Abstract
Coronavirus disease 2019 (COVID-19), a pandemic infection with profound effects on human society, has challenged our ability to control viral infections. Although at the beginning of the COVID-19 outbreak, the epidemic seemed controllable in Southern Iran, the disease presented a critical pattern as of May 2020. After a few months of the emergence of COVID-19, its severity and mortality increased dramatically. It has been proposed that antibodies produced during previous exposure to local circulating human coronaviruses or possibly severe acute respiratory syndrome coronavirus 2 might contribute to the development of more severe and lethal presentations of COVID-19 possibly by triggering antibody-dependent enhancement. The binding of virions complexed with antibodies to Fcγ receptors on the target cells initiates receptor-mediated signaling events, leading to enhanced expression of inflammatory cytokines and suppression of intracellular antiviral responses at the transcriptome level, followed by endocytosis of the virus and subsequent activation of immune cells. The activated immune cells might accumulate in the lung and promote cytokine storm and lymphopenia. Furthermore, the formation of immune complexes can promote complement activation and subsequent tissue damage. Although there are currently no clinical data to support this hypothesis, a better understanding of these immunopathologic phenomena and their relation to the disease course and severity might give insights into the development of the most efficient prophylactic and therapeutic approaches. This review demonstrates the critical pattern of COVID-19 in Southern Iran and highlights the possible interplay of factors leading to this condition.
Keywords: SARS-CoV-2, COVID-19, Spike protein, Angiotensin-converting enzyme 2 receptor, Antibody-dependent enhancement, Cytokine storm
Highlights of the Study
The pathogenicity of several viral infections is mediated by effector mechanisms of the immune system.
Antibodies produced during the previous exposure to seasonal coronaviruses or possibly severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are proposed to trigger antibody-dependent enhancement.
Antibody-dependent enhancement might lead to more severe and lethal illness upon infection with SARS-CoV-2, results in cytokine storm and tissue damage, and can modulate immune responses toward an inflammatory profile.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of coronavirus disease 2019 (COVID-19), an unprecedented pandemic that is currently the greatest concern of global health [1, 2, 3, 4]. This pandemic has affected every corner of the world, with 134,957,021 confirmed cases and 2,918,752 deaths globally as of 11 April 2021 [5]. COVID-19 is characterized by an asymptomatic, mild, or severe course; while most patients have asymptomatic or mild to moderate clinical symptoms of viral pneumonia, some patients experience life-threatening clinical illness due to acute respiratory distress syndrome, kidney injury, multiple organ dysfunction, and lung failure, eventually leading to death [6, 7, 8].
SARS-CoV-2 is a novel Betacoronavirus in the family Coronaviridae, with a positive single-stranded RNA genome and enveloped icosahedral capsid [9, 10, 11]. This virus shares 96.2% sequence identity to a bat coronavirus, suggesting interspecies transmissions [12, 13]. The viral genome encodes spike (S), nucleocapsid (N), membrane (M), and envelope (E) structural proteins and 16 nonstructural proteins [8, 14, 15, 16]. The S gene, encoding the most immunogenic viral protein, encounters the highest frequency of mutation and recombination, resulting in the evolution of different strains of SARS-CoV-2 [17, 18]. The S protein has 2 subunits: subunit 1 that contains a receptor-binding domain (RBD) and subunit 2. The RBD of spike protein binds with high affinity to angiotensin-converting enzyme 2 (ACE2) receptor, the main host cell receptor for SARS-CoV-2, and facilitates the entry of the virus into target cells, predominantly resulting in viral spreading into the heart, lungs, and gastrointestinal tract, that is, tissues that express high levels of ACE2 [8, 19, 20, 21]. Antibodies targeting the RBD of S protein are neutralizing antibodies, while antibodies against other parts of S protein tend not to neutralize the virus but to induce more severe disease upon secondary infection [22]. Nonneutralizing antibodies are produced commonly during the course of human coronavirus infection, such as MERS-CoV and SARS-CoV. Moreover, seasonal human coronaviruses may cause repeated infection with short-lived immunity [23, 24, 25].
