Abstract
Functional connectivity (FC) and resting-state network (RSN) analyses using functional magnetic resonance imaging (fMRI) have evolved into a growing field of research and have provided useful biomarkers for the assessment of brain function in neurological disorders. However, the underlying mechanisms of the blood oxygen level-dependant (BOLD) signal are not fully resolved due to its inherent complexity. In contrast, [18F]fluorodeoxyglucose positron emission tomography ([18F]FDG-PET) has been shown to provide a more direct measure of local synaptic activity and may have additional value for the readout and interpretation of brain connectivity. We performed an RSN analysis from simultaneously acquired PET/fMRI data on a single-subject level to directly compare fMRI and [18F]FDG-PET-derived networks during the resting state. Simultaneous [18F]FDG-PET/fMRI scans were performed in 30 rats. Pairwise correlation analysis, as well as independent component analysis (ICA), were used to compare the readouts of both methods. We identified three RSNs with a high degree of similarity between PET and fMRI-derived readouts: the default-mode-like network (DMN), the basal ganglia network and the cerebellar-midbrain network. Overall, [18F]FDG connectivity indicated increased integration between different, often distant, brain areas compared to the results indicated by the more segregated fMRI-derived FC. Additionally, several networks exclusive to either modality were observed using ICA. These networks included mainly bilateral cortical networks of a limited spatial extent for fMRI and more spatially widespread networks for [18F]FDG-PET, often involving several subcortical areas.
This is the first study using simultaneous PET/fMRI to report RSNs subject-wise from dynamic [18F]FDG tracer delivery and BOLD fluctuations with both independent component analysis (ICA) and pairwise correlation analysis in small animals. Our findings support previous studies, which show a close link between local synaptic glucose consumption and BOLD-fMRI-derived FC. However, several brain regions were exclusively attributed to either [18F]FDG or BOLD-derived networks underlining the complementarity of this hybrid imaging approach, which may contribute to the understanding of brain functional organization and could be of interest for future clinical applications.
Keywords: Resting-state networks, Functional connectivity, Dynamic [18F]FDG correlations, Connectomics, simultaneous PET/MRI
1. Introduction
Although the understanding of brain function has taken major leaps over the past several decades due to the development of novel imaging and computational modelling techniques, functional brain organization and its dysfunctions continue to be incompletely understood. Functional connectivity (FC) describes synchronous oscillations of brain activity in different parts of the brain and can be used to study its functional organization. Ogawa et al. introduced functional magnetic resonance imaging (fMRI), making use of the blood oxygenation level dependant (BOLD) signal to derive neural activity (Ogawa et al., 1990). This technique opened the possibility of acquiring FC using MRI (Biswal et al., 1995). FMRI-derived FC studies focus on characterizing functional resting-state networks (RSN) through temporal correlations of the BOLD signal between different brain regions (Fox et al., 2005; Greicius et al., 2003).
The default-mode network (DMN) is the most widely reported resting-state network (Raichle et al., 2001). The DMN is composed of a number of medial and lateral cortical regions characterized by decreased connectivity during tasks compared to the resting state. Hence, the DMN appears to be related to a certain default state of the brain during rest. The DMN is believed to play an essential role in brain organization, recollection, self-awareness and imagination (Raichle et al., 2001). The function of the DMN has been shown to be disrupted in several clinical populations, including Alzheimer’s disease and schizophrenia (Badhwar et al., 2017; Bluhm et al., 2007; Greicius et al., 2004; Mwansisya et al., 2017). In addition to the DMN, several other RSNs have been reported to involve other cortical areas (Damoiseaux et al., 2006). Additionally, RSNs have been identified in other brain regions, including the cerebellum and brainstem and subcortical areas such as the basal ganglia (Joel and Weiner, 1994). These networks encompass a variety of different structures and are therefore thought to be involved in a number of integrative processes, including perception, memory, attention, seizure suppression, executive control and emotion (Becerra et al., 2011; Cole et al., 2010). Due to their manifold roles, such networks are of interest for various clinical and research applications, such as diagnostics or drug development.
Although networks such as the DMN were initially assumed to be exclusive to the human brain, analogues of human RSNs have been observed in other species, including rats (Hutchison et al., 2010; Lu et al., 2012; Majeed et al., 2011; Zhang et al., 2010). The interpretation of the significance of the RSN continues to be a topic of scientific interest. However, it is widely established that numerous such networks are not only reproducible across species, individuals and ages but can also play a role as biomarkers, as they are disrupted in neurological disorders or by certain drugs (Barkhof et al., 2014). The field of research investigating disruptions of whole-brain or network-level FC has profited from the emergence of connectomics, which focus on defining the properties of the brain as a network (Bullmore and Sporns, 2009; Sporns et al., 2005). The framework of connectomics includes measures defining the functional organization of the brain to enable the analysis and detection of alterations to brain functionality (Sporns, 2010).
Although better established through BOLD-fMRI studies, the concept of functional connectivity between different brain regions first emerged using [18F]fluorodeoxyglucose positron emission tomography ([18]FDG-PET). Horwitz et al. showed relationships between the metabolism of different brain regions by computing correlations between regional subject series derived from static [18F]FDG-PET scans (Horwitz et al., 1984). Compared to the BOLD signal, which is driven by a still incompletely understood convolution of cerebral blood flow (CBF), cerebral blood volume (CBV) and cerebral metabolic rate of oxygen (CMRO2) (Buxton, 2012), [18F]FDG-PET may represent a more direct reflection of neural activity (Hahn et al., 2016; Lanzenberger et al., 2012). By operating on a different neurophysiological level, [18F]FDG correlations could potentially provide complementary information to BOLD-derived FC and thereby further help decipher the substrate of brain connectivity.
More recent studies focusing on [18F]FDG correlations have derived networks similar to those revealed by BOLD-fMRI (Di and Biswal, 2012). Upon further investigation by other groups, these networks acquired using [18F]FDG-PET have been shown to exhibit comparable organizational properties to BOLD-derived RSNs (Sanabria-Diaz et al., 2013; Seo et al., 2013b). Importantly, Savio and colleagues described several similar networks from simultaneously acquired FC and [18F]FDG-PET-derived connectivity in humans, suggesting the presence of a common substrate between both outputs (Savio et al., 2017). Recently, the same group has applied a similar methodology to investigate the effects of Alzheimer’s Disease on both metabolic and hemodynamic RSNs (Ripp et al., 2020). The authors showed that connectivity acquired using [18F]FDG-PET not only depicted partly complementary aspects of RSNs, but also exhibited increased specificity for Alzheimer’s Disease compared to FC derived from simultaneously acquired BOLD-fMRI, thereby emphasizing the value of [18F]FDG correlation analysis for diagnostics of neurodegenerative diseases.
All the studies mentioned above derived [18F]FDG correlations at inter-subject level from static PET scans. To enable subject-level computation of connectivity from [18F]FDG scans, dynamic PET acquisitions are employed to compute correlations between dynamic regional [18F]FDG tracer delivery fluctuations. Studies computing correlations between regional [18F]FDG dynamic PET time-courses are however few and the majority did not acquire PET simultaneously to BOLD-fMRI (Passow et al., 2015a; Tomasi et al., 2017). To the best of our knowledge, only two studies have reported the simultaneous acquisition of dynamic PET scans and fMRI to investigate correlations of dynamic [18F]FDG tracer delivery and BOLD fluctuations (Amend et al., 2019; Wehrl et al., 2013). These studies indicated a significant amount of complementary data by the two methods for both rest and task conditions.
The aim of the present study was to compare the resting-state outputs of dynamic [18F]FDG-PET and BOLD-fMRI by analysing different RSNs derived from [18F]FDG correlations and FC at the single-subject level using both pairwise correlation analysis and independent component analysis (ICA). This study builds on our prior work focused on analysing and comparing the effects indicated by BOLD-fMRI and [18F]FDG-PET following brain stimulation (Wehrl et al., 2013) and on defining the dynamics of tracer delivery driving [18F]FDG correlations (Amend et al., 2019) by elucidating the complementarity of the [18F]FDG correlations and FC readouts on a physiological level. We hypothesized that the common substrate of both outputs will enable the observation of similar organizational properties; however, we also expect a complementary input to be provided by [18F]FDG correlations. To the best of our knowledge, this is the first study investigating RSNs of both FC and subject-wise [18F]FDG correlations using simultaneous PET/MRI and employing both independent component analysis (ICA) and pairwise correlation analysis in small animals.
2. Materials and methods
2.1. Animals
Healthy male Lewis rats (n = 30) were ordered from Charles Rivers Laboratories (Sulzfeld, Germany). The animals used in the experiments had bodyweights of 292.2 ± 22.1 g, corresponding to an age of approximately 11 weeks. Rats were kept on a 12 h day-night cycle at a room temperature of 22 °C and 40–60% humidity. The rats received a standard diet and tap water ad libitum before and during the experimental period. All animals were fasted for 6 h prior to the scans. A separate cohort of three rats was used for determining physiological parameters over the course of the scans using pulse oximetry (please refer to Supplementary Information for details). All experiments were performed in accordance with the German federal regulations regarding the use and care of experimental animals and under approval by the local research administration.
Fifteen of the datasets were used in our previous study on the methodology of acquiring [18F]FDG and BOLD-fMRI resting-state functional connectivity using simultaneous acquisitions in rats (Amend et al., 2019).
2.2. Simultaneous PET/MRI experiments
The rats were placed into a knock-out box and 3% isoflurane in regular air was delivered to induce anaesthesia. The isoflurane concentration was then reduced to 2% for all subsequent preparation steps. First, the baseline capillary blood glucose levels were measured by puncturing the tail vein and the weights of the animals were examined. Subsequently, an intravenous catheter was placed into a tail vein using a 30 G needle for [18F]FDG administration. Finally, the animals were positioned onto a dedicated small-animal bed and introduced inside a PET/MRI scanner. The temperature was monitored and maintained at 36.5 °C using a rectal temperature probe and a feedback-controlled system, and the respiration rate was observed during the experiments. For the resting-state measurements, the isoflurane concentration was reduced to 1.3% in regular air over the course of the scan. At the end of each scan, the capillary blood glucose concentrations were re-evaluated. For additional data on the physiology of the animals under these experimental conditions, including heart rate, breathing rate and blood oxygenation, please refer to Supplementary Information.
Simultaneous PET/MRI measurements were performed over the course of 60 min. Magnetic resonance imaging was performed using a small-animal 7 T ClinScan system (Bruker BioSpin MRI, Ettlingen, Germany). A linearly polarized RF coil (Bruker) with a diameter of 72 cm was employed for transmission and a four-channel rat brain coil (Bruker) was used for reception. First, localizer scans were acquired to adjust the position of the rat brains to the centre of the PET/MRI field of view (FoV). Before performing further sequences, local field maps were acquired to optimize the local magnetic field homogeneity. Subsequently, T2 sequences were acquired as anatomical references (TR: 1800 ms, TE: 67.11 ms, 0.25 mm3 isotropic resolution, FoV: 40 × 32 × 32 mm, image size: 160 × 128 × 128 px, Rare factor: 28, averages: 1). Functional MRI scans were performed using a T2 * -weighted gradient echo EPI sequence (TE: 18 ms, TR: 2500 ms, 0.27 mm isotropic resolution, FoV 25 × 23 mm, image size: 92 × 85 × 20 px, slice thickness: 0.8 mm, 20 slices, 0.2 mm separation between slices). The fMRI scans were performed over the same 60 min time interval as the PET scans described below.
[18F]FDG-PET acquisitions were performed using a small-animal PET insert developed in cooperation with Bruker, a second generation with similar performance parameters as our in-house developed PET insert (Wehrl et al., 2013). Additionally, 31.6 ± 0.9 MBq of [18F]FDG diluted in NaCl was administered over a period of 30 s as a bolus shortly after the start of the PET/MRI acquisition. PET data were stored as list-mode files and reconstructed in 60 1-minute frames employing an in-house written ordered-subsets expectation maximization 2 (OSEM-2D) algorithm including random, decay, time-delay corrections and normalization. For a detailed description of [18F]FDG production, see the Supplementary Information.
The original raw dataset will be made available on Mendeley.
2.3. Data processing and analysis
Both the PET scans and the anatomical and functional MRI data were first realigned using Statistical Parametric Mapping 12 (SPM 12, Wellcome Trust Centre for Neuroimaging, University College London, London, United Kingdom) and checked for motion. The motion parameters were stored for fMRI denoising. Mean images of each data set were created and used to generate binary brain masks for the fMRI and PET scans using Analysis of Functional NeuroImages (AFNI, National Institute of Mental Health (NIMH), Bethesda, Maryland, USA). Additionally, brain masks were generated for the T2-weighted anatomical MRI scans. The respective masks were used to remove outer-cerebral tissues from all the PET, fMRI and T2-weighted anatomical MRI datasets. The remaining pre-processing steps were performed using SPM 12. First, all the realigned and skull-stripped fMRI and PET datasets were co-registered to their respective anatomical reference scans. Subsequently, using the anatomical images as a reference, all the datasets were normalized to the Schiffer rat brain atlas (Schiffer et al., 2006). Motion regression was applied to the normalized datasets using an in-house written script by regressing out the motion parameters acquired during the realignment step at the beginning of the pre-processing pipeline. Additionally, fMRI signal drifts were removed voxel-wise using linear and quadratic fits. Following motion regression, both the fMRI and PET datasets were smoothed using a 1.5 × 1.5 × 1.5 mm3 full-width-half-maximum (FWHM) Gaussian kernel as described previously (Wehrl et al., 2013), which is equivalent to the physical spatial resolution of the PET system used.
