Abstract
Objectives Previous studies have suggested that SARS-CoV-2 viral load, measured on upper respiratory tract samples at presentation to hospital using PCR Cycle threshold (Ct) value, has prognostic utility. However, these studies have not comprehensively adjusted for factors known to be intimately related to viral load. We aimed to evaluate the association between Ct value at admission and patient outcome whilst adjusting carefully for covariates.
Methods We evaluated the association between Ct value at presentation and the outcomes of ICU admission and death, in patients hospitalised during the first wave of the pandemic in Southampton, UK. We adjusted for covariates including age, duration of illness and antibody sero-status, measured by neutralisation assay.
Results 185 patients were analysed, with a median [IQR] Ct value of 27.9 [22.6–32.1]. On univariate analysis the Ct value at presentation was associated with the risk of both ICU admission and death. In addition, Ct value significantly differed according to age, the duration of illness at presentation and antibody sero-status. On multivariate analysis, Ct value was independently associated with risk of death (aOR 0.84, 95% CI 0.72–0.96; p = 0.011) but not ICU admission (aOR 1.04, 95% CI 0.93–1.16; p = 0.507). Neutralising antibody status at presentation was not associated with mortality or ICU admission (aOR 10.62, 95% CI 0.47–889; p = 0.199 and aOR 0.46, 95% CI 0.10–2.00; p = 0.302, respectively).
Conclusions SARS-CoV-2 Ct value on admission to hospital was independently associated with mortality, when comprehensively adjusting for other factors and could be used for risk stratification.
Keywords: Viral load, SARS-CoV-2, COVID-19, Prognosis, Antibodies
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in 2019 as a novel respiratory pathogen.1 In the 13 months since, it has spread around the world causing over 100 million confirmed cases and over 2.5 million deaths.2 The spectrum of disease associated with COVID-19 ranges from asymptomatic carriage to life threatening pneumonia and multi-organ failure. A number of socio-demographic factors, patient co-morbidities, clinical signs, and laboratory results have been identified as risk factors for severe disease and subsequent mortality.3, 4, 5, 6, 7 The utility of early risk stratification of patients may include the optimisation of resource allocation for clinical management. As treatments that are more effective become available, risk stratification may aid targeted therapeutic interventions.
The use of upper respiratory tract viral load as a prognostic marker in patients admitted to hospital has previously been explored for respiratory viruses such as influenza and RSV, with studies showing conflicting results.8, 9, 10 Studies undertaken during the first wave of the SARS-CoV-2 pandemic have suggested that the magnitude of viral load at presentation may be associated with clinical outcomes in hospitalised patients, leading to consideration for its use as a prognostic tool in this setting.11 , 12 However, several confounding patient factors have been identified which strongly influence SARS-CoV-2 viral load at the point of admission and have not always been considered or comprehensively controlled for in these early studies. These factors include patient age, duration of illness, and antibody sero-status at the point of presentation.13, 14, 15
We hypothesised that the association between viral load and clinical outcome seen in early studies may be dependant on other factors rather than independently associated. The aim of the study was therefore to evaluate the association between viral load, as measured by real-time PCR cycle threshold (Ct) value, and outcome whilst carefully adjusting for covariates including age, duration of illness and antibody sero-status.
Methods
Setting, study design and participants
Study data were collated from all SARS-CoV-2 positive patients in the point-of-care testing (POCT) arm of the CoV-19POC trial, a single centre study evaluating the impact of molecular POCT for SARS-CoV-2 in hospital.16 Inclusion criteria for these patients required that they were adults (>18 years) in the Emergency Department (ED) or Acute Medical Unit (AMU) with suspected COVID-19 who were recruited within 24 h of admission. The study was prospectively registered and approved, full details including inclusion/ exclusion criteria are available in the protocol.17
A combined nasal and pharyngeal swab was obtained from all participants and tested on the QIAstat-Dx PCR platform using the Respiratory SARS-CoV-2 Panel.18 , 19 The QIAstat-Dx analyser uses multiplexed real-time PCR. The SARS-CoV-2 gene targets in the panel are the ORF1b and E gene, with detection of either gene reported as a positive result. The lowest Ct value (i.e. highest viral load) for either target detected is displayed on the analyser.
All patients who were PCR positive after recruitment and remained in hospital were approached for a blood test within 24 h. Serum was separated on the day of collection and frozen at −80 °C.
Measurement of serum neutralising antibodies
Measurement of antibodies was performed at the Animal and Plant Health Agency (APHA), Weybridge, Surrey. Human blood samples were centrifuged, and the serum fraction was transferred to fresh tubes within a medical safety cabinet and inactivated by heat at 56 °C for 30 min. The virus neutralisation test was adapted from Loeffen, et al.20 In 96 well plate format, in quadruplicate, two-fold dilutions were made of the serum sample in virus growth media. 100 TCID50 of SARS-CoV-2 virus (2019-nCoV/Italy - INMI 1 [GISAID ID EPI_ISL_410,545]) was added to each well. Plates were sealed and incubated at 37 °C with 5% CO2 for 1 hour. Back titration of input virus was performed for each aliquot used by two-fold serial dilution in virus growth media. A negative control plate was also included. After incubation, a suspension of 5 × 104 Vero E6 cells were added to each well. Plates were sealed and incubated for 5 days at 37 °C with 5% CO2. Each well was visualised for cytopathic effect under a microscope. The titre of the virus and the samples were calculated using Spearman-Karber method and displayed as inhibitory concentration 50% (IC50). The limit of detection was 2.82 IC50 with all titre above this being considered positive.
Comparison of viral load (Ct value)
Cycle threshold (Ct) value is derived from the number of amplification cycles needed during real time PCR for sufficient gene amplification to produce a probe-based fluorescent signal that crosses a predefined threshold. The Ct value is inversely correlated to the quantitative viral load i.e. a low Ct value corresponds with a high viral load. For this study, viral load (Ct values) was categorised into three groups: high (Ct value ≤20), moderate (Ct value of ≥20.1 to 29.9) and low (Ct value of ≥30). These Ct value categories are similar to categories used in published studies.10 , 11
Data collection and outcome measures
Baseline data were collected prospectively at enrolment and outcome data collected retrospectively from patient case-notes and hospital information systems.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 9.0.1 (GraphPad Software, La Jolla, CA, USA), Python version 3.7 and R version 4.0.3. We compared baseline characteristics and outcomes of hospitalised patients with SARS-CoV-2 who had high (Ct <20), medium (Ct 20.1–29.9), and low (Ct >30) initial viral loads. For categorical variables difference in proportions were analysed using Fisher's exact test or Chi-squared test as appropriate. Continuous variables were analysed using Mann-Whitney U or Kruskall-Wallis test and are expressed as median and interquartile range [IQR]. Correlations between two continuous variables were analysed using a two-tail Spearman's rank correlation coefficient. A two-sided p-value of <0.05 was used for defining significance. 95% confidence intervals (CI) were calculated using the Clopper-Pearson exact method. Missing data is ≤3% unless otherwise stated in all analyses.
