Abstract
Approximately 20,000 patients per year are diagnosed with esophageal adenocarcinoma (EAC) and malignant pleural mesothelioma (MPM); fewer than 20% survive 5 years. Effective therapeutic strategies are limited although patients receive a combination of chemotherapeutics. These tumors harbor thousands of mutations that contribute to tumor development. Downstream of oncogenic driving mutations, altered tumor mitochondria promote resistance to apoptosis. Dynamic Bcl-2 homology-3 profiling (DBP) is a functional assay of live cells that identifies the mitochondrial proteins responsible for resistance to apoptosis. We hypothesized that DBP will predict which protein to target to overcome resistance thereby enhancing combinatorial therapy.
DBP predicted that targeting either Mcl-1 or Bcl-xL increases the efficacy of the chemotherapeutic agent, cisplatin, whereas targeting Bcl-2 does not. We performed these assays by treating EAC and MPM cells with a combination of Bcl-2 homology-3 (BH3) mimetics and cisplatin. Following treatments, we performed efficacy assessments including apoptosis assays, IC50 calculations, and generation of a combinatorial index. DBP confirmed that targeting mitochondria with BH3 mimetics alters the threshold of apoptosis. These apoptotic effects were abolished when the mitochondrial pathway was disrupted. We validated our findings by developing knockdown models of antiapoptotic proteins Mcl-1, Bcl-xL, and the mitochondrial effector proteins Bax/Bak. Knockdown of Mcl-1 or Bcl-xL recapitulated the results of BH3 mimetics. In addition, we report an approach for BH3 profiling directly from patient tumor samples. We demonstrate that the DBP assay on living tumor cells measures the dynamic changes of resistance mechanisms, assesses response to combinatorial therapy, and provides results in a clinically feasible time frame.
Introduction
Esophageal adenocarcinoma (EAC) and malignant pleural mesothelioma (MPM) have poor 5-year survival rates of 10% to 20% (1, 2). Effective therapy, especially targeted therapy, for EAC and MPM is limited (3, 4). Both are treated with doublet chemotherapy. Even when treatment leads to tumor responses, these cancers usually develop resistance. The common link between these cancers is chronic exposure to the environmental carcinogens, bile acid, and asbestos (5–7). These carcinogens induce thousands of mutations that account for their aggressive phenotypes (8–10). Whether treating with chemotherapy or targeting a specific mutation, these tumors circumvent death through other somatic mutations or bypass pathways to enable therapeutic resistance (11). Therefore, treatment strategies that target these resistance mechanisms are urgently needed (12).
Recently, we showed that chronic exposure of preneoplastic Barrett esophageal cells to bile salt induced malignant transformation through a mitochondrial mechanism termed, “minority MOMP” (mitochondrial outer membrane permeabilization; refs. 5, 13, 14). MOMP is the critical event mediated by the colocalization of the membrane channel proteins, Bax and Bak, to the outer mitochondrial membrane which releases cytochrome c (cyt c) to activate the caspase system resulting in cell death (15, 16). Minority MOMP activates the intrinsic pathway of the apoptotic machinery in roughly 8% of the cancer cells' mitochondria, which is not sufficient to result in cell death. Instead, minority MOMP promotes genomic instability, cellular transformation, and tumorigenesis (17–19). Because mitochondrial pathways are downstream of oncogenic driver proteins, targeting these pathways is a strategy that can overcome the mechanisms that enable tumor cell resistance (15, 20).
MOMP is a switch-like event regulated by the balance of antiapoptotic and proapoptotic B-cell lymphoma 2 (Bcl-2) family members (21). Toxic stimuli may make cells more susceptible to MOMP; however, cells may withstand stressful stimuli and avoid apoptosis by upregulating antiapoptotic proteins (22). During minority MOMP, we noted that the expression of the antiapoptotic protein, myeloid cell leukemia (Mcl)-1, doubled (6, 23). Mcl-1 is a critical antiapoptotic protein that prevents the activation of the intrinsic mitochondrial apoptotic pathway (24–27). Bcl-xL is also a clinically promising target because it is highly expressed in both EAC and MPM (5, 28). Mcl-1 and Bcl-xL may have redundant functions. Studies on Bcl-xL and Mcl-1 in EAC and MPM as the potential therapeutic targets are relatively limited (24, 25).
Bcl-2 homology-3 (BH3) profiling is a functional assay of live cells that identifies which of these family of proteins are responsible for resistance to apoptosis (29). The proapoptotic and antiapoptotic Bcl-2 proteins that regulate MOMP interact at hydrophobic BH3 domains (22). BH3 profiling measures the relative interactions of these proteins to determine whether a tumor cell is near the threshold to activate apoptosis (29–31). Dynamic BH3 profiling (DBP) is a live cell assay that measures changes in the BH3 profile before and after drug treatment to predict clinical response (32). Drugs that inhibit Bcl-2 proteins are a class of compounds termed “BH3 mimetics” (33). To be a BH3 mimetic, a drug must selectively inhibit an antiapoptotic protein, bind with high affinity, and induce MOMP in a Bak/Bax-dependent manner. Recently, Villalobos-Ortiz and colleagues developed a biochemical “tool kit” and utilized DBP to identify the appropriate BH3 mimetic in murine cells that overexpressed Bcl-2 proteins (34).
Experience with DBP in solid tumors is limited and, to our knowledge, reports of DBP in EAC and MPM do not exist. Ni Chonghaile and colleagues reported that BH3 profiling successfully predicted response to chemotherapy in cell culture models (31). Montero and colleagues noted that DBP predicted the emergence of resistance to multiple tyrosine kinase inhibitors (TKIs) in lung cancer cells (32). Bhola and colleagues reported that DBP predicts response in colon cancer (35). Interestingly, they found that apoptotic sensitizers differed between tumors of the same histology. Priming of the mitochondria in solid tumors, especially EAC and MPM, is difficult secondary to variability of antiapoptotic proteins per tumor, lack of response to single-agent therapy, and difficulty with ex vivo culture of solid tumors. To overcome these obstacles, we developed a proof-of-concept model wherein DBP can predict which BH3 mimetic will sensitize tumor cells to cisplatin. We hypothesized that DBP profiling is predictive of EAC and MPM response to combined treatment with BH3 mimetics and cisplatin.
Materials and Methods
Cell lines and treatments
EAC lines FLO-1, Eso26, and OE33 were purchased from the European Collection of Authenticated Cell Cultures via Sigma-Aldrich. Mesothelioma cell lines H28, H226, H2052, and H2452 were purchased from ATCC. All cell lines were authenticated yearly with HLA analysis and tested for Mycoplasma contamination every 6 months. Cells were cultured in RPMI (Thermo Fisher Scientific, 11875-093) with 10% FCS and 1% penicillin/streptomycin (Gibco No. 15140-122).
Cells were treated as described with Bcl-2 inhibitor, ABT199 (Apexbio; ref. 36), Bcl-xL inhibitor, A1155463 (MedChemExpress; ref. 37), or Mcl-1 inhibitor, S63845 (Active Biochem) and AZD-5991 (MedChemExpress; refs. 33, 38), and CDDP (APP Pharmaceuticals, NDC 63323-103-65).
Primary cells
Tumor samples were placed on ice in serum-free RPMI (Corning, 15-040-CV). Tissues were minced and dissociated using the human tumor dissociation kit (Miltenyi Biotec). Samples were either incubated at 37°C with gentle rocking for 1 hour or dissociated using the gentleMACS system following the manufacturer's protocol. The suspension was filtered through a 70-μm cell strainer (BD Falcon) and washed with PBS.
BH3 profiling and DBP
Intracellular BH3 profiling has been described in detail previously (29). Briefly, primary cells were stained with viability dye, LIVE/DEAD Fixable Aqua (Thermo Fisher Scientic), washed, then incubated with FcR blocking reagent (Miltenyi Biotec) and surface stained with phycoerythrin (PE)-conjugated antihuman epithelial cell adhesion molecule (EpCAM) antibody (clone HEA-125, Miltenyi Biotec) or PE-conjugated D2-40 antibody (clone NC-08, BioLegend) and common leukocyte antigen CD45 antibody (clone HI30, BioLegend). Peptide treatments and controls were mixed at 2× desired concentration in MEB2 buffer + 20 μg/mL digitonin (Sigma, D5628) and 50 μL of this combination was added per well in a 96-well, nonbinding plate (Celltreat, 229590).