Fifty-two regions of the Schiffer rat brain atlas were selected for region of interest (ROI)-based analysis (Schiffer et al., 2006) (for a list of regions of the Schiffer rat brain atlas, see the Supplementary Methods). The fMRI and [18F]FDG-PET time courses from these ROIs were extracted using the SPM toolbox Marseille Boîte À Région d’Intérêt (MarsBaR) (Matthew Brett, June 2–6, 2002). Prior to analysis, the extracted time courses underwent whole-brain normalization to account for global effects. Therefore, the values of each ROI at every time-point were divided by the whole-brain mean of the signal at the respective time-points. For each dataset, Pearson’s r correlation coefficients were computed between the normalized time courses of each pair of regions, resulting in correlation matrices of 52 × 52 entries for each dataset. Since the 52 self-correlations were set to zero, 2652 correlation coefficients were computed for each individual scan. Due to the findings in our previous study (Amend et al., 2019) on the inability to ensure temporal stability of the [18F]FDG correlation readout when applying a bolus protocol and the lack of a gold standard to determine the most suited period for the analysis, correlations were performed over the course of the entire scan to avoid potential bias by choosing a specific period.
For group-level analysis, Pearson’s r values were transformed into Fischer’s z-scores, and mean correlation matrices were generated for both fMRI and PET datasets (for a detailed description, please refer to the Supplementary Methods). Additionally, a second-level analysis was performed to test every edge for significance (one-sample t-test, p ≤ 0.05, corrected for multiple comparisons). All multiple comparison corrections presented in the manuscript were performed using Bonferroni-Holm.
To investigate the whole-brain organization of FC and [18F]FDG connectivity, three network measures were applied to the PET and fMRI-derived correlation matrices of each subject using the Brain Connectivity Toolbox (Rubinov and Sporns, 2010). First, for each individual correlation matrix, 500 randomized matrices with the same average degree were generated by rewiring each of the edges of the respective matrix. Ideal modularity (Reichardt and Bornholdt, 2006) and clustering coefficients (Onnela et al., 2005) were then computed for both the individual correlation matrices and all randomized matrices. To derive values representative of a null distribution the network metrics of the random matrices were averaged over the 500 iterations. Finally, small-world coefficients were derived for each matrix using the averaged randomized network metrics (for detailed descriptions of these methods, see the Supplementary Methods). As described previously, networks were defined as exhibiting small-world properties when the small-world coefficients were higher than 1 (Humphries and Gurney, 2008). To define the nodes and edges of the generated correlation matrices for further network analysis, a threshold was applied to achieve a sparsity value of 20%. Di et al. investigated metabolic connectivity matrices using thresholds between 4% and 18% sparsity (Di et al., 2017). For this application, a slightly more liberal threshold was chosen to avoid missing relevant edges since the data were acquired from anaesthetized animals, as opposed to humans in the mentioned study. Elsewhere, thresholds up to 40% sparsity have been reported for metabolic connectivity studies (Seo et al., 2013a).
To assess the similarity between group-averaged [18F]FDG correlations and FC readouts on whole-brain and RSN levels the correlation coefficients between the respective outputs were computed (Di et al., 2017). Therefore the Pearson’s correlation coefficient between the two sets of connectivity values was employed. The same calculations were also repeated for [18F]FDG correlations and FC readouts of each subject at whole-brain level, as well as and for the DMN, basal ganglia network and cerebellar-midbrain network. To report group-level mean correlation values between the [18F]FDG connectivity and FC of single subjects, the correlation coefficients were transformed to Fischer’s z-scores before averaging over all subjects.
Independent component analysis (ICA) was performed using Group ICA of the fMRI Toolbox (GIFT, MIALAB, University of New Mexico, Albuquerque, NM, USA). As described previously for small-animal datasets, 20 components were chosen for the ICA algorithm (Wehrl et al., 2013). The maps of the generated components were thresholded (z ≥ 1.96, corresponding to p ≤ 0.05) (Di et al., 2012). Voxel-wise Dice-Sörensen coefficients (DSCs) were computed between the components generated from the fMRI and PET datasets. Furthermore, DSCs were computed between each component and the 52 regions under consideration from the Schiffer brain atlas to determine the voxel-wise percentage contribution of each region to the respective components. By combining these measures with a visual inspection, similar components of the fMRI and PET datasets, as well as components exclusive to either one of the two datasets, were determined and quantified.
For the defined networks common to both BOLD-fMRI and [18F]FDG-PET, specific sets of nodes were isolated based on previous rat FC studies describing the regions contributing to each of these RSNs (Becerra et al., 2011). Using these nodes, unthresholded and thresholded (20%) mean correlation matrices were generated. The similarity of these RSNs for [18F]FDG and BOLD-fMRI using ROI-based methods was visualized through scatter plots and quantified using Pearson’s r. Additionally, common edges between FC and [18F]FDG correlations were derived from the thresholded matrices and illustrated using the BrainNet Viewer toolbox (Xia et al., 2013). For information on the regions included in each network, please refer to the Supplementary Methods. For the thresholded correlation matrices of each RSN, please refer to the Supplementary Results.
3. Results
3.1. Whole-brain connectivity assessment
First, the properties of the whole-brain [18F]FDG correlations and FC were assessed (Fig. 1). The analysis of an exemplary single subject is first depicted (Fig. 1A–F). The [18F]FDG and BOLD-fMRI correlation matrices were compared by computing their similarity using Pearson’s r. Additionally, clustering, modularity and small-worldness were quantified to derive the network properties of the investigated matrices. In the second part of the figure (Fig. 1G–L), the same analysis measures on the group level are shown.
Fig. 1.
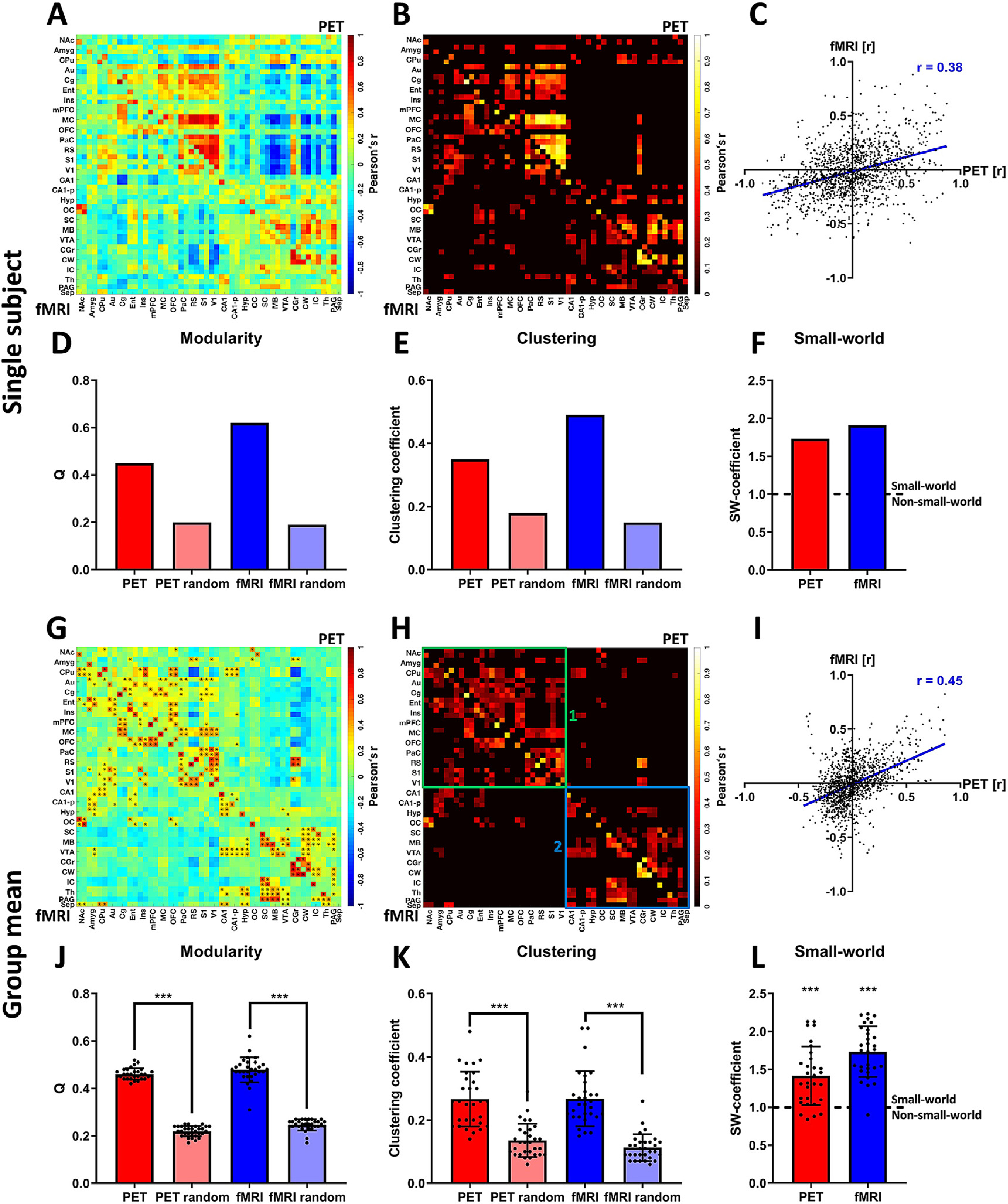
Assessment of whole-brain [18F]FDG correlations and FC. Panels (A)-(F) depict an exemplary analysis at the single-subject level: (A) PET/fMRI correlation matrix indicating [18F]FDG-PET-derived correlations (upper half of the matrix above the diagonal) and fMRI-derived FC (lower half of the matrix below the diagonal). (B) Thresholded PET/fMRI correlation matrix (20% sparsity) indicating similar areas of [18F]FDG connectivity and BOLD FC. (C) Scatter plot of all [18F]FDG correlations and FC values to assess the correlation of both outputs on the whole-brain level. (D) Ideal modularities of [18F]FDG connectivity and FC compared to the ideal modularity of respective randomized networks with the same average degrees. (E) Clustering coefficients of [18F]FDG correlation matrices and FC compared to the clustering coefficients of respective randomized matrices with the same average degrees. (F) Small-world coefficients of [18F]FDG connectivity and FC. The dotted line indicates the threshold (SW ≥ 1) for which networks are considered to have small-world properties. Panels (G)-(L) depict the analysis at the group-mean level: (G) Mean correlation matrix revealing similar patterns between [18F]FDG correlations (above diagonal) and FC (below diagonal). ∗ indicate significant correlations (p ≤ 0.05, corrected for multiple comparisons using Bonferroni-Holm). (H) Mean positive correlation matrix thresholded to a sparsity of 20% to define similar clusters between [18F]FDG correlations and FC. (1) Cluster indicating common [18F]FDG connectivity and FC in the cortex and in the anterior subcortical regions. (2) Cluster indicating common subcortical, midbrain and cerebellar [18F]FDG connectivity and FC. (I) Scatter plot of [18F]FDG connectivity and FC along with Pearson’s r coefficient to assess the correlation of both outputs. (J) Mean ideal modularity of [18F]FDG correlations and FC of every subject compared to respective randomized networks with the same average degree (paired-t-tests). (K) Mean clustering coefficients of [18F]FDG connectivity and FC of every subject compared to the respective randomized networks with the same average degree (paired-t-tests). (L) Small-world coefficients of [18F]FDG connectivity and FC of every subject showing small-world properties for the majority of subjects for both outputs. Both group means were significantly higher than 1 (p ≤ 0.001, one-sample t-tests). (*** represents a confidence interval of p ≤ 0.001; dotted line in (L) indicates threshold for which networks are considered to have a small-world organization (SW-coefficient ≥ 1) or not to exhibit such organization (SW-coefficient < 1), while *** represents a confidence of p ≤ 0.001 for the respective dataset being significantly higher than 1). FC = fMRI-derived functional connectivity, SW = small-world. For a list of abbreviations of all regions, please refer to Supplementary Table 1.
The whole-brain correlation matrix of the exemplary single-subject analysis (Fig. 1A) indicates its PET-derived interregional [18F]FDG correlations (upper half above the diagonal) and its fMRI-derived FC (lower half below the diagonal). This matrix was thresholded at 20% (Fig. 1B) to depict the common areas of [18F]FDG correlations and FC. The correlation between [18F]FDG connectivity and FC was also calculated and is shown as a scatter plot (Fig. 1C). For this exemplary subject, the correlation between the [18F]FDG and BOLD-fMRI readouts on the whole-brain level was r = 0.38 (p ≤ 0.001), and thus, it was highly significant (p ≤ 0.001). In addition, graph theory measures, including modularity (Fig. 1D), clustering (Fig. 1E) and small-worldness (Fig. 1F), were calculated for the exemplary subject, as well as for all other subjects. For this subject, both [18F]FDG correlations (small-world coefficient = 1.73) and FC (small-world coefficient = 1.91) exhibited small-world properties.
