Abstract
Post-embryonic organogenesis is critical for plant development. Underground, lateral roots (LR) form the bulk of mature root systems, yet the ontogeny of the LRP is not clear. Here, we use single-cell RNA-seq through the first four stages of LR formation in Arabidopsis. Our analysis leads to a model in which a single group of precursor cells, with different cell identity from their pericycle origins, rapidly reprograms and splits into a mixed ground tissue/stem cell niche fate and a vascular precursor fate. The ground tissue and stem cell niche fates soon separate and a subset of more specialized vascular cells form sucrose transporting phloem cells that appear to connect to the primary root. We did not detect cells resembling epidermis or root cap, suggesting that outer tissues may form later preceding LR emergence. At this stage, some remaining initial precursor cells form the primordium flanks while the rest create a reservoir of pluripotent cells able to replace the LR if damaged. Laser ablation of the central and lateral LRP regions shows that remaining cells restart the sequence of tissue initiation to form a LR. Our analysis reveals an ontological hierarchy for LR formation with an early and sequential split of main root tissues and stem cells.
Keywords: organogenesis, patterning, stem cells, single cell RNA-seq, cell fate, regeneration
SUMMARY
Transcriptomics of individual cells from the first stages of the LRP, when tissue initiation presumably first occurs, identified 7 early cell populations. Monitorization of these cell populations through confocal microscopy, genetics and laser ablation experiments revealed that the ontogeny of the LRP is organized and involves the assembly of cell lineages following conserved sequences within three developmental trajectories. We also identified the associated role of camalexin in one of the three developmental trajectories.
INTRODUCTION
Postembryonic plant organogenesis requires the recruitment of specific subsets of cells known as (organ) founder cells (Chandler, 2011; Van Norman et al, 2013). Root founder cells typically give rise to lateral roots (LR) (De Rybel et al., 2010; Motte et al., 2019; Perez-Garcia and Moreno-Risueno, 2018). The LR organogenic process initiates with asymmetric founder cell division and culminates with the formation of a new root meristem, thus generating all root cell types from a limited number of initial cells (Malamy and Benfey, 1997; Motte et al., 2019; Trinh et al., 2018). .
Careful characterization of LR primordium (LRP) formation in Arabidopsis has led to its classification in seven developmental and/or temporal stages and the identification of central and lateral growth domains (Malamy and Benfey, 1997), which were shown to emerge from non-deterministic cell division patterns (Lucas et al., 2013a; von Wangenheim et al., 2016). A gene regulatory network of LR formation (Lavenus et al., 2015) inferred from gravistimulated roots (Voß et al., 2015) showed dependency on AUXIN RESPONSE FACTOR (ARF) 7 for the formation of lateral growth domains of the LRP, while the central growth domain was defined by mutual inhibition between ARF7 and ARF5. PLETHORA (PLT) 3, PLT5, PLT7 and SCARECROW (SCR) factors have been shown to be expressed at the initial stages of LRP formation (Du and Scheres, 2017; Goh et al., 2016). PLT3, PLT5 and PLT7 are required for the initial formative divisions of the LRP central domain, which leads to the specification of the LR meristem tissues, activation of PLT1, PLT2 and PLT4 and the establishment of an auxin maximum. In turn, SCR initiates the specification of the quiescent center (QC) of the new LR. These findings support a major role for genetic components in LRP patterning indicating that cell lineages must form early during LR formation, with one of them leading to QC specification.
Organ formation has been widely studied during embryogenesis. Arabidopsis embryogenesis follows a highly ordered sequence of cell divisions forming stereotypic developmental stages. At the globular stage of embryogenesis the main tissue identities have been already specified as well as the hypophysis, the precursor cell of QC and of distal stem cells of the root (Möller et al., 2017; ten Hove et al., 2015). Thus, histogenesis in the Arabidopsis embryo was understood as the simultaneous formation of multiple cell types or lineages, although more recent experiments have established the outline of a possible hierarchy during embryo tissue formation (Smit et al., 2020).
Recently, high throughput single cell RNA Sequencing (sc-RNA-Seq) of the primary root meristem have deepened our knowledge of tissue formation (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shahan et al., 2020; Shulse et al., 2019; Zhang et al., 2019). These studies have addressed the developmental transitions taking place to generate tissues from the stem cell niche (SCN), indicating that during primary root growth, formation of most tissues occurs simultaneously. Tissue formation is also critical during regenerative processes. sc-RNA-Seq of root regeneration upon resection of the main meristem also suggests concurrent formation of the new tissues and SCN, with early transitioning cells showing mixed precursor identities (Efroni et al., 2016). Analysis of xylem identity through sc-RNA-Seq provided a molecular mechanism for cell identity transitions and differentiation showing the importance of the underlying gene regulatory circuit (Turco et al., 2019). Recent sc-RNA-Seq profiling LR initiation during root gravistimulation has identified novel regulators of pericycle reprogramming as well as of LRP growth and stemness (Gala et al., 2021). However, powerful sc-RNA-seq approaches have not yet been applied to capture the specific cells of the early LRP when tissue initiation presumably first occurs (Du and Scheres, 2017; Goh et al., 2016), and whether LR histogenesis follows any of the steps known from embryonic, adult, or regenerative root formation.
In this work, we have specifically isolated cells from early stages of the Arabidopsis LRP and performed sc-RNA-Seq. Our research has determined the initiation of six novel cell populations following three developmental branches from an initial pool of primordial cells. One of these branches involves the sequential formation of two cell populations with stem cell features and contributing to the specification of a new SCN. A second branch involves the formation of three vascular-like cell populations which might culminate in protopholem formation connecting the LRP and the primary root. Finally, a later third branch initiates a cell population associated with the primordium flanks and maintaining a reservoir of primordial/pluripotent cells able to regenerate the LR if damaged. Our work shows the orderly assembly of cell lineages during LR histogenesis, suggesting the existence of hierarchical cell states of formation during postembryonic development.
RESULTS
Transcriptional profiling of lateral root formation with single cell resolution
In order to investigate LR organogenesis, we took advantage of the ability of sc-RNA-Seq to comprehensively profile cell states at given stages of development. This technique has been shown to accurately recapitulate existing cell populations and predict developmental transitions in the primary root (Denyer et al., 2019; Jean-Baptiste et al., 2019; Ryu et al., 2019; Shahan et al., 2020; Shulse et al., 2019; Zhang et al., 2019). Most LR tissues are presumably initiated from stage I to IV as supported by genetic evidences (Du and Scheres, 2017; Goh et al., 2016). To specifically profile early stages (I-IV) of LR formation we used the 2 kb promoter of HOMEOBOX 53 (pHB53) transcription factor (TF) fused to the mCherry fluorescent protein and a nuclear localization signal (NLS), as this promoter specifically drives expression to early LRP cells (González-Grandío et al., 2017) (Fig 1A). LR formation occurs sequentially, following a shootward timing, and thus, LR are at different stages of formation in the same plant. We used 4 day-old roots, which under our growth conditions contained a mixture of LRP from stage I to IV. Based on LRP quantification and number of cells in each stage of LRP development, we estimated a proportion of cells of 21.8%, 20%, 24.1% and 34.1% from stage I to IV, respectively, in our samples. Thus, our samples represented LRs at different stages of development that would need to be deconvoluted (see below). We enzymatically digested cell walls, sorted 3 replicates of mCherry positive cells (using Fluorescent Activated Cell Sorting; Fig 1B) into 96-well plates, and generated cDNA libraries based on the Smart-Seq2 protocol (Picelli et al., 2014). We opted for the plate-based technique in order to provide deep per cell read profiles of the rare but highly targeted LR cells.
Figure 1. Distinctive cell populations can be identified through Single-Cell RNA sequencing (sc-RNA-seq) during early lateral root (LR) organogenesis.
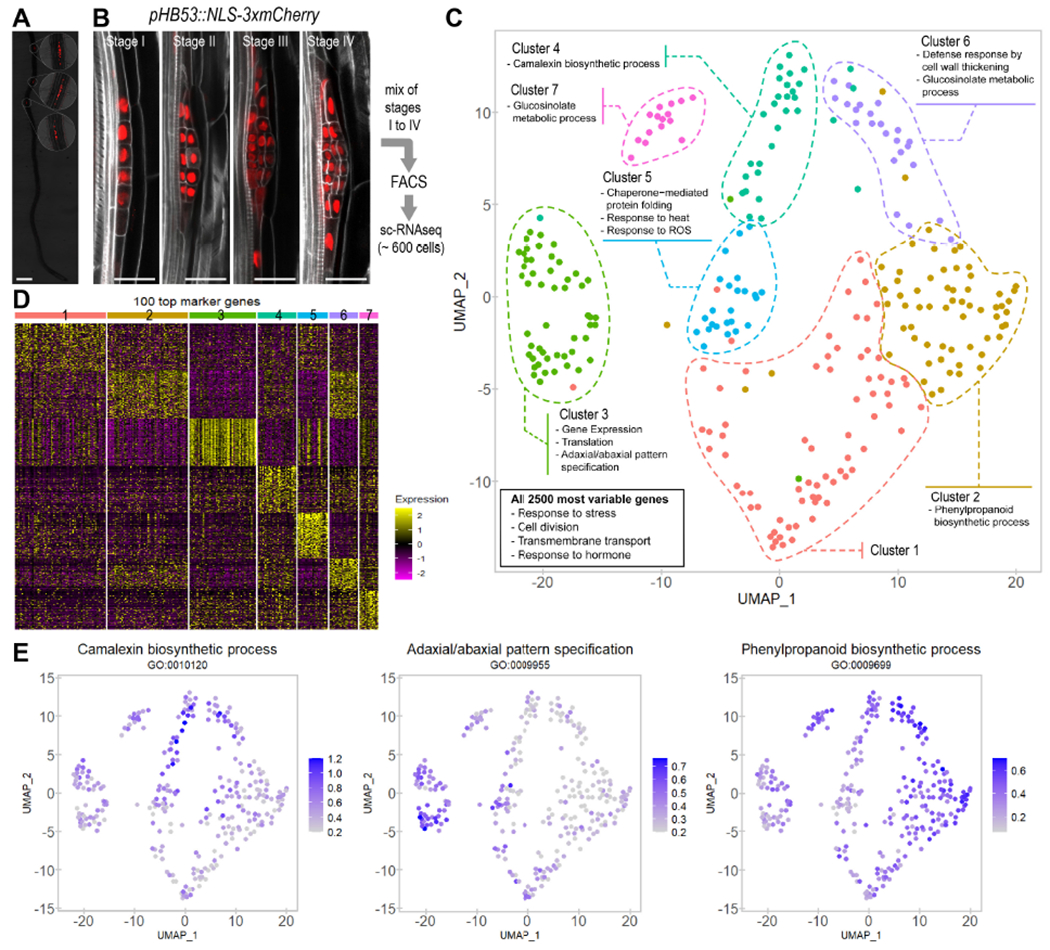
(A) Overlapping fluorescent and bright field images of a 4 day-old Arabidopsis seedling showing expression of the LR primordium (LRP) specific marker HOMEOBOX 53 (HB53) mCherry (pHB53::NLS-3xmCherry). Scale bar: 0.5 mm. Insets: 3.5x magnification. (B) Confocal images showing expression of pHB53::NLS-3xmCherry at the LR developmental stages profiled by sc-RNA-seq. Proportion of cells in each developmental stage is indicated. Stained cell walls are shown in a gray scale. Scale bars: 25 μm. (C) Uniform Manifold Approximation and Projection (UMAP) dimensional reduction of the highly informative LRP cells profiled through sc- RNA-Seq. Each dot corresponds to an individual cell. Cells were clustered in 7 populations and colored according to the legend. A summary of the GO terms enriched in the top 100 gene markers for each population is shown. Box: summary of enriched GO terms for the 2,500 most variable genes among all populations (D) Heat map showing scaled expression of the top 100 gene markers for each population in all individual cells. Cells were arranged by populations. (E) UMAP plots showing the average expression of the genes annotated in three representative GO terms enriched in population 4 (left), population 3 (center) and populations 6 and 2 (right).