Multivariate logistic regression modelling was performed for outcomes of in-hospital mortality and ICU admission, considering the pairwise interactions between Ct values, symptom duration and age in addition to other variables. For pairwise interactions, Ct values, age and symptom duration were centred around their means before computing the interaction terms.21 The covariates for all hospitalised COVID-19 patients were: age, BAME ethnicity, cardiovascular disease, asthma, COPD, chronic kidney disease, diabetes, National Early Warning Score 2, symptoms duration prior to admission, the presence of infiltrates on chest x-ray, white cell count, CRP, lymphocyte count, creatinine, urea, LDH, D-dimer, platelets, ferritin, troponin, and Ct value. Most of the variables considered for investigation were complete or near complete (>96% data completeness) with the exception of LDH (62%), D-dimer (74%) and troponin (75%). Missing data were imputed using K-nearest neighbour using the KNNImpute module of scikit-learn, Python 3.7. Multivariate logistic regression modelling and the calculation of adjusted odd ratios (aOR), their 95% confidence interval and p-values, were performed using R-studio version 1.4.1103. Interaction probing of significant interaction terms was performed using the interactions package of R. Additionally, Cox proportional hazard model to investigate cumulative risks of in-hospital admission by Ct values adjusted for age and other variables was performed using lifelines library of Python 3.7.
Neutralising antibody sero-status results were available for a subset of 99 patients. This subgroup was analysed using multivariate logistic regression, also testing for the three pairs of interaction of Ct-values, duration of illness in days and age. As the number of subjects of this sub-cohort was small, variable selection was performed based on univariable logistic regression to pick the top ten significant variables for multivariate logistic regression analysis in R-studio version 1.4.1103.
Results
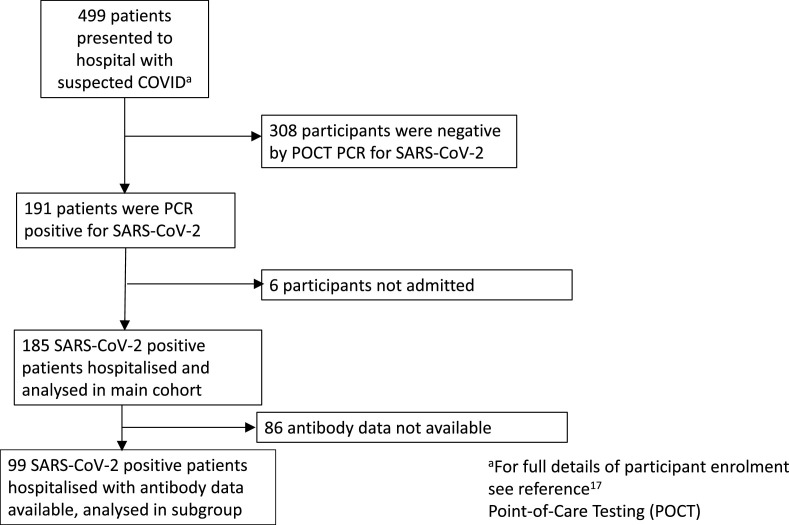
Patients were enroled between 20th March and 29th April 2020. We identified 185 patients hospitalised with COVID-19 (Fig. 1 ). These patients had a median [IQR] SARS-CoV-2 Ct value of 27.9 [22.6–32.1]. Of these 185 patients, 28 (15%) had a low Ct value (high viral load), 79 (43%) of 185 had a moderate Ct value (moderate viral load) and 78 (42%) of 185 had a high Ct value (low viral load). Baseline characteristics and clinical outcome are shown in Table 1 for all patients and according to Ct value category.
Fig. 1.
Flow of participants through the study.
Table 1.
Baseline characteristics, clinical, laboratory features and outcomes for all COVID-19 patients at admission combined (n = 185) and by Ct value (viral load).
| All patientsN = 185 | Ct Value (viral load) | p value a | p value b | ||||
| ≤20n = 28 | 20.1–29.9n = 79 | ≥30n = 78 | |||||
| Demographics | |||||||
| Age (years) | 65 [50–80] | 82 [51–92] | 69 [56–81] | 59 [48–73] | 0.002 | 0.013 | |
| Male | 101 (55%) | 16 (57%) | 39 (49%) | 46 (59%) | 0.461 | 0.839 | |
| Current Smoker | 7 (4%) | 1 (4%) | 2 (3%) | 4 (5%) | 0.694 | >0.999 | |
| Pregnant | 1 (1%) | 0 (0%) | 1 (1%) | 0 (0%) | 0.509 | >0.999 | |
| White Britishc | 129 (73%) | 24 (92%) | 57 (74%) | 48 (66%) | 0.031 | 0.016 | |
| BAMEd | 40 (23%) | 2 (8%) | 18 (24%) | 20 (28%) | 0.111 | 0.046 | |
| Co-morbidities | |||||||
| Hypertensionc | 74 (42%) | 13 (48%) | 36 (47%) | 25 (35%) | 0.260 | 0.529 | |
| Cardiovascular Diseasee | 53 (30%) | 13 (48%) | 20 (26%) | 20 (27%) | 0.076 | 0.038 | |
| Respiratory Disease (any) | 55 (31%) | 13 (50%) | 25 (32%) | 17 (23%) | 0.032 | 0.036 | |
| Asthmae | 30 (17%) | 8 (31%) | 12 (15%) | 10 (14%) | 0.116 | 0.050 | |
| COPD | 24 (13%) | 5 (19%) | 13 (17%) | 6 (8%) | 0.186 | 0.354 | |
| Chronic Kidney Disease | 17 (9%) | 3 (12%) | 10 (13%) | 4 (5%) | 0.269 | 0.720 | |
| Chronic Liver Diseasef | 9 (5%) | 3 (12%) | 2 (3%) | 4 (5%) | 0.197 | 0.129 | |
| Diabetes | 46 (26%) | 8 (30%) | 18 (23%) | 20 (27%) | 0.790 | 0.636 | |
| Active Malignancye | 9 (5%) | 1 (4%) | 3 (4%) | 5 (7%) | 0.682 | >0.999 | |
| Dementia | 23 (13%) | 7 (25%) | 7 (9%) | 9 (12%) | 0.096 | 0.059 | |
| Immunosuppressedc | 10 (6%) | 2 (7%) | 3 (4%) | 5 (7%) | 0.703 | 0.652 | |
| Clinical Features on presentation | |||||||
| Heart rate (beats/min) | 94 [82–109] | 92 [79–114] | 91 [80–109] | 98 [86–109] | 0.451 | 0.909 | |
| Respiratory Rate (breaths/min) | 25 [21–30] | 24 [20–28] | 26 [22–31] | 26 [21–30] | 0.462 | 0.223 | |
| Oxygen Saturations (%) | 96 [92–97] | 96 [93–98] | 96 [91–97] | 95 [93–96] | 0.390 | 0.172 | |
| Systolic Blood Pressure (mmHg) | 130 [120–145] | 131 [113–146] | 128 [117–149] | 134 [124–143] | 0.634 | 0.774 | |
| Temperature (⁰C) | 37.1 [36.6–38.1] | 37.0 [36.6–37.9] | 37.0 [36.5–38.1] | 37.2 [36.6–38.3] | 0.339 | 0.623 | |
| Temperature ≥37.8⁰C | 62 (34%) | 7 (25%) | 26 (35%) | 29 (37%) | 0.505 | 0.289 | |
| On supplemental oxygen | 82 (44%) | 12 (43%) | 35 (44%) | 35 (45%) | 0.983 | >0.999 | |
| NEWS2 Score | 6 [3–8] | 6 [3–8] | 5 [3–7] | 6 [3–8] | 0.892 | 0.709 | |