The whole-brain connectivity matrix (Fig. 1G) reveals the connectivity patterns of [18F]FDG-PET (above diagonal) and BOLD-fMRI (below diagonal) data on the group-mean level. All correlations were tested on group level for significance using multiple comparison correction (p ≤ 0.05, Bonferroni-Holm) and significant edges are indicated by asterisks. For an additional comparison between [18F]FDG correlations and FC involving only significant edges, please refer to Supplementary Figure 9. Here, a sparsity threshold of 20% was applied across all correlations to further compare the distribution of the strongest edges between [18F]FDG connectivity and FC (Fig. 1H). A number of similarities could be assessed visually from the whole-brain connectivity matrices. First, two large clusters with high densities of connections could be observed for both [18F]FDG connectivity and FC. The first cluster (indicated by green box (1) in Fig. 1I) was mainly composed of cortical regions, such as the auditory (Au), cingulate (Cg), entorhinal (Ent), motor (M1), orbitofrontal (OFC), parietal (PaC), retrosplenial (RS), primary sensory (S1) or visual (V1) cortices. Other regions included in this cluster were anterior subcortical areas, such as the nucleus accumbens (NAc), amygdala (Amyg) or caudate putamen (CPu). The remaining regions in the subcortical and posterior brain areas, including the superior and inferior colliculi (SC and IC, respectively), the midbrain (MB), the cerebellum white and grey matter (CW and CG), the thalamus (Th), the hypothalamus (Hyp), the periaquaeductal grey (PAG) and the septum (Sep), formed a separate cluster (indicated by the blue box (2) in Fig. 1H) with sparse connections to the anterior cluster mentioned above. The anterodorsal and posterior hippocampus (CA1 and CA1-p) exhibited connectivity to both clusters in the FC matrix; however, they were much less connected in the [18F]FDG readout. Observing the two separate clusters in more detail, further smaller clusters of dense and intense correlations could be observed for both outputs, including one composed of posterior cortical regions (RS, PaC, S1 and V1) and another of the cerebellum (CG and CW).
Quantitatively, the observed similarities translated into a Pearson’s r coefficient of 0.45 (p ≤ 0.001, Fig. 1I). The visual observations regarding modularity and clustering were quantified by computing afferent network measures (Fig. 1J–L). The ideal modularity Q differed significantly (p ≤ 0.001, paired t-tests) from the respective randomized groups of matrices for both [18F]FDG correlations (0.46 ± 0.02 for subject matrices and 0.22 ± 0.02 for randomized matrices) and FC (0.47 ± 0.05 for subject matrices and 0.25 ± 0.02 for randomized matrices). Similarly, clustering was significantly higher (p ≤ 0.001, paired t-tests) for [18F]FDG connectivity from the respective randomized groups (0.27 ± 0.09 for subject matrices and 0.14 ± 0.05 for randomized matrices), as well as for FC (0.27 ± 0.09 for subject matrices and 0.11 ± 0.04 for randomized matrices). Finally, the small-world properties of the outputs were computed using the small-world coefficient. For [18F]FDG connectivity, a mean value of 1.42 ± 0.39 was found. For FC, the mean small-world coefficient was 1.73 ± 0.34. Both datasets were significantly higher than 1 (p ≤ 0.001, one-sample t-test). On the individual level, one subject exhibited small-world coefficients lower than 1 for both [18F]FDG connectivity and FC, and three other subjects had small-world coefficients lower than 1 for [18F]FDG-PET.
3.2. Network-level connectivity analysis shows similar components for both readouts
Fig. 2 shows the analysis of a DMN-like network obtained for both modalities. The correlation matrices of [18F]FDG connectivity and FC and the respective edges obtained from applying a 20% sparsity threshold are depicted, and Pearson’s r correlation between both outputs is revealed. For ICA, the two similar components associated with the DMN are illustrated.
Fig. 2.
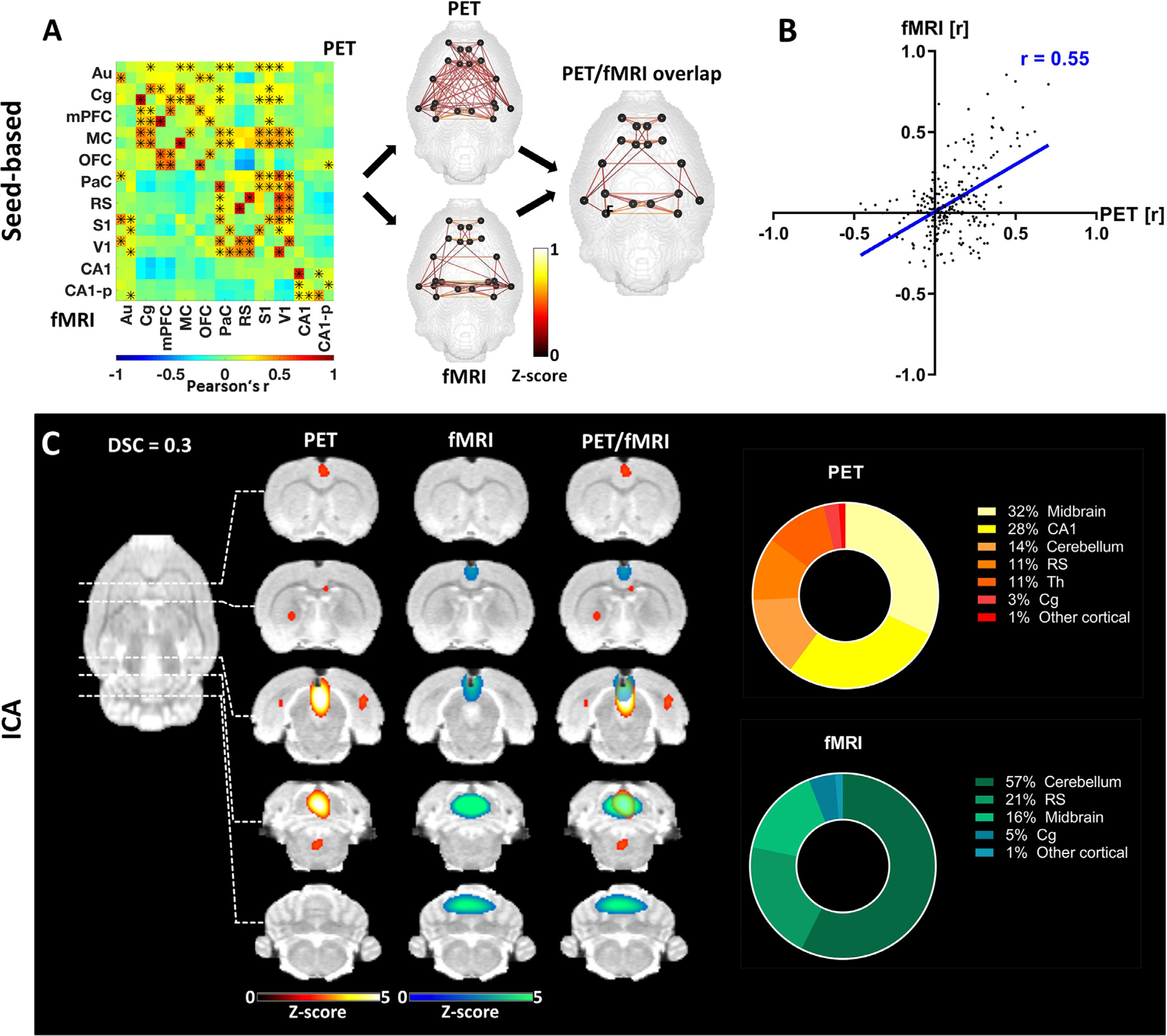
Pairwise correlation analysis and ICA-derived connectivity of the DMN-like network. (A) Mean connectivity matrix of the DMN for [18F]FDG-PET and BOLD-fMRI. The positive edges derived from a whole-brain sparsity threshold of 20% are depicted along with the common edges for both methods. * indicate significant correlations (p ≤ 0.05, corrected for multiple comparison using Bonferroni-Holm). (B) Correlation of [18F]FDG connectivity and FC within the DMN assessed using Pearson’s r. (C) ICA-derived components of [18F]FDG connectivity and FC comprising regions associated with the posterior DMN along with their overlap (z ≤ 1.96) and the respective contributions of different regions to the ICA-derived components. The results are reported at group-mean level (n = 30).). FC = fMRI-derived functional connectivity, ICA = independent component analysis, DSC = Dice-Sörensen coefficient. For a list of abbreviations of all regions, please refer to Supplementary Table 1.
The correlation matrices obtained from pairwise analysis of dynamic [18F]FDG tracer delivery fluctuations and FC of regions associated with the DMN revealed two clusters for both outputs (Fig. 2A). The first cluster included regions belonging to the anterior DMN, centred around the Cg. Other regions in this cluster were the mPFC, OFC and M1. The second cluster comprised posterior cortical regions, with lateral areas such as the PaC, S1 and V1, while the RS featured as a medial cortical region. The depiction of the common edges between both outputs yielded the observation that many edges were situated within the two respective clusters and fewer edges were situated between them. However, this observation was less pronounced for the [18F]FDG-PET readout. A Pearson’s r correlation coefficient of 0.55 (p ≤ 0.001) between [18F]FDG connectivity and FC within the DMN-like network was higher than that at the whole-brain level (Fig. 2B). Two components derived from the ICA of the two datasets were located around the posterior area of the DMN (Fig. 2C). The two cortical regions with the highest contributions to these components were the widely reported DMN hubs: the RS (11% in PET and 21% in fMRI) and Cg (3% in PET and 5% in fMRI). Other cortical regions contributed less than two percent of the signal in either component. However, large implications were observed from more posterior regions, such as the midbrain (32% in PET and 16% in fMRI) and the cerebellum (14% in PET and 57% in fMRI). Furthermore, CA1 (28%) and Th (11%) contributed to the signal in the PET-derived component.
Fig. 3 focuses on the connectivity within the basal ganglia network. The mean connectivity matrices for [18F]FDG and BOLD-fMRI are depicted, along with the edges derived from applying a 20% whole-brain sparsity threshold. To assess similar information for both outputs, the common edges are shown and Pearson’s r is calculated for the unthresholded outputs. Second, two components exhibiting a significant spatial distribution along the striatum are presented along with the quantification of the contributions of different regions to the respective components.
Fig. 3.
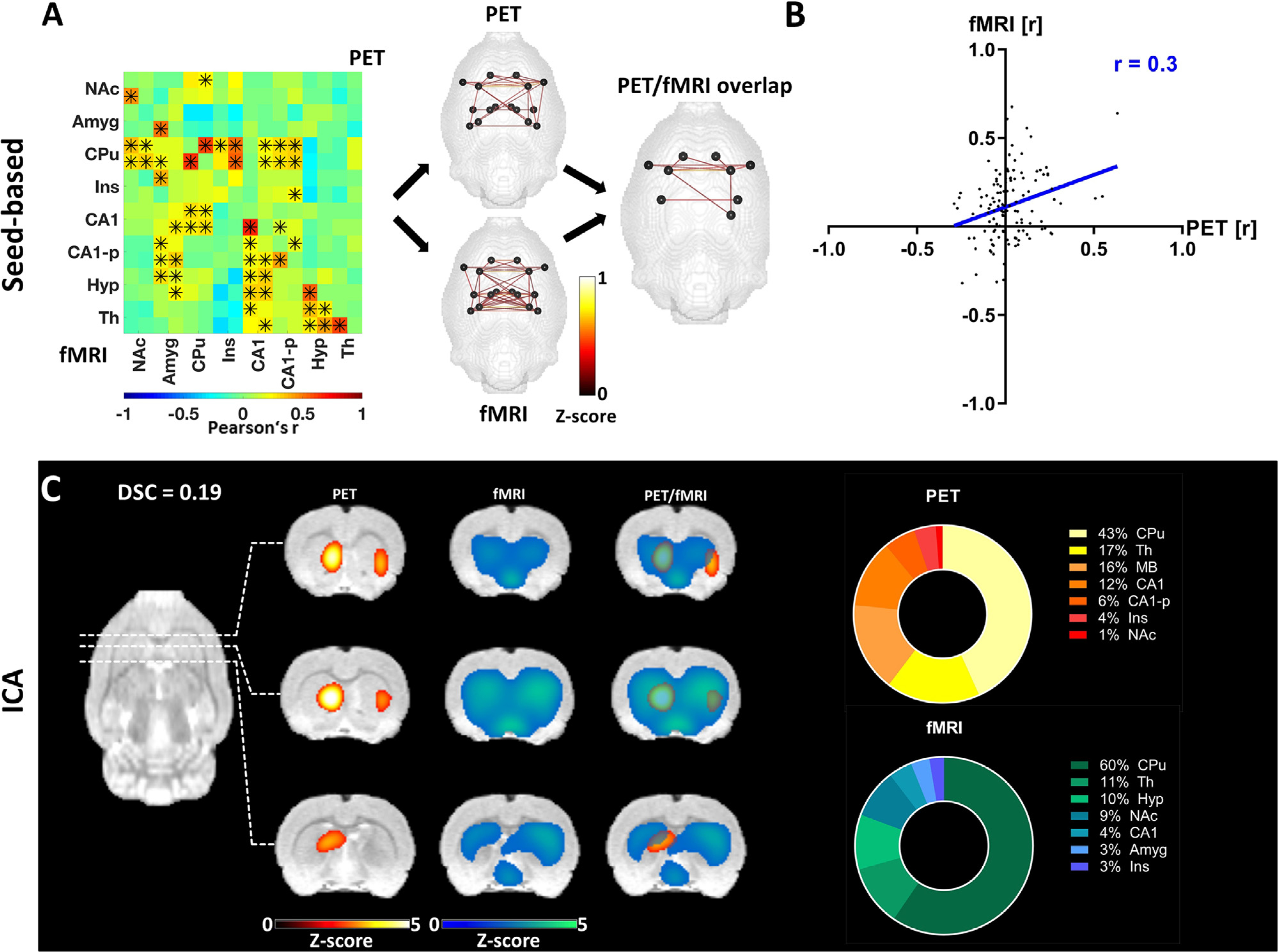
Pairwise correlation analysis and ICA-derived connectivity of the basal ganglia network. (A) Mean connectivity matrix of the basal ganglia network for [18F]FDG and BOLD-fMRI. The positive edges derived from a whole-brain sparsity threshold of 20% are depicted along with the common edges for both methods. * indicate significant correlations (p ≤ 0.05, corrected for multiple comparisons using Bonferroni-Holm). (B) Correlation of [18F]FDG connectivity and FC within the basal ganglia assessed using Pearson’s r. (C) ICA-derived components of [18F]FDG connectivity and FC comprising regions associated with the basal ganglia network along with their overlap (z ≥ 1.96) and the percentage contribution of different regions to the ICA-derived components. The results are reported at group-mean level (n = 30). FC = fMRI-derived functional connectivity, ICA = independent component analysis, DSC = Dice-Sörensen coefficient. For a list of abbreviations of all regions, please refer to Supplementary Table 1.
