Abstract
Objective
To assess the diagnostic performance of lateral flow immunochromatographic assays (LFAs) of 4 different manufacturers to identify SARS-CoV-2 antibodies (IgM, IgG, or total), comparing them with the nucleic acid amplification test (NAAT) or the clinical defined test (definite or probable SARS-CoV-2 infection, respectively).
Methods
One hundred nineteen serum samples were randomly selected by convenience and distributed in the following groups: (1) group with SARS-CoV-2 infection (n = 82; RT-qPCR positive [definite, n = 70] and probable [n = 12]); (2) other diseases (n = 27; other viruses identified [n = 8] and SARS of other etiologies [n = 19]); and (3) healthy control group (n = 10). LFAs of 4 manufacturers were compared: MedTest Coronavirus (COVID-19) IgG/IgM (MedLevensohn, Brazil); COVID-19 IgG/IgM ECO Test (Ecodiagnóstica, Brazil); Camtech COVID-19 IgM/IgG Rapid Test Kit (Camtech Diagnostics Pte Ltd, Singapore); and 1-Step COVID-19 Test for total antibodies (Guangzhou Wondfo Biotech Co., China).
Results
The 4 tests studied showed high diagnostic performance characteristics for the diagnoses of definite or probable SARS-CoV-2 infection. The best measures were for the Wondfo test: sensitivity (86.59%; 95% CI: 77.26–93.11%), specificity (100%; 90.51–100%), DOR (257; 60–1,008), LR+ (33.43; 4.82–231.85), LR− (0.13; 0.08–0.23), accuracy (90.76%; 84.06–95.29%), and Matthews correlation coefficient (MCC) 0.82. Although considering only the probable SARS-CoV-2 infection (PCR−) cases, all the kits studied showed limited values.
Conclusion
Our data demonstrate the excellent performance of LFA for the diagnoses of definite or probable SARS-CoV-2 infection. There was substantial heterogeneity in sensitivities of IgM and IgG antibodies among the different kits. LFA tests cannot replace molecular diagnostics but should be used as an additional screening tool.
Keywords: SARS-CoV-2, Serology, Rapid test, Lateral flow immunochromatographic assay
Highlights of the Study
Serological tests to detect SARS-CoV-2 antibodies aim to identify previous infection and confirm the presence of current infection.
Lateral flow immunochromatographic assays showed excellent diagnostic performance.
There was substantial heterogeneity in sensitivities of IgM and IgG antibodies between the kits.
Serology tests cannot replace molecular diagnostics, which is considered the gold standard, but should be used only as an additional screening tool.
Introduction
The SARS-CoV-2 pandemic presents important diagnostic challenges [1]. Serology tests to detect the presence of antibodies to SARS-CoV-2 aim to identify previous infection and to confirm the presence of current infection [1, 2]. SARS-CoV-2 serology testing relies on targeted antibodies binding to SARS-CoV-2-specific antigens. This study aimed to assess the diagnostic performance of lateral flow immunochromatographic assays (LFAs) of 4 different manufactures and compare them with the nucleic acid amplification test (NAAT, definite) or the clinical defined test (probable) in individuals with SARS-CoV-2 infection. Tests were selected based on their availability and approval by the National Health Surveillance Agency (ANVISA) in Brazil at the time of the study.
Materials and Methods
The samples consisted of serum specimens sent to the Virology Laboratory, Hospital de Clínicas, Universidade Federal do Parana (HC-UFPR), Brazil. Immunological and RT-qPCR for SARS-CoV-2 assays were performed in the HC-UFPR Virology Laboratory, which is certified by the Health Secretary of Paraná, Brazil. This study was approved under a waiver of informed consent by the HC-UFPR Institutional Review Board, Brazil.
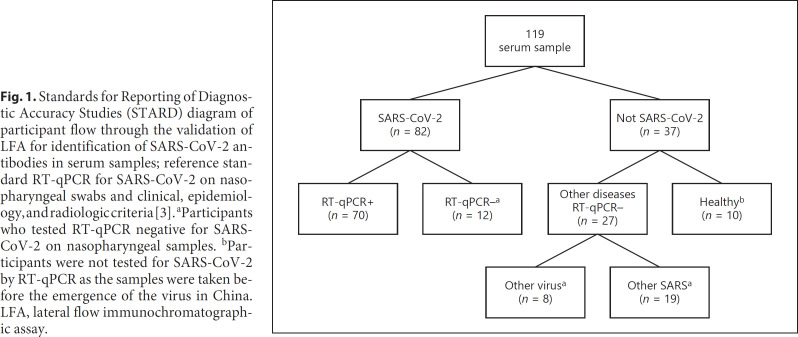
Patients admitted to the HC-UFPR between March 1 and August 7, 2020, were eligible if they had respiratory symptoms suspicious of COVID-19. The performances of the 4 different LFAs were evaluated in serum samples obtained on corresponding dates the respiratory samples were collected for the performance of NAAT. A total of 119 serum samples were randomly selected for convenience and distributed in the following groups (Fig. 1).
Fig. 1.
Standards for Reporting of Diagnostic Accuracy Studies (STARD) diagram of participant flow through the validation of LFA for identification of SARS-CoV-2 antibodies in serum samples; reference standard RT-qPCR for SARS-CoV-2 on nasopharyngeal swabs and clinical, epidemiology, and radiologic criteria [3]. aParticipants who tested RT-qPCR negative for SARS-CoV-2 on nasopharyngeal samples. bParticipants were not tested for SARS-CoV-2 by RT-qPCR as the samples were taken before the emergence of the virus in China. LFA, lateral flow immunochromatographic assay.
Group with SARS-CoV-2 (COVID-19, n = 82)
(a) Definite SARS-CoV-2 infection (n = 70): patients who tested positive for SARS-CoV-2 on RT-qPCR from nasopharyngeal samples. Participants admitted to the hospital COVID-19 unit or intensive care unit, n = 60 (86%); outpatients (n = 10; 14%); male, 38 (54%); median (IQR) age, 50 (38, 58.5) years; time after symptoms onset, 17 (12, 23) days. Two participants (2.9%) were asymptomatic.
(b) Probable SARS-CoV-2 infection (n = 12): patients who tested RT-qPCR negative for SARS-CoV-2 on nasopharyngeal samples but fulfilled the World Health Organization (WHO) clinical diagnostic case definitions for SARS-CoV-2 [3] (see online suppl. Table 1; see www.karger.com/doi/10.1159/000516776 for all online suppl. material). All participants were admitted in the hospital COVID-19 unit or intensive care unit; male, 6 (50%); median (IQR) age, 61.5 (47.5, 74.5) years; time after symptoms onset, 11 (7.5, 19) days (definite vs. probable, p = 0.046 and 0.118, respectively).