Neutralizing antibody responses against coronaviruses wane rapidly over time; therefore, the chance of reinfection is high [26]. However, despite short-lived neutralizing antibody responses in most cases of SARS and MERS, the probability of a secondary infection is extremely low due to the low prevalence of SARS-CoV and MERS-CoV in the community [25, 26]. On the contrary, the risk of reinfection in the case of SARS-CoV-2 seems to be more real [26]. SARS-CoV-2 infects a significant proportion of the general population due to the high circulation of the virus in the community. Furthermore, various SARS-CoV-2 strains have been isolated from infected persons. The RBD domain of the SARS-CoV-2 spike protein encounters critical mutations that might increase RBD-binding affinities of some of the mutated strains [18]. Pre-existing anti-RBD antibodies obtained from primary SARS-CoV-2 infection bind with reduced affinity to the RBD-mutated strain. The development of such mutated strains might exacerbate the COVID-19 pandemic with higher severity and mortality due to the reinfection of convalescent patients with the mutated strains [27]. Furthermore, protective neutralizing antibodies are serotype-specific and are therefore cross-reactive against other serotypes but cannot neutralize them [24].
The Critical Pattern of COVID-19 in Southern Iran
Since the emergence of the SARS-CoV-2 pandemic, Iran became one of the world's worst-affected countries. As of 11 April 2021, more than 2,049,078 confirmed cases of COVID-19 with 64,232 deaths have been documented in Iran [5]. Nevertheless, the number of infected cases is presumably much higher, resulting from asymptomatic cases of SARS-CoV-2 infection, symptomatic cases with undetectable viral load in the nasal and pharyngeal swabs, or lack of analysis due to limited resources. Moreover, part of symptomatic cases might not be detected due to low viral load. The high incidence of false-negative results is the greatest limitation of COVID-19 diagnostic tests, which is influenced by viral mutations, sampling time, improper sample types, inadequate handling, and improper transportation of samples [28, 29]. In addition to the transmissibility of COVID-19, high viral load is reported to correlate with worse disease outcomes, thus fatal symptomatic cases have higher viral loads than asymptomatic cases [30, 31, 32].
Southern Iran has been particularly heavily affected by COVID-19 and had a critical disease pattern as of May 2020, despite having a controllable epidemic pattern with very low severity and mortality in the first few months of COVID-19 emergence. At the beginning of the outbreak, severe course of SARS-CoV-2 infection and hospitalization were reported mostly among aged patients in Southern Iran. Children and young people were less likely to have symptomatic SARS-CoV-2 infection as than adults and the elderly [33]. Therefore, young infected subjects mainly remained undiagnosed, despite having a risk of infection similar to adults. Being deceived by mild or absent clinical symptoms, young people became drivers of SARS-CoV-2 transmission in society. After a few months of COVID-19 emergence, the severity and mortality of COVID-19 increased dramatically, and symptomatic cases and cases with severe clinical illness were increasingly reported among young people. The age distribution of the pandemic changed, and severe forms of COVID-19 and subsequently mortality were observed in all age-groups. Furthermore, the incidence of SARS-CoV-2 infection has been progressively increasing even in the hot humid weather of Southern Iran, while the transmissibility and stability of respiratory viral infections decrease with hot and humid weather conditions [34, 35].
Underlying Factors Responsible for the Critical Pattern of COVID-19
Multiple interplaying factors could have contributed to increasing the incidence of COVID-19 in Southern Iran. The use of face masks was initially thought to be ineffective in protecting against SARS-CoV-2 infection [36]. In the traditional society of Southern Iran, most young people live with their parents. This multigenerational cohabitation was believed to increase the risk of SARS-CoV-2 transmission among family members. Moreover, the positive cases were mostly in clusters of family members and relatives, indicating transmission of SARS-CoV-2 at family gatherings by asymptomatic or irresponsible infected persons. The government initially underestimated the threat of COVID-19 and perceived it as some sort of flu with cold-like symptoms. On the other hand, the general population did not take the disease seriously, and people were generally reluctant to adhere to the social restrictions enforced by the government. Furthermore, a full lockdown was not enforced due to economic reasons. Southern Iran is vital to the economy of the entire country, where business activities are essential. The repeated cycles of circulation of the virus within healthcare settings have been proposed as a significant contributor to enhancing the risk of COVID-19 in the community. Hospital-associated transmission has been suggested since a considerable proportion of the infected population is healthcare staff. Considering the role of the abovementioned factors in increasing the incidence of COVID-19 in Southern Iran, it can be presumed that SARS-CoV-2 has probably been circulating in the community for weeks before the first cases were detected. The virus had enough time to circulate and infect a significant proportion of the population, most of whom had asymptomatic infections and remained undiagnosed. Exclusively, hospitalized COVID-19 patients with severe presentations of COVID-19, of note older patients and those with major underlying diseases or comorbidities, were identified. Due to limited resources, there was no possibility of public screening and only those severely ill patients, their close contacts, and healthcare workers were tested for SARS-CoV-2 infection.
