient plasma, which is also correlated with BMI status. Despite the potential of chemerin as plasma biomarker based on current data, more analyses in a larger cohort or a case-control studies are needed to reduce the potential confounding factors such other inflammatory states. Nonetheless, it is possible that plasma chemerin can be informative when combined with existing practice tools for early detection and follow up ccRCC patients.
The implications of an adipokine regulating lipid metabolism and ferroptosis in ccRCC could have broad impact, as obesity is increasingly recognized as risk factor for a multitude of cancers. Before the recent advances in immunotherapy, the primary focus of cancer therapy had traditionally been on tumor and tumor microenvironmental phenotypes. Consideration of systemic mediators of transformation and progression has remained an overlooked attribute. While obesity driven metabolic mediators may be particularly relevant for highly lipid-laden cancers such as ccRCC and clear cell ovarian tumors, metabolic alterations in cancers are essentially universal. Targeting chemerin to block its downstream effects on FAO and ferroptosis is therefore a conceptually novel therapeutic approach to that possibly could be modeled in additional diseases.
METHODS
LEAD CONTACT AND MATERIALS AVAILABILITY
Further information and request for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Scott M. Welford (scott.welford@med.miami.edu). This study did not generate new unique reagents.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell Cultures.
786-O, UOK101, A498, and HEK293T cells were cultured in DMEM (Corning, Cat# 10-013-CV) containing 10% fetal bovine serum (FBS) (VWR, Cat# 89510-186). 769-P was cultured in RPMI (Gibco, Cat# 11875-093, 31800-022) containing 10% fetal bovine serum. HK-2 cell was cultured in Keratinocyte Serum Free Medium (K-SFM) (Invitrogen, Cat# 10724-011) supplemented with bovine pituitary extract (BPE) and human recombinant epidermal growth factor (EGF). 769-P, A-498, HEK293T and HK-2 were obtained from ATCC. 786-O and UOK-101 were gifts of Dr. Sandra Turcotte and Dr. W. Marston Linehan (National Cancer Institute, Bethesda), respectively. Cells were used for experiments within 10-15 passages from thawing. All the cells were tested to ensure they are mycoplasma free and they were authenticated by short tandem repeat profiling by Genetica DNA Laboratories.
Animal Studies.
All animal experiments were in compliance with National Institutes of Health guidelines and were approved by the Institutional Animal Care and Use Committee (IACUC) of the University of Miami Miller School of Medicine. Six-week-old female athymic nude mice (Charles River Laboratories) were used for xenograft studies. Mice were housed (3-5 mice per cage) on a 12-hour light/dark cycle in individually ventilated cages, and with ad libitum access to food and water. The experimental holding room had temperature (21.5°C set point) and humidity control (40% set point), and it was equipped with HEPA-filtered air. For subcutaneous tumor growth model, cells were pelleted and resuspended in a PBS/Matrigel Matrix (Corning, Cat# 356234) mix at 1:1 ratio. 2×106 cells in a 100 μL solution were injected subcutaneously into each flank. Once palpable tumors were established, digital calipers were used to measure tumor size twice weekly. Tumor volume was calculated using the equation V = ½ (length × width2). For orthotopic tumor growth assays, soft collagen pellets containing 1× 106 cells were implanted orthotopically under the capsules of the left kidney. Tumors were palpated and monitored twice weekly using IVIS bioluminescence imaging (Perkin Elmer). At the end of the experiments, blood was collected and both kidneys were harvested.
Patient Information.
Fresh normal-tumor tissues were obtained from nephrectomy cases at the University of Miami Hospital. Patients’ whole blood was also obtained and processed in the laboratory to obtain plasma. The samples were obtained under approvals from institutional research ethics review committees and with written informed patient consent under the institutional review board protocol 20140385.
METHOD DETAILS
RNA Isolation and Real Time Quantitative Polymerase Chain Reaction (qRT-PCR).
Total RNA was extracted using TRIzol reagent (Invitrogen, Cat# 15596018). The concentration and quality of RNA were determined using a NanoDrop 1000 spectrophotometer (Thermo). 500 ng of RNA was used to synthesize cDNA using the Quantabio qScript cDNA SuperMix (Quantabio, Cat# 95048-100) according to the manufacturer’s recommendations. The cDNA was diluted by 10-fold before used for qRT-PCR gene expression analysis. qRT-PCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Cat# 4367659) and SYBR Green primers (listed in Supplementary Table S3). The Ct values of the gene of interest was normalized to the Ct value of the housekeeping control, β-actin. The fold change was then calculated using the 2−ΔΔCt method.
Lentiviral transduction.
For shRNA knockdown models, HEK293T cells were transfected with a mixture of the lentiviral transfer plasmid of interest (Supplementary Table S4), psPAX2 and PVSVG using Lipofectamine 2000 transfection reagent (Invitrogen, Cat# 11668-019). shRNA GFP was used as a control. The media containing the viral particles was collected 24 hours post-transfection and filtered through 0.45 μm PVDF sterile syringe filter (Santa Cruz). Cells were infected with the lentiviral supernatant in the presence of 10 μg/mL of polybrene (Sigma-Aldrich, Cat# H9268). RARRES2 knockout cell lines were generated by cloning sgRNAs (Supplementary Table S5) into the lentiviral vector lentiCrisprv2 (Addgene), followed by lentivirus production in HEK293T. After puromycin selection, single cell clones were generated by limiting dilution in 96 well plates. Single-cell clones were expanded and lysates were collected to verify knockout by western blotting.
Protein Extraction and Western blotting.