Group with Other Diseases (n = 27)
RT-qPCR for SARS-CoV-2 on Nasopharyngeal Samples Negative
(a) Other viruses were identified (n = 8) by RT-qPCR on a nasopharyngeal swab tested against a respiratory virus panel: rhinovirus (n = 6) and coronavirus 229e/NL63 (1 case). One case was positive for Epstein-Barr virus and cytomegalovirus identified in blood.
(b) Severe acute respiratory syndromes: RT-qPCR for SARS-CoV-2 on respiratory samples was either negative and did not fulfill the WHO case definitions for SARS-CoV-2 [3] or other etiologies were identified (n = 19). Median (IQR) age was 54 (41, 76) years; male, 13 (48%) (definite vs. probable vs. other diseases, p = 0.078 and 0.851, respectively); time after symptoms onset, 4 (3, 8.5) days.
Group without Disease (Healthy Control Group, n = 10)
Ten serum samples from blood donors were collected in 2015 (HIV, HCV, HBV, HTLV I/II, syphilis, and Chagas disease negative). This group was not tested for SARS-CoV-2 by RT-qPCR as the samples were taken before the emergence of the virus in China [3].
The research was conducted ethically in accordance with the World Medical Association Declaration of Helsinki. This study was approved under a waiver of informed consent by the HC-UFPR Institutional Review Board, Brazil.
Lateral Flow Immunochromatographic Assays
The kits use capture reaction to detect SARS-CoV-2 IgM/IgG or total antibody in the samples, a 1-step method with results obtained within 15 min, using serum or plasma samples.
IgG, IgM, and Total Antibodies for SARS-CoV-2
The MedTest Coronavirus (COVID-19) IgG/IgM (MedLevensohn, Brazil), the COVID-19 IgG/IgM ECO Test (Ecodiagnóstica, Brazil), and the Camtech COVID-19 IgM/IgG Rapid Test Kit (Camtech Diagnostics Pte Ltd, Singapore) detect IgM and IgG. Samples were tested in parallel in the 4 assays. The tests were performed at room temperature according to the manufacturer's instructions. For all tests, the recommended sample volume of 10 μL serum was added to the specimen well on the individual test cassettes followed by the supplied buffer. The result was visually read after 10 min, by 2 researchers; in case of doubt, a third researcher checked it. For combination of the IgM and the IgG kit, the test card or cassette had 2 test lines (M and G lines) and a quality control line (C line). The M line was fixed with a monoclonal anti-human IgM antibody for detecting SARS-CoV-2 antibody, the G line was fixed with a reagent for detecting SARS-CoV-2 antibody, and the C line was fixed with a quality control antibody. Any visible band for IgG, IgM, or unspecified immunoglobulin was indicative for a positive result. The test card of the Wondfo kit has only 1 test line (T line) and a quality control line (C line). This kit does not differentiate IgM or IgG, and thus results were interpreted as positive or negative for SARS-CoV-2 antibody.
RT-qPCR for SARS-CoV-2
The RT-qPCR for SARS-CoV-2 was composed of dual positive results from a single NAAT targeting 2 different SARS-CoV-2 genes. Samples were collected with a rayon swab and transported immediately to the virology laboratory in a viral transport medium. Samples were taken from the oral cavity and subsequently from the nasal cavity using a nasopharyngeal rayon swab. We performed RT-qPCR using the XGEN-Master COVID-19 (XGEN) for qualitative detection of nucleic acid in RT-qPCR format-reverse transcription, followed by amplification of a conserved region of the ORF1ab and N genes for SARS-CoV-2 [4, 5], using specific primers and a fluorescence-labeled probe in respiratory samples. Specificity: 100% for SARS-CoV-2 (ORF1ab gene), 10 copies/reaction, with probability ≥95%. Sensitivity − SARS-CoV-2 (ORF1ab gene): 10 copies/reaction, with probability ≥95%. Sensitivity − SARS-CoV-2 (N gene): 50 copies/reaction, with probability ≥95%.
Statistical Analyses
The results were presented as the median (IQR) and number (n) and percentage, as appropriate. Categorical variables were compared between groups using the Fisher's exact test, and continuous variables were compared using the Mann-Whitney or Kruskal-Wallis test for nonparametric data, as appropriate. We performed the comparison of concordance and discordance proportions of the different kits with the McNemar test for paired nominal data. Results were considered significant at the 5% alpha level.
Clinical Performance Characteristics of the Tests
We evaluated, for each kit, the clinical performance of the LFA for SARS-CoV-2 IgM/IgG or total antibody (index test) in predicting the SARS-CoV-2 infection. The RT-qPCR for SARS-CoV-2 was the reference method. We analyzed separately the diagnostic performance for the detection of IgM and IgG antibodies in each test. For the calculation of SARS-CoV-2 IgG clinical performance, we included samples from patients who presented symptoms onset ≥8 days. If an IgG result was positive before the 8th day, we considered it true positive (n = 75). For IgM, we included all the samples collected.
The following clinical performance measures were calculated: sensitivity, specificity, accuracy (efficiency), positive and negative predictive values (PPV and NPV), Youden index [6], and positive and negative clinical utility index (CUI+ and CUI−). The CUI values were classified as follows: excellent, ≥ 0.81; good, ≥ 0.64; fair, ≥ 0.49; poor, ≤ 0.49; and very poor, ≤ 0.36 [7, 8]. We calculated the positive and negative likelihood ratio (LR+ and LR−) and diagnostic odds ratio (DOR), in which an LR+ value ≥10.0 indicates that a positive test almost confirmed the disease, a value of ∼6.0 indicates that the disease was present, and a value of ∼1.0 indicates that the test was not able to show whether the disease was present or not. An LR+ value ≤0.1 indicates that the disease was practically absent [9, 10]. The Matthews correlation coefficient (MCC) is a value between −1 and +1.
As the Wondfo test detects total antibodies, in order to compare its results with other tests, we evaluated the clinical performance results of all kits considering any positive results (IgM or IgG), and in a subsequent analysis, we evaluated them separately (IgM and IgG). Positive rates and levels of agreement between the kits were assessed using Cohen's kappa coefficients of agreement, which may be interpreted as follows: values ≤0 as indicating no agreement (i.e., purely random), 0.01–0.20 as none to slight, 0.21–0.40 as fair, 0.41–0.60 as moderate, 0.61–0.80 as substantial, and 0.81–1.00 as almost perfect agreement [11].