The mean connectivity matrices of the basal ganglia network revealed more widely distributed connectivity patterns for FC than for [18F]FDG correlations (Fig. 3A). The similarities of both outputs included pronounced connectivity between the CPu, Ins, CA1 and CA1-p. The quantification of similarity between both outputs using Pearson’s r had a value of 0.3 (p ≤ 0.001, Fig. 3B). Similar to the pairwise correlation approach, the ICA-derived components indicated an overlap in the anterior subcortical regions, especially in the striatum (Fig. 3C). The quantification of regional contributions confirmed that the CPu contributes extensively to the signal in both components (43% in PET and 60% in fMRI). Additionally, the second-highest contribution to both components was provided by the Th (17% for PET and 11% for fMRI). Other areas, such as the NAc, Ins and CA1, contributed to the signal in both components, while the Amyg and Hyp only featured in the fMRI output, and the MB exclusively featured in the PET-derived output.
Fig. 4 illustrates the connectivity in the cerebellar-midbrain network. First, the FC and [18F]FDG connectivity derived using the pairwise correlation approach are shown, and the respective outputs compared. Second, two components derived from both PET and fMRI are presented and the regional contributions to the signal of each component are quantified voxel-wise.
Fig. 4.
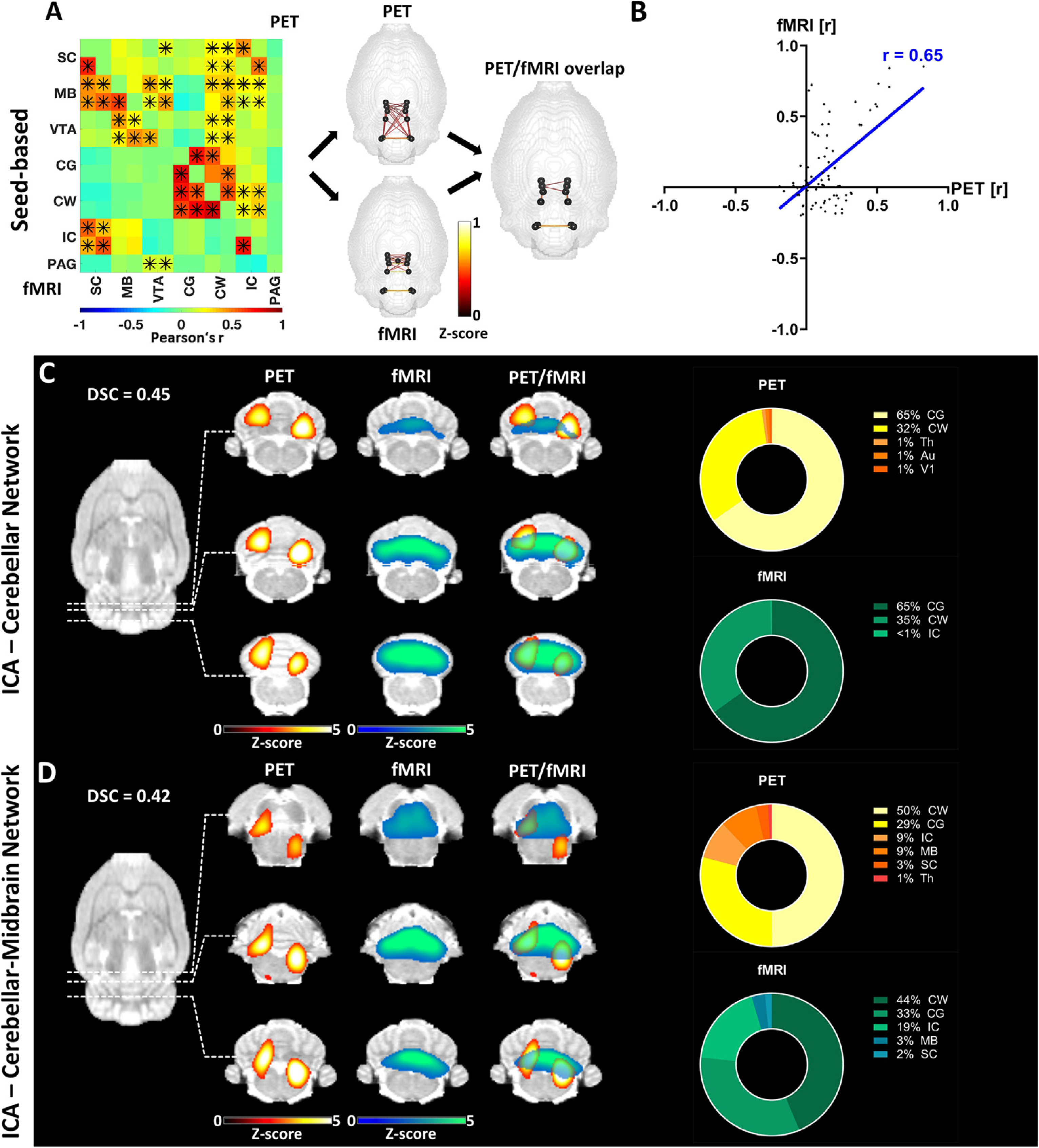
Pairwise correlation analysis and ICA-derived connectivity of the cerebellar-midbrain network. (A) Mean connectivity matrix of the cerebellar-midbrain network for [18F]FDG-PET and BOLD-fMRI. The positive edges derived from a whole-brain sparsity threshold of 20% are depicted along with the common edges for both methods. * indicate significant correlations (p ≤ 0.05, corrected for multiple comparisons using Bonferroni-Holm). (B) Correlation of [18F]FDG connectivity and FC within the cerebellar-midbrain network assessed using Pearson’s r. (C) ICA-derived components [18F]FDG-PET and BOLD-fMRI data (z ≥ 1.96) comprising cerebellar regions along with their overlap and the contribution of different regions to the respective components. (D) ICA-derived components of [18F]FDG-PET and BOLD-fMRI data (z ≥ 1.96) comprising cerebellar and midbrain regions along with their overlap and the contribution of different regions to the respective components. The results are reported at group-mean level (n = 30). FC = fMRI-derived functional connectivity, ICA = independent component analysis, DSC = Dice-Sörensen coefficient. For a list of abbreviations of all regions, please refer to Supplementary Table 1.
The connectivity in the cerebellar and midbrain areas of the brain revealed increased spatial distribution for [18F]FDG correlations (Fig. 4A). While both [18F]FDG correlations and FC could be found within the cerebellum (CG and CW) and the midbrain areas (MB, SC, IC, VTA), in contrast to [18F]FDG connectivity, no FC was detected between the midbrain regions and cerebellar regions. Nonetheless, a relatively high correlation of 0.65 (p ≤ 0.001) between the two unthresholded outputs of [18F]FDG-PET and BOLD-fMRI was quantified using Pearson’s r (Fig. 4B). Two separate components for the two areas were assessed using ICA. The cerebellar components (Fig. 4C) showed significant overlap, both having high contributions from the CG (65% in PET and 65% in fMRI) and the CW (32% in PET and 35% in fMRI). A further pair of components (Fig. 4D) was derived from both the midbrain and cerebellar areas. While the cerebellum provided the largest proportion of signal for both PET and fMRI, the MB also contributed to both components (9% in PET and 3% in fMRI) along with the IC (9% in PET and 19% in fMRI) and the SC (3% in PET and 2% in fMRI).
3.3. Single-subject correlations between hemodynamic and [18F]FDG-PET-derived RSNs
We calculated correlations between the single-subject readouts of the three reported RSNs. The values are presented in Fig. 5.
Fig. 5.
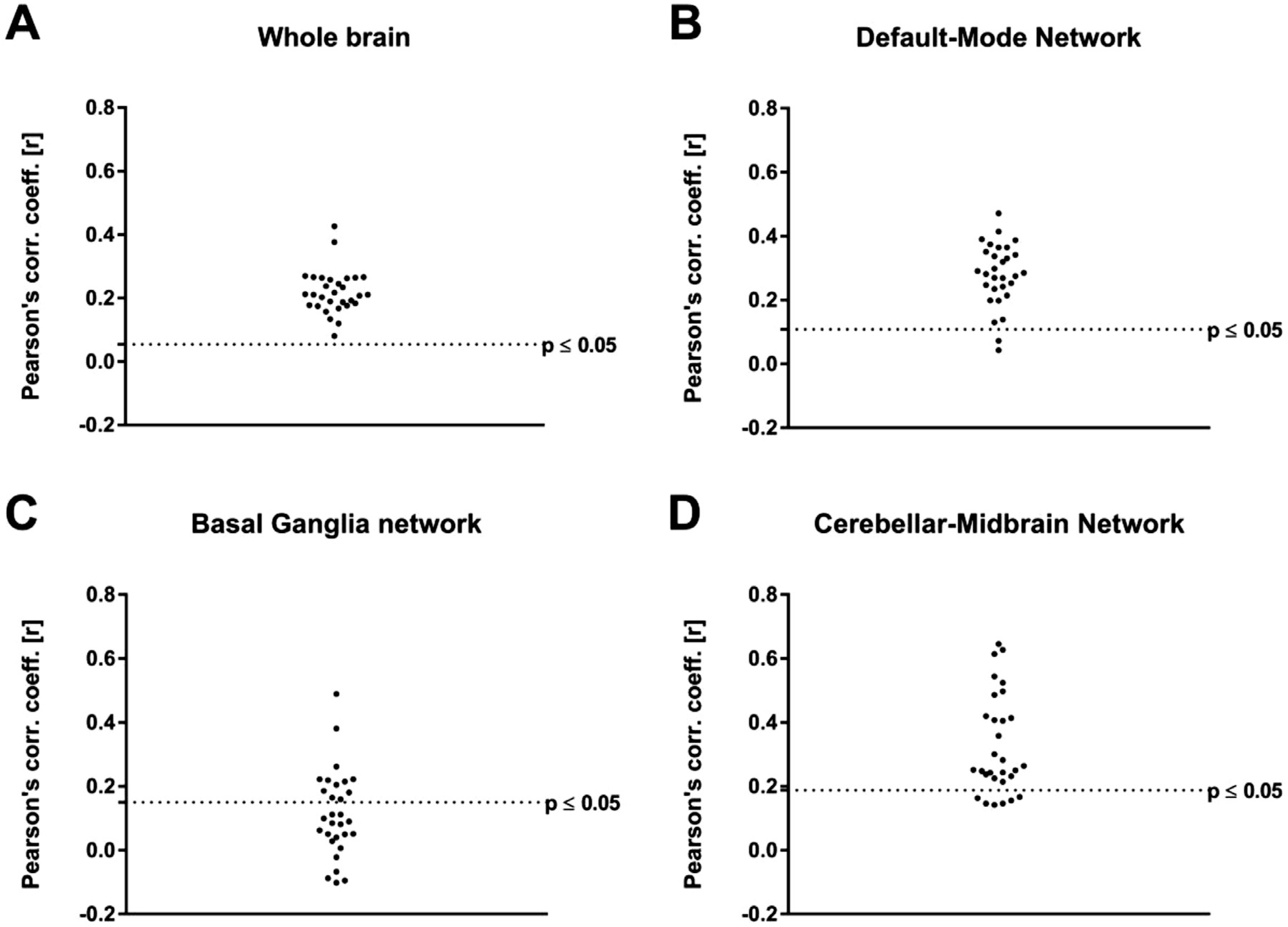
Correlations between the [18F]FDG and BOLD-fMRI readouts on single-subject level for every RSN. The subject-level correlations were tested for significance (A) at whole-brain level and in the (B) default-mode network, (C) basal ganglia network and (D) cerebellar-midbrain network with and without multiple comparison corrections. The dotted lines indicate significance without multiple comparison correction (p ≤ 0.05).
