Abstract
Pancreatic ductal adenocarcinoma (PDAC) is almost uniformly fatal and characterized by early metastasis. Oncogenic KRAS mutations prevail in 95% of PDAC tumors and co-occur with genetic alterations in the TP53 tumor suppressor in nearly 70% of patients. Most TP53 alterations are missense mutations that exhibit gain-of-function phenotypes that include increased invasiveness and metastasis yet the extent of direct cooperation between KRAS effectors and mutant p53 remains largely undefined. We show that oncogenic KRAS effectors activate cyclic AMP responsive element binding protein 1 (CREB1) to allow physical interactions with mutant p53 that hyperactivate multiple pro-metastatic transcriptional networks. Specifically, mutant p53 and CREB1 upregulate the pro-metastatic, pioneer transcription factor, FOXA1, activating its transcriptional network while promoting WNT/β-catenin signaling, together driving PDAC metastasis. Pharmacologic CREB1 inhibition dramatically reduced FOXA1 and β-catenin expression and dampened PDAC metastasis, identifying a new therapeutic strategy to disrupt cooperation between oncogenic KRAS and mutant p53 to mitigate metastasis.
Keywords: mutant p53, oncogenic KRAS, CREB1, metastasis, PDAC, FOXA1, pancreatic cancer, β-catenin, mouse model, Wnt/β-catenin
Introduction
The incidence of pancreatic ductal adenocarcinoma (PDAC) continues to rise with approximately 50% of patients presenting with distant metastatic disease. PDAC metastasis occurs early after tumor initiation and is a result of complex interplay between tumor cell-autonomous processes and cellular elements within the tumor microenvironment(1–3). Oncogenic KRAS promotes tumor initiation while the associated loss of key tumor suppressor genes accelerates the malignant progression of precursor pancreatic intraepithelial neoplasia (PanIN) lesions and promotes metastasis(4–7). Among affected tumor suppressors, TP53 is altered in 70% of PDAC patients and the majority of alterations are missense mutations that co-occur with KRAS mutations(7,8). The prevalence of co-occurring KRAS and TP53 mutations in PDAC suggests underlying mechanisms of cooperativity that drive tumor development and metastasis yet remain poorly defined. A deeper understanding of the complex interplay between oncogenic KRAS and mutant p53, the most commonly mutated human oncogene and tumor suppressor gene, respectively, may lead to therapies that reverse cooperative mechanisms and expose new therapeutic vulnerabilities.
Results
Mutant p53R172H promotes PDAC metastasis relative to p53 loss
The impact of mutant p53 on tumor initiation and metastasis has been largely studied through comparison of KPC (K, Kras; P, p53; C, Cre-recombinase) genetically engineered mouse models (GEMMs) of pancreatic cancer that incorporate conditional, oncogenic Kras (LOX-STOP-LOX(LSL)-KrasG12D) and mutant p53 alleles(9–11). Notably, these mutant p53 alleles are maintained as p53-null alleles due to a LSL-cassette which, when removed, permits expression of mutant p53 in a temporal and spatial manner(12). As such, cellular constituents of the tumor microenvironment (TME) remain heterozygous for wild-type (WT) p53. p53 heterozygosity in stromal cells affects tumor development in some cancers and may obfuscate the underlying roles of mutant p53 in PDAC due to contributions of stromal cells on PDAC biology(1,13–15). To express mutant p53R172H (corresponding to the human p53R175H hot spot mutation) only in tumor cells while preserving WT p53 functions in all stromal cells, we used a conditional knock-in mutant p53 allele that constitutively expresses WT p53 and recombines to mutant p53R172H (wild-type to mutant, wm), named p53wmR172H (Fig. 1A)(16). Cre-mediated recombination excises the WT p53 cDNA encoding exons 5–11 from the endogenous locus resulting in somatic mutant p53R172H expression. Extensive characterization of p53wmR172H/+ mice indicated a p53-dependent DNA damage response with activation of p53 transcriptional target genes and lack of a tumor phenotype (16). Thus, tissues without cre-expression, including the TME and immune system, remain WT for p53 without potential attendant effects on tumor development and progression.
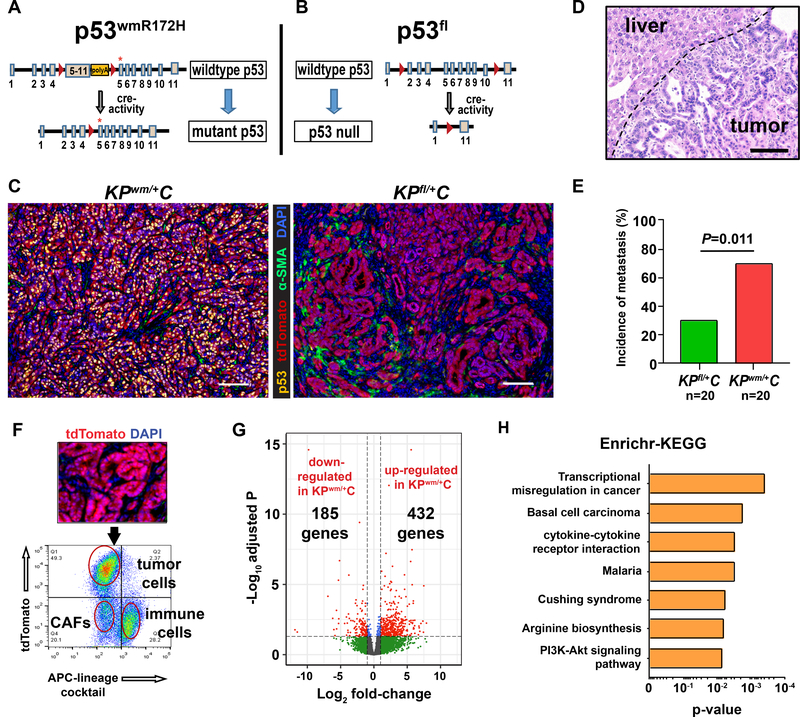
Figure 1. Mutant p53 augments PDAC metastasis.
A, The KPwm/+C mouse model incorporates a mutant p53 allele that converts from constitutive wildtype p53 to mutant p53R172H expression after cre-mediated recombination. Mutant p53R172H is somatically expressed in pancreatic epithelial cells and wildtype p53 is expressed in stromal cells present in the TME. B, The KPfl/+C mouse model incorporates a loss-of-function p53 allele with loxP sites flanking exons 2–11. Wildtype p53 is somatically deleted in pancreatic epithelial cells following cre-recombination and wildtype p53 is expressed in stromal cells present in the TME. C, Mutant p53 is expressed and stabilized in pancreatic tumor cells in KPwm/+C mice. Wildtype p53 expression is lost in pancreatic tumor cells in KPfl/+C mice. Scale bar, 250μM. D, Representative histology of a liver metastasis in a KPwm/+C mouse. Scale bar, 250μM. E, The incidence of liver or lung metastasis in KPwm/+C and KPfl/+C PDAC models. P value determined by unpaired two-tailed t-test. F, Single tumor cells from KPwm/+C and KPfl/+C mice were positively selected by FACS based on a tdTomato reporter. G, Volcano plot of genes differentially expressed in enriched PDAC cell populations in KPwm/+C (n=4) versus KPfl/+C (n=4) mice. H, Enrichr-KEGG analysis of KPwm/+C tumors demonstrates enriched transcriptional misregulation in cancer.
To dissect the cell autonomous functions of mutant p53 in PDAC biology, we generated LSL-KrasG12D; p53wmR172H/+; Pdx1-Cre (hereafter KPwm/+C) mice and compared them to LSL-KrasG12D; p53fl/+; Pdx1-Cre (KPfl/+C) mice in which WT p53 is somatically deleted (Fig. 1B). Some mice from both genotypes also contained the Rosa26-LSL-tdTomato reporter allele without effects on overall survival. PDAC tumors from KPwm/+C mice expressed and stabilized mutant p53R172H while tumors from KPfl/+C mice failed to express p53 (Fig. 1C). Although overall survival between these two somatic models were similar due to the burden of primary tumor, tumors in KPwm/+C mice resulted in a >2-fold incidence of metastatic lesions relative to KPfl/+C mice, reaffirming an in vivo pro-metastatic gain-of-function (GOF) phenotype of mutant p53R172H in PDAC (Fig. 1D, E; Supplementary Fig. 1A, B)(10,11). Interestingly, comparison of KPwm/+C and traditional KPLSLR172H/+C models failed to demonstrate differences in disease-specific survival despite differences in overall survival that approached significance, likely due to a higher incidence of extra-pancreatic malignancies in KPLSLR172H/+C mice (Supplementary Fig. 1C, D). PanINs and invasive tumoral compartments derived from KPfl/+C and KPwm/+C mice appeared grossly similar based on H&E staining (Supplementary Fig. 1E). Cell lines derived from tumors in KPwm/+C mice (hereafter, KPwmC) and recently-established human pancreatic adenocarcinoma tumor cell lines (hereafter, PATC) harbored p53 mutations and lost the WT p53 allele as confirmed by Sanger sequencing (Supplementary Fig. 1F). All tested PDAC cell lines recapitulated mutant p53-dependent, pro-invasive and pro-migratory phenotypes as measured by transwell assays following siRNA-mediated knockdown of mutant p53 (Supplementary Fig. 1G). Mutant p53R172H overexpression in KPflC cell lines lacking WT p53 alleles also increased PDAC cell migration/invasion by transwell assay measurements (Supplementary Fig. 1H). Collectively, these data indicate that somatic mutant p53R172H significantly increases PDAC metastasis in vivo as compared to somatic p53 deletion in a TME normalized for WT p53 expression.
Mutant p53 tumors are associated with a FOXA1 transcriptional signature
To identify mechanisms of mutant p53-mediated metastasis, advanced PDAC tumor cells from KPwm/+C and KPfl/+C mice expressing the tdTomato reporter were isolated using fluorescence-activated cell sorting (FACS) (Fig. 1F). This approach enriched for tdTomato(+) PDAC cells while excluding tdTomato(–) stromal and immune cells to maximize the signal-to-noise ratio of mutant p53-dependent transcriptional signatures present only in tumor cells. As mutant p53 is known to exert GOF through its effects on resident transcription factor activity, enriched tumor cells sorted from KPwm/+C (n=4) and KPfl/+C (n=4) mice underwent gene expression profiling with RNA-sequencing and pathway analysis to identify transcriptional programs associated with mutant p53R172H. To confirm pathways dysregulated in KPwm/+C tumors as relevant to human PDAC, we performed gene set enrichment analysis (GSEA) of KPwm/+C tumors which demonstrated enrichment in hallmark pathways observed in human PDAC (Supplementary Fig. 1I)(17). Overall, 432 genes were upregulated and 185 downregulated in KPwm/+C tumors (mutant p53 tumors) relative to KPfl/+C tumors (p53-null tumors) (Fig. 1G) (Supplementary data- Mutant p53 associated genes). Analysis using Erichr with embedded KEGG pathway analysis indicated “Transcriptional misregulation in cancer” (p=0.0004) and “Basal cell carcinoma” (p=0.0018) as the top activated pathways(18,19) (Fig. 1H), the latter due to the relative overexpression of various Wnt/β-catenin signaling elements and ligands.
