Abstract
Duck circovirus disease (DuCVD) caused by duck circovirus (DuCV) continues to spread in recent years, which brings serious harm to the poultry industry, so early diagnosis of DuCVD is of great significance for the prevention and control of this disease. Specific primers and probes for DuCV were designed in this study. Reverse primers and probes were modified at the 5′ ends with biotin and fluorescein, respectively, and they were combined with dipsticks labeled with biotin antibodies and fluorescein antibodies to establish a recombinase-aided amplification-lateral flow dipstick (RAA-LFD) assay for detection of duck circovirus. By using this method, the reaction products reached detectable levels in about 20 min as a result of rapid amplification at a constant temperature of 37℃. The detection results could be observed by dripping the reaction products onto the dipstick within 2 to 3 min. The RAA-LFD method has good specificity and high sensitivity (102 copies/μL). Compared with conventional polymerase chain reaction (PCR), RAA-LFD has no power limit on the testing instrument, and is easy to use, saving more time and manpower, so it is more suitable for clinical detection.
Key words: duck circovirus, recombinase-aided amplification, lateral flow dipstick
INTRODUCTION
Duck circovirus disease (DuCVD), as a newly discovered circovirus disease in the 21st century, is caused by duck circovirus (DuCV). DuCV was first discovered and recorded in 2003 (Hattermann et al., 2003). It is the smallest virus known to cause disease in ducks, and it has spread widely worldwide (Wu et al., 2019). This icosahedrally symmetrical, non-enveloped single-stranded circular DNA virus mainly infects duck immune tissues and organs, destroys humoral and cellular immune functions, causes immunosuppression, and predisposes to secondary infections (Bai et al., 2021). Developmental retardation, loss of feather, malnutrition and other signs often appear in animals infected with DuCV. In severe cases, asthma, anemia, decreased productivity and even death may occur. According to the clinical symptoms and pathological manifestations, it is not suitable to make an accurate judgment, and the detection of DuCV cannot be separated from the application of laboratory diagnostic technology. At present, 3 methods are often employed for the laboratory diagnosis of DuCV: scanning electron microscope (SEM) observation, enzyme-linked immunosorbent assay (ELISA) and molecular biological detection. SEM observation requires sophisticated instruments. ELISA can only be used to detect the presence of antibodies, so it is impossible to make early diagnosis. Molecular biological detection can be used to directly detect the presence of virus in duck target organs, and it is more meaningful in clinic.
There are a variety of molecular biological detection methods, which often rely on laboratory facilities and professional operation. However, there are few rapid methods suitable for clinical test at grassroots level. Up to now, the molecular biological detection methods for DuCV mainly include nucleic acid probe (NAP), PCR, real-time fluorescent quantitative PCR (RFQ-PCR), loop-mediated isothermal amplification (LAMP), etc.
Although the NAP assay had good stability and specificity in detecting DuCV, but it had high economic cost and required complicated operation (Zou et al., 2011). The RFQ-PCR method is highly sensitive to the detection of DuCV, but the required equipment is expensive (Liu et al., 2017). Whether it is conventional PCR, nested PCR or multiplex PCR, it requires complex experimental equipment and professional operators, and takes a long time, so it cannot meet the requirements of rapid detection of DuCV. The LAMP technique has the advantages of high specificity and low cost, but it requires reaction for 1 h at a constant temperature of 60 to 65℃ (Zhao et al., 2012). With the continuous development of molecular biotechnology, recombinase-aided amplification (RAA), as a new detection method, is increasingly favored by people. The technique has been used to detect a variety of viruses, such as avian influenza (Wang et al., 2020) and African Swine Fever (Fan et al., 2020).
This experiment combines the RAA technology with lateral flow test dipstick (LFD) coated with biotin antibodies and fluorescein antibodies to establish a visual detection method for rapid detection of DuCV, which provides an effective method for the prevention and control of DuCVD.
