Abstract
Purpose:
Ovarian cancer represents a major clinical hurdle for immune checkpoint blockade (ICB), with reported low patient response rates. We found that the immune checkpoint ligand PD-L2 is robustly expressed in patient samples of ovarian cancers and other malignancies exhibiting suboptimal response to ICB but not in cancers that are ICB sensitive. Therefore, we hypothesize that PD-L2 can facilitate immune escape from ICB through incomplete blockade of the PD-1 signaling pathway.
Experimental Design:
We engineered a soluble form of the PD-1 receptor (sPD-1) capable of binding and neutralizing both PD-L2 and PD-L1 with ×200 and ×10,000 folds improvement in binding affinity over wild-type PD-1 leading to superior inhibition of ligand-mediated PD-1 activities.
Results:
Both in vitro and in vivo analyses performed in this study demonstrated that the high-affinity sPD-1 molecule is superior at blocking both PD-L1– and PD-L2–mediated immune evasion and reducing tumor growth in immune-competent murine models of ovarian cancer.
Conclusions:
The data presented in this study provide justification for using a dual targeting, high-affinity sPD-1 receptor as an alternative to PD-1 or PD-L1 therapeutic antibodies for achieving superior therapeutic efficacy in cancers expressing both PD-L2 and PD-L1.
Translational Relevance.
Ovarian cancer is known to respond poorly to immune checkpoint blockade (ICB), with clinical trials reporting responses ranging from 6% to 22%. In this study, we found that the immune checkpoint ligand PD-L2 is associated with poor clinical response toward PD-1 inhibitors in ovarian cancer. By engineering the extracellular domain of PD-1, we developed a soluble PD-1 (sPD-1)-based fusion protein having superior binding affinities to both PD-L2 and PD-L1, relative to the wild-type receptor. Because improvement in the response rate of patients with ovarian cancer treated with ICB inhibitors is needed, the data presented in this study provide a preclinical justification for using engineered high-affinity sPD-1 as an alternative to PD-1 or PD-L1 therapeutic antibodies in cancers expressing both PD-L2 and PD-L1.
Introduction
Therapeutic inhibition of the PD-1 signaling pathway has proven to be an effective strategy in treating malignant diseases, showing unprecedented long-term durable responses in the clinic (1–3). In recent years, therapeutic antibodies against PD-1 and PD-L1 were approved for the treatment of various cancer types either as monotherapies or in combination with standard-of-care therapeutic regimens (4, 5). However, clinical responses vary significantly between patients and are thought to reflect the heterogeneous nature of individual tumors and their microenvironment (4, 6, 7). Ovarian cancer is known to respond poorly to immune checkpoint blockade (ICB) with clinical trials reporting responses ranging from 6% to 22% (8, 9). Clearly, there is a knowledge gap between our current understanding of immune regulation and how it can be manipulated to enhance the clinical efficacy of ICB therapies in ovarian cancer. Ongoing efforts to improve the response rate of ICB inhibitors are being pursued in the clinic by combining these inhibitors with cytotoxic drugs, targeted therapies, and radiation treatment (10–12). Despite these efforts, responses to ICB in ovarian cancer remain poor, even in combination settings (9), resulting in a paucity of ICB-approved therapies for ovarian cancer.
In contrast to the low response to ICB therapy, high-grade serous ovarian cancer was one of the first human cancers to have demonstrated a relationship between increased tumor-infiltrating lymphocytes (TIL) and improved patient survival (8, 13). Therefore, a major challenge remains to boost the clinical efficacy of ICB in ovarian cancers which possess low mutational burdens and are surrounded by an immunosuppressive tumor microenvironment (TME) comprised of regulatory T cells, macrophages, and PD-1 signaling (14). Strategies to overcome the immunosuppressive TME by antagonizing the PD-1 signaling pathway should facilitate enhanced TIL infiltration and activity, and enhance tumor control of ovarian cancer.
Mechanistically, most studies have exclusively focused on the role of PD-L1 on PD-1 signaling and have overlooked the consequences of PD-L2–mediated PD-1 activation in cancer. A second ligand to PD-1, PD-L2 is generally expressed at low levels on dendritic cells (DC), macrophages, and endothelial cells, suggesting that secreted factors in the TME upregulate PD-L2 expression (15, 16). For example, stimulation by pro-inflammatory cytokines such as IL4 can induce PD-L2 expression on both immune and stromal cells (17, 18). In addition, dysregulation of NFκB and STAT6 signaling can also increase PD-L2 expression on immune infiltrates (19).
Structural analysis suggests that PD-1 mechanistically engages its two ligands differently; binding to PD-L1 involves a conformational change, whereas binding to PD-L2 does not (20). This more direct mode of binding to PD-L2, where structural rearrangement is not required, is likely the molecular basis for the observed 6- to 10-fold stronger affinity relative to the PD-1/PD-L1 interaction (21). These differential affinities would drive PD-1 to preferentially engage with PD-L2 in the TME, providing a potential competitive advantage over therapeutic antibodies targeting PD-L1 or the PD-1 receptor itself.
In this study, we investigated the expression of PD-L2 and PD-L1 in ovarian cancer stratified according to tumor grade. To interrogate the importance of high-affinity PD-L2 binding to PD-1, we engineered a soluble form of the PD-1 receptor to have enhanced binding affinity to both PD-L1 and PD-L2. The resulting sPD-1–mutant clone has stronger affinity for PD-L1 (10,000-fold) and PD-L2 (200-fold) when compared with the wild-type interactions. We performed computational analysis of mutations identified from the combinatorial engineering efforts to demonstrate how changes both proximal and distal to the binding interface can modulate affinity. We determined that the enhancement in binding affinity translated into improved inhibition of PD-1 receptor activity in both in vitro and in vivo ovarian cancer models, and is superior to antibodies targeting PD-L1 and PD-L2. Taken together, these studies indicate that PD-L2 plays a critical role in promoting an immuno-suppressive TME that can be overcome with the use of a high-affinity sPD-1 mutant.
Materials and Methods
Study design
This study was designed to characterize structural, biochemical, functional, and therapeutic efficacy of sPD-1 with high binding affinity to both PD-L1 and PD-L2 both experimentally and therapeutically. All tumor microarray specimens were commercially purchased from US Biomax. A board-certified veterinarian pathologist performed the IHC scoring of the stained tumor microarray specimens. In vivo studies were conducted under the approval of Institutional Animal Care and Use Committee (IACUC) at Stanford University (Stanford, CA) and ChemPartner, Shanghai, P.R. China. Sample sizes for animal studies were determined on the basis of power calculations done on similar in vivo studies in previous studies. All animals were randomly assigned to treatment groups. Samples were not excluded from studies except for animals that required early termination due to illness that is unrelated to the study. Endpoints of experiments were defined in advance for each experiment. Tumor growth curves were presented for studies where tumor growth was measurable and Kaplan–Meier curves were used for orthotopic ovarian cancer models. Appropriate statistical analysis was used for each experimental study.
Cell lines
MC38, MC38 CRISPR cells, ID8, ID8 CRISPR cells, B16/OVA, MC38-hPD-L1, MC38-hPD-L2, Hep3B-hPD-L1, Hep3B-hPD-L2, and UPK10 cells were maintained in DMEM supplemented with 10% FBS and 1% antibiotics in a humidified 37°C, 5% CO2 incubator. Cells were trypsinized and passaged at 80% confluency. MC38- and Hep3B-derived cell lines were commercially obtained and authenticated through ChemPartner, Shanghai, P.R. China, before the commencement of the studies. Early passages of ID8 and UPK10 cell lines were obtained from Erinn Rankin and were authenticated by LabCorp in March 2021. PD-L1 (catalog no. sc-425636) and PD-L2 (catalog no. sc-425483) CRISPR KO construct were purchased through Santa Cruz Biotechnology and performed according to manufacturer's protocol.
In vivo tumor studies
All animal experiments were reviewed and approved by the Institutional Animal Care and Use Committee at Stanford University (Stanford, CA) and Animal Ethical Committee at ChemPartner, Shanghai. PD-L1 knockout (KO) mice on C57BL/6 background were kindly gifted by Dean W. Felsher (Stanford University, Stanford, CA). Female mice age 6–8 weeks were used for ID8 ovarian tumor studies. Mice were housed in a pathogen-free animal facility, and kept under constant temperature and humidity and controlled 12 hours light-dark cycles. For ID8 and UPK10 ovarian studies including ID8 PD-L1 CRISPR KO, PD-L2 CRISPR KO, and PD-L1/L2 CRISPR KO cells, 5 × 106 or 20 × 106cells were injected intraperitoneally or subcutaneously; animals were terminated upon the development of peritoneal ascites for survival analysis or once subcutaneous tumors reached ethical termination point. For all ID8 tumor growth studies, ID8 cells were suspended in 50% Matrigel (#356230) and injected subcutaneously.
Statistical analysis
The Pearson correlation was used for all correlation analysis of tumor specimens. IHC H score values were determined by a board-certified veterinarian pathologist. All tumor volume, survival, and quantification of in vivo IHC were conducted using GraphPad Prism software (GraphPad Software Inc). ANOVA with Tukey–Kramer test was used for comparing multiple treatment groups. P < 0.05 was considered significant. Repeated measure ANOVA was used for comparing multiple treatment groups measured over time. Statistical analysis of survival curves was conducted for the ID8 survival studies. A log-rank (Mantel–Cox) test was performed to compare mean survival among groups; P ≤ 0.05 was considered statistically significant.
Additional material and methods are available in the Supplementary Data.