On whole-brain level, all subjects correlated significantly (p ≤ 0.05) and all respective p-values survived multiple comparison correction using Bonferroni-Holm. The correlations ranged from 0.08 to 0.42 with a mean value of 0.22 ± 0.07. In the DMN, the correlations ranged from 0.08 to 0.42 with a mean value of 0.29 ± 0.10. 28 out of 30 subjects correlated significantly (p ≤ 0.05, uncorrected) and 26 out of the 30 p-values survived multiple comparison correction. In the basal ganglia network, correlations between [18F]FDG-PET and BOLD-fMRI connectivities on single-subject level were in the range of −0.1 and 0.49, the mean value being 0.11 ± 0.13. [18F]FDG connectivity and BOLD-derived FC correlated significantly for 12 out of 30 subjects before multiple comparison correction and for 3 out of 30 subjects after Bonferroni-Holm correction was applied. Finally, in the cerebellar-midbrain network the subject-level correlations of [18F]FDG connectivity and FC were in the range between 0.14 and 0.65 with an average of 0.35 ± 0.18. 24 out of 30 single-subject [18F]FDG correlation readouts and respective BOLD-fMRI outputs correlated significantly (p ≤ 0.05) before multiple comparison correction and 12 out of 30 survived Bonferroni-Holm correction.
3.4. Complementary networks can be found in fMRI and PET-derived connectivity outputs
Fig. 4 focuses on the components found exclusively in the fMRI-derived ICA. Here, four cortical components and one subcortical component are presented from the anterior to posterior areas of the brain. For a list of all 20 components found in the fMRI-derived ICA, see the Supplementary Results.
In the component comprising the anterior DMN, the Cg acted as a main hub, contributing 36% of the signal (Fig. 6A). Further anterior cortical areas with implications in this component were the mPFC (15%), M1 (14%) and OFC (10%). Subcortically, a significant signal could be found in the CPu (10%). Posteriorly, another main DMN hub, the RS, contributed 7% of the total signal, while 4% of the signal was derived from the S1 cortex.
Fig. 6.
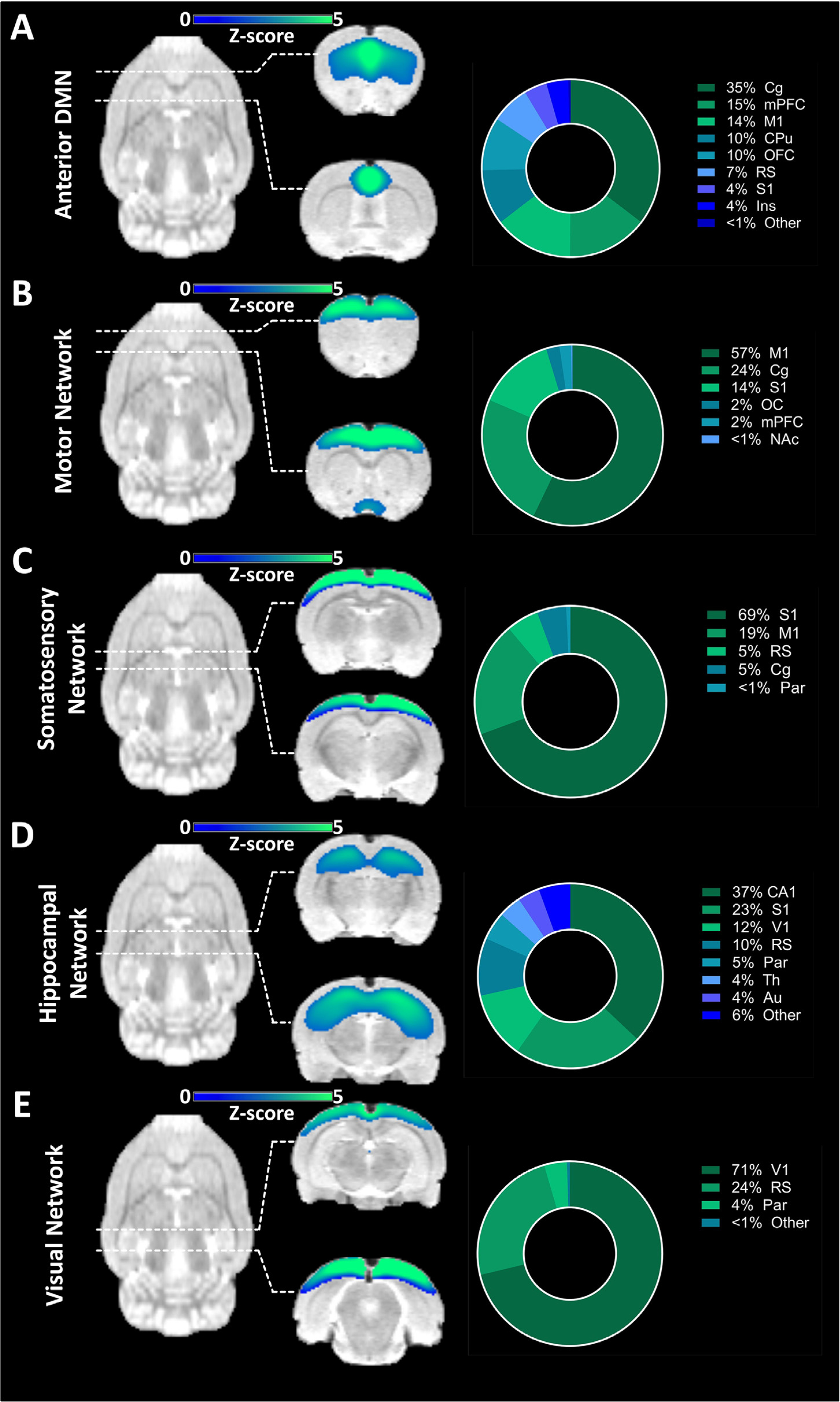
Components found exclusively in fMRI-derived ICA. (A) Component comprising regions associated with the anterior DMN along with the regional quantification of the derived signal. (B) Component comprising the motor network along with the regional quantification of the derived signal. (C) Component comprising the somatosensory network along with the regional quantification of the derived signal. (D) Component comprising regions of the hippocampal network along with the regional quantification of the derived signal. (E) Component comprising the visual network along with the regional quantification of the derived signal. The results are reported at group-mean level (n = 30). ICA = independent component analysis, DMN = default-mode like network. For a list of abbreviations of all regions, please refer to Supplementary Table 1.
Fifty-seven percent of a component representing the motor network (Fig. 6B) was found to be composed of M1, with further contributions from the Cg (24.%) and S1 (14%). The somatosensory network was derived to a proportion of 69% from S1 and 19% from M1 (Fig. 6C). The hippocampal network was mainly driven by CA1 (37%), while further signals could be found in cortical areas such as the S1, V1 and RS, as well as the Th (Fig. 6D). Finally, the visual network (Fig. 6E) was predominantly composed of V1 contributions (71%). Further contributions were provided by the RS (24%) and the Par (4%).
Three components found exclusively in the PET-derived ICA are depicted in Fig. 7. A list of all 20 components derived from the ICA applied to the PET data can be seen in the Supplementary Results.
Fig. 7.
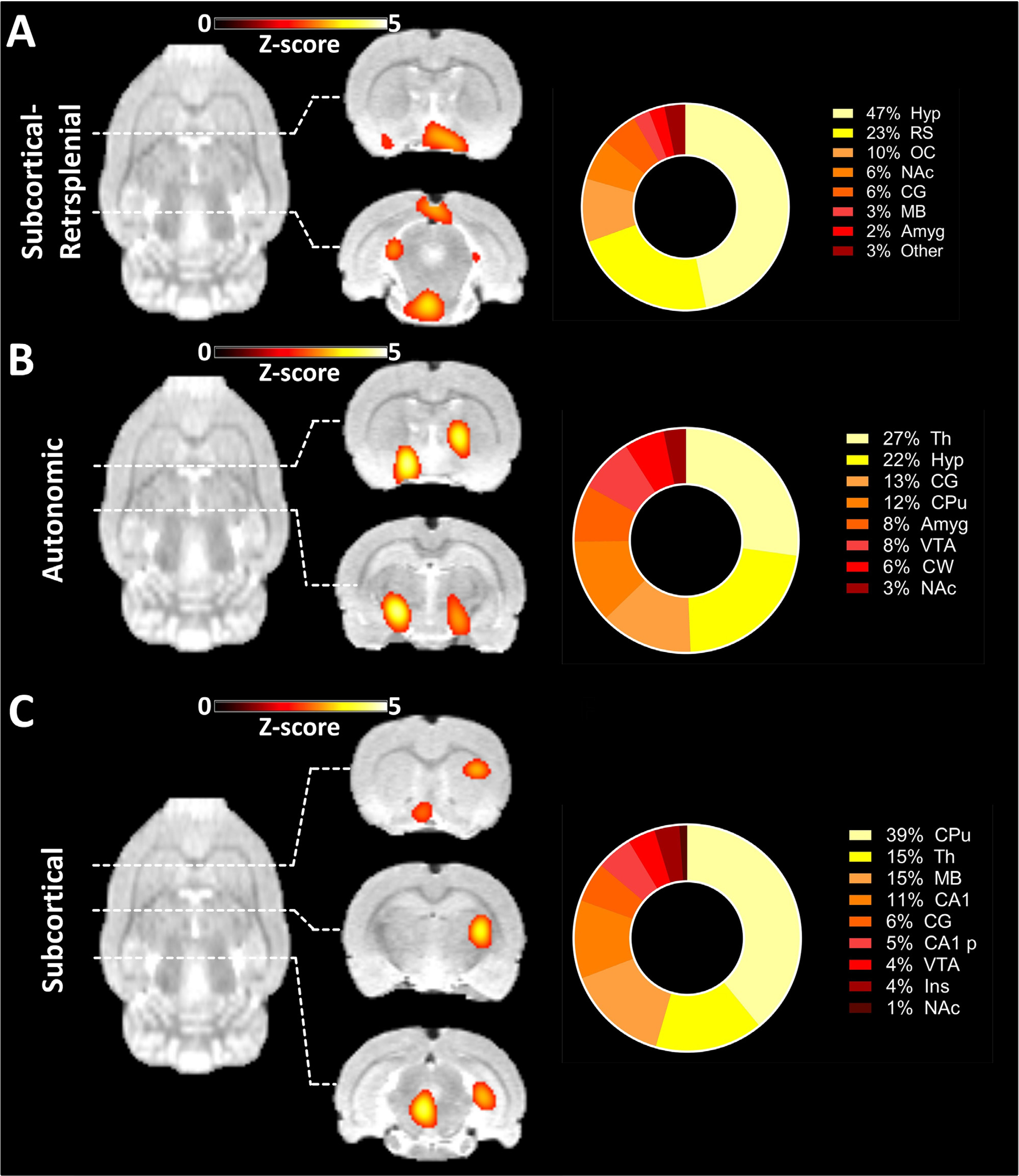
Components found exclusively in [18F]FDG-PET-derived ICA. (A) Component comprising the Hyp, RS, OC and other subcortical and posterior areas along with the regional quantification of the derived signal. (B) Component mainly composed of Th, Hyp and other subcortical areas along with the regional quantification of the derived signal. (C) Component comprising multiple subcortical areas along with the regional quantification of the derived signal. The results are reported at group-mean level (n = 30). ICA = independent component analysis. For a list of abbreviations of all regions, please refer to Supplementary Table 1.
The first component found exclusively in the PET-derived ICA (Fig. 7A) had the largest signal contributions from the Hyp (47%) and the RS (23%). Further signals could be detected in the OC (10%) and in anterior subcortical areas such as the NAc (6%) and the Amyg (2%). A smaller proportion of the signal was found posteriorly in the cerebellar and midbrain areas. A further component (Fig. 7B) was derived mainly from the Th (27%) and Hyp (22%). Smaller contributions were provided by other subcortical areas ranging from the anterior areas, including the CPu, NAc and Amyg, to the cerebellum. Finally, another spatially widely distributed subcortical component revealed the connectivity between the basal ganglia and the midbrain and the cerebellum (Fig. 7C). The majority of the signals stemmed from the CPu (39%), Th (15%), MB (15%) and CA1 (11%), with smaller implications from the CG, CA1-p, VTA, Ins and NAc.