We hypothesize that individuals with prior exposure to local circulating human coronaviruses experience more severe and lethal forms of the disease when they become infected with SARS-CoV-2. Moreover, most individuals naive to SARS-CoV-2 experience an asymptomatic or mild infection. However, these SARS-CoV-2 convalescent individuals might experience a severe and lethal illness upon reinfection with different strains of SARS-CoV-2 [27, 37]. According to this hypothesis, we speculate that the antibodies obtained from primary infections with circulating seasonal human coronaviruses or SARS-CoV-2 lead to more severe and lethal illness in secondary infection with SARS-CoV-2 possibly by triggering Fcγ receptor-mediated uptake of the virus. Also, our hypothesis for disease severity and fatality in older people is the repeated exposure to coronavirus infections in lifetime, which increases with age [35, 38, 39, 40]. Immunologic memory to closely related coronaviruses may induce antibody-dependent enhancement of infection and immune complex deposition [39, 41, 42, 43].
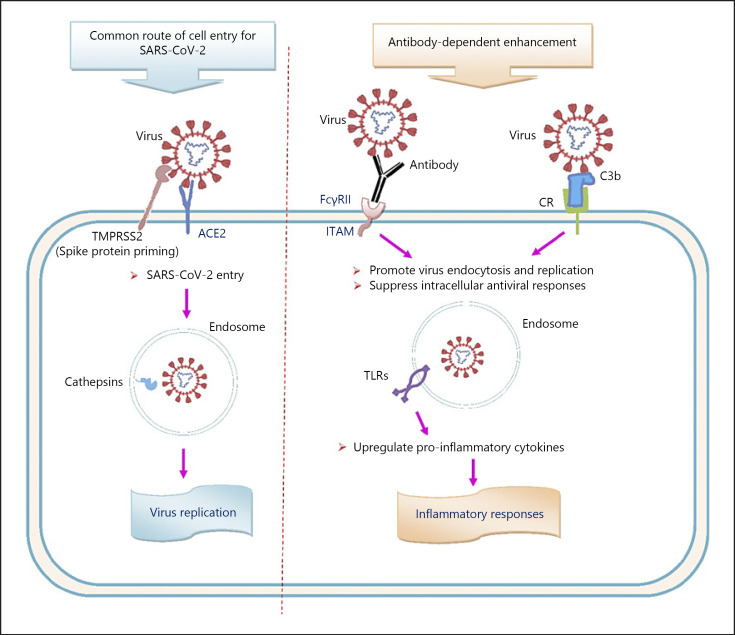
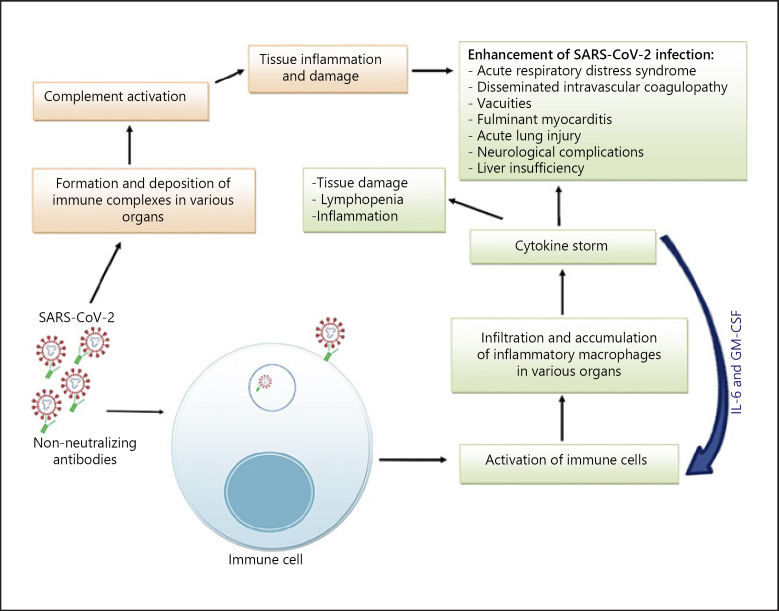
Secondary infection by related virus strains might lead to an exacerbated illness due to the presence of pre-existing cross-reactive nonneutralizing antibodies. The phenomenon in which nonneutralizing or sub-neutralizing antibodies facilitate viral cell entry by intermediation of Fcγ receptors (FcγRs) and/or complement receptors (CR) and lead to enhanced infection is termed antibody-dependent enhancement (ADE) (Fig. 1) [8, 27, 44, 45, 46, 47]. Binding of virions complexed with antibodies and/or complement fragments to FcγRII and/or CR on target cells initiates receptor-mediated signaling events, leading to enhanced expression of inflammatory cytokines and suppression of intracellular antiviral responses at the transcriptome level, followed by endocytosis of the virus and subsequent activation of immune cells. The activated immune cells might accumulate in the lung and promote cytokine storm and lymphopenia [22, 41, 48, 49, 50]. Furthermore, the formation of immune complexes can promote complement activation and subsequent tissue damage (Fig. 2) [41, 51, 52].