Results
The groups with definite and probable SARS-CoV-2 infection were comparable for time of start of symptoms, and for age, they were comparable with the group with other diseases, although different for age on PCR− and PCR+ groups.
Clinical Performance of LFAs
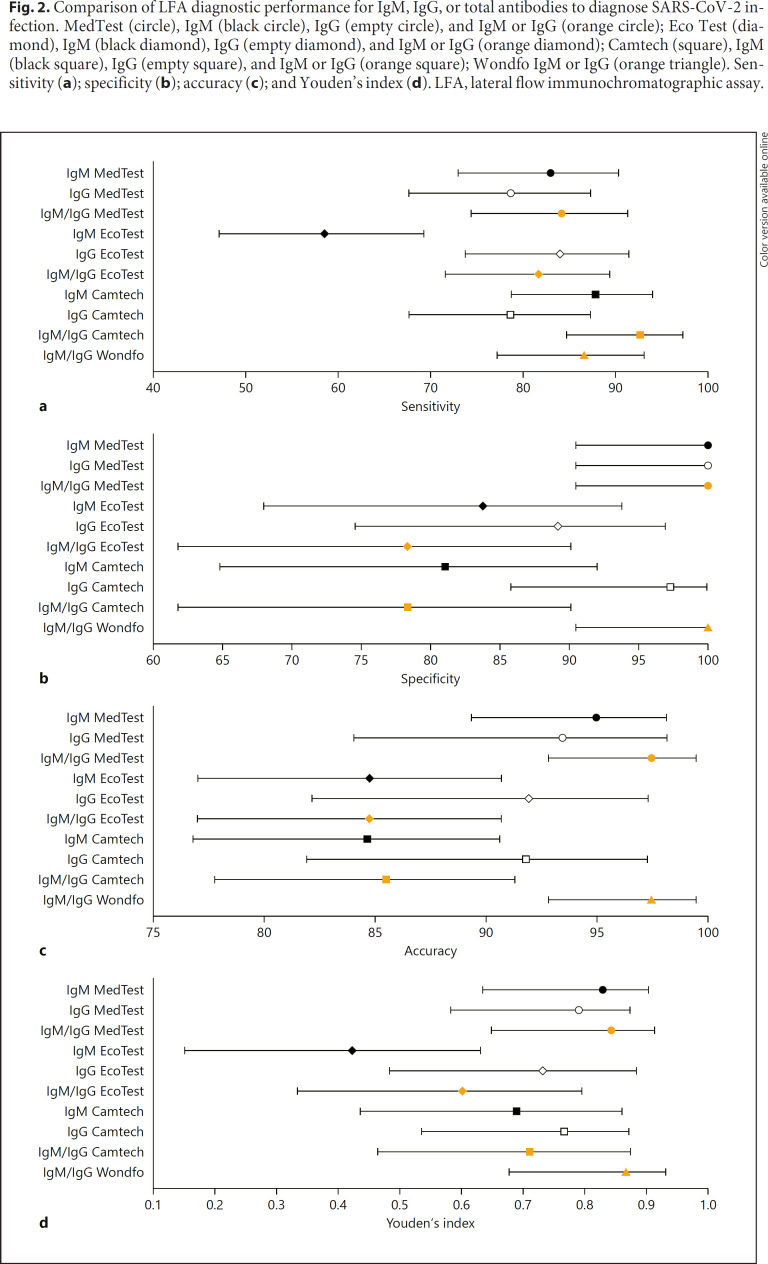
Overall, the analysis of the tests showed a good sensitivity; 1-Step Wondfo and MedTest presented the best results. In addition to the high sensitivity, they were the most accurate tests (Table 1). MedTest and Wondfo presented higher sensitivity, specificity, and accuracy, with less extended 95% confidence intervals (Fig. 2a–d).
Table 1.
Clinical performance characteristics of LFA in serum to detect SARS-CoV-2 antibodies
| IgM and/or IgG | MedTest | EcoTest | Camtech | Wondfo | ||||
|---|---|---|---|---|---|---|---|---|
| value | 95% CI | value | 95% CI | value | 95% CI | value | 95% CI | |
| Group PCR+/PCR– | ||||||||
| TP | 69 | 67 | 76 | 71 | ||||
| Sensitivity, % | 84.15 | 74.42; 91.28 | 81.71 | 71.63; 89.38 | 92.68 | 84.75; 97.27 | 86.59 | 77.26; 93.11 |
| Specificity, % | 100.00 | 90.51; 100.00 | 78.38 | 61.79; 90.17 | 78.38 | 61.79; 90.17 | 100.00 | 90.51; 100.00 |
| LR (+) | 32.50 | 4.68; 225.50 | 3.78 | 2.03; 7.04 | 4.29 | 2.31; 7.94 | 33.43 | 4.82; 231.85 |
| LR (−) | 0.16 | 0.10; 0.26 | 0.23 | 0.14; 0.38 | 0.09 | 0.04; 0.21 | 0.13 | 0.08; 0.23 |
| DOR | 203.00 | 46.80; 867.00 | 16.43 | 14.5; 18.53 | 47.67 | 57.75; 37.81 | 257.15 | 60.25; 1,008 |
| Youden's index | 0.84 | 0.65; 0.91 | 0.60 | 0.33; 0.80 | 0.71 | 0.47; 0.87 | 0.87 | 0.68; 0.93 |
| CUI+ | 0.84 | 0.77; 0.92 | 0.73 | 0.64; 0.83 | 0.84 | 0.77; 0.91 | 0.87 | 0.80; 0.94 |
| CUI– | 0.74 | 0.66; 0.82 | 0.52 | 0.40; 0.63 | 0.65 | 0.54; 0.76 | 0.77 | 0.69; 0.85 |
| PPV, % | 100.00 | − | 89.33 | 81.81; 93.98 | 90.48 | 83.68; 94.62 | 100.00 | − |
| NPV, % | 74.00 | 63.35; 82.41 | 65.91 | 54.28; 75.90 | 82.86 | 68.71; 91.41 | 77.08 | 66.00; 85.36 |
| Accuracy, % | 89.08 | 82.04; 94.05 | 80.67 | 72.42; 87.34 | 88.24 | 81.05; 93.42 | 90.76 | 84.06; 95.29 |
| MCC | 0.79 | − | 0.57 | − | 0.72 | − | 0.82 | − |
| Group PCR+ | ||||||||
| TP | 68 | 65 | 69 | 69 | ||||
| Sensitivity, % | 97.14 | 90.06; 99.65 | 92.86 | 84.11; 97.64 | 98.57 | 92.30; 99.96 | 98.57 | 92.30; 99.96 |
| Specificity, % | 100.00 | 90.51; 100.00 | 78.38 | 61.79; 90.17 | 78.38 | 61.79; 90.17 | 100.00 | 90.51; 100.00 |
| LR (+) | 37.37 | 5.40; 258.87 | 4.29 | 2.32; 7.96 | 4.56 | 2.47; 8.43 | 37.92 | 5.48; 262.56 |
| LR (−) | 0.03 | 0.01; 0.11 | 0.09 | 0.04; 0.22 | 0.02 | 0.00; 0.13 | 0.01 | 0.00; 0.10 |
| DOR | 1,245 | 540; 2,353 | 47.66 | 58.00; 36.18 | 228 | 0.00; 64.85 | 3,792 | 0.00; 2,625 |
| Youden's index | 0.97 | 0.81; 1.00 | 0.71 | 0.46; 0.88 | 0.77 | 0.54; 0.90 | 0.99 | 0.83; 1.00 |
| CUI+ | 0.97 | 0.94; 1.00 | 0.83 | 0.75; 0.91 | 0.88 | 0.82; 0.95 | 0.99 | 0.96; 1.00 |
| CUI– | 0.95 | 0.91; 0.99 | 0.67 | 0.56; 0.78 | 0.75 | 0.66; 0.86 | 0.97 | 0.94; 1.00 |
| PPV, % | 100.00 | − | 89.04 | 81.43; 93.77 | 89.61 | 82.35; 94.10 | 100.00 | − |