4. Discussion
Resting-state FC studies have been derived primarily from MRI data due to the high spatial and temporal resolution and relative ease of access to MR systems. The analysis of BOLD-fMRI RSN has become more streamlined in recent years, providing reliable quantitative measures of brain function. However, fMRI has some disadvantages, relying on the convoluted BOLD signal as an indirect reflection of neural activity via hemodynamic changes (Buxton, 2012), as well as a high sensitivity to artefacts such as the spin-history artefact already occurring at millimetre-range movements (Yancey et al., 2011). Recent advances in PET scanner technology with an improved temporal resolution allow functional PET imaging using [18F]FDG on the single-subject level (Villien et al., 2014). While [18F]FDG-PET directly measures the glucose consumption of cells, the BOLD signal is convoluted and weighted towards larger blood vessels (Wehrl et al., 2013). The recent advances in correlation analyses of [18F]FDG-PET data (Amend et al., 2019; Wehrl et al., 2013) performed in the same way as fMRI-derived FC using pairwise correlations or ICA have provided the opportunity to gain functional information using PET and enhance the understanding on functional connectivity. In this context, the availability of small-animal hybrid PET/MR systems provides the opportunity to simultaneously study functional RSNs with both modalities. Preclinical studies exhibit several advantages, including high subject cohort homogeneity and improved control over the experimental and living conditions of the cohort. Here, we present for the first time a comparison of subject-wise derived RSNs through the simultaneous acquisition of dynamic [18F]FDG-PET and fMRI data in rats.
In rats, fMRI-derived RSNs similar to those observed in humans have been reported by various groups (Becerra et al., 2011; Hutchison et al., 2010; Lu et al., 2012). Here, we report a DMN-like network, a basal ganglia network and a cerebellar-midbrain network observed using both BOLD-fMRI and [18F]FDG-PET, highlighting the common substrate of the two outputs. However, these networks were characterized by differences in the extent of their connectivity patterns, emphasizing the complementarity of the two methods. This aspect is further supported by a number of RSNs exclusive to either of the two outputs, indicated by ICA components associated with the anterior DMN, the motor network, the sensory network, the hippocampal network and the visual network for the fMRI dataset and a network comprising the retrosplenial cortex and several subcortical regions, one driven mainly by the hypothalamus and thalamus and one across several subcortical regions for the [18F]FDG-PET dataset.
4.1. Common RSNs for [18F]FDG-PET and BOLD-fMRI reveal differences regarding spatial extents
The DMN is one of the most widely reported RSNs in humans (Raichle et al., 2001). In rats, the DMN has been reported as being composed of an anterior and a posterior sub-component (Lu et al., 2012). This observation is confirmed by the present study. For FC, the pairwise correlation approach indicated an anterior cluster around the cingulate cortex and a posterior one anchored by the retrosplenial cortex with sparser connectivity between the two groups of regions. [18F]FDG correlations revealed a higher density of edges between the anterior and posterior parts of the DMN. ICA supported these findings: for fMRI, two separate anterior and posterior components associated with the DMN were observed. In contrast, only one component associated with the DMN was derived from the [18F]FDG-PET ICA. This component exhibited higher overlap with the posterior fMRI-derived DMN component than with the anterior component. Intriguingly, both the PET-derived and the fMRI-derived components comprised extensive contributions from cerebellar and midbrain areas. This finding can be explained by the fact that the midbrain and the cerebellum both cover a larger volume compared to the retrosplenial and cingulate cortices (see Supplementary Table 1). Additionally, recent reports in the literature indicated involvements of these areas in the DMN. In a human study, Habas et al. (Habas et al., 2009) found cerebellar, thalamic and midbrain contributions to the DMN, as indicated by our data. Other reports appear to converge towards the hypothesis that the cerebellum may contribute extensively to the DMN (Castellazzi et al., 2018; Savini et al., 2019), although its role has not been fully resolved. Studies have indicated its potential involvement in the elaboration of the past and future together with the retrosplenial cortex (Addis et al., 2007). The absence of the cerebellum from most reports could be a result of its exclusion due to a number of challenges it poses in fMRI, such as the risk of susceptibility artefacts (T2* effects) induced by nearby air-tissue interface in the ears (Batson et al., 2015; Becerra et al., 2011; Habas et al., 2009). In this respect, PET acquisitions are superior since they are not influenced by such artefacts. Nonetheless, both cerebellar and midbrain contributions to the DMN were also reported in rodents using fMRI (Becerra et al., 2011; Upadhyay et al., 2011). Our PET/fMRI data support the involvements of cerebellum and midbrain in the posterior DMN.
The second large-scale network featuring in the outputs of both modalities involved subcortical regions commonly termed the basal ganglia. The circuitry of this network is of particular interest since the basal ganglia have been shown to be chronologically amongst the first areas affected in several diseases, such as Parkinson’s disease, Huntington’s disease or dystonia (DeLong and Wichmann, 2007). Reports of this network in rats provide contrasting information regarding the integration of separate regions into a single network. Becerra et al. reported two components involving the basal ganglia in awake rats (Becerra et al., 2011). Other groups have observed different components separately involving the striatum, hippocampus or thalamic areas with no apparent connection between them (Hutchison et al., 2010; Majeed et al., 2011). For fMRI we found similar ICA components including separate hippocampal and thalamic circuitry (see component 7 in Supplementary Figure 3). Using the pairwise correlation approach, a more pronounced connectivity between the different structures could be observed for FC. For [18F]FDG, the pairwise correlation analysis showed decreased integration of the basal ganglia with pronounced connectivity predominantly in the anterior area around the striatum. This observation was forced by the ICA applied to the [18F]FDG-PET data. The highest overlap in regions associated with the basal ganglia network occurred between two components predominantly situated in the striatum.
The final network found to be similar for [18F]FDG correlations and BOLD-fMRI FC involved cerebellar-midbrain circuitry. Interestingly, novel evidence suggests an integration of these structures into a coherent network with the basal ganglia network presented above (Bostan and Strick, 2018). The analysis of cerebellar-midbrain circuitry enabled similar observations applied to the DMN described above; while [18F]FDG correlations suggested integration of midbrain and cerebellar circuitry, FC was pronounced within the cerebellum and the midbrain individually, but not between them. The performed ICA revealed one cerebellar component for both readouts and a second involving both cerebellum and midbrain. Interestingly, the cerebellar-midbrain component derived from [18F]FDG-PET also suggests increased spatial integration due to the more distributed signal with stronger contributions from the midbrain and thalamus.
In addition to the group-mean calculations, we conducted subject-wise correlation analyses between [18F]FDG and BOLD-fMRI readouts at whole-brain and RSN level, leading to two main conclusions. First, the subject [18F]FDG-PET and BOLD-fMRI FC readouts correlated for all RSNs and on whole-brain level in the majority of subjects. Significance testing of the single-subject correlations indicated differing extents of coherence between [18F]FDG correlations and FC depending on the RSN. Although the correlations between [18F]FDG-PET outputs and FC for certain subjects were as high as for the group-mean data, on average they were lower compared to group-mean readouts. Especially in the basal ganglia network, less than half of the subjects exhibited significant correlations between [18F]FDG-PET readouts and FC and merely three were significant following multiple comparison correction. These observations imply that there is a significant inter-subject variability in both [18F]FDG correlations and FC which is likely not entirely consistent between subjects. In spite of the single-subject variability, at the employed group size of 30 subjects the [18F]FDG-PET data and BOLD-fMRI FC converge to a higher correlation of the group-mean readouts. While inter-subject variability is a well-reported characteristic of resting-state fMRI (Chou et al., 2012) and appears to be similar for [18F]FDG-PET, it will be of interest for future studies to investigate direct relationships between the subject-level variance of the two readouts and to decipher their physiological significance. If the subject-wise variance of [18F]FDG correlations is able to partially explain that of FC, it may enhance the prospects of using connectivity as a tool for diagnosis or treatment monitoring in the future, especially in indications for which PET/MRI studies are more commonly performed, such as epilepsy (Shang et al., 2018).
4.2. Exclusive components derived from the BOLD-fMRI and [18F]FDG-PET datasets respectively reveal connectivity segregation and integration
The independent components found to be exclusive to either fMRI or [18F]FDG-PET appear to confirm the general trend suggested above. In general, the fMRI components can be described as being bilateral and having a limited anterior-posterior extent. Apart from the anterior DMN component, the other presented components were predominantly driven by a single brain area: the motor cortex, the sensory cortex, the hippocampus and the visual cortex. Such bilateral components were first described in rats by Hutchison et al. in 2010 (Hutchison et al., 2010).
Bilateral spatially limited components are observed much less frequently in the [18F]FDG-PET-derived ICA output. The first exclusive component presented principally included the hypothalamus, olfactory cortex and retrosplenial cortex. The retrosplenial cortex is a cortical region of a much larger relative extent in rodents than in humans; hence, its function in rats may be of increased importance. Previous studies have shown the connections of the hypothalamus to frontal cortical regions and the hippocampus, suggesting its role in cognition and decision-making (Nelson et al., 2014). Additionally, the hypothalamus it has been shown to be involved in spatial and mnemonic functions (Maguire, 2001; Vann et al., 2009). Since smell, which is processed in the olfactory cortex, is one of the most dominant senses in rodents and because the hypothalamus is known to play a major role in homoeostatic behaviours (Toni et al., 2004), this network may play a role in adapting to the environment by collecting cues via the sense of smell along with decision-making and navigation using the retrosplenial cortex and appropriate homoeostatic regulation through the hypothalamus. The autonomic network shown in Fig. 7B has been described in earlier BOLD-fMRI studies in awake rats (Becerra et al., 2011; Liang et al., 2011). Though the observation of this circuitry in the [18F]FDG-PET dataset may point towards the robustness of this method regarding anaesthesia, the interpretation of this network remains elusive. Finally, the spatially extensive component observed in Fig. 7C, comprised of basal ganglia, thalamus, midbrain and cerebellum, may represent a further hint towards an increased integration of these structures.
Our data strongly support the hypothesis that FC largely reflects interhemispheric connectivity between bilateral structures, while the [18F]FDG correlation output reflects a more integrated model of brain connectivity, revealing more extensive modules of functionally connected brain areas. Since many neurological disorders, such as AD and PD, have been shown to affect the brain on a large scale involving several interconnected brain regions distal from the main pathology, the readout of [18F]FDG-PET data will be a valuable tool to study brain connectivity changes in such diseases at early stages or to follow treatment responses.
4.3. Comparison of fMRI and [18F]FDG-PET connectivity imaging
Several studies have aimed to compare FC to brain metabolism metabolic connectivity or [18F]FDG correlations (Aiello et al., 2015; Di et al., 2017; Passow et al., 2015b; Savio et al., 2017; Tomasi et al., 2017; Wehrl et al., 2013). Aiello et al. compared brain metabolism to fMRI measures such as FC, regional homogeneity (ReHo) or fractional amplitude of low frequency fluctuations (fALFF) in humans and indicated correlations between the readouts ranging from 0.47 to 0.55 using Spearman’s ρ, which is in the same range as the correlation using Pearson’s r presented in our study (Aiello et al., 2015). On whole-brain level, our reported Pearson’s r correlation of 0.45 translates into a rank correlation of ρ = 0.44. Apart from the connectivity metric employed, the slightly lower values compared to those reported by Aiello might stem from the fact that, compared to the study by Aiello (Aiello et al., 2015), which only included the cortex, the present study was performed at whole-brain level. The overlaps between the networks we derived from BOLD-fMRI FC and [18F]FDG correlations are also in line with the values reported previously for a seed-based approach by Biswal et al. in humans (Di et al., 2017). With respect to RSNs, one study compared networks derived from simultaneous [18F]FDG-PET and fMRI acquisitions (Savio et al., 2017). Similarly to our data, the authors identified common components for fMRI and PET, as well as exclusive components for both outputs. Interestingly, the PET-exclusive components were also predominantly in subcortical brain areas (Savio et al., 2017), which is in line with our findings. However, none of the studies mentioned above derived [18F]FDG correlations from dynamic [18F]FDG-PET tracer delivery fluctuations. Two studies in humans have previously performed dynamic [18F]FDG-PET correlation analyses (Passow et al., 2015b; Tomasi et al., 2017), however without acquiring the fMRI data simultaneously, making the comparison of the respective outputs difficult. Previously, we have shown an interplay between fMRI and dynamic [18F]FDG-PET and the complementary information they provide in task-based and resting-state experiments (Wehrl et al., 2013). In the present study, we further support these findings with regard to RSNs.
As a general trend, both our study and the reports described above converge towards the conclusion that FC and [18F]FDG correlation readouts overlap partially while also offering significant complementary input. This is explained by the fact that while in essence, both methods are an indirect reflection of neural activity, the hemodynamic and [18F]FDG tracer fluctuations they respectively measure occur inherently at different time-scales (Amend et al., 2019; Rischka et al., 2018; Tomasi et al., 2017; Wehrl et al., 2013). Simultaneous [18F]FDG-PET/fMRI is therefore perfectly suited to delineate these two interconnected but distinct physiological readouts. BOLD-fMRI is able to capture hemodynamic changes using its second-range resolution and is complemented by [18F]FDG-PET reflecting glucose consumption using minute-range time-windows. This temporal mismatch likely contributes to the complementarity reported using BOLD-fMRI and [18F]FDG-PET in previous brain stimulation studies (Wehrl et al., 2013), as well as in the resting-state data reported here and elsewhere (Amend et al., 2019; Passow et al., 2015b; Tomasi et al., 2017).
Regarding the two types of analysis performed, several aspects need to be pointed out. First, while our ICA readout of the [18F]FDG data generally mirrors the reports of previous studies in rats (Wehrl et al., 2013), many components indicate focal, unilateral signal. Using different numbers of components may contribute towards solving this issue; however, one of the main reasons for this observation is the relatively low signal-to-noise PET data have at voxel level. Therefore, voxel-wise approaches such as ICA or voxel-wise seed-based correlations are likely to be less suited for [18F]FDG connectivity analysis compared to FC. Based on the reported data, we recommend the use of pair-wise correlation analysis including nodes of sufficient spatial extent to avoid particularly noisy signals.
