Abstract
Purpose:
Radiation-induced lung injury (RILI) is a progressive condition with an early phase (radiation pneumonitis) and a late phase (lung fibrosis). RILI may occur after partial-body ionizing radiation exposures or internal radioisotope exposure, with wide individual variability in timing and extent of lung injury. This study aimed to provide new insights into the pathogenesis and progression of RILI in the nonhuman primate (NHP) rhesus macaque model.
Methods and Materials:
We used an integrative approach to understand RILI and its evolution at clinical and molecular levels in 17 NHPs exposed to 10 Gy of whole-thorax irradiation in comparison with 3 sham-irradiated control NHPs. Clinically, we monitored respiratory rates, computed tomography (CT) scans, plasma cytokine levels, and bronchoalveolar lavage (BAL) over 8 months and lung samples collected at necropsy for molecular and histopathologic analyses using RNA sequencing and immunohistochemistry.
Results:
Elevated respiratory rates, greater CT density, and more severe pneumonitis with increased macrophage content were associated with early mortality. Radiation-induced lung fibrosis included polarization of macrophages toward the M2-like phenotype, TGF-β signaling, expression of CDKN1A/p21 in epithelial cells, and expression of α-SMA in lung stroma. RNA sequencing analysis of lung tissue revealed SERPINA3, ATP12A, GJB2, CLDN10, TOX3, and LPA as top dysregulated transcripts in irradiated animals. In addition to transcriptomic data, we observed increased protein expression of SERPINA3, TGF-β1, CCL2, and CCL11 in BAL and plasma samples.
Conclusions:
Our combined clinical, imaging, histologic, and transcriptomic analysis provides new insights into the early and late phases of RILI and highlights possible biomarkers and potential therapeutic targets of RILI. Activation of TGF-β and macrophage polarization appear to be key mechanisms involved in RILI.
Introduction
High doses of ionizing radiation exposure to the thorax during accidental or clinical events can disrupt normal lung biology substantially, leading to radiation-induced lung injury (RILI). The lung is a radiosensitive organ, and RILI may occur at single doses of ≥9.5 Gy. For fractionated therapeutic exposures, the mean lung dose (≥35.5 Gy), the volume of lung exposed to ≥20 Gy, and a higher total dose (≥45 Gy) are consistent predictors of radiation pneumonitis (RP).1–6 Nonuniform and partial body exposure are likely scenarios that could lead to significant lung damage.7 RILI is a progressive condition that has 2 phases: an early phase of RP, which includes inflammation, edema, and the proliferation of epithelial and endothelial cells, and a late phase of irreversible lung fibrosis (LF), with the accumulation of fibroblasts, myofibroblasts, and extracellular matrix remodeling. Persistent RILI leads to fibrosis, for which there is no treatment approved by the US Food and Drug Administration.8 Thus, RP and LF represent important clinical challenges for diagnosis, prognosis, and treatment.
Macrophages play an important role in instigating radiation-induced proinflammatory and profibrotic responses.9 However, their role in RILI and subsequent development of LF is not fully understood.10 TGF-β is a major regulator of fibrosis11 and is the most potent inducer of extracellular matrix synthesis. TGF-β promotes the development of myofibroblasts, which can arise from resident fibroblasts or circulating fibrocytes or via epithelial-mesenchymal transition (EMT).12 We hypothesized that macrophage polarization and increased TGF-β are important triggers for the progression of LF in the RILI nonhuman primate (NHP) model. Our data provide new insights into the pathogenesis and progression of RILI in the NHP model and a basis for investigating underlying mechanisms in RILI.
Methods and Materials
Animals and study design
For the present study, we used 17 rhesus macaques (median age, 4.76 years; median weight, 6.0 kg) given a single 10-Gy whole-thorax dose of ionization radiation at 4 Gy/min using a 6 MV linear accelerator (Methods E1) and 3 sham-irradiated, saline-treated controls (median age, 2.75 years; median weight, 3.6 kg). These animals were derived from prior studies of whole-thorax irradiation13 (Fig. E2). Our goal was to understand the intrinsic basis for varying individual responses to the same dose of irradiation.
Animals were studied for 8 months to assess clinical, pathologic, imaging, and molecular outcomes contributing to survival. Animals developing respiratory rates of >80 breaths per minute (the clinical endpoint) were euthanized and categorized as nonsurvivors. Animals surviving to the 35-week (8-month) experimental endpoint were categorized as survivors and were euthanized at the end of the study. Clinical observations, bronchoalveolar lavage (BAL) cytology/fluid samples, and computed tomography (CT) scans were taken at baseline and after irradiation at 2-month intervals throughout the study. After euthanasia, detailed necropsies were immediately performed, and lung tissue was collected for histopathologic and transcriptomic analysis. For transcriptomic analysis (RNA seq), 8 saline-treated irradiated animals were compared with the 3 sham-irradiated, saline-treated controls. Additional details are provided in the Methods E1, and a schematic of the experimental design is shown in Figure E1A.
Study approval
The protocol was approved by the Wake Forest University Institutional Animal Care and Use Committee. Wake Forest is accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. All procedures in this study were performed at Wake Forest University, in accordance with the Animal Welfare Act and other applicable federal regulations. All animals were screened for measles, simian retrovirus, filoviruses, simian immunodeficiency virus, simian-T-lymphotropic virus, and tuberculosis and were quarantined under Centers for Disease Control and Prevention guidelines before inclusion in the study. Supportive fluid therapy, analgesics (eg, ketoprofen), and symptomatic care were given as needed, based on clinical pathology abnormalities and clinical signs.
CT image analysis
CT scans were done before irradiation and every 2 months thereafter until the end of the study using a Toshiba Acquilion 32 slice scanner with a voxel resolution of 0.5 × 0.6 25 × 0.625 mm. Two animals reached their predefined euthanasia endpoint before the first 2-month CT scan; hence, 15 monkeys were used for CT analysis. We used Mimics Innovation Suite software, version 21.0.0.406 (Materialise), to quantify the percentage of the lung volume occupied by radio-opaque lung tissue and created a 3-dimensional reconstruction of abnormally dense lung tissue along with surrounding healthy tissue, using a modification of our previously published method13 to determine the percentage change in abnormally dense lung tissue by survival status, corrected for baseline. Additional details of CT procedures are described in Methods E1.
Histopathologic assessment, histochemical staining, and immunohistochemistry
At necropsy, 6 lung regions were consistently sampled from each animal, consisting of immediately adjacent fixed and frozen samples. Fixed samples were preserved in neutral 4% paraformaldehyde, embedded in paraffin blocks, and sectioned at 4 µm. An automated immunohistochemistry stainer, the Bond RX (Leica Biosystems, Buffalo Grove, IL), was used to stain slides. Sections were stained with hematoxylin and eosin (H&E) and Masson trichrome (MST), and immunohistochemistry staining was done for myofibroblasts (α-smooth muscle actin [α-SMA], Abcam, catalog # ab5694); macrophages (CD163, Thermo Scientific, catalog # MA1–82342; HAM56, MilliporeSigma, catalog # 279M-16); the M1 macrophage-specific transcription factor pSTAT1 (Cell Signaling Technology, Inc, catalog # 9167); the M2 macrophage-specific transcription factor CMAF-1 (Abcam, catalog # ab199424); the cell cycle arrest marker p21 (Abcam, catalog # ab109520); an epithelial marker (pancytokeratin, Dako/Agilent, catalog # M3515); and the type II pneumocyte marker SPC (Abcam, catalog # ab90716). Other details and a list of immunohistochemical markers are described in Methods E1 and Table E1.
Stained slides were scanned at 20 × magnification on an Olympus VS120 scanner. Slides stained with H&E were used for histopathologic assessment of pneumonitis severity. The histologic scoring criteria for pneumonitis are shown in Figure E4. Images of MST-stained slides were analyzed using the Visiopharm Integrator System image analysis software, version 6.5.0.2303 (Visiopharm, Horsholm, Denmark). The percentage of collagen area in the section was determined as percent collagen = (blue area/total area) × 100. Quantification details of fibrosis (MST), macrophage (HAM 56), M1 macrophages (CD163 + pSTAT 1), and M2 macrophages (CD163 + CMAF-1) are described in the Methods E1.
Cytokine analysis
Plasma and BAL supernatant samples from irradiated monkeys were assessed for 29 cytokines using the Monkey Cytokine Magnetic 29-Plex Panel (LPC0005M, Thermo Fisher); for TGF-β1, TGF-β2, and TGF-β3 using the U-PLEX TGF-β Combo NHP kit (catalog # K15243K, Meso Scale Diagnostics LLC) and Meso QuickPlex SQ120 (Meso Scale Diagnostics LLC); and for SERPINA3 using the Monkey Alpha1 Antichymotrypsin ELISA Kit (MBS7209859, MyBioSource) according to the manufacturers’ instructions (additional details are provided in Methods E1).