MATERIALS AND METHODS
Extraction of Viral Nucleic Acid
According to the instructions of Viral genomic DNA/RNA extraction kit (Tiangen Biotech Co., Ltd. Beijing, China), the DNA of DuCV, Duck enteritis virus (DEV), Muscovy duck parvovirus (MDPV), Fowl adenovirus (FAdV), Porcine circovirus (PCV), and the RNA of Duck hepatitis A virus (DHAV) were extracted (all viruses tested, isolated, identified and preserved by our laboratory). The corresponding cDNA was obtained by reverse transcription of extracted DNA and RNA virus, and stored at −20℃ for later use.
Design of Primers and Probes for RAA-LFD Detection of DuCV
By searching the complete genome sequences of DuCV in NCBI database, a pair of specific primers with a length of 30 to 35 bp and a probe with a length of 46 to 52 bp were designed for the conserved region (rep gene) according to the design principle of RAA primers (Li et al., 2019). The 5′ end of the reverse primer was modified with biotin, the 5′ end of the probe was modified with fluorescein, and the 3′ end of the probe was blocked by phosphorylation. A single-base gap was designed at a position 30 bp from the 5′ end, and it was modified with tetrahydrofuran (THF) residue. The designed primers and probes were synthesized by the reagent company (Sangon Biotech Co., Ltd. Shanghai, China).
The specific information of the primers and probes is as follows: DuCV-F (564-595bp): AAAGAGCCGTTATGCATTTGAATTTCCCGCCG, DuCV-R (686-717bp): Biotin-ACGGTCGGTAATTCTCAGCAAATCATCATACG, DuCV-T (605-655bp): FAM-ATTACAAACCACGCGGGAAGTGGTGGGACGGTTA/idSp/TCGGGAAATGACGTAG.
Development of RAA-LFD Method for Detecting DuCV
Using the DNA of DuCV as a template, a 50 μL reaction system was prepared: VI buffer (25 μL), ddH2O (16.5 μL), forward primers (10 μM, 2.1 μL), reverse primers (10 μM, 2.1 μL), template DNA (1.2 μL), and probe (0.6 μL), were mixed and moved to the basic reaction unit, and magnesium acetate (2.5 μL) was finally added to react 25 min at 39℃ in a RAA thermomixer (Qitian Gene Biological Technology Co., Ltd. Jiangsu. China). 10 μL of reaction product was dripped onto the detection area and the dipstick (Ustar Biological Co., Ltd. Hangzhou. China) was inserted into a 2 mL centrifuge tube containing 100 μL of buffer. After 2 to 3 min, the experimental results were observed and recorded. When there were 2 lines in the dipstick, one in the quality control area and the other in the test area, the result was positive. When there was only one red line in the quality control area, the result was negative.