Results
PD-L2 is abundantly expressed in human ovarian cancers, but not in ICB-sensitive bladder cancers
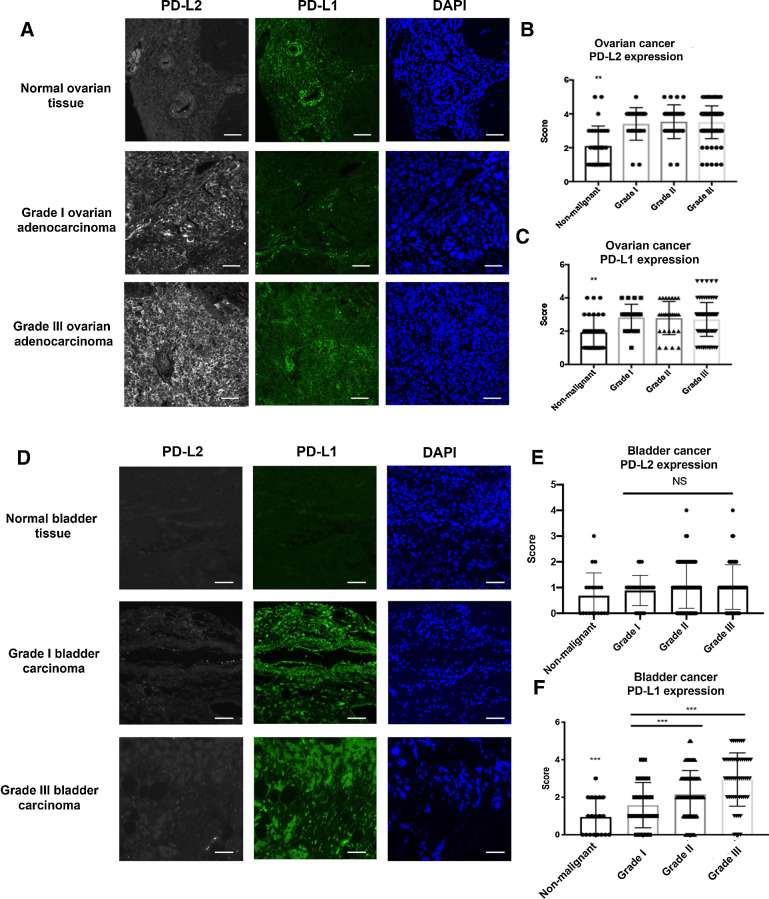
Although PD-L1 and PD-1 expression has been the subject of extensive study, less is known about the role of PD-L2 expression in cancer types that respond poorly to αPD-1 or αPD-L1 therapies (22, 23). To elucidate the clinical relevance of PD-L1 and PD-L2 expression in ovarian cancer, historically known to have a suboptimal clinical response to ICB, we analyzed the expression of both molecules in human cancer tissue microarrays (TMA; n = 156; Fig. 1A). The specificity of αPD-L1 and αPD-L2 antibodies was validated in human tonsillar tissue (Supplementary Fig. S1A). The spatial distribution and staining intensity of PD-L1 and PD-L2 expression were quantified and stratified according to tumor grades. In patients with ovarian cancer, the expression of both PD-L2 and PD-L1 are significantly elevated in the cancerous tissue compared with non-malignant specimens, but did not appear to change significantly with tumor grade (Fig. 1B and C). While Pearson correlation analysis showed a positive correlation between PD-L1 and PD-L2 expression in ovarian cancer specimens (Supplementary Fig. S1B), the expression pattern between PD-L1 and PD-L2 was nonoverlapping. In addition, we evaluated the same set of ovarian tumor microarrays with αPD-L1 and αPD-L2 antibodies that have been previously published as a means of checking the specificity of the expression (24, 25). In this comparison of antibodies for PD-L1 and PD-L2, we found similar staining patterns (Supplementary Figs. S2A and S2B) and scoring outcomes (Supplementary Figs. S2C–S2F), supporting the specificity and reliability of αPD-L1 and αPD-L2 antibodies used in this study. To evaluate whether high PD-L2 expression is a potential biomarker for low ICB responding cancer types, we also examined PD-L1 and PD-L2 expression in other cancers reported to have suboptimal clinical responses to ICB inhibitors (26–28) such as esophageal cancer (n = 72), gastric cancer (n = 76), and glioblastoma (n = 152). Similarly, high PD-L2 expression was also observed in tumor samples but not in non-malignant tissues (Supplementary Figs. S1C–S1E). In contrast, bladder cancer is an ICB-sensitive tumor type (29). Interestingly, the staining of bladder cancer TMA showed low PD-L2 staining, but robust PD-L1 ligand expression (n = 208; Fig. 1D–F). Furthermore, we surveyed a cohort of normal organ tissue to characterize the normal expression profile of PD-L1 and PD-L2. We found little overlap between PD-L1 and PD-L2 expression in different types of normal tissues (Supplementary Fig. S2G). The highest PD-L1 expression is found in spleen and stomach. Modest to low PD-L1 expression is observed in prostate, esophagus, liver, testis, small intestine, skin, bone, pancreas, rectum, and lung. No PD-L1 expression was detected in lymph node, thymus, cervix, or colon (Supplementary Fig. S2H). While PD-L2 expression was not found to be as ubiquitously expressed, high levels of PD-L2 expression were observed in lymph nodes, thymus, and testis, and low levels of expression were detected in small intestine, with minimal expression detected in other tissues (Supplementary Fig. S2G). We also co-stained normal tissue samples for PD-L1 and PD-L2 in EpCAM-positive cells (Supplementary Fig. S2H). Overall, expression of PD-L1 and PD-L2 shows distinct expression patterns in normal tissue and PD-L2 expression is more restricted in normal tissue.
Figure 1.
PD-L2 and PD-L1 expression in ovarian and bladder cancer. A, Representative images of ovarian cancer tissue microarray containing both normal and malignant samples (N = 156) stained with anti-human PD-L2 (gray), anti-human PD-L1 (green), and DAPI (blue) by fluorescent IHC. Scale bar 50 μm. The intensity of PD-L2 (B) and PD-L1 (C) were scored and stratified according to tumor grades. D, Bladder cancer tissue microarray was stained with anti-human PD-L2 (gray), anti-human PD-L1 (green), and DAPI (blue) by fluorescent IHC with representative images shown (N = 208). Scale bar 50 μm. E, PD-L2 expression in bladder cancer TMAs were scored and quantified according to tumor grade. F, PD-L1 expression in bladder cancer TMAs were scored and quantified according to tumor grade. Quantification data were plotted with mean and SD calculated. One-way ANOVA was used for analysis comparing the tumor grades. *, P < 0.05; **, P < 0.01; and ***, P < 0.001.
In summary, we found tumors that are poor responders to ICB has elevated levels of both PD-L1 and PD-L2, while ICB-sensitive tumors almost exclusively express PD-L1. On the basis of these observations, we hypothesize that resistance to ICB is in part attributed to elevated expression of both PD-L1 and PD-L2, thus inhibition of PD-1 signaling through blocking both PD-L1 and PD-L2 are required to enable effective ICB suppression.
Generation of soluble PD-1 decoy receptor with enhanced binding to PD-L1 and PD-L2
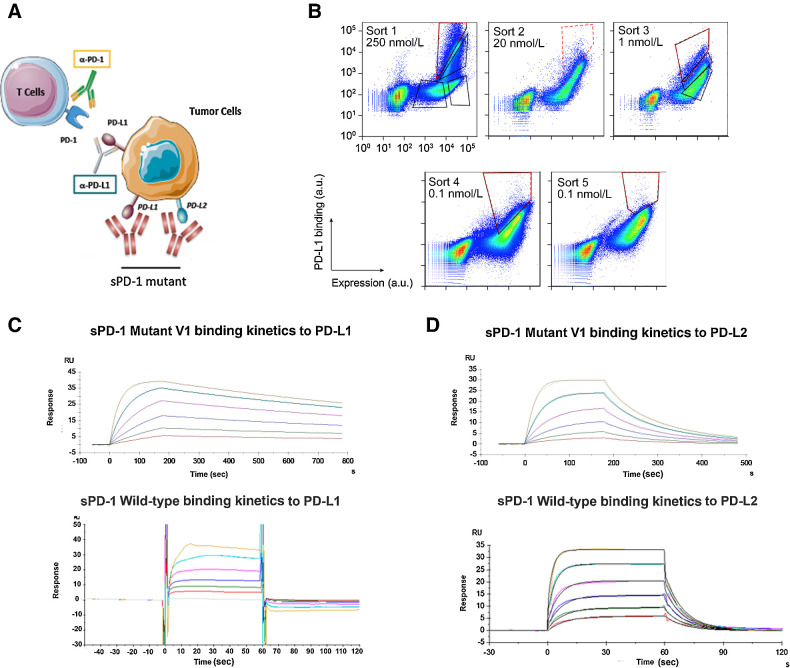
ICB is achieved by inhibiting the activation of PD-1 through the use of specific blocking antibodies. However, current avaliable PD-1 therapeutic antibodies are not optimized to prevent the interaction between PD-1 and PD-L2. Here, we propose to completely antagonize PD-1 signaling through generating an engineered soluble PD-1 decoy receptor (sPD-1). This approach exploits the native differences in binding affinities PD-1 has for its ligands, offers the ability to inhibit both interactions, and avoids chronically engaging T cells directly (Fig. 2A; ref. 30).
Figure 2.