RNA sequencing
Snap-frozen lung samples (immediately adjacent to the fixed samples) were selected from 3 lung regions from 8 irradiated animals and 2 regions from 3 control animals for RNA extraction and sequencing. Sequencing was done using an Illumina NextSeq 500 NGS (Illumina, San Diego, CA). Further details of RNA extraction, sequencing methods, and data analysis are described in Methods E1. Significant differentially expressed genes (DEGs) were conservatively defined as log2 fold change ratios ≥ ±1 and P < .05 after adjustment for false discovery (Benjamini-Hochberg). The NHP lung RNA-Seq data have been deposited in the Gene Expression Omnibus (GEO) Database at the National Center for Biotechnology Information (NCBI) under the accession number GSE174196.
Statistical analysis
All data were plotted, and descriptive analysis was done to assess normality and skew using GraphPad Prism 8. Statistical significance was determined using paired or unpaired Student t tests or Mann-Whitney U tests depending on sample distribution. For grouped or multivariate analysis, 1-way or 2-way analysis of variance or mixed-effects model (restricted or residual maximum likelihood) matching tests were done, followed by posttests (Tukey test or Wilcoxon signed rank test). The strength of correlations was determined by the Pearson test. For all experiments, a P value <.05 was considered statistically significant.
Results
Survival
The cumulative mortality rate at the study’s end was 52.9% (9 of 17). Nonsurvivors reached a predefined euthanasia endpoint, defined as having a respiratory rate of >80 respirations per minute, and were euthanized between 1.5 months (7.4 weeks) and 6 months (24.8 weeks) after irradiation (Fig. E1B-E1D).
CT imaging reveals greater lung injury in nonsurvivors relative to survivors
Increased CT density was a feature of RILI in both survivors and nonsurvivors. Figures 1A and 1B show CT images of a representative nonsurvivor and a survivor during the course of the study. Figures 1C and 1D show 3-dimensional reconstructions of abnormally dense radio-opaque lung tissue (gold) and surrounding normal lung tissue (blue) from the same 2 animals. The distribution of the lesions was patchy, and surprisingly, the location of opacities changed over time. The percentage of injured lung volume increased significantly in both nonsurvivors (Fig. 1E) and survivors (Fig. 1F) after irradiation, relative to baseline. However, the percentage of injured lung volume was significantly greater in nonsurvivors relative to survivors by 4 months after irradiation (Fig. 1G). There was a significant positive correlation between respiratory rates and the percentage of injured lung volume on CT scans, primarily driven by nonsurvivors (Fig. E3A and E3B).
Fig. 1.
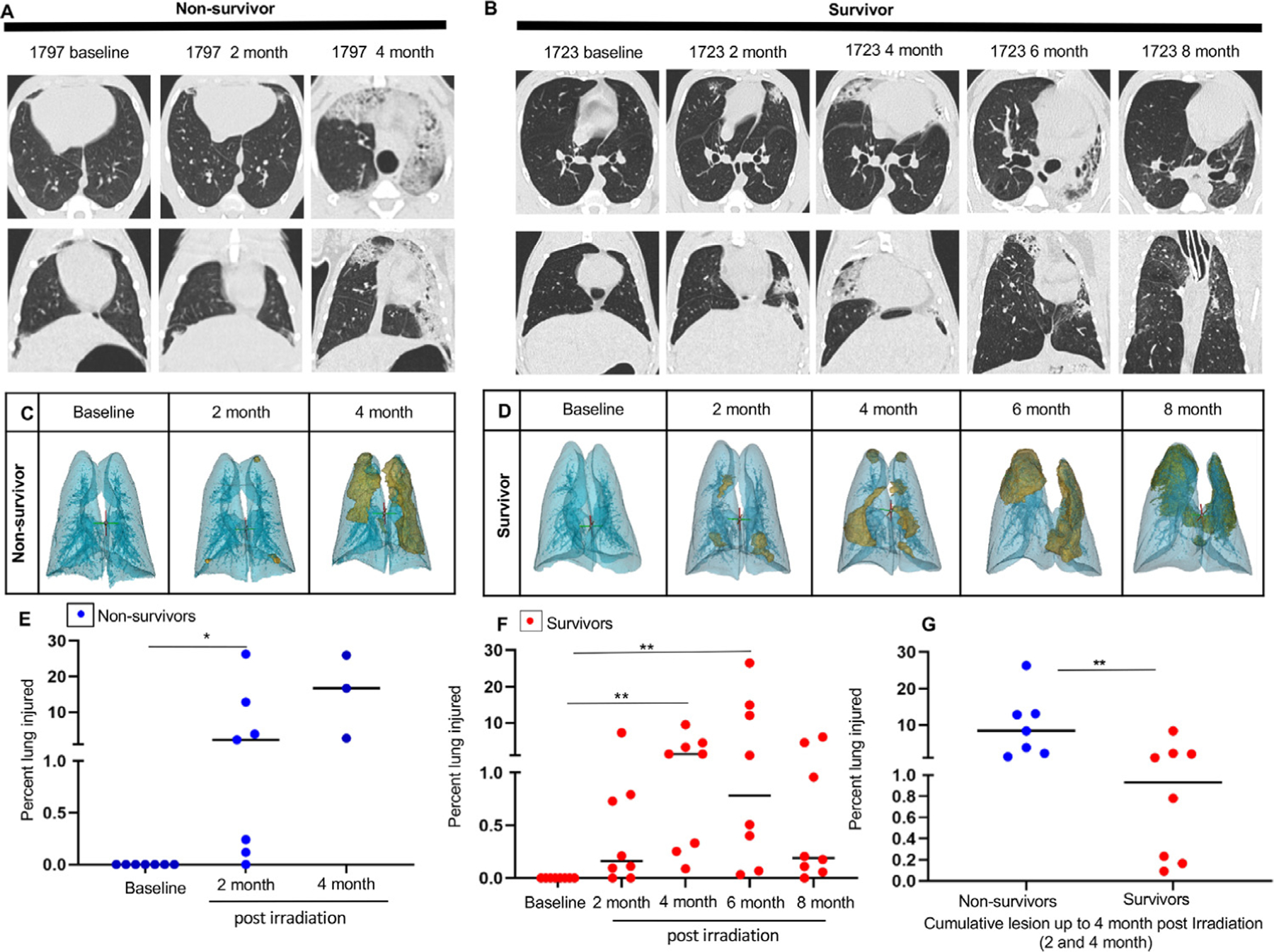
Postirradiation lung injury assessment using computed tomography. Computed tomography images (axial and coronal view) of a representative nonsurvivor (A) and survivor (B) at baseline and at the interval of 2 months until the clinical endpoint (4 months) or experimental endpoint (8 months). (C, D) Three-dimensional reconstruction of lung injury (gold) of the same representative nonsurvivor and survivor using Mimics software. (E) Percentage of injured lung volume of nonsurvivors at 2 months (n = 7) and 4 months (n = 3) relative to baseline. Results are expressed as the median and statistical significance was determined by the Wilcoxon signed rank test. (F) Percentage of injured lung volume of survivors at 2, 4, 6, and 8 months (n = 8) relative to baseline. Results are expressed as the median and statistical significance was determined by the Friedman test with a Dunn multiple comparison posttest. (G) Cumulative injured lung volume in survivors and nonsurvivors at 4 months. Results are expressed as the median. Statistical significance was determined by the Mann-Whitney test. For all data, *P < .05, **P < .01, and ***P < .001.
Histopathologic evaluation shows more severe pneumonitis in nonsurvivors
The average histologic pneumonitis score of each animal is shown in Figure 2A, where each bar represents 1 animal and dots represent 6 different regions of the lung. Monkeys at their clinical endpoints (nonsurvivors) developed moderate to severe pneumonitis. Animals euthanized at their experimental endpoints (survivors) had only minimal to mild pneumonitis by the study’s end (P < .05). Figures 2C to 2E show images of H&E-stained sections representative of a control nonirradiated lung, a nonsurvivor’s lung with a typical histologic pattern of RP, and a survivor’s lung, respectively. A significant positive correlation was observed between lung weights and pneumonitis scores (Fig. 2F), indicating the influence of pneumonitis on increased lung weights. HAM56 staining revealed a greater lung area occupied by macrophages at early clinical endpoints (nonsurvivors) relative to later endpoints (survivors) (Fig. 2G-2J). Together, these observations show that nonsurvivors developed significant pneumonitis predominated by macrophages, which was either not present or waned over time in survivors.
Fig. 2.