Fig. 1.
Potential routes of cell entry for SARS-CoV-2. TMPRSS2, transmembrane protease serine 2; ACE2, angiotensin-converting enzyme 2; FcγRII, Fcγ receptor II; CR, complement receptor; ITAM, immunoreceptor tyrosine-based activation motif; TLRs, toll-like receptors; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Fig. 2.
Potential pathways of antibody-dependent enhancement of SARS-CoV-2 infection. SARS-CoV-2 is recognized by cross-reactive nonneutralizing antibodies obtained from previous infections with circulating human coronaviruses. Virus-antibody complexes bind to immune cells and initiate receptor-mediated signaling events, followed by endocytosis of the virus and subsequent activation of immune cells. The activated immune cells, notably inflammatory macrophages, accumulate in various organs and promote cytokine storm, tissue damage, and lymphopenia, which contribute to the development of more severe presentations of COVID-19. On the other hand, IL-6 and GM-CSF induce monocyte activation and differentiation to inflammatory macrophages. Furthermore, the formation and deposition of immune complexes in various organs can promote complement activation and subsequent tissue damage. SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; COVID-19, coronavirus disease 2019.
ADE acts mainly through 2 pathways: FcγR-mediated ADE and complement-mediated ADE. In FcγR-mediated ADE, virus-antibody complexes bind to FcγRII on immune cells. The FcγRII activates signaling pathways by an immunoreceptor tyrosine-based activation motif to promote virus endocytosis and replication and to suppress the intracellular antiviral responses by decreasing antiviral transcription factors by inhibition of the STAT pathway. Besides, viral particles in the endosomes signal through Toll-like receptors to upregulate the production of pro-inflammatory cytokines and to activate immune cells [22, 53]. In complement-mediated ADE, following the activation of the complement cascade by virus-antibody complexes, complement-coated virions form and bind to CR on cells. Complement receptors activate signaling pathways to increase viral replication by suppressing intracellular antiviral responses [48, 54].
Hyperactivation of immune cells, notably inflammatory macrophages, triggers the excessive and prolonged inflammatory response known as cytokine storm [38]. Cytokine storm is mediated by increased levels of inflammatory mediators and cytokines, such as tumor necrosis factor-alpha, interferon-γ inducible protein 10, IL-1, IL-2, IL-6, IL-7, IL-8, IL-10, IL-18, macrophage colony-stimulating factor, granulocyte colony-stimulating factor, CCL2/monocyte chemotactic protein, CXCL10, and CCL3/macrophage inflammatory protein 1 alpha, which may promote acute lung injury, respiratory distress syndrome, disseminated intravascular coagulopathy, fulminant myocarditis, vacuities, multiple organ dysfunction, and many others [2, 3, 8, 38, 41, 55].
Lymphopenia is a predictor of disease severity and requirement for intensive care in COVID-19 patients. Lymphocyte apoptosis in response to high levels of inflammatory cytokines, lymphocyte lysis following the entry of SARS-CoV-2 into cells, inhibition of lymphocyte proliferation due to the interference by COVID-19 infection, and impairment of the cytotoxic activity of CD8+ T cells followed by the exhaustion of T cells have been proposed as the possible causes of lymphopenia. Nevertheless, the exact etiology remains unknown [56, 57].