| Duration of symptoms (days) | 6 [2–10] | 2 [0–7] | 6 [2–8] | 7 [3–10] | 0.005 | 0.008 | |
| Infiltrates on CXR | 146 (79%) | 20 (71%) | 64 (82%) | 62 (80%) | 0.492 | 0.310 | |
| Laboratory/POCT results on presentation | |||||||
| White blood cell count (109/L) | 7.2 [5.4–10.4] | 7.3 [5.8–14.0] | 7.4 [5.6–10.6] | 6.9 [5.1–10.0] | 0.422 | 0.247 | |
| CRP (mg/L) | 91 [44–152] | 109 [51–148] | 89 [52–152] | 55 [19–176] | 0.349 | 0.441 | |
| Lymphocyte Count (109/L) | 0.9 [0.7–1.2] | 0.9 [0.5–1.6] | 0.9 [0.6–1.2] | 1.0 [0.7–1.3] | 0.540 | 0.957 | |
| Creatinine (µmol/L) | 82 [64–111] | 100 [80–163] | 84 [60–113] | 77 [63–99] | 0.010 | 0.004 | |
| Urea (mmol/L) | 6.5 [4.8–10.9] | 11.5 [6.4–16.3] | 6.8 [5.1–10.4] | 5.7 [4.4–8.8] | 0.004 | 0.004 | |
| Neutrophil Count (109/L) | 5.5 [4–8.2] | 5.7 [4.3–11] | 5.6 [4.1–8.3] | 5.3 [3.9–7.6] | 0.448 | 0.366 | |
| LDHg (U/L) | 731 [518–998] | 505 [411–777] | 698 [503–1015] | 781 [597–1005] | 0.038 | 0.023 | |
| D-Dimerh (ng/L) | 478 [341–873] | 417 [305–953] | 573 [325–1023] | 452 [366–579] | 0.390 | 0.716 | |
| Platelets (109/L) | 230 [174–292] | 223 [154–272] | 230 [171–279] | 233 [178–321] | 0.276 | 0.266 | |
| Ferritini (µg/L) | 518 [239–1197] | 232 [62–1268] | 479 [183–1361] | 604 [326–1234] | 0.091 | 0.070 | |
| Troponinj (ng/ml) | 13 [5–45] | 49 [5–560] | 16 [7–46] | 11 [4–21] | 0.033 | 0.068 | |
| SARS-CoV-2 neutralising antibody positivek | 56 (57%) | 1 (8%) | 20 (49%) | 35 (78%) | <0.0001 | 0.0001 | |
| Ct Value (viral load) | 27.9 [22.6–32.1] | 16.7 [15.6–18.8] | 24.9 [22.6–27.3] | 32.5 [31.8–33.7] | <0.0001 | <0.0001 | |
| Outcome | |||||||
| Length of stayl,* (hours) | 194.9 [98.17–303.3] | 149.8 [11.33–313.4] | 220.4 [99.61–362] | 182.7 [106.1–264] | 0.299 | 0.248 | |
| Received supplemental oxygen | 149 (81%) | 18 (64%) | 66 (84%) | 65 (83%) | 0.062 | 0.035 | |
| Duration of received oxygenm, (hours) | 19.22 [8.00–64.01] | 12 [5.02–30.26] | 20.36 [9.46–113.50] | 19.13 [8.50–56.08] | 0.167 | 0.119 | |
| Received NIV | 36 (20%) | 4 (14%) | 19 (24%) | 13 (17%) | 0.381 | 0.607 | |
| Duration of NIVn, (hours) | 22.38 [9.28–42.25] | 27.83 [15.75–36.41] | 21.67 [5.98–54] | 25 [12.55–51.04] | 0.473 | 0.875 | |
| Received Intubation and Ventilation | 23 (12%) | 0 (0%) | 16 (20%) | 7 (9%) | 0.010 | 0.028 | |
| Duration of Intubation and Ventilationo, (hours) | 206.2 [113.7–482.2] | 0 [0–0] | 198.8 [119.2–487.3] | 229.8[88–482.2] | 0.922 | N/A | |
| Admitted to ICU | 36 (20%) | 2 (7%) | 22 (28%) | 12 (15%) | 0.029 | 0.117 | |
| Died within 30 days of admission | 41 (22%) | 14 (50%) | 21 (27%) | 6 (8%) | <0.0001 | 0.0004 | |
| Data are presented as number (%) and median [Inter-quartile range]. Abbreviations: BAME, Black Asian and minority ethnic; Ct, real-time PCR cycle threshold (a low Ct value represents a high viral load and vice versa);NEWS2, National Early Warning Score 2; POCT, Point-of-Care Test; CXR, Chest X-ray; COPD, Chronic Obstructive Pulmonary Disease; CRP, C Reactive Protein; LDH, Lactate dehydrogenase; NIV, non-invasive ventilation; ICU, intensive care unit. a: Across all Ct value groups (Kruskal–Wallis test, Chi Square). b: Between high viral load group (Ct ≤ 20) and other groups combined (Fisher's Exact, Mann Whitney U). C = 176; d = 174, e = 178, f = 177, g = 115, h = 100 (excluding d-dimers <230 or >5000), i = 143, j = 138, k = 99,l = 144, m = 149, n = 36, o = 23, *excluding those who died. | |||||||
The median [IQR] age of all patients was 65 [50–80] years and differed by Ct value; 82 [51–92] in those with a low Ct value, 69 [56–81] in those with a moderate Ct value and 59 [48–73] in those with a high Ct value, p = 0.002. Ct value was inversely correlated with age, (Spearman's rank correlation, r=−0.26; p = 0.0003). A higher proportion of patients in the low Ct value group were of White British ethnicity compared with the moderate and high Ct value groups; 24 (92%) of 26 versus 57 (74%) of 73 and 48 (66%) of 73 respectively, p = 0.031). The median [IQR] duration of symptoms prior to hospitalisation was 6 [2–10] days for all patients and differed by Ct value; 2 days [0–7], 6 days [2–8] and 7 days [3–10] for low, medium and high Ct value groups respectively, p = 0.005. Ct value (i.e. the inverse of viral load) was positively correlated with duration of illness, (Spearman's rank correlation, r=+0.25, p = 0.0005). Renal function, troponin and lactate dehydrogenase (LDH) on admission were significantly different across the three groups with median urea, creatinine, troponin and LDH levels being higher in those with lower Ct values (Table 1). Mortality was 41 (22%) of 185 overall. 14 (50%) of 28 patients died in the low Ct value group compared with 21 (27%) of 79 in the medium Ct value group and 6 (8%) of 78 in the high Ct value group, p<0.0001. 36 (20%) of 185 patients were admitted to ICU overall. 2 (7%) of 28 in the low Ct value group were admitted to ICU versus 22(28%) of 79 in the medium Ct value group and 12 (15%) of 78 in the high Ct value group, p = 0.029. No patient received any SARS-CoV-2 vaccine, immunomodulatory COVID-19 therapy, or antiviral for COVID-19 prior to collection of the nose and throat swabs.