Reads were processed using the R Seurat package (Butler et al., 2018; Satija et al., 2015). Quality control filters were applied to work with the more informative cells (see Material and Methods). As a result, we selected a total of 282 highly informative cells with 362,293 mean reads/cell (Fig S1A) and 4,415 mean transcripts/cell that corresponded to 22,557 genes from the Arabidopsis genome. During data processing, we regressed out variability related to total number of reads, the percentage of mitochondrial genes and the cell cycle stage of the cells. Thus, we ensured subsequent clustering was not affected by these unwanted sources of variation (Fig S1A-C). Many cells were detected to have recently divided or be dividing (phases G1 and G2/M based on expression of conserved cell cycle markers, Table S1) as expected for a LRP in formation which shows fast proliferation rates (Lucas et al., 2013a; von Wangenheim et al., 2016). In addition, cells collected by each replicate distributed homogenously, confirming reproducibility of our experimental design (Fig S1D).
A comprehensive classification of cell populations in the lateral root primordium
Unsupervised clustering based on the transcriptomic profiles of the selected cells resulted in the identification of differentiable populations. We represented their spatial distribution applying the dimensional reduction technique Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2018). To efficiently cluster cells in Seurat based on the diversity of the dataset, a resolution parameter can be adjusted. Clustering at very low resolutions differentiated two main populations, which suggests that at least two cell identities exist at early developmental stages (Fig S1E, F). Two additional clusters became distinguishable at a slightly higher resolution, suggesting a strong identity for cell populations eventually named as 3 and 5. Applying a resolution used in other root models (Ryu et al., 2019; Zhang et al., 2019) led to five different populations. Due to the highly dynamic nature of the LRP we used a less restrictive resolution (1.1) and identified 6 cell populations. Visual inspection allowed us to manually separate a seventh cluster (population 7) from population 4 as it was spatially separated (Fig 1C). Comparison of the expression levels of the top one hundred markers confirmed unique gene expression profiles for the seven populations identified (Fig 1D).
To investigate the functionality of these cell populations, we first analyzed the enriched GO terms in the 2,500 most variable genes (Table S2) among all populations. We found terms associated to cell division, response to stress and endogenous stimuli and transmembrane transport (Fig 1C, Fig S2A), as expected in a rapidly forming organ where signaling might mediate pattern formation. As LRP are known to accumulate auxin, which is required for their development (Benkova et al., 2003), we analyzed the expression of auxin polar transport genes, although auxin transport and response were not enriched GO terms. We found expression of both auxin influx- and eflux-transporter genes across all the populations (Fig S3A-B), which correlated well with a relatively ubiquitous auxin response in the early LRP cells (Fig S3C-D). In agreement with this observation, distribution of the auxin efflux transporters PINFORMED (PIN) 1 and PIN3 as well as DR5 reported auxin response did not show dramatic differences within LRP cells at very early stages of development (I and II) (Du and Scheres, 2017).
We also investigated GO enrichment in the top one hundred gene markers (Table S2) for each population and found specific GO terms, except for population 1 which did not show enrichment in any GO category (Fig 1C, Fig S2B). Population 4 was specifically enriched in camalexin metabolism (Fig 1C, E, Fig S2B), a phytoalexin related with pathogen response and which functions as a cell cycle regulator (Glawischnig, 2007). Population 3 showed enrichment in categories related to pattern specification, gene expression, chromatin remodeling and epigenetics marks (Fig 1C, E, Fig S2B-C). As cells in this population appeared to be very active transcriptionally and expressed patterning regulators, they might correspond to the central part of the LRP, since this region has been shown to very actively contribute to LRP formation (Lucas et al., 2013a). GO categories strongly represented in population 5 (Fig 1C, Fig S3B) could be grouped in heat shock response and protein folding, categories previously associated with stem cells (de Luis Balaguer et al., 2017). The GO terms associated with populations 2 and 6, related to callose deposition, cell wall thickening and phenylpropanoid metabolism such as lignin (Fig 1C, E, Fig S3B), which resembled developmental mechanisms observed in vascular tissues (Hellmann et al., 2018; Vatén et al., 2011; Xie et al., 2011).
Lateral root primordium cells share affinity to known root identities
As LR organogenesis leads to the formation of a root meristem (Trinh et al., 2018), we next explored whether the identified populations showed any quantitative affinities to root meristem cells. To systematically explore known cell identities (Table S3), we calculated the Index of Cell Identity (ICI) scores for our single cell dataset (Efroni et al., 2015). The majority of the cells in our dataset could not be assigned to a known identity with a significant p-value, which is consistent with the fact that the LRP is an organ forming de novo. Among the similarities found (Fig S4), we observed that cell populations 3 and 5 presented the higher scores for QC cells (~50 % of relative cell identity among all the identities found), while cell populations 1, 2, 4 and 6 had the highest vasculature scores. Pericycle identity scores were very low, which agrees with LRP cells rapidly reprogramming away from their original identity. These results are consistent with early cell identity acquisition during the LR organogenesis process, with cells gaining full root tissue identities during subsequent development.
Two lateral root primordium cell populations are enriched in stem cell like features and predict formation of the quiescent center and ground tissue lineages
As we found partial similarities to known root identities, we next used available root transcriptomes to further investigate the identity of the LRP cells. As population 5 was enriched in GO categories previously associated to stem cells (de Luis Balaguer et al., 2017), we further analyzed its similarity with transcriptomes related to root stem cells. We found enrichment in gene expression associated to cortex/endodermis initials, and thus cortex/endodermis initial markers (Clark et al., 2019) were specifically expressed in population 5 (Fig 2A). A requirement for fully functional proteins or proteostasis has been associated to stem cell identity (Yan et al., 2020), and we found that chaperones with enriched expression in the endodermis or the QC (Brady et al., 2007) had enriched expression in population 5 (Fig 2B). In addition, as population 5 had affinity to QC identity (Fig S4), it might represent a novel cell type transitioning to stem cells or to QC cells.
Figure 2. Two LRP cell populations are enriched in stem cell gene expression and give rise to cell lineages associated to endodermis/cortex initials and the quiescent center (QC).
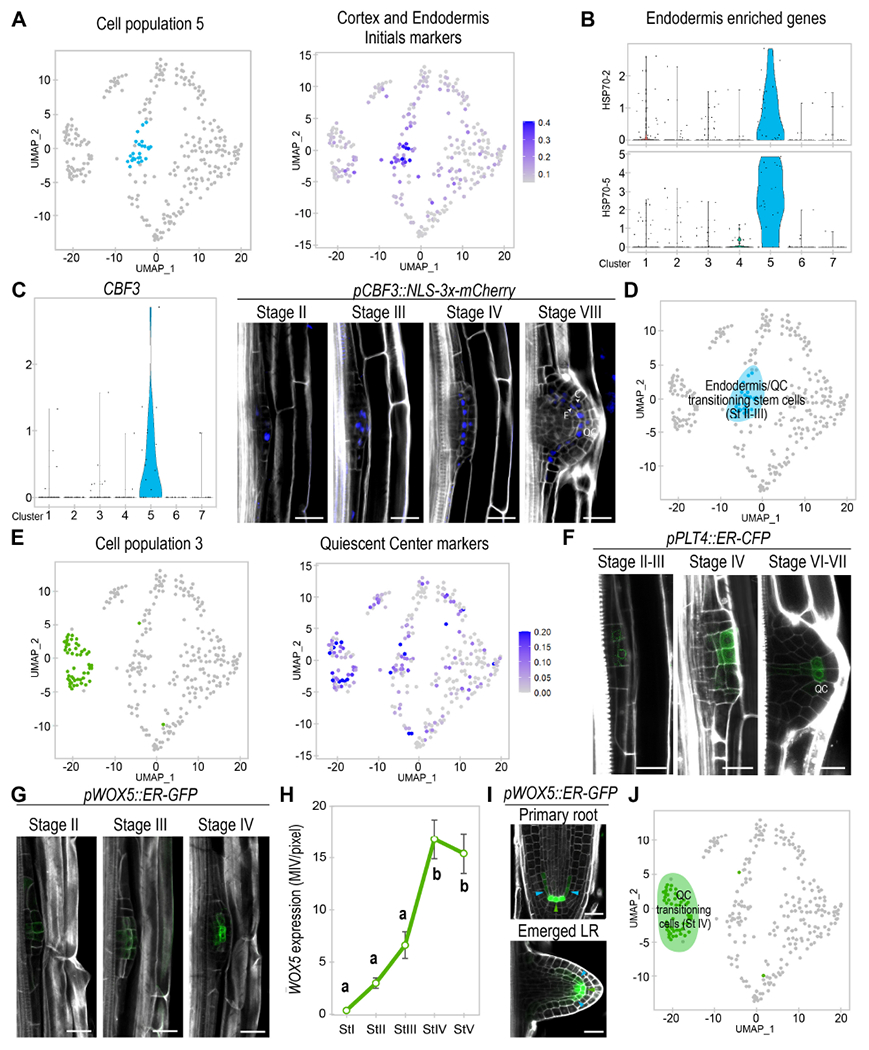
(A) Left, UMAP plot showing population 5 in pale blue. Right, UMAP representation of the average expression of cortex/endodermis initials marker genes. (B) Violin plot showing average expression of the endodermis enriched genes HEAT SHOCK PROTEIN (HSP) 70-2 and 70-5. (C) Left, violin plot showing expression of the population 5 biomarker CBF3. Right, confocal images of pCBF3::NLS-3x-mCherry (in blue) during LRP formation. (D) UMAP plot defining population 5 as Endodermis/QC-transitioning-stem-cells on a blue background. (E) Left, UMAP plot representing population 3 in green. Right, UMAP of QC markers average expression. (F-H) Confocal images showing (F) pPLT4:CFP and (G) pWOX5::ER-GFP expression (in green) and (H) quantification pWOX5 expression from stages I to V of LR formation. MIV/pixel: Mean Intensity Value/pixel. Different letters: p-value < 0.05 according to ANOVA and Tukey HSD post-hoc tests. n≥10 LRP/sample. (I) Confocal images of pWOX5::ER-GFP in the primary root and an emerged LR. Green and blue arrows: cells with higher and lower expression, respectively. (C, F, G, I) Stained cell walls are shown in a gray scale. E: endodermis, C: cortex, QC: Quiescent center. Scale bars: 25 μm. (J) UMAP plot defining population 3 as QC-transitioning-cells on a green background.
We next explored biomarker expression for population 5, and found the TF C-REPEAT BINDING FACTOR 3 (CBF3) to be enriched in this population (Fig 2C). We used the CBF3 promoter region to drive expression of a triple mCherry fluorescent protein fused to a NLS in a stably transformed plant line, and performed confocal laser microscopy. We first observed pCBF3::NLS-3xmCherry expression at stage II of LRP formation (Fig 2C), specifically, expression was observed at the central cells of the primordium on the external side. Analyses of subsequent stages of development showed that pCBF3::NLS-3xmCherry expression was gradually enriched in a row of cells that eventually corresponded to the endodermis/cortex lineage or the QC of the new LR. These results suggested that population 5 must be a transitioning type of stem cell with QC and endodermis features (Fig 2D).