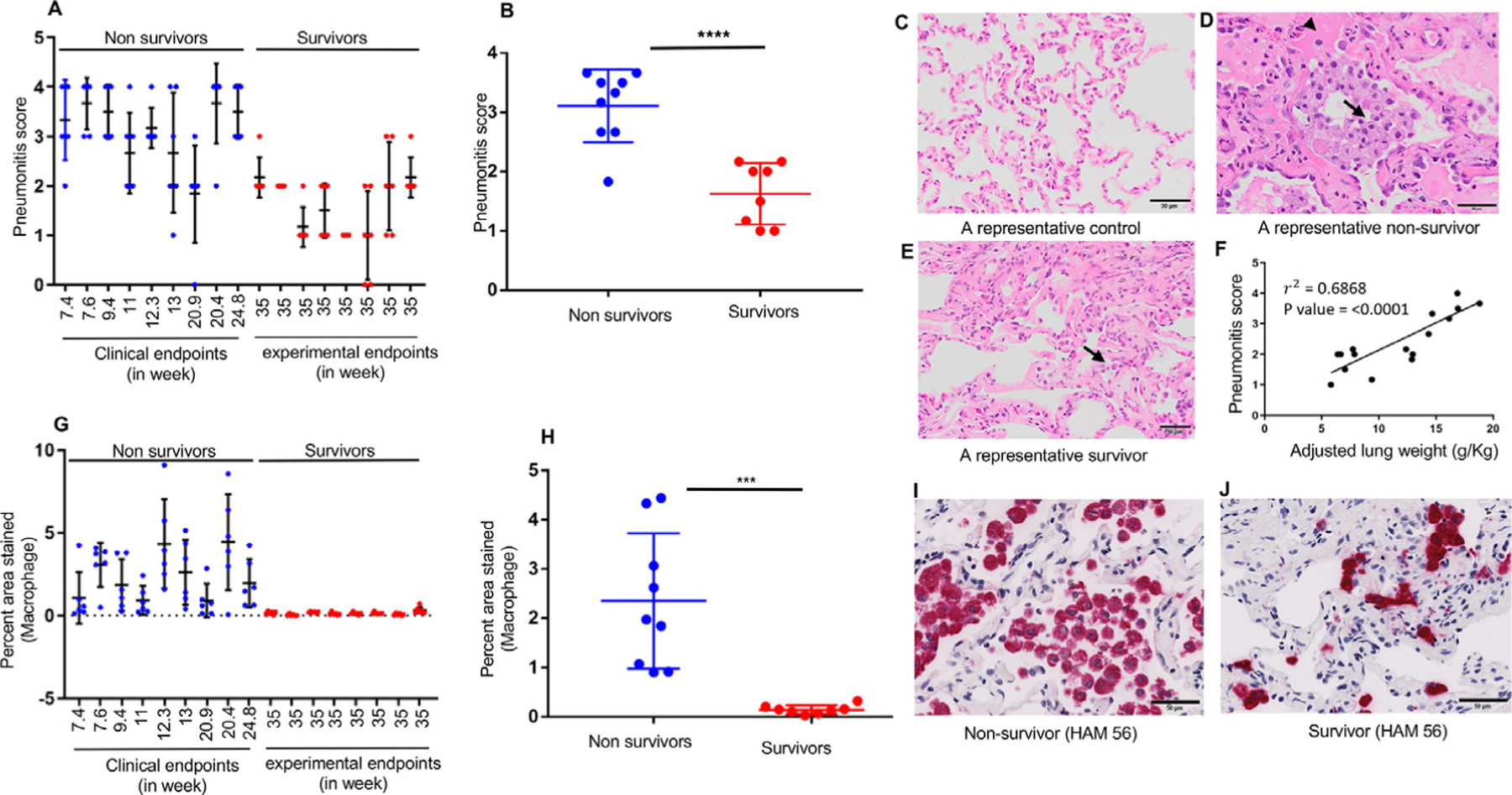
More severe pneumonitis dominated by macrophages in nonsurvivors relative to survivors at necropsy. (A) Pneumonitis scores of individual nonsurvivors (blue dots) and survivors (red dots). Dots represent 6 different lung regions in each animal. (B) Statistically significant differences in pneumonitis scores between nonsurvivors (n = 9) and survivors (n = 8). (C) A hematoxylin-and-eosin−stained photomicrograph of a control nonirradiated lung, (D) a representative nonsurvivor, and (E) a representative survivor showing infiltration of immune cells (arrow) and edema fluid (arrowhead) within the alveolar lumina. (F) A positive correlation between adjusted lung weights and pneumonitis scores of all irradiated animals. (G) M acrophage staining (HAM56, red stain) of individual nonsurvivors (blue dots) and survivors (red dots). The dots for each animal represent 6 different lung regions. (H) Macrophage staining (percentage of area stained) between nonsurvivors (n = 9) and survivors (n = 8). Results are expressed as the mean ± SD, and statistical significance was determined by an unpaired t test. For all data, *P < .05, **P < .01, and ***P < .001. (I, J) Representative images of a 13-week postirradiation nonsurvivor and a survivor, respectively, stained with the macrophage marker, HAM56, on the lung obtained at necropsy. Scale bars are 50 µm.
We observed a positive and significant correlation between the abnormally radiodense lung volume (quantified on the last CT scans corresponding to their endpoints) and the histopathologic pneumonitis scores in nonsurvivors (Fig. E3C). This correlation was not significant for survivors (Fig. E3D). These findings indicate that the inflammatory infiltrate was the major contributor to abnormally radiodense lung volume in nonsurvivors.
Irradiation induced significant fibrosis
Masson trichrome staining identified areas of collagen accumulation and fibrosis as early as 7.4 weeks after irradiation (Fig. 3A). Fibrosis was significantly greater in irradiated animals relative to nonirradiated controls (Fig. 3B) but did not differ between survivors and nonsurvivors. Representative MST-stained lung sections are shown in Figures 3C and 3D and Figure E5B.
Fig. 3.
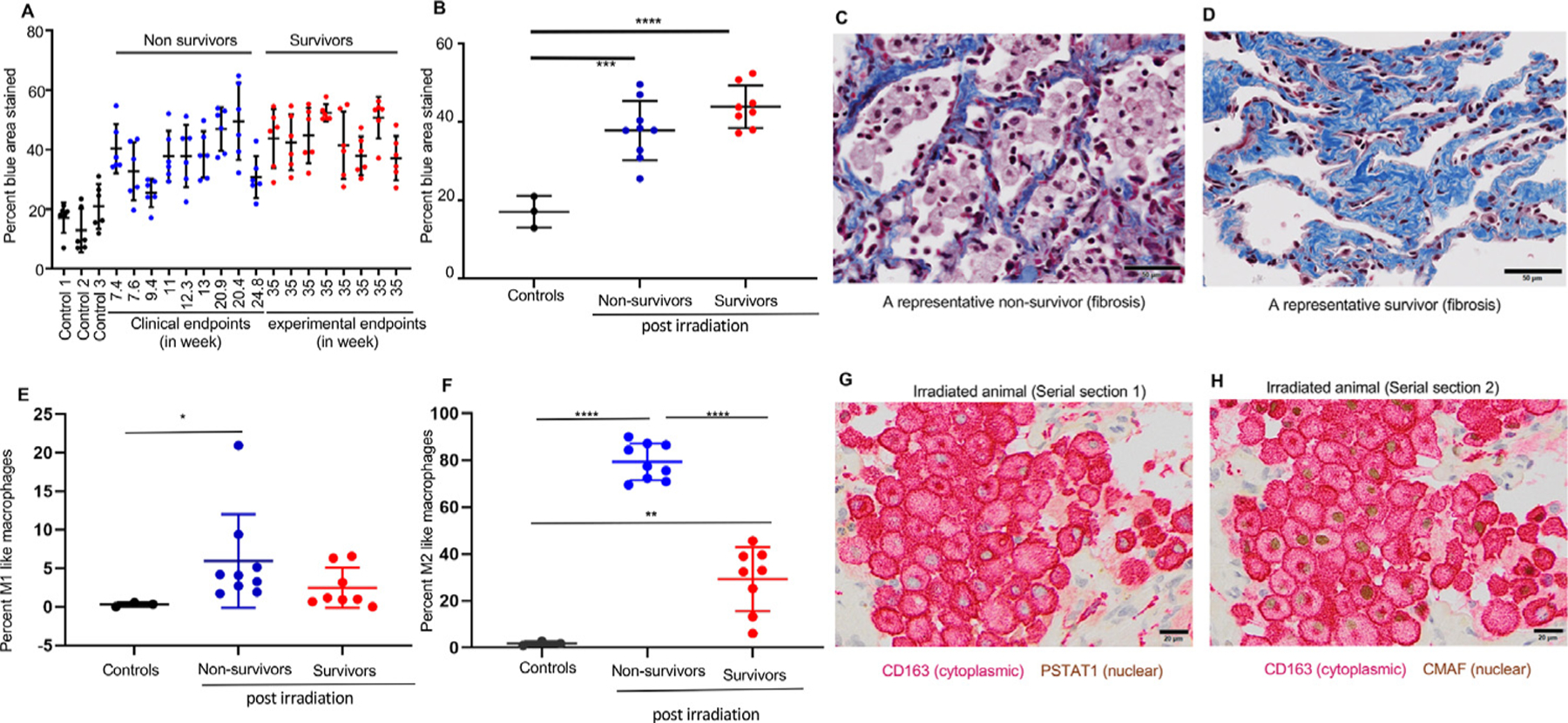
Quantitative assessment of the fibrosis and profibrotic macrophage (M2) phenotype in irradiated lungs. (A) Quantification of fibrosis as a percentage of area stained using Masson trichrome staining in individual controls (black bars), nonsurvivors (blue bars), and survivors (red bars). Six dots for each animal represent 6 different lung regions. (B) Average fibrotic area expressed as the mean ±SD between controls (n = 3), nonsurvivors (n = 9), and survivors (n = 9). Statistical significance was determined by 1-way analysis of variance with a Tukey multiple comparisons posttest. (C, D) Representative images of a postirradiation nonsurvivor and a survivor stained with Masson trichrome stain. Scale bars are 50 µm. (E, F) Quantification of M1-like (CD163 + PSTAT1) and M2-like (CD163 + CMAF) macrophages performed on serial lung sections of nonirradiated controls (n = 3) and irradiated monkeys (nonsurvivors, n = 9; survivors, n = 8) obtained at necropsy. Results for the percentage of M1 macrophages are expressed as medians ± interquartile ranges (IQRs); statistical significance was determined by the Mann-Whitney test. For M2, results are expressed as means ± SDs; statistical significance was determined by unpaired t tests. (G) Double immunostaining for macrophage marker CD163 (red) and M1-specific transcription factor PSTAT1 (brown) in a lung section 13 weeks after irradiation. (H) Double immunostaining for macrophage marker CD163 (red) and M2 specific transcription factor CMAF (brown) in a representative nonsurvivor postirradiated serial section of the same lung. For all data, *P < .05, **P < .01, and ***P < .001. Scale bars are 50 µm.