Neutralising antibody subgroup data
99 (54%) of 185 patients consented to venesection and had a serum sample taken. 56 (57%) of 99 patients had detectable neutralising antibodies against SARS-CoV-2. The baseline characteristics of patients who had serum neutralising antibodies measured are summarised in Table 2 .
Table 2.
Baseline demographic, clinical and laboratory features and outcomes for all COVID-19 patients with known SARS-CoV-2 neutralising antibody serological status combined (n = 99) and by antibody status.
| Demographics | All patients n = 99 | Antibody Negative n = 43 | Antibody Positive n = 56 | Difference(CI 95%)a | p valuea |
| Age (years) | 61 [48–74] | 68 [52–82] | 58 [45–70] | 10 (4 to 19) | 0.002 |
| Male (%) | 60 (61%) | 27 (63%) | 33 (59%) | −4% (−23% to 16%) | 0.836 |
| Current Smokerb | 4 (5%) | 3 (9%) | 1 (2%) | −7% (−16% to 2%) | 0.295 |
| Pregnant | 0 (0%) | 0 (0%) | 0 (0%) | N/A | N/A |
| White British | 63 (66%) | 34 (81%) | 29 (54%) | −27% (−46% to −8%) | 0.009 |
| BAME | 27 (28%) | 8 (19%) | 19 (35%) | 16% (−2% to 34%) | 0.110 |
| Co-morbidities | |||||
| Hypertensionc | 40 (42%) | 21 (50%) | 19 (36%) | −14% (−34% to 6%) | 0.210 |
| Cardiovascular Diseasec | 21 (22%) | 13 (30%) | 8 (15%) | −15% (−32% to 2%) | 0.135 |
| Respiratory Disease (any) | 25 (26%) | 18 (42%) | 7 (13%) | −44% (−64% to −24%) | 0.002 |
| Asthma | 16 (17%) | 10 (23%) | 6 (11%) | −12% (−27% to 3%) | 0.169 |
| COPD | 8 (8%) | 7 (16%) | 1 (2%) | −14% (−26% to −3%) | 0.021 |
| Chronic Kidney Disease | 7 (7%) | 4 (9%) | 3 (6%) | −4% (−14% to 7%) | 0.697 |
| Chronic Liver Disease | 3 (3%) | 1 (2%) | 2 (4%) | 1% (−6% to 8%) | >0.999 |
| Diabetes | 23 (24%) | 14 (33%) | 9 (17%) | −16% (−33% to 16%) | 0.095 |
| Active Malignancy | 7 (7%) | 3 (7%) | 4 (8%) | 1% (−10% to 11%) | >0.999 |
| Dementiac | 10 (11%) | 8 (19%) | 2 (4%) | −15% (−28% to −3%) | 0.021 |
| Immunosuppressedc | 5 (5%) | 1 (2%) | 4 (8%) | 5% (−4% to 14%) | 0.379 |
| Clinical Features on presentation | |||||
| Heart rate (beats/min) | 92 [82–109] | 90 [80–109] | 99 [85–109] | 9 (−2 to 14) | 0.141 |
| Respiratory Rate (breaths/min) | 25 [22–32] | 24 [20–30] | 26 [22–32] | 2 (−1 to 5) | 0.135 |
| Oxygen saturations (%) | 96 [92–97] | 96 [93–98] | 95 [92–96] | −1 (−2 to 0) | 0.143 |
| Systolic Blood Pressure (mmHg) | 133 [120–145] | 134 [122–146] | 133 [120–143] | −2 (−9 to 5) | 0.636 |
| Temperature (⁰C) | 37.3 [36.6–38.3] | 37.3 [36.7–38.2] | 37.3 [36.6–38.4] | 0.1 (−0.3 to 0.5) | 0.826 |
| Temperature ≥37.8⁰C | 38 (39%) | 15 (36%) | 23 (42%) | 6% (−14% to 26%) | 0.675 |
| On supplemental oxygen | 44 (44%) | 18 (42%) | 26 (46%) | −5% (−15% to 24%) | 0.687 |
| NEWS2 Score | 6 [4–7] | 5 [2–6] | 6 [4–8] | 1 (0 to 2) | 0.084 |
| Duration of symptoms (days) | 7 [4–10] | 6 [2–7] | 9 [6–11] | 3 (1 to 5) | 0.001 |
| Infiltrates on CXR | 86 (88%) | 36 (86%) | 50 (89%) | 4% (−10% to 17%) | 0.757 |
| Laboratory/POCT results on presentation | |||||
| White blood cell count (109/L) | 7.4 [5.5–11.2] | 6.3 [4.9–9.7] | 8.2 [5.7–11.4] | 1.9 (−0.2 to 2.5) | 0.139 |
| CRP (mg/L) | 108 [51–164] | 80 [19–174] | 124 [63–164] | 44 (−4 to 64) | 0.085 |
| Lymphocyte Count (109/L) | 1.0 [0.8–1.2] | 1.0 [0.8–1.4] | 1.0 [0.7–1.2] | −0.1 (−0.2 to 0.1) | 0.604 |
| Creatinine (µmol/L) | 85 [65–102] | 88 [76–121] | 81 [61–98] | −6.5 (−24 to 2) | 0.124 |
| Urea (mmol/L) | 6.3 [4.7–8.9] | 6.7 [5.0–11.2] | 5.8 [4.4–8.1] | −0.9 (−2.5 to 0.1) | 0.078 |
| Neutrophil Count (109/L) | 5.6 [3.9–8.6] | 4.5 [3.5–7.6] | 6.6 [4.4–9.7] | 2.1 (0.3 to 2.7) | 0.010 |
| LDHd (U/L) | 756 [534–1053] | 567 [429–759] | 927 [634–1173] | 360 (146 to 476) | 0.0001 |
| D-Dimere (ng/L) | 473 [334–927] | 443 [348–808] | 496 [327–973] | 53 (−125 to 170) | 0.876 |
| Platelets (109/L) | 241 [180–293] | 203 [164–269] | 253 [192–327] | 51 (7 to 76) | 0.019 |
| Ferritinf (µg/L) | 628 [287–1424] | 290 [121–621] | 1031 [537–1838] | 741 (291 to 911) | <0.0001 |
| Troponing (ng/ml) | 11 [5–26] | 13 [5–84] | 9 [5–19] | −4 (−27 to 1) | 0.101 |
| Ct Value (viral load) | 29.2 [23.2–32.3] | 24 [19.4–29.6] | 31.8 [28–33.3] | 7.8 (3.5 to 8.2) | <0.0001 |
| Outcome | |||||
| Length of stayh,* (hours) | 216.4 [120.6–339.8] | 262.4 [150.8–381.7] | 197.7 [100.1–296.2] | −64.74 (−133.7 to 31.85) | 0.204 |
| Received supplemental oxygen | 83 (84%) | 35 (81%) | 48 (86%) | 4% (−10% to 19%) | 0.592 |
