Abstract
Aims
To understand the impact of storage temperature on recovery of Staphylococcus aureus on sampling swabs. Staphylococcus aureus is a common cause of skin and soft tissue infections, but also causes a variety of life‐threatening diseases. With a large pool of asymptomatic carriers and transmission that can occur even through indirect contact, mitigation efforts have had limited success. Swab sampling, followed by culturing, is a cornerstone of epidemiological studies, however, S. aureus viability on swabs stored at different temperatures has not been characterized.
Methods and Results
We determined survival rates on swabs stored at five different temperatures. Samples stored at −70°C had no decay over time while samples stored at higher temperatures showed an exponential decay in viability. Mortality rates were greatest for swabs stored at 37°C. Survival at intermediate temperatures (−20 to 20·5°C) did not differ significantly, however, we observed more variation at higher temperatures.
Conclusions
To maximize recovery of S. aureus cells, samples should be stored at −70°C or processed for culturing without delay.
Significance and Impact of the Study
Epidemiological studies of bacterial diseases are typically limited to determination of pathogen presence/absence, yet quantitative assessments of pathogen load and genetic diversity can provide insights into disease progression and severity, likelihood of transmission and adaptive evolutionary potential. For studies of S. aureus where time or access to a microbiology laboratory may delay culturing, deep freezing or timely culturing will maximize the degree to which sampling results reflect source status.
Keywords: bacterial storage conditions, community sampling of S. aureus , determining bacterial concentration, microbial viability, storing bacterial samples, survival of bacteria on swabs, swab samples
Introduction
Staphylococcus aureus is a mesophilic Gram‐positive coccus‐shaped bacterium (Schleifer and Bell 2015), commonly associated with the epithelial tissues of humans and other animals. It is a common cause of skin, soft‐tissue, bone, joint, respiratory and endovascular infections in humans that occur when these bacteria penetrate the outer layers of skin or mucosa (Williams 1963). In 2017 in the United States, almost 120 000 bloodstream infections of S. aureus were recorded and 20 000 associated cases died (Kourtis et al. 2019). Treatment is increasingly more challenging due to the continuous emergence of strains resistant to methicillin and other beta‐lactam antibiotics, vancomycin, fluoroquinolones and other antibiotics (Chambers and DeLeo 2009; European Centre for Disease Prevention and Control 2019).
Historically, most clinical S. aureus infections were acquired in health care settings. However, during the past two decades, infections are increasingly acquired in community settings (Chambers and DeLeo 2009). This pathogen can be carried persistently or transiently in a large proportion of individuals without clinical manifestations (Williams 1963). In fact, colonization is prevalent in the nose of approximately one third of the healthy US population (Kluytmans et al. 1997; Wertheim et al. 2005). This wide distribution enables broad transmission, especially through transient and sporadic carriers, who experience frequent strain replacement (van Belkum et al. 1997; VandenBergh et al. 1999). In addition, horizontal gene transfer provides a mechanism for the spread of antibiotic resistances and virulence factors to new lineages (Fitzgerald et al. 2001). These characteristics emphasize the importance and challenge of understanding transmission in community settings (Zetola et al. 2005; Chambers and DeLeo 2009; Price et al. 2012; Grøntvedt et al. 2016; Pearson et al. 2019).
To better understand carriage and transmission of S. aureus in community settings, sampling strategies, which typically involve the use of swabs, must provide robust culture‐based isolation for downstream analyses for detection that accurately reflect carrier status (Williams 1963; Wertheim et al. 2005). Particularly for community‐based studies, storage conditions and elapsed time before culturing may play an important role on the viability of cells and therefore on successful culturing and accurate detection. The effectiveness of transport media for S. aureus kept for long periods of time has been widely studied (Morosini et al. 2006; Rishmawi et al. 2007; van Horn et al. 2008; Saegeman et al. 2011; Robinson et al. 2012). However, the use of transport media adds considerable expense to larger studies and precludes any downstream quantitative analyses as the bacteria will still replicate. The necessity of transport media has not been thoroughly evaluated.
In general, few studies have attempted to quantify the survival of S. aureus across different temperatures in the absence of media. Our understanding of S. aureus survival is based on studies of survival on frozen food (Tanaka et al. 1999; Casarin et al. 2009; Saklani et al. 2020), where viable cells could be detected for up to 2 months after storage without significant changes in concentration. At room temperature on a variety of substrates and conditions, S. aureus showed highly variable survival (Scott and Bloomfield 1990; Neely and Maley 2000; Wagenvoort et al. 2000; Noyce et al. 2006). However, almost no studies have quantified S. aureus survival in the absence of media and in the context of storage for down‐stream culture‐based isolation, which is critical to community‐based surveillance. In this study, we evaluated the longitudinal survival of five strains of S. aureus on swabs stored at five different temperatures.