Continuing our search for similarities between root meristematic tissues and LRP cells, we found enriched expression of QC related markers (Denyer et al., 2019) for population 3 and part of the Endodermis/QC-transitioning-stem-cells (Fig 2E), including the well-known QC regulator WUSCHEL RELATED HOMEOBOX 5 (WOX5) (Sarkar et al., 2007) and of the root stem cell regulator PLT4 (Galinha et al., 2007). Additional analyses showed that expression of SCN and other QC genes (Brady et al., 2007; Zhang et al., 2019) was also enriched in population 3 and part the Endodermis/QC-transitioning-stem-cells (Fig S5A), suggesting that population 3 would also be related to the formation of a new QC or SCN.
Next, we performed confocal laser microscopy using the markers pWOX5::ER-GFP and pPLT4::ER-CFP (Galinha et al., 2007; Sarkar et al., 2007). pPLT4::ER-CFP expression started at stage II/III of LRP formation. Prolonged exposure of pPLT4::ER-CFP showed signal in all central cells at stage II/III as previously described (Du and Scheres, 2017). However, we found that pPLT4::ER-CFP expression was enriched in several of the central cells on the external side (Fig 2F), which showed some overlap with pCBF3::NLS-3xmCherry (marking the Endodermis/QC-transitioning-stem-cells). pPLT4::ER-CFP expression became stronger at stage IV and remained restricted to the central cells of the LRP (Fig 2F), while at stage V/VI it mainly coincided with the QC of the new LR. Similarly, pWOX5::ER-GFP expression was also low and associated with the central cells of the LRP at stages II and III (Fig 2G). Similarly to pPLT4::ER-CFP, pWOX5::ER-GFP expression also greatly increased at stage IV (Fig 2H) remaining at the central area of the LRP and showing maximum expression in the cells predicted to become the new QC (Fig 2G). Inspection of pWOX5::ER-GFP expression pattern in the primary root and emerged LR (Fig 2I) showed high expression levels associated with the QC while low expression levels associated with stem cells such as the ground tissue initials and recently formed endodermis cells. Thus, we used pWOX5::ER-GFP to track initiation of QC identity. A peak in pWOX5::ER-GFP expression occurs at stage IV of LR emergency (Fig 2H), supporting the initiation of QC identity in population 3 at stage IV. In contrast, low pWOX5::ER-GFP expression appears to associate with stem cells, explaining the observed overlapping between pWOX5::ER-GFP and the Endodermis/QC transitioning stem cell biomarker at stage II/III. These results also support that some Endodermis/QC-transitioning-stem-cells might specialize to generate population 3 and eventually the new QC at stage IV. We define population 3 as QC-transitioning-cells (Fig 2J).
Three lateral root primordium cell populations reflect early vascular-like identity acquisition.
Further cell identity analysis showed that populations 1, 2 and 6 were enriched in early-stage vascular markers (Ryu et al., 2019) (Fig 3A). In addition, expression of known protophloem markers and functional regulators, such as OCTOPUS (OPS) and OBP2 as well as sieve element genes (Denyer et al., 2019; Miyashima et al., 2019; Ryu et al., 2019; Truernit et al., 2012) were enriched in population 1 (Fig 3B, S5B). The transcription factor NAC020, which is specifically expressed in developing phloem and is upstream of the master regulator of phloem development ALTERED PHOLEM DEVELOPMENT (APL) (Kondo et al., 2016), also showed enriched expression in population 1 (Fig 3B). The largest association of cluster 1 with protophloem marker genes in comparison with clusters 2 and 6 suggested that this cell population might be more developed or differentiated. As protophloem is one of the first vascular tissues undergoing differentiation to provide meristem loading (Furuta et al., 2014; Lucas et al., 2013b), these observations suggested that several LRP cells might rapidly specialize to assure water and nutrient supply from the primary root.
Figure 3. Different LRP cell populations show characteristic gene expression associated to vasculature development and one of them defines the cells adjacent to the phloem tissues of the primary root.
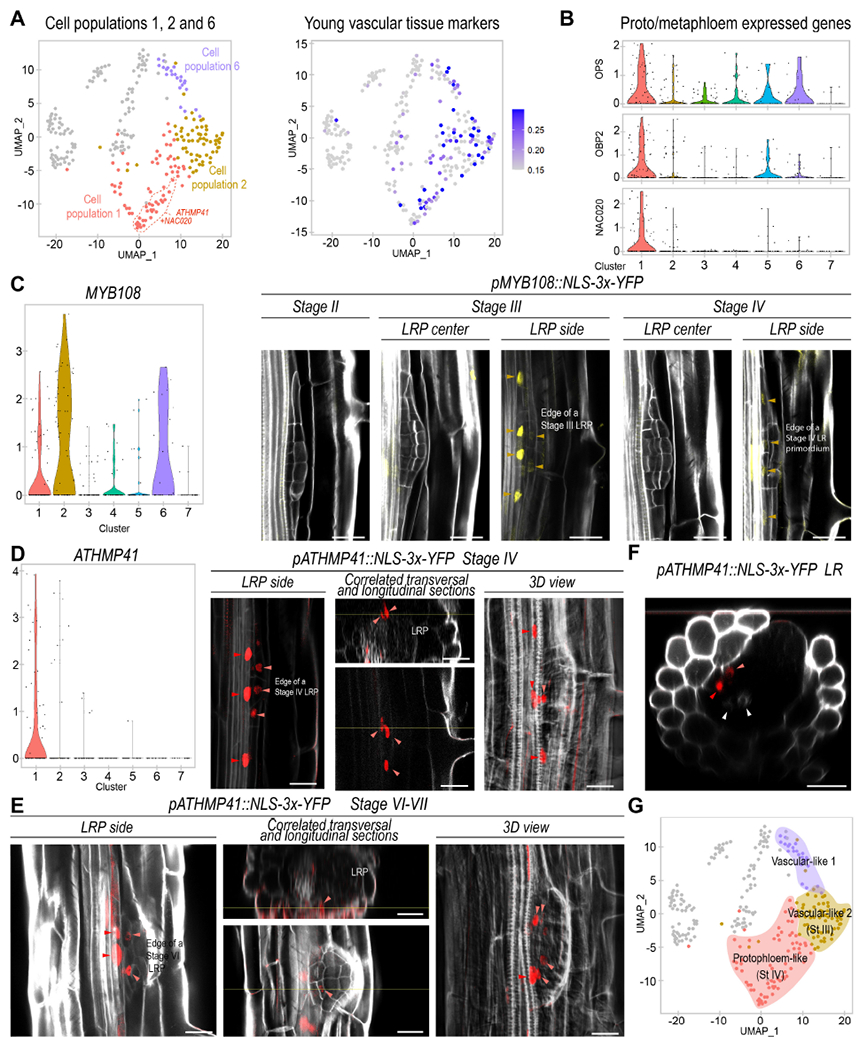
(A) Left, UMAP plot showing cell populations 1, 2 and 6 coded by colors. Cells with enriched expression of the population 1 biomarkers ATHMP41 and NAC020 are surrounded by a dashed red line Center, UMAP representation of the average expression of young vascular tissue markers. (B) Violin plot showing the average expression of protophloem and metaphloem representative genes: OCTOPUS (OPS), OBP2 and NAC020. (C) Left, violin plot showing expression of MYB108. Right, confocal images showing expression of pMYB108:NLS-3xYFP (in yellow) in a LRP (LRP) at stages II to IV. Orange arrows: pMYB108 marked cells. (D) Left, violin plot showing expression of ATHMP41. Right, confocal images showing expression of pATHMP41:NLS-3xYFP (in red) at stage IV of LR formation. (E-F) Images showing expression of pATHMP41:NLS-3xYFP during LR formation (D) and in a recently emerged LR (E). (D-F) Light red arrowheads: cells inside the LRP. Red arrowheads: primary root phloem. White arrowheads: primary root xylem. Stained cell walls are shown in a gray scale. Scale bars: 25 μm. (G) UMAP plot defining populations 6, 2 and 1 as vascular-like and protopholem-like cells on colored backgrounds.
To further explore the composition of the vascular identities, we identified as biomarkers the MYB108 and ATHMP41 genes. MYB108 showed enriched expression in population 2 while ATHMP41 was specifically expressed in population 1 (Fig. 3C, D). ATHMP41 encodes a metal transport protein, and intriguingly, this gene and the regulator of phloem development NAC020 showed enriched expression in a similar, coherent subset of cluster 1 (Fig 3A), suggesting that those cells might be more differentiated or constitute a cellular subtype. In agreement with this possibility, UMAP representations at very high resolutions led to the sub-clustering of population 1 (in subpopulations 1A, 1B and 1C) while the rest of populations remained largely unaltered (Fig S6A, B). Notably, expression of ATHMP41 and NAC020 was enriched in subpopulation 1A and absent in subpopulation 1C (Fig S6C).
We generated stable lines containing transcriptional fusions of the promoter regions of MYB108 and ATHMP41 to a triple yellow fluorescent protein (YFP) and a NLS. We monitored expression through confocal microscopy. We found consistent expression of pMYB108::NLS-3xYFP at stage III in cells adjacent to the vasculature of the primary root and located on the side of LRP (Fig 3C). pMYB108::NLS-3xYFP expression decreased at stage IV although it remained in similar locations of the LRP. Notably, pATHMP4::NLS-3xYFP expression started later, at stage IV (Fig 3D), while its expression pattern partially overlapped with that of pMYB108::NLS-3xYFP, and thus, some cells located on the side of the LRP base also showed pATHMP4::NLS-3xYFP expression at stage IV. The more restricted expression of pATHMP4::NLS-3xYFP correlates well with the predicted heterogeneity of cluster 1 and only a small portion of population 1 (or subpopulation 1A) being enriched in this biomarker expression. The cells expressing pATHMP4::NLS-3xYFP were adjacent to the phloem poles of the primary root, which also expressed pATHMP4::NLS-3xYFP (Fig 3D). At later stages of LR development pATHMP4::NLS-3xYFP expression remained adjacent to the phloem poles of the primary root (Fig 3E-F) suggesting that several cells within population 1 might early specify phloem to connect the flux of photosynthates and other potential metabolites to the LRP from the primary root.
The spatial and temporal expression pattern of pMYB108::NLS-3xYFP and pATHMP4::NLS-3xYFP support that some cells from population 2 might specialize to form population 1 and protophloem. Given that cell populations 1, 2 and 6 were arranged following a line in the UMAP representation, it appears that these cell populations could be part of a differentiation trajectory, with population 1 being more developed located at one end. We designated these cell populations as vascular-like-1, vascular-like-2 and protopholem-like (Fig 3G).
Establishing the ontogeny of the early lateral root primordium
Based on our analyses, the QC-transitioning-cells (stage IV) could be a specialized subset of the Endodermis/QC-transitioning-stem-cells (stage II), while the vascular-like populations might constitute a differentiation trajectory eventually leading to phloem specification. Analysis of enrichment in root meristematic markers (Ryu et al., 2019) showed that cells located in the UMAP representation at the end of these possible developmental processes (Fig 4A; following the colored arrows) had the highest enrichment, thus being the most similar to a root meristem. As culmination of the LRP developmental process is a root meristem, this analysis further supports that our data can be used to infer the developmental trajectories of the LRP cells.