| Duration of received oxygenh, (hours) | 23 [13–117.6] | 19.73 [12–112.1] | 30.43 [13.7–123.4] | 10.7 [−6 to 31] | 0.439 |
| Received NIV | 26 (26%) | 6 (14%) | 20 (36%) | 22% (4% to 39%) | 0.021 |
| Duration of received NIVi, (hours) | 32.63 [12.75–58.3] | 67.12 [28.24–124.5] | 24.43 [9.5–42.25] | −42.69 [−90.55 to 0.48] | 0.061 |
| Received Intubation and Ventilation | 19 (19%) | 7 (16%) | 12 (21%) | 5% (−11% to 21%) | 0.611 |
| Duration of Intubation and Ventilation, (hours)j | 206.2 [135.8–482.2] | 175.8 [135.8–424.9] | 285.8 [124.1–589.6] | 110 (−155.5 to 350.8) | 0.592 |
| Admitted to ICU | 30 (30%) | 10 (23%) | 20 (36%) | 12% (−6% to 31%) | 0.195 |
| Died within 30 days of admission | 16 (16%) | 13 (30%) | 3 (5%) | −25% (−40% to −10%) | 0.002 |
| Data are presented as number (%) and median [Inter-quartile range]. Abbreviations: BAME, Black Asian and minority ethnic; Ct, real-time PCR cycle threshold (a low Ct value represents a high viral load and vice versa); NEWS2, National Early Warning Score 2; CRP, C reactive protein; POCT, Point-of-Care Test; CXR, Chest X-ray; COPD, Chronic Obstructive Pulmonary Disease; LDH, Lactate dehydrogenase; NIV, Non-Invasive ventilation; ICU, intensive care unit. a: Comparing antibody positive and negative groups groups, (Fisher Exact, Mann Whitney U); b = 87; c = 95; d = 68; e = 60 (excluding d-dimers <230 or >5000); f = 80; g = 78; h = 83; i = 26; j = 19, *excluding those who died. | |||||
The median [IQR] age for those with neutralising antibodies detected was 58 [45–70] years compared to 68 [52–82] years in those without detectable antibodies (difference of 10 years, 95% CI 4–19; p = 0.002). A higher proportion of patients in the antibody negative group were of White British ethnicity compared with the antibody positive group, 34 (81%) of 96 versus 29 (54%) 96 (difference of −27%, 95%CI −46% to −8%; p = 0.009). There was a higher proportion of patients with underlying co-morbidity in the antibody negative group, included those with any pre-existing chronic respiratory disease, COPD and dementia (Table 2). Median [IQR] duration of symptoms prior to admission was longer in those with detectable neutralising antibodies, 9 [6–11] versus 6 [2–7] days (difference of 3 days, 95% CI 1–5; p = 0.001). The median neutrophil count on admission was significantly higher in the antibody positive group compared with the negative, 6.6 [4.4–9.7] vs 4.5 [3.5–7.6] (difference of 2.1, 95% CI 0.3 to 2.7; p = 0.010). Median [IQR] Ct value on admission was higher (i.e. viral load lower) in those with neutralising antibody detected, 31.8 [28–33.3] compared with those without, 24 [19.4–29.4] (difference of 7.8, 95% CI 3.5 to 8.2; p<0.0001). Mortality was lower in those with detectable neutralising antibodies, 3 (5%) of 56 compared to those without detectable antibody 13 (30%) of 43 (difference of −25%, 95% CI −40% to −10%; p = 0.002).
Multivariate analysis
Multiple logistic regression analysis for in-hospital mortality with 21 dependant covariates and three pairs of interaction terms (Table 3 ) showed that low Ct value (high viral load) at admission was associated with higher in-hospital mortality (adjusted odds ratio (aOR) of 0.84, 95% CI 0.72–0.96; p = 0.011). Longer duration of symptoms prior to admission was associated with lower in-hospital mortality (aOR 0.71, 95% CI 0.51–0.89; p = 0.011). Increasing age (aOR 1.16, 95% CI 1.07–1.28; p = 0.001) and the presence of cardiovascular and diabetes as co-morbidities were also associated with higher in-patient mortality (aOR 4.80, 95%CI 1.44–18.52, p = 0.014 and aOR 4.93, 95%CI 1.22–23.29; p = 0.031, respectively). The results also indicated a significant interaction of the two variables: symptom duration and age (aOR 1.02, 95% CI 1.01–1.04; p = 0.001), for in-hospital mortality outcome. The interaction probing results (Fig. S1a and b, supplementary appendix) demonstrated that symptom duration had a significantly different relationship to in-hospital mortality risk depending on the age of the patient, with younger patients having less probability of dying with increased duration of symptoms whilst the older age group has an increased risk of in-hospital mortality with longer symptom duration.
Table 3.
Logistic regression analysis for COVID-19 patients (n = 185), for in-hospital mortality and ICU admission.