Methods
Strains
Isolates were collected as part of an ongoing study of community carriage and transmission based in Yuma, AZ (Pearson et al. 2019). All isolates were collected using BD BBL CultureSwabs, cultured on CHROMAgar Staph aureus plates, and stored in 20% glycerol at −70°C (Pearson et al. 2019). Isolates used here (Table 1) were sequenced to determine multilocus sequence typing (MLST) genotype and none possessed the SCCmec mobile element conferring methicillin resistance. In order to represent a diverse group of strains, five isolates (termed isolates A, B, D, D and E) were selected within some of the common MLST genotypes of community isolates from Yuma, AZ: ST30 (two isolates), ST5, ST8 and ST45.
Table 1.
Sequence types and read accession numbers for the isolates used in this study
| Name for the isolates in this study | MLST genotype | Accession number |
|---|---|---|
| A | ST30 | SRX9078810 |
| B | ST5 | SRX9078814 |
| C | ST8 | SRX9078813 |
| D | ST30 | SRX9078812 |
| E | ST45 | SRX9078811 |
MLST, multilocus sequence typing.
Stock preparation
Glycerol stocks of strains collected from participants (Pearson et al. 2019) were streaked for isolation onto CHROMAgar Staph aureus plates and incubated at 37°C for 24 h. For each isolate, a single S. aureus colony was selected and streaked again on Tryptic Soy Agar (TSA) without dextrose plates and incubated for 24 h at 37°C. Four colonies from each re‐streaked TSA plate were transferred with a sterile plastic loop into 15 ml conical tubes containing 10 ml of sterile phosphate‐buffered saline (PBS) pH 7·4 and homogenized. Four colonies provided the approximate quantity of cells for our targeted initial concentration for inoculating swabs. Preliminary tests with isolate A showed little difference between the stock concentration of the 15 ml conical tubes and the concentration recovered immediately after inoculating a swab, suggesting high recovery potential and that enumeration on day 0 provided a good approximation of the inoculating concentration as well as a baseline value for determining longitudinal survival.
Storage treatments
On day 0, the same day that the stock solution was prepared, we placed 100 μl of stock solution onto each swab tip (sterile double swab BD BBL CultureSwab) using a micropipette. Each double swab was placed back into the storage tube and into a Ziploc bag. Each pair of swabs was processed at the same time and provided two independent replicates for each time and treatment point. Storage temperature treatments reflect ‘body temperature’ at 37°C (which varied from 36° to 37°C), ‘room temperature’ in a hermetic plastic box for protection against light and slight temperature changes (temperatures varied from 19·5 to 21·5°C with a mode of 20·5°C), ‘refrigerator’ at 5°C (varying from 3 to 6°C), ‘freezer’ at −20°C (undetermined variation) and ‘deep freezer’ at −70°C (varying from −69 to −70°C). We also performed an initial test of isolate A stored at 45°C, but no bacteria were recovered at or after 1 h, so we did not test any of the other isolates at this temperature.
Bacteria enumeration
To determine the concentration of S. aureus on each swab pair at time 0 and various time points after exposure to treatments, the tip of each swab was clipped off the shaft and placed in a microfuge tube containing 0.9ml of PBS and vortexed for 20 s. For accurate enumeration, we adjusted the concentration with a goal of yielding 30–300 colonies on, at least, one plate per sample. We did this by diluting the solution 1 : 10 followed by a 1 : 4 serial dilution and spreading 100ul on TSA plates using Drigalski spatulas. Plates were incubated at 37°C for approximately 24 h (except for isolate C that which was incubated for ~48 h due to slower growth).
The number of colonies on a plate was recorded for only those plates with 30–300 colonies as counts of <30 will be more influenced by small errors in dilution while counts of >300 will be difficult to count and contain colonies that are not fully separated. Our limit of quantification was therefore 30 colonies from the least diluted plate. A 10‐fold dilution series would not necessarily yield a plate in the countable range. We therefore used a 1 : 4 dilution series was as this was likely to yield one or two plates that were in the countable range, providing us with one or two measurements per swab. Cell concentration for each swab tip was calculated by multiplying the number of colonies by the dilution factor of each plate. Bacterial load represents the average cell concentration of each swab replicate in CFU/swab. We transformed bacterial load values to the natural log for posterior analyses.