Cells, in 50-μL MEB2, were incubated with peptides for 1 hour at room temperature then formaldehyde fixed. Anti-cyt-c antibody (BioLegend, 612308) was added at 1:2,000 final dilution at 4°C overnight. Viable EpCAM-positive (EpCAM+) or D2-40 positive (D2-40+) and CD45-negative cells were analyzed by multiparameter flow cytometry (MACSQuant, Miltenyi Biotec). Retained cyt c was measured and percent release was calculated from the MFI of cyt c stain normalized to that of wells containing 25 μmol/L alamethicin (ALM25; Enzo, BML-A150-0005) as 100% release control. For cultured lines, cells were harvested using TrypLE Express (Gibco, 12605-010), washed twice in PBS, and added at 5 × 104 cells per well in 50 μL of MEB2.
Lentiviral knockdowns
For double knockdowns, cells were transduced with commercially validated lentiviral short hairpin (shRNA) targeting Bak (Sigma NM_001188, TRCN0000033466), Bax (Sigma NM_004324, TRCN0000312625), or scrambled sequences (Sigma No. SHC002V) at multiplicity of infection (MOI) = 5. The bulk population was cloned by limiting dilution. Stable pools were expanded under puromycin (Sigma No. P9620-10ML) selection at 2 μg/mL. Individual clones were tested by qPCR and Western blot analysis to identify double knockdowns. Mcl-1 and Bcl-xL knockdowns were generated (Sigma No. NM_021960, TRCN0000194663, No. NM_001191. TRCM0000033499) using MOI = 5 and grown in 2 μg/mL puromycin.
Immunoblotting
Cell lysates (10 μg) were resolved on 4% to 20% SDS-polyacrylamide gels, electrotransferred onto nitrocellulose membranes, and blocked for 1 hour at 25°C in 5% nonfat dry milk/0.1% Tween. Blots were incubated with antibodies at 4°C overnight, washed in 0.05% Tween, and incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour. The blots were washed and immunocomplexes were detected using SuperSignal West Femto (Thermo Fisher Scientific No. 34095). Antibodies: Bak (ab32371, Abcam), Bax (ab32503, Abcam), Bcl-2 (sc-492, Santa Cruz Biotechnology), Bcl-xl (sc8392, Santa Cruz Biotechnology), and Mcl-1 (sc819, Santa Cruz Biotechnology).
IC50 and apoptosis assays
Cells treated as indicated were stained with annexin V and propidium iodide (PI) according to manufacturer's instructions (Takara, catalog No. 630110). Apoptotic cells were analyzed with FACS Caliber (Becton Dickinson) and AttuneNxT (Thermo Fisher Scientific). Apoptotic cell percentage corresponded to annexin V(+)/PI(−) cells. Data were analyzed using FlowJo 10.4.1 software.
IC50 curves were generated with 3,000 cells/well in a 96-well plate. Cells were treated overnight with ABT199, A1155463, or S63845. Cisplatin was added the next day at indicated concentrations. At 48 or 72 hours later, cell viability was assayed (CellTiter 96 AQueous One Solution, Promega).
Tissue arrays
Using array slides of normal tissue and tumor (Biomax), IHC staining was performed. Using a standardized IHC intensity-scoring schema, a third-party pathologist (Vitro Vivo) graded IHC staining as negative (0), low (25%), medium (50%) or high (≥75%).
Histologic analyses and IHC staining
Representative paraffin-embedded sections stained with hematoxylin–eosin (H&E) were analyzed by light microscopy. IHC analyses of paraffin-embedded tissue sections were performed using a VECTASTAIN ABC kit (Vector Laboratories Inc.). IHC staining for D2-40 (CellMarque Tissue Diagnostics, Mouse mAb, prediluted, 0.13 μg/mL, Roche) was performed using standard IHC techniques for tumor specimens fixed with paraformaldehyde (PFA) and paraffin embedded.
Immunofluorescence staining
Cells grown on coverslips were fixed in 4% PFA and permeabilized in PBS with 0.2% Triton X-100. After washing, cells were blocked in 3% BSA. Primary Mcl-1 (sc819, Santa Cruz Biotechnology), WT1 (NBP2-47858PE, NOVUS) conjugated with PE, pan-cytokeratin (sc-8018, Santa Cruz Biotechnology) conjugated with PE, Vimentin (sc-66002, Santa Cruz Biotechnology) conjugated with FITC antibodies and TOMM20 (ab209606, Abcam) antibodies conjugated with Alexa Fluor 647 were diluted in blocking solution and incubated overnight at 4°C. DAPI was used as internal control for immunofluorescence. Images were collected with a Zeiss 780 confocal microscope.
Statistical analysis
Quantitative data are presented as the mean ± SD. The Student t test compared means between two groups. One-way ANOVA followed by the post hoc Dunnett test compared means of more than two groups, and a multiple range least significant difference was used for intergroup comparisons. Survival curves were plotted by the Kaplan–Meier method, and compared by the log-rank test (*, # = P < 0.05; **, ## = P < 0.01). Statistical analyses were performed with Graph Pad Prism.
Results
Mcl-1 and Bcl-xL are highly expressed in EAC and MPM tumor samples and cell lines
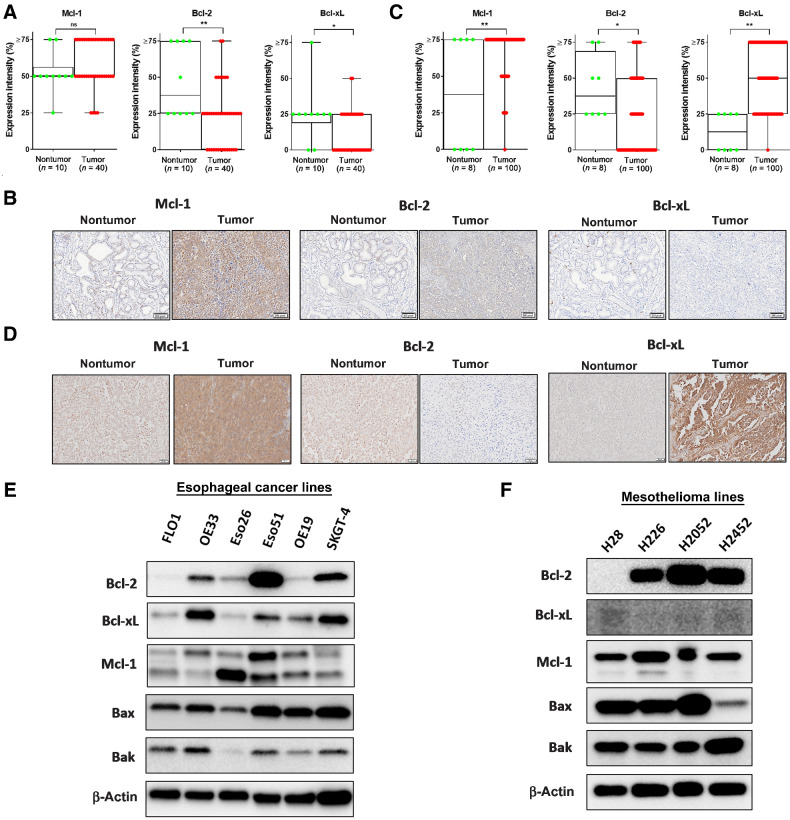
To investigate whether Bcl-2 family members are clinically relevant in human EAC and MPM, we examined the expression of antiapoptotic proteins, Mcl-1, Bcl-2, and Bcl-xL, by IHC staining in tissue microarray (TMA) slides. The TMA slides of 10 normal esophageal mucosa samples and 40 EAC tumors (Fig. 1A and B) and eight normal pleural and 100 MPM tumors (Fig. 1C and D) revealed >50% expression of Mcl-1 in more than 90% of tumor specimens in both histologies. Bcl-xL expression was higher in MPM compared with EAC tumor samples. Despite our hypothesis that function, not protein expression levels, are relevant for this mechanism, we assessed differences in survival based on expression from The Cancer Genome Atlas. As expected, expression levels did not correlate with survival (Supplementary Fig. S1).