We next sought to identify transcription factors, potentially misregulated through interactions with mutant p53R172H, responsible for the gene signatures identified in tumors from KPwm/+C mice. Using oPOSSUM-3.0, a computational system containing transcription factor binding sites (TFBS) from JASPAR, we examined the promoters of genes upregulated in mutant p53R172H tumors for overrepresented TFBS that would indicate aberrant activities of mutual transcription factors (Fig. 2A). Examination of overrepresented TFBS in the promoters of genes upregulated in mutant p53 PDAC tumors (KPwm/+C) revealed numerous candidate transcription factors whose activity is potentially affected by mutant p53: Sp1, Runx1, Foxa1, Klf4, Gfi (top five) (Fig. 2A).
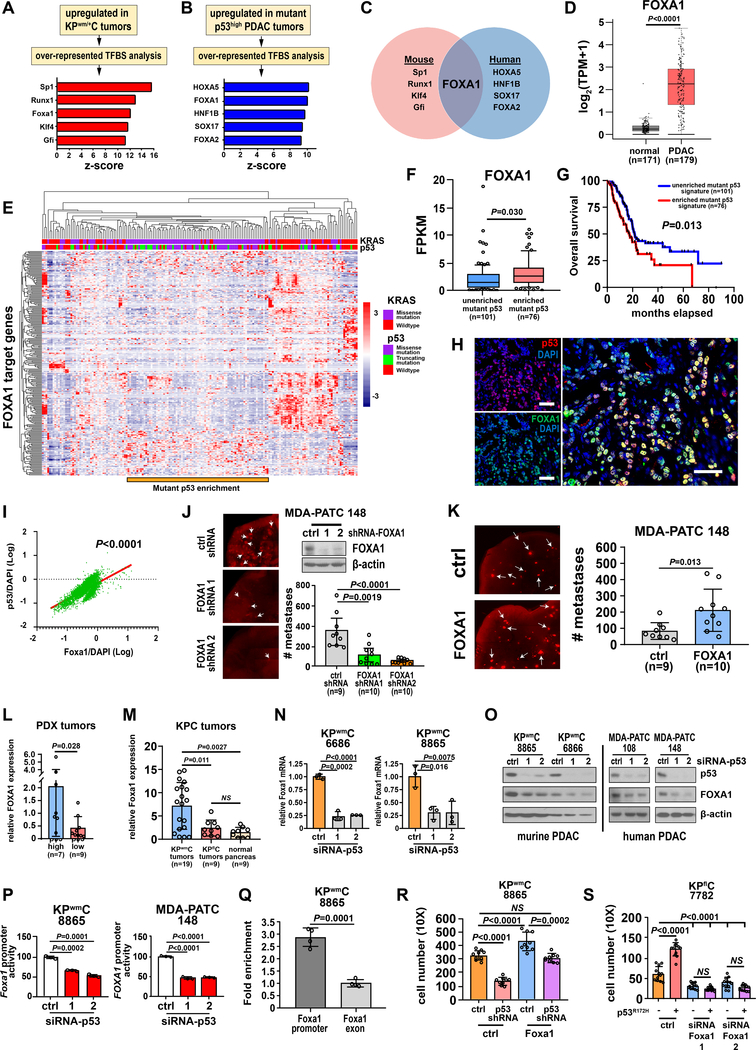
Figure 2. Mutant p53 is associated with a FOXA1 transcriptional signature and regulates FOXA1 to drive PDAC cell metastasis.
A, Overrepresented, conserved transcription factor binding site (TFBS) motifs in genes differentially upregulated in KPwm/+C (n=4) versus KPfl/+C (n=4) mice. B, Overrepresented, conserved TFBS motifs in genes differentially expressed in p53high (n=19) versus p53low (n=11) PDAC PDX tumors. C, Overlap of overrepresented TFBS motifs enriched in mutant p53 tumors in murine and human PDAC. D, Comparison of FOXA1 expression in normal pancreas and PDAC using the TCGA dataset. P value calculated by one-way ANOVA. E, Non-hierarchal clustering of genes upregulated in KPwm/+C tumors with FOXA1 binding motifs within the TCGA PDAC dataset. Relative to patients harboring truncating p53 mutations, patients with p53 missense mutations significantly clustered with oncogenic KRAS (Fisher exact test, p<0.0001). A distinct gene expression signature corresponds to this region as generated through listing FOXA1 target genes. F, Comparison of FOXA1 expression within and external to the region of mutant p53 enrichment. G, Overall survival of patients within and external to the region of mutant p53 enrichment that clusters with oncogenic KRAS. H, Representative spatial co-expression of mutant p53 and Foxa1 in KPwm/+C tumors as measured by confocal multicolor immunofluorescence (n=4 mice, 5 random fields/mouse). Scale bars, 50μM. I, Correlation of mutant p53 and Foxa1 expression as measured by Pearson correlation (p<0.0001). J, FOXA1 was knocked down in PDAC cells and metastasis measured by lung colonization metastasis assays. K, FOXA1 was overexpressed in PDAC cells and metastasis measured by lung colonization metastasis assays. L, Comparison of FOXA1 expression between p53high and p53low PDX models. M, Comparison of Foxa1 expression between tumors derived from KPwm/+C or KPfl/+C mice and normal murine pancreas. N and O, FOXA1 mRNA and protein levels after mutant p53 was silenced in murine and human PDAC. P, FOXA1 promoter reporter activity after mutant p53 knockdown. Q, Enrichment of mutant p53 occupancy at the Foxa1 promoter relative to a downstream exon. Pooled data from four independent experiments. R, Effect of Foxa1 overexpression on PDAC cell migration/invasion with and without mutant p53 knockdown. S, Effect of ectopic mutant p53 overexpression on PDAC cell migration/invasion with and without Foxa1 knockdown. (J, K, L, M, N, P, Q), Data are mean±s.d. and P values determined by unpaired two-tailed t-tests. (N, O, P), Data representative of at least three independent experiments, each in triplicate. (R, S), P value calculated by one-way ANOVA.
To orthogonally validate these murine transcription factors in human PDAC, we performed a similar analysis in a panel of clinically-relevant human PDAC patient-derived xenografts (PDXs). Thirty PDAC PDX tumors with accompanying whole-exome and RNA-sequencing data were identified that either strongly expressed mutant p53 (mutant p53high, n=19) or lacked p53 (p53low, n=11) by immunohistochemical analysis (Supplementary Fig. 2A, B). The presence of p53 missense mutations and the concomitant deletion of WT p53 were confirmed in p53high samples through analysis of variant allele frequencies in whole-exome sequencing data. Overrepresented TFBS associated with mutant p53 in PDAC PDX tumors included: HOXA5, FOXA1, HNF1B, SOX17 and FOXA2 (top five) (Fig. 2B). Comparison of overrepresented TFBS in murine and human PDAC tumors revealed the motif corresponding to FOXA1 as the sole common overrepresented motif in PDAC tumors that express mutant p53 (Fig. 2C).
FOXA1 is a pioneer transcription factor that modulates chromatin structure and transactivates thousands of target genes. Its relevance in breast and prostate cancer metastasis has been reported and FOXA1 has been linked to pancreatic cancer metastasis through enhancer reprogramming, independent of p53(20–22). To confirm the relevance of FOXA1 in human PDAC, we compared the expression of FOXA1 between normal pancreas and PDAC tumors using the human PDAC TCGA dataset. The average expression value of FOXA1 in PDAC patients was identified as increased >7-fold relative to the average expression in normal human pancreas (one-way ANOVA, p<0.0001) (Fig. 2D), indicating its general upregulation in pancreatic cancer. We next assessed the expression of all 258 FOXA1 target genes identified in KPwm/+C tumors within the human PDAC TGCA dataset and found clusters of patients with distinct transcriptional profiles (Fig. 2E). One patient cluster showed significant enrichment in patients with p53 missense mutations (Fisher exact test, p<0.0001) but not in p53 truncating mutations co-occurring with oncogenic KRAS (Fisher exact test, p=0.15), indicating the presence of a distinct FOXA1 transcriptional profile present only within mutant p53 PDAC tumors. Further analysis of FOXA1 expression within this mutant-p53 enriched patient cluster confirmed increased FOXA1 expression relative to other patients (Fig. 2F). Moreover, the survival of patients whose tumors exhibited this distinct FOXA1 transcriptional signature was significantly reduced compared to patients without it, affirming its biologic and prognostic significance (Fig. 2G). Notably, only the presence of p53 missense mutations, but not the specific sites of p53 missense mutations, was associated with this FOXA1 transcriptional signature, suggesting a general, global effect of mutant p53 not shared with truncating p53 mutations.
Mutant p53 transcriptionally regulates FOXA1 expression
Based on the overrepresentation of FOXA1 TFBS in murine PDAC tumors that express mutant p53 and the FOXA1 transcriptional signature identified in human PDAC tumors harboring p53 missense mutations, we hypothesized that mutant p53 alters the activity of FOXA1 to promote metastatic phenotypes. Accordingly, we initially sought to confirm the spatial co-expression of mutant p53 and Foxa1 in autochthonous KPwm/+C tumors. Immunofluorescent analysis of mutant p53 and Foxa1 in KPwm/+C tumors revealed >80% incidence of nuclear co-localization (Pearson correlation, p<0.0001), supporting our findings that Foxa1 levels correlate with the presence of mutant p53 in vivo (Fig. 2H, I). To test the biologic function of FOXA1 as it pertains to the metastatic phenotype identified in KPwm/+C mice, we performed shRNA-mediated FOXA1 knockdowns in a highly metastatic human PDAC cell line with a stably integrated RFP reporter, MDA-PATC 148, and performed lung colonization metastasis assays. Of note, MDA-PATC 148 cells were selected for this and other experiments due to their origin from a human hepatic PDAC metastasis harboring F134L p53 mutations (Supplementary Fig. 1F). This cell line therefore has pre-existing metastatic traits in which to test FOXA1 function while possessing a different and unique p53 missense mutation that would better support a broad effect of mutant p53 in PDAC metastasis, independent of missense site. FOXA1 knockdown resulted in a 67–85% reduction in the number of PDAC lung metastases relative to control and FOXA1 overexpression significantly increased the number lung metastases by >2.5-fold, indicating an important role in PDAC metastasis (Fig. 2J, K). Moreover, FOXA1 knockdown also demonstrated slight reductions in cell proliferation at 72hr. timepoints and a 25–50% reduction in anchorage-dependent clonogenic growth but not anchorage-independent growth, potentially indicating multiple roles in the metastatic cascade (Supplementary Fig. 2C–E).
Confident that mutant p53 and FOXA1 are effectors of PDAC metastasis, we next sought to validate differential FOXA1 expression levels in our model systems. FOXA1 expression was confirmed to be reduced in p53low PDX models and autochthonous tumors derived from KPfl/+C mice relative to p53high PDX models and KPwm/+C tumors, respectively (Fig. 2L, M). Given the strong correlations between mutant p53 and FOXA1 expression, we hypothesized that mutant p53 regulates FOXA1 to generate a distinct transcriptional signature that promotes PDAC metastasis. In addition, since analysis of upregulated genes containing Foxa1 motifs in KPwm/+C tumors relative to KPfl/+C tumors revealed 25 unique transcription factors, Foxa1 could potentially activate additional transcriptional networks that extend beyond its immediate primary network (Supplementary Fig. 2F). To test if mutant p53 directly binds to FOXA1 to increase its activity, we performed a series of co-immunoprecipitation experiments that failed to demonstrate direct interactions. We next tested if mutant p53 instead affects the expression of FOXA1, we performed siRNA-mediated knockdown of mutant p53 in murine and human PDAC cells and observed significant reductions in FOXA1 levels with a 70–77% reduction in FOXA1 expression similar to those measured in PDX p53low and KPfl/+C tumors (Fig. 2N, O). To further validate mutant p53 as a specific regulator of FOXA1, we transfected PDAC cell lines with a plasmid containing the FOXA1 promoter linked to a luciferase reporter and observed a 35–50% reduction in FOXA1 promoter reporter activity following mutant p53 knockdown (Fig. 2P).