Due to our exclusion criteria, those final swabs whose least diluted plate had less than 30 colonies, were not included in the analysis. Although the lack of quantifiable ending points deprives us of more data points, the inclusion of those low counts can be expected to contain significant error and thus could substantially affect the estimated rates. Moreover, those final plates with zero colonies would cause additional bias, as they do not represent the moment at which concentration intersected the X axis.
Statistical Analysis
Our goals for the statistical analysis were to determine the population decay rate of the isolates stored at different temperatures, and then to compare the decay rates across temperatures and isolates. To estimate exponential decay rates, we needed to conduct a linear regression of the natural log‐transformed bacterial load (CFU/swab) against time. In our analysis we also had to account for the following complexities: (i) each isolate had a unique starting concentration, and (ii) bacterial load per temperature per isolate was measured by one or more data point per swab duplicate. Therefore, we needed to allow for different intercepts between isolates and for nonindependent measurements from the same swabs.
We therefore constructed a linear mixed effects model, and chose to implement Bayesian inference using the packages rstan and rstanarm in the open‐source statistical software R, which interface with the Stan inference software (Goodrich et al. 2019; Stan Development Team 2019; R Core Team 2020). We chose a Bayesian framework due to the complexity of the model structure and because the Bayesian method allows us to better estimate the uncertainty in the group‐specific slopes and intercepts compared to frequentist methods (Goodrich et al. 2019). However, we emphasize that the choice to use the Bayesian approach does not change the statistical significance of the results.
The model implementation was of the form: log(Bacterial Load) ~ Time + (1|Isolate) + (1 | Swab.ID) + (0 + Time|Isolate:Temperature). This model thus allows for a random intercept that varies per Isolate and per Swab.ID. Then, the effect of time (i.e. the slope, the rate of decay) is allowed to vary per temperature, but the effect of temperature can also vary among isolates. The analysis results in posterior distributions of ‘random’ slopes that quantify the effect of each temperature on the decay rate of the bacteria, and these effects of temperature can vary across isolates. In the Stan model, we ran four independent chains for 2000 iterations each, discarding the first 1000 iterations as warm‐up. We used vague, default prior distributions for all model parameters.
Next, we compared whether the bacterial decay rates measurably differed across the temperature gradient. To do this, we conducted three post hoc comparisons of temperature groups, and averaged these effects across isolates. Specifically, we compared the decay rate of −70°C (we designate this as ‘group a’) to the average decay rate across −20°C through 20·5°C (group b); we compared −70 to 37°C (group c); and, finally, we compared the average decay rate across −20°C through 20·5°C to the decay rate at 37°C. This constituted three independent tests. To make these comparisons, we averaged the slope values at each posterior draw across the isolates, then calculated the differences in the slope estimates at each posterior draw. In other words, we calculated 4000 differences between each comparison group (a, b and c). Then, we considered the groups to be substantially different if the 95% credible interval of these differences did not include zero.
We report 95% credible interval for various statistics. These are calculated by randomly sampling from the posterior samples and then calculating the 95% quantile for these generated quantities. For instance, to get the 95% credible interval for the slope of isolate A at temperature 37°C, we have to randomly sample from the posterior of the global average slope, the random effect of isolate A and the random effect of temperature 37°C for isolate A.
Results
After treatment, almost all colonies showed typical morphological characteristics of S. aureus grown on TSA plates after 1 day of incubation: cream colour, circular, raised, with smooth margins and 1–5 mm in diameter. Occasionally, some colonies showed a more irregular shape, but reverted to typical morphologies when re‐streaked. Some colonies from swab samples stored in the refrigerator or freezer were particularly small, but reverted to typical size when re‐streaked. After any length of storage under any treatment temperature, isolate C grew slowly on TSA plates and yielded consistently small colonies. Increasing the incubation time for plates with isolate C to 48 h improved visualization of colonies without influencing colony number. Normal colony size was observed for this isolate after storage treatments when inoculated on CHROMAgar or re‐streaked on TSA from a fresh culture.
All five isolates survived best on swabs stored in the deep freezer at −70°C with almost no variation among isolates. We terminated this experiment at this temperature after 91 days for isolate A, 44 days for isolate B, 50 days for isolate C, 44 days for isolate D and 58 days for isolate E. At this temperature and for each endpoint, all concentrations for all isolates did not differ from the initial concentration as measured on day 0. For all other storage temperature conditions, we observed an exponential decrease in concentration over time. The last days that we were able to detect at least one colony from −20, 5, 20·5 and 37°C conditions are as follows: Isolate A: 21, 8, 7, 2 days; Isolate B: 17, 11, 11, 4 days; Isolate C: 15, 27, 10, 4 days; Isolate D: 17, 11, 11, 4 days; and Isolate E: 16, 13, 7, 0 days. These points were excluded from our analyses as they as they fell outside the quantifiable range of 30–300 cells.