There are currently no clinical data to support this hypothesis. The possibility of the development of more severe presentations of COVID-19 sustained by ADE, cytokine storm, and immune complex deposition, if confirmed, would have relevant implications for the management of the COVID-19 pandemic. A better understanding of these immunopathologic phenomena and their relation to disease course and severity might give us insights into the development of the most efficient prophylactic and therapeutic approaches. In addition to the immune-mediated pathogenesis, the severity of COVID-19 could be related to host genetic factors, such as mutations in the ACE2 protein, which are related to enhancing replication of the virus, as well as nonbiological factors [58, 59]. Notably, single-nucleotide polymorphisms in the genes related to immune responses, including cytokine genes and FcγR genes, can affect the severity and susceptibility of infectious diseases [49]. For example, FcγRIIa-R131 polymorphism is associated with SARS severity [27, 44]. The GCC haplotype in the IL10 promoter region is associated with the highest expression level of IL10 [49]. Inborn errors of Toll-like receptor 3- and interferon regulatory factor 7-dependent type I IFNs immunity are reported to correlate with life-threatening COVID-19, and patients with neutralizing autoantibodies against type I IFNs are at risk of developing critical COVID-19 [60, 61].
Given the rapid speed of SARS-CoV-2 transmission and life-threatening consequences of COVID-19 being observed over time, the global administration of efficient prophylactic vaccines seems to be the most affordable solution for controlling this pandemic. Moreover, pandemic management relies on prevention both at the individual and community levels [62]. Effective prevention can be achieved through promoting public health interventions, including face masks, handwashing, social distancing, public education, stay-at-home orders, workplace restrictions, and school closures, and establishing comprehensive screening programs for prompt identification and quarantining of infected subjects with the aim of breaking the cycle of SARS-CoV-2 transmission in the society [25]. Although prevention is a promising strategy to mitigate SARS-CoV-2 transmission, it may not suffice to eliminate this pandemic. In fact, offering regular SARS-CoV-2 testing to the general populations is impossible in resource-limited settings. Therefore, the specific focus should be on high-risk populations with the aim of protecting the vulnerable [63, 64]. Nevertheless, asymptomatic cases and those with undetectable viral load can be potentially considered as mobile infectious sources, contributing to SARS-CoV-2 spread [8]. Moreover, the socioeconomic impacts of social isolation and full lockdown and the afterward economic crisis are some other obstacles to manage this pandemic [65]. In these conditions, the development of potent prophylactic and therapeutic tools against COVID-19 is essential to drive away the rising wave of SARS-CoV-2 infection in Southern Iran.
Conclusion
Given previous data on the role of ADE in the pathogenicity of several viral infections, it is advisable to hypothesize a similar risk for COVID-19. Risk of ADE may be due to antibodies obtained from primary infections with circulating seasonal human coronaviruses or SARS-CoV-2. In addition, our hypothesis for disease severity and fatality in older people is the repeated exposure to coronavirus infections in the lifetime, which increases with age. Further thoughtful and rigorous research is needed to understand whether there is an association between ADE and rates of severity and mortality attributable to COVID-19.
Statement of Ethics
The author confirms that the ethical policies of the journal, as noted on the journal's author guidelines page, have been adhered to. No ethical approval was required as this is a review article with no original research data.
Conflict of Interest Statement
The authors declared that they do not have anything to disclose regarding funding or conflict of interest with respect to the manuscript.
Funding Sources
No funding was received for this review.
Author Contributions
Farshadpour F. and Taherkhani R. designed the study and drafted the manuscript; Taherkhani R. performed the literature review and edited the manuscript. All authors approved the final draft of the manuscript.
Acknowledgments
We extend our sincere gratitude to the staff of the Central Coronavirus Laboratory of Bushehr province who are supporting us during this difficult time.