The cells were washed with 1× ice-cold PBS (GE, Cat# SH30256.01) once before they were lysed in RIPA lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% NP-40; 50 mM NaF) and scraped from the plate on ice. The RIPA lysis buffer contained 1:1000 1× protease inhibitor cocktail (Sigma-Aldrich) and 1:100 1× phosphatase inhibitor (Sigma-Aldrich). The lysates were passed through a 16G needle 15 times before they were centrifuged at 4°C for 20 minutes at 20,000×g. The protein lysates were quantified using the Pierce BCA Protein Assay Kit (Thermo) according to the manufacturer’s protocols and absorbance was measured using GloMax Discover Microplate Reader (Promega). Between 50 and 100 μg of protein samples were mixed with 5× SDS Laemmli loading buffer and boiled at 95°C for 5 minutes. Proteins were separated using 4-20% SDS-PAGE gels (Bio-Rad) and transferred onto PVDF membrane (Millipore). The membrane was blocked with TBS-T supplemented with 5% non-fat dry milk primary antibody for 45 minutes at room temperature rocking before blotting with primary antibodies at 4°C overnight. The membrane was washed in TBS-T 3 times with 5 minutes each and blotted in secondary antibodies at room temperature for 2 hours. An Azure 300 chemiluminescent western blot imaging system (Azure biosystems) was used to visualize the membrane. The antibodies used were listed in Supplementary Table S6.
Enzyme-linked Immunosorbent Assay (ELISA) for Plasma Chemerin.
Plasma were diluted 1:1000 before incubated in a high-binding microplate (Santa Cruz) coated with capture antibody according to manufacturer’s instructions (R&D, Cat# DY2324). The samples were washed, and biotin-conjugated detection antibody was added into the plate wells for further incubation. Next, HRP-conjugated streptavidin was added, followed by substrate A/B 1:1 mixture. Finally, the catalytic reaction was terminated by adding the stop solution and the absorbance was measured at 450 nm using GloMax Discover Microplate Reader (Promega).
Cell Proliferation Assay.
Cells were plated at a density of 100,000 cells in triplicate wells of a 6-well plate, supplemented with DMEM containing 10% FBS. Cells were trypsinized and were counted using the Cell Counter Countess II (Life Technologies) at days 3, 6, 9 and 12.
Cell Viability Assay.
After treatment with monoclonal antibodies, 786-O cells were incubated with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenylterazolium bromide (MTT, Sigma-Aldrich) to assess cell survival. Briefly, the cells were plated at density of 1000 cells/plate in 96-well plates and treated with different concentrations of monoclonal antibodies (IgG or anti-RARRES2) for indicated incubation times (2 days, 4 days and 6 days). MTT solution was diluted 5-fold in DMEM media and incubated in 37°C incubator for 4 hours. The media was carefully removed and the precipitate was dissolved in DMSO. The solution later immediately quantified at 450 nm using the GloMax Discover Microplate Reader (Promega).
EdU incorporation assay.
Cells were plated on coverslips at 1×104 cells in 12-well plate and incubated with 10 μM EdU solution overnight at 37°C, 5% CO2. After incubation, the cells were fixed with 3.7% formaldehyde/PBS for 15 minutes at room temperature and washed with 1 mL of 3% BSA/PBS twice. 1 mL of 0.5% TritonX-100 in PBS was added for 20 minutes at room temperature to permeabilize the cells. Next, cells were incubated with 0.5 mL of Click-iT reaction cocktail (Invitrogen, Cat# C10337) prepared according to manufacturer’s instructions, at room temperature for 30 minutes and away from direct light. DAPI (Millipore, Cat# 268298) was added later for DNA staining. Coverslips were removed from wells and visualized using the z-stack of an Olympus FV1000 confocal scanning microscope. The images were analyzed using Fiji software (ImageJ).
Clonogenic survival assay.
The cells were plated in triplicate in 6cm plates (100 cells/well) with complete DMEM media to measure survival by colony forming unit (CFU). The colonies were counted after 2 weeks and the experiment was repeated twice.
siRNA transfection.
In one tube, the stock siRNA was diluted in serum-free medium and DharmaFECT transfection reagent (Horizon) was added to another tube. The content of each tube was gently mixed and incubated for 5 minutes at room temperature. The contents of both tubes were mixed and incubated for 20 minutes at room temperature. The culture media were removed from the plate and 2.5mL of serum-free medium was added together with the transfection medium. The cells were incubated at 37°C in 5% CO2 for 5 hours before changing the media to complete media with 10% FBS. Non-targeting Control siRNAs (siRISC) and KLF6 targeting siRNA (Horizon) were used.
Flow Cytometry.
Cells were stained with fluorescein isothiocyanate (FITC)-conjugated annexin V and propidium iodide (ChemCruz, Cat# sc-3541) using the Alexa Fluor 488 Annexin V Kit (Invitrogen, Cat# V13245) according to the manufacturer’s protocol. The cells were later analyzed using an LSR II cytometer instrument (BD Biosciences) and analyzed by FACS DIVA 8.1 software (BD Biosciences). The percentage of dead cells in measured by combining cells with annexin V-positive and annexin V/PI-double-positive.
BODIPY Lipid Droplet Imaging.
Cells were plated on round glass coverslips of 12-well plates and incubated with 1 mL of DMEM supplemented with 10% FBS. The medium was removed and cells were washed with room temperature PBS, fixed with 4% formaldehyde for 30 minutes and incubated with 300 μL of 1uM BODIPY 493/503 (Life Technologies, Cat# D3922) for 15 minutes, protected from direct light. 1 μg/mL of DAPI (Millipore) was added for nucleus staining. The z stack images were acquired using Olympus FV1000 confocal scanning microscope and analyzed using Fiji software (ImageJ).
Lipid Reactive Oxygen Species.