M2-like macrophages predominate in irradiated lungs
Inflammatory M1-like macrophages coexpressing CD163 and PSTAT1 were only significantly elevated in nonsurvivors (P < .05) (Fig. 3E). M2-like (profibrotic) macrophages coexpressing CD163 and CMAF were significantly higher in both irradiated groups (P = .0021) and more abundant in nonsurvivors relative to both controls and survivors (both P = .001) (Fig. 3F). A representative lung section from a nonsurvivor (13 weeks after irradiation) shows lung macrophages (CD163) negative for the M1-specific transcription factor (PSTAT1) (Fig. 3G), whereas macrophages in the corresponding serial section were positive for the M2-specific transcription factor (CMAF) (Fig. 3H).
Transcriptional profiling of lung tissues in survivors, nonsurvivors, and controls
Unsupervised principal-components analysis of RNA seq clustering data revealed 3 different gene expression patterns in controls, nonsurvivors, and survivors (Fig. 4A). A total of 2644 genes were differentially expressed (upregulated or downregulated) in nonsurvivors versus controls and 680 genes in survivors versus controls. Significant DEGs were conservatively defined as log2 fold change ratios ≥ ± 1 and P < .05 after adjustment for false discovery. Of the 2644 DEGs of nonsurvivors, 2202 differed from controls only in nonsurvivors, and 238 of 680 DEGs differed from controls only in survivors. A total of 422 DEGs were shared between nonsurvivors and survivors (Fig. 4B). Ingenuity pathway analysis (IPA) of nonsurvivor-specific DEGs (2202) revealed hepatic fibrosis as the top canonical pathway, with TGF-β as the main upstream regulator (Fig. 4B and 4C); the top upregulated transcripts included MMP3, DCSTAMP, GH1, PLA2G2D, and EREG, and the most downregulated transcripts included SLC6A4, MS4A15, SEC14LC3, SCGB3A1, and AWAT2 (Fig. 4D). Pathways related to cell division and cell cycle were prominent in IPA analysis of survivor-specific DEGs (238). Their top upregulated transcripts were ALB, ANKRD7, SPINK1, HCN4, and FAIM2, and downregulated transcripts involved KCN5, BRINP1, ESSRG, NUF2, and RACGAP1 (Fig. 4E).
Fig. 4.
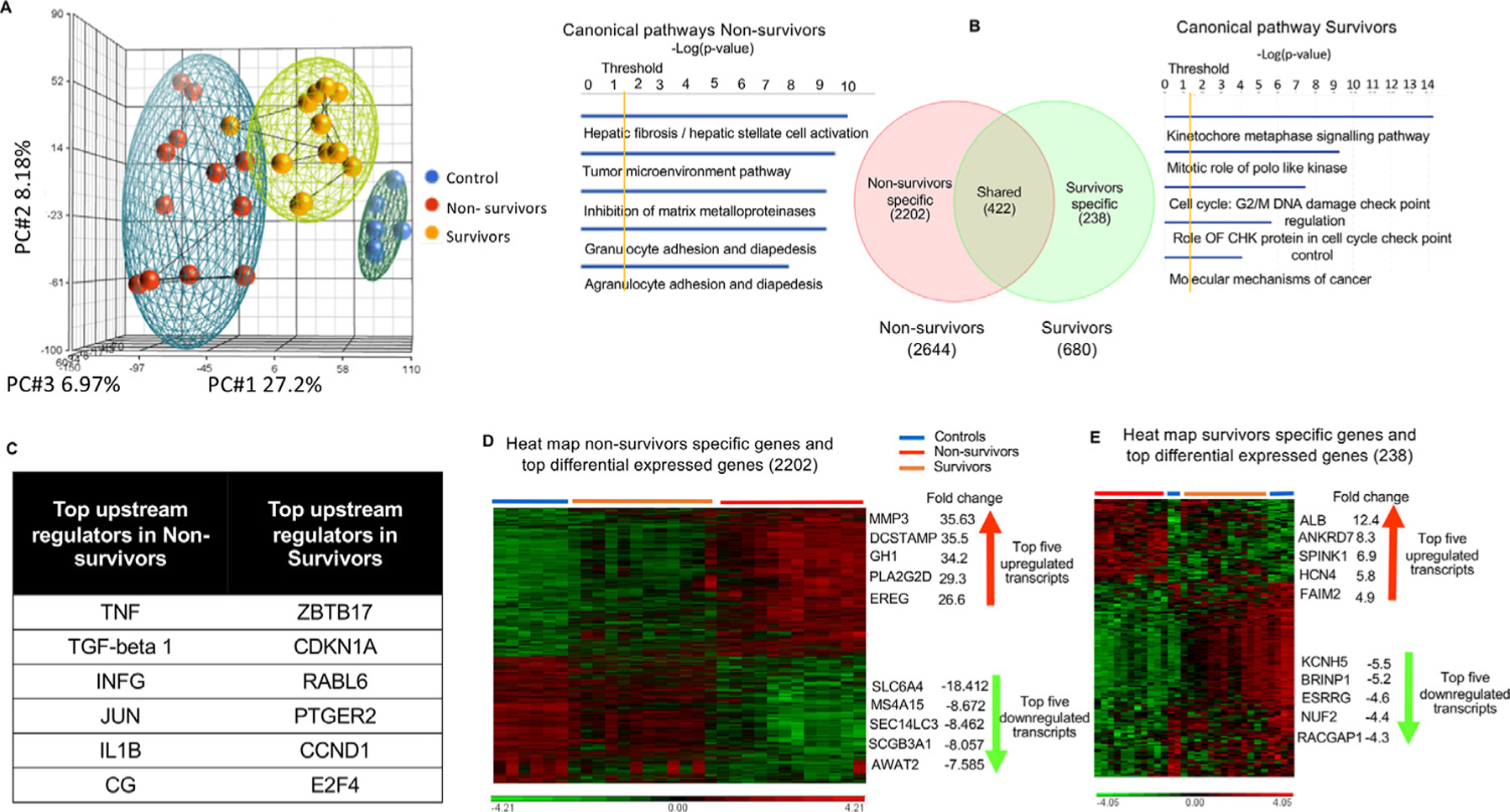
Specific lung transcriptomic analysis (RNA sequencing) in survivors and nonsurvivors. (A) Unsupervised principal component analysis of primate lung RNA sequencing data shows clear clustering into controls, nonsurvivors, and survivors (blue = controls, red = nonsurvivors, and yellow = survivors). (B) Venn diagram of differentially expressed transcripts by group comparisons. Values indicate the number of transcripts with significant differential expression (P < .05) across that contrast: nonsurvivors versus controls (2644); survivors versus controls (680); nonsurvivor-specific (2202), survivor-specific (238), and shared transcripts (422); along with canonical pathways elicited by nonsurvivor- and survivor-specific differentially expressed genes (DEGs) using ingenuity pathway analysis. (C) Upstream regulators (using Ingenuity Pathway Analysis) of nonsurvivor- and survivor-specific DEGs are illustrated by the Venn diagram. (D, E) Hierarchical clustering combined with a heat map of nonsurvivor- and survivor-specific DEGs shows clear differentiation between groups, listed with their top 5 upregulated and downregulated transcripts. Significant differentially expressed genes (DEGs) were conservatively defined as log2 fold change ratios ≥±1 and P values <.05 after adjustment for false discovery.
A unique subset of 101 genes out of 422 DEGs shared between survivors and nonsurvivors (Fig. 5A) showed significant differential expression in all 3 groups (control, survivors, and nonsurvivors). These 101 genes differentially affected nonsurvivors, survivors, and controls and were used to identify common target molecules and mechanisms underlying RILI leading to fibrosis. IPA analysis of these 101 DEGs reflected functions associated with cancer, organismal injury, cellular movements, immune cell trafficking, and inflammatory responses (Fig. E6A). The top upregulated genes among the 101 DEGs included SERPINA3, ATP12A, LRRN4, GJB2, and CLDN10, and the most downregulated genes were PNMT, TOX3, FAM107A, RXRG, and LPA (Fig. 5B and 5C). Angiotensinogen, ILA1, IL1B, and TGF-β1 were major upstream regulators (Fig. 5D). Associated target transcripts of TGF-β1 upstream regulators (SERPINA3, GJB2, CCL2, CCL11, and CDKN1A/p21) revealed by IPA are shown in Figure E6B and Table E2.