At the highest temperature, we observed lowest survival rates. All five isolates showed the most rapid decline at 37°C (Fig. 1). This was especially pronounced for isolate E, which showed no growth at any time point, the first of which was at 24 h (Fig. 1). At 37°C, none of the other four isolates (A–D) were cultivable after day 4 (Fig. 1).
Figure 1.
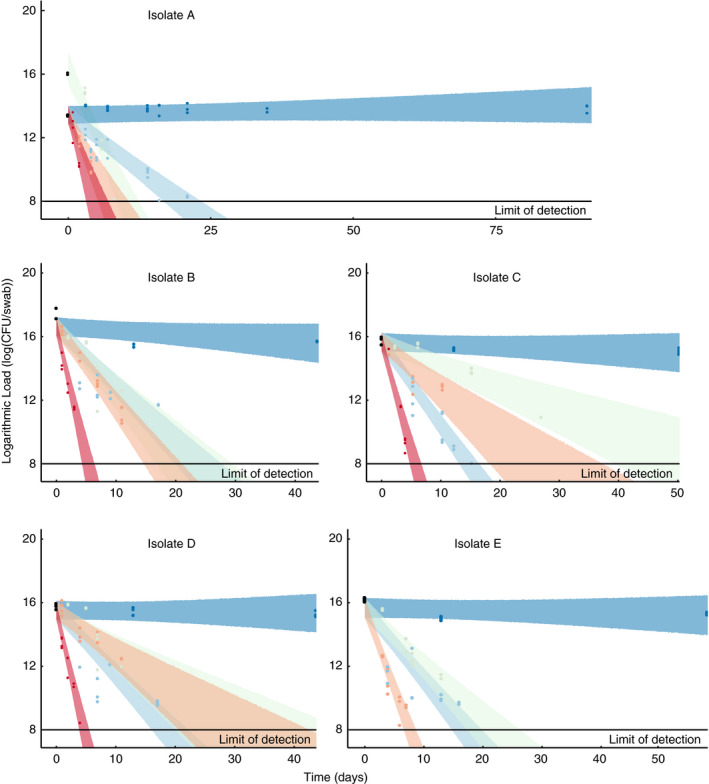
Concentration over time for five isolates on swabs stored at five different temperatures. Each panel plots the concentration dynamics of a different isolate (Table 1) at the five storage temperatures. Black points are the initial concentration recovered from swabs at time 0. All swabs across treatments were inoculated with the same stock and thus have the same initial concentration with the exception of isolate A at 5°C. The coloured ribbons show the 95% credible interval in the model predictions of bacterial concentration over time for each isolate and temperature treatment. Our statistical analyses allowed for us to vary the time points at which we measured survival. This was important as it was impossible to process all strains at the same time. This also, allowed us to assess the decay at −70°C over a longer time period. Temperature: ( ) −70°C, (
) −70°C, ( ) −20°C, (
) −20°C, ( ) 5°C, (
) 5°C, ( ) 20·5°C, (
) 20·5°C, ( ) 37°C.
) 37°C.
The statistical analysis revealed that the rate of bacterial decay increased with storage temperature (Fig. 2). Quantitatively this led to a more negative slope as temperature increased (i.e. a larger negative impact of temperature). We found that the −70°C storage temperature did not lead to measurable decay; the slope was not substantially different than zero (median (95% credible interval in the difference): −0·009 (−0·019, 0·002)). The −70°C storage condition (group a in Fig. 2) had a measurably lower decay rate compared to the average across −20, 5 and 20·5°C (group b in Fig. 2), where 95% credible interval of the difference did not include zero (median (95% CI): 0·46 (0·42, 0·49)); and separately, the −70°C group had lower decay compared to 37°C (group c in Fig. 2; median (95% CI): 1·48 (1·29, 1·67)). Furthermore, the mid‐range storage conditions of group b (−20 to 20·5°C) had a measurably lower decay rate compared to 37°C (median (95% CI) in the difference: 1·03 (0·83 1·21)). Qualitatively, we can see that isolates within the group b storage conditions (−20, 5 and 20·5°C) had similar decay rates; however, as temperature increased, the variation between isolates became more pronounced, and some isolates had substantially higher decay rates at 5 and 20·5°C (Fig. 2). Indeed, isolate E had the lowest survival rate at 20·5°C and was the only isolate that did not even survive for 24 h (the first time point) at 37°C (Fig. 2).
Figure 2.