To better determine if mutant p53-mediated regulation of FOXA1 is dependent on specific sites of p53 mutation, three different p53 hotspot missense mutant constructs were ectopically expressed in KPflC cells (p53R172H, p53R245W, p53R270H) and resulted in significant 30–260% increases in Foxa1 expression (Supplementary Fig. 2G). Moreover, orthotopic xenotransplantation of KPflC-p53R172H cells into nude mice resulted in an approximate 70% increase in Foxa1(+) nuclei by immunohistochemistry (Supplementary Fig. 2H). Finally, to test if mutant p53 directly regulates FOXA1 through binding to its promoter, we performed chromatin immunoprecipitation-qPCR (ChIP-qPCR) assays and detected an approximate 3-fold enrichment of mutant p53 occupancy within the FOXA1 promoter relative to an intra-exonic control (Fig. 2Q). Collectively, these data indicate that mutant p53 regulates FOXA1 expression directly at the FOXA1 promoter irrespective of p53 missense mutation.
Mutant p53 modulates the FOXA1 transcriptional signature to drive PDAC metastasis
Since mutant p53 transcriptionally regulates FOXA1, we next sought to determine if the Foxa1 signature identified in KPwm/+C tumors is sensitive to functional modulation by mutant p53 or Foxa1. We focused our evaluation on Foxa1 target genes that are transcription factors as these would potentially activate additional downstream transcriptional networks, amplifying the effects of mutant p53 and recapitulate metastatic phenotypes. We selected five transcription factors with Foxa1 motifs within promoter regions (Irf4, Zbtb16, cFos, Gas7, Runx1) that were upregulated in KPwm/+C tumors as putative Foxa1 target genes and measured expression changes following mutant p53 or Foxa1 knockdown. Among the 2 murine and 2 human PDAC cell lines tested for these 5 FOXA1 target genes, we observed near complete concordance (19/20 samples) in target gene reductions following knockdown of mutant p53 or FOXA1 (Supplementary Fig. 3A). Similarly, these same five FOXA1 target genes were downregulated following knockout of mutant p53 yet upregulated to at least control levels after enforced FOXA1 overexpression (Supplementary Fig. 3B). Taken together, these data indicate that the transcriptional effects of mutant p53 and FOXA1 are nearly identical vis-à-vis quantitative FOXA1 target gene responses and that the FOXA1 transcriptional signature is mutant p53-dependent.
We next sought to similarly relate the dependence of metastatic phenotypes observed in KPwm/+C mice to mutant p53 and Foxa1. Initial transwell experiments were performed in which siRNA-mediated knockdown of FOXA1 in human and murine PDAC cell lines resulted in a 50–90% reduction in migration/invasion (Supplementary Fig. 3C), phenocopying our results in vivo and after mutant p53 knockdown (Supplementary Fig. 1H). Likewise, overexpression of Foxa1 in p53-null KPflC cells resulted in a significant increase in PDAC cell migration (Supplementary Fig. 3D). To firmly link mutant p53 to the promotion of migratory phenotypes through alterations in FOXA1 expression, we performed a series of epistasis experiments to rescue reduced PDAC cell migration following mutant p53 or Foxa1 knockdown with overexpression of Foxa1 or mutant p53 protein, respectively. Following shRNA-mediated knockdown of mutant p53, Foxa1 overexpression effectively rescued PDAC cell migration and invasion to control levels (Fig. 2R). Likewise, ectopic expression of mutant p53 following knockdown of Foxa1 failed to result in increased PDAC cell migration and invasion, indicating the pro-migratory and pro-invasive effects of mutant p53 to be dependent on Foxa1 (Fig. 2S). We therefore conclude that mutant p53 transcriptionally upregulates FOXA1 expression/activity, a resultant FOXA1 transcriptional signature, and that the pro-metastatic effects of mutant p53 are FOXA1-dependent.
Oncogenic KRAS effectors activate CREB1
An important next question was how mutant p53 interacts with the transcriptional complex at the FOXA1 promoter to drive its expression. Although the transcriptional regulation of FOXA1 remains largely undefined, previous work has linked FOXA1 with the transcription factor, CREB1, to cooperatively activate genes that mediate the G1/S phase transition in prostate cancer(23). We hypothesized that CREB1 might also regulate FOXA1 and evaluated CREB1 as a potential transcriptional regulator of FOXA1. Indeed, siRNA-mediated knockdown of Creb1 resulted in significantly reduced Foxa1 levels, expression and promoter activity (Fig. 3A–C). Of note, previous reports have indicated a requirement of CREB1 to be phosphorylated at serine 133 (p-CREB1S133) for attendant transcriptional activity as mediated by the MAPK/MEK, PI3K/Akt and PKA signaling pathways(24–26). To test the dependency of CREB1 activation on serine 133 phosphorylation, we overexpressed wildtype CREB1 or mutant CREB1 containing a serine-to-alanine 133 mutation (CREB1S133A) that precludes CREB1 phosphorylation in PDAC cells. Despite equivalent total CREB1 levels, PDAC cells with CREB1S133A demonstrated significantly reduced p-CREB1S133 levels with corresponding reductions in Foxa1 expression and promoter reporter activity, supporting a substantial and specific role of p-CREB1S133 in FOXA1 regulation (Fig. 3D–F). Accordingly, spatial co-expression analysis of p-Creb1 and Foxa1 in autochthonous KPwm/+C tumors revealed significant spatial co-expression (Pearson correlation, p<0.0001) (Fig. 3G, H).
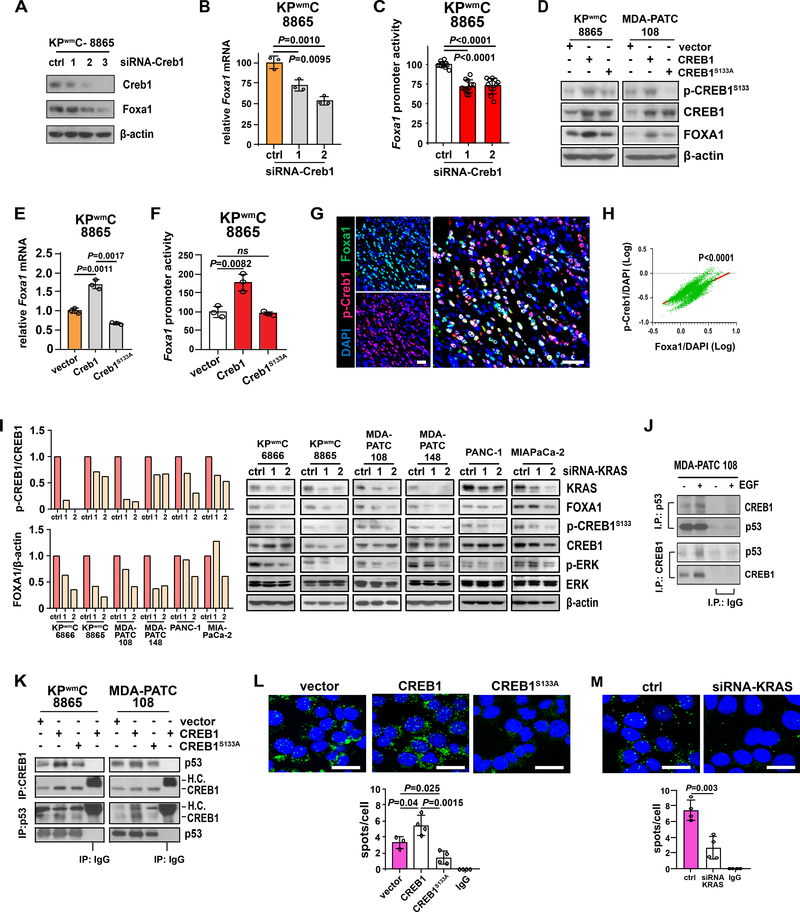
Figure 3. Oncogenic KRAS effectors enable mutant p53 binding to CREB1 and FOXA1 upregulation.
A, Foxa1 levels following Creb1 knockdown. B and C, Foxa1 expression and promoter reporter activity following Creb1 knockdown. D, Wildtype CREB1 or phosphorylation-resistant CREB1S133A mutant were transfected into PDAC cells and FOXA1 levels measured by western blot. E and F, The effect of wildtype Creb1 or Creb1S133A mutant overexpression on Foxa1 expression and Foxa1 promoter reporter activity. G, Representative spatial co-expression of p-Creb1 and Foxa1 in KPwm/+C tumors as measured by multicolor immunofluorescence (n=4 mice, 5 random fields/mouse). Scale bars, 50μM. H, Correlation of p-Creb1 and Foxa1 expression as measured by Pearson correlation (p<0.0001). I, p-CREB1S133 and FOXA1 levels following KRAS knockdown. J, Reciprocal co-immunoprecipitation of mutant p53 and CREB1, with and without EGF treatment. K, Reciprocal co-immunoprecipitation between CREB1 and mutant p53 following overexpression of wildtype CREB1 or the phosphorylation-resistant mutant, CREB1S133A. L, The effect of CREB1S133 phosphorylation on mutant p53 and CREB1 binding interactions as measured by proximity ligation assay (PLA). M, The effect of KRAS knockdown on mutant p53 and CREB1 binding interactions as measured by PLA. A-F, L, M. Representative of at least three independent experiments performed in triplicate. I-K, Representative of at least three independent experiments. (B, C, E, F, L, M) Data are mean±s.d. and P values determined by unpaired two-tailed t-tests. (L, M) Scale bars, 20μM.
The MAPK/MEK and PI3K/Akt pathways are robustly activated in PDAC and are considered key effectors of oncogenic KRAS. We therefore reasoned that sustained oncogenic signaling from mutant KRAS might serve as a dominant driver of CREB1 phosphorylation at serine 133 through effector pathways, thereby promoting FOXA1 expression. Indeed, KRAS knockdown in a panel of murine and human PDAC cell lines resulted in marked reductions in p-CREB1S133 and FOXA1 levels, confirming a significant role in CREB1 phosphorylation and activation (Fig. 3I). Moreover, to test if the activation of CREB1 is largely driven by KRAS effectors, we treated PDAC cells with pharmacologic inhibitors against MEK (AZD6244, U0126, trametinib), PI3K (pictilisib) and PKA (H89) and observed reductions in p-CREB1S133 levels (Supplementary Fig. 4A–C). Among these, pharmacologic inhibition of MEK produced the greatest reductions in p-CREB1S133 levels even following treatment with epidermal growth factor (EGF), a potent stimulator of the MAPK/MEK pathway and CREB1 phosphorylation (Supplementary Fig. 4B, C). Since p-CREB1S133 leads to CREB1 activation and the selective inhibition of KRAS effectors, specifically MAPK/MEK inhibition, largely prevents CREB1 serine 133 phosphorylation, we concluded that oncogenic KRAS largely drives CREB1 activation in the context of PDAC and contributes to FOXA1 expression.