Figure 1.
Expression levels of Bcl-2 protein family members in patient tissue and EAC and MPM cell lines. A, EAC samples were categorized into four levels of expression patterns, 10 normal esophageal tissues were compared with 40 tumor samples. B, Representative samples from IHC analysis of EAC samples microarray showing examples of expression of Bcl-2, Bcl-xL, and Mcl-1 (scale bar = 50 μmol/L). C, MPM samples were categorized into four levels of expression patterns; eight normal tissues were compared with 100 tumor samples. D, Representative samples from IHC analysis of MPM samples microarray showing examples of expression of Bcl-2, Bcl-xL, and Mcl-1. E, Immunoblot of Bcl-2 family expression in EAC lines. F, Immunoblot of Bcl-2 family expression in MPM cell lines.
Although most cells undergo MOMP, not all exhibit this mechanism. Therefore, we confirmed Bax and Bak expression by immunoblots and found both proteins in all EAC and MPM cell lines tested (Fig. 1E and F). In addition, the antiapoptotic function of Bcl-2, Bcl-xL, and Mcl-1 is often redundant. When we tested the protein expression levels, the expression differed significantly, yet at least one of the proteins was present in each line. In summary, these results show that the necessary proteins are present in EAC and MPM tissue arrays and cell lines for the intrinsic mitochondrial apoptotic pathway to function. Therefore, pursuing this mechanism to overcome treatment resistance is a clinically relevant strategy.
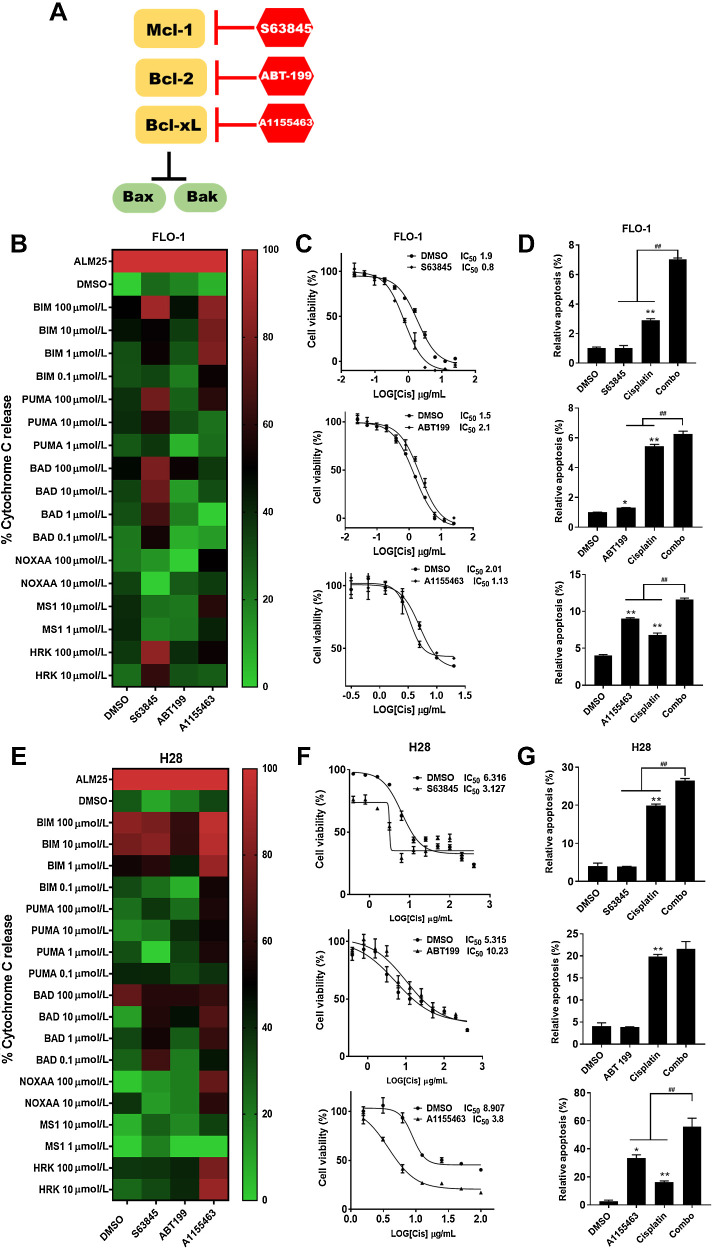
Targeting Mcl-1 or Bcl-xL selectively primes mitochondria and sensitizes EAC and MPM to cisplatin
To determine whether targeting Bcl-2 family proteins sensitized cells to chemotherapeutics, we chose the well-established BH3 mimetics, S63845, A1155463, and ABT-199, which selectively target Mcl-1, Bcl-xL, and Bcl-2, respectively (Fig. 2A). We performed DBP on EAC cells, FLO-1 and Eso26, treated with these drugs versus DMSO control (Fig. 2B). The outcome measurement of DBP is cyt c release. On the heatmap, green represents a lack of cyt c release and red represents 100% release. When the cells were treated with S63845 that targets Mcl-1, cyt c release increased noted by the BAD peptide showing red with S63845 compared with green with DMSO. This effect also was noted for Eso26, but not as robust. Similarly, treatment with A1155463 resulted in increased cyt c release. In contrast, ABT-199 targeting Bcl-2 showed no differences in the cyt c release compared with DMSO. These findings suggest that targeting Mcl-1 or Bcl-xL, but not Bcl-2, primed the mitochondria.
Figure 2.
Mcl-1 and Bcl-xL are involved in mitochondrial priming to enhance response to conventional treatment. A, Interaction of Bcl-2 proteins and BH3 mimetics: the function of Bcl-xL, Bcl-2 and Mcl-1 is to prevent the activation of Bax and Bak. A1155463 inhibits Bcl-xL, ABT‐199 inhibits Bcl-2 and S63845 inhibits Mcl-1. B, Heatmaps showing DBP in FLO-1 cells with 1.0 μmol/L A1155463, S63845, or ABT-199 pretreatment for 24 hours. C, cells were treated with different concentrations of CDDP (0.02–25 μg/mL) for 48 hours after pretreatment for 24 hours with 1.0 μM A1155463, S63845, or ABT-199; the IC50 values of CDDP on FLO-1 cells are determined. D, After pretreatment with 1.0 μM A1155463, S63845, or ABT199 for 24 hours, the apoptotic ratio in cells at 48 hours after treatment with CDDP were measured by annexin V flow cytometry; cells positive for annexin V staining were counted as apoptotic cells (*, P < 0.05; **, P < 0.01 for treatment vs. DMSO; ##, P < 0.01 for combination treatment vs. A1155463, ABT-199 or S63845 single treatment). E, Heatmap showing DBP in H28 cells with 1.0 μM A1155463, 5.0 μmol/L S63845 or ABT-199 pretreatment for 24 hours. F, Cells were treated with different concentrations of CDDP (0.4–400 μg/mL) for 48 hours after pretreatment for 24 hours with 1.0 μM A1155463, 5.0 μmol/L S63845 or ABT-199; the IC50 values of CDDP on H28 cells are determined. G, After pretreatment with 1.0 μmol/L A1155463, 5.0 μM S63845 or ABT199 for 24 hours, the apoptotic ratio in cells at 48 hours after treatment with CDDP were measured by annexin V flow cytometry; cells positive for annexin V staining were counted as apoptotic cells (*, P < 0.05; **, P < 0.01 for treatment vs. DMSO; ##P < 0.01 for combination treatment vs. A1155463, ABT-199, or S63845 single treatment).