Fig. 5.
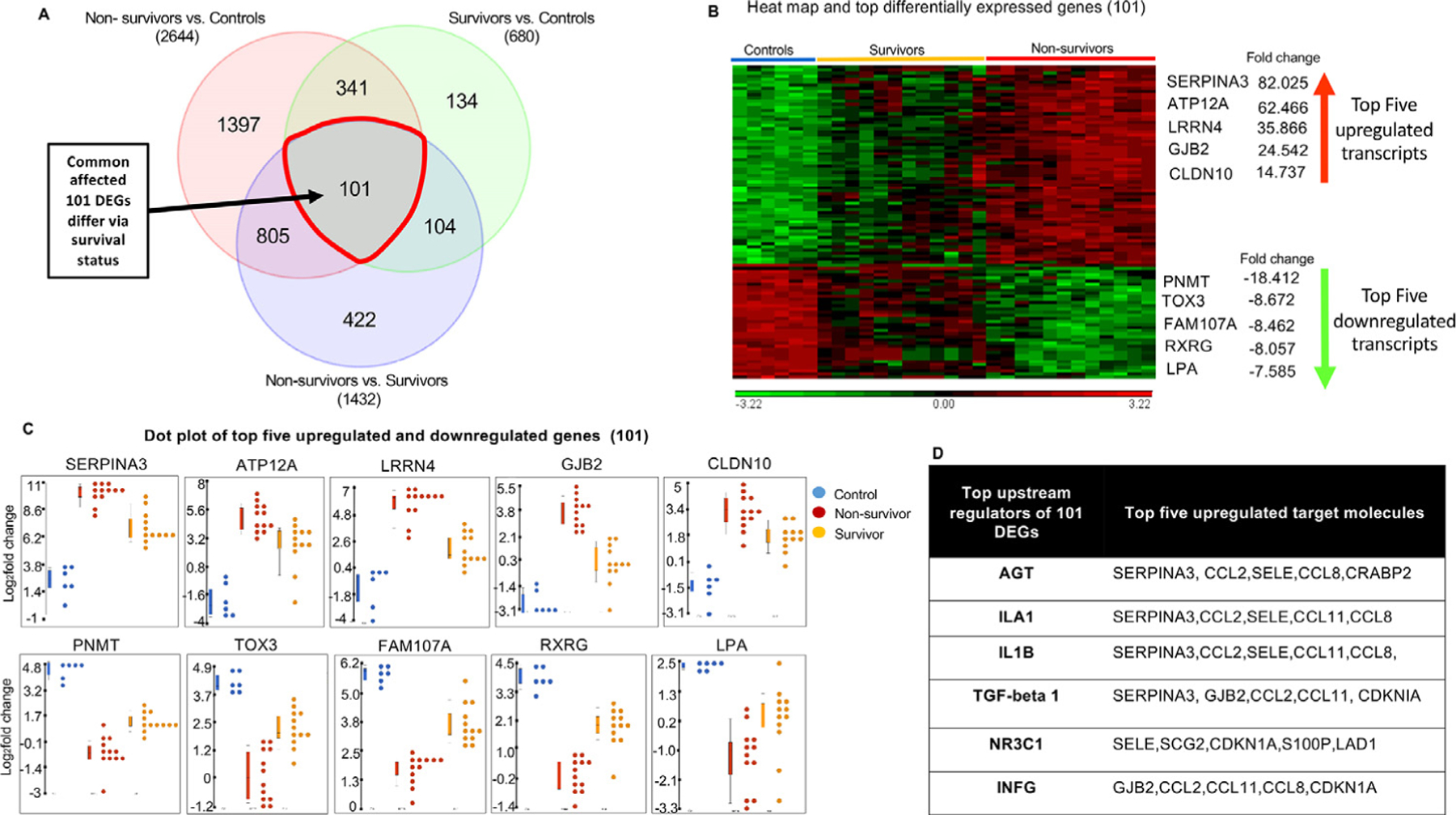
Transcriptomic analysis of 101 differentially expressed genes (DEGs), a subset of shared genes. (A) A Venn diagram of differentially expressed transcripts by group comparisons highlights 101 DEGs, a unique subset of shared genes. (B) A hierarchical clustering heat map shows clear differentiation between controls, nonsurvivors, and survivors, listed with their top 5 upregulated and downregulated transcripts. (C) Dot plots of the top 5 upregulated and downregulated genes, showing differing patterns by outcome. (D) Upstream regulators (using Ingenuity Pathway Analysis) and top target transcripts. Significant DEGs were conservatively defined as log2 fold change ratios ≥±1 and P values <.05 after adjustment for false discovery.
Cytokine analysis revealed significantly increased levels of CCL2 and CCL11 in plasma at 2.3, 3.4, and 4.3 weeks and at necropsy (Fig. 6A and 6B) and a significant increase in TGF-β1 in plasma at necropsy in the irradiated group relative to baseline (Fig. 6C). Cytokines CCL2 and CCL11 were most relevant to transcriptomic analysis and were most significantly elevated among the other 29 measured cytokines (Table E4). We also observed increased levels of SERPINA3 protein in BAL fluid at 9.4 weeks after irradiation (Fig. 6D) and increased TGF-β1 relative to baseline (Fig. 6E). Analysis of BAL cells revealed a significant increase in eosinophil percentage at 9.4 weeks after irradiation relative to baseline, irrespective of survival status (Fig. E7B).
Fig. 6.
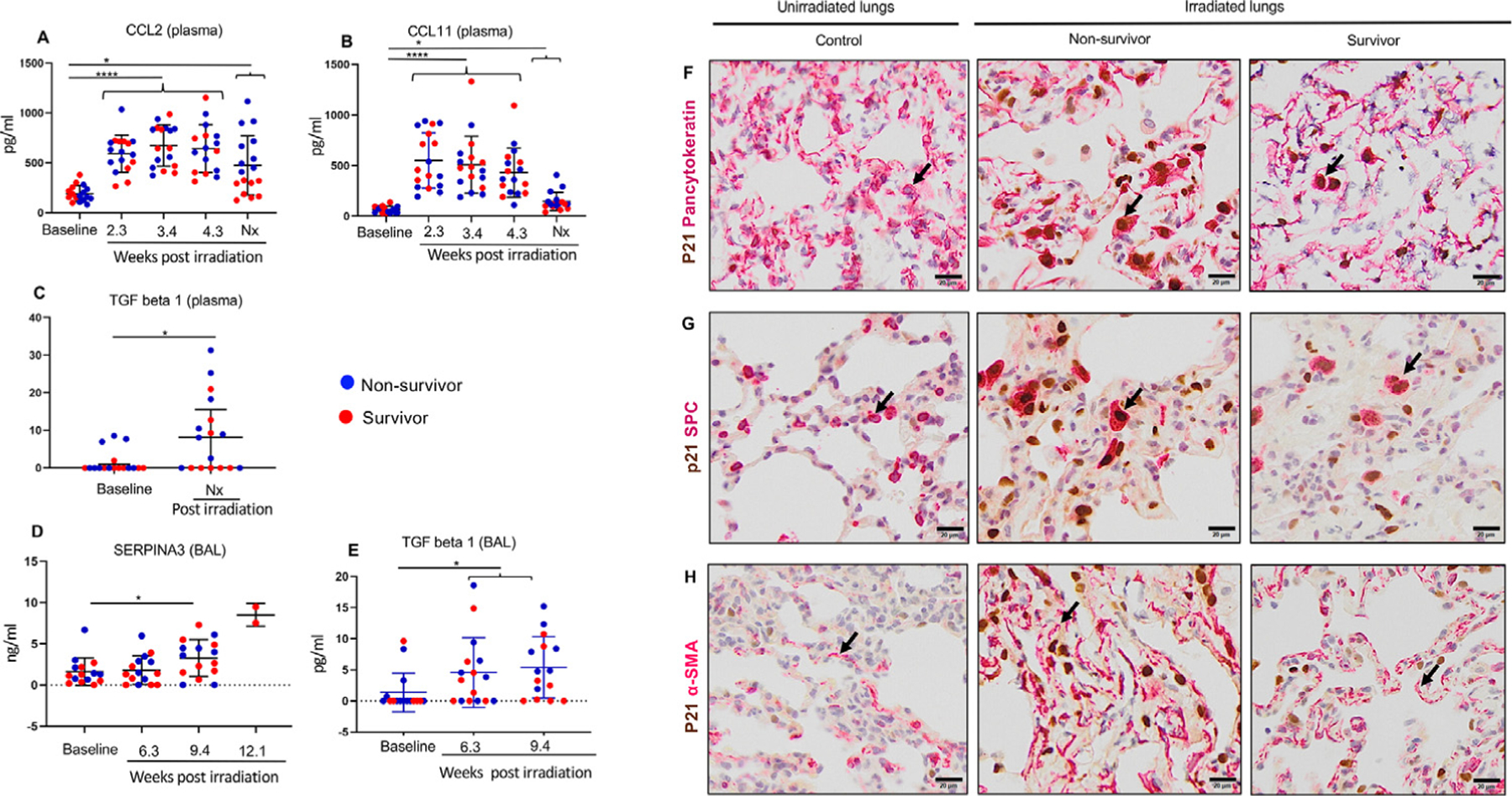
Protein expression of TGF-β1 and its associated transcripts. (A, B) Mean plasma values of CCL2 and CCL11 measured at 2.3 weeks (n = 17), 3.4 weeks (n = 17), 4.3 weeks (n = 16, n=17), and at necropsy (n = 17) after irradiation relative to baseline (n = 17). Results are expressed as means ± SDs, and statistical significance was determined by mixed-effects analysis, RM (repeated measures) 1-way analysis of variance, and Tukey multiple-comparison posttests. (C) TGF-β1 levels measured in plasma at necropsy (n = 17) relative to baseline (n = 17). Results are expressed as medians ± interquartile ranges (IQRs), and the Wilcoxon matched-pairs signed rank test was used for statistical significance. (D) Mean SERPINA3 levels detected via enzyme-linked immunosorbent assay in bronchoalveolar lavage (BAL) fluid (n = 15) collected at 3.6 weeks, 9.4 weeks, and 12.1 weeks (n = 2) after irradiation, relative to baseline. Results are expressed as means ± SDs, and statistical significance was determined by t tests. (E) TGF-β1 levels assessed on BAL fluid collected at 3.6 weeks (n = 16) and 9.4 weeks (n = 15) after irradiation, relative to baseline (n = 16). Results are expressed as medians ± IQRs, and the Wilcoxon matched-pairs signed rank test was used for statistical significance. (F) Upper panel, double staining (arrow) for epithelial marker, cyto-keratin (cytoplasmic), and cell cycle arrest marker p21 (nuclear). (G) Middle panel, double staining (arrow) for alveolar type II pneumocyte marker SPC (cytoplasmic) and p21 (nuclear). (H) Lower panel, smooth-muscle actin α-SMA (cytoplasmic, arrow) and p21 (nuclear) using representative lung sections of a control, nonsurvivor, and survivor. Scale bars are 50 µm.