Optimization of Reaction Conditions for RAA-LFD Detection of DuCV
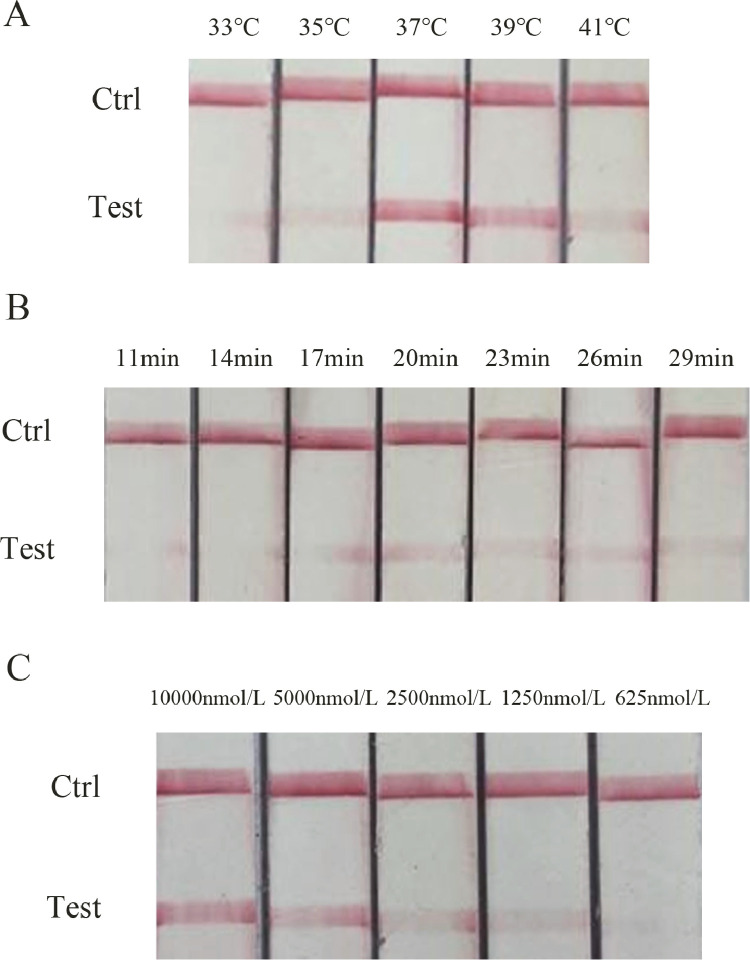
When the conditions of VI buffer and magnesium acetate in the RAA reaction system remain unchanged, the reaction temperature (33, 35, 37, 39, 41℃), reaction time (11, 14, 27, 20, 23, 26, 29 min) and primer concentration (625, 1,250, 2,500, 5,000, 10,000 nmol/L) for RAA test were optimized, respectively, and the RAA amplification products were detected by LFD, to find the conditions with shortest reaction time, lowest reaction temperature and lowest primer concentration which could provide clear results
Specificity Test for RAA-LFD Detection of DuCV
Using the DNA of DuCV, DEV, MDPV, FAdV, PCV, and the cDNA of DHAV as template and ddH2O as negative control, RAA-LFD test was carried out under optimized conditions to observe whether red line appeared in the detection area and verify the specificity of the reaction.
Sensitivity Test for RAA-LFD Detection of DuCV
Preparation of recombinant plasmid: using DuCV as template, a pair of primers with length of 20 to 25 bp (F: CACCGGAAAGAGCCGTTATGCATT, R: CGGGTAACGGTCGGTAATTCTCAGC) were designed and synthesized according to the principle of PCR primer design. The reaction system (50 μL): 2 × Taq Mix (25 μL), template DNA (3 μL), forward primer (10 μM, 1 μL), reverse primer (10 μM, 1 μL), ddH2O (20 μL). The reaction conditions: pre-denaturation at 94℃ for 5 min, annealing at 55℃ for 30 s, extension at 72℃ for 30 s, 34 cycles, and extension at 72℃ for 5 min. The purified target fragment was ligated to pMD20-T vector (Takara Biomedical Technology Co., Ltd. Beijing. China) and introduced into DH5α competent cells. After blue-white screening, the white colonies were selected and cultured overnight to extract plasmids for sequencing by the reagent company (Sangon Biotech Co., Ltd. Shanghai, China).
The number of DNA copies per unit volume of plasmid was calculated as follows:
Plasmid copy number (copies/L) = [plasmid concentration (g/μL) × 6.02 × 1023]/[total fragment length (bp) × 660 g/mol], in which total fragment length=vector length (bp)+target fragment length (bp).
The plasmid was diluted to 1 × 109 copies/μL−1 × 10° copies/μL, and stored at −20℃. Different concentrations of plasmids were used as templates for detection by RAA-LFD, conventional PCR and RFQ-PCR, control to compare the sensitivity of different detection methods.
RAA-LFD detection was carried out according to the optimized reaction conditions, with 1 × 105 copies/μL−1 × 10° copies/μL plasmid (1 μL) as template.