| NPV, % | 94.87 | 82.52; 98.64 | 85.29 | 71.02; 93.21 | 96.67 | 80.44; 99.51 | 97.37 | 84.09; 99.62 |
| Accuracy, % | 98.13 | 93.41; 99.77 | 87.85 | 80.12; 93.37 | 91.59 | 84.63; 96.08 | 99.07 | 94.90; 99.98 |
| MCC | 0.96 | − | 0.73 | − | 0.81 | − | 0.98 | − |
| Group PCR– | ||||||||
| TP | 01 | 02 | 07 | 02 | ||||
| Sensitivity, % | 8.33 | 0.21; 38.48 | 16.67 | 2.09; 48.41 | 58.33 | 27.67; 84.83 | 16.67 | 2.09; 48.41 |
| Specificity, % | 100.00 | 90.51; 100.00 | 78.38 | 61.79; 90.17 | 78.38 | 61.79; 90.17 | 100.00 | 90.51; 100.00 |
| LR (+) | 5.57 | 0.55; 56.78 | 0.77 | 0.19; 3.14 | 2.70 | 1.24; 5.87 | 8.36 | 0.95; 73.88 |
| LR (−) | 0.92 | 0.77; 1.09 | 1.06 | 0.78; 1.44 | 0.53 | 0.27; 1.06 | 0.83 | 0.65; 1.07 |
| DOR | 6.05 | 0.71; 52.02 | 0.73 | 0.24; 2.18 | 5.09 | 4.59; 5.54 | 10.07 | 1.46; 69.05 |
| Youden's index | 0.08 | −0.09; 0.39 | −0.05 | −0.36; 0.39 | 0.37 | −0.11; 0.75 | 0.17 | −0.07; 0.48 |
| CUI+ | 0.08 | 0.00; 0.55 | 0.03 | 0.00; 0.31 | 0.27 | 0.00; 0.60 | 0.17 | 0.00; 0.61 |
| CUI– | 0.77 | 0.69; 0.84 | 0.58 | 0.47; 0.70 | 0.67 | 0.56; 0.78 | 0.79 | 0.71; 0.87 |
| PPV, % | 100.00 | − | 20.00 | 5.77; 50.49 | 46.67 | 28.67; 65.57 | 100.00 | − |
| NPV, % | 77.08 | 73.93; 79.96 | 74.36 | 68.14; 79.72 | 85.29 | 74.41; 92.04 | 78.72 | 74.18; 82.65 |
| Accuracy, % | 77.55 | 63.38; 88.23 | 63.27 | 48.29; 76.58 | 73.47 | 58.92; 85.05 | 79.59 | 65.66; 89.76 |
| MCC | 0.25 | − | −0.05 | − | 0.34 | − | 0.36 | − |
LFA, lateral flow immunochromatographic assay; TP, true positive; PPV, positive predictive value; NPV, negative predictive value; LR, likelihood ratio; DOR, diagnostic odds ratio (the higher the DOR value, the better the test); CUI, clinical utility index; MCC, Matthews correlation coefficient.
Fig. 2.
Comparison of LFA diagnostic performance for IgM, IgG, or total antibodies to diagnose SARS-CoV-2 infection. MedTest (circle), IgM (black circle), IgG (empty circle), and IgM or IgG (orange circle); Eco Test (diamond), IgM (black diamond), IgG (empty diamond), and IgM or IgG (orange diamond); Camtech (square), IgM (black square), IgG (empty square), and IgM or IgG (orange square); Wondfo IgM or IgG (orange triangle). Sensitivity (a); specificity (b); accuracy (c); and Youden's index (d). LFA, lateral flow immunochromatographic assay.
Considering IgG, IgM, or total antibodies for the diagnoses of definite or probable SARS-CoV-2 infection, the 4 tests studied showed high diagnostic performance characteristics. The higher sensitivity was for Camtech; accuracy was similar for MedTest, Camtech, and Wondfo. Diagnostic specificity was 100% for the MedTest and Wondfo. LR+ was >30 for the MedTest and Wondfo, indicating a positive test, almost confirming the SARS-CoV-2 infection although it was limited for the Eco Test and Camtech. The highest DOR was seen in the Wondfo test.
For the PCR+ cases, the sensitivity and accuracy were higher for all the kits studied. Although for the probable SARS-CoV-2 infection (RT-qPCR negative) all the kits showed a limited value for total antibody, IgM, and IgG, a high specificity was seen for IgM and IgG.