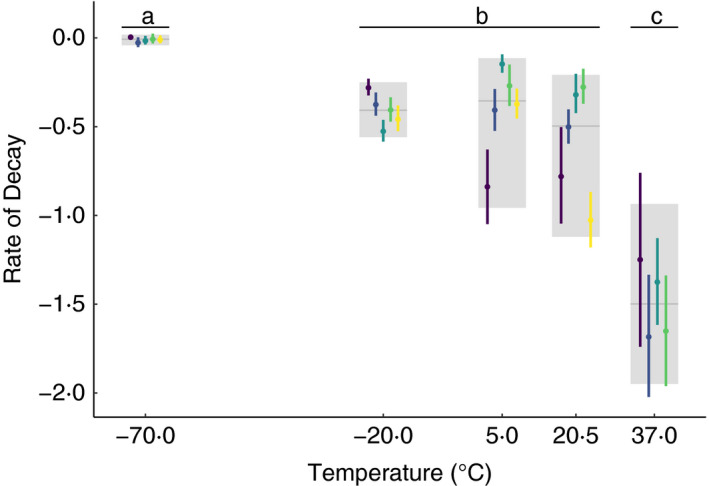
Rates of decay for each isolate at each storage temperature. The median (point) and 95% credible interval (error lines) of the decay rates (slope) for each temperature and isolate combination are shown in colour while the median and 95% credible interval of decay rate averaged across isolates for each temperature is shown in grey, as estimated by the random effects in the statistical model. Isolates: ( ) A, (
) A, ( ) B, (
) B, ( ) C, (
) C, ( ) D, (
) D, ( ) E.
) E.
Discussion
Survivability of S. aureus on a sampling substrate is a critical consideration for study design and for surveillance and downstream genomic analyses of S. aureus from community samples. In our study, we found that survival decays exponentially over time when samples are stored on swabs outside of a deep freezer, the rate of decay increased substantially at higher storage temperatures, and we provide empirical findings to quantify these dynamics. Samples exposed to 37°C experienced the lowest survival rate, while the best recoveries occurred after storage under the coldest condition (−70°C). In fact, at −70°C, there was no measurable decay in survival even after several months of storage without media. Intermediate temperatures (−20 to 20·5°C) showed moderate decay in survival over time although there was variation across isolates, with some isolates showing substantial differences in their decay rates at 5 and 20·5°C. Our results are broadly consistent with other studies that showed highest survival at the lowest temperatures (Tsvetkov and Shishkova 1982; Polo et al. 2017).
In some bacteria, low temperatures can induce cell damage due to ice crystal formation or other types of injuries which decrease the number of cultivable cells, however, a variety of stress strategies at different temperatures can prevent or reduce such damage (Wesche et al. 2009). Our results suggest that S. aureus can effectively survive storage at −70°C for long periods of time with no storage or transport media. At the other tested temperatures, we observed an exponential decay in survivability. S. aureus has been recovered at high doses even after 2 months at −20°C on substrates such as seafood (Saklani et al. 2020), possibly aided by cryoprotectant properties contained in many foods (Wesche et al. 2009) which were absent in our experiments.
Colonies from samples stored in the refrigerator (5°C) and especially freezer (−20°C) showed some variation in size and shape, that indicates that cold or freezing conditions could induce metabolic changes in S. aureus cells (Wesche et al. 2009; Onyango et al. 2012; Suo et al. 2018). As a result, although many cells could still be viable, not all would be immediately cultivable under the same incubation conditions (Watson et al. 1998), and this might explain the relatively low recovery at −20°C versus −70°C observed in our study. Our methods allowed us to quantify cultivable bacteria incubated for 24 h, but additional tests would be needed to estimate the total survival of viable cells.
Swabs stored at room temperature showed that S. aureus can be quantified with our method after 7 or 11 days, which is similar to the study of Neely and Maley (2000), who detected the presence of recoverable S. aureus cells after approximately 2 weeks for most of their isolates stored at room temperature on polyester (Neely and Maley 2000). However, in the Neely and Maley (2000) study, two of the six isolates survived for 40 and 56 days, illustrating the strain‐level variation of survival in response to temperature. Additionally, differences in results across studies may be influenced by different methods and differences in relative humidity for storage conditions. S. aureus can survive longer at lower relative humidity levels (Strasters and Winkler 1966; Kramer et al. 2006; Coughenour et al. 2011), but our samples contained 100 µl of PBS on each of two swabs that were enclosed in a single small tube. Survival may also be influenced by the load of bacteria on a surface (Watson et al. 1998; Warnke et al. 2014), where lower inocula were associated with lower survival rates (Watson et al. 1998; Warnke et al. 2014). We did not observe such a phenomenon, as the similarity of our initial concentrations across isolates precluded our ability to detect an influence of cell density on survival.