Figure 4. An early primordial cell population gives rise to three developmental trajectories during LR organogenesis.
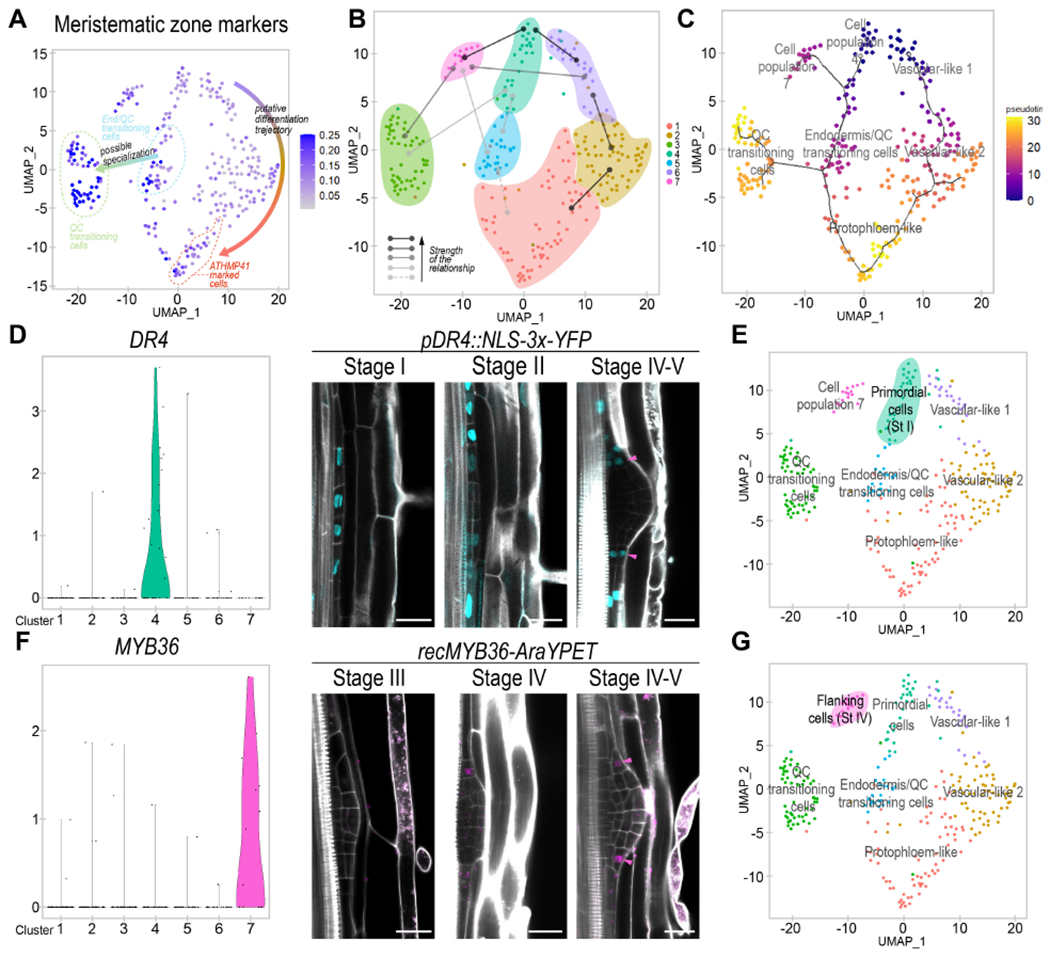
(A) UMAP plot showing the average expression of meristematic root markers. (B) Illustrative figure representing the results obtained from the Partition-based Graph Abstraction (PAGA) analysis. Lines: relationships among populations. Strength of the relationships is displayed according to the legend. (C) UMAP dimensional reduction plot of the pseudotemporal trajectories obtained by Monocle 3. Dark line: predicted pseudotemporal trajectories. White dots: root nodes. Dot color: pseudotime value for each cell. (D) Left, violin plot showing expression of the population 4 biomarker DR4. Right, confocal images showing the expression of pDR4:NLS-3xYFP (in pale blue) during LRP formation. (E) UMAP plot defining population 4 as primordial cells on a bluish green background. (F) Left, violin plot showing the expression of the population 7 biomarker MYB36. Right, confocal images showing expression of the MYB36-AraYPET recombineering line (in magenta) during LRP formation. (D, F) Magenta arrowheads show cells in the same relative position of the LRP. Stained cell walls are shown in gray scale. Scale bars: 25 μm. (G) UMAP plot defining population 7 as flanking cells on a magenta background.
To investigate the ontogeny of the seven cell populations identified, we performed connectivity and pseudotime analyses. We first used Partition-based Graph Abstraction (PAGA), which estimates connectivity of manifold partitions (Wolf et al., 2019). We observed very strong connectivity between populations 4 and 7, and 4 and the populations proposed to constitute a vasculature developmental trajectory (6, 2 and 1) (Fig 4B, Fig S7A). This analysis supports that the vasculature related populations could have originated from population 4 and form a developmental trajectory. A lower threshold showed moderate connectivity between population 4 and the Endodermis/QC-transitioning-stem-cells among other possible connections. As a whole, the PAGA analysis suggested that population 4 might constitute a primordial cell population that would give rise to most of the LRP cells.
Next, we performed a pseudotime ordering of the cells using Monocle 3 (Cao et al., 2019). We rooted the pseudotemporal trajectory on populations 4 and vascular-like-1 based on the PAGA analysis, and their lower expression of root meristematic markers. We observed a predicted trajectory from vascular-like-1 to vascular-like-2 to protopholem-like (Fig 4C) coinciding with the proposed vasculature developmental trajectory. Population 4 appeared in direct connection with the Endodermis/QC-transitioning-stem-cells, and these cells with the QC-transitioning-cells, defining a stem cell developmental trajectory, which was in agreement with some of our previous observations. Population 7 defined a third trajectory that also started from population 4. Finally, this analysis also predicted a trajectory between the Endodermis/QC-transitioning-stem-cells and the protopholem-like population, which could relate with the fact that the former population showed certain vasculature features (Fig S5B). Analysis of ground tissue formation during embryogenesis shows that ground tissue initials express vascular markers (Smit et al., 2020). As based on our analysis Endodermis/QC-transitioning-stem-cells would give rise to the LR ground tissue, it is possible that partially overlapping ground tissue and vasculature identities could be a common feature related to ground tissue initiation.
Cell population 4 was very intriguing as we had not found expression of any known regulators or gene markers of the primary root and might constitute a primordial cell population giving rise to the rest of the LRP cells. As pHB53::NLS-3xmCherry starts to be expressed at the onset of founder cell division, this population could represent dividing founder cells or their daughters at stage I, although we did not expect to capture founder cell division consistently given that this process occurs within ~3 hours (Goh et al., 2012). Comparison of biomarkers with previous LR initiation datasets (De Smet et al., 2008; Parizot et al., 2010) showed that population 4 had the closest association (out of the 7) with dividing founder cells as expected, although only ~1/3 of the biomarkers were putative founder cell division genes (Fig S7B). Based on this analysis, this population most likely corresponds to the founder cell daughters at stage I. We defined cell population 4 as primordial cells (Fig 4E).
Next, we constructed a transcriptional fusion of the promoter region of the identified biomarker gene DR4 (Fig 4D) encoding a putative protease inhibitor, to drive expression of the YFP fused to a NLS and performed confocal laser microscopy. We found expression of pDR4::NLS-3xYFP at stage I of the LR formation (Fig 4D), in agreement with the primordial cells being a precursor state. To better understand the relationship between the primordial cells and the other LRP cells, we analyzed pDR4::NLS-3xYFP expression at subsequent stages of LRP development. At stage II the dividing central cells of LRP showed weak expression of pDR4::NLS-3xYFP as compared with the flanking cells. At later stages, we found that pDR4::NLS-3xYFP expression was maintained on the sides of the LRP suggesting that cells with a slow proliferation rate might preserve the initial identity of this population. Comparison of our dataset with the LR gene regulatory network (Lavenus et al., 2015) inferred from gravistimulated roots (Voß et al., 2015) showed that the primordial cells had the greatest dependency on ARF regulation (Fig S7C). Notably, ARF7 regulation associates with initiation of the founder cell daughters and establishment of the LRP lateral zones. In addition, some ARF7 targets are expressed in the sides of the LRP base from stage IV to emergence (Lavenus et al., 2015), which is in agreement with the primordial cells being maintained on the sides of the LRP base. In contrast, cell fate transitions of the primordial cells associated with fast division of the LRP central cells to give rise to the Endodermis/QC-transitioning-stem-cells and QC-transitioning-cells, which also correlated well with the QC-transitioning-cells showing no dependency on ARF7 regulation (Fig S7C).
Analysis of the characteristics features of population 7 revealed the enriched expression of MYB36 (Fig S5C). MYB36 is a transcription factor expressed in the flanking regions of the LRP from stage V to emergence and defining the boundaries between proliferating and arrested cells (Fernández-Marcos et al., 2017). Given the predicted developmental trajectory from the primordial cells to cell population 7, we used MYB36 as biomarker gene (Fig 4F) to investigate the spatial and temporal relationships between these two cell populations. We performed confocal laser microscopy of the AraYPET recombineering construct (Liberman et al., 2015), detecting signal in several cells of the LRP flanking regions at stage IV/V. Importantly, cells in these flanking regions which showed MYB36-AraYPET expression also showed DR4-YFP expression (Fig 4D, F, magenta arrows), supporting that population 7 could derive from a subset of the primordial cells as predicted by the developmental trajectory. We defined population 7 as flanking cells (Fig 4G).
Functional relationships among primordial cells, endodermis/QC-transitioning-cells and QC-transitioning-cells support a stem cell developmental trajectory
Our confocal analyses on marker expression demonstrated stage-dependent initiation for each cell population, which spatially associated within predicted developmental trajectories (Fig 5A). Analysis of expression of the top 100 biomarkers for each population in the LR formation dataset generated by gravistimulation (Voß et al., 2015) showed that early identities such as primordial cells and vascular-like-1 cells peaked at early time points, while late identities such as flanking cells and QC-transitioning-cells went up later, coinciding, respectively, with stages I and IV as expected (Fig S7D). Primordial cell biomarker expression also went up at later times confirming our observations that this identity is maintained on the sides during formation of the LRP base. Endodermis/QC-transitioning-stem-cell identity peaked at stage II differing at later stages, while vascular-like-2 and protopholem-like identities were partially recapitulated, which could be caused by the fact that these populations share gene expression with the endodermis and the protopholem of the primary root and this dataset profiles whole roots (Voß et al., 2015). All these spatial temporal associations support initiation of new identities following a conserved sequence.
Figure 5. Functional relationships among primordial cells, endodermis/QC-transitioning-cells and QC-transitioning-cells support a stem cell developmental trajectory.