Engineering sPD-1 mutants with superior binding affinity to PD-L1 and PD-L2. A, Conceptual illustration of sPD-1 mutant inhibiting the PD-1 signaling. B, Representative flow cytometry dot plots showing clone selection pressure and gating strategies used for isolating high-affinity PD-L1 binding clones. Clones with the strongest binding to PD-L1 were selected and stringency was increased across sort rounds by decreasing the concentration of PD-L1 used to label the library. Gates used to collect the top 1% to 3% of the library are shown (red, dashed line). C, Binding kinetics of sPD-1 mutant version 1 (top) and wild-type PD-1 (bottom) binding to human PD-L1 was determined by surface plasmon resonance system. Each curve represents a single concentration of the analyte. D, Binding kinetics of sPD-1 mutant version 1 (top) and wild-type PD-1 (bottom) binding to human PD-L2 was determined by surface plasmon resonance system. Each curve represents a single concentration of the analyte.
To develop a high-affinity sPD-1 decoy receptor capable of neutralizing both PD-L1 and PD-L2, mutations were randomly introduced into the wild-type PD-1 gene using error-prone PCR as decribed previously (31, 32). The resulting library was displayed on the yeast cell surface and sorted by FACS to isolate clones with enhanced binding to hPD-L1 Fc. After five rounds of sorting, the enriched library displayed improved binding to hPD-L1 Fc (Fig. 2B). Sequence analysis of the sort five products revealed the emergence of several concensus mutations, most notably A132V (Supplementary Table S1). Despite the fact that significant diversity remained, to combinatorially explore the sequence space encompassed by the enriched mutations, sequences from sort five were shuffled and the wild-type PD-1 gene containing the A132V point mutation was added to the shuffle to help dilute out any neutral mutations. To increase stringency, monomeric hPD-L1 was used in place of hPD-L1 Fc, and a combination of equilibrium binding and kinetic off-rate sorts were employed to further impart selective pressure. Following six rounds of sorting, sequence analysis showed strong convergence at seven positions (Supplementary Table S2; Supplementary Fig. S3A), suggesting this core set of mutations was driving the observed improvement in binding to hPD-L1.
To characterize the engineered variants further, a sequence containing the concensus mutations was made recombinantly (sPD-1V1). Interestingly, one of the mutations, N116S, removes an N-glycosylation site so a second variant (sPD-1V2) was made without this mutation in an effort to preserve the native posttranslational modifications. The sPD-1 variants were fused to the Fc domain of human IgG4 to improve plasma exposure, reduce the rate of renal clearance, and increase protein stability and solubility. The S228P mutation was included in the Fc backbone to stabilize the hinge region and prevent Fab-arm exchange (33). The affinities of both sPD-1V1 and sPD-1V2 to human PD-L1 and PD-L2 were quantitatively measured using surface plasmon resonance. sPD-1V1 displayed an approximately 10,000-fold and 200-fold improvement in binding to hPD-L1 and hPD-L2, respectively, compared with wild-type PD-1 (Table 1). Kinetic analysis of the binding revealed that a majority of the improvement was due to a slower dissociation rate (koff; Fig. 2C and D). Importantly, sPD-1V2 showed similar binding, suggesting that removal of the glycosylation at N116 has little impact on binding (Supplementary Figs. S3B and S3C).
Table 1.
The apparent binding affinity between sPD-1 wild-type Fc, sPD-1V1 Fc, and sPD-1V2 Fc in co-complex with human PD-L1 and PD-L2.
| Analyte | Ligand | ka (1/ms) | kd (1/second) | Kd (mol/L) |
|---|---|---|---|---|
| Human PD-L1 | sPD-1 WT | N/A | N/A | 2.743E-6 |
| sPD-1V1 | 2.038E+6 | 6.941E-4 | 3.406E-10 | |
| sPD-1V2 | 2.139E+6 | 7.500E-4 | 3.420E-10 | |
| Human PD-L2 | sPD-1 WT | 1.811E+5 | 0.1029 | 5.471E-7 |
| sPD-1V1 | 4.366E+6 | 0.01288 | 2.949E-9 | |
| sPD-1V2 | 4.479E+6 | 0.01524 | 3.402E-9 |
Computational based modeling simulations to identify the structural basis for high-affinity binding
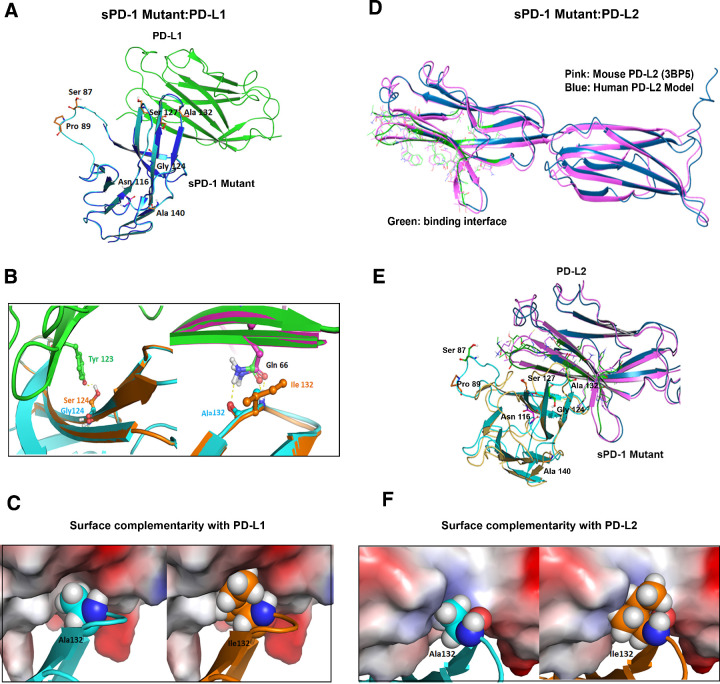
To determine the structural basis for increased binding affinity of the sPD-1 variants, we performed computational based model simulations. Wild-type human PD-1/PD-L1 complex structures (PDB ID: 4ZQK and 5IUS) were used as backbones for mapping amino acid mutations identified in the sPD-1 mutants. The sequence alignment for both hPD-1 structures no. 4ZQK and no. 5IUS is shown in Supplementary Fig. S4A. The crystal structure of sPD-1 and hPD-L1 are shown in blue and green, respectively, with the identified mutations shown in orange sticks (Fig. 3A). We identified residues G124S, S127V, and A132I as mutations within the binding interface; S87G, P89 L, and A140V are distal from the binding interface. The modeling revealed mutations G124S and A132I each make one additional hydrogen bond with Tyr123 and Gln66 of PD-L1, respectively (Fig. 3B, left and right). In addition, mutation A132I on sPD-1 resulted in an increase in the protein surface complementarity with hPD-L1 from 0.72 to 0.85 (Fig. 3C; Supplementary Fig. S4B). To further investigate how these mutations influence the overall binding between sPD-1 mutations and hPD-L1, the variation of the binding affinity and complex stability for each individual mutation were calculated and listed in Table 2. We also calculated the variation in binding affinity and complex stability for grouped mutations within and outside of the binding interface. A higher negative value is indicative of increased binding affinity and stability (Table 2). Overall, the interface mutations contribute to enhanced binding affinity and protein stability, and the mutations outside of the binding interface lead to increased stability.
Figure 3.
Computational modeling–based structural analysis of sPD-1 mutants in co-complex with human PD-L1 and PD-L2. A, Overlay of human PD-1 model in complex with human PD-L1 (PDBID:4ZQK) are shown in blue and green, respectively. The model of PD-1 with missing loop fixed is shown in cyan, residues mutated on sPD-1 are shown in orange sticks, and the N-glycosylation site (Asn116) is shown in magenta sticks. B, Left, The sPD-1 mutation G124S (orange) makes a hydrogen bond with PD-L1 Tyr123 (green and red sticks). Wild-type PD-1 structure is shown in cyan. Right, Mutation A132I makes one more hydrogen bond with PD-L1 Gln166. Wild-type PD-1 structure is shown in cyan and mutated PD-1 structure is shown in orange. Crystal PD-L1 structure is shown in pink and PD-L1 co-complex with sPD-1 mutant is shown in green. C, Comparison of surface complementarity of mutation A132I. PD-L1 binding site is marked to reveal electrostatic potential surface. Red indicates negative electrostatic potential, blue indicates positive electrostatic potential, and gray indicates hydrophobic regions. Wild-type PD-1 is shown in cyan cartoon and balls (left); mutated PD-1 is shown in orange cartoon and balls (right). D, Alignment of proposed human PD-L2 with mouse PD-L2 (PDBID: 3BP5). The interface residues are marked green within the dark blue human PD-L2 model and the crystal structure of mouse PD-L2 labeled pink. E, The overlay of proposed human PD-1 with mutations (cyan) in complex with human PD-L2 (dark blue) and mouse PD-1 (yellow) in complex with mouse PD-L2 (pink). F, Comparison of surface complementarity of mutation A132I with PD-L2 with same color annotation as C.
Table 2.
Variations within the binding affinity and protein stability (in kcal/mol) for each mutation in co-complex with hPD-L1.
| Residue | Original | Mutated | d Affinity | d Stability (solvated) |
|---|---|---|---|---|
| 87 | SER | GLY | 0.94 | 5.56 |
| 89 | PRO | LEU | 0 | −7.06 |
| 116 | ASN | SER | −0.02 | 0.82 |
| 124 | GLY | SER | −4.27 | −10.24 |
| 127 | SER | VAL | −0.12 | −1.82 |
| 132 | ALA | ILE | −10.94 | −11.9 |
| 140 | ALA | VAL | −0.77 | −25.89 |
Given that the hPD-L2 protein structure was not resolved until very recently (34), a model of the hPD-L2 was simulated based on available mPD-L2 structures. The sequence identity between hPD-L2 and mPD-L2 is 72% (Supplementary Fig. S4C). The overlay of the proposed hPD-1/PD-L2 model and murine PD-1/PD-L2 crystal structure (PDB ID: 3BP5) is shown in Fig. 3D and Supplementary Fig. S4D. The proposed co-complex structure between sPD-1 mutations with the proposed human PD-L2 is shown in Fig. 3E. Compared with the proposed wild-type hPD-1/hPD-L2 co-complex, mutation A132I created an additional hydrogen bond with PD-L2, resulting in increased surface complementarity (Fig. 3F; Supplementary Fig. S4E). The calculation of the binding affinity variation and protein stability was also performed for single and grouped mutations within and outside of the binding interface (Table 3). Importantly, the A140V mutation, in sPD-1, which lies outside of the binding interface, is a significant contributor to the overall stability of the sPD-1/PD-L1, PD-L2 co-complexes. This observation further emphasizes the importance of a nonbiased screening approach for identifying mutations outside of a binding interface for improving the overall stability of the protein–protein interaction.