The cells were plated at 50,000 cells per well in 24-well plate. 10uM BODIPY-581/591 C11 (Invitrogen, Cat# D3861) and 1ug/mL DAPI (Millipore) were added to the media and incubated for 1 hour at 37°C, 5% CO2. Cells were imaged using Olympus FV1000 confocal scanning microscope and analyzed using Fiji software (ImageJ). 9 randomly selected images per condition were selected for analysis.
Oil Red O (ORO) Lipid Droplet Staining.
For tissue sections staining, the tissues were cryoprotected by incubating in 30% sucrose/PBS solution. After the tissues sank to the bottom of the vial, the tissues were embedded in OCT (Fisher Healthcare). Embedded tissue blocks were sectioned at 10 μm thickness and stored at −80°C until ready for staining. Before ORO staining, the frozen sections were air dried and fixed in 10% neutral buffered formalin (Sigma-Aldrich, Cat# HT501128). After fixing in 100% propylene glycol (VWR, Cat# 0575) twice, sections were stained with freshly made ORO solution (Sigma-Aldrich, Cat# O0625) followed by staining for hematoxylin. Then, the slides were washed with distilled water and mounted using glycerin jelly (Sigma-Aldrich). The sections were visualized using a VS120 Virtual Slide Microscope scanner (Olympus). For cell staining, the cells plated in 12-well plates in triplicate were rinsed with PBS twice, and fixed with 10% formaldehyde for 1 hour. They were then rinsed with 60% isopropanol for 5 minutes, followed by ORO staining (3 mg/mL) for 4 minutes and the ORO solution was removed. After the cells were air dried, 250 μL of isopropanol was added to the wells and the eluted solution was measured at 510nm. Next, the cells were stained with sulforhodamine B as previously described, and measured at 564 nm using GloMax Discover Microplate Reader (Promega), as previously described (60).
BN-PAGE analysis.
Blue native-polyacrylamide gel electrophoresis (BN-PAGE) was performed as previously described (61). Briefly, 2x106 shGFP control and shRARRES2 cells were collected, resuspended in PBS and permeabilized with 2 mg/mL digitonin. Permeabilized cells were pelleted and resuspended in 100 μl buffer containing 1.5 M aminocaproic acid, 50 mM Bis-Tris pH 7.0. Mitochondrial proteins were extracted in the presence of 1% lauryl maltoside (LM), followed by clarification spin at 22,000 g for 30 minutes at 4°C. Native PAGE™ Novex® 3-12% Bis-Tris protein gels (Life Technologies) were loaded with 60-80 μg of total LM cell extracts. After electrophoresis, proteins were transferred to PDVF membranes and used for immunoblotting. Western blot was performed using primary antibodies raised against the following human OXPHOS subunits: NDUFA9 (Abcam, ab14713), CORE2 (UQCRC2, Abcam, ab14745), COX1 (Abcam, ab14705), SDHA (Protein tech, 14865-1-AP), ATP5 (ATP5A, Abcam, ab14748). In parallel, 60 μg aliquots of the same samples were resolved in a denaturing 12% SDS-PAGE and probed with a primary antibody against VDAC as loading control. Peroxidase-conjugated anti-mouse and anti-rabbit IgGs were used as a secondary antibodies (Rockland).
Polarographic analysis of cellular respiration.
Endogenous cell respiration was measured polarographically at 37°C using a Clark-type electrode from Hansatech Instruments (Norfolk, United Kingdom). Substrate-driven respiration was assayed in digitonin-permeabilized cultured cells as reported (62). Briefly, trypsinized cells were washed with permeabilized-cell respiration buffer (PRB) containing 0.3 M mannitol, 10 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 0.5 mM EGTA, 1 mg/ml BSA and 10 mM KH3PO4 (pH 7.4). The cells were resuspended at ~4 × 106 cells/ml in 0.5 ml of the same buffer air-equilibrated at 37 °C and supplemented with 10 units of hexokinase and 2 mM ADP. The cell suspension was immediately placed into the polarographic chamber to measure endogenous respiration. Cells were then permeabilized with digitonin (3.4 μg/106 cells) and complex I substrates glutamate and malate (to 2 mM each final concentration) were added to measure state 3 or ADP-stimulated (coupled) respiration. Oligomycin (to 1 μM final concentration) was successively added to inhibit ATP synthesis, and non-phosphorylating condition (state 4 or leak respiration) was measured. Mitochondrial respiration was uncoupled by 1 μL incremental addition of 0.1 M CCCP to reach maximal oxygen consumption. Lastly 3 μL of an 80 mM KCN solution were finally added to assess the specificity of the assay. Oxygen consumption rate were measured as nmol O2 consumed/min, normalized by number of cells and expressed as percentage of the shGFP control endogenous respiration.
ATP Measurement.
The relative cellular ATP concentration was measured using Cell Titer-Glo Luminescent reagent (Promega) according to manufacturer’s protocol. Briefly, an equal volume of the single-one-step reagent was added to each well of cells and incubated at room temperature for 15 minutes rocking. The luminescence was quantified using GloMax Discover Microplate Reader (Promega). The ATP level is normalized to cell count from an additional plate with same setup.
Extracellular Flux Analysis.
Prior to the Seahorse XF96 Cell Mito Stress Test assay, the optimal cell density was determined to be 2000 cells per well. A day before the experiment, the cells were plated in the Seahorse XF96 Cell Culture Microplate (Agilent 102601-100) using DMEM. The sensor cartridge was hydrated in Seahorse XF Calibrant at 37°C in a non-CO2 incubator overnight. The medium (Agilent 103575-100) was supplemented with 1 mM pyruvate, 2 mM glutamine and 10 mM glucose before adjusting the pH to be 7.4. 20 μL of 15 μM stock Oligomycin, 22 μL of 5 μM stock FCCP and 25 μL of 5 μM stock Rotenone/Antimycin A, were added to Port A, B and C of the microplate respectively. After replacing the cell media in the cell culture microplate with warmed assay medium using a multichannel pipette, the microplate was placed in a 37°C non-CO2 incubator for 45 hours. The assay was run using the Agilent Seahorse XF96 Analyzer (Seahorse Biosciences) and the assay parameters (including oxygen consumption rate) were exported from the Seahorse XF Stress Test Report Generator.