References
- 1.Alanagreh L, Alzoughool F, Atoum M. The human coronavirus disease COVID-19: its origin, characteristics, and insights into potential drugs and its mechanisms. Pathogens. 2020 Apr 29;9((5)):331. doi: 10.3390/pathogens9050331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo YR, Cao QD, Hong ZS, Tan YY, Chen SD, Jin HJ, et al. The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) outbreak: an update on the status. Mil Med Res. 2020 Mar 13;7((1)):11. doi: 10.1186/s40779-020-00240-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nikolich-Zugich J, Knox KS, Rios CT, Natt B, Bhattacharya D, Fain MJ. SARS-CoV-2 and COVID-19 in older adults: what we may expect regarding pathogenesis, immune responses, and outcomes. GeroScience. 2020 Apr;42((2)):505–14. doi: 10.1007/s11357-020-00186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vabret N, Britton GJ, Gruber C, Hegde S, Kim J, Kuksin M, et al. Immunology of COVID-19: current state of the science. Immunity. 2020 Jun 16;52((6)):910–41. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.WHO . WHO coronavirus disease (COVID-19) dashboard. [updated 11 April 2021]. Available from: https://covid19.who.int/?gclid=CjwKCAjw19z6BRAYEiwAmo64LXM8cRxNccb9fSrb3jI3kULSSk2ji9fVnDHiXvfJDNIAeeaeara4GxoC4hoQAvD_BwE. [Google Scholar]
- 6.Chen D, Xu W, Lei Z, Huang Z, Liu J, Gao Z, et al. Recurrence of positive SARS-CoV-2 RNA in COVID-19: a case report. Int J Infect Dis. 2020 Apr;93:297–9. doi: 10.1016/j.ijid.2020.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gavriatopoulou M, Korompoki E, Fotiou D, Ntanasis-Stathopoulos I, Psaltopoulou T, Kastritis E, et al. Organ-specific manifestations of COVID-19 infection. Clin Exp Med. 2020;20:493–506. doi: 10.1007/s10238-020-00648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siracusano G, Pastori C, Lopalco L. Humoral immune responses in COVID-19 patients: a window on the state of the art. Front Immunol. 2020;11:1049. doi: 10.3389/fimmu.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bahrami A, Ferns GA. Genetic and pathogenic characterization of SARS-CoV-2: a review. Future Virol. 2020 [Google Scholar]
- 10.Pal M, Berhanu G, Desalegn C, Kandi V. Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2): an update. Cureus. 2020 Mar 26;12((3)):e7423. doi: 10.7759/cureus.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ugurel OM, Ata O, Turgut-Balik D. An updated analysis of variations in SARS-CoV-2 genome. Turk J Biol. 2020;44((3)):157–67. doi: 10.3906/biy-2005-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ceraolo C, Giorgi FM. Genomic variance of the 2019-nCoV coronavirus. J Med Virol. 2020 May;92((5)):522–8. doi: 10.1002/jmv.25700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malaiyan J, Arumugam S, Mohan K, Gomathi Radhakrishnan G. An update on the origin of SARS-CoV-2: despite closest identity, bat (RaTG13) and pangolin derived coronaviruses varied in the critical binding site and O-linked glycan residues. J Med Virol. 2020 Jul 7;93((1)):499–505. doi: 10.1002/jmv.26261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J Med Virol. 2020 Apr;92((4)):2249–23. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satarker S, Nampoothiri M. Structural proteins in severe acute respiratory syndrome coronavirus-2. Arch Med Res. 2020 May 25;51:482–91. doi: 10.1016/j.arcmed.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshimoto FK. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020 Jun;39((3)):198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen B, Tian EK, He B, Tian L, Han R, Wang S, et al. Overview of lethal human coronaviruses. Signal Transduct Target Ther. 2020 Jun 10;5((1)):89. doi: 10.1038/s41392-020-0190-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ou J, Zhou Z, Zhang J, Lan W, Zhao S, Wu J, et al. RBD mutations from circulating SARS-CoV-2 strains enhance the structural stability and human ACE2 affinity of the spike protein. bioRxiv. 2020 [Google Scholar]
- 19.Bibiana SOF, Vargas-Pinilla P, Amorim CEG, Sortica VA, Bortolini MC. ACE2 diversity in placental mammals reveals the evolutionary strategy of SARS-CoV-2. Genet Mol Biol. 2020;43((2)):e20200104. doi: 10.1590/1678-4685-GMB-2020-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury MA, Hossain N, Kashem MA, Shahid MA, Alam A. Immune response in COVID-19: a review. J Infect Public Health. 2020 Jul 14;13:1619–29. doi: 10.1016/j.jiph.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, et al. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol. 2020;58((6)):e00461–20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki A, Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020 Jun;20((6)):339–41. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shokri S, Mahmoudvand S, Taherkhani R, Farshadpour F. Modulation of the immune response by Middle East respiratory syndrome coronavirus. J Cell Physiol. 2019 Mar;234((3)):2143–51. doi: 10.1002/jcp.27155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorse GJ, Donovan MM, Patel GB. Antibodies to coronaviruses are higher in older compared with younger adults and binding antibodies are more sensitive than neutralizing antibodies in identifying coronavirus-associated illnesses. J Med Virol. 2020 May;92((5)):512–7. doi: 10.1002/jmv.25715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirkcaldy RD, King BA, Brooks JT. COVID-19 and postinfection immunity: limited evidence, many remaining questions. JAMA. 2020 May 11;323((22)):2245–6. doi: 10.1001/jama.2020.7869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kellam P, Barclay W. The dynamics of humoral immune responses following SARS-CoV-2 infection and the potential for reinfection. J Gen Virol. 2020 Aug;101((8)):791–7. doi: 10.1099/jgv.0.001439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulrich H, Pillat MM, Tárnok A. Dengue fever, COVID-19 (SARS-CoV-2), and antibody-dependent enhancement (ADE): a perspective. Cytometry A. 2020 Jul;97((7)):662–7. doi: 10.1002/cyto.a.24047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu E. COVID-19 testing: challenges, limitations and suggestions for improvement. 2020. [Google Scholar]
- 29.Liu G, Rusling JF. COVID-19 antibody tests and their limitations. ACS Sens. 2021;6((3)):593. doi: 10.1021/acssensors.0c02621. [DOI] [PubMed] [Google Scholar]
- 30.Fajnzylber J, Regan J, Coxen K, Corry H, Wong C, Rosenthal A, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11((1)):5493–9. doi: 10.1038/s41467-020-19057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawasuji H, Takegoshi Y, Kaneda M, Ueno A, Miyajima Y, Kawago K, et al. Transmissibility of COVID-19 depends on the viral load around onset in adult and symptomatic patients. PloS One. 2020;15((12)):e0243597. doi: 10.1371/journal.pone.0243597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukagoshi H, Shinoda D, Saito M, Okayama K, Sada M, Kimura H, et al. Relationships between viral load and the clinical course of COVID-19. Viruses. 2021;13((2)):304. doi: 10.3390/v13020304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Felsenstein S, Hedrich CM. SARS-CoV-2 infections in children and young people. Clin Immunol. 2020;220:108588. doi: 10.1016/j.clim.2020.108588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eccles R. An explanation for the seasonality of acute upper respiratory tract viral infections. Acta Otolaryngol. 2002 Mar;122((2)):183–91. doi: 10.1080/00016480252814207. [DOI] [PubMed] [Google Scholar]
- 35.Cegolon L, Pichierri J, Mastrangelo G, Cinquetti S, Sotgiu G, Bellizzi S, et al. Hypothesis to explain the severe form of COVID-19 in Northern Italy. BMJ Glob Health. 2020 Jun;5((6)):e002564. doi: 10.1136/bmjgh-2020-002564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feng S, Shen C, Xia N, Song W, Fan M, Cowling BJ. Rational use of face masks in the COVID-19 pandemic. Lancet Respir Med. 2020;8((5)):434–6. doi: 10.1016/S2213-2600(20)30134-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lafaie L, Celarier T, Goethals L, Pozzetto B, Grange S, Ojardias E, et al. Recurrence or relapse of COVID-19 in older patients: a description of three cases. J Am Geriatr Soc. 2020 Jul 7 doi: 10.1111/jgs.16728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gomez-Rial J, Rivero-Calle I, Salas A, Martinon-Torres F. Role of monocytes/macrophages in Covid-19 pathogenesis: implications for therapy. Infect Drug Resist. 2020;13:2485–93. doi: 10.2147/IDR.S258639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sedokani A, Feizollahzadeh S. Plasmapheresis, anti-ACE2 and anti-FcγRII monoclonal antibodies: a possible treatment for severe cases of COVID-19. Drug Desi Devel Ther. 2020;14:2607–11. doi: 10.2147/DDDT.S262491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22((2)):72–3. doi: 10.1016/j.micinf.2020.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Felsenstein S, Herbert JA, McNamara PS, Hedrich CM. COVID-19: immunology and treatment options. Clin Immunol. 2020 Jun;215:108448. doi: 10.1016/j.clim.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fierz W, Walz B. Antibody dependent enhancement due to original antigenic sin and the development of SARS. Front Immunol. 2020;11:1120. doi: 10.3389/fimmu.2020.01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar R, Gupta N, Kodan P, Mittal A, Soneja M, Wig N. Is there antibody-dependent enhancement in SARS Coronavirus 2? J Family Med Prim Care. 2020;9((5)):2589–90. doi: 10.4103/jfmpc.jfmpc_540_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bournazos S, Gupta A, Ravetch JV. The role of IgG Fc receptors in antibody-dependent enhancement. Nat Rev Immunol. 2020 Aug 11;20:633–43. doi: 10.1038/s41577-020-00410-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Negro F. Is antibody-dependent enhancement playing a role in COVID-19 pathogenesis? Swiss Med Wkly. 2020;150((1516)):w20249. doi: 10.4414/smw.2020.20249. [DOI] [PubMed] [Google Scholar]