Other work has indicated that mutant p53 mediates PDAC metastasis through upregulation of the receptor tyrosine kinase (RTK), platelet derived growth factor receptor-β (PDGFR-β)(11). To evaluate the potential contributions of RTK signaling and oncogenic KRAS on CREB1 phosphorylation and resultant FOXA1 upregulation, we treated PDAC cells with epidermal growth factor (EGF) to activate an RTK upstream of KRAS, or PDGF in the presence and relative absence of oncogenic KRAS. Although EGF or PDGF treatment increased p-CREB1S133 and FOXA1 levels/promoter activity in the setting of oncogenic KRAS, following knockdown of oncogenic KRAS, only treatment with PDGF resulted in a slight increase in p-CREB1S133 and FOXA1 levels in 1 of 2 tested KRAS siRNA sequences without detectable changes in FOXA1 promoter reporter activity (Supplementary Fig. 4D, E). Taken together, these results indicate that oncogenic KRAS effectors predominantly mediate CREB1S133 phosphorylation and upregulate FOXA1 levels to drive PDAC metastasis.
Mutant p53 binds activated CREB1 to upregulate FOXA1
Mutant p53 GOF is largely imparted through interactions with transcription factors that modulate their transcriptional activity. After determining that mutant p53 and activated CREB1 individually regulate FOXA1, we next assessed if mutant p53 and CREB1 might together interact to transcriptionally regulate FOXA1 in PDAC. We therefore tested if mutant p53 binds to CREB1 or its activated form, p-CREB1S133, to enhance its transcriptional activity at the FOXA1 promoter. Reciprocal co-immunoprecipitation experiments between mutant p53 and CREB1 revealed clear binding between these two proteins that was enhanced with EGF treatment, indicating a potential requirement for p-CREB1S133 to enable pulldown (Fig. 3J). To test if interactions between mutant p53 and CREB1 were indeed dependent on p-CREB1S133, we performed reciprocal co-immunoprecipitation experiments between mutant p53, WT CREB1 and CREB1S133A. Indeed, despite equivalent immunoprecipitated CREB1 levels, significantly less mutant p53 was pulled down in the presence of CREB1S133A relative to WT CREB1 and significantly less CREB1 was pulled down with mutant p53 in the presence of CREB1S133A (Fig. 3K). These results were further confirmed through proximity ligation assays that demonstrated a 73% reduction in proximity between mutant p53 and CREB1 in the presence of CREB1S133A (Fig. 3L). As we previously determined KRAS effectors to serve as a dominant driver of CREB1 phosphorylation at serine 133, we next sought to determine if oncogenic KRAS significantly enabled interactions between mutant p53 and CREB1. Following siRNA-mediated KRAS knockdown, an approximate 64% reduction in mutant p53 and CREB1 proximity were observed, indicating dominant and permissive roles of oncogenic KRAS in binding between mutant p53 and CREB1 (Fig. 3M).
FOXA1 activates WNT/β-catenin signaling
After establishing that oncogenic KRAS effectors and mutant p53 consort to upregulate FOXA1, we next sought to better understand how FOXA1 promotes PDAC metastasis. While genes upregulated in mutant p53 tumors were enriched in Foxa1 TFBS relative to p53-null tumors, IPA analysis of these differentially expressed genes also revealed β-catenin as a dominant upstream regulator (Fig 4A, B). Furthermore, Enrichr analysis indicated “Basal cell carcinoma” as a top enriched pathway due to the presence of Wnt/β-catenin signaling genes differentially upregulated in KPwm/+C tumors (Fig. 1H). Taken together, these data suggest that Foxa1 and β-catenin activity are both enriched in mutant p53 PDAC tumors and might potentially function together to drive PDAC metastasis.
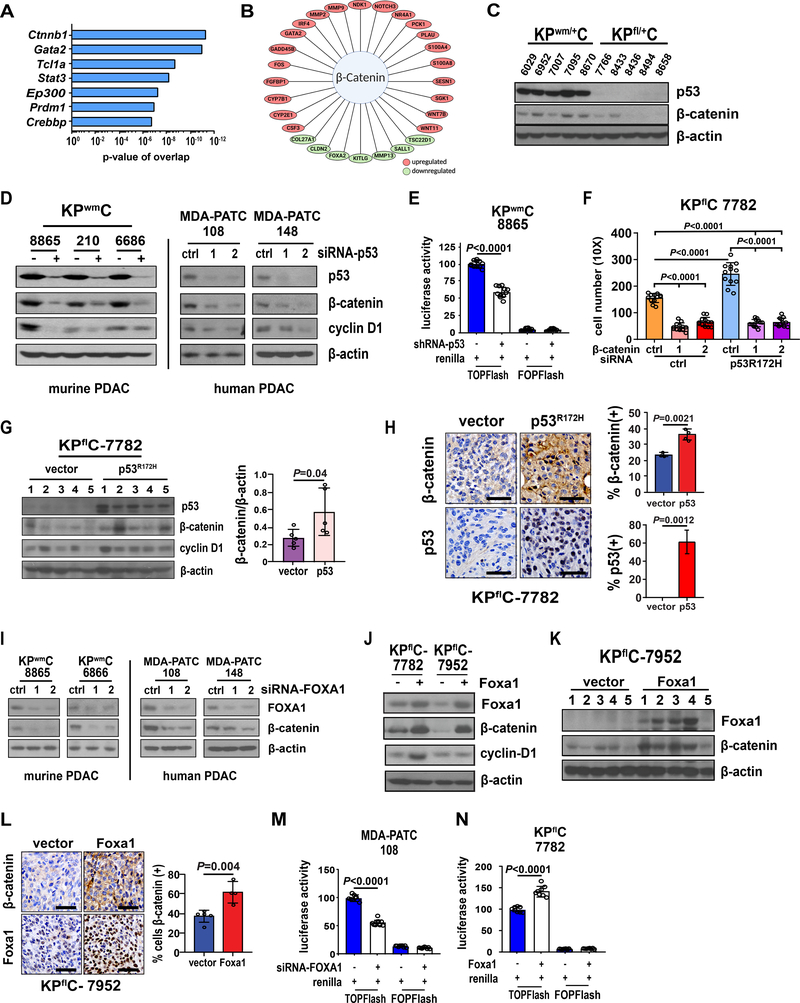
Figure 4. Mutant p53 promotes β-catenin stabilization and activity through FOXA1.
A, IPA analysis results of top upstream regulators of genes differentially expressed in tumors derived from KPwm/+C mice relative to KPfl/+C mice. B, Enriched transcription factors associated with Wnt/β-catenin signaling in KPwm/+C tumors. C, β-catenin levels measured in tumors derived from KPwm/+C and KPfl/+C mice. D, Effect of mutant p53 knockdown on β-catenin levels in murine and human PDAC. E, β-catenin activity following mutant p53 knockdown as measured by the TOPFlash luciferase reporter system (TLRS). F, The effect of β-catenin knockdown on PDAC cell migration/invasion, with and without mutant p53R172H. G, β-catenin levels following ectopic mutant p53R172H expression in p53 null, KPflC cells (KPflC-p53R172H) and heterotopic engraftment into immunodeficient mice. H, Immunohistochemical evaluation and quantification of β-catenin in KPflC (n=5) and KPflC-p53R172H (n=5) tumors. I, The effect of FOXA1 knockdown on β-catenin levels in murine and human PDAC. J, Levels of β-catenin following Foxa1 overexpression in two p53-null, KPflC cell lines (KPflC-Foxa1). K, β-catenin levels were measured in whole tumor lysates following engraftment of KPflC-Foxa1 cells into nude mice (n=5) along with vector controls (n=5). L, Immunohistochemical evaluation and quantification of Foxa1 in KPflC (n=5) and KPflC-Foxa1 (n=5) tumors. M, β-catenin activity (TLRS) following FOXA1 knockdown. N, β-catenin activity (TLRS) following FOXA1 overexpression. (E, G, H, L, M, N), Data are mean±s.d. and P values determined by unpaired two-tailed t-tests. (F), P value calculated by one-way ANOVA.
To determine if FOXA1 and β-catenin signatures are potentially interrelated, we first sought to confirm enriched Wnt/β-catenin signaling in tumors from KPwm/+C mice relative to KPfl/+C mice through measurement of total β-catenin levels. Overall, lysates generated from randomly selected tumors in KPwm/+C mice demonstrated higher levels of total β-catenin relative to tumors from KPfl/+C mice (Fig. 4C). To test if mutant p53 affects β-catenin levels, knockdown of mutant p53 in murine and human PDAC cell lines resulted in reduced β-catenin levels with a concomitant 40% reduction in β-catenin activity as confirmed by the TOPFLASH reporter system (Fig. 4D, E). Decreased protein levels of the β-catenin target gene, cyclin D1, were also observed, indicating that mutant p53 potentially stimulates β-catenin activity (Fig. 4D).
To test if β-catenin contributes to the prometastatic phenotype observed in mutant p53 tumors, we performed epistasis experiments to rescue reduced PDAC cell migration following β-catenin knockdown with overexpression of mutant p53R172H. After shRNA-mediated knockdown of β-catenin, mutant p53R172H overexpression failed to rescue PDAC cell migration and invasion, indicating an important role downstream of mutant p53 (Fig. 4F). Importantly, knockdown of mutant p53 did not result in reduction of β-catenin mRNA levels, indicating a role for mutant p53 in the stabilization of β-catenin protein (Supplementary Fig. 5A). To further confirm mutant p53 regulation of β-catenin in vivo, mutant p53R172H was ectopically produced in a p53-null, KPflC cell line and orthotopically transplanted into immunodeficient mice. Relative to vector controls, β-catenin levels increased by 2-fold in lysates from KPflC-p53R172H tumors (Fig. 4G). In parallel, immunohistochemical analysis of β-catenin levels in KPflC-p53R172H tumors also demonstrated gross increases in β-catenin(+) nuclei relative to vector controls (Fig. 4H). Collectively, these data corroborate our IPA analysis indicating enriched β-catenin stability and activity in mutant p53 tumors and demonstrate that mutant p53 promotes the stabilization and activity of β-catenin in PDAC.
Since mutant p53 transcriptionally upregulates FOXA1 and stabilizes β-catenin, we hypothesized that enhanced FOXA1 expression and activity promotes β-catenin stability with both transcriptional networks potentially serving as effectors of mutant p53-mediated metastasis. Accordingly, siRNA-mediated knockdown of Foxa1 resulted in reduced β-catenin levels and overexpression of Foxa1 in KPflC PDAC cells (KPflC-Foxa1) resulted in significant increases in β-catenin levels (Fig. 4I, J). This was confirmed in protein lysates and immunohistochemical analyses of xenografted tumors generated from KPflC-Foxa1 cells and corresponding vector controls (Fig. 4K, L). Overexpression of Foxa1 in KPflC cell lines also resulted in an approximate 43% increase in β-catenin activity and knockdown of Foxa1 resulted in a 44% reduction in β-catenin activity (Fig. 4M, N).
As cyclin D1 is only one β-catenin target gene used as a surrogate screen for β-catenin activity, we next assessed if the activation of other canonical Wnt/β-catenin target genes were concordantly modulated by mutant p53 and Foxa1. Indeed, 6 additional Wnt/β-catenin target genes were concordantly downregulated following knockdown of mutant p53 or Foxa1, supporting the activation of Wnt/β-catenin co-transcriptional networks by a mutant p53-FOXA1 signaling axis (Supplementary Fig. 5B).