Observed cell densities of S. aureus in the nares of human carriers fluctuate over time (Burian et al. 2010; Szafrańska et al. 2019), and S. aureus abundance in the anterior nares is lower for women compared to men (Liu et al. 2015). Staphylococcus aureus can range from less than 104 to 107 16S copies per swab sample (Liu et al. 2015), which is approximately 1 order of magnitude less than the concentrations of our initial mixes. Our results suggest that a highly concentrated sample from a person left at room temperature for a week would still be detectable via culture. Conversely, a low‐concentration swab may yield false negative results after only a few days of storage. Our data therefore suggest that if sampling swabs cannot be stored at −70°C, cultivation efforts should commence as soon as possible, given the exponential decay. While we saw no statistically significant difference between average survival rates at −20, 5 and 20·5°C, the increased variation in survival rates among isolates was noticeable and argues for storage at colder temperatures to avoid bias in the detection of certain isolates. In general, our data suggest that transport medium is not necessary if samples are stored at −70°C or if culturing occurs within a day or two of collection and samples have not been kept at high temperatures.
Our methodology was intended to quantify the dynamics of microbial decay, but not to empirically assess the last day of survival of our isolates in our particular experiments, although this can be predicted from our statistical model. With this aim, we reduce counting and sampling errors by including only plates with 30–300 colonies. We further reduce errors surrounding our estimation by using a 1 : 4 dilutions which are more likely to yield one to two plates in the countable range per replicate. Finally, our use of paired swabs provides the potential of two replicates for each point. Despite these precautions, variation in our estimates precludes us from observing any statistically significant differences in survival rates between −20, 5 and 20°C. If such differences actually exist, additional swab replicates and plate replicates would further reduce variation in cell concentration estimations.
In summary, our results suggest that transport medium is not always necessary for field‐based sampling, but reinforce the need for appropriate storage or rapid processing of swab samples with S. aureus when stored in the absence of media. Samples stored at −70°C had almost no decay over time while S. aureus at 37°C had the lowest survival rates. We did not observe any statistically significant differences in survival rates between −20, 5 and 20°C. However, we observed more variation among isolates stored at higher temperatures.
Author contributions
DP and TP were involved in study conception. DP, JM, MM, SK, HH, EC and TP designed the experiments. DP, JM, RT, CH and TP provided data, materials and funding. DP, JM and TP analysed the data and drafted the paper. All authors were involved in critical revisions and approval of final manuscript.
Acknowledgements
This work was funded by NAU Southwest Health Equities Research Collaborative (NIH/NIMHD U54, Grant #NIH U54MD012388) and an NIH/NIAID grant (R15AI156771).
References
- van Belkum, A., Riewarts Eriksen, N.H., van Marly Sijmons, W., Leeuwen, M., van den Bergh, J., Kluytmans, F.E. and Verbrugh, H. (1997) Coagulase and protein A polymorphisms do not contribute to persistence of nasal colonisation by Staphylococcus aureus . J Med Microbiol 46, 222–232. [DOI] [PubMed] [Google Scholar]
- Burian, M., Wolz, C. and Goerke, C. (2010) Regulatory adaptation of Staphylococcus aureus during nasal colonization of humans. PLoS One 5, e10040. 10.1371/journal.pone.0010040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casarin, L.S., Tondo, E.C., Klein, M.P. and Brandelli, A. (2009) Survival of Escherichia coli, Staphylococcus aureus and Salmonella enteritidis in frozen chicken hamburger. J Muscle Foods 20(4), 478–488. [Google Scholar]
- Chambers, H.F. and DeLeo, F.R. (2009) Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol 7(9), 629–641. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughenour, C., Stevens, V. and Stetzenbach, L.D. (2011) An evaluation of methicillin‐resistant Staphylococcus aureus survival on five environmental surfaces. Microb Drug Resist 17, 457–461. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control (2019) Surveillance of antimicrobial resistance in Europe 2018. ECDC Stockholm.