Table 3.
Variations within the binding affinity and protein stability (in kcal/mol) for each mutation in co-complex with hPD-L2.
| Residue | Original | Mutated | d Affinity | d Stability (solvated) |
|---|---|---|---|---|
| 87 | SER | GLY | 0 | −0.68 |
| 89 | PRO | LEU | 0.28 | −4.71 |
| 116 | ASN | SER | 0 | 0.85 |
| 124 | GLY | SER | −3.06 | 6.47 |
| 127 | SER | VAL | 1.65 | −4.42 |
| 132 | ALA | ILE | −9.47 | −0.55 |
| 140 | ALA | VAL | 0.25 | −11.76 |
sPD-1 mutants disrupt PD-L1 and PD-L2 binding to PD-1, resulting in T-cell activation
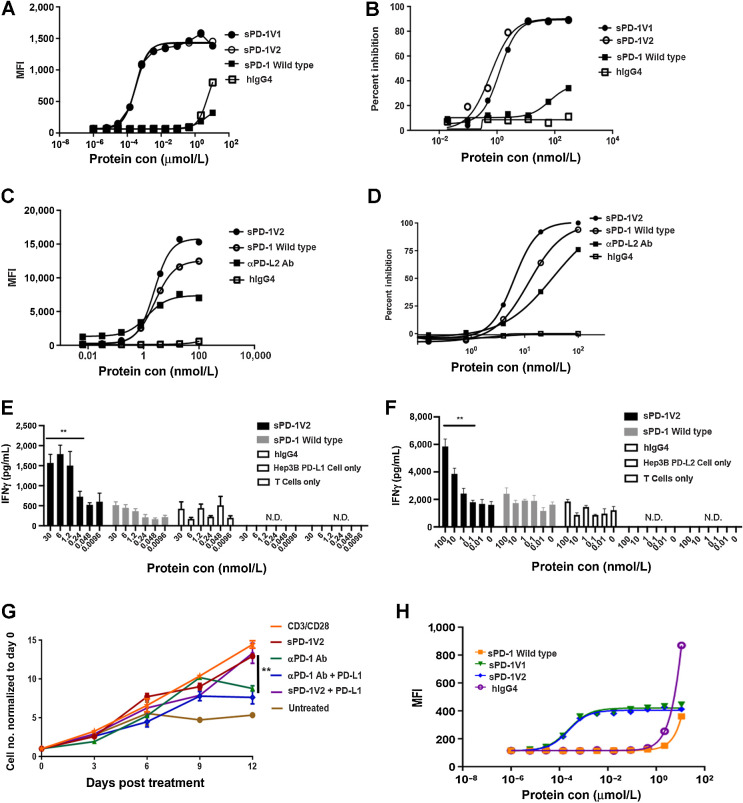
To demonstrate that the sPD-1 mutants can disrupt the interactions between human PD-L1 or PD-L2 and human PD-1 at the cellular level, we assessed the binding activity of sPD-1 mutants to PD-L1 in a murine cell line with a knock-in human PD-L1. Because our attempts to generate a knock-in hPD-L1 in murine ovarian cancer–derived ID8 and UPK10 cells were not successful, we performed our studies in murine MC38 hPD-L1 knock-in cells (Supplementary Fig. S5C). In addition, we also generated human Hep3B cells stably overexpressing hPD-L1. In the MC38 hPD-L1 knock-in cells, wild-type sPD-1 binds to hPD-L1 with low affinity and minimal mean fluorescent intensity (MFI) signal was detected up to 10 μmol/L (Fig. 4A). In contrast, similar binding affinities for both sPD-1V1 and sPD-1V2 to hPD-L1 with similar magnitudes of enhancement, further supporting the conclusion that the mutation of Asn116 is not critical for the enhancement of binding. To further examine the ability of sPD-1 mutants to block binding between endogenously expressed hPD-L1 and PD-1, we performed receptor blocking assays using sPD-1V1 and sPD-1V2, wild-type sPD-1, and hIgG4 controls. Both sPD-1 mutants showed up to 90% inhibition of PD-L1–mediated receptor binding toward PD-1 when compared with wild-type sPD-1 and hIgG4 (Fig. 4B). To demonstrate that the sPD-1 mutant is capable of binding to PD-L2 and blocking PD-L2 mediated PD-1 activation, we overexpressed human PD-L2 in MC38 cells (MC38-hPD-L2) and human hepatocellular carcinoma Hep3B cells (Hep3B-hPD-L2) to generate stable cell lines (Supplementary Fig. S5B). Because little difference in binding affinity was detected between sPD-1V1 and sPD-1V2, we decided to focus on sPD-1V2 (without the Asn116 mutation) for the rest of our studies. The sPD-1V2 exhibits strongest binding toward endogenous hPD-L2–expressing MC38 cells compared with wild-type sPD-1 or αPD-L2 antibody (25% and 50% reduction in MFI signal, respectively) at the same concentrations (Fig. 4C). The sPD-1V2 mutant also showed enhanced ability to block PD-L2–mediated activation of PD-1 in receptor blocking assays (Fig. 4D). The binding analysis was also repeated in Hep3B-hPD-L2 cell with similar outcomes (Supplementary Fig. S5A). We then investigated whether the sPD-1V2 can functionally inhibit PD-1 signaling in activated T cells. Human peripheral blood mononuclear cells (PBMC) were collected from healthy donors and incubated with Hep3B-hPD-L1 or Hep3B-hPD-L2 cells. The effect of increasing concentrations of wild-type PD-1, sPD-1V2, and hIgG4 on T-cell activity was assessed by measuring INFγ secretion. We found that sPD-1V2 increased INFγ in a concentration-dependent manner by blocking endogenous PD-L1 and PD-L2 binding to PD-1 (Fig. 4E and F). In addition, we observed that prolonged direct stimulation of T-cell activity through inhibition of PD-1 receptors by αPD-1 antibodies can potentially affect T-cell viability over time (Fig. 4G). We hypothesized that sPD-1V2 could overcome this shortcoming by targeting cancer cells expressing high levels of PD-L1 and PD-L2 but not directly interacting with T cells as a possible solution to preserve T-cell longevity and functionality. To test this hypothesis, activated human T cells (stimulated with CD3 and CD28) were treated with either αPD-1 antibody or sPD-1V2 in the presence or absence of PD-L1. Continuous treatment with αPD-1, but not sPD-1V2, inhibited CD3/CD28 stimulated T-cell proliferation (Fig. 4G). These observations suggest that sPD-1V2 is capable of binding and blocking PD-L1 and PD-L2 binding to PD-1, resulting in stimulation of T-cell activity while preserving T-cell viability and functionality. Because the PD-1 molecule is well conserved between murine and human genomes and human PD-1 is known to interact with mouse PD-L1 (35), we validated the binding of sPD-1V2 to mouse PD-L1 in wild-type parental MC38 cells, which only express mouse PD-L1 and PD-L2. An approximate 4-fold increase in binding signal was detected with sPD-1V2 compared with wild-type sPD-1 (Fig. 4H). This species cross-reactivity is important as it allows us to test the sPD-1V2 in a syngenic manner in murine models of malignancies.
Figure 4.
sPD-1 mutants demonstrated superior capability in blocking PD-L1– and PD-L2–mediated activities in a ligand-dependent manner without affecting T-cell viability. A, FACS-based binding analysis of wild-type PD-1, hIgG4, and sPD-1V1 and V2 to MC38 cells with human PD-L1 knock-in (MC38-hPD-L1). A–D are all presented as one of the two independent experiments shown here with individual points representing the mean of two technical repeats. B, Cell-based receptor-blocking assay showing inhibition of Hep3B-hPD-L1 binding to biotin conjugated PD-1 wild-type in competition with sPD-1 wild-type, hIgG4, and sPD-1 mutants. C, Binding of wild-type PD-1, αPD-L2 antibodies (Ab), hIgG4, and sPD-1V2 to MC38-hPD-L2. D, Cell-based receptor-blocking assay showing inhibition of Hep3B-hPD-L2 binding to biotin conjugated PD-1 wild-type in competition with sPD-1 wild-type, αPD-L2 antibody, hIgG4, and sPD-1V2. E, T-cell activation in the presence of Hep3B-hPD-L1 cells when incubated with sPD-1V2, sPD-1 wild-type, hIgG4 control, Hep3B-PD-L1 cell only, and T cell only control. IFNγ levels measured as a marker of T-cell activation. Error bars represent the mean and SD of technical triplicate. Experiment was conducted twice with PBMCs isolated from different donors. F, T-cell activation in the presence of Hep3B-hPD-L2 cells upon incubation with sPD-1V2, sPD-1 wild-type, and hIgG4, Hep3B-PD-L2 cell only and T cell only control. T-cell activity is measured by IFNγ level. Error bars represent the mean and SD of technical triplicate. Experiment was conducted twice independently with PBMC isolated from different donors. G, T-cell proliferation over-time in the presence of sPD-1V2 and αPD-1 antibody with or without hPD-L1 added. H, Binding kinetics between sPD-1V2 and mouse PD-L1 in MC38 parental cells. Each data point represents the mean and SD of technical duplicate. Experiment was repeated with T cells isolated from a second donor. Statistical analysis was conducted with one-way ANOVA for comparing between treatment groups and repeated ANOVA for changes over time. *, P < 0.05; **, P < 0.01.