| In-hospital mortality | ICU admission | |||
| aOR (95% CI) | p-value | aOR (95% CI) | p-value | |
| Age (years) | 1.16 (1.07–1.28) | 0.001 | 0.96 (0.92–1.00) | 0.042 |
| BAME Ethnicity | 0.64 (0.04–6.05) | 0.718 | 1.65 (0.48–5.62) | 0.419 |
| Cardiovascular disease | 4.80 (1.44–18.52) | 0.014 | 1.00 (0.24–3.79) | 0.997 |
| Asthma | 1.08 (0.18–6.23) | 0.929 | 1.58 (0.38–6.00) | 0.512 |
| COPD | 0.08 (0.01–0.44) | 0.008 | 0.87 (0.11–4.86) | 0.879 |
| Chronic Kidney disease | 0.24 (0.03–1.49) | 0.145 | 0.58 (0.01–16.36) | 0.777 |
| Diabetes | 4.93 (1.22–23.29) | 0.031 | 0.53 (0.13–1.90) | 0.344 |
| NEWS2 Score | 1.19 (0.95–1.52) | 0.141 | 1.05 (0.84–1.30) | 0.692 |
| Symptoms duration prior to admission, (days) | 0.71 (0.51–0.89) | 0.011 | 1.06 (0.95–1.18) | 0.301 |
| Infiltrates on CXR | 1.59 (0.37–7.30) | 0.537 | 4.97 (0.66–113.29) | 0.188 |
| White cell count (109/L) | 1.06 (0.89–1.27) | 0.531 | 1.08 (0.93–1.25) | 0.326 |
| CRP (mg/L) | 1.00 (1.00–1.01) | 0.312 | 1.01 (1.00–1.02) | 0.012 |
| Lymphocyte count (109/L) | 0.84 (0.50–1.09) | 0.296 | 0.71 (0.22–1.12) | 0.478 |
| Creatinine (µmol/L) | 1.01 (1.00–1.03) | 0.297 | 1.01 (1.00–1.03) | 0.546 |
| Urea (mmol/L) | 0.94 (0.82–1.07) | 0.367 | 0.97 (0.79–1.12) | 0.774 |
| LDH (U/L) | 1.00 (1.00–1.00) | 0.346 | 1.00 (1.00–1.00) | 0.204 |
| D-dimer (ng/L) | 1.00 (1.00–1.00) | 0.924 | 1.00 (1.00–1.00) | 0.584 |
| Platelets (109/L) | 1.00 (0.99–1.00) | 0.507 | 1.00 (0.99–1.00) | 0.125 |
| Ferritin (µg/L) | 1.00 (1.00–1.00) | 0.731 | 1.00 (1.00–1.00) | 0.176 |
| Troponin (ng/ml) | 1.00 (1.00–1.00) | 0.346 | 1.00 (0.99–1.00) | 0.637 |
| Ct value (viral load) | 0.84 (0.72–0.96) | 0.011 | 1.04 (0.93–1.16) | 0.507 |
| Interaction terms | ||||
| Ct Value x age | 1.00 (1.00–1.01) | 0.325 | 1.01 (1.00–1.01) | 0.040 |
| Symptoms days x age | 1.02 (1.01–1.04) | 0.001 | 1.00 (0.99–1.00) | 0.725 |
| Ct Value x symptom days | 1.01 (0.99–1.03) | 0.498 | 1.00 (0.99–1.02) | 0.729 |
| Data presented as adjusted odds ratios (aOR). Abbreviations: BAME, Black, Asian and minority ethnic; COPD, Chronic Obstructive Pulmonary Disease; NEWS2, National Early Warning Score 2; CXR, Chest X-Ray; CRP, C reactive protein; LDH, Lactate dehydrogenase; Ct, real-time PCR cycle threshold (a low Ct value represents a high viral load and vice versa). | ||||
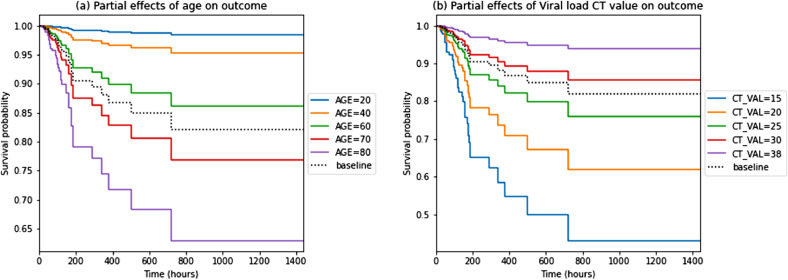
The time-based analysis using Cox proportional hazard model (Fig. S2, Table S1, supplementary appendix) revealed a higher viral load (i.e. lower Ct value) to be independently associated with higher risk of in-hospital mortality (hazard ratio (HR) of 0.89, 95% CI 0.84–0.96; p = 0.001). Additionally, increasing age (HR 1.06, 95% CI 1.02–1.10; p = 0.006), NEWS2 score (HR 1.18, 95%CI 1.02–1.36; p = 0.028), creatinine (HR 1.00, 95% CI 1.00–1.01; p = 0.035) and the presence of cardiovascular co-morbidities (HR 3.38, 95% CI 1.46–7.81; p = 0.004) were all associated with higher in-hospital mortality. Fig. 2 shows partial effects Kaplan-Meier graphs of age and viral load at admission and the probability of survival outcomes, for all hospitalised patients.
Fig. 2.
a and b. Partial effects Kaplan-Meier graphs of age (a) and viral load (b) at admission and the probability of survival outcomes for all hospitalised patients (n = 185).
Multivariate logistic regression for the outcome of ICU admission on the same set of covariates (Table 3) showed that younger age (aOR 0.96, 95% CI 0.92–1.00; p = 0.042) and higher CRP (aOR 1.01, 95%CI 1.00–1.02; p = 0.012) were independently associated with higher risk of ICU admission. The results also indicated a significant interaction of the two covariates; Ct value and age, for ICU admission outcome (aOR 1.01, 95%CI 1.00–1.01; p = 0.040). The interaction probing results (Fig. S1c and d, supplementary appendix) indicated that Ct values had slightly different effects on ICU admission depending on the age of the patients, but the magnitude of the increased risk or decreased risk associated with Ct values was minimal.
In the subset of patients with known neutralising antibody sero-status (n = 99), the presence of antibodies was found not to be significantly protective against in-hospital mortality in both the multivariate logistic regression analysis on covariates listed in Table 4 (aOR 10.62, 95% CI 0.47–889; p = 0.199) and the time-based analysis (HR 0.93, 95% CI 0.14–5.99; p = 0.936), shown in Tables 4 and S2, supplementary appendix. Increasing age (aOR 1.52, 95%CI 1.15–2.64; p = 0.041) and CRP (aOR 1.03, 95%CI 1.01–1.06; p = 0.047) showed a significant association with higher risk of in-hospital mortality for this sub-group. The presence of neutralising antibodies was found not to be protective against ICU admission (aOR 0.46, 95% CI 0.10–2.00; p = 0.302).
Table 4.
Logistic regression analysis for COVID-19 patients with known neutralising antibody serological status (n = 99) for in-hospital mortality and ICU admission.