Mutant p53 regulates Wnt/β-catenin stabilization through FOXA1-mediated CK1α downregulation
As with mutant p53 knockdown, FOXA1 overexpression did not affect β-catenin mRNA levels, indicating intermediary processes that stabilize β-catenin rather than affect gene transcription. β-catenin stabilization is the sum product of complex signals that balance β-catenin sequestration, release and degradation by the β-catenin destruction complex(27,28). Specifically, β-catenin levels are regulated by kinases that sequentially phosphorylate β-catenin at Ser45 (CK1α) followed by Ser33/37/Thr41 (GSK3), marking it for degradation by the β-catenin destruction complex(27,28). To identify the mechanism through which FOXA1 stabilizes β-catenin, we hypothesized that FOXA1 inhibits such key phosphorylation events, thereby maintaining β-catenin levels and activity. To test if FOXA1 affects β-catenin phosphorylation, we knocked down FOXA1 and observed significantly increased levels of phospho-β-catenin at Ser45 (p-β-cateninS45) and concomitant reductions in β-catenin levels (Supplementary Fig. 5C). CK1α, a serine-threonine kinase, phosphorylates β-catenin at Ser45 and measurement of CK1α levels following FOXA1 knockdown revealed increases in CK1α levels with a 3–8-fold increase in expression (Supplementary Fig. 5D). Likewise, Foxa1 overexpression in KPflC cell lines resulted in the marked reduction of Ck1α levels following orthotopic tumor formation (Supplementary Fig. 5E). Ectopic overexpression of FOXA1 in a KPwmC cell line also resulted in 8–9-fold reductions in p-β-cateninS45 levels with concomitant >2-fold increases in total β-catenin levels, much of which were reversed by ectopic CK1α expression (Supplementary Fig. 5F).
As expected, since CREB1 positively regulates FOXA1, introduction of CREB1S133A resulted in a 260% increase in CK1α expression with concomitant reductions in Wnt/β-catenin target gene expression (Supplementary Fig. 5G, H). Likewise, siRNA experiments targeting mutant p53 or FOXA1 and ectopic mutant p53 expression in 3 different KPflC cell lines were performed to identify concordant changes in CK1α, cyclin D1 and Wnt/β-catenin target gene expression, confirmatory of our defined mechanism (Supplementary Fig.5I–K). Collectively, our data indicate that mutant p53R172H interacts with p-CREB1S133 to upregulate FOXA1 expression which, in turn, down-modulates CK1α, reduces p-β-cateninS45 levels and mitigates β-catenin degradation, resulting in the activation of β-catenin target genes. Such co-activation of the FOXA1 and β-catenin-associated transcriptional networks, as enabled by oncogenic KRAS effectors and mutant p53, drive pro-metastatic processes that result in PDAC metastasis.
Therapeutic CREB1 inhibition reduces PDAC metastasis
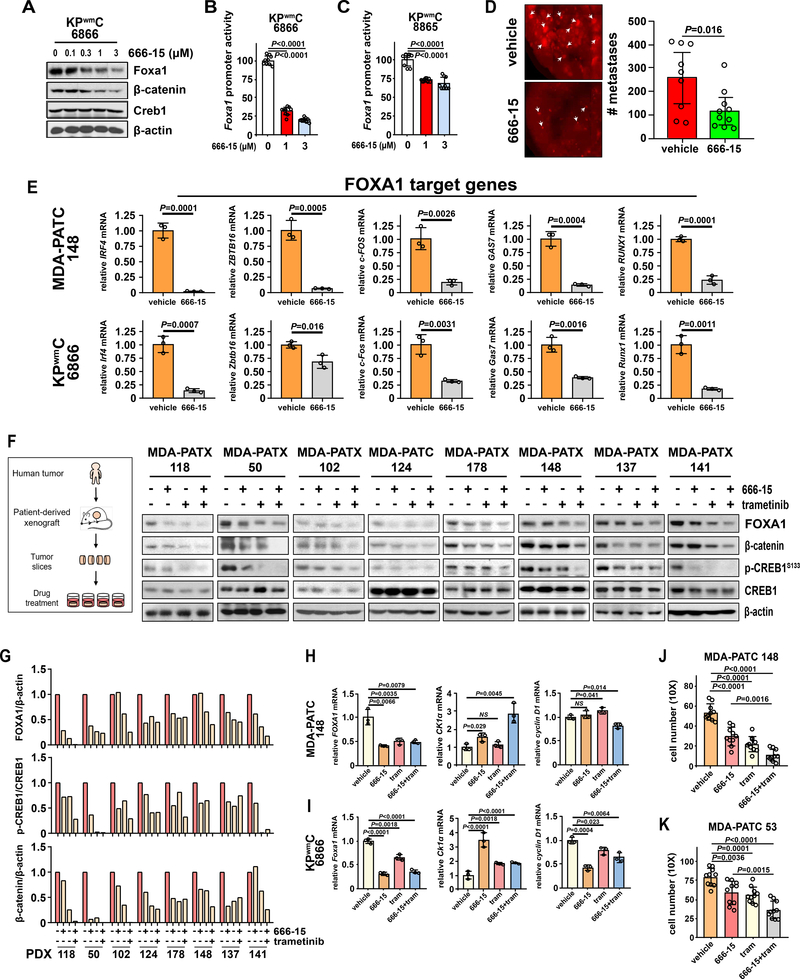
We have identified CREB1 as a point of convergence between oncogenic KRAS effectors and mutant p53 that upregulates FOXA1 and PDAC metastasis. Accordingly, we hypothesized that pharmacologic inhibition of CREB1 with a potent and specific CREB1 inhibitor, 666–15, would minimize the downstream activation of FOXA1/β-catenin signaling, thereby inhibiting PDAC metastasis(29,30). 666–15 is a bioavailable, potent, and selective CREB1 inhibitor that is well tolerated in vivo with ample pharmacokinetic data and demonstrated anticancer activity using once daily dosing (29,30). To test if 666–15 affects Foxa1 and β-catenin levels, PDAC cells were treated with 666–15 and western analysis revealed dose-dependent reductions in Foxa1 and β-catenin levels along with a 27–68% reduction in Foxa1 activity (Fig 5A–C). We next tested if pharmacologic inhibition of CREB1 would reduce PDAC metastasis using conventional tail vein metastasis assays. Following transfection with RFP reporter plasmids, human PDAC cells were injected into the tail veins of nude mice and were treated with either vehicle or 666–15 (10mg/kg/day) for 5 days. Treatment with 666–15 significantly abrogated the number of formed lung metastases relative to vehicle, confirming CREB1 inhibition as a viable approach to reduce PDAC metastasis (Fig. 5D). Likewise, treatment of PDAC cells with 666–15 resulted in the reduced expression of all tested FOXA1 target genes, supporting our mechanism while providing evidence of pharmacologic inhibition of FOXA1 activity (Fig. 5E).
Figure 5. Therapeutic CREB1 inhibition limits PDAC metastasis.
A, Foxa1 and β-catenin levels following treatment with a CREB1 inhibitor (666–15). B and C, The effect of 666–15 treatment on Foxa1 promoter reporter activity. D, The effect of pharmacologic CREB1 inhibition on PDAC metastasis as determined by lung colonization metastasis assays in host mice treated with vehicle (n=9) or 666–15 (n=9). P values determined by unpaired two-tailed t-tests. E, The effect of 666–15 treatment on FOXA1 target gene activation in murine and human PDAC. F, The effects of single and combinatorial treatment with 666–15 and trametinib (MEK inhibitor) on FOXA1, β-catenin, and p-CREB1S133 levels in 8 unique PDAC PDX models. G, Quantification of FOXA1 and p-CREB1S133 levels in 8 unique PDAC PDX models following treatment with 666–15 and/or trametinib. H and I, FOXA1, CK1α and cyclin D1 expression following treatment with a CREB1 inhibitor (666–15), MEK inhibitor (trametinib) or combination in human and murine PDAC. J and K, The effects of treatment with a CREB1 inhibitor (666–15), MEK inhibitor (trametinib) or combination on PDAC cell migration/invasion as measured by transwell assays. (A-C, E, H-K) Representative data from at least three independent experiments performed in triplicate. Data are mean±s.d. P values determined by unpaired two-tailed t-tests.
Since our data implicate MEK1/2 as the predominant KRAS effector responsible for the activation of CREB1 through phosphorylation at ser133, we further reasoned that co-treatment with a MEK inhibitor (trametinib) and CREB1 inhibitor (666–15) might result in improved therapeutic efficacy. To test the combinatorial effects of trametinib and 666–15 on CREB1 phosphorylation and further validate our derived mechanism within a relevant biological system, we treated freshly resected PDAC PDX tumors using our live tissue sensitivity assay (LTSA) system(31). Serial tumor sections from 8 unique, freshly resected PDAC PDX tumors were treated with vehicle, 666–15, trametinib or both drugs for 24 hours and tumor lysates were generated to measure FOXA1, p-CREB1S133 and β-catenin levels (Fig 5F). As expected, CREB1 and MEK inhibition reduced FOXA1, p-CREB1S133 and β-catenin levels with the largest reduction evident in the dual treatment group (Fig. 5F, G). Expected effects of 666–15 and trametinib treatment on FOXA1, CK1α and cyclin D1 expression were confirmed in PDAC cell lines by RT-qPCR (Fig. 5H, I). The individual and combined effects of trametinib and 666–15 treatment on PDAC cell migration/invasion were functionally confirmed by transwell migration and invasion (Fig. 5J, K). Taken together, pharmacologic CREB1 inhibition with 666–15 significantly reduced PDAC metastasis in vivo and co-inhibition of CREB1 and MEK significantly reduced p-CREB1S133A, FOXA1 and β-catenin levels in clinically relevant models of PDAC.
Discussion
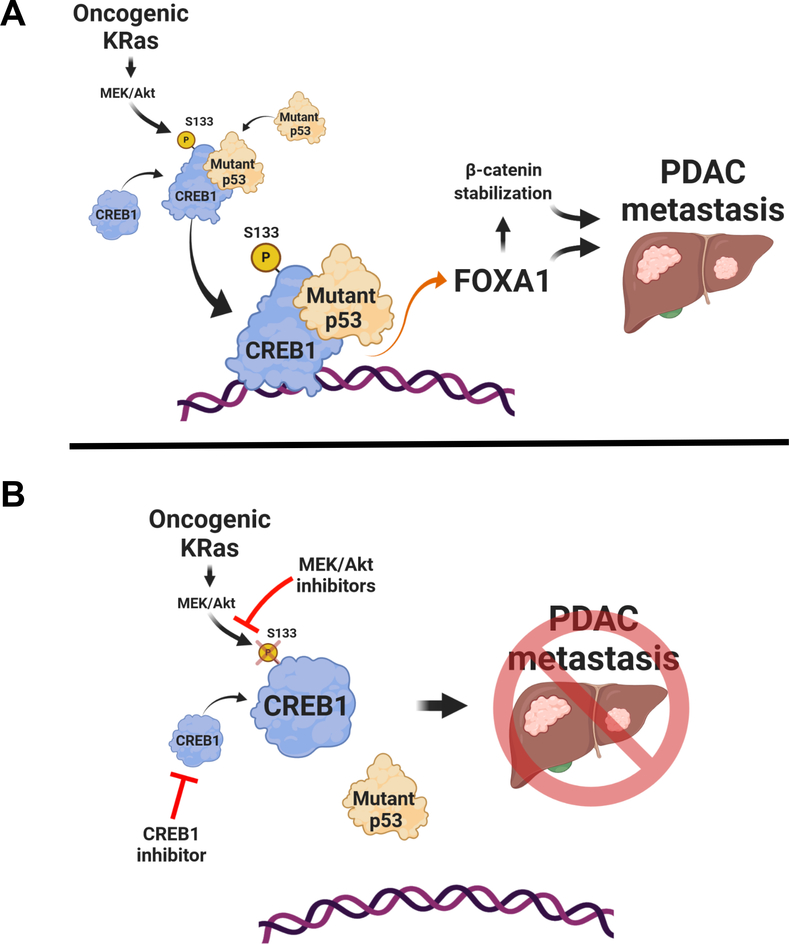
Our work details a novel mechanism through which mutant p53 exerts GOF to drive PDAC metastasis that is conditional on CREB1S133 phosphorylation, mediated largely through the oncogenic KRAS-RAF-MEK-MAPK effector pathway. The activation of CREB1 by oncogenic KRAS effectors and resultant binding to mutant p53 to upregulate FOXA1 expression represents a unique mechanism whereby mutant p53 directly cooperates with an oncogenic signaling axis prevalent in the vast majority of PDAC patients (Fig 6A). Moreover, the enhanced activities of CREB1, FOXA1 and β-catenin result in the activation of many downstream target genes, vastly amplifying the oncogenic effects of mutant KRAS and p53 in a KRAS-dependent manner. To our knowledge, this is the first report to describe direct and dependent cooperation between the most commonly mutated oncogene and tumor suppressor gene in human cancer and PDAC that result in the expansive activation of pro-metastatic transcriptional networks. Most importantly, decoupling this cooperative node between oncogenic KRAS effectors and mutant p53 mitigates PDAC metastasis and combinatorial prevention of CREB1S133 phosphorylation and/or direct CREB1 inhibition represents a novel therapeutic approach to target dominant genetic driver pathways in PDAC (Fig. 6B).