PCR system (25 μL): 2 × Taq Mix (12.5 μL) (Vazyme Biotech Co., Ltd. Nanjing. China), template (1 × 107 copies/μL−1 × 10° copies/μL plasmid, 1.0 μL), forward primers (10 μM, 0.5 μL), primers (the same primers used to prepare recombinant plasmids) (10 μM, 0.5 μL), ddH2O (10.5 μL). Predenaturation at 94℃ for 5 min, annealing at 55℃ for 30 s, extension at 72℃ for 60 s, 34 cycles, and extension at 72℃ for 5 min. The resulting PCR product was detected by agarose gel electrophoresis, and the results were observed in the gel imaging system.
RFQ-PCR reaction system (25 μL) (primers same as those in conventional PCR): TB Green Premix Dimer Eraser (2X) (12.5 μL) (Takara Biomedical Technology Co., Ltd. Beijing. China), ddH2O (10 μL), primers (primers ibid) (10 μM, 0.75 μL), template (1 × 106 copies/μL−1 × 10° copies/μL plasmid, 1 μL). The reaction conditions: pre-denaturation at 95℃ for 30 s, denaturation at 95℃ for 5 s, annealing at 55℃ for 30 s, extension at 72℃ for 30 s, 40 cycles.
The PCR and RFQ-PCR methods used in this experiment have been repeatedly verified by our laboratory.
Clinical Sample Detection
The infected ducks came from 5 duck farms in northern China and showed symptoms of hair loss, growth retardation and decreased feeding intake, and were clinically diagnosed as suspected duck circovirus disease. The 40 samples of bursa of diseased ducks were taken as disease material, and the virus nucleic acid was extracted according to the instructions of DNA/RNA nucleic acid extraction kit (Tiangen Biotech Co., Ltd. Beijing, China). RAA-LFD, RFQ-PCR and conventional PCR were used for test, and the coincidence rates of the 3 methods were compared.
RESULTS AND DISCUSSION
Optimization of RAA-LFD Reaction Conditions
The brightness of detection line obviously changed with temperature, and was clearer at 37℃ (Figure 1A). With the extension of reaction time at 37℃, the detection line became clear gradually, and an obvious detection line could be seen at 20 min (Figure 1B). The RAA-LFD reaction was carried out at 37℃ for 20 min, and the detection line did not show color when the primer concentration was 625 nmol/L. With the increase of primer concentration, the detection line became brighter and an obvious red line could be seen at 1,250 nmol/L, so 1,250 nmol/L was chosen as the lowest primer concentration (Figure 1C). Finally, the optimal reaction condition of RAA-LFD was determined as follows: primer concentration (1,250 nmol/L), reaction temperature (37℃), reaction time (20 min).
Figure 1.
Optimization of reaction conditions for RAA-LFD detection of DuCV. (A) Optimization of RAA-LFD reaction temperature. (B) Optimization of RAA-LFD reaction time. (C) Optimization of RAA-LFD primer concentration. Abbreviations: DuCV, duck circovirus; RAA-LFD, recombinase-aided amplification-lateral flow dipstick.
RAA-LFD can be used to amplify a large number of target fragments in a short time, and it only takes 20 min for the reaction products to reach detectable levels. Compared with RFQ-PCR, it shortens detection time and realizes rapid nucleic acid amplification test under nonlaboratory conditions. At the same time, RAA has no strict requirements for temperature, reaction can be carried out at 35 to 41℃, and there is no need to buy thermocyclers or other expensive experimental equipment, which is more cost-effective compared with both LAMP and PCR.
Specificity Test of RAA-LFD in the Detection of DuCV
Two red lines that meet the positive criteria could be observed on the dipstick only for the RAA amplification products with DuCV as template, and only one quality control line appeared on the dipstick for other viruses. It was proved that the RAA-LFD detection method had good specificity in the detection of DuCV and there was no cross reaction with other avian viruses.