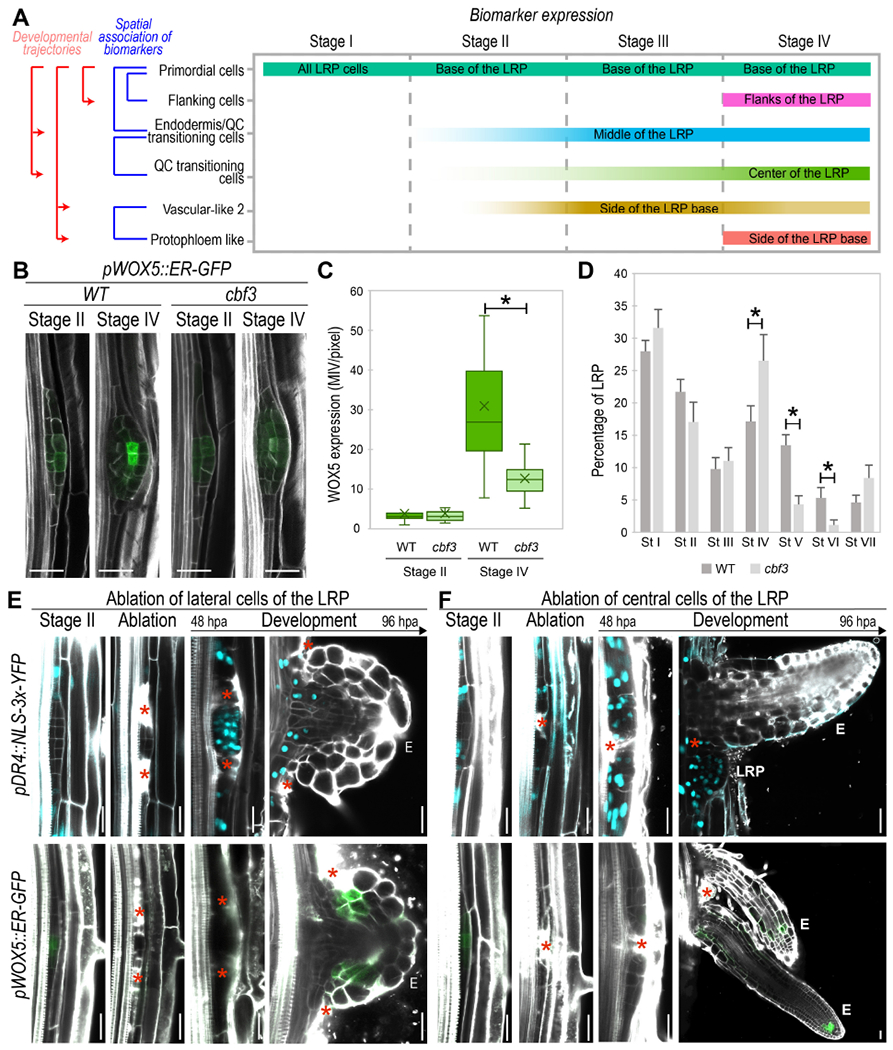
(A) Schematic representation of LRP cell populations initiation based on confocal microscopy analyses of biomarkers. Intensity of the color bar represents the intensity of the biomarker expression. (B-C) Confocal images showing (B) pWOX5::ER-GFP expression in the WT and in cbf3 mutant at St II and IV of LRP formation and (C) its quantification at 6 days post imbibition (dpi). Bars: boxplot whiskers. Crosses: average. MIV/pixel: Mean Intensity Value/pixel. *: p-value < 0.05 by ANOVA and Bonferroni post-hoc test. n≥ 15 LRP/sample. (D) Quantification of LRP stages in the WT and cbf3 mutant at 6 dpi. *: p-value < 0.05 by ANOVA and Bonferroni tests. n≥ 25 roots/sample. (E-F) Confocal images showing the expression of pDR4::NLS::3xYFP (in cyan) and pWOX5::ER-GFP (in green) following laser ablation of LRP lateral (E) or central (F) cells at stage II in 5 dpi roots, hpa: hours post ablation. Red asterisks: sites of ablation. E: emerged LR, LRP: Lateral root primordium. (B, E, F) Stained cell walls are shown in gray scale. Scale bars: 25 μm.
To further test LR tissue ontogeny we focused on the stem cell developmental trajectory. Based on this trajectory the Endodermis/QC-transitioning-stem-cells should give rise to the QC-transitioning-cells. Analysis of the mutant for the Endodermis/QC-transitioning-stem-cells marker (cbf3) showed a reduction in pWOX5::ER-GFP expression at stage IV, whereas no change was observed at stage II as compared with control roots (Fig 5B-C). As high pWOX5::ER-GFP expression at stage IV associated with specification of the QC-transitioning-cells, the observed reduction in pWOX5::ER-GFP expression in cbf3 indicates altered specification of this cell population. When we analyzed LRP formation in cbf3, we observed abnormal accumulation of LRP at stage IV while LRP in the subsequent developmental stages V and VI decreased; confirming that initiation of QC-transitioning-cells from Endodermis/QC-transitioning-stem-cells is required for LRP development.
Next, we performed laser ablation of the LRP central and lateral regions at stage II in plants carrying the primordial cell population marker (pDR4::NLS-3x-YFP) and pWOX5::ER-GFP. Ablation of the LRP lateral regions, resulted in inactivation of pWOX5::ER-GFP and activation of pDR4::NLS-3x-YFP expression in the remaining cells to resume LRP formation, and eventually, to express pWOX5::ER-GFP again (Fig 5E). Thus, ablation of the lateral regions of the LRP appeared to restart the sequence of identity initiation predicted by the stem cell developmental trajectory to form a new LR. Ablation of the LRP central region caused fast division of the primordial cells in the laterals to form two adjacent LRP or LRs, which indicates that all primordial cells have similar formative capacities regardless of their position or origins. In agreement with the proposed stem cell developmental trajectory, cells marked with pDR4::NLS-3x-YFP did not show pWOX5::ER-GFP expression (Fig 5F) (i.e. if they correspond to the large daughter cells or are immediately derived from them).
Overall, these results show the functional relevance of the Endodermis/QC-transitioning-stem-cells, a previously unknown cell population, and the existence of a stem cell developmental trajectory that initiates cell identities following a conserved interconnected sequence.
The primordial cells constitute a pluripotent cell reservoir and associate with camalexin metabolite
As ablation of the central side of the LRP resulted in division of the remaining primordial cells to form new LRs, we wondered if the primordial cells might maintain their identity and functionality after LR emergence. We observed pDR4::NLS-3x-YFP expression at the base of emerged LR (Fig 6A). Furthermore, expression of the founder cell marker pSKP2Bs::NLS-3x-mCherry (Perianez-Rodriguez et al., 2021) was also found at the base of emerged LRs, suggesting that primordial cells remained and might be functional. Next, we excised LRs and observed that new LRs emerged from the root regions containing the pDR4::NLS-3x-YFP and pSKP2Bs::NLS-3x-mCherry marked cells (Fig 6B), indicating that primordial cells appear to form new LRs when the initial LR was damaged.
Figure 6. The primordial cells constitute a reservoir for pluripotent cells and associate with camalexin metabolite.
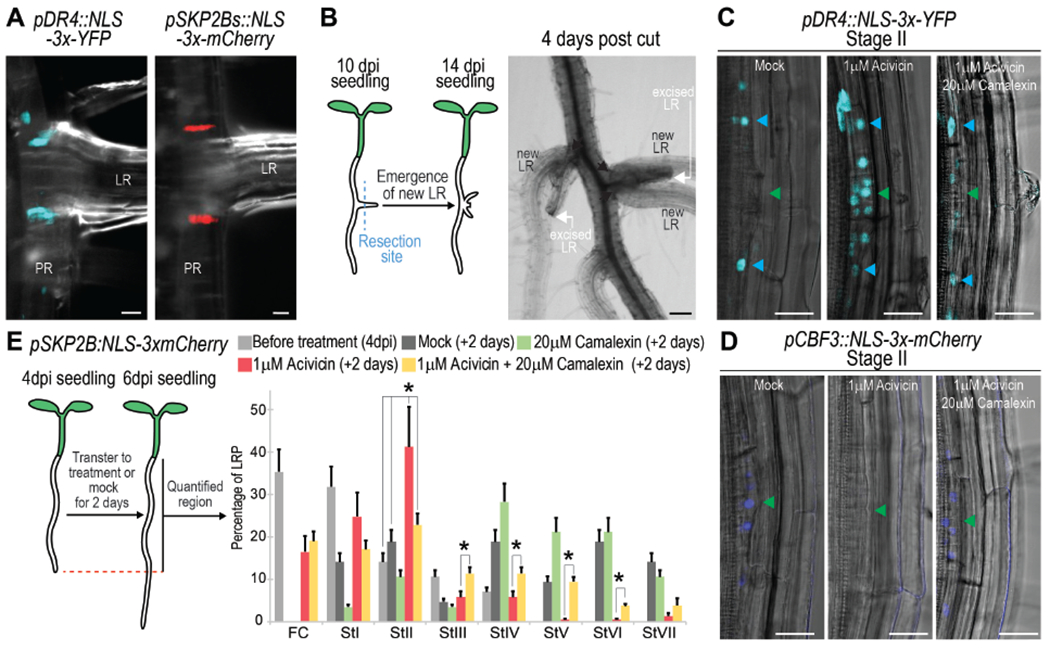
(A) Confocal images showing expression of the marker for the primordial cells pDR4:NLS-3xYFP and the founder cell marker pSKP2Bs:NLS-3xmCherry at the base of emerged LRs. (B) Left, cartoon illustrating experiment. Right) Bright field images showing formation of new LRs after excision of emerged LRs. Black arrows: sites of emergence. (C-D) Confocal images showing expression of (C) pDR4:NLS-3xYFP or (D) pCBF3:NLS-3xmCherry in 4 dpi roots treated with 1μM acivicin or 1μM acivicin and 20 μM camalexin for 2 days. Acivicin is a camalexin biosynthesis inhibitor. Green/cyan arrows: central/lateral LRP regions, respectively. (E) Acivin and camalexin regulate LRP formation. Left, cartoon illustrating experiment. Right, quantification of LRP stages following treatments. *: p-value < 0.05 by ANOVA and Bonferroni tests. n≥20 roots/condition. (A-C, D) Scale bars: 25 μm (A,C,D) and 125 μm (B).
The camalexin biosynthesis pathway was a GO term enriched in the primordial cells (Fig 1C, E), so we decided to further explore this association. When plants were treated with acivicin, which has been shown to inhibit camalexin biosynthesis in Arabidopsis (Su et al., 2011), we found activation of pDR4::NLS-3x-YFP expression in the central cells of the LRP at stage II, while pCBF3::NLS-3x-mCherry expression was turned off (Fig 6C, D). Supplementation of roots with both acivicin and camalexin restored pDR4::NLS-3x-YFP expression to the LRP sides and pCBF3::NLS-3x-mCherry expression to the central region of the LRP. When we analyzed LRP formation in roots treated with acivicin, we observed that LRP did not develop normally accumulating at stage II. However, this developmental inhibition could be partially reversed when the roots were additionally supplemented with camalexin (Fig 6E), which suggest that initiation of Endodermis/QC-transitioning-stem-cells at stage II is critical for LRP development. Finally, as activating pDR4::NLS-3x-YFP expression is accompanied by turning off pCBF3::NLS-3x-mCherry and vice versa (Fig 6C, D) it can be inferred that primordial and Endodermis/QC-transitioning-stem-cell identities are part of a conserved sequence, further supporting the stem cell developmental trajectory and the associated role of camalexin.
DISCUSSION
Our research has identified seven cell populations formed during the early stages of LR organogenesis. Five out of them constitute completely novel cell types, while the QC-transitioning-cells represent a precursor state of the LR QC and the primordial cells likely correspond to the stage I founder cell daughters. Interestingly, our results show the same identity for all these cells, although they are formed by a division morphologically asymmetric. We have also determined the spatial localization of six of these cell populations within the LRP and their temporal order of formation, which is hierarchical and conserved. We also established functional relationships among several of them that validate one of the three inferred developmental trajectories. Our research reveals the ontogeny of LRP tissue initiation with an unprecedented level of resolution. Furthermore, we demonstrate functionality of the primordial cells at the sides of LRP base to regenerate new LRs following laser ablation and LR resection. We also identified novel transcriptomic features of the primordial cells that led to the identification of camalexin as a new LRP developmental regulator associated to initiation of Endodermis/QC-transitioning-stem-cells.