Figure. 6. Mutant p53 cooperates with oncogenic KRAS effectors to generate metastatic phenotypes.
A, Model depicting KRAS effectors phosphorylating CREB1 to enable binding and hyperactivation by mutant p53. Consequently, FOXA1 is upregulated and β-catenin is stabilized, leading to pro-metastatic phenotypes. B, Reductions in CREB1 phosphorylation through MEK inhibition, coupled with direct CREB1 inhibition with 666–15, blocks FOXA1 upregulation and β-catenin stabilization, inhibiting PDAC metastasis.
Our findings also contextualize a major observation in the PDAC mutational landscape as it pertains to p53 and PDAC biology. Specifically, p53 is the only major tumor suppressor in PDAC to predominantly undergo missense mutation rather than loss of protein expression like in the case of CDKN2A or SMAD4. This observation suggests the presence of biologic advantages to p53 missense mutations that result in stabilized, though altered, mutant p53 proteins relative to the absolute loss of protein. Indeed, the prevalence of stable, mutant p53 proteins in human cancers has long supported mutant p53 GOF activities that increase tumor cell proliferation, drug resistance, and migration/invasion, among other activities(32,33). In particular, the high incidence of co-occurring oncogenic KRAS and missense p53 mutations in PDAC suggests the underlying presence of cooperative mechanisms that promote tumor growth and progression. Convergent, cooperative hubs between mutant KRAS and p53, as we identified at CREB1, may at least partially account for the prevalence and dominance of these two driver genes within the mutational landscape of human cancers. Moreover, such cooperation may produce phenotypes with superior fitness features relative to cells that do not form stable mutant p53 protein products, even extending beyond the metastatic phenotype observed in our study. Additional studies are necessary to define the extent and impact of combinatorial therapies directed against CREB1 to further exploit this therapeutic vulnerability in PDAC.
Another important aspect of mutant p53 biology apparent in our work is the promiscuous nature of mutant p53 to engage potent transcription factors, in a multiplexed fashion, to magnify its oncogenic effects. Through the hyperactivation of CREB1, mutant p53 likely affects many CREB1 target genes and associated cellular processes. In parallel, through mutant p53/CREB1-mediated upregulation of FOXA1, many FOXA1 target genes are also aberrantly activated in concert with the co-transcriptional activities of β-catenin. Such multiplexing of transcriptional networks amplifies the downstream effects of mutant p53, effectively converting a single, mutated tumor suppressor gene into a bona fide oncoprotein that fuels PDAC metastasis. Importantly, since FOXA1 is a pioneer transcription factor, mutant p53 may also promote the expression of pro-metastatic genes through increased chromatic accessibility, indirectly regulating genes without physically occupying promoter regions. Efforts to determine the cistrome of mutant p53 through ChIP analyses, therefore, may not entirely capture the gene repertoire recruited by mutant p53 to curate metastatic phenotypes. Further elucidation of essential target genes responsible for pro-metastatic traits within activated FOXA1 and β-catenin transcriptional networks may drive additional clinical therapeutics to mitigate PDAC metastasis, increase surgical resection rates, and improve patient outcomes.
Methods
Mice.
Previously characterized KRasLSL-G12D(ref. (34)), Trp53flox/flox(ref. (35)), Trp53wmR172H(ref. (16)), Pdx1-cre (ref. (5)), Rosa26LSL-tdTomato (ref. (36)) were bred and crossed to generate KPC mice exclusively in the MD Anderson animal facility. All mice were bred and maintained in a C57/bl6 background with roughly equal numbers of male and female mice examined in the study. 10–12 week-old female nude mice (Envigo; Athymic Nude-Foxn1 nu) were used for cell-lined based xenotransplantation experiments. All mice were monitored daily by Investigators and staff of the MD Anderson Veterinary Medicine and Surgery Department. Any mice exhibiting signs of physical distress or undue discomfort were immediately euthanized. All animal studies and associated procedures were reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at MD Anderson Cancer Center (protocols 00000985-RN02 00001089-RN02).
Cell Lines.
Primary murine PDAC cell lines (KPwmC, KPflC) were generated from mice of corresponding genotypes (KPwm/+C and KPfl/+C mice) through mechanical digestion of pancreatic tumors and subsequent tissue culture in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All primary human PDAC cell lines (MDA-PATC108, 148, 102, 124) were isolated from patient derived xenograft tumors in our laboratory at various times between 2010–2016 (37). Other PDAC cell lines (MIA-PaCa-2, BxPC-3 and PANC1) were obtained from the American Type Culture Collection (ATCC) in 2020. All cells were verified by DNA fingerprinting at the Characterized Cell Line Core Facility at MD Anderson Cancer Center. All cells were cultured in DMEM supplemented with 10% FBS. All cell lines were routinely confirmed negative for mycoplasma every 2 months by PCR, last tested 12/3/2020. Cells were passaged no longer than 12 to 20 times prior to the completion of experiments.
Orthotopic and heterotopic xenotransplantation of tumor cells.
KPC or human PATC tumor cells were suspended in 1X phosphate-buffered saline solution (PBS) at a concentration of 1×105 cells/25μL with one volume of Matrigel (Corning) and kept on ice. 25 μL were injected into the subcutaneous flank or mouse pancreas in 10–12-week-old nude mice. After 3 weeks, all mice were euthanized, and tumors harvested for analysis.
Lung colonization metastasis assays.
PDAC cells were first transduced with pcDH-EF1-MCS-RFP (CSB1 system bioscience), passaged and then transduced with nontargeting control shRNA or shRNA targeting FOXA1. Following confirmation of FOXA1 knockdown, 1 × 106 cells/100 μl PBS from control shRNA or shRNA-FOXA1 were injected into the tail veins of nude mice. 3 weeks later, hepatic and pulmonary metastases were identified and enumerated based on the detection of RFP(+) lesions using the EVOS® FL cell imaging system (ThermoFisher). For studies involving treatment with 666–15 (Tocris), PDAC cells transduced with pcDH-EF1-MCS-RFP were treated with 666–15 (1μM) for 5 days and 1 × 106 cells/100 μl were subsequently injected into mouse tail veins. Following tail vein injection, 666–15 (10 mg/kg) was dissolved in 1% N-2-methylpyrrolidone (NMP) and 5% Tween-80 solution and administered by intraperitoneal injection once a day for 5 days. Control mice were treated with identical volumes of vehicle without 666–15.
Genomic DNA extraction, PCR amplification and sequencing
Cell pellets from 1X106 cells were collected and 300 μl of DNA lysis buffer (10 mM Tris, 100 mM NaCl, 10 mM EDTA, 0.5% SDS, 0.4 mg/ml proteinase K) was added. Cell mixtures were incubated at 55°C for 2hr. followed by centrifugation at 13,000 rpm for 10 min. The supernatant was then combined with 300μL isopropanol, mixed and centrifuged at 13,000 rpm for 10 min. The DNA pellet was then dissolved in 300 μl of double-distilled water. 1 μL of DNA solution was used for each genotyping PCR reaction. PCR reactions were performed on a T100™ Thermal Cycler (Bio-Rad). Primer pairs used to amplify the exon-5 of human and mouse p53 genes are listed in supplementary materials and methods. PCR fragments were gel-purified with a DNA-gel extraction kit (Qiagen) and quantified with the NanoDrop™ 2000 (Thermo Fisher). Recovered PCR fragments were Sanger sequenced at MD Anderson Advanced Technology Genomic Core. Sequencing was performed on ABI 3730XL DNA analyzer (Thermo Fisher), and the results were analyzed with KB™ BaseCaller software v5.4 (Thermo Fisher). The DNA sequences were visualized using Chromos software (Technelysium Pty Ltd.) and mutations were confirmed through alignment with wild type gene references using online Basic local Alignment Search Tool (BLAST) from the National Center for Biotechnology Information.
Transwell migration/invasion assay.
BioCoat Matrigel invasion chambers with 8.0μm PET membrane in 24-well plates (354480, Corning) were thawed and 400 μl of serum-free DMEM was added to the upper chamber for rehydration. After incubating at 37°C for 2 hours, 1 × 105 KPC cells or 2×105 MDA-PATC cells were resuspended in 100 μl of serum-free DMEM and added to the upper chamber. In the lower chamber, 800 μl of DMEM with 10% FBS was deposited as a chemoattractant. After 16 hours of 37°C incubation, the upper well membrane was scrubbed with a cotton swab soaked in PBS to remove non-invaded cells. The lower membrane of the well was carefully rinsed with PBS; invaded cells were fixed with 100% cold methanol and stained in the dark with 0.05 mg/ml 4,6-diamidino-2-phenylindole (DAPI) (BD Biosciences) for 10 min. Images of cells in each chamber were obtained with a fluorescence microscope (Olympus IX71) and invading cells were enumerated in eight random fields (10X magnification) per chamber by ImageJ software (NIH).
Lentiviral shRNA-mediated gene silencing.
shRNA glycerol stocks contained pGIPZ-GFP expressing shRNA against mouse Trp53 and human FOXA1 (Open Biosystems) were obtained from the MDACC Functional Genomic Core (Supplementary Materials and Methods). The sequences and the RNAi consortium clone IDs for shRNAs used are: pGIPZ-GFP-shRNA-Trp53: 5’-CACTACAAGTACATGTGTA-3’ (Dharmacon, v3LHS_646511), pGIPZ-GFP-shRNA-Trp53: 5’- CGCGCCATGGCCATCTACA-3’ (Dharmacon, v2LHS_287039), pGIPZ-GFP-shRNA-hFOXA1: 5’-CTCCGTATATTTACATAAC-3’ (Dharmacon, V2LHS16780) and pGIPZ-GFP-shRNA-hFOXA1:5’-GCAATACTCTTAACCATAA-3’ (Dharmacon, V2LHS16813). GIPZ lentiviral non-targeting shRNA were used as controls. Lentiviral shRNA plasmids were packaged into lentiviral particles and infected into PDAC cell lines. Stably integrated shRNAs were selected by adding puromycin (2 μg/ml for human cell lines and 4 μg/ml for mouse cell lines) (Invitrogen) to culture medium for 3 days. These pools of colonies were isolated and screened for Trp53 or FOXA1 expression by Western blot analysis. Sequences and RNAi consortium clone IDs are also listed in supplementary materials and methods.