4.4. Study limitations
There are several limitations to our study. First, fasting, a standard procedure for [18F]FDG-PET scans, has been recently shown to impact fMRI-derived FC in mice (Tsurugizawa et al., 2019). Future studies are required to reveal the impact of fasting on the relationship between FC and [18F]FDG correlations. Interestingly, the above-mentioned study reported an increase in retrosplenial FC being possibly associated with food-seeking. The [18F]FDG-PET-exclusive component in our study comprising the retrosplenial cortex and hypothalamus may be related to this type of behaviour.
Additionally, the choice of isoflurane has been shown to influence rs-FC readouts (Paasonen et al., 2018; Williams et al., 2010). However, we kept isoflurane at a level previously indicated as feasible for FC acquisition in rodents (Hutchison et al., 2014) to minimize its effects. Importantly, previous studies evaluated interhemispheric FC as a measure of the effects of different anaesthesia regimes and dosages (Jonckers et al., 2014). The bilateral ICs, as well as the strong interhemispheric FC revealed by the pairwise correlation analysis for FC indicate isoflurane was at sufficiently low levels. Furthermore, isoflurane has been shown to impact [18F]FDG uptake (Spangler-Bickell et al., 2016). For fMRI studies, a cocktail of isoflurane and medetomidine has been shown to have the most reduced impact on FC readout in rodents (Paasonen et al., 2018). Unfortunately, previous reports (Wehrl et al., 2013) have shown that the effects of medetomidine on [18F]FDG metabolism are significantly more pronounced than those of isoflurane. Concerning the interpretation of our data, previous studies have reported a regionally differentiated impact of isoflurane on brain metabolism, cortical and thalamic [18F]FDG uptake being most affected with more conserved metabolism in cerebellar and midbrain areas (Park et al., 2017; Spangler-Bickell et al., 2016). This may explain a certain bias of our [18F]FDG readout towards the latter structures compared to FC, especially in the ICA analysis. Therefore, effects on the chosen anaesthesia cannot be excluded on the analysis performed in this study and the described network architecture is valid for this type of anaesthesia. Ultimately, whether and to which extent the choice of anaesthesia does influence the readout remains to be elucidated in future studies. On a general note, due to its inherent combination of two imaging modalities, finding an anaesthesia protocol suited for animal PET/fMRI will be one of the major future challenges of similarly designed studies. For an additional analysis and brief discussion on the [18F]FDG uptake in the present study compared to previous reports in conscious and unconscious animals, please refer to Supplementary Information.
Furthermore, we discussed our findings in the context of previous studies performed in rodents but also in humans. While many RSNs have been shown to be reproducible and comparable amongst species, some differences cannot be excluded and should be kept in mind when directly comparing findings between species, in addition to the role played by the choice of anaesthesia.
Additionally, although potential benefits have been delineated (Li et al., 2019; Murphy and Fox, 2017), global normalization or global signal regression (GSR) is a topic of debate in rs-fMRI due to its decrease of sensitivity and induction of artificial negative correlations (Chuang et al., 2019). Due to the latter, we refrained from interpreting the negative correlations observed in our data. Nonetheless, global normalization is a standard method in [18F]FDG-PET and previous studies investigating [18F]FDG correlations using dynamic [18F]FDG-PET reported global normalization as the method of choice (Amend et al., 2019; Passow et al., 2015b). Employing the same method for fMRI enabled an increased coherence between the analysis pipelines of the two datasets. Previous studies focused on rodent fMRI preprocessing and signal cleaning have shown that many approaches which are considered standard for human rs-fMRI cannot be easily translated to small animals (Chuang et al., 2019). Nonetheless, while further studies are clearly required to elucidate the effects of different preprocessing pipelines on both FC and [18F]FDG correlations, to investigate the effect of global signal removal on the computation of FC we also employed an alternative approach recently described by Chuang et al. (Chuang et al., 2019) which did not involve global signal removal. Our comparison shows that global signal removal does not majorly affect our findings which could also be reproduced without applying this method (please refer to Supplementary Results).
Finally, as shown in our previous publication (Amend et al., 2019), one of the main drawbacks of inferring [18F]FDG correlations using a bolus protocol is the inherent inability to achieve stable tracer kinetics. Since the data in the mentioned publication (Amend et al., 2019) indicated the importance of k1 for the readout, the analysis was performed over the course of the entire scan. Hence, it cannot be excluded that by choosing this approach the readout may be to a certain extent biased towards flow effects and when performing correlation analyses the resulting correlation coefficients may be higher compared to analyses performed over later periods of the scan. To this extent, infusion protocols may offer a more accurate subject-level reflection of the metabolic connectivity described on group level by previous studies (Horwitz et al., 1984; Ripp et al., 2020). Additionally, due to the potential bias of using the entire scans in bolus protocols towards tracer flow, results in pathological cohorts with inherent vasculature alterations should also be interpreted with caution when comparing them to data of healthy controls. However, major advantages of choosing the entire period of the scan include the straightforward implementation and the avoidance of user bias of choosing a specific scan period for the analysis. Although infusion protocols do enable a stable readout after achieving stable kinetics (Amend et al., 2019) and are fast becoming more widespread (Rischka et al., 2018; Villien et al., 2014), research on this topic is still at an incipient stage and infusion protocols are thus far still not widely established. Another drawback of infusion protocols is the period required to reach pseudo-equilibrium, an aspect particularly important for scans involving the use of anaesthesia. Therefore, at this stage bolus protocols represent a viable option which can be easily implemented for similarly designed studies both in a clinical and preclinical setting.
The main advantages of the performed study were the simultaneous PET/fMRI acquisitions and dynamic PET analysis. Our data reveal that simultaneously acquired PET data driven by their sub-pico-molar sensitivity (Price, 2001) complement resting-state fMRI network readouts. The use of simultaneous PET/fMRI data acquisition is essential to ensure a temporal coherence between the observations made on both physiological levels (Judenhofer et al., 2008; Wehrl et al., 2013). Dynamic PET is required for inferring subject-wise [18F]FDG correlations from the time series of each subject, rather than on the group level from subject series of one single image per subject. Hence, dynamic PET acquisition is a prerequisite for potential future clinical applications.
5. Conclusion
Deriving [18F]FDG correlations from dynamic [18F]FDG-PET data provides important complementary information to BOLD-fMRI results regarding brain connectivity. Our data reveal a common substrate of both outputs with similar RSNs observed using the two methods. However, the added value of [18F]FDG-PET was apparent both from the extent of the similar networks observed from both datasets and from the components exclusive to the [18F]FDG-PET dataset. It is also important to note that [18F]FDG reflects only one PET tracer amongst many; therefore, PET offers a multitude of possibilities to study different facets of brain metabolism and physiology. Our results indicate the potential of [18F]FDG correlations to reflect a model of brain connectivity with enhanced integration of different brain areas by revealing several large-scale networks, including the cortical, subcortical, midbrain and cerebellar regions. These findings may be of interest for a basic understanding of brain functionality, as well as for studying and developing therapies for neurodegenerative and psychiatric diseases.
Supplementary Material
Acknowledgements
We thank PD Dr. Gerald Reischl (Department of Preclinical Imaging and Radiopharmacy, Werner Siemens Imaging centre, Eberhard Karls University Tuebingen) for producing the [18F]FDG tracer. Additionally, we acknowledge Dr. Julia Mannheim, Dr. Rebecca Rock, Ines Herbon and Stacy Huang (Department of Preclinical Imaging and Radiopharmacy, Werner Siemens Imaging centre, Eberhard Karls University Tuebingen) for their administrative and technical support. This study is also part of the PhD thesis of Tudor Ionescu.
Funding
The research was funded by the German Ministry of Education and Research (BMBF, Grant No. 01GQ1415) to BJP and HFW, the Werner Siemens Foundation and the National Institute of Health (NIH R01 DA038895) to BBB.
Footnotes
Declaration of Competing Interest
The authors declare no conflict of interest.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2021.118045.
Data availability
The original dataset will be made available on Mendeley.
References
- Addis DR, Wong AT, Schacter DL, 2007. Remembering the past and imagining the future: common and distinct neural substrates during event construction and elaboration. Neuropsychologia 45, 1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiello M, Salvatore E, Cachia A, Pappata S, Cavaliere C, Prinster A, Nicolai E, Salvatore M, Baron JC, Quarantelli M, 2015. Relationship between simultaneously acquired resting-state regional cerebral glucose metabolism and functional MRI: a PET/MR hybrid scanner study. Neuroimage 113, 111–121. [DOI] [PubMed] [Google Scholar]
- Amend M, Ionescu TM, Di X, Pichler BJ, Biswal BB, Wehrl HF, 2019. Functional resting-state brain connectivity is accompanied by dynamic correlations of application-dependent [(18)F]FDG PET-tracer fluctuations. Neuroimage 196, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badhwar A, Tam A, Dansereau C, Orban P, Hoffstaedter F, Bellec P, 2017. Resting-state network dysfunction in Alzheimer’s disease: a systematic review and meta-analysis. Alzheimers Dement. (Amst.) 8, 73–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkhof F, Haller S, Rombouts SA, 2014. Resting-state functional MR imaging: a new window to the brain. Radiology 272, 29–49. [DOI] [PubMed] [Google Scholar]
- Batson MA, Petridou N, Klomp DWJ, Frens MA, Neggers SFW, 2015. Single Session Imaging of Cerebellum at 7 Tesla: obtaining Structure and Function of Multiple Motor Subsystems in Individual Subjects. PLoS ONE 10, e0134933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra L, Pendse G, Chang PC, Bishop J, Borsook D, 2011. Robust reproducible resting state networks in the awake rodent brain. PLoS ONE 6, e25701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Yetkin FZ, Haughton VM, Hyde JS, 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn. Reson. Med. 34, 537–541. [DOI] [PubMed] [Google Scholar]
- Bluhm RL, Miller J, Lanius RA, Osuch EA, Boksman K, Neufeld RW, Theberge J, Schaefer B, Williamson P, 2007. Spontaneous low-frequency fluctuations in the BOLD signal in schizophrenic patients: anomalies in the default network. Schizophr. Bull. 33, 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostan AC, Strick PL, 2018. The basal ganglia and the cerebellum: nodes in an integrated network. Nat. Rev. Neurosci. 19, 338–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullmore E, Sporns O, 2009. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat. Rev. Neurosci. 10, 186–198. [DOI] [PubMed] [Google Scholar]
- Buxton RB, 2012. Dynamic models of BOLD contrast. Neuroimage 62, 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellazzi G, Bruno SD, Toosy AT, Casiraghi L, Palesi F, Savini G, D’Angelo E, Wheeler-Kingshott CAMG, 2018. Prominent changes in cerebro-cerebellar functional connectivity during continuous cognitive processing. Front. Cell. Neurosci. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou Y.-h., Panych LP., Dickey CC., Petrella JR., Chen N.-k., 2012. Investigation of long-term reproducibility of intrinsic connectivity network mapping: a resting-state fMRI study. Am. J. Neuroradiol. 33, 833–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang KH, Lee HL, Li Z, Chang WT, Nasrallah FA, Yeow LY, Singh K, 2019. Evaluation of nuisance removal for functional MRI of rodent brain. Neuroimage 188, 694–709. [DOI] [PubMed] [Google Scholar]
- Cole MW, Pathak S, Schneider W, 2010. Identifying the brain’s most globally connected regions. Neuroimage 49, 3132–3148. [DOI] [PubMed] [Google Scholar]
- Damoiseaux JS, Rombouts SA, Barkhof F, Scheltens P, Stam CJ, Smith SM, Beckmann CF, 2006. Consistent resting-state networks across healthy subjects. Proc. Natl. Acad. Sci. U. S. A. 103, 13848–13853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLong MR, Wichmann T, 2007. Circuits and circuit disorders of the basal ganglia. Arch. Neurol. 64, 20–24. [DOI] [PubMed] [Google Scholar]