- 46.Wan Y, Shang J, Sun S, Tai W, Chen J, Geng Q, et al. Molecular mechanism for antibody-dependent enhancement of coronavirus entry. J Virol. 2020 Feb 14;94((5)):94. doi: 10.1128/JVI.02015-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yager EJ. Antibody-dependent enhancement and COVID-19: moving toward acquittal. Clin Immunol. 2020 Aug;217:108496. doi: 10.1016/j.clim.2020.108496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willey S, Aasa-Chapman MM, O'Farrell S, Pellegrino P, Williams I, Weiss RA, et al. Extensive complement-dependent enhancement of HIV-1 by autologous non-neutralising antibodies at early stages of infection. Retrovirology. 2011 Mar 14;8:16. doi: 10.1186/1742-4690-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smatti MK, Al Thani AA, Yassine HM. Viral-induced enhanced disease illness. Front Microbiol. 2018;9:2991. doi: 10.3389/fmicb.2018.02991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kadkhoda K. COVID-19: are neutralizing antibodies neutralizing enough? Transfusion. 2020 Jul;60((7)):1602–3. doi: 10.1111/trf.15897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burton DR, Walker LM. Rational vaccine design in the time of COVID-19. Cell Host Microbe. 2020 May 13;27((5)):695–8. doi: 10.1016/j.chom.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Felsenstein S, Hedrich CM. COVID-19 in children and young people. Lancet Rheumatol. 2020 Sep;2((9)):e514–16. doi: 10.1016/S2665-9913(20)30212-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Siracusano G, Pastori C, Lopalco L. Humoral immune responses in COVID-19 patients: a window on the state of the art. Front Immunol. 2020;11:1049. doi: 10.3389/fimmu.2020.01049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smatti MK, Al Thani AA, Yassine HM. Viral-induced enhanced disease illness. Front Microbiol. 2018;9:2991. doi: 10.3389/fmicb.2018.02991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jafarzadeh A, Chauhan P, Saha B, Jafarzadeh S, Nemati M. Contribution of monocytes and macrophages to the local tissue inflammation and cytokine storm in COVID-19: lessons from SARS and MERS, and potential therapeutic interventions. Life Sci. 2020 Sep 15;257:118102. doi: 10.1016/j.lfs.2020.118102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tavakolpour S, Rakhshandehroo T, Wei EX, Rashidian M. Lymphopenia during the COVID-19 infection: what it shows and what can be learned. Immunol Lett. 2020 Sep;225:31–2. doi: 10.1016/j.imlet.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Terpos E, Ntanasis-Stathopoulos I, Elalamy I, Kastritis E, Sergentanis TN, Politou M, et al. Hematological findings and complications of COVID-19. Am J Hematol. 2020 Jul;95((7)):834–47. doi: 10.1002/ajh.25829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hou Y, Zhao J, Martin W, Kallianpur A, Chung MK, Jehi L, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020 Jul 15;18((1)):216. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.LoPresti M, Beck DB, Duggal P, Cummings DAT, Solomon BD. The role of host genetic factors in coronavirus susceptibility: review of animal and systematic review of human literature. Am J Hum Genet. 2020 Sep 3;107((3)):381–402. doi: 10.1016/j.ajhg.2020.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020 Oct 23;370((6515)) doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Q, Bastard P, Liu Z, Le Pen J, Moncada-Velez M, Chen J, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020 Oct 23;370((6515)) doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Javed M, Javed F, Ergin HE, Maung TZ, Khan S. Do COVID-19 and SARS gene complexities and variations help overcome the knowledge gap? Cureus. 2020;12((6)):e8439. doi: 10.7759/cureus.8439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Taherkhani R, Farshadpour F. Global elimination of hepatitis C virus infection: progresses and the remaining challenges. World J Hepatol. 2017 Nov 28;9((33)):1239–52. doi: 10.4254/wjh.v9.i33.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taherkhani R, Farshadpour F. Lurking epidemic of hepatitis C virus infection in Iran: a call to action. World J Hepatol. 2017 Aug 28;9((24)):1040–2. doi: 10.4254/wjh.v9.i24.1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, et al. The socio-economic implications of the coronavirus pandemic (COVID-19): a review. Int J Surg. 2020 Jun;78:185–93. doi: 10.1016/j.ijsu.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]