Short interfering RNA (siRNA)-mediated gene silencing.
siRNAs used in this study (see supplementary materials and methods) are as follows: mouse Trp53 (sigma, NM_001127233), human TP53 (sigma, NM_000546), mouse FOXA1 (sigma, NM_008259), human FOXA1 (sigma, NM_004496), mouse KRAS (sigma, NM_021284), human KRAS (sigma, NM_004985) and mouse CREB1 (sigma, NM_001037726). MISSION® siRNA Transfection Reagent (Sigma) was used to transfect siRNA into PDAC cell lines. 1×105 cells were seeded in a 6-well plate. siRNA and transfection reagent were mixed per manufacturer’s instructions and added into 200μl serum-free medium for 15 min. The transfection mixture was then combined with 2ml fresh culture medium and added to cell-bearing wells. Effective gene silencing was confirmed 24–48 hours post-transfection by qPCR or western blot relative to control.
Gene expression plasmids.
Trp53R172H was expressed in KPC cell lines using pBABE-puro-Trp53R172H previously generated by our group(38). Plasmid was packaged into retrovirus particles and infected into KPC cell lines. PDAC cells with stable integration of plasmid were selected by adding G418 (500 μg/ml) to the culture medium for 5–6 days. Colonies were pooled and screened for p53 expression by Western blot analysis. pLenti-C-mGFP-P2A-puro-FOXA1 (murine) ORFs (Origene) were packaged into lentiviral particles and infected into PDAC cell lines. PDAC cells with stable integration of plasmid were selected with puromycin (4 μg/ml) for 3 days. Pools of colonies were screened for increased FOXA1 expression by Western blot analysis. pCMV vectors expressing mouse CREB1 or CREB1S133A ORFs (Clontech) were transfected into PDAC cells using Lipofectamine 2000 (Invitrogen). Likewise, a pLC-Flag-Puro vectors expressing CK1α (murine) ORFs were transfected into PDAC cells using Lipofectamine 2000 (Invitrogen). Expected changes in CREB1, CREB1S133A, CK1α levels were confirmed by western blot 24 hours after transfection.
Luciferase Reporter Gene Assays.
PDAC cells (10,000) were plated in 100μL of medium containing 10% FBS per well in a white-walled 96-well plate (Perkin Elmer). After 24hrs., cells were co-transfected using Lipofectamine 2000 transfection reagent (Invitrogen) according to the manufacturer’s instructions with 150 ng of TOPFLASH Firefly (M50) reporter 41 (Addgene) and 50 ng of pRL-SV40P Renilla (Addgene) constructs, or 150ng of FOPFLASH Firefly (M51) reporter 41 (Addgene) and 50 ng of pRL-SV40P Renilla. After 24 hrs. transfection, cells with or without treatment were detected using the Dual-Luciferase® Reporter Assay System (Promega) according to the manufacturer’s instructions. To control for transfection efficiency, Firefly luciferase levels were normalized to Renilla luciferase levels to generate a measurement of relative luciferase units. For FOXA1 promoter activity reporter assays, the FOXA1 promoter (murine) sequence spanning 1619 bp (1355 bp upstream and 263bp downstream of the transcription start site) was cloned into the lentivirus promoter reporter plasmid pEZX-LvPG02 (GeneCopoeia) through EcoRI and BamHI restrictions enzyme sites and verified with Sanger sequencing. The FOXA1 reporter plasmid and negative control (NEG-LVPG02) were packaged into lentiviral particles and infected into PDAC cells. Cells with stable integration of plasmid were selected with puromycin for 3 days. Stable cell lines were then transfected with siRNA, ORFs or treated with small molecule inhibitors for 24 hours or treated with EGF and PDGF-BB for 30 minutes. After transfection or treatment, luminescence in media or cell lysates was measured using the Pierce gaussia luciferase glow assay kit (ThermoFisher).
Proximity ligation assay.
The Duo-link In Situ-Fluorescence kit was used according to the manufacturer’s instructions (Sigma-Aldrich). PDAC cells were grown on eight chamber slides (Corning), fixed in cold 100% methanol, and permeabilized using 0.1% Triton-X-100. After 1 hour of blocking solution incubation, cells were incubated overnight with primary antibodies against p53(CM-5) and CREB1(D-4) at 4°C (see supplementary materials and methods). The cells were subsequently incubated with PLA plus and minus probes for mouse and rabbit and incubated with ligation-ligase solution for 60 min at 37 °C followed by amplification-polymerase solution. Slide were mounted with DAPI solution and images were obtained using a confocal microscope (Nikon A1). Fluorescent signals per cell were enumerated in four random fields (60X magnification) and normalized with total cell number by ImageJ software (NIH).
Immunoprecipitation and Immunoblotting.
Techniques performed as previously described(39). For immunoprecipitation, cells were lysed with RIPA buffer containing protease and phosphatase inhibitors (Santa Cruz). p53 (clone DO-1, Santa Cruz) and CREB-1(D-4) primary antibodies (supplementary materials and methods) were bound to 40 μl of Protein A/G plus-Agarose (Santa Cruz). Immunoprecipitations were performed overnight at 4°C using equal amounts of total protein and primary antibody. Beads were subsequently washed four times and heated to 95°C for 5 min in 30 μl of sample buffer. For immunoblotting, equal amounts of protein from each sample were subjected to 9.5% SDS-PAGE and transferred to a polyvinylidene membrane (Millipore). The membrane was blocked with 5% skim milk in tris-buffered saline with 0.1% Tween 20 for 1hr. and incubated with the following primary antibodies, as indicated: β-catenin (E-5), Phospho-β-Catenin (Ser33/37/Thr41), Vinculin, PARP, p53 (DO-1), FOXA1, CREB-1 (E306), Phospho-CREB (Ser133), Cyclin D1, CK1α, p44/42 MAPK (Erk1/2), Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204), AKT, phosphor-AKT, K-Ras and β-Actin primary antibodies (supplementary materials and methods). The membrane was then incubated with horseradish peroxidase–conjugated secondary antibodies (ThermoFisher), and the bands were visualized using Western Lightning Plus-ECL (PerkinElmer).
Immunohistochemistry
All mouse tissues were rinsed with PBS and immediately placed into zinc-buffered formalin (Polysciences) for fixation. PDX tumors were fixed with 10% formalin at the time of harvest from host mice. All tissues were paraffin-embedded and sectioned at 4 microns for staining and histological analysis. For staining, slides bearing paraffinized tissue sections were deparaffinized, rehydrated and antigen retrieval was performed in citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0). Sections were treated with 3% H2O2, blocked with Fc receptor blocker (Innovex) and incubated with blocking buffer (2% Goat serum for rabbit primary antibodies or 2% horse serum for mouse primary antibodies) for 1 hour. Tissue sections were then incubated overnight at 4°C with primary antibodies against p53 (DO-1 for PDX tissues; CM-5 for mouse tissues), FOXA1, β-catenin (clone E-5) and CK1α (supplementary materials and methods). Biotinylated secondary antibody kits (VECTASTAIN® ABC kit) was used to develop slides per the manufacturer’s instructions (Vector Labs). Sections were counterstained with hematoxylin and eosin. Images were visualized using automated multispectral imaging microscope (Nikon Eclipse Ci). DAB signal quantification was observed in five random fields (40X magnification) and normalized with total cell number using ImageJ software (NIH).
Immunofluorescence
Paraffin-embedded sections of tumors were deparaffinized, rehydrated and antigen retrieval performed in citrate buffer (10 mM sodium citrate, 0.05% Tween 20, pH 6.0). Slide were blocked in PBS containing 5% fish gelatin (VWR) for 1 hour. Tissue sections were incubated overnight at 4°C with primary antibodies (supplementary materials and methods). Tissue sections were then stained with secondary antibodies conjugated with Alexa Fluor 488 (1:200 dilution) or AlexaFluor 555 (1:200 dilution) (Thermo Fisher Scientific). Coverslips were mounted by mounting medium containing DAPI (Vectashield, H-2000). Images were acquired using a Nikon 80i upright wide field fluorescence microscope with Elements software. The fluorescence intensity of each fluorophore was quantified in five random fields (20X magnification) and normalized with DAPI using NIS-Elements AR4.40.00 software. The correlation of fluorescent intensity was calculated by GraphPad Prism 8.
Quantitative Real-Time PCR (RT-qPCR)
RNA was prepared using TRIzol Reagent (Invitrogen) and purified using the RNeasy mini kit (Qiagen). cDNA was synthesized using the iScript reverse transcription supermix kit (Bio-RAD). Quantitative PCR was performed with the StepOne Real-Time PCR System (Bio-RAD) and analyzed using StepOne Software v2.2.1 (Bio-RAD). mRNA levels were calculated using the ΔCt method and normalized by large ribosomal protein (RPLPO). qPCR primer sequences are listed in supplementary materials and methods.
In Vitro Drug Treatment
1×105 KPwmC cells were seeded in a 6-well plate and cultured in DMEM supplemented with 10% FBS 37°C for 24 hours. Media was then changed to DMEM serum-free medium overnight and subsequently treated with 666–15 (Tocris) at 0, 0.1, 0.3, 1 and 3μM at 37°C for 2 hours. Alternatively, cells were treated with 1μM MEK inhibitors (AZD6244, U0126, trametinib), 1μM PI3K inhibitor (pictilisib) or 10μM PKA inhibitor (H89) in 10% FBS culture medium for 30 minutes. After treatment, cells were gently washed with cold PBS twice and then collected and stored at −80°C. Western blot analysis was carried out and β-actin was used as a loading control.
Ex Vivo Tissue Slice Culture and Drug Treatment
Tissue cores were generated with disposable 3-mm biopsy punches (Integra Miltex) from PDX tumors and immediately placed in Belzer UW® Cold Storage Solution supplemented with 2% Penicillin-Streptomycin-Neomycin (PSN). Tissue cores were embedded in 1% low melting-point agarose gel (Sigma) and cut into 200 μm slices with the Krumdieck Tissue Slicer (Alabama Research and Development). The tissue slices were sequentially arrayed in 24-well plates with 400μl DMEM medium supplemented with 10% fetal bovine serum (FBS) and 2% PSN and incubated in a humidified 37°C incubator with 5% CO2. The plates were seated on a platform shaker at 150 RPM. After 2 hours incubation, tissue slices were treated with or without 666–15 (1 μM) and trametinib (1 μM) along with an additional 100ul medium, totaling 500 μl medium per well. The plates were returned to the incubator/shaker and cultured for 24 hours. Tissue slices were then harvested, and protein lysates generated.