Cell cycle arrest in a variety of lung cells, including alveolar type II pneumocytes
In this study, irradiation induced significant fibrosis and TGF-β expression. Ingenuity pathway analysis of the 101 DEGs also revealed CDKN1A/p21 among the top upregulated target molecules of TGF-β (Fig. 5D and Fig. E6B). We observed abundant CDKN1A/p21 staining in epithelial and alveolar type 2 (SPC) cells in nonsurvivors and survivors in comparison with nonirradiated controls (Fig. 6F and 6G), indicating cell cycle inhibition. Alpha smooth muscle actin (α-SMA) staining was also identified in irradiated animals in contrast to controls, indicating a profibrotic myofibroblast phenotype (Fig. 6H).
Discussion
In this study, we provide, to our knowledge, the first long-term in vivo characterization of RILI in the NHP model in conjunction with quantitative CT analysis. Our findings are consistent with prior work.2 In addition, we assessed gene-expression patterns in lung tissue at the RNA level and confirmed the protein expression of target molecules in bronchoalveolar lavage and plasma samples to expand our understanding of the time course of disease in RILI. We observed a 52.9% cumulative mortality rate at the experimental endpoint, which is similar to a previous report of 62.5% in an established whole-thorax lung irradiation NHP model.2 Nonsurvivors developed more severe pneumonitis, as evidenced by increased CT opacities and higher respiratory rates, and the degree of injury revealed by CT imaging corresponded to histopathologic pneumonitis scoring. Previous studies in humans have suggested that after radiation therapy, severe RP is symptomatic and often includes dyspnea and coughing with or without mild fever and chest discomfort.14 Our data show that even survivors with no elevated respiratory rates (Fig. E1C and E1D) have CT evidence of lung injury (Fig. 1F).
Histologic data showed severe pneumonitis predominated by macrophage infiltration in the animals euthanized at their clinical endpoints (nonsurvivors, 7–24 weeks) relative to animals euthanized at a later experimental endpoint (survivors, 35 weeks). Both survivors and nonsurvivors, however, developed significant fibrosis relative to controls (Fig. 3B). Similar histologic evidence of increased fibrosis has been reported in other NHP model studies.2 The mild pneumonitis accompanied by significant fibrosis in survivors at their experimental endpoints was likely due to persistent injury,15 and their CT scans showed evidence of pneumonitis in the same time frame as nonsurvivors. Similarly, in human patients, the establishment of a fibrotic phase from pneumonitis using CT scans has been reported between 9 months and 2 years after the completion of radiation therapy.14 In our study, the predominance of M2-like macrophages in both survivors and nonsurvivors could be an important contributor to activation of TGF-β pathways, along with other stressed cell types expressing p21.16
Our RNA sequencing data highlight non−survival-specific, survival-specific, and common gene signatures that differed via survival status. Observed upregulation of MMP3 in 2202 nonsurvivor-specific genes has also been reported in idiopathic pulmonary fibrosis (IPF) and in bleomycin-induced lung fibrosis, suggesting a role in extracellular matrix remodeling and EMT in RILI. Increased serum levels of MMP3 are associated with shortened survival time in patients with IPF.17 Another upregulated gene, DCSTAMP (dendritic cells-specific transmembrane protein), has been previously shown to promote macrophage infiltration and fusion through the CCL2 chemotaxis.18 We also observed increased expression of epiregulin in nonsurvivors. Epiregulin belongs to the epidermal growth factor family and has an important role in inflammation, wound healing, tissue repair, and cell-growth regulation in acute lung injury.19,20 Our results showed downregulation of solute carrier family 6 member 4 (SLC6A4) in nonsurvivors’ lungs. This gene encodes for serotonin neurotransmitter transporter, also known as 5HTT/SERT, which has an important role in the uptake of extracellular 5-HT. Changes in serotonin levels have been observed in various pulmonary disorders, including pulmonary edema and fibrosis.21,22 These gene-expression patterns in nonsurvivors indicate striking ongoing inflammatory and profibrotic processes leading to increased mortality. Several DEGs specific to survivors (SPINK1, FAIM2, KCNH5, NUF2, and RACGAP1) highlighted processes related to cell-cycle regulation, growth, proliferation, invasion, and migration, indicating a more repair-oriented response to the same radiation insult.
Among the 101 common DEGs, SERPINA3 was the top upregulated transcript both in nonsurvivors and survivors (Fig. 5B and 5C), and we observed elevated levels of SERPINA3 in the BAL fluid of the irradiated group at 9.4 weeks after irradiation, relative to baseline (Fig. 6D), which could be an early indicator for extracellular matrix remodeling. Hence, SERPINA3 is a potential biomarker of extracellular matrix remodeling locally (BAL) in irradiated lungs. Our findings are in line with previous studies showing that SERPINA3 is a biomarker of extracellular matrix remodeling in various diseases,23 including idiopathic pulmonary fibrosis.24 SERPINA3 is also a urinary biomarker of acute renal injury in the chronic progression of renal fibrosis.25 The second most upregulated gene was ATP12A (ATPase H+/K+ transporting nongastric alpha 2 subunit), whose upregulation is associated with cystic fibrosis.26 Other upregulated genes included LRRN4 (leucine rich repeat neuronal 4), GJB2/Connexin 26 (gap junction protein beta 2), and CLDN10 (claudin-10); their increased expression has been observed in acute, chronic lung injury and in EMT.27–30 Key genes among downregulated genes were TOX3 and LPA (lipoprotein A). Downregulation of the TOX3 gene is associated with increased EMT.31 Although LPA is an independent risk factor for cardiovascular disease, its plasma levels have been shown to be reduced in patients affected by chronic obstructive pulmonary disease compared with healthy individuals.32
Our IPA analysis of 101 DEGs also indicated angiotensinogen and TGF-β as key upstream regulators. Dysregulation of renin angiotensin system (RAS) in RILI is well known, and a number of investigations have studied various drugs that target RAS.33 Our observation that TGF-β was a key factor in RILI provides further evidence that TGF-β signaling has an important role in the early and late phases of RILI. We validated the corresponding protein expression in BAL fluid, plasma, and tissues collected at necropsy. Most of the aforementioned genes described in nonsurvivors (2202) and survivors (238) and the unique set of 101 common DEGs have been implicated in general lung injury, inflammation, EMT, and fibrotic processes such as IPF, bleomycin-induced fibrosis, and renal fibrosis. Their dysregulation in irradiated lungs indicates their likely role as therapeutic targets and biomarkers of early and late RILI.