The ontogeny of the LRP is organized and involves the hierarchical assembly of cell lineages
Based on the spatial localization of the cell populations, the order of formation and the developmental branch to which they belong (i.e. the capacity of the cell populations to give rise or derive from other cell populations); we propose a model for LRP patterning during organogenesis (Fig 7A). In this model the spatial organization of the populations in layers would be determined by the position of the precursor population. Thus, the vascular-like-1 cells (on the inside) and the Endodermis/QC-transitioning-stem-cells (on the outside) would be derived at stage II-III from the primordial cells. Undivided or slow dividing cells on the sides would remain as primordial cells, whereas several of them would specialize at stage IV into the flanking cells to eventually establish the growth boundaries of the LRP. Some of the Endodermis/QC-transitioning-stem-cells in the central region would form the QC-transitioning-cell at stage IV to subsequently generate a QC and maybe other SCN cells. The remaining Endodermis/QC-transitioning-stem-cells would establish the ground tissue lineage.
Figure 7. Model for LRP patterning during organogenesis.
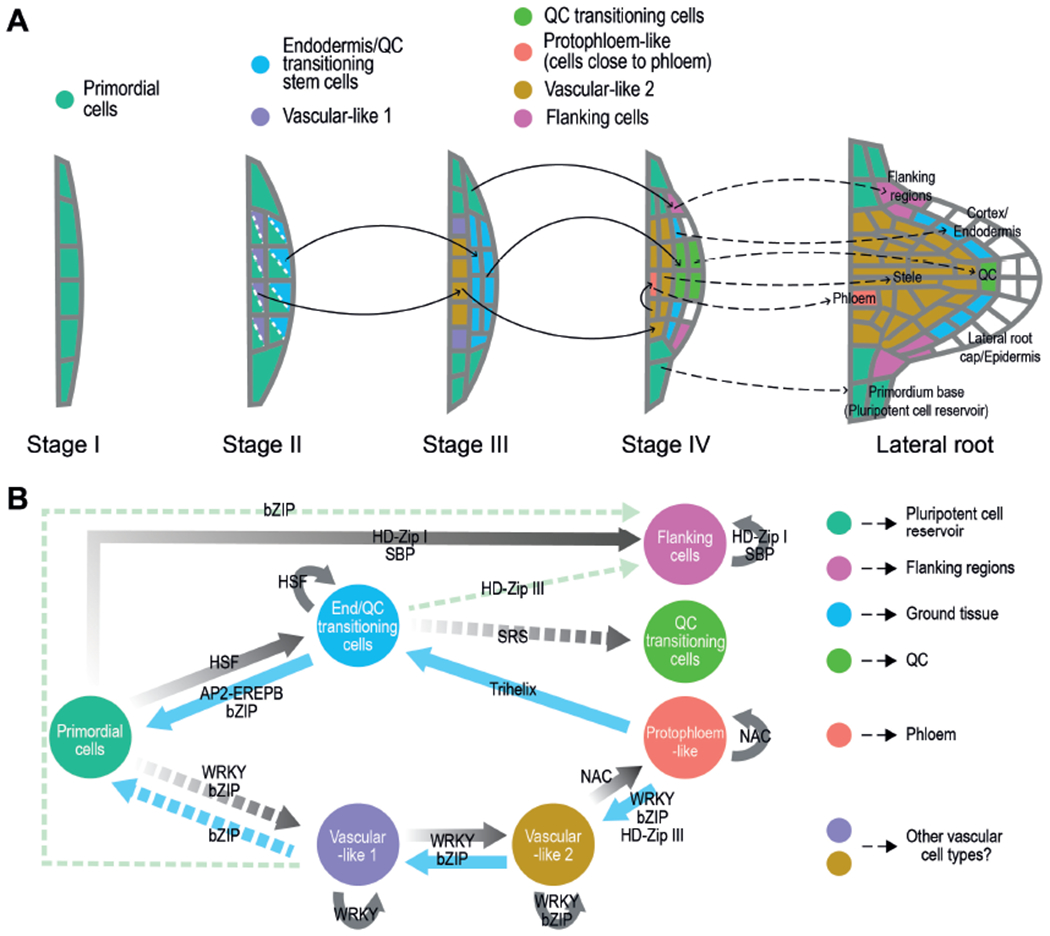
(A) The localization of the different cell populations is represented by colors as indicated. The model is based on the capacity of the populations to give rise or derive from other cell populations (black arrows) based on their spatial localization, sequential order of formation and developmental branch. The dashed arrows indicate subsequent tissue or region of the LR would be originated by each cell population based on localization of biomarkers at stage IV or later if evaluated. In our model LR external layers (epidermis and lateral root cap) would be formed after stage IV of LR formation. (B) Hypothetical regulation among the different LRP cell populations based on PAGA and pseudotime analyses (Figure 4B and C) and enrichment in Transcription Factor (TF) families and TF binding sites (TFBS) (table S4). Continuous lines: relationships predicted by PAGA, Monocle and TF/TFBS enrichment analyses. Discontinuous lines: relationship predicted by the TF/TFBS enrichment analysis and either PAGA or Monocle. Gray lines: regulation through a developmental trajectory. Note that all the TF families putatively mediating transitions also self-regulate the newly formed population (except SRS). Blue lines: additional regulation within a developmental trajectory. Pale green lines: regulation between different developmental trajectories.
Based on this model, the LRP regulator SCR (Goh et al., 2016) would be expressed in the Endodermis/QC-transitioning-stem-cells (a novel cell type), likely contributing to initiate the QC-transitioning-cells to specify a new QC. In agreement with the proposed additional pathways leading to QC specification (Goh et al., 2016), we found CBF3 to specifically regulate initiation of the QC-transitioning-cells. In addition, the PLT factors (Du and Scheres, 2017) could also belong to the same stem cell developmental trajectory. PLT3/5/7 could regulate initiation of the Endodermis/QC-transitioning-stem-cells and PLT1/2/4 initiation of the QC-transitioning-cells, which could require additional positional signals such as auxin. Future experiments might investigate the role of ARF7 in primordial cell fate specification and its maintenance after LR emergence.
In the vascular development branch (Fig 7A), vascular-like-1 would initiate vascular-like-2 cells at stage III, while at stage IV, some of these would have transitioned to protopholem-like cells. Intriguingly, the cells putatively becoming protophloem would be the cells adjacent to the phloem tissues of the primary root, which anticipates vasculature connection and suggests that signals from primary root tissues might induce their specialization. In agreement with this idea, during grafting, genes related to vascular tissue formation are activated above and below the graft junction, which is part of a recognition mechanism that leads to reconnection of vasculature tissues (Melnyk et al., 2018). Interestingly, grafting does not involve establishment of stem cells, and neither the vasculature developmental trajectory in the LRP shows stem cell features.
Our analyses did not identify any cell population with affinity to epidermis or lateral-root-cap cells, suggesting that these tissues might be formed at later developmental stages, possibly from QC-transitioning-cells according to their localization or other undetermined cells. Further supporting this idea, expression of the columella and epidermis regulators FEZ and WEREWOLF, respectively, was observed at developmental stages close to LR emergence (Du and Scheres, 2017).
Regulation of the developmental trajectories of the LRP
In addition to the identification of CBF3 as regulator of LRP ontogeny, our research also has the potential of identifying other novel molecular interactions. As a proof of concept, we analyzed enrichment of TF families and TF binding sites (TFBS) in the seven cell populations of the early LRP. We found at least one specific family of TF to be enriched in a population and its corresponding TFBS to be enriched in the temporally subsequent (or precursor) population (Fig 7B, Table S4). Thus, it can be inferred the putative role of class I HD-ZIP TFs mediating the transition from primordial to the flanking cells, which could lead to MYB36 activation, or that of HD-ZIP III TFs and SHI RELATED SEQUENCE (SRS) proteins mediating the transition from Endodermis/QC-transitioning-stem-cells to QC-transitioning-cells, among others. Interestingly, SRS proteins regulate auxin biosynthetic genes favoring the local induction of auxin maxima (Eklund et al., 2010), which would be in agreement with an expected auxin maximum present at the future domain of the LR QC.
In some other cases, the same family of TF was enriched along an entire developmental trajectory, such as the WRKY TFs (Fig 7B). WRKY18 was specifically expressed in the primordial cells while subsequent vascular-like-1/2 were enriched in WRKY binding sites and in other WRKY TFs. WRKY18 has been shown to form a complex with histone acetytransferases to activate sugar response genes (Chen et al., 2019) and in addition sugar response has been associated to vascular reconnection (Melnyk et al., 2018) suggesting that sugar or nutrient response might be involved in the specialization of the vasculature trajectory. In addition, NAC TFs such as NAC020 might mediate the transition from vascular-like-2 to protopholem-like cells. NAC020 is specifically expressed in protopholem-like cells and is upstream of the regulator of phloem development APL (Kondo et al., 2016). Moreover, we observed enrichment of the vasculature regulators PHLOEM EARLY DOF TFs (PEAR) (Miyashima et al., 2019) in the protopholem-like cells. PEAR genes have been shown to activate expression of HD-ZIP III TFs, and we found HD-ZIP III TFs to be enriched in the same cells. As HD-ZIP III are negative regulators of vasculature proliferation (Miyashima et al., 2019) their activation might facilitate the specification of protopholem-like cells.
Final Remarks
Organ formation in plants involves the specification of new cell identities and their organization in tissues or growth domains. Previous research in tissue ontogeny showed limited hierarchy or no obvious order of formation. Our findings provide an insight into a possible tissue formation mechanism which involves the hierarchical assembly or initiation of tissues, thus shedding light on the long-term question of how plants conduct developmental programs to establish cell fate while growing or developing structures.
MATERIALS AND METHODS
Plant material and growth conditions
Arabidopsis thaliana Columbia-0 (Col-0) accession was the genetic background used in this study. Seeds were surfaced-sterilized upon exposition to Cl2 gas and stratified in sterile water at 4°C in darkness during 2 days. After stratification, seeds were transferred to petri dishes containing half-Murashige & Skoog (MS) medium with 1% sucrose and 10 g/L Plant Agar (Duchefa). Acivicin (Thermo Fisher, Waltham, Massachusetts, USA) and Camalexin (Avantor, Radnor, Pennsylvania, USA) were supplemented to the growth media at the indicated concentrations. Arabidopsis seedlings were vertically grown in chambers under a 16/8 photoperiod at 22°C. pWOX5::ER-GFP (Sarkar et al., 2007), pPLT4:: ER-CFP (Du and Scheres, 2017), RecMYB36::AraYPET (Liberman et al., 2015) and pSKP2Bs::NLS-3x-mCherry (Perianez-Rodriguez et al., 2021) lines were also used in this study.