Immunohistochemistry staining.
The paraffin-embedded ccRCC tumor microarray (KD601) was purchased from US Biolab that contains clear cell renal cell carcinoma tissue with matched cancer adjacent kidney tissue, 2 cores per case. The slides were baked for 60 min at 60°C, deparaffinized in xylene, and rehydrated through graded concentrations of ethanol in water. The slides were then subjected to antigen retrieval. Immunohistochemistry staining was performed using BOND-III Fully Automated IHC and ISH Stainer according to the manufacturer’s instructions (Leica Biosystems, Wetzlar, Germany). Anti-rabbit polyclonal human chemerin (29-67) (H-002-52, Phoenix Pharmaceuticals) was used at 1:200 dilution, and other reagents for IHC were provided by Bond™ Polymer Refine Detection (DS9800) kit. The microarray sections were left to air-dry, mounted with permanent mounting medium and coverslipped. The bright field images were acquired by an automated VS200 Research Slide Scanner (Olympus) at 40× magnification. The images were subsequently viewed using the OlyVIA software (Olympus). The intensity of the staining was quantified using Image J (63).
Sodium palmitate preparation and supplementation.
To prepare BSA-palmitate conjugate, sodium palmitate (Thermo, Cat# P0500) was added to a NaCl solution and heated in a water bath while stirring. The hot palmitate solution was mixed with BSA solution, 5mL at a time. The final volume was adjusted with 150 mM NaCl for 1 mM palmitate solution and pH was adjusted to 7.4. Sodium palmitate solution was added to cell at final concentration 500 μM and incubated for 24 hours.
RNA-sequencing.
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Cat# 74104) according to the manufacturer’s instructions. Before library construction, all samples were assessed for RNA purity via 260/280 ratio using Nanodrop, then RNA integrity and potential DNA contamination using agarose gel electrophoresis, and RNA integrity again using the Agilent Bioanalyzer 2100. mRNA was purified from total RNA using poly-T-oligo-attached magnetic beads and the mRNA were fragmented randomly by addition of fragmentation buffer. For NEB library preparation using NEBNext® Ultra™ RNA Library Prep Kit for Illumina®, briefly, the first strand cDNA was synthesized using random hexamer primer and M-MuLV Reverse Transcriptase (RNase H-) and second strand cDNA synthesis was subsequently performed using DNA polymerase I and RNase H. The double-stranded cDNA was then purified using AMPure XP beads and the remaining overhangs of the purified double-stranded cDNA are converted into blunt ends via exonuclease/polymerase activities. After adenylation of 3’ ends of DNA fragments, NEBNext Adaptor with hairpin loop structure was ligated to prepare for hybridization. In order to select cDNA fragments of preferentially 150~200 bp in length, the library fragments are purified with AMPure XP system (Beckman Coulter, Beverly, USA). Finally, the final library was obtained by PCR amplification and purification of PCR products by AMPure XP beads. The libraries were pooled in equimolar amounts and sequenced in paired end 150bp reactions on the Illumina NovaSeq 6000.
Untargeted lipidomics and metabolomics.
Cell samples were extracted using the modified folch biphasic extraction procedure (64). Briefly, for metabolomics, 20 μL of internal standard mixture were added to cell pellets of 10 million cells. Protein quantification were done for all the samples for pre-normalization. Ice cold methanol (80%) were used for global metabolome extraction. The supernatant was collected after centrifugation, transferred to a new tube and dried under nitrogen. The dried sample was reconstituted in 0.1% formic acid in water (70 μL). For lipidomics, 20 μL of 10x diluted internal standard mixture (stock solution of 50 ppm, w: v) was added. Samples were extracted using ice cold 4:2:1 chloroform:methanol:water (v:v:v), and the organic phase was collected, dried down under nitrogen flow, and reconstituted in 75 μL of isopropanol plus 1 μL of injection standard mixture (100 ppm, w:v). They were used for global lipidome extraction. Metabolomics and lipidomics samples ran separately and for each sequence, solvent blanks, extraction blanks (without internal standard), neat quality controls (QCs, blanks with internal standards), pooled samples QCs were also prepared for evaluation of extraction and data collection efficiency. High-pressure liquid chromatography coupled high-resolution tandem mass spectrometry (LC-HRMS/MS) were used for data collection. Chromatographic separation was achieved using reverse phase chromatography (Dionex Ultimate 3000 RS UHLPC system, Thermo Scientific, San Jose, CA, USA) with a Waters C18-pfp column (Ace, 100 x 2.1 mm, 2 μM) for metabolomics and with a Waters Acquity C18 BEH column maintained at 30˚C (2.1 × 100 mm, 1.7 μm particle size, Waters, Milford, MA, US) for lipidomics. In the case of metabolomics, the gradient consisted of solvent A (0.1% FA in H2O) and solvent B (Acetonitrile), both with 10 mM ammonium formate and 0.1% formic acid. The flow rate was 350 μL/min. The column temperature was maintained at 25°C. In the case of lipidomics, the gradient consisted of solvent A (60:40 acetonitrile:water) and solvent B (90:8:2 isopropanol:acetonitrile:water), both with 10 mM ammonium formate and 0.1% formic acid. The flow rate was 500 μL/min. The column temperature was maintained at 50°C. Samples were analyzed in positive and negative electrospray ionization on a ThermoScientific Q-Exactive mass spectrometry (Thermo Scientific, San Jose, CA). Data-dependent (ddMS2-top5) MS/MS data were obtained on pooled samples per group for identification purposes. In addition, full-scan data was acquired for all the samples without MS/MS for comparing metabolite or lipid intensities across groups.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification and statistical analysis.