sPD-1V2 demonstrates strong antitumor efficacy in syngenic ovarian and other murine cancer models
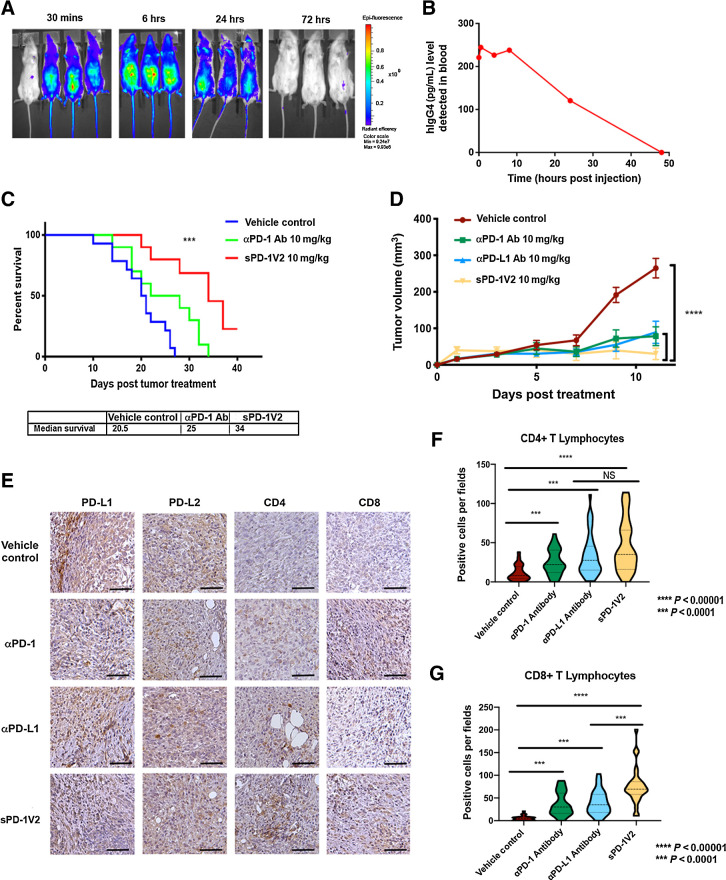
The therapeutic potency of sPD-1V2 was evaluated in multiple syngenic mouse tumor models. To evaluate the pharmacokinetics of sPD-1V2 in vivo, a single 10 mg/kg intravenous dose of fluorescently labeled sPD-1V2 was injected into mice and imaging was done at 30 minutes, 6 hours, 24 hours, and 72 hours after treatment (Fig. 5A). To monitor sPD-1V2 levels over time, we used both changes in fluorescence as assessed by whole body imaging as well as ELISA assays to detect the human IgG4 component of sPD-1V2 (Fig. 5B). Both fluorescent images and ELISA analysis indicated that the half-life of the sPD-1V2 molecule is approximately 24 hours, providing the rationale for in vivo dosing at 10 mg/kg every 48 hours.
Figure 5.
sPD-1V2 inhibits tumor growth in mouse tumor models of ovarian cancer. A, Bio-distribution of sPD-1V2 labeled with Alexa Fluor 488 after a single dose of 10 mg/kg were imaged over time (N = 3 with 1 vehicle control). B, Biodistribution of sPD-1V2 in mouse serum after a single dose of molecule at 10 mg/kg detected with ELISA against human IgG4. Each data point represents the mean of two animals collected at the same timepoint. C, Kaplan–Meier survival plot of C57BL/6 mice orthotopically inoculated with ID8 mouse ovarian tumor cells treated with vehicle control (N = 14), anti-mouse αPD-1 blocking antibody 10 mg/kg (N = 10), and sPD-1V2 10 mg/kg (N = 10). Animals terminated upon the development of ascites. Median survival of each experimental group listed below. D, Subcutaneous UPK10 ovarian tumor growth over time in C57BL/6 mice treated with vehicle control, sPD-1V2, αPD-1 antibody, and αPD-L1 antibody (N = 7). E, The expression of PD-L1, PD-L2, CD4+, and CD8+ cells in UPK10 tumors post-treatment was analyzed by IHC staining. Scale bar 50 μm. F, Violin plot of CD4+ T lymphocyte infiltration into UPK10 ovarian tumors post treatment. G, Violin quantitative plot of CD8+ T lymphocyte infiltration into UPK10 ovarian tumors post treatment. Statistical analysis was conducted using one-way ANOVA for comparing between treatment groups and repeated ANOVA for changes over time. Kaplan–Meier estimator was calculated for survival curves. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To determine the efficacy of sPD-1V2 compared with an αPD-1 antibody, we inoculated C57BL/6 mice with ID8 mouse ovarian cancer cells intraperitoneally and treated with α-mouse PD-1 antibody or sPD-1V2 every 48 hours. Treatment with sPD-1V2 significantly prolonged overall survival compared with the vehicle control group (median survival of 34 vs. 20.5 days). The αPD-1–treated group showed an intermediate response between the control and sPD-1V2–treated groups (Fig. 5C). A subset of animals from each treatment group was sacrificed on day 20 after treatment and analyzed for PD-L1, PD-L2, CD4, and CD8 expression immunohistochemically (Supplementary Fig. S6A). Tumors in all groups stained positive for PD-L1 and PD-L2 expression, but significantly higher numbers of CD4+ and CD8+ T cells were observed infiltrating the tumors treated with sPD-1V2 compared with the vehicle control or αPD-1–treated groups (Supplementary Figs. S6B and S6C). A second syngenic murine ovarian cancer model UPK10 was tested. Animals were treated with sPD-1V2, αPD-1, or αPD-L1 antibodies. A significant reduction in tumor growth was observed in all three treated groups, but tumor reduction was most pronounced in sPD-1V2–treated animals (Fig. 5D). UPK10 tumors showed a positive correlation between antitumor activity of sPD-1V2 and increased CD4+ and CD8+ T-cell infiltration. While TILs were observed in both αPD-1– and αPD-L1–treated groups, they were elevated to a higher level in sPD-1V2–treated tumors (Fig. 5E–G). No significant changes in animal body weight were observed throughout the experiment (Supplementary Fig. S6D). The therapeutic efficacy of sPD-1V2 was also studied in additional tumor models. MC38-hPD-L1 colorectal cancer cells were inoculated subcutaneously in C57BL/6 mice and treated with sPD-1V2 or αPD-L1 therapeutic antibody (atezolizumab). A significant reduction in tumor growth was observed in both sPD-1V2 and αPD-L1 (atezolizumab) treated tumors (Supplementary Figs. S7A and S7C). Compared with αPD-L1 treatment, tumors treated with the sPD-1V2 showed superior efficacy in suppressing tumor growth and prolonged survival with no significant changes in body weight (Supplementary Figs. S7B and S7D). The ability for sPD-1V2 to inhibit PD-L2–mediated tumor growth of MC38 tumors overexpressing hPD-L2 was evaluated and compared with αPD-1 antibody pembrolizumab treatment. Significant delays in tumor growth were observed in tumor-bearing mice treated with sPD-1V2 or pembrolizumab. Although statistically not significant, there was a trend toward stronger inhibition of tumor growth in the sPD-1V2–treated group compared with the pembrolizumab-treated group (Supplementary Fig. S7E). Again, the antitumor activity of sPD-1V2 was tested in B16/OVA melanoma model and resulted in reduced tumor growth compared with vehicle-treated groups (Supplementary Fig. S7F). TILs were analyzed in mice bearing B16/OVA (Supplementary Fig. S7G) and MC38 (Supplementary Figs. S7H and S7I) tumors treated with sPD-1V2 or αPD-1 antibody. While both treatments resulted in increased infiltration of CD4+ and CD8+ cytotoxic T cells, the most significant changes were observed in the sPD-1V2–treated groups. Tumor-infiltrating natural killer cells and macrophages were also analyzed, but no significant differences were detected between groups (Supplementary Figs. S7J and S7K). Collectively, these studies demonstrated enhanced infiltration of TILs and strong antitumor efficacy of sPD-1V2 in multiple syngenic mouse tumor models.