Protoplast isolation and sc-RNAseq
The biological samples were extracted from plants expressing the pHB53:NLS-3xmCherry construct at 4 days of growth (i.e. post-imbibition) after stratification. These roots showed 49%, 22%, 17% and 12% of LRP from stages I to IV, which we estimated corresponded to 21.8%, 20%, 24.1% and 34.1% of cells in each stage, respectively. For each independent replicate, the primary root tip and the aerial part of ~400 plants were removed and the remaining roots were subjected to 1.5 hours of protoplasting as detailed in (Efroni et al., 2016). The cell suspension containing mCherry positive cells was sorted on a FACSAria II (Becton Dickenson) into 96-well plates using the single-cell sort mode. Three replicates were processed on a Bravo robotic system (Agilent) and a Mantis (Formulatrix) using the cDNA construction and barcoding protocol detailed in (Efroni et al., 2016) to construct cDNA libraries. Libraries were generated based on the Smart-Seq2 protocol (Picelli et al., 2014) using the Nextseq 500 Mid Output 150 V2.5 kit and run on the Illumina Nextseq 500. This approach yielded 573 single cells, which represent over 5-fold coverage of all the cells contained in LRP from stages I to IV. Reads were trimmed using Trimmomatics with the following settings: Illumina clip: 2:30:10, Crop: 48, Sliding window: 4:5, Minleng: 36 and then mapped to the Arabidopsis genome reference using Star, with default settings, using the -- quantMode GeneCounts argument to generate counts in Star (Efroni et al., 2016). The data was deposited in GEO under the accession number GSE161970.
Generation of a single-cell gene expression matrix
Raw counts processing and subsequent analysis were performed in R (https://www.R-project.org/) using Seurat (v.3.1) (Butler et al., 2018; Satija et al., 2015) unless indicated. Cut-off parameters for the 573 sequenced cells required at least 1,000 total reads, between 200 and 8000 transcripts and between 0.1 and 6% of mitochondrial genes per cell. In addition, we removed transcripts expressed in fewer than 3 cells. The expression data were log normalized to generate an expression matrix reflecting the expression of 22,557 transcripts across 282 cells, with 362,293 mean reads/cell and 4415 mean transcripts/cell. Cell cycle stage was estimated for each cell based on expression of G2/M and S phase markers (Table S1) using the Seurat function CellCycleScoring() which assigns a score. For subsequent clustering the expression matrix data was modeled using the Seurat function ScaleData() to regress out the variability for the total number of reads, percentage of mitochondrial genes and the cell cycle stage. To regress out the cell cycle stage of the cells, the relationship between gene expression and the S and G2M cell cycle scores was calculated. The scaled residuals of remaining genes were used for subsequent dimensional reduction, avoiding clustering cells based on their cell cycle stage, reads or mitochondrial genes. Using the expression matrix, genes differentially expressed among clusters (p value<0.01, log-fold-change ≥ 0.25, present in ≥ 25% cells) were identified as biomarkers. Protoplasting-induced genes (Birnbaum, 2003) were excluded from transcriptomic analyses.
Data analysis and clustering
Dimensionality reduction by Principal Component Analysis (PCA) was performed using the top 2,500 most variable features in Seurat (v.3.1) (Table S2). The top 5 principal components were determined as informative using the Elbow plot and Jack Straw plot in Seurat and used as input for clustering and visualization using Uniform Manifold Approximation and Projection (UMAP) (Becht et al., 2018). Clusters were identified using Seurat at different resolutions as indicated. Heatmaps were performed in Seurat.
Assessment of cell identity and gene expression analysis
The Index of Cell Identity was calculated in R according to gene expression in the Root Map (Brady et al., 2007; Efroni et al., 2016). Spec values were also calculated in R using the following parameters: cuts = 4 and distshape = 1. Biomarkers for known cell types were selected according to previous studies (Brady et al., 2007; Clark et al., 2019; Denyer et al., 2019; Efroni et al., 2016; Ryu et al., 2019) as specified in Table S3. Specific identity features in our dataset was performed by analyzing the average expression of known cell type biomarkers: QC markers from (Brady et al., 2007; Denyer et al., 2019), SCN markers (cluster 12 enriched genes) from (Zhang et al., 2019), CEI markers from (Clark et al., 2019), young vascular markers from (Ryu et al., 2019) and protophloem markers from (Denyer et al., 2019; Ryu et al., 2019). Meristematic markers were those in (Huang and Schiefelbein, 2015).
GO Enrichment analysis
GO enrichment analyses were performed in g:Profiler (Raudvere et al., 2019) using a p-value < 0.001 to detect the GO terms enriched in the 2,500 most variable genes and the top 100 markers for each cluster. Visualization of GO enrichment in UMAP plots was performed using the average expression of all the genes included in those GO terms according to the TAIR database.
Pseudotime analyses
Pseudotemporal relationships among single cells were assessed by two different methods: Partition-Based Graph Abstraction (PAGA) (Wolf et al., 2019) and Monocle 3 (Cao et al., 2019). Clustering from Seurat was used as the initial input, so identities were not re-established by these methods.
TF and TF binding sites (TFBS) enrichment
TF and TFBS enrichment was performed using Agris and Athamap databases, respectively (Davuluri et al., 2003; Steffens, 2004). As input for TF and TFBS search, we used the top 100 markers for each population and the 500 bp upstream and 50 bp downstream of each gene. Enrichment was calculated as the ratio between the TFs found in each population and those in the set of the 2,500 most variable genes or those provided by Athamap. Enriched TFs or TFBSs were those with a ratio ≥ 1.5 (Table S4).
Selection, cloning and transformation of marker genes
The 2Kb promoter of HB53 (AT5G66700) was amplified by PCR (Phusion High-Fidelity DNA Polymerase Thermo Fisher Scientific) using the primers 5′-GTTCGTTGCCCACACATTACT-3′ and 5′-TTTCTCTCTCTAGTTTTTCAGAC-3′. The identification of markers up-regulated in each cluster as compared to the rest of cells was performed in Seurat (v.3.1). The promoter region of selected marker genes were amplified by PCR (Phusion High-Fidelity DNA Polymerase, Thermo Fisher Scientific) using the primers 5′-CTCCTTGACTTACAGACCAAAC-3′ and 5′-GGTGGCCTTCATGATTGTTTC-3′ for DR4 (AT1G73330), 5′-ACTTCTTTGCTTCACATAAGTTAAAAGTCA-3′ and 5′- TGATCAGAAGAGTACTCTGTTTCAAGA-3′ for CBF3 (AT4G25480) and 5′- GTGCCCTGGAATCTAACATT −3′ and 5′-CTTGTTGATTTTATCTTTGACTT-3′ for ATHMP41 (AT4G39700). Constructs cloning was performed using MultiSite Gateway Three- Fragment Vector Construction Kit (Invitrogen) to fuse promoter regions to a nuclear localization sequence and 3 repeats of the Yellow (YFP) or mCherry fluorescent protein genes in the dpGreen BarT vector as indicated. Constructs were transformed into Col-0 background by floral dip method. Homozygous lines were selected among the T2 progeny in ammonium glufosinate (Merck).
Microscopy and regeneration assays
4-6 days old seedlings from homozygous lines were stained with 10 mg/mL Propidium Iodide (PI) (Sigma-Aldrich) or 1 mg/mL SCRI Renaissance 2200 dye (SR2200) (Renaissance Chemicals) and imaged in a Leica SP8 laser-scanning confocal microscope (Leica) using a hybrid detector counting mode and the following settings: YFP or AraYPET (excitation 514 nm, acquisition 524 - 544 nm), mCherry (excitation 561 nm, acquisition 600 - 650 nm), SR2200 (excitation 405 nm, acquisition 430 - 450 nm) and PI (excitation 561 nm, acquisition 600 - 630 nm). Quantification of fluorescent signal was performed in Leica LAS AF Lite software as pixel density. Laser ablation of cells was performed using pulses of lasers of 405 nm and 458 nm for 1 minute. Fluorescence microscopy was performed in Flumazone (Leica M205FA adapted with Hamamatsu EMCCD X2 camera). LRs resection was performed using a scalpel in Flumazone.
Statistical analysis
Statistical differences were detected using R. Homoscedastic groups were analyzed using Analysis of variance (ANOVA) and Tukey HSD or Bonferroni post-hoc tests for one or more factors, respectively. Significant differences were collected with 5% level of significance.
Supplementary Material
ACKNOWLEDGMENTS
This work was funded by Ministerio de Economía y Competitividad (MINECO) of Spain and ERDF (grants BFU2016-80315-P and PID2019-111523GB-I00 to M.A.M.-R), by Comunidad de Madrid (CM) and Universidad Politécnica de Madrid (UPM (grant APOYO_JOVENES_2Y36R7_20_TRG6W7 (Plant_Stem) to P.P.-G.) and by National Institutes of Health (grant #1R35GM136362) and National Science Foundation (grant #1934388) to K.D.B. L.S.-R. and A.S.-C. were supported by FPI contracts (BES-2017-080155 and BES-2014-068852, respectively) from MINECO, P.P.-G. by Programa Atraccion Talento from CM (2017-T2/BIO-3453) and J.C. by a Juan de la Cierva Contract from MINECO (FJCI-2016-28607). We thank Dr. C. Manzano for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, and Newell EW (2018). Dimensionality reduction for visualizing single-cell data using UMAP. Nature Biotechnology 37:38–44. [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, and Friml J (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602. [DOI] [PubMed] [Google Scholar]
- Birnbaum K (2003). A Gene Expression Map of the Arabidopsis Root. Science 302:1956–1960. [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, and Benfey PN (2007). A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318:801–806. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566:496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Xu X, Xu D, Zhang H, Zhang C, and Li G (2019). WRKY18 and WRKY53 Coordinate with HISTONE ACETYLTRANSFERASE1 to Regulate Rapid Responses to Sugar. Plant physiology 180:2212–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark NM, Buckner E, Fisher AP, Nelson EC, Nguyen TT, Simmons AR, de Luis Balaguer MA, Butler-Smith T, Sheldon PJ, Bergmann DC, et al. (2019). Stem-cell-ubiquitous genes spatiotemporally coordinate division through regulation of stem-cell-specific gene networks. Nature Communications 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davuluri RV, Sun H, Palaniswamy SK, Matthews N, Molina C, Kurtz M, and Grotewold E (2003). AGRIS: Arabidopsis Gene Regulatory Information Server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC bioinformatics 4:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luis Balaguer MA, Fisher AP, Clark NM, Fernandez-Espinosa MG, Möller BK, Weijers D, Lohmann JU, Williams C, Lorenzo O, and Sozzani R (2017). Predicting gene regulatory networks by combining spatial and temporal gene expression data in Arabidopsis root stem cells. Proceedings of the National Academy of Sciences 114:E7632–E7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rybel B, Vassileva V, Parizot B, Demeulenaere M, Grunewald W, Audenaert D, Van Campenhout J, Overvoorde P, Jansen L, Vanneste S, et al. (2010). A novel aux/IAA28 signaling cascade activates GATA23-dependent specification of lateral root founder cell identity. Current biology : CB 20:1697–1706. [DOI] [PubMed] [Google Scholar]
- De Smet I, Vassileva V, De Rybel B, Levesque MP, Grunewald W, Van Damme D, Van Noorden G, Naudts M, Van Isterdael G, De Clercq R, et al. (2008). Receptor-like kinase ACR4 restricts formative cell divisions in the Arabidopsis root. Science 322:594–597. [DOI] [PubMed] [Google Scholar]