Previous studies have reported that TGF-β1 induces the expression of CCL11 (eotaxin1) in human airway fibroblasts,34 and eotaxin 1 promotes the recruitment of eosinophils and subsequent development of fibrosis in bleomycin-induced pulmonary injury.35 We observed elevated plasma protein levels of CCL11 (Fig. 6B) and an increased percentage of eosinophils in BAL cytospins (Fig. E7B). Similarly, human studies have reported increased eosinophils in blood and BAL fluid after radiation therapy.36,37 Radiation may produce eosinophilic pneumonitis in the early proliferative and fibrogenesis phase of RILI, and cytokines such as TGF-β38 and TNF-α39 are chemotactic for eosinophils.
Stress signals induced by TGF-β can cause cell-cycle arrest by stabilizing p21 in a Smad-dependent40,41 or Smadindependent42,43 manner. Our observation of positive immunostaining for CDKN1A/p21 in multiple cell types, including epithelial cells (Fig. 6F-6H), is consistent with the finding of a heterogeneous cell population of senescent cells in radiation-injured lungs in mice.44 Cell-cycle arrest due to sustained injury may contribute to the progression of fibrosis via releasing profibrotic cytokines from senescent cells, which is a feature of the cellular senescence in RILI.16,45,46 TGF-β has an important role in the transformation of various cell types (resident fibroblasts, circulating fibrocytes, EMT, and endothelial EMT) to myofibroblasts, which are associated with excessive extracellular matrix deposition and fibrosis. We believe this was the case in our study, as supported by positive α-SMA staining and fibrosis data.
Significant differences exist between this experimental model of RILI in rhesus monkeys and RILI in humans. In particular, we used a single dose (10 Gy) of irradiation to the whole thorax, whereas in humans, only a targeted volume of lung is exposed and doses are usually fractionated. However, the 10-Gy dose used in this study was higher than the single large estimated dose of 9.3 Gy in a human retrospective study that caused RP in approximately 50% of patients.1,47 The overall objective of the current study was to understand individual differences in the course of RILI and to find new molecular targets in animals exposed to a single high thoracic dose of ionization radiation, which mimics the scenario of accidental nonuniform or partial-body exposure to irradiation.7 However, this information can be applied to a clinical situation such as single-fraction stereotactic body radiation therapy (SF-SBRT), in which a single high dose is prescribed to target a tumor. Although SF-SBRT is highly focused, there is some likelihood of developing lung toxicity in the surrounding tissue. However, because this is a relatively new field, literature regarding SF-SBRT safety and efficacy is rare. In a clinical trial, the incidence of RP after a single dose of 10 Gy and a fractionated total dose of 12 Gy (total-body irradiation) was comparable.48,49 Clinical studies of RILI are hampered by comorbidities such as COVID-19 and concurrent chemotherapy. Studies in NHP whole-thorax lung irradiation models offer an opportunity to explore possible translatable biomarkers and molecular targets in the absence of such confounders.
This study did not address the effects of sex or age on RILI. Female C57BL/6 mice are marginally more sensitive to RILI than males.50 Human and NHP studies have not shown an influence of sex in radioresponsiveness, but data are limited.51,52 Human studies show that older patients are at greater risk of developing RILI after irradiation.51 In this study, the median age of NHP nonsurvivors was 3.76 years at irradiation, whereas the median age of survivors was 5.7 years at irradiation; both of these are “adolescent” ages in rhesus monkeys, which have a typical lifespan of 30 to 35 years. The median body weight of nonsurvivors (5.95 kg) and survivors (7.17 kg) at irradiation was not significantly different. The age and body-weight distribution of studied animals is shown in Table E3. Future studies with more advanced-aged animals will likely show an effect of age.
In addition to the lung, the radiation field in this study included the distal trachea, esophagus, thymus, heart, and other adjacent tissues, including the superior portion of the liver and stomach. However, histologic examination of the trachea, esophagus, thymus, and liver in field and out of field did not show any evidence of cellular injury, inflammation, or fibrosis aside from thymic degeneration in animals dying early (data not shown). The co-irradiation of lung and heart can cause cardiac and pulmonary injury via different mechanisms. In general, the heart is less sensitive to doses <15 Gy as compared to the lungs soon after irradiation.53 Hence, the 10-Gy dose likely did not cause major heart toxicity in short-term experimentation. However, short- and long-term effects of radiation on the heart cannot be disregarded and require further investigation. Postirradiation changes in heart pathology and heart function are part of an ongoing separate study.
A possible confounder in this study was the inclusion of animals treated with superoxide dismutase (SOD) mimetic drugs. We did not see any effect of SOD mimetic treatment on study outcomes; however, the possibility of an effect at the molecular level cannot be ruled out. Therefore, for RNA expression studies, we used only irradiated and unirradiated animals that were not treated with drugs. We believe these data provide useful and new insights to the understanding of RILI.
Conclusion
Our data enhance understanding of RILI in nonsurvivors and survivors in an NHP model and support the role of TGF-β activation and macrophage polarization in RILI. RNA sequencing highlights the role of SERPINA3, ATP12A, GJB2, CLDN10, TOX3, and LPA as possible biomarkers and potential therapeutic targets of RILI. Their differential expression in nonsurvivors and survivors correlated with the severity of the disease. The observed upregulation of TGF-β1 is consistent with prior reports in showing the increased expression of TGF-β1’s associated target transcripts (SERPINA3, CCL11, CCL2, p21) at the mRNA and protein level, indicating not only the role of active TGF-β signaling but also the role of these molecules in the early proliferative phase of RP (nonsurvivors) and the late phase of fibrogenesis and remodeling (survivors).
Supplementary Material
Acknowledgments—
We are grateful to the members of Cline Laboratory for their valuable suggestions and technical expertise; to Ms Jean Gardin, Ms Stacey Combs, Ms Chrystal Bragg, Mr Matt Dwyer, and Mr Michael Adam Benedict; to Russell O’Donnell for technical support; and to the Register Laboratory (Mr J.D. Bottoms, Mr Vincent Yao, and Ms Maryanne Post) for biomarker support. We thank Dr Ashley A. Weaver for the valuable advice about using Mimics software. We are grateful for the advice of Drs Cristina Furdui, Doris M. Brown, Timothy Howard, and Isabel L. Jackson. We acknowledge use of the Comparative Pathology Core Laboratory and Bioinformatics Core of the Molecular Genetics and Genomics Program, supported by a Cancer Center Support Grant (P30CA012197) from the National Cancer Institute and a North Carolina Biotechnology Center Grant (2015-IDG-1006).
This research has been supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, through the Radiation Countermeasures Center of Research Excellence (U19 AI67798/Core D), Department of Defense grants DOD W81XWH-15–1-0574, NIH/ORIP T32 OD010957, and NIH/NCI P30CA012197, North Carolina Biotechnology Center Grant 2015-IDG-1006, and the Wake Forest Department of Pathology.
Footnotes
Disclosures: none.
Research data are stored in an institutional repository and will be shared upon request to the corresponding author.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijrobp.2021.03.058.