The Wondfo and MedTest showed the best clinical diagnostic performance characteristics with a high LR (+), which indicates that a positive test almost confirmed the disease, and a high Youden's index, DOR, accuracy, and MCC (Table 1). Concerning the detection of IgM for the diagnoses of definite or probable SARS-CoV-2 infection, MedTest and Camtech displayed higher sensitivity and accuracy. All the kits showed high diagnostic specificity for IgM detection (Table 2). Concerning the detection of IgG for SARS-CoV-2, the sensitivity was low, although it was specific; the MedTest and Camtech showed similar diagnostic performance characteristics (Table 3).
Table 2.
Clinical performance characteristics of LFA in serum to detect SARS-CoV-2 IgM antibodies
| IgM | MedTest | EcoTest | Camtech | |||
|---|---|---|---|---|---|---|
| value | 95% CI | value | 95% CI | value | 95% CI | |
| Group PCR+/PCR– | ||||||
| TP | 68 | 48 | 72 | |||
| Sensitivity, % | 82.93 | 73.02; 90.34 | 58.54 | 47.12; 69.32 | 87.80 | 78.71; 93.99 |
| Specificity, % | 100.00 | 90.51; 100.00 | 83.78 | 67.99; 93.81 | 81.08 | 64.84; 92.04 |
| LR (+) | 32.04 | 4.62; 222.32 | 3.61 | 1.70; 7.68 | 4.64 | 2.37; 9.09 |
| LR (−) | 0.17 | 0.11; 0.28 | 0.49 | 0.37; 0.66 | 0.15 | 0.08; 0.27 |
| DOR | 188.47 | 42.00; 794.00 | 7.37 | 4.60; 11.64 | 30.93 | 29.63; 33.67 |
| Youden's index | 0.83 | 0.64; 0.90 | 0.42 | 0.15; 0.63 | 0.69 | 0.44; 0.86 |
| CUI+ | 0.83 | 0.75; 0.91 | 0.52 | 0.39; 0.65 | 0.80 | 0.72; 0.88 |
| CUI– | 0.73 | 0.64; 0.81 | 0.40 | 0.29; 0.51 | 0.61 | 0.50; 0.72 |
| PPV, % | 100.00 | − | 88.89 | 79.00; 94.45 | 91.14 | 84.01; 95.27 |
| NPV, % | 72.55 | 62.12; 80.98 | 47.69 | 40.47; 55.02 | 75.00 | 62.18; 84.55 |
| Accuracy, % | 88.24 | 81.05; 93.42 | 66.39 | 57.15; 74.78 | 85.71 | 78.12; 91.45 |
| MCC | 0.78 | − | 0.39 | − | 0.68 | − |
| Group PCR+ | ||||||
| TP | 67 | 46 | 66 | |||
| Sensitivity, % | 95.71 | 87.98; 99.11 | 65.71 | 53.40; 76.65 | 94.29 | 86.01; 98.42 |
| Specificity, % | 100.00 | 90.51; 100.00 | 83.78 | 67.99; 93.81 | 81.08 | 64.84; 92.04 |
| LR+ | 36.83 | 5.32; 255.17 | 4.05 | 1.91; 8.59 | 4.98 | 2.55; 9.73 |
| LR– | 0.04 | 0.01; 0.13 | 0.41 | 0.29; 0.58 | 0.07 | 0.03; 0.18 |
| DOR | 921 | 532; 1,962 | 9.87 | 6.59; 14.81 | 71.14 | 85.00; 54.05 |
| Youden's index | 0.96 | 0.79; 0.99 | 0.50 | 0.21; 0.71 | 0.75 | 0.51; 0.91 |
| CUI+ | 0.96 | 0.92; 1.00 | 0.58 | 0.45; 0.71 | 0.85 | 0.78; 0.93 |
| CUI– | 0.93 | 0.87; 0.98 | 0.47 | 0.36; 0.58 | 0.72 | 0.61; 0.82 |
| PPV, % | 100.00 | − | 88.46 | 78.33; 94.21 | 90.41 | 82.84; 94.85 |
| NPV, % | 92.50 | 80.30; 97.39 | 56.36 | 47.55; 64.79 | 88.24 | 74.09; 95.16 |
| Accuracy, % | 97.20 | 92.02; 99.42 | 71.96 | 62.45; 80.22 | 89.72 | 82.35; 94.76 |
| MCC | 0.94 | − | 0.47 | − | 0.77 | − |
| Group PCR– | ||||||
| TP | 01 | 02 | 06 | |||
| Sensitivity, % | 8.33 | 0.21; 38.48 | 16.67 | 2.09; 48.41 | 50.00 | 21.09; 78.91 |
| Specificity, % | 100.00 | 90.51; 100.00 | 83.78 | 67.99; 93.81 | 81.08 | 64.84; 92.04 |
| LR+ | 5.57 | 0.55; 56.78 | 1.03 | 0.24; 4.43 | 2.64 | 1.10; 6.34 |
| LR– | 0.92 | 0.77; 1.09 | 0.99 | 0.74; 1.33 | 0.62 | 0.34; 1.11 |
| DOR | 6.05 | 0.714; 52.09 | 1.04 | 0.32; 3.33 | 4.26 | 3.33; 5.71 |
| Youden's index | 0.08 | −0.09; 0.39 | 0.01 | −0.30; 0.42 | 0.31 | −0.14; 0.71 |
| CUI+ | 0.08 | 0.00; 0.55 | 0.04 | 0.00; 0.35 | 0.23 | 0.0; 0.57 |
| CUI– | 0.77 | 0.69; 0.85 | 0.63 | 0.53; 0.74 | 0.68 | 0.57; 0.78 |
| PPV, % | 100.00 | − | 25.00 | 7.17; 58.98 | 46.15 | 26.33; 67.27 |
| NPV, % | 77.08 | 73.93; 79.96 | 75.61 | 69.88; 80.56 | 83.33 | 73.55; 89.99 |
| Accuracy, % | 77.55 | 63.38; 88.23 | 67.35 | 52.46; 80.05 | 73.47 | 58.92; 85.05 |
| MCC | 0.25 | − | 0.01 | − | 0.30 | − |
LFA, lateral flow immunochromatographic assay; TP, true positive; PPV, positive predictive value; NPV, negative predictive value; LR, likelihood ratio; DOR, diagnostic odds ratio (the higher the DOR value, the better the test); CUI, clinical utility index; MCC, Matthews correlation coefficient.
Table 3.