- Denyer T, Ma X, Klesen S, Scacchi E, Nieselt K, and Timmermans MCP (2019). Spatiotemporal Developmental Trajectories in the Arabidopsis Root Revealed Using High-Throughput Single-Cell RNA Sequencing. Developmental cell 48:840–852.e845. [DOI] [PubMed] [Google Scholar]
- Du Y, and Scheres B (2017). PLETHORA transcription factors orchestrate de novo organ patterning during Arabidopsis lateral root outgrowth. Proceedings of the National Academy of Sciences of the United States of America 114:11709–11714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Ip P-L, Nawy T, Mello A, and Birnbaum KD (2015). Quantification of cell identity from single-cell gene expression profiles. Genome Biology 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efroni I, Mello A, Nawy T, Ip P-L, Rahni R, DelRose N, Powers A, Satija R, and Birnbaum KD (2016). Root Regeneration Triggers an Embryo-like Sequence Guided by Hormonal Interactions. Cell 165:1721–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eklund DM, Ståldal V, Valsecchi I, Cierlik I, Eriksson C, Hiratsu K, Ohme-Takagi M, Sundström JF, Thelander M, Ezcurra I, et al. (2010). The Arabidopsis thaliana STYLISH1 Protein Acts as a Transcriptional Activator Regulating Auxin Biosynthesis. The Plant cell 22:349–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Marcos M, Desvoyes B, Manzano C, Liberman LM, Benfey PN, del Pozo JC, and Gutierrez C (2017). Control of Arabidopsis lateral root primordium boundaries by MYB36. New Phytologist 213:105–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta KM, Yadav SR, Lehesranta S, Belevich I, Miyashima S, Heo J.o., Vaten A, Lindgren O, De Rybel B, Van Isterdael G, et al. (2014). Arabidopsis NAC45/86 direct sieve element morphogenesis culminating in enucleation. Science 345:933–937. [DOI] [PubMed] [Google Scholar]
- Gala HP, Lanctot A, Jean-Baptiste K, Guiziou S, Chu JC, Zemke JE, George W, Queitsch C, Cuperus JT, and Nemhauser JL (2021). A single cell view of the transcriptome during lateral root initiation in Arabidopsis thaliana. The Plant cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, and Scheres B (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057. [DOI] [PubMed] [Google Scholar]
- Glawischnig E (2007). Camalexin. Phytochemistry 68:401–406. [DOI] [PubMed] [Google Scholar]
- Goh T, Joi S, Mimura T, and Fukaki H (2012). The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 139:883–893. [DOI] [PubMed] [Google Scholar]
- Goh T, Toyokura K, Wells DM, Swarup K, Yamamoto M, Mimura T, Weijers D, Fukaki H, Laplaze L, Bennett MJ, et al. (2016). Quiescent center initiation in the Arabidopsis lateral root primordia is dependent on the SCARECROW transcription factor. Development 143:3363–3371. [DOI] [PubMed] [Google Scholar]
- González-Grandío E, Pajoro A, Franco-Zorrilla JM, Tarancón C, Immink RGH, and Cubas P (2017). Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proceedings of the National Academy of Sciences 114:E245–E254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellmann E, Ko D, Ruonala R, and Helariutta Y (2018). Plant Vascular Tissues—Connecting Tissue Comes in All Shapes. Plants 7:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, and Schiefelbein J (2015). Conserved Gene Expression Programs in Developing Roots from Diverse Plants. The Plant cell 27:2119–2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean-Baptiste K, McFaline-Figueroa JL, Alexandre CM, Dorrity MW, Saunders L, Bubb KL, Trapnell C, Fields S, Queitsch C, and Cuperus JT (2019). Dynamics of Gene Expression in Single Root Cells of Arabidopsis thaliana. The Plant cell 31:993–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo Y, Nurani AM, Saito C, Ichihashi Y, Saito M, Yamazaki K, Mitsuda N, Ohme-Takagi M, and Fukuda H (2016). Vascular Cell Induction Culture System Using Arabidopsis Leaves (VISUAL) Reveals the Sequential Differentiation of Sieve Element-Like Cells. The Plant cell 28:1250–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavenus J, Goh T, Guyomarc’h S, Hill K, Lucas M, Voss U, Kenobi K, Wilson MH, Farcot E, Hagen G, et al. (2015). Inference of the Arabidopsis lateral root gene regulatory network suggests a bifurcation mechanism that defines primordia flanking and central zones. The Plant cell 27:1368–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman LM, Sparks EE, Moreno-Risueno MA, Petricka JJ, and Benfey PN (2015). MYB36 regulates the transition from proliferation to differentiation in the Arabidopsis root. Proceedings of the National Academy of Sciences 112:12099–12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas M, Kenobi K, von Wangenheim D, Vobeta U, Swarup K, De Smet I, Van Damme D, Lawrence T, Peret B, Moscardi E, et al. (2013a). Lateral root morphogenesis is dependent on the mechanical properties of the overlaying tissues. Proceedings of the National Academy of Sciences of the United States of America 110:5229–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas WJ, Groover A, Lichtenberger R, Furuta K, Yadav S-R, Helariutta Y, He X-Q, Fukuda H, Kang J, Brady SM, et al. (2013b). The Plant Vascular System: Evolution, Development and FunctionsF. Journal of Integrative Plant Biology 55:294–388. [DOI] [PubMed] [Google Scholar]
- Malamy JE, and Benfey PN (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124:33–44. [DOI] [PubMed] [Google Scholar]
- Melnyk CW, Gabel A, Hardcastle TJ, Robinson S, Miyashima S, Grosse I, and Meyerowitz EM (2018). Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proceedings of the National Academy of Sciences 115:E2447–E2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashima S, Roszak P, Sevilem I, Toyokura K, Blob B, Heo J.-o., Mellor N, Help-Rinta-Rahko H, Otero S, Smet W, et al. (2019). Mobile PEAR transcription factors integrate positional cues to prime cambial growth. Nature 565:490–494. [DOI] [PubMed] [Google Scholar]
- Möller BK, ten Hove CA, Xiang D, Williams N, López LG, Yoshida S, Smit M, Datla R, and Weijers D (2017). Auxin response cell-autonomously controls ground tissue initiation in the earlyArabidopsisembryo. Proceedings of the National Academy of Sciences 114:E2533–E2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motte H, Vanneste S, and Beeckman T (2019). Molecular and Environmental Regulation of Root Development. Annual Review of Plant Biology 70:465–488. [DOI] [PubMed] [Google Scholar]
- Parizot B, De Rybel B, and Beeckman T (2010). VisuaLRTC: A New View on Lateral Root Initiation by Combining Specific Transcriptome Data Sets. Plant physiology 153:34–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Garcia P, and Moreno-Risueno MA (2018). Stem cells and plant regeneration. Developmental Biology 442:3–12. [DOI] [PubMed] [Google Scholar]
- Perianez-Rodriguez J, Rodriguez M, Marconi M, Bustillo-Avendaño E, Wachsman G, Sanchez-Corrionero A, De Gernier H, Cabrera J, Perez-Garcia P, Gude I, et al. (2021). An auxin-regulable oscillatory circuit drives the root clock in Arabidopsis. Science Advances 7:eabd4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picelli S, Faridani OR, Björklund ÅK, Winberg G, Sagasser S, and Sandberg R (2014). Full-length RNA-seq from single cells using Smart-seq2. Nature protocols 9:171–181. [DOI] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, and Vilo J (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Research 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KH, Huang L, Kang HM, and Schiefelbein J (2019). Single-Cell RNA Sequencing Resolves Molecular Relationships Among Individual Plant Cells. Plant physiology 179:1444–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar AK, Luijten M, Miyashima S, Lenhard M, Hashimoto T, Nakajima K, Scheres B, Heidstra R, and Laux T (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446:811–814. [DOI] [PubMed] [Google Scholar]
- Satija R, Farrell JA, Gennert D, Schier AF, and Regev A (2015). Spatial reconstruction of single-cell gene expression data. Nature Biotechnology 33:495–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahan R, Hsu C-W, Nolan TM, Cole BJ, Taylor IW, Vlot AHC, Benfey PN, and Ohler U (2020). A single cell Arabidopsis root atlas reveals developmental trajectories in wild type and cell identity mutants. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulse CN, Cole BJ, Ciobanu D, Lin J, Yoshinaga Y, Gouran M, Turco GM, Zhu Y, O’Malley RC, Brady SM, et al. (2019). High-Throughput Single-Cell Transcriptome Profiling of Plant Cell Types. Cell Reports 27:2241–2247.e2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit ME, Llavata-Peris CI, Roosjen M, van Beijnum H, Novikova D, Levitsky V, Sevilem I, Roszak P, Slane D, Jürgens G, et al. (2020). Specification and regulation of vascular tissue identity in the Arabidopsis embryo. Development 147:dev186130. [DOI] [PubMed] [Google Scholar]
- Steffens NO (2004). AthaMap: an online resource for in silico transcription factor binding sites in the Arabidopsis thaliana genome. Nucleic Acids Research 32:368D–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su T, Xu J, Li Y, Lei L, Zhao L, Yang H, Feng J, Liu G, and Ren D (2011). Glutathione-Indole-3-Acetonitrile Is Required for Camalexin Biosynthesis in Arabidopsis thaliana. The Plant cell 23:364–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Hove CA, Lu KJ, and Weijers D (2015). Building a plant: cell fate specification in the early Arabidopsis embryo. Development 142:420–430. [DOI] [PubMed] [Google Scholar]
- Trinh CD, Laplaze L, and Guyomarc’h S (2018). Lateral Root Formation: Building a Meristem de novo.847–890. [Google Scholar]
- Truernit E, Bauby H, Belcram K, Barthelemy J, and Palauqui JC (2012). OCTOPUS, a polarly localised membrane-associated protein, regulates phloem differentiation entry in Arabidopsis thaliana. Development 139:1306–1315. [DOI] [PubMed] [Google Scholar]
- Turco GM, Rodriguez-Medina J, Siebert S, Han D, Valderrama-Gómez MÁ, Vahldick H, Shulse CN, Cole BJ, Juliano CE, Dickel DE, et al. (2019). Molecular Mechanisms Driving Switch Behavior in Xylem Cell Differentiation. Cell Reports 28:342–351.e344. [DOI] [PubMed] [Google Scholar]
- Vatén A, Dettmer J, Wu S, Stierhof Y-D, Miyashima S, Yadav, Shri R, Roberts, Christina J, Campilho A, Bulone V, Lichtenberger R, et al. (2011). Callose Biosynthesis Regulates Symplastic Trafficking during Root Development. Developmental cell 21:1144–1155. [DOI] [PubMed] [Google Scholar]
- von Wangenheim D, Fangerau J, Schmitz A, Smith RS, Leitte H, Stelzer EH, and Maizel A (2016). Rules and Self-Organizing Properties of Post-embryonic Plant Organ Cell Division Patterns. Current biology : CB 26:439–449. [DOI] [PubMed] [Google Scholar]
- Voß U, Wilson MH, Kenobi K, Gould PD, Robertson FC, Peer WA, Lucas M, Swarup K, Casimiro I, Holman TJ, et al. (2015). The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nature Communications 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf FA, Hamey FK, Plass M, Solana J, Dahlin JS, Göttgens B, Rajewsky N, Simon L, and Theis FJ (2019). PAGA: graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biology 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie B, Wang X, Zhu M, Zhang Z, and Hong Z (2011). CalS7 encodes a callose synthase responsible for callose deposition in the phloem. The Plant Journal 65:1–14. [DOI] [PubMed] [Google Scholar]
- Yan P, Ren J, Zhang W, Qu J, and Liu G-H (2020). Protein quality control of cell stemness. Cell Regeneration 9:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T-Q, Xu Z-G, Shang G-D, and Wang J-W (2019). A Single-Cell RNA Sequencing Profiles the Developmental Landscape of Arabidopsis Root. Molecular Plant 12:648–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