Efficacy of sPD-1V2 in PD-L2–driven murine models of ovarian cancer
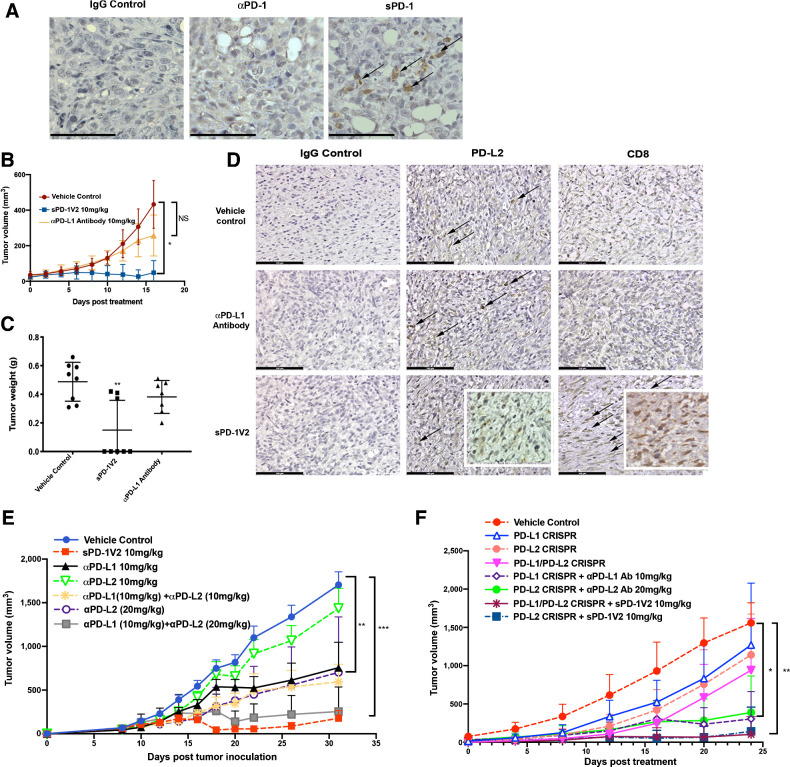
To demonstrate the requirement for PD-L2 in facilitating PD-1–mediated tumor growth, we genetically ablated PD-L1 in ID8 ovarian tumor cells and MC38 colorectal cells using a CRISPR-CAS9 approach and implanted these tumor cells in PD-L1 KO mice, completely eliminating PD-L1 expression in both the tumor and the host (Fig. 6A; Supplementary Fig. S8). While MC38 PD-L1 KO tumor failed to form tumors in PD-L1 KO mice, ID8 PD-L1 KO tumor cells did develop tumors when higher number of tumor cells were implanted. In the ID8 PD-L1 KO-derived tumors, the inhibition of PD-L2 interaction with PD-1 through administering sPD-1V2 led to significant tumor regression; as expected tumors treated with αPD-L1 antibody remained largely unresponsive (Fig. 6B and C). When ID8 tumors were stained for both PD-L2 and CD8 expression, all tumors regardless of treatment groups expressed PD-L2. However, only tumors treated with sPD-1V2 exhibited infiltrating CD8+ T cells (Fig. 6D). These results highlight the importance of blocking PD-L2–mediated activation of PD-1 to suppress immune evasion.
Figure 6.
sPD-1V2 treatment suppresses tumor growth in ovarian cancer models that are PD-L2 dependent. A, ID8 PD-L1 CRSIPR tumors inoculated into PD-L1 KO mice. Tumor stained for PD-L1 and PD-L2 expression. Scale bar 50 μm. B, Subcutaneous ID8 PD-L1 CRSIPR tumor growth in C57BL/6 PD-L1 KO mice. Mice were treated with vehicle control, anti-mouse αPD-L1 blocking antibody, and sPD-1V2 (N = 8). C, Total tumor weight of ID8 PD-L1 CRISPR KO tumors at the time of termination. D, IHC staining of PD-L1 and PD-L2 in ID8 PD-L1 CRISPR KO tumors treated with vehicle control, anti-mouse αPD-L1 blocking antibody 10 mg/kg, and sPD-1V2 10 mg/kg. Scale bar 100 μm. E, ID8 ovarian tumor growth kinetics upon treatment with sPD-1V2 10 mg/kg, αPD-L1 antibody (10 mg/kg), αPD-L2 (10 mg/kg and 20 mg/kg), and the combination of αPD-L1 and αPD-L2 Abs (N = 5 for all experimental groups). F, Tumor growth kinetic of ID8 CRISPR PD-L1 and/or PD-L2 ovarian tumor treated with αPD-L1, αPD-L2 therapeutic antibodies, or sPD-1V2 to evaluate the tumor and/or stromal contribution of PD-L1, PD-L2 in ovarian tumor (N = 5 for all experimental groups). Statistical analysis used are one-way ANOVA for comparing between treatment groups and repeated ANOVA for changes over time. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We performed a tumor study where αPD-L1 and αPD-L2 antibodies were administered either individually or simultaneously in mice bearing ID8 syngenic ovarian tumors to determine whether combined treatment with αPD-L1 and αPD-L2 mAbs could achieve similar antitumor activity when compared with sPD-1V2 treatment. Treatment with αPD-L1 or αPD-L2 individually resulted in partial inhibition of tumor growth, whereas the combination of αPD-L1 and αPD-L2 treatment lead to enhanced antitumor activities (Fig. 6E). As a technical point, treatment with αPD-L2 alone at 10 mg/kg failed to show significant tumor inhibition, but αPD-L2 antibodies tested at a higher concentration (20 mg/kg) as a monotherapy or in combination with αPD-L1 did indeed decrease tumor growth, suggesting a dose-dependent effect (Fig. 6E). To further investigate the role of PD-L2 in evading immune suppression, we again used a CRISPR-CAS9 approach to generate PD-L2 CRISPR KO and PD-L1/L2 double CRISPR KO in ID8 cells that were implanted into female syngenic C57BL/6 mice. There were no significant differences observed in tumor growth between PD-L1 or PD-L2 individual KO ID8 tumors, double PD-L1/PD-L2 CRISPR KO ID8 tumors, and the control group, suggesting a significant contribution of stromal-derived PD-L1 and PD-L2 (Fig. 6F). This is consistent with previous reports that the endogenous contribution of PD-L1 and PD-L2 is necessary for promoting ID8 tumor growth (36). Therefore, host inhibition of PD-L1 and PD-L2 in ID8 tumors are required to achieve maximal inhibition of the PD-1 signaling. In the same study, systemic inhibition of PD-L1, PD-L2, or both were achieved through treating tumor-bearing animals with αPD-L1 antibody, αPD-L2 antibody, or sPD-1V2. Specifically, mice bearing PD-L1 CRISPR KO ID8 tumors were treated with αPD-L1 antibody, mice bearing PD-L2 CRISPR KO ID8 tumors were treated with αPD-L2 antibody, and mice bearing double CRISPR PD-L1/L2 KO ID8 tumors were treated with sPD-1V2 to neutralize both PD-L1 and PD-L2. In addition, a separate cohort of mice bearing CRISPR PD-L2 KO ID8 tumors were treated with sPD-1V2 molecules to evaluate the effect of systemic inhibition of PD-L1 and PD-L2 on the growth of PD-L2 CRISPR KO ID8 tumors. We found that the inhibition of both PD-1 ligands either with the combination of αPD-L1 and αPD-L2 antibodies or sPD-1V2 treatment markedly reduced tumor growth in all PD-L1 and/or PD-L2 CRISPR KO ID8 ovarian tumor models, with the most significant inhibition observed in sPD-1V2–treated groups (Fig. 6F).
In summary, we propose that high PD-L2 expression in human ovarian cancer is a contributing factor to poor response toward ICB. To test this hypothesis, we developed an engineered soluble PD-1 decoy receptor sPD-1V2 that blocks the binding of PD-L1 and PD-L2 to PD-1 with enhanced binding affinity. Structural and biological analysis of the high-affinity sPD-1V2 supports the hypothesis that inhibiting both PD-L1 and PD-L2 can neutralize PD-1 more effectively than blocking antibodies to PD-1 or PD-L1. Therefore, affinity-enhanced sPD-1V2 represents a new therapeutic approach for treating ovarian tumors and/or other tumors expressing PD-L1 and/or PD-L2.
Discussion
To date, the single-agent activities of αPD-1 and αPD-L1 therapeutic antibodies in ovarian cancer have being disappointing (26, 28). In the recent JAVELIN Ovarian 100 trial evaluating avelumab (αPD-L1 antibody) in combination with or following platinum-based chemotherapy in 998 previously untreated patients, there was not a significant improvement in patient response. Similarly, suboptimal clinical outcomes have also been reported in brain and gastrointestinal malignancies (26, 27, 37, 38). The immunosuppressive TME in these cancer types is particularly challenging to overcome, because tumor- and stromal-derived PD-L1 and PD-L2 promote immune evasion, preventing the activation of TILs (39, 40), therefore, complete inhibition of PD-1 by neutralizing both of its ligands is necessary to boost T cell–mediated killing. Clinically, most ICB inhibits PD-1 by blocking PD-1 or PD-L1. However, PD-L2 still remains therapeutically unexplored in the context of ICB with only one clinical candidate, AMP-224, comprising a Fc fusion with the extracellular domain of PD-L2 to act as trap for the PD-1 receptor (41). The lack of therapeutic molecules targeting PD-L2 is predominantly based on the belief that the tissue expression of PD-L2 is low and irrelevant during cancer progression. It is also widely believed that blocking PD-1 receptor therapeutically is sufficient to inhibit the interaction between PD-1 receptor and both of its ligands. However, emerging clinical and biological evidence suggest that in some cancer types, PD-L2 expression is induced by tumors and the TME to promote survival and immune evasion (42–44). In addition, PD-L2 binds to PD-1 receptor at 6- to 10-fold higher binding affinity compared with PD-L1, which potentially can act as a significant competitor to therapeutic αPD-1 antibodies for receptor binding and engagement, leading to diminished efficacy.