To determine whether the results of DBP are phenotypically relevant, we asked whether priming the mitochondria changed the effects of cisplatin (CDDP) on the cells. We used CDDP because it is the most common backbone of chemotherapy regimens for EAC and MPM. We performed IC50 calculations for FLO-1 and noted that at the IC50 level, S63845 and A1155463, but not ABT-199, increased the sensitivity to CDDP (Fig. 2C). With an annexin V assay to determine whether BH3 mimetics enhanced apoptosis with CDDP, we noted that combining S63845 or A1155463 with CDDP significantly increased apoptosis when compared with CDDP alone (Fig. 2D). In contrast, ABT-199 in combination with CDDP was not different from CDDP alone. To further characterize apoptotic cell morphologic changes, we used Hoechst 33342 to stain pyknotic nuclei, which revealed that the relative apoptotic cells are consistent with the annexin V analysis (Supplementary Fig. S2). To determine whether the effects of S63845 where synergistic with CDDP, we performed a combinatorial index (CI) evaluation. We use the method of Chou and Talalay to quantify synergy between S63845 and CDDP (39). The CI at different dose levels that caused growth suppression (fraction affected) revealed synergy between S63845 and CDDP at most doses in two cell lines. In contrast, only one of the eight doses showed synergy between ABT199 and CDDP (Supplementary Fig. S3). We repeated the experiments in Eso26 and noted similar findings (Supplementary Fig. S4A–S4C).
EAC and MPM have different embryologic origins, different cell surface IHC markers, and different sensitivities to drug treatment. Therefore, we repeated these experiments in the two MPM cell lines, H28 and H2452. S63845 or A1155463 increased mitochondrial cyt c release in the MPM cell line, H28, whereas ABT-199 did not (Fig. 2E). Similar to EAC lines, the IC50 level decreased and relative apoptosis increased with S63845 or A1155463 but not ABT-199 (Fig. 2F and G; Supplementary Fig. S2). Of note, the mitochondrial priming was reduced with both S63845 and A1155463, however, A1155463 was particularly effective noted by a high percentage of black and red on the heatmap. In addition, the CI revealed synergy between S63845 and CDDP in H28 (Supplementary Fig. S3B). Similar results were noted with H2452 (Supplementary Fig. S4E and S4F). Of note, the IC50 levels of MPM were substantially higher compared to EAC cells which is expected given that MPM response to therapy is particularly poor (Supplementary Fig. S5).
Together, these results indicate that DBP was predictive of synergistic responses of Bcl-2 family inhibition with CDDP. The results of DBP are phenotypically relevant given that treatment with S63845 or A1155463 and CDDP, the effects of CDDP were enhanced. These results are specific given that when we treated the cells with ABT-199 and CDDP, no enhancement of CDDP effects were noted. In summary, DBP can predict which antiapoptotic protein(s) to target to overcome therapeutic resistance.
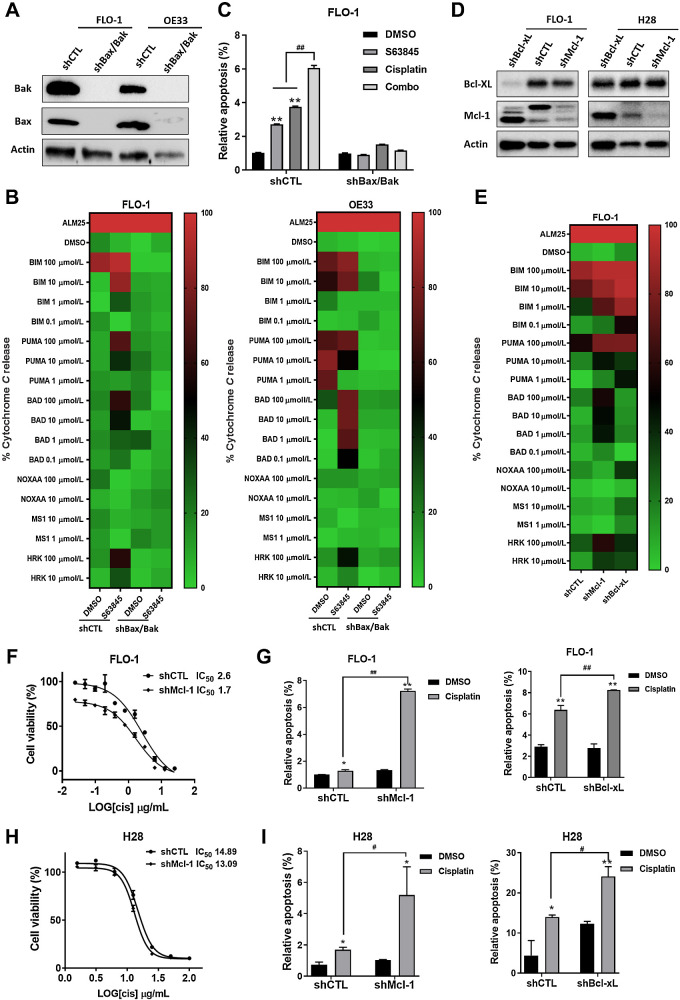
BH3 mimetic enhances the effects of CDDP through the intrinsic mitochondrial apoptotic pathway
As mentioned previously, a BH3 mimetic must induce MOMP in a Bak/Bax-dependent manner. Therefore, we tested whether the results were abrogated when this pathway is disrupted. We developed double knockdowns (DKDs) of Bax and Bak in two EAC cell lines, FLO-1 and OE33, by lentiviral transduction of shRNA (shBax/Bak; Fig. 3A; Supplementary Fig. S6). We performed a DBP with the shBax/Bak versus nontargeted control (shCTL). cyt c release was completely abrogated in the shBak/Bak cells (Fig. 3B). When the shCTL cells were treated with S63845, cyt c release occurred. As further support, the heatmap with shBax/Bak displayed green with every peptide, which suggests that no baseline level of MOMP occurred without these proteins.
Figure 3.
BH3 mimetic enhances the effects of CDDP through the intrinsic mitochondrial apoptotic pathway. A, Stable DKD of Bax and Bak (shBax/Bak) or scrambled control (shCTL) were generated in FLO-1 and OE33 cells by transfecting shRNA lentiviruses. The protein levels of Bax and Bak expression were analyzed by Western blot analysis. B, Heatmap shows DBP in FLO-1 and OE33 cells treated with or without S63845 for 24 hours. C, FLO-1 cells were pretreated with S63845 for 24 hours; the apoptotic ratio in DKD and shCTL cells at 48 hours after treatment with CDDP measured by annexin V flow cytometry; cells positive for annexin V staining were counted as apoptotic cells (**, P < 0.01 for treatment vs. DMSO; ##, P < 0.01 for combination treatment vs. S63845 single treatment). D, Stable knockdown of Mcl-1 (shMcl-1), Bcl-xL (shBcl-xL) or scrambled control (shCTL) were generated by transfecting shRNA lentiviruses. The protein levels of Mcl-1 and Bcl-xL expression were analyzed by Western blot analysis. E, Heatmap shows DBP in shMcl-1, shBcl-xL, or shCTL FLO-1 cells. F, The IC50 values of CDDP with shMcl-1 or shCTL FLO-1 cells are determined. G, The apoptotic ratio in shMcl-1, shBcl-xL, or shCTL FLO-1 cells after treatment with CDDP for 48 hours, measured by annexin V flow cytometry; cells positive for annexin V staining were counted as apoptotic cells. H, The IC50 values of CDDP with shMcl-1 or shCTL H28 cells are determined. I, The apoptotic ratio in shMcl-1, shBcl-xL, or shCTL H28 cells after treatment with CDDP for 48 hours, measured by annexin V flow cytometry; cells positive for annexin V staining were counted as apoptotic cells. (**, P < 0.01 for CDDP versus DMSO; ##, P < 0.01 for shMcl-1, shBcl-xL vs. scrambled control).