- Fitzgerald, J.R., Sturdevant, D.E., Mackie, S.M., Gill, S.R. and Musser, J.M. (2001) Evolutionary genomics of Staphylococcus aureus: insights into the origin of methicillin‐resistant strains and the toxic shock syndrome epidemic. Proceedings of the National Academy of Sciences of the USA 98, 8821–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich, B., Gabry, J., Ali, I. and Brilleman, S. (2019) Rstanarm: Bayesian applied regression modeling via Stan. R Package Version 2. https://mc‐stan.org/rstanarm
- Grøntvedt, C.A., Elstrøm, P., Stegger, M., Skov, R.L., Andersen, P.S., Larssen, K.W., Urdahl, A.M.et al. (2016) Methicillin‐resistant Staphylococcus aureus CC398 in humans and pigs in Norway: a ‘One Health’ perspective on introduction and transmission. Clin Infect Dis 63, 1431–1438. 10.1093/cid/ciw552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kluytmans, J., van Belkum, A. and Verbrugh, H. (1997) Nasal carriage of Staphylococcus aureus: epidemiology, underlying mechanisms, and associated risks. Clin Microbiol Rev 10, 505–520.http://cmr.asm.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis, A.P., Hatfield, K., Baggs, J., Mu, Y., See, I., Epson, E., Nadle, J., Kainer, M.et al. (2019) Vital signs: Epidemiology and recent trends in methicillin‐resistant and in methicillin‐susceptible Staphylococcus aureus bloodstream infections‐United States. https://www.cdc.gov/mmwr [DOI] [PMC free article] [PubMed]
- Kramer, A., Schwebke, I. and Kampf, G. (2006) How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect Dis 6, 130. 10.1186/1471-2334-6-130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.M., Price, L.B., Hungate, B.A., Abraham, A.G., Larsen, L.A., Christensen, K., Stegger, M., Skov, R.et al. (2015) Staphylococcus aureus and the ecology of the nasal microbiome. Sci Adv 1(5), e1400216. 10.1126/sciadv.1400216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morosini, M.‐I., Loza, E., Gutiérrez, O., Almaraz, F., Baquero, F. and Cantón, R. (2006) Evaluation of 4 swab transport systems for the recovery of ATCC and clinical strains with characterized resistance mechanisms. Diagn Microbiol Infect Dis 56, 19–24. 10.1016/j.diagmicrobio.2006.02.011 [DOI] [PubMed] [Google Scholar]
- Neely, A.N. and Maley, M.P. (2000) Survival of Enterococci and Staphylococci on hospital fabrics and plastic. J Clin Microbiol 38, 724–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noyce, J.O., Michels, H. and Keevil, C.W. (2006) Potential use of copper surfaces to reduce survival of epidemic meticillin‐resistant Staphylococcus aureus in the healthcare environment. J Hosp Infect 63, 289–297. [DOI] [PubMed] [Google Scholar]
- Onyango, L.A., Hugh Dunstan, R., Gottfries, J., von Eiff, C. and Roberts, T.K. (2012) Effect of low temperature on growth and ultra‐structure of Staphylococcus Spp. PLoS One 7(1), e29031. 10.1371/journal.pone.0029031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, T., Barger, S.D., Lininger, M., Wayment, H., Hepp, C., Villa, F., Tucker‐Morgan, K.et al. (2019) Health disparities in Staphylococcus aureus transmission and carriage in a border region of the United States based on cultural differences in social relationships: protocol for a survey study. JMIR Res Protocols 8, e14853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo, L., Mañes‐Lázaro, R., Olmeda, I., Cruz‐Pio, L.E., Medina, S.F. and Pardo, I. (2017) Influence of freezing temperatures prior to freeze‐drying on viability of yeasts and lactic acid bacteria isolated from wine. J Appl Microbiol 122, 1603–1614. 10.1111/jam.13465 [DOI] [PubMed] [Google Scholar]
- Price, L.B., Stegger, M., Hasman, H., Aziz, M., Larsen, J., Andersen, P.S., Pearson, T. and Waters, A.E. et al. (2012) Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. MBio 3, e00305. 10.1128/mBio.00305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2020) R: a language and environment for statistical computing. Vienna, Austria: Foundation for Statistical Computing. https://www.R‐project.org/ [Google Scholar]