Unlike PD-L1, which is expressed in abundance on a variety of both malignant and normal cells, it is thought that basal PD-L2 expression is low on tumor cells and is restricted to very few immune cell types of the myeloid lineage. DCs and macrophages produce Th2 cytokines such as IL4 and drive PD-L2 expression, leading to T-cell suppression and immune escape (43). In addition, T cells can induce PD-L2 through IL4-mediated signaling, which can also result in inhibition of T-cell activity (43, 45). In the context of cancer, studies have reported an increase in PD-L2 expression in both cancer and tumor-associated stromal components upon malignant transformation, which contribute to immune evasion (44). The data presented here suggest that high PD-L2 expression is often associated with cancers with low clinical response toward αPD-1 and αPD-L1 therapies. In particular, ovarian cancers show elevated PD-L2 in tumor tissue and surrounding stroma, with undetectable levels in normal ovarian tissue. This finding supports the concept that PD-L2 expression responds to stimuli in the TME and is inducible during tumor progression. Therefore, inhibition of both PD-L1 and PD-L2 in cancer is important for maximal blockade of the PD-1 signaling pathway.
The increased binding affinity between PD-L2 and PD-1 compared with PD-L1 and PD-1 presents the possibility that abundant PD-L2 can undermine the therapeutic activity of αPD-1 antibodies through competing for PD-1 receptor binding. Our strategy to overcome this potential affinity competition was to develop a soluble PD-1 decoy receptor molecule engineered with high-affinity binding to both PD-L1 and PD-L2. Unlike directed mutagenesis approaches that only mutate residues at the ligand-receptor binding interface, our unbiased mutation strategy identified three mutations within the binding interface of sPD-1, as well as four mutations distal to it. In particular, the mutation A140V located outside of the binding interface resulted in improved overall stability to the PD-1/PD-L1 and PD-1/PD-L2 co-complexes, illustrating the importance of searching beyond the direct binding interface when seeking to optimize an interaction. Interestingly, while several other studies have generated high-affinity binding sPD-1 clones, none have reported mutations at A140, nor analyzed their molecules for binding toward PD-L2 (34, 46, 47). Compared with the binding affinity of commercially available αPD-L1 antibody atezolizumab (TENCENTRIQ), our lead candidate sPD-1V2 has enhanced binding affinity to PD-L1 (sPD-1V2 KD = 3.41 × 10–10 mol/L vs. atezolizumab KD = 9.96 × 10–09 mol/L) under similar experimental conditions (30). In addition, sPD-1V2 has enhanced binding affinity (KD = 3.4 × 10–09) to PD-L2, whereas atezolizumab does not bind to PD-L2 (48). Therefore, sPD-1V2 is the first ligand trap that has been demonstrated to have enhanced binding capabilities to both PD-1 ligands.
Biologically, we tested sPD-1V2 in two syngenic ovarian cancer models. We found that the sPD-1V2 out-performed both αPD-L1 and αPD-1 antibodies in syngenic models of ovarian and other murine cancer models, displaying improved CD4+ and CD8+ TIL infiltration. More importantly, when PD-L1 was genetically ablated from both the cancer cells and the host, only sPD-1V2, but not αPD-L1 antibody–treated animals exhibited antitumor activity. Interestingly, while we developed both MC38 PD-L1 CRISPR KO murine colorectal cancer model and ID8 CRISPR KO murine ovarian cancer model, the MC38 PD-L1 KO model failed to form tumors, which is consistent with previous reports that MC38 tumor growth largely relies on host PD-L1 expression (36). Therefore, the antitumor activity we observed in the MC38 in vivo study is likely to be primarily driven by PD-L1–mediated signaling. In contrast, the ID8 CRISPR PD-L1, PD-L2 single or double KO tumor models did form tumors, although tumor growth was delayed compared with the wild-type ID8 model, indicating that both tumor or host derived PD-L1 and PD-L2 can drive tumor growth and immune evasion.
In summary, we identified PD-L2 as a critical factor associated with poor clinical response toward PD-1 inhibitors in ovarian cancer. Given that improving the response rate of ICB is an urgent unmet need for patients with ovarian cancer, the data presented in this study provide justification for using a dual targeting, high-affinity sPD-1 decoy receptor as an alternative to PD-1 or PD-L1 therapeutic antibodies for achieving superior therapeutic efficacy in cancers expressing both PD-L2 and PD-L1.
Authors' Disclosures
Y.R. Miao reports other support from AKSO Biopharmaceutical Inc outside the submitted work; in addition, Y.R. Miao has a patent for PCT/US2019/050742 pending and licensed to AKSO Biopharmaceutical Inc and a patent for US16/785332 pending and licensed to AKSO Biopharmaceutical Inc. K.N. Thakkar reports a patent for sPD-1 variant-Fc fusion proteins issued and licensed to AKSO Biopharmaceutical Inc, Leland Stanford Junior University. M.S. Kariolis reports a patent for sPD-1 variant-Fc fusion protein pending and licensed to AKSO Biopharmaceutical Inc and a patent for “Receptor-based antagonists of the programmed cell death (PD-1) pathway” issued and licensed to AKSO Biopharmaceutical Inc; M.S. Kariolis also has stock equity in Aravive Biologics Inc. and AKSO Biopharmaceutical Inc. A.J. Giaccia reports other support from AKSO Biopharmaceutical Inc and Aravive Biologics Inc. during the conduct of the study; in addition, A.J. Giaccia has a patent for Stanford University pending and licensed to AKSO Biopharmaceutical Inc. No disclosures were reported by the other authors.
Supplementary Material
Supplementary data, material methods and table
Acknowledgments
This work was supported by NIH grants CA67166, CA197713, and CA198291; a grant from MRC UK; the Silicon Valley Foundation; the Sydney Frank Foundation, and the Kimmelman Fund. A.J. Giaccia is the principle investigator for all grants listed above. The authors would like to thank Hannah Elizabeth Skolnik for technical support on in vivo studies.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Authors' Contributions
Y.R. Miao: Conceptualization, resources, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. K.N. Thakkar: Conceptualization, resources, data curation, software, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing. J. Qian: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. M.S. Kariolis: Data curation, formal analysis, validation, investigation, methodology, writing–review and editing. W. Huang: Data curation, software, formal analysis, validation, investigation, visualization, methodology. S. Nandagopal: Data curation, formal analysis, validation, investigation, methodology. T.T.C. Yang: Data curation, software, formal analysis, validation, investigation, methodology. A.N. Diep: Data curation, formal analysis, validation, investigation, visualization, writing–review and editing. G.M. Cherf: Data curation, software, validation, investigation, methodology. Y. Xu: Conceptualization, data curation, validation, investigation, methodology. E.J. Moon: Conceptualization, resources, data curation, software, validation, investigation, methodology. Y. Xiao: Resources, data curation, software, formal analysis, validation, investigation, visualization, methodology. H. Alemany: Conceptualization, data curation, software, formal analysis, validation, investigation, methodology. T. Li: Resources, data curation, formal analysis, validation, investigation, methodology. W. Yu: Data curation, software, formal analysis, validation, investigation, project administration. B. Wei: Data curation, validation. E.B. Rankin: Resources, data curation, formal analysis, validation, investigation, methodology. A.J. Giaccia: Conceptualization, resources, data curation, supervision, funding acquisition, validation, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387:1540–50. [DOI] [PubMed] [Google Scholar]
- 2. Larkin J, Minor D, D'Angelo S, Neyns B, Smylie M, Miller WH Jr, et al. Overall survival in patients with advanced melanoma who received nivolumab versus investigator's choice chemotherapy in CheckMate 037: a randomized, controlled, open-label phase III trial. J Clin Oncol 2018;36:383–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schachter J, Ribas A, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017;390:1853–62. [DOI] [PubMed] [Google Scholar]
- 4. Wang C, Thudium KB, Han M, Wang XT, Huang H, Feingersh D, et al. In vitro characterization of the anti-PD-1 antibody nivolumab, BMS-936558, and in vivo toxicology in non-human primates. Cancer Immunol Res 2014;2:846–56. [DOI] [PubMed] [Google Scholar]
- 5. Rodriguez CP, Wu Q, Voutsinas J, Fromm JR, Jiang X, Pillarisetty VG, et al. A phase II trial of pembrolizumab and vorinostat in recurrent metastatic head and neck squamous cell carcinomas and salivary gland cancer. Clin Cancer Res 2020;26:837–45. [DOI] [PubMed] [Google Scholar]
- 6. Varga A, Piha-Paul S, Ott PA, Mehnert JM, Berton-Rigaud D, Morosky A, et al. Pembrolizumab in patients with programmed death ligand 1-positive advanced ovarian cancer: analysis of KEYNOTE-028. Gynecol Oncol 2019;152:243–50. [DOI] [PubMed] [Google Scholar]