Clinical performance characteristics of LFA in serum to detect SARS-CoV-2 IgG antibodies
| IgG | MedTest | EcoTest | Camtech | |||
|---|---|---|---|---|---|---|
| value | 95% CI | value | 95% CI | value | 95% CI | |
| Group PCR+/PCR– | ||||||
| TP | 59 | 63 | 59 | |||
| Sensitivity, % | 78.67 | 67.68; 87.29 | 84.00 | 73.72; 91.45 | 78.67 | 67.68; 87.29 |
| Specificity, % | 100.00 | 90.51; 100.00 | 89.19 | 74.58; 96.97 | 97.30 | 85.84; 99.93 |
| LR+ | 30.39 | 4.37; 211.13 | 7.77 | 3.06; 19.71 | 29.11 | 4.20; 201.92 |
| LR– | 0.21 | 0.14; 0.33 | 0.18 | 0.11; 0.30 | 0.22 | 0.14; 0.34 |
| DOR | 144 | 31.21; 639 | 43.17 | 27.82; 65.70 | 132.32 | 30; 594 |
| Youden's index | 0.79 | 0.58; 0.87 | 0.73 | 0.48; 0.88 | 0.76 | 0.54; 0.87 |
| CUI+ | 0.79 | 0.70; 0.88 | 0.79 | 0.70; 0.88 | 0.77 | 0.68; 0.87 |
| CUI– | 0.70 | 0.61; 0.79 | 0.65 | 0.55; 0.76 | 0.67 | 0.58; 0.77 |
| PPV, % | 100.00 | − | 94.03 | 86.13; 97.56 | 98.33 | 89.48; 99.76 |
| NPV, % | 69.81 | 59.96; 78.12 | 73.33 | 61.80; 82.38 | 69.23 | 59.22; 77.71 |
| Accuracy, % | 85.71 | 77.84; 91.61 | 85.71 | 77.84; 91.61 | 84.82 | 76.81; 90.90 |
| MCC | 0.74 | − | 0.70 | − | 0.72 | − |
| Group PCR+ | ||||||
| TP | 58 | 62 | 57 | |||
| Sensitivity, % | 87.88 | 77.51; 94.62 | 93.94 | 85.20; 98.32 | 86.36 | 75.69; 93.57 |
| Specificity, % | 100.00 | 90.51; 100.00 | 89.19 | 74.58; 96.97 | 97.30 | 85.84; 99.93 |
| LR+ | 33.84 | 4.88; 234.75 | 8.69 | 3.44; 21.97 | 31.95 | 4.61; 221.41 |
| LR– | 0.12 | 0.06; 0.23 | 0.07 | 0.03; 0.18 | 0.14 | 0.08; 0.26 |
| DOR | 282 | 81.33; 1,020.00 | 124 | 115; 122 | 228 | 7.62; 850 |
| Youden's index | 0.88 | 0.68; 0.95 | 0.83 | 0.60; 0.95 | 0.84 | 0.62; 0.94 |
| CUI+ | 0.88 | 0.81; 0.95 | 0.88 | 0.81; 0.95 | 0.85 | 0.77; 0.93 |
| CUI– | 0.82 | 0.75; 0.90 | 0.80 | 0.71; 0.88 | 0.78 | 0.70; 0.86 |
| PPV, % | 100.00 | − | 93.94 | 85.98; 97.51 | 98.28 | 89.16; 99.75 |
| NPV, % | 82.22 | 70.72; 89.85 | 89.19 | 76.02; 95.55 | 80.00 | 68.50; 88.04 |
| Accuracy, % | 92.23 | 85.27; 96.59 | 92.23 | 85.27; 96.59 | 90.29 | 82.87; 95.25 |
| MCC | 0.85 | − | 0.83 | − | 0.81 | − |
| Group PCR– | ||||||
| TP | 01 | 01 | 02 | |||
| Sensitivity, % | 11.11 | 0.28; 48.25 | 11.11 | 0.28; 48.25 | 22.22 | 2.81; 60.01 |
| Specificity, % | 100.00 | 90.51; 100.00 | 89.19 | 74.58; 96.97 | 97.30 | 85.84; 99.93 |
| LR+ | 7.09 | 0.71; 71.11 | 1.03 | 0.13; 8.12 | 8.22 | 0.83; 80.97 |
| LR– | 0.89 | 0.71; 1.12 | 1.00 | 0.77; 1.29 | 0.80 | 0.56; 1.14 |
| DOR | 7.97 | 1.00; 63.49 | 1.03 | 0.17; 6.30 | 10.28 | 1.48; 71.03 |
| Youden's index | 0.11 | −0.09; 0.48 | 0.003 | −0.25; 0.45 | 0.20 | −0.11; 0.60 |
| CUI+ | 0.11 | 0.00; 0.65 | 0.02 | 0.00; 0.35 | 0.15 | 0.00; 0.64 |
| CUI– | 0.82 | 0.75; 0.90 | 0.72 | 0.62; 0.81 | 0.82 | 0.74; 0.89 |
| PPV, % | 100.00 | − | 20.00 | 3.07; 66.38 | 66.67 | 16.88; 95.17 |
| NPV, % | 82.22 | 78.59; 85.35 | 80.49 | 76.14; 84.21 | 83.72 | 78.32; 87.98 |
| Accuracy, % | 82.61 | 68.58; 92.18 | 73.91 | 58.87; 85.73 | 82.61 | 68.58; 92.18 |
| MCC | 0.30 | − | 0.003 | − | 0.31 | − |
LFA, lateral flow immunochromatographic assay; TP, true positive; PPV, positive predictive value; NPV, negative predictive value; LR, likelihood ratio; DOR, diagnostic odds ratio (the higher the DOR value, the better the test); CUI, clinical utility index; MCC, Matthews correlation coefficient.
Two subjects (2.9%) were asymptomatic and were positive for SARS-CoV-2 RT-qPCR from nasopharyngeal samples; Wondfo was positive in both patients; however, only one of them tested positive for IgM in the MedTest and Camtech. IgG was not positive in all assays performed. The other subject tested positive for both IgM and IgG in all assays, but the Camtech test was positive only for IgG.
Analytical Specificity (Cross-Reaction)
For the Wondfo and MedTest IgM and IgG and the Camtech IgG, no cross-reaction was identified, and the analytical specificity was 100%. However, for the Eco Test, the analytical specificity for IgG or IgM was 80% each. For IgM, 1 sample cross-reacted with rhinovirus and 2 samples with Tb. Concerning IgG, 1 sample cross-reacted with rhinovirus and 2 samples with sepsis. For Camtech, there was a cross-reaction of IgM with 4 samples (2 samples with rhinovirus, 1 community-acquired pneumonia, and 1 pleural Tb). The analytical specificity was 73.30%.