Unless indicated in the figure legend, all experiments were performed at least in triplicate and results were presented as mean ± standard error of mean (SEM) of absolute values or percentages of control. All the statistical analyses performed were described in each figure legend. Statistical p values were obtained by application of the appropriate statistical tests using the Prism 8 program. P-values lower than 0.05 were considered significant (*p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001; ns, not significance).
RNA-sequencing data analysis.
The raw FASTQ data were processed through an in-house bioinformatics pipeline including quality and adapter trimming with TrimGalore version 0.6.1 (https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), alignment to the human genome hg38/GRCh38 and gene quantification with the STAR algorithm v2.5.2 (65). The differential expression analysis was performed using the quasi-likelihood function implemented in edgeR (66). Gene set enrichment analysis was performed using fully unfiltered gene list in the GSEA desktop application (http://software.broadinstitute.org/gsea/downloads.jsp) (67,68).
Single cell RNA-sequencing data analysis.
Single-cell RNA-sequencing data was assessed from Young et al. and analyzed as previously described (19). Briefly, in order to remove low quality cells, only single cell libraries with at least 1000 features with greater than 1 count per million were retained and cells with high fraction of counts from mitochondrial genes (indicating dying cells) were removed. In addition, genes with expression less than 1 count per million in at least 2 cells per tumor were excluded from analysis. After concatenating cell by gene count matrices of all samples into a single matrix and normalized, the relationship between cells was visualized in two dimensions through Uniform Manifold Approximation and Projection (UMAP) plots. Each cluster was assigned to respective cell identity by marker differential gene expressions described in previous literatures. The data supplement from Young et al. defined tissue of origin, and was used to stratify during several different analyses. Our analysis followed the primary workflow of the Seurat pipeline (69,70). According to the authors, the data was normalized by dividing the total number of unique molecular identifiers (UMIs) in each cell to obtain the sequencing depth before transforming to a log scale for each cell using the Seurat (http://satijalab.org/seurat/) NormalizeData function: (19)
x: UMI count matrix
g: gene
c: cell
F: Seurat “scale.factor” (default value 10,000)
Next, COMBAT (19) was used in batch correction step to regress out the variability introduced by individual 10× channels. All the entries which were 0 before batch correction were maintained to be at 0 after batch correction to prevent imputing expression from genes with no expression. The data was re-normalized so that it was consistent with being derived from an expression vector with the sum to 1 after such correction:
The threshold for mitochondrial genes is set at 5%.
Lipidomics and metabolomics data analysis.
For lipidomics data analysis, LipidMatch Flow was used for file conversion, peak picking (implementing MZMine 2 (71), blank filtration (72,73), lipid annotation (74), and combining positive and negative datasets. LipidMatch Flow software and tutorials (including video tutorials) can be found at http://secim.ufl.edu/secim-tools/. LipidMatch was used to annotate ions using data dependent MS/MS analysis. For metabolomics data analysis, metabolite identification was performed with MZmine 2.0 and matching metabolite retention time and m/z value to an internal library of over 1000 metabolites. Metabolic pathway analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database by matching metabolite sets with human metabolome (https://www.genome.jp/kegg/pathway.html). Metabolite set enrichment (fold enrichment) were investigated using MetaboAnalyst (open source R package).
DATA AND CODE AVAILABILITY
This study did not generate new unique reagents. The RNA -sequencing data was deposited in GEO database GSE159716. The lipidomics and metabolomics data were deposited in MetaboLights MTBL2435.
Supplementary Material
STATEMENT OF SIGNIFICANCE.
Identification of a HIF-dependent adipokine that prevents fatty acid oxidation and causes escape from ferroptosis highlights a critical metabolic dependency unique in the clear cell subtype of kidney cancer. Targeting lipid metabolism via inhibition of a soluble factor is a promising pharmacological approach to expand therapeutic strategies for ccRCC patients.
ACKNOWLEDGEMENTS
We thank M. Linehan for UOK101 cells. This work was supported in part by National Institutes of Health: R01CA254409, P30CA240139, R01DK120679, P50AA024333, and P01HL147823; U24DK097209 to Southeast Center for Integrated Metabolomics (SECIM) at the University of Florida.
Footnotes
Conflict of interests: The authors declare no potential conflict of interests.