To confirm that results of DBP were functionally relevant, the cells were treated with S63845 and CDDP. If S63845 enhances the efficacy of CDDP via the intrinsic pathway, loss of Bax and Bak should abrogate apoptosis. Treatment of shBax/Bak cells with S63845 and CDDP alone or in combination resulted in complete loss of apoptosis compared with controls (Fig. 3C). The lack of cyt c release in the DBP and the loss of apoptosis without Bax and Bak indicate that S63845 kills the cells through the intrinsic mitochondrial apoptotic pathway.
In addition to induction of MOMP in a Bax/Bak-dependent manner, a BH3 mimetic must selectively inhibit an antiapoptotic protein. Therefore, we established Mcl-1 and Bcl-xL knockdowns (shMcl-1, shBcl-xL) in FLO-1 cells by lentiviral transduction to determine whether these knockdowns phenocopied the effects of Mcl-1 and Bcl-xL inhibition (Fig. 3D). The DBP revealed cyt c release from the shMcl-1 and shBcl-xL cells was similar compared with drug treatment (Fig. 3E). Again, to determine whether the cellular phenotype was similar, both shMcl-1and shBcl-xL cells were treated with CDDP. The IC50 level decreased with shMcl-1 versus shCTL (Fig. 3F and H). Apoptosis significantly increased in the shMcl-1 or shBcl-xL cells (Fig. 3G and I). These results show that drug treatment and protein knockdown of Mcl-1 or Bcl-xL have similar effects. As further support for the crucial role for Mcl-1, we tested Mcl-1 localization to the mitochondria and its inhibitory effect on MOMP. Mcl-1 localized primarily to the mitochondria of FLO-1 and H28 cells confirmed by its colocalization with mitochondrial marker TOMM20 (Supplementary Fig. S7). In summary, these results indicate that the BH3 mimetics specifically enhances the effects of CDDP through the intrinsic mitochondrial apoptotic pathway in an Mcl-1 or Bcl-xL–dependent manner.
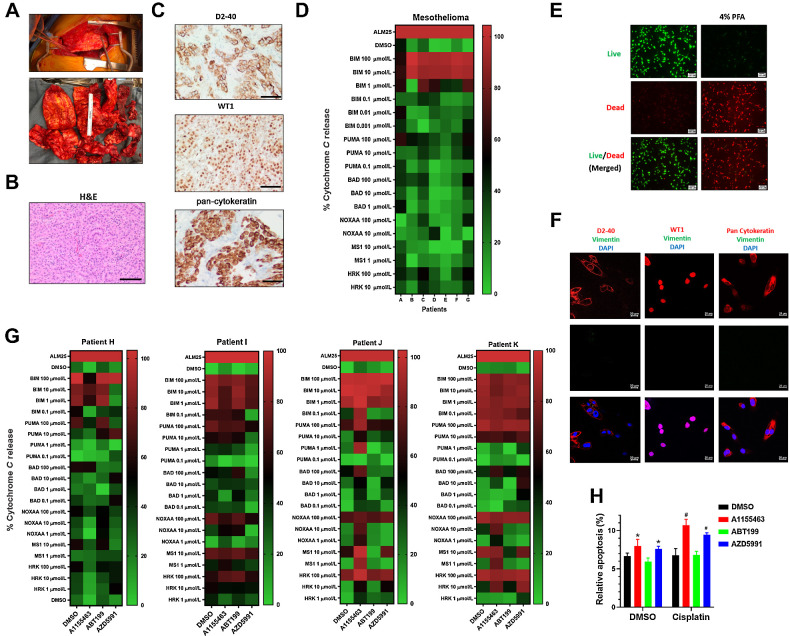
DBP in mesothelioma tumor specimens reveals patient-specific responses
Given that targeting antiapoptotic proteins may enhance the efficacy of standard therapeutics, we assessed whether DBP could be performed directly from fresh EAC and MPM tumor specimens. To our knowledge, DBP has not been performed in MPM or EAC, therefore, we first asked whether BH3 profiling could be performed in these histologies and whether the profiles differed between patients.
For patients with MPM, we performed operations called pleurectomy and decortications (Fig. 4A). Specimens were sent to pathology for pathologic confirmation of the laboratory findings. Both H&E and IHC are required for diagnosis; IHC revealed high expression of D2-40, WT-1, and pan-cytokeratin (Fig. 4B and C). The D2-40 protein enables FACS analysis to identify tumors from the stromal background which were performed on single-cell suspensions for live, CD45−, D2-40+ cells to generate BH3 profiles (Fig. 4D). We observed that patients A, C, and G showed a strong response to BAD and HRK BH3 peptides in addition to the ubiquitous sensitizer, PUMA. Patients B and E appear to be poorly primed, given the weak response to BH3 peptides, which suggests resistance to Bcl-2 antagonism. In summary, these results indicate that BH3 profiles can be developed directly from MPM specimens.
Figure 4.
DBP in mesothelioma tumor specimens reveals patient-specific responses. A, Top, shows gross findings of the MPM during surgery. Bottom, shows the appearance of mesothelioma once removed. B, H&E staining of the resected specimen shows epithelioid component with tubule formation and solid architecture (scale bar = 200 μmol/L). C, Representative results of D2-40, pan-cytokeratin, and WT1 IHC staining on MPM specimens establish the diagnosis and correlate to surface markers used for FACS analysis (scale bar = 200 μmol/L). D, Heatmap showing BH3 profiling in seven patients reveal patient-specific profiles. E, A LIVE/DEAD assay performed in ex vivo cultured tumor cells after surgery 48 hours with 4% PFA fixed cells for positive control. Green fluorescence denotes viable cells stained with calcein-AM, while reddish-orange fluorescence represents dead cells stained with ethidium homodimer. F, Immunofluorescence double staining of pan-cytokeratin, D2-40, and WT1 (red) and vimentin (green) in ex vivo cultured patient K tumor cells (scale bar = 200 μmol/L). G, DBP performed in four patient tumors with 0.2 μmol/L A1155463, AZD5991, and ABT-199 for 20 hours ex vivo. H, An annexin V assay reveals that pretreatment with 0.2 μmol/L A1155463, AZD5991, and ABT199 for 24 hours prior to 48 hours of CDDP treatment primed the mitochondria as noted by the apoptotic ratios. Cells positive for annexin V staining were counted as apoptotic cells (*, P < 0.05 for treatment vs. DMSO; #, P < 0.05 for combination treatment vs. A1155463, ABT-199 or S63845 single treatment).
After performing BH3 profiles, we asked whether DBP can be performed in MPM samples when targeting the antiapoptotic proteins with A1155463, ABT-199, and AZD-5991. Given that AZD-5991 is an Mcl-1 inhibitor that is currently in clinical trials, we switched from the cell culture model with S638452 to AZD-5991 to translate this work into clinical trials. We performed LIVE/DEAD staining at 48 hours post-surgery and noted that the majority of cells were alive (Fig. 4E). Next, we performed IHC revealing high expression of WT-1, D2-40 and pan-cytokeratin, which confirms that the experiments are performed on MPM tumor cells (Fig. 4F). Finally, we performed DBP in MPM tumors with A1155463, ABT-199, and AZD-5991 (Fig. 4G). The DBP indicate that mitochondrial priming in MPM is feasible and that the patterns differ between patients. Patient H showed about 50% cyt c release with AZD5991 but otherwise had relatively poor priming with BH3 mimetic. In contrast, patients J and K appear Bcl-xL–dependent as noted by a marked response to A1155463 with NOXAA, MS1, and HRK peptides. Patient K also had a >50% cyt c release with the BAD protein when treated with AZD5991 whereas patients I and J were poorly primed with AZD5991. To further confirm the predicted drug's efficacy in patient K, we combined the BH3 mimetics with CDDP which revealed significant increases in apoptosis with A1155463 or AZD-5991 and CDDP compared with ABT-199 or DMSO control (Fig. 4H).