| In-hospital mortality | ICU admission | |||
| aOR (95% CI) | p-value | aOR (95% CI) | p-value | |
| Age (years) | 1.52 (1.15–2.64) | 0.041 | 0.92 (0.85–0.97) | 0.005 |
| Cardiovascular disease | N/A | – | 1.40 (0.18–9.31) | 0.732 |
| NEWS2 score | 1.13 (0.72–1.92) | 0.604 | 1.22 (0.96–1.59) | 0.113 |
| Symptom duration prior to admission (days) | 0.51 (0.16–0.99) | 0.137 | 0.94 (0.81–1.06) | 0.320 |
| Infiltrates on CXR | 0.25 (0.01–3.48) | 0.330 | >10 (0.9-∞) | 0.988 |
| White blood cell count (109/L) | 0.98 (0.65–1.46) | 0.901 | 1.28 (1.05–1.61) | 0.022 |
| CRP (mg/L) | 1.03 (1.01–1.06) | 0.047 | N/A | – |
| Lymphocyte count (109/L) | 0.75 (NA-1.15) | 0.343 | 0.44 (0.09–0.86) | 0.220 |
| Urea (mmol/L) | N/A | – | 1.13 (0.98–1.36) | 0.153 |
| Platelets (109/L) | 0.99 (0.98–1.01) | 0.418 | N/A | – |
| Neutralising antibody status | 10.62 (0.47–889) | 0.199 | 0.46 (0.10–2.00) | 0.302 |
| Ct value (viral load) | 0.61 (0.28–1.01) | 0.104 | 1.09 (0.93–1.29) | 0.320 |
| Interaction terms | ||||
| Ct Value x age | 1.02 (0.99–1.05) | 0.214 | 1.01 (1.00–1.02) | 0.075 |
| Symptoms days x age | 1.03 (1.00–1.08) | 0.096 | 1.00 (1.00–1.01) | 0.287 |
| Ct Value x symptom days | 1.03 (0.98–1.09) | 0.278 | 1.02 (0.99–1.05) | 0.235 |
| Data presented as adjusted odds ratios (aOR). Abbreviations: N/A, Not applicable, not included as covariate in the model; NEWS2, National Early Warning Score 2; CXR, Chest X-Ray; CRP, C reactive protein; Ct, real-time PCR cycle threshold (a low Ct value represents a high viral load and vice versa); ∞, infinity. Multiple regression analysis of each outcome was performed on a subset of covariates used for the whole cohort, where the ten most significant ones from univariate analyses with the outcome were chosen as dependant covariates. | ||||
Discussion
Our study demonstrates that in a large cohort of hospitalised COVID-19 patients, SARS-CoV-2 viral load at presentation was independently associated with mortality, even when comprehensively adjusting for other variables. This finding is in keeping with other studies and strengthens the case for utilising admission Ct value to assist with risk stratification alongside clinical judgement, in newly hospitalised patients.11 , 12 The clinical course of COVID-19 is unpredictable and utilising Ct value measurement as a predictor of mortality may allow more effective directed escalation to closely monitored clinical areas. The prognostic role of including Ct admission values within other validated severity mortality scoring systems such as ISARIC 4C, is unknown and would need further prospective studies to evaluate.3 Ct value-based risk stratification may also be useful for therapeutic decisions particularly once more effective antiviral therapies are developed and available, as well as informing patients and their families of their own potential prognosis.
Contrary to previous studies, we found that viral load on admission was not independently associated with the risk of admission to the intensive care unit. The interaction analysis revealed that changes in age was the major factor influencing admission to ICU, suggesting that the decision to be admitted to ICU in COVID-19 patients was strongly influenced by patient factors related to age, such as comorbidity and frailty.
The interaction effect of age on outcomes and duration of illness could be hypothesised to be due to older patients having less physiological reserves, and therefore being admitted earlier in their duration of illness and having higher viral loads in the upper respiratory tract22. Older age is a known strong risk factor in predicting mortality23 and therefore, age, duration of symptoms, and viral load are all likely to be strongly inter-related.
In addition to the benefits of clinical risk stratification, measurement of Ct value at presentation may be useful to guide infection prevention measures and prevent nosocomial outbreaks. Although direct evidence for the risk of transmissibility according to viral load (as measured by real time PCR) is lacking, several studies have demonstrated that the ability to culture SARS-CoV-2 diminishes with reducing viral load suggesting that patients presenting with low viral loads are unlikely to be infectious to others.24, 25, 26 For example, a patient who presents to hospital with a very high SARS-CoV-2 Ct value, may be judged in the right clinical circumstances to have resolving infection and to no longer represent a high risk of infection to others. PCR-based point-of-care testing (POCT) has already been shown to improve infection control measures in hospitals by reducing the number of bed moves and improving time to arrival in definitive SARS-CoV-2 positive or negative areas.16
Analysis of the subgroup of patients who were tested for the presence of neutralising antibodies did not demonstrate a significant protective benefit against mortality in those with detectable antibodies at presentation. This is consistent with the results of a previous study evaluating antibody sero-status, viral load and 30 day mortality.27 This suggests that the measurement of neutralising antibodies at presentation to hospital is not helpful for prognostication.
The strengths of this study include the robust statistical methodology, which comprehensively adjusted for known variables associated with SARS-CoV-2 viral load and clinical outcome. The heterogeneity of individuals in our study allows the results to be widely generalisable to other acute hospitals, due to patients presenting with a wide spectrum of COVID-19 illness severity and duration. To our knowledge, we are the first study to adjust for neutralising antibody serological status when evaluating the association between viral load at admission and clinical outcome. The limitations of our study include: the relatively small sample size in the antibody subgroup which may explain the non-significant association between Ct value and outcome28 and being a single centre study, In addition, this study was performed during the first wave of COVID-19 pandemic before antiviral and immunomodulatory therapies were approved for use, which may now influence outcomes and potentially, viral load. Some patients were entered into COVID-19 therapeutics trials, but we do not know what each patient received. We acknowledge that SARS-CoV-2 Ct values are an aid to clinical decision making, and do not replace clinical judgement and also that Ct values do not represent a consistent quantity of virus across different PCR assays and gene targets. Finally, this study was completed before the emergence of new variant SARS-CoV-2 strains and before vaccination was widely introduced and it is uncertain what effects these factors may have on the relationship between viral load and outcome. Further studies should be undertaken to evaluate this.
In conclusion SARS-CoV-2 viral load measured at the point of hospitalisation was associated with the risk of death even after adjustment for age, duration of illness and neutralising antibody sero-status. Measurement of Ct value at admission may be useful for risk stratification.
CRediT authorship contribution statement
Alex R Tanner: Conceptualization, Formal analysis, Data curation, Writing – original draft, Writing – review & editing. Hang Phan: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Nathan J Brendish: Conceptualization, Data curation, Writing – original draft, Writing – review & editing. Florina Borca: Data curation, Writing – review & editing. Kate R Beard: Writing – review & editing, Writing – original draft. Stephen Poole: Data curation, Writing – original draft, Writing – review & editing. Tristan W Clark: Conceptualization, Visualization, Data curation, Writing – original draft, Writing – review & editing.
Declaration of Competing Interest
TWC has received speaker fees, honoraria, travel reimbursement, and equipment and consumables free of charge for the purposes of research outside of this submitted study, from BioFire diagnostics LLC and BioMerieux. TWC has received consultancy fees from Synairgen research Ltd, Randox laboratories Ltd and Cidara therapeutics. He a member of an advisory board for Roche and a member of two independent data monitoring committees for trials sponsored by Roche. He has acted as the UK chief investigator for an IMP study sponsored by Janssen. KRB has received honoraria from Randox laboratories Ltd. All other authors have no competing interests to declare.
Acknowledgments
Funding
University Hospital Southampton NHS Foundation Trust.
Acknowledgements
We would like to acknowledge and gives thanks to all the patients who kindly participated in this study and to all the clinical staff at University Hospital Southampton who cared for them. We would also like to acknowledge the NIHR Southampton Clinical Research Facility (CRF) laboratory, project and nursing teams; the UHSFT research nursing team; and the NIHR Southampton Biomedical Research Centre staff and project teams for their support in the set-up of this study. We thank Dr Ashley Banyard, Animal and Plant Health Agency (APHA), Weybridge, Surrey, for his teams work on the serological antibody measurement.
Data sharing
Following publication of major outputs all anonymised data will be made available on request to the corresponding author for appropriate, ethically approved research.
This report is independent research supported by the National Institute for Health Research (NIHR Post Doctorial Fellowship, Dr Tristan Clark, PDF 2016–09–061). The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research or the Department of Health.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jinf.2021.08.003.