REFERENCES
- 1.Zhang Z, Lin E, Zhuang H, Xie L, Feng X, Liu J, et al. Construction of a novel gene-based model for prognosis prediction of clear cell renal cell carcinoma. Cancer Cell Int 2020;20:27 doi 10.1186/s12935-020-1113-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cui W, Luo W, Zhou X, Lu Y, Xu W, Zhong S, et al. Dysregulation of Ketone Body Metabolism Is Associated With Poor Prognosis for Clear Cell Renal Cell Carcinoma Patients. Front Oncol 2019;9:1422 doi 10.3389/fonc.2019.01422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7–34 doi 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 4.Padala SA, Barsouk A, Thandra KC, Saginala K, Mohammed A, Vakiti A, et al. Epidemiology of Renal Cell Carcinoma. World J Oncol 2020;11(3):79–87 doi 10.14740/wjon1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garje R, An J, Greco A, Vaddepally RK, Zakharia Y. The Future of Immunotherapy-Based Combination Therapy in Metastatic Renal Cell Carcinoma. Cancers 2020;12(1):143 doi 10.3390/cancers12010143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linehan WM, Schmidt LS, Crooks DR, Wei D, Srinivasan R, Lang M, et al. The Metabolic Basis of Kidney Cancer. Cancer Discov 2019;9(8):1006–21 doi 10.1158/2159-8290.Cd-18-1354. [DOI] [PubMed] [Google Scholar]
- 7.Ackerman D, Tumanov S, Qiu B, Michalopoulou E, Spata M, Azzam A, et al. Triglycerides Promote Lipid Homeostasis during Hypoxic Stress by Balancing Fatty Acid Saturation. Cell Rep 2018;24(10):2596–605 e5 doi 10.1016/j.celrep.2018.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan SK, Welford SM. Lipid in Renal Carcinoma: Queen Bee to Target? Trends Cancer 2020;6(6):448–50 doi 10.1016/j.trecan.2020.02.017. [DOI] [PubMed] [Google Scholar]
- 9.Kaelin WG Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell 2008;30(4):393–402 doi 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 10.Sanchez DJ, Simon MC. Genetic and metabolic hallmarks of clear cell renal cell carcinoma. Biochim Biophys Acta Rev Cancer 2018;1870(1):23–31 doi 10.1016/j.bbcan.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kwon TJ, Ro JY, Mackay B. Clear-cell carcinoma: an ultrastructural study of 57 tumors from various sites. Ultrastruct Pathol 1996;20(6):519–27. [DOI] [PubMed] [Google Scholar]
- 12.Melone MAB, Valentino A, Margarucci S, Galderisi U, Giordano A, Peluso G. The carnitine system and cancer metabolic plasticity. Cell death & disease 2018;9(2):228 doi 10.1038/s41419-018-0313-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Serra D, Mera P, Malandrino MI, Mir JF, Herrero L. Mitochondrial fatty acid oxidation in obesity. Antioxidants & redox signaling 2013;19(3):269–84 doi 10.1089/ars.2012.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du W, Zhang L, Brett-Morris A, Aguila B, Kerner J, Hoppel CL, et al. HIF drives lipid deposition and cancer in ccRCC via repression of fatty acid metabolism. Nature communications 2017;8(1):1769 doi 10.1038/s41467-017-01965-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seidell JC, Halberstadt J. The global burden of obesity and the challenges of prevention. Ann Nutr Metab 2015;66 Suppl 2:7–12 doi 10.1159/000375143. [DOI] [PubMed] [Google Scholar]
- 16.da Costa WH, Moniz RR, da Cunha IW, Fonseca FP, Guimaraes GC, de Cassio Zequi S. Impact of renal vein invasion and fat invasion in pT3a renal cell carcinoma. BJU Int 2012;109(4):544–8 doi 10.1111/j.1464-410X.2011.10366.x. [DOI] [PubMed] [Google Scholar]
- 17.Helfer G, Wu QF. Chemerin: a multifaceted adipokine involved in metabolic disorders. J Endocrinol 2018;238(2):R79–r94 doi 10.1530/joe-18-0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tun HW, Marlow LA, von Roemeling CA, Cooper SJ, Kreinest P, Wu K, et al. Pathway signature and cellular differentiation in clear cell renal cell carcinoma. PloS one 2010;5(5):e10696 doi 10.1371/journal.pone.0010696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young MD, Mitchell TJ, Vieira Braga FA, Tran MGB, Stewart BJ, Ferdinand JR, et al. Single-cell transcriptomes from human kidneys reveal the cellular identity of renal tumors. Science (New York, NY) 2018;361(6402):594–9 doi 10.1126/science.aat1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landolt L, Eikrem O, Strauss P, Scherer A, Lovett DH, Beisland C, et al. Clear Cell Renal Cell Carcinoma is linked to Epithelial-to-Mesenchymal Transition and to Fibrosis. Physiological reports 2017;5(11) doi 10.14814/phy2.13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goralski KB, Jackson AE, McKeown BT, Sinal CJ. More Than an Adipokine: The Complex Roles of Chemerin Signaling in Cancer. International Journal of Molecular Sciences 2019;20(19):4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buechler C, Feder S, Haberl EM, Aslanidis C. Chemerin Isoforms and Activity in Obesity. International journal of molecular sciences 2019;20(5) doi 10.3390/ijms20051128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ribeiro RA, Bonfleur ML, Batista TM, Borck PC, Carneiro EM. Regulation of glucose and lipid metabolism by the pancreatic and extra-pancreatic actions of taurine. Amino Acids 2018;50(11):1511–24 doi 10.1007/s00726-018-2650-3. [DOI] [PubMed] [Google Scholar]