Analytical Interference
Analytical interference is defined as the effect of a substance present in the sample that alters the correct value of the result. There was hemolysis in 1 sample in the group with definite SARS-CoV-2 infection. This sample tested positive for IgM, IgG, and total antibodies by all the 4 kits in the group with RT-qPCR positive. The agreement between LFA kits is shown in online suppl. Table 2.
Discussion
In this study, we report the performance characteristics of the LFA for IgG, IgM, and total antibody anti-SARS-CoV-2 from 3 different manufacturers for IgG and IgM and one for total antibody anti-SARS-CoV-2. We used sera from hospitalized adult patients with definite (RT-qPCR proved) and probable (clinical, epidemiologic, and radiologic criteria, with RT-qPCR negative) SARS-CoV-2 infection. The diagnosis of SARS-CoV-2 infection involves collecting the correct specimen at the right time. IgM antibodies usually appear after the 7th day of infection, indicating acute infection. However, IgG antibodies appear from the 14th day on. However, the usefulness of monitoring IgM and discriminating them from IgG is largely debated [12].
Overall, the LFA to identify SARS-CoV-2 antibodies (IgM, IgG, or total) in the definite and probable cases showed high sensitivity, specificity, and PPV. This indicates that a positive result is often seen in those with SARS-CoV-2 infection, and a negative result is often seen in those without the infection. The very high PPV that indicates a false positive is rare, which suggests this can be a potential confirmatory test. A high CUI+ suggested LFAs are excellent for case finding and a CUI− is good for screening. The overall value of this single test for combined screening and case finding is good. The clinical performance characteristics presented higher values considering only the definite SARS-CoV-2 cases determined by positive RT-qPCR. However, for probable cases, the clinical performance characteristics showed limited values. Overall, the manufacturers' DOR and MCC of IgM were higher than IgG, and the diagnostic sensitivity and specificity were similar for IgG and IgM.
In the present study, the Wondfo test presented the best clinical performance characteristic, followed by the MedTest. Our findings are in accordance with previous studies, in which the Wondfo test was one of the 2 best-performing LFAs of the 10 evaluated, with pooled estimates of sensitivity (84.8%; 95% confidence interval [CI] = 81.4–87.8%) and specificity (99.0%; 95% CI = 97.8–99.7%) [13, 14]. However, the Wondfo test has the advantage of not differentiating between IgM and IgG. For IgM and IgG, the best results of clinical performance characteristic were with the MedTest. The Youden's index, accuracy, and MCC of IgG were similar for the 3 manufacturers.
Test performance characteristics as provided by manufacturers were similar with those observed in our study [15]. In the present study, the median time after the symptoms appeared was 11 and 17 days in the groups with definite and probable SARS-CoV-2. We observed that the positivity of immunological tests for SARS-CoV-2 diagnosis is time dependent. Patients with at least 8 days of symptoms presented a higher sensitivity [16, 17]. Antibody-mediated immunity in SARS-CoV-2-specific IgM and IgG is detectable in the serum between 7 and 14 days after the onset of the symptoms, respectively. The SARS-CoV-2 virus RNA peak occurs at 3–5 days after exposure; virus RNA inversely correlated with neutralizing antibody titers. In the acute phase of the disease, nucleic acid detection of SARS-CoV-2 in respiratory samples was greater for antibody detection in the diagnosis of COVID-19 [16, 18, 19, 20, 21, 22]. After that period, tests that detect the presence of specific antibodies are recommended [12, 23]. Antibody tests could play a useful role in the detection of previous SARS-CoV-2 infection if applied 15 or more days after the onset of symptoms. The persistence of antibody rises is currently unknown, as there are very little data beyond 35 days after symptoms onset [1] as well as about the extension of protection of neutralizing antibodies against subsequent infection with the virus [24]. The sensitivity of LFA was 11.1% from the 1st to the 7th day after the onset of the symptoms, 92.9% from the 8th to the 14th days, and 96.8% from day 15 after the onset of the symptoms [25].
Our study will add to the previous ones [1, 13, 25, 26, 27] as SARS-CoV-2 infection cases were diagnosed based on RT-qPCR and clinical radiologic criteria, that is, definite and probable SARS-CoV-2 cases, respectively [3]; 15% of the samples included in the SARS-CoV-2 group were probable infection. Although these cases fulfill the clinical and radiological criteria for SARS-CoV-2 infection, some cases could be miss enrolled.
The main limitation of this study is the small number of samples and the lack of blinding of the index test and reference standard; the majority patients were admitted in the hospital, which represents clinically more severe patients. Only 14% were outpatients, and of these, only 2 were asymptomatic. Therefore, it is unclear if the results could be expanded to asymptomatic or oligosymptomatic persons or even used on serological community surveys. Only adults were included; reports on the dynamics and detection of SARS-CoV-2 antibodies in children are lacking and require urgent attention.
Conclusion
Our data demonstrate excellent performance of the antibody assays studied. We observed substantial heterogeneity in sensitivities of IgM and IgG antibodies between manufacturers. The high specificity of LFAs may contribute to rapidly confirm the presence of SARS-CoV-2 infection and accelerate decision-making in emergency rooms and routing to appropriate hospital wards. Nonetheless, these LFA tests cannot replace molecular diagnostics in acute-care settings but should only be used as an additional screening tool when the improvement of hospital logistics is expected, and their limitations are carefully considered.
Statement of Ethics
This study was approved under a waiver of informed consent by the HC-UFPR Institutional Review Board, Brazil (no. 30578620.70000.0008).
Conflictof Interest Statement
The authors have no conflicts of interest to declare.
Funding Sources
The authors did not receive any funding.
Author Contributions
Conception of the study − S.M. de Almeida, M. B. Nogueira, and S.M. Raboni; study design − S.M. de Almeida, M. B. Nogueira, and S.M. Raboni; data acquisition − S. M. de Almeida, R. N. Spalanzani, M.B. Nogueira, B. Sanada, B.M. Cavalli, I. Rotta, G. R.A. Takahashi, L.A. Pereira, F.B. Moreira, C.L.T. Dino, M.E. Graf, B.M.M. de Almeida, N. R. Domino, G. Genelhoud, L.B. Bueno, and S.M. Raboni; data analysis − S.M. de Almeida, R.N. Spalanzani, M.B. Nogueira, B. Sanada, B.M. Cavalli, I. Rotta, G.R.A. Takahashi, L. A. Pereira, F.B. Moreira, C.L.T. Dino, M.E. Graf, B. M.M. de Almeida, N.R. Domino, G. Genelhoud, L. B. Bueno, and S. Raboni; data interpretation − S.M. de Almeida, M. B. Nogueira, and S.M. Raboni; drafting of the manuscript − S.M. de Almeida, M.B. Nogueira, and S.M. Raboni; final approval of the manuscript − S.M. de Almeida, R.N. Spalanzani, M.B. Nogueira, B. Sanada, B.M. Cavalli, I. Rotta, G.R.A. Takahashi, L. A. Pereira, F.B. Moreira, C.L.T. Dino, M.E. Graf, B. M.M. de Almeida, N.R. Domino, G. Genelhoud, L. B. Bueno, and S. Raboni.