Appendix. Supplementary materials
References
- 1.Chan J.F.W., Kok K.H., Zhu Z., Chu H., Kai-Wang To K., Yuan S. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9(1):221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organisation. WHO Coronavirus disease (COVID-19) dashboard. Available at https://covid19.who.int/table. Accessed January 30, 2021.
- 3.Gupta R.K., Harrison E.M., Ho A., Docherty A.B., Knight S.R., Smeden M. Development and validation of the ISARIC 4C deterioration model for adults hospitalised with COVID-19: a prospective cohort study. Lancet Respir Med. 2021;2600(20) doi: 10.1016/S2213-2600(20)30559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grasselli G., Zangrillo A., Zanella A., Antonelli M., Cabrini L., Castelli A. Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the lombardy region, Italy. JAMA. 2020;323(16):1574. doi: 10.1001/jama.2020.5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brendish N.J., Poole S., Naidu V.V., Mansbridge C.T., Norton N., Borca F. Clinical characteristics, symptoms and outcomes of 1054 adults presenting to hospital with suspected COVID-19: a comparison of patients with and without SARS-CoV-2 infection. J Infect. 2020;81(6):937–943. doi: 10.1016/j.jinf.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Izcovich A., Ragusa M.A., Tortosa F., Marzio M.A.L., Agnoletti C., Bengolea A. Prognostic factors for severity and mortality in patients infected with COVID-19: a systematic review. PLoS ONE. 2020;15:1–30. doi: 10.1371/journal.pone.0241955. 11 November. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de la Calle C., Lalueza A., Mancheño-Losa M., Maestro-de la Calle G., Lora-Tamayo J., Arrieta E. Impact of viral load at admission on the development of respiratory failure in hospitalized patients with SARS-CoV-2 infection. Eur J Clin Microbiol Infect Dis. 2021;40(6):1209–1216. doi: 10.1007/s10096-020-04150-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee N., Chan M.C.W., Lui G.C.Y., Li R., Wong R.Y.K., Yung I.M.H. High viral load and respiratory failure in adults hospitalized for respiratory syncytial virus infections. J Infect Dis. 2015;212(8):1237–1240. doi: 10.1093/infdis/jiv248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee N., Chan P.K.S., Hui D.S.C., Rainer T.H., Wong E., Choi K.W. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis. 2009;200(4):492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lalueza A., Folgueira D., Muñoz-Gallego I., Trujillo H., Laureiro J., Hernández-Jiménez P. Influence of viral load in the outcome of hospitalized patients with influenza virus infection. Eur J Clin Microbiol Infect Dis. 2019;38(4):667–673. doi: 10.1007/s10096-019-03514-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Magleby R., Westblade L.F., Trzebucki A., Simon M.S., Rajan M., Park J. Impact of SARS-CoV-2 viral load on risk of intubation and mortality among hospitalized patients with Coronavirus disease. Clin Infect Dis. 2019;2020:1–14. doi: 10.1093/cid/ciaa851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pujadas E., Chaudhry F., McBride R., Richter F., Zhao S., Wajnberg A. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2(1):e13–e22. doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Euser S., Aronson S., Manders I., Lelyveld S., Herpers B., Sinnige J. SARS-CoV-2 viral load distribution reveals that viral loads increase with age: a retrospective cross-sectional cohort study. MedRxiv. 2021 doi: 10.1093/ije/dyab145. 2021.01.15.21249691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J. SARS-CoV-2 neutralizing antibody LY-CoV555 in outpatients with Covid-19. N Engl J Med. 2021;384(3):229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brendish N.J., Poole S., Naidu V.V., Mansbridge C.T., Norton N.J., Wheeler H. Clinical impact of molecular point-of-care testing for suspected COVID-19 in hospital (COV-19POC): a prospective, interventional, non-randomised, controlled study. Lancet Respir Med. 2020;8(12):1192–1200. doi: 10.1016/S2213-2600(20)30454-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.T.W. Clark. Evaluating the clinical impact of routine molecular point-of-care testing for COVID-19 in adults presenting to hospital: A prospective, interventional, non-randomised, controlled study (CoV-19POC) [Protocol]. 2020. 3 June. Available at https://eprints.soton.ac.uk/439309/2/CoV_19POC_Protocol_v2_0_eprints.pdf. Accessed February 7, 2021.
- 18.Parčina M., Schneider U.V., Visseaux B., Jozić R., Hannet I., Lisby J.G. Multicenter evaluation of the QIAstat respiratory panel-a new rapid highly multiplexed PCR based assay for diagnosis of acute respiratory tract infections. PLoS ONE. 2020;15(3):1–12. doi: 10.1371/journal.pone.0230183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Visseaux B., Hingrat Q.L., Collin G., Bouzid D., Lebourgeois S., Pluart D.L. Evaluation of the qiastat-dx respiratory sars-cov-2 panel, the first rapid multiplex PCR commercial assay for sars-cov-2 detection. J Clin Microbiol. 2020;58(8):1–5. doi: 10.1128/JCM.00630-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loeffen W., Quak S., Boer-Luijtze E., Hulst M., Wim W.P., Bouwstra R. Development of a virus neutralisation test to detect antibodies against Schmallenberg virus and serological results in suspect and infected herds. Acta Vet Scand. 2012;54:44. doi: 10.1186/1751-0147-54-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aiken L.S., West S.G., Reno R.R. Newbury Park (Calif.): Sage publications; 1991. Multiple regression : testing and interpreting interactions. [Google Scholar]
- 22.Cevik M., Kuppalli K., Kindrachuk J., Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- 23.Knight S.R., Ho A., Pius R., Buchan I., Carson G., Drake T.M. Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO clinical characterisation protocol: development and validation of the 4C mortality score. BMJ. 2020:m3339. doi: 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bullard J., Dust K., Funk D., Strong J.E., Alexander D., Garnett L. Predicting Infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663–2666. doi: 10.1093/cid/ciaa638. Doi. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walsh K.A., Jordan K., Clyne B., Rohde D., Drummond L., Byrne P. SARS-CoV-2 detection, viral load and infectivity over the course of an infection. J Infect. 2020;81(3):357–371. doi: 10.1016/j.jinf.2020.06.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scola B.L., Bideau MLe, Andreani J., Hoang V.T., Grimaldier C., Colson P. Viral RNA load as determined by cell culture as a management tool for discharge of SARS-CoV-2 patients from infectious disease wards. Eur J Clin Microbiol Infect Dis. 2020;39(6):1059–1061. doi: 10.1007/s10096-020-03913-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bryan A., Fink S.L., Gattuso M.A., Pepper G., Chaudhary A., Wener M.H. SARS-CoV-2 viral load on admission is associated with 30-day mortality. Open Forum Infect Dis. 2020;7(12):1–5. doi: 10.1093/ofid/ofaa535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hauck W.W., Donner A. Wald's test as applied to hypotheses in logit analysis. J Am Stat Assoc. 1977;72(360a):851–853. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.