- 24.Lee B, Mahmud I, Marchica J, Dereziński P, Qi F, Wang F, et al. Integrated RNA and metabolite profiling of urine liquid biopsies for prostate cancer biomarker discovery. Scientific reports 2020;10(1):3716 doi 10.1038/s41598-020-60616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flores-Romero H, Ros U, Garcia-Saez AJ. A lipid perspective on regulated cell death. Int Rev Cell Mol Biol 2020;351:197–236 doi 10.1016/bs.ircmb.2019.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Pratt DA, Tallman KA, Porter NA. Free radical oxidation of polyunsaturated lipids: New mechanistic insights and the development of peroxyl radical clocks. Acc Chem Res 2011;44(6):458–67 doi 10.1021/ar200024c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swenson SA, Moore CM, Marcero JR, Medlock AE, Reddi AR, Khalimonchuk O. From Synthesis to Utilization: The Ins and Outs of Mitochondrial Heme. Cells 2020;9(3) doi 10.3390/cells9030579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Antonicka H, Leary SC, Guercin GH, Agar JN, Horvath R, Kennaway NG, et al. Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple, early-onset clinical phenotypes associated with isolated COX deficiency. Hum Mol Genet 2003;12(20):2693–702 doi 10.1093/hmg/ddg284. [DOI] [PubMed] [Google Scholar]
- 29.Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. Embo j 2007;26(6):1713–25 doi 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang H, Liu C, Zhao Y, Gao G. Mitochondria regulation in ferroptosis. Eur J Cell Biol 2020;99(1):151058 doi 10.1016/j.ejcb.2019.151058. [DOI] [PubMed] [Google Scholar]
- 31.Syafruddin SE, Rodrigues P, Vojtasova E, Patel SA, Zaini MN, Burge J, et al. A KLF6-driven transcriptional network links lipid homeostasis and tumour growth in renal carcinoma. Nature communications 2019;10(1):1152 doi 10.1038/s41467-019-09116-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu B, Ackerman D, Sanchez DJ, Li B, Ochocki JD, Grazioli A, et al. HIF2alpha-Dependent Lipid Storage Promotes Endoplasmic Reticulum Homeostasis in Clear-Cell Renal Cell Carcinoma. Cancer Discov 2015;5(6):652–67 doi 10.1158/2159-8290.Cd-14-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoo W, Noh KH, Ahn JH, Yu JH, Seo JA, Kim SG, et al. HIF-1alpha expression as a protective strategy of HepG2 cells against fatty acid-induced toxicity. Journal of cellular biochemistry 2014;115(6):1147–58 doi 10.1002/jcb.24757. [DOI] [PubMed] [Google Scholar]
- 34.Rajandram R, Perumal K, Yap NY. Prognostic biomarkers in renal cell carcinoma: is there a relationship with obesity? Transl Androl Urol 2019;8(Suppl 2):S138–s46 doi 10.21037/tau.2018.11.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee CH, Woo YC, Wang Y, Yeung CY, Xu A, Lam KS. Obesity, adipokines and cancer: an update. Clin Endocrinol (Oxf) 2015;83(2):147–56 doi 10.1111/cen.12667. [DOI] [PubMed] [Google Scholar]
- 36.Stone TW, McPherson M, Gail Darlington L. Obesity and Cancer: Existing and New Hypotheses for a Causal Connection. EBioMedicine 2018;30:14–28 doi 10.1016/j.ebiom.2018.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierzbicki PM, Klacz J, Kotulak-Chrzaszcz A, Wronska A, Stanislawowski M, Rybarczyk A, et al. Prognostic significance of VHL, HIF1A, HIF2A, VEGFA and p53 expression in patients with clear-cell renal cell carcinoma treated with sunitinib as first-line treatment. International journal of oncology 2019;55(2):371–90 doi 10.3892/ijo.2019.4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gan CL, Heng DYC. New insights into the obesity paradox in renal cell carcinoma. Nat Rev Nephrol 2020;16(5):253–4 doi 10.1038/s41581-020-0264-y. [DOI] [PubMed] [Google Scholar]
- 39.Parker AS, Lohse CM, Cheville JC, Thiel DD, Leibovich BC, Blute ML. Greater body mass index is associated with better pathologic features and improved outcome among patients treated surgically for clear cell renal cell carcinoma. Urology 2006;68(4):741–6 doi 10.1016/j.urology.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 40.Hakimi AA, Furberg H, Zabor EC, Jacobsen A, Schultz N, Ciriello G, et al. An epidemiologic and genomic investigation into the obesity paradox in renal cell carcinoma. J Natl Cancer Inst 2013;105(24):1862–70 doi 10.1093/jnci/djt310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shin WJ, Zabel BA, Pachynski RK. Mechanisms and Functions of Chemerin in Cancer: Potential Roles in Therapeutic Intervention. Front Immunol 2018;9:2772 doi 10.3389/fimmu.2018.02772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bozaoglu K, Curran JE, Stocker CJ, Zaibi MS, Segal D, Konstantopoulos N, et al. Chemerin, a novel adipokine in the regulation of angiogenesis. J Clin Endocrinol Metab 2010;95(5):2476–85 doi 10.1210/jc.2010-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gimm T, Wiese M, Teschemacher B, Deggerich A, Schodel J, Knaup KX, et al. Hypoxia-inducible protein 2 is a novel lipid droplet protein and a specific target gene of hypoxia-inducible factor-1. FASEB J 2010;24(11):4443–58 doi 10.1096/fj.10-159806. [DOI] [PubMed] [Google Scholar]
- 44.Bersuker K, Hendricks JM, Li Z, Magtanong L, Ford B, Tang PH, et al. The CoQ oxidoreductase FSP1 acts parallel to GPX4 to inhibit ferroptosis. Nature 2019;575(7784):688–92 doi 10.1038/s41586-019-1705-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang WH, Ding CC, Sun T, Rupprecht G, Lin CC, Hsu D, et al. The Hippo Pathway Effector TAZ Regulates Ferroptosis in Renal Cell Carcinoma. Cell Rep 2019;28(10):2501–8.e4 doi 10.1016/j.celrep.2019.07.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumar JD, Aolymat I, Tiszlavicz L, Reisz Z, Garalla HM, Beynon R, et al. Chemerin acts via CMKLR1 and GPR1 to stimulate migration and invasion of gastric cancer cells: putative role of decreased TIMP-1 and TIMP-2. Oncotarget 2019;10(2):98–112 doi 10.18632/oncotarget.26414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J-J, Yin H-K, Guan D-X, Zhao J-S, Feng Y-X, Deng Y-Z, et al. Chemerin suppresses hepatocellular carcinoma metastasis through CMKLR1-PTEN-Akt axis. British journal of cancer 2018;118(10):1337–48 doi 10.1038/s41416-018-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vr Wittamer, Franssen J-D, Vulcano M, Mirjolet J-Fo, Le Poul E, Migeotte I, et al. Specific Recruitment of Antigen-presenting Cells by Chemerin, a Novel Processed Ligand from Human Inflammatory Fluids. Journal of Experimental Medicine 2003;198(7):977–85 doi 10.1084/jem.20030382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rourke JL, Muruganandan S, Dranse HJ, McMullen NM, Sinal CJ. Gpr1 is an active chemerin receptor influencing glucose homeostasis in obese mice. J Endocrinol 2014;222(2):201–15 doi 10.1530/joe-14-0069. [DOI] [PubMed] [Google Scholar]