- Di X, Biswal BB Alzheimer’s Disease Neuroimaging, I., 2012. Metabolic brain covariant networks as revealed by FDG-PET with reference to resting-state fMRI networks. Brain Connect 2, 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di X, Gohel S, Thielcke A, Wehrl HF, Biswal BB Alzheimer’s Disease Neuroimaging, I., 2017. Do all roads lead to Rome? A comparison of brain networks derived from inter-subject volumetric and metabolic covariance and moment-to-moment hemodynamic correlations in old individuals. Brain Struct Funct 222, 3833–3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MD, Snyder AZ, Vincent JL, Corbetta M, Van Essen DC, Raichle ME, 2005. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc. Natl. Acad. Sci. U. S. A. 102, 9673–9678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Krasnow B, Reiss AL, Menon V, 2003. Functional connectivity in the resting brain: a network analysis of the default mode hypothesis. Proc. Natl. Acad. Sci. U. S. A. 100, 253–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greicius MD, Srivastava G, Reiss AL, Menon V, 2004. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc. Natl. Acad. Sci. U. S. A. 101, 4637–4642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habas C, Kamdar N, Nguyen D, Prater K, Beckmann CF, Menon V, Greicius MD, 2009. Distinct cerebellar contributions to intrinsic connectivity networks. J. Neurosci. 29, 8586–8594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn A, Gryglewski G, Nics L, Hienert M, Rischka L, Vraka C, Sigurdardottir H, Vanicek T, James GM, Seiger R, Kautzky A, Silberbauer L, Wadsak W, Mitterhauser M, Hacker M, Kasper S, Lanzenberger R, 2016. Quantification of Task-Specific Glucose Metabolism with Constant Infusion of 18F-FDG. J. Nucl. Med. 57, 1933–1940. [DOI] [PubMed] [Google Scholar]
- Horwitz B, Duara R, Rapoport SI, 1984. Intercorrelations of glucose metabolic rates between brain regions: application to healthy males in a state of reduced sensory input. J. Cereb. Blood Flow Metab. 4, 484–499. [DOI] [PubMed] [Google Scholar]
- Humphries MD, Gurney K, 2008. Network ‘small-world-ness’: a quantitative method for determining canonical network equivalence. PLoS ONE 3, e2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Hutchison M, Manning KY, Menon RS, Everling S, 2014. Isoflurane induces dose-dependent alterations in the cortical connectivity profiles and dynamic properties of the brain’s functional architecture. Hum. Brain Mapp. 35, 5754–5775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchison RM, Mirsattari SM, Jones CK, Gati JS, Leung LS, 2010. Functional networks in the anesthetized rat brain revealed by independent component analysis of resting-state FMRI. J. Neurophysiol. 103, 3398–3406. [DOI] [PubMed] [Google Scholar]
- Joel D, Weiner I, 1994. The organization of the basal ganglia-thalamocortical circuits: open interconnected rather than closed segregated. Neuroscience 63, 363–379. [DOI] [PubMed] [Google Scholar]
- Jonckers E, Delgado y Palacios R, Shah D, Guglielmetti C, Verhoye M, Vander Linden A, 2014. Different anesthesia regimes modulate the functional connectivity outcome in mice. Magn. Reson. Med. 72, 1103–1112. [DOI] [PubMed] [Google Scholar]
- Judenhofer MS, Wehrl HF, Newport DF, Catana C, Siegel SB, Becker M, Thielscher A, Kneilling M, Lichy MP, Eichner M, Klingel K, Reischl G, Widmaier S, Rocken M, Nutt RE, Machulla HJ, Uludag K, Cherry SR, Claussen CD, Pichler BJ, 2008. Simultaneous PET-MRI: a new approach for functional and morphological imaging. Nat. Med. 14, 459–465. [DOI] [PubMed] [Google Scholar]
- Lanzenberger R, Kranz GS, Haeusler D, Akimova E, Savli M, Hahn A, Mitterhauser M, Spindelegger C,Philippe C, Fink M, Wadsak W, Karanikas G, Kasper S, 2012. Prediction of SSRI treatment response in major depression based on serotonin transporter interplay between median raphe nucleus and projection areas. Neuroimage 63, 874–881. [DOI] [PubMed] [Google Scholar]
- Li J, Kong R, Liégeois R, Orban C, Tan Y, Sun N, Holmes AJ, Sabuncu MR, Ge T, Yeo BTT, 2019. Global signal regression strengthens association between resting-state functional connectivity and behavior. Neuroimage 196, 126–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Z, King J, Zhang N, 2011. Uncovering intrinsic connectional architecture of functional networks in awake rat brain. J. Neurosci. 31, 3776–3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zou Q, Gu H, Raichle ME, Stein EA, Yang Y, 2012. Rat brains also have a default mode network. Proc. Natl. Acad. Sci. U. S. A. 109, 3979–3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire EA, 2001. The retrosplenial contribution to human navigation: a review of lesion and neuroimaging findings. Scand. J. Psychol. 42, 225–238. [DOI] [PubMed] [Google Scholar]
- Majeed W, Magnuson M, Hasenkamp W, Schwarb H, Schumacher EH, Barsalou L, Keilholz SD, 2011. Spatiotemporal dynamics of low frequency BOLD fluctuations in rats and humans. Neuroimage 54, 1140–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthew Brett J-LA, Valabregue, Romain, Poline, Jean-Baptiste, 2002. Region of interest analysis using an SPM toolbox. 8th Internation Conference on Functional Mapping of the Human Brain. [Google Scholar]
- Murphy K, Fox MD, 2017. Towards a consensus regarding global signal regression for resting state functional connectivity MRI. Neuroimage 154, 169–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwansisya TE, Hu A, Li Y, Chen X, Wu G, Huang X, Lv D, Li Z, Liu C, Xue Z, Feng J, Liu Z, 2017. Task and resting-state fMRI studies in first-episode schizophrenia: a systematic review. Schizophr. Res. 189, 9–18. [DOI] [PubMed] [Google Scholar]
- Nelson AJ, Hindley EL, Haddon JE, Vann SD, Aggleton JP, 2014. A novel role for the rat retrosplenial cortex in cognitive control. Learn. Mem. 21, 90–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa S, Lee TM, Kay AR, Tank DW, 1990. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl. Acad. Sci. U. S. A. 87, 9868–9872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onnela J-P., Saramäki J., Kertész J., Kaski K., 2005. Intensity and coherence of motifs in weighted complex networks. Phys. Rev. E 71, 065103. [DOI] [PubMed] [Google Scholar]
- Paasonen J, Stenroos P, Salo RA, Kiviniemi V, Gröhn O, 2018. Functional connectivity under six anesthesia protocols and the awake condition in rat brain. Neuroimage 172, 9–20. [DOI] [PubMed] [Google Scholar]
- Park TY, Nishida KS, Wilson CM, Jaiswal S, Scott J, Hoy AR, Selwyn RG, Dardzinski BJ, Choi KH, 2017. Effects of isoflurane anesthesia and intravenous morphine self-administration on regional glucose metabolism ([18F]FDG-PET) of male Sprague-Dawley rats. Eur. J. Neurosci. 45, 922–931. [DOI] [PubMed] [Google Scholar]
- Passow S, Specht K, Adamsen TC, Biermann M, Brekke N, Craven AR, Ersland L, Gruner R, Kleven-Madsen N, Kvernenes OH, Schwarzlmuller T, Olesen R, Hugdahl K, 2015a. A close link between metabolic activity and functional connectivity in the resting human brain. EJNMMI Phys 2, A78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passow S, Specht K, Adamsen TC, Biermann M, Brekke N, Craven AR, Ersland L, Gruner R, Kleven-Madsen N, Kvernenes OH, Schwarzlmuller T, Olesen RA, Hugdahl K, 2015b. Default-mode network functional connectivity is closely related to metabolic activity. Hum. Brain Mapp. 36, 2027–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price P, 2001. PET as a potential tool for imaging molecular mechanisms of oncology in man. Trends Mol. Med. 7, 442–446. [DOI] [PubMed] [Google Scholar]
- Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, Shulman GL, 2001. A default mode of brain function. Proc. Natl. Acad. Sci. U. S. A. 98, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt J, Bornholdt S, 2006. Statistical mechanics of community detection. Phys. Rev. E 74, 016110. [DOI] [PubMed] [Google Scholar]
- Ripp I, Stadhouders T, Savio A, Goldhardt O, Cabello J, Calhoun V, Riedl V, Hedderich D, Diehl-Schmid J, Grimmer T, Yakushev I, 2020. Integrity of neurocognitive networks in dementing disorders as measured with simultaneous PET/fMRI. J. Nucl. Med.. [DOI] [PubMed] [Google Scholar]
- Rischka L, Gryglewski G,Pfaff S, Vanicek T, Hienert M, Klobl M, Hartenbach M, Haug A, Wadsak W, Mitterhauser M, Hacker M, Kasper S, Lanzenberger R, Hahn A, 2018. Reduced task durations in functional PET imaging with [(18)F]FDG approaching that of functional MRI. Neuroimage 181, 323–330. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O, 2010. Complex network measures of brain connectivity: uses and interpretations. Neuroimage 52, 1059–1069. [DOI] [PubMed] [Google Scholar]
- Sanabria-Diaz G, Martinez-Montes E, Melie-Garcia L Alzheimer’s Disease Neuroimaging, I., 2013. Glucose metabolism during resting state reveals abnormal brain networks organization in the Alzheimer’s disease and mild cognitive impairment. PLoS ONE 8, e68860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savini G, Pardini M, Castellazzi G, Lascialfari A, Chard D, D’Angelo E, Gandini Wheeler-Kingshott CAM, 2019. Default mode network structural integrity and cerebellar connectivity predict information processing speed deficit in multiple sclerosis. Front. Cell. Neurosci. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savio A, Funger S, Tahmasian M, Rachakonda S, Manoliu A, Sorg C, Grimmer T, Calhoun V, Drzezga A, Riedl V, Yakushev I, 2017. Resting-state networks as simultaneously measured with functional MRI and PET. J. Nucl. Med. 58, 1314–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiffer WK, Mirrione MM, Biegon A, Alexoff DL, Patel V, Dewey SL, 2006. Serial microPET measures of the metabolic reaction to a microdialysis probe implant. J. Neurosci. Methods 155, 272–284. [DOI] [PubMed] [Google Scholar]
- Seo EH, Lee DY, Lee J-M., Park J-S., Sohn BK., Lee DS., Choe YM., Woo JI., 2013a. Whole-brain functional networks in cognitively normal, mild cognitive impairment, and Alzheimer’s disease. PLoS ONE 8, e53922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EH, Lee DY, Lee JM, Park JS, Sohn BK, Lee DS, Choe YM, Woo JI, 2013b. Whole-brain functional networks in cognitively normal, mild cognitive impairment, and Alzheimer’s disease. PLoS ONE 8, e53922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang K, Wang J, Fan X, Cui B, Ma J, Yang H, Zhou Y, Zhao G, Lu J, 2018. Clinical Value of Hybrid TOF-PET/MR Imaging–Based Multiparametric Imaging in Localizing Seizure Focus in Patients with MRI-Negative Temporal Lobe Epilepsy. American Journal of Neuroradiology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spangler-Bickell MG, de Laat B, Fulton R, Bormans G, Nuyts J, 2016. The effect of isoflurane on 18F-FDG uptake in the rat brain: a fully conscious dynamic PET study using motion compensation. EJNMMI Res. 6, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, 2010. Networks of the Brain. The MIT Press. [Google Scholar]
- Sporns O, Tononi G, Kotter R, 2005. The human connectome: a structural description of the human brain. PLoS Comput. Biol. 1, e42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi DG, Shokri-Kojori E, Wiers CE, Kim SW, Demiral SB, Cabrera EA, Lindgren E, Miller G, Wang GJ, Volkow ND, 2017. Dynamic brain glucose metabolism identifies anti-correlated cortical-cerebellar networks at rest. J. Cereb. Blood Flow Metab. 37, 3659–3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toni R, Malaguti A, Benfenati F, Martini L, 2004. The human hypothalamus: a morpho-functional perspective. J. Endocrinol. Invest. 27, 73–94. [PubMed] [Google Scholar]
- Tsurugizawa T, Djemai B, Zalesky A, 2019. The impact of fasting on resting state brain networks in mice. Sci. Rep. 9, 2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyay J, Baker SJ, Chandran P, Miller L, Lee Y, Marek GJ, Sakoglu U, Chin CL, Luo F, Fox GB, Day M, 2011. Default-mode-like network activation in awake rodents. PLoS ONE 6, e27839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vann SD, Aggleton JP, Maguire EA, 2009. What does the retrosplenial cortex do? Nat. Rev. Neurosci. 10, 792–802. [DOI] [PubMed] [Google Scholar]
- Villien M, Wey H-Y., Mandeville JB., Catana C., Polimeni JR., Sander CY., Zürcher NR., Chonde DB., Fowler JS., Rosen BR., Hooker JM., 2014. Dynamic functional imaging of brain glucose utilization using fPET-FDG. Neuroimage 100, 192–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wehrl HF, Hossain M, Lankes K, Liu CC, Bezrukov I, Martirosian P, Schick F, Reischl G,Pichler BJ, 2013. Simultaneous PET-MRI reveals brain function in activated and resting state on metabolic, hemodynamic and multiple temporal scales. Nat. Med. 19, 1184–1189. [DOI] [PubMed] [Google Scholar]
- Williams KA, Magnuson M, Majeed W, LaConte SM, Peltier SJ, Hu X, Keilholz SD, 2010. Comparis on of α-chloralose, medetomidine and isoflurane anesthesia for functional connectivity mapping in the rat. Magn. Reson. Imaging 28, 995–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M, Wang J, He Y, 2013. BrainNet Viewer: a network visualization tool for human brain connectomics. PLoS ONE 8, e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yancey SE, Rotenberg DJ, Tam F, Chiew M, Ranieri S, Biswas L, Anderson KJT, NicoleBaker S, Wright GA, Graham SJ, 2011. Spin-history artifact during functional MRI: potential for adaptive correction. Med. Phys. 38, 4634–4646. [DOI] [PubMed] [Google Scholar]
- Zhang N, Rane P, Huang W, Liang Z, Kennedy D, Frazier JA, King J, 2010. Mapping resting-state brain networks in conscious animals. J. Neurosci. Methods 189, 186–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original dataset will be made available on Mendeley.