References
- 1.Van Dyk J, Keane TJ, Kan S, Rider WD, Fryer CJ. Radiation pneumonitis following large single dose irradiation: A re-evaluation based on absolute dose to lung. Int J Radiat Oncol Biol Phys 1981;7:461–467. [DOI] [PubMed] [Google Scholar]
- 2.Garofalo M, Bennett A, Farese AM, et al. The delayed pulmonary syndrome following acute high-dose irradiation: a rhesus macaque model. Health Phys 2014;106:56–72. [DOI] [PubMed] [Google Scholar]
- 3.Angel E, Yaghmai N, Jude CM, et al. Dose to radiosensitive organs during routine chest CT: Effects of tube current modulation. AJR Am J Roentgenol 2009;193:1340–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh VK, Newman VL, Berg AN, MacVittie TJ. Animal models for acute radiation syndrome drug discovery. Expert Opin Drug Discov 2015;10:497–517. [DOI] [PubMed] [Google Scholar]
- 5.Kwa SL, Lebesque JV, Theuws JC, et al. Radiation pneumonitis as a function of mean lung dose: An analysis of pooled data of 540 patients. Int J Radiat Oncol Biol Phys 1998;42:1–9. [DOI] [PubMed] [Google Scholar]
- 6.Mehta V Radiation pneumonitis and pulmonary fibrosis in non-small-cell lung cancer: Pulmonary function, prediction, and prevention. Int J Radiat Oncol Biol Phys 2005;63:5–24. [DOI] [PubMed] [Google Scholar]
- 7.Mahmood J, Jelveh S, Calveley V, Zaidi A, Doctrow SR, Hill RP. Mitigation of lung injury after accidental exposure to radiation. Radiat Res 2011;176:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanania AN, Mainwaring W, Ghebre YT, Hanania NA, Ludwig M. Radiation-induced lung injury: Assessment and management. Chest 2019;156:150–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duru N, Wolfson B, Zhou Q. Mechanisms of the alternative activation of macrophages and non-coding RNAs in the development of radiation-induced lung fibrosis. World J Biol Chem 2016;7:231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meziani L, Deutsch E, Mondini M. Macrophages in radiation injury: A new therapeutic target. Oncoimmunology 2018;7 e1494488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caja L, Dituri F, Mancarella S, et al. TGF-β and the tissue microenvironment: Relevance in fibrosis and cancer. Int J Mol Sci 2018;19:1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park HR, Jo SK, Jung U. Ionizing radiation promotes epithelial-to-mesenchymal transition in lung epithelial cells by TGF-β-producing M2 macrophages. In Vivo 2019;33:1773–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cline JM, Dugan G, Bourland JD, et al. Post-irradiation treatment with a superoxide dismutase mimic, MnTnHex2-PyP(5+), mitigates radiation injury in the lungs of non-human primates after whole-thorax exposure to ionizing radiation. Antioxidants (Basel) 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benveniste MF, Gomez D, Carter BW, et al. Recognizing radiation therapy-related complications in the chest. Radiographics 2019;39:344–366. [DOI] [PubMed] [Google Scholar]
- 15.Fumagalli M, Rossiello F, Clerici M, et al. Telomeric DNA damage is irreparable and causes persistent DNA-damage-response activation. Nat Cell Biol 2012;14:355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tominaga K, Suzuki HI. TGF-β signaling in cellular senescence and aging-related pathology. Int J Mol Sci 2019;20:5002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pardo A, Cabrera S, Maldonado M, Selman M. Role of matrix metalloproteinases in the pathogenesis of idiopathic pulmonary fibrosis. Respir Res 2016;17:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helming L, Gordon S. Molecular mediators of macrophage fusion. Trends Cell Biol 2009;19:514–522. [DOI] [PubMed] [Google Scholar]
- 19.Riese DJ 2nd, Cullum RL. Epiregulin: Roles in normal physiology and cancer. Semin Cell Dev Biol 2014;28:49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finigan JH, Downey GP, Kern JA. Human epidermal growth factor receptor signaling in acute lung injury. Am J Respir Cell Mol Biol 2012;47:395–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tamarapu Parthasarathy P, Galam L, Huynh B, et al. MicroRNA 16 modulates epithelial sodium channel in human alveolar epithelial cells. Biochem Biophys Res Commun 2012;426:203–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goolaerts A, Roux J, Ganter MT, et al. Serotonin decreases alveolar epithelial fluid transport via a direct inhibition of the epithelial sodium channel. Am J Respir Cell Mol Biol 2010;43:99–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aslam MS, Yuan L. Serpina3n: Potential drug and challenges, mini review. J Drug Target 2020;28:368–378. [DOI] [PubMed] [Google Scholar]
- 24.Chen H, Fang X, Zhu H, et al. Gene expression profile analysis for different idiopathic interstitial pneumonias subtypes. Exp Lung Res 2014;40:367–379. [DOI] [PubMed] [Google Scholar]
- 25.Sanchez-Navarro A, Mejia-Vilet JM, Perez-Villalva R, et al. SerpinA3 in the early recognition of acute kidney injury to chronic kidney disease (CKD) transition in the rat and its potentiality in the recognition of patients with CKD. Sci Rep 2019;9:10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scudieri P, Musante I, Caci E, et al. Increased expression of ATP12A proton pump in cystic fibrosis airways. JCI Insight 2018;3 e123616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simon BA, Easley RB, Grigoryev DN, et al. Microarray analysis of regional cellular responses to local mechanical stress in acute lung injury. Am J Physiol Lung Cell Mol Physiol 2006;291:L851–L861. [DOI] [PubMed] [Google Scholar]
- 28.Johnson LN, Koval M. Cross-talk between pulmonary injury, oxidant stress, and gap junctional communication. Antioxid Redox Signal 2009;11:355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, Qin G, Luo M, et al. Reciprocal positive regulation between Cx26 and PI3K/Akt pathway confers acquired gefitinib resistance in NSCLC cells via GJIC-independent induction of EMT. Cell Death Dis 2015;6:e1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ip YC, Cheung ST, Lee YT, Ho JC, Fan ST. Inhibition of hepatocellular carcinoma invasion by suppression of claudin-10 in HLE cells. Mol Cancer Ther 2007;6:2858–2867. [DOI] [PubMed] [Google Scholar]
- 31.Jiang B, Chen W, Qin H, et al. TOX3 inhibits cancer cell migration and invasion via transcriptional regulation of SNAI1 and SNAI2 in clear cell renal cell carcinoma. Cancer Lett 2019;449:76–86. [DOI] [PubMed] [Google Scholar]
- 32.Basili S, Ferroni P, Vieri M, et al. Lipoprotein(a) serum levels in patients affected by chronic obstructive pulmonary disease. Atherosclerosis 1999;147:249–252. [DOI] [PubMed] [Google Scholar]
- 33.Medhora M, Gao F, Jacobs ER, Moulder JE. Radiation damage to the lung: Mitigation by angiotensin-converting enzyme (ACE) inhibitors. Respirology 2012;17:66–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wenzel SE, Trudeau JB, Barnes S, et al. TGF-beta and IL-13 synergistically increase eotaxin-1 production in human airway fibroblasts. J Immunol 2002;169:4613–4619. [DOI] [PubMed] [Google Scholar]
- 35.Huaux F, Gharaee-Kermani M, Liu T, et al. Role of Eotaxin-1 (CCL11) and CC chemokine receptor 3 (CCR3) in bleomycin-induced lung injury and fibrosis. Am J Pathol 2005;167:1485–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakayasu H, Shirai T, Tanaka Y, Saigusa M. Chronic eosinophilic pneumonia after radiation therapy for squamous cell lung cancer. Respir Med Case Rep 2017;22:147–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim JH, Nam HS, Kim HJ, et al. Migratory eosinophilic alveolitis caused by radiation therapy. J Thorac Dis 2015;7:E117–E121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luttmann W, Franz P, Matthys H, Virchow JC Jr. Effects of TGF-beta on eosinophil chemotaxis. Scand J Immunol 1998;47:127–130. [DOI] [PubMed] [Google Scholar]
- 39.Nam HS, Lee SY, Kim SJ, et al. The soluble tumor necrosis factor-alpha receptor suppresses airway inflammation in a murine model of acute asthma. Yonsei Med J 2009;50:569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramachandra M, Atencio I, Rahman A, et al. Restoration of transforming growth factor Beta signaling by functional expression of smad4 induces anoikis. Cancer Res 2002;62:6045–6051. [PubMed] [Google Scholar]
- 41.Moustakas A, Kardassis D. Regulation of the human p21/WAF1/Cip1 promoter in hepatic cells by functional interactions between Sp1 and Smad family members. Proc Natl Acad Sci U S A 1998;95:6733–6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim GY, Mercer SE, Ewton DZ, Yan Z, Jin K, Friedman E. The stress-activated protein kinases p38 alpha and JNK1 stabilize p21 (Cip1) by phosphorylation. J Biol Chem 2002;277:29792–29802. [DOI] [PubMed] [Google Scholar]
- 43.Han SU, Kim HT, Seong DH, et al. Loss of the Smad3 expression increases susceptibility to tumorigenicity in human gastric cancer. Oncogene 2004;23:1333–1341. [DOI] [PubMed] [Google Scholar]
- 44.Soysouvanh F, Benadjaoud MA, Dos Santos M, et al. Stereotactic lung irradiation in mice promotes long-term senescence and lung injury. Int J Radiat Oncol Biol Phys 2020;106:1017–1027. [DOI] [PubMed] [Google Scholar]
- 45.Freund A, Orjalo AV, Desprez PY, Campisi J. Inflammatory networks during cellular senescence: Causes and consequences. Trends Mol Med 2010;16:238–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beach TA, Groves AM, Johnston CJ, Williams JP, Finkelstein JN. Recurrent DNA damage is associated with persistent injury in progressive radiation-induced pulmonary fibrosis. Int J Radiat Biol 2018;94:1104–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Abratt RP, Morgan GW, Silvestri G, Willcox P. Pulmonary complications of radiation therapy. Clin Chest Med 2004;25:167–177. [DOI] [PubMed] [Google Scholar]
- 48.Thomas ED, Clift RA, Hersman J, et al. Marrow transplantation for acute nonlymphoblastic leukemic in first remission using fractionated or single-dose irradiation. Int J Radiat Oncol Biol Phys 1982;8:817–821. [DOI] [PubMed] [Google Scholar]
- 49.Carruthers SA, Wallington MM. Total body irradiation and pneumonitis risk: A review of outcomes. Br J Cancer 2004;90:2080–2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson IL, Baye F, Goswami CP, et al. Gene expression profiles among murine strains segregate with distinct differences in the progression of radiation-induced lung disease. Dis Model Mech 2017;10:425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kong FM, Wang S. Nondosimetric risk factors for radiation-induced lung toxicity. Semin Radiat Oncol 2015;25:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thrall KD, Mahendra S, Jackson MK, Jackson W 3rd, Farese AM, MacVittie TJ. A comparative dose-response relationship between sexes for mortality and morbidity of radiation-induced lung injury in the rhesus macaque. Health Phys 2019;116:354–365. [DOI] [PubMed] [Google Scholar]
- 53.Ghobadi G, Hogeweg LE, Faber H, et al. Quantifying local radiation-induced lung damage from computed tomography. Int J Radiat Oncol Biol Phys 2010;76:548–556. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