- 50.Huang C, Dai XY, Cai JX, Chen J, Wang BB, Zhu W, et al. A Screened GPR1 Peptide Exerts Antitumor Effects on Triple-Negative Breast Cancer. Mol Ther Oncolytics 2020;18:602–12 doi 10.1016/j.omto.2020.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pachynski RK, Wang P, Salazar N, Zheng Y, Nease L, Rosalez J, et al. Chemerin Suppresses Breast Cancer Growth by Recruiting Immune Effector Cells Into the Tumor Microenvironment. Front Immunol 2019;10:983 doi 10.3389/fimmu.2019.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pachynski RK, Wang P, Salazar N, Zheng Y, Nease L, Rosalez J, et al. Chemerin Suppresses Breast Cancer Growth by Recruiting Immune Effector Cells Into the Tumor Microenvironment. Frontiers in Immunology 2019;10(983) doi 10.3389/fimmu.2019.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin PC, Giannopoulou EG, Park K, Mosquera JM, Sboner A, Tewari AK, et al. Epigenomic alterations in localized and advanced prostate cancer. Neoplasia 2013;15(4):373–83 doi 10.1593/neo.122146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alholle A, Brini AT, Gharanei S, Vaiyapuri S, Arrigoni E, Dallol A, et al. Functional epigenetic approach identifies frequently methylated genes in Ewing sarcoma. Epigenetics 2013;8(11):1198–204 doi 10.4161/epi.26266. [DOI] [PubMed] [Google Scholar]
- 55.Jacenik D, Fichna J. Chemerin in immune response and gastrointestinal pathophysiology. Clinica Chimica Acta 2020;504:146–53 doi 10.1016/j.cca.2020.02.008. [DOI] [PubMed] [Google Scholar]
- 56.Liao LM, Weinstein SJ, Pollak M, Li Z, Virtamo J, Albanes D, et al. Prediagnostic circulating adipokine concentrations and risk of renal cell carcinoma in male smokers. Carcinogenesis 2013;34(1):109–12 doi 10.1093/carcin/bgs322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Spyridopoulos TN, Petridou ET, Dessypris N, Terzidis A, Skalkidou A, Deliveliotis C, et al. Inverse association of leptin levels with renal cell carcinoma: results from a case-control study. Hormones (Athens) 2009;8(1):39–46 doi 10.14310/horm.2002.1220. [DOI] [PubMed] [Google Scholar]
- 58.Choi SH, Chun SY, Kim TH, Kwon TG. Identifying the emerging role of adipokine as a diagnostic and prognostic biomarker of renal cell carcinoma. Urol Oncol 2016;34(6):259.e15–9 doi 10.1016/j.urolonc.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 59.Lu Z, Liang J, He Q, Wan Q, Hou J, Lian K, et al. The serum biomarker chemerin promotes tumorigenesis and metastasis in oral squamous cell carcinoma. Clin Sci (Lond) 2019;133(5):681–95 doi 10.1042/cs20181023. [DOI] [PubMed] [Google Scholar]
- 60.Vichai V, Kirtikara K. Sulforhodamine B colorimetric assay for cytotoxicity screening. Nat Protoc 2006;1(3):1112–6 doi 10.1038/nprot.2006.179. [DOI] [PubMed] [Google Scholar]
- 61.Diaz F, Barrientos A, Fontanesi F. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using blue native gel electrophoresis. Curr Protoc Hum Genet 2009;Chapter 19:Unit19.4 doi 10.1002/0471142905.hg1904s63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Barrientos A, Fontanesi F, Diaz F. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr Protoc Hum Genet 2009;Chapter 19:Unit19.3 doi 10.1002/0471142905.hg1903s63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Crowe AR, Yue W. Semi-quantitative Determination of Protein Expression using Immunohistochemistry Staining and Analysis: An Integrated Protocol. Bio Protoc 2019;9(24) doi 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 1957;226(1):497–509. [PubMed] [Google Scholar]
- 65.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1):15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139–40 doi 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 2005;102(43):15545–50 doi 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 2003;34(3):267–73 doi 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 69.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36(5):411–20 doi 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive Integration of Single-Cell Data. Cell 2019;177(7):1888–902.e21 doi 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pluskal T, Castillo S, Villar-Briones A, Oresic M. MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 2010;11:395 doi 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirpich AS, Ibarra M, Moskalenko O, Fear JM, Gerken J, Mi X, et al. SECIMTools: a suite of metabolomics data analysis tools. BMC Bioinformatics 2018;19(1):151 doi 10.1186/s12859-018-2134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patterson R, Kirpich A, Koelmel J, Kalavalapalli S, Morse A, Cusi K, et al. Improved experimental data processing for UHPLC–HRMS/MS lipidomics applied to nonalcoholic fatty liver disease. Metabolomics 2017;13(11):142. [Google Scholar]
- 74.Koelmel JP, Kroeger NM, Ulmer CZ, Bowden JA, Patterson RE, Cochran JA, et al. LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data. BMC Bioinformatics 2017;18(1):331 doi 10.1186/s12859-017-1744-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate new unique reagents. The RNA -sequencing data was deposited in GEO database GSE159716. The lipidomics and metabolomics data were deposited in MetaboLights MTBL2435.