DBP in EAC specimens
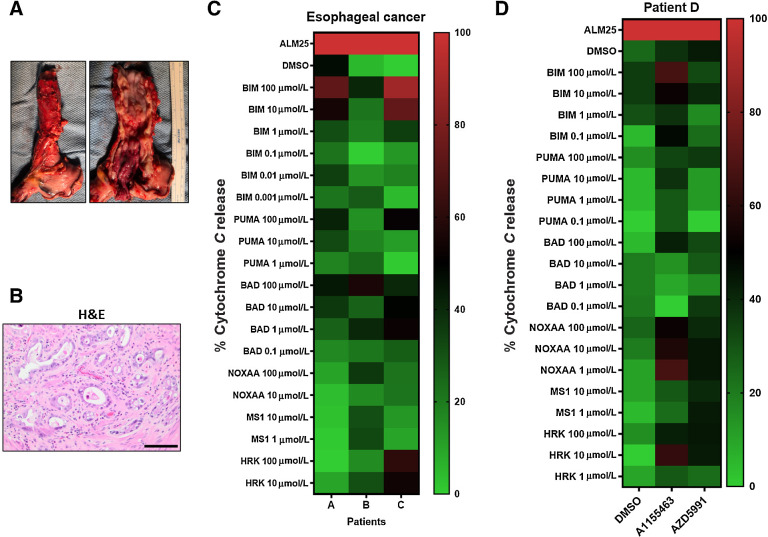
EAC is an epithelial tumor, whereas MPM is a mesenchymal tumor; therefore, we established whether the methods developed for MPM would apply to EAC. Unlike MPM, patients with EAC receive neoadjuvant chemoradiotherapy prior to resection (Fig. 5A). Therefore, tumor specimens are radiated and contain a high level of fibrosis. Regardless of this treatment, biopsies were sent for confirmation by standard H&E stains (Fig. 5B). After dissociated into single-cell suspensions, gating identified single, live, CD45− population, and EpCAM+ cells. BH3 profiles were performed in three patients with EAC, which revealed variable patterns of cyt c release (Fig. 5C). In contrast to MPM, the radiated specimens would not survive 48 hours of ex vivo culture to perform DBP. However, one patient had metastatic EAC to the pleura, which was biopsied and DBP was performed on this nonirradiated tumor. In the DBP of this patient, both A1155463 and AZD5991 primed mitochondria noted by about 50% increases in cyt c release. These results suggest that this patient is sensitive to both Bcl-xL and Mcl-1 inhibition (Fig. 5D). Our results reveal the DBP may be successful in patients with EAC; however, pretreatment biopsies will be required given that irradiated specimens do not survive ex vivo.
Figure 5.
BH3 profiling in patients with EAC. A, An EAC of the distal esophagus after surgery. Left, The gross appearance of the intact esophagus. Right, The mucosal surface with the tumor. B, H&E staining of the resected specimen shows atypical cytologic features including increased nuclear/cytoplasmic ratio, pleomorphism, prominent nucleoli, and intraluminal necrotic debris (scale bar = 200 μmol/L). C, Heatmap showing BH3 profilings in three patients with EAC. D, DBP with 0.2 μmol/L A1155463 and AZD5991 for 20 hours ex vivo from a metastatic pleural implant that was not radiated with the primary esophageal tumor.
Discussion
Even when EAC and MPM tumors respond to systemic chemotherapy, resistance eventually develops in nearly all patients. Resistance develops regardless of whether patients receive standard chemotherapy or targeted therapy. Standard pathologic assessment and even molecular assessments of solid tumors have produced significant advancements in cancer treatment. Yet, in vivo, cancer cells are dynamic and overcome most therapeutic strategies, even those that were initially successful. Current molecular diagnostic assays do not account for cellular plasticity, a common feature underlying tumor heterogeneity in situ (40). The dynamic nature of these mechanisms permits tumor cells to adapt to therapy and acquire resistance. To account for the dynamic cellular changes, bioassays should measure changes in the cell when exposed to therapeutics. Measurement of molecular changes after therapy is particularly important in tumors that are no longer responding to therapy. In addition, EAC and MPM rarely respond to single-agent therapy; therefore, an ideal bioassay also should assess the response to multiple agents. Finally, an effective bioassay must produce results rapidly enough for clinical decisions in patients whose tumors often progress quickly. DBP has the potential to achieve these goals. Therefore, we sought to develop a proof-of-concept model with DBP as a bioassay of living tumor cells in MPM and EAC that accounts for the dynamic changes in the resistance mechanisms, assesses response to combinatorial therapy, and provides results in a clinically feasible time frame.
Given that DBP has promise in predicting response to therapy, we asked whether DBP predicts enhanced efficacy of BH3 mimetics in combination with standard therapy. Ni Chonghaile and colleagues reported that BH3 profiling successfully predicted response to chemotherapy in cell culture (31). The authors observed that BH3 profiling predicted initial response to chemotherapy, which suggested that BH3 profiling is likely an actionable bioassay in solid tumors. Montero and colleagues advanced these findings when they noted that DBP predicted the emergence of resistance to multiple TKIs in lung cancer cells (32). They showed that DBP performed at progressive timepoints correlated with emergence of resistance in lung cancer cells treated with TKIs over several months. Bhola and colleagues advanced these findings with solid tumors by screening patient-derived xenografts (PDXs) from colon cancer with multiple therapeutics. They noted that responses were highly patient specific. Despite these advances, whether DBP could predict targets that enhanced the response to standard therapy is unknown. Because EAC and MPM are rarely treated with single agents, we asked whether DBP could foretell which resistance protein to target in combination with CDDP treatment. We chose BH3 mimetics that directly target the antiapoptotic proteins in the intrinsic mitochondrial apoptotic pathway as a proof-of-concept approach. This pathway is downstream of mutant driver proteins and therefore, should be effective regardless of mutational burden. We chose CDDP because it is the most commonly used chemotherapeutic in both EAC and MPM. We observed that the BH3 mimetics that altered the DBP predicted which drug enhances efficacy of CDDP. We noted that targeting Mcl-1 or Bcl-xL, but not Bcl-2, showed alterations in the DBP. Subsequently, we noted that targeting Mcl-1 or Bcl-xL increased the efficacy of CDDP whereas targeting Bcl-2 did not. These results show that DBP is predictive of combinatorial therapy and that directly targeting the antiapoptotic proteins is a feasible approach. This approach will help identify a multidrug treatment strategy for treatment refractory EAC and MPM.
Considering that BH3 mimetics appear effective with CDDP, we queried whether these effects were specific to the intrinsic mitochondrial apoptotic pathway. Villalobos-Ortiz and colleagues developed a biochemical “tool kit” which is a methodology to ensure that the most selective and potent BH3 mimetics are identified (34). They used DBP in conjunction with annexin V/Hoechst viability testing to ensure that the BH3 mimetics functioned as predicted on the basis of their antiapoptotic target. They defined a BH3 mimetic based on selectively inhibiting an antiapoptotic protein, binding with high affinity, and inducing MOMP in a Bak/Bax-dependent manner. They overexpressed Bcl-2 antiapoptotic proteins in murine cells and treated with a panel of putative BH3 mimetics. However, whether DBP will predict which antiapoptotic protein to target in solid tumor cells that do not overexpress these proteins is unknown. Therefore, we asked whether the effects of combinatorial therapy with BH3 mimetics and CDDP occurred by selectively targeting these antiapoptotic proteins. By knocking down the effector proteins, Bax and Bak, we observed complete elimination of apoptosis in these cells. Next, to determine whether the BH3 mimetics were specific for Mcl-1 or Bcl-xL as predicted by the DBP, we knocked down Mcl-1 or Bcl-xL and observed the same effects as blocking with a drug. Taken together, by blocking effector proteins and Mcl-1 and Bcl-xL, we observed that the combined effects were specific for the intrinsic mitochondrial apoptotic pathway.