Comparison of Sensitivity of Three Methods for Detecting DuCV
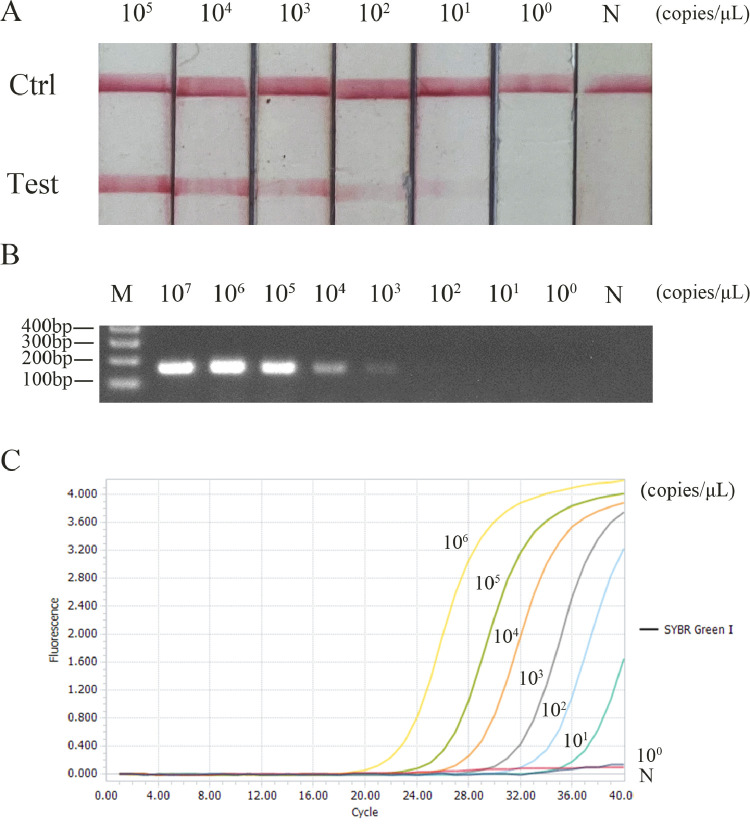
After being diluted in a certain proportion, the plasmid was detected by RAA-LFD, conventional PCR and RFQ-PCR, respectively. The results showed that the LDL of DuCV was 102 copies/μL for RAA-LFD (Figure 2A), 104 copies/μL for conventional PCR (Figure 2B) and 10 copies/μL for RFQ-PCR (Figure 2C). The Sensitivity of RAA-LFD can reach 102 copies/μL, which is 100 times higher than that of conventional PCR.
Figure 2.
Comparison of sensitivity of three methods for detecting DuCV. (A) Sensitivity of RAA-LFD in detecting DuCV. (B) Sensitivity of PCR in detecting DuCV. (C) Sensitivity of RFQ-PCR in detecting DuCV. Abbreviations: DuCV, duck circovirus; M, marker; N, negative control; PCR, polymerase chain reaction; RAA-LFD, recombinase-aided amplification-lateral flow dipstick.
Clinical Sample Detection
RAA-LFD, RFQ-PCR and conventional PCR were used to detect the bursa of Fabricius in 40 ducks suspected to be infected with DuCV. The results showed that 31 samples were positive and 9 samples were negative by RAA-LFD method. Conventional PCR method detected 28 positive strains and 12 negative strains. Using PCR as the benchmark, the clinical detection coincidence rate of RAA-LFD detection method reached 92.50%. There were 33 positive strains and 7 negative strains detected by RFQ-PCR. Using RFQ-PCR as the benchmark, the clinical detection coincidence rate of RAA-LFD detection method reached 95.00%.