RNA-sequencing and analysis
Murine- Raw RNA-sequencing reads were mapped to the mouse mm9 assembly reference genome using tophat and bowtie bioinformatics tools(40). The read count was generated with HTseq-count(41). Differential gene expression analysis was performed with DESeq (a R/Bioconductor package) using adjusted p-value < 0.05 as the significance cutoff(42). Pathway analysis and upstream regulatory elements were performed with Enrichr-KEGG and IPA (Ingenuity Pathway Analysis) developed by Ingenuity, Inc. Significantly differentially expressed genes were used as the input gene list for the IPA core analysis. Enriched transcription factor binding site analyses were performed with oPOSSUM-3.0. Human- Transcriptional profiling was carried out by NYGC. RNA libraries were prepared with the Agilent TruSeq Stranded kit and sequenced on the HiSeq2500, with paired end 50 bp reads at ~100M reads per sample. Raw reads were aligned to joint index (hg19+mm10) with STAR aligner. Reads unambiguously mapping to the mouse genome were filtered and unmapped and mapped reads to the human genome were aligned to hg19 using STAR aligner. Quality control of aligned reads was performed using RSeQC and Picard. Gene quantification was performed using featureCounts and gene annotation was performed using Gencode 19. RSEM was used for gene and transcript quantification. Enriched transcription factor binding site analyses were performed with oPOSSUM-3.0. Clinical information and datasets related to genetic alterations of KRAS and p53 from the TCGA PAAD cohort were acquired via cBioPortal website (https://www.cbioportal.org/). Simple somatic mutation dataset and raw read counts for the TCGA PAAD cohort were downloaded from the ICGC data portal (https://dcc.icgc.org/). RNA-Seq datasets related to gene expression values (FPKM) of 177 TCGA PAAD cohorts was downloaded from the GDC data portal website (https://portal.gdc.cancer.gov/). An unsupervised hierarchical cluster heatmap based on FPKM values was generated using the heatmap package (version 1.0.12). Differential gene expression (DGE) analysis was performed using DESeq2 package (version 3.12) on R platform (version 3.6.3). Correlation plots were generated using ggplot2 package (version 3.3.2) and correlation analyses was performed using Pearson correlation test. Boxplots and Kaplan-Meier Curve were generated and tested for statistical significance using GraphPad Prism 8 software.
Whole exome sequencing
Whole exome sequencing was carried out by the New York Genome Center (NYGC). DNA libraries were prepared with the Agilent SureSelect XT capture (51Mb, v4) and sequenced on the HiSeq2500, with paired end 125 bp reads to ~125x coverage. Data analysis was carried out the NYGC standard processing pipeline (NYGC_Somatic_Pipelines_v4.pdf).
Statistical Analysis
All data are represented as mean ± s.d. Graphpad 8.0 Prism was used to perform all statistical analyses. Significance of differences between groups was evaluated by Student’s t-test or ANOVA. p < 0.05 was considered significant. Pearson correlation coefficient was used to analyze p53 and FOXA1 expression or FOXA1 and CREB1 expression in mouse tissues.
Data and code availability
RNA-sequencing data is deposited to the Gene Expression Omnibus (GEO). The accession numbers for the RNA-seq transcriptome data for human and mouse experiments reported in this paper is GSE158221. All other data may be request from the corresponding author upon reasonable request.
Supplementary Material
Statement of Significance.
Oncogenic KRAS and mutant p53 are the most commonly mutated oncogene and tumor suppressor gene in human cancers yet direct interactions between these genetic drivers remain undefined. We identified a cooperative node between oncogenic KRAS effectors and mutant p53 that can be therapeutically targeted to undermine cooperation and mitigate metastasis.
Acknowledgements
This work was supported by NIH/NCI K08CA218690 (M.P.K.), American College of Surgeons Faculty Research Fellowship (M.P.K.), NIH P01CA117969 (M.P.K., Y.K., B.D.), NIH/NCI R01CA82577 (G.L.) and the NIH T32 CA 009599 (T.G.H.). A.M. is supported by P01CA117969. B.L. is supported by CPRIT RP17002. We acknowledge Dr. Paulucci-Holthauzen at the BSRB Microscopy Facility for training and support and the NIH shared instrumentation grant (1S10OD024976-01) for supporting the confocal microscope. The Cancer Center Support Grant (P30CA016672) supported all Core Facilities including the Flow Cytometry and Cellular Imaging Core Facility (FCCICF), the Functional Genomics Core, and the Advanced Technology Genomics Core. We acknowledge Jared Burks and Wendy Schober in the FCCICF for their assistance and support. We acknowledge the gracious support of the Richard K. Lavine Pancreatic Fund, the Ben and Rose Cole Charitable Pria Foundation, and the Skip Viragh Foundation for their generous support and dedication to pancreatic cancer research. Images were created with BioRender.com.
Footnotes
Disclosure statement: The authors declare no potential conflicts of interest.
References
- 1.Ozdemir BC, Pentcheva-Hoang T, Carstens JL, Zheng X, Wu CC, Simpson TR, et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell 2015;28(6):831–3 doi 10.1016/j.ccell.2015.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012;148(1–2):349–61 doi 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rhim AD, Oberstein PE, Thomas DH, Mirek ET, Palermo CF, Sastra SA, et al. Stromal elements act to restrain, rather than support, pancreatic ductal adenocarcinoma. Cancer Cell 2014;25(6):735–47 doi 10.1016/j.ccr.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aguirre AJ, Bardeesy N, Sinha M, Lopez L, Tuveson DA, Horner J, et al. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev 2003;17(24):3112–26 doi 10.1101/gad.1158703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hingorani SR, Petricoin EF, Maitra A, Rajapakse V, King C, Jacobetz MA, et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003;4(6):437–50. [DOI] [PubMed] [Google Scholar]
- 6.Muzumdar MD, Dorans KJ, Chung KM, Robbins R, Tammela T, Gocheva V, et al. Clonal dynamics following p53 loss of heterozygosity in Kras-driven cancers. Nat Commun 2016;7:12685 doi 10.1038/ncomms12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Connor AA, Denroche RE, Jang GH, Lemire M, Zhang A, Chan-Seng-Yue M, et al. Integration of Genomic and Transcriptional Features in Pancreatic Cancer Reveals Increased Cell Cycle Progression in Metastases. Cancer Cell 2019;35(2):267–82 e7 doi 10.1016/j.ccell.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cancer Genome Atlas Research Network. Electronic address aadhe, Cancer Genome Atlas Research N. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell 2017;32(2):185–203 e13 doi 10.1016/j.ccell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani SR, Wang L, Multani AS, Combs C, Deramaudt TB, Hruban RH, et al. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005;7(5):469–83 doi 10.1016/j.ccr.2005.04.023. [DOI] [PubMed] [Google Scholar]
- 10.Morton JP, Timpson P, Karim SA, Ridgway RA, Athineos D, Doyle B, et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc Natl Acad Sci U S A 2010;107(1):246–51 doi 10.1073/pnas.0908428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weissmueller S, Manchado E, Saborowski M, Morris JPt, Wagenblast E, Davis CA, et al. Mutant p53 drives pancreatic cancer metastasis through cell-autonomous PDGF receptor beta signaling. Cell 2014;157(2):382–94 doi 10.1016/j.cell.2014.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Olive KP, Tuveson DA, Ruhe ZC, Yin B, Willis NA, Bronson RT, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 2004;119(6):847–60 doi 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Hill R, Song YR, Cardiff RD, Van Dyke T. Selective evolution of stromal mesenchyme with p53 loss in response to epithelial tumorigenesis. Cell 2005;123(6):1001–11 doi 10.1016/j.cell.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 14.Kalluri R The biology and function of fibroblasts in cancer. Nat Rev Cancer 2016;16(9):582–98 doi 10.1038/nrc.2016.73. [DOI] [PubMed] [Google Scholar]
- 15.Saison-Ridinger M, DelGiorno KE, Zhang T, Kraus A, French R, Jaquish D, et al. Reprogramming pancreatic stellate cells via p53 activation: A putative target for pancreatic cancer therapy. PLoS One 2017;12(12):e0189051 doi 10.1371/journal.pone.0189051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y, Xiong S, Liu B, Pant V, Celii F, Chau G, et al. Somatic Trp53 mutations differentially drive breast cancer and evolution of metastases. Nat Commun 2018;9(1):3953 doi 10.1038/s41467-018-06146-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science 2008;321(5897):1801–6 doi 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353–D61 doi 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28(1):27–30 doi 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adams EJ, Karthaus WR, Hoover E, Liu D, Gruet A, Zhang Z, et al. FOXA1 mutations alter pioneering activity, differentiation and prostate cancer phenotypes. Nature 2019;571(7765):408–12 doi 10.1038/s41586-019-1318-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell 2002;9(2):279–89 doi 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 22.Roe JS, Hwang CI, Somerville TDD, Milazzo JP, Lee EJ, Da Silva B, et al. Enhancer Reprogramming Promotes Pancreatic Cancer Metastasis. Cell 2017;170(5):875–88 e20 doi 10.1016/j.cell.2017.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang C, Wang L, Wu D, Chen H, Chen Z, Thomas-Ahner JM, et al. Definition of a FoxA1 Cistrome that is crucial for G1 to S-phase cell-cycle transit in castration-resistant prostate cancer. Cancer Res 2011;71(21):6738–48 doi 10.1158/0008-5472.CAN-11-1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science 1999;286(5443):1358–62 doi 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 25.Du K, Montminy M. CREB is a regulatory target for the protein kinase Akt/PKB. J Biol Chem 1998;273(49):32377–9 doi 10.1074/jbc.273.49.32377. [DOI] [PubMed] [Google Scholar]
- 26.Xing J, Ginty DD, Greenberg ME. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 1996;273(5277):959–63 doi 10.1126/science.273.5277.959. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee A, Dhar N, Stathos M, Schaffer DV, Kane RS. Understanding How Wnt Influences Destruction Complex Activity and beta-Catenin Dynamics. iScience 2018;6:13–21 doi 10.1016/j.isci.2018.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell 2017;169(6):985–99 doi 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 29.Xie F, Li BX, Kassenbrock A, Xue C, Wang X, Qian DZ, et al. Identification of a Potent Inhibitor of CREB-Mediated Gene Transcription with Efficacious in Vivo Anticancer Activity. J Med Chem 2015;58(12):5075–87 doi 10.1021/acs.jmedchem.5b00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li BX, Gardner R, Xue C, Qian DZ, Xie F, Thomas G, et al. Systemic Inhibition of CREB is Well-tolerated in vivo. Sci Rep 2016;6:34513 doi 10.1038/srep34513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roife D, Dai B, Kang Y, Perez MVR, Pratt M, Li X, et al. Ex Vivo Testing of Patient-Derived Xenografts Mirrors the Clinical Outcome of Patients with Pancreatic Ductal Adenocarcinoma. Clin Cancer Res 2016;22(24):6021–30 doi 10.1158/1078-0432.CCR-15-2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kastenhuber ER, Lowe SW. Putting p53 in Context. Cell 2017;170(6):1062–78 doi 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 2014;25(3):304–17 doi 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, et al. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev 2001;15(24):3243–8 doi 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marino S, Vooijs M, van Der Gulden H, Jonkers J, Berns A. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev 2000;14(8):994–1004. [PMC free article] [PubMed] [Google Scholar]
- 36.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010;13(1):133–40 doi 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, Truty MA, Kang Y, Chopin-Laly X, Zhang R, Roife D, et al. Extracellular lumican inhibits pancreatic cancer cell growth and is associated with prolonged survival after surgery. Clin Cancer Res 2014;20(24):6529–40 doi 10.1158/1078-0432.CCR-14-0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong S, Tu H, Kollareddy M, Pant V, Li Q, Zhang Y, et al. Pla2g16 phospholipase mediates gain-of-function activities of mutant p53. Proc Natl Acad Sci U S A 2014;111(30):11145–50 doi 10.1073/pnas.1404139111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chou RH, Wang YN, Hsieh YH, Li LY, Xia W, Chang WC, et al. EGFR modulates DNA synthesis and repair through Tyr phosphorylation of histone H4. Dev Cell 2014;30(2):224–37 doi 10.1016/j.devcel.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 2012;7(3):562–78 doi 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 2015;31(2):166–9 doi 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol 2010;11(10):R106 doi 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-sequencing data is deposited to the Gene Expression Omnibus (GEO). The accession numbers for the RNA-seq transcriptome data for human and mouse experiments reported in this paper is GSE158221. All other data may be request from the corresponding author upon reasonable request.