- 7. Wang BC, Zhang ZJ, Fu C, Wang C. Efficacy and safety of anti-PD-1/PD-L1 agents vs chemotherapy in patients with gastric or gastroesophageal junction cancer: a systematic review and meta-analysis. Medicine 2019;98:e18054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Imai Y, Hasegawa K, Matsushita H, Fujieda N, Sato S, Miyagi E, et al. Expression of multiple immune checkpoint molecules on T cells in malignant ascites from epithelial ovarian carcinoma. Oncol Lett 2018;15:6457–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pietzner K, Nasser S, Alavi S, Darb-Esfahani S, Passler M, Muallem MZ, et al. Checkpoint-inhibition in ovarian cancer: rising star or just a dream? J Gynecol Oncol 2018;29:e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tran L, Allen CT, Xiao R, Moore E, Davis R, Park SJ, et al. Cisplatin alters antitumor immunity and synergizes with PD-1/PD-L1 inhibition in head and neck squamous cell carcinoma. Cancer Immunol Res 2017;5:1141–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rini BI, Plimack ER, Stus V, Gafanov R, Hawkins R, Nosov D, et al. Pembrolizumab plus axitinib versus sunitinib for advanced renal-cell carcinoma. N Engl J Med 2019;380:1116–27. [DOI] [PubMed] [Google Scholar]
- 12. Tree AC, Jones K, Hafeez S, Sharabiani MTA, Harrington KJ, Lalondrelle S, et al. Dose-limiting urinary toxicity with pembrolizumab combined with weekly hypofractionated radiation therapy in bladder cancer. Int J Radiat Oncol Biol Phys 2018;101:1168–71. [DOI] [PubMed] [Google Scholar]
- 13. Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, et al. Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 2003;348:203–13. [DOI] [PubMed] [Google Scholar]
- 14. Luo Z, Wang Q, Lau WB, Lau B, Xu L, Zhao L, et al. Tumor microenvironment: the culprit for ovarian cancer metastasis? Cancer Lett 2016;377:174–82. [DOI] [PubMed] [Google Scholar]
- 15. Ishida M, Iwai Y, Tanaka Y, Okazaki T, Freeman GJ, Minato N, et al. Differential expression of PD-L1 and PD-L2, ligands for an inhibitory receptor PD-1, in the cells of lymphohematopoietic tissues. Immunol Lett 2002;84:57–62. [DOI] [PubMed] [Google Scholar]
- 16. Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. PD-L2 is a second ligand for PD-1 and inhibits T cell activation. Nat Immunol 2001;2:261–8. [DOI] [PubMed] [Google Scholar]
- 17. Lesterhuis WJ, Punt CJ, Hato SV, Eleveld-Trancikova D, Jansen BJ, Nierkens S, et al. Platinum-based drugs disrupt STAT6-mediated suppression of immune responses against cancer in humans and mice. J Clin Invest 2011;121:3100–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yamazaki T, Akiba H, Iwai H, Matsuda H, Aoki M, Tanno Y, et al. Expression of programmed death 1 ligands by murine T cells and APC. J Immunol 2002;169:5538–45. [DOI] [PubMed] [Google Scholar]
- 19. Liang SC, Latchman YE, Buhlmann JE, Tomczak MF, Horwitz BH, Freeman GJ, et al. Regulation of PD-1, PD-L1, and PD-L2 expression during normal and autoimmune responses. Eur J Immunol 2003;33:2706–16. [DOI] [PubMed] [Google Scholar]
- 20. Lazar-Molnar E, Yan Q, Cao E, Ramagopal U, Nathenson SG, Almo SC. Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2. Proc Natl Acad Sci U S A 2008;105:10483–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Youngnak P, Kozono Y, Kozono H, Iwai H, Otsuki N, Jin H, et al. Differential binding properties of B7-H1 and B7-DC to programmed death-1. Biochem Biophys Res Commun 2003;307:672–7. [DOI] [PubMed] [Google Scholar]
- 22. Derks S, Nason KS, Liao X, Stachler MD, Liu KX, Liu JB, et al. Epithelial PD-L2 expression marks Barrett's esophagus and esophageal adenocarcinoma. Cancer Immunol Res 2015;3:1123–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pujade-Lauraine E, Fujiwara K, Dychter SS, Devgan G, Monk BJ. Avelumab (anti-PD-L1) in platinum-resistant/refractory ovarian cancer: JAVELIN Ovarian 200 Phase III study design. Future Oncol 2018;14:2103–13. [DOI] [PubMed] [Google Scholar]
- 24. Rimm DL, Han G, Taube JM, Yi ES, Bridge JA, Flieder DB, et al. A prospective, multi-institutional, pathologist-based assessment of 4 immunohistochemistry assays for PD-L1 expression in non-small cell lung cancer. JAMA Oncol 2017;3:1051–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Inoue Y, Yoshimura K, Mori K, Kurabe N, Kahyo T, Mori H, et al. Clinical significance of PD-L1 and PD-L2 copy number gains in non-small-cell lung cancer. Oncotarget 2016;7:32113–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Groot J, Penas-Prado M, Alfaro-Munoz KD, Hunter K, Pei B, O'Brien B, et al. Window-of-opportunity clinical trial of pembrolizumab in patients with recurrent glioblastoma reveals predominance of immune-suppressive macrophages. Neuro Oncol 2020;22:539–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pfizer. Avelumab in previously untreated patients with epithelial ovarian cancer (JAVELIN OVARIAN 100). Available from: https://www.pfizer.com/avelumab-previously-untreated-patients-epithelial-ovarian-cancer-javelin-ovarian-100.
- 28. Pfizer. EMD Serono and Pfizer provide update on phase III JAVELIN Gastric 100 Trial. Available from: https://www.pfizer.com/news/press-release/press-release-detail/emd_serono_and_pfizer_provide_update_on_phase_iii_javelin_gastric_100_trial.
- 29. Fradet Y, Bellmunt J, Vaughn DJ, Lee JL, Fong L, Vogelzang NJ, et al. Randomized phase III KEYNOTE-045 trial of pembrolizumab versus paclitaxel, docetaxel, or vinflunine in recurrent advanced urothelial cancer: results of >2 years of follow-up. Ann Oncol 2019;30:970–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang F, Qi X, Wang X, Wei D, Wu J, Feng L, et al. Structural basis of the therapeutic anti-PD-L1 antibody atezolizumab. Oncotarget 2017;8:90215–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kariolis MS, Miao YR, Jones DS 2nd, Kapur S, Mathews II, Giaccia AJ, et al. An engineered Axl ‘decoy receptor’ effectively silences the Gas6-Axl signaling axis. Nat Chem Biol 2014;10:977–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kariolis MS, Miao YR, Diep A, Nash SE, Olcina MM, Jiang D, et al. Inhibition of the GAS6/AXL pathway augments the efficacy of chemotherapies. J Clin Invest 2017;127:183–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Silva JP, Vetterlein O, Jose J, Peters S, Kirby H. The S228P mutation prevents in vivo and in vitro IgG4 Fab-arm exchange as demonstrated using a combination of novel quantitative immunoassays and physiological matrix preparation. J Biol Chem 2015;290:5462–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tang S, Kim PS. A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery. Proc Natl Acad Sci U S A 2019;116:24500–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burova E, Hermann A, Waite J, Potocky T, Lai V, Hong S, et al. Characterization of the anti-PD-1 antibody REGN2810 and its antitumor activity in human PD-1 knock-in mice. Mol Cancer Ther 2017;16:861–70. [DOI] [PubMed] [Google Scholar]
- 36. Lin H, Wei S, Hurt EM, Green MD, Zhao L, Vatan L, et al. Host expression of PD-L1 determines efficacy of PD-L1 pathway blockade-mediated tumor regression. J Clin Invest 2018;128:805–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Moehler M, Ryu MH, Dvorkin M, Lee KW, Coskun HS, Wong R, et al. Maintenance avelumab versus continuation of first-line chemotherapy in gastric cancer: JAVELIN Gastric 100 study design. Future Oncol 2019;15:567–77. [DOI] [PubMed] [Google Scholar]
- 38. Kudo T, Hamamoto Y, Kato K, Ura T, Kojima T, Tsushima T, et al. Nivolumab treatment for oesophageal squamous-cell carcinoma: an open-label, multicentre, phase 2 trial. Lancet Oncol 2017;18:631–9. [DOI] [PubMed] [Google Scholar]
- 39. Nazareth MR, Broderick L, Simpson-Abelson MR, Kelleher RJ Jr, Yokota SJ, Bankert RB. Characterization of human lung tumor-associated fibroblasts and their ability to modulate the activation of tumor-associated T cells. J Immunol 2007;178:5552–62. [DOI] [PubMed] [Google Scholar]
- 40. Messal N, Serriari NE, Pastor S, Nunes JA, Olive D. PD-L2 is expressed on activated human T cells and regulates their function. Mol Immunol 2011;48:2214–9. [DOI] [PubMed] [Google Scholar]
- 41. NCI. AMP-224, a PD-1 inhibitor, with stereotactic body radiation therapy in metastatic colorectal cancer. Available from: https://clinicaltrials.gov/ct2/show/NCT02298946. [DOI] [PMC free article] [PubMed]
- 42. Huber S, Hoffmann R, Muskens F, Voehringer D. Alternatively activated macrophages inhibit T-cell proliferation by Stat6-dependent expression of PD-L2. Blood 2010;116:3311–20. [DOI] [PubMed] [Google Scholar]
- 43. Lesterhuis WJ, Steer H, Lake RA. PD-L2 is predominantly expressed by Th2 cells. Mol Immunol 2011;49:1–3. [DOI] [PubMed] [Google Scholar]
- 44. Yearley JH, Gibson C, Yu N, Moon C, Murphy E, Juco J, et al. PD-L2 expression in human tumors: relevance to anti-PD-1 therapy in cancer. Clin Cancer Res 2017;23:3158–67. [DOI] [PubMed] [Google Scholar]
- 45. Hebenstreit D, Wirnsberger G, Horejs-Hoeck J, Duschl A. Signaling mechanisms, interaction partners, and target genes of STAT6. Cytokine Growth Factor Rev 2006;17:173–88. [DOI] [PubMed] [Google Scholar]
- 46. Li Y, Liang Z, Tian Y, Cai W, Weng Z, Chen L, et al. High-affinity PD-1 molecules deliver improved interaction with PD-L1 and PD-L2. Cancer Sci 2018;109:2435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Maute RL, Gordon SR, Mayer AT, McCracken MN, Natarajan A, Ring NG, et al. Engineering high-affinity PD-1 variants for optimized immunotherapy and immuno-PET imaging. Proc Natl Acad Sci U S A 2015;112:E6506–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Deng R, Bumbaca D, Pastuskovas CV, Boswell CA, West D, Cowan KJ, et al. Preclinical pharmacokinetics, pharmacodynamics, tissue distribution, and tumor penetration of anti-PD-L1 monoclonal antibody, an immune checkpoint inhibitor. MAbs 2016;8:593–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data, material methods and table