- Rishmawi, N., Ghneim, R., Kattan, R., Ghneim, R., Zoughbi, M., Abu‐Diab, A., Turkuman, S.et al. (2007) Survival of fastidious and nonfastidious aerobic bacteria in three bacterial transport swab systems. J Clin Microbiol 45, 1278–1283. 10.1128/JCM.02110-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, G.L., Harris, A.D., Morgan, D.J., Pineles, L., Belton, B.M., Kristie, J. and Johnson, J.K.(2012) Survival of methicillin‐resistant Staphylococcus aureus and vancomycin‐resistant Enterococcus Spp. for an extended period of transport. J Clin Microbiol 50, 2466–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saegeman, V., Flamaing, J., Muller, J., Peetermans, W.E., Stuyck, J. and Verhaegen, J. (2011) Clinical evaluation of the copan ESwab for methicillin‐resistant Staphylococcus aureus detection and culture of wounds. Eur J Clin Microbiol Infect Dis 30, 943–949. 10.1007/s10096-011-1178-1 [DOI] [PubMed] [Google Scholar]
- Saklani, P., Lekshmi, M., Nayak, B.B. and Kumar, S. (2020) Survival of methicillin‐resistant Staphylococcus aureus in fish and shrimp under different storage conditions. J Food Prot 83, 844–848. 10.4315/JFP-19-546 [DOI] [PubMed] [Google Scholar]
- Schleifer, K.‐H. and Bell, J.A. (2015) Staphylococcus. In Bergey’s Manual of Systematics of Archaea and Bacteria ed. William, W. pp. 1–43. Chichester, UK: John Wiley and Sons. Ltd. 10.1002/9781118960608.gbm00569 [DOI] [Google Scholar]
- Scott, E. and Bloomfield, S.F. (1990) The survival and transfer of microbial contamination via cloths, hands and utensils. J Appl Bacteriol 68, 271–278. [DOI] [PubMed] [Google Scholar]
- Stan Development Team (2019) RStan: The R Interface to Stan. R Package Version 2.19.1. https://mc‐stan.org/
- Strasters, K.C. and Winkler, K.C. (1966) Viability of hospital Staphylococci in air. Bacteriological Reviews 30, 674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suo, B., Yang, H., Wang, Y., Lv, H., Li, Z., Xu, C. and Ai, Z. (2018) Comparative proteomic and morphological change analyses of Staphylococcus aureus during resuscitation from prolonged freezing. Front Microbiol 9. 10.3389/fmicb.2018.00866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szafrańska, A.K., Junker, V., Steglich, M. and Nübel, U. (2019) Rapid cell division of Staphylococcus aureus during colonization of the human nose. BMC Genom 20. 10.1186/s12864-019-5604-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, Y., Ishino, G., Matsuba, T., Takayama, H. and Ishida, S. (1999) Survival of bacteria at a subfreezing temperature (‐1°C). Yonago Acta Med 42, 147–152. [Google Scholar]
- Tsvetkov, T. and Shishkova, I. (1982) Studies on the effects of low temperatures on lactic acid bacteria. Cryobiology 19, 211–214. [DOI] [PubMed] [Google Scholar]
- Van Horn, K.G., Audette, C.D., Sebeck, D. and Tucker, K.A. (2008) Comparison of the Copan ESwab system with two amies agar swab transport systems for maintenance of microorganism viability. J Clin Microbiol 46, 1655–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBergh, M.F.Q., Yzerman, EdPF, van Belkum, A., Boelens, H.A.M., Sijmons, M. and Verbrugh, H.A. (1999) Follow‐up of Staphylococcus aureus nasal carriage after 8 years: Redefining the persistent carrier state. J Clin Microbiol 37, 3133–3140. 10.1128/JCM.37.10.3133-3140.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenvoort, J.H.T., Sluijsmans, W. and Penders, R.J.R. (2000) Better environmental survival of outbreak vs. sporadic MRSA isolates. J Hosp Infect 45, 231–234. [DOI] [PubMed] [Google Scholar]
- Warnke, P., Frickmann, H., Ottl, P. and Podbielski, A. (2014) Nasal screening for MRSA: different swabs – different results! PLoS One 9, 10.1371/journal.pone.0111627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson, S.P., Clements, M.O. and Foster, S.J. (1998) Characterization of the starvation‐survival response of Staphylococcus aureus . J Bacteriol 180, 1750–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertheim, H.F.L., Melles, D.C., Vos, M.C., van Leeuwen, W., van Belkum, A., Verbrugh, H.A. and Nouwen, J.L. (2005) The Role of nasal carriage in Staphylococcus aureus Infections. Lancet Infect Dis 5, 751–762. [DOI] [PubMed] [Google Scholar]
- Wesche, A.M., Gurtler, J.B., Marks, B.P. and Ryser, E.T. (2009) Stress, sublethal injury, resuscitation, and virulence of bacterial foodborne pathogens. J Food Prot 72(5), 1121–1138. [DOI] [PubMed] [Google Scholar]
- Williams, R.E.O. (1963) Healthy carriage of Staphylococcus aureus: its prevalence and importance. Bacteriol Rev 27, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetola, N., Francis, J.S., Nuermberger, E.L. and Bishai, W.R. (2005) Community‐Acquired Meticillin‐Resistant Staphylococcus aureus: an emerging threat. Lancet Infect Dis 5, 275–286. [DOI] [PubMed] [Google Scholar]