Supplementary Material
Supplementary data
References
- 1.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Spijker R, Taylor-Phillips S, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev. 2020;6:CD013652. doi: 10.1002/14651858.CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caliendo AM, Gilbert DN, Ginocchio CC, Hanson KE, May L, Quinn TC, et al. Better tests, better care: improved diagnostics for infectious diseases. Clin Infect Dis. 2013 Dec;57((Suppl 3)):S139. doi: 10.1093/cid/cit578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Public health surveillance for COVID-19. WHO/2019-nCoV 2020. /SurveillanceGuidance/2020.7; Last accessed, June 9, 2020.
- 4.Silveira MF, Barros AJD, Horta BL, Pellanda LC, Victora GD, Dellagostin OA, et al. Population-based surveys of antibodies against SARS-CoV-2 in Southern Brazil. Nature Medicine. 2020 doi: 10.1038/s41591-020-0992-3. Available from: www.nature.com/naturemedicine. [DOI] [PubMed] [Google Scholar]
- 5.Food and Drug Administration New York SARS-CoV-2 Real-time RT-PCR diagnostic panel. 2020 Available from: . Last accessed, March 9, 2020. [Google Scholar]
- 6.Galen RS, Gambino SR. New York: Wiley & Sons; 1975. Beyond normality, the predictive value and efficiency of medical diagnoses; p. p. 237. [Google Scholar]
- 7.Mitchell AJ. The clinical significance of subjective memory complaints in the diagnosis of mild cognitive impairment and dementia: a meta-analysis. Int J Geriatr Psychiatry. 2008 Nov;23((11)):1191–202. doi: 10.1002/gps.2053. [DOI] [PubMed] [Google Scholar]
- 8.Mitchell AJ. Sensitivity x PPV is a recognized test called the clinical utility index (CUI+) Eur J Epidemiol. 2011 Mar;26((3)):251–2. doi: 10.1007/s10654-011-9561-x. [DOI] [PubMed] [Google Scholar]
- 9.McGee S. Simplifying likelihood ratios. J Gen Intern Med. 2002 Aug;17((8)):646–9. doi: 10.1046/j.1525-1497.2002.10750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Akobeng AK. Understanding diagnostic tests 2: likelihood ratios, pre- and post-test probabilities and their use in clinical practice. Acta Paediatr. 2007 Apr;96((4)):487–91. doi: 10.1111/j.1651-2227.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 11.McHugh ML. Interrater reliability: the kappa statistic. Biochem Med. 2012;22((3)):276–82. [PMC free article] [PubMed] [Google Scholar]
- 12.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19. Immunity. 2020 Jun;52((6)):910–41. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitman JD, Hiatt J, Mowery CT, Shy BR, Yu R, Yamamoto TN, et al. Test performance evaluation of SARS-CoV-2 serological assays. MedRxiv. 2020 [Google Scholar]
- 14.Pellanda LC, Wendland EM, McBride AJA, Tovo-Rodrigues L, Ferreira MRA, Dellagostin OA, et al. Sensitivity and specificity of a rapid test for assessment of exposure to SARS-CoV-2 in a community-based setting in Brazil. MedRxiv. 2020 [Google Scholar]
- 15.Castro R, Luz PM, Wakimoto MD, Veloso VG, Grinsztejn B, Perazzo H. COVID-19: a meta-analysis of diagnostic test accuracy of commercial assays registered in Brazil. Braz J Infect Dis. 2020;24((2)):180–7. doi: 10.1016/j.bjid.2020.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J, Yuan Q, Wang H, Liu W, Liao X, Su Y, et al. Antibody responses to SARSCoV- 2 in patients of novel coronavirus disease 2019. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, et al. Antibody detection and dynamic characteristics in patients with coronavirus disease 2019. Clin Infect Dis. 2020 Nov;71((8)):1930–4. doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, et al. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020 Jul 28;71((15)):778–85. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cassaniti I, Novazzi F, Giardina F, Salivaro F, Sachs M, Perlini S, et al. Performance of vivadiag COVID-19 IgM/IgG rapid test is inadequate for diagnosis of COVID-19 in acute patients referring to emergency room department. J Med Virol. 2020 March;92:1724–7. doi: 10.1002/jmv.25800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.To KK, Tak O, Tsang Y, Leung W, Tam AR, Wu T, et al. Articles temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020 May;20((5)):565–74. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Long Q, Deng H, Chen J, Hu J, Liu B, Liao P, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. MedRxiv. 2020 [Google Scholar]
- 22.Li Z, Yi Y, Luo X, Xiong N, Liu Y, Li S, et al. Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis. J Med Virol. 2020 Sep;92((9)):1518–24. doi: 10.1002/jmv.25727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peeling RW, Wedderburn CJ, Garcia PJ, Boeras D, Fongwen N, Nkengasong J, et al. Serology testing in the COVID-19 pandemic response. Lancet Infect dis. 2020 Sept;20:e245–9. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Cruz RJ, Currier AW, Sampson VB. Laboratory testing methods for novel severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2) Front Cell Dev Biol. 2020 Jun;8:468. doi: 10.3389/fcell.2020.00468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Y, Li X, Yang G, Fan J, Tang Y, Zhao J, et al. Serological immunochromatographic approach in diagnosis with SARS-CoV-2 infected COVID-19 patients. J Infect. 2020 July;81:e28–32. doi: 10.1016/j.jinf.2020.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Geurtsvan Kessel CH, Okba NMA, Igloi Z, Bogers S, Embregts CWE, Laksono BM, et al. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat Commun. 2020 Jul;11:3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flower B, Brown JC, Simmons B, Moshe M, Frise R, Penn R, et al. Clinical and laboratory evaluation of SARS-CoV-2 lateral flow assays for use in a national COVID-19 seroprevalence survey. Thorax. 2020;75:1082–8. doi: 10.1136/thoraxjnl-2020-215732. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data