To determine whether this approach is clinically feasible in MPM and EAC, we performed DBP directly from patient tumors. The majority of DBP have been performed on nonsolid tumors such as leukemia and lymphoma. Leukemia and lymphoma are conducive to single-cell suspensions when isolated from the blood. In contrast, solid tumors have a highly developed microenvironment that contains stromal cells, neovascular cells, immune cells as well as actual tumor cells. Bhola and colleagues reported DBP from two colon cancer resections (35). They dissociated the tumors, treated for 24 hours, and performed a DBP on EpCAM+ cells (31). Analysis directly from the tumor cells is critical because the other cells and especially the immune cells will generate DBP that will result in misleading information. Therefore, we isolated the CD45− cells to avoid performing DBP on immune cells and stained with D2-40 and EpCAM, respectively. Prior to the DBP, we performed BH3 profiles on 10 patients' samples that revealed substantial heterogeneity between patients regardless of histology. This heterogeneity is consistent with the report by Bhola and colleagues who tested 11 drugs on each of the two colon cancer specimens and noted substantial differences in priming between the two specimens despite the same histology. After the initial BH3 profiles, we performed DBP on the MPM tumors which revealed different responses to BH3 mimetics per patient. These findings highlight the requirement for precision-based medicine with an effective bioassay such as DBP to overcome resistance in these tumors.
Our results show that DBP can screen drug combinations for synergistic effects. In addition, we generated BH3 profiles in 10 patients and DBP in four patients with two different solid tumors, but several limitations exist. Bhola and colleagues report high-throughput DBP to screen a larger number of drugs in PDX models (35). Although the PDX model generates reliable results, production of a PDX per patient sample requires several months, which is not a clinically feasible. Developing the high-throughput technique directly from patient samples is a future direction that is critical for clinical translation. Another critical limitation is post-therapy DBP. DBP was not feasible after tumor cells were irradiated, therefore, results from pretreatment biopsies will be required for tumors that are treated with radiation.
EAC and MPM contain thousands of mutations per cell; therefore, targeting one mutation or mutant pathway is overcome by redundant mutations that enable cancer cell growth. Targeting mitochondrial pathways holds promise because mitochondria regulate cell death through the intrinsic pathway and serve as a critical center for multiple metabolic pathways essential for tumor growth. However, this approach requires live cell assays that measure the alterations in mitochondria when treated with therapeutics. Therefore, a dynamic bioassay is necessary for this strategy to be successful. DBP achieves all of these goals. In summary, DBP is a bioassay of living tumor cells that measures changes in resistance mechanisms, predicts response to combinatorial therapy, and produces results rapidly enough for clinical feasibility. Our future goal is to develop this technology to generate clinically meaningful results within 7 days of tumor biopsy (Fig. 6).
Figure 6.
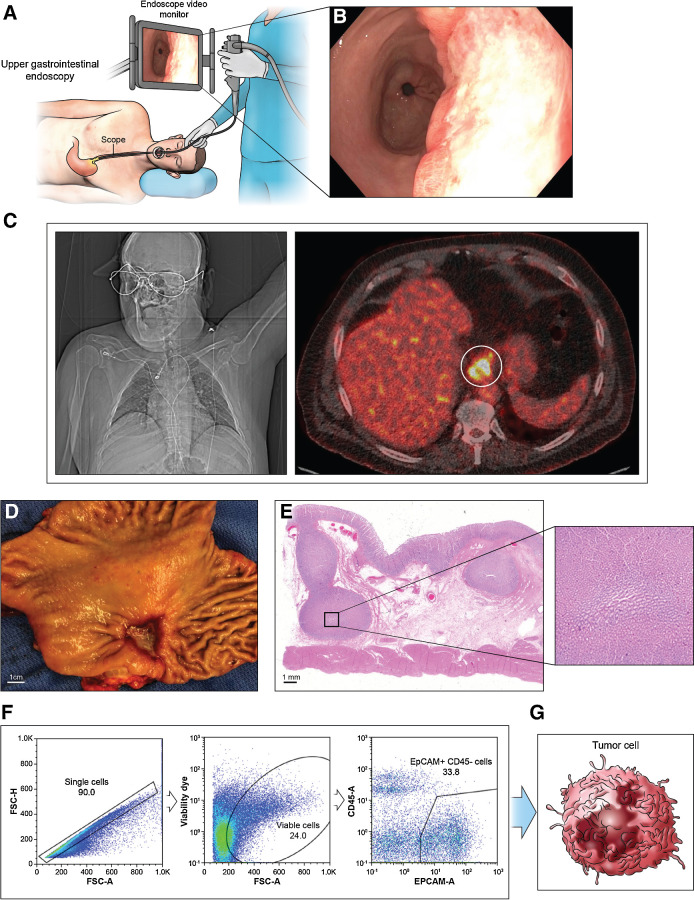
Schema of cancer cell isolation from patient. A, An endoscopy procedure involves inserting a long, flexible tube (endoscope) down the throat and into the esophagus. A camera on the end of the endoscope shows the esophagus. B, Endoscopic view of EAC. C, Positron emission tomography of EAC shows high glucose uptake. D, Appearance of esophagus after surgery. E, H&E staining of the resected specimen. A magnified view of black-boxed area reveals large tumor nest. F and G, Flow cytometry analysis of cancer cell subsets isolates single cancer cell suspensions labeled with cell surface markers produce distinguishable BH3 profiles upon gating.
Authors' Disclosures
B.M. Burt reports grants from Momotaro-Gene and AstraZeneca outside the submitted work. No disclosures were reported by the other authors.
Acknowledgments
This project was supported by the Michael E. DeBakey Department of Surgery, Faculty Research Award, and the Roderick D. MacDonald Research Fund at Baylor St. Luke's Medical Center. This project was supported by Cytometry and Cell Sorting Core at Baylor College of Medicine with funding from CPRIT Core Facility Support Award (CPRIT-RP180672), NIH (P30 CA125123 and S10 RR024574), and expert assistance from Joel M. Sederstrom. H.C. Hodges is a CPRIT Scholar supported by grants from CPRIT (RR170036), NIH (R35GM137996), Gabrielle's Angel Foundation, the V Foundation (V2018-003), and Mark Foundation for Cancer Research (20-024-ASP). This project was supported by Pathology and Histology Core (HTAP) at Baylor College of Medicine with funding from the Dan L. Duncan Comprehensive Cancer Center NIH grant [P30 Cancer Center Support Grant (NCI-CA125123)] and expert assistance of Patricia Castro.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Note: Supplementary data for this article are available at Molecular Cancer Therapeutics Online (http://mct.aacrjournals.org/).
Authors' Contributions
Deborah R. Surman: Conceptualization, data curation, formal analysis, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing. Yuan Xu: Conceptualization, data curation, formal analysis, supervision, validation, investigation, methodology, writing–original draft, writing–review and editing. Min-Jung Lee: Conceptualization, resources, formal analysis. Jane Trepel: Resources, formal analysis, investigation, methodology, writing–original draft. Kate Brown: Formal analysis, investigation, methodology. Maheshwari Ramineni: Data curation, formal analysis, methodology. Taylor G. Splawn: Data curation, methodology, writing–review and editing. Laurence P. Diggs: Validation, investigation, methodology, writing–original draft. H. Courtney Hodges: Conceptualization, resources, formal analysis, writing–original draft, writing–review and editing. Jeremy L. Davis: Resources, data curation, investigation, writing–original draft. Hyun-Sung Lee: Resources, formal analysis, funding acquisition, investigation, writing–original draft. Bryan M. Burt: Resources, supervision, validation, investigation. Robert Taylor Ripley: Conceptualization, resources, formal analysis, supervision, funding acquisition, methodology, writing–original draft, project administration, writing–review and editing.