In this study, the conserved gene (rep gene) of DuCV was used as the target gene, and specific primers and probes were designed for RAA reaction. Using RAA combined with LFD, the naked-eye observation of amplification products could be completed in 2 to 3 min, and a rapid, simple and visual RAA-LFD detection method was established. RAA-LFD is expected to become a new means of pathogen detection to be widely used in veterinary hospitals and farms at grassroots level, which is of great significance for the prevention and control of animal diseases.
Acknowledgments
ACKNOWLEDGMENTS
This work was supported by Hebei Province Agricultural Quality Development Key Generic Technology Targeted Project (21326617D), Key R&D Plan of Science and Technology Department of Hebei Province (20326621D) from Science and Technology Department, Hebei Province, China, and Open Fund of State Key Laboratory of Veterinary Etiological Biology (BYSWX2021KFKT03) from Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China
We acknowledge funding by Hebei Province Agricultural Quality Development Key Generic Technology Targeted Project (21326617D), Key R&D Plan of Science and Technology Department of Hebei Province (20326621D) from Science and Technology Department, Hebei Province, China, and Open Fund of State Key Laboratory of Veterinary Etiological Biology (BYSWX2021KFKT03) from Lanzhou Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
DISCLOSURES
The authors declare that they have no competing interests, the manuscript has not been published or submitted to other journals previously.
REFERENCES
- Bai B., Zhang W.X., He W.T., Zhao J.J., Gong F.P., Wang G.P., Yuan J.F. Epidemiological investigation of duck circovirus in Guangdong from 2018 to 2020. Chin. J. Anim. Infect. Dis. 2021;29:80–84. [Google Scholar]
- Fan X.X., Lin L., Gang Y.Y., Liu Y.T., Liu C.J., wang Q.H., Dong Y.Q., Wang S.J., Chi T.Y., Song F.F., Sun C.Y., Wang Y.L., Ha D.C., Zhao Y., Bao J.Y., Wu X.D., Wang Z.L. Front. Microbiol. 2020;11:1696. doi: 10.3389/fmicb.2020.01696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattermann K, Schmitt C, Soike D, Mankertz A. Cloning and sequencing of duck circovirus (DuCV) Arch Virol. 2003;148:2471–2480. doi: 10.1007/s00705-003-0181-y. [DOI] [PubMed] [Google Scholar]
- Li X.N., Shen X.X., Li M.H., Qi J.J., Wang R.H., Duan Q.X., Zhang R.Q., Fan T., Bai X.D., Fan G.H. Applicability of duplex real time and lateral flow strip reverse-transcription recombinase aided amplification assays for the detection of Enterovirus 71 and Coxsackievirus A16. Virol. 2019;16:166–176. doi: 10.1186/s12985-019-1264-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L.F., Li Z.B., Si X.K. Establishment and preliminary application of real-time fluorescence quantitative PCR for detection of duck circovirus. Chin. Poult. 2017;39:15–19. [Google Scholar]
- Wang W.J., Li X.Y., Zhang P., Liu J.R., Yao S.S., Wang C.G., Zhang T. Establishment of visual detection method for avian influenza RT-RAA lateral flow dipstick. Chin. J. Vet. Med. 2020;40:2159–2163. [Google Scholar]
- Wu Z.C., Xia X.J., Li H.R., Jiang S.J., Wang X. Tandem repeat sequence of duck circovirus serves as downstream sequence element to regulate viral gene expression. Vet. Microbiol. 2019;239 doi: 10.1016/j.vetmic.2019.108496. [DOI] [PubMed] [Google Scholar]
- Zhao G.Y., Xie Z.X., Xie L.J., Pang Y.S., Deng X.W., Liu J.B., Fan Q. Establishment of LAMP visual detection method for duck circovirus. Chin. J. Anim. Quarant. 2012;29:24–26. [Google Scholar]
- Zou J.F., Liu S.N., Lei Z., Sun H., Wei Z., Wang X., Jiang S.J. Preparation and application of duck circovirus nucleic acid probe. Chin. J. Vet. Med. 2011;31:1578–1581. [Google Scholar]