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 2. Rusch VW, Giroux D, Kennedy C, Ruffini E, Cangir AK, Rice D, et al. Initial analysis of the international association for the study of lung cancer mesothelioma database. J Thorac Oncol 2012;7:1631–9. [DOI] [PubMed] [Google Scholar]
- 3. Kindler HL, Ismaila N, Armato SG, Bueno R, Hesdorffer M, Jahan T, et al. Treatment of malignant pleural mesothelioma: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 2018;36:1343–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kong CY, Kroep S, Curtius K, Hazelton WD, Jeon J, Meza R, et al. Exploring the recent trend in esophageal adenocarcinoma incidence and mortality using comparative simulation modeling. Cancer Epidemiol Biomarkers Prev 2014;23:997–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xu Y, Surman DR, Diggs L, Xi S, Gao S, Gurusamy D, et al. Bile acid-induced "Minority MOMP" promotes esophageal carcinogenesis while maintaining apoptotic resistance via Mcl-1. Oncogene 2020;39:877–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bajpai M, Aviv H, Das KM. Prolonged exposure to acid and bile induces chromosome abnormalities that precede malignant transformation of benign Barrett's epithelium. Mol Cytogenet 2012;5:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rake C, Gilham C, Hatch J, Darnton A, Hodgson J, Peto J. Occupational, domestic and environmental mesothelioma risks in the British population: a case-control study. Br J Cancer 2009;100:1175–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Y, Feingold PL, Surman DR, Brown K, Xi S, Davis JL, et al. Bile acid and cigarette smoke enhance the aggressive phenotype of esophageal adenocarcinoma cells by downregulation of the mitochondrial uncoupling protein-2. Oncotarget 2017;8:101057–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Palanca-Wessels MC, Barrett MT, Galipeau PC, Rohrer KL, Reid BJ, Rabinovitch PS. Genetic analysis of long-term Barrett's esophagus epithelial cultures exhibiting cytogenetic and ploidy abnormalities. Gastroenterology 1998;114:295–304. [DOI] [PubMed] [Google Scholar]
- 11. Huo X, Juergens S, Zhang X, Rezaei D, Yu C, Strauch ED, et al. Deoxycholic acid causes DNA damage while inducing apoptotic resistance through NF-kappaB activation in benign Barrett's epithelial cells. Am J Physiol Gastrointest Liver Physiol 2011;301:G278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feingold PL, Surman DR, Brown K, Xu Y, McDuffie LA, Shukla V, et al. Induction of thioredoxin-interacting protein by a histone deacetylase inhibitor, entinostat, is associated with DNA damage and apoptosis in esophageal adenocarcinoma. Mol Cancer Ther 2018;17:2013–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brokatzky D, Dörflinger B, Haimovici A, Weber A, Kirschnek S, Vier J, et al. A non-death function of the mitochondrial apoptosis apparatus in immunity. EMBO J 2019;38:e100907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu Y, Surman DR, Ripley RT. Minority MOMP: a toxic, slow demise. Oncotarget 2020;11:3559–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gillies LA, Kuwana T. Apoptosis regulation at the mitochondrial outer membrane. J Cell Biochem 2014;115:632–40. [DOI] [PubMed] [Google Scholar]
- 16. Westphal D, Dewson G, Czabotar PE, Kluck RM. Molecular biology of Bax and Bak activation and action. Biochim Biophys Acta 2011;1813:521–31. [DOI] [PubMed] [Google Scholar]
- 17. Ichim G, Lopez J, Ahmed SU, Muthalagu N, Giampazolias E, Delgado ME, et al. Limited mitochondrial permeabilization causes DNA damage and genomic instability in the absence of cell death. Mol Cell 2015;57:860–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tait SWG, Parsons MJ, Llambi F, Bouchier-Hayes L, Connell S, Muñoz-Pinedo C, et al. Resistance to caspase-independent cell death requires persistence of intact mitochondria. Dev Cell 2010;18:802–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Tang HL, Tang HM, Mak KH, Hu S, Wang SS, Wong KM, et al. Cell survival, DNA damage, and oncogenic transformation after a transient and reversible apoptotic response. Mol Biol Cell 2012;23:2240–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tait SW, Green DR. Mitochondria and cell death: outer membrane permeabilization and beyond. Nat Rev Mol Cell Biol 2010;11:621–32. [DOI] [PubMed] [Google Scholar]
- 21. Singh R, Letai A, Sarosiek K. Regulation of apoptosis in health and disease: the balancing act of BCL-2 family proteins. Nat Rev Mol Cell Biol 2019;20:175–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Certo M, Moore VDG, Nishino M, Wei G, Korsmeyer S, Armstrong SA, et al. Mitochondria primed by death signals determine cellular addiction to antiapoptotic BCL-2 family members. Cancer Cell 2006;9:351–65. [DOI] [PubMed] [Google Scholar]
- 23. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646–74. [DOI] [PubMed] [Google Scholar]
- 24. Perciavalle RM, Opferman JT. Delving deeper: MCL-1's contributions to normal and cancer biology. Trends Cell Biol 2013;23:22–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Williams MM, Lee L, Hicks DJ, Joly MM, Elion D, Rahman B, et al. Key survival factor, Mcl-1, correlates with sensitivity to combined Bcl-2/Bcl-xL blockade. Mol Cancer Res 2017;15:259–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ertel F, Nguyen M, Roulston A, Shore GC. Programming cancer cells for high expression levels of Mcl1. EMBO Rep 2013;14:328–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rezaei Araghi R, Bird GH, Ryan JA, Jenson JM, Godes M, Pritz JR, et al. Iterative optimization yields Mcl-1-targeting stapled peptides with selective cytotoxicity to Mcl-1-dependent cancer cells. Proc Natl Acad Sci U S A 2018;115:E886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Arulananda S, O'Brien M, Evangelista M, Harris TJ, Steinohrt NS, Jenkins LJ, et al. BCL-XL is an actionable target for treatment of malignant pleural mesothelioma. Cell Death Discov 2020;6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ryan J, Montero J, Rocco J, Letai A. iBH3: simple, fixable BH3 profiling to determine apoptotic priming in primary tissue by flow cytometry. Biol Chem 2016;397:671–8. [DOI] [PubMed] [Google Scholar]
- 30. Potter DS, Letai A. To prime, or not to prime: that is the question. Cold Spring Harb Symp Quant Biol 2016;81:131–40. [DOI] [PubMed] [Google Scholar]
- 31. Chonghaile TN, Sarosiek KA, Vo T-T, Ryan JA, Tammareddi A, Moore VDG, et al. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science 2011;334:1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Montero J, Sarosiek KA, DeAngelo JD, Maertens O, Ryan J, Ercan D, et al. Drug-induced death signaling strategy rapidly predicts cancer response to chemotherapy. Cell 2015;160:977–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kotschy A, Szlavik Z, Murray J, Davidson J, Maragno AL, Le Toumelin-Braizat G, et al. The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature 2016;538:477–82. [DOI] [PubMed] [Google Scholar]
- 34. Villalobos-Ortiz M, Ryan J, Mashaka TN, Opferman JT, Letai A. BH3 profiling discriminates on-target small molecule BH3 mimetics from putative mimetics. Cell Death Differ 2020;27:999–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bhola PD, Ahmed E, Guerriero JL, Sicinska E, Su E, Lavrova E, et al. High-throughput dynamic BH3 profiling may quickly and accurately predict effective therapies in solid tumors. Sci Signal 2020;13:eaay1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Souers AJ, Leverson JD, Boghaert ER, Ackler SL, Catron ND, Chen J, et al. ABT-199, a potent and selective BCL-2 inhibitor, achieves antitumor activity while sparing platelets. Nat Med 2013;19:202–8. [DOI] [PubMed] [Google Scholar]
- 37. Leverson JD, Phillips DC, Mitten MJ, Boghaert ER, Diaz D, Tahir SK, et al. Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci Transl Med 2015;7:279ra40. [DOI] [PubMed] [Google Scholar]
- 38. Tron AE, Belmonte MA, Adam A, Aquila BM, Boise LH, Chiarparin E, et al. Discovery of Mcl-1-specific inhibitor AZD5991 and preclinical activity in multiple myeloma and acute myeloid leukemia. Nat Commun 2018;9:5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Chou TC, Talalay P. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 1984;22:27–55. [DOI] [PubMed] [Google Scholar]
- 40. Smith EA, Hodges HC. The spatial and genomic hierarchy of tumor ecosystems revealed by single-cell technologies. Trends Cancer 2019;5:411–25. [DOI] [PMC free article] [PubMed] [Google Scholar]